Significance
Cells need to rapidly control their cell cycle profile, gene expression, or protein stability in response to the activation of stress-response pathways. One such type of stress results from plasma membrane damage, which can arise because of physical damage or pathogen invasion. Here, we report that plasma membrane stress inhibits polarized cell growth and DNA replication through a novel cell cycle checkpoint pathway in Saccharomyces cerevisiae. To our knowledge, this is the first study to link a plasma membrane signaling pathway with the cell cycle and DNA replication control, which is relevant to plasma membrane repair and the role of stress and cell proliferation control in higher eukaryotes.
Keywords: cdc6, mck1, cell wall integrity, sic1, DNA replication
Abstract
Cellular wound healing or the repair of plasma membrane/cell wall damage (plasma membrane damage) occurs frequently in nature. Although various cellular perturbations, such as DNA damage, spindle misalignment, and impaired daughter cell formation, are monitored by cell cycle checkpoint mechanisms in budding yeast, whether plasma membrane damage is monitored by any of these checkpoints remains to be addressed. Here, we define the mechanism by which cells sense membrane damage and inhibit DNA replication. We found that the inhibition of DNA replication upon plasma membrane damage requires GSK3/Mck1-dependent degradation of Cdc6, a component of the prereplicative complex. Furthermore, the CDK inhibitor Sic1 is stabilized in response to plasma membrane damage, leading to cell integrity maintenance in parallel with the Mck1-Cdc6 pathway. Cells defective in both Cdc6 degradation and Sic1 stabilization failed to grow in the presence of plasma membrane damage. Taking these data together, we propose that plasma membrane damage triggers G1 arrest via Cdc6 degradation and Sic1 stabilization to promote the cellular wound healing process.
It is common for cells to suffer from various attacks to their plasma membrane and cell wall, such as physical damage, pathogen invasion, and various environmental perturbations (1). Maintaining structured barriers between the internal and external environment of a cell, upon such challenges as those listed, is critical for the integrity of genetic materials. Therefore, it is advantageous to study cellular wound healing in a simple, genetically tractable model organism such as yeast.
The budding yeast, Saccharomyces cerevisiae, responds to cell wall and plasma membrane damage using an evolutionary conserved signaling cascade known as the cell wall integrity (CWI) pathway. The CWI pathway includes a Rho-type GTPase (Rho1) (2, 3) and protein kinase C (Pkc1) (4), both conserved plasma membrane repair proteins, and also includes part of the mitogen-activated protein (MAP) kinase cascade (5). One major consequence of the CWI pathway activation is transcriptional activation of enzymes required for cell wall synthesis (6). The CWI pathway can be studied using chemicals such as sodium dodecyl sulfate (SDS), which perturbs the plasma membrane. Previously, Kono et al. established a laser damage assay that specifically creates acute local damage to the yeast membrane (7). Using this assay, the authors showed that daughter cell growth is temporarily arrested by Pkc1-dependent degradation of cell-polarity regulators (7). Although the growth arrest was suggested to be triggered by plasma membrane damage, how the cell cycle progression was modulated upon local membrane damage remains to be understood.
The cell cycle is a series of events that leads to genome duplication and cell division, producing two daughter cells. In most eukaryotes, cells commit to division in G1, at which point the cells integrate internal and external cues to determine cell fates (8). In budding yeast, the restriction point is called START (9). During pre-Start, cells arrest in response to mating pheromone, whereas post-Start cells are committed to one cell cycle progression (10). G1 progression is triggered by the G1 cyclin Cln3/CDK complex, which phosphorylates and inactivates Whi5, an inhibitor of transcription factor Swi4/Swi6 (SBF) (11). SBF and MBF, an additional transcription factor complex, then activate the transcription of two additional G1 cyclins, Cln1 and Cln2 (10, 12). Cln1 and Cln2 compose a positive feedback circuit via the activation of transcription factors SBF and MBF (13, 14), triggering a genome-wide transcriptional change that promotes the G1/S transition (15, 16). Subsequently, the CDK inhibitor Sic1 is phosphorylated by the G1 cyclins, ubiquitinated, and degraded in a SCF (Skp1–Cullin–F-box)-dependent manner (17, 18). DNA replication is triggered upon Sic1 degradation. DNA replication origins are licensed for subsequent firing by assembly of prereplicative complexes (pre-RCs). Each pre-RC contains the Mcm2-7 helicase loaded onto origin DNA through sequential binding of Orc1-6, Cdc6, and Cdt1 (19). At the onset of S-phase, the pre-RC is acted on by two protein kinases, Dbf4-Cdc7 (DDK) and S-phase Cyclin-Cdk (S-CDK), and then converts into the Cdc45-Mcm2-7-GINS (CMG) replicative DNA helicase (20). This step involves DDK-dependent loading of Sld3 and CDK-dependent assembly of the preloading complex, consisting of Sld2, Dpb11, GINS, and DNA polymerase ε (20, 21). In yeast, three pre-RC components (Cdc6, Mcm2-7, and the ORC) are phosphorylated by Cyclin/CDK to prevent a second round of DNA replication, thereby inhibiting DNA rereplication (22–28).
Cdc6 protein degradation ensures DNA replication once and only once during the cell cycle by which Cdc6 requires phosphorylation by CDK for degradation (29). Previously, we demonstrated that the yeast GSK-3 kinase, Mck1, phosphorylates Cdc6 at the GSK-3 consensus site, at Thr-39 and Thr-368, promoting degradation of Cdc6 to guarantee genome integrity (30). Cdc6-Thr368 phosphorylation requires a priming phosphorylation at Ser372 by Cyclin/CDK (31). This double phospho-degron at Cdc6-T368-S372 created by Mck1 and CDK serves as a binding site for the SCF F-box protein, Cdc4 (31).
The yeast Mck1 is a serine/threonine protein kinase homologous to mammalian glycogen synthase kinase-3 (GSK-3) (32, 33). Mck1 also plays a role in the stress response, such that mck1 deletion cells are sensitive to hot and cold temperatures (34), benomyl (34), and osmotic stress (35). Mck1 also stimulates calcineurin signaling (36–38), and binds stress-response elements to activate transcription (38). We recently showed that GSK-3–dependent Cdc6 degradation plays a role in genome integrity maintenance when cells are exposed to DNA damage (31). Thus, Mck1 ensures proper DNA replication, prevents DNA damage, and maintains genome integrity by inhibiting Cdc6 (31).
Here we provide evidence that plasma membrane damage activates a novel cell cycle checkpoint in G1 through Mck1-depedent Cdc6 degradation and Sic1 stabilization.
Results
Plasma Membrane Stress Inhibits S-Phase Entry.
