Abstract
The metalloregulatory protein ArsR, which offers high affinity and selectivity toward arsenite, was overexpressed in Escherichia coli in an attempt to increase the bioaccumulation of arsenic. Overproduction of ArsR resulted in elevated levels of arsenite bioaccumulation but also a severe reduction in cell growth. Incorporation of an elastin-like polypeptide as the fusion partner to ArsR (ELP153AR) improved cell growth by twofold without compromising the ability to accumulate arsenite. Resting cells overexpressing ELP153AR accumulated 5- and 60-fold-higher levels of arsenate and arsenite than control cells without ArsR overexpression. Conversely, no significant improvement in Cd2+ or Zn2+ accumulation was observed, validating the specificity of ArsR. The high affinity of ArsR allowed 100% removal of 50 ppb of arsenite from contaminated water with these engineered cells, providing a technology useful to comply with the newly approved U.S. Environmental Protection Agency limit of 10 ppb. These results open up the possibility of using cells overexpressing ArsR as an inexpensive, high-affinity ligand for arsenic removal from contaminated drinking and ground water.
Arsenic (As) is an extremely toxic metalloid that adversely affects human health. Both highly toxic trivalent arsenite [As(III)] and less-toxic pentavalent arsenate [As(V)] have been associated with increased risk of skin, kidney, lung, and bladder cancer (12). The toxicity of arsenic is attributed to the substitution of As(V) for phosphate, affinity of As(III) for protein thiol groups, and protein-DNA and DNA-DNA cross-linking (20). Arsenic enters the water supply primarily from geochemical sources, such as the mining of arsenopyrite gold ores (16), which constitute about one-third of world gold reserves. Additional contaminations arise from anthropomorphic sources such as arsenical-containing herbicides, pesticides, and the widely used wood preservative chromated copper arsenic (17, 21). Untreated, highly toxic arsenic effluent has been disposed of in rivers and ended up in groundwater. An estimated 20 million Americans consume water containing arsenic at levels presenting a potentially fatal cancer risk (9). Worldwide, 57 million people have been exposed to arsenic through contaminated wells in Bangladesh (18). These incidents again serve as a reminder of the toxic consequences of arsenic mobilization and the needs for efficient removal of arsenic in aquatic systems (19).
Because of the toxicity, the regulatory limit on arsenic in the United States is currently set at 10 ppb. There are a variety of methods currently available for removal of arsenic from contaminated water. Conventional technologies, such as coagulation, do not discriminate between arsenic and other elements and involve alteration of the water chemistry and addition of other chemicals (7). Current technologies, such as activated alumina sorption, polymeric anion exchange, and polymeric ligand exchange (6), are more effective for As(V) than As(III), and most commonly used methods require prior oxidation of all of the As(III) to As(V) (10, 30). Biological methods are gaining momentum because of their potential in providing a cost-effective technology for heavy metal remediation. One emerging technology that has received more attention in recent years is the development of biosorbents with high affinity and specificity. Unfortunately, very few reports on the use of microorganisms for arsenic removal have been demonstrated except for nonspecific binding to the cell walls (26). These biosorbents, however, generally lack the high affinity and specificity. Although arsenic hyperresistant organisms have been isolated from arsenic-contaminated sites (5, 15), their use for arsenic bioremediation has not been reported.
It is well established that many microorganisms survive in the presence of toxic metals or metalloids by inducing the expression of an array of resistance proteins. The highly specific nature of these resistance mechanisms is the result of a cleverly designed genetic circuit that is tightly controlled by specific metalloregulatory proteins. The Escherichia coli chromosomal ars operon, for example, confers resistance to arsenicals and antimonials (31). The regulatory protein of the operon, ArsR, has been shown to be a trans-acting repressor that senses environmental As(III) (32). The ArsR protein contains a very specific binding site toward As(III) and can discriminate effectively against phosphate, sulfate, cobalt, and cadmium (28). ArsR has a strikingly high affinity, as even 10−15 M As(III) could induce the ars promoter (22). Although the high affinity and specificity of ArsR have been exploited for development of whole-cell biosensors (22), no one has taken advantage of ArsR for arsenic remediation. In this work, we present a new method for the selective removal of arsenic by using engineered E. coli cells overexpressing ArsR. The resulting E. coli strain was endowed with the ability to bind arsenic with high affinity and selectivity similar to that exhibited by ArsR.
