Abstract
Transcription factors R2R3MYB family have been associated with the control of secondary metabolites, development of structures, cold tolerance and response to biotic and abiotic stress, among others. In recent years, genomes of Rosaceae botanical family are available. Although this information has been used to study the karyotype evolution of these species from an ancestral genome, there are no studies that treat the evolution and diversity of gene families present in these species or in the botanical family. Here we present the first comparative study of the R2R3MYB subfamily of transcription factors in three species of Rosaceae family (Malus domestica, Prunus persica and Fragaria vesca). We described 186, 98 and 86 non-redundant gene models for apple, peach and strawberry, respectively. In this research, we analyzed the intron–exon structure and genomic distribution of R2R3MYB families mentioned above. The phylogenetic comparisons revealed putative functions of some R2R3MYB transcription factors. This analysis found 44 functional subgroups, seven of which were unique for Rosaceae. In addition, our results showed a highly collinearity among some genes revealing the existence of conserved gene models between the three species studied. Although some gene models in these species have been validated under several approaches, more research in the Rosaceae family is necessary to determine gene expression patterns in specific tissues and development stages to facilitate understanding of the regulatory and biochemical mechanism in this botanical family.
Keywords: Peach, Strawberry, Apple, R2R3MYB transcription factors
| Specifications | |
|---|---|
| Organism | Prunus persica, Malus domestica and Fragaria vesca |
| Sex | Not applicable |
| Sequencer or array type | Genome sequences |
| Experimental factors | R2R3MYB gene models of three Rosaceae genomes |
| Experimental features | Gene models of R2R3MYB transcription factors were obtained from functional annotation of each genome project for apple, strawberry and peach. Sequences identified were checked for R2R3 domains through InterProscan. Intron – exon structure was determined using the information available for each genome project. To determine the relationship across Rosaceae genomes and predicted R2R3MYB we analyzed the synteny and collinearity of these genes between the genomes. A Phylogenetic analysis and functional diversity study of R2R3MYB genes in Rosaceae were realized. |
| Consent | Not applicable |
| Sample source location |
https://phytozome.jgi.doe.gov/pz/portal.html https://www.arabidopsis.org/ |
1. Introduction
Global sequencing of genomes is an invaluable tool for plant research. Many genomes have been sequenced, facilitating the understanding of the physiology, biochemistry, genetics and evolution of a great number of species [1], [2], [3]. Sequencing of Arabidopsis genome (The Arabidopsis Genome Initiative, 2000) marked the beginning of plant genomics. However, this provided only partial support for the study of phylogenetically distant species. This led to the sequencing of agronomic important species such as rice [4], tomato [3], potato [5] and grape [6], among others. In recent years there has been a general interest to sequence different genomes of the botanical family Rosaceae; the sequences of apple [7], strawberry [8] and peach [9] are currently available. Each of these species is a specific genome model for the subfamilies Malaideae, Rosoideae and Amygdeloideae, respectively, which allows more precise understanding of the biochemical and physiological processes which occur in each of these taxa [1].
The genomes of Fragaria (x = 7), Prunus (x = 8) and Malus (x = 17) evolved from a common ancestor with x = 9 [10]. The evolutionary mechanisms involved in the speciation have been studied with molecular markers [11] and later, more precisely, by the comparative study of the sequenced genomes [2]. These studies determined that the genus Malus underwent a whole duplication event (WGD), and that Prunus is the genus with the most conserved karyotype compared to the ancestral karyotype configuration. In addition, high co-linearity between Pyrus and Malus, and high macrosynteny between Fragaria and Prunus was described [11].
In spite of this knowledge about genome evolution, there are no analyses which explain the physiological and biochemical differences among these genera. In recent years, studies in grape [12], maize [13], soybean [14] and tomato [15] have attempted to identify the physiological effects of acquisition, duplication, conservation and deletions of the R2R3MYB family of transcription factors. This family has been associated with the control of secondary metabolites [16], [17], [18], [19], development of structures [20], opening and closing of stomata [21], cold tolerance [22] phosphorous capture [23], hormone synthesis and cell signaling biotic [24], [25] and abiotic stress [22], among others.
The MYB transcription factors are characterized by the presence of the MYB domain, which is composed of four imperfectly repeated sequences (R). Each R domain has about 52 amino acids which may form three α-helix structures [26]. The MYB family is classified into four subgroups according to the number of R domains present in the protein; so far R1, R2R3, R3 and R4 have been described. The R2R3MYB subfamily is the largest in plants; it is characterized by two types of R domain in the N-terminal end and usually by a transcription activator or repressor in the C-terminal end [26].
To improve our understanding of the gene subfamily of R2R3MYB transcription factors in the sequenced genomes of the three most important species of the Rosaceae family: Malus domestica, Prunus persica and Fragaria vesca, we describe the expansion and conservation of clades of the R2R3MYB gene subfamily in each taxon. We also incorporate a functional analysis based on the clustering of R2R3MYB genes of Arabidopsis, evolutionarily more recent than Rosaceae family. We determine the chromosome distribution of the predicted models and putative paralog gene models. Our results provide the first comprehensive analysis of the gene structure, gene distribution, gene duplication and putative function of the R2R3MYB family encoded in the Rosaceae genome.
