Abstract
Background
Xanthophylls are oxygenated carotenoids and fulfill critical roles in plant growth and development. In plants, two different types of carotene hydroxylases, non-heme di-iron and heme-containing cytochrome P450, were reported to be involved in the biosynthesis of xanthophyll. Citrus fruits accumulate a high amount of xanthophylls, especially β,β-xanthophylls. To date, however, the roles of carotene hydroxylases in regulating xanthophyll content and composition have not been elucidated.
Results
In the present study, the roles of four carotene hydroxylase genes (CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C) in the biosynthesis of xanthophyll in citrus fruits were investigated. Phylogenetic analysis showed that the four citrus carotene hydroxylases presented in four distinct clusters which have been identified in higher plants. CitHYb was a non-heme di-iron carotene hydroxylase, while CitCYP97A, CitCYP97B, and CitCYP97C were heme-containing cytochrome P450-type carotene hydroxylases. Gene expression results showed that the expression of CitHYb increased in the flavedo and juice sacs during the ripening process, which was well consistent with the accumulation of β,β-xanthophyll in citrus fruits. The expression of CitCYP97A and CitCYP97C increased with a peak in November, which might lead to an increase of lutein in the juice sacs during the ripening process. The expression level of CitCYP97B was much lower than that of CitHYb, CitCYP97A, and CitCYP97C in the juice sacs during the ripening process. Functional analysis showed that the CitHYb was able to catalyze the hydroxylation of the β-rings of β-carotene and α-carotene in Escherichia coli BL21 (DE3) cells. Meanwhile, when CitHYb was co-expressed with CitCYP97C, α-carotene was hydroxylated on the β-ring and ε-ring sequentially to produce lutein.
Conclusions
CitHYb was a key gene for β,β-xanthophyll biosynthesis in citrus fruits. CitCYP97C functioned as an ε-ring hydroxylase to produce lutein using zeinoxanthin as a substrate. The results will contribute to elucidating xanthophyll biosynthesis in citrus fruits, and provide new strategies to improve the nutritional and commercial qualities of citrus fruits.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0840-2) contains supplementary material, which is available to authorized users.
Keywords: β-Cryptoxanthin, Flavedo, Juice sacs, Lutein, Satsuma mandarin
Background
Carotenoids are a diverse group of pigments widely distributed in nature that provide distinct colors to fruits and flowers, and fulfill critical roles in plant growth and development [1–4]. In nature, more than 700 carotenoids have been identified and divided into two groups: carotenes and xanthophylls. Carotenes are liner or cyclic hydrocarbons, and xanthophylls are oxygenated derivatives of carotenes, such as lutein, β-cryptoxanthin, zeaxanthin, and astaxanthin. In higher plants, xanthophylls play an important role in the photosynthesis and photoprotection. They are structural elements of the photosynthetic apparatus, and the xanthophyll cycle (lutein, zeaxanthin, and antheraxanthin) protects plants from the damage of high light irradiation by dissipating excess light energy [5–8]. In addition, xanthophylls can be oxidatively cleaved in a site-specific manner, producing different apocarotenoids with important metabolic functions, such as plant hormones, pigments, as well as aroma and scent compounds [9–13]. Xanthophylls are not only important to the plants themselves, but also beneficial to human health. Epidemiological studies suggested that xanthophylls, such as lutein, β-cryptoxanthin, and astaxanthin, were effective to prevent eye diseases, certain cancers and inflammation because of their high antioxidant activity [14–21].
In plants, two different types of carotene hydroxylases, non-heme di-iron carotene hydroxylase and heme-containing cytochrome P450-type carotene hydroxylase, are involved in the biosynthesis of xanthophyll. Non-heme di-iron carotene hydroxylase (also called BCH, HYD, or HYb) efficiently catalyzes the hydroxylation of the β-rings of β-carotene (Fig. 1). In some plants species, it has been reported that two members of non-heme di-iron carotene hydroxylase existed, which had similar functions but tissue-specific expression patterns [22–25]. In Arabidopsis, a double-null mutation of BCH1 and BCH2 led to a significant decrease of β,β-xanthophylls [24, 26]. Recently, three heme-containing cytochrome P450-type carotene hydroxylases (CYP97A3, CYP97B3, and CYP97C1) have been identified in Arabidopsis. As shown in Fig. 1, CYP97C1 encoded by the LUT1 locus in Arabidopsis is responsible for the ε-ring hydroxylation [27, 28]. CYP97C1 is a key enzyme for the biosynthesis of lutein, and its activity can not be replaced by other carotene hydroxylases. In tomato, up-regulation of CYP97C11 led to an increase in the content of lutein in leaves. In contrast, when CYP97C11 was down-regulated, lutein was almost absent (0.8 %) in tomato leaves [29]. CYP97A3 encoded by LUT5 locus exhibits a major activity towards the β-ring of α-carotene, and a minor activity towards the β-rings of β-carotene in Arabidopsis [28, 30] (Fig. 1). Quinlan et al. [31] reported that OsCYP97A4 interacted with OsCYP97C2 in maize protoplasts, and the synergistic interaction between OsCYP97A4 and OsCYP97C2 drove the formation of lutein. Unlike CYP97A and CYP97C, the roles of CYP97B in the xanthophyll biosynthesis are still poorly studied. It has been suggested that CYP97B3 might be able to hydroxylate the β-rings of β-carotene and α-carotene in Arabidopsis [32, 33]. However, in the quadruple mutant (bch1, bch2, cyp97c1, and cyp97a3) that contained only CYP97B3, xanthophylls did not accumulated, indicating that CYP97B might not be an important enzyme for carotene hydroxylation [8, 24].
Fig. 1.

Xanthophyll biosynthetic pathway in Arabidopsis. LCYb, lycopene β-cyclase; LCYe, lycopene ε-cyclase; BCH1/2, β-ring hydroxylase1/2; CYP97A3, heme-containing cytochromes P450 monooxygenases A3; CYP97B3 heme-containing cytochromes P450 monooxygenases B3; CYP97C1, heme-containing cytochrome P450 monooxygenases C1; ZEP, zeaxanthin epoxidase
Citrus fruits accumulate a high amount of xanthophylls, especially β,β-xanthophylls, which account for up to 90 % of total carotenoids [34, 35]. In the previous studies, carotenoid metabolism has been extensively investigated in the fruits of different citrus varieties [11, 34–38]. Meanwhile, some key carotenoid metabolic genes have been isolated and their functions were deeply investigated in citrus fruits [9, 11, 12, 37, 39]. To date, however, the roles of carotene hydroxylases in regulating carotenoid content and composition are still unclear in citrus. In the present study, the changes in the expression of four carotene hydroxylase genes (CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C) were investigated in the flavedo and juice sacs during the ripening process. In addition, to elucidate their roles in xanthophyll biosynthesis, functional analyses of the four carotene hydroxylase genes were conducted in Escherichia coli cells accumulating with different carotenoids. The results present in this study will contribute to further elucidating the mechanism of carotenoid accumulation in citrus fruits, and provide new insights into enhancing the nutritional and commercial qualities of citrus fruits.
