Significance
Efflux-based drug resistance complicates the treatment of infectious diseases and cancers. Cellular exposure to ecological toxins or reactive metabolites may influence the conservation and activity of efflux pumps. Yet, for most such systems, we know very little about their natural substrates and the signals controlling pump expression. Using diverse approaches, including a chip-based method of electrochemical detection, we show that the endogenous and reactive antibiotic 5-methylphenazine-1-carboxylate (5-Me-PCA) is transported by the efflux pump MexGHI-OpmD in Pseudomonas aeruginosa. Furthermore, we demonstrate that 5-Me-PCA activates expression of its cognate transporter and that it is required for normal P. aeruginosa biofilm morphogenesis. Our results provide insight into mechanisms of self-resistance and determinants of multicellular behavior in this major cause of biofilm-based infections.
Keywords: phenazine, RND efflux, MexGHI-OpmD, biofilm, antibiotic
Abstract
Redox-cycling compounds, including endogenously produced phenazine antibiotics, induce expression of the efflux pump MexGHI-OpmD in the opportunistic pathogen Pseudomonas aeruginosa. Previous studies of P. aeruginosa virulence, physiology, and biofilm development have focused on the blue phenazine pyocyanin and the yellow phenazine-1-carboxylic acid (PCA). In P. aeruginosa phenazine biosynthesis, conversion of PCA to pyocyanin is presumed to proceed through the intermediate 5-methylphenazine-1-carboxylate (5-Me-PCA), a reactive compound that has eluded detection in most laboratory samples. Here, we apply electrochemical methods to directly detect 5-Me-PCA and find that it is transported by MexGHI-OpmD in P. aeruginosa strain PA14 planktonic and biofilm cells. We also show that 5-Me-PCA is sufficient to fully induce MexGHI-OpmD expression and that it is required for wild-type colony biofilm morphogenesis. These physiological effects are consistent with the high redox potential of 5-Me-PCA, which distinguishes it from other well-studied P. aeruginosa phenazines. Our observations highlight the importance of this compound, which was previously overlooked due to the challenges associated with its detection, in the context of P. aeruginosa gene expression and multicellular behavior. This study constitutes a unique demonstration of efflux-based self-resistance, controlled by a simple circuit, in a Gram-negative pathogen.
Although efflux pumps are notorious for transporting clinically relevant antibiotics, there is great interest in identifying the natural substrates and inducers of these systems (1, 2). In some Gram-positive Streptomyces species, known for their production of small-molecule antibiotics and pigments, genes for antibiotic biosynthetic enzymes are found in large clusters, which can also contain genes encoding efflux pumps and regulatory proteins that control expression of adjacent loci. Efflux pumps have been implicated in the transport of, and resistance to, antibiotics such as actinorhodin, daunorubicin, and oxytetracycline in their streptomycete producers (3–6). In several cases, either the antibiotic itself, an intermediate in antibiotic synthesis, or compounds with structural similarities to the antibiotic have been demonstrated or inferred to be inducers of pump expression (3, 4, 7, 8).
For most well-studied efflux pumps from Gram-negative bacteria, on the other hand, the hypothesized natural or ecologically relevant substrates remain mysterious. Recent evidence suggests that Pseudomonas aeruginosa MexAB-OprM and MexEF-OprN transport intercellular signals and/or intermediates formed during their synthesis (9, 10). Two systems in enteric bacteria—the VexAB pump of Vibrio cholerae and the Acr(Z)AB-TolC pump of Escherichia coli—have been linked to protection from accumulated metabolic intermediates and fatty acids and bile salts found in the gut (11–14). In the enteric systems, regulators have been identified that mediate induction of these pumps in response to their substrates (11, 15). However, discrete mechanistic connections between activators, regulators, and efflux pumps have not been clarified in most cases, especially for endogenously produced substrates.
P. aeruginosa is an opportunistic pathogen that causes biofilm-based infections with devastating effects (16). This Gram-negative bacterium synthesizes phenazines, redox-cycling antibiotics that affect redox homeostasis and gene expression in both the producer and hosts (17–22). Several years ago, our group showed that pyocyanin (5-N-methyl-1-hydroxyphenazine), the most well-known P. aeruginosa phenazine (23), activates the redox-active transcription factor SoxR and thereby induces expression of a small regulon that includes the RND (resistance-nodulation-division) efflux pump-encoding operon mexGHI-opmD (hereafter referred to as “mex”) (24, 25). As this was, to our knowledge, the first demonstration of SoxR homolog activation by an endogenous metabolite, it led to a reevaluation of the physiological activators of SoxR in E. coli, where previous studies had led to a long-standing model of SoxR activation by superoxide (typically generated by xenobiotics) (26–29). Although still under discussion in the community (30–33), the current prevailing view is that endogenously produced or exogenously added redox-cycling compounds activate SoxR by direct oxidation of its iron–sulfur cluster and that the SoxR targets, or targets of transcription factors controlled by SoxR, function to protect cells from at least some of these inducing compounds (28, 34).
We recognized the potential of the P. aeruginosa phenazine/SoxR/MexGHI-OpmD system as a model for regulation of endogenous natural product transport and self-protection in a Gram-negative producer of antibiotics. Initial characterizations have implicated MexGHI-OpmD in the export of several types of compounds: xenobiotics, including the antibiotic norfloxacin and the heterocyclic dye acriflavine (35); a precursor or derivative of the Pseudomonas quinolone signal (PQS) (36); and uncharacterized red pigments thought to be derived from 5-methylphenazine-1-carboxylic acid (5-Me-PCA) (37). 5-Me-PCA, a reactive compound with a higher redox potential than the well-studied P. aeruginosa phenazines, has not been fully purified from wild-type P. aeruginosa cultures, although various observations indicate that it is the intermediate between phenazine-1-carboxylate (PCA) and pyocyanin in the phenazine biosynthetic pathway (38–40). Among these are the facts that the red phenazine aeruginosin A (5-methyl-7-amino-1-carboxyphenazinium betaine) is overproduced by P. aeruginosa phzS mutants (41, 42), which lack the hydroxylase required to convert 5-Me-PCA to pyocyanin (43, 44), and that E. coli expressing phzM converts PCA into a red pigment (42). Interestingly, the gene required for production of 5-Me-PCA from PCA, phzM, is located next to the mex operon on the P. aeruginosa chromosome (Fig. 1A), an arrangement that is common for genes encoding (i) efflux pumps and (ii) biosynthetic enzymes that generate efflux pump substrates (8). This genetic organization prompts the hypothesis that 5-Me-PCA is the natural substrate of MexGHI-OpmD.
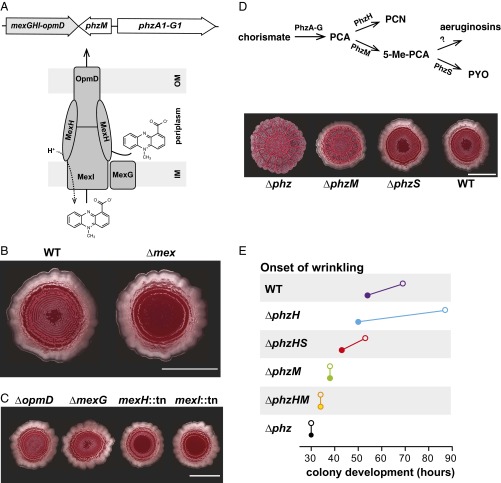
Fig. 1.
MexGHI-OpmD and specific P. aeruginosa phenazines contribute to P. aeruginosa PA14 colony development. (A) Illustrations depicting the genomic organization of the mex operon and predicted subunit arrangement of the encoded efflux pump. (B) PA14 wild type (WT) and ∆mex mutant colonies grown for 65 h in the colony morphology assay. (Scale bar, 1 cm.) (C) Mex component mutant colonies grown for 65 h. (Scale bar, 1 cm.) (D) 5-Me-PCA, but not pyocyanin, is required for P. aeruginosa PA14 WT colony morphology. (Top) Abbreviated P. aeruginosa phenazine biosynthetic pathway. PCN, phenazine-1-carboxamide; PYO, pyocyanin. (Bottom) Images of 3-d-old colony biofilms grown on colony morphology assay medium. (Scale bar, 1 cm.) (E) Onset of wrinkling in colonies of phenazine biosynthetic mutant (or wild-type) strains (closed circles) and their ∆mex counterparts (open circles). Colony images and wrinkling data are representative of three independent experiments.
In this study, we investigated the role of MexGHI-OpmD in P. aeruginosa PA14 during colony biofilm morphogenesis on agar-solidified medium, a process strongly influenced by phenazine production, and during growth in liquid culture. These analyses used diverse approaches including an electrochemical method for specific detection of naturally produced 5-Me-PCA released from multicellular PA14 communities. Results of these studies suggest that 5-Me-PCA and the structurally similar, synthetic compound phenazine methosulfate (PMS) are substrates of MexGHI-OpmD. Our observations also indicate that, among the phenazines produced by P. aeruginosa, 5-Me-PCA and 5-Me-PCA derivatives other than pyocyanin lead to the highest induction of mex and that this operon is required for 5-Me-PCA resistance. We present results supporting the hypothesis that 5-Me-PCA is the primary physiological substrate of MexGHI-OpmD and demonstrate a role for this efflux pump in P. aeruginosa self-resistance to a relatively high-potential and reactive product.
Results
MexGHI-OpmD and 5-Me-PCA Play Roles in P. aeruginosa PA14 Colony Morphogenesis.
