Abstract
Copy number variants (CNVs) at the Breakpoint 1 to Breakpoint 2 region at 15q11.2 (BP1-2) are associated with language-related difficulties and increased risk for developmental disorders in which language is compromised. Towards underlying mechanisms, we investigated relationships between single nucleotide polymorphisms (SNPs) across the region and quantitative measures of human brain structure obtained by magnetic resonance imaging of healthy subjects. We report an association between rs4778298, a common variant at CYFIP1, and inter-individual variation in surface area across the left supramarginal gyrus (lh.SMG), a cortical structure implicated in speech and language in independent discovery (n = 100) and validation cohorts (n = 2621). In silico analyses determined that this same variant, and others nearby, is also associated with differences in levels of CYFIP1 mRNA in human brain. One of these nearby polymorphisms is predicted to disrupt a consensus binding site for FOXP2, a transcription factor implicated in speech and language. Consistent with a model where FOXP2 regulates CYFIP1 levels and in turn influences lh.SMG surface area, analysis of publically available expression data identified a relationship between expression of FOXP2 and CYFIP1 mRNA in human brain. We propose that altered CYFIP1 dosage, through aberrant patterning of the lh.SMG, may contribute to language-related difficulties associated with BP1-2 CNVs. More generally, this approach may be useful in clarifying the contribution of individual genes at CNV risk loci.
Introduction
Rare multi-gene copy number variants (CNVs) are well established to increase risk for neurodevelopmental disorders, but translational efforts have been hindered by a limited understanding of underlying mechanisms. The four gene region between breakpoints 1 and 2 (BP1-2) at 15q11.2 is interesting in this regard in that deletions are associated with increased risk for epilepsy and schizophrenia [1–6], and the reciprocal duplications may be relevant to autism [7–9]. Language appears to be compromised in a large number of CNV carriers, in that a meta-analysis of clinically ascertained BP1-2 CNV carriers determined that speech delay was present in 92% and 49% of deletion and duplication subjects, respectively (8). Similarly, separate work found evidence for a significant association between deletion status and self-reported reading difficulties (e.g. “Did you experience any difficulties in reading in elementary school?”) [4]. Described below is work aimed at providing molecular insights into how genetic variation at the BP1-2 region may contribute to disease-related variation in human brain structure.
Recent work found a relationship between gene dosage at BP1-2 and structural variation at multiple disease associated brain regions [4]. Through analysis of magnetic resonance imaging (MRI) data from BP1-2 deletion, duplication, and CNV neutral subjects, investigators identified linear relationships between gene dosage and volume of the left insula (lh.insula), right anterior cingulate cortex (rh.ACC), corpus callosum (CC), left temporal white matter (lh.tempWM), and the left supramarginal gyrus (lh.SMG). The volumes of the lh.insula, rh.ACC, lh.SMG, and lh.tempWM were reduced in deletion carriers and increased in duplication carriers, whereas the opposite directionality was observed for the CC. Interestingly, people with schizophrenia are reported to show reduced volumes of the insula, CC, and lh.tempWM [10–12], as well as abnormal connectivity of the rh.ACC [13]. Structural and functional alterations of the lh.SMG have been implicated in speech and language [14,15]. Taken together, these data support a model whereby altered gene dosage at BP1-2 influences regional brain development and increases risk for disease. We hypothesized that common regulatory variants at the locus might be associated with similar effects in healthy individuals, and that their identification might clarify mechanisms of disease.
The BP1-2 region at 15q11.2 contains four genes (TUBGCP5, CYFIP1, NIPA2, and NIPA1); each has interesting connections to neurodevelopment. TUBGCP5 encodes a protein involved in regulation of chromosomal segregation [16–18]. Although not yet studied with regard to disease, it is directly regulated by GSK3beta, implicated in schizophrenia [19–21]. More is known about the adjacent gene that encodes CYFIP1. Binding to RAC1 activates the Wave Regulatory Complex (WRC) and initiates cytoskeletal remodeling [22,23]. A separate interaction between CYFIP1 and the fragile X mental retardation protein (FMRP) results in the repression of eIF4E-mediated translation [24,25]. Knockdown or overexpression of CYFIP1 impacts neuronal morphology, brain development and function [26–29] and common CYFIP1 regulatory variants have been associated with schizophrenia and autism [2,28,30]. Separate studies looking at NIPA2, a third gene in the region, identified rare functional variants in epilepsy patients but not controls [31,32] and found a schizophrenia associated single nucleotide polymorphism (SNP) upstream of the gene [2]. Moreover, investigation of the relationship between regional gene expression and behavior in individuals with Prader-Willi Syndrome found strong correlations with NIPA2 [33]. NIPA1, immediately downstream of NIPA2, has been implicated in axonal growth [34]. Related, is that dominant negative mutations in NIPA1 cause hereditary spastic paraplegia [35].
In this study, examination of a discovery cohort identified SNPs associated with structural variation at brain regions known to be sensitive to gene dosage at the BP1-2 region. The top hit, upstream of CYFIP1, was likewise associated with variation in lh.SMG surface area in a large independent cohort. Consistent with a gene dosage effect, interrogation of publically available expression data revealed a relationship between genotype at this variant and CYFIP1 levels in human brain. Additional in silico analyses we performed suggest that this association with CYFIP1 expression may be mediated by allele-dependent regulation by FOXP2, a transcription factor (TF) implicated in speech and language [36–38]. Results provide independent support for previous work identifying a relationship between genetic variation at BP1-2 and human brain structure, and point to CYFIP1 as a likely contributor. The novel link to FOXP2 provides convergent evidence for involvement of the BP1-2 region in language. The approach used here could be useful in understanding risk mechanisms at other disease-related CNVs.
