Abstract
The autophagy–lysosomal pathway (ALP) is involved in the degradation of long-lived proteins. Deficits in the ALP result in protein aggregation, the generation of toxic protein species, and accumulation of dysfunctional organelles, which are hallmarks of Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), and prion disease. Decades of research have therefore focused on enhancing the ALP in neurodegenerative diseases. More recently, transcription factor EB (TFEB), a major regulator of autophagy and lysosomal biogenesis, has emerged as a leading factor in addressing disease pathology. We review the regulation of the ALP and TFEB and their impact on neurodegenerative diseases. We also offer our perspective on the complex role of autophagy and TFEB in disease pathogenesis and its therapeutic implications through the examination of prion disease.
Subtypes and Machinery of Autophagy
Autophagy or autophagocytosis is derived from the Greek words auto, ‘self’, and phagein, ‘to eat’. This so-called self-eating refers to a basic catabolic mechanism/degradation pathway that involves the delivery of cytoplasmic cargo to the lysosome. Autophagy is typically categorized into three groups based on the particular physiological role of the process, the mode of cargo delivery, and the pathway. These groups are as follows: macroautophagy (see Glossary; herein referred to as autophagy), chaperone-mediated autophagy, and microautophagy. Despite mechanistic differences between the three groups, all are induced by similar stimuli such as environmental stress, nutrient starvation, oxidative stress, and infection. In addition, all converge on the lysosomal pathway, wherein TFEB plays a key role as the major regulator of lysosomal biogenesis (and autophagy) [1,2].
Autophagy is particularly important in non-dividing neurons. Mice deficient in autophagy exhibit neuronal accumulation of aggregate-prone proteins and neurodegeneration, demonstrating the crucial role of autophagy in neuronal homeostasis [3,4]. The role of the autophagy–lysosomal pathway (ALP) in neurodegenerative disease has been extensively studied in recent years, with defects in the pathway being strongly associated with disease [5]. In particular, late-onset neurodegenerative diseases such as PD, HD, and AD are characterized by the accumulation of intracellular aggregates in the brain, and clearance of these aggregates is typically associated with improvement of symptoms [6]. Thus, correcting ALP defects and enhancing the activity of the pathway are appealing therapeutic interventions. Since its discovery and characterization in 2009, TFEB, a master regulator of the ALP, has been widely demonstrated to ameliorate pathology in these diseases [7–11], as well as in models of spinal and bulbar muscular atrophy and in lysosomal storage disorders (LSDs) when activated [12–15]. The broad applicability of TFEB and its role in linking the different forms of autophagy make it an exceedingly attractive therapeutic target. We provide here a general description of the ALP and its impact on neurodegenerative disease, followed by in-depth coverage of TFEB. Learning from prion disease, we conclude by offering our perspective on the two-sided nature of the ALP and how it may influence disease pathogenesis and therapeutic development.
Macroautophagy
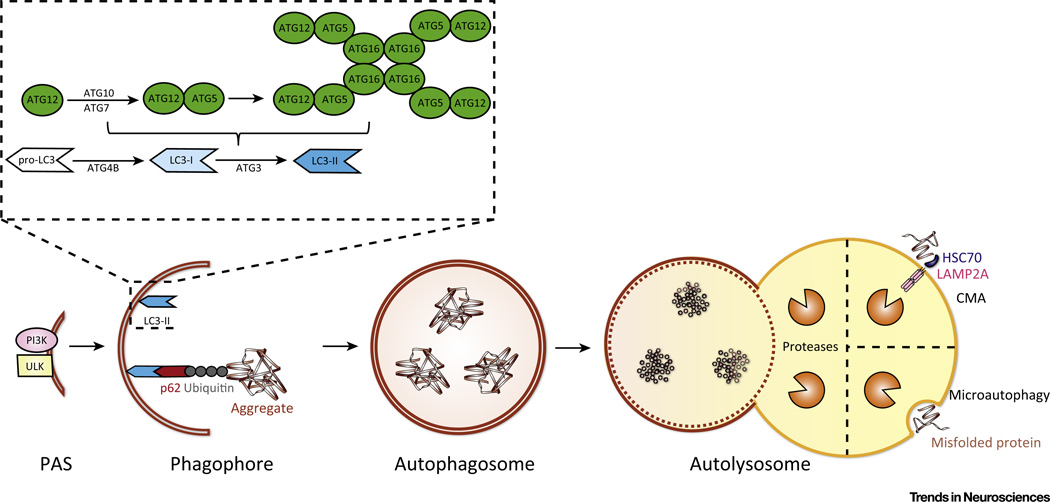
Autophagy is the major pathway for bulk protein degradation. More than 30 autophagy-related genes (ATGs) have been identified in yeast, with the majority having homologs in mammals [16]. These genes function at different stages of autophagy including initiation, elongation, maturation, and fusion with the lysosome (Box 1 for detailed mechanism). Autophagy is initiated by the formation of a crescent-shaped double-membrane structure known as the phagophore. The autophagosome membrane could potentially be derived from multiple sources including the cytoplasmic membrane, mitochondria, endoplasmic reticulum (ER), or Golgi apparatus [17–20]. In elongation, the double-membrane phagophore grows in size and utilizes two ubiquitin-like conjugation systems for elongation of pre-autophagosomal structures and generation of the autophagosome marker LC3-II. Following formation of the complete autophagosomal structure, the autophagosome can fuse with other endosomes or with the lysosome to form autolysosomes. In the autolysosome, contents are degraded by lysosomal enzymes (Figure 1).
Box 1. The Machinery of Macroautophagy.
Initiation
Two major protein complexes, ULK (UN51-like Ser/Thr kinases complex) and the class III phosphatidylinositol 3-kinase (PI3K) complex, are recruited to the phagophore assembly site (PAS). The ULK complex contains ULK1/2, FAK family kinase interacting protein of 200 kDa (FIP200), and ATG13 [102]. Another equivalent autophagy initiation complex is the PI3K complex, also called the Beclin 1 complex, which contains vacuolar protein sorting 34 (Vps34), p150 (Vps15), Beclin 1 (ATG6), and Barkor (ATG14) [103]. The activity of the class III PI3K is enhanced by dissociation of Beclin 1 from Bcl-2 allowing Beclin 1 to interact with Vps34 [104]. Phosphatidylinositol 3-phosphate [PI(3)P], the product of class III PI3K, is highly enriched on the surfaces of the phagophore and autophagosome [105]. PI(3)P is also thought to be a scaffold molecule to recruit other ATG proteins to the PAS to aid in autophagosome formation.
Elongation
Two ubiquitin-like conjugation systems are featured. The first system involves the linkage of ATG12 to ATG5 through interactions with ATG7 and ATG10. The ATG12–ATG5 conjugate then interacts with ATG6 to form the ATG12–ATG5–ATG16 complex, which is responsible for elongation of the pre-autophagosomal structures [106,107]. In the second ubiquitin-like conjugation system, the precursor of microtubule-associated protein 1 light chain 3 (LC3 or ATG8) is cleaved by ATG4B at its C-terminal domain to generate LC3-I [108]. LC3-I is then conjugated to phosphatidylethanolamine (PE) via interaction with ATG3 and ATG7 to form LC3-II [109,110]. The ATG12–ATG5–ATG16 complex can also display ubiquitin-like conjugation activity for the formation of PE-conjugated LC3-II. This final product specifically localizes to the autophagosome from elongation to lysosomal fusion, and is a well-known protein marker of autophagosomes [108].