In published work, we have used a colony morphology assay to study the contributions of SoxR, SoxR targets [including the mexGHI-opmD operon (mex)], and phenazines to P. aeruginosa PA14 biofilm development. In this assay, a 10-μL droplet of a PA14 cell suspension is used to inoculate agar-solidified medium containing the dye Congo red, which binds to biofilm matrix (19, 37). Over several days of incubation at room temperature, wild-type PA14 first forms a smooth colony. Concentric ridges appear on the colony surface starting on day 3. We have previously reported that the initiation of wrinkling is delayed in PA14 ∆mex mutant colonies relative to those of the wild type (Fig. 1B), which we interpret as a phenazine-toxicity effect, as this mutant also shows an extended lag phase in liquid culture but only if the mutant can also produce phenazines (37).
MexI, MexH, and OpmD are inner membrane, periplasmic, and outer membrane proteins (Fig. 1A), respectively, that bear homology to their respective counterparts in other RND-family efflux pumps. MexG is an anomalous cytoplasmic membrane component with unknown function. We tested mutants with disruptions in each gene of the mex operon and found that MexH and MexI were required for wild-type colony morphology, whereas OpmD and MexG were not (Fig. 1C). Complementation of the mexH transposon-insertion strain by chromosomal restoration of the wild-type allele showed that this phenotype could be attributed to disruption of the mex operon (SI Appendix, Fig. S1). The dispensability of OpmD is consistent with prior observations that outer membrane components of RND efflux pumps are interchangeable; given that the P. aeruginosa genome encodes several of these proteins, heterologous complementation by other porins is likely (45).
We have also previously reported that a PA14 mutant unable to produce PCA and therefore all phenazines (∆phz) exhibits a unique colony morphology phenotype in that it begins to wrinkle on day 2, forming disorganized ridges in the colony center that are surrounded by radial spokes emanating from the center (Fig. 1D) (19, 37). As phenazines have been shown to support intracellular redox balancing and survival in electron acceptor-limited PA14 cultures (17, 46), we interpret the ∆phz phenotype as a response to oxidant limitation that increases the colony surface-to-volume ratio and access to oxygen for resident cells. Phenazines in wild-type biofilms that accept electrons from cells in anoxic biofilm subzones could (i) diffuse into oxic zones to react with oxygen and/or (ii) transfer electrons to other phenazines repeatedly until they are ultimately transferred to oxygen in the oxic zone. Either of these scenarios would allow cells in anoxic biofilm subzones to use phenazines as mediators to access electron acceptors that are not available in the immediate environment (19).
We tested whether 5-Me-PCA or other PA14 phenazines make differential contributions to biofilm morphogenesis by growing mutants with disruptions in the phenazine biosynthetic pathway in the colony morphology assay. The combinations of phenazines produced by these mutants can be discerned from the biosynthetic pathway depicted in Fig. 1D. [Additional derivatives that may be produced as side products of this pathway, such as phenazine-1,6-dicarboxylic acid and 1-hydroxyphenazine, are not represented (42, 47).] The ∆phzM biofilm exhibited similar wrinkling but less spreading compared with the ∆phz biofilm, suggesting that PCA and/or PCN inhibit spreading but that they are not sufficient for wild-type colony development (Fig. 1D). Surprisingly, the ∆phzS colony biofilm, which produced 5-Me-PCA in addition to PCA and PCN, showed a wild-type morphology. This result was unexpected because the product of PhzS, pyocyanin, is the best studied P. aeruginosa phenazine and supports survival in liquid-culture models under oxygen-limited conditions considered relevant for biofilms (17, 46). The ∆phzS colony morphology phenotype observed here reveals the importance of 5-Me-PCA production in biofilm morphogenesis.
The mex operon is adjacent to the phzM gene (Fig. 1A), encoding the N-methyltransferase presumed to convert PCA to 5-Me-PCA (43), in the P. aeruginosa genome, suggesting that 5-Me-PCA is a substrate of MexGHI-OpmD. We created combinatorial mutants lacking the mex operon and genes involved in phenazine biosynthesis and examined their development in the colony morphology assay (Fig. 1E). We found that the deletion of mex leads to delayed wrinkling solely in strains capable of producing 5-Me-PCA (i.e., strains that contain the phenazine biosynthetic enzymes PhzA-G and PhzM) (43, 44).
MexGHI-OpmD Contributes to Fitness in Shaken Liquid Cultures When N-Methylated Phenazines Are Produced.
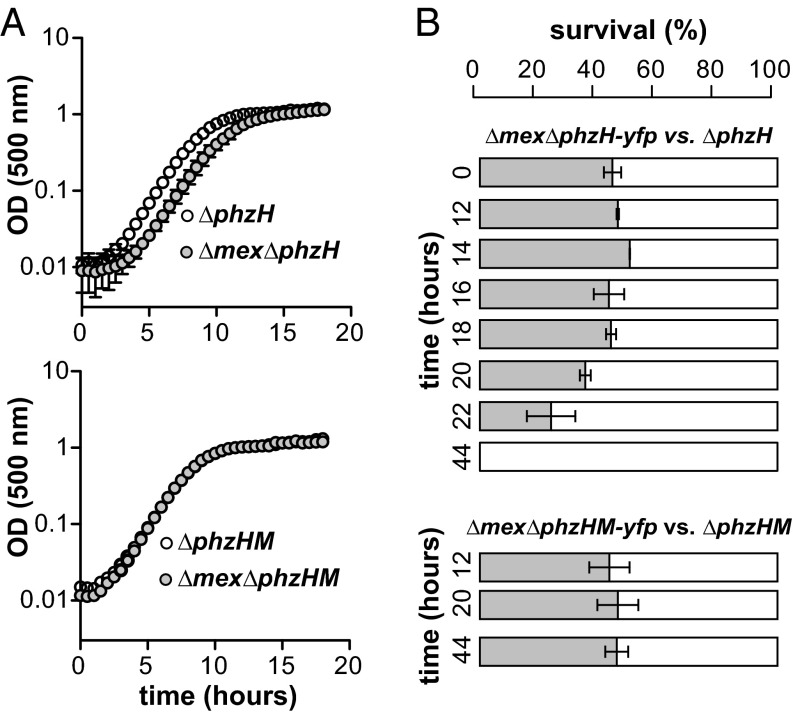
To further examine the role of MexGHI-OpmD in the context of 5-Me-PCA production, we grew combinatorial mutants affected in phenazine biosynthesis and MexGHI-OpmD activity in liquid cultures, individually and in competition. All mutants examined in these assays were derived from the ∆phzH parent strain, which is incapable of producing PCN and therefore may shunt more PCA toward PhzM-catalyzed N-methylation (48). In pure cultures, we found that the ∆mex∆phzH mutant exhibited an extended lag phase relative to the ∆phzH mutant, whereas the ∆mex∆phzHM mutant exhibited growth identical to that of ∆phzHM (Fig. 2A). In competition assays, the ∆mex∆phzH mutant showed a fitness disadvantage when competed against ∆phzH, whereas the ∆mex∆phzHM mutant showed no disadvantage when competed against ∆phzHM. A time course of colony-forming unit (cfu) counts confirmed that the benefit of MexGHI-OpmD arises specifically in late stationary phase in strains with an intact phzM gene (Fig. 2B). In these experiments, phenazine production became detectable by HPLC analysis at the 12-h time point, ∼6 h before the disadvantage of the ∆mex∆phzH mutant became detectable by cfu counts. Control experiments competing each strain against itself and reversing the strain tagging (using the YFP label on the opposite strain) further supported this conclusion (SI Appendix, Fig. S2). These results suggest that 5-Me-PCA and/or its derivative is a substrate of MexGHI-OpmD and that the accumulation of this compound is toxic to PA14.
Fig. 2.
MexGHI-OpmD is required for wild-type fitness when N-methylated phenazines are produced. (A) Growth of PA14 mutant cultures in liquid LB medium. Cultures were inoculated with samples from stationary-phase precultures. (B) Percentage of the population represented by strains with intact or deleted mex genes after coculturing for the time indicated in liquid 1% tryptone medium. Phenazines were first detected in the culture at 12 h. Error bars indicate SD of the mean for three replicates.
The Synthetic Phenazine PMS Modulates PA14 Colony Development at Subinhibitory Concentrations.