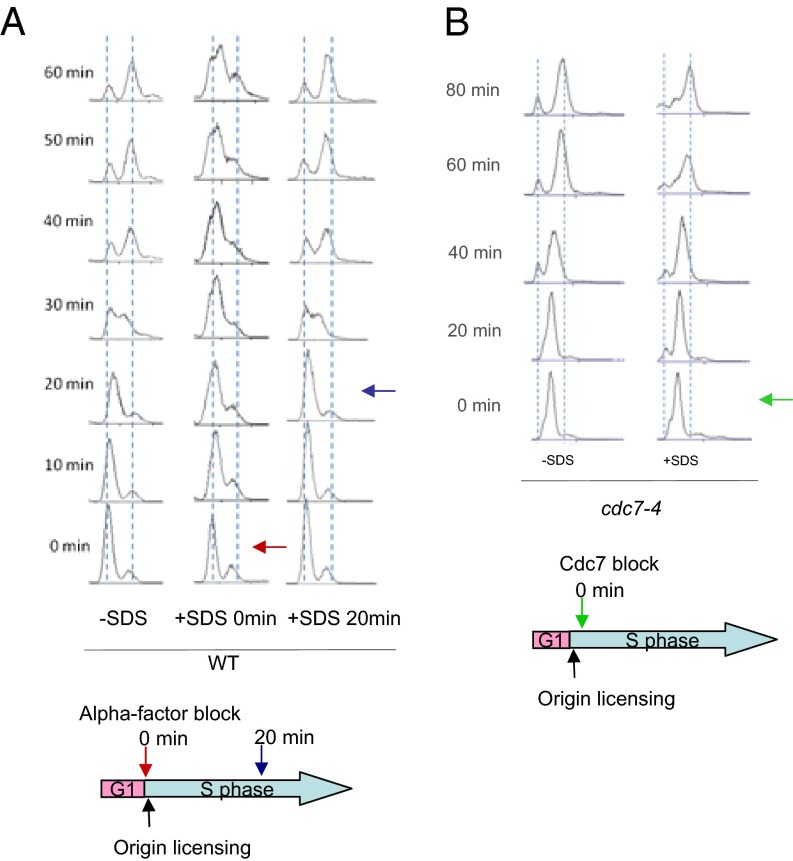
First, we analyzed the cell cycle profile upon plasma membrane stress. Cells were synchronized in G1 by α-factor and then released into media with or without SDS treatment. Wild-type cells, without SDS treatment, entered S-phase 30 min after G1 release (Fig. 1A, Left column). In contrast, S-phase entry was significantly delayed with SDS treatment (Fig. 1A, Center column). However, cell cycle progression was completely normal when cells were treated with SDS after the origins have been fired, 20 min after G1 release (Fig. 1A, Right column). This finding indicates that S-phase entry is inhibited in response to plasma membrane damage. To test which DNA replication step is affected by SDS treatment, we used a cdc7-4 mutant to arrest the cell cycle. Cdc7 binds to Dbf4 to make the DDK complex, which then phosphorylates and activates Mcm4 to initiate DNA replication after origin licensing (39, 40). cdc7-4 is a DDK temperature-sensitive mutant that arrests the cell cycle during G1-S phase, after pre-RC formation but with an inactive helicase not yet able to unwind DNA (41). When cells were treated with SDS upon cdc7-4 block and release, the cell cycle proceeded normally (Fig. 1B), indicating that SDS affects the DNA replication step before DDK is activated, probably at pre-RC formation.
Fig. 1.
S-phase progression is inhibited in response to SDS. (A) Wild-type cells were arrested and released in G1-phase by α-factor. The cell cycle progression was monitored by FACS analysis in the presence or absence of 0.0075% SDS added at time 0 (red arrow) or 20 min after release (blue arrow). (B) First, cdc7-4 cells were arrested in G1-phase by α-factor at 23 °C and then released at 37 °C, the restrictive temperature, to block the cells after pre-RC formation but with an inactive helicase. Cells were then released into YPD media at 23 °C and collected every 20 min to monitor the cell cycle progression by FACS analysis in the presence or absence of SDS 0.0075% added at time 0 (green arrow). The diagrams below show at what point of the cycle that the cells were in when SDS was added.
S-cyclin/CDK Activity Is Inhibited in Response to Plasma Membrane Damage.
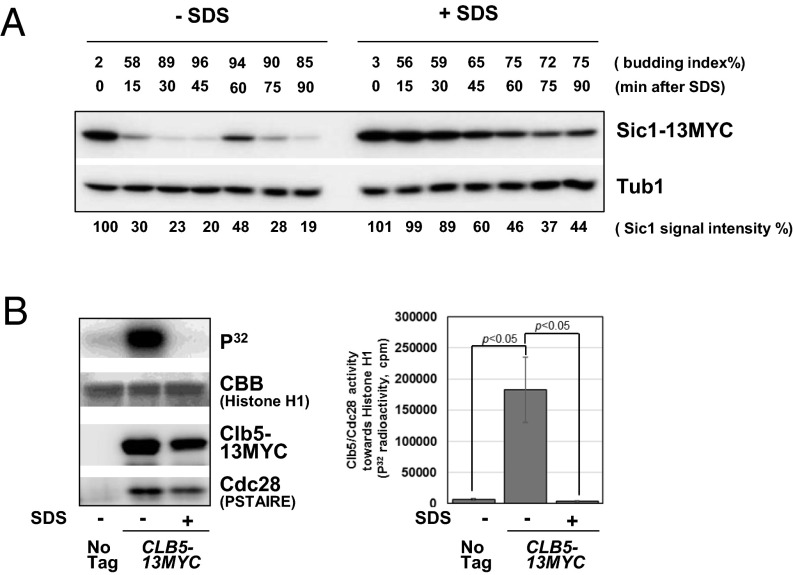
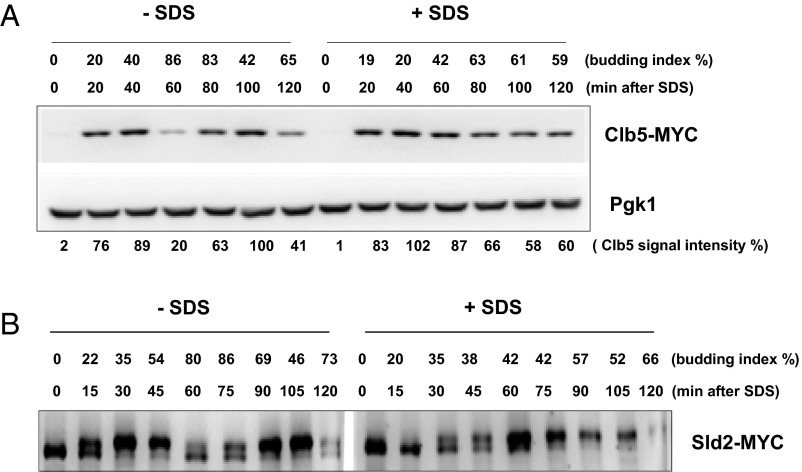
Next, we examined cell cycle regulators to test if they play a role in G1 arrest induced by membrane damage. We investigated Sic1 protein expression, the S-phase CDK inhibitor, during cell cycle arrest induced by plasma membrane stress. Sic1 is rapidly degraded at the onset of the G1/S transition in untreated cells and is expressed again 60 min later during the next G1 phase (Fig. 2A, Left) (18, 42). In contrast, we found the Sic1 protein levels to be more stable in the presence of SDS throughout the time course (Fig. 2A, Right). Next, we examined protein levels of the S-phase cyclin, Clb5, under plasma membrane stress. Under normal conditions, Clb5 is degraded by the APCcdc20 complex during mitosis (43), which we observed 60 min after α-factor block and release in untreated cells (Fig. S1A, Left). Clb5 expression was observed again 80 min later during the next cell cycle. Under SDS treatment, Clb5 was continuously expressed throughout the time course (Fig. S1A, Right). Because of stable Sic1 protein level under SDS treatment, CDK activity might be inhibited despite continual Clb5 expression. To test this possibility, we examined the phosphorylation status of a CDK substrate, Sld2, a DNA replication protein phosphorylated by Clb5 (44). In a normal cell cycle progression, Sld2 is phosphorylated during G1/S-phase and dephosphorylated after S-phase (Fig. S1B, Left). In contrast, the timing of Sld2 phosphorylation was delayed in cells treated with SDS (Fig. S1B, Right). This delay indicates that CDK activity is inhibited in response to plasma membrane stress, which leads to S-phase delay; this was further tested by in vitro kinase assay using Clb5/CDK purified from S-phase cells treated with SDS. We found that CDK activity is greatly reduced after SDS treatment (Fig. 2B).
Fig. 2.
CDK is inhibited through Sic1 stabilization upon SDS treatment. (A) Cells expressing Sic1-13myc (BY4741) from the genomic locus were synchronized by α-factor, and then released into fresh YPD medium in the presence or absence of 0.02% SDS to induce plasma membrane damage. Protein samples were collected every 15 min to observe Sic1-13myc levels by Western blotting. Tub1 was used as a loading control. The budding index is shown as percent of budded cells. (B) CLB5-13MYC cells (BY4741) were synchronized in G1 phase by α-factor. The cell cycle block was released and SDS was added to the media. Samples were collected at 20 min after the release. Clb5-13myc was immunopurified using anti-myc (9E10) antibody. Kinase reactions were performed with Histone H1 as a substrate. Total Histone H1 was visualized by Coomassie staining. Western blotting was performed to show the total amount of Clb5 and Cdc28. The graph indicates the averages of four independent experiments. Error bars represent SEM. Statistical significance was determined by Student’s t test.
Fig. S1.
Clb5 is stabilized after SDS treatment. (A) CLB5-MYC cells were synchronized during G1 phase by α-factor. The cell cycle block was released and SDS was added to the media. Samples were collected every 20 min for protein extractions. Each time point was subjected to Western blot analysis. Pgk1 was used as a loading control. Budding index is shown as percent of budded cells. (B) SLD2-MYC cells were used to monitor the Sld2 phosphorylation status upon G1 block and release as described in A.
