Octopamine acts as a metabolic sensor that links environmental nutrient signals to energy homeostasis in C. elegans.
Keywords: Octopamine, C. elegans, DAF-12, lipid hydrolysis, starvation
Abstract
Starvation is probably the most common stressful situation in nature. In vertebrates, elevation of the biogenic amine norepinephrine levels is common during starvation. However, the precise role of norepinephrine in nutrient deprivation remains largely unknown. We report that in the free-living nematode Caenorhabditis elegans, up-regulation of the biosynthesis of octopamine, the invertebrate counterpart of norepinephrine, serves as a mechanism to adapt to starvation. During nutrient deprivation, the nuclear receptor DAF-12, known to sense nutritional cues, up-regulates the expression of tbh-1 that encodes tyramine β-hydroxylase, a key enzyme for octopamine biosynthesis, in the RIC neurons. Octopamine induces the expression of the lipase gene lips-6 via its receptor SER-3 in the intestine. LIPS-6, in turn, elicits lipid mobilization. Our findings reveal that octopamine acts as an endocrine regulator linking nutrient cues to lipolysis to maintain energy homeostasis, and suggest that such a mechanism may be evolutionally conserved in diverse organisms.
INTRODUCTION
Biogenic amines modulate a variety of physiological functions in animals (1). Octopamine, a functional analogue of mammalian norepinephrine, has been implicated in the responses associated with the absence of food in invertebrate species, such as insects and worms. In fruit flies Drosophila melanogaster, octopamine promotes locomotor activity and foraging behavior in response to starvation (2, 3). In the free-living nematode Caenorhabditis elegans, exogenous application of octopamine can inhibit egg laying and pharyngeal pumping, which are normally promoted by the presence of food (4, 5). Thus, it seems that octopamine can mimic a starvation-like signal. In some species of vertebrates, elevation of norepinephrine levels has been observed under starvation conditions (6–9). However, the precise role of octopamine or norepinephrine in nutrient deprivation remains largely unknown.
C. elegans can undergo physiological and behavioral changes to cope with nutrient deprivation. For instance, starvation alters C. elegans pharyngeal muscle function to enhance ingestion when food becomes available again (10). Meanwhile, L4 and young adult worms can maintain energy balance through increases in lipid hydrolysis by a variety of lipases during starvation (11–13). For example, starvation mediates the induction of genes involved in converting fat stores into energy production, thus allowing worms to survive (14). Similarly, the endoplasmic reticulum proteins IRE-1 and HSP-4 recognize nutritional changes in response to starvation, resulting in up-regulation of lipase genes fil-1 and fil-2 (11). These lipases, in turn, break down fat storage, thus maintaining whole-body energy homeostasis. In addition, the lysosomal lipases, such as LIPL-1 and LIPL-3, promote the fat mobilization through lipophagy, which is tightly regulated by two transcription factors, MXL-3 and HLH-30 (12). When food is available, MXL-3 inhibits the expression of these lysosomal lipase genes. Following fasting, HLH-30 elicits the activation of lipophagy by up-regulating lysosomal lipase and autophagy genes.
Endocrine systems regulate a variety of physiological processes in response to environmental cues. In C. elegans, the nuclear receptor DAF-12 acts as a dietary and environmental sensor to orchestrate diverse aspects of development, metabolism, and reproduction. The regulation of DAF-12 is tightly linked to its steroidal ligands Δ4- and Δ7-dafachronic acids (DAs). Favorable environments, such as an abundant food supply, activate DAF-9, a cytochrome P450 enzyme that catalyzes the last step of DA production from cholesterol (15). By binding with DA, DAF-12 promotes reproductive development under favorable conditions. When nematodes detect environmental stresses, such as food limitation, these pathways are inhibited, leading to reduced production of DA (16, 17). In times of food scarcity, the production of the ligand DA is inhibited, leading to a shift of the liganded DAF-12 (DAF-12/DA) toward the unliganded DAF-12/DIN-1 complex (16, 18). The fact that DAF-12/DIN-1 is dominant during starvation implicates a role of this complex in regulation of starvation responses.
Here, we uncovered that disruption of tbh-1, a gene coding for tyramine β-hydroxylase that is required for octopamine biosynthesis (4), resulted in enhanced susceptibility to starvation. Octopamine promoted a lipase LIPS-6–mediated lipid mobilization via its receptor SER-3 in the intestine. Finally, our results demonstrated that the expression of tbh-1 was up-regulated by the DAF-12/DIN-1 complex.
RESULTS
Up-regulation of octopamine biosynthesis is required for starvation resistance
It has been shown that the octopamine signaling is activated after food deprivation in C. elegans (19). In C. elegans, tbh-1 encodes tyramine β-hydroxylase, which catalyzes the conversion of tyramine to octopamine (4). We thus determined the expression of tbh-1 using transgenic animals carrying the tbh-1 promoter fused to green fluorescent protein (GFP). We found that the expression of Ptbh-1::gfp was markedly up-regulated after worms were starved for 12 hours, compared with worms fed with laboratory food Escherichia coli OP50 during this time (Fig. 1A). These results were confirmed by determining the expression of tbh-1 using quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 1B). To determine the octopamine content in worms, we used liquid chromatography–tandem mass spectrometry (LC-MS/MS). We found that the octopamine levels were higher in worms after 12 hours of starvation than in well-fed worms (Fig. 1C and fig. S1).
Fig. 1. Biosynthesis of octopamine is up-regulated during starvation.
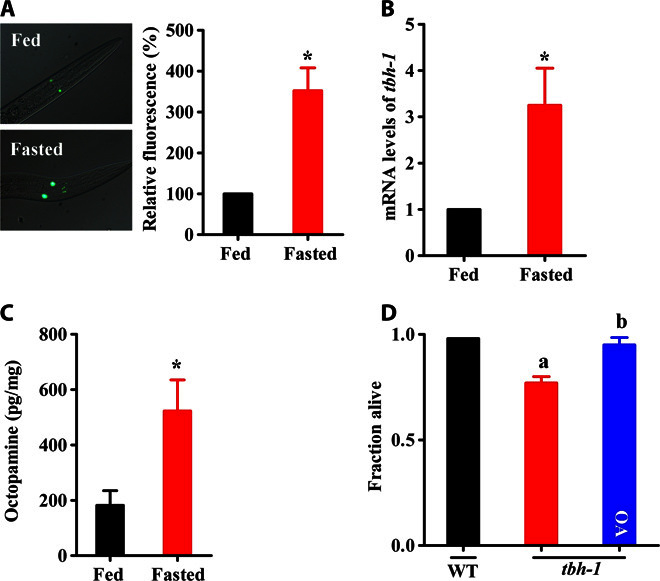
(A) Expression of Ptbh-1::gfp was up-regulated in starved worms relative to well-fed worms (in the presence of E. coli OP50). The right panel shows quantification of GFP levels. (B) The mRNA levels of tbh-1 were determined by qRT-PCR. (C) The levels of octopamine in starved worms were higher than those in well-fed worms. Results are means ± SD of three experiments. *P < 0.05 versus well-fed worms. (D) Survival rate of tbh-1(n3247) worms was lower than that of wild-type (WT) worms after 3 days of starvation. Exogenous octopamine (OA; 1 mM) markedly restored the survival of tbh-1(n3247) worms. Results are means ± SD of three experiments. aP < 0.05, tbh-1(n3247) versus WT worms; bP < 0.05, tbh-1 + octopamine versus tbh-1.