Materials and Methods
Discovery Cohort: Subjects, MRI, and Genotyping
The Einstein Institutional Review Board approved this study and subject recruitment. All participants gave informed consent prior to participation. 100 unrelated adults without any self-reported physical disability, psychiatric diagnosis, or first degree relatives with a psychiatric diagnosis were recruited for MRI imaging (S1 Table). 81 of these individuals were Caucasian. Saliva was collected using Oragene OG-500 kits (DNA Genotek). T1-weighed anatomical images were acquired using a Philips 3T Achieva Quasar TX scanner with 32 channel SENSE head coils. An MPRAGE protocol with the following parameters was used: TR/TE = 8.187/3.724 ms, TFE prepulse inversion time = 900ms, flip angle = 8°, 220 axial slices (1.0 mm thick) with 256x256 acquisition matrix. 101 HapMap SNPs with a minor allele frequency (MAF) ≥15% mapped to the BP1-2 interval (chr15:22783636–23085559; Hg19). Tagger was used to define 57 tag-SNPs (r2≥0.8, CEU/TSI populations from Hapmap Version 3, Release 2; S1 Fig and S2 Table) [39]. DNA was extracted using QuickGene SP kits (Kurabo) and a QuickGene 610 instrument (Autogen). Integrity was verified via electrophoresis and concentration measured using a NanoDrop 2000 (Thermo). Primers for Sequenom genotyping were designed for 56/57 SNPs that could be multiplexed [40]. Genotype quality was evaluated by visual examination of clustering, testing for Hardy-Weinberg equilibrium, and retyping of random subjects. 52 SNPs remained after quality control.
Validation Cohort—Subjects, MRI, and Genotyping
Subjects were healthy adults of Dutch descent included in the Nijmegen Brain Imaging Genetics cohort [41,42]. None reported any neurological or psychiatric history. Research was approved by the CMO Region Arnhem-Nijmegen ethics committee. All participants gave written informed consent prior to participation. Imaging data was available or generated for 1276 subjects (all Caucasian) typed for each of rs12102024 and rs1051288, and 2621 individuals (2383 Caucasian) typed for rs4778298 (S3 Table). T1-weighted anatomical scans were acquired for previous work using a 1.5T or 3T Siemens scanner. rs12102024 and rs1051288 genotypes, obtained by Affymetrix GeneChip SNP 6.0 array, were likewise generated previously [43]. rs4778298 was genotyped for this study at the Max Planck Institute in Nijmegen using a custom KASP assay (LGC Genomics). The assay was validated by Sanger sequencing of randomly-selected samples.
Image processing
Scans were processed using FreeSurfer 5.3 [44,45]. For discovery cohort subjects, segmentation was confirmed by inspection of a midline saggital section. Volumes, surface areas, and thicknesses for all regions in the Desikan-Killany atlas [46] were acquired. Analyses were, however, tightly restricted to the lh.insula, rh.ACC, CC, lh.temp.WM, and lh.SMG, regions of interest (ROIs) shown previously to be sensitive to CNVs at BP1-2 [4]. Volume of the rh.caudalanteriorcingulate was taken to represent the rh.ACC, and the volumes of five individually segmented callosal subregions (anterior, mid anterior, central, mid posterior, and posterior) as the CC. lh.tempWM was defined as white matter within the: left bank of the superior temporal sulcus, left entorhinal cortex, left fusiform gyrus, left inferior-temporal cortex, left middle-temporal cortex, left parahippocampal cortex, left superior-temporal cortex, and left temporal pole. Because FreeSurfer does not output surface area or thickness of subcortical regions, the CC and lh.temp.WM were only included for analysis of volume.
Statistical Analyses
In the discovery cohort, associations of individual SNP-ROI pairs were examined using R [47] according to the following linear regression model: Volume of ROI = α + β1(variant) + β2(age) + β3(gender) + β4(ethnicity) + β5(handedness) + β6(total brain volume) + ε, in which α is the intercept and ε random error. For analysis of surface area and cortical thickness, total brain surface area and mean cortical thickness of the whole brain were used for β6, respectively. To assess the aggregate effect of individual SNPs across ROIs, the sum of chi-square statistics (X20) was calculated and its significance (ppermutation corrected < 0.05) was evaluated by permutation with 10,000 replicates (X2n). By aggregating effects across ROIs for each SNP, we were able to perform only one test for each anatomical trait (volume, surface area, and thickness), improving power to detect any effects present. In the validation cohort, three SNP-ROI pairs were tested in PLINK [48] under a linear regression model as above, but scanner field strength was included and handedness omitted. Handedness was not included as a covariate in our analysis of data from the validation cohort. Left handed individuals made up less than 5% of subjects and previously published work in this cohort saw no relationship between handedness and any regional measure of surface area [49].
Gene Expression
Normalized expression data (Human Exon 1.0 ST, Affymetrix) for 1231 human brain samples from 134 individuals (16–102 at death; mean age 59) and corresponding genotypes (Omni 1M Immunochip, Illumina) were downloaded from BRAINEAC [50]. The relationship between expression (averaged across brain regions) and genotype was modeled using linear regression to identify expression quantitative trait loci (eQTL) variants. No correction for multiple comparisons was performed because only one probe (Affymetrix 3583676), specific for the 5’ UTR of CYFIP1short, was tested. For evaluating the relationship between levels of CYFIP1short and candidate TFs, Pearson's correlation coefficients were calculated, and significance assessed using an F-test based on a lack-of-fit sum-of-squares method.
TF Binding
50 bps of genomic sequence on either side of each CYFIP1short eQTL variant was extracted from the UCSC browser (Hg19). Sequences (1 per allele) were screened against human Position Weight Matrices (PWMs) in JASPAR, a database of TF binding profiles, using the Transcription Factor Binding Site Tools package [51,52]. Matches were defined as having a relative score ≥0.8. For differential binding, information content ≥1 at the variable base within the PWM was required.