Plasma Membrane Damage Inhibits DNA Replication Through Cdc6 Degradation.
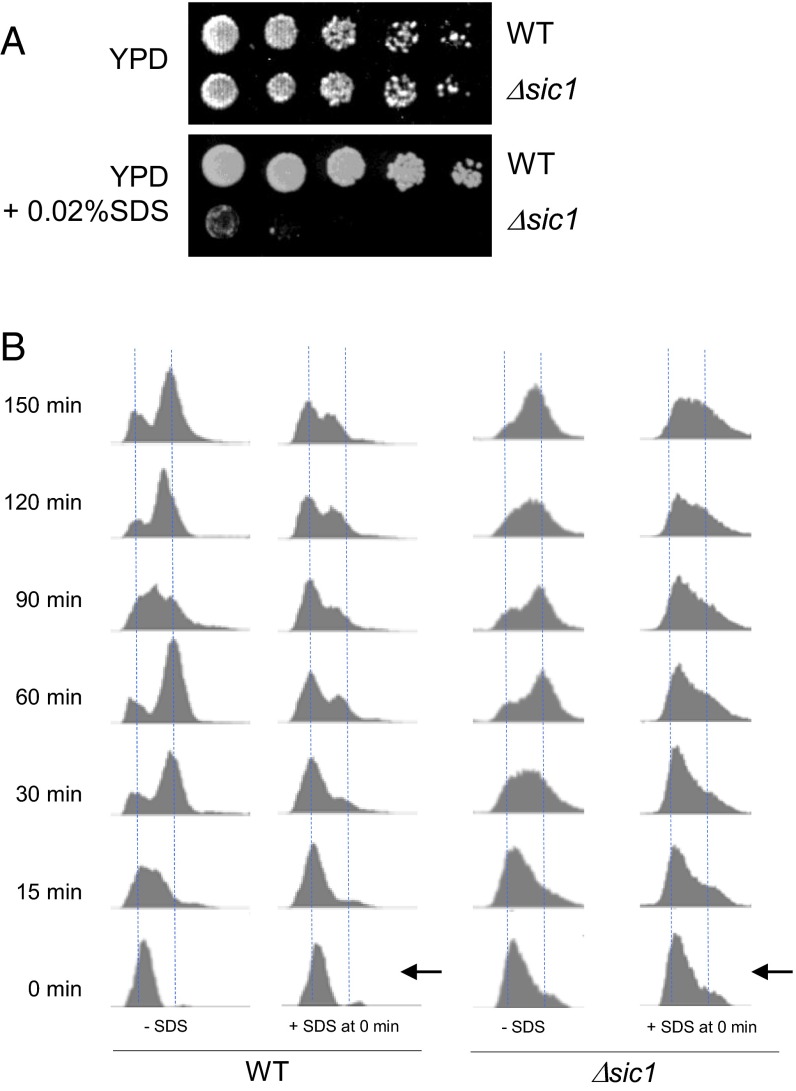
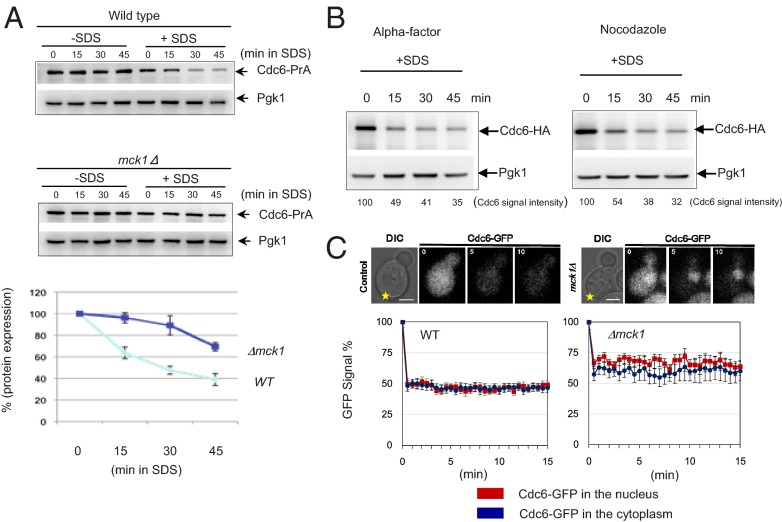
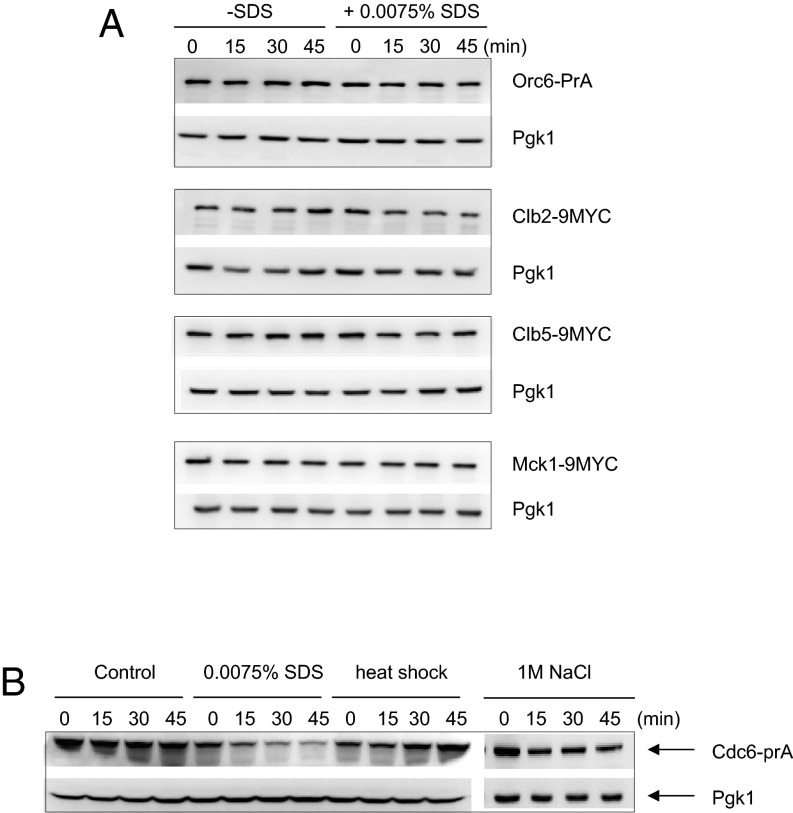
Sic1 protein levels were more stable in response to SDS (Fig. 2A). The Sic1 deletion strain, Δsic1, was sensitive to SDS, indicating that Sic1 may play a role in G1 arrest upon SDS treatment (Fig. S2A). We tested if Δsic1 rescues the S-phase delay caused by SDS and observed that the cell cycle arrest was sustained (Fig. S2B), indicating that Sic1 is dispensable for G1 arrest induced by SDS treatment. This result is consistent with previous findings that Sic1 degradation is not a cause but a consequence of commitment to the cell cycle progression (45). Next, we considered the possibility that the cell cycle arrest upon SDS treatment is a result of pre-RC disassembly, because the cell cycle progression had no effect when SDS was added after pre-RC formation (Fig. 1B). We analyzed protein levels of pre-RC components after SDS treatment and observed that only Cdc6 was degraded in response to membrane damage triggered by SDS (Fig. 3A and Fig. S3A). Next, we monitored Cdc6 levels after different stress treatments. In contrast to Cdc6 degradation under SDS treatment (plasma membrane stress), we found that Cdc6 was stable at 40 °C (heat shock) or in the presence of 1 M NaCl (osmotic stress) (Fig. S3B). Previously, we showed that Cdc6 degradation is mediated by Mck1 during an unperturbed cell cycle or during a DNA damage response (30, 31). Δmck1 deletion cells are sensitive to various stress stimuli, including plasma membrane stress (Fig. 4B), which suggests a link between Mck1-dependent Cdc6 degradation and membrane stress. Therefore, we examined the possibility that Mck1 promotes Cdc6 degradation in the presence of membrane damage. Indeed, Cdc6 degradation was rescued in Δmck1 deletion cells, indicating that Cdc6 degradation in response to plasma membrane stress requires Mck1 (Fig. 3A).
Fig. S2.
