Abstract
Allergen-specific IgE (allergic sensitization) plays a central role in the pathogenesis of allergic disease. We performed the first large-scale genome wide association study (GWAS) of allergic sensitization in 5,789 affected individuals and 10,056 controls and followed up the top SNP from 26 loci in 6,114 affected individuals and 9,920 controls. We increased the number of susceptibility loci with genome-wide significant association to allergic sensitization from three to 10, including SNPs in or near TLR6, C11orf30, STAT6, SLC25A46, HLA-DQB1, IL1RL1, LPP, MYC, IL2 and HLA-B. All the top-SNPs were associated with allergic symptoms in an independent study. Risk variants at these 10 loci were estimated to account for at least 25% of allergic sensitization and allergic rhinitis. Understanding the molecular mechanisms underlying these associations may provide novel insight into the etiology of allergic disease.
Allergic sensitization is defined as the presence of allergen-specific immunoglobulin E (IgE) against common environmental antigens.1 Allergen-specific IgE plays a pivotal role in pathogenesis of allergic reactions by binding allergens and initiating the immunological processes leading to allergic inflammation, and IgE-mediated sensitivity is considered to be central to the initiation of the allergy-associated (atopic) diseases rhinitis, asthma and eczema.
The effort to identify genes for atopic diseases is limited both by the heterogeneity of these diseases2 and by diagnostic uncertainty. An alternative approach is to study an intermediate phenotype, such as allergic sensitization. This is likely to be more closely related to a specific physiological mechanism, and thereby to the genetic substrate, and is clearly defined from objective, standardized assessments. It may therefore be a more powerful phenotype in genetic studies. Insight into the genetics of sensitization will also increase understanding of the shared genetic basis of sensitization and atopic diseases, and thereby their casual relationship.
Allergic sensitization has an estimated heritability of 0.40-0.85.3, 4 Heritability seems higher for the general tendency to sensitization (sensitization against any allergen) than for sensitization against one specific allergen.5 A large number of candidate genes has been reported6 but few have been firmly established. The first GWAS on allergic sensitization7 identified three putative susceptibility loci (5q22.1 near TMEM232, 11q13.5 near C11ORF30 and the HLA-region), but considered only sensitization to a single allergen (grass), was based on a modest sample size, and did not include replication.
We conducted a two-stage meta-analysis of GWAS of allergic sensitization including data from 16 studies from the EAGLE and AAGC consortia. Sensitization status was assessed objectively by either elevated levels of allergen-specific IgE in blood or a positive skin reaction after puncture of the skin through a droplet of allergen extract (skin prick test, SPT) and included sensitization against common food and inhalant allergens (Supplementary Tables 1, 2 and 3).
The stage one discovery phase included 5,789 individuals who were sensitized (cases) and 10,056 who were not sensitized (controls). In silico and de novo replication of the top-SNP from 26 loci included up to 6,114 independent cases and 9,920 independent controls, all of European descent.
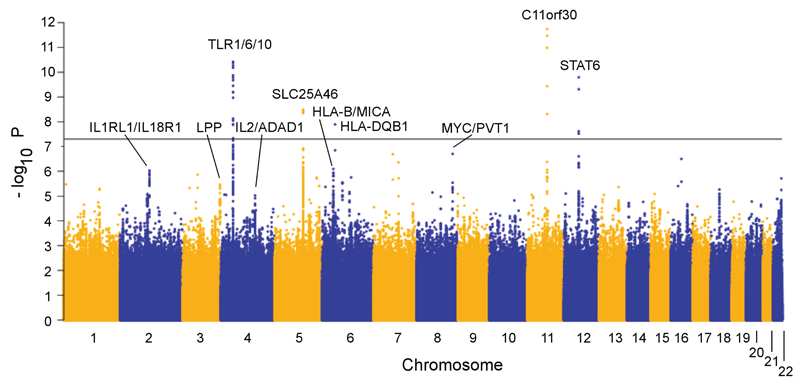
In stage 1, results were meta-analyzed after applying study-specific genomic control adjustment. There was minimal overall evidence for population stratification or other unaccounted biases on the overall results (λGC = 1.01). We detected an excess of association signals beyond those expected by chance (Supplementary Fig. 1) with SNPs in five regions reaching genome-wide significance (P < 5 × 10-8) (Fig. 1, Table 1). These five loci and 21 with suggestive evidence of association (P < 1 × 10-5), were selected for replication in stage two.
Figure 1. Manhattan plot for the discovery genome-wide association meta-analysis.