Results
Common genetic variation at BP1-2 is associated with inter-individual differences in brain structure
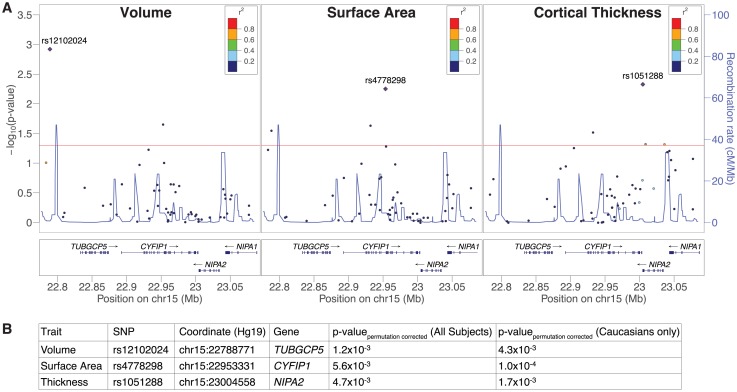
We performed targeted genetic association analyses in a discovery cohort (S1 Table). 52 tag-SNPs from the BP1-2 region were genotyped (S2 Table), and the relationships to volume, surface area, and thickness at predefined ROIs examined under an additive model. Hypothesizing that common regulatory variants within the interval would give rise to effects like those seen in rare CNV carriers, we restricted analyses to ROIs shown previously to be sensitive to BP1-2 dosage [4]. Significant associations were observed (Fig 1A and S4 Table). The strongest signal (rs4778298 at CYFIP1; ppermutation corrected = 5.6x10-3; Fig 1B) became more pronounced after limiting analysis to Caucasians (ppermutation corrected = 1.0x10-4; Fig 1B). Finally, even if an overly stringent Bonferonni correction is used to account for the 3 endpoints examined, p-values of 1.7x10-2 and 3.3x10-4 are observed. Counter to our initial hypothesis that common regulatory variants might uniformly impact all BP1-2 sensitive brain regions, effects were attributable to individual ROIs (Fig 2A and S5 Table). A post-hoc permutation analysis, accounting for all 52 tag-SNPs and ROIs, revealed a significant association between rs4778298 and lh.SMG surface area in Caucasians (ppermutation corrected = 7.5x10-3; see S6 Table). Linear effects at the CC and lh.SMG were observed for rs12102024 and rs4778298, respectively, consistent with an effect of gene dosage (Fig 2B).
Fig 1. Common variant genotypes at 15q11.2 arssociated with differences in regional cortical volume, thickness, and surface area in a discovery cohort.
(A) Aggregate -log p-values for single nucleotide polymorphisms (SNPs) are plotted against genomic position. P-values reflect the relationships between genotype in the discovery cohort and volume (left), surface area (middle), or thickness (right) across all regions of interest (ROIs). Significant associations (ppermutation corrected<0.05) appear above the horizontal red line. Linkage disequilibrium of top hits (diamonds) to typed variants is specified, and estimated recombination rates plotted (blue lines). r2 in the legend indicates the correlation between genotype at each tag-SNP (circles) and genotype at the top hit (diamond). (B) Top aggregate p-values and their SNP descriptions for each trait examined in the full cohort and the Caucasian subset are summarized.
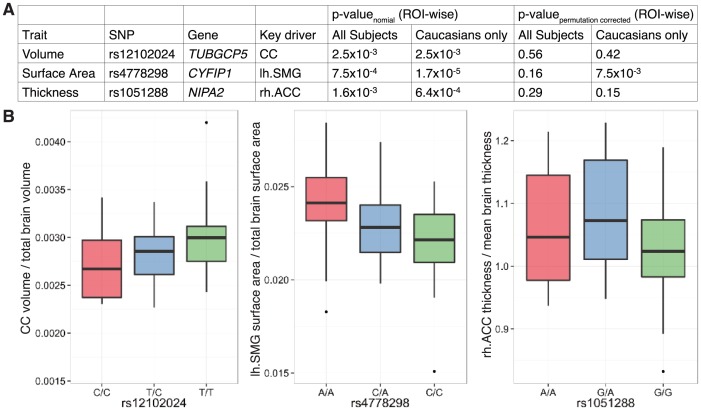
Fig 2. Individual regions of interest (ROIs) drive associations observed in the discovery cohort.
(A) Pairwise analyses of the top signal for each of volume, surface area, and thickness from the discovery cohort determined that aggregate effects were attributable to individual ROIs (and see S5 Table). Post-hoc permutation testing, accounting for all 52 tag-SNPs and ROIs examined, found a significant association between rs4778298 genotype and the surface area of the left supramarginal gyrus (lh.SMG) in Caucasian subjects (ppermutation corrected = 7.5x10-3). (B) Linear effects at the CC and lh.SMG were observed for rs12102024 and rs4778298, respectively, consistent with an effect of allele dosage. rh.ACC and CC correspond to right anterior cingulate cortex and corpus callosum, respectively.
Relationship between SNP genotype at CYFIP1 and left supramarginal gyrus surface area in an independent population
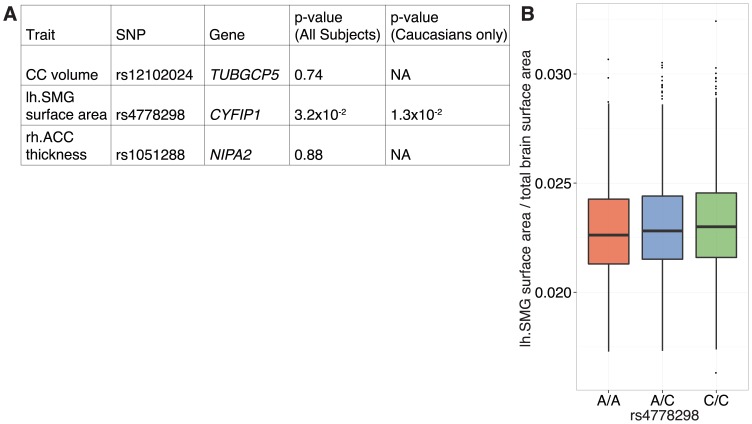
To assess the generalizability of our findings, we analyzed the top three hits in an independent cohort of healthy adults [43]. Imaging data was available for 1276 subjects typed for each of rs12102024 and rs1051288, and 2621 individuals typed for rs4778298 (S3 Table). We saw no signal at rs12102024 or rs1051288, but did see an effect at rs4778298, the top hit in our discovery cohort (pnominal = 3.2x10-2; Fig 3). As above, the effect became stronger after restricting the analysis to Caucasians and survived correction for multiple comparisons (pnominal = 1.3x10-2; pbonferroni corrected = 4.9x10-2; Fig 3). A linear relationship between genotype and lh.SMG surface area was also observed as above, although in contrast to the discovery cohort the ‘A’ allele was seen to be associated with a relative reduction.
