Abstract
The human T-cell leukaemia virus type 1 and type 2 (HTLV-1/HTLV-2) antisense proteins HBZ and APH-2 play key roles in the HTLV lifecycles and persistence in the host. Nuclear Factors Associated with double-stranded RNA (NFAR) proteins NF90/110 function in the lifecycles of several viruses and participate in host innate immunity against infection and oncogenesis. Using GST pulldown and co-immunoprecipitation assays we demonstrate specific novel interactions between HBZ/APH-2 and NF90/110 and characterised the protein domains involved. Moreover we show that NF90/110 significantly enhance Tax mediated LTR activation, an effect that was abolished by HBZ but enhanced by APH-2. Additionally we found that HBZ and APH-2 modulate the promoter activity of survivin and are capable of antagonising NF110-mediated survivin activation. Thus interactions between HTLV antisense proteins and the NFAR protein family have an overall positive impact on HTLV infection. Hence NFARs may represent potential therapeutic targets in HTLV infected cells.
Keywords: HTLV, NFAR, HBZ, APH-2, Survivin, YM155, Tax1, Tax2
1. Background
Human T-cell leukaemia virus type 1 (HTLV-1) is the causative agent of two distinct pathologies; adult T-cell leukaemia (ATL), an aggressive malignancy of CD4+/CD25+T-lymphocytes, and a chronic neurodegenerative disorder termed HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) (Poiesz et al., 1980; Hinuma et al., 1981; Kaplan et al., 1990). HTLV-2 is closely related to HTLV-1 however, despite sharing similar genetic organisation and expression strategies, HTLV-2 has not been linked to the development of ATL or any malignancy (Cavallari et al., 2013; Bender et al., 2012; Ciminale et al., 2014; Rende et al., 2012). Instead infection is associated with non-malignant lymphocyte proliferation, elevated platelet counts and milder neurological disorders (Araujo and Hall, 2004; Roucoux and Murphy, 2004; Bartman et al., 2008).
HTLV-1 and -2 encode the regulatory proteins Tax1 and Tax2 which are pivotal to HTLV-1 pathogenesis (Higuchi and Fujii, 2009). Interaction of Tax1 with CREB/ATF and p300/CBP complexes enhances their affinity for cAMP responsive elements within the viral long terminal repeats (LTRs) and hence potently activates viral gene expression (Zhao and Giam, 1991; Suzuki et al., 1993; Jiang et al., 1999). Moreover activation of NF-κB pathways by Tax1 plays central roles in T-cell transformation (Currer et al., 2012; Grassmann et al., 2005) and in the development of leukaemia/lymphoma in Tax1 transgenic animals (Hasegawa et al., 2006). ATL cells display constitutive NF-κB activation despite the loss of Tax1 expression due to genetic and epigenetic modification of the 5′LTR (Matsuoka and Jeang, 2007; Nasr et al., 2011), indicating that while Tax1 is critical for T-cell transformation, additional viral/cellular interactions are responsible for the later stages of leukaemogenesis.
The antisense strand of the HTLV-1 genome encodes a regulatory protein, termed HTLV-1 bZIP factor (HBZ) (Gaudray et al., 2002). HBZ is recognised as a key player in HTLV-1 pathogenesis as its constitutive expression in HTLV-1 infected cells and ATL cells appears to be fundamental to ATL and HAM/TSP progression (Saito et al., 2009; Satou et al., 2006, 2011). HBZ often exhibits opposing effects compared to Tax1; for instance, the interaction between HBZ and CREB prevents association of CREB to CRE present in the HTLV-1 LTR resulting in inhibition of HTLV-1 gene expression (Gaudray et al., 2002). Moreover, HBZ modulates both the AP-1 (Basbous et al., 2003; Thebault et al., 2004; Matsumoto et al., 2005) and canonical NF-κB (Zhao et al., 2009) pathways in an opposing manner to Tax1. While HBZ expression is dispensable for viral replication and cellular immortalisation, it plays an important role in the maintenance of infection, evidenced by its ability to enhance T-cell proliferation and promote viral persistence (Arnold et al., 2006; Arnold et al., 2008). Transgenic expression of HBZ in a murine model induces increased T-cell proliferation in addition to the development of T-cell lymphomas and chronic inflammation, suggesting a key role in HTLV-1 pathogenesis (Satou et al., 2011).
HTLV-2 also exploits 3′LTR transcription to express its antisense protein, APH-2 (antisense protein of HTLV-2) (Halin et al., 2009), however little is known about the role of APH-2 in HTLV-2 infection. Similarly to HBZ, APH-2 inhibits Tax2-mediated LTR trans-activation through its interaction with CREB (Halin et al., 2009; Yin et al., 2012). APH-2 can also modulate the AP-1 pathway; APH-2 interacts with c-Jun, JunB and JunD resulting in stimulation of their transcriptional activity, which stands in contrast to HBZ (Marban et al., 2012). Unlike HBZ, APH-2 is not capable of promoting T-cell proliferation in vitro, while lymphocytosis in HTLV-2-infected individuals is not attributed to APH-2. Remarkably inoculation of APH-2 knockout mutant viruses in a rabbit model results in increased proviral loads and immunological responses compared to wild-type HTLV-2-infected rabbits (Yin et al., 2012), suggesting that the expression of APH-2 may in fact hinder the spread of HTLV-2 infection, likely contributing to its decreased pathogenicity compared to HTLV-1.
The NFAR protein family are double-stranded RNA binding proteins encoded by the ilf3 gene, which by differential splicing produces at least five distinct mRNA species encoding proteins that differ primarily in their C-terminus (Reichman et al., 2003). NFAR proteins are particularly renowned for their roles in regulating cellular and viral gene expression at both transcriptional and post-transcriptional levels, having critical functions in mRNA stability, export and translational events (Reichman et al., 2003; 2002; Corthesy and Kao, 1994; Kao et al., 1994; Shim et al., 2002; Kuwano et al., 2010; Shi et al., 2007). More recently it has been established that the NFAR family may also play a role in cancer progression. Transcriptional and post-transcriptional regulation of several key cancer genes by NFAR proteins, particularly the anti-apoptotic factor survivin, is associated with oncogenesis (Vumbaca et al., 2008; Hu et al., 2013; Nakamura et al., 2012).
In addition to their role in tumourigenesis, NFAR proteins play a key role in host innate antiviral defence, primarily due to their direct interaction with the interferon-inducible dsRNA-dependent protein kinase (PKR) (Saunders et al., 2001). PKR is induced by viral invasion and its activation causes suppression of viral and cellular translation (Galabru and Hovanessian, 1987; Hovanessian, 1989; Rhoads, 1993). NFARs are known substrates of PKR and recent studies have demonstrated that during viral infection NFARs are retained on the polysomes where they are capable of interacting with viral mRNA and suppressing viral mRNA translation in an EIF2α-independent manner (Harashima et al., 2010). Additionally NFAR proteins appear to moonlight as regulators of numerous clinically significant viral lifecycles including HIV-1, hepatitis C virus, hepatitis B virus, influenza, adenovirus, ebola and dengue viruses. As a result of both their nucleic acid and protein binding capabilities, NFARs can regulate these viral lifecycles at multiple levels, resulting in enhancement or inhibition of viral amplification, depending on the virus in question (Wang et al., 2009; Isken et al., 2007; Agbottah et al., 2007; Urcuqui-Inchima et al., 2006, 2011; Shabman et al., 2011; Gomila et al., 2011; Gwizdek et al., 2004).
Given that NFARs are associated with a broad spectrum of viral infections, it led us to speculate whether NFARs may regulate the HTLV lifecycles. The aim of this study was to characterise the role of NFARs in HTLV infection, namely through comparative interaction and functional studies encompassing HBZ and APH-2. This study highlights that HBZ and APH-2 are novel interacting partners of the NFAR family and indicate that these interactions impact on viral and cellular gene expression.
2. Material and methods
2.1. Yeast two-hybrid screen
The yeast two-hybrid screen was performed by Myriad Pharmaceuticals, Salt Lake City, UT, USA. Full length HBZ or APH-2 were used as bait to screen T-cell, brain and stem cell cDNA libraries for possible interactions. Positive “hits” were confirmed by nutritional and colorimetric selection.
2.2. Cell culture
293T and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Life Technologies) supplemented with 10% foetal bovine serum (FBS) (Gibco, Life Technologies). Jurkat, HTLV-1-infected cell lines, MT2 and C91-PL, and ATL cell lines ATL-CR, ATL-TH and were maintained in RPMI 1640 medium (Gibco, Life Technologies) containing 10% FBS and 100 μg/ml penicillin-streptomycin (Gibco, Life Technologies). The two ATL cell lines were established from Brazilian patients with ATL and were previously described (Miyatake et al., 2013). To establish these cell lines, PBMCs isolated from patients with ATL were cultured in the presence of recombinant IL-2. After long-term culture, they acquired IL-2 independence. The HTLV-2-infected cell line, Mo, was cultured in RPMI 1640 medium containing 20% FBS and 100 μg/ml penicillin-streptomycin (Gibco, Life Technologies). Cells were cultured under standard tissue culture conditions.
2.3. Plasmid constructs
The expression plasmids encoding GST-APH-2, GFP-APH-2, FLAG-HBZ and FLAG-APH-2, FLAG-APH-2ΔncbZIP and FLAG-APH-2 (1–93) were previously described (Marban et al., 2012). To obtain the GST-HBZ construct, we generated an HBZ PCR product using the pcDNA-HBZ-Myc-His (kindly provided by Dr. Jean-Michel Mesnard, Université de Montpellier, France) as a template. The HBZ PCR product was then digested with BamHI/EcoRI and cloned into the pGEX-2 T (GE Healthcare). The NF110a-His construct used in GST pulldowns was generated by firstly amplifying an NF110a cDNA fragment from pcDNA3.1-NF110a by PCR and cloning into pBAD/myc-His C vectors using EcoRI. The pFLAG-HBZ mutants pFLAG-HBZΔAD, pFLAG-HBZΔCD and pFLAG-HBZΔbZIP were constructed using site-directed mutatgenesis (Phusion® Site-Directed Mutatgenesis kit, Thermo Scientific) using pFLAG-HBZ as a template and the primers: 5′-CAGCGACGGGCTGAGGAG - 3′ and 5′-AAGCTTGTCGTCATCGTCTTTG - 3′ for pFLAG-HBZΔAD; 5′-CAGGAGCGCCGTGAGCG - 3′ and 5′-CTGCTTTCTCCGGGCAAC - 3′ for pFLAG-HBZΔCD; 5′-GAATTCATCGATAGATCTGATATCGGTA - 3′ and 5′-CTGCTTTCTCCGGGCAAC - 3′ for pFLAG-HBZΔbZIP. The NFAR-encoding plasmids, pcDNA3.1-NF90a and pcDNA3.1-NF110a were kindly provided by Prof. Glen Barber, University of Miami, FL, USA. The NF110a mutants NF110a-167-394 and NF110a-398-603 were generated by cloning the respective cDNAs as EcoRI fragments obtained from pcDNA3.1-NF110a into pCAGGS vectors. The pEGFP-HBZ-Sp1 was a kind gift from Dr. Jean-Michel Mesnard, Université de Montpellier, France. HTLV-1-LTR-luc, HTLV-2-LTR-luc, pCAGGS-Tax-1-His and pCAGGS-Tax2B-His were previously described (McCabe et al., 2013; Sheehy et al., 2006). The HTLV-1 (pACH) and HTLV-2 (pH6neo) proviral clones were kindly provided by Prof. Lee Ratner, Washington University, St. Louis, MO, USA and Prof. Patrick Green, Ohio State University, Columbus, OH, USA, respectively. The human survivin promoter pLuc-Hsp-281 luciferase reporter construct was available from CH3 BioSystems™.