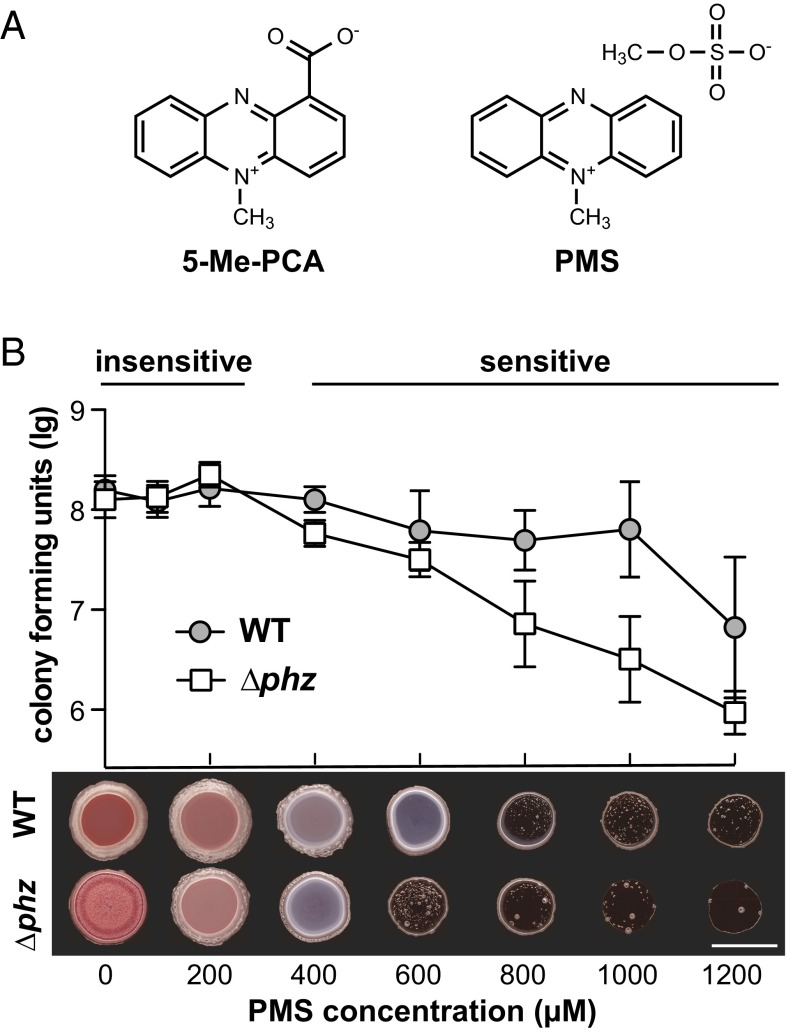
5-Me-PCA is a reactive compound that is difficult to purify and maintain in the laboratory (39, 40). Previous studies of 5-Me-PCA metabolism have therefore used the structurally similar, synthetic phenazine PMS (49) (Fig. 3A). We wondered whether, at subinhibitory concentrations, PMS would have an effect similar to that of 5-Me-PCA in the colony morphology assay. We grew wild-type and ∆phz biofilms on agar-solidified media containing a range of concentrations of PMS, then resuspended the biofilms and plated for cfus. PMS at 100–200 µM, a range comparable to concentrations observed for natural phenazines (18), did not adversely affect the viability of cells in ∆phz colonies (Fig. 3B). At 200 µM, PMS promoted smooth colony formation in the ∆phz mutant. This result shows that low, noninhibitory concentrations of PMS can substitute for endogenously produced phenazines in preventing the wrinkled colony morphology characteristic of the phenazine-nonproducing mutant. However, at higher PMS concentrations, we observed an effect on biofilm coloration (less Congo red binding) and a drop in cfus that was more pronounced in ∆phz. The increased resistance of the wild type (compared with ∆phz) to higher concentrations of PMS may arise from preinduction of self-resistance mechanisms by endogenous phenazines in cultures used to inoculate PMS plates.
Fig. 3.
PMS modulates PA14 colony morphology at subinhibitory concentrations. (A) Structures of 5-Me-PCA and PMS. (B, Top) Quantification of cells (expressed as log10 of cfus) present in PA14 biofilms grown on media containing various concentrations of PMS. Error bars represent SD of the mean for three replicates. (Bottom) Images of colonies grown for 2 d on media containing PMS as indicated, representative of three independent experiments. (Scale bar, 1 cm.)
Select PA14 SoxR Regulon Mutants Show Sensitivity to PMS.
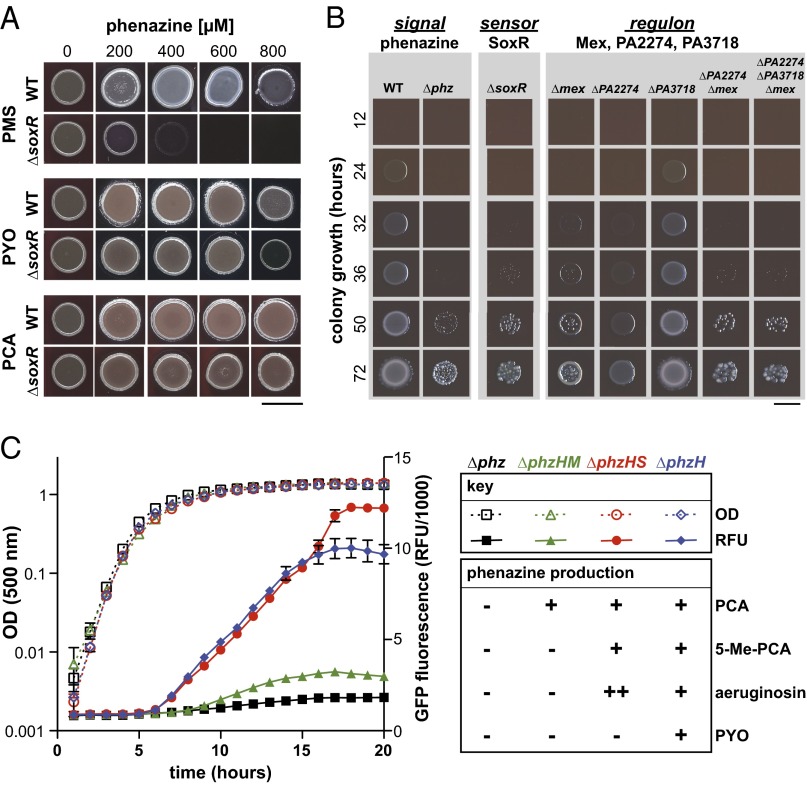
In a recent study examining the physiology of the SoxR-mediated response in P. aeruginosa, the phenazines pyocyanin, PCA, and PMS were found to activate SoxR-dependent gene expression (32). However, a P. aeruginosa ∆soxR mutant did not show increased sensitivity to pyocyanin compared with the wild type in a toxicity assay. Here, we further characterized the SoxR-mediated response to individual phenazines by first growing the P. aeruginosa PA14 wild type and a ∆soxR mutant on agar-solidified media containing various concentrations of pyocyanin, PCA, and PMS. We also tested sensitivity to paraquat, historically recognized as a SoxR inducer in E. coli (26, 27). Paraquat is a broadly toxic, synthetic redox-cycling compound that generates intracellular reactive oxygen species. We found that the ∆soxR mutant showed little or no sensitivity to PCA, pyocyanin, and paraquat but a strong sensitivity to PMS (Fig. 4A and SI Appendix, Fig. S3A). Wild-type PA14 showed no sensitivity to PMS.
Fig. 4.
N-methylated phenazines other than pyocyanin are toxic to SoxR-regulon and ∆phz mutants and maximally induce expression of the mexGHI-opmD operon. (A) Growth of PA14 WT and ∆soxR mutant on media containing various concentrations of PMS, PYO, or PCA. (Scale bar, 5 mm.) (B) Growth of SoxR-regulon mutants on medium containing 600 µM PMS. (Scale bar, 1 cm.) Images are representative of three independent experiments. (C) Strains that contain deletions in phenazine biosynthetic genes and express GFP under the control of the SoxR-dependent mex promoter were grown in liquid MOPS–LB medium. Open symbols represent OD values, whereas closed symbols represent GFP fluorescence values. Relative amounts of each phenazine produced are indicated in the Right panel.
Next, we tested which of the SoxR-target loci contribute to PMS resistance by growing various mutants on agar-solidified medium containing this phenazine (Fig. 4B). P. aeruginosa SoxR regulates three targets: the mexGHI-opmD operon (PA4205-PA4208) (mex); PA2274, which encodes a putative monooxygenase; and PA3718, which encodes a major facilitator superfamily (MFS) transporter (24) (ORF numbers refer to those from P. aeruginosa PAO1). The individual ∆mex and ∆PA2274 mutants showed growth defects, but these were less severe than that of ∆soxR. Growth of the ∆PA3718 mutant was identical to that of the wild type. The defect of the ∆mex∆PA2274 double mutant was more severe than that of the individual ∆mex and ∆PA2274 mutants. Growth phenotypes of the ∆mex∆PA2274 double mutant and the triple mutant lacking all SoxR targets were identical to each other and to the growth phenotype of the ∆soxR mutant, demonstrating that the combination of mex and PA2274 is sufficient for wild-type PMS resistance. Finally, ∆phz showed a growth delay in this assay that was comparable to that of the ∆soxR mutant (Fig. 4B). Because ∆phz does not display a growth delay in the absence of PMS (SI Appendix, Fig. S4), we speculate that SoxR is induced by endogenous phenazines in liquid cultures and thereby confers protection when these cultures are spotted on a PMS-containing medium. The sensitivities of SoxR target mutants were also demonstrated by a filter disk assay for PMS-dependent growth inhibition of ∆phz strains in which soxR or SoxR target genes were also deleted (SI Appendix, Fig. S3B). Notably, ∆PA2274 had a defect when grown in the presence of PMS but not on PMS-free medium (Fig. 4B and SI Appendix, Fig. S3B versus Fig. S4) (37).
PhzM, but Not PhzS, Is Necessary and Sufficient for Full Induction of mexGHI-opmD.
The roles of MexGHI-OpmD in supporting PA14 biofilm morphogenesis and in protection from N-methylated phenazines raise the question of whether 5-Me-PCA and possibly also its derivatives other than pyocyanin specifically activate SoxR and thereby stimulate mex expression. To test this question, we moved a reporter construct that expresses GFP under the control of the mex promoter into strains with deletions in various phenazine biosynthetic loci. Analysis of GFP expression during growth in liquid cultures (Fig. 4C and SI Appendix, Fig. S5) and colony biofilms (SI Appendix, Fig. S6) showed increased levels specifically in strains with intact phzM genes, indicating that phzM is necessary and sufficient for full induction of mex. These results support a simple, elegant model in which the PhzM product(s) directly induces expression of its cognate transporter.
