Benthic deep-sea archaea are mainly controlled by temperature, whereas bacteria are controlled by food availability.
Keywords: deep-sea macroecology, benthic bacteria, archaea, environmental drivers, global change
Abstract
Bacteria and archaea dominate the biomass of benthic deep-sea ecosystems at all latitudes, playing a crucial role in global biogeochemical cycles, but their macroscale patterns and macroecological drivers are still largely unknown. We show the results of the most extensive field study conducted so far to investigate patterns and drivers of the distribution and structure of benthic prokaryote assemblages from 228 samples collected at latitudes comprising 34°N to 79°N, and from ca. 400- to 5570-m depth. We provide evidence that, in deep-sea ecosystems, benthic bacterial and archaeal abundances significantly increase from middle to high latitudes, with patterns more pronounced for archaea, and particularly for Marine Group I Thaumarchaeota. Our results also reveal that different microbial components show varying sensitivities to changes in temperature conditions and food supply. We conclude that climate change will primarily affect deep-sea benthic archaea, with important consequences on global biogeochemical cycles, particularly at high latitudes.
INTRODUCTION
Deep-sea sediments, which are the largest ecosystems on Earth, covering ca. 65% of the surface of the globe and ca. 95% of the ocean sea floor, control global biogeochemical cycles (1, 2). Life in the deep-sea sediments is largely constrained by the limited and episodic food supply from the water column, possibly supplemented by benthic chemoautotrophic production (1, 3). Here, total benthic biomass is dominated by prokaryotes (up to more than 90%), and bacteria and archaea play a pivotal role in C production (either heterotrophic or chemoautotrophic), nutrient cycling, and energy transfer to the higher trophic levels (1, 4). Deep-sea prokaryotes are concentrated in surface sediments, where their abundance per unit of volume can be 103 to 106 times higher than in the dark portion of the water column and in the subsea-floor biosphere (5, 6). Using a modeling approach, the global prokaryotic biomass (reported as “bacteria”) on the sea floor has been estimated in the order of ca. 35 Mt C (7).
Although regional-scale studies reported the presence of significant depth-related patterns of benthic prokaryotic abundance and biomass (8, 9), meta-analyses conducted at global scale suggest that their abundance and biomass remain rather constant with increasing water depth (7, 10, 11). Because food supply to deep-sea benthos changes with depth, latitude, biogeographic region, and related productivity, the lack of depth-related patterns at global scale suggests that prokaryotes are dependent not only on the quality and quantity (or dilution) of organic matter available in the deep sea (12–18) but also on other factors acting at different spatial scales. In particular, high deep-sea biomasses have been reported in highly productive oceanic regions (for example, the North Atlantic and upwelling and polar regions), whereas the oligotrophic conditions support biomass values of one to two orders of magnitude lower (for example, Central Pacific and the deep Mediterranean Sea) (7, 19). Recent studies based on the metabolic theory provided evidence that the relative influence of chemical and thermal energy on deep-sea organisms varies considerably across levels of biological organization and that thermal energy has a major effect at lower levels of biological organization (20). Moreover, benthic deep-sea prokaryotes can be controlled by viral infections able to abate >80% of biomass production (11). We also know that archaeal abundance increases beneath 1000-m depth and can equal bacterial abundance in the deeper portion of the water column and in subsurface deep-sea sediments. However, the factors explaining such shifts in the relative importance of archaea and bacteria are still largely unknown (5, 6, 21–23). Overall, our knowledge of the patterns and drivers controlling the distributions of bacteria and archaea in deep-sea sediments is completely insufficient to fully comprehend their ecology and response to multiple stressors, including the effects of global change predicted for the coming decades.
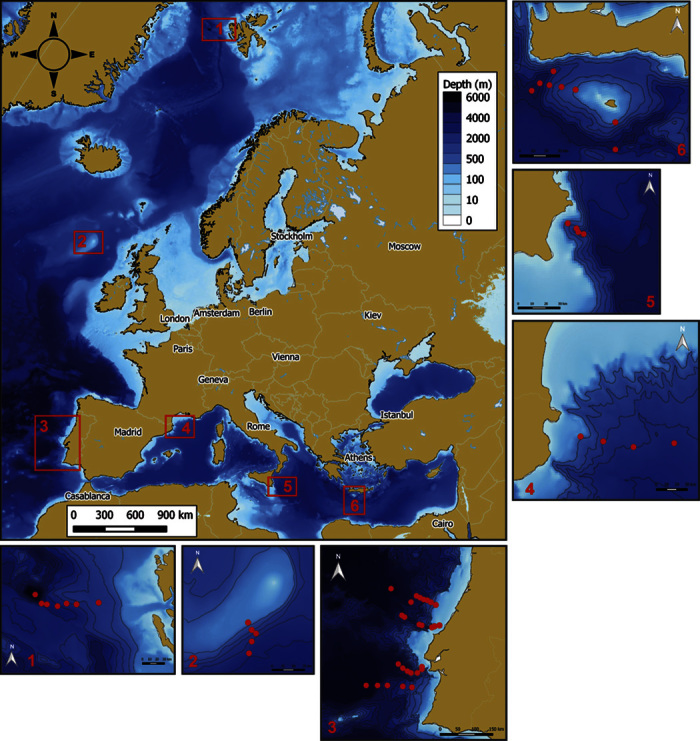
Here, we investigated the macroecology of bacteria and archaea in surface deep-sea sediments by means of an intensive and highly replicated macroscale sampling strategy. Two hundred twenty-eight samples were collected from 58 benthic deep-sea sites, spanning from 34°N to 79°N (Fig. 1) and encompassing different physical-chemical and trophic characteristics (for example, from the Arctic to the North Atlantic to the Mediterranean basin, with temperature deltas >10°C at the same depths), and at depths ranging from ca. 400 to 5570 m.
Fig. 1. Sampling areas and locations of the sampling stations.
The present study was designed to provide new insights into the following: (i) the factors driving the distribution of benthic bacteria and archaea and, within the domain Archaea, of Marine Group I (MG-I) Thaumarchaeota and MG-II Euryarchaeota, which represent the dominant archaeal groups in surface deep-sea sediments (24, 25), and (ii) the different sensitivity of bacteria and archaea to changing trophic and thermohaline conditions to forecast their potential responses to global changes.
Our results point out the increasing importance of benthic archaea at high latitudes. Our findings also reveal that bacteria and archaea respond with different sensitivity to temperature shifts and changes in food supply, showing a higher sensitivity of archaea, and particularly of the MG-I Thaumarchaeota, to changes in thermal energy. Our results suggest that global climate change will have important consequences on the structure and distribution of deep-sea prokaryotic assemblages, with profound implications on global biogeochemical cycles.
RESULTS
Environmental setting
Bottom water temperature across the different sampling regions investigated ranged from −0.90° to 14.17°C (varying from −0.90° to −0.51°C in the Arctic Ocean, from 2.0° to 12.05°C in the Atlantic Ocean, and from 13.02° to 14.17°C in the Mediterranean Sea; table S1). Salinity at the water-sediment interface ranged from 34.82 to 34.92 in the Arctic Ocean, from 34.09 to 36.29 in the Atlantic Ocean, and from 38.44 to 38.85 in the Mediterranean Sea (table S1).
Trophic characteristics of the sediments were investigated in terms of total phytopigment content, biopolymeric C concentrations (as the sum of C equivalents of proteins, carbohydrates, and lipid content of the sediments), and organic C fluxes to the sea floor. These variables represent excellent proxies of the food supply and availability of organic substrates for heterotrophic metabolism in the deep sea (18, 26, 27). Total phytopigment concentrations ranged, on average, from 12.8 ± 2.9 to 16.8 ± 2.8 μg g−1 in the Arctic and Atlantic oceans, respectively (table S1). Biopolymeric C concentrations were higher in the Arctic Ocean (3.6 ± 0.6 mg C g−1) than in the Atlantic Ocean and Mediterranean Sea (1.7 ± 0.2 and 1.6 ± 0.2 mg C g−1, respectively). Organic C fluxes to the deep-sea floor were two- to threefold higher in the Arctic and Atlantic oceans (on average, 21.5 ± 5.6 and 26.9 ± 4.4 mg C m−2 day−1, respectively) than in the Mediterranean Sea (9.9 ± 2.2 mg C m−2 day−1; table S1).