2. Results and discussion
2.1. Identification of R2R3MYB genes in Rosaceae
Gene models of R2R3MYB transcription factors were obtained from functional annotation of each genome project for apple, strawberry and peach. Sequences identified were checked for R2R3 domains through InterProscan (V.5.14), removing sequences with incomplete R2 or R3 domains. Previously, 244; 157; 126; 121; 109 and 108 R2R3MYB genes have been described in maize [13], soybean [14], Arabidopsis [16], tomato [15], rice [27] and grape [12], respectively. Our study identified 186, 98 and 86 non-redundant gene models for apple, peach and strawberry, respectively. Both the largest number of predicted genes in sequenced genomes of plants and the greatest number of predicted transcription factors have been reported in apple; more than 400 transcription factors of the MYB family have been identified [7]. Cao et al. mentioned that 222 and 121 models correspond to apple and peach R2R3MYB subfamily, respectively [28]. Our study reduced these values to 16% for apple and 30% for peach updating the number of R2R3MYB transcription factors available in each genome. By contrast, in strawberry only 187 MYB models have been identified [8], and we identified the smallest R2R3MYB subfamily (86 models) described in Rosacea family up to now.
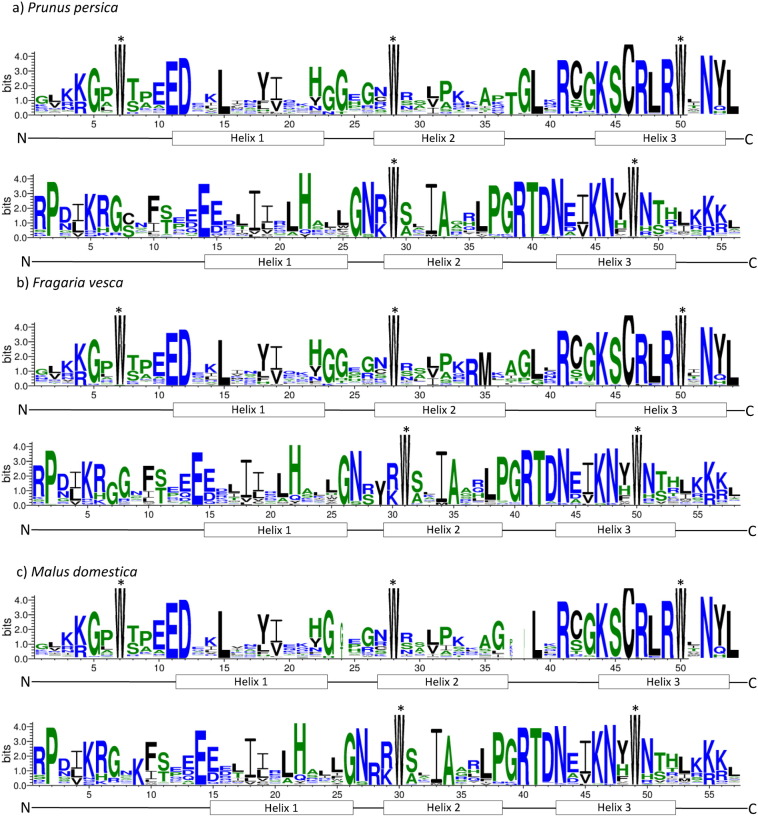
To determine both homologous domain sequences and amino acid frequency in each position within R2 and R3 domains, we performed a multiple alignment analysis for each domain in each species under study. In Fig. 1 we can observe different amino acids frequency for each position of the R2 and R3 domains. Our results confirm the conserved nature of each of these domains. All gene families have three conserved tryptophans in the R2 domain, while only two are present in the R3 domain, where the first tryptophan is always replaced by a hydrophobic amino acid, which is consistent with that was observed in other studies of this gene family such as cucumber [29] and tomato [15].
Fig. 1.
R2 and R3 MYB repeats are highly conserved across all R2R3MYB domain proteins. Sequence logos of the R2 (top) and R3 (bottom) MYB repeats are based on conserved alignments for Prunus persica (a), Fragaria vesca (b) and Malus domestica (c). The overall height of each stack indicates the conservation of the sequence at that position, whereas the height of letters within each stack represents the relative frequency of the corresponding amino acid. Asterisk indicates the position of the conserved amino acid that are identical among R2R3MYB proteins.
2.2. Intron – exon structure of the Rosaceae R2R3MYB family and genomic distribution
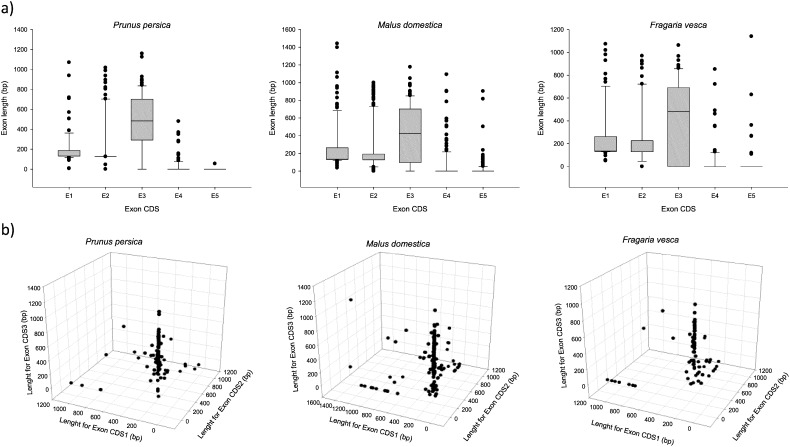
We found 4, 7.5 and 8% of single exon genes for peach, apple and strawberry, respectively. Exon 1 and 2 usually encode almost the entire R2R3 domain, presenting both exons a uniform size range through the botanical family. In peach, the first exon has an average size of 202 bp, while it reaches 246 bp and 253 bp for strawberry and apple, respectively. Furthermore, exon 2 achieved average size values of 241, 261 and 271 bp for peach, apple and strawberry, respectively (Fig. 2a). Regardless of the species analyzed, exon 3 is the one that has the greatest diversity in sequence length (strawberry, 44–1062 bp; apple, 52–1176 bp; peach, 56–1159 bp). Diversity in sequence length leads into a source of functional divergence due to amino acid changes in motifs and domains. Considering length of the first three exons, we have identified a common pattern of gene diversity in Rosaceae (Fig. 2b) with similar results to those obtained in tomato [15], Arabidopsis and grape [12].