Sic1 is dispensable for G1 arrest caused by SDS. (A) BY4741 background yeast cells with indicated genotypes were serially diluted at fourfold on YPD or YPD +0.02% SDS and incubated for 4 d to test the viability. (B) Wild-type cells and Δsic1 cells in BY4741 background were arrested and released in G1 phase by α-factor. Cell cycle progression was monitored by FACS analysis in the presence or absence of SDS 0.02% added at time 0 after the release.
Fig. 3.
Cdc6 is degraded in response to plasma membrane stress. (A) CDC6-prA or Δmck1 CDC6-prA cells were grown to log-phase. Samples were collected every 15 min in the presence or absence of 0.0075% SDS. Protein was extracted from each time point for Western blotting to detect Cdc6-prA or Pgk1 as a loading control. The same experiment was repeated three times and the signal was quantified to show the average with SD. (B) GAL-CDC6 cells were incubated in galactose-containing media first. The cell cycle was arrested in G1 by α-factor or in mitosis by nocodazole. The cell cycle block was 86% in G1 and 83% in mitosis. Samples were collected every 15 min in the presence or absence of 0.0075% SDS. Protein was extracted from each time point and subjected to Western blotting. Pgk1 was used as a loading control. (C) Wild-type or Δmck1 cells were damaged with a laser at the location marked with a star. The Cdc6-GFP signal was monitored by live-cell imaging using a fluorescence microscope. The numbers in the upper left corner indicate the time after laser damage (min). (Scale bars, 2 µm.) The graphs show the mean GFP signal intensity after background subtraction (control cells, n = 15; mck1Δ cells, n = 11). The error bars represent SEM.
Fig. S3.
Cdc6 is degraded in response to SDS. (A) ORC6-prA, CLB2-9MYC, CLB5-9MYC, or MCK1-9MYC cells were grown to log-phase first, and then treated with 0.0075% SDS. Cells were collected every 15 min and protein extracts from each time point were subjected to Western blotting. (B) Cdc6-prA cells were grown to log-phase first, and were treated with either 0.0075% SDS at 40 °C or 1 M NaCl. Cells were collected every 15 min and protein extracts from each time point were subjected to Western blotting.
Fig. 4.
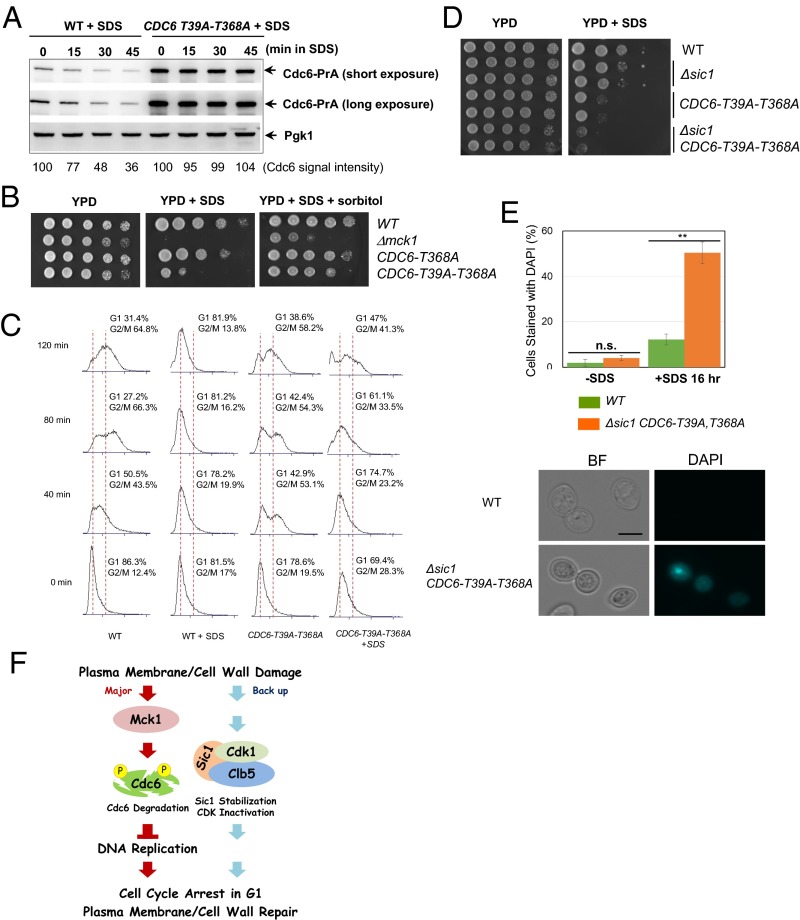
Phosphorylation of Cdc6-T39 and T368 is required for Cdc6 degradation and cell cycle arrest after plasma membrane damage. (A) CDC6-prA or CDC6-T39A-T368A-prA cells were grown to log-phase. SDS at 0.0075% concentration was added to the media, and samples were collected every 15 min. Protein was extracted from each time point for Western blotting to detect Cdc6-prA or Pgk1 as a loading control. (B) Indicated W303 background yeast strains were serially diluted at fivefold, and spotted on YPD, YPD containing 0.005% SDS, or YPD containing 0.005% SDS plus 1 M sorbitol plates. Plates were incubated for 2–3 days. (C) Indicated strains were grown in YPD and were synchronized in G1 phase by α-factor. The G1 block was released into nocodazole-containing YPD media plus 0.0075% SDS at time 0. Samples were collected every 40 min to monitor cell cycle progression via FACS analysis. (D) Indicated strains were serially diluted at 10-fold and plated on YPD with or without 0.0075% SDS plates. (E) Indicated strains were grown in YPD with or without 0.0075% SDS for 16 h. Cells were washed once with YPD and then stained with DAPI to visualize cells with a ruptured plasma membrane under a fluorescence microscope. Averages of three independent experiments are shown. n > 100 per each experiment. Error bars, SD **P < 0.01 (Student’s t test). (Scale bar, 5 μm.) The images are representations of DAPI stained cells treated with SDS. (F) A model of a novel cell cycle checkpoint activated by plasma membrane damage.
To further understand how Cdc6 is degraded upon plasma membrane perturbation in respect to the cell cycle, Cdc6 protein degradation was monitored using cells synchronized in G1 with low CDK activity or in mitosis with high CDK activity. Cdc6 was degraded after SDS treatment when cells were arrested during G1 phase by α-factor, suggesting that CDK activity is not required for Cdc6 degradation under membrane stress (Fig. 3B, Left). We also observed Cdc6 degradation during mitotic arrest induced by the nocodazole treatment (Fig. 3B, Right), suggesting that Cdc6 degradation takes place independent of the cell cycle stages. We conclude that Cdc6 does not require CDK activity for its degradation in response to plasma membrane damage.
Next, the subcellular localization of Cdc6-GFP was monitored under a time-lapse microscope after local plasma membrane laser damage (Fig. 3C). Both the control and Δmck1 cells show an immediate drop in GFP signal that we suspect to be an artifact, possibly because of pH change or protein loss through membrane leakage or photo bleaching. After the initial decline, the Cdc6-GFP signal for both genotypes became relatively stable; however, the GFP signal in the Δmck1 cells stabilized with a higher intensity than in the wild-type cells. The Δmck1 cells retained a nuclear Cdc6-GFP signal even after laser damage, confirming the above results that Mck1 is required for Cdc6 degradation after plasma membrane damage (Fig. 3A). Thus, Cdc6 is selectively degraded in a Mck1-dependent manner after membrane damage.
Mck1-Dependent Cdc6 Phosphorylation at T39 and T368 Are Responsible for DNA Replication Inhibition Induced by Plasma Membrane Stress.
Previously, we showed that Mck1 phosphorylates Cdc6 at T39 and T368 (30, 31). Motivated by these findings, we tested if the Cdc6 phosphorylation mutant, Cdc6-T39A-T368A, is stabilized upon SDS stress. The levels of Cdc6-T39A-T368A protein, even after SDS treatment, remained unchanged (Fig. 4A), similar to wild-type Cdc6 protein in Δmck1 cells (Fig. 3A), suggesting that SDS-dependent Cdc6 degradation requires phosphorylation at T39 and T368 by Mck1.