Abundances of total prokaryotes, bacteria, and archaea
Total prokaryotic abundances were high at all depths, from the shelf break down to abyssal sediments (fig. S1), and did not show significant bathymetric patterns. On average, the highest total prokaryotic abundance was observed in the sediments of the Arctic Ocean (9.92 ± 1.16 × 108 cells g−1), followed by sediments of the Atlantic Ocean and Mediterranean Sea (both 4.8 × 108 cells g−1; fig. S1). The sum of archaea and bacteria determined by catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) analyses accounted for ca. 75% of total prokaryotic abundances determined by SYBR Green I. Such a discrepancy can be due to the lack of probe specificity toward all bacterial and archaeal taxa inhabiting benthic deep-sea ecosystems (table S2). Data from CARD-FISH were compared for their consistency with results of the quantitative polymerase chain reaction (qPCR) on either bacteria or archaea (fig. S2). Bacteria dominated over archaea in almost all of the investigated sites.
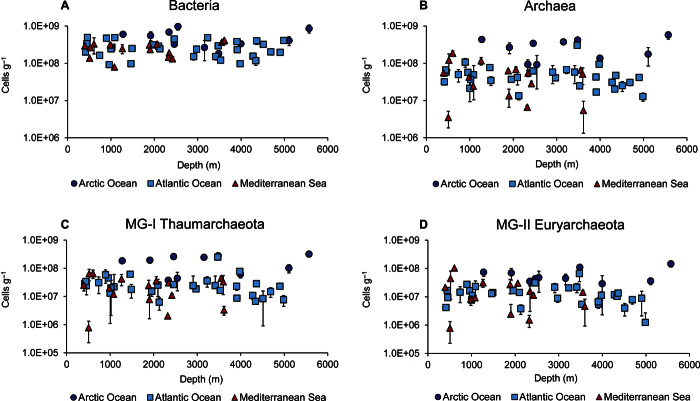
Consistent with results obtained for total prokaryotic abundance, the abundances of bacteria, archaea, MG-I Thaumarchaeota, and MG-II Euryarchaeota did not show significant bathymetric patterns for all of the oceanic regions investigated (Fig. 2, A to D). Significant differences in all microbial components were found among the different oceanic regions, but not among areas within the same oceanic region (that is, between the two areas of the Atlantic Ocean or among the three areas of the Mediterranean Sea; table S3). In particular, bacterial abundances were significantly higher in Arctic sediments (on average, 5.17 ± 0.83 × 108 cells g−1) than in the Atlantic and Mediterranean sediments (on average, 2.69 ± 0.23 × 108 and 2.61 ± 0.25 × 108 cells g−1, respectively; Fig. 2A). The abundances of archaea in the Arctic sediments (2.91 ± 0.52 × 108 cells g−1) were fivefold higher than in the Atlantic and Mediterranean sediments (on average, 0.53 ± 0.01 × 108 and 0.57 ± 0.12 × 108 cells g−1; Fig. 2B). Archaea accounted, on average, for ca. 11 to 13% of the total prokaryotic abundance in Atlantic and Mediterranean sediments and ca. 31% in Arctic sediments.
Fig. 2. Bathymetric patterns of the different microbial components in surface deep-sea sediments.
(A to D) Depth-related patterns of the abundances of bacteria (A), archaea (B), MG-I Thaumarchaeota (C), and MG-II Euryarchaeota (D) in the top 1 cm of sediments collected in the different oceanic regions. Means and SDs (n = 3) are reported.
Within the domain Archaea, the abundance of MG-I Thaumarchaeota in the sediments of the Arctic Ocean (1.72 ± 0.33 × 108 cells g−1) was significantly higher than in the Atlantic Ocean or in the Mediterranean Sea (0.33 ± 0.08 and 0.26 ± 0.05 × 108 cells g−1, respectively; Fig. 2C and table S3). Similarly, the abundance of MG-II Euryarchaeota was significantly higher in the Arctic Ocean (0.63 ± 0.12 × 108 cells g−1) than in the Atlantic Ocean and Mediterranean Sea (0.17 ± 0.03 × 108 and 0.22 ± 0.07 × 108 cells g−1, respectively; Fig. 2D and table S3). On average, Thaumarchaeota accounted for ca. 53% of the total archaeal abundance, whereas Euryarchaeota accounted for ca. 29%. The highest contribution of Euryarchaeota was observed in the sediments of the Atlantic Ocean and Mediterranean Sea (on average, ca. 30%), followed by benthic deep-sea ecosystems of the Arctic Ocean (25%).
Considering the whole data set, bacterial and archaeal abundances in the top 1 cm of the sediments investigated were significantly correlated (Pearson correlation, n = 174, r = 0.356, P < 0.01); they were positively related with trophic conditions (as biopolymeric C concentrations in the sediment; Fig. 3A) and inversely related with bottom water temperature (Fig. 3B).
Fig. 3. Benthic bacterial and archaeal abundances in relation to food availability and bottom water temperature.
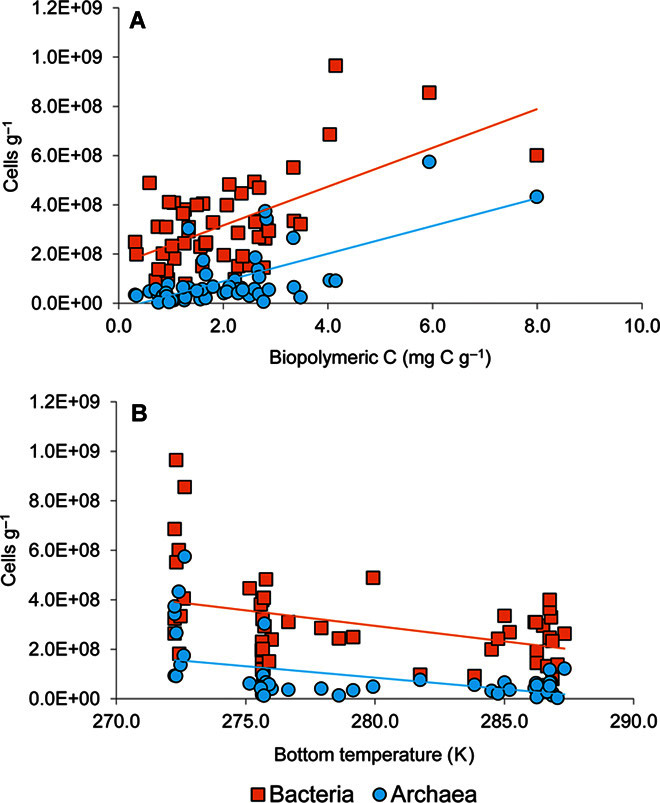
(A and B) Relationships between benthic bacterial and archaeal abundances and food availability (expressed as biopolymeric C concentrations in the top 1 cm of sediments) (A) and between benthic bacterial and archaeal abundances and bottom water temperature (B). (A) The color-coded fitted curves show the correlations for bacterial abundances (Y = 8 × 107 X + 2 × 108; n = 54; R2 = 0.360; P < 0.01) and archaeal abundances (Y = 6 × 107 X − 2 × 107; n = 54; R2 = 0.432; P < 0.01) versus food availability. (B) The color-coded fitted curves show the correlations for bacterial abundances (Y = −1 × 107 X + 4 × 109; n = 53; R2 = 0.151; P < 0.05) and archaeal abundances (Y = −9 × 106 X + 3 × 109; n = 53; R2 = 0.194; P < 0.05) versus bottom water temperature.