Fig. 2.
Exon length distribution analysis of Rosaceae R2R3MYB genes. a) Exon length (Exon 1–Exon 5) values were analyzed using Box plot depicted by SigmaPlot 10.0 on Prunus persica, Malus domestica and Fragaria vesca, respectively. Each box represents the exon size range in which 50% of the values for a particular exon are grouped. The median is represented as a continuous line. b) First, second and third exon lengths in Prunus persica, Malus domestica and Fragaria vesca, respectively.
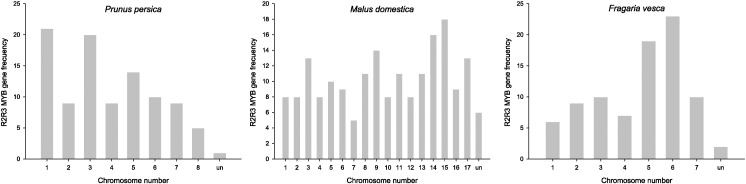
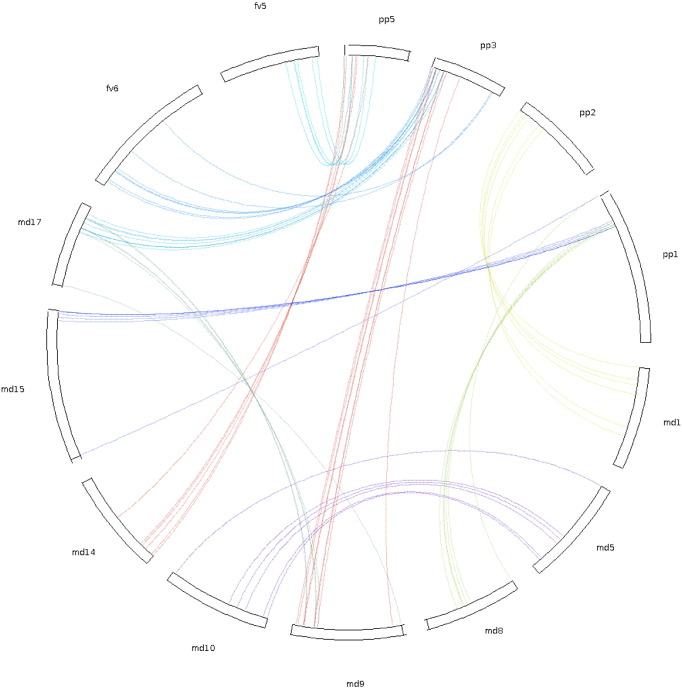
Using available genome position reference for each gene model, it was possible to determine that R2R3MYB genes are distributed across all chromosomes (Fig. 3 and Fig. S1). Using this information, we proceeded to obtain specific evolutionary relationships between both species and gene models through synteny analysis. Our results, considering gene model position and sequence similarity, determined that 105 genes are collinear between species (29.09%), and ten syntenic blocks were found (Table S1). Apple chromosomes 9 and 17, as well as 5 and 10, have gene models highly collinear coming from a segmental duplication event. In addition, apple chromosomes 9 and 17 are highly collinear with peach chromosome 3 and strawberry chromosome 6, respectively. In turn, peach chromosomes 1 and 2 are collinear with apple chromosomes 8 and 1, respectively (Fig. 4), being consistent with what was reported by Illa et al. [10].
Fig. 3.
Chromosome locations of R2R3MYB transcription factors at Prunus persica, Malus domestica and Fragaria vesca. Chromosomal position of each R2R3MYB was mapped according to each genome information.
Fig. 4.
Schematic representation of R2R3MYB genes synteny in Rosaceae family. The chromosome number is indicated at the top of each chromosome pp.: Prunus persica, md: Malus domestica and fv: Fragaria vesca. Color lines suggest duplicated R2R3MYB gene pairs between chromosomes.