Table 1. Discovery and replication results of the 10 loci associated with allergic sensitization.
| Discovery | Replication | Combined | Replication in study of allergy symptoms | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | SNP/effect allele | Nearest gene | Eff. all. freq. | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | Het P/I2 (%) | P-category** |
| 11q13.5 | rs2155219/t | C11orf30 | 0.47 | 1.20 (1.14-1.24) | 1.8 × 10-12 | 1.15 (1.09, 1.21) | 1.1 × 10-7 | 1.18 (1.13, 1.22) | 1.4 × 10-18 | 0.57/0 | +++ |
| 12q13.3 | rs1059513/t | STAT6 | 0.90 | 1.34 (1.22-1.47) | 1.6 × 10-10 | 1.24 (1.13, 1.37) | 1.0 × 10-5 | 1.30 (1.21, 1.39) | 1.0 × 10-14 | 0.25/17 | + |
| 5q22.1 | rs10056340/t | SLC25A46 | 0.83 | 0.82 (0.77-0.88) | 3.2 × 10-9 | 0.83 (0.77, 0.90) | 4.4 × 10-6 | 0.83 (0.78, 0.87) | 5.2 × 10-14 | 0.93/0 | +++ |
| 6p21.32 | rs6906021/t | HLA-DQB1 | 0.55 | 0.86 (0.82-0.91) | 1.3× 10-8 | 0.87 (0.81, 0.93) | 4.8 × 10-5 | 0.87 (0.83, 0.90) | 2.2 × 10-12 | 0.51/0 | +++ |
| 2q12.1 | rs3771175/a | IL1RL1/IL18R1 | 0.14 | 0.79 (0.72-0.88) | 9.1× 10-6 | 0.83 (0.77, 0.90) | 1.1 × 10-6 | 0.83 (0.78, 0.88) | 4.9 × 10-11 | 0.39/5 | +++ |
| 4p14 | rs17616434/t | TLR1/6/10 | 0.78 | 1.24 (1.16-1.32) | 3.8× 10-11 | 1.22 (1.15, 1.31) | 3.8 × 10-10 | 1.23 (1.18, 1.29) | 5.2 × 10-11* | 0.04/40 | +++ |
| 3q28 | rs9865818/a | LPP | 0.59 | 0.88 (0.84-0.93) | 3.4 × 10-6 | 0.90 (0.85, 0.94) | 2.2 × 10-5 | 0.89 (0.86, 0.92) | 2.7 × 10-10 | 0.49/0 | ++ |
| 8q24.21 | rs4410871/t | MYC/PVT1 | 0.28 | 1.16 (1.09-1.23) | 2.0 × 10-7 | 1.12 (1.05, 1.20) | 6.7 × 10-4 | 1.14 (1.09, 1.19) | 5.4 × 10-10 | 0.45/1 | + |
| 4q27 | rs17454584/a | IL2/ADAD1 | 0.74 | 0.88 (0.83-0.93) | 9.5 × 10-6 | 0.86 (0.80, 0.92) | 1.4 × 10-5 | 0.87 (0.83, 0.91) | 5.5 × 10-10 | 0.74/0 | ++ |
| 6p21.33 | rs6932730/t | HLA-B/MICA | 0.82 | 1.18 (1.10-1.26) | 7.9 × 10-7 | 1.10 (1.02, 1.18) | 0.0075 | 1.14 (1.09, 1.20) | 4.2 × 10-8 | 0.26/16 | ++ |
P value for discovery and combined analysis is in bold if genome-wide significant (P < 5 × 10-8)
Het P: Heterogeneity P for Cochrane’s Q statistic
The P value was calculated by random effects model due to evidence of heterogeneity between studies (Heterogeneity P for Cochrane’s Q statistic < 0.05, I2 > 25%)
P value levels are categorized for the replication in an independent study on allergic symptoms8: + (significant after Bonferroni correction for the 10 genome-wide significant loci, P < 0.005,), ++ (P < 1 × 10-4), +++ (genome-wide significant, P < 5 × 10-8)
In stage two, 10 of the 26 loci showed evidence for association (P < 0.05) and directional consistency with sensitization status. All 10 loci reached genome-wide significance in the combined meta-analysis (Table 1, Supplementary Table 4, Supplementary Fig. 2 and 3). There was no statistically significant effect modification by age, assessment method (SPT or blood IgE) or sex after adjustment for multiple comparisons.
All 10 genome-wide significant SNPs showed evidence for association (P < 0.005) with self-reported allergy symptoms in an independent companion study8 based on more than 53,000 individuals (Table 1 and Supplementary Table 5). This suggests that these loci not only increase the risk of sensitization but also that of allergic disease.
