Summary
Listeria monocytogenes is a Gram-positive environmental bacterium that lives within soil but transitions into a pathogen upon contact with a mammalian host. The transition of L. monocytogenes from soil dweller to cytosolic pathogen is dependent upon secreted virulence factors that mediate cell invasion and intracellular growth. PrsA1 and PrsA2 are secreted bacterial lipoprotein chaperones that contribute to the folding of proteins translocated across the bacterial membrane; PrsA2 is required for L. monocytogenes virulence, whereas the function of PrsA1 remains to be determined. We have solved an X-ray crystal structure of PrsA1 and have used this model to guide comparison structure-based mutagenesis studies with PrsA2. Targeted mutagenesis of PrsA2 demonstrates that oligomerization of PrsA2 as well as molecular features of the foldase domain are required for protein secretion and virulence, whereas a functional role was uncovered for PrsA1 in bacterial resistance to alcohol. Interestingly, PrsA2 membrane localization is not required for all PrsA2-dependent activities, suggesting that the lipoprotein retains function when released from the bacterial cell. PrsA chaperones are thus multifaceted proteins with distinct domains adapted to accommodate the functional needs of a diverse array of secreted substrates.
Keywords: post-translocation chaperone, bacterial pathogenesis, PPIase, foldase, secretion, PrsA
Introduction
The transport of proteins across lipid-rich bacterial membranes is a fundamental process that is required for the assembly of complex structures such as pili and flagella as well as for nutrient acquisition, cell wall synthesis and division, and bacterial attachment to environmental surfaces. For Gram-positive bacteria, protein secretion requires translocation across a single membrane coupled with protein folding in the externally exposed environment of the membrane-cell wall interface. The Gram-positive cell wall consists of multiple glycan chains cross-linked by short peptides which together form the peptidoglycan while also containing teichoic and lipotechoic acids that represent a high density of negative charge as well as a capacity to bind cationic molecules (Weidenmaier & Peschel, 2008). Bacteria therefore must be capable of correctly folding translocated proteins in a highly charged and potentially fluctuating environment, while those proteins destined for release face the additional challenge of translocation across the thick Gram-positive peptidoglycan cell wall.
Listeria monocytogenes (Lm) is a Gram-positive bacterium that has developed a capacity for survival under a wide range of environmental conditions. The bacterium lives as a saprophyte in decaying plant matter, but maintains the ability to transition into an intracellular pathogen upon the consumption of contaminated food by a human host (Freitag et al., 2009). Lm typically causes mild forms of disease in healthy individuals, however in immunocompromised patients and the elderly, bacterial infections can lead to meningitis, meningoencephalitis, and brain abscesses (Czuprynski, 2005, Drevets & Bronze, 2008). Lm is also capable of crossing the placenta of pregnant woman to infect the developing fetus, leading to stillbirth and abortion (Drevets & Bronze, 2008, Guerrero et al., 2012). The ability of Lm to survive in diverse and challenging environments outside of the human host has resulted in the bacterium becoming a frequent contaminant of food processing facilities, resulting in some of the largest and most deadly food recalls in the United States (Silk et al., 2013, Johnson, 2014).
Central to the ability of Lm to enter a mammalian host and establish an infection is the coordinated secretion of a variety of protein virulence factors. Surface proteins such as internalin A (InlA) and internalin B (InlB) contribute to host cell invasion (Bierne et al., 2007, Disson & Lecuit, 2013), while the secreted pore-forming hemolysin listeriolysin O (LLO) (Koster et al., 2014) and phospholipases mediate vacuole membrane lysis, facilitating bacterial entry into the cytosol. Surface expressed ActA directs host actin-assembly and enables Lm movement through the host cytosol and into adjacent cells (Yeung et al., 2007, O’Neil et al., 2009, Boujemaa-Paterski et al., 2001). Additional secreted or membrane bound proteins contribute to nutrient acquisition within host cells as well as modulation of host innate immune responses (Chaudhuri et al., 2013, Chico-Calero et al., 2002, Sauer et al., 2011, Woodward et al., 2010). Many of these secreted proteins and virulence factors are dependent upon a post-translocation secretion chaperone known as PrsA2 for folding and activity (Alonzo & Freitag, 2010, Alonzo et al., 2009, Alonzo et al., 2011, Zemansky et al., 2009).
PrsA2 and PrsA-like proteins are present in a variety of Gram-positive bacteria where they assist in the folding and secretion of proteins with diverse functions (Jacobs et al., 1993, Cahoon & Freitag, 2015). They are structurally similar to periplasmic secretion chaperones found in Gram-negative bacteria, in that they consist of a central foldase region that helps stabilize unfolded protein chains and a peptidyl-prolyl cis-trans isomerase (PPIase) domain of the parvulin family (Rahfeld et al., 1994, Jakob et al., 2015, Bitto & McKay, 2002, Clantin et al., 2010, Kale et al., 2011). Moreover, they are tethered to the membrane by a lipid-modified N-terminal cysteine so they may act upon substrates at the membrane interface as they are secreted (Baumgartner et al., 2007, Vitikainen et al., 2004, Bitto & McKay, 2002). The activity of PrsA proteins is linked to a variety of physiological phenotypes, including resistance to osmotic stress and extremes of pH, cell-wall integrity, motility, membrane protein folding, and virulence in pathogenic species (Behrens-Kneip, 2010, Cahoon & Freitag, 2015, Hyyrylainen et al., 2010).
Lm has two known PrsA isoforms, PrsA1 and PrsA2 (Alonzo et al., 2009). PrsA1 currently has no known function, and PrsA2 is required for swimming motility, stress resistance, cell wall integrity, and virulence (Alonzo & Freitag, 2010, Alonzo et al., 2009, Alonzo et al., 2011, Cahoon & Freitag, 2015, Zemansky et al., 2009). Although PrsA proteins are homologous at the sequence and structural level, their functions are non-redundant suggesting a level of substrate specificity (Cahoon & Freitag, 2015). This is exemplified by Bacillus subtilis PrsA that is required for normal growth conditions (Vitikainen et al., 2001), whereas Lm PrsA1 and PrsA2 are dispensable and instead appear to function in an environmental or host specific manner (Alonzo & Freitag, 2010). Furthermore, Lm PrsA1 and PrsA2 share 58% sequence identity and 75% homology, yet PrsA1 cannot substitute for PrsA2 when prsA1 is expressed from the prsA2 promoter (Alonzo et al., 2009). Additionally, PrsA orthologues from several species complement for only a select number of phenotypes associated with the loss of prsA2 in Lm (Cahoon & Freitag, 2015). PrsA orthologues restore swimming motility and bacterial growth under extremes of pH, but fail to complement bacterial resistance to antibiotics, lysozyme, and most notably virulence in a mouse model of infection (Cahoon & Freitag, 2015).
Given these observations, we hypothesized that there may exist subtle structural differences between PrsA proteins that account for the recognition of different protein substrates, and thus for different functional roles. To address this, we have taken a structural approach and solved an X-ray crystal structure of Lm PrsA1 and have used this as a basis to model Lm PrsA2. Based on the comparison of these models and with other known PrsA-family structures, we generated a number of site-directed mutants corresponding to conserved and differing structural features. Here we uncover a role for Lm PrsA1 in bacterial resistance to alcohol, and assign distinct functions to PrsA2 domains that contribute to substrate recognition and aspects of protein activity crucial for bacterial virulence.
Results
X-ray crystal structure of PrsA1 and comparison with B. subtilis PrsA
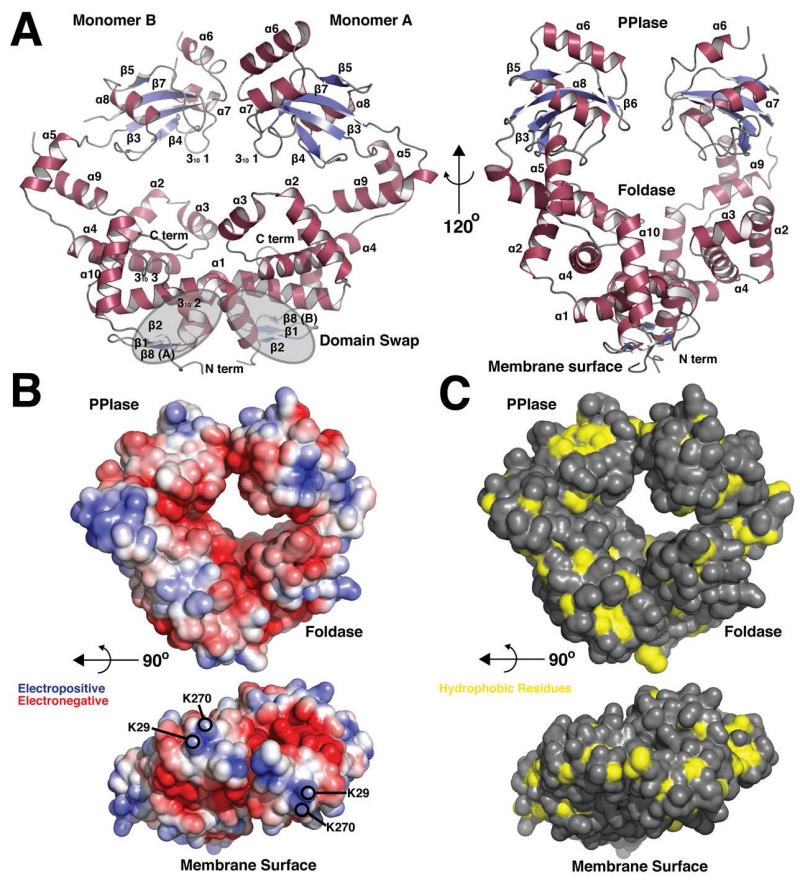
The x-ray crystal structure shows PrsA1 to be a homodimer primarily composed 10 α-helices per monomer (Figure 1A, Table 1). The architecture reveals two general domains, a peptidyl-prolyl isomerase (PPIase) domain (residues 137-228) and a ‘foldase’ domain that consists of both the previously identified N and C terminal domain elements (Alonzo et al., 2011). The foldase, or N-C domain, is formed from the dimerization of PrsA1 with the interface stabilized by a domain swap of the N-terminal segment of helix α1 and the formation of a β-sheet consisting of strands β1, β2 from one monomer and strand β8 from the other monomer (Figure 1A). It is these two β-sheets that are expected to face and potentially interact with the bacterial membrane surface. The foldase domain also contains two mobile ‘arm’ segments (Clantin et al., 2010) comprised of helices α2, α3, and α4 that protrude from the protein surface. This domain architecture is conserved across this structural family of molecular chaperones as it is also observed in Par27 from Bordetella pertussis that aids in the secretion of filamentous hemagglutinin (Clantin et al., 2010), the periplasmic chaperone PEB4 from Campylobacter jejuni (Kale et al., 2011) and PrsA from Bacillus subtilis (Jakob et al., 2015) (Figure 2). Moreover, in this protein family the PPIase domain exhibits rotational freedom relative to the foldase domain, a property which likely serves to aid in the recognition of diverse substrates and to catalyze protein folding (Clantin et al., 2010). To this effect, the PPIase of monomer A in our crystal structure is well resolved and stabilized by crystal packing interactions, whereas the PPIase of monomer B extends into a solvent channel and is poorly resolved. The PPIase of monomer B exhibits higher average B-factors and poor electron density, with residues 229-233 that link the PPIase to the foldase showing no density at a contour level of 1.0 rmsd or 0.19 e/Å3 (Figure 1A and Figure S1A). Furthermore, the conformation of the PrsA1 monomers is non-equivalent, with the PPIase domains displaying a shift of approximately 8 Å relative to each other when the foldase segments are aligned (Figure S1B). Although dynamic, in the trapped conformation of this crystal form the active site faces of the PPIase domains that consist of strands β3, β4, β5, β6, β7 and helix α8 point down and inward towards the foldase domain, with helix 310 1 coming into close proximity of the arm domain of the foldase (helix α2) (Figure 1A).