2.3. Phylogenetic analysis and functional diversity of R2R3MYB genes in Rosaceae family
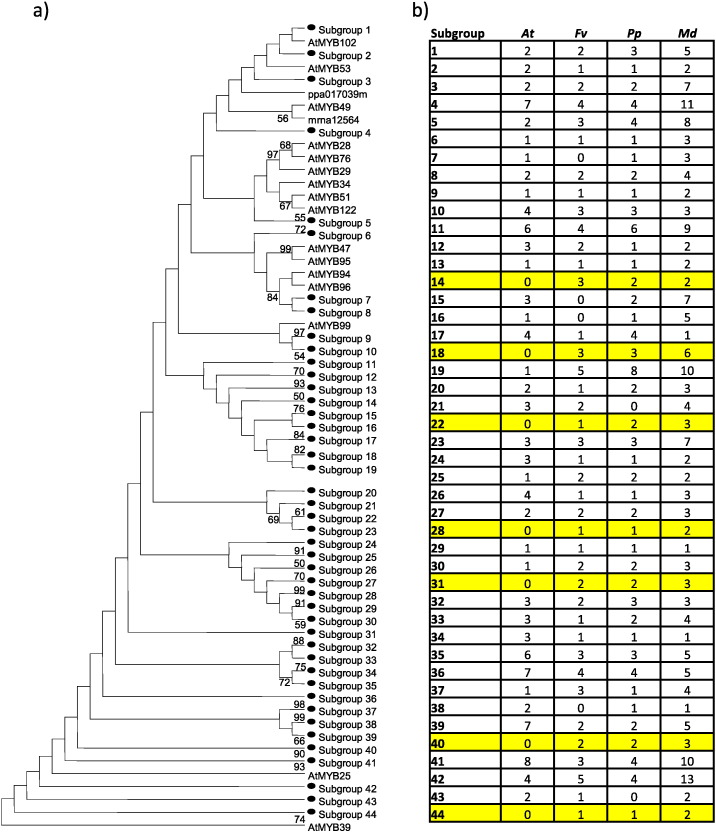
To identify new clades, expansions and genic conservation, we constructed phylogenetic tree using the neighbor-joining algorithm. Neighbor-joining tree considered all putative genes identified in each genome and all R2R3MYB genes described in Arabidopsis (Fig. 5; Fig. S2). The association in clades and subclades of gene models and described genes, has facilitated the identification of possible functions of candidate genes [12], allowing the recognition of similar genes with cooperative or redundant functions. These analyses allowed us the identification of 44 functional clusters suggested by R2R3MYB Arabidopsis sequences and, additionally, this strategy allowed us the identification of seven new clusters, identified as subgroup 14 (14S); subgroup 18 (18S); subgroup 22 (22S); subgroup 28 (28S); subgroup 31 (31S); subgroup 40 (40S) and subgroup 44 (44S). Furthermore, gene models inside of each syntenic block mentioned above (Table S1) were found in different subgroups, suggesting that the evolution of this family was through a mechanism different from tandem duplication.
Fig. 5.
a) Phylogenetic relationships and subgroup designations in R2R3MYB proteins from Rosaceae. 44 subgroups were identified in the neighbor-joining tree analysis. 125 protein sequences of Arabidopsis (At), 86 from strawberry (Fv), 98 from peach (Pp) and 186 from apple (Md) were used. Not condensed neighbor-joining tree is showed in Supplementary Fig. 2. b) Membership of each subgroup is described in the table. Highlighted subgroups are conformed only by Rosaceae sequences.
a) Phylogenetic relationships and subgroup designations in R2R3MYB proteins from Rosaceae. 44 subgroups were identified in the neighbor-joining tree analysis. 125 protein sequences of Arabidopsis (At), 86 from strawberry (Fv), 98 from peach (Pp) and 186 from apple (Md) were used. Not condensed neighbor-joining tree is showed in Supplementary Fig. 2. b) Membership of each subgroup is described in the table. Highlighted subgroups are conformed only by Rosaceae sequences.
2.3.1. Regulation of plant development and cell destiny
By RNA-seq, gene models proposed in the apple genome have been widely validated, allowing the study of transcriptional profile of primary roots (radicles) [30], auxin-responsive genes [31] and regulatory genes in different stages of fruit development [32]. Specifically, Legay et al. [33] validated by RNA-seq the following R2R3MYB transcription factors: MDP0000655330 (S16), MDP0000787808 (S10), MDP0000149535 (S8), MDP0000320772 and MDP0000228252 (S3), MDP0000197283 (S1), MDP0000291518 and MDP0000852158 (S41), MDP0000322479 (S23), MDP0000157506 (S30). These transcription factors are expressed in apple exocarp, where some of them are expressed differentially in fruit with russet defect characterized by the accumulation of suberin at the inner part of the cell wall of the outer epidermal cell layers. On the other hand, several gene models of R2R3MYB transcription factors have been validated in peach [34], [35], [36], [37].
Arabidopsis has many control points in the development of male reproductive structures, which is reflected in the number of MYB factors that participate in this process. It has been reported that AtMYB21, AtMYB24, AtMYB57 (S34) and AtMYB108 (S35) are expressed during pollen and stamen maturation. Factor clustered in subgroup 34 responds to gibberellin to induce jasmonate synthesis [39]; AtMYB24 and AtMYB108 have redundant functions [40] and are downstreamed from the regulator AtMYB21 [20]. Studies in Arabidopsis have shown that AtMYB103 (S25) is closely associated with tapetum, anthers and exine development [41]. Although male structures are highly conserved in angiosperm evolution, we found fewer gene models of subgroup 34 in Rosaceae than the genes described in Arabidopsis.
AtMYB61 (S26) has been reported to be a transcription factor associated with photomorphogenic control, mucilage deposition, stomatal aperture, xylem formation and carbon translocation to the roots, among others [42], [43], [44], [45]. This group also includes transcription factors AtMYB50, AtMYB55 and AtMYB86, for which no associated functions have been found yet. The number of apple gene models found in subgroup 26 was similar to that described for Arabidopsis; however, only one gene was found for peach and strawberry, respectively. In Arabidopsis, there was probably an event of genic sub-functionalization with respect to peach and strawberry, while apple may have conserved these genes after global genome duplication.
Arabidopsis transcription factor AtMYB5 was found in subgroup 16 (16S). This factor participates in trichome morphogenesis and mucilage synthesis [46]. In this subgroup, we only found one gene model for peach and three for apple. Gene model MDP0000655330 has been previously described in russeted cultivars for apple, characterized by a slightly rough skin phenotype [33].