Next we examined whether Cdc6 degradation is required for viability in the presence of SDS. Indeed, cells with Cdc6-T39A-T368A failed to grow in the presence of SDS, similar to Δmck1 cells (Fig. 4B, Center). Moreover, the marginal growth phenotype of Cdc6-T39A-T368A cells and Δmck1 cells was suppressed by the addition of the plasma membrane-stabilizing reagent, sorbitol (46) (Fig. 4B, Right), suggesting that the cell lethality in Δmck1 and CDC6-T39A-T368A cells in the presence of SDS was because of cell lysis. We tested if the stabilized Cdc6 bypasses the S-phase delay caused by SDS. CDC6-T39A-T368A cells entered mitosis 40 min after α-factor block and release and showed normal cell cycle progression when cells were untreated (Fig. 4C). Wild-type cells arrested their cell cycle during G1-phase during SDS treatment (Fig. 4C). Conversely, the CDC6-T39A-T368A cells treated with SDS progressed through S-phase and entered mitosis 80 min after α-factor release (Fig. 4C). G1-phase in the CDC6-T39A-T368A cells was greatly reduced despite the presence of SDS (Fig. 4C). We also detected an increase in the sub-G1 population in CDC6-T39A-T368A cells treated with SDS after 120 min, suggesting that Cdc6 stabilization after plasma membrane damage causes cell lysis. Thus, upon plasma membrane damage, Mck1-dependent phosphorylation/degradation of Cdc6 is required for cell cycle arrest before the G1/S-transition.
When CDC6-T39A-T368A mutations are combined with Δsic1, the SDS sensitivity was exacerbated (Fig. 4D), suggesting that the Sic1 stabilization and Cdc6 degradation were genetically parallel to maintain cell viability upon SDS treatment. What is the consequence in Δsic1 CDC6-T39A-T368A cells after plasma membrane damage? To test whether cells lacking both Cdc6 degradation and Sic1 stabilization mechanisms undergo cell lysis, we examined the plasma membrane’s integrity after SDS treatment by using DAPI, as it only stains cells with a ruptured membrane (47). In wild-type untreated cells DAPI does not permeate the cell membrane, therefore there was no DAPI staining observed (Fig. 4E). In wild-type cells treated with SDS, 12% of cells showed plasma membrane rupture via DAPI staining (Fig. 4E). In contrast, we found that 50% of Δsic1 CDC6-T39A-T368A cells underwent plasma membrane rupture under SDS treatment (Fig. 4E). Thus, continued DNA synthesis and cell cycle progression in the presence of plasma membrane damage induces plasma membrane rupture and cell lysis, eventually leading to cell death.
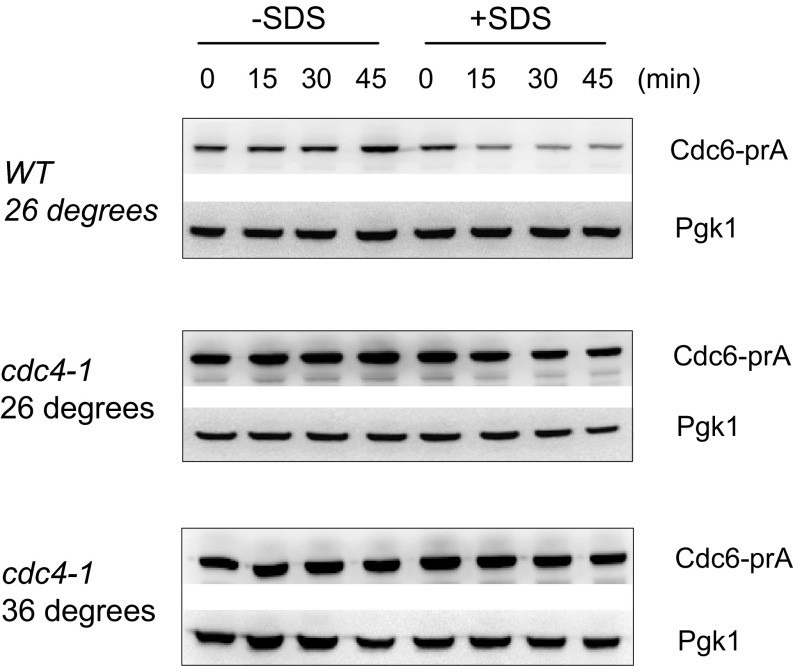
Cdc6 degradation is mediated through the SCFCdc4 complex (22–24). Previously we showed that methyl methanesulfonate (MMS)-induced Cdc6 degradation was rescued when Cdc4 was defective (31). To test if Cdc6 degradation induced by SDS is also mediated through Cdc4, we monitored the Cdc6 protein levels in a cdc4-1 mutant. Cdc6 was stabilized in the presence of SDS when the cdc4-1 temperature-sensitive mutant was incubated at the nonpermissive temperature (36 °C) (Fig. S4). Thus, Cdc4 is likely to be involved in the degradation of Cdc6 during plasma membrane damage.
Fig. S4.
Cdc6 protein degradation by SDS is mediated through SCFCdc4. CDC6-prA (WT) or CDC6-prA cdc4-1 (cdc4-1) cells were arrested during mitosis by nocodazole first at the permissive temperature, 26 °C. Cells were then incubated at either 26 °C or 36 °C for 1.5 h, and samples were collected every 15 min with or without SDS treatment. Protein was extracted from each time point and subjected to Western blotting to visualize Cdc6-prA. Anti-Pgk1 antibody was used as a loading control.
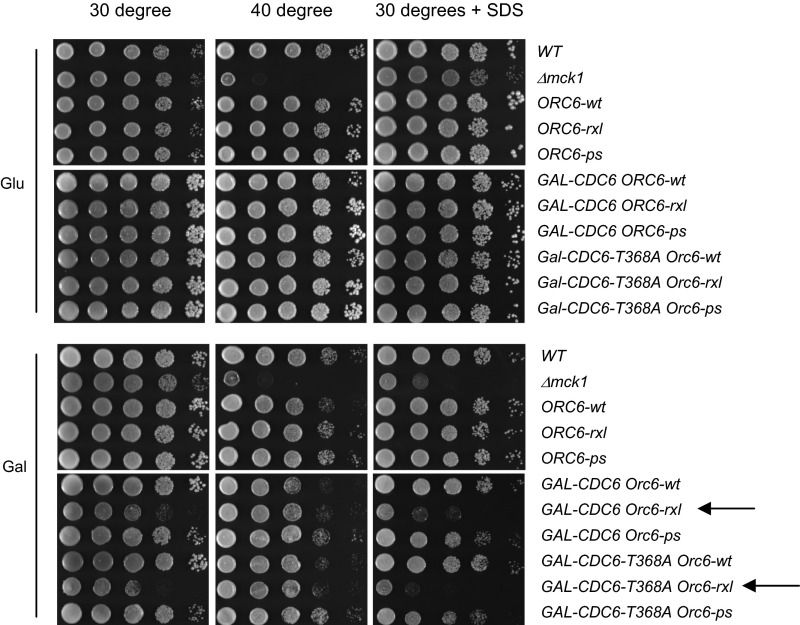
Our results imply that inhibition of DNA replication is crucial when cells encounter plasma membrane damage. Aberrant DNA replication under membrane stress may lead to cell death because cells have no chance to repair membrane damage. To assess this hypothesis, we tested if cells that rereplicate DNA are sensitive to plasma membrane damage induced by SDS. DNA rereplicating cells such as ORC6-rxl GAL-CDC6 or ORC6-rxl GAL-CDC6-T368A, grown on galactose-containing plates, were sensitive to SDS, but not to high temperatures (Fig. S5). These results are consistent with our understanding that continuous DNA replication in the presence of plasma membrane damage leads to lethality. This genetic evidence supports the biological importance of DNA replication control under plasma membrane stress.
Fig. S5.
DNA replication mutants are sensitive to SDS. Strains with indicated genotypes were serially diluted at tenfold on YPD or YPG plates. The plates were incubated at 30 °C, 40 °C, or 30 °C using plates containing SDS. Strains indicated by arrows induce DNA rereplication in the presence of galactose.
Discussion
In this work, we revealed a novel cell cycle checkpoint activated by plasma membrane damage. This checkpoint arrests the cell cycle after Start but before DNA replication initiation. DNA replication is inhibited upon plasma membrane damage, primarily via Mck1-dependent Cdc6 degradation. In addition to the primary pathway, Sic1 stabilization also plays a role in CDK inhibition, to promote plasma membrane healing and protect cells from cell lysis (Fig. 4F).