Maturation and Lysosome Fusion
Following the completion of elongation and formation of a complete bubble-like structure, the autophagosome can move bidirectionally along microtubules, mediated by kinesin and dynein motor proteins [111,112]. During transport along the microtubules, autophagosomes first fuse with endosomes to generate amphisomes, followed by fusion with lysosomes to form autolysosomes. These fusion reactions are mediated by several membrane protein complexes such as the soluble NSF attachment protein receptor (SNAREs) [113]. After fusion with the lysosome, contents of the autophagosome and its inner membrane are digested by lysosomal enzymes.
Figure 1. Simplified Diagram of Autophagy Pathways.
Macroautophagy is initiated by recruitment of ULK and PI3K complex to the phagophore assembly site (PAS). The ULK complex is composed of ULK1/2, FIP200, and ATG13. The PI3K complex is made up of Vps34, Vps15, Beclin 1 (ATG6), and Barkor (ATG14). In elongation, two ubiquitin-like conjugating systems are involved. In the first, ATG12 is linked to ATG5 through ATG7 and ATG10, then the ATG5–ATG12 conjugate forms a complex with ATG16. In the second ubiquitin-like reaction, LC3 is cleaved at its C-terminus by ATG4B to form LC3-I. LC3-I is then conjugated to phosphatidylethanolamine (PE) by ATG7 and ATG3 to form LC3-II. The ATG12–ATG5–ATG16 complex can also conjugate PE to LC3-I to form LC3-II. LC3-II localizes to the autophagosome and remains on the membrane from elongation to lysosome fusion. In selective autophagy, LC3-II binds to p62, which delivers ubiquitinated protein aggregates for autophagic degradation. At completion of elongation, a complete bubble-like autophagosome surrounding the cargo has formed. The autophagosome can travel along microtubules via motor proteins, and eventually fuses with the lysosome to form an autolysosome. Within the autolysosome, the lysosomal proteases degrade the cargo and inner membrane of the former autophagosome. Chaperone-mediated autophagy (CMA) differs from macroautophagy in that HSC70 binds to protein substrates containing a KFERQ motif. The substrate–HSC complex interacts with LAMP-2A which multimerizes in response to binding to form an active transport complex through which substrates pass into the lysosome after unfolding. With microautophagy, the lysosome directly engulfs cytoplasmic cargo, such as protein aggregates and lysosomal proteases degrade the material.
Selective Autophagy
Selective autophagy ensures proper organelle, macromolecule, and protein turnover in the cytosol. Selective autophagy may arise under stress conditions that result from protein aggregation or damaged mitochondria, thus corresponding to the terms aggrephagy and mitophagy, respectively [21,22].
Aggrephagy refers to the specific degradation of protein aggregates. Several reports have demonstrated that ubiquitin tagging of aggregated proteins is the first step of aggrephagy [23]. The recognition of ubiquitinated substrates depends on adaptor proteins such as p62/sequestesome (SQSTM) 1, neighbor of BRCA1 gene 1 (NBR1), and optineurin (OPTN) which all share similar cargo-binding domains. Through the direct interaction of adaptors with LC3 or the membrane of the autophagosome, aggregates are selectively delivered to the autophagosome (Figure 1) [24].
Mitophagy refers to the selective degradation of damaged mitochondria by autophagy. Among the several pathways regulating mitophagy, the PTEN-induced putative kinase (PINK) 1–Parkin pathway is the best characterized. PINK1 is a mitochondrial Ser/Thr kinase that monitors mitochondrial function. Damaged mitochondria cannot maintain sufficient membrane potential, inducing retention of PINK1 on the outer membrane of the mitochondria. PINK1 then recruits Parkin, an E3 ubiquitin ligase. After ubiquitination, damaged mitochondria are selectively recognized by adaptor proteins and engulfed by the autophagosome [25].
Chaperone-Mediated Autophagy (CMA)
CMA is a multistep process characterized by substrate proteins containing a KFERQ motif that is recognized by a cytosolic chaperone heat-shock cognate protein of 70 kDa (HSC70). The substrate–HSC complex then interacts with lysosome-associated membrane protein type 2A (LAMP-2A), which serves as the receptor protein. Binding of the substrate triggers the assembly of LAMP-2A multimers, which function as an active transport complex through which substrates pass to enter the lysosome after unfolding [26].
Microautophagy
Microautophagy is characterized by direct lysosomal engulfment of cytoplasmic cargo. Although the mechanism and relationship to disease largely remain unclear, microautophagy is known to be triggered by conditions similar to those that drive autophagy and CMA, such as starvation, nitrogen deprivation, and rapamycin treatment. This observation suggests that the three types of autophagy are likely regulated by similar signaling pathways. Several reports have also implied that membrane fusion systems in autophagy, such as Rab and ESCRT I/III, also contribute to microautophagy [27,28].
Regulatory Pathways of the ALP
Mammalian (or Mechanistic) Target of Rapamycin (mTOR)
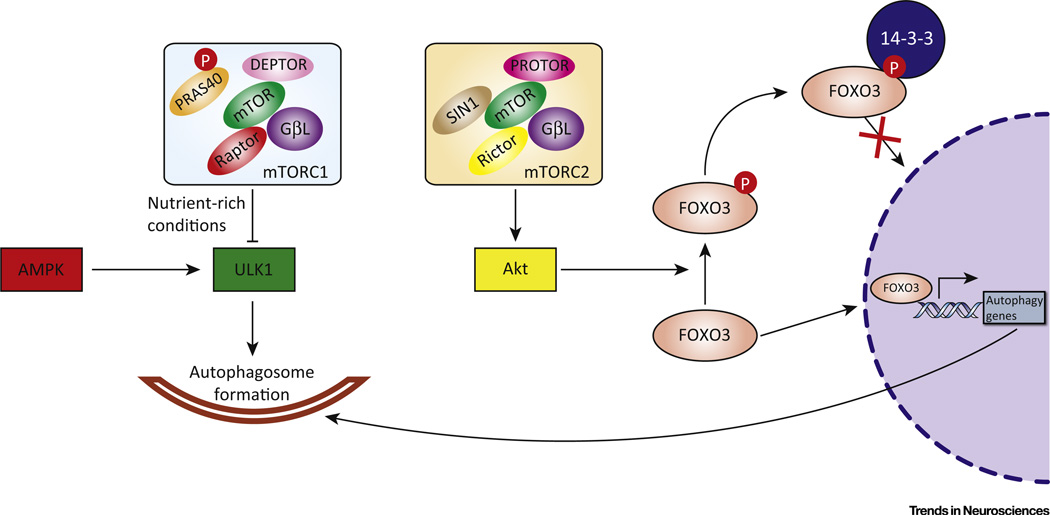
The best-studied regulator of mammalian autophagy is mTOR (Figure 2). Through mTOR, we have several well-defined pathways of the regulation of autophagy (Box 2).
Figure 2. mTOR Regulatory Pathways of the Autophagy–Lysosomal Pathway (ALP).
mTORC1 is composed of mTOR, regulatory associated protein of mTOR (Raptor), G protein β-subunit-like protein (GβL), proline-rich Akt substrate of 40 kDa (PRAS40), and DEP domain-containing mTOR-interacting protein (Deptor). PRAS40 is phosphorylated (P) by Akt and dissociates from Raptor to activate mTORC1. Activated mTORC1 (in nutrient-rich conditions or in response to growth factors, etc.) phosphorylates ULK1 to inhibit its role in autophagosome formation. AMPK, however, phosphorylates ULK1 at different sites to activate autophagy. mTORC2 is composed of mTOR, rapamycin-insensitive companion of mTOR (Rictor), GβL, stress-activated protein kinase-interacting protein (SIN) 1, and protein observed with Rictor (PROTOR). mTORC2 can also participate in autophagy regulation through the FOXO3 pathway. mTORC2 phosphorylates Akt, followed by Akt phosphorylation of FOXO3. Phosphorylated FOXO3 binds to 14-3-3 protein, which retains it in the cytoplasm, preventing activation of autophagy gene transcription.