MATERIALS AND METHODS
Bacterial strains and molecular techniques.
E. coli JM109 {Δ(lac-proAB) glnV44 e14- gyrA96 recA1 endA1 thi hsdR17 [F′ traD36 proA+ B+ lacIq (lacZ)M15]} was used in all cloning steps. E. coli BLR(DE3) (Novagen, Madison, Wis.), a common strain used for high-level expression from the T7 promoter, was used for protein expression and arsenic-binding experiments. All molecular biology techniques were carried out as described previously (23).
Expression vectors for ArsR and the elastin-like protein (ELP)-ArsR fusion.
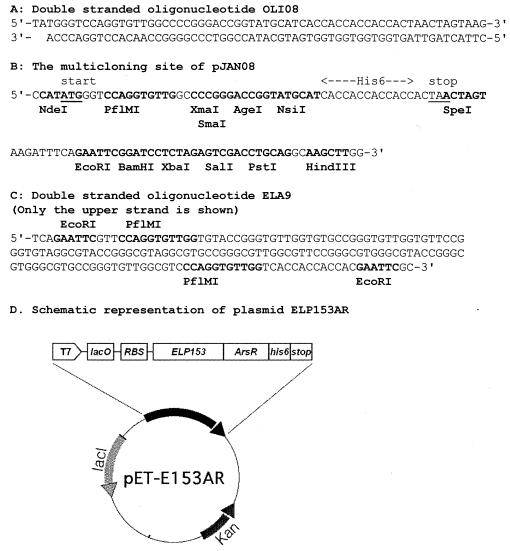
Plasmid pMal-c2x (New England BioLabs, Beverly, Mass.) was digested with PflMI, blunt ended by removing the single-stranded overhangs with mung bean nuclease, and religated, thus replacing the PflMI site with a new NdeI site. The resulting plasmid (6,645 bp) was digested with NdeI and XmnI, and a 3,972-bp fragment was ligated with a synthetic linker OLI08 (Fig. 1A), creating the 4,037-bp vector pJAN08. This vector was constructed to facilitate subcloning by providing the start and stop codons, a number of additional restriction sites, and a C-terminal hexahistidine tag (Fig. 1B).
FIG. 1.
Oligonucleotide and cloning sequences used for the construction of expression vectors.
To construct an expression vector for ArsR, the arsR gene was retrieved from plasmid pETR (donation by B. P. Rosen, Wayne State University, Detroit, Mich.) by PCR using the primers arsfor, 5′-GCACCGGTGGCATGTCATTTCTGTTACCC-3′, and arsback, 5′-CCTCTGCAGTTAACTGCAAATGTTCTT-3′. The amplified fragment was digested with AgeI and PstI and inserted into the corresponding sites in pJAN08, creating p08-ArsR. To express ArsR under control of a T7 promoter, the NdeI-PstI fragment of p08-ArsR was cloned directly into the expression vector pET38b+ (Novagen) to create pETAR.