Fig 3. Linear association between common variant genotype at CYFIP1 and surface area across the left supramarginal gyrus (lh.SMG) in an independent cohort.
(A) An association between rs4778298 genotype and lh.SMG surface area was observed (pnominal = 3.2x10-2) in a large independent population (n = 1276 for rs12102024 and rs1051288; n = 2621 for rs4778298). Similar to what we observed in our discovery cohort, this effect became stronger on analysis of Caucasian subjects alone and survived correction for multiple comparisons (pnominal = 1.3x10-2; pbonferroni corrected = 4.9x10-2). (B) As in the discovery cohort, a linear relationship between genotype and lh.SMG surface area was observed. Notably, however, directionality was the reverse of what was seen previously. rh.ACC and CC correspond to right anterior cingulate cortex and corpus callosum, respectively.
Common variant signal associated with CYFIP1 levels in human brain
Inspection of the genomic region surrounding rs4778298 determined that it was 2.2 kb upstream of the transcriptional start for a CYFIP1 isoform encoding a 95 kDa protein (CYFIP1short; Fig 4A). To test the hypothesis that rs4778298 genotype is associated with variation in mRNA levels for CYFIP1short, we turned to BRAINEAC, a resource integrating genotype and expression data for human brain samples [50]. Analyses identified a significant linear relationship between rs4778298 genotype and CYFIP1short levels (Affymetrix Probe 3583676; p = 7.2x10-5; S7 Table). rs4778298-C was associated with increased mRNA levels. Additional eQTL variants in high linkage disequilibrium (LD) with rs4778298 (r2>0.9; S7 Table) were also identified. No relationship between genotype at any of these SNPs and levels of TUBGCP5, NIPA2, or NIPA1 was observed (p>0.05).
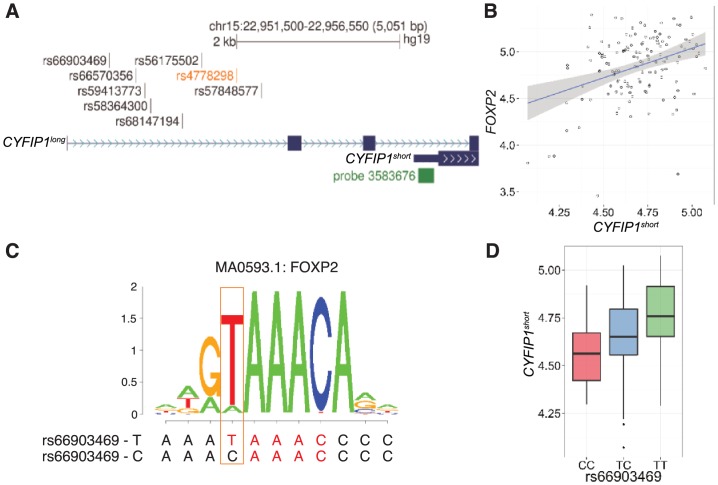
Fig 4. Evidence for genotype-dependent activation of CYFIP1 by FOXP2.
(A) rs4778298 (orange) associated with variation across the surface area of the left supramarginal gyrus is 2.2 kb upstream of CYFIP1short. Seven linked variants (r2>0.9), and an expression probe specific for CYFIP1short (Affymetrix 3583676), are shown. (B) A significant positive association between levels of FOXP2 and CYFIP1 is seen in human brain (r2 = 0.36; p = 2.5x10-5). (C) rs66903469-T is predicted to bind FOXP2 with 11.8-fold greater affinity than the alternate C allele. MA0593.1 corresponds to the position weight matrix for FOXP2 defined in JASPAR. (D) rs66903469-T (r2 with rs4778298-C of 0.93), is strongly associated with increased levels of CYFIP1short (p = 2.9x10-5).
Evidence for genotype-dependent activation of CYFIP1 by FOXP2
We sought next to understand how genetic variation upstream of CYFIP1short might impact expression and perhaps brain structure. Beginning with TFs in JASPAR for which binding in human was characterized (n = 127), we identified a subset of twenty-eight whose expression in brain was correlated with CYFIP1short (pbonferroni corrected<0.05; S8 Table). Ten of these were predicted to bind in a genotype dependent manner to one or more of the eight CYFIP1short eQTL variants we identified (S9 Table). After removing candidate TFs on the basis of logical inconsistencies (e.g. negatively correlated with CYFIP1short expression but having a binding site containing the allele associated with increased expression), or where binding was unlikely to be sensitive to genotype (see methods; S9 Table), only E2F4 and FOXP2 remained. FOXP2 is particularly interesting, as it is well recognized as an important contributor to human speech and language [36–38]. We see a positive relationship between FOXP2 and CYFIP1short levels (r2 = 0.36; p = 2.5x10-5; Fig 4B). Consistent with these findings, the rs66903469-T allele predicted to bind FOXP2 with 11.8-fold greater affinity than the alternate C allele (Fig 4C) is associated with higher levels of CYFIP1short (p = 2.9x10-5; Fig 4D and S9 Table).