The fact that starvation enhanced the biosynthesis of octopamine led us to investigate its role in starvation resistance. We found that tbh-1(n3247) worms had lower survival rates after 3 days of starvation (Fig. 1D). Exogenous application of octopamine (1 mM) significantly restored the survival of tbh-1(n3247) worms to a similar extent as that of wild-type worms. Collectively, these results suggest that octopamine, which is elevated in starvation, promotes worm survival under nutrient deprivation.
Octopamine mediates lipid mobilization by up-regulating lips-6 expression during starvation
Next, we investigated the molecular basis of octopamine function in starvation resistance. Like mammals, C. elegans consumes stored lipid droplets in response to nutrient deprivation (11–13). Consistent with these data, we found that the quantity of lipid droplets in the intestine, as judged by Oil Red O staining, was reduced by approximately 60% in wild-type worms after 1 day of starvation (Fig. 2A). In contrast, a mutation in tbh-1(n3247) did not affect lipid hydrolysis in starved worms. However, the quantity of lipid droplets in tbh-1(n3247) worms was comparable to that in wild-type worms in the presence of E. coli OP50. Furthermore, we extracted total lipids, separated them by thin-layer chromatography (TLC) (fig. S2), and quantified triacylglycerol (TAG) content by gas chromatography (GC)–MS. We found that the TAG content was significantly lower in wild-type than in tbh-1(n3247) worms after starvation (Fig. 2B). Application of octopamine promoted lipid hydrolysis in starved tbh-1(n3247) worms (both fed and fasted states) and well-fed wild-type worms (Fig. 2, A and B). Finally, overexpression of tbh-1 in the RIC neurons enhanced lipid hydrolysis in well-fed wild-type worms (fig. S3). These results suggest that TBH-1/octopamine signaling mediates lipid hydrolysis.
Fig. 2. Octopamine promotes lipid hydrolysis by up-regulating the expression of the lipase gene lips-6.
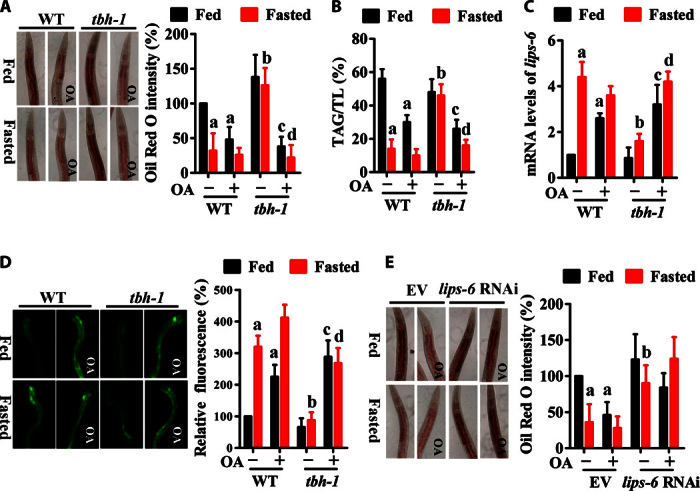
(A and B) Lipid mobilization was regulated by octopamine. (A) The quantity of lipid droplets determined by Oil Red O staining in WT worms was significantly lower than that in tbh-1(n3247) worms after 24 hours of starvation. Octopamine (1 mM) treatment promoted lipid hydrolysis in either starved tbh-1(n3247) worms or well-fed WT worms. The right panel represents relative Oil Red O intensity. (B) The relative TAG contents were determined by GC-MS. Results are means ± SD of three experiments. TL, total lipid. (C and D) The mRNA levels of lips-6 (C) and the expression of Plips-6::gfp (D) were up-regulated by octopamine after 12 hours of starvation. Results are means ± SD of three experiments. aP < 0.05 versus well-fed WT worms; bP < 0.05 versus starved WT worms; cP < 0.05 versus well-fed tbh-1(n3247) worms; dP < 0.05 versus starved tbh-1(n3247) worms. (E) Lipid hydrolysis was suppressed by lips-6 RNAi after 24 hours of starvation. However, exogenous octopamine (1 mM) failed to restore lipid hydrolysis in lips-6 (RNAi) worms. The right panel represents relative Oil Red O intensity. Results are means ± SD of three experiments. aP < 0.05 versus well-fed worms + empty vector (EV); bP < 0.05 versus starved worms + empty vector.
It has been shown that the serotonin [5-hydroxytryptamine (5-HT)]/octopamine signaling elicits lipid hydrolysis despite food abundance (20). The function of octopamine and its receptor SER-6 is to maintain 5-HT synthesis. 5-HT production in the ADF neurons activates the chloride channel MOD-1 in the URX neurons, leading to up-regulation of the lipase gene atgl-1 in the intestine via the nuclear receptor NHR-76. ATGL-1, in turn, mediates lipid hydrolysis in well-fed worms. Consistent with the observations from Lee et al. (21), the mutation in atgl-1(tm3116) also led to lipid accumulation in starved worms (fig. S4A). However, we found that nutrient deprivation did not alter either the mRNA level of atgl-1 or the expression of Patgl-1::atgl-1::GFP (fig. S4, B and C). Meanwhile, the mutation in tbh-1(n3247) did not alter the expression of Patgl-1::atgl-1::GFP during starvation (fig. S4C), suggesting that octopamine does not regulate atgl-1 expression. Octopamine treatment does not alter the expression of Patgl-1::atgl-1::GFP in well-fed worms (20). In any case, our results suggest that ATGL-1 is not involved in octopamine-induced lipid hydrolysis during starvation.
A previous study using microarray analysis indicated that starvation significantly up-regulated the expression of three TAG lipases (fil-1, fil-2, and lips-6), and suggested that these three genes are probably involved in lipid hydrolysis (11). We thus investigated the role of these TAG lipases on the lipid mobilization mediated by the TBH-1/octopamine signaling. We found that, although the expression of fil-1, fil-2, and lips-6 was up-regulated by starvation, only lips-6 was inhibited by a mutation in tbh-1(n3247) (Fig. 2C and fig. S5). Using transgenic animals that express Plips-6::gfp, we observed that lips-6 was expressed in the intestine. Starvation significantly increased the expression of Plips-6::gfp in wild-type, but not in tbh-1(n3247), worms (Fig. 2D). Furthermore, the addition of octopamine restored the mRNA levels of lips-6 as well as the expression of Plips-6::gfp in tbh-1(n3247) mutants (both fed and fasted states) and well-fed wild-type worms. To test the role of lips-6 in lipid hydrolysis, we knocked down lips-6 using RNA interference (RNAi). We found that endogenous lips-6 expression was markedly ablated by RNAi (fig. S6). Knockdown of lips-6 significantly suppressed lipid hydrolysis during starvation (Fig. 2E), whereas overexpression of lips-6 promoted lipid hydrolysis in well-fed worms (fig. S7). However, application of octopamine failed to restore lipid hydrolysis in worms subjected to lips-6 RNAi. Together, these results suggest that the TBH-1/octopamine signaling promotes lipid hydrolysis through LIPS-6.