Subgroup 5 (5S) contains transcription factors AtMYB106/NOK and AtMYB16/MIXTA. These transcription factors participate in trichome development, and the latter determines the form of petals epidermal cells [47]. Recently, ppa023143m has been validated and characterized as a positive regulator of trichome formation in peach fruit, generating the difference among nectarine and peach fruits [37]. Nevertheless, our data indicate that there are a large number of genes in each of the three studied genomes, which reveals strong control over the development of these structures in Rosaceae.
2.3.2. Control of primary and secondary metabolism
Transcription factors AtMYB28, AtMYB29, AtMYB34, AtMYB51, AtMYB76 and AtMYB122 are associated with the synthesis of glucosinolates [48], [49], [50], [51] and always formed a monophyletic clade without interacting directly with any Rosaceae gene model. This clade is related to the evolution of the order Brassicaleae as a function of the adaptive response to herbivory [52].
We obtain consistent and conserved clades with factors AtMYB58 and AtMYB63 (S4), both associated with lignin synthesis during the formation of the secondary cell wall in plant cells [53]. Lignin biosynthesis implies highly complex transcriptional networks [54]. Secondary Wall-Associated NAC Domain Protein 1 (SND1) is a key transcription factor for the formation of the secondary wall, including cellulose and lignin synthesis. AtMYB83 and AtMYB46 are direct targets of SND1, showing functional redundancy in the secondary wall formation cascade [18]. AtMYB46 would also regulate AtMYB52 and AtMYB63 [55], which are transcriptional activators of lignin biosynthesis regulated by homologs of SND1 [53]. AtMYB54 (S41), AtMYB61 (S26) and AtMYB69 (S41) also participate in secondary wall synthesis, in biosynthetic genes activation and in lignification control [40].
The high number of gene models related to these genes may be justified by the structural diversity observed in trees (peach and apple) compared to herbaceous species (strawberry). We must consider the total genome duplication in apple as well as physiological differences; for example, pit formation in peach, where R2R3MYB genes had to change their spatial and temporal expresion patterns to give origin to this particular structure.
Regulation of phenylpropanoid pathway, and specifically, flavonoid pathway, has been well described [26]. The AtMYB11, AtMYB12 and AtMYB111 genes (S12) of Arabidopsis are related to the production of flavonol glycosides [56]. This clade is conserved with respect to Arabidopsis in the Rosaceae; there were two or three gene models in each analyzed genome (Fig. S2). In peach, ppa004560m (MYB15), ppa010277m (MYB16) and ppa010716m (MYB111) have already been described [34]. While MYB15 is part of the flavonol glycosides clade (S4).
The repressor group (S11) has been extensively described in Arabidopsis. Jin et al. [57] and Preston et al. [58] found two protein domains associated with this clade, which were also identified in our alignment. High level of regulation by this subgroup in the phenylpropanoid pathway has been described in Arabidopsis. AtMYB32 mutants decrease the expression levels of DFR (dihydrofavonol-4 reductase) and LDOX (leucoanthocianidine dioxygenase) enzymes [57], while AtMYB4 has been associated with the production of cinnamate-4 hydroxylase and photoprotector compounds in response to UV radiation [59], [60]. In Rosaceae, studies of repressor MYB transcription factors are scarce. However, FaMYB1 is strongly implicated in the suppression of anthocyanins and the accumulation of flavonols, probably by the regulation of the UFGT (UDP glucose: flavonoid-3-O-glucosyltransferase) and LDOX enzymes [61].
Many studies have been performed to determine genes homologous to AtMYB75/PAP1 and AtMYB90/PAP2 in Rosaceae. These are specific activators of the flavonoid pathway which, when overexpressed, stimulate the production of anthocyanins [62]. Several studies in apple determined the allelic variants and the regulatory zone of gene MYB10 of Rosaceae [17], [19], [63], [64], [65]. This gene is strongly involved in the regulation of structural genes of the metabolic pathway associated with anthocyanin synthesis and thus directly determines their accumulation. In our alignments, genes AtMYB75/PAP1, AtMYB90/PAP2, AtMYB113 and AtMYB114 always formed a specific clade for Arabidopsis. In peach, the regulation of flavonoids pathway have been extensively characterized revealing tandem duplication of MYB10 (linkage group 3): ppa026640m (MYB10.1), ppa016711m (MYB10.2) and ppa020385m (MYB10. 3) [36]. Zhou et al. [38] have also validated these gene models-integrating to the analysis- the three transcription factors belonging to the linkage group 6. However, these models do not correspond to R2R3MYB; therefore they have been excluded from our analysis.
Genes AtMYB5 (S16) and AtMYB123/TT2 (S19) showed partially redundant functions in tannin synthesis in Arabidopsis seeds [66]. Ravaglia et al. [34] have described ppa023768m (MYB123) and ppa009439m (MYBPA1/subgroup 14). Our study proposes a total of 5, 8 and 10 gene models for MYB123-like, and 3, 2, and 2 for MYBPA1-like in strawberry, peach and apple, respectively. In the genome sequence of strawberry there is an expansion of the clade associated with AtMYB123 [8]. We determined that this expansion occurs in the botanical family probably due, among other reasons, to a response in the formation of fleshy fruit structures which accumulate large amounts of these compounds in the first development stages. It should be noted that neither apples nor strawberries are true fruits, thus the events of gene duplication, which occurred in these subfamilies, would also have facilitated the accumulation of anthocyanins and tannins in other structures.