Figure 1. Overall Structure and molecular surface properties of PrsA1.
(A) Cartoon representation of PrsA1 (residues 22-294). Secondary structure elements are displayed in red (α-helix) and blue (β-strand). Grey highlights indicate the domain swap region at the dimer interface. The PPIase domain and Foldase domain are labeled, in addition to the site of contact with the bacterial membrane. Each secondary structure element is labeled in by number order from N to C terminus (α = α-helix, β = β-strand) (B) Surface electrostatic potential generated by the Adaptive Poisoon-Boltzmann Software (APBS). Red represents electronegative and blue electropositive. The contour is −5 to 5 kT/e. The lower panel shows the surface facing the bacterial membrane with lysine residues highlighted. (C) Solvent accessible hydrophobic residues (A, G, I, L, M, F, V) on the surface of PrsA1 are displayed in yellow, all other residues in grey. The lower panel shows the surface facing the bacterial membrane.
Table 1. Crystallographic Data and Refinement Statistics.
| Native | SeMet (2 crystals) | |
|---|---|---|
| Data Collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 65.3, 84.0, 114.8 | 65.2, 85.1, 116.4 |
| α,β,γ (°) | 90.0, 90.0, 90.0 | 90, 90, 90 |
| Wavelength | 0.97941 | 0.97872 |
| Resolution (Å) | 67.0 – 2.1 | 68.7 – 2.3 |
| Rmeas | 7.0 (108) | 17.2 (106) |
| CC(1/2) | 99.9 (70.1) | 99.4 (73.9) |
| I/σ/ | 17.4 (1.9) | 9.0 (1.5) |
| Completeness (%) | 99.5 (99.3) | 99.4 (97.8) |
| Redundancy | 5.7 (5.8) | 6.2 (3.7) |
| Refinement | ||
| Resolution (Å) | 67.0 – 2.1 | 19.9 – 2.3 |
| No. reflections | 213078 | 342463 |
| No. unique reflections | 37408 | 55605 |
| Rwork/Rfree | 19.2/24.0 | |
| No. atoms | ||
| Protein | 4095 | |
| Water | 279 | |
| B-factors | ||
| Protein | 23.1 | |
| Water | 48.0 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.017 | |
| Bond angles (°) | 1.707 |
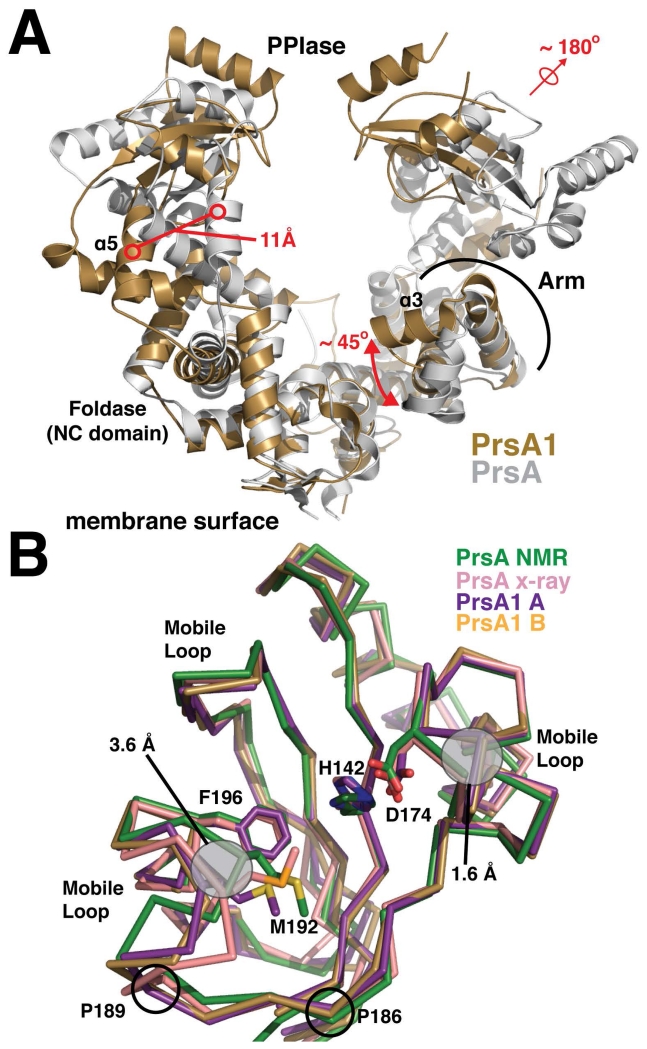
Figure 2. Structural alignment of the PrsA1 and PrsA structures.
(A) Listeria monocytogenes PrsA1 (gold) and Bacillus subtilis PrsA (PDB accession code 4WO7) (grey) are shown. The structures are aligned by an overlay of the foldase (NC) domains. Significant structural differences are highlighted in red. (B) The PPIase domains from each monomer of PrsA1 (purple, gold) are aligned with the PrsA PPIase from Bacillus subtilis (green and pink, PDB accession codes 1ZK6 and 4WO7). The side chains of several conserved catalytic residues are drawn in stick representation for comparison with conformational differences outlined in grey. The approximate regions experiencing dynamic motion as indicated from the PrsA NOE relaxation data are marked as mobile loops. The position of conserved proline residues found in both PrsA1 and PrsA2, but not in PrsA are indicated.
To further explore the general structure features of PrsA1, we also examined the solvent accessible surface of the protein. The charge properties of PrsA1 were calculated and plotted using the adaptive poisson-boltzmann solver (APBS) (Dolinsky et al., 2007, Baker et al., 2001) (Figure 1B) and the positions of hydrophobic residues mapped (Figure 1C). As shown in Figure 1B, PrsA1 has several charged patches. Most notably, the internal self-facing regions of the chaperone are electronegative and the outer surfaces tend towards electropositive. Furthermore, if the model is rotated 90° to observe the surface facing the membrane we observe two electropositive patches formed by lysine residues (Figure 1B). Residue K29 is conserved in both PrsA1 and PrsA2, whereas in PrsA2 the surface lysine K270 is switched to position K268.
When examining the surface of PrsA1 for areas of hydrophobicity, there is no significant clustering of hydrophobic amino-acids on the surface (Figure 1C). The face opposite the active site of the PPIase shows the most notable hydrophobic region of PrsA1. Likewise, the face of PrsA1 that faces the membrane does not exhibit a hydrophobic patch. In this construct, the N-terminal 21 residues are truncated for solubility, as they largely hydrophobic and comprise the signal sequence that serves to target PrsA1 and PrsA2 to the bacterial surface (Forster & Marquis, 2012).
Although Lm PrsA1 shares the same general fold as Bacillus subtilis PrsA, there are structural variations that may help to explain their functional differences (Cahoon & Freitag, 2015) (Figure 2). Overall we observe that our crystal form has trapped a different conformation of the PrsA chaperone. Compared to Lm PrsA1, one PPIase domain of the Bacillus PrsA has been rotated approximately 180° away from the foldase pocket. Additionally, helix α5 that connects the other PPIase domain is shifted just over 11Å (Figure 2A). The PPIase domains of Lm PrsA1 face each other with their active sites pointed towards the foldase pocket making a ‘closed’ conformation. In the Bacillus protein the PPIase domains are non-equivalent and make an intermediate ‘open’ conformation, similar to the motion observed in the SAXS analysis of Par27 (Clantin et al., 2010).
In considering the foldase domain of Lm PrsA1 and PrsA, there are several important differences. We observe that both the Lm PrsA1 and the PrsA foldase are electronegative (Figure S2A and S2B), however the surface of the foldase domain of Bacillus PrsA is more hydrophobic than that seen in our crystal structure with a clear hydrophobic patch in the arm domain made by residues L66, L73, and L82 (Jakob et al., 2015) (Figure S2C). Additionally, helix α3 on the arm domain of Lm PrsA1 that contains the differing hydrophobic patch is rotated 45° up towards the PPIase and inward making the foldase pocket width several angstroms smaller (Figure 2A). However, as the arm is dynamic (Clantin et al., 2010) and the Bacillus PrsA conformation would clash with the Lm PrsA1 crystal packing we cannot rule out that they could adopt each other’s displayed conformations. We do observe that the conformation of the Lm PrsA1 arm is identical in both the monomers and is stabilized by a small hydrophobic core consisting of F88, L92, and F102, making this contrasting conformation likely the dominant fold.
Although the PPIase domain is homologous to structures of the parvulin family (Tossavainen et al., 2006, Jakob et al., 2015) [Z-score of 17 as analyzed by the Dali sever (Holm & Rosenstrom, 2010)], there are distinctions most notably in conformation of the region from G164 to E194 that bears catalytic residue M192 (Figure 2B). Comparing the Bacillus NMR structure to Lm PrsA1 where we observe a shift of 3.6Å outward that slightly expands the observed active site (Figure 2B). These and other loop regions in the PrsA PPIase domain are shown to be mobile, experiencing dynamics as indicated by 1H-15N heteronuclear NOE relaxation data (Tossavainen et al., 2006). The differences could be due to crystal packing of the x-ray structure. However, the PrsA1 PPIase of both monomers display the same conformation and are still non-equivalent to the PrsA X-ray structure (Figure 2B). To account for this, both Lm PrsA1 and PrsA2 have two proline residues in this loop region (P186 and P189, W and K in PrsA respectively) (Figure 2B). As prolines have less conformational freedom and impose rigidity in the peptide backbone, this may serve to restrict the PrsA1/2 PPIase conformations as compared to the Bacillus subtilis PrsA structure, potentially effecting substrate recognition.
Modeling of PrsA2 based on PrsA1 structure
Although PrsA1 and PrsA2 are homologous (57% sequence identity), they share limited functional overlap in Lm (Alonzo & Freitag, 2010, Alonzo et al., 2009, Cahoon & Freitag, 2015). To further dissect differences between the two chaperones, the structure of PrsA1 was analyzed using ConSurf (Ashkenazy et al., 2010, Landau et al., 2005) to plot regions of surface conservation in the PrsA protein family and plot regions of similarity to PrsA2. Aside from the PPIase domain, the family appears generally diverse (Figure S3A) which agrees with the observation that deletion of PrsA-like proteins exhibit different phenotypes indicative of varying functional roles (Alonzo et al., 2009, Hyyrylainen et al., 2010). This is further underscored by the observation that PrsA orthologues cannot fully complement for all Lm PrsA2 activities when expressed in Lm, consistent with distinct protein substrate specificity (Cahoon & Freitag, 2015). When comparing PrsA1 directly to PrsA2, we generally observe a mostly random distribution of residue substitutions on the surface (Figure S3B). However, there are critical differences and similarities in both the PPIase domain and the foldase domain (Figure 3).
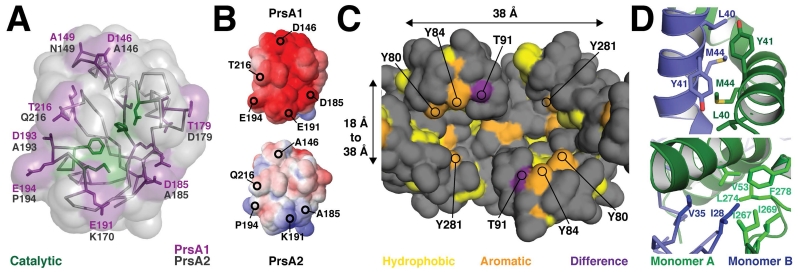
Figure 3. Comparison of the molecular properties of the PrsA1 and PrsA2 secretion chaperones.