MexGHI-OpmD Supports Release of Red 5-Me-PCA Derivatives.
In an earlier study, Aendekerk et al. (36, 50) characterized P. aeruginosa PAO1 mutants with disruptions in mexI and opmD. These mutants showed extended lag phases in liquid culture compared with the wild type, consistent with our previously published (37) and present work, and were defective in the production of 2-heptyl-3-hydroxy-4-quinolone (i.e., PQS). The growth phenotypes of combinatorial PAO1 mexI mutants with defects in PQS biosynthesis led Aendekerk et al. to implicate MexGHI-OpmD in the export of the PQS precursor anthranilate or a toxic derivative thereof (36). However, we measured the levels of PQS in P. aeruginosa PA14 wild type and ∆mex mutant whole colonies and found that PQS production in our system was unaffected by deletion of mexGHI-opmD (SI Appendix, Fig. S7A). We also observed no deficiency in pyocyanin production in the mex mutant of PA14 compared with wild-type PA14, whereas Aendekerk et al. reported that a mex mutant of PAO1 was markedly deficient in pyocyanin release. The discrepancies in these results may have arisen from strain- or condition-dependent effects.
Wild-type PA14 colonies grown on 1% tryptone, 1% agar medium release an uncharacterized combination of red compounds that likely includes the phenazines aeruginosin A and aeruginosin B (5-methyl-7-amino-1-carboxy-3-sulfophenazinium betaine) (38, 39, 41, 42). Red phenazine production requires PhzM and increases when PhzS is not functional, suggesting that these compounds are produced from 5-Me-PCA (42). We prepared aqueous extracts from the agar underlying ∆phzH and ∆phzHM colonies and determined the emission maximum of PhzM-dependent red phenazines to be 620 nm (when excited at 510 nm) (SI Appendix, Fig. S7 B and C). We then used this method to quantify relative levels of red phenazines produced by these mutants and the ∆phzH∆mex mutant. Consistent with our previously reported results (37), disrupting mex led to a marked decrease in the amount of red phenazine released from colonies (SI Appendix, Fig. S7D). These results implicate mex in the transport of 5-Me-PCA and/or 5-Me-PCA-derived red phenazines.
MexGHI-OpmD Supports Release of 5-Me-PCA.
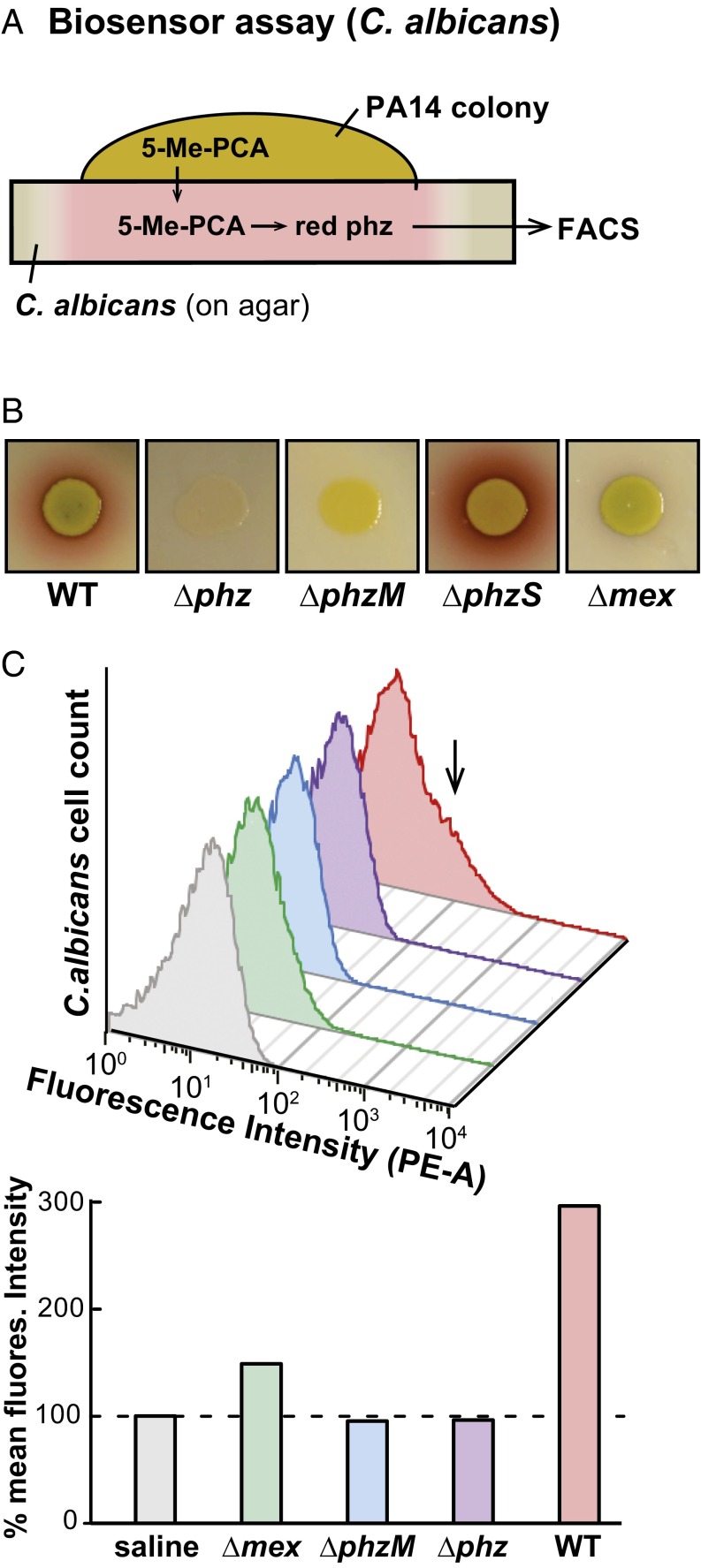
To determine whether 5-Me-PCA itself is a substrate of MexGHI-OpmD, we first used a 5-Me-PCA bioassay in which P. aeruginosa biofilms are grown on top of Candida albicans lawns. Under this condition, C. albicans takes up 5-Me-PCA, which is then modified to yield a red, fluorescent derivative thought to largely arise from covalent linkage of 5-Me-PCA to amine residues on C. albicans intracellular proteins (Fig. 5A) (49, 51). Although wild-type PA14 stimulated the formation of red pigments in C. albicans, ∆mex did not (Fig. 5B). Mutants representing individual MexGHI-OpmD subunits produced results that correlated with their colony morphotypes (SI Appendix, Fig. S8 and Fig. 1C), and complementation of the mexH transposon-insertion strain showed that its phenotype could also be attributed to disruption of the mex operon (SI Appendix, Fig. S8). The inability of ∆mex to release 5-Me-PCA was confirmed by recovering C. albicans cells after coculture with wild type and various P. aeruginosa mutants in this assay and using flow cytometry to count cells exhibiting increased 5-Me-PCA–dependent fluorescence. The mean fluorescence intensity of C. albicans cells cocultured with ∆mex was 75% lower than that of C. albicans cells cocultured with the wild type (Fig. 5C).
Fig. 5.
A C. albicans-based bioassay reveals a role for MexGHI-OpmD in 5-Me-PCA release. (A) Setup diagram of the C. albicans-based biosensor assay for 5-Me-PCA production. (B) PhzM-dependent red phenazine formation is visible when P. aeruginosa PA14 WT and ∆phzS colonies are grown on top of C. albicans lawns, and this coloration is not evident for ∆phz, ∆phzM, and ∆mex colonies. Colonies (∼8 mm in diameter) were imaged after 72 h (24 h at 30 °C and 48 h at room temperature). (C) Fluorescence-activated cell sorting of C. albicans cells cocultured with P. aeruginosa PA14 strains. The arrow indicates increased fluorescence intensity in the phycoerythrin channel (PE-A) consistent with 5-Me-PCA production. Representative data are shown.
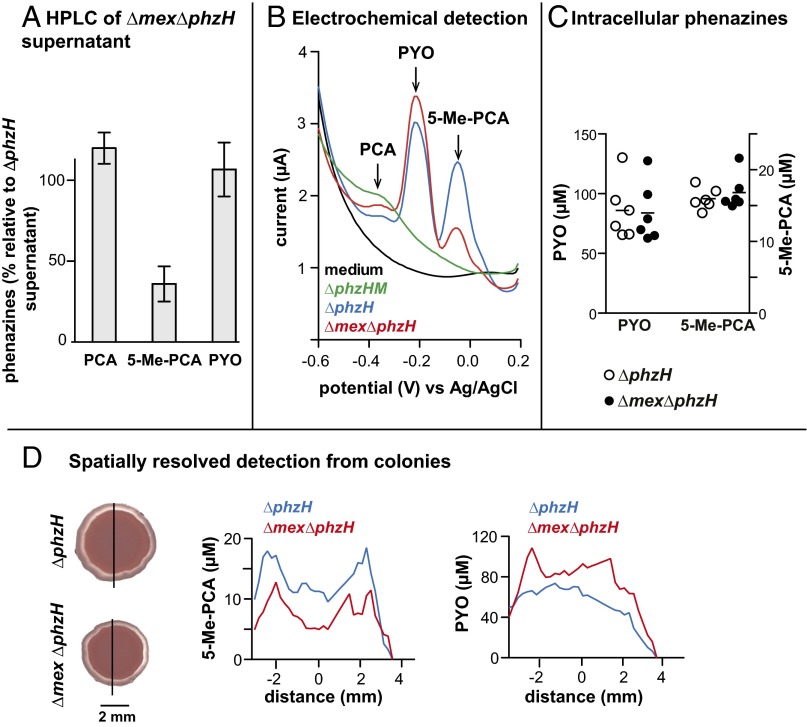
We next endeavored to directly measure 5-Me-PCA release from various strains. Due to its reactivity and polarity, detection of this compound in supernatants and extracts from bacterial cultures has been technically challenging (39, 40). We developed an HPLC-based method for 5-Me-PCA detection and compared liquid culture supernatants from the ∆phzH mutant (expected to produce 5-Me-PCA at levels higher than the wild type) to those obtained from the ∆phzHM mutant (which cannot produce 5-Me-PCA). 5-Me-PCA was identified as a unique peak found specifically in the ∆phzH mutant, eluting at 6 min (SI Appendix, Fig. S9 A and B). Applying our HPLC assay to quantification of phenazines released by the ∆phzH and ∆mex∆phzH mutants, we detected a decrease in 5-Me-PCA release in the absence of Mex (Fig. 6A).