The abundances of bacteria, archaea, MG-I Thaumarchaeota, and MG-II Euryarchaeota in the sediments significantly increased from middle to high latitudes, considering either the whole data set or data selected at similar depths (that is, considering only sites at ca. 1000- or 2000-m depth; Fig. 4, A and B).
Fig. 4. Changes of the different benthic microbial components in relation to latitude.
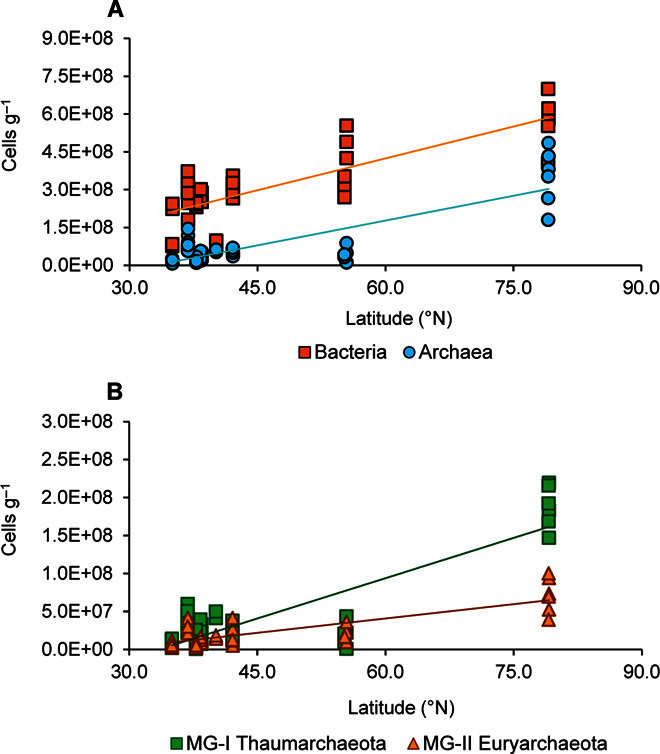
(A and B) Latitudinal patterns of bacterial and archaeal abundances (A) and MG-I Thaumarchaeota and MG-II Euryarchaeota abundances (B) in the top 1 cm of sediments collected in the different oceanic regions at similar depths. The color-coded fitted curves show the correlations for bacteria (Y = 8.42 × 106 X − 8.1 × 107; n = 45; R2 = 0.679; P < 0.01), archaea (Y = 6.56 × 106 X − 2.2 × 108; n = 45; R2 = 0.679; P < 0.01), MG-I Thaumarchaeota (Y = 3.54 × 106 X − 1.2 × 108; n = 45; R2 = 0.747; P < 0.01), and MG-II Euryarchaeota (Y = 1.26 × 106 X − 3.5 × 107; n = 45; R2 = 0.629; P < 0.01).
The abundances of all investigated microbial components significantly decreased with increasing depth along the vertical profile of the sediment [analysis of variance (ANOVA), P < 0.01; Fig. 5, A to D]. However, bacterial abundance decreased to a larger extent than archaeal abundance with increasing depth in the sediment, as also revealed by the increase of the archaeal-to-bacterial abundance ratio (on average, from 0.3 in the 0- to 1-cm sediment layer to 0.5 in the 10- to 15-cm layer). The MG-I Thaumarchaeota to MG-II Euryarchaeota abundance ratio decreased with increasing depth in the sediment (on average, from 1.8 to 1.2 at 0- to 1-cm and 10- to 15-cm depth, respectively).
Fig. 5. Distribution of the different microbial components along the vertical profiles of the sediments.
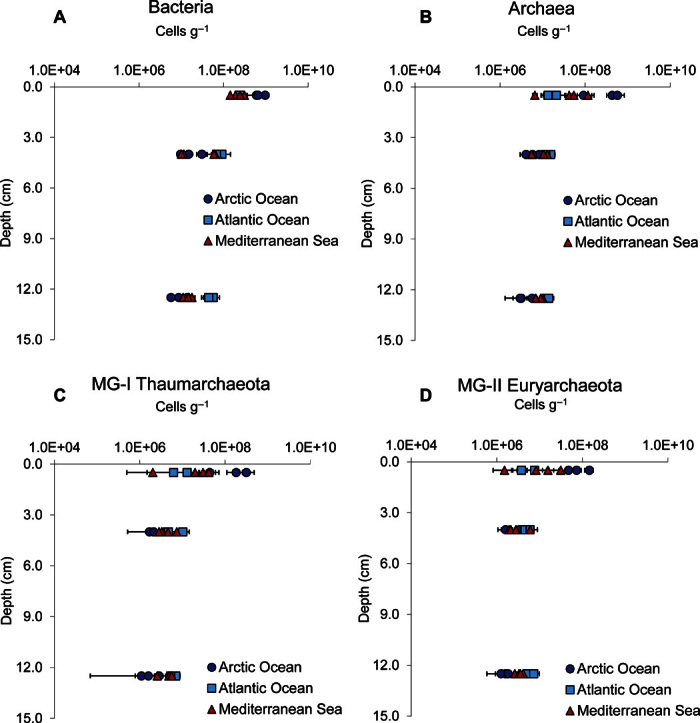
(A to D) Abundances of bacteria (A), archaea (B), MG-I Thaumarchaeota (C), and MG-II Euryarchaeota (D) along the vertical profiles of sediments (down to 15-cm depth) collected in the different oceanic regions. Means and SDs (n = 3) are reported.
Effects of the environmental variables on microbial components
The results of the regression tree analysis highlighted that bacterial distribution in all oceanic regions was primarily controlled by the availability of trophic resources, which overall explained 30% of the variance (Fig. 6). Bacterial distribution, especially at high latitudes, was also dependent on temperature (explaining 7% of the variance), whereas the food quality (as protein-to-carbohydrate ratio) explained 7% of the variance of the bacterial abundance in the Atlantic sediments. The output of the regression tree analysis provided evidence that archaeal abundances were dependent on bottom water temperatures (explaining 28% of the variance; Fig. 7), especially in the sediments of the polar region. In the middle-latitude ecosystems, benthic archaeal abundances were also controlled by the quantity and quality of trophic resources (explaining 17 and 11% of the variance, respectively). In all oceanic regions investigated, bottom water temperature was the only factor driving the distribution of MG-I Thaumarchaeota (29% of the variance; fig. S3), whereas food quantity and quality controlled the distribution of MG-II Euryarchaeota (explaining together 41% of the variance; fig. S4).
Fig. 6. Output of the regression tree analysis carried out to identify environmental factors explaining the distribution of bacterial abundances in the top 1 cm of sediments collected in the different oceanic regions.
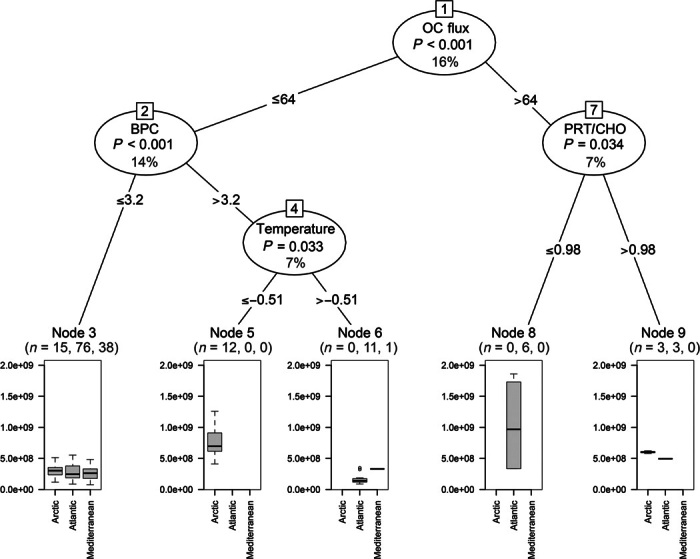
The significance level and the percentage of the explained variance of each predictor variable are reported along with the number of sampling sites for each oceanic region at the terminal nodes. OC flux, organic C that reaches the sea floor through particle sinking (expressed as mg C m−2 day−1); BPC, biopolymeric C concentrations in the sediment (expressed as mg C g−1); PRT/CHO, protein-to-carbohydrate ratio in the sediment (adimensional).