(A) The active site surface of the PPIase domain is shown with the sequence differences to PrsA2 highlighted in purple (PrsA1) and black (PrsA2). (B) Electrostatic surface of the PPIase of PrsA1 (top) and PrsA2 model calculated using Modeller (bottom). Surface electrostatic potential was generated by (APBS). Electronegative potential is shown in red and electropositive potential in blue. The contour is −5 to 5 kT/e. (C) View looking down into the Foldase domain pocket from the PPIase domain(s). The approximate size of the surface is indicated in Angstroms as 38 Å long, 18 Å to 38 Å in width, and 20 Å deep as measured from the base to the top of the ‘Arm.’ Hydrophobic residues (A, G, I, L, M, F, V) are colored in yellow, aromatic residues (Y, W, but not F) in orange, and the sequence difference between PrsA1 and PrsA2 in purple (T91 and V91 respectively). (D) Several residues at the dimer interface and that contribute to the domain swap are displayed in stick representation. Monomer A is displayed in green and monomer B in blue.
Structural comparison of the PPIase domains of PrsA1 and PrsA2
To probe for differing structural properties between the PrsA1 and PrsA2 PPIase domain, we created a PrsA2 homology model with the program Modeller (Eswar et al., 2006) using the PrsA1 monomer A as a template. Alignment of the structures reveals several sequence substitutions on the surface of the PPIase domain (Figure 3A and Figure S3C). The core catalytic residues at the center of the active site are conserved (H142, D174, M192, and F196), however there are eight substitutions that encircle that active site on the outer face of the domain (Figure 3A). The general trend of these variations appears to make the PPIase active site surface of PrsA2 less charged or polar relative to PrsA1. This is evident as several acidic residues are swapped for alanines and a proline (D146, D185, D193, and E194). Additionally, the surface electrostatic properties of both models were calculated using APBS (Dolinsky et al., 2007, Baker et al., 2001). In agreement with the observed sequence variation, the PrsA1 PPIase domain is highly electronegative in contrast to the PrsA2 PPIase domain that appears largely uncharged with a small electropositive surface segment (Figure 3B). Notably, the residue variation at positions 185 and 191 (D185 to A185 and E191 to K191) has the effect of creating an opposite surface charge at least on part of the molecule. In contrast, a surface patch of amino acid variations on the face opposing the PPIase active site have a less concerted pattern and do not appear to relay a significant change in surface properties (Figure S3C).
Molecular comparison of the PrsA1 and PrsA2 foldase domains
The foldase domain forms a large pocket on the surface of PrsA1 with the arm segments extending up towards the PPIase domain (Figure 1A and Figure 3C). The pocket is approximately 38 Å long (S251 A to S251 B), 18 Å wide at the bottom of the cavity (S94 A to N95 B), 38 Å at its widest (S87 A to Y84 B), and almost 20 Å deep (bottom of the cavity to the top of the arm domain) (Figure 3C). However, given the dynamic properties of the arm segments the functional size of the foldase pocket is likely variable (Clantin et al., 2010). It is this cavity that is expected to bind a protein substrate during secretion to encourage proper folding, and has been shown to be necessary for chaperone activity (Bitto & McKay, 2002, Kale et al., 2011).
To explore the chemical properties of the foldase cavity, we plotted the solvent accessible hydrophobic and aromatic residues (Tyr and Trp), and also calculated surface electrostatics. As shown in Figure 3C, the foldase cavity has several hydrophobic and aromatic residues suggesting that it can accommodate the partially folded chains of globular proteins. However, compared to PEB4 the foldase domain is significantly less hydrophobic (Kale et al., 2011). This is further exemplified by the surface charge properties as calculated by APBS (Dolinsky et al., 2007, Baker et al., 2001). Here we observe the PrsA1 foldase to be highly electronegative (Figure S2C). Additionally, upon examination of sequence conservation (Figure S3D) the PrsA family members are divergent, supportive of the theme that these chaperones have their own subset of substrates. Conversely, when we compare the Lm PrsA1 to PrsA2 we observe that the interior foldase pocket is largely homologous with the most significant variation at position 91 (T in PrsA1, V in PrsA2) (Figure 3C). However, the general chemical properties of the variant side chains are preserved and largely involve substitution by homologous residues, for example S for T and Q for N. It is also worth noting that a patch of surface residues on the outside face of the arm domain shows significant variation (Figure S3E).
In addition to the slight variation of the foldase surface, an interesting feature of the PrsA1 and PrsA2 foldase cavity is the high representation of tyrosine residues. The cavity is lined with tyrosines, with two of these residues (Y80 and Y84) on the mobile arm domain (Figure 3C). Tyrosine residues are typically enriched in protein:protein interfaces (Moreira et al., 2007), which is demonstrated by PrsA1 as these aromatic residues are oriented to point the phenol side chain directly into the cavity where they presumably can participate in backbone hydrogen bonding and potentially van der waal interactions with a substrate. The variation at position 91 is also on the dynamic arm segment and represents a chemical change from polar in PrsA1 (T91) to hydrophobic in PrsA2 (V91). Given that this residue is surface adjacent to the conserved residues Y80 and Y81 it is tempting to speculate that this residue may impart a level of substrate specificity between PrsA1 and PrsA2.
As the foldase domain is formed from a dimerization event we also examined in detail how the dimer is stabilized. The interface consists of the packing of helix α1 from one monomer against the other and is stabilized by extensive hydrophobic interactions (residues 27 to 31 and to 41 to 60), which includes part of the kinked segment of helix α1 (residues 37 to 47) (Figure 1 and Figure 3D). The N-terminal segment of helix α1 preceding the kink displays the tightest packing interaction where the interaction of the side chains of L40, Y41, and M44 from each monomer is observed. The domain swap also consists of residue I28 from one monomer tucked into a hydrophobic pocket formed by several residues from the other monomer (Figure 1 and Figure 3D). When compared to PrsA2, the highlighted residues at the interface are conserved, with the exception of the substitution of valine in PrsA2 for isoleucine at position 28.
Selection of residues and variant controls for comparative mutational analysis
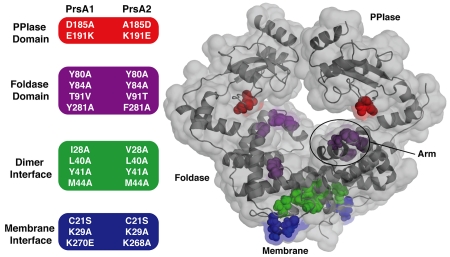
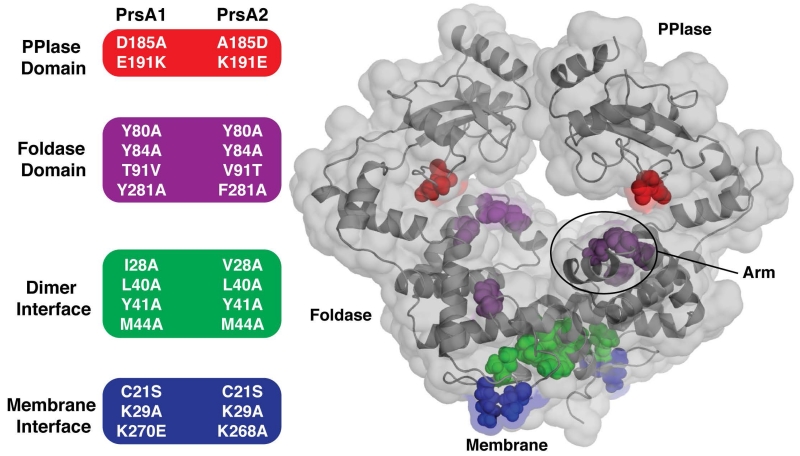
Based on our structural analysis of Lm PrsA1 relative to Lm PrsA2, we targeted a number of amino acid residues based on predicted domain function in both Lm proteins to probe how the structural features of the PrsA2 chaperone correlate to observable phenotypes related to protein secretion, membrane and cell wall stress, and virulence (Alonzo et al., 2009, Cahoon & Freitag, 2015) (Figure 4 and Figure S4). To explore the PPIase domain, residues at positions 185 and 191 were swapped between PrsA1 and PrsA2 to change the surface charge surrounding the active site (Figure 3B, Figure 4). For the foldase, the conserved tyrosines lining the pocket were selected as we hypothesized that they may disrupt general function (Figure 3C, Figure 4). Positions 80 and 281 were of special consideration as they are usually aromatic or hydrophobic in related chaperones. Additionally, residue 91 was swapped (T91V, V91T) in PrsA1 and PrsA2 respectively to test if this residue helps to discriminate substrates. As dimerization is required for activity in vitro (Jakob et al., 2015), we mutated the hydrophobic residues at the interface to alanine to inhibit oligomerization (Figure 3D, Figure 4). Given that Lm PrsA proteins are tethered to the membrane by lipid modification of residue C21 (Baumgartner et al., 2007), yet also appear to be released from the membrane (Cahoon & Freitag, 2014) we created a C21S mutant that would lack lipid modification (Figure 4). As it is a common feature of membrane-associated proteins to contain positively charged residues at the membrane interface to favorably interact with the negative head-groups of phospholipids (Lovering et al., 2007, Lovering et al., 2010), we also mutated K29 and K268/270 (Figure 1B, Figure 4).
Figure 4. Summary of PrsA1 and PrsA2 variants tested in vivo.
(Left) The residues selected for substitution in both PrsA1 and PrsA2 are grouped and color-coded by domain and/or structural feature. (Right) Transparent surface representation of PrsA1 with selected variant residues drawn as color-coded spheres. PPIase domain substitutions (red), Foldase domain substitutions (purple), dimer interface substitutions (green), and membrane interface substitutions (blue).
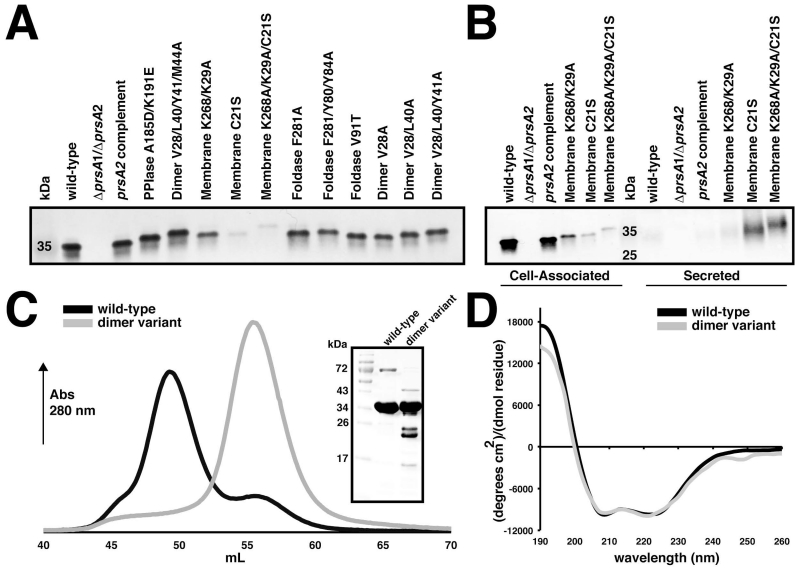
The genes encoding the PrsA1 and PrsA2 variants were expressed from their native promoters at an ectopic chromosomal integration site (Lauer et al., 2002) in a Lm ΔprsA1/ΔprsA2 or ΔprsA2 background. We examined the activity of PrsA1 variants in Lm ΔprsA1/ΔprsA2 strains so as to be able to clearly distinguish PrsA1 variant activity without background contributions from PrsA2, given that PrsA2 activity often predominates over PrsA1. As a control, the mutant strains were verified for normal patterns of growth (data not shown) and for expression levels of Lm PrsA proteins compared to wild-type and a ΔprsA2 complemented with prsA2. Similar protein levels and cellular localization of PrsA2 mutant alleles were observed, with the exception of variants that were designed to disrupt association with the cell membrane (Figure 4, Figure 5A). These variants were predominant in supernatant fractions versus cell-associated fractions, particularly for strains expressing the PrsA2 C21S mutation (Figure 5B). Mutation of the lysine residues (K29 and K268E) disrupted membrane association, though significantly less in comparison to the C21S mutation (Figure 5B). Verification of PrsA1 expression levels proved unsuccessful with either an anti-PrsA2 antibody that binds PrsA1 in vitro (Alonzo et al., 2011) or by adding a C-terminal his-tag to facilitate detection (data not shown). This was not unexpected, as we have observed that PrsA1 as well as other select PrsA orthologues expressed in Lm are produced at very low levels despite retaining the ability to complement for a number of PrsA2-associated functions (Cahoon & Freitag, 2015).