To provide insight into the molecular mechanisms underlying each of the 10 associations, we searched for cis –acting expression quantitative trait loci (eQTLs) using gene expression data obtained from six cell types or tissues (white blood cells [WBC], lymphoblastoid cell lines [LCLs], whole blood and adipose tissue, B cells and monocytes9). For nine of the 10 loci, the sentinel SNP was associated (P<0.001) with the expression of one or more nearby (+/- 1 Mb) genes (Supplementary Table 6, 7 and 8). Furthermore, for all 10 loci, we identified variants located within ENCODE-predicted regulatory regions that were in high LD (r2 > 0.8) with the sentinel SNP (Supplementary Table 9). Together, these results suggest that for the ten loci the underlying causal variant(s) is likely to influence DNA transcription and/or mRNA degradation. Using data from the 1000 Genomes Project, we also identified missense variants in high LD with the sentinel SNP in TLR1 at 4p14 and in MICA at 6p21.32 (Supplementary Table 10).
The 8q24 region does not include any gene previously implicated in allergic disease or related traits. The lead SNP, rs4410871, is located in the PVT1 transcript, downstream of the MYC gene. MYC is a transcription factor involved in multiple cellular processes - including cell growth, differentiation and apoptosis of lymphocytes - and has an established role for several cancers, including B-cell malignancies.10 PVT1 is a non-coding, RNA transcription factor hosting several miRNAs,11 which have been suggested to be as important as MYC for T-cell activation.12 The same variant (rs4410871) was reported to associate with multiple sclerosis, with opposite direction of effect.13
The 11q13.5 and 5q22.1 loci do not harbor any genes with an established role in the immune system. However, both loci were identified recently by a GWAS of allergic sensitization 7 and the 11q13.5 locus seems to have pleiotropic effects of importance for development of both atopic 7, 14, 15 and autoimmune disease.16, 17
The 4p14 locus harbors the genes encoding Toll-like receptors 6, 1 and 10. This locus was associated with asthma and IgE levels in candidate gene studies.18–20 Toll-like receptors are pattern recognition molecules involved in innate immunity and immune responses to microbial exposure.21 The top SNP at this locus showed evidence for heterogeneity between studies. This may indicate effect modification from environmental factors and pinpoint a focus for future gene-environment interaction studies.
The 6 remaining genome-wide significant loci all imply genes with an established role in the immune system: STAT6,22 IL1RL1,23, 24 BCL6,25 IL2,26 HLA-DQB16, 27 and HLA-B/MICA.28–30 (Supplementary Note)
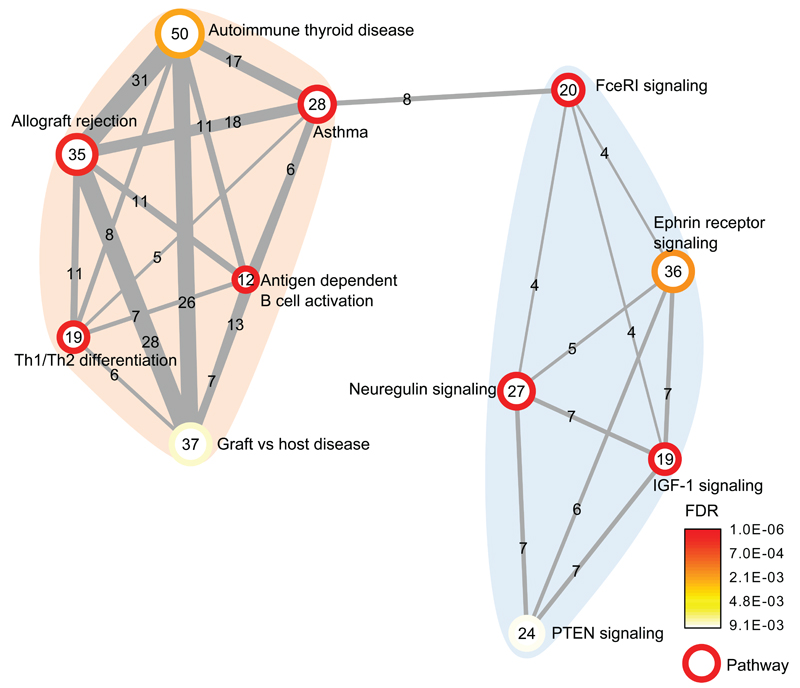
To explore biological connections and potentially identify new pathways associated with allergic sensitization, we applied a gene set enrichment analysis (MAGENTA)31 to the discovery stage GWAS results and identified significant enrichment of 11 partially overlapping gene sets. (Fig. 2 and Supplementary Table 11) These results suggest two cohesive clusters of biologically related gene sets. The largest cluster includes gene sets related to immune function, all sharing multiple genes, suggesting a broad overlap between autoimmunity and allergy (Supplementary Fig. 4). The second cluster includes potential novel sensitization pathways such as PTEN signalling, Ephrin receptor signalling, and IGF1-signaling, all characterized by involvement in cell growth and cancer. Three of 19 SNPs not in LD (r2 < 0.1) with the loci identified in our discovery GWAS (Supplementary Table 12) (in PTPN11, IL5 and CD86) showed evidence for association (P < 0.05) with self-reported allergy in the companion paper (Hinds et al.), suggesting that these are true, albeit weaker, allergy susceptibility loci.