Box 2. Regulation of the ALP via mTOR.
mTOR, a serine/threonine kinase, is the best-studied regulator of mammalian autophagy. This kinase is the catalytic subunit of mTOR complexes. Based on structural differences, mTOR complexes are classified as mTOR complex (mTORC) 1 and mTORC2. mTORC1 is composed of mTOR, regulatory associated protein of mTOR (Raptor), G protein β-subunit-like protein (GβL), proline-rich Akt substrate of 40 kDa (PRAS40), and DEP domain-containing mTOR-interacting protein (DEPTOR). mTORC2 is composed of mTOR, rapamycin-insensitive companion of mTOR (Rictor), GβL, stress-activated protein kinase-interacting protein (SIN) 1, and protein observed with Rictor (PROTOR) [114]. mTORC1 responds to several input signals including nutrients, growth factors, and cellular energy status [115]. The lysosomal surface is the site of activation for mTORC1, where it is bound and activated by Rag GTPases [36]. The V-ATPase complex on the lysosome is involved in amino acid sensing and interacts with Rag GTPases and Ragulator in an amino acid-dependent manner [37]. Amino acids activate Rags, with Ragulator mediating docking and activation of Rags followed by the physical binding of mTORC1 [116]. These aforementioned components are known as the LYNUS machinery. Under nutrient-rich conditions, mTORC1 phosphorylates ULK1 at Ser757 to inhibit ULK1 activity and autophagosome formation. By contrast, AMP-activated protein kinase (AMPK) competitively phosphorylates ULK1 at Ser317 and Ser777 to promote ULK1 activity and autophagy (see Figure 2 in main text). Although mTORC1 is the major sensor of nutrition and growth factor signals, autophagy can also be regulated by mTORC2 through the mTORC2–Akt–FoxO3 signaling pathway [117]. FoxO3 is a transcription factor that is activated in starvation conditions and promotes the transcription of genes that regulate autophagy induction [118]. Here, mTORC2 phosphorylates Akt at Ser473, and Akt in turn phosphorylates FoxO3 at Thr32, which induces cytosolic retention of FoxO3 through 14-3-3 binding, and thus transcriptionally inhibits autophagy (see Figure 2 in main text) [119]. A recent report also revealed that mTORC2 inhibits CMA through lysosomal Akt modulation of the assembly/disassembly of the LAMP-2A translocation complex, indicating that mTOR may mediate multiple autophagy pathways [120].
Role of TFEB in the ALP
Coordination of the multiple steps in the ALP and the need to adapt this process to different physiological and pathological conditions requires the existence of a master regulator. One such global regulator of the ALP is TFEB, a member of the MiT family of transcription factors that also includes TFE3, TFEC, and MITF (Box 3) [29]. TFEB is a basic helix-loop-helix leucine zipper transcription factor originally associated with renal carcinoma [30]. More recently, TFEB was identified as the transcription factor that binds to a promoter motif responsible for coordinating the expression of lysosomal genes, or the CLEAR element [1]. The coordinated lysosomal expression and regulation (CLEAR) network consists of genes involved in processes such as autophagy, lysosomal biogenesis, lysosomal exocytosis, endocytosis, and membrane repair [31]. TFEB was identified as a main player in coordinating autophagy through positively regulating autophagosome formation and autophagosome–lysosome fusion [2]. In addition, it enhances cellular clearance through lysosomal exocytosis, a process mediated by activation of the lysosomal Ca2+ channel MCOLN1 [12]. Thus, TFEB positively regulates genes key to cellular degradative pathways [1,31].
Box 3. The MiT Family of Transcription Factors.
This family of transcription factors consists of MITF, TFEB, TFE3, and TFEC. These basic helix-loop-helix leucine zipper transcription factors all bind to a similar E-box core sequence (CANNTG) as homodimers or heterodimers in combination with other family members [121,122]. Recent work demonstrates that TFE3 and MITF are also regulated by mTORC1 and promote the transcription of similar gene networks to those regulated by TFEB [61,123,124]. These overlapping roles in mammals indicate redundancy in this family of transcription factors, suggesting the possibility of compensation and/or differential expression in various cell types. With respect to murine CNS expression, all four family members are expressed, with TFEB and TFE3 mRNA being expressed at higher levels [125]. For all MiT family members, expression levels in microglia are particularly high, likely due to their role as phagocytic cells [125]. The significance of this differential expression in CNS cell types warrants further investigation.
Regulation of TFEB
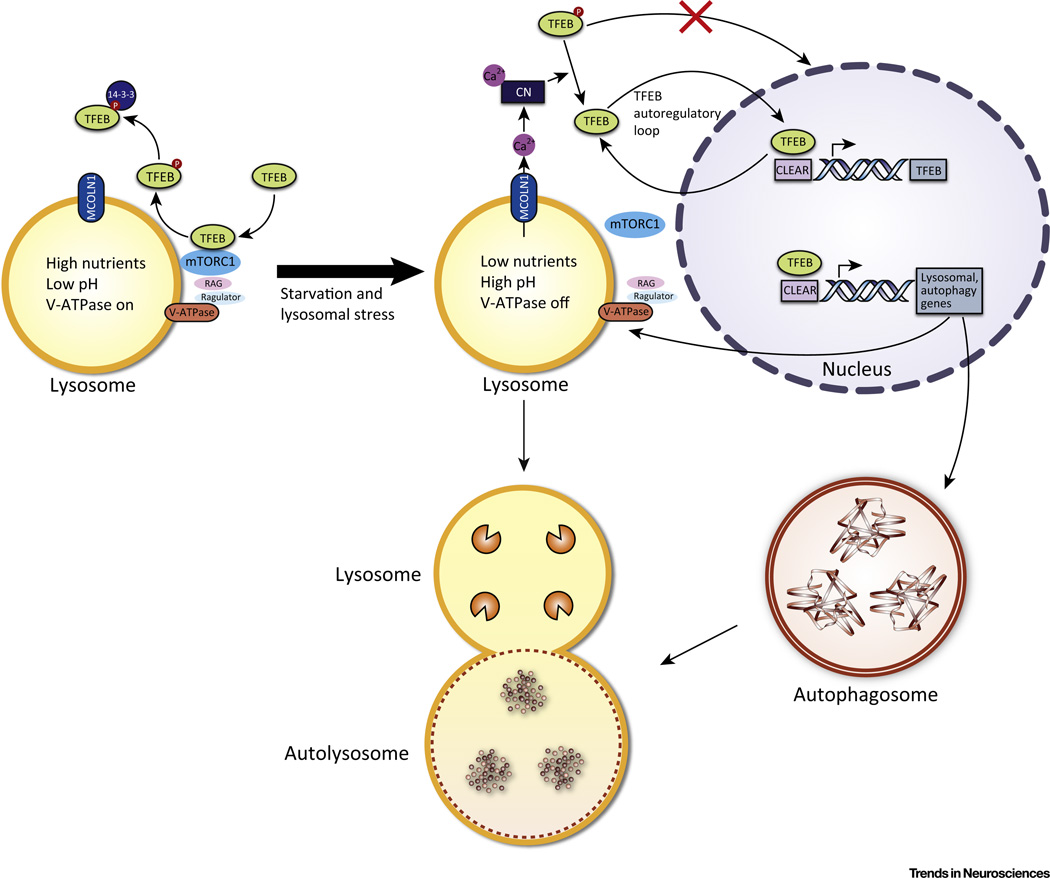
TFEB is an important, major regulator of the ALP, with its own regulation being a complex combination of autoregulatory networks and positive feedback loops. Crucially, the cell maintains homeostasis in stress conditions through lysosomal status triggering the transcription of genes necessary to correct perturbations, also known as lysosomal adaptation. Through such communication between the lysosome and nucleus, TFEB regulates metabolism and cellular clearance (Figure 3).