Figure 5. Mutant strain and protein variant control assays.
(A) Cell associated protein profiles for PrsA2 variants expressed in the ΔprsA2/ΔprsA1 background. (B) Secretion profiles of PrsA2 membrane variants in the ΔprsA2/ΔprsA1 background. Bacterial pellets were collected at mid-log phase to detect cell associated protein expression, whereas secreted proteins were precipitated from the media. In both cases PrsA2 variants were visualized by western blot with anti-PrsA2 antibody. (C) Gel-filtration profile of wild-type PrsA1 (black) and the PrsA1 dimer variant (V28/L40/Y41/M44A). Dimerized PrsA1 elutes at ~ 49 mL and monomeric PrsA1 at ~ 55 mL. The inset shows purified wild-type and variant PrsA1 separated by SDS-PAGE gel visualized by Coomassie stain. (D) Circular dichroism spectra of PrsA1 wild-type and the dimer variant plotted as mean residue ellipticity.
To verify that our proposed mutations disrupted PrsA dimer formation, we purified the four-fold PrsA1 dimer variant (I28/L40/Y41/M44A) and tested its ability to dimerize compared to the wild-type protein. As shown in Figure 5C, when the proteins were assayed by size-exclusion chromatography the dimer variant persisted in the monomeric state whereas the wild-type was in an equilibrium that favored the dimer. Moreover, analysis by circular dichroism confirmed that the dimer variant was not unfolded and that it adopts essentially the same helical structure as the wild-type (Figure 5D). As an additional control, the PrsA1 PPIase variant (D185A/E191K) was also tested for dimerization to demonstrate that the substitution of residues located outside of the hydrophobic dimerization interface did not interfere with dimer formation (Figure S5). As an internal control (and as will be demonstrated), we note that every variant has the ability to complement at the wild-type level for one or more PrsA associated phenotypes and exhibits a different pattern of functional loss. This demonstrates that generally active PrsA variants were being expressed for each mutant strain and suggests that our selected mutations likely interfere in the interaction with a subset of protein substrates (Figures 6-10).
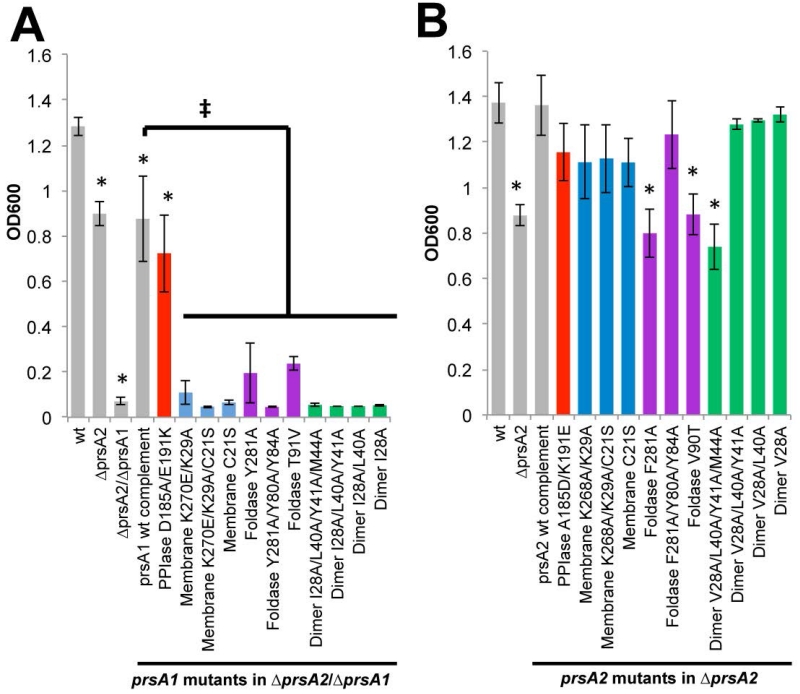
Figure 6. The role of PrsA1 and PrsA2 in resistance to ethanol.
Bacterial growth is shown as the optical density at 600nm (OD600) of strains inoculated from a saturated culture into liquid broth containing 4% ethanol (EtOH) and grown overnight. (A) prsA1 mutants in the ΔprsA2/ΔprsA1 background and (B) prsA2 mutants in the ΔprsA2 background are shown. The grey bars represent the reference strains and the variants are color-coded as Figure 5. Error bars represent the standard error of the mean of 4 independent cultures where P≤0.05 by two-tailed Student’s T-test when compared to Lm wt or prsA1 complement as indicated by an asterisks or ‡, respectively.
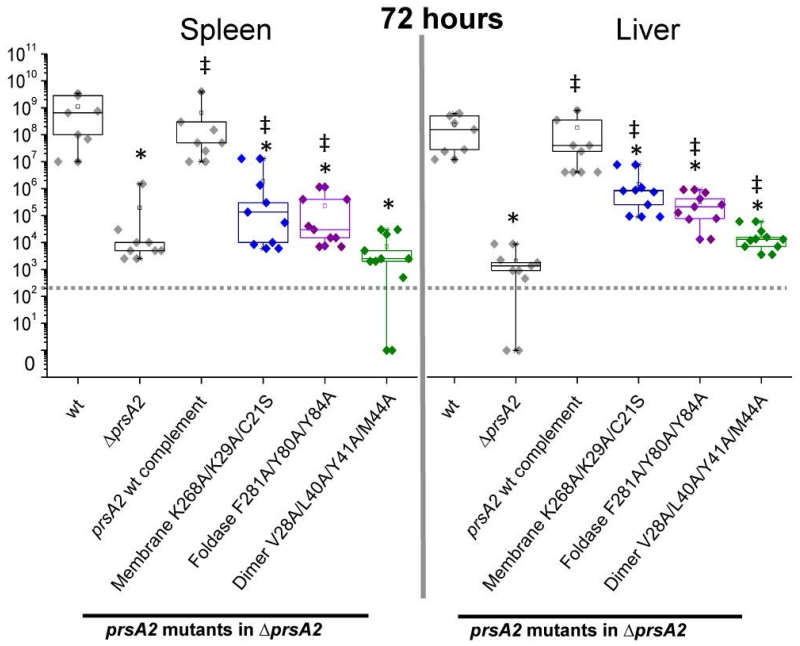
Figure 10. Lm prsA2 mutant virulence phenotypes.
Mice were intravenously infected with 2×104 colony forming units (CFUs) with the indicated reference strains (grey), membrane interface mutant (blue), foldase mutant (purple), and dimer mutant (green). At 72 hours post-infection bacterial burdens were determined for spleens and livers. Box plots are shown where each point represents one mouse. A dotted line indicates the limit of detection. Asterisks and ‡ indicate statistical significance of P≤0.05, by two-tailed Wilcoxon Rank-Sum Test when compared to Lm wt and ΔprsA2, respectively.
Phenotypic analyses of PrsA1 and PrsA2 variants in bacterial resistance to ethanol
Ethanol attacks lipid bilayers and cellular proteins, which impairs multiple functions and elicits a stress response (Silveira et al., 2004, Konopasek et al., 2000). Given Lm PrsA1 and PrsA2 are molecular chaperones located at the membrane-cell wall interface, we assessed the need of these proteins for survival in the presence of 4% ethanol (Figure 6). Both PrsA1 and PrsA2 were found to contribute to ethanol resistance as indicated by comparisons of the growth of wild type, ΔprsA2, and ΔprsA2/ΔprsA1 strains in the presence of ethanol. However, this is the first identified phenotype for which the contribution of PrsA1 appears to dominate over that of PrsA2. Comparison of strains expressing the various prsA1 mutants to the prsA1 wild-type complemented strain indicated that only amino acid substitutions within the PPIase domain were tolerated with respect to functional complementation of ethanol resistance, indicating that PrsA1/PrsA2-specific amino acid variations that alter surface charge near the PPIase active site do not significantly affect aspects of chaperone function relating to ethanol stress (Figure 6A). Substitution of amino acids within the foldase domain (F281A and V90T) or those that most dramatically affected dimerization (V28A/L40A/Y80/M44A) eliminated complementation by prsA1 mutants in comparison to the wild-type allele (Figure 6A). A contribution by PrsA2 to resistance to ethanol stress was apparent and required foldase and dimerization activities (Figure 6B). Interestingly, while single mutations within the foldase domain disrupted PrsA2 activity, the PrsA2 foldase variant containing three amino acid substitutions exhibited wild type levels of activity (Figure 6B). While the reason for this increased activity is unclear, we speculate that the additional hydrophobicity provided by the substituted alanine methyl side-chains in the mobile arm may allow for more, albeit non-specific, interactions with an unfolded peptide chain (Figure 6B). Taken together, these data demonstrate that PrsA1 and PrsA2 contribute to bacterial resistance to ethanol, potentially by stabilizing ethanol susceptible substrates or proteins important for repairing membrane damage. Furthermore, the ability to confer this resistance depends upon proper membrane interaction (K29/K270A), a polar foldase pocket (T91V, and tyrosine variants), and dimerization.