Discussion
Towards insight into how genetic variation at the 15q11.2 BP1-2 region may influence disease risk, we examined the relationship between tag-SNPs spanning the interval and aspects of regional brain structure. We report a common variant 2 kb upstream of CYFIP1 associated with the surface area of the lh.SMG, a brain region implicated in language. A relationship between this genotype and lh.SMG surface area was also observed in an independent population composed of 2621 individuals. Consistent with a model whereby the effect is mediated by altered gene expression, in silico analyses revealed that genotype at this SNP, and at seven other variants in high LD, is associated with variation in CYFIP1short mRNA levels in human brain. Differential expression may be mediated by FOXP2, a TF implicated in speech and language [36–38], as it is predicted to bind one of these eQTL variants in an allele-dependent fashion. Supportive of this idea, we report a correlation between mRNA levels for FOXP2 and CYFIP1short in human brain.
Although we see a relationship between rs4778298 genotype in two independent cohorts, the direction of the effect differs between the populations. The ‘A’ allele is associated with increased lh.SMG surface area in the discovery cohort, but a decrease in the validation cohort. While we cannot rule out the possibility that this result may be a false positive, the linearity of the observed effects that recapitulate those for rare CNVs at BP1-2 argue against this [4]. Moreover, as described below, the link to FOXP2 provides biological plausibility for the observed association. The difference in directionality we observed in the two cohorts we looked at may be the result of genetic interactions with variants at the BP1-2 locus or elsewhere in the genome [53,54]. Multiple CYFIP1 regulatory variants independent from rs4778298 / rs66903469 have been associated with increased risk for schizophrenia and autism [2,28,30,55]. Further underscoring complexity at the locus, in one of these studies no association with schizophrenia was observed when the CYFIP1 regulatory variant rs4778334 was considered in isolation, but a clear signal was seen when the effect of a second eQTL variant at the ACTR2 gene encoding an interacting protein was taken into account [28]. Importantly, directionality was reversed as a function of this second genotype, with rs4778334-C seen to be protective in rs268864-AA individuals but found to be risk associated in rs268864-GG subjects. The difference in directionality we observed between the two populations we looked at could also be related to other variables. For example, parent of origin effects in abnormalities of synaptic transmission and behavior in Cyfip1+/- have been reported [56]. Similarly, socioeconomic factors are known to be associated with the regional surface area of numerous brain regions including those involved in language [57]. The relatively small number of subjects within the discovery cohort may also be relevant. Although relatively little is known about CYFIP1 function in mammalian brain development, early knockdown of Cyfip1 in mouse results in inappropriate outward migration of radial glia (RG) from the ventricular zone [28]. Our findings are consistent with this result, in that separate work in human suggests that RG are important regulators of cortical surface area [58,59]. Our results also provide potential insights into mechanisms that may mediate these effects. Full length CYFIP1 binds both RAC1 and FMRP, whereas CYFIP1short is truncated and binds only FMRP [22,25]. All else being equal, less CYFIP1short would result in fewer FMRP binding sites and increase competition between FMRP and RAC1 for CYFIP1long. Reduced RAC1 binding would be expected to dampen WRC activity and in turn interfere with RG migration, a hypothesis that could be tested experimentally. Such experiments, together with studies in additional cohorts, might also shed light on regional specificity. As it stands, it’s unclear whether the signal we observed at rs4778298 / rs66903469 is specific for the lh.SMG or alternatively a result of insufficient power. Results from these investigations will provide new insights into the relationship between genetic variation at the BP1-2 region and brain patterning.
The novel relationship we describe between FOXP2 and CYFIP1 is likewise intriguing given involvement of FOXP2 in the production and comprehension of speech and language. A point mutation disrupting the DNA-binding domain of FOXP2 was seen to segregate with a severe speech and language disorder in the multi-generational KE family [37,60], and screening of unrelated individuals with similar problems identified additional subjects harboring FOXP2 mutations [61]. Experimental work will be required to establish that FOXP2 binding at rs66903469 is genotype-dependent and that variation here is sufficient to alter CYFIP1short levels, efforts that may be complicated by temporal and regional specificity. Nevertheless, the previously unknown relationship between FOXP2 and CYFIP1short provide new insight into language-related aspects of human brain development. Moreover, in keeping with our model that the lh.SMG may be sensitive to FOXP2 activity, an fMRI study determined that KE mutation carriers show hypo-activity at this region during a word retrieval task [62].
Although we did not evaluate directly the relationship between common variation at CYFIP1 and language ability in this study, our results are consistent with the hypothesis that language impairment in BP1-2 deletion carriers [4,8] may be attributable to reduced CYFIP1 acting on the lh.SMG. With regard to idiopathic disease, a meta-analysis points to a reduced volume of lh.SMG in dyslexic children prior to the onset of reading impairment [63,64]. Similarly, adults with dylexia show under-activation of the lh.SMG in a number of fMRI studies [65–67]. In terms of function, the lh.SMG as a part of the dorsal lexical pathway is known to be important in transforming written language components (graphemes) into phonological objects (phonemes) [68,69]. Another function of the lh.SMG is the integration of auditory and sensory inputs before forwarding signals to other brain regions relevant to speech articulation [70,71]. Based on these findings, evaluation of phonological processing and speech articulation in BP1-2 CNV carriers is warranted. Separate studies characterizing the relationships between common variation at the locus and language related outcomes in healthy individuals might complement these efforts.
Lastly, our study points to overlap between the effect of rare and common genetic variation at BP1-2 locus. Given practical difficulties assembling cohorts where all individuals harbor the same rare variant, and progress establishing large cohorts with both MRI and genetic data, the approach described here may be useful in understanding the mechanisms underlying risk for disease at other multi-gene CNVs.
Supporting Information
Haploview was used to generate a schematic representation of the interval and define LD structure. Genes within the interval are illustrated (TUBGCP5, CYFIP1, NIPA2, and NIPA1). Below this, SNPs are represented by short vertical lines. LD blocks appear as black triangles.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
Thanks to the subjects whose participation enabled this work. Thanks also to the following core facilities at Einstein: Human Phenotyping, Molecular Cytogenetics, Neurogenomics, and Translational Neuroimaging. This work makes use of the Brain Imaging Genetics database, first established in Nijmegen in 2007. This resource is now part of Cognomics (www.cognomics.nl), a joint initiative by researchers at the Donders Centre for Cognitive Neuroimaging, the Human Genetics and Cognitive Neuroscience departments of the Radboud University Medical Centre, and the Max Planck Institute for Psycholinguistics in Nijmegen. We thank Sarah Graham for assistance with KASP genotyping and Adam Auton for helpful discussion.