2.4. Transfections
293T cells were transiently transfected using Lipofectamine™ 2000 (Life Technologies) according to the manufacturer’s guidelines. Transfections of HeLa cells were performed using Turbofect™ (Thermo Scientific) following the manufacturer’s instructions. The HTLV-1 and HTLV-2 proviral clones were transfected into HeLa cells using PolyFect® as per manufacturer’s protocol. The overall DNA concentration was normalised for all transfections by use of the relevant parent plasmid.
2.5. Western blotting and antibodies
Cellular lysates were subjected to SDS-PAGE and transferred to PVDF membranes (GE Healthcare) using standard procedures. Membranes were probed using the SNAP i.d.™ system (Merck Millipore), using the indicated antibodies. The antibodies were used in Western blotting and immunofluorescence procedures were as follows: anti-FLAG (Sigma-Aldrich, F7425), anti-ILF3 (Abcam, ab92355), anti-His (Clontech 631212), anti-α-tubulin (Abcam, ab7291), anti-survivin (R&D Systems, AF886), anti-HTLV-1/2 p24 (Zeptometrix 75/4.21.11), Alexa Fluor 594-conjugated goat anti-rabbit IgG (Life Technologies, A-11012) and Alexa Fluor 488-conjugated goat anti-mouse IgG (Life Technologies, A-11001). Densitometry analysis of western blot protein bands was performed using LI-COR Image Studio™ Lite software.
2.6. GST pulldown assays
Glutathione-S-transferase (GST) and GST-fusion proteins were expressed and purified from Escherichia coli (E.coli) BL21 as previously described (Marban et al., 2012). The NF110a protein was expressed in E.coli Top10F and purified using a nickel resin (QIA-GEN). For the pulldown assays, GST and GST-fusion proteins were immobilised onto Gluthathione Sepharose™ 4 Fast Flow resin (GE Healthcare) overnight. Following incubation, purified NF110a was incubated with GST and GST-fusion proteins for a further 24 h. Resins were washed in a GST wash buffer (0.5% Triton® X-100 in PBS) and bound proteins were eluted from the beads using GST elution buffer (50 mM Tris–HCl pH 8.0 containing 10 mM reduced glutathione). Interactions were analysed by Western blotting using an anti-ILF3 antibody and Coomassie Brilliant Blue staining.
2.7. Co-immunoprecipitations
293T cells were transfected with the relevant amounts of expression constructs as indicated in the individual using Lipofectamine™ 2000 (Life Technologies) according to the manufacturer’s guidelines. Transfected cells were incubated for 24 h and lysed in a buffer containing 1X TBS, 0.005M EDTA, 1% Triton® X-100 supplemented with protease inhibitors (Complete Protease Inhibitor Cocktail EDTA-free, Roche) or RIPA buffer containing 50 mM Tris–HCl pH8, 150 mM NaCl, 1% Triton® X-100, 0.1% SDS, 0.5% sodium deoxycholate supplemented with protease inhibitors as indicated in Cellular lysates were subjected to co-immunoprecipitation with an anti-FLAG M2 resin (Sigma-Aldrich) overnight at 4 °C. The beads were then washed 3 times in the relevant buffer. Co-immunoprecipitations were analysed by western blot using anti-FLAG, anti-His and anti-ILF-3 antibodies.
2.8. Immunofluorescence
HeLa cells were seeded onto chamber slides and transiently transfected with the indicated expression vectors using Turbofect™ transfection reagent (Thermo Scientific) according to the manufacturer’s protocol. Transfected cells were incubated for 24 h. The HTLV-1 (pACH) and HTLV-2 (pH6neo) proviral clones were transfected into HeLa cells using PolyFect transfection reagent (QIAGEN) as per the manufacturer’s recommendations and incubated for 48 h. All cells were fixed with 4% paraformaldehyde for 10 min at room temperature (RT), and permeabilised using 0.5% Triton® X-100/PBS for 10 min at RT. All slides were blocked in 0.02% Tween 20/5% FBS/10% goat serum in PBS for 1 h at room temperature. To stain endogenous NFARs, slides were incubated with an anti-ILF-3 antibody for 2 h followed by Alexa Flour 594-conjugated goat anti-rabbit IgG for 1 h at room temperature. HTLV-1 and HTLV-2 p24 capsid gene products were detected by incubating the cells with an anti-HTLV-1/2 p24 antibody (Zeptometrix 75/4.21.11) for 2 h at RT, followed by incubation with Alexa Fluor 488-conjugated goat anti-mouse IgG for one hour at RT. Nuclei were stained using DAPI (Sigma-Aldrich) and slides were mounted in ProLong Gold Anti-Fade (Life Technologies). Images were obtained by use of a Zeiss AxioImager MI fluorescent microscope and Axiocam HR camera.
2.9. Knockdown of NFARs by siRNA
The control and NFAR-specific siRNA duplexes targeting both NF90 and NF110 were: 5′-AAGCCACTGATGCTATTGGGC-3′ and were purchased from QIAGEN. Briefly, approximately 40,000 HeLa cells were seeded into individual wells of a 24-well plate and transfected with 40 pmol siRNA using Lipofectamine™ RNAiMAX according to the manufacturer’s recommendations. 72 h post-siRNA transfection, the cellular lysates were prepared and knockdown was validated for each experiment by Western blot analysis using anti-ILF3 and anti-α-tubulin antibodies.
2.10. Luciferase reporter gene assays
293T or HeLa cells were transfected with either HTLV-1-LTR-Luc, HTLV-2-LTR-Luc or human survivin promoter pLuc-Hsp-281 and different combinations of expression vectors as indicated using Lipofectamine™ 2000 (Life Technologies) or Turbofect™ (Thermo Scientific), respectively. Transfected cells were lysed in Cell Culture Lysis Reagent and luciferase assays were performed as previously described using the Luciferase® Assay System (Pro-mega) (Sheehy et al., 2006). Luciferase values were normalised for all samples with the respective protein concentrations obtained by performing a BCA assay (Pierce™ BCA Protein Assay Kit, Thermo Scientific).
2.11. YM155 treatments
The survivin inhibitor YM155 was purchased from Calbiochem. 1 × 106 MT2 or Mo cells were seeded into a 24-well plate and treated with 100 nM of YM155 or DMSO as a control for 48 h. 293T cells were seeded into individual wells of a 6-well dish and treated with 100 nM YM155 or DMSO for 24 h. Cells were harvested for various analytical procedures.
2.12. p19gag ELISA assays
The concentration of HTLV matrix p19gag protein was quantified in the cell culture supernatants from MT2 and Mo cells collected 48 h post-YM155 treatment using a commercially available ELISA kit (Retrotek HTLV1/2 p19 antigen, Zeptometrix), following the manufacturer’s recommendations.
3. Results
3.1. Screening of HTLV-2 APH-2-interacting proteins using the yeast two-hybrid system
In contrast to HTLV-1 HBZ, very little is known about the role of APH-2 in HTLV-2 infection. In order to investigate the function of APH-2, we employed a yeast two-hybrid approach to screen cDNA libraries to identify potential APH-2 interactions. One of the most common interactors identified from that study was cDNAs corresponding to NF110a, encoded by the ilf3 gene [GenBank: NM_012218]. Alternative splicing of the ilf3 gene produces at least five distinct mRNA species encoding individual protein isoforms (Reichman et al., 2003; Duchange et al., 2000) (Fig. 1A). The predominant protein isoforms have approximate molecular masses of 90 kDa and 110 kDa, termed NF90 and NF110, respectively. NF90 and NF110 exhibit significant homology having identical N-terminal and central regions, but divergent C-termini. Differential splicing events give rise to the insertion of a four amino acid sequence (NVQK), separating NF90 and NF110 into two further isoforms; NF90b and NF110b, while it is absent in NF90a and NF110a (Patino et al., 2015). A further NFAR isoform is referred to as NF90c or NF90ctv (NF90 C-terminal variant), however it is presently unclear whether this variant is an artefact of cloning or if it is a bona fide, naturally occurring variant in vivo (Reichman and Mathews, 2003). NF90 and NF110 possess several important functional domains, notably two double-strand RNA binding domains (DRBDs) that facilitate their interaction with RNA species, in addition to nuclear export and localisation signals (NES and NLS, respectively) which permit their nuclear-cytoplasmic shuttling (Masuda et al., 2013). The C-terminally extended NF110 isoforms bear a GQSY-rich motif which is absent from NF90 isoforms.
Fig. 1.
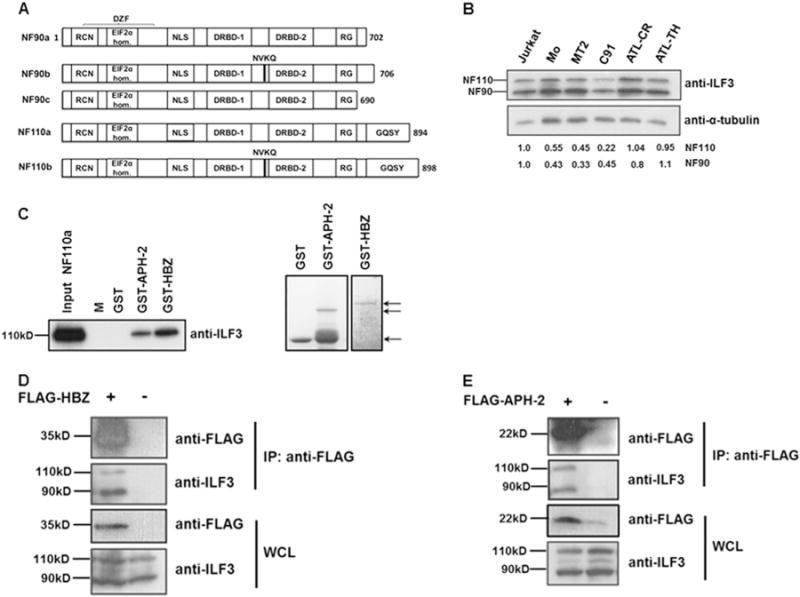
HTLV-1 HBZ and HTLV-2 APH-2 interact with NFARs in vitro and in vivo. (A) Schematic representation of NFAR isoforms. The functional domains are indicated and are as follows: RCN: region containing NES; EIF2α homology: region homologous to EIF2α; DZF: double zinc finger; NLS: nuclear localisation signal; DRBD-1/-2: double-stranded RNA binding domain −1 and −2; RG: arginine/glycine rich domain; GQSY: GQSY-rich region. (B) Expression levels of endogenous NF90 (90 kD) and NF110 (110 kD) in various cell lines. Equal amounts of whole cell extracts from Jurkat, HTLV-1 transformed cell lines, MT2 and C91, a HTLV-2 transformed cell line, Mo and ATL cell lines, ATL-TH and ATL-CR were electrophoresed on an 8% polyacrylamide gel and immunoblotted with antibodies directed against ILF3 which detects both NF90 and NF110, in addition to α-tubulin antibodies. For densitometry analysis of protein bands the individual signals for NF90 and NF110 in all cell lines were quantified and normalised against the signal for α-tubulin. To determine fold changes normalised values for HTLV and ATL cell lines were expressed relative to normalised values for uninfected Jurkat cells. The figures shown beneath the immunoblot denote the intensities of NF90 and NF110 relative to Jurkat cells (C) GST pulldown assays were performed by incubating purified NF110a with GST, GST-APH-2 or GST-HBZ immobilised on GST resin. The eluates were analysed by immunoblot with anti-ILF-3 antibodies (left panel) and coomassie blue staining (right panel). NF110a input corresponds to 20% of total NF110a loaded to each pulldown. M indicates protein molecular-weight marker. The arrows indicate purified GST, GST-APH-2 and GST-HBZ used in the pulldown assays. (D–E) HBZ and APH-2 interact with endogenous NF90 and NF110 in vivo. 293T cells seeded in 60 mm cell culture dishes were transiently transfected with 8 μg pFLAG-HBZ (D) or pFLAG-APH-2 (E) expression vectors as indicated. Immunoprecipitations were performed using an anti-FLAG M2 resin and precipitates were analysed by western blot using anti-FLAG and anti-ILF3 antibodies. IP: immunoprecipitation; WCL: whole cell lysate.