Octopamine mediates lipid mobilization via its receptor SER-3
There are three octopamine receptors: SER-3, SER-6, and OCTR-1 (22). We thus tested whether these receptors are involved in lipid hydrolysis during starvation. A mutation in ser-3(ad1774), but not ser-6(tm2146) or octr-1(ok371), significantly inhibited lipid hydrolysis (Fig. 3, A and B). Likewise, the mRNA levels of lips-6 were also reduced in ser-3(ad1774), but not ser-6(tm2146) or octr-1(ok371), worms during starvation (Fig. 3C). Furthermore, the addition of octopamine failed to restore lipid hydrolysis and the mRNA levels of lips-6 in ser-3(ad1774) mutants (fig. S8, A and B).
Fig. 3. Octopamine promotes lipid hydrolysis via its receptor SER-3.
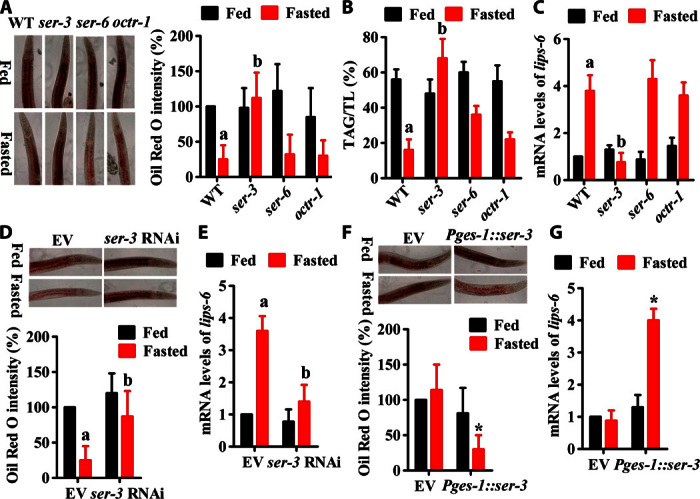
(A and B) Lipid hydrolysis was inhibited in ser-3(ad1774), but not ser-6(tm2146) or octr-1(ok371), mutants after 24 hours of starvation. (A) Oil Red O staining. The right panel represents relative Oil Red O intensity. (B) The relative TAG contents were determined by GC-MS. Results are means ± SD of three experiments. (C) The mRNA levels of lips-6 were also reduced in ser-3(ad1774), but not ser-6(tm2146) or octr-1(ok371), worms during starvation. Results are means ± SD of three experiments. aP < 0.05 versus well-fed WT worms; bP < 0.05 versus starved WT worms. (D and E) Intestinal-specific knockdown of ser-3 by RNAi reduced lipid hydrolysis (D) and the expression of lips-6 (E). Results are means ± SD of three experiments. aP < 0.05 versus well-fed worms + empty vector; bP < 0.05 versus starved worms + empty vector. (F and G) Expression of ser-3 under the control of the intestinal ges-1 markedly restored lipid hydrolysis (F) and the expression of lips-6 (G) in ser-3(ad1774) mutants. Results are means ± SD of three experiments. *P < 0.05 versus empty vector.
ser-3 is expressed in neurons in the head and tail, muscles, and intestine (23). To determine tissue-specific activities of SER-3 in the regulation of lipid hydrolysis, we knocked down ser-3 by RNAi in the neurons, muscle, and intestine. However, we found that neither neuronal- nor muscular-specific RNAi of ser-3 had an effect on lipid hydrolysis (fig. S9, A and C) and the expression of lips-6 (fig. S9, B and D). In contrast, intestinal-specific knockdown of ser-3 by RNAi reduced lipid hydrolysis and the expression of lips-6 (Fig. 3, D and E). Furthermore, expression of ser-3 under the control of the intestinal-specific ges-1 promoter (24) markedly restored lipid hydrolysis and the expression of lips-6 in ser-3(ad1774) mutants (Fig. 3, F and G). As expected, ser-3(ad1774) mutants exhibited reduced rates of survival after 3 days of starvation (fig. S10). Expression of ser-3 in the intestine markedly suppressed the sensitivity of ser-3(ad1774) mutants to starvation. These results suggest that octopamine promotes lipid hydrolysis through its receptor SER-3.
The expression of tbh-1 is regulated by the DAF-12/DIN-1 complex upon starvation
It is believed that the production of DAs by DAF-9 converts DAF-12 to a liganded state under favorable growth conditions, whereas DAF-12 is mostly unliganded under harsh environmental conditions such as limited food supply and overcrowding (16, 18). In the tbh-1 promoter, there are three AGTACA hexamer elements (−4139, −3193, and −2711 upstream of ATG), which are putative DAF-12 binding sites (Fig. 4A) (25). Here, we investigated whether DAF-12/DIN-1 regulates octopamine biosynthesis after nutrient deprivation. We found that mutations in daf-12(rh61rh411) or din-1(dh127) reduced both the expression of Ptbh-1::gfp (Fig. 4B) and the mRNA level of tbh-1 in starved worms (Fig. 4C). Consistently, the octopamine levels in daf-12(rh61rh411) or din-1(dh127) mutants were significantly lower than those in wild-type worms after starvation (Fig. 4D).
Fig. 4. DAF-12/DIN-1 up-regulates the expression of tbh-1 during starvation.
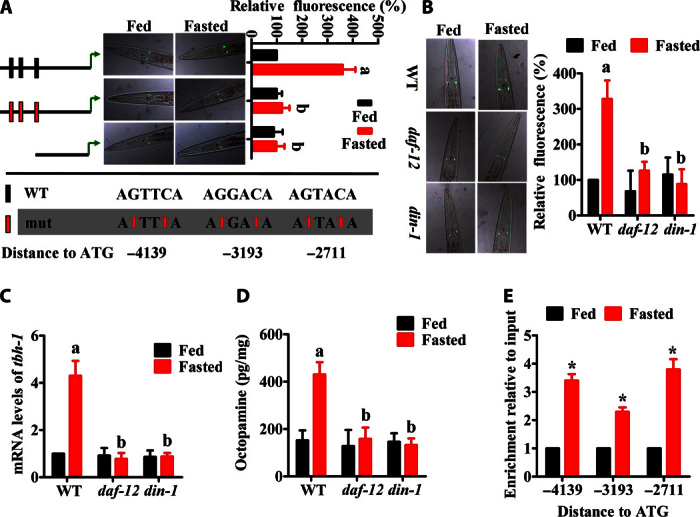
(A) tbh-1 promoters with three AGTACA hexamer elements (−4139, −3193, and −2711 upstream of ATG) for DAF-12. Mutagenesis or deletion of these elements led to a significant reduction in Ptbh-1::gfp expression during food-deprived conditions. (B to D) A mutation in daf-12(rh61rh411) or din-1(dh127) inhibited the expression of Ptbh-1::gfp expression (B) and reduced the mRNA levels of tbh-1 (C) and the levels of octopamine (D) in starved worms. The right part shows quantification of GFP levels (B). Results are means ± SD of three experiments. aP < 0.01 versus well-fed WT worms; bP < 0.05 versus starved WT worms. (E) The three putative DAF-12 binding sites in the promoter regions of tbh-1 were detected by ChIP with anti-GFP antibody. ChIP results were quantitated by qPCR. *P < 0.05 versus well-fed worms.