Data Availability
The full discovery dataset has been included as supplemental data (see S6 Table). To protect participant privacy the Max Planck Institute Institutional Review Board does not allow data to be shared publicly. Requests for data may be sent to Dr. Fisher (Simon.Fisher@mpi.nl). Expression and transcription factor data are available from UK Expression Consortium (http://www.braineac.org) and JASPAR (http://jaspar.genereg.net) database.
Funding Statement
Work was supported by an Einstein New Investigator Award (www.einstein.yu.edu), an IDDRC pilot (P30HD071593, www.nichd.nih.gov), and a subcontract from an Autism Center of Excellence (9R01 MH100027, www.nimh.nih.gov) to BSA. Support for RAN was provided through the Training Program in Cellular and Molecular Biology and Genetics (T32GM007491, www.nigms.nih.gov). The Cognomics Initiative (www.cognomics.nl/) is supported by participating groups in Nijmegen and funds from the Biobanking and Biomolecular Resources Research Infrastructure Netherlands (www.bbmri.eu), the Hersenstichting Nederland (www.hersenstichting.nl), and the Netherlands Organisation for Scientific Research (www.nwo.nl/en). TG, AV and SEF are supported by the Max Planck Society (www.mpi.nl). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stefansson H, Rujescu D, Cichon S, Pietiläinen OPH, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455: 232–6. 10.1038/nature07229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao Q, Li T, Zhao X, Huang K, Wang T, Li Z, et al. Rare CNVs and Tag SNPs at 15q11.2 Are Associated With Schizophrenia in the Han Chinese Population. Schizophr Bull. 2012; 10.1093/schbul/sbr197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Kovel CGF, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133: 23–32. 10.1093/brain/awp262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. Nature Publishing Group; 2014;505: 361–6. 10.1038/nature12818 [DOI] [PubMed] [Google Scholar]
- 5.Mullen SA, Carvill GL, Bellows S, Bayly M a, Trucks H, Lal D, et al. Copy number variants are frequent in genetic generalized epilepsy with intellectual disability. Neurology. 2013;81: 1507–14. 10.1212/WNL.0b013e3182a95829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jähn JA, von Spiczak S, Muhle H, Obermeier T, Franke A, Mefford HC, et al. Iterative phenotyping of 15q11.2, 15q13.3 and 16p13.11 microdeletion carriers in pediatric epilepsies. Epilepsy Res. Elsevier B.V.; 2014;108: 109–116. 10.1016/j.eplepsyres.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 7.Chaste P, Sanders SJ, Mohan KN, Klei L, Song Y, Murtha MT, et al. Modest impact on risk for autism spectrum disorder of rare copy number variants at 15q11.2, Specifically Breakpoints 1 to 2. Autism Res. 2014;7: 355–362. 10.1002/aur.1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnside RD, Pasion R, Mikhail FM, Carroll AJ, Robin NH, Youngs EL, et al. Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: a susceptibility region for neurological dysfunction including developmental and language delay. Hum Genet. 2011;130: 517–28. 10.1007/s00439-011-0970-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Zwaag B, Staal WG, Hochstenbach R, Poot M, Spierenburg HA, de Jonge M V, et al. A co-segregating microduplication of chromosome 15q11.2 pinpoints two risk genes for autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153B: 960–6. 10.1002/ajmg.b.31055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makris N, Goldstein JM, Kennedy D, Hodge SM, Caviness VS, Faraone S V, et al. Decreased volume of left and total anterior insular lobule in schizophrenia. Schizophr Res. 2006;83: 155–71. 10.1016/j.schres.2005.11.020 [DOI] [PubMed] [Google Scholar]
- 11.Arnone D, McIntosh AM, Tan GMY, Ebmeier KP. Meta-analysis of magnetic resonance imaging studies of the corpus callosum in schizophrenia. Schizophr Res. 2008;101: 124–132. 10.1016/j.schres.2008.01.005 [DOI] [PubMed] [Google Scholar]
- 12.Bora E, Fornito A, Radua J, Walterfang M, Seal M, Wood SJ, et al. Neuroanatomical abnormalities in schizophrenia: A multimodal voxelwise meta-analysis and meta-regression analysis. Schizophr Res. Elsevier B.V.; 2011;127: 46–57. 10.1016/j.schres.2010.12.020 [DOI] [PubMed] [Google Scholar]
- 13.Guo W, Liu F, Liu J, Yu L, Zhang J, Zhang Z, et al. Abnormal causal connectivity by structural deficits in first-episode, drug-naive schizophrenia at rest. Schizophr Bull. 2015;41: 57–65. 10.1093/schbul/sbu126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liégeois F, Mayes A, Morgan A. Neural Correlates of Developmental Speech and Language Disorders: Evidence from Neuroimaging. Curr Dev Disord reports. 2014;1: 215–227. 10.1007/s40474-014-0019-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sliwinska MW, Khadilkar M, Campbell-Ratcliffe J, Quevenco F, Devlin JT. Early and sustained supramarginal gyrus contributions to phonological processing. Front Psychol. 2012;3: 161 10.3389/fpsyg.2012.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy SM, Preble a M, Patel UK, O’Connell KL, Dias DP, Moritz M, et al. GCP5 and GCP6: two new members of the human gamma-tubulin complex. Mol Biol Cell. 2001;12: 3340–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izumi N, Fumoto K, Izumi S, Kikuchi A. GSK-3β regulates proper mitotic spindle formation in cooperation with a component of the γ-tubulin ring complex, GCP5. J Biol Chem. 2008;283: 12981–12991. 10.1074/jbc.M710282200 [DOI] [PubMed] [Google Scholar]
- 18.Xiong Y, Oakley BR. In vivo analysis of the functions of gamma-tubulin-complex proteins. J Cell Sci. 2009;122: 4218–27. 10.1242/jcs.059196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36: 131–7. 10.1038/ng1296 [DOI] [PubMed] [Google Scholar]