We firstly analysed the expression levels of NF90 and NF110 in two HTLV-1 chronically infected cell lines (MT2 and C91), a HTLV-2 chronically infected cell line (Mo) and two ATL cell lines (ATL-CR and ATL-TH) compared to control uninfected Jurkat cells (Fig. 1B). The ATL-CR and ATL-TH cell lines were established from patients with ATL and viral gene expression is silenced in these cells (Miyatake et al., 2013, 2015). The status of HBZ expression in the ATL-CR and ATL-TH cell lines was not investigated. While NF90 and NF110 proteins were abundantly expressed in all the cell lines tested, densitometry analysis of western blot bands revealed lower or equivalent levels of expression of both proteins compared to the HTLV uninfected Jurkat cell line.
3.2. HTLV-1 HBZ and HTLV-2 APH-2 interact with NFAR members in vitro and in vivo
To verify the findings of the yeast two-hybrid screen, we firstly sought to confirm the interaction between APH-2 and NF110a. Additionally we investigated whether the HTLV-1 antisense protein HBZ may also interact with NF110a. To this extent we performed GST pulldown assays, using GST, GST-APH-2, GST-HBZ and NF110a-His purified from E.coli (Fig. 1C). Our results illustrate that NF110a-His binds both GST-APH-2 and GST-HBZ but not GST, indicating direct specific physical interactions between these proteins in vitro.
We next investigated whether HBZ and APH-2 interact with endogenous NFARs in vivo. To this extent we transfected 293T cells with either FLAG-HBZ or FLAG-APH-2 constructs. Cellular lysates were subjected to co-immunoprecipitation using an anti-FLAG M2 resin and resultant interactions were analysed by western blotting using anti-FLAG and anti-ILF3 antibodies. We demonstrate that endogenous NF90/NF110 can be precipitated in the presence of but not in the absence of HBZ expression (Fig. 1D). Furthermore we report a specific interaction between APH-2 and endogenous NF90/NF110 (Fig. 1E). Thus these data demonstrate specific interactions between HBZ and APH-2 and endogenous NF90 and NF110 in mammalian cells.
3.3. The central domain of HBZ and the N-terminus of APH-2 are involved in the interaction with endogenous NF90 and NF110
We next sought to ascertain the domains in HBZ or APH-2 that mediate their interaction with endogenous NF90 and NF110. HBZ contains three interaction domains, namely an activation domain (AD), a central domain (CD) and a basic leucine zipper motif (bZIP). These functional domains are responsible for the ability of HBZ to bind numerous cellular factors which in turn leads to modulation of certain cellular signalling pathways (Zhao and Matsuoka, 2012). To determine the domain of HBZ that binds endogenous NF90 and NF110, we transfected 293T cells with plasmids encoding full length HBZ or the indicated HBZ deletions (Fig. 2A). Protein complexes from transfected lysates were precipitated using an anti-FLAG M2 resin and interactions were analysed by western blotting using antibodies directed against the FLAG epitope and ILF3. Our data shows that HBZ proteins lacking the central domain failed to interact with NF90 or NF110, leading us to conclude that the central domain of HBZ is responsible for mediating its interaction with NFAR members (Fig. 2B).
Fig. 2.
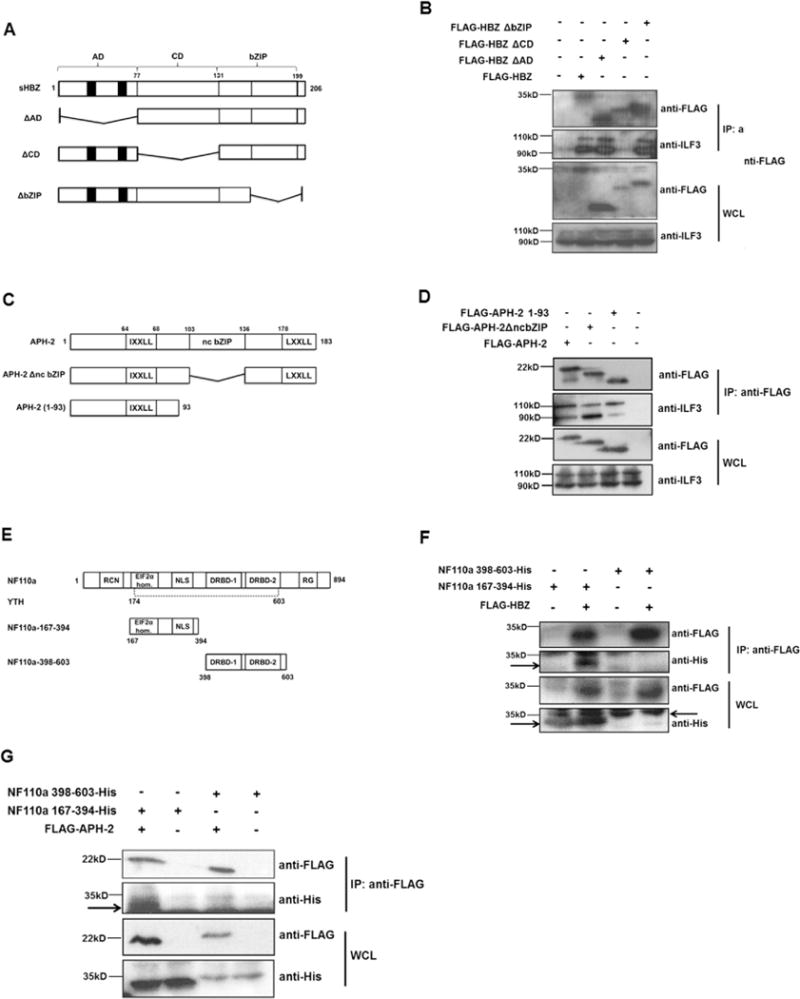
Deletion mapping of HBZ, APH-2 and NF110a interaction domains. (A) Schematic of HBZ and deletion mutants used in co-immunoprecipitations. Functional domains are: AD: activation domain; CD: central domain; bZIP: basic leucine zipper. Black boxes represent the LXXLL motifs. (B) NFARs interact with HBZ central domain. 293T cells were seeded into 60 mm cell cultures dishes and transfected with 8 μg of pFLAG-HBZ or pFLAG-HBZΔAD and 10 μg of pFLAG-HBZΔCD or pFLAG-HBZΔbZIP as indicated. 24 hours post-transfection cells were lysed in RIPA buffer containing 50 mM Tris–HCl, 150 mM NaCl, 1% Triton® X-100, 0.1% SDS, 0.5% sodium deoxycholate. An anti-FLAG M2 resin was used to precipitate protein complexes from lysates overnight. Interactions were analysed by western blot using anti-FLAG and anti-ILF3 antibodies. (C) Schematic of APH-2 and mutants used in co-immunoprecipitations. Functional domains are: IXXLL: LXXLL-like motif; ncbZIP: non-canonical basic leucine zipper domain; LXXLL: LXXLL motif. (D) 293T cells were seeded into 60 mm cell cultures dishes and transfected with 8 mg of full length APH-2 or the indicated mutants. Lysates from transfected cells were prepared using a buffer containing 1X TBS, 0.005M EDTA, 1% Triton-X 100 24 h post-transfection and subjected to immunoprecipitation using FLAG M2 resin. Precipitates were analysed by western blot using anti-FLAG and anti-ILF-3 antibodies. (E) Schematic representation of NF110a and mutants used in co-immunoprecipitations. Mutants are: NF110a-167-394 incorporating the EIF2α-homology region and NLS; 398–603 incorporating two double-stranded RNA-binding domains (DRBD-1/-2). Y2H represents the minimum domain in NF110a required for interaction with APH-2 obtained from yeast two-hybrid screen. (F and G) HBZ and APH-2 interact with NF110a 167-394. 293T cells were seeded into 60 mm cell cultures dishes and co-transfected with 2 μg pCAGGS-NF110a-167-394-His or 8 μg pCAGGS-NF110a-398-603-His and 4 μg pFLAG-HBZ (F) or pFLAG-APH-2 (G) as indicated. Cells were lysed in 1X TBS, 0.005M EDTA, 1% Triton-X 100 buffer as above. Interactions were analysed by immunoblotting using anti-FLAG and anti-His antibodies. Arrows indicate specific protein bands.
Similar experiments were performed to determine the region of APH-2 that binds endogenous NF90 and NF110. APH-2 harbours a non-conventional bZIP domain (ncbZIP), as it contains seven instead of six amino acids between the sixth and the seventh leucine. In addition APH-2 contains an LXXLL and an IXXLL motif located in its C-and N-termini, respectively (Halin et al., 2009). Two deletion mutants were used in the APH-2 deletion analysis studies; APH-2ΔncbZIP lacks the non-conventional bZIP domain, while APH-2 1–93 represents the N-terminal portion of APH-2 that contains its IXXLL motif (Fig. 2C). 293T cells were transfected with the indicated plasmids and an anti-FLAG M2 resin precipitated the interacting proteins from transfected lysates before being investigated by western blot analysis. Our results demonstrate that the non-canonical bZIP domain of APH-2 does not mediate its interaction with endogenous NF90 and NF110, and that the N-terminal region of APH-2, namely amino acids 1–93 is sufficient for these interactions (Fig. 2D).