To express ArsR as a fusion to ELP, the desired ELP sequence was constructed by concatenation of a basic DNA monomer coding for the VPGVG (13) sequence of ELP. Two oligonucleotides were annealed, and the overhangs were filled in with Pfu polymerase to create the 179-bp DNA monomer ELA9 (Fig. 1C). ELA9 was digested with EcoRI and inserted into the EcoRI site of pBluescript SK(+) (Stratagene, La Jolla, Calif.). Larger quantities of ELA9 were then generated by PCR, using the primers KS, 5′-TCGAGGTCGACGGTATC-3′, and SK, 5′-CGCTCTAGAACTAGTGGATC-3′ [flanking the EcoRI site in the original pBluescript SK(+)] and the plasmid pBluescript-ELA9 as the template. The 239-bp product was digested with PflMI, and the resulting 130-bp fragment was concatenated with T4 DNA ligase at a high DNA concentration (60 μg/ml). The resulting high-molecular-weight (>10-kb) DNA fragments were partially digested with PflMI to yield products of the required sizes, which were then inserted into the PflMI site of pJAN08 to provide an in-frame fusion with the start codon. One plasmid containing 153 repeats of VPGVG (17 copies of the DNA monomer) was designated p08-ELP153 and was used for future work. The resulting ELP sequence was excised as an NdeI-XmaI fragment and inserted into corresponding sites of p08-ArsR, creating p08-ELP153-ArsR. The open reading frame coding for the ELP-ArsR (ELP153AR) fusion was then transferred into the expression vector pET38b+ (Novagen) as an NdeI-PstI fragment.
Protein expression, purification, and characterization.
E. coli BLR(DE3) cells containing different expression vectors were grown for 2 days in Terrific broth (TB) (23) at 30°C. Cells were harvested, washed, resuspended in TB74S buffer (50 mM Tris, 150 mM NaCl; pH 7.4) to an optical density at 600 nm of ∼10, and lysed with a French press. Production of ELP153AR and ArsR was verified by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gel (14), followed by Coomassie blue staining (23).
Arsenic accumulation by growing cells.
Cells were grown in TB as described above in the presence of 10 μM sodium arsenite. One-milliliter samples were taken at various time points for cell mass determination and for arsenic analysis. Samples were harvested in a microcentrifuge and resuspended in fresh TB74S buffer, and the supernatant was carefully removed by centrifugation. The cell pellet was then air dried for 2 days at room temperature and dissolved in 200 μl of concentrated nitric acid for 2 days at room temperature. As a control, cells harboring pET38b+ were used for comparison.
Arsenic and heavy metal accumulation by resting cells.
Cells were grown in TB and harvested as described above. Cells were resuspended in prewarmed TB74S buffer (30°C) to a final concentration of 2.2 mg/ml (dry cell weight) unless stated otherwise. Sodium arsenite, sodium arsenate, cadmium chloride, or zinc chloride was added as described in each experiment. Cells harboring pET38b+ without ArsR overexpression were used as a control for comparison. To study the effect of ionic strength, various amounts of sodium chloride were added as indicated. Samples were agitated at 30°C for 1 h or the indicated time period, after which 1 ml of the suspension was used for analysis.
To evaluate the efficiency of resting cells to remove low levels of As(III), contaminated water spiked with 50 ppb of As(III) was incubated with different amounts of resting cells and the percentage of As(III) removed was determined after 1 h of incubation.
Determination of metal content.
Arsenic was determined by atomic absorption spectrophotometry (Shimadzu AA6701) at 193.7 nm using a pyrolytically coated graphite furnace tube with an inserted L'vov platform. Then, 0.054% palladium and 0.011% magnesium were added as a matrix modifier prior to loading of the diluted samples. All other parameters used were as recommended by the manufacturer (Shimadzu). Zinc and cadmium were determined by atomic absorption spectrophotometry at 213.9 and 228.8 nm, respectively, in the flame atomization mode.
RESULTS
Overexpression of ArsR in E. coli and bioaccumulation of arsenite.