Figure 3. Regulation and Activity of TFEB.
In conditions of normal nutrient availability, no lysosomal stress, and no V-ATPase inhibition, the V-ATPase, Ragulator, and Rag GTPases form an active complex that binds to mTORC1 at the lysosomal surface and activates it. mTORC1 then phosphorylates (P) TFEB which sequesters it in the cytoplasm. Phosphorylated TFEB is also bound by 14-3-3 protein in the cytoplasm. In conditions of starvation, lysosomal stress, or V-ATPase inhibition, the Rag GTPases are turned off, which releases mTORC1 from the lysosomal surface and inactivates it. Because mTORC1 can no longer phosphorylate TFEB, the unphosphorylated TFEB translocates to the nucleus to bind to the CLEAR sequence of its target genes, leading to the upregulation of autophagy and lysosomal genes. The TFEB gene also has numerous CLEAR sequences in its promoter, and thus TFEB upregulates its own expression in an autoregulatory loop. Another positive feedback loop deals with MCOLN1, a lysosomal calcium efflux channel that is a transcriptional target of TFEB. MCOLN1 creates a microdomain of high Ca2+ concentration near the lysosomal surface. The higher Ca2+ concentration then activates the phosphatase calcineurin (CN), which dephosphorylates TFEB, promoting its activation.
Under normal conditions, TFEB is located in the cytoplasm. However, conditions of stress, such as starvation or lysosomal dysfunction, result in TFEB nuclear translocation, where it promotes the transcription of its target genes [1,2]. TFEB activity and nuclear translocation correlate with its phosphorylation state [2]. In particular, Ser142 and Ser211 were identified as being phosphorylated in nutrient-rich conditions, and mutations of these serines to alanines resulted in significantly increased nuclear localization of TFEB [2,32]. Two kinases are known to phosphorylate TFEB at these sites: ERK2 [2] and mTORC1 [32–35]. In addition to phosphorylation, members of the 14-3-3 protein family bind to phosphorylated TFEB to sequester it in the cytoplasm [33,34]. When TFEB is cytoplasmic, it can be located both in the cytosol and on the lysosomal surface, where it can interact with mTORC1 [32,33]. TFEB regulation by mTORC1 provides an illustration of where the lysosome, sensing nutrient status through the lysosome nutrient-sensing (LYNUS) machinery, signals to the nucleus to adapt to conditions of starvation or plenty. Amino acids and the V-ATPase interact to regulate Rag GTPases, which then activate mTORC1 by translocating it to the lysosomal surface [36,37]. On the lysosomal surface, mTORC1 binds to TFEB, facilitating its phosphorylation and cytoplasmic sequestration in nutrient-rich conditions [32]. Inhibiting mTORC1 with Torin 1 results in TFEB nuclear accumulation [32]. In addition, knockdown of the Rag GTPases, part of the lysosomal nutrient-sensing machinery that signal to mTORC1, leads to nuclear translocation of TFEB even in the presence of nutrients [32]. Finally, cellular conditions that are known to inactivate mTORC1, such as stress, starvation, and lysosomal inhibition, also result in TFEB nuclear localization and activation of its target genes [32–34]. In addition to being a major regulator of autophagy–lysosomal genes, TFEB also regulates its own expression through a starvation-induced autoregulatory loop [38]. The TFEB promoter contains multiple CLEAR elements to which the transcription factor binds to induce further TFEB expression [38]. TFEB is regulated by another positive feedback loop where a TFEB transcriptional target, the calcium channel MCOLN1, leads to TFEB dephosphorylation and nuclear translocation through calcineurin [39]. In starvation or high energy consumption, Ca2+ is released from the lysosome through MCOLN1, creating a Ca2+ microdomain near the lysosome [39]. This local increase in Ca2+ leads to the activation of the phosphatase calcineurin, which dephosphorylates TFEB, resulting in TFEB nuclear translocation and the transcription of target genes [39].
ALP and TFEB in Neurodegeneration
Huntington's Disease
HD is a neurodegenerative disease caused by expansion of a CAG trinucleotide repeat in the first exon of the huntingtin (HTT) gene. This results in a protein that contains an expanded polyglutamine (polyQ) tract, leading to misfolding into a pathogenic conformation. Despite the relatively simple genetic cause of HD, the etiology of the disease is vastly unknown. HTT is a large protein with ubiquitous cellular localization. Its normal function remains poorly defined, complicating understanding of the role of mutant HTT (mHTT) in HD. However, recent studies suggest that non-mutant HTT binds to p62 to facilitate its autophagy receptor function in selective autophagy, in addition to promoting autophagy by interacting with ULK1 to avoid negative regulation via mTORC1 [40]. Additional work suggests that the structural similarity between HTT and ATG11 allows HTT to serve as an ATG11-like scaffold protein for selective autophagy [41]. Previous findings of empty autophagosomes and defects in cargo sequestration in HD support the newly proposed role of nonmutant HTT in selective autophagy [42].
HTT is also thought to be a CMA substrate, given its three KFERQ-like motifs. Normal HTT degradation is enhanced by phosphorylation-regulated CMA [43]. Indeed, HTT degradation is increased by overexpression of HSC70 or LAMP2A and is decreased by knockdown of these genes in cellular models [43]. However, mHTT is less efficiently phosphorylated and degraded, a problem compounded by age-related decreases in lysosomal function [43]. More recent work has used a fusion molecule containing polyglutamine binding peptide 1 and HSC70 binding motifs to specifically target mHTT through its expanded polyQ tract for CMA-mediated degradation [44].
In addition to the loss of normal HTT function in HD, mHTT aggregates could provide a gain of function phenotype through their high affinity for other proteins. In particular, Beclin 1 and mTOR are sequestered in mHTT inclusions bodies [45,46]. In both cases, enhancing autophagy through Beclin 1 overexpression or rapalogue CCI-779 treatment promotes degradation of mHTT aggregates [45,46]. Furthermore, enhancing the ALP through TFEB overexpression has vast therapeutic effects. TFEB RNA levels and several of its target genes are reduced in HD mice, suggesting a deficiency in the ALP [7]. In vitro studies have shown that enhancing the ALP through TFEB overexpression reduces HTT protein aggregation in cells expressing huntingtin with the polyglutamine expansion [1,7]. TFEB was also identified as the downstream mediator and transcriptional target of PGC-1∝, which was shown to improve neurological function when overexpressed in a mouse model of HD [7]. In addition, the deficits in TFEB and its target genes were corrected with PGC-1∝ overexpression [7].
In general, enhancing autophagy in HD has proved beneficial. Animal models of HD treated with drugs targeting the ALP, for example CCI-779, rilmenidine, and trehalose, have been demonstrated to have therapeutic effects [46–48]. Enhancing the ALP through TFEB overexpression also has marked beneficial effects in ameliorating HD in mammalian cells and mouse models [1,7].