3.4. The N-terminal domain of NF110a binds HBZ and APH-2
We also wished to determine the protein domains in NF110a that mediate its interaction with HBZ and APH-2 in mammalian cells. Data obtained from our yeast two-hybrid screen indicates that the minimum domain in NF110a required for the interaction between APH-2 and NF110a lies between amino acids 174–603 (Fig. 2E). Based on this evidence we generated two different expression constructs encompassing either the N-terminal EIF2α homology region and the nuclear localisation signals (NLS) (NF110a-167-394), or the C-terminal double-stranded RNA binding domains −1 and −2 (NF110a-398-603), respectively (Fig. 2E). These NF110a mutated plasmids were co-transfected together with either HBZ or APH-2 expression plasmids into 293T cells and lysates were immunoprecipitated using an anti-FLAG M2 resin. Western blot analyses of precipitated complexes show that the N-terminal region of NF110a containing the EIF2α-homology region and the NLS is responsible for binding to not only HBZ but also APH-2 (Fig. 2F and G).
3.5. Subcellular localisation of HTLV antisense proteins and NFARs
While NF90 and NF110 are principally nuclear-localising proteins, several lines of evidence suggest that they shuttle between the nuclear and cytoplasmic compartments as a result of specific cellular stimuli (Harashima et al., 2010; Larcher et al., 2004; Pfeifer et al., 2008; Parrott et al., 2005). Next we sought to examine the subcellular localisation of HBZ, APH-2 and endogenous NFARs in HeLa cells (Fig. 3A). HeLa cells were transfected with expression vectors encoding GFP, GFP-HBZ or GFP-APH-2 (green). Endogenous NFARs were detected using an anti-ILF3 antibody that detects both NF90 and NF110, followed by Alexa Fluor 594 staining (red). Nuclei were stained using DAPI (blue) and the subcellular distribution of these proteins was visualised by fluorescence microscopy. GFP exhibited diffuse staining throughout the cells examined, and as has been previously demonstrated NFARs were localised in the nucleolar compartment, in addition to diffusely in the nucleoplasm (Viranaicken et al., 2011) (Fig. 3A; Panel 1). GFP-HBZ was localised exclusively in the nuclear compartment, and exhibited its characteristic speckled pattern (Fig. 3A; panel 2) (Hivin et al., 2005). GFP-APH-2 distribution was predominantly nuclear and accumulated in a granular pattern; however we did observe some staining outside of the nucleus which is in agreement with a previously report (Fig. 3A; panel 3) (Halin et al., 2009). Despite having determined that HBZ and APH-2 interact with endogenous NFARs by co-immunoprecipitation assays, NFARs do not appear to colocalise to the same nuclear region as the HTLV antisense proteins (Fig. 3A; merged panels). Moreover the expression HBZ and APH-2 did not result in redistribution of NFARs to the cytoplasm.
Fig. 3.
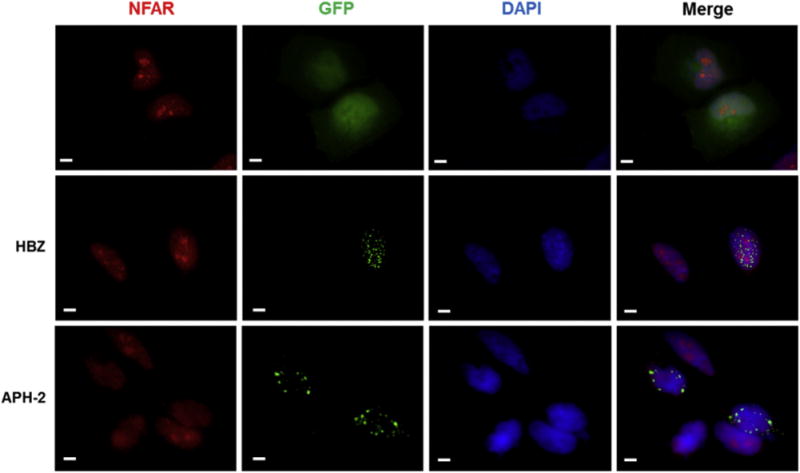
Subcellular localisation of HTLV antisense proteins and endogenous NFARs in HeLa cells. (A) Immunofluorescence experiments were carried out on HeLa cells transfected with 750 ng of pEGFP, pEGFP-APH-2, pEGFP-HBZ (green). Endogenous NFARs were detected using anti-ILF3 antibodies, followed Alexa Fluor 594® staining (red). Location of nuclei was determined using of DAPI (blue). Scale bar denotes 10 μm.
3.6. NFARs are required for HTLV-1 and HTLV-2 LTR transactivation
Previous studies have demonstrated that NF90 and NF110 have the ability to alter the activity of various cellular and viral promoters in transfected mammalian cells (Reichman et al., 2003, 2002; Shi et al., 2007; Agbottah et al., 2007). Thus to gain some insight into the functionality of NFARs in the HTLV lifecycles, we sought to investigate whether NFARs affect basal or Tax-mediated HTLV-1 or -2 LTR activation. In terms of HTLV-1, we observed that NFAR knockdown resulted in approximately 70% reduction in both basal (Fig. 4A) and Tax1-mediated LTR activation, suggesting that NFARs are required for HTLV-1 gene transcription (Fig. 4B). Moreover HBZ-mediated inhibition of basal or Tax1 LTR activation was further reduced to approximately 10% in NFAR knockdown cells indicating that the combined effect of HBZ expression and NFAR inactivation reduces basal and Tax1 mediated LTR activation by 90%. Thus these data suggest that HBZ may target NFARs as a means of inhibiting Tax1-mediated HTLV-1 LTR activation. Similar trends were observed for HTLV-2 LTR activity, where repression of NFARs inhibited basal (Fig. 4C) and Tax2-mediated LTR activity by approximately 50–70% (Fig. 4D). In control siRNA-treated cells, APH-2 inhibits basal LTR activity by approximately 35% (Fig. 4C), while Tax-mediated LTR transactivation was inhibited by approximately 20% by APH-2 (Fig. 4D). We observed that, in contrast to HBZ, NFAR knockdown had little or no effect on the ability of APH-2 to inhibit basal or Tax2-mediated LTR activation (Fig. 4C and D), suggesting that unlike HBZ, NFAR proteins may not be targeted by APH-2 to repress HTLV-2 LTR activation. We also investigated whether ectopic expression of NF90a or NF110a in the absence or presence of HBZ or APH-2 may modulate Tax-mediated transactivation of the HTLV-1 and -2 LTRs (Fig. 4E and F). Overall our results indicate that overexpression of NF90a and NF110a substantially upregulates luciferase activity driven by the activation of the HTLV LTRs, however a number of disparities were observed between the viral promoters. Firstly, NF90a and NF110a are more potent activators of Tax1-mediated HTLV-1 gene transcription compared to HTLV-2 (6–8 fold compared to 2–3 fold, respectively) (Fig. 4E lanes 2 and 4 compared to Fig. 4F lanes 2 and 4). Secondly, NF110a appears to promote HTLV-1 LTR transactivation to a greater extent than NF90a, whereas the opposite was observed for HTLV-2 (Fig. 4E lanes 2 and 4 compared to Fig. 4F lanes 2 and 4). Interestingly, stark differences in NFAR-mediated LTR activation were observed in the presence of the respective HTLV antisense proteins. We observed that HBZ has the ability to strongly repress NFAR-mediated HTLV-1 LTR transactivation to almost undetectable levels (Fig. 4E, lanes 3 and 5). Conversely, expression of APH-2 resulted in a synergistic upregulation of NFAR-mediated HTLV-2 LTR activity (Fig. 4F, lower panels, lanes 3 and 5).
Fig. 4.
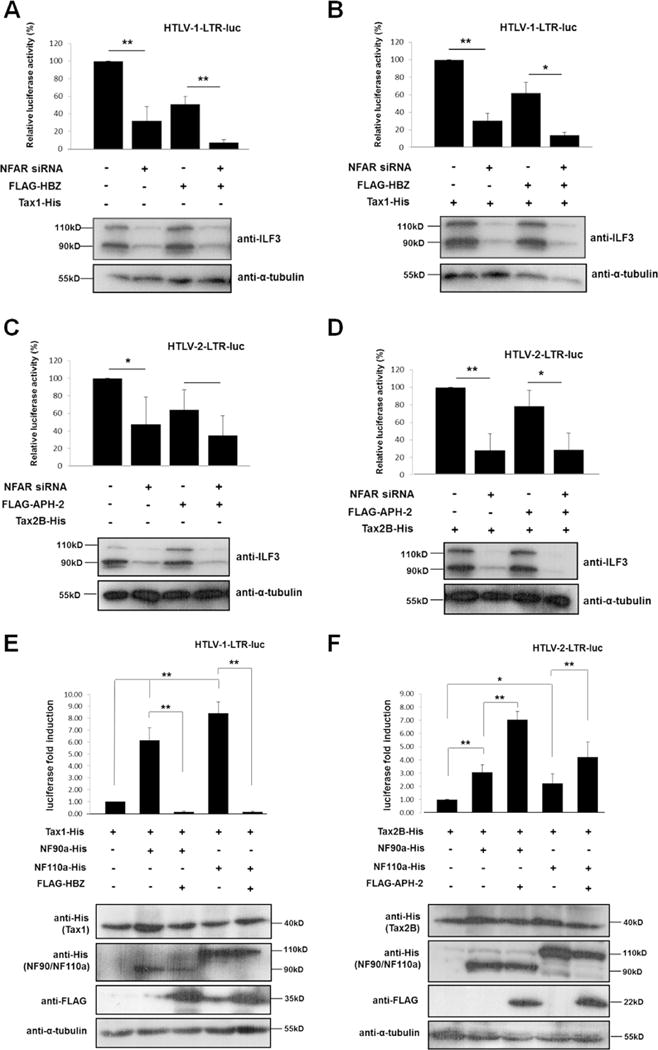
NFARs are required for HTLV-1 and HTLV-2 basal and Tax-mediated LTR activation. (A–D) Effect of NFARs on basal and Tax-mediated LTR transactivation. Approximately 40,000 HeLa cells were seeded into individual wells of 24-well cell culture dishes and transfected with 40 pmol of NFAR-specific or control siRNA. 24 h post-siRNA transfection, HeLa cells were co-transfected with 100 ng HTLV-1 or HTLV-2 LTR luciferase reporter constructs, 20 ng pCAGGS-Tax1 or -Tax2B, and 50 ng pFLAG-HBZ or -APH-2 as indicated. Luciferase assays were performed on lysates 72 hours post-siRNA transfection and were normalised against protein concentration. The average of three independent experiments is shown. Results are plotted as percentage luciferase activation relative to the control, which was assigned the arbitrary value of 100%. Error bars represent standard deviation. Asterisk (*) or (**) indicates statistical significance of p = <0.05 or p = <0.01 respectively obtained by two-tailed Student’s t-test. Shown beneath is NFAR knockdown and α-tubulin expression in cells used in luciferase assays assessed by subjecting normalised cellular lysates to western blot analysis using anti-ILF3 and anti-α-tubulin antibodies. (E–F) Effect of NF90/NF110 ectopic expression on HTLV-1 and HTLV-2 LTR transactivation. 293T cells seeded into individual wells of 6-well cell culture dishes were co-transfected with 500 ng HTLV-1 or HTLV-2 LTR luciferase reporters, 100 ng pCAGGS-Tax1 or -Tax2B, 2 μg pcDNA3.1-NF90a or -NF110a and 3 μg FLAG-HBZ or -APH-2 as indicated. Luciferase activities were measured 48 h post-transfection and normalised against protein concentration. The average of three independent experiments is shown and values are expressed as fold luciferase change relative to the control which was assigned the arbitrary value of 1. Error bars represent the standard deviation. Asterisk (*) or (**) indicates statistical significance of p = <0.05 or p = <0.01 respectively obtained by two-tailed Student’s t-test. Levels of NF90a, NF110a, Tax1, Tax2B, HBZ, APH-2 and α-tubulin expression in the cellular lysates used in the luciferases assays was determined by western blot analysis and are shown beneath.