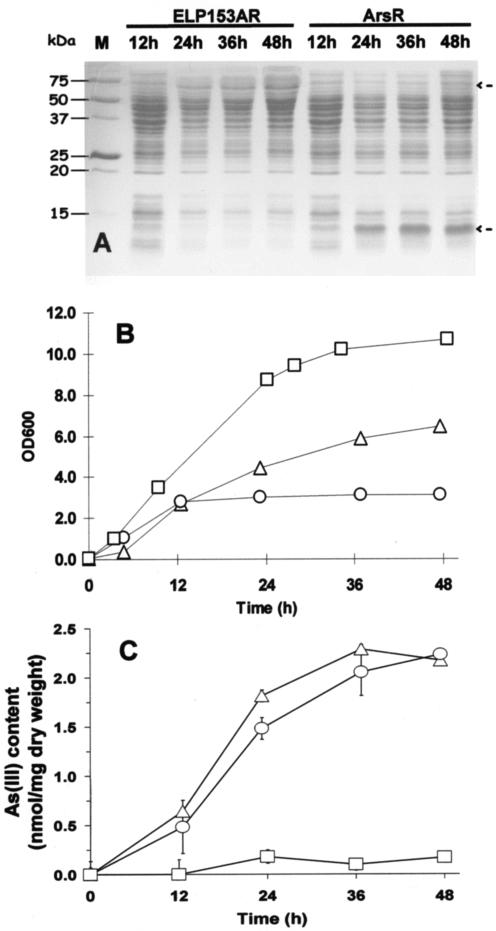
The arsR gene was placed under control of a strong T7 promoter in p08-ArsR, allowing high-level synthesis of ArsR without isopropyl-β-d-thiogalactopyranoside addition (Fig. 1D). Overexpression of ArsR was demonstrated by detection of a strong band corresponding to the expected size of ArsR (Fig. 2A). The ability of engineered cells to accumulate As(III) was investigated by the addition of 10 μM sodium arsenite to the growth medium. Only a modest level of As(III) accumulation occurred at 12 h, when ArsR expression was low (Fig. 2A and C). However, the level of accumulation increased by fourfold at 24 h, when the ArsR expression level was substantially increased. The specific As(III) content was 13-fold higher than the control without ArsR expression after 48 h. This level of As(III) accumulation (2.3 nmol/mg [dry weight]) is also higher than the value reported for arsenic accumulation (1.47 nmol/mg [dry weight]) by E. coli cells producing phytochelatins (PCs) (25).
FIG. 2.
Bioaccumulation of arsenite. (A) Expression of ELP153AR and ArsR. Samples were harvested at different time points, and the total cellular proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12.5% polyacrylamide). The desired proteins are marked with arrows. (B and C) Growth profiles (B) and arsenite contents (C) of E. coli BLR(DE3) cells expressing ArsR (○), ELP153AR (▵), or no protein (□).
High-level expression of ArsR was apparently toxic to the cells, as a threefold reduction in the final cell density was observed (Fig. 2B). This reduction did not appear to be the toxic effect of As(III), as the same reduction was observed with or without As(III). It has been shown that the addition of a fusion partner could alleviate the toxic effect of protein overexpression (8). To test this possibility, an ELP coding for 153 repeats of VPGVG, which has been shown to decrease single-chain Fv fragment toxicity (13), was fused to ArsR to generate an ELP-ArsR fusion (ELP153AR). Production of ELP153AR was again detected under the same growth conditions (Fig. 2A). More importantly, the final cell density was increased by more than twofold (Fig. 2B). Although the level of expression was lower than that of ArsR, the profile and the level of As(III) accumulation were very similar to those of cells overexpressing ArsR (Fig. 2C). This may be attributed to the inefficient As(III) uptake, which limits the overall accumulation of As(III). Owing to the similar level of As(III) accumulation and the improved cell growth, cells expressing ELP153AR were subjected to further investigation.
Arsenite accumulation by resting cells.