Results
Isolation and characterization of carotene hydroxylase genes in citrus fruits
In order to identify the carotene hydroxylase genes in citrus, we performed blast searches in the Citrus clementina v.10 and Citrus sinesis v 1.1 genome databases (http://www.phytozome.net/) using the sequences of Arabidopsis BCH1, BCH2, CYP97A3, CYP97B3, and CYP97C1 as queries, respectively. Four carotene hydroxylase genes (HYb, CYP97A, CYP97B, and CYP97C) were identified in citrus genome database. In our previous study, CitHYb was isolated from Satsuma mandarin (Accession number: AB114653), while the information on CYP97A, CYP97B, and CYP97C in citrus fruits was completely unknown. In the present study, the full-length cDNAs of CYP97A, CYP97B, and CYP97C were isolated from Satsuma mandarin by RT-PCR using the primers designed within 5′ and 3′ UTRs according to the sequences obtained from the citrus genome database. The sequences of CYP97A, CYP97B, and CYP97C were named as CitCYP97A, CitCYP97B, and CitCYP97C, and submitted to the NCBI database (Accession numbers: CitCYP97A, LC143646; CitCYP97B, LC143647; CitCYP97C, LC143648). Phylogenetic analysis showed that the four citrus carotene hydroxylases presented in four distinct clusters which have been identified in higher plants (Fig. 2). CitHYb was a non-heme di-iron carotene hydroxylase, and its nucleotide sequence contained 936 bp, encoding a putative protein of 311 amino acids with a predicted molecular of 34.7 kDa. CitCYP97A, CitCYP97B, and CitCYP97C were heme-containing cytochrome P450-type carotene hydroxylases. The nucleotide sequence of CitCYP97A contained 1839 bp, and encoded a putative protein of 612 amino acids with a predicted molecular of 68.4 kDa. The nucleotide sequence of CitCYP97B contained 1749 bp, and encoded a putative protein of 582 amino acids with a predicted molecular of 65.2 kDa. The nucleotide sequence of CitCYP97C contained 1641 bp, and encoded a putative protein of 546 amino acids with a predicted molecular of 61.6 kDa. A chloroplastic transit peptide with different lengths was predicted in the N-terminal region of the proteins of CitHYb (62 amino acids), CitCYP97A (37 amino acids), CitCYP97B (50 amino acids), and CitCYP97C (18 amino acids).
Fig. 2.
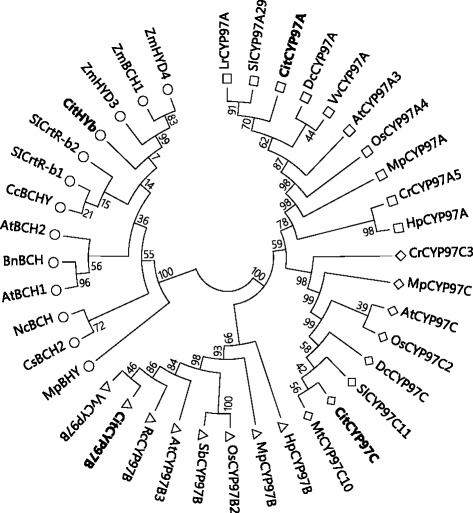
Phylogenetic analysis of carotene hydroxylases. The Neighbor-joining phylogenetic tree was constructed based on the alignment of the deduced amino acid sequences of carotene hydroxylases using MEGA6 software
Changes in carotenoid contents and expression of carotene hydroxylase genes in the flavedo during the ripening process
In the present study, carotenoids were extracted from citrus fruits during the ripening process, and the changes in carotenoid content and composition were analyzed by HPLC. In the flavedo, the contents of β-carotene, α-carotene and lutein decreased rapidly from August, and then kept at a low level during the ripening process (Fig. 3a). The content of β-cryptoxanthin, the major carotenoid in Satsuma mandarin, increased significantly during the ripening process, and reached 49.7 μg g−1 in December. In addition, the contents of zeaxanthin, all-trans-violaxanthin and cis-violaxanthin also gradually increased, and as a result β,β-xanthophyll (sum of β-cryptoxanthin, zeaxanthin, all-trans-violaxanthin, and cis-violaxanthin) accumulated massively during the ripening process (Fig. 3b). Gene expression results showed that the expression of CitHYb increased gradually from September, which was consistent with the accumulation of β,β-xanthophyll during the ripening process (Fig. 4a). The expression of CitCYP97A decreased rapidly to a low level in October, and then increased with a peak in November during the ripening process. The expression of CitCYP97C increased with two peaks in September and November, respectively. Similarly to CitCYP97C, the expression of CitCYP97B increased gradually with a peak in September.
Fig. 3.

Carotenoid accumulation in the citrus fruits during the ripening process. a Changes in the colors of citrus fruits during the ripening process. b Changes in the carotenoid content in the flavedo during the ripening process. c Changes in the carotenoid content in the juice sacs during the ripening process. The results shown are the mean ± SE for triplicate samples. β-Car, β-carotene; β-Cry, β-cryptoxanthin; Zea, zeaxanthin; T-vio, all-trans-violaxanthin; C-vio, 9-cis-violaxanthin; α-Car, α-carotene; Lut, lutein; β,β-Xanthophyll, sum of β-Cry, Zea, T-vio and C-vio. A, August; S, September; O, October; N, November; D, December
Fig. 4.

Changes in the expression of carotene hydroxylase genes in the flavedo (a) and juice sacs (b) during the ripening process. The results shown are the mean ± SE for triplicate samples. The mRNA levels were analyzed by TaqMan real-time quantitative RT-PCR. Real-time RT-PCR amplification of 18S rRNA was used to normalize the expression of the genes under identical conditions. A, August; S, September; O, October; N, November; D, December
Changes in carotenoid contents and expression of carotene hydroxylase genes in the juice sacs during the ripening process
In the juice sacs, the content of β-carotene decreased rapidly to an extremely low level in October, while the contents of β-cryptoxanthin and zeaxanthin increased significantly during the ripening process (Fig. 3c). The contents of α-carotene and lutein increased gradually in the juice sacs during the ripening process. Gene expression results showed that the expression of CitHYb increased in the juice sacs during the ripening process, which was in parallel with the accumulation of β,β-xanthophyll (Fig. 4b). The expression of CitCYP97A, CitCYP97B, and CitCYP97C increased with a peak in October and November, respectively. In addition, the expression level of CitCYP97B was much lower than that of CitCYP97A and CitCYP97C in the juice sacs during the ripening process (Fig. 4b).