3.7. Regulation of HTLV replication by NFARs
Previous reports have established that NFARs can either promote or impede viral replication. For instance, NFAR protein members negatively regulate HIV-1, influenza and Ebola propagation (Wang et al., 2009; Agbottah et al., 2007; Urcuqui-Inchima et al., 2006; Pfeifer et al., 2008; Krasnoselskaya-Riz et al., 2002), whereas hepatitis C virus, adenovirus and dengue virus exploit NFAR function to aid their amplification (Isken et al., 2007; Gomila et al., 2011; Gwizdek et al., 2004). Therefore we wished to elucidate whether NFARs may also regulate HTLV-1 and HTLV-2 replication. Given that the antiviral properties of NFARs mediated through the PKR pathway are strongly related to NFAR redistribution and sequestration in the cytoplasm following infection, we initially sought to determine the possible impact of HTLV infection on the localisation of endogenous NFARs. To this extent we transfected HeLa cells with either the HTLV-1 (pACH) or HTLV-2 (pH6neo) proviral clones and stained the cells for endogenous NFARs (Fig. 5A). We found that HTLV-1 and HTLV-2 p24gag capsid proteins exhibited the expected punctate localisation throughout the cytoplasm (Fig. 5A, panels 1 and 2) and the presence of HTLV-1 or HTLV-2 infection did not result in redistribution of NFARs from the nucleus to the cytoplasm (Fig. 5A panels 1 and 2). Given that NFARs are retained in the nucleus during HTLV infection, this analysis indicates that NFARs are not participating in PKR-mediated innate immune responses against HTLVs.
Fig. 5.
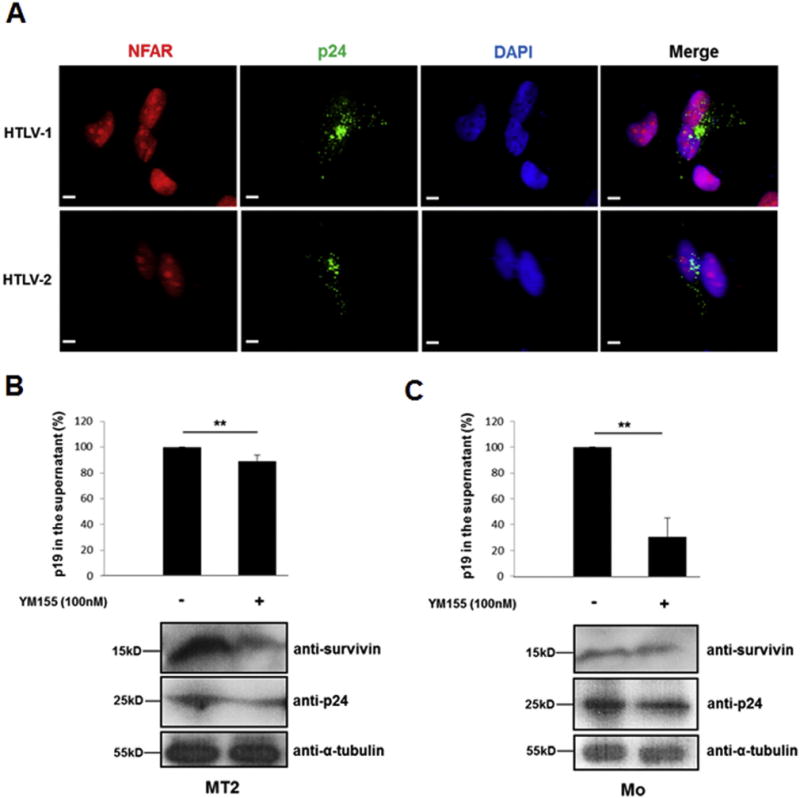
Regulation of HTLV replication by NFARs. (A) HTLV infection does not induce NFAR relocalisation. HeLa cells were transfected with 750 ng HTLV proviral clones pACH (HTLV-1) or pH6neo (HTLV-2) (green). HTLV p24 capsid proteins were detected using an antibody against HTLV-2 p24, followed by Alexa Flour 488® staining. Endogenous NFARs were stained as in Fig. 3A. (B–C) YM155 inhibits HTLV replication. MT2 and Mo cells were treated with 100 nM of YM155 (control: 0.2% DMSO). 48 h post-YM155 treatment, cellular supernatants were collected and cells were lysed in RIPA buffer. P19gag levels in the supernatants were quantified by ELISA and cellular lysates were immunoblotted with anti-p24gag, anti-survivin and anti-α-tubulin antibodies. The average of three independent experiments ± standard deviation is shown. Data are represented as percentage change compared to the P19gag levels obtained for YM155-untreated cells which was set arbitrarily to 100%. Asterix (**) indicates statistical significance of p = <0.01 obtained by two-tailed Student’s t-test.
In order to investigate the effect of NFARs on HTLV replication we initially sought to knockdown NFARS in the HTLV-1 infected cell line MT2 and the HTLV-2 infected cell line Mo using siRNA or shRNA. However despite repeated attempts we failed to achieve reproducible knockdown of NFAR proteins in HTLV infected cells using these methods. As an alternative strategy, we decided to utilize the small molecule survivin inhibitor, YM155, which was shown in previous studies to bind NF110 and inhibit its ability to activate survivin expression (Nakamura et al., 2012; Nakahara et al., 2007; Yamauchi et al., 2012). We hypothesised that even though YM155 does not knockdown NFARs per se, it inhibits it’s activity and as such represents a valid means of investigating the role of NFARs in HTLV replication. To this end we incubated HTLV-1 (MT2) or HTLV-2 (Mo) infected cells with YM155 and quantified p19gag levels in the supernatants by ELISA from YM155-treated and untreated cells. Our results indicate that YM155 treatment resulted in not only reduced survivin expression (Fig. 5B and C, lower panels) but also inhibited the production of viral particles released from Mo cells by almost 70% (Fig. 5C) while the effect was considerably less pronounced in MT2 cells, which exhibited a marginal 12% decrease of p19gag (Fig. 5B). We also analysed intracellular levels of p24gag from YM155-treated and untreated cells and found that consistent with the ELISA results, YM155 treatment of MT2 and Mo cells resulted in a reduction of p24gag levels (Fig. 5B and C, lower panels). Thus these results suggest that NFARs may promote HTLV replication which may be due to their ability to activate survivin expression and repress apoptosis.
3.8. HTLV-1 HBZ and HTLV-2 APH-2 regulate survivin expression
Survivin is a member of the inhibitor of apoptosis (IAP) protein family that is frequently highly expressed in many cancer cell types and positively correlated with tumour progression (Coumar et al., 2013). Based on studies showing that survivin expression is elevated in HTLV-1-infected cell lines and samples from ATL patients (Kamihira et al., 2001; Che et al., 2006) and our findings that HBZ and APH-2 interact with NFAR proteins, which are known survivin activators (Nakamura et al., 2012; Yamauchi et al., 2012), we sought to investigate whether the HTLV antisense proteins may play a role in NF110-mediated regulation of survivin expression. We initially determined the effect of HBZ or APH-2 on survivin promoter activity. Using luciferase reporter constructs that contain −1257 (data not shown) or −281 regions of the survivin promoter relative to the transcription start site (Fig. 6A), we can show that HBZ, and to a lesser extent APH-2, activate the survivin promoter in a dose-dependent manner (Fig. 6B and C). Similarly to previously published data (Nakamura et al., 2012), we observed an approximate 10-fold increase in survivin promoter activity in the presence of overexpressed NF110a compared to basal levels using either −281 to +30 (Fig. 6D) or −1257 to +30 (data not shown) survivin promoter constructs. This suggests that the minimum promoter sequence required for activation of the survivin promoter by NF110a spans −281 base pairs relative to the transcription start site, as has been previously reported (Nakamura et al., 2012). We found that HBZ and APH-2 inhibit NF110a-mediated transactivation of the survivin promoter, with HBZ having a significantly greater inhibitory effect compared to APH-2 (Fig. 6D). Given that HBZ and APH-2 regulate survivin gene activation, we next wished to determine whether this regulation resulted in regulation of survivin protein levels (Fig. 6E). Our results reveal that ectopic expression of HBZ and APH-2 results in only a minor increase in survivin protein expression. Altogether these results clearly show that HBZ and to a lesser extent APH-2 regulate survivin gene expression and both can down regulate NF110a-mediated survivin expression at a transcriptional level.
Fig. 6.