Fig. 6.
MexGHI-OpmD is required for wild-type export of 5-Me-PCA by PA14. (A) HPLC-based detection of phenazines in ΔmexΔphzH culture supernatants compared to those of ΔphzH. Cultures were grown in liquid MOPS–LB medium and sampled at an OD (500 nm) of ∼4. Error bars represent SD of the mean for three replicates. (B) Square-wave voltammogram of supernatants from early-stationary phase MOPS–LB liquid cultures. (C) Quantification of intracellular phenazines from PA14 grown in liquid MOPS-LB and sampled at an OD (500 nm) of ∼3. Concentrations were adjusted to total extracted protein. Horizontal lines represent mean values. The phenazine PCA was not detected. (D, Left) Day 1 ∆phzH and ∆mex∆phzH colonies used for on-chip electrochemical phenazine detection. The line indicates the region for which phenazines were quantified. (Right) Quantification of phenazines released by colonies on the IC chip.
As 5-Me-PCA is considered a reactive intermediate of phenazine biosynthesis (44), it does not accumulate in wild-type cultures but might be expected to accumulate in our ∆phzHS mutant, which is unable to convert 5-Me-PCA or its precursor, PCA, into pyocyanin or PCN, respectively. We conducted a growth experiment in liquid culture to examine changes in 5-Me-PCA over time. Liquid cultures of the ∆phzHS mutant exhibit a lime-green color in the early and midstationary phase (SI Appendix, Fig. S10C). Chemically synthesized 5-Me-PCA and the 5-Me-PCA analog PMS are yellow (52), and PMS takes on a bright green color upon reduction (53). Accumulation of a reduced form of 5-Me-PCA could occur under the reducing conditions prevailing in the high-cell density, oxygen-limited conditions of stationary phase cultures; if reduced 5-Me-PCA is green, this could account for the coloration of ∆phzHS cultures. We performed chloroform extractions on culture supernatants taken over the course of ∆phzHS growth and used HPLC to follow the distribution of phenazines in the aqueous and organic phases. The concentration of 5-Me-PCA in aqueous extracts peaked at the 9-h time point (when culture supernatants are green) and decreased over the rest of the experiment. Consistent with the decrease in 5-Me-PCA at time points when supernatants are red, we propose that the red color arises from modification of the 5-Me-PCA (SI Appendix, Fig. S10C). In samples taken after 8 h, we also observed a compound in the organic phase that eluted at the same time as 5-Me-PCA, with an absorbance spectrum that differed slightly from that of 5-Me-PCA (SI Appendix, Fig. S10 A, B, and D). The presence of MOPS [3-(N-morpholino)propanesulfonic acid] in our medium held the pH at 7, indicating that the appearance of the compound in the organic phase could not be attributed to pH-dependent effects on 5-Me-PCA similar to those described previously (40). The appearance of the hydrophobic phenazine derivative may be influenced by other aspects of the chemistry of the culture medium (i.e., oxygen availability) or regulation of uncharacterized phenazine transformations.
The defect in 5-Me-PCA release displayed by ∆mex (Figs. 5C and 6A) indicates that deleting mex either (i) disrupts transport of 5-Me-PCA by MexGHI-OpmD or (ii) indirectly affects total 5-Me-PCA synthesis, leading to decreased extracellular levels. To distinguish between these possibilities, we sought to measure intracellular phenazine concentrations. Due to the chemical complexity of cell lysates, we could not use HPLC to detect phenazines in these samples and instead used square wave voltammetry (SWV), a linear potential sweep voltammetry that measures the redox current of a working electrode as its potential is swept relative to a reference electrode (48). Currents are sampled in a manner that reduces the effect of charging currents. First, we verified that SWV applied to culture supernatants could detect differences in 5-Me-PCA release between the ∆phzH and ∆mex∆phzH mutants (Fig. 6B). Next, we used SWV to measure phenazines in lysates and found that intracellular 5-Me-PCA levels in ∆mex∆phzH were comparable to or slightly higher than those in ∆phzH (Fig. 6C), even though extracellular 5-Me-PCA levels are lower in ∆mex∆phzH relative to ∆phzH. We note that electrochemical detection methods show that the ∆mex∆phzH mutant also releases more pyocyanin than the ∆phzH mutant, suggesting a shunt of accumulated 5-Me-PCA toward PhzS-catalyzed hydroxylation (37) (Fig. 6 B and D). These results support a model in which mex deletion does not affect 5-Me-PCA synthesis but rather its transport out of the cell.
We attempted detection of 5-Me-PCA release from colonies by conducting HPLC analysis of extracts from the underlying agar-solidified growth medium. However, we were unable to detect 5-Me-PCA using this method (SI Appendix, Fig. S9C), possibly due to degradation or modification during the prolonged agar extraction protocol. Thus, we sought to demonstrate a role for MexGHI-OpmD in 5-Me-PCA release from colonies by using an integrated circuit (IC) chip, an electrochemical metabolite sensing platform recently developed by our group (48, 54). This technique enables direct detection of phenazines as they are released from the biofilm using the same SWV-based electrochemical detection modality described earlier, eliminating the need for agar processing and extraction steps that may allow conversion of 5-Me-PCA to other derivatives before HPLC analysis (SI Appendix, Fig. S11). In a significant advantage over many other methods, it also provides information about the spatial distribution of metabolites released from intact multicellular samples. Electrochemical imaging showed that 5-Me-PCA was localized to a region near the edges of both the ∆phzH and ∆mex∆phzH biofilms, with less total 5-Me-PCA detected in ∆mex∆phzH (Fig. 6D). The specific localization of 5-Me-PCA, in comparison with the more even distribution of pyocyanin, suggests that this patterning is relevant for the unique effect of 5-Me-PCA on colony morphology (Fig. 1D). Furthermore, the dependence of colony biofilm 5-Me-PCA release on MexGHI-OpmD activity is consistent with the delayed-wrinkling phenotypes exhibited by ∆mex mutants that produce 5-Me-PCA (Fig. 1E).
Discussion
The opportunistic pathogen P. aeruginosa produces phenazines, antibiotics that contribute to virulence not only by directly damaging host tissues but also by enhancing pseudomonad fitness during host colonization (18, 21, 22, 46, 55). In particular, P. aeruginosa is well known for production of the bright blue phenazine pyocyanin, which contributes to the coloration of sputum and pus associated with infections. Pyocyanin is one of several redox-active compounds that have been shown to activate the redox-sensing transcription factor SoxR (25, 32). In P. aeruginosa, SoxR induces a small regulon that includes the mexGHI-opmD (mex) operon (24, 25). We examined the physiological role of MexGHI-OpmD, the RND efflux pump encoded by mex. We found that deleting soxR or mex had little to no effect on the sensitivity of P. aeruginosa PA14 to pyocyanin and the precursor phenazine PCA but rather rendered PA14 sensitive to the synthetic phenazine PMS (Fig. 4 A and B).
PMS is structurally similar to 5-Me-PCA, an elusive, implied intermediate in the pyocyanin biosynthetic pathway (44). Zheng et al. recently detected 5-Me-PCA in pure P. aeruginosa cultures for the first time using a mutant that overproduces PhzM and reported a redox potential for 5-Me-PCA of +129 mV (vs. NHE at pH 7.0; two-electron transfer) (40). Before the Zheng et al. study, the highest redox potential measured for a pseudomonad phenazine had been that of pyocyanin, which is –40 mV (vs. NHE at pH 7.0). The remarkably high redox potential of 5-Me-PCA has implications for our interpretations of its physiological effects. On one hand, this property could make its intracellular accumulation, as would be predicted in ∆mex, a harmful condition that leads to oxidation of intracellular metabolites and redox-sensitive proteins. This effect may be responsible for the altered colony development (i.e., persistent smoothness or wrinkling delayed beyond the timing observed for the wild type) and liquid-culture growth and competition defects observed in ∆mex strains (Fig. 1, Fig. 2, and SI Appendix, Fig. S2). On the other hand, the redox potential of 5-Me-PCA would make it a favorable mediator for extracellular electron shuttling by cells in electron acceptor-limited biofilm microenvironments: The potential of 5-Me-PCA could theoretically support ATP generation when coupled to the oxidation of electron donors such as succinate in anoxic regions of the biofilm but is also low enough to allow oxidation of 5-Me-PCA by oxygen in oxic biofilm subzones. Our previous work indicates that extracellular electron shuttling by endogenous P. aeruginosa phenazines has a beneficial effect in that it promotes the relatively smooth morphotype of the wild type (19). Indeed, our results suggest that 5-Me-PCA is the primary phenazine responsible for wild-type PA14 colony morphogenesis (Fig. 1D), consistent with previous chemical complementation results in which PCA more effectively inhibited wrinkling and spreading of ∆phz colonies than pyocyanin (56) (phzM is intact in the ∆phz mutant, allowing for conversion of added PCA to 5-Me-PCA). We speculate that 5-Me-PCA supports redox balancing for cells in anoxic biofilm subzones and may contribute to survival in the host.