Figure 2. Gene set enrichment map for the 11 significant gene sets from the MAGENTA analysis.
Vertices depict pathways and edges depict the mutually overlapping genes, suggesting that significant pathways are highly overlapping.
To further highlight biological connections between genes located within the 10 genome-wide significant loci, we applied the GRAIL pathway analysis tool32 and identified biologically connected genes related to immune function. Interestingly, we found that STAT6, TLR6, IL18RAP and HLA-DQB are inter-related, and expression of these genes are associated with the top SNPs, suggesting their involvement in a common mechanism that is likely to be causally associated with sensitization (Supplementary Fig. 5).
We tested the potential role of the 10 sensitization loci in determining asthma or eczema risk by testing for association with these diseases in two large published GWAS.33, 34 There was significant association with asthma with directional consistency for 9 out of the 10 SNPs (Supplementary Table 13). This overlap was considerably stronger than the previously reported minimal overlap between total IgE levels and asthma associated gene variants33 consistent with a potentially causal role of sensitization in the pathogenesis of asthma. For eczema the associations were weaker but with consistent direction of effect (Supplementary Table 14).
Our analysis also provided the opportunity to investigate whether previously reported susceptibility loci for atopic diseases and related intermediate phenotypes are associated with allergic sensitization (Supplementary Tables 15 and 16). Many reported atopy-genes were associated with sensitization. However, several strong total IgE-loci, such as FCER1A, HLA-A and HLA-DRB, as well as strong asthma loci, such as the 17q12-21 locus and IL-33, were only weakly or not significantly associated with sensitization, suggesting that these loci increase asthma risk by mechanisms other than allergic sensitization.
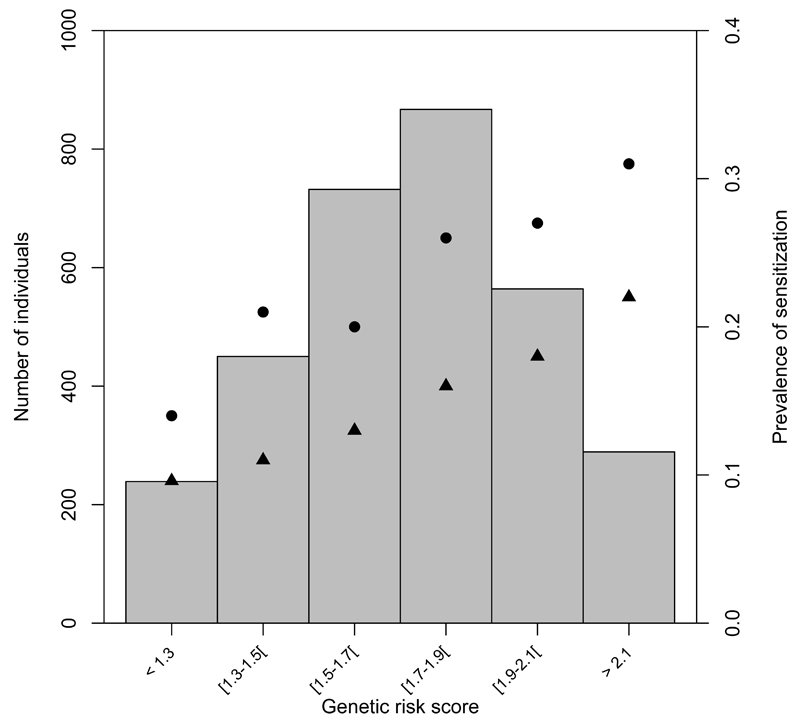
Lastly, we estimated the proportion of allergic sensitization in the general population caused by the 10 loci (Supplementary Table 17). A conservative PARF estimate above 25% was obtained by considering the 10% of the population with lowest risk score as the “unexposed” group. Results were similar for allergic rhinitis, which is the strongest clinical correlate of sensitization. The capacity to predict individual risk was limited (Supplementary notes). The sensitization prevalence by genetic risk score is depicted in Fig. 3 showing approximately 2 times higher prevalence of sensitization in the 9,2% of the population with highest genetic risk score compared to the 7,6% of the population with lowest risk score.
Figure 3. Combined impact of risk alleles from the 10 genome-wide significant loci on prevalence of allergic sensitization and allergic rhinitis (hay fever) in the population-based Health2006 replication study.