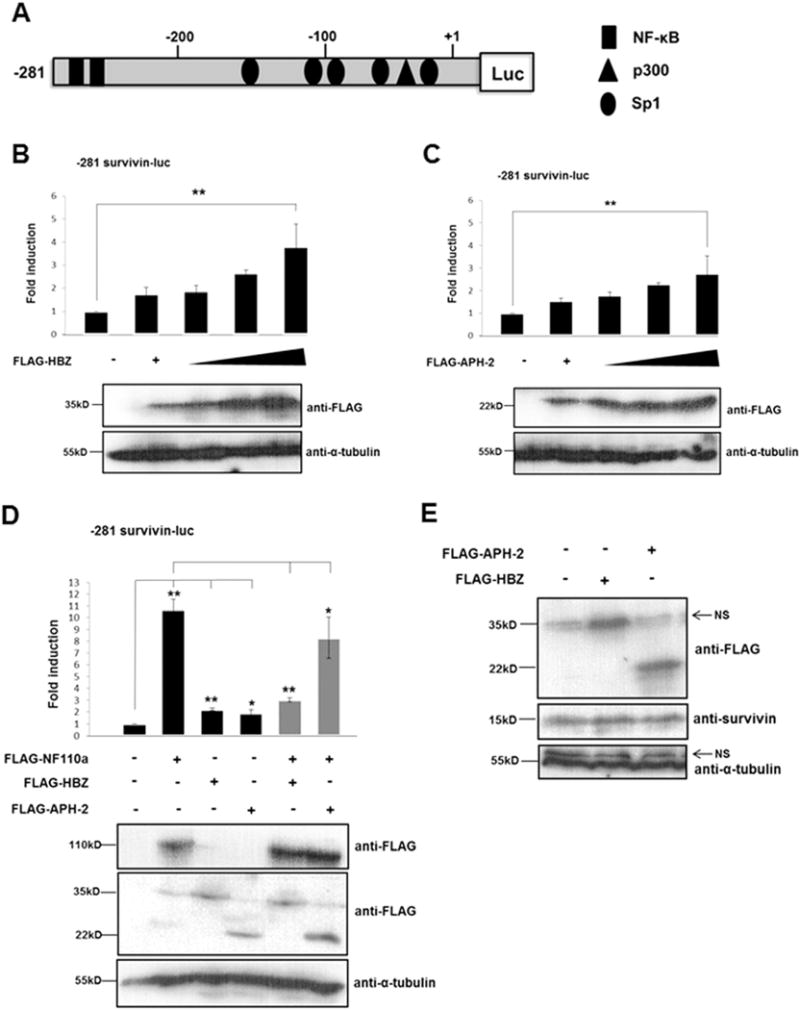
HTLV-1 HBZ and HTLV-2 APH-2 regulate survivin gene expression. (A) Schematic representation of the survivin promoter luciferase reporter constructs used in (B–D). This shows the approximate location of Sp1, NF-κB and p300 transcription factors adapted from (Kawakami et al., 2005; Banerjee et al., 2008; Mityaev et al., 2008). (B–C) HBZ and APH-2 regulate survivin promoter activity. 293T cells were co-transfected with pLuc-Hsp-281 to +30 survivin promoter luciferase reporter constructs and increasing amounts of pFLAG-HBZ or pFLAG-APH-2 as indicated. Luciferase assays were performed on cellular lysates 48 h post-transfection. Data are represented as the fold change survivin induction relative to the control, which was assigned the arbitrary value of 1. The average of three independent experiments ± standard deviation is shown. Asterisk (**) indicates statistical significance at a level of p < 0.01 obtained by two-tailed Student’s t-test. (D) HBZ and APH-2 inhibit NF110a-mediated survivin promoter transactivation. 293 T cells were cotransfected with pLuc-Hsp-281 to +30 survivin promoter luciferase reporter constructs and FLAG-NF110a, FLAG-HBZ or FLAG-APH-2 expression constructs as indicated. Luciferase assays were performed as in (B–C). The average of three independent experiments 7 standard deviation is shown. Results are represented as the fold change survivin induction relative to the control, which was assigned the arbitrary value of 1. The asterisk (*) or (**) indicates statistical significance at a level of p < 0.05 or p < 0.01, respectively obtained using a two-tailed Student’s t-test. (D) HBZ and APH-2 promote survivin protein expression. 293T cells were seeded into individual wells of a 6-well cell culture dish and transfected with 5 μg of pFLAG-HBZ or pFLAG-APH-2. 48 hours post-transfection the cellular lysates were prepared using RIPA buffer containing 50 mM Tris–HCl, 150 mM NaCl, 1% Triton® X-100, 0.1% SDS, 0.5% sodium deoxycholate. Total protein levels in the lysates were measured using a BCA assay. Equal amounts of the whole cell extracts were electrophoresed on 12% and 15% polyacrylamide gels, followed by immunoblot using anti-FLAG, anti-survivin and anti-tubulin antibodies.
4. Discussion
The NFAR proteins belong to the double stranded RNA-binding protein (DRBP) family of proteins and were first identified as factors involved in transcriptional and post-transcriptional gene regulation (Reichman et al., 2003; Shim et al., 2002; Shi et al., 2007; Reichman and Mathews, 2003; Pfeifer et al., 2008). Recent studies have shown that while these proteins are involved in host antiviral innate immune responses, their abnormal expression in several cancers is linked to severity of disease, indicating their diverse roles in health and disease. In the present study, we investigated physical and functional interactions between the HTLV antisense proteins HBZ and APH-2 and NFAR proteins.
Using GST pulldown and co-immunoprecipitation assays we show that HBZ and APH-2 specifically interact with NF90/110 in vivo and in vitro, thus confirming the interactions obtained from the yeast two-hybrid system. We demonstrate that the N-terminal region of NF110a comprising amino acids 167–394 and not the C-terminal 398–603 encompassing the RNA-binding domains, is responsible for its binding to not only HBZ but also APH-2. We acknowledge that the relative expression level of the 398–603 mutant is inherently lower than that of the 167–394 mutant, and it could therefore be argued that the higher expression of the 167–394 mutant promoted its interaction with HBZ and APH-2. However given the results from several independent experiments we are satisfied that amino acids 167–394 of NF110a are at least sufficient for its interaction with the HTLV antisense proteins.
HBZ harbours several domains that are responsible for its association with numerous cellular factors (Zhao and Matsuoka, 2012). We show here that the central domain of HBZ facilitates its interaction with endogenous NF90 and NF110. We accept that the expression level of the CD mutant is lower than wild type HBZ or the other mutants however this result coincides with a previous report (Satou et al., 2011), whereby the authors conclude an interaction between Foxp3 and the HBZ CD despite the inherently lower relative expression of the CD domain. Recent studies have shown that the central domain of HBZ is responsible for suppressing the transcriptional activation of the pro-apoptotic gene Bim, by binding and sequestering its critical transcriptional activator, FoxO3a, equating to novel function of the central domain in ATL cell proliferation and oncogenesis (Tanaka-Nakanishi et al., 2014). The central domain contains two of the three nuclear localisation signals required to facilitate the distinct nuclear localisation exhibited by HBZ (Hivin et al., 2005). Even though we found that NFAR proteins bind HBZ through the central domain which contains its nuclear localisation signals, NFARs do not affect the nuclear localisation of HBZ as seen in our immunofluorescence data. We report that while the non-canonical bZIP domain of APH-2 is not involved in the in vivo interaction with NF90 and NF110, the region between amino acids 1–93 which contains the IXXLL motif mediates this interaction. Of the limited studies conducted on APH-2, this is the first study to show that the N-terminus of APH-2 mediates its interaction with cellular factors.
While NFARs are predominantly nuclear/nucleolar localising proteins, they may also display cytoplasmic localisation during specific cellular events, particularly in response to viral infection (Harashima et al., 2010). For instance, the association of NF90, NF110 and NF45 with mRNA ribonucleoprotein (RNP) complex in the nucleus facilitates mRNA export and translation in the cytoplasm from where they are reshuttled back to the nucleus. However in the event of viral infection and phosphorylation by PKR, NFARs are retained on the polysomes where they are capable of interacting with viral mRNA and suppressing viral mRNA translation (Harashima et al., 2010). Our immunofluorescence studies indicate that NFARs are retained in the nucleus in the presence of HBZ, APH-2 expression or HTLV-1 or HTLV-2 infection suggesting that NFARs do not participate in innate immune responses against HTLV. Despite clearly showing that NFAR proteins interact with HBZ and APH-2 both in vitro and in vivo, detailed computational analysis of fluorescence microscopy images (data not shown) revealed that NFARs did not localise to the same distinct foci occupied by HBZ or APH-2. The reasons for the lack of colocalisation are unclear but may be related to disparities in the cell lines employed for co-immunopreciptions and immunofluorescence or differences in the experimental conditions used in both assays.
Several studies have shown that NFARs can both positively and negatively regulate transcription from several viral promoters in mammalian cells. For instance, NF90 inhibits transcription from the major late 1 promoter of adenovirus, whereas it activates transcription from the cytomegalovirus (CMV) and SV40 promoters (Reichman et al., 2002; Saunders et al., 2001). Moreover, NF90 binds the TAR element present in HIV-1 transcripts and in turn disrupts Tat-TAR interaction, resulting in suppression of Tat-mediated transcriptional activation of the HIV-1 LTR (Agbottah et al., 2007). In this study, the combination of knockdown and overexpression studies clearly demonstrates that NFARs are required for activation of both the HTLV-1 and -2 LTRs. Even though the mechanisms involved have not been determined, these data suggest that NFARs may cooperate with the core HTLV transcriptional machinery including CREB, ATF, SRF and CBP/p300 complexes to enhance activation of HTLV promoters. NF110 was previously shown to be present in a transcription regulation complex with p300 at the β-globin locus (Karmakar et al., 2010). Moreover recent studies have shown that NFAR proteins function as transcriptional coactivators and physically interact with coactivators such as CREB, SRF and ATF-1 (Nakadai et al., 2015), thus it is possible that NF110 may cooperate with the p300 complex to activate HTLV gene expression. We show that HBZ expression together with NFAR knockdown results in a cumulative repression of Tax-mediated LTR activity by approximately 90%. The ability of APH-2 to repress basal and Tax2B-mediated LTR was unaffected by NFAR knockdown. These results suggest that HBZ but not APH-2 may target NFARs in part to inhibit HTLV LTR transcription. The disparities in the levels of LTR repression by HBZ or APH-2 in the presence of reduced levels of NFAR expression might be related to the fact that HBZ inhibits LTR activation by forming hetrodimers with CREB/ATF-1 and p300/CBP complexes (Gaudray et al., 2002; Lemasson et al., 2007; Hagiya et al., 2011; Clerc et al., 2008; Cook et al., 2011), while APH-2 only binds CREB (Halin et al., 2009; Yin et al., 2012). Our overexpression studies illustrate that HBZ strongly represses NFAR-mediated LTR activation, again indicating that NFARs may be targeted by HBZ to repress HTLV-1 gene transcription. Conversely, the opposite was true for APH-2 in relation to HTLV-2. These results suggest that in the presence of high levels of NFAR expression, which is likely the case in HTLV-infected cells, the propensity of HBZ to inhibit LTR transactivation dominates over NFAR’s ability to transactivate the promoter, whereas the repressive effect of APH-2 is unable to counteract NFAR-mediated LTR activation. This observation is in accordance with a previous report showing that the inhibitory potential APH-2 is significantly attenuated compared to HBZ (Yin et al., 2012). Although we are unable to explain the reasons for the contrasting functions of the antisense proteins under these conditions, HBZ and APH-2 have been previously shown to exert distinct functional effects, for instance on the on the AP-1 pathway. Several studies have elucidated that HBZ represses the transcriptional activation of several AP-1 family members (Basbous et al., 2003; Matsumoto et al., 2005; Hivin et al., 2007; Clerc et al., 2009), whereas APH-2 potentiates their activation (Marban et al., 2012). Altogether these studies demonstrate the role of NFARs in HTLV gene transcription, and highlight the differential functional capacities of the HTLV antisense proteins relative to LTR transactivation.
The association of NFARs with certain viral nucleic acids or proteins has been shown to impact either positively or negatively on viral replication. The interaction between the hepatitis C virus NS5a protein and NFAR members triggers the formation of a loop structure within the HCV genome that has been shown to be indispensable for viral replication (Isken et al., 2007). Conversely, knockdown of NF90 expression results in significant enhancement of influenza A polymerase activity and viral replication (Wang et al., 2009). A lack of relocalisation of NFARs from the nucleus to the cytoplasm was observed in de novo HTLV infected cells which suggest that NFARs do not play a role in innate immune responses to HTLV infection. We did however observe that treatment of HTLV infected cells with YM155, which has been shown to suppress the ability of NF110 to activate survivin gene expression (Gwizdek et al., 2004), results in reduced viral replication in MT2 and Mo cells. This is consistent with previous studies reports showing that YM155 causes apoptosis of ATL cells (Mitobe et al., 2015; Chen et al., 2013). As YM155 inhibits NF110-mediated survivin expression, this effect could be due to induction of apoptosis in treated cells, however further analysis would be required to confirm this. Another possibility is that YM155 may suppress HTLV replication by inhibiting NFAR-mediated activation of the viral LTRs as we have shown in this study. Overall these results support the view that NF110 promotes the survival of HTLV infected cells which, together with our data showing that NFARs activate HTLV transcription, suggests that these proteins enhance the survival of infected cells and hence viral persistence.