The N-methylation reaction catalyzed by PhzM is unique to P. aeruginosa among the phenazine-producing pseudomonad species studied to date (57) and has important consequences for phenazine chemistry. In P. aeruginosa, the phenazine PCA, which has a carboxylate group, is converted into the zwitterionic 5-Me-PCA. Further conversion to pyocyanin allows for tautomerization to the uncharged form, which increases hydrophobicity and the potential of the compound to cross lipid membranes at neutral pH without the need for a membrane transporter. This property of pyocyanin would agree with our observation that ∆mex mutants exhibit a blue-green coloration after prolonged (5 d) coculture on C. albicans lawns, suggesting that deletion of ∆mex does not prevent release of pyocyanin. Simple chloroform extractions indicate that pyocyanin and PCA partition readily into the organic phase whereas 5-Me-PCA does not (SI Appendix, Fig. S12). As 5-Me-PCA is zwitterionic and reactive under physiological conditions (pH 7), it constitutes a likely substrate of transmembrane efflux pumps, and we have demonstrated that MexGHI-OpmD is a 5-Me-PCA transporter that confers protection from the toxic effects of this unusually reactive phenazine on its producer (Figs. 2, 5, and 6).
In many antibiotic-producing organisms, resistance mechanisms include derivatization of the antibiotic (7); by analogy, conversion of 5-Me-PCA to the more lipid-soluble pyocyanin could constitute a means of protection rather than a targeted effort to produce this most notorious phenazine. Our systematic analyses of the effects of various phenazine biosynthetic defects revealed an interesting relationship between phenazine chemistry and mex physiology. Delayed colony wrinkling resulting from the mex deletion and strong induction of mex expression are two phenotypes that were both observed specifically in strains with an intact PhzM gene (i.e., strains that produce 5-Me-PCA such as ∆phzH and ∆phzHS) (Figs. 1E and 4C). We have also observed that deleting mex increases expression of the Pmex reporter even further (i.e., increases activation of SoxR) in both the ∆phzH and ∆phzHS backgrounds (SI Appendix, Fig. S13). These results imply that delayed colony wrinkling and SoxR induction arise from common intracellular effects and highlight the unique physiology of N-methylated phenazine production.
In this study, we identified 5-Me-PCA as a natural substrate of the RND efflux transporter MexGHI-OpmD. This pump therefore confers resistance to xenobiotic compounds, such as synthetic antibiotics and dyes (35), and an endogenous P. aeruginosa product (Fig. 2). Of the several RND pumps encoded by the P. aeruginosa genome, MexGHI-OpmD is distinguished by the organization of its encoding locus. The mexAB-oprM, mexEF-oprN, and mexXY operons each encode one inner membrane component, one periplasmic component, and an optional outer membrane component, all of which play canonical roles in the functions of RND efflux pumps (2). The mexGHI-opmD operon, however, encodes an additional membrane-associated component (MexG) with unknown functionality. Although we found that MexG was not required for normal export of 5-Me-PCA, we speculate that this protein may act to modulate the specificity of MexHI in a manner similar to that of the AcrZ accessory protein (14). In experiments examining the resistance of E. coli MG1655 ∆acrB and ∆acrZ mutants to antibiotics commonly used in human medicine, Hobbs et al. showed that AcrZ is required for full resistance to a relatively hydrophilic subset of the antibiotics that are transported by AcrB. It would be interesting to test whether AcrZ is involved in resistance to the fatty acids and bile salts among the suggested ecological substrates of AcrB, particularly considering that acrZ is regulated by a transcription factor that is sensitive to these compounds (14, 15). Studies elucidating the ecologically relevant or endogenous inducers and substrates of efflux pumps will ultimately help us to better understand their evolution, maintenance in diverse genomes, and critical roles in the physiology of pathogens and malignant host cells (58–60).
Methods
Strains and Culture Conditions.
Strains used in this study are listed in SI Appendix, Table S1. For preculturing and genetic manipulation, bacteria were routinely grown at 37 °C in Lysogeny Broth (LB) with shaking at 250 rpm (Forma Orbital Shaker, Thermo Scientific) or on LB solidified with agar (15 g/L) (61). MOPS-LB (50 mM MOPS buffer, 43 mM NaCl, 93 mM NH4Cl, 2.2 mM KH2PO4, 1 mM MgSO4·7H2O, 3.6 µM FeSO4·7H2O, 20 mM glucose, 20% LB) was used for experiments requiring a semidefined medium. Basal “colony morphology assay medium,” used for phenazine sensitivity and colony development studies, contains 1% tryptone, 1% agar, 40 µg/mL Congo red, and 20 µg/mL Coomassie blue. For selection during genetic manipulation, gentamicin was added to the medium at 15 µg/mL for E. coli or 100 µg/mL for P. aeruginosa.
Strain Construction.
Plasmids and primers used in this study are listed in SI Appendix, Tables S2 and S3.
Markerless deletions in specific loci were generated by homologous recombination as described previously (18). Deletion plasmids were generated using the yeast gap repair method as described previously (18, 62). The same approach was used to complement the transposon insertion mutant mexH::tn, with the modification that plasmid pLD1574, instead of pMQ30, was used to replace the disrupted allele with the wild-type sequence.
Strains constitutively expressing eyfp from the PA1/04/03 promoter, for use in competition experiments, were generated using the pAKN69-eYFP plasmid (63), which was mobilized into PA14 via triparental conjugation. Stationary phase cultures of the donor (BW29427 containing pAKN69-eYFP), recipient, and helper (β2155) strains were mixed in equal ratios, spotted on LB agar containing diaminopimelic acid, and incubated at room temperature overnight. The cells were scraped off the agar using a pipette tip, washed with LB, and plated on selective medium (with 70 μg/mL gentamicin). Resistant transconjugants were screened by PCR for insertion of the reporter construct at the neutral attTn7 site on the PA14 chromosome, and expression of the reporter was verified by fluorescence scanning (Typhoon FLA7000, GE Healthcare).
Strains expressing GFP under the control of Pmex were generated by homologous recombination. First, we created a plasmid for homologous recombination into the neutral attB site on the PA14 chromosome. Regions ∼1 kb in size on either side of this locus were amplified using PA14 genomic DNA as a template, whereas gfp was amplified from pYL122 (64). The fragments were cloned into pMQ30 using homologous recombination in Saccharomyces cerevisiae InvSc1, creating pLD2477. Next, a region ∼500 bp upstream of the mexG start site was amplified and cloned into pLD2477 using SpeI and XhoI, giving pLD2524. This plasmid was moved into various strain backgrounds using the BW29427 donor strain (see SI Appendix, Table S1). Stationary phase cultures of the donor and recipient strains were mixed in 4:1 ratios, spotted on LB agar containing diaminopimelic acid, and incubated at room temperature overnight. The cells were scraped from the agar using a pipette tip, washed with LB, and plated on selective medium. Single recombinants (containing the genomically inserted pLD2524) were grown in LB to exponential phase and then plated on LB agar containing no NaCl and 10% sucrose. Double recombinants (that resolved the plasmid but retained Pmex-GFP) were verified by PCR and fluorescence scanning.
Colony Morphology Assay.
Ten microliters of overnight precultures were spotted on colony morphology assay medium (described earlier; 60 mL in a 9-cm square plate) and incubated at 25 °C, >95% humidity. The colonies were imaged daily using a Keyence VHX-1000 digital microscope. Time-lapse movies of colony development were taken using an iPod running Lapse It software. Images of colonies growing in a humidified chamber in the dark were taken every 15 min. An LED light that was programmed to turn on every 15 min (LabVIEW software) was used to illuminate the colonies for photographing.
Growth Curves.
Overnight precultures were diluted 1:100 in fresh LB and grown to exponential phase. The exponential phase cultures were diluted to an optical density (OD) at 500 nm of 0.05 in LB medium and grown for 24 h. The cultures were then diluted 1:200 in 1 mL LB. Two hundred microliters of the diluted cultures were transferred to a sterile 96-well plate (Greiner Bio-One, 655001) and incubated at 37 °C with continuous shaking on the “fast” setting in a BioTek Synergy 4 plate reader.
Competition Experiments.
Overnight precultures were diluted 1:100 in LB and grown to exponential phase. The exponential phase cultures were centrifuged at 2,152 × g for 2 min, washed, resuspended in 1% tryptone, and mixed in 50:50 ratios. Cultures were grown at 37 °C with shaking at 250 rpm. At indicated time points, cultures were diluted to 10−7–10−4 and spread on 1% tryptone, 1% agar plates.
PMS Sensitivity.
Liquid precultures of each strain to be tested were grown overnight in LB medium. Ten microliters of these precultures were spotted on agar-solidified medium containing 600 µM PMS and incubated at 25 °C in the dark. Colonies were imaged at indicated time points using a Canon CanoScan 5600F scanner. For quantification of cfus in colonies, 10 µL of overnight LB precultures were spotted on colony morphology assay medium containing different concentrations of PMS. After 2 d of growth, each colony was scraped from the agar using a razor blade and added to a 1.5 mL screw-cap tube containing 1 mL LB and 0.5 g·1.4-mm acid-washed zirconium beads (OPS Diagnostics, LLC). The colonies were homogenized at the highest setting using a Mini Beadbeater (Biospec Products) for 3 min and diluted in LB. Ninety-microliter aliquots of 10−4, 10−6, and 10−7 dilutions were spotted on 1% tryptone, 1% agar plates and incubated overnight at 37 °C before colony enumeration.