In conclusion, 186, 98 and 86 non-redundant gene models were identified for apple, peach and strawberry, respectively. Phylogenetic comparisons grouped gene models into 44 functional subgroups, seven of which were unique for Rosaceae and syntenic analysis determinted 10 syntenic block and 105 collinear gene models across the studied species. Although some gene models in peach and apple have been validated under several approaches, more research in the Rosaceae family is necessary to determine gene expression patterns in specific tissues and development stages for each gene model identified in this study.
3. Materials and methods
3.1. Search for MYBR2R3 transcription factors and sequence conservation analysis
R2R3MYB transcription factors annotated (predicted) for each genome project were downloaded from Phytozome database (www.phytozome.net) [67]. In order to confirm the presence of R2R3MYB domain and verify the reliability of the candidate sequences, all sequences were analyzed using InterProScan V.5.14 [68]. Only those which had complete R2R3 domains were used in order to make the search more precise and obtain models with greater probability of being expressed, thus avoiding pseudogenes. Sequences which share the same chromosome position but are given different names were considered as redundant in the counts of total numbers of MYB transcription factors. On the other hand, 125 Arabidopsis R2R3MYB protein sequences were downloaded from TAIR Arabidopsis genome (http://www.Arabidopsis.org/). All IDs of selected sequences from Phytozome and TAIR databases are shown in the Supplementary Table 2.
To analyze the features of the R2R3MYB domain of each genome, the sequences of R2 and R3 MYB repeats were aligned with clustalW. Then, multiple alignment sequences were submitted to WEBLOGO website (http://weblogo.berkeley.edu/logo.cgi).
3.2. Intron – exon structure of the Rosaceae R2R3MYB family and genomic distribution
Intron – exon structure was determined using the information available for each genome project. GFF files (General Feature Format files) of each genome were used to extract the exact position of all intron and exons for each predicted gene model. Then, number and length of introns and exons were calculated. Linkage groups were constructed using the software Mapchart 2.2; while all the graphs were constructed using SigmaPlot version 10.0.
To determine the relationship across Rosaceae genomes and predicted R2R3MYB we analyzed the synteny and collinearity of these genes between the genomes. For this, we used the MCScan software [69]. This software is an algorithm to scan multiple genomes or subgenomes to identify putative homologous chromosomal regions, and then align these regions using genes as anchors, in our case, R2R3MYB transcription factors.
3.3. Phylogenetic analysis and functional diversity of R2R3MYB genes in Rosaceae
Using the dataset generated for each species we performed a phylogenetic analysis along with the 125 R2R3MYB genes described for Arabidopsis (www.arabidopsis.org) [70]. Alignments were performed with a BLOSUM matrix (gap opening penalty 25 and gap extension penalty 1) using the ClustalW algorithm (based on the AlignX module) in the MEGA6 program. Phylogenetic analyses used the neighbor-joining algorithm. Confidence in the nodes was estimated by bootstrap sampling with 1000 repetitions. Classification of the R2R3MYB genes was performed according to their phylogenetic relationships with their corresponding Arabidopsis R2R3MYB genes.
The following are the supplementary data related to this article.
Relationship across Rosaceae genomes and predicted R2R3MYB based on R2R3MYB gene models collinearity.
Linkage groups and chromosomal positioning of the gene models found peach (a), strewberry (b) and apple (c).
Phylogenetic relationships and subgroup designations in R2R3MYB protiens from Rosaceae. The neighbor-joining tree includes 126 protien sequences of Arabidopsis (At), 86 from strewberry (Fv), 99 from peach (Pp) and 186 from apple (Md).
Supplementary material.
Conflict of interest
All authors read the manuscript and declare no conflict of interests.
Acknowledgments
This work was funded by the Comisión Nacional de Ciencia y Tecnología, CONICYT. Scholarships Doctorado en Chile 2009 No. 21090118; Apoyo a la Tesis Doctoral 2011 No. 24110179; Tesis en la Industria - Consorcio tecnológico de la Fruta No. 781211008.