(a-c) For each individual a genetic risk score was calculated by applying the per-allele risk estimates from the replication sets, to the number of higher-risk alleles of each of the 10 genome-wide significant loci (one SNP per locus). The risk-score thus represents an index of the number of weighted risk alleles. Along the × axis, individuals in each risk score interval are shown, and the prevalence of sensitization/hay fever in each interval is plotted (y axis on right). The histogram (y axis on left) represents the number of individuals in each risk score interval. (a) Sensitization measured by Skin Prick Test. (b) Sensitization measured by Specific IgE in blood (circles and triangles depict sensitization prevalence with an IgE level cut off of 0.35 IU/mL and 3.5 IU/mL respectively). (c) Hayfever.
Several of the identified loci for sensitization are also reported susceptibility loci for autoimmune disease (Supplementary Table 18). We observed examples both of opposing direction of effect (for multiple sclerosis at the MYC/PVT1 locus) and same direction of effect (for diabetes and Crohn’s disease at the IL2 and C11orf30 loci) suggesting that the Th1/Th2 paradigm35 does not adequately explain the relationship between these disorders.
In conclusion, we increased the number of established susceptibility loci for allergic sensitization from three to 10, with a substantial population attributable risk fraction. Understanding the molecular mechanisms underlying the association with these sequence variants may provide novels insight into the etiology of allergy and other immune diseases.
URLs
MACH, http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G-PhaseI-Interim.html; HaploReg, http://www.broadinstitute.org/mammals/haploreg/haploreg.php
Methods
Phenotype definition
We studied the general tendency of specific IgE production without restriction by assessment method or allergen. Cases were defined as individuals with objectively measured sensitization against at least one of the allergens tested for in the respective studies, and controls were defined as individuals who were not sensitized against any of the allergens tested for (Supplementary Tables 1 and 2). We included sensitization assessed by skin reaction after puncture of the skin through a droplet of allergen extract (skin prick test, SPT) and/or by elevated levels of circulating allergen-specific IgE in blood. The SPT cut off level was 3 mm larger than the negative control for cases, and below 1 mm for controls. In order to optimize case specificity and the correlation between methods we chose a high cut off of specific IgE level for cases (3.5 IU/mL) and a low cut off for controls (0.35 IU/mL). We combined data from children and adults, but chose a lower age limit of 6 years since sensitization status at lower ages shows poorer correlation with sensitization later in life both due to transient sensitization and frequent development of sensitization during late childhood.
Discovery analysis
The discovery analysis was based on 5,798 individuals who were sensitized (cases) and 10,056 controls of European descent from 10 studies. The individual studies were approved by the local research ethics committees, and informed consent was obtained from all participants when necessary Details on sample recruitment and phenotypes and summary details for each collection are given in the Supplementary Notes and in Supplementary Table 1 and 2. Genome-wide genotyping was performed independently in each cohort with the use of various standard genotyping technologies (Supplementary Notes and Supplementary Table 3). Imputation was independently conducted for each study with reference to HapMap phase 2 or 3 CEU genotypes, and association analysis was performed using logistic regression models based on an expected allelic dosage model for SNPs, adjusting for ancestry-informative principal components as necessary. SNPs with MAF <1% and/or poor imputation quality (r2 <0.3, if using the 24 imputation algorithm MACH, or proper info <0.4, if using the IMPUTE imputation algorithm) were excluded. After genomic control at the level of the individual studies, we meta-analysed association data for ˜2.5 million imputed and genotyped autosomal SNPs using additive model fixed-effects meta-analysis as implemented in METAL (version 2010-08-01). For a given SNP, study-specific effect size estimates (transformed to Z-scores by METAL) were weighted inversely with their variance.
Replication analysis
We selected for replication the most strongly associated SNP in each of the 26 most strongly associated loci in the discovery meta-analysis (all with P < 1 × 10-5 in stage I; Table 1 and Supplementary Table 4). These SNPs were analyzed using in silico data from 6 studies with GWAS-data not included in the discovery meta-analysis and de novo genotyping data from 3 additional studies (Supplementary Tables 1, 2 and 3). The largest possible replication sample size amounted to 6,114 cases and 9,920 controls from 9 studies, all of European descent. Association analyses were again conducted for each study using a logistic regression model with similar covariate adjustments, based on expected allelic dosage for the in silico studies and allele counts in the de novo genotyping study. We applied a threshold of P < 5 × 10-8 for genome-wide significance.
Association and Manhattan plots
The Locuszoom tool was used to make association plots with 200kb flanking regions.36 Manhattan plots were plotted with the R Bioconductor software.