Sensitivity to Different Phenazines and Paraquat.
Phenazines were added to the colony morphology assay medium from stock solutions after autoclaving. Stock solutions of PCA (Apexmol Technology) and pyocyanin (Cayman Chemical) were dissolved in DMSO, whereas PMS (MP Biomedicals) and paraquat (Acros Organics) were dissolved in water. The stocks were diluted so that equal volumes of water or DMSO were added to the medium.
Liquid precultures of each strain to be tested were grown overnight in LB medium. Two microliters of overnight cultures were spotted on agar-solidified media containing the indicated concentrations of phenazines and incubated at 25 °C, >95% humidity in the dark. After 24 h of growth, colonies were imaged using a Keyence VHX-1000 digital microscope.
Quantification of GFP Fluorescence in Pmex Reporter Experiments.
For quantification of mex expression in liquid cultures, MOPS–LB overnight precultures were diluted to an OD (500 nm) of 0.01 in MOPS–LB medium in 96-well plates. Plates were incubated at 37 °C with continuous shaking on the fast setting in a BioTek Synergy 4 plate reader. GFP fluorescence was detected by excitation at 480 nm and measurement of emission at 510 nm.
For quantification of mex expression in colony biofilms, LB overnight precultures were used as inocula for biofilms grown on 1% tryptone, 1% agar at 25 °C, >95% humidity. Colonies were scanned using a Typhoon FLA7000 model scanner (GE Healthcare). GFP fluorescence was detected by excitation at 473 nm and measurement of emission at 520 nm. Fluorescence data were processed using ImageJ.
Extraction and Detection of PQS Produced by Colonies.
Ten microliters of stationary phase LB cultures were spotted on polycarbonate membrane filters (Whatman 110606; pore size, 0.2 µm) on colony morphology assay medium and incubated at 25 °C, >95% humidity. After 4 d of growth, the filter paper and attached colony were removed from the agar plate with forceps and placed inside a 20 mL scintillation vial containing 1 mL 0.015% acetic acid/ethyl acetate. The PQS was extracted into the ethyl acetate overnight in the dark at room temperature. The extract was dried using a vacuum centrifuge for 20 min at 60 °C. The dried extract was resuspended in 200 µL MOPS buffer (pH 7.3) and analyzed using a BioTek Synergy 4 plate reader. The solution was excited at 340 nm, and the emitted fluorescence was measured from 340 to 800 nm in steps of 10 nm.
Extraction and Detection of Red Phenazines Produced by Colonies.
Ten microliters of overnight LB precultures were spotted on solidified medium containing 1% tryptone, 1% agar and incubated at 25 °C, >95% humidity. After 2 d of growth, the colonies were scraped from the agar using a metal spatula, and the agar was placed in a 15-mL conical tube and chopped. Red phenazines were extracted into 2 mL of water in the dark at room temperature. Five hundred microliters of extract was removed and centrifuged at 16873 × g for 2 min. The supernatant was filtered through a cellulose acetate membrane (Corning Life Sciences Plastic 8161; pore size, 0.22 µm), and a 200-µL aliquot was analyzed using a BioTek Synergy 4 plate reader. The solution was excited at 510 nm, and the emitted fluorescence was measured from 540 to 800 nm in steps of 10 nm.
C. albicans Bioassay.
C. albicans was grown in yeast extract peptone dextrose medium overnight at 30 °C. Seven hundred microliters of the culture were spread onto yeast extract peptone dextrose medium 1.5% agar plates and incubated at 30 °C for 48 h. Five microliters of LB-grown cultures of P. aeruginosa PA14 wild type and mutants of interest were spotted onto C. albicans lawns and incubated 24 h at 30 °C (51), unless otherwise noted. The cocultures were imaged with a Canon CanoScan 5600F scanner. The intracellular fluorescence of C. albicans cultured with or without P. aeruginosa was analyzed as previously described with few modifications (49). Briefly, fungal cells were sampled and suspended into sterile saline solution (final volume 1 mL) and fixed with 2% paraformaldehyde for 15 min on ice. Cell solutions were washed twice with 1 mL PBS and scanned by flow cytometry in a BD Canto instrument using the R-phycoerythrin (PE-A) channel (excitation, 488 nm). Approximately 105 cells were evaluated for fluorescence. Analyses were performed for two independent coculture assays.
Phenazine Detection by HPLC.
P. aeruginosa precultures were grown overnight, diluted 1:50 in liquid LB, and grown to exponential phase. The cells were centrifuged at 2,152 × g for 2 min, resuspended in MOPS–LB, and diluted to OD (500 nm) of 0.05 in 50 mL MOPS–LB. Cultures were grown at 37 °C with shaking at 250 rpm, and sampling times for individual experiments are indicated in figure legends. Samples were centrifuged at maximum speed, and the supernatant was filtered through a cellulose acetate membrane (pore size, 0.22 µm). A 50-µL sample was loaded on an analytical Waters Symmetry C-18 reverse-phase column (5 µm particle size, 4.6 × 150 mm2). HPLC-based separation was carried out using a gradient of water–0.01% TFA (pH 3.01; solvent A) to acetonitrile–0.01% TFA (solvent B) at a flow rate of 0.4 mL·min−1 using the following protocol: linear gradient from 0% to 15% solvent B for 2 min, linear gradient to 83% solvent B for 20 min, linear gradient from 83% to 100% solvent B for 10 min, and finally a linear gradient to 0% solvent B for 5 min. The total method time was 38 min. The retention times for 5-Me-PCA, pyocyanin, and PCA were 6 min, 13 min, and 25 min, respectively.
Phenazine Detection by SWV.
Overnight P. aeruginosa precultures were diluted 1:50 in LB and grown to exponential phase. The cells were centrifuged at 2,152 × g for 2 min, resuspended in MOPS–LB, and diluted to an OD500 of 0.05 in 50 mL MOPS–LB. Stationary-phase cells were centrifuged at maximum speed, and the supernatant was collected. The SWV signal consisted of the response to a series of forward and reverse voltage steps. The current value at the end of each voltage step was sampled, and the current sample after a forward voltage step was subtracted from the current sample after a reverse voltage step. The resulting signal was plotted as a function of applied potentiostat voltage.
Phenazines released from colonies were detected and quantified by using an IC chip as described in ref. 54.
Cell Lysis for Quantification of Intracellular Phenazines.
Each PA14 culture was grown in 500 mL of MOPS–LB medium to an OD (500 nm) of 3.1 and centrifuged at 6,000 × g for 10 min. Supernatant was removed and the pellet was resuspended in 20 mL MOPS–LB medium. Cells were lysed by two rounds of processing in a Sim-Aminico (Spectronic Instruments) French press at 1,250 lbs/in2.
Supplementary Material
Acknowledgments
We thank Yu-Cheng Lin for constructing plasmid pLD2524; Arie Zask and Nicholas Jacobs for discussions and critical reading of the manuscript; and Jessie Scott and Emily Dolben for capturing images in the yeast biosensor assay. H.S. and K.S. were supported by IGERT Fellowship Award 0801530. L.K. was supported by the Amgen Scholars Program at Columbia University/Barnard College. Y.Z. was supported by a fellowship from the Chinese Scholarship Council. This work was also funded by NSF Award 1353553 (to K.S. and L.E.P.D.), NIH Grant R01AI103369 (to L.E.P.D.), a diversity supplement for the aforementioned NIH grant (to B.L.F.), Columbia University’s Bridge Program (B.L.F.), and NIH Grants R01GM108492 and R01AI091702 (to D.A.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600424113/-/DCSupplemental.