Contributions of PrsA1 and PrsA2 functional domains to cell wall integrity
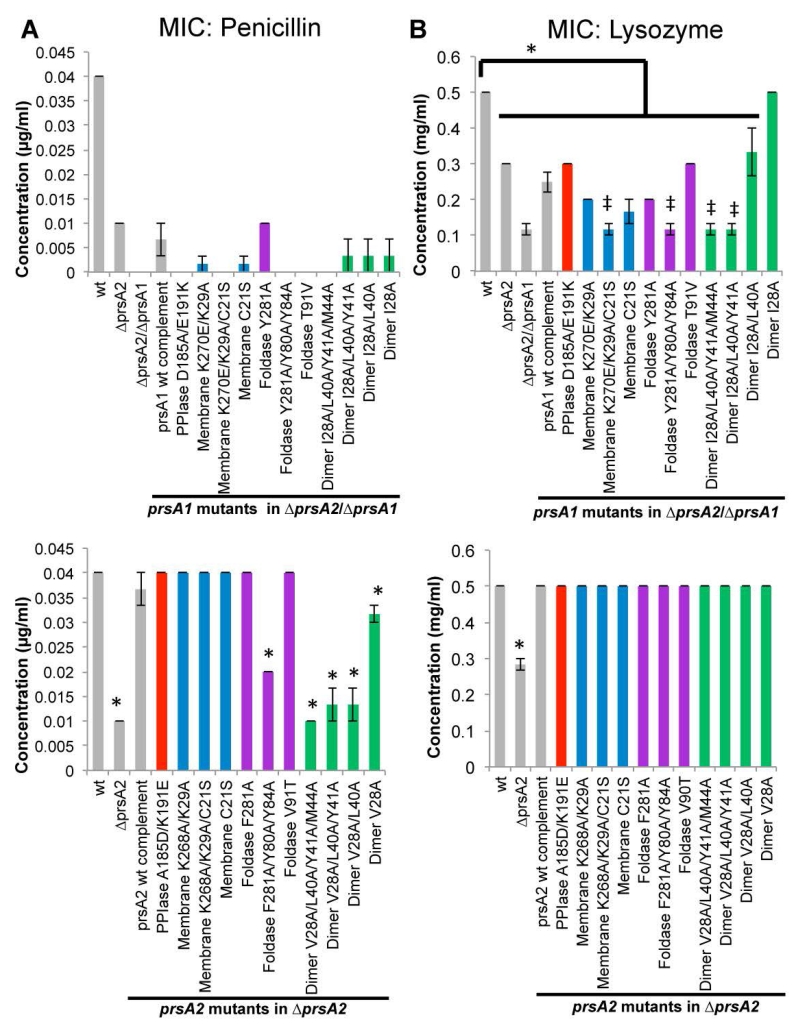
Lm ΔprsA2 strains exhibit increased sensitivity to penicillin, which inhibits peptidoglycan transpeptidation reactions (Alonzo et al., 2011, Cahoon & Freitag, 2015) and the mutants exhibit reduced levels of several penicillin binding proteins (PBPs) (Alonzo & Freitag, 2010). In addition, these strains show increased sensitivity to lysozyme, which catalyzes the hydrolysis of peptidoglycan linkages (Cahoon & Freitag, 2015, Forster et al., 2011). Therefore, we assessed the contributions of PrsA1 and PrsA2 domains to cell wall biosynthesis and integrity by determining the minimum inhibitory concentration (MIC) to penicillin and lysozyme for strains expressing wild-type and targeted mutant proteins (Figure 7). As shown in Figure 7A, PrsA2 is the major contributor to penicillin resistance, however in its absence PrsA1 can compensate to a limited extent. Penicillin resistance mediated by PrsA1 requires its native PPIase suggesting that an electrostatic interaction is necessary for substrate recognition. In addition, nearly every amino acid substitution within a PrsA1 functional domain (with the exception of the Y281A foldase substitution) reduced the already modest compensation provided by PrsA1 for penicillin resistance when prsA1 was expressed from its native promoter. Resistance mediated by PrsA1 to lysozyme appeared somewhat more permissive with respect to substitutions within PrsA1 domains, as single and double amino acid substitutions were tolerated. However both the tyrosine residues of the foldase and dimerization appear necessary for function (Figure 7B). In contrast, resistance to penicillin associated with PrsA2 required protein dimerization and only single aromatic residue substitutions were tolerated within the foldase pocket, again stressing the importance of the foldase aromatic residues. Substitutions within the PPIase domain and membrane binding regions had little effect on resistance (Figure 7B). It has previously been demonstrated that strains expressing PrsA2 variants that completely lack the PPIase domain exhibit increased sensitivity to penicillin (Alonzo et al., 2011). Taken together, these results indicate that the PPIase domain itself is important for activity while remaining functionally tolerant of amino acid substitutions. All PrsA2 variants restored resistance to lysozyme (Fig. 7B) thereby demonstrating that not all PrsA2 functions associated with the cell wall require the same domains or molecular surfaces for substrate recognition.
Figure 7. The contributions of PrsA1 and PrsA2 to cell wall integrity.
The minimum inhibitory concentration (MIC) to prevent growth was determined by bacterial inoculation into broth containing dilutions of the indicated cell wall active antibiotics. (A) The resistance to inhibition of peptidoglycan transpeptidation as measured by the MIC of penicillin. (B) The resistance to hydrolysis of peptidoglycan as measured by the MIC of lysozyme. The top of each panel are prsA1 mutants in the ΔprsA2/ΔprsA1 background and the bottom of each panel are prsA2 mutants in the ΔprsA2 background. The grey bars represent the reference strains and the variants are color-coded as Figure 4. Error bars represent the standard error of the mean of 4 independent cultures where P≤0.05 by two-tailed Student’s T-test when compared to Lm wt or prsA1 complement as indicated by an asterisks or ‡, respectively.
PrsA2 domain function relating to osmotic stress and extreme pH environments
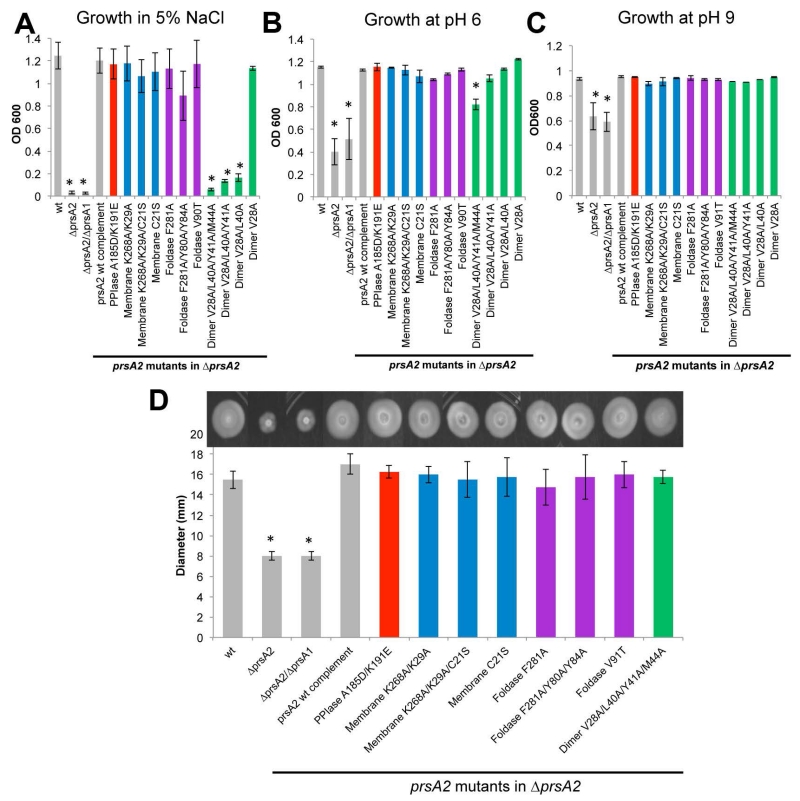
As an environmental pathogen adapted to life under a diverse array of conditions, Lm has evolved to withstand exposure to high osmolarity and a broad range of pH (Chaturongakul et al., 2008). This profound stress resistance is dependent upon functional PrsA2 (Cahoon & Freitag, 2015). Following challenge with 5% NaCl, Lm strains expressing PrsA2 variants with substitutions designed to reduce dimer formation were no longer capable of surviving osmotic stress, whereas all other variants maintained activity similar to the wild-type (Figure 8A). In contrast, the PrsA2 variants exhibited pH tolerance with the exception of the four-fold substituted dimer variant. This variant shows drastically reduced dimer formation in vitro (Figure 5C), which appears to correlate to a loss of activity at low pH (Figure 8B and 8C). The ability of PrsA2 chaperone function to tolerate a wide-range of amino acid substitutions suggests that the protein substrates engaged by PrsA2 in response to pH stress may not require a single specific molecular surface for recognition and folding, or that the domains of PrsA2 may be able to compensate for each other for substrate interactions necessary for low pH resistance. These observations of PrsA2 variant function with respect to low pH are consistent with the ability of many PrsA orthologues, including PrsA1, to complement for PrsA2 in extreme pH conditions (Cahoon & Freitag, 2015). Similarly, all PrsA2 variants retained functional complementation of swimming motility (Figure 8D), an activity that was also complemented by multiple PrsA orthologues (Cahoon & Freitag, 2015) and which may reflect an indirect role of PrsA2 for motility.
Figure 8. The contribution of prsA2 mutants to cellular physiology.
Shown is the growth (OD600) of strains inoculated from a saturated culture into liquid broth containing 5% NaCl (A), a pH of 6 (B), and a pH of 9 (C). (D) Swimming motility of the prsA2 mutants. Motility was measured as the diameter of the swimming colony. Diameters are indicated as the average of 4 swimming colonies from 4 independent experiments. The grey bars represent the reference strains and the variants are color-coded as Figure 4. Error bars represent the standard error of the mean of 4 independent experiments where P≤0.05 by two-tailed Student’s T-test when compared to Lm wt as indicated by asterisks.
The contribution of PrsA2 structural features to Lm virulence
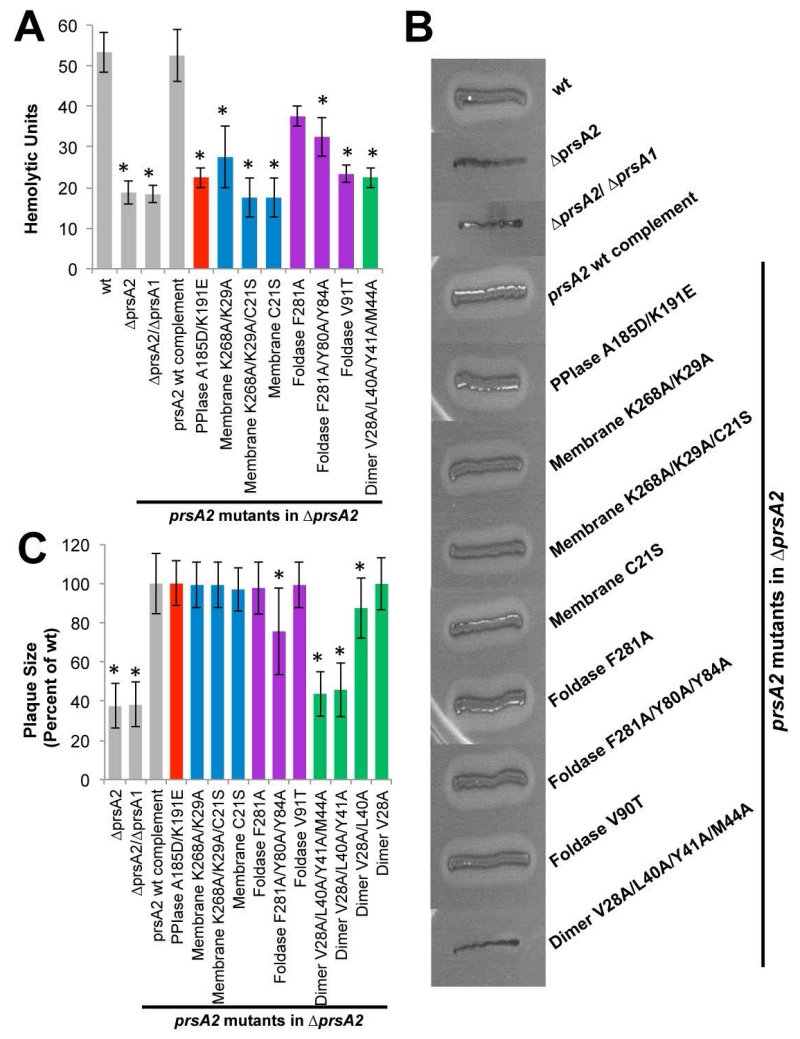
PrsA2 is necessary for the proper folding and secretion of several virulence factors including listeriolysin O (LLO) and phospholipase (PC-PLC) and is required for bacterial virulence in a mouse models of Lm infection (Alonzo & Freitag, 2010, Alonzo et al., 2011, Cahoon & Freitag, 2015, Forster et al., 2011, Zemansky et al., 2009). When we examined the supernatants derived from strains expressing the PrsA2 variants for evidence of the secretion of active LLO, we found that all substitutions with the exception of the single F281A foldase substitution resulted in reduced LLO activity (Figure 9A). This suggests a role for both the electrostatic surface of the PPIase and the presence of a hydrophobic residue in the dynamic arm domain (Foldase V91T) in the recognition and folding of LLO. This is one of the few phenotypes associated thus far with these specific amino acid substitution mutants and demonstrates that a very specific molecular surface is likely required to recognize and fold LLO. The requirement for the electrostatic surface of the PPIase domain was surprising, given that we had earlier demonstrated that this entire domain can be removed without a dramatic effect on secreted LLO activity (Alonzo et al., 2011) and that substitutions are tolerated within this domain for other activities (Figure 7 and 8). It would thus appear that having an altered PPIase domain is more detrimental to PrsA2 activity as it relates to LLO activity than the complete absence of the domain. In contrast, all PrsA2 variants were capable of restoring secreted PC-PLC activity except for the dimer variant (Figure 9B).
Figure 9. The effect of prsA2 mutants of the secretion of known virulence determinants.