Functional Annotation
To help identify the potential causal gene(s) underlying each of the ten associations, we searched for cis eQTLs using gene expression results obtained from “in house” datasets of four cell types or tissues: white blood cells, lymphoblastoid cell lines (LCLs), whole blood and adipose tissue. ALSPAC: mRNA quantified expression data were collected for 947 unrelated ALSPAC participants from lymphoblastoid cell lines (LCLs) established from blood samples taken when the children were 9 years old. All cell lines were from a first passage, cells were harvested when confluent, washed in phosphate buffered saline and frozen in RNA Later (Ambion, Life Technologies, Grand Island, NY), RNA was extracted using Qiagen’s Rneasy extraction kit and amplified using Ambion’s Illumina TotalPrep 96 RNA amplification kit. Expression was surveyed using the Illumina HG-12 bead arrays and each individual run with 2 replicates. Expression data were normalized by quantile normalization between replicates and then median normalization across individuals was taken forward for further analysis. KORA: The expression analysis in this study was based on whole blood samples of the KORA (Cooperative Heath Research in the Region of Augsburg) F4 participants aged 62 to 81 years.37 Gene expression profiling was performed from whole blood using the Illumina HumanHT-12 v3 BeadChip as described elsewhere.38 DeCODE: The expression analyses were based on white blood cells (N=745) and adipose tissue (N=600) from unselected Icelandic individuals. Gene expression profiling was performed as previously described.39 B-cells and monocyte eQTL study: Expression quantitative trait analysis was performed on gene expression data from CD14+ monocytes (n=282) and CD19+ B-cells that were positively selected from freshly purified peripheral blood mononuclear cell fractions as previously described.40 Genotyping data for these European individuals was performed using HumanOmniExpress-12v1.0 BeadChips (Illumina), with whole genome imputation being performed using IMPUTE2 against the 1KG phase I integrated variant set (March 2012). Expression data was obtained using HumanHT-12 v4 BeadChips (Illumina), with variance stabilizing transformation and robust spline normalization between chips being performed using the lumi R package.41 To identify putative cis eQTL, linear analysis was performed using Matrix eQTL,42 using a 1Mb window between SNP and probe and using the first 15 principal components of the expression data as covariates.
Furthermore, we used publicly available bioinformatic resources to annotate putative associations with gene expression and coding SNPs. We annotated SNPs in linkage disequilibrium (r2>0.8, within 2Mb) with the top 10 associated SNPs based on 1000G Interim Phase I data (b37) of 381 Europeans. Minor and major alleles, MAF and the corresponding LD were computed using the haplotype data downloaded from MACH. Chromosomal positions were compared against UCSC dbSNP132 (hg19) for alternative SNP ID (e.g. chr4:38774337 to rs111437368). The GHS-Express database of Zeller et al.9 was used to identify SNPs associated with variant in gene expression levels (eQTL); RefGene tract of UCSC was used for gene annotation. Regulatory elements were annotated by HaploReg in terms of predicted ENCODE chromatin state (promoter and enhancer histone modification signals) and DNaseI hypersensitivity.
Population attributable risk fractions
(PARFs) were estimated from 3 large, general-population samples: B58C (age 44-45 years), Health2006 (age 18-69 years), and ALSPAC (age 7.5 years). Health2006 and the B58C sub-sample used for this purpose were part of the replication stage only while ALSPAC was both part of both the discovery and replication stage. Within each cohort, a genetic risk score was calculated by applying the pooled per-allele coefficients (lnORs) from the replication sets, to the number of higher-risk alleles of each of the 10 genome-wide significant loci (one SNP per locus). PARFs were estimated in two ways. Firstly, the prevalence of sensitization at a genetic risk score of zero (ie no higher risk alleles for any of the 10 SNPs) was predicted, with 95% confidence intervals, from the intercept term in a logistic regression model with risk score as a continuous explanatory variable. The PARF was then derived (with 95% confidence intervals) by expressing the difference between the observed prevalence and predicted (“unexposed”) prevalence as a percentage of the observed prevalence. Secondly, since there were no individuals observed with zero risk alleles, the prevalence of sensitization among individuals in the lowest decile of the genetic risk score was estimated in a similar manner, and used to derive a PARF on the assumption that this 10% of the population were “unexposed”. This estimate has the advantage that it does not predict beyond the bounds of the data, but its results are conservative compared to the first method. PARFs were estimated for sensitization measured by SPT and specific IgE (cut off 0.35 and 3.5 IU/mL) and allergic rhinitis.