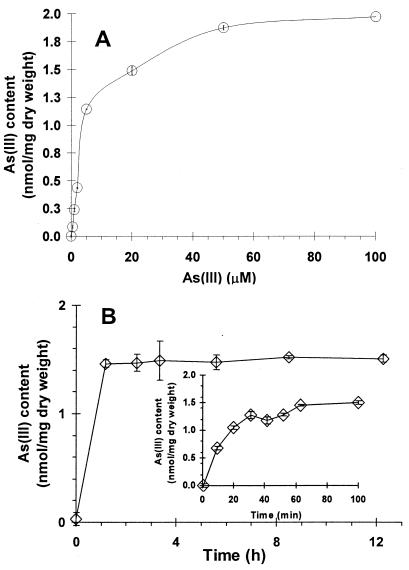
One potential utility of this technology is the use of resting cells as affinity ligands for arsenic removal. To investigate whether resting cells could accumulate As(III) to a similar level as growing cells, the binding profile was determined over a range of As(III) concentrations. Overnight-grown cells were resuspended in buffer to a concentration of 2.2 mg (dry weight)/ml, and the level of As(III) that accumulated after 1 h was determined at various As(III) concentrations (Fig. 3A). The binding level increased quickly at low concentrations and reached a plateau of 1.9 nmol/mg (dry weight) around 50 μM arsenite. This maximum accumulation level was virtually the same as the value obtained with growing cultures. This represented a 60-fold increase in As(III) accumulation, as resting cells not expressing ELP153AR bound virtually no As(III) (0.03 nmol/mg [dry weight]) under this condition.
FIG. 3.
As(III) bioaccumulation by E. coli BLR(DE3) cells harboring pET-E153AR. (A) As(III) binding isotherm. As(III) binding was determined at various concentrations after 1 h of incubation. (B) Time profile of As(III) uptake by resting cells. Resting cells were resuspended in Tris buffer (pH 7.4) containing 20 μM As(III) and incubated for the indicated lengths of time. Data shown are the mean values (± standard deviations) obtained from three independent experiments.
To determine the rate of As(III) accumulation, a time course experiment was conducted in the presence of 20 μM arsenite. As shown in Fig. 3B, As(III) accumulation occurred very rapidly, with maximum accumulation within 1 h. The same level of As(III) accumulation was maintained over a 12-h period, indicating that As(III) was stably bound to the cells. This rapid accumulation rate and good stability suggest that these resting cells may be suitable as an efficient ligand for arsenic removal in contaminated water.
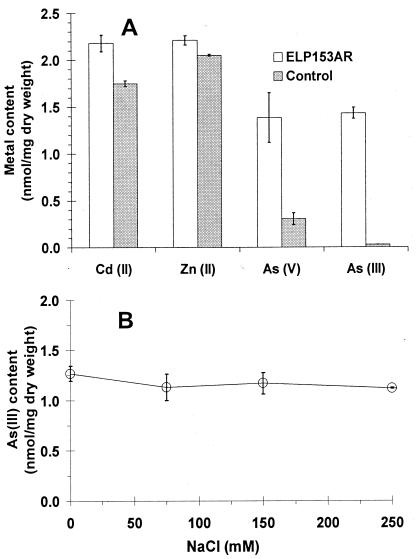
Binding selectivity.
To demonstrate the selectivity of ELP153AR toward arsenic, similar binding experiments were conducted with arsenite, arsenate, cadmium, and zinc. For cadmium and zinc, no significant increase in accumulation was observed compared to control cells (Fig. 4A). Similar to As(III), a significant increase in arsenate accumulation was observed; however, the extent of improvement was only fivefold. Although ArsR is known to be specific for As(III), removal of As(V) from the supernatant may occur through the initial conversion of As(V) into As(III) by the arsenate reductase and the subsequent sequestration by ArsR.
FIG. 4.
Arsenic selectivity. (A) The intracellular metal or As contents of resting cells with or without overexpression of ELP153AR (control) were determined after 1 h of incubation with either As(III) (10 μM), As(V) (10 μM), cadmium chloride (25 μM), or zinc chloride (65 μM). (B) Effect of sodium ion on As(III) binding. Resting cells were resuspended in Tris buffer (pH 7.4) containing 10 μM As(III), and the indicated concentration of NaCl was added. As(III) binding was determined after 1 h of incubation. Data shown are the mean values (± standard deviations) obtained from three independent experiments.
Many whole-cell sorbents for heavy metals are sensitive to the Na+ commonly found in contaminated waters (11). To investigate whether the presence of Na+ has any effect on whole-cell binding of As(III), binding experiments were performed at increasing concentrations of sodium chloride. As shown in Fig. 4B, no decrease in As(III) binding was observed at any Na+ concentration tested up to 250 mM.