Fig. 7. Output of the regression tree analysis carried out to identify environmental factors explaining the distribution of archaeal abundances in the top 1 cm of sediments collected in the different oceanic regions.
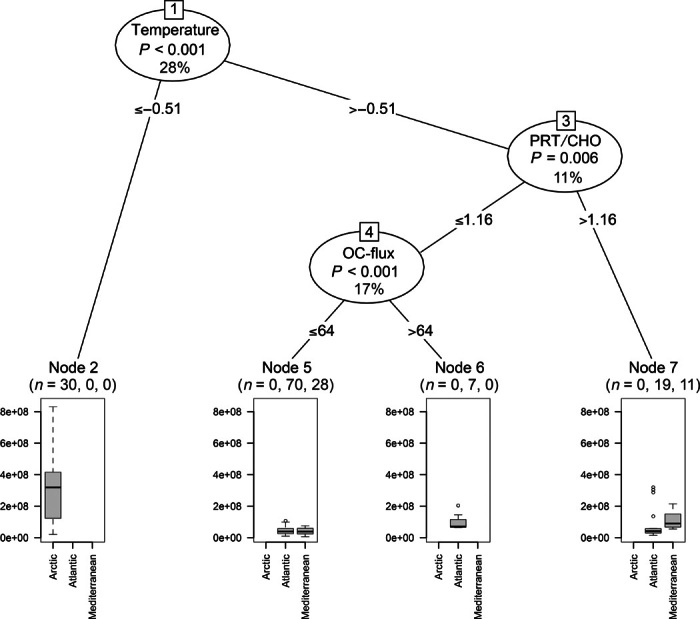
The significance level and the percentage of the explained variance of each predictor variable are reported along with the number of sampling sites for each oceanic region at the terminal nodes. OC flux, organic C that reaches the sea floor through particle sinking (expressed as mg C m−2 day−1); PRT/CHO, protein-to-carbohydrate ratio in the sediment (adimensional).
DISCUSSION
The presence of archaea in deep-sea sediments was documented for the first time more than 15 years ago (28), and all available studies conducted to date on surface marine sediments (almost exclusively at shallow depths) reported that archaea provide an almost negligible contribution to total prokaryotic abundance (from <1 to 6%) (29–32). Only recently, the quantitative importance of archaea has been reevaluated in subsurface sediments (down to >300 m below the sediment surface) (5, 19, 33–36), but no attempts have been made yet to analyze these benthic components in a macroecological perspective.
Results presented here provide evidence that the relative importance of archaea in surface deep-sea sediments of different biogeographic regions is significantly higher (range, 11 to 31%) than the one reported from all continental shelf ecosystems investigated to date (29–32). Because we used protocols here that were largely used previously, such differences cannot be due to methodological biases. We are aware that the reliability of CARD-FISH analysis to quantify the abundances of bacteria and archaea in benthic ecosystems depends on an array of factors, including the following: (i) the procedure used for the dislodgement of the cells from the sediment, (ii) the methodology used to permeabilize the cell wall, (iii) the selection of specific probes, and (iv) the hybridization conditions (22, 23, 32). Here, we have carefully taken into account all of these factors to optimize the procedure for the determination of bacterial and archaeal abundances in deep-sea sediments. In addition, we performed qPCR analyses on the same samples where CARD-FISH analyses were conducted, and we found that the two methods provided consistent results.
Meta-analyses on benthic prokaryotic abundances (reported as bacteria) at global scale showed the lack of bathymetric patterns (7, 10). Here, using the most consistent data set available so far (that is, same sampling strategy, same sampling equipment, and same protocols for the analyses; use of multiple independent approaches), we provide evidence of the lack of depth-related patterns for either bacterial and archaeal abundance in all oceanic regions investigated.
Conversely to terrestrial ecosystems, photosynthetic primary production in the open oceans increases from equatorial/tropical toward higher latitudes (37). Such a gradient, influencing the magnitude of organic carbon export from the surface waters to the deep sea floor through sinking particles, is expected to play an important role in controlling the metabolism and standing stocks of benthic assemblages in the deep sea (1).
Here, we show that bacterial and archaeal abundances covaried, suggesting that the two prokaryotic domains are partly controlled by the same factors. For instance, the abundances of either bacteria or archaea were significantly related with food availability in the sediment (that is, as biopolymeric C concentrations). Similarly, a positive relationship between trophic conditions and richness of bacterial taxa has been previously reported in surface deep-sea sediments (38).
Our data provide unprecedented evidence that the abundances of both benthic bacteria and archaea in the deep sea increased from middle to high latitudes. Indeed, the highest abundances of both prokaryotic domains were reported in Arctic sediments, which were characterized by a higher availability of trophic resources. These results hold true either considering only the abundances at equal depths (for example, at 1000- or 2000-m depth) or pooling together all sampling depths (that is, 228 benthic samples).
We also show that the abundances of the two dominant groups of the archaeal domain, MG-I Thaumarchaeota and MG-II Euryarchaeota, increased from middle to high latitudes but to a larger extent for MG-I Thaumarchaeota. An increasing abundance of Thaumarchaeota (previously defined as marine Crenarchaeota Group I) toward high latitudes (from 5°N to 65°N) has been reported in ammonia-rich deep-water masses of the North Atlantic (39). This could be explained by the fact that higher concentrations of ammonia (that is, higher remineralization activity by heterotrophic components) can sustain higher chemoautotrophic production rates of Thaumarchaeota through ammonia oxidation processes (3, 39).
Here, an increase of the archaeal-to-bacterial abundance ratio with increasing depth in the sediment was observed. This pattern has been described for subsurface sediments (5), and our results refine these findings also for the top 15 cm of the sediment. The subsurface sediments are poorer than surface sediments in terms of food and energy resources (for example, labile organic compounds, electron acceptors, dissolved substrates, and metabolites) so that the increase of the relative importance of archaea with increasing depth in the sediment could be due to a higher efficiency to grow under these limiting conditions (40). However, further manipulative and in situ studies are needed to support this hypothesis.
Previous studies, using random forest model applied to a data set compiled from literature, reported that primary productivity predictors (that is, sea surface temperature, irradiance, mixed layer depth, phytoplankton biomass and growth rates, and chlorophyll- and carbon-based primary production) and water depth explained a large fraction of the variance of the distribution of benthic prokaryotic abundance and biomass of the global oceans (7). These findings provide further support of the role of surface primary production processes and related fluxes of particulate organic carbon to the sea floor in driving the distribution of benthic prokaryotic standing stocks worldwide. The authors also hypothesized that an important portion of benthic prokaryotic standing stocks could come from prokaryotes attached to particles sinking on the ocean floor. However, the supply of prokaryotes adsorbed onto sinking particles to the deep sea (on average, 2.5 to 4.1 × 108 cells m−2 day−1 at 1000- and 4700-m depth) (41) is orders of magnitude lower than prokaryotes in situ produced on the ocean floor (0.3 to 1.4 × 1012 cells m−2 day−1; obtained by combining prokaryotic abundance and their turnover rates contextually determined) (11).