References
- 1.Shulaev V., Korban S.S., Sosinski B., Abbott A.G., Aldwinckle H.S., Folta K.M., Iezzoni A., Main D., Arús P., Dandekar A.M., Lewers K., Brown S.K., Davis T.M., Gardiner S.E., Potter D., Veilleux R.E. Multiple models for Rosaceae genomics. Plant Physiol. 2008;147:985–1003. doi: 10.1104/pp.107.115618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung S., Cestaro A., Troggio M., Main D., Zheng P., Cho I., Folta K.M., Sosinski B., Abbott A., Celton J.M., Arús P., Shulaev V., Verde I., Morgante M., Rokhsar D., Velasco R., Sargent D.J. Whole genome comparisons of Fragaria, Prunus and Malus reveal different modes of evolution between Rosaceous subfamilies. BMC Genomics. 2012;13:129. doi: 10.1186/1471-2164-13-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zovine M., Latché A., Rousseau C., Regad F., Pech J.C., Philippot M. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J., Hu S., Wang J., Wong G.K.S., Li S., Liu B. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 5.Xu X., Pan S., Cheng S., Zhang B., Mu D., Ni P. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 6.Jaillon O., Aury J.M., Noel B., Policriti A., Clepet C., Casagrande A. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 7.Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A. The genome of the domesticated apple (Malus × domestica Borkh.) Nat. Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 8.Shulaev V., Sargent D.J., Crowhurst R.N., Mockler T.C., Folkerts O., Delcher A.L. The genome of woodland strawberry (Fragaria vesca) Nat. Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmad R., Parfitt D., Fass J., Ogundiwin E., Dhingra A. The International Peach Genome Initiative. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013;45:487–494. doi: 10.1038/ng.2586. [DOI] [PubMed] [Google Scholar]
- 10.Illa E., Sargent D.J., Girona E.L., Bushakra J., Cestaro A., Crowhurst R., Pindo M., Cabrera A., van der Knaap E., Iezzoni A., Gardiner S., Velasco R., Arús P., Chagné D., Troggio M. Comparative analysis of rosaceous genomes and the reconstruction of a putative ancestral genome for the family. BMC Evol. Biol. 2011;11:9. doi: 10.1186/1471-2148-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilanova S., Sargent D., Arús P., Monfort A. Synteny conservation between two distantly-related Rosaceae genomes: Prunus (the stone fruits) and Fragaria (the strawberry) BMC Plant Biol. 2008;8:67. doi: 10.1186/1471-2229-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matus J.T., Aquea F., Arce-Johnson P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008;8:83. doi: 10.1186/1471-2229-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du H., Fen B.R., Yang S.S., Huang Y.B., Tang Y.X. The R2R3-MYB transcription factor gene family in maize. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du H., Yang S.S., Liang Z., Feng B.R., Liu L., Huang Y.B., Tang Y.X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106. doi: 10.1186/1471-2229-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao P., Li Q., Li J., Wang L., Ren Z. Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol. Gen. Genomics. 2014;289:1183–1207. doi: 10.1007/s00438-014-0879-4. [DOI] [PubMed] [Google Scholar]
- 16.Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 17.Takos A.M., Jaffé F.W., Jacob S.R., Bogs J., Robinson S.P., Walker A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy R.L., Zhong R., Ye Z.H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009;50:1950–1964. doi: 10.1093/pcp/pcp139. [DOI] [PubMed] [Google Scholar]
- 19.Espley R.V., Brendolise C., Chagné D., Kutty-Amma S., Green S. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009;21:168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandaokar A., Browse J. MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 2009;149:851–862. doi: 10.1104/pp.108.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cominelli E., Galbiati M., Vavasseur A., Conti L., Sala T., Vuylsteke M., Leonhardt N., Dellaporta S.L., Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., Zhao G., Jia J., Liu X., Kong X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 2012;63:203–214. doi: 10.1093/jxb/err264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai X., Wang Y., Yang A., Zhang W.H. OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol. 2012;159:169–183. doi: 10.1104/pp.112.194217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vailleau F., Danie X., Tronchet M., Montillet J.L., Triantaphylidès C., Roby D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. PNAS. 2002;99:10179–10184. doi: 10.1073/pnas.152047199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi H., Cui R., Hu B., Wang X., Zhang S., Liu R., Dong H. Overexpression of transcription factor AtMYB44 facilitates Botrytis infection in Arabidopsis. Physiol. Mol. Plant Pathol. 2011;76:90–95. [Google Scholar]
- 26.Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Yanhui C., Xiaoyuan Y., Kun H., Meihua L., Jigang L., Zhaofeng G. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 28.Cao Z.H., Zhang S.Z., Wang R.K., Zhang R.F., Hao Y.J. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q., Zhang C., Li J., Wang L., Ren Z. Genome-wide identification and characterization of R2R3MYB family in Cucumis sativus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0047576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen R., Djozgic H., Rieger B., Rapp S., Schmidt E.R. Columnar apple primary roots share some features of the columnar-specific gene expression profile of aerial plant parts as evidenced by RNA-Seq analysis. BMC Plant Biol. 2015;15:34. doi: 10.1186/s12870-014-0356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devoghalaere F., Doucen T., Guitton B., Keeling J., Payne W., Ling T.J., Ross J.J., Hallett I.C., Gunaseelan K., Dayatilake G., Diak R., Breen K.C., Tustin D.S., Costes E., Chagné D., Schaffer R.J., David K.M. A genomics approach to understanding the role of auxin in apple (Malus × domestica) fruit size control. BMC Plant Biol. 2012;12:7. doi: 10.1186/1471-2229-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai Y., Dougherty L., Cheng L., Zhong G.Y., Xu K. Uncovering co-expression gene network modules regulating fruit acidity in diverse apples. BMC Genomics. 2015;16:612. doi: 10.1186/s12864-015-1816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Legay S., Guerriero G., Deleruelle A., Lateur M., Evers D., André C.M., Hausman J.F. Apple russeting as seen through the RNA-seq lens: strong alterations in the exocarp cell wall. Plant Mol. Biol. 2015;88:21–40. doi: 10.1007/s11103-015-0303-4. [DOI] [PubMed] [Google Scholar]