Functional analysis of carotene hydroxylase genes in E. coli cells
In the present study, the cDNAs of CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C without transcript peptides were cloned into pRSF-2 Ek/LIC vector, respectively. The four recombinant plasmids were transformed into β-carotene-accumulating E. coli BL21 (DE3) cells, as well as α-carotene- and β-carotene-accumulating E. coli BL21 (DE3) cells, respectively. Carotenoids were extracted from bacteria and analyzed by HPLC. When CitHYb was expressed in the β-carotene-accumulating E. coli BL21 (DE3) cells, the peaks of β-cryptoxanthin and zeaxanthin were observed (Fig. 5b). When CitHYb was expressed in the α-carotene- and β-carotene-accumulating E. coli BL21 (DE3) cells, a monohydroxylated intermediate, zeinoxanthin, was also detected, except for β-cryptoxanthin and zeaxanthin (Fig. 6b). In contrast to CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C did not exhibit any carotene hydroxylation activity in the β-carotene-accumulating E. coli BL21 (DE3) cells, or α-carotene- and β-carotene-accumulating E. coli BL21 (DE3) cells (Figs. 5c, d, e and 6c, d, e).
Fig. 5.

HPLC analysis of carotenoids in β-carotene-accumulating E. coli BL21 (DE3) cells transformed with pRSF-2 Ek/LIC-CitHYb (b), pRSF-2 Ek/LIC-CitCYP97A (c), pRSF-2 Ek/LIC-CitCYP97B (d), and pRSF-2 Ek/LIC-CitCYP97C (e). Carotenoids extracted from the suspension cultures of β-carotene-accumulating E. coli BL21 (DE3) cells with pRSF-2 (empty vector) were used as control (a). β-Car, β-carotene; β-Cry, β-cryptoxanthin; Zea, zeaxanthin
Fig. 6.
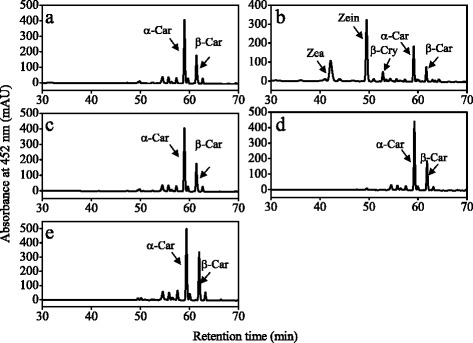
HPLC analysis of carotenoids in α-carotene and β-carotene-accumulating E. coli BL21 (DE3) cells transformed with pRSF-2 Ek/LIC-CitHYb (b), pRSF-2 Ek/LIC-CitCYP97A (c), pRSF-2 Ek/LIC-CitCYP97B (d), and pRSF-2 Ek/LIC-CitCYP97C (e). Carotenoids extracted from the suspension cultures of α-carotene and β-carotene-accumulating E. coli BL21 (DE3) cells with pRSF-2 (empty vector) were used as control (a). β-Car, β-carotene; β-Cry, β-cryptoxanthin; Zea, zeaxanthin; Zein, zeinoxanthin
To further investigate the functions of carotene hydroxylase genes of citrus, we co-transformed CitCYP97C with CitHYb, CitCYP97A, and CitCYP97B, respectively. The recombinant plasmids were expressed in the α-carotene- and β-carotene-accumulating E. coli BL21 (DE3) cells. As shown in Fig. 7b, when CitHYb and CitCYP97C were co-expressed, the monohydroxylated zeinoxanthin, which was produced by CitHYb, was further converted to lutein by CitCYP97C. However, when CitCYP97A and CitCYP97C were co-expressed or CitCYP97B and CitCYP97C were co-expressed, no hydroxylated carotene was detected in the α-carotene- and β-carotene-accumulating E. coli BL21 (DE3) cells (Fig. 7c, d).
Fig. 7.

HPLC analysis of carotenoids in α-carotene and β-carotene-accumulating E. coli BL21 (DE3) cells co-transformed with pCDF-2 Ek/LIC-CitCYP97C and pRSF-2 Ek/LIC-CitHYb (b), pCDF-2 Ek/LIC-CitCYP97C and pRSF-2 Ek/LIC-CitCYP97A (c), pCDF-2 Ek/LIC-CitCYP97C and pRSF-2 Ek/LIC-CitCYP97B (d). Carotenoids extracted from the suspension cultures of α-carotene and β-carotene-accumulating E. coli BL21 (DE3) cells with pRSF-2 (empty vector) were used as control (a). β-Car, β-carotene; β-Cry, β-cryptoxanthin; Zea, zeaxanthin; Zein, zeinoxanthin; Lut, lutein
Discussion
Isolation and characterization of carotene hydroxylase genes in citrus fruits
Carotene hydroxylases were the key enzymes responsible for xanthophyll biosynthesis in plants. Two different types of carotene hydroxylases, non-heme di-iron carotene hydroxylase and heme-containing cytochrome P450-type carotene hydroxylase, have been identified in higher plants. In the present study, the roles of four carotene hydroxylase genes (CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C) in regulating xanthophylls biosynthesis were investigated in citrus fruits. As shown in Fig. 2, the four carotene hydroxylase genes were clustered in distinct groups. CitHYb was a non-heme di-iron carotene hydroxylase, while CitCYP97A, CitCYP97B, and CitCYP97C were heme-containing cytochrome P450-type carotene hydroxylases. It has been reported that two or more HYb genes existed in some plant species, such as pepper, tomato, and Arabidopsis [23, 25, 40–42]. However, only one HYb was identified and isolated from citrus fruits, and it showed approximately 70 % identities to Arabidopsis BCH1 and BCH2 at the amino acid level. In the previous studies, the changes in the expression of HYb were extensively investigated in citrus fruits during the ripening process and under different environmental conditions [11, 34, 35, 43, 44]. In the juice sacs, different expression levels of CitHYb led to distinct carotenoid compositions between Satsuma mandarin and Valencia orange [34]. In contrast to CitHYb, the roles of heme-containing cytochrome P450-type carotene hydroxylases in regulating carotenoid accumulation in citrus fruits are completely unknown. In this study, it was the first time to isolate CitCYP97A, CitCYP97B, and CitCYP97C from the citrus fruits. Phylogenetic analysis suggested that CitCYP97A was more closely related to CitCYP97C than to CitCYP97B. CitCYP97B contained three insertions in the amino acid sequence compared with CitCYP97A and CitCYP97C, and shared around 42 % amino acid identity with CitCYP97A and CitCYP97C (Additional file 1: Figure S1). It has been reported that CYP97B3 in Arabidopsis was an uncharacterized cytochrome P450 monooxygenase, and the three amino acid insertions differentiated CYP97B from CYP97A and CYP97C [33, 45]. In addition, a transit peptide was predicted in the N-terminal reign of proteins encoded by CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C, which suggested that the four carotene hydroxylases of citrus were able to import into plastids. As most carotenoids are synthesized and stored in plastids, the location of CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C within plastid allows them to catalyze the reaction of carotene hydroxylation.