Here, we found that both bacterial and archaeal abundances significantly decreased with increasing bottom water temperature. This applies even when the “warmer” Mediterranean samples were excluded from the analysis.
The regression tree analysis conducted in the present study revealed that different environmental drivers were responsible for the distribution of bacteria and archaea. For instance, in all oceanic regions investigated, bacterial abundances were primarily controlled by food availability, thus suggesting a prevalence of heterotrophic metabolism. Conversely, archaeal abundances were mostly driven by changes in temperature of bottom waters. In particular, the distribution of archaea in the polar region was exclusively dependent on bottom water temperatures, whereas in the other oceanic regions, the quantity and quality of food resources played a more important role. Because archaea are abundant in diverse low-temperature environments throughout the globe (42), our findings suggest a thermal adaptation of benthic archaea to colder deep-sea ecosystems, possibly due to the specific characteristics of their cell membranes and proteins (42–44).
Our findings also revealed that MG-I Thaumarchaeota and MG-II Euryarchaeota displayed a different sensitivity toward changes in the main environmental drivers identified. In particular, MG-I Thaumarchaeota, which represent the dominant component of archaeal assemblages in surface deep-sea sediments, were apparently very sensitive to changes in temperature at all latitudes investigated (see the results of the regression tree analysis). Conversely, the abundance of MG-II Euryarchaeota was dependent on the quantity and quality of organic food resources, supporting the hypothesis of a heterotrophic (or mixotrophic) metabolism in Euryarchaeota (45). The different sensitivity of these two archaeal groups to the main environmental drivers might favor their coexistence by reducing niche overlap and competition for available resources.
Overall, our results suggest that the archaeal contribution to the total benthic microbial biomass in surface deep-sea sediments worldwide could be far larger than previously hypothesized, especially at high latitudes. Combining worldwide estimates of prokaryotic abundances, obtained by dividing their global biomass previously reported (7) by C content per cell (10 to 20 fg C per cell) (11, 19), with the average contribution of archaea to total prokaryotes that we found (on average, 20%), we estimated that archaeal abundance on the ocean floor is ca. 0.4 to 0.7 × 1027. Such archaeal abundances once projected on the top 15 cm of deep-sea sediments worldwide are equivalent to ca. 5 to 10% of the total abundance of pelagic archaea in the global oceans (21). Because our findings indicate that >50% of archaea in the surface sediments are accounted for by MG-I Thaumarchaeota, we suggest that this component can have a crucial role in biomass production and biogeochemical cycles of benthic deep-sea ecosystems at a global scale.
Global changes are predicted to have stronger effects at the high latitudes, particularly in the Northern Hemisphere (46), with expected increase of temperature and decrease of salinity due to freshening caused by ice melting. The response of the different deep-sea microbial components to altered thermohaline and trophic conditions due to global change or climate-driven episodic events has been, so far, largely unknown (47, 48). However, the results reported in this study suggest that changes in temperature and trophic characteristics could significantly influence the abundance and structure of benthic prokaryotic assemblages and provide evidence of a potentially major impact on archaea and prokaryotic assemblages at high latitudes.
The findings presented here contribute to our understanding of the functioning of deep-sea ecosystems and of the expected changes in the structure of prokaryotic assemblages in response to changes in environmental variables. These results will contribute to better forecast the potential consequences of climate changes on benthic processes and biogeochemical cycles in the largest ecosystem on Earth.
MATERIALS AND METHODS
Study areas
The investigated regions display important differences in terms of thermohaline characteristics. The wide variability of temperature and salinity values in the northeast Atlantic Ocean are due to differences in the thermohaline conditions of water masses at different depths. In particular, at latitudes comprising 53°N to 56°N (that is, including area 2 investigated), these water masses include the following: (i) the Antarctic Bottom Waters (at a depth >3000 m), (ii) the Northeast Atlantic Deep Waters (at depths from 2500 to 3000), (iii) the Labrador Sea Waters (from 1500- to 2000-m depth), (iv) Subarctic Intermediate Waters (from 800- to 1000-m depth, (v) the Mediterranean Waters (from 700- to 800-m depth), and (vi) the Eastern North Atlantic Waters (in the upper 700 m). The water masses in the northeast Atlantic Ocean at latitudes comprising 38°N to 46°N (that is, including area 3 investigated) include the following: (i) the Lower Deep Waters (mainly composed of Antarctic Bottom Waters), at depths >4000 m (that is, across the abyssal plains); (ii) the North Atlantic Deep Waters (from 4000- and 2200-m depth); (iii) the Labrador Sea Waters, circulating regionally toward the southwest between 2200- and 1500-m depth; (iv) the Mediterranean Outflow Waters (from 1500- to 600-m depth), flowing along the Iberian margin to the Bay of Biscay; and (v) the North Atlantic Central Waters, in the top 600-m depth. The peculiar thermohaline characteristics of the Mediterranean Sea (high temperature and high salinity, when compared to the Atlantic waters) are due to the local climate, which, in this semi-enclosed basin, determines a deficit of the water budget.
The Mediterranean Sea is characterized by three major water layers: (i) the surface waters flowing eastward and northward and (ii) the intermediate and (iii) deep waters flowing westward and filling more than 70% of the whole Mediterranean basin. The thermohaline circulation is driven by seasonal variations of salinity and by changes in the surface water temperature. The Levantine Intermediate Waters flowing from the eastern to the western basin at 300- to 600-m depth can be identified through their high-salinity values. Surface water cooling during winter in the Adriatic region and in the northwest Mediterranean is also responsible for the formation of dense waters, ventilating the deep Mediterranean water masses and leading to the formation of the Eastern Mediterranean Deep Waters and the Western Mediterranean Deep Waters.
Sampling design
Sediment samples were collected from 58 deep-sea sites in the Arctic Ocean (Hausgarten region, ca.120 km west of Spitsbergen, an island of the Svalbard archipelago in Norway), in two areas of the northeast Atlantic Ocean (offshore of northeast Ireland and west Iberia), and in three areas of the Mediterranean Sea (North Western, Central, and Eastern Mediterranean). The areas investigated included only open-ocean sites and continental-margin systems and excluded specific hot-spot ecosystems (that is, deep-water coral sites, cold seeps, and hydrothermal vents). This allowed comparisons of similar systems covering >96% of the deep-sea surface. At all of the sampling sites, the sediments were collected using a multiple corer (MaxiCorer; inside diameter, 9.0 cm; depth penetration, >20 cm). Sediment samples from replicate sediment cores (n = 3) were collected from independent multiple-corer deployments at each deep-sea site (see the Supplementary Materials for details).
Environmental and trophic characteristics
Bottom water temperature and salinity were measured by a conductivity-temperature-depth profiler. Chlorophyll a and phaeopigments were extracted from sediment samples using 90% acetone (24 hours in the dark, at 4°C) and then analyzed fluorometrically (49). After centrifugation (800g), the supernatant was used to determine the functional chlorophyll a and thereafter acidified with 0.1 N HCl to estimate the amount of phaeopigments (49). Total phytopigments were defined as the sum of chlorophyll a and phaeopigments, and their concentrations were reported as μg g−1. The concentrations of proteins, carbohydrates, and lipids in the sediment were determined spectrophotometrically and expressed as bovine serum albumin, glucose, and tripalmitine equivalents, respectively (49). Biopolymeric C concentrations in the sediments were obtained by the sum of the carbohydrate, protein, and lipid concentrations converted into carbon equivalents (using the conversion factors of 0.40, 0.49, and 0.75 mg C mg−1, respectively) (26).