Removal of arsenite from contaminated water.
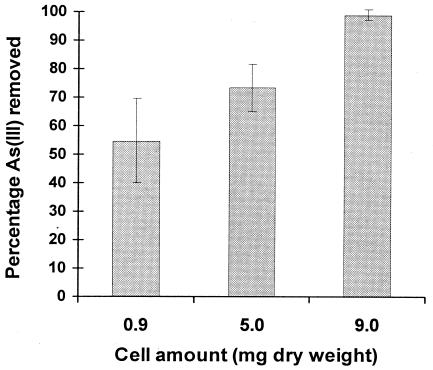
According to the estimate of the U.S. Environmental Protection Agency, the newly promulgated arsenic maximum contaminant level of 10 ppb from 50 ppb in drinking water would require corrective action for over 4,000 water supply systems serving an approximate population of 20 million (9). To utilize the recombinant cells as an efficient ligand for arsenic treatment would necessitate removal beyond the required regulatory limit. To investigate this possibility, resting cells were incubated with water spiked with 50 ppb of arsenite, and the percent removal was determined after 1 h. At a cell loading of 0.9 mg (dry weight)/ml, only 54% of the added As(III) was removed from the supernatant (Fig. 5). Increasing the cell loading resulted in increased efficiencies, with 98% of the added As(III) removed at a cell loading of 9 mg (dry weight)/ml. The high efficiency of arsenic removal along with the high selectivity toward arsenic suggest that these engineered cells may be very useful for arsenic removal from municipal water sources.
FIG. 5.
Evaluation of arsenic binding by E. coli BLR(DE3) cells harboring pET-E153AR. Water spiked with 50 ppb of arsenite was incubated with different amounts of resting cells, and the percentage of arsenite removed was determined after 1 h of incubation. Data shown are the mean values (± standard deviations) obtained from three independent experiments.
DISCUSSION
The use of plant, algae, or microorganisms as biosorbents for heavy metal removal from contaminated water is well documented (15, 24, 26). In recent years, enhanced biosorbents have been engineered by genetic incorporation of metal-binding peptides such as metallothioneins (29) or synthetic PCs (2, 3) in different bacteria or yeasts. Arsenic, a metalloid, on the other hand, does not bind well to these peptides. The only exception is PCs, which are naturally occurring metal-binding peptides found in plants and yeast (27). The presence of a γ bond between glutamic acid and cysteine in PCs perhaps allows the required coordination for thiol-As complexation. Expression of PC synthase in E. coli resulted in production of PCs and enhanced accumulation of arsenate (25). However, this strategy lacked selectivity, as the engineered cells also demonstrated enhanced binding to Cd2+, Zn2+, Pb2+, and Cu2+.
The use of a metalloregulatory protein as a specific metal-binding ligand has recently been reported for enhanced mercury removal (1). The success of this strategy is based on the physiological role of these metalloregulatory proteins as specific sensors toward the metals of interest (4). E. coli confers resistance to arsenic by a specific efflux pump controlled by an arsenite-inducible repressor, ArsR (31). In this work, both growing and resting cells overexpressing ArsR accumulated 13- to 60-fold-higher levels of As(III) and As(V) than control cells without ArsR overexpression. The level of arsenic accumulation (∼2.2 to 1.47 nmol/mg [dry weight]) is also higher than that reported for cells producing PC (25). In contrast to PCs, the physiological role of ArsR as an As(III) sensor molecule allows a high degree of specificity and affinity towards arsenic, as no significant improvement in Cd2+ or Zn2+ accumulation was observed. This represents the first demonstration of enhanced intracellular accumulation of both As(III) and As(V) by a genetically engineered microorganism. The increased accumulation suggests that the high levels of ArsR could compete with the efflux pump for the available intracellular As(III), thus rendering the bound As(III) in nontoxic forms by sequestration. It is also possible that the overexpression of ArsR results in repression of the ars operon, reducing the efflux efficiency and further increasing the intracellular sequestration of As(III) by ArsR. More importantly, the high affinity of these engineered cells toward As enables 98% removal of 50 ppb of As(III) from contaminated water. Therefore, overexpression of ArsR could be a promising strategy to increase the cellular accumulation of arsenic and provide a highly selective ligand for bioremediation processes. As a result of the selectivity and affinity, the use of these engineered cells as arsenic-binding ligands, either in biofilm reactors or biofilters, may be an effective alternative to chemical ligands for treatment of contaminated water.