Changes in carotenoid contents and expression of carotene hydroxylase genes in citrus fruits during the ripening process
In the previous studies, non-heme di-iron carotene hydroxylases were isolated from different plant species, and their roles in the carotenoid biosynthesis have been characterized [23–25, 46]. In Arabidopsis, BCH1 and BCH2 primarily catalyzed the hydroxylation of the β-rings of β-carotene [22, 47]. Du et al. [48] reported that DSM2 gene (BCH) controlled the biosyntheses of zeaxanthin and ABA, and conferred drought and oxidative stress resistance in rice. In citrus fruits, massive accumulation of xanthophylls, especially β,β-xanthophylls, occurred during the ripening process. In the present study, the results showed that the expression of CitHYb increased gradually in the flavedo and juice sacs during the ripening process (Fig. 4). The increase in the expression of CitHYb was well in agreement with the accumulation of β,β-xanthophyll in the flavedo and juice sacs (Fig. 3). This result was consistent with the findings of Pons et al. [49], in which inhibiting the expression of Csβ-CHX (HYb) by RNA interference led to a significant increase of β-carotene (up to 36-fold) and a decrease of β,β-xanthophylls in sweet orange. Thus, it was suggested that CitHYb was a key gene for β,β-xanthophyll biosynthesis in citrus fruits.
In contrast to non-heme di-iron carotene hydroxylase, heme-containing cytochrome P450-type carotene hydroxylases preferentially hydroxylated the β- and α-rings of α-carotene, yielding lutein in Arabidopsis [30]. Quinlan et al. [31] found that a synergistic interaction existed between rice OsCYP97A4 and OsCYP97C2, which was required to drive the biosynthesis of lutein. In the present study, gene expression results showed that the expression of CitCYP97A and CitCYP97C increased with a peak in November in juice sacs, which might lead to an increase of lutein during the ripening process (Figs. 3c and 4b). In the flavedo, the expression of CitCYP97A decreased rapidly from August, which was in parallel with the reduction of lutein in the green stage. In the orange stage (from October), the expression of CitCYP97A and CitCYP97C increased with a peak in November, while the content of lutein decreased to a low level in the flavedo (Figs. 3b and 4a). In citrus fruits, a change from β,ε-carotenoid accumulation (α-carotene and lutein) to β,β-carotenoid accumulation (β-carotene, β-cryptoxanthin, zeaxanthin, all-trans-violaxanthin, and cis-violaxanthin) was observed in the flavedo during the ripening process, accompanying the disappearance of CitLCYe transcripts and the increase in CitLCYb transcripts. We previously reported that the expression of CitLCYe decreased rapidly to a low level in the orange stage in the flavedo of Satsuma mandarin [34, 39]. Thus, it was suggested that the level of lutein in the orange stage was mainly controlled by CitLCYe instead of CitCYP97A and CitCYP97C in the flavedo.
CYP97B is another member of CYP97 family with carotene hydroxylation activity. CYP97B3 of Arabidopsis exhibited a potential hydroxylation activity towards β-carotene and α-carotene [32, 33]. However, it remains controversial whether CYP97B is involved in the xanthophyll biosynthesis, because xanthophylls did not accumulate in the Arabidopsis quadruple mutant (bch1, bch2, cyp97c1, and cyp97a3) with only CYP97B3 [8, 24]. In the present study, the results showed that the expression level of CitCYP97B in the juice sacs was much lower than that of CitHYb, CitCYP97A, and CitCYP97C, indicating that CitCYP97B might not be a key gene for carotene hydroxylation in the juice sacs of citrus fruits (Fig. 4b).
Functional analysis of carotene hydroxylase genes in E. coli cells
In citrus, it is difficult to investigate the gene functions in the transgenic fruits because of its long juvenile phase that delays fruiting for 5–15 years [50]. As an alternative, E. coli cells that accumulate different carotenoids have been identified to be an efficient platform for investigating the functions of citrus carotenoid metabolic genes [11, 39, 43]. In the present study, we used the β-carotene-accumulating E. coli BL21 (DE3) cells and α-carotene- and β-carotene-accumulating E. coli BL21 (DE3) cells to investigate the functions of the four carotene hydroxylase genes of citrus. The results showed that CitHYb catalyzed the hydroxylation of the β-rings of β-carotene and α-carotene in E. coli BL21 (DE3) cells. When CitHYb was expressed in β-carotene-accumulating E. coli BL21 (DE3) cells, β-carotene was converted to β-cryptoxanthin and zeaxanthin, which supported the finding that CitHYb was a key gene for β,β-xanthophyll accumulation in citrus fruits (Fig. 5b). In Arabidopsis, it was reported that CYP97A3 and CYP97C1 were involved in lutein biosynthesis through hydroxylation of β- and ε-rings of α-carotene, respectively [24, 31]. However, in the absence of CYP97A the biosynthesis of lutein was not completely blocked. In Arabidopsis and rice, mutant of CYP97A only reduced around 20 % lutein compared with WT, which indicated that other carotene hydroxylases must also be able to catalyze hydroxylation of α-carotene on the β-ring [30, 51]. In the present study, we found that CitHYb participated in the biosynthesis of lutein. It converted α-carotene to zeinoxanthin, and the zeinoxanthin was further hydroxylated by CitCYP97C to produce lutein. Interestingly, CitCYP97C exhibited ε-ring hydroxylation activity only when it was co-expressed with CitHYb. Moreover, zeinoxanthin was the substrate for CitCYP97C instead of α-carotene, because α-cryptoxanthin, a monohydroxylated α-carotene on the ε-ring, was not detected in E. coli cells that transformed with CitCYP97C (Fig. 6e). These results suggested that α-carotene was hydroxylated on the β-ring and ε-ring sequentially to produce lutein in citrus fruits. A similar result was also reported in Arabidopsis, tomato and liverwort [29, 30, 52].
In contrast to the CitHYb, the carotene hydroxylation activities of CitCYP97A and CitCYP97B were not detected in E. coli BL21 (DE3) cells. In higher plants, only rice OsCYP97A4 was reported to exhibit β-ring hydroxylation activity in E. coli cells [31]. Amino acid sequence analysis showed that CitCYP97A shared 70 % similarity with OsCYP97A4, and the conserved oxygen-binding and heme-binding domains detected in CitCYP97A were identical to those of OsCYP97A4 (Additional file 2: Figure S2). In addition, we tested the activity of the full-length CitCYP97A, and optimized the culturing temperature and IPTG concentration as suggested in the study of Quinlan [45] (data not shown). Unfortunately, we could not detect any hydroxylation activity of CitCYP97A in E. coli cells. Similarly, attempts to assay the carotene hydroxylation activity of Arabidopsis CYP97A3 and liverwort MpCYP97A in E. coli cells were also failed [30, 52]. Whereas, mutant studies showed that Arabidopsis CYP97A3 (LUT5 locus) exhibited a major hydroxylation activity towards the β-ring of α-carotene, and a minor activity towards the β-rings of β-carotene [30]. In orange carrot, a deficient CYP97A3 allele caused the accumulation of α-carotene and a high α-/β-carotene ratio [53]. The research of Pons et al. [49] suggested that a second carotene hydroxylase might be present in sweet orange, because inhibiting the expression of Csβ-CHX (HYb) by RNA interference only caused a slight decrease in xanthophylls. In our present study, the gene expression results showed that CitCYP97A expressed in the flavedo and juice sacs during the ripening process. Meanwhile, the changes in CitCYP97A expression were consistent with the accumulation of lutein in the juice sacs (during the ripening stage) and flavedo (in the green stage), which indicating that CitCYP97A might be involved in the biosynthesis of lutein in citrus fruits. However, the mechanism that CitCYP97A regulates xanthophyll biosynthesis in citrus fruits seems to be more complicated, and some co-factors that are absent in E. coli cells might be needed for CitCYP97A to exert its activities. In the future research, further identification of the co-factors for heme-containing cytochrome P450-type carotene hydroxylases will contribute to elucidating the role of CitCYP97A in carotenoid accumulation.