Organic C fluxes that reached the sea floor through particle sinking were estimated on the basis of the net photosynthetic primary production extracted from the ocean productivity database (www.science.oregonstate.edu/ocean.productivity/index.php), derived from a C-based productivity model algorithm (50), and referred to the same sampling periods when the sediments were collected and the water column depth (51). The following equation was used
where JCorg (mg C m−2 day−1) is the flux of organic C, PP is the net photosynthetic primary production (mg C m−2 day−1), and z is the water depth (m). This model takes into consideration the degradation processes occurring to organic particles during their sinking, thus allowing to estimate the amount of organic material that actually reaches the surface of the deep-sea floor.
Prokaryotic abundance
Prokaryotic abundance was determined after cell detachment from the sediment using pyrophosphate (final concentration, 5 mM) and ultrasound treatment (three times, 1 min each) (11). The samples were then diluted with sterile and 0.2-μm prefiltered formaldehyde solution (final concentration, 2%); following filtering onto 0.2-μm pore size filters, the samples were stained using SYBR Green I. The filters were analyzed by epifluorescence microscopy (×1000 magnification). For each slide, at least 10 fields were observed, and a total of at least 400 cells were counted. Prokaryotic abundances were normalized to sediment dry weight after desiccation (60°C, 24 hours).
Quantification of bacteria, archaea, MG-I Thaumarchaeota, and MG-II Euryarchaeota
The analysis of the prokaryotic assemblage structure (bacteria versus archaea and MG-I Thaumarchaeota and MG-II Euryarchaeota) was performed by CARD-FISH (32, 52), following the procedure optimized for deep-sea sediments (22). Briefly, sediment samples were centrifuged (16,000g for 5 min), washed with phosphate-buffered saline (PBS), then centrifuged (16,000g for 5 min), and resuspended in PBS/96% ethanol. The samples were then treated with ultrasounds (three times, 1 min each), properly diluted, and then filtered onto 0.2-μm polycarbonate membrane filters. The filters were dipped in low–gelling point agarose [0.1% (w/v) in Milli-Q water], dried on a glass petri dish at 37°C, and dehydrated in 95% ethanol. Cell wall permeabilization was optimized by incubation at 37°C with lysozyme for bacteria or proteinase K for archaea. After Milli-Q washing and incubation in 10 mM HCl (room temperature, 20 min), the filters were washed again with Milli-Q water, dehydrated in 95% ethanol, dried, and hybridized with oligonucleotide horseradish peroxidase–labeled probes targeting bacteria, archaea, MG-I Thaumarchaeota, and MG-II Euryarchaeota (table S2). Such probes were selected for consistency with previous studies investigating archaeal and bacterial dynamics in deep-sea sediments conducted worldwide (23, 53). Hybridization (35°C for bacteria and 46°C for archaea) was performed for 2 hours. Then, the filters were transferred into preheated washing buffer, placed in PBS buffer (pH 7.6, 0.05% Triton X-100), and incubated at room temperature for 15 min. After removal of the buffer, the samples were incubated for 30 min in the dark at 37°C for Cy3-tyramide signal amplification. Filters were then observed under epifluorescence microscopy. All analyses were performed in triplicate. The data were normalized to sediment dry weight after desiccation (60°C, 24 hours).
Comparative analyses on the same sediment samples were based on qPCR, using the TaqMan technology, targeting 16S ribosomal RNA (rRNA) genes of bacteria and archaea. To remove potential biases due to the presence of extracellular DNA, sediment samples were pretreated with a chemical-physical procedure before the extraction of DNA by in situ cell lysis procedure (49, 54). The DNA was then extracted and purified by using the UltraClean Soil DNA Isolation Kit according to the manufacturer’s instructions. The quantification of archaeal and bacterial 16S rRNA genes was performed using primers and probes selectively targeting archaea (forward primer, 5′-GYGCASCAGKCGMGAAW-3′; reverse primer, 5′-GGACTACVSGGGTATCTAAT-3′; probe, 5′-TGYCAGCCGCCGCGGTAAHACCVGC-3′) (55) or bacteria (forward primer, 5′-TCCTACGGGAGGCAGCAGT-3′; reverse primer, 5′-GGACTACCAGGGTATCTAATCCTGTT-3′; probe, 5′-CGTATTACCGCGGCTGCTGGCAC-3′) (56). These primers and probes were selected for consistency with previous studies investigating archaeal and bacterial dynamics in deep-sea sediments conducted worldwide (23, 57). Hydrolysis probes for qPCR TaqMan assay contained a fluorescent reporter dye (6-FAM) in 5′, and a Black Hole Quencher 1 (BHQ-1) in the 3′ position. The amplification reactions were performed in a final volume of 15 μl in film-sealed optical 96-well qPCR plates. Each reaction contained the iQ Supermix (Bio-Rad), 0.8/0.2 μM (for archaeal) or 0.1/0.1 μM (for bacterial) primers/probes, respectively, and 1 μl of template DNA. According to the standard MIQE (Minimum Information for Publication of Quantitative Real-Time PCR Experiments) guidelines for best practice in qPCR analyses (58), calibration curves were included in all reactions, with a five-log10 linear dynamic range, from 2.5 × 10−1 to 2.5 × 10−5 pg of template 16S rDNA from Escherichia coli (for bacteria) or Methanocaldococcus jannaschii (for archaea). The limit of detection of the assay was ca. 25 and 30 16S rRNA gene copies per reaction, for bacteria and archaea, respectively. To test for possible inhibition of qPCR, reactions were run in triplicate, using undiluted aliquots of DNA obtained from the different sediment samples, in addition to running all sample extracts in serial 10-fold dilutions. The log-linear relationship between quantification cycle (Cq) and the dilution factor was obtained in all of the samples by using the 10-fold dilution, allowing us to exclude possible bias due to qPCR inhibition (59, 60). The optimal concentration of primers and probes and the thermal cycles were set following temperature and primer/probe concentration gradient tests. The adopted thermal cycling conditions were as follows: 3 min at 95°C, followed by 40 cycles for 15 s at 95°C, 1 min at 60°C for bacteria; 3 min at 95°C, followed by 40 cycles for 15 s at 95°C, 5 min at 57°C for archaea. The presence of a single PCR product of the expected length size was checked using 1% agarose gel electrophoresis. The iQ5 Optical System 2.1 Software was used to calculate Cq, efficiency (E), and R2 values for each reaction. Results from qPCR runs were used in this study if E > 98% and < 102%, and R2 > 0.99 (even more stringent than MIQE guidelines); samples from the same incubation experiment were always analyzed within the same qPCR run, thus ensuring technical consistency. All samples, standards, and negative controls were analyzed in triplicate qPCR reactions. Any run for which negative controls were positive was excluded and conducted again. The number of copies of 16S rRNA genes was calculated on the basis of the template length in base pairs (bp) and an average weight of 650 daltons bp−1.
Statistical analyses
To test for differences in the investigated microbial variables among oceanic regions and sampling areas, ANOVA [using permutational multivariate analysis of variance (PERMANOVA)] was carried out. Before analysis, the homogeneity of variance was checked using the Cochran test on appropriately transformed data, whenever necessary. For those data sets for which the transformation did not allow to obtain homogeneous variances, a more conservative level of significance was considered. When significant differences were encountered, a post hoc comparison test (at α = 0.05) was also carried out. To identify factors influencing the abundances of bacteria, archaea, MG-I Thaumarchaeota, and MG-II Euryarchaeota, regression tree analyses were carried out (61). In particular, temperature, salinity, and quantity and quality of organic matter were used as predictor variables. Conditional inference procedures were applied at recursive binary portioning to solve two problems of exhaustive search procedures: overfitting and selection bias toward covariates with many possible splits (62). The conditional inference trees were constructed with cquad-type test statistics and α = 0.05 with Bonferroni correction. Regression tree analysis was performed using “party” package version 1.0-25 in free statistical environment R (R version 3.2.2) (63).