To determine the importance of the consensus DAF-12 binding sequences, we generated a transcriptional reporter consisting of GFP under the control of the tbh-1 promoter, which contains the AGTACA hexamer elements. Mutagenesis or deletion of these elements resulted in a significant reduction in Ptbh-1::gfp expression during starvation (Fig. 4A). Finally, chromatin immunoprecipitation (ChIP)–qPCR analysis demonstrated that binding to the three elements of tbh-1 promoter was markedly increased in worms under starvation conditions compared to well-fed worms (Fig. 4E and table S1). Together, these results indicate that the DAF-12/DIN-1 signaling is crucial for tbh-1 expression under starvation conditions.
The DAF-12/DIN-1 complex is involved in lipid mobilization
To investigate whether the DAF-12/DIN-1 complex is involved in starvation resistance, we first determined its effect on lipid hydrolysis. Mutations in daf-12(rh61rh411) or din-1(dh127) significantly inhibited lipid hydrolysis during starvation compared with starved wild-type worms (Fig. 5A). Meanwhile, the expression of Plips-6::gfp and the mRNA levels were substantially reduced in daf-12(rh61rh411) or din-1(dh127) worms compared to wild-type animals (Fig. 5, B and C). However, application of octopamine markedly restored lipid hydrolysis and the expression of lips-6 in daf-12(rh61rh411) or din-1(dh127) worms (both fed and fasted states). These results suggest that the DAF-12/DIN-1 complex regulates lipid mobilization via octopamine.
Fig. 5. DAF-12/DIN-1 confers resistance to starvation.
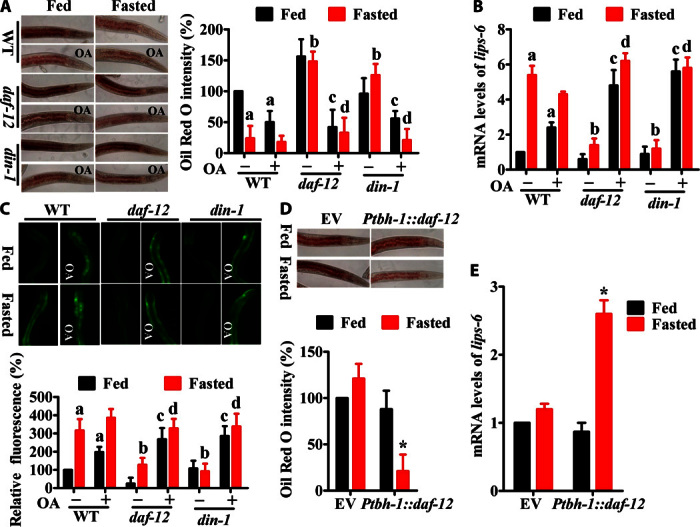
(A) Mutation in daf-12(rh61rh411) or din-1(dh127) suppressed lipid hydrolysis determined by Oil Red O staining after 24 hours of starvation. Application of octopamine (1 mM) restored fasting-induced breakdown. The right panel represents relative Oil Red O intensity. Results are means ± SD of three experiments. (B and C) Expression of lips-6 in WT, daf-12(rh61rh411), and din-1(dh127) worms after 12 hours of starvation. (B) lips-6 mRNA levels. (C) Expression of Plips-6::gfp. The right part shows quantification of GFP levels. Results are means ± SD of three experiments. aP < 0.05 versus well-fed WT worms; bP < 0.05 versus starved WT worms; cP < 0.05 versus well-fed daf-12(rh61rh411) or din-1(dh127) worms; dP < 0.05 versus starved daf-12(rh61rh411) or din-1(dh127) worms. (D and E) Expression of daf-12 under the control of the tbh-1 promoter restored lipid hydrolysis (D) and the expression of lips-6 (E) in daf-12(rh61rh411) animals. The lower panel represents relative Oil Red O intensity (D). Results are means ± SD of three experiments. *P < 0.05 versus empty vector.
Because tbh-1 is mainly expressed in the RIC neurons (4), we hypothesized that DAF-12 carried out its role in lipid mobilization in the same neurons upon starvation. Indeed, expression of daf-12 under the control of the tbh-1 promoter markedly restored lipid hydrolysis and the expression of lips-6 in daf-12(rh61rh411) animals (Fig. 5, D and E). These results suggest that up-regulation of octopamine biosynthesis is one of the mechanisms underlying the DAF-12/DIN-1 complex–mediated lipid hydrolysis.
DISCUSSION
Using a combination of tissue-autonomous metabolic adjustments and hormonal signals to coordinate the mobilization of nutrients is a common physiological response upon nutrient depletion (6–9). Here, we characterized an endocrine mechanism in coupling nutrient cues to lipid metabolism to maintain energy homeostasis in C. elegans (Fig. 6). When encountering food limitation, the nutrient-sensing nuclear receptor DAF-12 perceives a state of starvation and consequently augments the biosynthesis of octopamine in the neuronal cells. The biogenic amine activates its receptor SER-3 in the intestine to induce hydrolysis of lipid droplets for maintaining energy balance.
Fig. 6. A proposed model for octopamine-mediated starvation resistance.
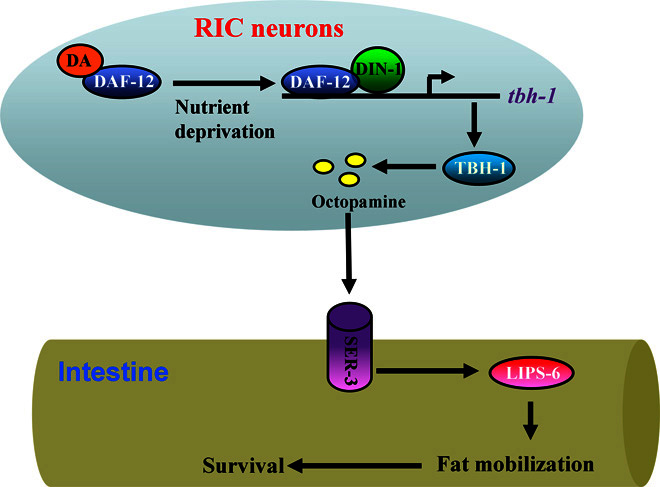
Under nutrient deprivation, the liganded DAF-12 shifts the equilibrium to the unliganded DAF-12/DIN-1 complex. The latter promotes the octopamine biosynthesis by up-regulating the expression of tbh-1 in the RIC neurons. Octopamine, in turn, activates its receptor SER-3 to up-regulate the expression of the lipase gene lips-6 in the intestine. Concomitantly, LIPS-6 contributes to lipid hydrolysis to maintain energy balance.