Conclusion
In the present study, the roles of the four carotene hydroxylase genes (CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C) in regulating xanthophyll biosynthesis were investigated in citrus fruits. The results showed that CitHYb was a key gene for β,β-xanthophyll biosynthesis in citrus fruits. Functional analysis showed that the CitHYb was able to hydroxylate the β-rings of β-carotene and α-carotene in E. coli BL21 (DE3) cells. Meanwhile, when CitHYb was co-expressed with CitCYP97C, α-carotene was hydroxylated on the β-ring and ε-ring sequentially to produce lutein. In addition, we detected the expression of CitCYP97A in citrus fruits during the ripening process, and the changes in its expression were consistent with the accumulation of lutein in the juice sacs (during the ripening stage) and flavedo (in the green stage), which indicating that CitCYP97A might be involved in the biosynthesis of lutein in citrus fruits. The results presented in this study will contribute to further elucidating the mechanism of carotenoid biosynthesis in citrus fruits, and provide new strategies to improve carotenoid composition of citrus fruits.
Methods
Plant material
Satsuma mandarin (Citrus unshiu Marc.) cultivated at the Fujieda Farm of Shizuoka University (Shizuoka, Japan) were used as materials. Fruit samples were collected periodically from August to December. The flavedo and juice sacs were separated from sampled fruits, immediately frozen in liquid nitrogen, and kept at −80 °C until used.
Extraction and determination of carotenoids
The identification and quantification of carotenoids were conducted according to the methods described by Ma et al. [11]. Pigments were extracted from the samples using a hexane:acetone:ethanol (2:1:1 [v/v]) solution containing 0.1 % (w/v) 2,6-di-tert-butyl-4-methylphenol and 10 % (w/v) magnesium carbonate basic. After the organic solvents had been completely evaporated, the extracts containing carotenoids esterified to fatty acids were saponified with 20 % (w/v) methanolic KOH. Water-soluble extracts were then removed by adding NaCl-saturated water. The pigments repartitioned into the diethylether phase were recovered and evaporated to dryness. Subsequently, the residue was redissolved in 5 mL of a TBME: methanol (1:1 [v/v]) solution. An aliquot (20 μL) was separated by a reverse-phase HPLC system (Jasco, Tokyo, Japan) fitted with a YMC Carotenoid S-5 column of 250- × 4.6-mm-i.d. (Waters, Milford, MA) at a flow rate of 1 mL min−1. The eluent was monitored by a photodiode array detector (MD-2015, Jasco). The carotenoid concentration was estimated by the standard curves and expressed as milligrams per gram fresh weight [34]. Carotenoid quantification was performed in three replicates.
Isolation and sequence analysis of carotene hydroxylase genes
Total RNA was extracted from the flavedo of Satsuma mandarin fruits according to the method described by Ikoma et al. [54]. First-strand cDNA was synthesized from 2 μg of total RNA using TaqMan Reverse Transcription Reagents (Applied Biosystems). The full-length cDNAs of CitCYP97A, CitCYP97B, and CitCYP97C were amplified by RT-PCR using the primers designed within 5′ and 3′ UTRs according to the sequences obtained from the citrus genome database (Additional file 3: Table S1). The amplified cDNAs were cloned into the TOPO TA vector and sequenced using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) with an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
The alignment of CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C was created using CLUSTAL W (http://www.clustal.org). The Neighbor-joining phylogenetic tree was constructed based on the alignment of the deduced amino acid sequences of carotene hydroxylases using MEGA6 software [55]. Accession numbers are: Arabidopsis AtBCH1, AY113923; Arabidopsis AtBCH2, AY117225; Brassica napus BnBCH, EF026098; Coffea canephora CcBCHY, DQ157165; Citrus CitHYb1, AB114653; Crocus sativus CsBCH2, AY579207; Lycopersicon esculentum SICrtR-b1, Y14809; Lycopersicon esculentum SICrtR-b2, Y14810; Marchantia polymorpha MpBHY, AB981062; Narcissus tazetta var. chinensis NcBCH, JN625263; Zea mays ZmBCH1, GQ131287; Zea mays ZmHYD3, AY844958; Zea mays ZmHYD4, AY844956; Arabidopsis AtCYP97A3, NM_102914; Chlamydomonas reinhardtii CrCYP97A5, EF587911; Citrus CitCYP97A, LC143646; Daucus carota DcCYP97A3, JQ655297; Haematococcus pluvialis HpCYP97A, JX308236; Lycopersicon esculentum SlCYP97A29, EU849605; Lycium ruthenicum LrCYP97A, KF957714; Marchantia polymorpha MpCYP97A, AB981063; Oryza sativa OsCYP97A4, AK068163; Vitis vinifera VvCYP97A, XP_002279984; Arabidopsis AtCYP97C1, AY424805; Citrus CitCYP97C, LC143648; Chlamydomonas reinhardtii CrCYP97C3, EF587910; Daucus carota DcCYP97C, ABB52076; Lycopersicon esculentum SlCYP97C11, EU849604; Medicago truncatula MtCYP97C10, ABC59096; Marchantia polymorpha MpCYP97C, AB981065; Oryza sativa OsCYP97C2, AK065689; Arabidopsis AtCYP97B3, NM_117600; Citrus CitCYP97B, LC143647; Haematococcus pluvialis HpCYP97B, JX272918; Marchantia polymorpha MpCYP97B, AB981064; Oryza sativa OsCYP97B2, XM_015771315; Ricinus communis RcCYP97B, XP_002520583; Sorghum bicolor SbCYP97B, XP_002451628; Vitis vinifera VvCYP97B, XP_002266883. Predictions of transit peptides of CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C were carried out using TargetP.
Total RNA extraction and real-time quantitative RT-PCR
Total RNA was extracted from the flavedo and juice sacs of Satsuma mandarin at different stages according to the method described by Ikoma et al. [54]. The total RNA was cleaned up using the RNeasy Mini Kit (Qiagen) with on-column DNase digestion. The reverse transcription (RT) reaction was performed with 2 μg of purified RNA and a random hexamer at 37 °C for 60 min using TaqMan Reverse Transcription Reagents (Applied Biosystems).
TaqMan MGB probes and sets of primers for CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C were designed with the Primer Express software (Additional file 4: Table S2). For the endogenous control, the TaqMan Ribosomal RNA Control Reagents VIC Probe (Applied Biosystems) was used. TaqMan real-time PCR was carried out with the TaqMan Universal PCR Master Mix (Applied Biosystems) using ABI PRISM 7300 (Applied Biosystems) according to the manufacturer’s instructions. Each reaction contained 900 nM of the primers, 250 nM of the TaqMan MGB Probe, and template cDNA. The thermal cycling conditions were 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. The levels of gene expression were analyzed with ABI PRISM 7300 Sequence Detection System Software (Applied Biosystems) and normalized with the results of 18S ribosomal RNA. Real-time quantitative RT-PCR was performed in three replicates for each sample.