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for useful suggestions. Funding: This study was conducted in the framework of the European funded projects HERMIONE (Hotspot Ecosystem Research and Man’s Impact On European Seas) (contract no. 226354) and MIDAS (Managing Impacts of Deep-sea Resource Exploitation) (grant agreement no. 603418) and of the Flagship Project RITMARE—The Italian Research for the Sea—coordinated by the Italian National Research Council and funded by the Italian Ministry of Education, University and Research within the National Research Program. Author contributions: R.D., C.C., and A.D. planned the project; M.M. performed the laboratory and part of the fieldwork; A.D., C.C., M.M., and R.D. performed the data analysis; R.D., M.M., C.C., and A.D. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/4/e1500961/DC1
Supplementary Materials and Methods
table S1. Details of the station locations, temperature, and salinity of bottom waters, total phytopigment (CPE) and biopolymeric C (BPC) concentrations, and protein-to-carbohydrate ratio (PRT/CHO) in surface sediments (0 to 1 cm), net photosynthetic primary production (NPP), and organic C fluxes (OC fluxes).
table S2. Output of the in silico analysis dealing with the coverage of probes targeting 16S rRNA used in the present study.
table S3. Statistical analysis testing for differences in the distribution of the different microbial components.
fig. S1. Depth-related patterns of total prokaryotic abundances (obtained using SYBR Green I) in the top 1 cm of sediments collected in the different oceanic regions.
fig. S2. Comparison of the abundances of bacteria and archaea obtained by CARD-FISH, with the number of 16S rDNA copies of bacteria and archaea obtained in surface sediments of different oceanic regions.
fig. S3. Output of the regression tree analysis carried out to identify environmental factors explaining the distribution of MG-I Thaumarchaeota in the top 1 cm of sediments collected in the different oceanic regions.
fig. S4. Output of the regression tree analysis carried out to identify environmental factors explaining the distribution of MG-II Euryarchaeota in the top 1 cm of sediments collected in the different oceanic regions.
REFERENCES AND NOTES
- 1.Danovaro R., Snelgrove P. V. R., Tyler P., Challenging the paradigms of deep-sea ecology. Trends Ecol. Evol. 29, 465–475 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Canals M., Puig P., de Madron X. D., Heussner S., Palanques A., Fabres J., Flushing submarine canyons. Nature 444, 354–357 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Molari M., Manini E., Dell’Anno A., Dark inorganic carbon fixation sustains the functioning of benthic deep-sea ecosystems. Global Biogeochem. Cycles, 27, 212–221 (2013). [Google Scholar]
- 4.Jørgensen B. B., Boetius A., Feast and famine—Microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5, 770–781 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Lipp J. S., Morono Y., Inagaki F., Hinrichs K.-U., Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454, 991–994 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Biddle J. F., Lipp J. S., Lever M. A., Lloyd K. G., Sørensen K. B., Anderson R., Fredricks H. F., Elvert M., Kelly T. J., Schrag D. P., Sogin M. L., Brenchley J. E., Teske A., House C. H., Hinrichs K.-U., Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U.S.A. 103, 3846–3851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei C.-L., Rowe G. T., Escobar-Briones E., Boetius A., Soltwedel T., Caley M. J., Soliman Y., Huettmann F., Qu F., Yu Z., Pitcher C. R., Haedrich R. L., Wicksten M. K., Rex M. A., Baguley J. G., Sharma J., Danovaro R., MacDonald I. R., Nunnally C. C., Deming J. W., Montagna P., Lévesque M., Weslawski J. M., Wlodarska-Kowalczuk M., Ingole B. S., Bett B. J., Billett D. S. M., Yool A., Bluhm B. A., Iken K., Narayanaswamy B. E., Global patterns and predictions of seafloor biomass using random forests. PLOS One 5, e15323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danovaro R., Marrale D., Dell’Anno A., Della Croce N., Tselepides A., Fabiano M., Bacterial response to seasonal changes in labile organic matter composition on the continental shelf and bathyal sediments of the Cretan Sea. Prog. Oceanogr. 46, 345–366 (2000). [Google Scholar]
- 9.Quéric N.-V., Soltwedel T., Arntz W. E., Application of a rapid direct viable count method to deep-sea sediment bacteria. J. Microbiol. Meth. 57, 351–367 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Rex M. A., Etter R. J., Morris J. S., Crouse J., McClain C. R., Johnson N. A., Stuart C. T., Deming J. W., Thies R., Avery R., Global bathymetric patterns of standing stock and body size in the deep-sea benthos. Mar. Ecol. Prog. Ser. 317, 1–8 (2006). [Google Scholar]
- 11.Danovaro R., Dell’Anno A., Corinaldesi C., Magagnini M., Noble R., Tamburini C., Weinbauer M., Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454, 1084–1087 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Boetius A., Ferdelman T., Lochte K., Bacterial activity in sediments of the deep Arabian Sea in relation to vertical flux. Deep-Sea Res. Pt. II 47, 2835–2875 (2000). [Google Scholar]
- 13.Turley C., Bacteria in the cold deep-sea benthic boundary layer and sediment–water interface of the NE Atlantic. FEMS Microbiol. Ecol. 33, 89–99 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Turley C. M., Dixon J. L., Bacterial numbers and growth in surficial deep-sea sediments and phytodetritus in the NE Atlantic: Relationships with particulate organic carbon and total nitrogen. Deep-Sea Res. Pt. I 49, 815–826 (2002). [Google Scholar]
- 15.Deming J. W., Carpenter S. D., Factors influencing benthic bacterial abundance, biomass, and activity on the northern continental margin and deep basin of the Gulf of Mexico. Deep-Sea Res. Pt. II 55, 2597–2606 (2008). [Google Scholar]
- 16.Giovannelli D., Molari M., d’Errico G., Baldrighi E., Pala C., Manini E., Large-scale distribution and activity of prokaryotes in deep-sea surface sediments of the Mediterranean Sea and the adjacent Atlantic Ocean. PLOS One 8, e72996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arrieta J. M., Mayol E., Hansman R. L., Herndl G. J., Dittmar T., Duarte C. M., Dilution limits dissolved organic carbon utilization in the deep ocean. Science 348, 331–333 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Dell’Anno A., Corinaldesi C., Danovaro R., Virus decomposition provides an important contribution to benthic deep-sea ecosystem functioning. Proc. Natl. Acad. Sci. U.S.A. 112, E2014–E2019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kallmeyer J., Pockalny R., Adhikari R. R., Smith D. C., D’Hondt S., Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 109, 16213–16216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClain C. R., Allen A. P., Tittensor D. P., Rex M. A., Energetics of life on the deep seafloor. Proc. Natl. Acad. Sci. U.S.A. 109, 15366–15371 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karner M. B., DeLong E. F., Karl D. M., Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Molari M., Manini E., Reliability of CARD-FISH procedure for enumeration of Archaea in deep-sea surficial sediments. Curr. Microbiol. 64, 242–250 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Lloyd K. G., May M. K., Kevorkian R. T., Steen A. D., Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor. Appl. Environ. Microbiol. 79, 7790–7799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durbin A. M., Teske A., Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ. Microbiol. 13, 3219–3234 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Orcutt B. N., Sylvan J. B., Knab N. J., Edwards K. J., Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pusceddu A., Dell’Anno A., Fabiano M., Danovaro R., Quantity and bioavailability of sediment organic matter as signatures of benthic trophic status. Mar. Ecol. Prog. Ser. 375, 41–52 (2009). [Google Scholar]
- 27.Dell’Anno A., Danovaro R., Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science 309, 2179 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Vetriani C., Jannasch H. W., MacGregor B. J., Stahl D. A., Reysenbach A.-L., Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol. 65, 4375–4384 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahm K., Berninger U.-G., Abundance, vertical distribution, and community structure of benthic prokaryotes from permanently cold marine sediments (Svalbard, Arctic Ocean). Mar. Ecol. Prog. Ser. 165, 71–80 (1998). [Google Scholar]
