Summary
Clearance of entangled DNA from the anaphase mid-region must accurately proceed in order for chromosomes to segregate with high fidelity. Loss of Taz1 (fission yeast ortholog of human TRF1/TRF2) leads to stalled telomeric replication forks that trigger telomeric entanglements; the resolution of these entanglements fails at ≤20°C. Here, we investigate these entanglements and their promotion by the conserved replication/repair protein Rif1. Rif1 plays no role in taz1Δ fork stalling. Rather, Rif1 localizes to the anaphase mid-region and regulates the resolution of persisting DNA structures. This anaphase role for Rif1 is genetically separate from the role of Rif1 in S/G2, though both roles require binding to PP1 phosphatase, implying spatially and temporally distinct Rif1-regulated phosphatase substrates. Rif1 thus acts as a double-edged sword. Although it inhibits the resolution of taz1Δ telomere entanglements, it promotes the resolution of non-telomeric ultrafine anaphase bridges at ≤20°C. We suggest a unifying model for Rif1’s seemingly diverse roles in chromosome segregation in eukaryotes.
Keywords: telomere, anaphase, chromosome segregation, Rif1, Taz1, ultrafine anaphase bridge, phosphatase, replication fork stalling
Graphical Abstract

Highlights
-
•
Telomere replication problems lead to DNA entanglements persisting in anaphase
-
•
Rif1 specifically acts in anaphase to inhibit entanglement resolution
-
•
Anaphase Rif1 function promotes the resolution of non-telomeric entanglements
Zaaijer et al. show that the conserved DNA replication and repair protein Rif1 promotes resolution of non-telomeric DNA entanglements at anaphase, but further entwines entanglements between dysfunctional telomeres. This anaphase role is distinct from Rif1’s activities in S phase and G2 but shares the requirement for Rif1 to interact with protein phosphatase 1.
Introduction
By the time a cell divides, all chromosomes need to be equally partitioned between the two separated chromatid masses; DNA persisting in the middle of an anaphase cell threatens the integrity of the inherited genome. Nonetheless, a growing number of studies have revealed the tenacious presence of DNA in the anaphase mid-region, implying a dramatic last-minute scramble by the cell to clear the region and ensure faithful genomic segregation. In particular, challenges during S phase can result in entwined sister DNA strands, catenated duplexes, or strand invasion intermediates. Failure to resolve any of these structures results in chromosome entanglements.
Chromosomal entanglements can manifest as fine DNA-containing strands persisting between segregating DNA masses, referred to as ultrafine anaphase bridges or UFBs (Baumann et al., 2007, Chan et al., 2007). UFBs are distinct from anaphase bridges comprising whole lagging chromosomes, which often originate from merotelic attachments in which single centromeres are attached and pulled simultaneously to both poles (reviewed in Gregan et al., 2011); in addition, whereas lagging chromosomes are clearly coated with histones, the latter have rarely been detected on UFBs (Chan and Hickson, 2011). Mammalian UFBs are generally visualized by immunofluorescence using antibodies against proteins implicated in UFB resolution. Most commonly, UFBs originate from centromeric DNA and may represent loops arising from defective replication of the repeated centromeric units or catenated centromeric duplex sister chromatids; such centromeric UFBs are seen in unperturbed cells in early anaphase and do not impact wild-type (WT) cell viability (Baumann et al., 2007, Chan et al., 2007). These UFBs are decorated by PICH (Plk1-interacting protein) (Baumann et al., 2007) and the BLM helicase, and their levels are increased on treatment with topoisomerase II inhibitors (Baumann et al., 2007, Chan et al., 2007). A second type of UFB, thought to originate from persisting late replication intermediates, is characterized by the additional presence of the FANCD2 and FANCI DNA repair proteins at sites flanking the UFBs, as well as RPA along the intervening stretch (Chan and Hickson, 2011, Chan et al., 2009). UFBs of this class are rare, but increase in frequency in cells treated with replication inhibitors. This type of UFB can generate a DNA double-strand break (DSB) response in the daughter nuclei during the ensuing G1 phase, as indicated by formation of 53BP1 foci (Chan et al., 2009, Lukas et al., 2011).
Although PICH constitutes a robust marker for mammalian UFBs (Baumann et al., 2007), no PICH ortholog has been identified in Saccharomyces cerevisiae or Schizosaccharomyces pombe. Nonetheless, UFB-like structures have been reported in S. pombe strains experiencing replication stalling at lacO/I arrays that serve as bidirectional barriers to replication fork progression (Sofueva et al., 2011); such arrays are clearly visible as UFBs. Formation and removal of these UFBs is Rad51 independent, as is also true for mammalian UFBs (Chan et al., 2009). In S. cerevisiae, UFBs also have been suggested to originate from induced fork barriers and are coated by RPA, Dpb11TopBP1, and Sgs1BLM (Germann et al., 2014, Sofueva et al., 2011).