Acknowledgments
This work was funded by grants from the U.S. EPA (R829606) and NSF (BES0331416).
REFERENCES
- 1.Bae, W., C. Wu, J. Kostal, A. Mulchandani, and W. Chen. 2003. Enhanced mercury biosorption by bacterial cells with surface-displayed MerR. Appl. Environ. Microbiol. 69:3176-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, W., R. Mehra, A. Mulchandani, and W. Chen. 2001. Genetic engineering of Escherichia coli for enhanced bioaccumulation of mercury. Appl. Environ. Microbiol. 67:5335-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, W., W. Chen, A. Mulchandani, and R. Mehra. 2000. Enhanced bioaccumulation of heavy metals by bacterial cells displaying synthetic phytochelatins. Biotechnol. Bioeng. 70:518-523. [DOI] [PubMed] [Google Scholar]
- 4.Bontidean, I., C. Berggren, G. Johansson, E. Csorgi, B. Mattiasson, J. R. Lloyd, K. J. Jakeman, and N. L. Brown. 1998. Detection of heavy metal ions at femtomolar levels using protein-based biosensors. Anal. Chem. 70:4162-4169. [DOI] [PubMed] [Google Scholar]
- 5.Canovas, D., C. Duran, N. Rodriguez, R. Amils, and V. De Lorenzo. 2003. Testing the limits of biological tolerance to arsenic in a fungus isolated from the River Tinto. Environ. Microbiol. 5:133-138. [DOI] [PubMed] [Google Scholar]
- 6.Chwirka, J. D., B. M. Thomson, and J. M. Stomp. 2000. Removing arsenic from groundwater. J. Am. Water Works Assoc. 92:79-88. [Google Scholar]
- 7.Clifford, D., S. Subramonian, and T. Sorg. 1998. Removing dissolved inorganic contaminants from water. Environ. Sci. Technol. 20:1072-1080. [Google Scholar]
- 8.Davis, G. D., C. Elisee, D. M. Newham, and R. G. Harrison. 1999. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol. Bioeng. 65:382-388. [PubMed] [Google Scholar]
- 9.DeMarco, M. J., A. K. SenGupta, and J. E. Greenleaf. 2003. Arsenic removal using a polymeric/inorganic hybrid sorbent. Water Res. 37:164-176. [DOI] [PubMed] [Google Scholar]
- 10.Driehaus, W., R. Seith, and M. Jekel. 1995. Oxidation of arsenic(III) with manganese oxides in water treatment. Water Res. 29:297-305. [Google Scholar]
- 11.Gang, M. A., and D. Langmuir. 1974. Controls on heavy metals in surface and ground waters affected by coal mine drainage, p. 39-69. In Proceedings of the 5th Symposium on Coal Mine Drainage Research. National Coal Mine Association, Washington, D.C.
- 12.Karagas, M. R., T. D. Tosteson, J. Blum, J. S. Morris, J. A. Baron, and B. Klaue. 1998. Design of an epidemiologic study of drinking water arsenic exposure and skin and bladder cancer risk in a U.S. population. Environ. Health Perspect. 106:1047-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, J.-Y., A. Mulchandani, and W. Chen. 2003. An immunoassay for atrazine using tunable immunosorbent. Anal. Biochem. 322:251-256. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Ma, L. Q., K. M. Komar, C. Tu, W. Zhang, Y. Cai, and E. D. Kennelley. 2001. A fern that hyperaccumulates arsenic. Nature 409:579. [DOI] [PubMed] [Google Scholar]
- 16.Macy, J. M., K. Nunan, K. D. Hagen, D. R. Dixon, P. J. Harbour, M. Cahill, and L. I. Sly. 1996. Chrysiogenes arsenatis gen. nov., sp. nov., a new arsenate respiring bacterium isolated from gold mine wastewater. Int. J. Syst. Bacteriol. 46:1153-1157. [DOI] [PubMed] [Google Scholar]
- 17.National Research Council. 1977. Arsenic. National Academy of Sciences, Washington, D.C.