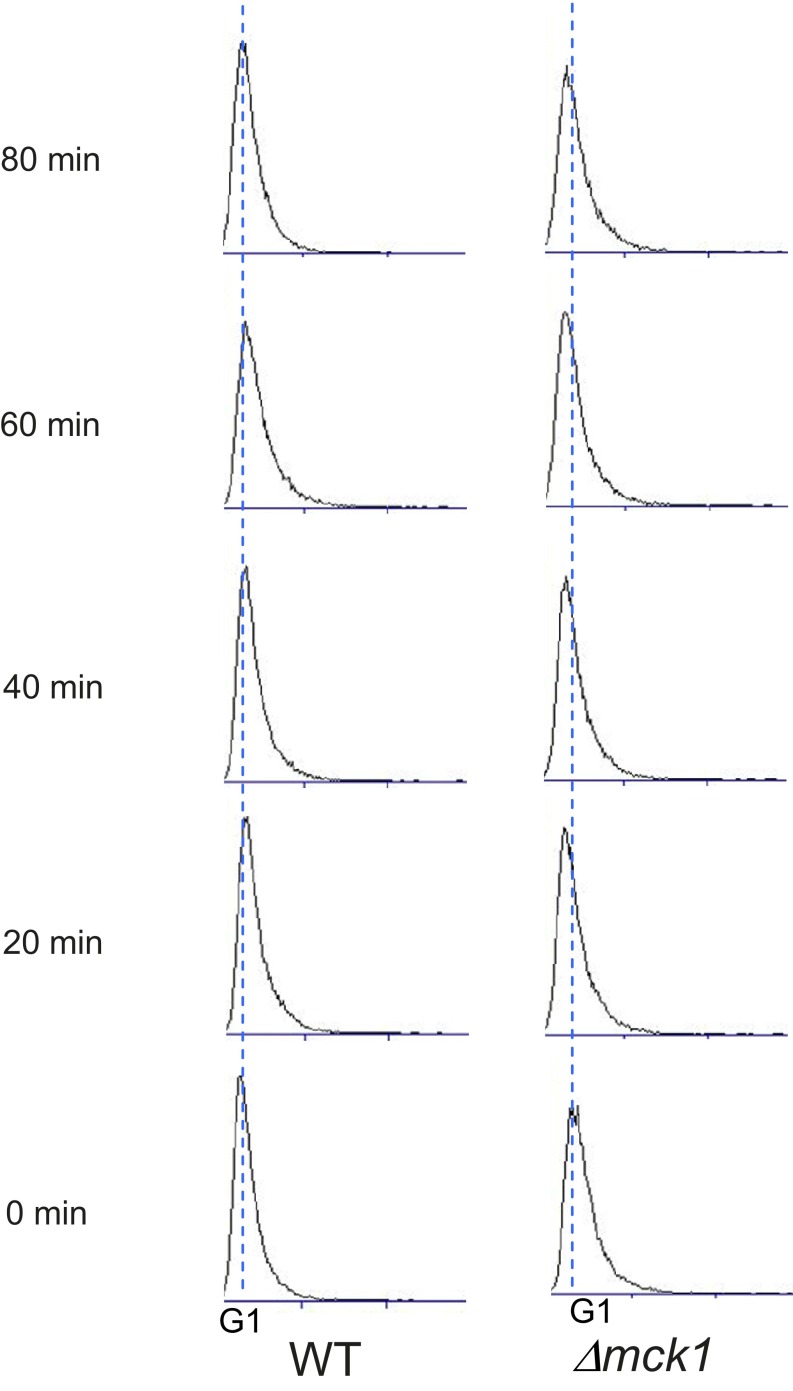
What is the molecular mechanism of Cdc6 degradation caused by membrane stress? Cdc6 phosphorylation at T368 by Mck1 requires a priming phosphorylation at S372 by CDK. Considering that CDK activity is inhibited by Sic1 in response to plasma membrane stress, it is possible that Cdc6 uses a different priming kinase other than CDK that enables cells to promote Cdc6 degradation independently from cell cycle progression but in response to plasma membrane stress. In fact, Cdc6 was still degraded in G1 cells when CDK activity is low (Fig. 3B). It is of interest to study if the kinases involved in the CWI pathway phosphorylate Cdc6 at its priming site, which allows Cdc6 to create a “stress-responsive” phosphodegron to ensure timely protein degradation in response to stress. It is worth noting that Δmck1 cells did not rescue G1 arrest induced by SDS (Fig. S6), which is probably because of other unknown functions of Mck1 during the G1-S transition. Furthermore CDC6-T39A-T368A mutant cells partially but not completely rescued the G1 arrest (Fig. 4C), which indicates that there might be a Mck1-independent Cdc6 regulation mechanism involved in the G1 arrest caused by SDS. It is also possible that other DNA replication proteins and steps are regulated during S-phase in response to stress.
Fig. S6.
WT or Δmck1 cells were arrested and released in G1-phase by α-factor. The cell cycle progression was monitored by FACS analysis in the presence of SDS 0.0075% added at time 0 upon release.
What is known about DNA replication control in response to stress in general? DNA replication is linked to the osmotic stress-response pathway. Hog1 (stress-activated protein kinase, SAPK) phosphorylates Mrc1 to delay origin firing in response to osmotic stress, resulting in the maintenance of genome integrity in yeast (48). Mrc1 phosphorylation by Hog1 is independent of that by the DNA damage checkpoint; therefore, Hog1 and Mrc1 play a role in a novel S-phase checkpoint upon osmotic stress (48). Taken together, these data show that cells use distinct kinases and downstream cell cycle targets to respond to various environmental stresses and maintain genome integrity.
The sustained Cdc6 expression in CDC6-T39A-T368A mutants led to S-phase entry even in the presence of membrane damage caused by SDS, leading to cell lethality (Fig. 4 B and D). A CDC6-T368A single mutation did not cause severe cell growth defects in the presence of SDS (Fig. 4B). It might be interesting to study the role of the Cdc6-T39 phosphorylation site, which might have prominent role in the stress response.
Our results also explain previous observations that cells defective in SCF function (cdc53-1 and cdc34-2 mutants) show SDS sensitivity (49). First, Cdc6 phosphorylation is recognized by the SCFCDC4 complex for its degradation (22, 23, 29, 31), suggesting that the SCF mutants cause Cdc6 stabilization. Furthermore, Sic1 is also targeted for its protein degradation by the SCF complex (18). Therefore, Sic1 is ectopically accumulated in cells defective in SCF function. Thus, SCF function seems to be critical for cell cycle arrest as well as for initiating a wound-healing response.
The sensitivity of the CDC6-T39A-T368A mutant to SDS was more severe than that in Δsic1, indicating that Cdc6 degradation might be the primary mechanism to arrest the cell cycle during G1 (Fig. 4D). DNA rereplication could trigger chromosome instability, which is a hallmark of tumorigenesis in higher eukaryotes (50). The DNA replication protein Cdc6 has to be degraded after origin licensing to prevent DNA rereplication in yeast (24, 51–53). In human, Cdc6 is overexpressed in brain tumors (54), lung carcinomas (55), and lymphomas (56), indicating the importance of Cdc6 protein levels in tumorigenesis. An ectopic expression of CDC6 induces DNA replication in quiescent cells (57) and DNA rereplication in tumor cells (58). To our knowledge, this is the first evidence suggesting that plasma membrane damage promotes a checkpoint-like mechanism inhibiting DNA replication. Given that the players involved in plasma membrane damage-dependent inhibition of DNA replication are evolutionarily conserved, the cell cycle and DNA replication control under plasma membrane stress should be investigated in higher eukaryotes. Thus, linking the stress response to DNA replication and cell cycle control will offer insights into the mechanism for control of cancers.
Materials and Methods
Plasmids and Strains.
Standard methods were used for mating, tetrad dissection, and transformation. All strains listed in Table S1 are congenic with W303 unless noted.
Table S1.