Parkinson's Disease
PD is a neurodegenerative disorder affecting dopaminergic neurons in the substantia nigra. A major hallmark of PD is the accumulation of proteinaceous cytoplasmic inclusions (Lewy bodies) in nigral dopaminergic neurons. The main component of Lewy bodies is misfolded and aggregated ∝-synuclein. As in other neurodegenerative disease, the misfolding and accumulation of protein aggregates correlates with the progression of PD pathogenesis. Studies indicate that ∝-synuclein accumulation is cytotoxic and blocks ER–Golgi vesicular trafficking, which may hinder autophagy [49]. Rab1, a protein important in ER–Golgi vesicular trafficking, was linked to macroautophagy regulation, with Rab1 overexpression rescuing ∝-synuclein-induced autophagy impairment [49]. In addition to affecting autophagy, PD-related ∝-synuclein mutations cause CMA dysfunction, resulting in ∝-synuclein accumulation and the disturbance of neuronal survival factor myocyte enhancer factor 2D [50]. Interestingly, carriers of mutations in genes associated with the LSDs Gaucher and Niemann–Pick A disease have an increased risk of developing PD, underscoring the importance of lysosomal degradation pathways in the disease [51,52]. Loss of glucocerebrosidase, the enzyme mutated in Gaucher's disease, results in accumulation of ∝-synuclein and neurotoxicity in cellular models [53]. In addition, ∝-synuclein inhibits normal glucocerebrosidase activity in neurons, suggesting a positive feedback loop in cases of mutation and a possible role in sporadic PD [53]. A mouse model expressing mutant glucocerebrosidase results in ∝-synuclein pathology and memory aberrations [54]. Restoring normal glucocerebrosidase activity ameliorates ∝-synuclein pathology and behavioral deficits in ∝-synuclein mice [55]. Furthermore, lysosomal deficiency is evident in other in vitro models and rodent models of PD, as well as in human PD [8,56]. Overexpressing TFEB or inducing its nuclear translocation erases this deficit and ameliorates ∝-synuclein pathology [8,56,57]. In postmortem human PD brains, nuclear TFEB is significantly reduced in the midbrain [8]. Interestingly, TFEB colocalizes with ∝-synuclein in Lewy body-containing nigral neurons in both human PD brains and in a rodent model of PD [8]. Furthermore, ∝-synuclein was found to bind to TFEB through coimmunoprecipitation, sequestering the transcription factor in the cytoplasm and preventing its activation [8]. Studies of TFEB in PD have also examined using pharmacological activation of TFEB as a potential therapy [56,57]. Trehalose, a disaccharide, is able to activate TFEB, evidenced by its nuclear translocation upon treatment, and results in reduced cell death in MPP+ intoxicated cells [56]. In addition, 2-hydroxypropyl-β-cyclodextrin also activates TFEB and enhances clearance of ∝-synuclein aggregates in human neuroglioma cells [57,58]. Finally, rapamycin, an FDA-approved drug, which activates autophagy through inhibition of mTOR (and thus stimulation of TFEB function) has been used in vitro and in vivo to reduce ∝-synuclein pathology [8,56].
While the aforementioned studies focus on the clearance of ∝-synuclein aggregates, defects in the handling of damaged mitochondria play a major role in PD pathogenesis. Up to 10% of PD cases are linked to genetic mutations. One of the well-known genes causing PD is SNCA, encoding ∝-synuclein, with two other genes associated with early-onset recessive PD being PARK2 and PARK6, encoding Parkin and PINK1, respectively. Parkin and PINK1 have crucial roles in mitophagy, as evidenced by Parkin and Pink1 knockout and mutant mouse models [59]. The hypothesis developed from these studies suggests that the accumulation of dysfunctional mitochondria due to failed mitophagy could play a significant role in PD pathogenesis. New therapeutic strategies have investigated restoring PINK1 kinase activity in loss of function mutants through the ATP analog kinetin triphosphate [60]. In addition, TFEB has recently been shown to play a role in mitophagy [61]. During this process, TFEB translocates to the nucleus and becomes transcriptionally active in a PINK1- and Parkin-dependent manner [61]. Furthermore, another recent study suggests that the reciprocal positive feedforward TFEB–PGC1∝ signaling pathway plays a crucial role in the clearance of damaged mitochondria and mitochondrial biogenesis in mice with a Q311X Parkin mutation [62]. In this case, TFEB–PGC1∝ signaling was reduced by the increased levels of PARIS, which is less efficiently bound and ubiquitinated by mutant Parkin [62]. This recently discovered role for TFEB in mitochondrial quality control expands the therapeutic potential of TFEB in PD beyond the ALP-dependent clearance of ∝-synuclein aggregates.
AD and Other Tauopathies
AD is the most common form of neurodegenerative disease. It is characterized by extracellular amyloid plaques consisting of β-amyloid peptides (Aβ) and by intracellular neurofibrillary tangles (NFTs) composed of hyperphosphorylated tau protein. AD can have direct genetic origins, such as mutations in amyloid precursor protein (APP) and presenilin (PS) 1 and 2. Loss of function or AD-related mutations in PS1 have been shown to result in failure of V-ATPase V0a1 subunit maturation and assembly of the V-ATPase complex, which is necessary for lysosomal acidification and protease activity [63,64]. Alternatively, another study demonstrates impaired lysosomal fusion capacity and accumulation of endomembranes in PS deficiency as a result of defective lysosomal calcium storage/release [65]. In addition, postmortem studies on human samples have revealed the accumulation of autophagosomes, multivesicular bodies, and autolysosomes in dystrophic neurites [66]. Furthermore, impairment of the ALP has been indicated in AD [5]. Specifically, the ALP has been shown to regulate APP turnover and Aβ metabolism [67–69], in addition to tau protein degradation [10,70–74].
The beneficial effect mediated by TFEB has been demonstrated in multiple mouse models of AD addressing Aβ and tau pathology [9–11,75]. With respect to tau, enhancing the ALP through exogenous TFEB expression in the brain dramatically reduces tau pathology, neurodegeneration, and behavioral deficits in the rTg4510 mouse model [10]. The proposed mechanism of action in this study operates via a TFEB–PTEN–Akt–mTOR–TFEB feedback regulatory loop, where TFEB induces the ALP through upregulation of PTEN and inhibition of Akt and mTOR, which further activates TFEB [10]. For amyloid pathology, TFEB expression in astrocytes facilitates a reduction in Aβ plaque pathology in the APP/PS1 mouse model by the proposed mechanism of astrocyte uptake and lysosomal degradation of Aβ [9]. In addition, intracranial stereotaxic injection of adeno-associated virus (AAV)–TFEB under the control of a cytomegalovirus (CMV) promoter in APP/PS1 mice leads to a reduction of APP, Aβ production, and plaque deposition via enhanced flux of APP through the endosome–lysosome pathway, resulting in lysosomal degradation of holo-APP [11]. Finally, the effect of glycogen synthase kinase 3 inhibition on APP degradation in vitro is thought to be mediated by enhancing the ALP through increasing TFEB nuclear localization and transcriptional activity [75].
While the beneficial effect of exogenous TFEB expression in these AD mouse models is clear, the role of endogenous TFEB in AD progression is less well defined. miR128 is a microRNA identified to be upregulated in the hippocampus of AD patients [76] and in monocytes derived from late-onset AD (LOAD) patients [77]. Of note, miR128 targets TFEB, leading to its downregulation [1,8,77]. In the case of LOAD patient-derived monocytes, lysosomal enzymes and Aβ degradative capacity are reduced compared to healthy donors, likely owing to a decrease in TFEB transcripts and nuclear localization caused by miR128 upregulation [77]. By contrast, patient-derived fibroblasts containing the familial AD PS1 mutation A246E display lysosomal alkalization and decreased cathepsin D activity, resulting in upregulation of ALP genes, in particular TFEB [78]. Furthermore, a study of PS conditional knockout mice failed to find defects in lysosomal acidification, but lysosomal genes containing the CLEAR sequence were transcriptionally upregulated [79]. A working hypothesis is that defects in membrane protein processing that result from PS/γ-secretase absence could result in protein accumulation in the lysosome, causing lysosomal stress and activating TFEB, much like LSDs and chloroquine treatment [1,15]. In addition, the 5 ×FAD mouse model, which contains five familial mutations related to AD (including two mutations in PS1), demonstrates transcriptomic upregulation of TFEB targets [80]. In these cases, TFEB is activated in response to lysosomal dysfunction, but is insufficient to address accumulating autophagosomes containing partially degraded material. However, in the absence of such a response to lysosomal stress the disease might progress even more rapidly, making the upregulation of endogenous TFEB and its targets a line of defense that is ultimately overwhelmed. Thus, exogenously induced expression of TFEB would be necessary to ameliorate the disease.