- 18.Nickson, R., J. McArthur, W. Burgess, K. M. Ahmed, P. Ravenscroft, and M. Rahman. 1998. Arsenic poisoning of Bangladesh groundwater. Nature 395:338. [DOI] [PubMed] [Google Scholar]
- 19.Nickson, R. T., J. M. McArthur, P. Ravenscroft, W. G. Burgess, and K. M. Ahmed. 2000. Mechanism of arsenic release to groundwater, Bangladesh and West Bengal. Appl. Geochem. 15:403-413. [Google Scholar]
- 20.Norman, N. C. 1998. Chemistry of arsenic, antimony, and bismuth. J. Natl. Cancer Inst. 40:453-463. [Google Scholar]
- 21.Peryea, F. J., and R. Kammereck. 1997. Phosphate-enhanced movement of arsenic out of lead arsenate-contaminated topsoil and through uncontaminated subsoil. Water Air Soil Pollut. 93:243-254. [Google Scholar]
- 22.Ramanathan, S., W. Shi, B. P. Rosen, and S. Daunert. 1997. Sensing antimonite and arsenite at the subattomole level with genetically engineered bioluminescent bacteria. Anal. Chem. 69:3380-3384. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Sar, P., and S. D'Souza. 2001. Biosorptive uranium uptake by a Pseudomonas strain: characterization and equilibrium studies. J. Chem. Technol. Biotechnol. 76:1286-1294. [Google Scholar]
- 25.Sauge-Merke, S., S. Cuine, P. Carrier, C. Lecomte-Pradines, D.-T. Luu, and G. Peltier. 2003. Enhanced toxic metal accumulation in engineered bacterial cells expressing Arabidopsis thaliana phytochelatin synthase. Appl. Environ. Microbiol. 69:490-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Say, R., N. Yilmaz, and A. Denizli. 2003. Biosorption of cadmium, lead, mercury, and arsenic ions by the fungus Penicillium purpurogenum. Sep. Sci. Technol. 38:2039-2053. [Google Scholar]
- 27.Schmoger, M. E. V., M. Oven, and E. Grill. 2000. Detoxification of arsenic by phytochelatins in plants. Plant Physiol. 122:793-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott, D. L., S. Ramanathan, W. Shi, B. P. Rosen, and S. Daunert. 1997. Genetically engineered bacteria: electrochemical sensing systems for antimonite and arsenite. Anal. Chem. 69:16-20. [DOI] [PubMed] [Google Scholar]
- 29.Sousa, C., P. Kotrba, T. Ruml, A. Cebolla, and V. de Lorenzo. 1998. Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. J. Bacteriol. 180:2280-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkie, J. A., and J. G. Hering. 1998. Rapid oxidation of geothermal arsenic(III) in streamwaters of the eastern Sierra Nevada. Environ. Sci. Technol. 32:657-662. [Google Scholar]
- 31.Xu, C., and B. P. Rosen. 1997. Dimerization is essential for DNA binding and repression by the ArsR metalloregulatory protein of Escherichia coli. J. Biol. Chem. 272:15734-15738. [DOI] [PubMed] [Google Scholar]
- 32.Xu, C., W. Shi, and B. P. Rosen. 1996. The chromosomal arsR gene of Escherichia coli encodes a trans-acting metalloregulatory protein. J. Biol. Chem. 271:2427-2432. [DOI] [PubMed] [Google Scholar]