- 30.Llobet-Brossa E., Rosellò-Mora R., Amann R., Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64, 2691–2696 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravenschlag K., Sahm K., Amann R., Quantitative molecular analysis of the microbial community in marine arctic sediments (Svalbard). Appl. Environ. Microbiol. 67, 387–395 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishii K., Mußmann M., MacGregor B. J., Amann R., An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microb. Ecol. 50, 203–213 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Schippers A., Köweker G., Höft C., Teichert B. M., Quantification of microbial communities in forearc sediment basins off Sumatra. Geomicrobiol. J. 27, 170–182 (2010). [Google Scholar]
- 34.Schippers A., Kock D., Höft C., Köweker G., Siegert M., Quantification of microbial communities in subsurface marine sediments of the Black Sea and off Namibia. Front. Microbiol. 3, 16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jorgensen S. L., Hannisdal B., Lanzén A., Baumberger T., Flesland K., Fonseca R., Øvreås L., Steen I. H., Thorseth I. H., Pedersen R. B., Schleper C., Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl. Acad. Sci. U.S.A. 109, E2846–E2855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie S., Lipp J. S., Wegener G., Ferdelman T. G., Hinrichs K.-U., Turnover of microbial lipids in the deep biosphere and growth of benthic archaeal populations. Proc. Natl. Acad. Sci. U.S.A. 110, 6010–6014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P., Primary production of the Biosphere: Integrating terrestrial and oceanic components. Science 281, 237–240 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Bienhold C., Boetius A., Ramette A., The energy–diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J. 6, 724–732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varela M. M., Van Aken H. M., Sintes E., Herndl G. J., Latitudinal trends of Crenarchaeota and Bacteria in the meso- and bathypelagic waters of the eastern North Atlantic. Environ. Microbiol. 10, 110–124 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Valentine D. L., Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 5, 316–323 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Vanucci S., Dell’Anno A., Pusceddu A., Fabiano M., Lampitt R. S., Danovaro R., Microbial assemblages associated with sinking particles in the Porcupine Abyssal Plain (NE Atlantic Ocean). Prog. Oceanogr. 50, 105–121 (2001). [Google Scholar]
- 42.Cavicchioli R., Cold-adapted archaea. Nat. Rev. Microbiol. 4, 331–343 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Cavicchioli R., Thomas T., Curmi P. M. G., Cold stress response in Archaea. Extremophiles 4, 321–331 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Feller G., Gerday C., Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 1, 200–208 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Lloyd K. G., Schreiber L., Petersen D. G., Kjeldsen K. U., Lever M. A., Steen A. D., Stepanauskas R., Richter M., Kleindienst S., Lenk S., Schramm A., Jørgensen B. B., Predominant archaea in marine sediments degrade detrital proteins. Nature 496, 215–218 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Hoegh-Guldberg O., Bruno J. F., The impact of climate change on the world’s marine ecosystems. Science 328, 1523–1528 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Danovaro R., Dell’Anno A., Fabiano M., Pusceddu A., Tselepides A., Deep-sea ecosystem response to climate changes: The eastern Mediterranean case study. Trends Ecol. Evol. 16, 505–510 (2001). [Google Scholar]
- 48.Mora C. , Wei C.-L., Rollo A., Amaro T., Baco A. R., Billett D., Bopp L., Chen Q., Collier M., Danovaro R., Gooday A. J., Grupe B. M., Halloran P. R., Ingels J., Jones D. O. B., Levin L. A., Nakano H., Norling K., Ramirez-Llodra E., Rex M., Ruhl H. A., Smith C. R., Sweetman A. K., Thurber A. R., Tjiputra J. F., Usseglio P., Watling L., Wu T., Yasuhara M., Biotic and human vulnerability to projected changes in ocean biogeochemistry over the 21st century. PLOS Biol. 11, e1001682 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R. Danovaro, Methods for the study of deep-sea sediments, their functioning and biodiversity (CRC Press, UK, 2010). [Google Scholar]
- 50.Behrenfeld M. J., Boss E., Siegel D. A., Shea D. M., Carbon-based ocean productivity and phytoplankton physiology from space. Global Biogeochem. Cycles 19, GB1006 (2005). [Google Scholar]
- 51.W. H. Berger, K. Fischer, C. Lai, G. Wu, Ocean Productivity and Organic Carbon Flux. I. Overview and Maps of Primary Production and Export Production (University of California, San Diego, CA, 1987). [Google Scholar]
- 52.Teira E., Reinthaler T., Pernthaler A., Pernthaler J., Herndl G. J., Combining catalyzed reporter deposition-fluorescence in-situ hybridization and microautoradiography to detect substrate utilization by Bacteria and Archaea in the deep ocean. Appl. Environ. Microbiol. 70, 4411–4414 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schippers A., Neretin L. N., Kallmeyer J., Ferdelman T. G., Cragg B. A., Parkes R. J., Jørgensen B. B., Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433, 861–864 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Corinaldesi C., Tangherlini M., Luna G. M., Dell’Anno A., Extracellular DNA can preserve the genetic signatures of present and past viral infection events in deep hypersaline anoxic basins. P. Roy. Soc. B-Biol. Sci. 281, 20133299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takai K., Horikoshi K., Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66, 5066–5072 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nadkarni M. A., Martin F. E., Jacques N. A., Hunter N., Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148, 257–266 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Schippers A., Neretin L. N., Quantification of microbial communities in near-surface and deeply buried marine sediments on the Peru continental margin using real-time PCR. Environ. Microbiol. 8, 1251–1260 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., Wittwer C. T., The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Cao Y., Griffith J. F., Dorevitch S., Weisberg S. B., Effectiveness of qPCR permutations, internal controls and dilution as means for minimizing the impact of inhibition while measuring Enterococcus in environmental waters. J. Appl. Microbiol. 113, 66–75 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Lloyd K. G., MacGregor B. J., Teske A., Quantitative PCR methods for RNA and DNA in marine sediments: Maximizing yield while overcoming inhibition. FEMS Microbiol. Ecol. 72, 143–151 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Dèath G., Fabricius K. E., Classification and regression trees: A powerful yet simple technique for ecological data analysis. Ecology 81, 3178–3192 (2000). [Google Scholar]
- 62.Hothorn T., Hornik K., Zeileis A., Unbiased recursive partitioning: A conditional inference framework. J. Comput. Graph. Stat. 15, 651–674 (2006). [Google Scholar]
- 63.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2013); www.R-project.org/ [accessed January 2016].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/4/e1500961/DC1
Supplementary Materials and Methods
table S1. Details of the station locations, temperature, and salinity of bottom waters, total phytopigment (CPE) and biopolymeric C (BPC) concentrations, and protein-to-carbohydrate ratio (PRT/CHO) in surface sediments (0 to 1 cm), net photosynthetic primary production (NPP), and organic C fluxes (OC fluxes).
table S2. Output of the in silico analysis dealing with the coverage of probes targeting 16S rRNA used in the present study.
table S3. Statistical analysis testing for differences in the distribution of the different microbial components.
fig. S1. Depth-related patterns of total prokaryotic abundances (obtained using SYBR Green I) in the top 1 cm of sediments collected in the different oceanic regions.
fig. S2. Comparison of the abundances of bacteria and archaea obtained by CARD-FISH, with the number of 16S rDNA copies of bacteria and archaea obtained in surface sediments of different oceanic regions.
fig. S3. Output of the regression tree analysis carried out to identify environmental factors explaining the distribution of MG-I Thaumarchaeota in the top 1 cm of sediments collected in the different oceanic regions.
fig. S4. Output of the regression tree analysis carried out to identify environmental factors explaining the distribution of MG-II Euryarchaeota in the top 1 cm of sediments collected in the different oceanic regions.