Functional analysis of the carotene hydroxylases in E. coli cells
The cDNAs of CitHYb, CitCYP97A, CitCYP97B, and CitCYP97C without transit peptide were cloned into the pRSF-2 Ek/LIC vector or pCDF-2 Ek/LIC vector, respectively. In the previous study, two recombinant plasmids pET-CitLCYb1 and pET-CitLCYb1 + CitLCYe were constructed, and transformed into E. coli BL21 (DE3) cells harboring a lycopene biosynthetic palsmid pACCRT-EIB, respectively [39, 56]. The co-transformation of pACCRT-EIB with pET-CitLCYb1 or pET-CitLCYb1 + CitLCYe led to β-carotene accumulation or α-carotene and β-carotene-accumulation in the E. coli BL21 (DE3) cells. In the present study, the recombinant plasmids pRSF-2-CitHYb, pRSF-2-CitCYP97A, pRSF-2-CitCYP97B, and pRSF-2-CitCYP97C were transformed into the β-carotene-accumulating, as well as α-carotene and β-carotene-accumulating E. coli BL21 (DE3) cells. To investigate the interactions among the carotene hydroxylases of citrus, we co-expressed pCDF-2-CitCYP97C with pRSF-2-CitHYb, pRSF-2-CitCYP97A, and pRSF-2-CitCYP97B in the α-carotene and β-carotene-accumulating E. coli BL21 (DE3) cells, respectively. After induction with 0.05 M isopropyl β-D-thiogalactoside (IPTG) for 2 d at 27 °C, carotenoids were extracted from E. coli cells. Cultures of E. coli cells were centrifuged at 5,000 g for 10 min and the bacterial pellet was washed twice with Tris–HCl (pH 8.0). The pellet was dried using vacuum freeze drying and stored at −20 °C until the HPLC analysis. The freeze-ground material was extracted with a mixture of chloroform and methanol (2:1 [v/v]) until all the color was removed from the E. coli cells. The carotenoid extracts were reduced to dryness by rotary evaporation, and then dissolved in the methyl tert-butyl ether: methanol (1:1 [v/v]) solution containing 0.1 % butylated hydroxyl toluene. α-Carotene, β-carotene, β-cryptoxanthin, zeaxanthin, and lutein were identified by comparing their specific retention times and absorption spectra with the authentic standards (Kato et al. 2004). The identification of zeinoxanthin was conducted using the methods described by Meléndez-Martínez et al. [57]. For each carotene hydroxylase gene, three replicates were conducted using different colonies in the functional analysis in E. coli cells.
Statistical analysis
All values are shown as the mean ± SE for three replicates. The data were analyzed, and Tukey’s HSD test (at P < 0.05) was used to compare the treatment means.
Abbreviations
CYP, cytochrome P450; HYb, β-ring hydroxylase; LCYb, lycopene β-cyclase; LCYe, lycopene ε-cyclase; ZEP, zeaxanthin expoxidase.
Acknowledgments
We thank Professor Norihiko Misawa for providing the pACCRT-EIB plasmid (Research Institute for Bioresources and Biotechnology, Ishikawa Prefectural University, Nonoichi Ishikawa, Japan).
Funding
This work was supported by Grant-in-Aid for Young Scientists B (16 K18649).
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional files.
Authors’ contributions
GM and MK conceived and designed the study. WY and IT conducted the experiments and collected the data. NI and MO analyzed the data. KY and MY provided plant materials and reagents. LZ and MK prepared the manuscript. All authors read and approved the manuscript.
Completing interests
The authors declare that they have no completing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
Three sequence insertions in CitCYP97B compared with that of CitCYP97A and CitCYP97C. (DOCX 38 kb)
Alignment of deduced amino acid sequences of CitCYP97A and OsCYP97A4. The alignment was created using CLUSTAL W (http://www.clustal.org). (DOCX 153 kb)
Primer sequences used for isolating full-length cDNAs of CitCYP97A, CitCYP97B, CitCYP97C. (DOCX 12 kb)
Primer sequences and TaqMan MGB Probes used for the quantitative RT-PCRs of carotene hydroxylase genes. (DOCX 38 kb)
Contributor Information
Gang Ma, Email: agma@ipc.shizuoka.ac.jp.
Lancui Zhang, Email: archo@ipc.shizuoka.ac.jp.
Witchulada Yungyuen, Email: witchu.y@gmail.com.
Issei Tsukamoto, Email: 09tsukamoto@gmail.com.
Natsumi Iijima, Email: 7tsumikan@gmail.com.
Michiru Oikawa, Email: m.oikawa.0610@gmail.com.
Kazuki Yamawaki, Email: abkyama@ipc.shizuoka.ac.jp.
Masaki Yahata, Email: amyahat@ipc.shizuoka.ac.jp.
Masaya Kato, Email: amkato@ipc.shizuoka.ac.jp.