Here, our results demonstrate that the biosynthesis of octopamine is up-regulated during starvation. The molecular mechanism underlying transcriptional regulation of tbh-1 in C. elegans remains largely unknown. A recent study has demonstrated that the activation of CREB [adenosine 3′,5′-monophosphate (cAMP) response element–binding protein]–regulated transcription coactivator–1 (CRTC-1) by CREB leads to a 1.5-fold increase in tbh-1 expression in well-fed worms (26). Promoter analysis reveals that tbh-1 gene contains the putative CREB binding site in its promoter, implicating that CRTC-1/CRH-1 may directly regulate transcription of tbh-1. Our data indicate that the DAF-12/DIN-1 complex is involved in the induction of tbh-1 expression in response to starvation. We show that the tbh-1 promoter contains three AGTACA hexamer elements, previously identified as a DAF-12 binding site (25). 5′-Deletion and ChIP analyses demonstrate that the three elements are required for induction of tbh-1 expression under starvation conditions. Thus, DAF-12/DIN-1 represents a novel pathway to regulate tbh-1 expression and octopamine levels during starvation.
It has been shown that starvation extends life span in C. elegans (27–29). Here, our results demonstrate that starvation increases the levels of octopamine, and the mutation in tbh-1(n3247) markedly reduces survival of worms after nutrient deprivation, suggesting that octopamine is essential for resistance to starvation. Recently, Burkewitz et al. (26) have reported that transgenic worms carrying constitutively active AMPK catalytic subunit increases life span probably by reducing octopamine levels under ad libitum feeding conditions, implicating that octopamine negatively modulates life span. Unexpectedly, these authors found that worms harboring null mutations in either tdc-1(n3421) or tbh-1(n3247), both of which lack octopamine, have a slightly shorter life span than wild-type animals. The most parsimonious explanation for these conflicting observations is that octopamine plays a complicated role in life span under different physiological conditions (fed or fasted state) or genetic backgrounds.
It has been shown that octopamine released from RIC neurons mediates a starvation response, leading to activation of the transcription factor CREB (19, 23). This starvation response is required for the octopamine receptor SER-3 in SIA neurons. Although different regulatory mechanisms have evolved for nutrient mobilization, the ability to regulate lipid consumption is a fundamental process upon starvation. It has been shown that fasting or dietary restriction elicits the expression of the lipase gene lips-6 in worms (11, 30). Here, we show that octopamine mediates lipid hydrolysis by up-regulating lips-6, permitting C. elegans to survive starvation. However, octopamine exhibits the action via its receptor SER-3 in the intestine, suggesting that octopamine also regulates a starvation response in peripheral tissues. Because the elevated circulating concentrations of norepinephrine are reported in vertebrates during starvation (6–9), the role of norepinephrine in energy balance is likely to be conserved.
Norepinephrine, which is associated with chronic stress, has been identified as an etiological factor in various types of cancer (31–33). Norepinephrine via β-adrenergic receptors modulates expression of metastasis-associated genes involved in angiogenesis, inflammation, tissue invasion, and epithelial-mesenchymal transition (31, 34). Because cancer cells are often hypersensitive to starvation, characterization of the role for norepinephrine in energy balance may provide mechanistic insights into caloric restriction for cancer treatment.
MATERIALS AND METHODS
Nematode strains
The C. elegans strains were cultured under standard conditions and fed E. coli OP50 (35). Wild-type animals were C. elegans Bristol N2. The strains tbh-1(n3247), daf-12(rh61rh411), din-1(dh127), hjIs67[atgl-1p::atgl-1::gfp], octr-1(ok371), and TU3401 [sid-1(pk3321)V; uIs69V] for neuronal-specific RNAi and NR350 [rde-1(ne219)v; kzIs20] for muscular-specific RNAi were provided by the Caenorhabditis Genetics Center. The strain MGH170 for intestinal-specific RNAi (sid-1(qt9); Is[vha-6pr::sid-1]; Is[sur-5pr::GFPNLS]), the strain that expresses Ptbh-1::gfp, ser-3(ad1774), ser-6(tm2146), and atgl-1(tm3116) worms were provided by G. Ruvkun (Massachusetts General Hospital, Harvard Medical School), H. R. Horvitz (Massachusetts Institute of Technology), Z. Wu (Huazhong University of Science and Technology), and B. Liang (Kunming Institute of Zoology, Chinese Academy of Sciences), respectively. Mutants were backcrossed three times into the N2 strain used in the laboratory.
Construction of lips-6 RNAi
To generate a clone directed against lips-6, a 1042–base pair (bp) fragment was amplified from genomic DNA by PCR using the primers 5′-GTGTGCTGATGTGACTGC-3′ (forward) and 5′-ATGCCAACTATCCACGAC-3′ (reverse). The fragment was TA-cloned into a Hind III– and Kpn I–linearized L4440 feeding vector, as in the RNAi library (36).
Construction of transgenic strains
The vector expressing Plips-6::gfp was generated by subcloning a 3.2-kb promoter fragment of lips-6 into an expression vector (pPD95.75). Pdaf-12::daf-12-gfp was generated by subcloning a 5.3-kb daf-12 promoter fragment (corresponding to nucleotides −5313 to −1 relative to the translational start site) into an expression vector (pPD95.75) into which the daf-12 complementary DNA (cDNA) was fused at its 3′ end in-frame with GFP and the 3′ untranslated region (UTR) of unc-54. Ptbh-1::daf-12-gfp was generated by subcloning a 4.6-kb tbh-1 promoter fragment (corresponding to nucleotides −4613 to −1 relative to the translational start site) into the pPD95.75 vector into which the daf-12 cDNA was fused at its 3′ end in-frame with GFP and the 3′UTR of unc-54. cDNAs encoding daf-12, tbh-1, ser-3, and lips-6 were chemically synthesized and obtained from Generay Biotech Co. The tbh-1 minimal promoter vector was generated by a 2.7-kb promoter fragment (corresponding to nucleotides −2710 to −1 relative to the translational start site) and inserted into a pPD95.75 reporter vector. The deleted fragment of tbh-1 promoter was chemically synthesized (Generay Biotech) and subcloned into a pPD95.75 reporter vector. Ptbh-1::daf-12::gfp and Ptbh-1::tbh-1::gfp were generated by subcloning a 4.6-kb tbh-1 promoter fragment (corresponding to nucleotides −4613 to −1 relative to the translational start site) into the pPD95.75 vector into which the daf-12 or tbh-1 cDNA was fused at its 3′ end in-frame with GFP and the 3′UTR of unc-54. Pges-1::ser-3::gfp and Pges-1::lips-6::gfp were generated by subcloning a 2.9-kb ges-1 promoter fragment (corresponding to nucleotides −2963 to −1 relative to the translational start site) into the pPD95.75 vector into which the ser-3 or lips-6 cDNA was fused at its 3′ end in-frame with GFP and the 3′UTR of unc-54. These constructs were co-injected with the marker plasmid pRF4 containing rol-6(su1006) into the gonads of wild-type, daf-12(rh61rh411), or ser-3(ad1774) worms by standard techniques. The transgenic worms were confirmed before assay (37).