- 34.Ravaglia D., Espley R.V., Henry-Kirk R., Andreotti C., Ziosi V., Hellens R.P., Costa G., Allan A.C. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013;13:68. doi: 10.1186/1471-2229-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y., Mao Y., Liu H., Yu F., Li S., Yin T.M. Transcriptome analysis of differentially expressed genes relevant to variegation in peach flowers. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahim M.A., Busatto N., Trainotti L. Regulation of anthocyanin biosynthesis in peach fruits. Planta. 2014;240:913–929. doi: 10.1007/s00425-014-2078-2. [DOI] [PubMed] [Google Scholar]
- 37.Vendramin E., Pea G., Dondini L., Pacheco I., Dettori M.T., Gazza L., Scalabrin S., Strozzi F., Tartarini S., Bassi D., Verde I., Rossini L. A unique mutation in a MYB gene cosegregates with the nectarine phenotype in peach. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y., Zhou H., Lin-Wang K., Vimolmangkang S., Espley R.V., Wang L., Allan A.C., Han Y. Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol. 2014;14:388. doi: 10.1186/s12870-014-0388-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lippold F., Sanchez D.H., Musialak M., Schlereth A., Scheible W.R., Hincha D.K., Udvardi M.K. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 2009;149:1761–1772. doi: 10.1104/pp.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez M.D., Urbez C., Perez-Amador M.A., Carbonell J. Characterization of constricted fruit (ctf) mutant uncovers a role for AtMYB117/LOF1 in ovule and fruit development in Arabidopsis thaliana. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penfield S., Meissner R.C., Shoue D.A., Carpita N.C., Bevan M.W. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant. 2001;13:2777–2791. doi: 10.1105/tpc.010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman L.J., Perazza D.E., Juda L., Campbell M.M. Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and darphotomorphogenic components of the det3 mutant phenotype. Plant J. 2004;37:239–250. doi: 10.1046/j.1365-313x.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- 44.Liang Y.K., Dubos C., Dodd I.C., Holroyd G.H., Hetherington A.M., Campbell M.M. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 45.Romano J.M., Dubos C., Prouse M.B., Wilkins O., Hong H., Poole M., Kang K.Y., Li E., Douglas C.J., Western T.L., Mansfield S.D., Campbell M.M. AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol. 2012;195:774–786. doi: 10.1111/j.1469-8137.2012.04201.x. [DOI] [PubMed] [Google Scholar]
- 46.Sun Y., Zhou Q., Zhang W., Fu Y., Huang H. ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta. 2002;214:694–702. doi: 10.1007/s004250100673. [DOI] [PubMed] [Google Scholar]
- 47.Baumann K., Perez-Rodriguez M., Bradley D., Venail J., Bailey P., Jin H., Koes R., Roberts K., Martin C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development. 2007;134:1691–1701. doi: 10.1242/dev.02836. [DOI] [PubMed] [Google Scholar]
- 48.Gigolashvili T., Berger B., Mock H.P., Müller C., Weisshaar B., Flügge U.I. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- 49.Gigolashvili T., Yatusevich R., Berger B., Müller C., Flügge U.I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;51:247–261. doi: 10.1111/j.1365-313X.2007.03133.x. [DOI] [PubMed] [Google Scholar]
- 50.Gigolashvili T., Engqvist M., Yatusevich R., Müller C. U.I. Flügge HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 2008;177:627–642. doi: 10.1111/j.1469-8137.2007.02295.x. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y., Yan X., Chen S. Bioinformatic analysis of molecular network of glucosinolate biosynthesis. Comput. Biol. Chem. 2011;35:10–18. doi: 10.1016/j.compbiolchem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Grubb C.D., Abel S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006;11:89–100. doi: 10.1016/j.tplants.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 53.Zhou J., Lee C., Zhong R., Ye Z.H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21(1):248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Q., Dixon R.A. Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 2011;16:227–233. doi: 10.1016/j.tplants.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 55.Ko J.H., Kim W.C., Han K.H. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 2009;60:649–665. doi: 10.1111/j.1365-313X.2009.03989.x. [DOI] [PubMed] [Google Scholar]
- 56.Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., Niehaus K., Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weissharr B., Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Preston J., Wheeler J., Heazlewood J., Li S.F., Parish R.W. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 2004;40:979–995. doi: 10.1111/j.1365-313X.2004.02280.x. [DOI] [PubMed] [Google Scholar]
- 59.Hemm M.R., Herrmann K.M., Chapple C. AtMYB4: a transcription factor general in the battle against UV. Trends Plant Sci. 2001;6:135–136. doi: 10.1016/s1360-1385(01)01915-x. [DOI] [PubMed] [Google Scholar]
- 60.Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aharoni A., De Vos C.H., Wein M., Sun Z., Greco R. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001;28(3):319–332. doi: 10.1046/j.1365-313x.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- 62.Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2393. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ban Y., Honda C., Hatsuyama Y., Igarashi M., Bessho H., Moriguchi T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007;48:958–970. doi: 10.1093/pcp/pcm066. [DOI] [PubMed] [Google Scholar]
- 64.Espley R.V., Hellens R.P., Putterill J., Stevenson D.E., Kutty-Amma S., Allan A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin-Wang K., Bolitho K., Grafton K., Kortstee A., Karunairetnam S., McGhie T.K., Espley R.V., Hellens R.P., Allan A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gonzalez A., Mendenhall J., Huo Y., Lloyd A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 2009;325:412–421. doi: 10.1016/j.ydbio.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 67.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Miltros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., Kissinger J.C., Paterson A.H. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huala E., Dickerman A.W., Garcia-Hernandez M., Weems D., Reiser L., LaFond F. The Arabidopsis information resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relationship across Rosaceae genomes and predicted R2R3MYB based on R2R3MYB gene models collinearity.
Linkage groups and chromosomal positioning of the gene models found peach (a), strewberry (b) and apple (c).
Phylogenetic relationships and subgroup designations in R2R3MYB protiens from Rosaceae. The neighbor-joining tree includes 126 protien sequences of Arabidopsis (At), 86 from strewberry (Fv), 99 from peach (Pp) and 186 from apple (Md).
Supplementary material.