(A) Hemolytic activity of prsA2 mutants. Bacterial culture supernatant dilutions were assessed for the ability to lyse sheep red blood cells (RBCs) in vitro. The reciprocal of the dilution that resulted in 50% RBC lysis (hemolytic units) was determined for 4 independent experiments. (B) Phospholipase activity of the prsA2 mutants. Phospholipase activity was determined by the incubation of strains on Listeria selective agar plates containing lecithin that when hydrolyzed produces a zone of opacity surrounding the bacterial streak. (C) Intracellular growth and cell to cell spread of prsA2 mutants. A representative plaque assay is shown where monolayers of mouse L2 fibroblasts were infected with the indicated Lm strains and plaque formation was determined in the presence of gentamicin 72 hours post-infection. At least 25 plaques were measured in 3 independent experiments for all strains. Measurements represent plaque size with respect to Lm wt (set at 100%). Grey bars represent the reference strains and the variants are color-coded as Figure 4. Error bars represent the standard error of the mean where P≤0.05 by two-tailed Student’s T-test when compared to Lm wt as indicated by asterisks.
The formation of zones of clearing, or plaques, within monolayers of Lm-infected L2 fibroblast cells has been used as a measure of the ability of Lm to infect cell monolayers, escape from host cell vacuoles, and spread from cell to cell (Sun et al., 1990). Given that it has been previously demonstrated that only a fraction of LLO activity is required for efficient escape from host cell vacuoles (Freitag & Portnoy, 1994), and that despite reduced levels of secreted LLO activity Lm mutants lacking prsA2 appear to mediate efficient vacuole escape (Alonzo et al., 2011), we assessed the ability of strains expressing PrsA2 variants to mediate plaque formation in fibroblast monolayers. While most amino acid substitutions within PrsA2 reduced secreted LLO activity, many of these mutants remained capable of restoring intracellular growth and cell-to-cell spread to strains infecting fibroblast monolayers (Figure 9C). Notable exceptions were strains expressing PrsA2 variants containing amino acid substitutions designed to reduce dimer formation (triple and quadruple substitution mutants prsA2 V28A L40A Y41A and prsA2 V28A L40A Y41A M44A). Substitution of the aromatic residues within the foldase domain (prsA2 Y80A Y84A F281A) seemed to also play a minor role (Figure 9C).
Based on the ability of different targeted mutations to affect specific aspects of PrsA2-associated activities, we assessed the influence of selected mutations to interfere with PrsA2 functions required for virulence in a mouse model of infection. Selected mutants included the triple membrane association variant (prsA2 C21S K29A K268A), an aromatic residue foldase variant (prsA2 Y80A Y84A F281A), and the four substitution dimer variant (prsA2 V28A L40A Y41A M44A) for function within the host. Mutant strains bearing these protein variants were compared to both the wild-type and wild-type prsA2 complemented strains for bacterial burdens in the spleens and livers of infected mice (Figure 10). Strains expressing the PrsA2 mutant with substitutions within the dimerization domain exhibited levels of virulence attenuation essentially equivalent to strains that completely lack prsA2 (Figure 10). This virulence defect appeared consistent with observations that the dimer variant exhibited reduced activity for most PrsA2-associated phenotypes, illustrating the fundamental role of dimerization for PrsA2 activity. Alteration of PrsA2 membrane association also significantly reduced bacterial virulence to mutants, although not to the same extent as inhibition of dimer formation, especially within the liver (Fig. 10). Substitution of the foldase aromatic residues also significantly reduced virulence, strongly suggesting a requirement for foldase activity and directly relating the importance of this specific molecular surface to virulence (Figure 10). These experiments suggest that while individual PrsA2 substrates may only require a subset of PrsA2 functional domains, the complex coordination of protein secretion and cell wall integrity required for successful bacterial infection of a mammalian host imposes extensive demands on all aspects of PrsA2 activity.
Discussion
The folding and secretion of active protein molecules across the bacterial membrane is fundamental to the ability of Lm to survive under a variety of environmental conditions, including the ability of the bacterium to establish a replication niche within an infected host. Secreted chaperones of the PrsA family have been implicated in the folding and activity of diverse secreted proteins in Gram-positive bacteria (Cahoon & Freitag, 2014), and we have focused our analyses on the PrsA1 and PrsA2 chaperones of Lm to gain a clearer understanding of how these proteins contribute to both Lm physiology and pathogenesis in addition to the general process of Gram-positive protein secretion. To this end, we have solved an X-ray crystal structure of Lm PrsA1 and used this model as a template to correlate specific molecular features to observable PrsA-dependent phenotypes. With this approach, we have been able to identify important residues within the multiple functional domains of PrsA1 and PrsA2, and to distinguish the requirements of PrsA2 membrane association, foldase, and dimerization for several distinct PrsA2-associated activities. While some PrsA2-associated activities tolerated multiple amino acid substitutions within PrsA2, the complex requirements of host infection require the full activity of all PrsA2 domains. These studies are the first to demonstrate functional residues outside of the PPIase catalytic domain, and to demonstrate the complexity of substrate interactions mediated by this family of secreted chaperones. In addition to these findings, we have successfully identified at least one role for Lm PrsA1, that being its contribution to ethanol resistance.
Structural analysis of PrsA1 and the use of PrsA1 to model PrsA2 highlighted several differences in the molecular structure between the two chaperones, consistent with the idea that these proteins have non-redundant substrates. Significant structural differences include the extreme charge difference of the PrsA1 and PrsA2 PPIase domains and a few substitutions within the foldase pocket including residue T91 (Figure 3). Although we have yet to discover a phenotype that solely depends upon PrsA1, we do observe that most of the resistance conferred to challenge by ethanol is mediated by PrsA1 (Figure 6). Furthermore the activity of PrsA1 was dependent upon both dimerization and the polar nature of the foldase pocket (Y80, Y84, and T91), but not on the surface charge of the PPIase. This suggests that the PPIase acts equally well (or not at all) regardless of charge in this case and that the tyrosine residues within the foldase pocket contribute to substrate selection. As a role in ethanol resistance is the first phenotype associated with this chaperone, and given that ethanol disrupts lipid bilayers, it is possible that PrsA1 may play a principal role in general maintenance of the Lm membrane.
Evidence for potentially over-lapping chaperone function can be observed in the ability of Lm PrsA1 to contribute to the integrity of the peptidoglycan layer and to contribute along with PrsA2 to resistance to beta-lactam antibiotics (Figure 7A). For PrsA1, cell wall related activities appear dependent upon the charged PPIase surface, dimerization, and on foldase residue T91, whereas cell wall related activities for PrsA2 require dimerization and the conserved aromatics present within the foldase pocket. These differences in what are required for PrsA1 and PrsA2 cell wall-related activities argue that the distinctions in the molecular surface between PrsA1 and PrsA2 contribute to substrate recognition. PrsA1 activity significantly contributes to bacterial resistance to lysozyme, playing a role nearly equal in magnitude to that of PrsA2 (Figure 7B). However, where all strains expressing variants of PrsA2 remained resistant to lysozyme, a clear residue dependency was detectable with PrsA1. Strains expressing PrsA1 depended upon the foldase aromatic residues for activity, suggesting that each PrsA isoform recognizes potentially distinct protein substrates to maintain the cell wall. Additionally, as PrsA proteins have been reported to interact directly with PBPs (Hyyrylainen et al., 2010), it is tempting to speculate that residues Y80, Y84, and F281 of both PrsA1 and PrsA2 directly interact with nascent PBPs as they are secreted to direct proper folding. Given that PrsA1 may have a substantial role in the general activities of membrane and peptidoglycan integrity, perhaps the function of PrsA1 is to lessen the total substrate burden in environments where Lm PrsA2 is needed for virulence-associated substrates.
The present studies have made it possible to correlate molecular features of PrsA2 directly to virulence and substrate recognition. Most importantly, and in agreement with in vitro activity assays (Jakob et al., 2015), dimerization of PrsA proteins is a major determinant of activity. We see that the ability to dimerize is required for virulence (Figure 5C and Figure 10), and indeed, dimerization was required for most PrsA2-associated phenotypes including the ability to grow in environments of osmotic stress (Figure 8), challenge by beta-lactams (Figure 7), secretion of active LLO and PC-PLC, and intracellular growth and cell to cell spread (Figure 9). It is interesting to note however, that the dimer mutant was still active in some cases, such as the ability to maintain growth a pH 9 (Figure 8) as well as support swimming motility (Figure 9). As some structurally related PrsA-like chaperones are known to function as monomers, such as Campylobacter jejuni Cj1298 (Kale et al., 2011), this suggests that the PPIase and partial foldase pocket can be sufficient for a subset of PrsA2 functions. Alternatively, as PrsA2 is tethered to the membrane it may still be able to form a dimer on the membrane, albeit less efficiently and therefore substrate localization may be able to direct and rescue chaperone activity.
It appears that the ring of aromatic residues that line the PrsA2 foldase pocket (Y80, Y84, and F281) is a critical structural feature for virulence and recognition and folding of many PrsA2 substrates (Figure 10). The tyrosine residues likely stabilize folding protein chains through hydrogen bonding with the peptide backbone, and support overall folding by hydrophobic and van der waal interactions with their aromatic side chain. Furthermore, the hydrophobic character of V91 (T91 in PrsA1) appears to participate directly in substrate recognition and discrimination between the isoforms. Residue V91 is required for the secretion of the virulence factor LLO (Figure 9) and ethanol resistance mediated by PrsA2 (Figure 6). Residues T91 in PrsA1 is required for ethanol resistance (Figure 6) and beta-lactam resistance (Figure 7). As position 91 is not required for all activities of both chaperones, this suggests it necessary for a subset of substrates for each protein. Additionally, it may allow PrsA1 to recognize protein substrates required for full beta-lactam resistance that are not substrates for PrsA2. Overall these observations agree with the dimerization requirement, as oligomerization is what forms the foldase pocket. However, as mutation of the foldase pocket did not disrupt all the same activities as the dimer mutations (Figure 6B, 8A, 9) this reinforces that residues Y80, Y84, F281, and V91 are needed to recognize subsets of protein substrates.
As Lm PrsA1 and PrsA2 are lipoproteins and are thought to work primarily at the membrane interface, it was interesting to observe that membrane localization was not a strict requirement for activity. Similar to the foldase mutants, localization was required for full virulence but its deletion also resulted in an intermediate phenotype. Moreover, membrane localization is dispensable for resistance to lysozyme, osmotic stress and extreme pH environments, and surprisingly secretion of flagellin and PC-PLC (Figure 8D and Figure 9B). These observations support evidence that PrsA2 is secreted from the bacterial cell as part of its normal function (Cahoon & Freitag, 2014). Alternatively, it is possible that the transient association of PrsA2 membrane localization variants that occurs during the process of secretion may be sufficient for partial complementation of chaperone activity. Additionally, correct interaction with the membrane as mediated through an electropositive surface is important for chaperone activity (K29, K268/270 mutants). These mutants remained tethered but could not function in the secretion of LLO (Figure 9), and also disrupted the ability of PrsA1 to function (Figure 6A).