RNA interference
The clones of genes for RNAi were from the Ahringer library (38). RNAi feeding experiments were performed on synchronized L1 larvae at 20°C for 40 hours.
Starvation survival analysis
Synchronized populations of worms were cultivated on nematode growth medium (NGM) plates in the presence of E. coli OP50 at 20°C until the young adult stage. They were then transferred to NGM agar plates containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), amphotericin B (0.25 μg/ml), and 5′-fluoro-2′-deoxyuridine (75 μg/ml) at 20°C. The number of living worms was counted at 3 days of starvation. Three plates were performed per assay, and all experiments were performed three times.
Detection of octopamine levels
The worms were homogenized with perchloric acid as previously described (4) with modifications. Briefly, 60 to 80 mg of worms was homogenized with a pestle in 500 μl of 0.3 M perchloric acid and centrifuged at 13,000 rpm × 30 min at 4°C to remove the insoluble residue. The supernatant was filtered through a 0.22-μm centrifugal filter and freeze-dried in a VirTis freeze dryer. The resulting powders were dissolved in 50 μl of 0.3 M perchloric acid. Samples were analyzed by LC-MS/MS on a Shimadzu LC-20ADvp pump equipped with an API 3200 mass spectrometer (Applied Biosystems/MDS Sciex). Samples were injected into a Shim-pack XR-ODS (3.0 mm inside diameter × 75 mm) column at a flow rate of 0.4 ml/min and a temperature of 40°C. The analytes were eluted with a mobile phase comprising water with 0.4% formic acid (solvent A) and acetonitrile (solvent B). Gradient program of B: 0 min, 5%; 1.0 min, 5%; 4.0 min, 80%; 4.5 min, 80%; 4.6 min, 5%; 6.0 min, 5%. The mass spectrometer was operated under the following conditions: nebulizer gas, 60 psi; curtain gas, 20 psi; auxiliary gas, 55 psi; ion spray voltage, 5.0 kV; and temperature, 65°C. The detection mode was multiple reaction monitoring, positive. The octopamine concentrations were calculated on the basis of standard curves (1 to 100 ng of octopamine, Sigma) and expressed as picogram of octopamine per milligram of worm wet weight.
Quantitative RT-PCR
Total RNA was isolated from worms with TRIzol Reagent (Invitrogen). Random-primed cDNAs were generated by reverse transcription of the total RNA samples with SuperScript II (Invitrogen). A real-time PCR analysis was conducted using SYBR Premix Ex Taq (Takara) on a Roche LightCycler 480 System (Roche Applied Science). ama-1 was used as an internal control. The primers used for PCR were as follows: tbh-1, 5′-AAGAAGAACCAGCCGCTCT-3′ (forward) and 5′-TGCTCATTTCCAGGTGTCAC-3′ (reverse); fil-1, 5′-TGATGTTGCTGGTCCATTG-3′ (forward) and 5′-CCCTGAGTTCCATAGTCTGCT-3′ (reverse); fil-2, 5′-TGGTTATTCATCTACTCCCTGC-3′ (forward) and 5′-TCGTCGGCTTTACTCCAAGT-3′ (reverse); lips-6, 5′-ACTGCTGATTTCGTCGTAGGT-3′ (forward) and 5′-CTCGTGGTTCCTTCCTTCA-3′ (reverse); ama-1, 5′-TCCTGACCCAAAGAACACGGT-3′ (forward) and 5′-ATCCACCTGCTCCTCCTGAG-3′ (reverse).
Oil Red O staining
Oil Red O staining was performed as previously described (39). Briefly, after the worms were washed in phosphate-buffered saline (PBS) buffer, they were suspended in 120 μl of PBS, to which an equal volume of 2× MRWB buffer (160 mM KCl, 40 mM NaCl, 14 mM Na2EGTA, 1 mM spermidine-HCl, 0.4 mM spermine, 30 mM Na-Pipes at pH 7.4, 0.2% β-mercaptoethanol) was added. Then, the worms were immediately frozen in liquid nitrogen and subjected to three freeze/thaw cycles. After they were washed with the PBS buffer, they were dehydrated in 60% isopropyl alcohol for 10 min and stained with filtered Oil Red O solution [60% Oil Red O stock solution (isopropanol, 5 mg/ml)/40% water] overnight. Photographs of 15 worms were taken using a Nikon E800 fluorescence microscope. Anterior intestinal cell areas were selected to determine Oil Red O intensities. Relative intensities were quantified using ImageJ software [National Institutes of Health (NIH)]. All experiments were performed three times.
Quantitative analysis of TAG
TAG was determined using the method previously described (40). Briefly, approximately 10,000 worms were collected and washed repeatedly with M9 buffer until bacteria were removed. The worm pellets were incubated with 5 ml of ice-cold chloroform/methanol (1:1) overnight at −20°C with occasional vortexing. A solution of 0.2 M H3PO4 and 1 M KCl was added to the samples, resulting in phase separation of the organic and aqueous phases. The organic phase was collected and dried under dry nitrogen and then resuspended in chloroform. The samples were loaded, and TLC plates were developed two-thirds of the way up the plate in the solvent system: hexane/diethyl ether/acetic acid (80:20:2). After the plate was sprayed with 0.005% primuline, the lipids were visualized under ultraviolet light. The spots corresponding to TAG and the major phospholipids (PLs) were scraped for GC-MS analysis to determine the relative levels of the TAG and PL fractions.
Fluorescence microscopic analysis of GFP-labeled worms
After the young adult worms were starved for 12 hours, they were mounted in M9 buffer on microscope slides. The worms were grown on E. coli OP50 at the same time as a control. The slides were imaged using a Nikon E800 fluorescence microscope. Fluorescence intensity was quantified by using the ImageJ software (NIH). Three plates of about 30 animals per plate were tested per assay, and all experiments were performed three times independently.
ChIP-qPCR
ChIP was performed as described previously (41). Briefly, young adult worms were grown in NGM plates in the presence or absence of E. coli OP50 for 24 hours. The worms were then collected and washed repeatedly with M9 buffer until bacteria were removed. The samples were resuspended in 47 ml of M9 buffer and 2.8 ml of 37% formaldehyde solution, and crosslinked for 30 min with rotation at 100 rpm. The worms were then washed with 50 ml of 100 mM tris (pH 7.5) to quench the formaldehyde solution, and washed twice with 50 ml of M9 buffer and once with 10 ml of FA buffer [50 mM Hepes/KOH (pH 7.5), 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 150 mM NaCl] supplemented with protease inhibitors (Complete Mini Protease Inhibitor Cocktail Tablets, Roche). Worms were then collected by centrifugation and were stored at −80°C.