Yeast strains used in this study
| Strain | Genotype | Source |
| RUY508 | MATa bar1 ura3 leu2 his3 trp1 can1-100 | A.E.I. (Brooklyn College, Brooklyn, NY) |
| BCY574 | MATa cdc7-4 trp ura leu ade his | J. Diffley (41) |
| YKK2253 | MATa SIC1-13MYC::HIS3 his3 leu2 met15, ura3 | K.K. (Nagoya City University, Nagoya, Japan) |
| YKK2400 | MATa CLB5-13MYC::HIS3 his3 leu2 met15 ura3 | K.K. |
| RUY267 | MATa CLB5-9MYC::TRP ade2 ura3 leu2 his3 trp1 can1-100 | VAY106 (F. Cross, The Rockefeller University, New York) |
| BCY366 | MATa bar SLD2-9MYC::TRP ADE2 | Present study |
| BCY355 | MATa bar1::hisG ade2 ura3 leu2 his3 trp1 can1-100 | DOM0090 (D. Morgan, University of California, San Francisco) |
| BCY077 | MATa bar1 CDC6-prA::HIS3 ade2 ura3 leu2 his3 trp1 can1-100 | A.E.I. |
| BCY078 | MATa bar1 mck1::KanMX CDC6-prA::HIS3 ade2 ura3 leu2 his3 trp1 can1-100 | A.E.I. |
| BCY461 | MATa CDC6-GFP::HIS3 ura3 leu2 his3 trp1 can1-100 | A.E.I. |
| BCY463 | MATa mck1::KanMX CDC6-GFP::HIS3 ura3 leu2 his3 trp1 can1-100 | A.E.I. |
| BCY234 | MATa bar1 URA3::GAL-CDC6(s) ADE2 | A.E.I. |
| BCY581 | MATa bar1 CDC6-T39A-T368A-prA::HIS3 ade2 ura3 leu2 his3 trp1 can1-100 | A.E.I. |
| BCY615 | MATa bar1 cdc7-4 CDC6-T39A-T368A-prA::HIS3 ade2 ura3 leu2 his3 trp1 can1-100 | Present Study |
| BCY061 | MATa mck1::KanMX ade2 ura3 leu2 his3 trp1 can1-100 | A.E.I. |
| BCY639 | MATa sic1::KanMX CDC6-T39A-T368A-prA::HIS3 ade2 ura3 leu2 his3 trp1 can1-100 | Present study |
| YKK1404 | MATa sic1::KanMX his3 leu2 met15 ura3 | K.K. |
| RUY315 | MATa ORC6-prA::HIS3 ade2 ura3 leu2 his3 trp1 can1-100 | A.E.I. |
| RUY266 | MATa CLB2-9MYC::TRP ade2 ura3 leu2 his3 trp1 can1-100 | VAY100 (F. Cross) |
| BCY101 | MATa bar1 MCK1-9MYC:TRP ade2 ura3 leu2 his3 trp1 can1-100 | A.E.I. |
| BCY282 | MATa cdc4-1 CDC6-prA::HIS3 ADE ade2 ura3 leu2 his3 trp1 can1-100 | A.E.I. |
| RUY004 | MATa bar1 LEU2::ORC6-wt lys2 ade2 ura3 leu2 his3 trp1 can 1 | GW185-3-2 [S. Bell (60)] |
| RUY005 | MATa bar1 LEU2::ORC6-rxl lys2 ade2 ura3 leu2 his3 trp1 can 1 | GW186-3-2 [S. Bell (60)] |
| RUY006 | MATa bar1 LEU2::ORC6-ps lys2 ade2 ura3 leu2 his3 trp1 can 1 | GW187-3-2 [S. Bell (60)] |
| BCY466 | MATa LEU2::ORC6-wt URA3::GAL-CDC6 ade2 ura3 leu2 his3 trp1 can1-100 | Present study |
| BCY467 | MATa LEU2::ORC6-rxl URA3::GAL-CDC6 ade2 ura3 leu2 his3 trp1 can1-100 | Present study |
| BCY468 | MATa LEU2::ORC6-ps URA3::GAL-CDC6 ade2 ura3 leu2 his3 trp1 can1-100 | Present study |
| BCY469 | MATa LEU2::ORC6-wt URA3::GAL-CDC6-T368A ade2 ura3 leu2 his3 trp1 can1-100 | Present study |
| BCY470 | MATa LEU2::ORC6-rxl URA3::GAL-CDC6-T368A ade2 ura3 leu2 his3 trp1 can1-100 | Present study |
| BCY471 | MATa LEU2::ORC6-ps URA3::GAL-CDC6-T368A ade2 ura3 leu2 his3 trp1 can1-100 | Present study |
Cell Cycle Block and Release.
Log-phase cells (OD = 0.3–0.4) were blocked by incubating for 2 h with the addition of α-factor (10−7 M). Cells were washed three times with YPD and then placed into fresh YPD media to release the block. For the cdc7-4 strain, cells were incubated to log-phase at 23 °C and blocked with α-factor, as described above. The temperature was shifted to 36 °C for 1 h upon α-factor release to create an additional block. The cell cycle was released again by shifting the temperature back to 23 °C. To induce mitotic block, cells were treated with nocodazole at the concentration of 15 μg/mL for 2 h.
SDS Experiment.
SDS at the concentration of 0.0075% was used unless indicated. BY4741 is less sensitive to SDS and was treated at 0.02% in Fig. 2.
Western Blotting.
Cells were lysed by agitation in SDS sample buffer with glass beads using a FastPrep (MP Biomedicals) for 20 s, twice, at speed 6. The protein was separated by SDS/PAGE with 10% (wt/vol) polyacrylamide gel. Western blot analysis was performed using an anti-cMYC antibody 9E10 (Sigma) at 1:2,000 dilution, anti-cMYC antibody A-14 (Santa Cruz) at 1:1,000 dilution, anti-Cdc2 (PSTAIRE) antibody (Santa Cruz) at 1:1,500 dilution, anti-Pgk1 antibody (Life Technologies) at 1:2,000 dilution as a loading control, anti-Tub1 antibody (AbD Serotec) at 1:5,000 dilution as an additional loading control. Protein-A–tagged proteins were probed using HRP-conjugated anti-rabbit IgG antibody (Sigma) at 1:5,000 dilution.
Fluorescence Microscopy.
The laser-damage experiment was performed as previously described (7), with modifications as follows. A yeast culture grown overnight was refreshed and incubated for an additional 2–6 h until OD600 reached 0.1–0.3. Cells were then spotted onto an agarose bed (SD medium + 1.2% agarose) on glass slides. An LSM 780 NLO confocal laser scanning microscopy (Carl Zeiss) was used to create the damage and to monitor the fluorescent and bright-field images. Signal quantification was performed using FIJI software.
Kinase Assay.
Cdc28/Clb5-13myc was immunopurified using anti-myc (9E10) antibody. Kinase reactions were performed as described previously (59). Histone H1 (Roche) was used as a substrate.
Plasma Membrane Integrity Assay.
Yeast strains were grown to early-log phase (0.1–0.2 OD600) in YPD media, transferred to YPD supplemented with 0.0075% SDS for 16 h. OD600 = 1.5 equivalent of cells were collected and washed once with YPD, then resuspended in YPD including DAPI. Cells were then observed under a fluorescence microscope.
Acknowledgments
We thank Atsuya Nishiyama, Nube Ramon, and Hsin-I Wen for technical support. This study was supported in part NIH Grant 5SC3GM105498 (to A.E.I.); PSC CUNY enhanced award (to A.E.I.); and JSPS Grants 25112520 (to K.K.) and 26250027 (to M.N.). A portion of this work was conducted in the Institute of Transformative Bio-Molecules at Nagoya University, supported by the Japan Advanced Plant Science Network.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.J.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523824113/-/DCSupplemental.