References
- 1.Martín JF, Casqueiro J, Liras P. Secretion systems for secondary metabolites: How producer cells send out messages of intercellular communication. Curr Opin Microbiol. 2005;8(3):282–293. doi: 10.1016/j.mib.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Martinez JL, et al. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev. 2009;33(2):430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y, Willems A, Au-Yeung C, Tahlan K, Nodwell JR. A two-step mechanism for the activation of actinorhodin export and resistance in Streptomyces coelicolor. MBio. 2012;3(5):e00191-12. doi: 10.1128/mBio.00191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guilfoile PG, Hutchinson CR. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc Natl Acad Sci USA. 1991;88(19):8553–8557. doi: 10.1073/pnas.88.19.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W, Sharma M, Kaur P. The DrrAB efflux system of Streptomyces peucetius is a multidrug transporter of broad substrate specificity. J Biol Chem. 2014;289(18):12633–12646. doi: 10.1074/jbc.M113.536136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohnuki T, Katoh T, Imanaka T, Aiba S. Molecular cloning of tetracycline resistance genes from Streptomyces rimosus in Streptomyces griseus and characterization of the cloned genes. J Bacteriol. 1985;161(3):1010–1016. doi: 10.1128/jb.161.3.1010-1016.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cundliffe E, Demain AL. Avoidance of suicide in antibiotic-producing microbes. J Ind Microbiol Biotechnol. 2010;37(7):643–672. doi: 10.1007/s10295-010-0721-x. [DOI] [PubMed] [Google Scholar]
- 8.Mak S, Xu Y, Nodwell JR. The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol Microbiol. 2014;93(3):391–402. doi: 10.1111/mmi.12689. [DOI] [PubMed] [Google Scholar]
- 9.Minagawa S, et al. RND type efflux pump system MexAB-OprM of Pseudomonas aeruginosa selects bacterial languages, 3-oxo-acyl-homoserine lactones, for cell-to-cell communication. BMC Microbiol. 2012;12:70. doi: 10.1186/1471-2180-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamarche MG, Déziel E. MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline) PLoS One. 2011;6(9):e24310. doi: 10.1371/journal.pone.0024310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor DL, Ante VM, Bina XR, Howard MF, Bina JE. Substrate-dependent activation of the Vibrio cholerae vexAB RND efflux system requires vexR. PLoS One. 2015;10(2):e0117890. doi: 10.1371/journal.pone.0117890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz C, Levy SB. Regulation of acrAB expression by cellular metabolites in Escherichia coli. J Antimicrob Chemother. 2014;69(2):390–399. doi: 10.1093/jac/dkt352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zgurskaya HI, Nikaido H. Bypassing the periplasm: Reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci USA. 1999;96(13):7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc Natl Acad Sci USA. 2012;109(41):16696–16701. doi: 10.1073/pnas.1210093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol. 2003;48(6):1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 16.Tolker-Nielsen T. Pseudomonas aeruginosa biofilm infections: From molecular biofilm biology to new treatment possibilities. APMIS Suppl. 2014;(138):1–51. doi: 10.1111/apm.12335. [DOI] [PubMed] [Google Scholar]
- 17.Price-Whelan A, Dietrich LE, Newman DK. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189(17):6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Recinos DA, et al. Redundant phenazine operons in Pseudomonas aeruginosa exhibit environment-dependent expression and differential roles in pathogenicity. Proc Natl Acad Sci USA. 2012;109(47):19420–19425. doi: 10.1073/pnas.1213901109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietrich LE, et al. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195(7):1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glasser NR, Kern SE, Newman DK. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol. 2014;92(2):399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun. 2004;72(7):4275–4278. doi: 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rada B, Leto TL. Pyocyanin effects on respiratory epithelium: Relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 2013;21(2):73–81. doi: 10.1016/j.tim.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villavicencio RT. The history of blue pus. J Am Coll Surg. 1998;187(2):212–216. doi: 10.1016/s1072-7515(98)00137-9. [DOI] [PubMed] [Google Scholar]
- 24.Palma M, et al. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect Immun. 2005;73(5):2958–2966. doi: 10.1128/IAI.73.5.2958-2966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006;61(5):1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 26.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87(16):6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsaneva IR, Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172(8):4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu M, Imlay JA. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol. 2011;79(5):1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci USA. 1996;93(19):10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imlay J, Gu M. Many plants and bacteria excrete redox-cycling compounds. Free Radic Biol Med. 2011;50(12):1814–1815. doi: 10.1016/j.freeradbiomed.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 31.Shin JH, Singh AK, Cheon DJ, Roe JH. Activation of the SoxR regulon in Streptomyces coelicolor by the extracellular form of the pigmented antibiotic actinorhodin. J Bacteriol. 2011;193(1):75–81. doi: 10.1128/JB.00965-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheplock R, Recinos DA, Mackow N, Dietrich LE, Chander M. Species-specific residues calibrate SoxR sensitivity to redox-active molecules. Mol Microbiol. 2013;87(2):368–381. doi: 10.1111/mmi.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh AK, Shin JH, Lee KL, Imlay JA, Roe JH. Comparative study of SoxR activation by redox-active compounds. Mol Microbiol. 2013;90(5):983–996. doi: 10.1111/mmi.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietrich LE, Kiley PJ. A shared mechanism of SoxR activation by redox-cycling compounds. Mol Microbiol. 2011;79(5):1119–1122. doi: 10.1111/j.1365-2958.2011.07552.x. [DOI] [PubMed] [Google Scholar]
- 35.Sekiya H, et al. Functional cloning and characterization of a multidrug efflux pump, mexHI-opmD, from a Pseudomonas aeruginosa mutant. Antimicrob Agents Chemother. 2003;47(9):2990–2992. doi: 10.1128/AAC.47.9.2990-2992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aendekerk S, et al. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology. 2005;151(Pt 4):1113–1125. doi: 10.1099/mic.0.27631-0. [DOI] [PubMed] [Google Scholar]
- 37.Dietrich LE, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321(5893):1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flood ME, Herbert RB, Holliman FG. Pigments of Pseudomonas species. V. Biosynthesis of pyocyanin and the pigments of Ps. aureoaciens. J Chem Soc Perkin 1. 1972;4(4):622–626. doi: 10.1039/p19720000622. [DOI] [PubMed] [Google Scholar]
- 39.Hansford GS, Holliman FG, Herbert RB. Pigments of Pseudomonas species. IV. In vitro and in vivo conversion of 5-methylphenazinium-1-carboxylate into aeruginosin A. J Chem Soc Perkin 1. 1972;1:103–105. doi: 10.1039/p19720000103. [DOI] [PubMed] [Google Scholar]
- 40.Zheng H, et al. Redox metabolites signal polymicrobial biofilm development via the NapA oxidative stress cascade in Aspergillus. Curr Biol. 2015;25(1):29–37. doi: 10.1016/j.cub.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abu EA, et al. Cyclic voltammetric, fluorescence and biological analysis of purified aeruginosin A, a secreted red pigment of Pseudomonas aeruginosa PAO1. Microbiology. 2013;159(Pt 8):1736–1747. doi: 10.1099/mic.0.065235-0. [DOI] [PubMed] [Google Scholar]
- 42.Mavrodi DV, et al. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183(21):6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsons JF, et al. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry. 2007;46(7):1821–1828. doi: 10.1021/bi6024403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenhagen BT, et al. Crystal structure of the pyocyanin biosynthetic protein PhzS. Biochemistry. 2008;47(19):5281–5289. doi: 10.1021/bi702480t. [DOI] [PubMed] [Google Scholar]
- 45.Li XZ, Barré N, Poole K. Influence of the MexA-MexB-oprM multidrug efflux system on expression of the MexC-MexD-oprJ and MexE-MexF-oprN multidrug efflux systems in Pseudomonas aeruginosa. J Antimicrob Chemother. 2000;46(6):885–893. doi: 10.1093/jac/46.6.885. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Kern SE, Newman DK. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J Bacteriol. 2010;192(1):365–369. doi: 10.1128/JB.01188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu N, et al. Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 8):1403–1413. doi: 10.1107/S0907444913008354. [DOI] [PubMed] [Google Scholar]
- 48.Bellin DL, et al. Integrated circuit-based electrochemical sensor for spatially resolved detection of redox-active metabolites in biofilms. Nat Commun. 2014;5:3256. doi: 10.1038/ncomms4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morales DK, et al. Antifungal mechanisms by which a novel Pseudomonas aeruginosa phenazine toxin kills Candida albicans in biofilms. Mol Microbiol. 2010;78(6):1379–1392. doi: 10.1111/j.1365-2958.2010.07414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aendekerk S, Ghysels B, Cornelis P, Baysse C. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology. 2002;148(Pt 8):2371–2381. doi: 10.1099/00221287-148-8-2371. [DOI] [PubMed] [Google Scholar]
- 51.Gibson J, Sood A, Hogan DA. Pseudomonas aeruginosa-Candida albicans interactions: Localization and fungal toxicity of a phenazine derivative. Appl Environ Microbiol. 2009;75(2):504–513. doi: 10.1128/AEM.01037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phelan VV, et al. Impact of a transposon insertion in phzF2 on the specialized metabolite production and interkingdom interactions of Pseudomonas aeruginosa. J Bacteriol. 2014;196(9):1683–1693. doi: 10.1128/JB.01258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaugg WS. Spectroscopic characteristics and some chemical properties of N-methylphenazinium methyl sulfate (phenazine methosulfate) and pyocyanine at the semiquinoid oxidation level. J Biol Chem. 1964;239:3964–3970. [PubMed] [Google Scholar]
- 54.Bellin DL, et al. Electrochemical camera chip for simultaneous imaging of multiple metabolites in biofilms. Nat Commun. 2016;7:10535. doi: 10.1038/ncomms10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunter RC, et al. Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am J Respir Cell Mol Biol. 2012;47(6):738–745. doi: 10.1165/rcmb.2012-0088OC. [DOI] [PubMed] [Google Scholar]
- 56.Ramos I, Dietrich LE, Price-Whelan A, Newman DK. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res Microbiol. 2010;161(3):187–191. doi: 10.1016/j.resmic.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol. 2006;44:417–445. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- 58.Gupta S, et al. Efflux inhibition with verapamil potentiates bedaquiline in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58(1):574–576. doi: 10.1128/AAC.01462-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alnaseri H, et al. Inducible expression of a resistance-nodulation-division-type efflux pump in Staphylococcus aureus provides resistance to linoleic and arachidonic acids. J Bacteriol. 2015;197(11):1893–1905. doi: 10.1128/JB.02607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fletcher JI, Haber M, Henderson MJ, Norris MD. ABC transporters in cancer: More than just drug efflux pumps. Nat Rev Cancer. 2010;10(2):147–156. doi: 10.1038/nrc2789. [DOI] [PubMed] [Google Scholar]
- 61.Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol. 2004;186(3):595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O’Toole GA. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol. 2006;72(7):5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lambertsen L, Sternberg C, Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 2004;6(7):726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- 64.Lequette Y, Greenberg EP. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187(1):37–44. doi: 10.1128/JB.187.1.37-44.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.