Approximately 0.5 ml of worms was resuspended in 2 ml of FA buffer plus protease inhibitors (two tablets of protease inhibitors, 250 μl of 100 mM phenylmethylsulfonyl fluoride, 50 μl of 1 M dithiothreitol in 50 ml of FA buffer). After sonication in ice water, the sample was transferred to microfuge tubes and centrifuged at 13,000g for 15 min at 4°C. The protein concentration of the supernatant was measured by the Bradford method using bovine serum albumin as the standard. Extract containing approximately 3 mg of total protein was added to a microfuge tube, and the volume was brought to 500 μl with FA buffer plus protease inhibitors. Then, 25 μl of 20% Sarkosyl solution was added, and the tube was spun at 13,000g for 5 min at 4°C. The supernatant was then collected, and 10% of the extract was removed and stored at −20°C for future use as input DNA. Anti-GFP or control immunoglobulin G antibodies (15 μg) were added to the extract and rotated at 4°C overnight. Then, 25 μl of protein G conjugated to Sepharose beads was added to each ChIP sample and rotated at 4°C for 2 hours. Each ChIP sample was washed four times with 1 ml of FA buffer. After the washes, the beads were suspended in one bed volume of FA buffer, and 40 μl of the bead slurry was added to each ChIP sample and rotated at 4°C for 2 hours. The beads were then washed twice at 4°C in 1 ml of FA buffer and once in FA buffer with 1 M NaCl. The beads were collected between each wash. FA buffer (1 ml) with 500 mM NaCl was added to the beads and rotated for 10 min. The beads were then washed in TEL buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM tris-HCl, pH 8.0) for 10 min and twice in TE for 5 min. To elute the immunocomplexes, 150 μl of elution buffer (1% SDS in TE with 250 mM NaCl) was added and incubated at 65°C for 15 min, with brief vortexing every 5 min. The beads were spun down at 2500g for 2 min, and the supernatant was transferred to a new tube. The elution was repeated, and supernatants were combined four times. At this time, input samples were thawed. Two microliters of ribonuclease (10 mg/ml) was added to each sample, and then the samples were incubated at room temperature for 2 hours. The samples were incubated with 250 μl of elution buffer and 1 μl of proteinase K (20 mg/ml) for 2 hours at 55°C. All samples were transferred to 65°C for 12 hours to reverse crosslinks. The DNA was extracted by a DNA purification kit (BioTeke Corp.). A 5-μl aliquot of the input DNA was then run on a 2% agarose gel to check the extent of shearing, with an expected range between 200 and 800 bp. The immunoprecipitated DNA was detected by qPCR. All ChIP experiments were performed with three replicates.
Statistics
The statistical significance of differences in gene expression, survival rate, and fluorescence intensity was assessed with one-way analysis of variance (ANOVA), followed by a Student-Newman-Keuls test. Data were analyzed using SPSS 11.0 software (SPSS Inc.).
Supplementary Material
Acknowledgments
We are grateful to J. Xu (McMaster University) and Z. An (University of Texas Health Science Center at Houston) for their critical reading of this manuscript. We thank the Caenorhabditis Genetics Center and H. R. Horvitz, G. Ruvkun, Z. Wu, and B. Liang for worm strains. Funding: This work was supported in part by a grant (2013CB127500) from the National Basic Research Program of China (973) (to K.-Q.Z.) and a grant from the National Natural Science Foundation of China (311171365) (to C.-G.Z.). Y.-C.M. is the recipient of Outstanding PhD Training Fund of Yunnan University (2014YUN05). Author contributions: J.T. and Y.-C.M. performed all of the experiments, analyzed the data, and helped write the manuscript. Z.-S.Y. performed octopamine content analysis. C.-G.Z. and K.-Q.Z. designed the experiments, interpreted the results, and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data used to obtain the conclusions in this paper are presented in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/5/e1501372/DC1
fig. S1. LC-MS/MS detection of octopamine contents in worms.
fig. S2. TLC analysis of total lipids extracted from worms.
fig. S3. Overexpression of tbh-1 enhances lipid hydrolysis in well-fed wild-type worms.
fig. S4. Starvation does not alter the expression of atgl-1.
fig. S5. A mutation in tbh-1(n3247) does not affect the expression of fil-1 or fil-2 in starved worms.
fig. S6. Knockdown of lips-6 leads to reduced lips-6 expression.
fig. S7. Overexpression of lips-6 in the intestine promotes lipid hydrolysis in well-fed worms.
fig. S8. Octopamine fails to restore lipid hydrolysis and expression of lips-6 in ser-3(ad1774) mutants during starvation.
fig. S9. SER-3 in the neurons and muscle is not involved in lipid hydrolysis during starvation.
fig. S10. SER-3 is involved in resistance to starvation.
table S1. ChIP-qPCR analysis of the tbh-1 promoter.