- 20.Freyberg Z, Ferrando SJ, Javitch JA. Roles of the Akt/GSK-3 and Wnt signaling pathways in schizophrenia and antipsychotic drug action. Am J Psychiatry. 2010;167: 388–96. 10.1176/appi.ajp.2009.08121873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hur E-M, Zhou F-Q. GSK3 signalling in neural development. Nat Rev Neurosci. 2010;11: 539–51. 10.1038/nrn2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, et al. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem. 1998;273: 291–5. Available: http://www.ncbi.nlm.nih.gov/pubmed/9417078 [DOI] [PubMed] [Google Scholar]
- 23.Schenck A, Bardoni B, Langmann C, Harden N, Mandel JL, Giangrande A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38: 887–898. 10.1016/S0896-6273(03)00354-4 [DOI] [PubMed] [Google Scholar]
- 24.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134: 1042–54. 10.1016/j.cell.2008.07.031 [DOI] [PubMed] [Google Scholar]
- 25.Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98: 8844–9. 10.1073/pnas.151231598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bozdagi O, Sakurai T, Dorr N, Pilorge M, Takahashi N, Buxbaum JD. Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS One. 2012;7: e42422 10.1371/journal.pone.0042422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oguro-Ando A, Rosensweig C, Herman E, Nishimura Y, Werling D, Bill BR, et al. Increased CYFIP1 dosage alters cellular and dendritic morphology and dysregulates mTOR. Mol Psychiatry. 2014; 10.1038/mp.2014.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon K-J, Nguyen HN, Ursini G, Zhang F, Kim N-S, Wen Z, et al. Modeling a Genetic Risk for Schizophrenia in iPSCs and Mice Reveals Neural Stem Cell Deficits Associated with Adherens Junctions and Polarity. Cell Stem Cell. Elsevier Inc.; 2014;15: 79–91. 10.1016/j.stem.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Rubeis S, Pasciuto E, Li KW, Fernández E, Di Marino D, Buzzi A, et al. CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron. 2013;79: 1169–82. 10.1016/j.neuron.2013.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Tao Y, Song F, Sun Y, Ott J, Saffen D. Common Regulatory Variants of CYFIP1 Contribute to Susceptibility for Autism Spectrum Disorder (ASD) and Classical Autism. Ann Hum Genet. 2015; 329–340. 10.1111/ahg.12121 [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y, Zhang Y, Zhang P, Sang T, Zhang F, Ji T, et al. NIPA2 located in 15q11.2 is mutated in patients with childhood absence epilepsy. Hum Genet. 2012;131: 1217–24. 10.1007/s00439-012-1149-3 [DOI] [PubMed] [Google Scholar]
- 32.Xie H, Zhang Y, Zhang P, Wang J, Wu Y, Wu X, et al. Functional Study of NIPA2 Mutations Identified from the Patients with Childhood Absence Epilepsy. PLoS One. 2014;9: e109749 10.1371/journal.pone.0109749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bittel DC, Kibiryeva N, Butler MG. Expression of 4 genes between chromosome 15 breakpoints 1 and 2 and behavioral outcomes in Prader-Willi syndrome. Pediatrics. 2006;118: e1276–83. 10.1542/peds.2006-0424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang HTH, Edwards TL, Wang X, Connell JW, Davies RJ, Durrington HJ, et al. The hereditary spastic paraplegia proteins NIPA1, spastin and spartin are inhibitors of mammalian BMP signalling. Hum Mol Genet. 2009;18: 3805–21. 10.1093/hmg/ddp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rainier S, Chai J-H, Tokarz D, Nicholls RD, Fink JK. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6). Am J Hum Genet. 2003;73: 967–71. 10.1086/378817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher SE, Scharff C. FOXP2 as a molecular window into speech and language. Trends Genet. 2009;25: 166–177. 10.1016/j.tig.2009.03.002 [DOI] [PubMed] [Google Scholar]
- 37.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413: 519–523. 10.1038/35097076 [DOI] [PubMed] [Google Scholar]
- 38.Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359: 2337–45. 10.1056/NEJMoa0802828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21: 263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 40.Ross P, Hall L, Smirnov I, Haff L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16: 1347–51. 10.1038/4328 [DOI] [PubMed] [Google Scholar]
- 41.Franke B, Vasquez AA, Veltman JA, Brunner HG, Rijpkema M, Fernández G. Genetic variation in CACNA1C, a gene associated with bipolar disorder, influences brainstem rather than gray matter volume in healthy individuals. Biol Psychiatry. 2010;68: 586–8. 10.1016/j.biopsych.2010.05.037 [DOI] [PubMed] [Google Scholar]
- 42.Guadalupe T, Zwiers MP, Teumer A, Wittfeld K, Vasquez AA, Hoogman M, et al. Measurement and genetics of human subcortical and hippocampal asymmetries in large datasets. Hum Brain Mapp. 2014;35: 3277–89. 10.1002/hbm.22401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guadalupe T, Zwiers MP, Wittfeld K, Teumer A, Vasquez AA, Hoogman M, et al. Asymmetry within and around the human planum temporale is sexually dimorphic and influenced by genes involved in steroid hormone receptor activity. Cortex. 2014;2: 1–15. 10.1016/j.cortex.2014.07.015 [DOI] [PubMed] [Google Scholar]
- 44.Reuter M, Schmansky NJ, Rosas HD, Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage. 2012;61: 1402–18. 10.1016/j.neuroimage.2012.02.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9: 179–94. 10.1006/nimg.1998.0395 [DOI] [PubMed] [Google Scholar]
- 46.Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31: 968–80. 10.1016/j.neuroimage.2006.01.021 [DOI] [PubMed] [Google Scholar]
- 47.R Core Team. R: A Language and Environment for Statistical Computing. Available: http://www.r-project.org/ [Google Scholar]