Chaperones with structural similarity to PrsA are ubiquitous in bacterial species, and the elucidated crystal structures appear to show a general trend that this family functions as homo-dimers stabilized by a domain swap that serves to form the pocket of the foldase, with the PPIase domain being rotationally independent (Figure 1) (Jakob et al., 2015, Kale et al., 2011, Clantin et al., 2010, Bitto & McKay, 2002). However, although the architecture is conserved and some chaperone functions are general, each protein has evolved specific molecular features for a subset of substrates. For example, our mutational analyses indicate that both the surface charge of the Lm PrsA2 PPIase and hydrophobic nature of residue V91 on the dynamic arm are important for the secretion of active LLO, but not for resistance to osmotic stress or extreme pH environments (Figure 8 and Figure 9). It is interesting that the N-C or foldase domain in this protein family is largely responsible for most chaperone activity as it can complement for deletion strains, yet the conserved PPIase domain is dispensable (Alonzo et al., 2011). Indeed, complete removal of the PrsA2 PPIase domain or swapping of the PrsA2 PPIase domain with that of PrsA1 had little effect on secreted LLO activity (Alonzo et al., 2011); however alteration of individual amino acids within the domain did reduce the secretion of active LLO. It is possible that substitutions within the PPIase domain results in an inactive domain that impedes foldase activity by making non-productive interactions or steric hindrance that inhibits the folding process, whereas complete loss of the domain removes this burden. This scenario is likely, as the work with B. subtilis PrsA shows that both the PPIase and foldase (NC domain) can make contacts with a small peptide substrate simultaneously (Jakob et al., 2015). In contrast, as the pocket of the foldase diverges heavily between species not only in sequence but also in molecular surface, ranging from very hydrophobic in PEB4 (Kale et al., 2011) to very polar in Lm PrsA1 (Figure S2B), these results demonstrate that the foldase domain is the major determinant of substrate discrimination.
Our work here has provided an in depth exploration of the molecular features of the Lm PrsA chaperones and has provided a framework for Lm specific drug discovery. Given the critical role for PrsA2 in cell wall integrity, antibiotic resistance, and virulence, the inhibition of PrsA2 function would provide an effective means of reducing Lm pathogenicity while increasing its sensitivity to drugs that target the cell wall. Given the accessibility of PrsA family members at the membrane-cell wall interface, these chaperones may represent promising drug targets for a variety of Gram-positive pathogens.
Experimental Procedures
Bacterial strains, plasmids, and media
Bacterial strains used in this study are listed in Supplemental Table 1. Lm 10403S is the wild-type (wt) strain and Lm 10403S containing an erythromycin resistance gene (erm) in place of the prsA2 coding sequences is referred to as ΔprsA2 (Alonzo et al., 2009), while ΔprsA1 transduced with ΔprsA2::erm is ΔprsA2/ΔprsA1 (Alonzo & Freitag, 2010). Either ΔprsA2 or ΔprsA2/ΔprsA1 was used for complementation with the designated prsA2 or prsA1 mutant allele. Escherichia coli One Shot TOP10 (Invitrogen), SM10, and S17 (a kind gift from N. Cianciotto, Northwestern University) were used as host strains for recombinant plasmids. Luria broth (LB) and brain heart infusion (BHI) medium were used for growth in of E. coli and Lm, respectively. The integration plasmid pPL2 (Lauer et al., 2002) was used for genetic complementation.
Protein Expression and Purification
For protein expression, PrsA1 residues 22-294 (coding region lacking the N terminal 21 amino acid secretion signal sequence) were cloned as a C-terminal 6-his tag in the vector pQE60 (Alonzo & Freitag, 2010). BL21(DE3) cells containing the PrsA1 pQE60 expression vector were grown in LB media at 37°C until an OD of > 0.6 at 600 nm at which point a final concentration of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added. The temperature was reduced for 20°C and cultures allowed to grow overnight. Cells were harvested by centrifugation, lysed with an Emulsiflex-C5 (Avestin), and lysate cleared by centrifugation at 16,000 rpm for 30 minutes. Lysates were passed over a nickel NTA gravity column (Pierce) and washed with 50 column volumes of chilled buffer (50 mM Tris pH 7.5, 500 mM NaCl, 25 mM imidazole). Protein was eluted with 5 column volumes elution buffer (50 mM Tris pH 7.5, 500 mM NaCl, 500 mM imidazole) and concentrated for injection onto an SD75 16/60 superdex gel filtration column (GE Healthcare) at 4°C. Fractions were collected via AKTA (GE Healthcare) in a final buffer of 20 mM Tris pH 7.5, 100 mM NaCl, 1 mM betamercaptoethanol (β-ME). Selenomethionine labeled protein was produced using metabolic inhibition. Briefly cells were grown in M9 media at 37°C then the temperature reduced to 20°C with the addition of 0.05 g/L seleomethionine, leucine, valine, proline, and 0.1 g/L lysine, threonine, phenylalanine. Cells were allowed to grow for an additional 30 minutes before induction with 1 mM IPTG.
Protein Crystallization
PrsA1 was concentrated to 20 mg/mL for initial screening in commercially available conditions with a Tecan Freedom Evo 200 robot at the University of Illinois at Chicago Research Resources Center High Through-put facility. The crystallization conditions were 26 mg/mL PrsA1 with a 1:1 mixture of 30% MPD (2-methyl-2,4-pentadiol) 100 mM Na Acetate pH 4.5. Crystals were grown by sitting drop vapor diffusion at 4°C with micro-seeding using SeedBead (Hampton Research) required for optimal growth.
Data collection and refinement
Diffraction data was collected at the Advanced Photon Source at Argonne National Laboratories as part of the LS-CAT, Sector 21. Protein crystals were prepared by soaking in mother liquor at 25% MPD plus 25% PEG 4000 and then flash freezing in liquid nitrogen. After mounting, annealing for 10 seconds was required. Data was processed using XDS (Kabsch, 2010) and phases determined by single anomalous dispersion (SAD) on data collected near the selenium peak using both the Phenix package (Adams et al., 2010) and Sharp/Autosharp (Bricogne et al., 2003, Vonrhein et al., 2007). The initial model was further built and refined using Coot (Emsley et al., 2010), Refmac5 (Murshudov et al., 1997) from the CCP4 suite of programs (Winn et al., 2011), and TLS refinement (Painter & Merritt, 2006). The final model as an R/Rfree of 19.3/23.5% with 99% of residues in the allowed region of the ramachandran plot. The coordinates and structure factors (code 5HTF) have been deposited to the Protein Data Bank, Research Collaboratory for Structural bioinformatics, Rutgers University, New Brunswick, NJ (www.pdb.org).
Protein variant expression assays
To observe the cell associated and secreted protein expression profiles for the PrsA1 and PrsA2 variants in the ΔprsA2/ΔprsA1 and ΔprsA2 backgrounds, respectively, strains were grown to mid-log phase and collected at equivalent densities. For cell-associated proteins the bacterial pellet was isolated by centrifugation. Secreted proteins were precipitated from the culture medium using trichloroacetic acid (TCA) with a final concentration of 10%. Equivalent sample volumes of cell-associated or secreted material were re-suspended in sample buffer containing β-mercaptoethanol, boiled, and separated by SDS-PAGE gel. Proteins were transferred to nitrocellulose and visualized by western-blot using a polyclonal anti-PrsA2 antibody.
Protein dimer assay
The PrsA1 dimer variant (I28/L40/Y41/M44A) was expressed and purified using the wild-type protocol. To assay for the ability to form a dimer, the wild-type and variant proteins were subjected to size exclusion chromatography using an SD75 16/600 column (GE Healthcare). The column was equilibrated in 20 mM HEPES pH 7.5, 100 mM NaCl, 5 mM β-ME and both proteins were injected at a concentration of 60 μM at a total of 2 mg, as determined by absorbance at 280 nm. To assay the purity of the samples, the isolated material was separated by 15% SDS-PAGE gel and proteins stained by Coomassie dye for visualization.
Circular Dichroism Spectroscopy
Purified PrsA1 wild-type and PrsA1 dimer variant (I28/L40/Y41/M44A) were diluted into 20 mM KHPO4 at pH 7.5 in a 0.2 cm pathlength cuvette. Spectra were collected on a Jasco-815 spectropolarimeter from a wavlength of 260 nm to 190 nm. Background spectra of the PrsA1 buffer diluted into 20 mM KHPO4 was subtracted from the data. To plot the data as mean residue ellipticity, a mean residue weight of 111 was used with protein concentrations of 0.05 mg/mL for wild-type and 0.02 mg/mL for the dimer variant.
Construction of ΔprsA2 and ΔprsA2/ΔprsA1 strains containing mutant prsA2 and prsA1 alleles
Genomic DNA from Lm 10403S was used to amplify prsA2 and prsA1, where forward primers were designed with a SacI restriction site and reverse primers were designed with an XmaI site (Table S2). PCR amplified products were cloned into pPL2, which integrates into a single neutral site with the Lm chromosome and maintains single copy expression (Lauer et al., 2002). Site directed mutants were generated using a kit and primers were designed as specified by the manufacture (QuikChange Lightning, Agilent Technologies) (Table S2). Plasmids were propagated in E. coli One Shot TOP10 cells, sequenced (UIC Research Resources Center Core Genomics Facility), and transformed into E. coli S17 or SM10 cells, then subsequently introduced into the Lm ΔprsA2 or Lm ΔprsA2/ΔprsA1 strain by conjugation.
Growth assays in ethanol, at high osmolarity, and at acidic and basic pH
Growth assays were performed as follows: 2μL of a saturated overnight culture was inoculated into 2 mL BHI liquid broth containing 4% ethanol, 5% NaCl w/v or broth at pH 6 or pH 9 (where HCl or NaOH was used to obtain the respective pH). These cultures were grown overnight at 37°C with agitation and growth was measured as a function of optical density (OD660nm).
Swimming motility assay
For swimming motility, mid-log phase (OD600nm~0.8) bacterial BHI broth cultures (2μL) were inoculated into soft BHI agar (0.3%) plates and grown at 37°C for 24 hours and subsequently at 25°C for 24 hours. Then motility was measured as the diameter of the spreading colony.
Determination of antibiotic minimum inhibitory concentration
The minimum inhibitory concentration (MIC) to prevent bacterial growth was determined following 2 μL inoculation from mid-log phase (OD600nm ~0.8) cultures into 2 ml BHI broth in 4 ml polypropylene tubes containing dilutions of penicillin G or lysozyme. Cultures were grown at 37°C with agitation for 16 hours followed by the MIC determination based on the complete inhibition of bacterial growth.
Hemolysin assays
Hemolytic activity was measured for strains as previously described (Cahoon & Freitag, 2015). Briefly, strains were grown overnight in LB, diluted 1:10 in fresh LB and grown for 5 hours. Normalized culture supernatants were serially diluted into PBS pH 5 containing 1 mM DTT (PBS-DTT) and incubated at 37°C for 30 min. Then 100μL of a 1:5 dilution of PBS-DTT washed sheep red blood cells (RBCs) was added and incubated for 30 minutes at 37°C. Bacterial supernatant/RBCs mixtures were pelleted by centrifugation and the supernatant dilution resulting in 50% RBC lysis was determined by visual inspection of the pellet.
L2 plaque assays
Plaque assays were conducted as previously described (Sun et al., 1990). Briefly, monolayers of L2 fibroblasts in 6-well culture dishes were infected at an MOI of 30:1 for 1 hour. Then infected monolayers were washed three times with PBS pH 7 and overlaid with DMEM/agarose containing 10μg/ml gentamicin to kill extracellular bacteria. Plaques were measured at 72 hours with a micrometer.
Detection of phospholipase activity
To detect phospholipase activity, Brilliance Listeria Agar with differential supplement containing lecithin (Oxoid) was used. An opaque halo is produce around the bacterial streak upon Lm phospholipase hydrolysis of lecithin in the medium. Colonies were struck onto the medium and incubated for 24 hours at 37°C followed by visual inspection of the zone of opacity surrounding the bacterial streaks.
Intravenous mouse infections
Animal procedures were approved by the UIC Animal Care Committee and were conducted in the Biological Resources Laboratory. Saturated overnight bacterial cultures were diluted 1:20 in BHI broth and grown to an OD600nm ~0.6 at 37°C. Bacteria were normalized to 6 × 108 CFU/ml, washed twice with PBS pH 7, diluted, and re-suspended in PBS pH 7 to a final concentration of 1 × 105 CFU/ml. Seven to nine week old female Swiss Webster mice (Charles River Laboratories) were injected with 200 μL containing 2 × 104 CFU bacteria by tail vein injection. Livers and spleens of infected animals were collected at 72 hours post infection. Organs were homogenized and 10-fold serial dilutions were plated for the determination of total CFUs.