REFERENCES AND NOTES
- 1.Verlinden H., Vleugels R., Marchal E., Badisco L., Pflüger H.-J., Blenau W., Broeck J. V., The role of octopamine in locusts and other arthropods. J. Insect Physiol. 56, 854–867 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Koon A. C., Ashley J., Barria R., DasGupta S., Brain R., Waddell S., Alkema M. J., Budnik V., Autoregulatory and paracrine control of synaptic and behavioral plasticity by octopaminergic signaling. Nat. Neurosci. 14, 190–199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z., Yu Y., Zhang V., Tian Y., Qi W., Wang L., Octopamine mediates starvation-induced hyperactivity in adult Drosophila. Proc. Natl. Acad. Sci. U.S.A. 112, 5219–5224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkema M. J., Hunter-Ensor M., Ringstad N., Horvitz H. R., Tyramine Functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron 46, 247–260 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Horvitz H. R., Chalfie M., Trent C., Sulston J. E., Evans P. D., Serotonin and octopamine in the nematode Caenorhabditis elegans. Science 216, 1012–1014 (1982). [DOI] [PubMed] [Google Scholar]
- 6.McKnight K. A., Rupp H., Dhalla K. S., Beamish R. E., Dhalla N. S., Biphasic changes in heart performance with food restriction in rats. J. Appl. Physiol. (1985) 87, 1909–1913 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Zauner C., Schneeweiss B., Kranz A., Madl C., Ratheiser K., Kramer L., Roth E., Schneider B., Lenz K., Resting energy expenditure in short-term starvation is increased as a result of an increase in serum norepinephrine. Am. J. Clin. Nutr. 71, 1511–1515 (2000). [DOI] [PubMed] [Google Scholar]
- 8.De Pedro N., Delgado M. J., Gancedo B., Alonso-Bedate M., Changes in glucose, glycogen, thyroid activity and hypothalamic catecholamines in tench by starvation and refeeding. J. Comp. Physiol. B 173, 475–481 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Goldstein J. L., Zhao T.-j., Li R. L., Sherbet D. P., Liang G., Brown M. S., Surviving starvation: Essential role of the ghrelin-growth hormone axis. Cold Spring Harb. Symp. Quant. Biol. 76, 121–127 (2011). [DOI] [PubMed] [Google Scholar]
- 10.You Y.-j., Kim J., Cobb M., Avery L., Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 3, 237–245 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jo H., Shim J., Lee J. H., Lee J., Kim J. B., IRE-1 and HSP-4 contribute to energy homeostasis via fasting-induced lipases in C. elegans. Cell Metab. 9, 440–448 (2009). [DOI] [PubMed] [Google Scholar]
- 12.O’Rourke E. J., Ruvkun G., MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 15, 668–676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKay R. M., McKay J. P., Avery L., Graff J. M., C. elegans: A model for exploring the genetics of fat storage. Dev. Cell 4, 131–142 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Gilst M. R., Hadjivassiliou H., Yamamoto K. R., A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. U.S.A. 102, 13496–13501 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Motola D. L., Cummins C. L., Rottiers V., Sharma K. K., Li T., Li Y., Suino-Powell K., Xu H. E., Auchus R. J., Antebi A., Mangelsdorf D. J., Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell 124, 1209–1223 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Fielenbach N., Antebi A., C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22, 2149–2165 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wollam J., Antebi A., Sterol regulation of metabolism, homeostasis, and development. Annu. Rev. Biochem. 80, 885–916 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ao W., Gaudet J., Kent W. J., Muttumu S., Mango S. E., Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science 305, 1743–1746 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Suo S., Culotti J. G., Van Tol H. H. M., Dopamine counteracts octopamine signalling in a neural circuit mediating food response in C. elegans. EMBO J. 28, 2437–2448 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noble T., Stieglitz J., Srinivasan S., An integrated serotonin and octopamine neuronal circuit directs the release of an endocrine signal to control C. elegans body fat. Cell Metab. 18, 672–684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J. H., Kong J., Jang J. Y., Han J. S., Ji Y., Lee J., Kim J. B., Lipid droplet protein LID-1 mediates ATGL-1-dependent lipolysis during fasting in Caenorhabditis elegans. Mol. Cell. Biol. 34, 4165–4176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mills H., Wragg R., Hapiak V., Castelletto M., Zahratka J., Harris G., Summers P., Korchnak A., Law W., Bamber B., Komuniecki R., Monoamines and neuropeptides interact to inhibit aversive behaviour in Caenorhabditis elegans. EMBO J. 31, 667–678 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suo S., Kimura Y., Van Tol H. H. M., Starvation induces cAMP response element-binding protein-dependent gene expression through octopamine–Gq signaling in Caenorhabditis elegans. J. Neurosci. 26, 10082–10090 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Libina N., Berman J. R., Kenyon C., Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115, 489–502 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Shostak Y., Van Gilst M. R., Antebi A., Yamamoto K. R., Identification of C. elegans DAF-12-binding sites, response elements, and target genes. Genes Dev. 18, 2529–2544 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burkewitz K., Morantte I., Weir H. J. M., Yeo R., Zhang Y., Huynh F. K., Ilkayeva O. R., Hirschey M. D., Grant A. R., Mair W. B., Neuronal CRTC-1 governs systemic mitochondrial metabolism and lifespan via a catecholamine signal. Cell 160, 842–855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cypser J. R., Tedesco P., Johnson T. E., Hormesis and aging in Caenorhabditis elegans. Exp. Gerontol. 41, 935–939 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinkove D., Halstead J. R., Gems D., Divecha N., Long-term starvation and ageing induce AGE-1/PI 3-kinase-dependent translocation of DAF-16/FOXO to the cytoplasm. BMC Biol. 4, 1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang C., Avery L., Systemic regulation of starvation response in Caenorhabditis elegans. Genes Dev. 23, 12–17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palgunow D., Klapper M., Döring F., Dietary restriction during development enlarges intestinal and hypodermal lipid droplets in Caenorhabditis elegans. PLOS One 7, e46198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakroborty D., Sarkar C., Basu B., Dasgupta P. S., Basu S., Catecholamines regulate tumor angiogenesis. Cancer Res. 69, 3727–3730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thaker P. H., Han L. Y., Kamat A. A., Arevalo J. M., Takahashi R., Lu C., Jennings N. B., Armaiz-Pena G., Bankson J. A., Ravoori M., Merritt W. M., Lin Y. G., Mangala L. S., Kim T. J., Coleman R. L., Landen C. N., Li Y., Felix E., Sanguino A. M., Newman R. A., Lloyd M., Gershenson D. M., Kundra V., Lopez-Berestein G., Lutgendorf S. K., Cole S. W., Sood A. K., Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma. Nat. Med. 12, 939–944 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Fitzgerald P. J., Is norepinephrine an etiological factor in some types of cancer? Int. J. Cancer 124, 257–263 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Cole S. W., Sood A. K., Molecular pathways: Beta-adrenergic signaling in cancer. Clin. Cancer Res. 18, 1201–1206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timmons L., Fire A., Specific interference by ingested dsRNA. Nature 395, 854 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Mello C., Fire A., DNA transformation. Methods Cell Biol. 48, 451–482 (1995). [PubMed] [Google Scholar]
- 38.Kamath R. S., Ahringer J., Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Soukas A. A., Kane E. A., Carr C. E., Melo J. A., Ruvkun G., Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 23, 496–511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi X., Li J., Zou X., Greggain J., Rødkær S. V., Færgeman N. J., Liang B., Watts J. L., Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J. Lipid Res. 54, 2504–2514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong M., Niu W., Lu Z. J., Sarov M., Murray J. I., Janette J., Raha D., Sheaffer K. L., Lam H. Y. K., Preston E., Slightham C., Hillier L. W., Brock T., Agarwal A., Auerbach R., Hyman A. A., Gerstein M., Mango S. E., Kim S. K., Waterston R. H., Reinke V., Snyder M., Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLOS Genet. 6, e1000848 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/5/e1501372/DC1
fig. S1. LC-MS/MS detection of octopamine contents in worms.
fig. S2. TLC analysis of total lipids extracted from worms.
fig. S3. Overexpression of tbh-1 enhances lipid hydrolysis in well-fed wild-type worms.
fig. S4. Starvation does not alter the expression of atgl-1.
fig. S5. A mutation in tbh-1(n3247) does not affect the expression of fil-1 or fil-2 in starved worms.
fig. S6. Knockdown of lips-6 leads to reduced lips-6 expression.
fig. S7. Overexpression of lips-6 in the intestine promotes lipid hydrolysis in well-fed worms.
fig. S8. Octopamine fails to restore lipid hydrolysis and expression of lips-6 in ser-3(ad1774) mutants during starvation.
fig. S9. SER-3 in the neurons and muscle is not involved in lipid hydrolysis during starvation.
fig. S10. SER-3 is involved in resistance to starvation.
table S1. ChIP-qPCR analysis of the tbh-1 promoter.