- 48.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M a R, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guadalupe T, Willems RM, Zwiers MP, Arias Vasquez A, Hoogman M, Hagoort P, et al. Differences in cerebral cortical anatomy of left- and right-handers. Front Psychol. 2014;5: 261 10.3389/fpsyg.2014.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. Nature Publishing Group; 2014;17: 1418–1428. 10.1038/nn.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42: 142–147. 10.1093/nar/gkt997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandelin A, Höglund A, Lenhard B, Wasserman WW. Integrated analysis of yeast regulatory sequences for biologically linked clusters of genes. Funct Integr Genomics. 2003;3: 125–134. 10.1007/s10142-003-0086-6 [DOI] [PubMed] [Google Scholar]
- 53.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460: 748–52. 10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey a J, et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. Molecular Autism; 2012;3: 9 10.1186/2040-2392-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waltes R, Duketis E, Knapp M, Anney RJL, Huguet G, Schlitt S, et al. Common variants in genes of the postsynaptic FMRP signalling pathway are risk factors for autism spectrum disorders. Hum Genet. 2014;133: 781–792. 10.1007/s00439-013-1416-y [DOI] [PubMed] [Google Scholar]
- 56.Chung L, Wang X, Zhu L, Towers A, Kim IH, Jiang Y-H. Parental origin impairment of synaptic functions and behaviors in cytoplasmic FMRP interacting protein 1 (Cyfip1) deficiency mice. Brain Res. Elsevier; 2015;1: 1–11. 10.1016/j.brainres.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noble KG, Houston SM, Brito NH, Bartsch H, Kan E, Kuperman JM, et al. Family income, parental education and brain structure in children and adolescents. Nat Neurosci. 2015;18: 773–778. 10.1038/nn.3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nonaka-Kinoshita M, Reillo I, Artegiani B, Martínez-Martínez MÁ, Nelson M, Borrell V, et al. Regulation of cerebral cortex size and folding by expansion of basal progenitors. EMBO J. 2013;32: 1817–28. 10.1038/emboj.2013.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ecker C, Murphy D. Neuroimaging in autism—from basic science to translational research. Nat Rev Neurol. Nature Publishing Group; 2014;10: 82–91. 10.1038/nrneurol.2013.276 [DOI] [PubMed] [Google Scholar]
- 60.Fisher SE, Vargha-Khadem F, Watkins KE, Monaco AP, Pembrey ME. Localisation of a gene implicated in a severe speech and language disorder. Nat Genet. 1998;18: 168–170. 10.1038/ng0298-168 [DOI] [PubMed] [Google Scholar]
- 61.Graham SA, Fisher SE. Decoding the genetics of speech and language. Curr Opin Neurobiol. Elsevier Ltd; 2013;23: 43–51. 10.1016/j.conb.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 62.Liégeois F, Baldeweg T, Connelly A, Gadian DG, Mishkin M, Vargha-Khadem F. Language fMRI abnormalities associated with FOXP2 gene mutation. Nat Neurosci. 2003;6: 1230–1237. 10.1038/nn1138 [DOI] [PubMed] [Google Scholar]
- 63.Linkersdörfer J, Lonnemann J, Lindberg S, Hasselhorn M, Fiebach CJ. Grey matter alterations co-localize with functional abnormalities in developmental dyslexia: an ALE meta-analysis. PLoS One. 2012;7: e43122 10.1371/journal.pone.0043122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raschle NM, Chang M, Gaab N. Structural brain alterations associated with dyslexia predate reading onset. Neuroimage. 2011;57: 742–9. 10.1016/j.neuroimage.2010.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Temple E. Brain mechanisms in normal and dyslexic readers. Curr Opin Neurobiol. 2002;12: 178–83. Available: http://www.ncbi.nlm.nih.gov/pubmed/12015234 [DOI] [PubMed] [Google Scholar]
- 66.Dufor O, Serniclaes W, Sprenger-Charolles L, Démonet J-F. Top-down processes during auditory phoneme categorization in dyslexia: a PET study. Neuroimage. 2007;34: 1692–707. 10.1016/j.neuroimage.2006.10.034 [DOI] [PubMed] [Google Scholar]
- 67.Ruff S, Cardebat D, Marie N, Démonet JF. Enhanced response of the left frontal cortex to slowed down speech in dyslexia: an fMRI study. Neuroreport. 2002;13: 1285–9. Available: http://www.ncbi.nlm.nih.gov/pubmed/12151788 [DOI] [PubMed] [Google Scholar]
- 68.Price CJ. A review and synthesis of the first 20years of PET and fMRI studies of heard speech, spoken language and reading. Neuroimage. Elsevier Inc.; 2012;62: 816–847. 10.1016/j.neuroimage.2012.04.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price CJ. The anatomy of language: a review of 100 fMRI studies published in 2009. Ann N Y Acad Sci. 2010;1191: 62–88. 10.1111/j.1749-6632.2010.05444.x [DOI] [PubMed] [Google Scholar]
- 70.Hickok G. Computational neuroanatomy of speech production. Nat Rev Neurosci. 2012;13: 135–45. 10.1038/nrn3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Golfinopoulos E, Tourville JA, Guenther FH. The integration of large-scale neural network modeling and functional brain imaging in speech motor control. Neuroimage. 2010;52: 862–74. 10.1016/j.neuroimage.2009.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Haploview was used to generate a schematic representation of the interval and define LD structure. Genes within the interval are illustrated (TUBGCP5, CYFIP1, NIPA2, and NIPA1). Below this, SNPs are represented by short vertical lines. LD blocks appear as black triangles.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The full discovery dataset has been included as supplemental data (see S6 Table). To protect participant privacy the Max Planck Institute Institutional Review Board does not allow data to be shared publicly. Requests for data may be sent to Dr. Fisher (Simon.Fisher@mpi.nl). Expression and transcription factor data are available from UK Expression Consortium (http://www.braineac.org) and JASPAR (http://jaspar.genereg.net) database.