Pathway enrichment analyses
MAGENTA pathway analysis
To explore potentially new pathways associated with allergic sensitisation in the full GWAS dataset, we applied an adaption of gene set enrichment analysis (meta-analysis gene-set enrichment of variants associations, MAGENTA version 2).31 Briefly, each gene in the genome is mapped to a single SNP with the lowest P value within a 110 Kb upstream or 40kb downstream window. Gene p-values are corrected for confounding factors such as gene size, LD patterns, SNP density, and other genetic factors. The adjusted P values are ranked and the observed number of genes in a given pathway above a given significance threshold (95th percentile) is calculated. This statistic is compared with 10000 randomly permuted pathways of identical size to generate an empirical GSEA P value for each pathway. In our study, individual pathways that reached a false discovery rate < 0.01 were deemed significant, and, unless otherwise stated, results for the 95-percentile cut-off analysis are reported.
To identify the degree of overlap between gene sets, we plotted an enrichment map of the significant pathways. An enrichment map is a network where vertices represent pathways, vertex size is proportional to the number of genes, and edge width represents the number of genes shared by two pathways. Vertices are colour coded according to enrichment FDR. To further identify which significant pathways are central, we plotted a bipartite network of significant genes (adjusted P value < 0.0273) connected to their corresponding pathways.
GRAIL pathway analysis
GRAIL32 was run with gene size correction enabled and based on PubMed abstracts up to December 2006. We specified genes at each of the 10 validated loci explicitly by considering all genes within a 1 mega base flanking regions of either side of the given lead SNP. VIZ-GRAIL tool43 was used for visualization of the GRAIL results.
Supplementary Material
Acknowledgements
The full list of acknowledgements for each study is in the Supplementary Note.
Footnotes
Author Contributions
Study level data analysis: K.B., T.H.P. (meta-analysis and systems biology analyses), R. Granell (meta-analysis), D.P.S., A.C.A. (systems biology analyses), A.L., J.A.C., N.M.W., M.S., M. Kerkhof, B.K.B., M. Kaakinen, P. Sleimann, G.T., K. Schramm, E.K.-M., A.S., L.C., C.S.T., B.P.F. , R. Gupta, M.B.. S.W., H.H., D.S.P., A.C., G.H.K., N.T.
Study design: K.B., M.C.M., T.H.P., D.P.S., A.L., B.K.B., E.K.-M., A.S., C.F.R., G.W.M., S.C.D., P.E., M.J.A., J.C.K., R. Gupta, P.J.T., J.N.H., M.B., S.W., H.H., J. Heinrich, D.S.P., A.C., C.E.P., M.-R.J., G.H.K., N.T., M.A.F., H.B., A.J.H.
Writing paper: K.B., T.H.P., R. Granell, D.P.S., A.C.A., A.L., J.A.C., G.H.K., N.T., M.A.F., H.B., A.J.H.
Data collection: K.B., M.C.M., D.P.S., A.L., I.J., B.K.B., U.T., A.S., J. Hui, E.H.W., D.L.D., G.J., L.P., C.F.R., J.P., E.T., A.L.H., S.D., L.L.H., C.H., A.J., B.P.F. , J.C.K., M.J.A., P.J.T., P.H., P. Sly, M.B., H.H., K. Stefansson, J. Heinrich, D.S.P., A.C., M.-R.J., G.H.K., H.B., A.J.H.
Genotyping: K.B., A.L., J.A.C., I.J., P. Sleimann, U.T., S.B., E.K.-M., A.S., B.S.P., C.M.T.T., S.M.R., W.L.M., G.W.M., L.L.H., J.P.K., M.W., H.H., K. Stefansson, A.C., C.E.P., M.-R.J., G.H.K., H.B.
Revising and reviewing paper: K.B., M.C.M., T.H.P., R. Granell, D.P.S., A.C.A., A.L., J.C., N.M.W., M.S., M. Kerkhof, I.J., B.K.B., M. Kaakinen, P. Sleimann, G.T., U.T., K. Schramm, S.B., E.K.-M., A.S., B.S.P., L.C., J. Hui, E.H.W., C.M.T.T., D.L.D., G.J., S.M.R., W.L.M., L.P., C.F.R., J.P., C.S.T., E.T., G.W.M., A.L.H., S.C.D., L.L.H., C.H., J.P.K., P.E., A.J., M.W., M.J.A., B.P.F., J.C.K., R. Gupta, P.J.T., P.H., P. Sly, J.N.H., M.B., S.W., H.H., K. Stefansson, J. Heinrich, D.S.P., A.C., C.E.P., M.R.J., G.H.K., N.T., M.A.F., H.B., A.J.H.
Competing financial interests
I.J., G.T., U.T. and K. Stefansson are employees of deCODE genetics. The remaining authors declared no competing interests of relevance to this paper.