Elevated levels of ilf3 expression are reported in non-small cell lung carcinoma, ovarian cancer and nasopharyngeal carcinoma (Guo et al., 2008, 2012; Fung et al., 2000; Zhu and Yu, 2010). Moreover the intensity of nuclear NF110 staining in breast tumour specimens was correlated with disease severity and knockdown of NFARs in a breast cancer animal model significantly diminished breast tumour outgrowth and lung metastasis (Hu et al., 2013). Therefore these studies, as well as their role in survivin regulation suggest an important link between NFARs and oncogenesis. Our studies demonstrate that HBZ, and to a lesser extent APH-2 significantly upregulates survivin expression. To our knowledge this is the first report showing that APH-2 can regulate survivin, though both Tax1 (Kawakami et al., 2005; Banerjee et al., 2008), and recently, HBZ (Mitobe et al., 2015) have been demonstrated to modulate survivin. As elevated levels of survivin are linked to the pathogenesis of HTLV-1, our study coupled with the work of Mitobe et al. Mitobe et al., (2015) strongly supports a role for HBZ in survivin regulation. In that study (Mitobe et al., 2015), the authors employed methodologies that distinguished between HBZ RNA and protein activity on survivin gene transcription. Their analysis indicates that HBZ RNA functions as an anti-apoptotic factor by upregulating survivin expression, whereas HBZ protein predominantly modulates the transcription of immune-related genes and promotes proliferation and apoptosis leading to the suggestion that HBZ RNA likely compensates for the apoptosis-promoting effects of HBZ protein. Even though our study did not discriminate between HBZ RNA or protein, our data demonstrates that HBZ and APH-2 regulate survivin expression through the −281 to +30 region of the survivin promoter, which encompasses the region −84 to +34 bp recently reported to be involved in survivin activation by HBZ RNA (Mitobe et al., 2015). Hence we suspect that the effect of HBZ on survivin expression that we observe may be due to HBZ RNA rather than its protein product. Our finding that HBZ, and to a lesser extent APH-2 antagonise the ability of NF110a to enhance survivin expression was unexpected given that all proteins individually can activate survivin expression. This raises the possibility that HBZ/APH-2 may compete with NF110a for promoter binding or alternatively the physical interaction between these proteins renders NF110a unavailable to optimally activate the promoter.
5. Conclusions
In conclusion we have identified the NFAR family as novel interaction partners of the HTLV antisense proteins HBZ and APH-2. We demonstrate that NFAR proteins enhance the activity of Tax proteins and HTLV replication. Additionally we show that HBZ and APH-2 modulate survivin expression, as well as demonstrating that they are capable of antagonising NF110-mediated survivin activation. Thus interactions between HTLV antisense proteins and the NFAR protein family have an overall positive impact on HTLV infection and may contribute to the survival of infected cells.
Acknowledgments
We would like to thank Prof. P Green for the generous gift of the HTLV-2 proviral clone pH6neo, as well as Prof. G Barber and Prof. M Matthews for kindly providing the NFAR constructs, and Dr Jean-Michel Mesnard for providing the GFP-HBZ vectors. We thank Prof. R Mahieux and members of his group for assistance with proviral clone work. We are grateful to Jonathan Dean for support preparing the manuscript and to Elena Woods for assistance with fluorescence microscopy analysis.
Funding
This work was supported by the National Virus Reference Laboratory (NVRL), University College Dublin, Ireland and NIH grants CA10073 and CA63417.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ Contributions
Conceived and designed the study: JM, NS, WWH. Performed the experiments: JM. Contributed to particular reagents: LR. Wrote the paper: JM. Assisted in drafting the manuscript: NS, WWH.
References
- Araujo A, Hall WW. Human T-lymphotropic virus type II and neurological disease. Ann Neurol. 2004;56(1):10–19. doi: 10.1002/ana.20126. [DOI] [PubMed] [Google Scholar]
- Arnold J, et al. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood. 2006;107(10):3976–3982. doi: 10.1182/blood-2005-11-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J, et al. Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation. Blood. 2008;112(9):3788–3797. doi: 10.1182/blood-2008-04-154286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbottah ET, et al. Nuclear Factor 90(NF90) targeted to TAR RNA inhibits transcriptional activation of HIV-1. Retrovirology. 2007;4:41. doi: 10.1186/1742-4690-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C, et al. Temporal regulation of HTLV-2 expression in infected cell lines and patients: evidence for distinct expression kinetics with nuclear accumulation of APH-2 mRNA. Retrovirology. 2012;9:74. doi: 10.1186/1742-4690-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman MT, et al. Long-term increases in lymphocytes and platelets in human T-lymphotropic virus type II infection. Blood. 2008;112(10):3995–4002. doi: 10.1182/blood-2008-05-155960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbous J, et al. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J Biol Chem. 2003;278(44):43620–43627. doi: 10.1074/jbc.M307275200. [DOI] [PubMed] [Google Scholar]
- Banerjee P, et al. Human T-cell lymphotropic virus type 1 infection of CD34 + hematopoietic progenitor cells induces cell cycle arrest by modulation of p21(cip1/waf1) and survivin. Stem Cells. 2008;26(12):3047–3058. doi: 10.1634/stemcells.2008-0353. [DOI] [PubMed] [Google Scholar]
- Cavallari I, et al. Fine tuning of the temporal expression of HTLV-1 and HTLV-2. Front Microbiol. 2013;4:235. doi: 10.3389/fmicb.2013.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciminale V, et al. HTLV-1 and HTLV-2: highly similar viruses with distinct oncogenic properties. Front Microbiol. 2014;5:398. doi: 10.3389/fmicb.2014.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currer R, et al. HTLV tax: a fascinating multifunctional co-regulator of viral and cellular pathways. Front Microbiol. 2012;3:406. doi: 10.3389/fmicb.2012.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthesy B, Kao PN. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J Biol Chem. 1994;269(32):20682–20690. [PubMed] [Google Scholar]
- Coumar MS, et al. Treat cancers by targeting survivin: just a dream or future reality? Cancer Treat Rev. 2013;39(7):802–811. doi: 10.1016/j.ctrv.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Che XF, et al. Overexpression of survivin in primary ATL cells and sodium arsenite induces apoptosis by down-regulating survivin expression in ATL cell lines. Blood. 2006;107(12):4880–4887. doi: 10.1182/blood-2005-08-3423. [DOI] [PubMed] [Google Scholar]
- Clerc I, et al. An interaction between the human T cell leukemia virus type 1 basic leucine zipper factor (HBZ) and the KIX domain of p300/CBP contributes to the down-regulation of tax-dependent viral transcription by HBZ. J Biol Chem. 2008;283(35):23903–23913. doi: 10.1074/jbc.M803116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR, Polakowski N, Lemasson I. HTLV-1 HBZ protein deregulates interactions between cellular factors and the KIX domain of p300/CBP. J Mol Biol. 2011;409(3):384–398. doi: 10.1016/j.jmb.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc I, et al. Propensity for HBZ-SP1 isoform of HTLV-I to inhibit c-Jun activity correlates with sequestration of c-Jun into nuclear bodies rather than inhibition of its DNA-binding activity. Virology. 2009;391(2):195–202. doi: 10.1016/j.virol.2009.06.027. [DOI] [PubMed] [Google Scholar]
- Chen J, et al. Markedly additive antitumor activity with the combination of a selective survivin suppressant YM155 and alemtuzumab in adult T-cell leukemia. Blood. 2013;121(11):2029–2037. doi: 10.1182/blood-2012-05-427773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchange N, et al. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene. 2000;261(2):345–353. doi: 10.1016/s0378-1119(00)00495-9. [DOI] [PubMed] [Google Scholar]
- Fung LF, et al. Differential gene expression in nasopharyngeal carcinoma cells. Life Sci. 2000;67(8):923–936. doi: 10.1016/s0024-3205(00)00684-6. [DOI] [PubMed] [Google Scholar]
- Grassmann R, Aboud M, Jeang KT. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene. 2005;24(39):5976–5985. doi: 10.1038/sj.onc.1208978. [DOI] [PubMed] [Google Scholar]
- Gaudray G, et al. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J Virol. 2002;76(24):12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galabru J, Hovanessian A. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J Biol Chem. 1987;262(32):15538–15544. [PubMed] [Google Scholar]
- Gomila RC, Martin GW, Gehrke L. NF90 binds the dengue virus RNA 3′ terminus and is a positive regulator of dengue virus replication. PLoS One. 2011;6(2):e16687. doi: 10.1371/journal.pone.0016687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C, et al. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J Biol Chem. 2004;279(2):884–891. doi: 10.1074/jbc.M306808200. [DOI] [PubMed] [Google Scholar]
- Guo NL, et al. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin Cancer Res. 2008;14(24):8213–8220. doi: 10.1158/1078-0432.CCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, et al. Correlations among ERCC1, XPB, UBE2I, EGF, TAL2 and ILF3 revealed by gene signatures of histological subtypes of patients with epithelial ovarian cancer. Oncol Rep. 2012;27(1):286–292. doi: 10.3892/or.2011.1483. [DOI] [PubMed] [Google Scholar]
- Hinuma Y, et al. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci USA. 1981;78(10):6476–6480. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Fujii M. Distinct functions of HTLV-1 Tax1 from HTLV-2 Tax2 contribute key roles to viral pathogenesis. Retrovirology. 2009;6:117. doi: 10.1186/1742-4690-6-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, et al. Thymus-derived leukemia-lymphoma in mice transgenic for the Tax gene of human T-lymphotropic virus type I. Nat Med. 2006;12(4):466–472. doi: 10.1038/nm1389. [DOI] [PubMed] [Google Scholar]
- Halin M, et al. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2-mediated transcription. Blood. 2009;114(12):2427–2438. doi: 10.1182/blood-2008-09-179879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, et al. Interleukin enhancer-binding factor 3 promotes breast tumor progression by regulating sustained urokinase-type plasminogen activator expression. Oncogene. 2013;32(34):3933–3943. doi: 10.1038/onc.2012.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanessian AG. The double stranded RNA-activated protein kinase induced by interferon: dsRNA-PK. J Interferon Res. 1989;9(6):641–647. doi: 10.1089/jir.1989.9.641. [DOI] [PubMed] [Google Scholar]
- Harashima A, Guettouche T, Barber GN. Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes Dev. 2010;24(23):2640–2653. doi: 10.1101/gad.1965010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hivin P, et al. Nuclear localization of HTLV-I bZIP factor (HBZ) is mediated by three distinct motifs. J Cell Sci. 2005;118(Pt 7):1355–1362. doi: 10.1242/jcs.01727. [DOI] [PubMed] [Google Scholar]