References
- 1.Sonnemann KJ, Bement WM. Wound repair: Toward understanding and integration of single-cell and multicellular wound responses. Annu Rev Cell Dev Biol. 2011;27:237–263. doi: 10.1146/annurev-cellbio-092910-154251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bickle M, Delley PA, Schmidt A, Hall MN. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 1998;17(8):2235–2245. doi: 10.1093/emboj/17.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delley PA, Hall MN. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J Cell Biol. 1999;147(1):163–174. doi: 10.1083/jcb.147.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levin DE, Fields FO, Kunisawa R, Bishop JM, Thorner J. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell. 1990;62(2):213–224. doi: 10.1016/0092-8674(90)90360-q. [DOI] [PubMed] [Google Scholar]
- 5.Nonaka H, et al. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14(23):5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics. 2011;189(4):1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kono K, Saeki Y, Yoshida S, Tanaka K, Pellman D. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell. 2012;150(1):151–164. doi: 10.1016/j.cell.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 8.Johnson A, Skotheim JM. Start and the restriction point. Curr Opin Cell Biol. 2013;25(6):717–723. doi: 10.1016/j.ceb.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartwell LH. Saccharomyces cerevisiae cell cycle. Bacteriol Rev. 1974;38(2):164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doncic A, Falleur-Fettig M, Skotheim JM. Distinct interactions select and maintain a specific cell fate. Mol Cell. 2011;43(4):528–539. doi: 10.1016/j.molcel.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costanzo M, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117(7):899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 12.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12(5):1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrews BJ, Herskowitz I. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989;342(6251):830–833. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- 14.Cross FR, Hoek M, McKinney JD, Tinkelenberg AH. Role of Swi4 in cell cycle regulation of CLN2 expression. Mol Cell Biol. 1994;14(7):4779–4787. doi: 10.1128/mcb.14.7.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol Cell. 2011;43(4):515–527. doi: 10.1016/j.molcel.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14(8):518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nash P, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414(6863):514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 18.Verma R, Feldman RM, Deshaies RJ. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997;8(8):1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 20.Araki H. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol. 2010;22(6):766–771. doi: 10.1016/j.ceb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 21.Boos D, Frigola J, Diffley JF. Activation of the replicative DNA helicase: Breaking up is hard to do. Curr Opin Cell Biol. 2012;24(3):423–430. doi: 10.1016/j.ceb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Perkins G, Drury LS, Diffley JF. Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 2001;20(17):4836–4845. doi: 10.1093/emboj/20.17.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drury LS, Perkins G, Diffley JF. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16(19):5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol Biol Cell. 1999;10(10):3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labib K, Diffley JF, Kearsey SE. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat Cell Biol. 1999;1(7):415–422. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen VQ, Co C, Irie K, Li JJ. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr Biol. 2000;10(4):195–205. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411(6841):1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 28.Vas A, Mok W, Leatherwood J. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol Cell Biol. 2001;21(17):5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drury LS, Perkins G, Diffley JF. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr Biol. 2000;10(5):231–240. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- 30.Ikui AE, Rossio V, Schroeder L, Yoshida S. A yeast GSK-3 kinase Mck1 promotes Cdc6 degradation to inhibit DNA re-replication. PLoS Genet. 2012;8(12):e1003099. doi: 10.1371/journal.pgen.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Zain A, Schroeder L, Sheglov A, Ikui AE. Cdc6 degradation requires phosphodegron created by GSK-3 and Cdk1 for SCFCdc4 recognition in Saccharomyces cerevisiae. Mol Biol Cell. 2015;26(14):2609–2619. doi: 10.1091/mbc.E14-07-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dailey D, et al. Novel yeast protein kinase (YPK1 gene product) is a 40-kilodalton phosphotyrosyl protein associated with protein-tyrosine kinase activity. Mol Cell Biol. 1990;10(12):6244–6256. doi: 10.1128/mcb.10.12.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puziss JW, Hardy TA, Johnson RB, Roach PJ, Hieter P. MDS1, a dosage suppressor of an mck1 mutant, encodes a putative yeast homolog of glycogen synthase kinase 3. Mol Cell Biol. 1994;14(1):831–839. doi: 10.1128/mcb.14.1.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shero JH, Hieter P. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1) Genes Dev. 1991;5(4):549–560. doi: 10.1101/gad.5.4.549. [DOI] [PubMed] [Google Scholar]
- 35.Griffioen G, Swinnen S, Thevelein JM. Feedback inhibition on cell wall integrity signaling by Zds1 involves Gsk3 phosphorylation of a cAMP-dependent protein kinase regulatory subunit. J Biol Chem. 2003;278(26):23460–23471. doi: 10.1074/jbc.M210691200. [DOI] [PubMed] [Google Scholar]
- 36.Hilioti Z, et al. GSK-3 kinases enhance calcineurin signaling by phosphorylation of RCNs. Genes Dev. 2004;18(1):35–47. doi: 10.1101/gad.1159204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizunuma M, Hirata D, Miyaoka R, Miyakawa T. GSK-3 kinase Mck1 and calcineurin coordinately mediate Hsl1 down-regulation by Ca2+ in budding yeast. EMBO J. 2001;20(5):1074–1085. doi: 10.1093/emboj/20.5.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirata Y, Andoh T, Asahara T, Kikuchi A. Yeast glycogen synthase kinase-3 activates Msn2p-dependent transcription of stress responsive genes. Mol Biol Cell. 2003;14(1):302–312. doi: 10.1091/mbc.E02-05-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463(7277):113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24(1):101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bousset K, Diffley JF. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12(4):480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verma R, et al. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278(5337):455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 43.Shirayama M, Tóth A, Gálová M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402(6758):203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- 44.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445(7125):281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 45.Cross FR, Schroeder L, Bean JM. Phosphorylation of the Sic1 inhibitor of B-type cyclins in Saccharomyces cerevisiae is not essential but contributes to cell cycle robustness. Genetics. 2007;176(3):1541–1555. doi: 10.1534/genetics.107.073494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verna J, Lodder A, Lee K, Vagts A, Ballester R. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94(25):13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kapuscinski J. DAPI: A DNA-specific fluorescent probe. Biotech Histochem. 1995;70(5):220–233. doi: 10.3109/10520299509108199. [DOI] [PubMed] [Google Scholar]
- 48.Duch A, et al. Coordinated control of replication and transcription by a SAPK protects genomic integrity. Nature. 2013;493(7430):116–119. doi: 10.1038/nature11675. [DOI] [PubMed] [Google Scholar]
- 49.Varelas X, Stuart D, Ellison MJ, Ptak C. The Cdc34/SCF ubiquitination complex mediates Saccharomyces cerevisiae cell wall integrity. Genetics. 2006;174(4):1825–1839. doi: 10.1534/genetics.106.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol. 2007;19(6):663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calzada A, Sánchez M, Sánchez E, Bueno A. The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J Biol Chem. 2000;275(13):9734–9741. doi: 10.1074/jbc.275.13.9734. [DOI] [PubMed] [Google Scholar]
- 52.Archambault V, Ikui AE, Drapkin BJ, Cross FR. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol Cell Biol. 2005;25(15):6707–6721. doi: 10.1128/MCB.25.15.6707-6721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Honey S, Futcher B. Roles of the CDK phosphorylation sites of yeast Cdc6 in chromatin binding and rereplication. Mol Biol Cell. 2007;18(4):1324–1336. doi: 10.1091/mbc.E06-06-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohta S, et al. Cdc6 expression as a marker of proliferative activity in brain tumors. Oncol Rep. 2001;8(5):1063–1066. doi: 10.3892/or.8.5.1063. [DOI] [PubMed] [Google Scholar]
- 55.Karakaidos P, et al. Overexpression of the replication licensing regulators hCdt1 and hCdc6 characterizes a subset of non-small-cell lung carcinomas: Synergistic effect with mutant p53 on tumor growth and chromosomal instability—Evidence of E2F-1 transcriptional control over hCdt1. Am J Pathol. 2004;165(4):1351–1365. doi: 10.1016/S0002-9440(10)63393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinyol M, et al. Unbalanced expression of licensing DNA replication factors occurs in a subset of mantle cell lymphomas with genomic instability. Int J Cancer. 2006;119(12):2768–2774. doi: 10.1002/ijc.22146. [DOI] [PubMed] [Google Scholar]
- 57.Cook JG, et al. Analysis of Cdc6 function in the assembly of mammalian prereplication complexes. Proc Natl Acad Sci USA. 2002;99(3):1347–1352. doi: 10.1073/pnas.032677499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaziri C, et al. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell. 2003;11(4):997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 59.Niida H, et al. Depletion of Chk1 leads to premature activation of Cdc2-cyclin B and mitotic catastrophe. J Biol Chem. 2005;280(47):39246–39252. doi: 10.1074/jbc.M505009200. [DOI] [PubMed] [Google Scholar]
- 60.Wilmes GM, et al. Interaction of the S-phase cyclin Clb5 with an 'RXL' docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004;18(9):981–991. doi: 10.1101/gad.1202304. [DOI] [PMC free article] [PubMed] [Google Scholar]