References
- 1.Cunningham FX, Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:557–83. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- 2.Havaux M. Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci. 1998;3:147–51. doi: 10.1016/S1360-1385(98)01200-X. [DOI] [Google Scholar]
- 3.Cazzonelli CI, Pogson BJ. Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010;15:266–74. doi: 10.1016/j.tplants.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Nisar N, Li L, Lu S, Khin NC, Pogson BJ. Carotenoid Metabolism in Plants. Mol Plant. 2015;8:68–82. doi: 10.1016/j.molp.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Niyogi KK, Björkman O, Grossman AR. The roles of specific xanthophylls in photoprotection. Proc Natl Acad Sci U S A. 1997;94:14162–7. doi: 10.1073/pnas.94.25.14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croce R, Weiss S, Bassi R. Carotenoid-binding sites of the major light-harvesting complex II of higher plants. J Biol Chem. 1999;274:29613–23. doi: 10.1074/jbc.274.42.29613. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, et al. Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature. 2004;428:287–92. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 8.Fiore A, Dall’Osto L, Cazzaniga S, Diretto G, Giuliano G, Bassi R. A quadruple mutant of Arabidopsis reveals a β-carotene hydroxylation activity for LUT1/CYP97C1 and a regulatory role of xanthophylls on determination of the PSI/PSII ratio. BMC Plant Biol. 2012;12:50. doi: 10.1186/1471-2229-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato M, Matsumoto H, Ikoma Y, Okuda H, Yano M. The role of carotenoid cleavage dioxygenases in the regulation of carotenoid profiles during maturation in citrus fruit. J Exp Bot. 2006;57:2153–64. doi: 10.1093/jxb/erj172. [DOI] [PubMed] [Google Scholar]
- 10.Walter MH, Floss DS, Strack D. Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta. 2010;232:1–17. doi: 10.1007/s00425-010-1156-3. [DOI] [PubMed] [Google Scholar]
- 11.Ma G, Zhang LC, Matsuta A, Matsutani K, Yamawaki K, Yahata M, et al. Enzymatic formation of β-citraurin from β-cryptoxanthin and zeaxanthin by carotenoid cleavage dioxygenase4 in the flavedo of citrus fruit. Plant Physiol. 2013;163:682–95. [DOI] [PMC free article] [PubMed]
- 12.Rodrigo MJ, Alquézar B, Alós E, Lado J, Zacarías L. Biochemical bases and molecular regulation of pigmentation in the peel of Citrus fruit. Sci Hortic. 2013;163:46–62. doi: 10.1016/j.scienta.2013.08.014. [DOI] [Google Scholar]
- 13.Ahrazem O, Rubio-Moraga A, Berman J, Capell T, Christou P, Zhu C, Gómez-Gómez L. The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2016;209:650–63. doi: 10.1111/nph.13609. [DOI] [PubMed] [Google Scholar]
- 14.Cerhan JR, Saag KG, Merlino LA, Mikuls TR, Criswell LA. Antioxidant micronutrients and risk of rheumatoid arthritis in a cohort of older women. Am J Epidemiol. 2003;157:345–54. doi: 10.1093/aje/kwf205. [DOI] [PubMed] [Google Scholar]
- 15.Khachik F, London E, de Moura FF, Johnson M, Steidl S, Detolla L, et al. Chronic ingestion of (3R,3′R,6′R)-lutein and (3R,3′R)-zeaxanthin in the female rhesus macaque. Invest Ophthalmol Vis Sci. 2006;47:5476–86. doi: 10.1167/iovs.06-0194. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi M, Hamamoto R, Uchiyama S, Ishiyama K, Hashimoto K. Anabolic effects of bee pollen cistus ladaniferus extract on bone components in the femoral-diaphyseal and -metaphyseal tissues of rats in vitro and in vivo. J Health Sci. 2006;52:43–9. doi: 10.1248/jhs.52.43. [DOI] [Google Scholar]
- 17.Sugiura M, Nakamura M, Ogawa K, Ikoma Y, Ando F, Shimokata H, Yano M. Dietary patterns of antioxidant vitamin and carotenoid intake associated with bone mineral density: findings from post-menopausal Japanese female subjects. Osteoporos Int. 2011;22:143–52. doi: 10.1007/s00198-010-1239-9. [DOI] [PubMed] [Google Scholar]
- 18.Takayanagi K, Morimoto S, Shirakura Y, Mukai K, Sugiyama T, Tokuji Y, Ohnishi M. Mechanism of visceral fat reduction in Tsumura Suzuki obese, diabetes (TSOD) mice orally administered β-cryptoxanthin from Satsuma mandarin oranges (Citrus unshiu Marc) J Agric Food Chem. 2011;59:12342–51. doi: 10.1021/jf202821u. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi M. Role of carotenoid β-cryptoxanthin in bone homeostasis. J Biomed Sci. 2012;19:36. doi: 10.1186/1423-0127-19-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iskandar AR, Liu C, Smith DE, Hu KQ, Choi SW, Ausman LM, Wang XD. β-Cryptoxanthin restores nicotine-reduced lung SIRT1 to normal levels and inhibits nicotine-promoted lung tumorigenesis and emphysema in A/J mice. Cancer Prev Res (Phila) 2013;6:309–20. doi: 10.1158/1940-6207.CAPR-12-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pouchieu C, Galan P, Ducros V, Latino-Martel P, Hercberg S, Touvier M. Plasma carotenoids and retinol and overall and breast cancer risk: a nested case–control study. Nutr Cancer. 2014;66:980–8. doi: 10.1080/01635581.2014.936952. [DOI] [PubMed] [Google Scholar]
- 22.Tian L, DellaPenna D. Characterization of a second carotenoid beta-hydroxylase gene from Arabidopsis and its relationship to the LUT1 locus. Plant Mol Biol. 2001;47:379–88. doi: 10.1023/A:1011623907959. [DOI] [PubMed] [Google Scholar]
- 23.Galpaz N, Ronen G, Khalfa Z, Zamir D, Hirschberg J. A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell. 2006;18:1947–60. doi: 10.1105/tpc.105.039966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Smith JJ, Tian L, DellaPenna D. The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol. 2009;50:463–79. doi: 10.1093/pcp/pcp005. [DOI] [PubMed] [Google Scholar]
- 25.D’Ambrosio C, Stigliani AL, Giorio G. Overexpression of CrtR-b2 (carotene beta hydroxylase 2) from S. lycopersicum L. differentially affects xanthophyll synthesis and accumulation in transgenic tomato plants. Transgenic Res. 2011;20:47–60. doi: 10.1007/s11248-010-9387-4. [DOI] [PubMed] [Google Scholar]
- 26.Tian L, Magallanes-Lundback M, Musetti V, DellaPenna D. Functional analysis of beta- and epsilon-ring carotenoid hydroxylases in Arabidopsis. Plant Cell. 2003;15:1320–32. doi: 10.1105/tpc.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D. The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid epsilon-ring hydroxylation activity. Proc Natl Acad Sci U S A. 2004;101:402–7. doi: 10.1073/pnas.2237237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiore A, Dall’Osto L, Fraser PD, Bassi R, Giuliano G. Elucidation of the beta-carotene hydroxylation pathway in Arabidopsis thaliana. FEBS Lett. 2006;580:4718–22. doi: 10.1016/j.febslet.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 29.Stigliani AL, Giorio G, D’Ambrosio C. Characterization of P450 carotenoid beta- and epsilon-hydroxylases of tomato and transcriptional regulation of xanthophyll biosynthesis in root, leaf, petal and fruit. Plant Cell Physiol. 2011;52:851–65. doi: 10.1093/pcp/pcr037. [DOI] [PubMed] [Google Scholar]
- 30.Kim J, DellaPenna D. Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid beta-ring hydroxylase CYP97A3. Proc Natl Acad Sci U S A. 2006;103:3474–9. doi: 10.1073/pnas.0511207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan RF, Shumskaya M, Bradbury LM, Beltrán J, Ma C, Kennelly EJ, Wurtzel ET. Synergistic interactions between carotene ring hydroxylases drive lutein formation in plant carotenoid biosynthesis. Plant Physiol. 2012;160:204–14. doi: 10.1104/pp.112.198556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JE, Punja ZK, Douglas CJ. Co-expression of Arabidopsis thaliana cytochrome P450 enzymes and NADPH-cytochrome P450 reductase in Escherichia coli: testing the function of candidate β-carotene hydroxylases. In: P450 Systems and Regulation, Proceedings of 14th International Conference on Cytochromes P450, Dallas, TX. Italy: Medimond S.r.l; 2005. p. 115–20.