- Hagiya K, et al. ATF3, an HTLV-1 bZip factor binding protein, promotes proliferation of adult T-cell leukemia cells. Retrovirology. 2011;8:19. doi: 10.1186/1742-4690-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hivin P, et al. The HBZ-SP1 isoform of human T-cell leukemia virus type I represses JunB activity by sequestration into nuclear bodies. Retrovirology. 2007;4:14. doi: 10.1186/1742-4690-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, et al. Nuclear factors are involved in hepatitis C virus RNA replication. RNA. 2007;13(10):1675–1692. doi: 10.1261/rna.594207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, et al. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol Cell Biol. 1999;19(12):8136–8145. doi: 10.1128/mcb.19.12.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JE, et al. The risk of development of HTLV-I-associated myelopathy/tropical spastic paraparesis among persons infected with HTLV-I. J Acquir Immune Defic Syndr. 1990;3(11):1096–1101. [PubMed] [Google Scholar]
- Kao PN, et al. Cloning and expression of cyclosporin A-and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem. 1994;269(32):20691–20699. [PubMed] [Google Scholar]
- Kuwano Y, et al. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2010;38(1):225–238. doi: 10.1093/nar/gkp861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnoselskaya-Riz I, et al. Nuclear factor 90 mediates activation of the cellular antiviral expression cascade. AIDS Res Hum Retrovir. 2002;18(8):591–604. doi: 10.1089/088922202753747941. [DOI] [PubMed] [Google Scholar]
- Kamihira S, et al. Aberrant expression of caspase cascade regulatory genes in adult T-cell leukaemia: survivin is an important determinant for prognosis. Br J Haematol. 2001;114(1):63–69. doi: 10.1046/j.1365-2141.2001.02902.x. [DOI] [PubMed] [Google Scholar]
- Karmakar S, et al. A multiprotein complex necessary for both transcription and DNA replication at the beta-globin locus. EMBO J. 2010;29(19):3260–3271. doi: 10.1038/emboj.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H, et al. Transcriptional activation of survivin through the NF-kappaB pathway by human T-cell leukemia virus type I tax. Int J Cancer. 2005;115(6):967–974. doi: 10.1002/ijc.20954. [DOI] [PubMed] [Google Scholar]
- Larcher JC, et al. Ilf3 and NF90 associate with the axonal targeting element of Tau mRNA. FASEB J. 2004;18(14):1761–1763. doi: 10.1096/fj.04-1763fje. [DOI] [PubMed] [Google Scholar]
- Lemasson I, et al. Human T-cell leukemia virus type 1 (HTLV-1) bZIP protein interacts with the cellular transcription factor CREB to inhibit HTLV-1 transcription. J Virol. 2007;81(4):1543–1553. doi: 10.1128/JVI.00480-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7(4):270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- Matsumoto J, et al. HTLV-1 HBZ suppresses AP-1 activity by impairing both the DNA-binding ability and the stability of c-Jun protein. Oncogene. 2005;24(6):1001–1010. doi: 10.1038/sj.onc.1208297. [DOI] [PubMed] [Google Scholar]
- Marban C, et al. Interplay between the HTLV-2 Tax and APH-2 proteins in the regulation of the AP-1 pathway. Retrovirology. 2012;9:98. doi: 10.1186/1742-4690-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatake Y, et al. Protective roles of epithelial cells in the survival of adult T-cell leukemia/lymphoma cells. Am J Pathol. 2013;182(5):1832–1842. doi: 10.1016/j.ajpath.2013.01.015. [DOI] [PubMed] [Google Scholar]
- McCabe A, et al. The four and a half LIM family members are novel interactants of the human T-cell leukemia virus type 1 Tax oncoprotein. J Virol. 2013;87(13):7435–7444. doi: 10.1128/JVI.00070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, et al. NF90 in posttranscriptional gene regulation and microRNA biogenesis. Int J Mol Sci. 2013;14(8):17111–17121. doi: 10.3390/ijms140817111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatake Y, et al. Anchorage-dependent multicellular aggregate formation induces CD44 high cancer stem cell-like ATL cells in an NF-kappaB- and vimentin-dependent manner. Cancer Lett. 2015;357(1):355–363. doi: 10.1016/j.canlet.2014.11.055. [DOI] [PubMed] [Google Scholar]
- Mitobe Y, et al. HTLV-1 bZIP factor RNA and protein impart distinct functions on T-cell proliferation and survival. Cancer Res. 2015;75(19):4143–4152. doi: 10.1158/0008-5472.CAN-15-0942. [DOI] [PubMed] [Google Scholar]
- Mityaev MV, et al. Functional significance of a putative sp1 transcription factor binding site in the survivin gene promoter. Biochemistry. 2008;73(11):1183–1191. doi: 10.1134/s0006297908110035. [DOI] [PubMed] [Google Scholar]
- Nasr R, et al. Controversies in targeted therapy of adult T cell leukemia/lymphoma: ON target or OFF target effects? Viruses. 2011;3(6):750–769. doi: 10.3390/v3060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, et al. Interleukin enhancer-binding factor 3/NF110 is a target of YM155, a suppressant of survivin. Mol Cell Proteomics. 2012;11(7):M111 013243. doi: 10.1074/mcp.M111.013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara T, et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007;67(17):8014–8021. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- Nakadai T, et al. The RNA binding complexes NF45-NF90 and NF45-NF110 associate dynamically with the c-fos gene and function as transcriptional coactivators. J Biol Chem. 2015;290(44):26832–26845. doi: 10.1074/jbc.M115.688317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poiesz BJ, et al. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77(12):7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino C, Haenni AL, Urcuqui-Inchima S. NF90 isoforms, a new family of cellular proteins involved in viral replication? Biochimie. 2015;108:20–24. doi: 10.1016/j.biochi.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Pfeifer I, et al. NFAR-1 and −2 modulate translation and are required for efficient host defense. Proc Natl Acad Sci USA. 2008;105(11):4173–4178. doi: 10.1073/pnas.0711222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott AM, et al. RNA binding and phosphorylation determine the intracellular distribution of nuclear factors 90 and 110. J Mol Biol. 2005;348(2):281–293. doi: 10.1016/j.jmb.2005.02.047. [DOI] [PubMed] [Google Scholar]
- Rende F, et al. Comparison of the genetic organization, expression strategies and oncogenic potential of HTLV-1 and HTLV-2. Leuk Res Treat. 2012;2012:876153. doi: 10.1155/2012/876153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roucoux DF, Murphy EL. The epidemiology and disease outcomes of human T-lymphotropic virus type II. AIDS Rev. 2004;6(3):144–154. [PubMed] [Google Scholar]
- Reichman TW, et al. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J Mol Biol. 2003;332(1):85–98. doi: 10.1016/s0022-2836(03)00885-4. [DOI] [PubMed] [Google Scholar]
- Reichman TW, Muniz LC, Mathews MB. The RNA binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol Cell Biol. 2002;22(1):343–356. doi: 10.1128/MCB.22.1.343-356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads RE. Regulation of eukaryotic protein synthesis by initiation factors. J Biol Chem. 1993;268(5):3017–3020. [PubMed] [Google Scholar]
- Reichman TW, Mathews MB. RNA binding and intramolecular interactions modulate the regulation of gene expression by nuclear factor 110. RNA. 2003;9(5):543–554. doi: 10.1261/rna.2181103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, et al. The trans-activator tax of human T-cell leukemia virus type 1 (HTLV-1) interacts with cAMP-responsive element (CRE) binding and CRE modulator proteins that bind to the 21-base-pair enhancer of HTLV-1. Proc Natl Acad Sci USA. 1993;90(2):610–614. doi: 10.1073/pnas.90.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, et al. In vivo expression of the HBZ gene of HTLV-1 correlates with proviral load, inflammatory markers and disease severity in HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP) Retrovirology. 2009;6:19. doi: 10.1186/1742-4690-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, et al. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci USA. 2006;103(3):720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, et al. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 2011;7(2):e1001274. doi: 10.1371/journal.ppat.1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J, et al. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol Cell. 2002;10(6):1331–1344. doi: 10.1016/s1097-2765(02)00730-x. [DOI] [PubMed] [Google Scholar]
- Shi L, et al. NF90 regulates inducible IL-2 gene expression in T cells. J Exp Med. 2007;204(5):971–977. doi: 10.1084/jem.20052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, et al. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and −2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem. 2001;276(34):32300–32312. doi: 10.1074/jbc.M104207200. [DOI] [PubMed] [Google Scholar]
- Shabman RS, et al. DRBP76 associates with Ebola virus VP35 and suppresses viral polymerase function. J Infect Dis. 2011;204(Suppl 3):S911–S918. doi: 10.1093/infdis/jir343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy N, et al. Functional analysis of human T lymphotropic virus type 2 Tax proteins. Retrovirology. 2006;3:20. doi: 10.1186/1742-4690-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thebault S, et al. HBZ interacts with JunD and stimulates its transcriptional activity. FEBS Lett. 2004;562(1–3):165–170. doi: 10.1016/S0014-5793(04)00225-X. [DOI] [PubMed] [Google Scholar]
- Tanaka-Nakanishi A, et al. HTLV-1 bZIP factor suppresses apoptosis by attenuating the function of FoxO3a and altering its localization. Cancer Res. 2014;74(1):188–200. doi: 10.1158/0008-5472.CAN-13-0436. [DOI] [PubMed] [Google Scholar]
- Urcuqui-Inchima S, et al. Nuclear Factor 90, a cellular dsRNA binding protein inhibits the HIV Rev-export function. Retrovirology. 2006;3:83. doi: 10.1186/1742-4690-3-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urcuqui-Inchima S, et al. Production of HIV particles is regulated by altering sub-cellular localization and dynamics of Rev induced by double-strand RNA binding protein. PLoS One. 2011;6(2):e16686. doi: 10.1371/journal.pone.0016686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vumbaca F, et al. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol Cell Biol. 2008;28(2):772–783. doi: 10.1128/MCB.02078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viranaicken W, et al. L-Ilf3 and L-NF90 traffic to the nucleolus granular component: alternatively-spliced exon 3 encodes a nucleolar localization motif. PLoS One. 2011;6(7):e22296. doi: 10.1371/journal.pone.0022296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, et al. Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. J Virol. 2009;83(16):7850–7861. doi: 10.1128/JVI.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, et al. Human T-cell leukemia virus type 2 antisense viral protein 2 is dispensable for in vitro immortalization but functions to repress early virus replication in vivo. J Virol. 2012;86(16):8412–8421. doi: 10.1128/JVI.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, et al. Sepantronium bromide (YM155) induces disruption of the ILF3/p54(nrb) complex, which is required for survivin expression. Biochem Biophys Res Commun. 2012;425(4):711–716. doi: 10.1016/j.bbrc.2012.07.103. [DOI] [PubMed] [Google Scholar]
- Zhao LJ, Giam CZ. Interaction of the human T-cell lymphotrophic virus type I (HTLV-I) transcriptional activator Tax with cellular factors that bind specifically to the 21-base-pair repeats in the HTLV-I enhancer. Proc Natl Acad Sci USA. 1991;88(24):11445–11449. doi: 10.1073/pnas.88.24.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, et al. Human T-cell leukemia virus type 1 bZIP factor selectively suppresses the classical pathway of NF-kappaB. Blood. 2009;113(12):2755–2764. doi: 10.1182/blood-2008-06-161729. [DOI] [PubMed] [Google Scholar]
- Zhao T, Matsuoka M. HBZ and its roles in HTLV-1 oncogenesis. Front Microbiol. 2012;3:247. doi: 10.3389/fmicb.2012.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Yu JJ. Gene expression patterns in the histopathological classification of epithelial ovarian cancer. Exp Ther Med. 2010;1(1):187–192. doi: 10.3892/etm_00000030. [DOI] [PMC free article] [PubMed] [Google Scholar]