Reference List
- 1.Johansson SG, et al. Revised nomenclature for allergy for global use: Report of the Nomenclature Review Committee of the World Allergy Organization, October 2003 12. J Allergy Clin Immunol. 2004;113:832–836. doi: 10.1016/j.jaci.2003.12.591. [DOI] [PubMed] [Google Scholar]
- 2.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease 10. Lancet. 2008;372:1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 3.Los H, Postmus PE, Boomsma DI. Asthma genetics and intermediate phenotypes: a review from twin studies 1. Twin Res. 2001;4:81–93. doi: 10.1375/1369052012191. [DOI] [PubMed] [Google Scholar]
- 4.Thomsen SF, Ulrik CS, Kyvik KO, Ferreira MA, Backer V. Multivariate genetic analysis of atopy phenotypes in a selected sample of twins. Clin Exp Allergy. 2006;36:1382–1390. doi: 10.1111/j.1365-2222.2006.02512.x. [DOI] [PubMed] [Google Scholar]
- 5.Thomsen SF, van der SS, Kyvik KO, Backer V. A study of asthma severity in adult twins. Clin Respir J. 2012;6:228–237. doi: 10.1111/j.1752-699X.2011.00273.x. [DOI] [PubMed] [Google Scholar]
- 6.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 7.Ramasamy A, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 8.Hinds D. A meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;99:99. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeller T, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 11.Huppi K, et al. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. 2008;6:212–221. doi: 10.1158/1541-7786.MCR-07-0105. [DOI] [PubMed] [Google Scholar]
- 12.Beck-Engeser GB, et al. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4. doi: 10.1186/1742-4690-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esparza-Gordillo J, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet. 2009;41:596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- 15.Ferreira MA, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kormann MS, et al. Toll-like receptor heterodimer variants protect from childhood asthma. J Allergy Clin Immunol. 2008;122:86–92. doi: 10.1016/j.jaci.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 19.Lazarus R, et al. TOLL-like receptor 10 genetic variation is associated with asthma in two independent samples. Am J Respir Crit Care Med. 2004;170:594–600. doi: 10.1164/rccm.200404-491OC. [DOI] [PubMed] [Google Scholar]
- 20.Reijmerink NE, et al. Toll-like receptors and microbial exposure: gene-gene and gene-environment interaction in the development of atopy. Eur Respir J. 2011;38:833–840. doi: 10.1183/09031936.00099210. [DOI] [PubMed] [Google Scholar]
- 21.Tesse R, Pandey RC, Kabesch M. Genetic variations in toll-like receptor pathway genes influence asthma and atopy. Allergy. 2011;66:307–316. doi: 10.1111/j.1398-9995.2010.02489.x. [DOI] [PubMed] [Google Scholar]
- 22.Potaczek DP, Kabesch M. Current concepts of IgE regulation and impact of genetic determinants. Clin Exp Allergy. 2012;42:852–871. doi: 10.1111/j.1365-2222.2011.03953.x. [DOI] [PubMed] [Google Scholar]
- 23.Gudbjartsson DF, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 24.Savenije OE, et al. Interleukin-1 receptor-like 1 polymorphisms are associated with serum IL1RL1-a, eosinophils, and asthma in childhood. J Allergy Clin Immunol. 2011;127:750–756. doi: 10.1016/j.jaci.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 25.Ichii H, et al. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol. 2002;3:558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 26.Christensen U, et al. Family based association analysis of the IL2 and IL15 genes in allergic disorders. Eur J Hum Genet. 2006;14:227–235. doi: 10.1038/sj.ejhg.5201541. [DOI] [PubMed] [Google Scholar]
- 27.Howell WM, Holgate ST. HLA genetics and allergic disease. Thorax. 1995;50:815–818. doi: 10.1136/thx.50.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin AM, et al. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-Hom variant. Proc Natl Acad Sci U S A. 2004;101:4180–4185. doi: 10.1073/pnas.0307067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strange A, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moffatt MF, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paternoster L, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat Genet. 2012;44:187–192. doi: 10.1038/ng.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabin RL, Levinson AI. The nexus between atopic disease and autoimmunity: a review of the epidemiological and mechanistic literature. Clin Exp Immunol. 2008;153:19–30. doi: 10.1111/j.1365-2249.2008.03679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathmann W, et al. Incidence of Type 2 diabetes in the elderly German population and the effect of clinical and lifestyle risk factors: KORA S4/F4 cohort study. Diabet Med. 2009;26:1212–1219. doi: 10.1111/j.1464-5491.2009.02863.x. [DOI] [PubMed] [Google Scholar]
- 38.Mehta D, et al. Impact of common regulatory single-nucleotide variants on gene expression profiles in whole blood. Eur J Hum Genet. 2012 doi: 10.1038/ejhg.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 40.Fairfax BP, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 42.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raychaudhuri S. VIZ-GRAIL: visualizing functional connections across disease loci. Bioinformatics. 2011;27:1589–1590. doi: 10.1093/bioinformatics/btr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.