Based on recent studies in AD, HD, and PD, the role of TFEB in neurodegenerative disease may possibly be two-sided. On the one hand, TFEB inhibition or cytoplasmic sequestration could result in deficiency in the ALP, worsening the accumulation of pathologic aggregates. On the other hand, TFEB could be upregulated in response to lysosomal stress resulting from a preexisting insult such as mutation or dysfunction in other components of the ALP; however, it remains unable to compensate for increasing disease pathology. In both scenarios, the net output is TFEB insufficiency and should therefore be offset by enhanced TFEB activity.
Contrasting Roles of ALP in Neurodegeneration? Lessons from Prion Disease
While enhancing the ALP offers attractive therapeutic potential for clearing misfolded proteins common to many neurodegenerative diseases, understanding the ALP in physiological and disease context is crucial. An important lesson in the complexity of autophagy may be gained from examining prion disease, a unique infectious and fatal neurodegenerative disorder. In the protein-only hypothesis of prion disease, the cellular prion protein, PrPC, is converted to the pathogenic conformation PrPSc. Monomeric PrPSc forms the so-called ‘seed’ responsible for disease progression, where additional monomeric PrPSc is recruited to the two terminal ends to form an amyloid fiber [81]. The amyloid fiber can be fragmented to form more seeds, which then spread to neighboring cells to trigger a new round of fiber growth [81]. This hypothesis successfully interprets the infectious nature of prion disease. Although this pathological spreading was initially viewed to be unique to prion disease, recent studies revealed that other neurodegenerative diseases, AD and PD in particular, exhibit similar seeding and spreading phenomena [82–87].
In several prion models, autophagy is activated through both mTOR and Beclin 1 signaling pathways [88,89], and ALP activators such as rapamycin and trehalose decrease PrPSc in vitro and in vivo [90,91]. Paradoxically, the antimalarial drug quinacrine, which functions as an autophagy–lysosomal degradation inhibitor, has been shown to have therapeutic effects in prion disease models [92]. Although a randomized, double-blind, placebo-controlled clinical trial failed to show a positive effect on survival [93], the quinacrine studies beg the question if PrPSc seed formation requires autophagy–lysosomal function. During PrPSc fiber growth, fragmentation of large aggregates into smaller seeds, as exemplified by sonication in vitro, increases the number of terminals where monomeric PrPSc can be recruited, enhancing the spread of disease [94]. In this regard, it is tempting to speculate that insufficient autophagy–lysosomal degradation could potentially serve as an ‘innate sonication machine’ to produce more seeds and to exacerbate disease progression. Thus, quinacrine could function to inhibit seed formation by blocking the ALP. Remarkably, protein truncation by autophagy–lysosomal degradation has been shown to occur in AD- and PD-relevant conditions, with the truncated proteins having a higher propensity to form amyloid fibers [95,96]. Similarly to N-terminal truncation of PrPSc by the lysosome [97], APP and APP-processing enzymes BACE1 and γ-secretase are detected inside the autophagosome [98]. Tau is selectively cleaved by CMA, which generates a highly amyloidogenic tau fragment [70]. In PD, lysosomal cathepsin D has been shown to generate C-terminal truncation of ∝-synuclein [99], and inhibition of autophagy and CMA decreases seeding induced ∝-synuclein aggregation [100]. The increase in seeds would not only promote protein aggregation in a cell-autonomous manner but also promote the propagation of disease pathology, which has recently become an established mechanism in AD and PD progression, paralleling prion disease [82–86].
Concluding Remarks
Overwhelming evidence supports a crucial role of the ALP in the degradation of misfolded proteins that accumulate in numerous neurodegenerative disorders. Enhancing the ALP thus holds therapeutic promise in the treatment of neurodegenerative diseases. TFEB has emerged as a potent activator of the ALP by coordinating autophagy induction with lysosomal biogenesis. The fact that TFEB can be modulated by phosphorylation and activated through kinase inhibition, which is potentially druggable, makes it an attractive therapeutic target. However, as essential cellular degradative machinery, the level and duration of ALP and/or TFEB activation need to be tightly regulated. Constitutive activation of MiT/TFE proteins is hypothesized to drive metabolic reprogramming in pancreatic malignancy, and TFEB/TFE3 overexpression is a hallmark of some renal carcinomas [30,101]. Conversely, inefficient activation may not only be therapeutically ineffective but also has the potential for worsening neurodegenerative disease, chiefly by increasing aggregate formation and propagation. Studies of the ALP in prion disease and of endogenous TFEB suggest complexity in how this pathway functions in disease. Thus, in-depth understanding of TFEB and the ALP under various physiological and pathological conditions at the molecular, cellular, and functional levels will be necessary to decipher their role in disease pathogenesis and to inform therapeutic development (see Outstanding Questions).
Outstanding Questions.
Are there druggable targets via small molecules or others that activate TFEB and could potentially translate to human therapeutics? If such therapeutics are developed, what are the potential consequences of long-term therapeutic activation of the ALP that will be necessary to treat neurodegenerative diseases?
Do the autophagic defects observed in the various neurodegenerative diseases represent a causal factor or a compensatory response to disease progression? Could the answer potentially vary depending on disease type (and precise etiology)? In particular, what is the mechanistic explanation for the opposing status of TFEB activation in different neurodegenerative diseases, despite markedly similar features in terms of inhibition of autophagic flux and accumulation of autophagosomes (e.g., AD vs PD and HD)?
Are lysosomal defects/dysfunction a common consequence of aging that function as a common causative agent in neurodegenerative diseases?
Given the complexity of the role of autophagy in prion disease, could inhibition of autophagy play a role in therapeutic development to reduce seeding and prevent propagation in AD and PD? How will future therapeutics address the balance between the beneficial and detrimental roles of autophagy in the prion-like diseases?
Does the role and response of the ALP to neurodegenerative disease vary in a cell type-dependent manner? Can we modulate disease progression via astrocytes, microglia, etc. without directly targeting neurons? Could enhancing the ALP in these cell types ameliorate disease pathology without potentially enhancing seeding and propagation?
Trends.
Recent work has continued to build on the importance of the ALP in the degradation of misfolded proteins that accumulate in many neurodegenerative diseases, and on elucidating further mechanisms of how disease features interact with the pathway.
Enhancing the ALP in neurodegenerative diseases continues to be a focal point of therapeutic development. In particular, TFEB has emerged as a potent activator of the ALP by coordinating autophagy induction with lysosomal biogenesis, and its activation has successfully ameliorated disease in mouse models of various neurodegenerative disorders.
Recent studies reveal that AD and PD exhibit a seeding and spreading phenomenon similar to that of prion disease. This phenomenon has driven the field to focus on the mechanics of seeding and spreading, and the role that autophagy may play.
Acknowledgments
We thank members of the laboratories of H.Z. and A.B. for stimulating discussions. Results presented from the our laboratory were supported by R01 grants from the National Institutes of Health (NIH) (NS093652, AG020670, and NS076117) and the Belfer Neurodegeneration Consortium to H.Z., and the Italian Telethon Foundation, the Beyond Batten Disease Foundation, the European Research Council, and the NIH (R01 NS 078072-01A1) to A.B.
Glossary
- Autophagy–lysosomal pathway (ALP)
autophagy is a conserved degradation pathway. Three types of autophagy transport substrates to the lysosome where lysosomal enzymes degrade substrates.