Statistical analyses
For data represented as a bar graph, a two-tailed Student’s T-test was used for statistical analysis where P≤0.05 and error bars represent the standard error of the mean. For data represented as box plots, a two-tailed Wilcoxon Rank-Sum Test was used where P≤0.05.
Supplementary Material
Acknowledgements
We thank members of the Freitag laboratory and the UIC Positive Thinking group for helpful discussions. We also thank Kiira Ratia at the UIC Research Resources High throughput screening facility for help with initial crystal screening and Bernard D. Santarsiero for support in x-ray data collection. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). This work was supported by NIH grants R01 AI083241 and AI083241-03S1 to NEF. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the funding source.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo F, 3rd, Freitag NE. Listeria monocytogenes PrsA2 is required for virulence factor secretion and bacterial viability within the host cell cytosol. Infect immun. 2010;78:4944–4957. doi: 10.1128/IAI.00532-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo F, 3rd, Port GC, Cao M, Freitag NE. The posttranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect Immun. 2009;77:2612–2623. doi: 10.1128/IAI.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo F, 3rd, Xayarath B, Whisstock JC, Freitag NE. Functional analysis of the Listeria monocytogenes secretion chaperone PrsA2 and its multiple contributions to bacterial virulence. Mol Microbiol. 2011;80:1530–1548. doi: 10.1111/j.1365-2958.2011.07665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nuc Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Nat Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner M, Karst U, Gerstel B, Loessner M, Wehland J, Jansch L. Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J Bacteriol. 2007;189:313–324. doi: 10.1128/JB.00976-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens-Kneip S. The role of SurA factor in outer membrane protein transport and virulence. Int J Med Microbiol. 2010;300:421–428. doi: 10.1016/j.ijmm.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Bierne H, Sabet C, Personnic N, Cossart P. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes infect. 2007;9:1156–1166. doi: 10.1016/j.micinf.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Bitto E, McKay DB. Crystallographic structure of SurA, a molecular chaperone that facilitates folding of outer membrane porins. Structure. 2002;10:1489–1498. doi: 10.1016/s0969-2126(02)00877-8. [DOI] [PubMed] [Google Scholar]
- Boujemaa-Paterski R, Gouin E, Hansen G, Samarin S, Le Clainche C, Didry D, Dehoux P, Cossart P, Kocks C, Carlier MF, Pantaloni D. Listeria protein ActA mimics WASp family proteins: it activates filament barbed end branching by Arp2/3 complex. Biochem. 2001;40:11390–11404. doi: 10.1021/bi010486b. [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta crystallographica. Section D, Biological crystallography. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Cahoon LA, Freitag NE. Listeria monocytogenes virulence factor secretion: don’t leave the cell without a chaperone. Front Cellular Infect Microbiol. 2014;4:13. doi: 10.3389/fcimb.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon LA, Freitag NE. Identification of Conserved and Species-Specific Functions of the Listeria monocytogenes PrsA2 Secretion Chaperone. Infect Immun. 2015;83:4028–4041. doi: 10.1128/IAI.00504-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul S, Raengpradub S, Wiedmann M, Boor KJ. Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol. 2008;16:388–396. doi: 10.1016/j.tim.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S, Gantner BN, Ye RD, Cianciotto NP, Freitag NE. The Listeria monocytogenes ChiA chitinase enhances virulence through suppression of host innate immunity. mBio. 2013;4:e00617–00612. doi: 10.1128/mBio.00617-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico-Calero I, Suarez M, Gonzalez-Zorn B, Scortti M, Slaghuis J, Goebel W, T. European Listeria Genome Consortium. Vazquez-Boland JA. Hpt, a bacterial homolog of the microsomal glucose- 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci U S A. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clantin B, Leyrat C, Wohlkonig A, Hodak H, Ribeiro Ede A, Jr., Martinez N, Baud C, Smet-Nocca C, Villeret V, Jacob-Dubuisson F, Jamin M. Structure and plasticity of the peptidyl-prolyl isomerase Par27 of Bordetella pertussis revealed by X-ray diffraction and small-angle X-ray scattering. J Struct Biol. 2010;169:253–265. doi: 10.1016/j.jsb.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Czuprynski CJ. Listeria monocytogenes: silage, sandwiches and science. Animal Health Res Rev. 2005;6:211–217. doi: 10.1079/ahr2005111. [DOI] [PubMed] [Google Scholar]
- Disson O, Lecuit M. In vitro and in vivo models to study human listeriosis: mind the gap. Microbes Infect. 2013;15:971–980. doi: 10.1016/j.micinf.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nuc Acids Res. 2007;35:W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets DA, Bronze MS. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS immunol Med Microbiol. 2008;53:151–165. doi: 10.1111/j.1574-695X.2008.00404.x. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Cur Protocols Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5: Unit 5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster BM, Marquis H. Protein transport across the cell wall of monoderm Gram-positive bacteria. Mol Microbiol. 2012;84:405–413. doi: 10.1111/j.1365-2958.2012.08040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster BM, Zemansky J, Portnoy DA, Marquis H. Posttranslocation chaperone PrsA2 regulates the maturation and secretion of Listeria monocytogenes proprotein virulence factors. J Bacteriol. 2011;193:5961–5970. doi: 10.1128/JB.05307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag NE, Port GC, Miner MD. Listeria monocytogenes - from saprophyte to intracellular pathogen. Nat Rev Microbiol. 2009;7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag NE, Portnoy DA. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol Microbiol. 1994;12:845–853. doi: 10.1111/j.1365-2958.1994.tb01070.x. [DOI] [PubMed] [Google Scholar]
- Guerrero MLF, Torres R, Mancebo B, Gonzalez-Lopez JJ, Gorgolas M, Jusdado JJ, Roblas RF. Antimicrobial treatment of invasive non-perinatal human listeriosis and the impact of the underlying disease on prognosis. Clin Microbiol Infec. 2012;18:690–695. doi: 10.1111/j.1469-0691.2011.03616.x. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nuc Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyyrylainen HL, Marciniak BC, Dahncke K, Pietiainen M, Courtin P, Vitikainen M, Seppala R, Otto A, Becher D, Chapot-Chartier MP, Kuipers OP, Kontinen VP. Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis. Molecular microbiology. 2010;77:108–127. doi: 10.1111/j.1365-2958.2010.07188.x. [DOI] [PubMed] [Google Scholar]
- Jacobs M, Andersen JB, Kontinen V, Sarvas M. Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequences. Mol Microbiol. 1993;8:957–966. doi: 10.1111/j.1365-2958.1993.tb01640.x. [DOI] [PubMed] [Google Scholar]
- Jakob RP, Koch JR, Burmann BM, Schmidpeter PA, Hunkeler M, Hiller S, Schmid FX, Maier T. Dimeric Structure of the Bacterial Extracellular Foldase PrsA. J Biol Chem. 2015;290:3278–3292. doi: 10.1074/jbc.M114.622910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NB. CDC National Health Report: Leading Causes of Morbidity and Mortality and Associated Behavioral Risk and Protective Factors-United States, 2005-2013 (vol 63, pg 3, 2014) MMWR. 2014;63:1015–1015. [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale A, Phansopa C, Suwannachart C, Craven CJ, Rafferty JB, Kelly DJ. The virulence factor PEB4 (Cj0596) and the periplasmic protein Cj1289 are two structurally related SurA-like chaperones in the human pathogen Campylobacter jejuni. J Biol Chem. 2011;286:21254–21265. doi: 10.1074/jbc.M111.220442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopasek I, Strzalka K, Svobodova J. Cold shock in Bacillus subtilis: different effects of benzyl alcohol and ethanol on the membrane organisation and cell adaptation. Biochimica et biophysica acta. 2000;1464:18–26. doi: 10.1016/s0005-2736(99)00240-0. [DOI] [PubMed] [Google Scholar]
- Koster S, van Pee K, Hudel M, Leustik M, Rhinow D, Kuhlbrandt W, Chakraborty T, Yildiz O. Crystal structure of listeriolysin O reveals molecular details of oligomerization and pore formation. Nat Commun. 2014;5 doi: 10.1038/ncomms4690. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nuc Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering AL, de Castro LH, Lim D, Strynadka NC. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science. 2007;315:1402–1405. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- Lovering AL, Lin LY, Sewell EW, Spreter T, Brown ED, Strynadka NC. Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis. Nat Struct Mol Biol. 2010;17:582–589. doi: 10.1038/nsmb.1819. [DOI] [PubMed] [Google Scholar]
- Moreira IS, Fernandes PA, Ramos MJ. Hot spots--a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica. Section D, Biological crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- O’Neil HS, Forster BM, Roberts KL, Chambers AJ, Bitar AP, Marquis H. The propeptide of the metalloprotease of Listeria monocytogenes controls compartmentalization of the zymogen during intracellular infection. J Bacteriol. 2009;191:3594–3603. doi: 10.1128/JB.01168-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. [Google Scholar]
- Rahfeld JU, Rucknagel KP, Schelbert B, Ludwig B, Hacker J, Mann K, Fischer G. Confirmation of the existence of a third family among peptidyl-prolyl cis/trans isomerases. Amino acid sequence and recombinant production of parvulin. FEBS Lett. 1994;352:180–184. doi: 10.1016/0014-5793(94)00932-5. [DOI] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-Ethyl-N-Nitrosourea-Induced Goldenticket Mouse Mutant Reveals an Essential Function of Sting in the In Vivo Interferon Response to Listeria monocytogenes and Cyclic Dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk BJ, Mahon BE, Griffin PM, Gould H, Tauxe RV, Crim SM, Jackson KA, Gerner-Smidt P, Herman KM, Henao OL. Vital Signs: Listeria Illnesses, Deaths, and Outbreaks - United States, 2009-2011. MMWR. 2013;62:448–452. [PMC free article] [PubMed] [Google Scholar]
- Silveira MG, Baumgartner M, Rombouts FM, Abee T. Effect of adaptation to ethanol on cytoplasmic and membrane protein profiles of Oenococcus oeni. Applied Environ Microbiol. 2004;70:2748–2755. doi: 10.1128/AEM.70.5.2748-2755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun AN, Camilli A, Portnoy DA. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 1990;58:3770–3778. doi: 10.1128/iai.58.11.3770-3778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossavainen H, Permi P, Purhonen SL, Sarvas M, Kilpelainen I, Seppala R. NMR solution structure and characterization of substrate binding site of the PPIase domain of PrsA protein from Bacillus subtilis. FEBS Lett. 2006;580:1822–1826. doi: 10.1016/j.febslet.2006.02.042. [DOI] [PubMed] [Google Scholar]
- Vitikainen M, Lappalainen I, Seppala R, Antelmann H, Boer H, Taira S, Savilahti H, Hecker M, Vihinen M, Sarvas M, Kontinen VP. Structure-function analysis of PrsA reveals roles for the parvulin-like and flanking N- and C-terminal domains in protein folding and secretion in Bacillus subtilis. J Biol Chem. 2004;279:19302–19314. doi: 10.1074/jbc.M400861200. [DOI] [PubMed] [Google Scholar]
- Vitikainen M, Pummi T, Airaksinen U, Wahlstrom E, Wu H, Sarvas M, Kontinen VP. Quantitation of the capacity of the secretion apparatus and requirement for PrsA in growth and secretion of alpha-amylase in Bacillus subtilis. J Bacteriol. 2001;183:1881–1890. doi: 10.1128/JB.183.6.1881-1890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung PSM, Na YJ, Kreuder AJ, Marquis H. Compartmentalization of the broad-range phospholipase C activity to the spreading vacuole is critical for Listeria monocytogenes virulence. Infect Immun. 2007;75:44–51. doi: 10.1128/IAI.01001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemansky J, Kline BC, Woodward JJ, Leber JH, Marquis H, Portnoy DA. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J Bacteriol. 2009;191:3950–3964. doi: 10.1128/JB.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.