- 33.Kim JE, Cheng KM, Craft NE, Hamberger B, Douglas CJ. Over-expression of Arabidopsis thaliana carotenoid hydroxylases individually and in combination with a β-carotene ketolase provides insight into in vivo functions. Phytochemistry. 2010;71:168–78. doi: 10.1016/j.phytochem.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Kato M, Ikoma Y, Matsumot H, Sugiura M, Hyodo H, Yano M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol. 2004;134:824–37. doi: 10.1104/pp.103.031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodrigo MJ, Marcos JF, Zacarías L. Biochemical and molecular analysis of carotenoid biosynthesis in flavedo of orange (Citrus sinensis L.) during fruit development and maturation. J Agric Food Chem. 2004;52:6724–31. doi: 10.1021/jf049607f. [DOI] [PubMed] [Google Scholar]
- 36.Rodrigo MJ, Zacarías L. Effect of postharvest ethylene treatment on carotenoid accumulation and the expression of carotenoid biosynthetic genes in the flavedo of orange (Citrus sinensis L. Osbeck) fruit. Postharvest Biol Technol. 2007;43:14–22. doi: 10.1016/j.postharvbio.2006.07.008. [DOI] [Google Scholar]
- 37.Ríos G, Naranjo MA, Rodrigo MJ, Alós E, Zacarías L, Cercós M, Talón M. Identification of a GCC transcription factor responding to fruit colour change events in citrus through the transcriptomic analyses of two mutants. BMC Plant Biol. 2010;10:276. doi: 10.1186/1471-2229-10-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X, Chen C, Yu Q, Gady A, Yu Y, Liang G, Gmitter FG., Jr Comparison of carotenoid accumulation and biosynthetic gene expression between Valencia and Rohde Red Valencia sweet oranges. Plant Sci. 2014;227:28–36. doi: 10.1016/j.plantsci.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, Ma G, Shirai Y, Kato M, Yamawaki K, Ikoma Y, Matsumoto H. Expression and functional analysis of two lycopene β-cyclases from citrus fruits. Planta. 2012;236:1315–25. doi: 10.1007/s00425-012-1690-2. [DOI] [PubMed] [Google Scholar]
- 40.Bouvier F, Keller Y, d’Harlingue A, Camara B. Xanthophyll biosynthesis: molecular and functional characterization of carotenoid hydroxylases from pepper fruits (Capsicum annuum L.) Biochim Biophys Acta. 1998;1391:320–8. doi: 10.1016/S0005-2760(98)00029-0. [DOI] [PubMed] [Google Scholar]
- 41.Diretto G, Welsch R, Tavazza R, Mourgues F, Pizzichini D, Beyer P, Giuliano G. Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol. 2007;7:11. doi: 10.1186/1471-2229-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, Farre G, Naqvi S, Breitenbach J, Sanahuja G, Bai C, Sandmann G, Capell T, Christou P, Zhu C. Cloning and functional characterization of the maize carotenoid isomerase and β-carotene hydroxylase genes and their regulation during endosperm maturation. Transgenic Res. 2010;19:1053–68. doi: 10.1007/s11248-010-9381-x. [DOI] [PubMed] [Google Scholar]
- 43.Alquézar B, Zacarías L, Rodrigo MJ. Molecular and functional characterization of a novel chromoplast-specific lycopene β-cyclase from Citrus and its relation to lycopene accumulation. J Exp Bot. 2009;60:1783–97. doi: 10.1093/jxb/erp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Ma G, Kato M, Yamawaki K, Takagi T, Kiriiwa Y, Ikoma Y, Matsumoto H, Yoshioka T, Nesumi H. Regulation of carotenoid accumulation and the expression of carotenoid metabolic genes in citrus juice sacs in vitro. J Exp Bot. 2012;63:871–86. doi: 10.1093/jxb/err318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan RF, Jaradat TT, Wurtzel ET. Escherichia coli as a platform for functional expression of plant P450 carotene hydroxylases. Arch Biochem Biophys. 2007;458:146–57. doi: 10.1016/j.abb.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang HM, To KY, Lai HM, Jeng ST. Modification of flower colour by suppressing β-ring carotenehydroxylasegenes in Oncidium. Plant Biol (Stuttg) 2016;18:220–9. doi: 10.1111/plb.12399. [DOI] [PubMed] [Google Scholar]
- 47.Sun Z, Gantt E, Cunningham Jr FX. Cloning and functional analysis of the β-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem. 1996;271:24349–52. [DOI] [PubMed]
- 48.Du H, Wang N, Cui F, Li X, Xiao J, Xiong L. Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol. 2010;154:1304–18. doi: 10.1104/pp.110.163741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pons E, Alquézar B, Rodríguez A, Martorell P, Genovés S, Ramón D, Rodrigo MJ, Zacarías L, Peña L. Metabolic engineering of β-carotene in orange fruit increases its in vivo antioxidant properties. Plant Biotechnol J. 2014;12:17–27. doi: 10.1111/pbi.12112. [DOI] [PubMed] [Google Scholar]
- 50.Peña L, Cervera M, Fagoaga C, Romero J, Ballester A, Soler N, Pons E, Rodríguez A, Peris J, Juárez J, Navarro L. Citrus. In C Kole, TC Hall. Citrus. In: Kole C, Hall TC, editors. Compendium of Transgenic Crop Plants: Tropical and Subtropical Fruits and Nuts. Oxford, UK: Blackwell Publishing; 2008. p. 1–62.
- 51.Lv MZ, Chao DY, Shan JX, Zhu MZ, Shi M, Gao JP, Lin HX. Rice carotenoid β-ring hydroxylase CYP97A4 is involved in lutein biosynthesis. Plant Cell Physiol. 2012;53:987–1002. doi: 10.1093/pcp/pcs041. [DOI] [PubMed] [Google Scholar]
- 52.Takemura M, Maoka T, Misawa N. Biosynthetic routes of hydroxylated carotenoids (xanthophylls) in Marchantia polymorpha, and production of novel and rare xanthophylls through pathway engineering in Escherichia coli. Planta. 2015;241:699–710. doi: 10.1007/s00425-014-2213-0. [DOI] [PubMed] [Google Scholar]
- 53.Arango J, Jourdan M, Geoffriau E, Beyer P, Welsch R. Carotene hydroxylase activity determines the levels of both α-carotene and total carotenoids in orange carrots. Plant Cell. 2014;26:2223–33. doi: 10.1105/tpc.113.122127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikoma Y, Yano M, Ogawa K, Yoshioka T, Xu ZC, Hisada S, Omura M, Moriguchi T. Isolation and evaluation of RNA from polysaccharide-rich tissues in fruit for quality by cDNA library construction and RT-PCR. J Jpn Soc Hortic Sci. 1996;64:809–14. doi: 10.2503/jjshs.64.809. [DOI] [Google Scholar]
- 55.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Misawa N, Shimada H. Metabolic engineering for the production of carotenoids in non-carotenogenic bacteria and yeasts. Journal of Biotechnol. 1998;59:169–81. doi: 10.1016/S0168-1656(97)00154-5. [DOI] [PubMed] [Google Scholar]
- 57.Meléndez-Martínez AJ, Britton G, Vicario IM, Heredia FJ. Identification of zeinoxanthin in orange juices. J Agric Food Chem. 2005;53:6362–7. doi: 10.1021/jf050370c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the results of this article are included within the article and its additional files.