- Autophagy-related genes (ATGs)
a class of genes encoding proteins facilitating the process of the ALP.
- Chaperone-mediated autophagy
the specific degradation of protein substrates with a pentapeptide KFERQ motif.
- Coordinated lysosomal expression and regulation (CLEAR) network
the lysosomal gene network regulated by TFEB. TFEB binds to a consensus motif known as the CLEAR sequence in the promoter region to promote transcription of these genes. The CLEAR sequence consists of a palindromic 10 bp GTCACGTGAC motif, typically within 200 bp of the transcription start-site, and can be a single sequence or arranged in tandem with multiple copies.
- Lysosome nutrient-sensing (LYNUS) machinery
located on the lysosomal surface, this responds to lysosomal amino acid content by signaling to the nucleus, often via TFEB activation or inactivation. LYNUS consists of the mTORC1 complex, the V-ATPase complex, Rag GTPases, Ragulator, and Rheb. The V-ATPase complex senses amino acids and interacts with Rags and Ragulator. Rag GTPases physically bind to mTORC1 on the lysosomal surface and activate it. Activated mTORC1 phosphorylates TFEB to retain it in the cytoplasm. During starvation, mTORC1 is released from the LYNUS machinery and becomes inactive. mTORC1 can no longer phosphorylate TFEB, and unphosphorylated TFEB can then translocate to the nucleus to activate its target genes.
- Lysosomal adaptation
the concept that lysosomal genes are regulated in such a way to provide optimal lysosomal function in various physiological and pathological conditions. TFEB is one such global regulator of the lysosomal gene network that responds to stress, starvation, and nutrient abundance, among other stimuli.
- Macroautophagy
functions within the cell to degrade old or damaged organelles, as well as long-lived or aggregated proteins too large to be handled by the proteasome. Basal autophagy functions in maintenance of the healthy cell.
- Microautophagy
the direct engulfment of cytoplasmic cargo by the lysosome.
- Prion
a term used to describe an infectious protein particle. Prions can self-propagate and spread from one cell to another in a manner similar to a virus.
- Seeding model
a hypothetical model to describe prion replication. Seeds are oligomeric disease-associated forms of prion protein (PrPSc) that recruit monomeric cellular prion protein (PrPC) to form amyloid fibers. The amyloid fiber is fragmented to oligomers again to form more seeds and accelerate prion propagation.
- Selective autophagy
ensures proper organelle, macromolecule, and protein turnover in the cytosol. The process is also thought to arise in conditions of stress, specifically degrading protein aggregates and damaged mitochondria.
References
- 1.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 2.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 4.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 5.Menzies FM, et al. Compromised autophagy and neuro-degenerative diseases. Nat. Rev. Neurosci. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto A, Simonsen A. The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol. Dis. 2011;43:17–28. doi: 10.1016/j.nbd.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsunemi T, et al. PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 2012;4:142ra197. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decressac M, et al. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E1817–E1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao Q, et al. Enhancing astrocytic lysosome biogenesis facilitates Ab clearance and attenuates amyloid plaque pathogenesis. J. Neurosci. 2014;34:9607–9620. doi: 10.1523/JNEUROSCI.3788-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polito VA, et al. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol. Med. 2014;6:1142–1160. doi: 10.15252/emmm.201303671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao Q, et al. Neuronal-targeted TFEB accelerates lysosomal degradation of APP, reducing Abeta generation and amyloid plaque pathogenesis. J. Neurosci. 2015;35:12137–12151. doi: 10.1523/JNEUROSCI.0705-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medina DL, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell. 2011;21:421–430. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes CJ, et al. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat. Neurosci. 2014;17:1180–1189. doi: 10.1038/nn.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spampanato C, et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 2013;5:691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song W, et al. TFEB regulates lysosomal proteostasis. Hum. Mol. Genet. 2013;22:1994–2009. doi: 10.1093/hmg/ddt052. [DOI] [PubMed] [Google Scholar]
- 16.Mizushima N, et al. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 17.Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 18.Ravikumar B, et al. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng J, Klionsky DJ. The Golgi as a potential membrane source for autophagy. Autophagy. 2010;6:950–951. doi: 10.4161/auto.6.7.13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamark T, Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int. J. Cell Biol. 2012;2012:736905. doi: 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K, Klionsky DJ. Mitochondria removal by autophagy. Autophagy. 2011;7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogov V, et al. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawamura N, et al. Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat. Commun. 2012;3:1071. doi: 10.1038/ncomms2069. [DOI] [PubMed] [Google Scholar]
- 28.Sahu R, et al. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rehli M, et al. Cloning and characterization of the murine genes for bHLH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics. 1999;56:111–120. doi: 10.1006/geno.1998.5588. [DOI] [PubMed] [Google Scholar]
- 30.Kuiper RP, et al. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum. Mol. Genet. 2003;12:1661–1669. doi: 10.1093/hmg/ddg178. [DOI] [PubMed] [Google Scholar]
- 31.Palmieri M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 32.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martina JA, et al. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roczniak-Ferguson A, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pena-Llopis S, et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30:3242–3258. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sancak Y, et al. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+ - ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Settembre C, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medina DL, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rui YN, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochaba J, et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. U.S.A. 2014;111:16889–16894. doi: 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Vicente M, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson LM, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi L, et al. The role of chaperone-mediated autophagy in huntingtin degradation. PLoS ONE. 2012;7:e46834. doi: 10.1371/journal.pone.0046834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata M, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 46.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 47.Rose C, et al. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington's disease. Hum. Mol. Genet. 2010;19:2144–2153. doi: 10.1093/hmg/ddq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka M, et al. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat. Med. 2004;10:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 49.Winslow AR, et al. α-Synuclein impairs macroautophagy: implications for Parkinson's disease. J. Cell Biol. 2010;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Q, et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gan-Or Z, et al. Genotype–phenotype correlations between GBA mutations and Parkinson disease risk and onset. Neurology. 2008;70:2277–2283. doi: 10.1212/01.wnl.0000304039.11891.29. [DOI] [PubMed] [Google Scholar]
- 52.Gan-Or Z, et al. The p.L302P mutation in the lysosomal enzyme gene SMPD1 is a risk factor for Parkinson disease. Neurology. 2013;80:1606–1610. doi: 10.1212/WNL.0b013e31828f180e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mazzulli JR, et al. Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146:37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sardi SP, et al. CNS expression of glucocerebrosidase corrects alpha-synuclein pathology and memory in a mouse model of Gaucher-related synucleinopathy. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12101–12106. doi: 10.1073/pnas.1108197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sardi SP, et al. Augmenting CNS glucocerebrosidase activity as a therapeutic strategy for parkinsonism and other Gaucher-related synucleinopathies. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3537–3542. doi: 10.1073/pnas.1220464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dehay B, et al. Pathogenic lysosomal depletion in Parkinson's disease. J. Neurosci. 2010;30:12535–12544. doi: 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kilpatrick K, et al. Genetic and chemical activation of TFEB mediates clearance of aggregated alpha-synuclein. PLoS ONE. 2015;10:e0120819. doi: 10.1371/journal.pone.0120819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song W, et al. 2-Hydroxypropyl-beta-cyclodextrin promotes transcription factor EB-mediated activation of autophagy: implications for therapy. J. Biol. Chem. 2014;289:10211–10222. doi: 10.1074/jbc.M113.506246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hertz NT, et al. A neo-substrate that amplifies catalytic activity of Parkinson's-disease-related kinase PINK1. Cell. 2013;154:737–747. doi: 10.1016/j.cell.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nezich CL, et al. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 2015;210:435–450. doi: 10.1083/jcb.201501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siddiqui A, et al. Mitochondrial quality control via the PGC1alpha–TFEB signaling pathway is compromised by parkin Q311X mutation but independently restored by rapamycin. J. Neurosci. 2015;35:12833–12844. doi: 10.1523/JNEUROSCI.0109-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JH, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JH, et al. Presenilin 1 maintains lysosomal Ca2+ homeostasis via TRPML1 by regulating vATPase-mediated lysosome acidification. Cell Rep. 2015;12:1430–1444. doi: 10.1016/j.celrep.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coen K, et al. Lysosomal calcium homeostasis defects, not proton pump defects, cause endo-lysosomal dysfunction in PSEN-deficient cells. J. Cell Biol. 2012;198:23–35. doi: 10.1083/jcb.201201076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nixon RA, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 67.Pickford F, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J. Clin. Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rohn TT, et al. Depletion of Beclin-1 due to proteolytic cleavage by caspases in the Alzheimer's disease brain. Neurobiol. Dis. 2011;43:68–78. doi: 10.1016/j.nbd.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang DS, et al. Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer's disease ameliorates amyloid pathologies and memory deficits. Brain. 2011;134:258–277. doi: 10.1093/brain/awq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kruger U, et al. Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol. Aging. 2012;33:2291–2305. doi: 10.1016/j.neurobiolaging.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 72.Schaeffer V, et al. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain. 2012;135:2169–2177. doi: 10.1093/brain/aws143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caccamo A, et al. mTOR regulates tau phosphorylation and degradation: implications for Alzheimer's disease and other tauopathies. Aging Cell. 2013;12:370–380. doi: 10.1111/acel.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ozcelik S, et al. Rapamycin attenuates the progression of tau pathology in P301S tau transgenic mice. PLoS ONE. 2013;8:e62459. doi: 10.1371/journal.pone.0062459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parr C, et al. Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-beta precursor protein. Mol. Cell. Biol. 2012;32:4410–4418. doi: 10.1128/MCB.00930-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 77.Tiribuzi R, et al. miR128 up-regulation correlates with impaired amyloid beta(1–42) degradation in monocytes from patients with sporadic Alzheimer's disease. Neurobiol. Aging. 2014;35:345–356. doi: 10.1016/j.neurobiolaging.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 78.Coffey EE, et al. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer's disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience. 2014;263:111–124. doi: 10.1016/j.neuroscience.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X, et al. A role for presenilins in autophagy revisited: normal acidification of lysosomes in cells lacking PSEN1 and PSEN2. J. Neurosci. 2012;32:8633–8648. doi: 10.1523/JNEUROSCI.0556-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Landel V, et al. Temporal gene profiling of the 5×FAD transgenic mouse model highlights the importance of microglial activation in Alzheimer's disease. Mol. Neurodegener. 2014;9:33. doi: 10.1186/1750-1326-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aguzzi A, Sigurdson CJ. Antiprion immunotherapy: to suppress or to stimulate? Nat. Rev. Immunol. 2004;4:725–736. doi: 10.1038/nri1437. [DOI] [PubMed] [Google Scholar]
- 82.Stohr J, et al. Purified and synthetic Alzheimer's amyloid beta (Abeta) prions. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11025–11030. doi: 10.1073/pnas.1206555109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanders DW, et al. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron. 2014;82:1271–1288. doi: 10.1016/j.neuron.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iba M, et al. Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer's-like tauopathy. J. Neurosci. 2013;33:1024–1037. doi: 10.1523/JNEUROSCI.2642-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clavaguera F, et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luk KC, et al. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jaunmuktane Z, et al. Evidence for human transmission of amyloid-beta pathology and cerebral amyloid angiopathy. Nature. 2015;525:247–250. doi: 10.1038/nature15369. [DOI] [PubMed] [Google Scholar]
- 88.Xu Y, et al. Activation of the macroautophagic system in scrapie-infected experimental animals and human genetic prion diseases. Autophagy. 2012;8:1604–1620. doi: 10.4161/auto.21482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Joshi-Barr S, et al. De novo prion aggregates trigger autophagy in skeletal muscle. J. Virol. 2014;88:2071–2082. doi: 10.1128/JVI.02279-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cortes CJ, et al. Rapamycin delays disease onset and prevents PrP plaque deposition in a mouse model of Gerstmann–Straussler–Scheinker disease. J. Neurosci. 2012;32:12396–12405. doi: 10.1523/JNEUROSCI.6189-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aguib Y, et al. Autophagy induction by trehalose counter-acts cellular prion infection. Autophagy. 2009;5:361–369. doi: 10.4161/auto.5.3.7662. [DOI] [PubMed] [Google Scholar]
- 92.Doh-Ura K, et al. Lysosomotropic agents and cysteine protease inhibitors inhibit scrapie-associated prion protein accumulation. J. Virol. 2000;74:4894–4897. doi: 10.1128/jvi.74.10.4894-4897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Geschwind MD, et al. Quinacrine treatment trial for sporadic Creutzfeldt–Jakob disease. Neurology. 2013;81:2015–2023. doi: 10.1212/WNL.0b013e3182a9f3b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Castilla J, et al. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 95.Wischik CM, et al. Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4506–4510. doi: 10.1073/pnas.85.12.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Michell AW, et al. The effect of truncated human alpha-synuclein (1–120) on dopaminergic cells in a transgenic mouse model of Parkinson's disease. Cell Transplant. 2007;16:461–474. doi: 10.3727/000000007783464911. [DOI] [PubMed] [Google Scholar]
- 97.Caughey B, et al. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J. Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu WH, et al. Macroautophagy – a novel beta-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sevlever D, et al. Cathepsin D is the main lysosomal enzyme involved in the degradation of alpha-synuclein and generation of its carboxy-terminally truncated species. Biochemistry. 2008;47:9678–9687. doi: 10.1021/bi800699v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsujimura A, et al. Lysosomal enzyme cathepsin B enhances the aggregate forming activity of exogenous alpha-synuclein fibrils. Neurobiol. Dis. 2014;73C:244–253. doi: 10.1016/j.nbd.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 101.Perera RM, et al. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jung CH, et al. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fan W, et al. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc. Natl. Acad. Sci. U.S.A. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 105.Obara K, Ohsumi Y. Dynamics and function of PtdIns(3)P in autophagy. Autophagy. 2008;4:952–954. doi: 10.4161/auto.6790. [DOI] [PubMed] [Google Scholar]
- 106.Shao Y, et al. Stimulation of ATG12–ATG5 conjugation by ribonucleic acid. Autophagy. 2007;3:10–16. doi: 10.4161/auto.3270. [DOI] [PubMed] [Google Scholar]
- 107.Fujioka Y, et al. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J. Biol. Chem. 2010;285:1508–1515. doi: 10.1074/jbc.M109.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Satoo K, et al. The structure of Atg4B–LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Metlagel Z, et al. Structural insights into E2–E3 interaction for LC3 lipidation. Autophagy. 2014;10:522–523. doi: 10.4161/auto.27594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nath S, et al. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 2014;16:415–424. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ravikumar B, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 112.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 113.Itakura E, et al. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 114.Zoncu R, et al. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jewell JL, et al. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bar-Peled L, et al. Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lamming DW, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 119.Tzivion G, et al. FoxOtranscription factors; regulation by AKT and 14-3-3 proteins. Biochim. Biophys. Acta. 2011;1813:1938–1945. doi: 10.1016/j.bbamcr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 120.Arias E, et al. Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Mol. Cell. 2015;59:270–284. doi: 10.1016/j.molcel.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hemesath TJ, et al. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 122.Pogenberg V, et al. Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev. 2012;26:2647–2658. doi: 10.1101/gad.198192.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Martina JA, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 2014;7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ploper D, et al. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E420–E429. doi: 10.1073/pnas.1424576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]