As highly repetitive genomic regions, telomeres are notorious for challenging replication (Barefield and Karlseder, 2012, Miller et al., 2006, Sfeir et al., 2009). The telomere binding protein Taz1 mitigates the tendency of telomere repeats to impede replication in S. pombe (Miller et al., 2006); this ability to promote telomere replication has been shown to be a conserved role for the mammalian Taz1 ortholog, TRF1 (Sfeir et al., 2009). Stalled RFs in taz1Δ cells are aberrantly processed upon sumoylation and activation of the RecQ helicase Rqh1 (a BLM ortholog), rendering them unable to restart (Rog et al., 2009). These irreversibly stalled forks lead to a number of phenotypes, including sub-telomeric hyper-recombination, immediate telomere loss on telomerase inactivation, and cold-sensitive viability. Hence, at temperatures of ≤20°C, taz1Δ cells show associations between the separating chromatin masses that fail to resolve at mitosis; we refer to these taz1Δ DNA bridges as telomere entanglements. These entanglements are distinct from telomere fusions, as the absence of Lig4, the ligase required for non-homologous end joining (NHEJ), fails to rescue their occurrence (Miller and Cooper, 2003); moreover, they are not products of Rad51-dependent homologous recombination, as taz1Δ cold sensitivity is not rescued by Rad51 inhibition. The origin of telomere entanglements in stalled RFs is further illustrated by comparing cells lacking Taz1 versus cells lacking Rap1, a Taz1-binding protein. While taz1Δ and rap1Δ strains share several phenotypes (including elongated telomeres, loss of telomere position effect, and susceptibility to NHEJ-mediated fusions during G1 arrest), rap1Δ telomeres do not suffer replication fork stalling and therefore do not experience entanglement or reduced viability in colder environments (Miller et al., 2005, Miller et al., 2006). A phenomenon reminiscent of taz1Δ telomere entanglements, in which NHEJ-independent telomere associations are seen between mitotic chromosomes, has been observed in mammalian TRF1-deficient cells experiencing compromised telomere replication (Muñoz et al., 2009, Ohishi et al., 2014, Sfeir et al., 2009). Moreover, stalled telomeric RFs provoked by the depletion of the ReqQ helicase WRN, or aphidicolin treatment, generate UFBs (Barefield and Karlseder, 2012). Hence, telomere disentanglement represents a universal challenge to genome stability.
Rif1 is a highly conserved and multifaceted protein, first identified as a budding yeast telomere length regulator (Hardy et al., 1992). ScRif1 associates with telomeres via Rap1 binding (Hardy et al., 1992, Shi et al., 2013); it also appears to bind the single-strand (ss) overhangs or ss/ds junctions at telomeres (Xu et al., 2010), where it plays a role in limiting 5′ telomeric resection and preventing cell-cycle arrest (Xue et al., 2011). Although mammalian Rif1 has not been implicated in telomere length regulation, it controls the choice of DNA repair pathway at internal DSBs and dysfunctional telomeres, favoring NHEJ by inhibiting resection of 5′ ends (Chapman et al., 2013, Di Virgilio et al., 2013, Escribano-Díaz et al., 2013, Silverman et al., 2004, Xu and Blackburn, 2004, Zimmermann et al., 2013). Moreover, both yeast and mammalian Rif1 play key roles in replication initiation, as well as in the replication timing program; in the absence of Rif1, many late-firing origins fire early and vice versa (Cornacchia et al., 2012, Hayano et al., 2012, Lian et al., 2011, Yamazaki et al., 2013). The roles of yeast Rif1 in replication have been linked to interaction of protein phosphatase 1 (PP1) with the SILK/RvXF motif in the N-terminal region of Rif1 (Davé et al., 2014, Mattarocci et al., 2014). The presence of PP1 at S. pombe origins opposes phosphorylation of the replicative MCM helicases by the Dbf4-dependent kinase (DDK). Phosphorylation of Rif1 in S phase promotes the release of PP1, allowing DDK-mediated phosphorylation to trigger origin firing (Davé et al., 2014). Finally, consistent with an additional function for Rif1 beyond the foregoing S- and interphase-based functions, mammalian Rif1 has been observed in the mid-region between separating chromatin masses at anaphase (Hengeveld et al., 2015, Xu and Blackburn, 2004).
Remarkably, deletion of rif1+ suppresses the cold sensitivity of taz1Δ cells, implicating Rif1 as a causative factor in telomere entanglements (Miller et al., 2005). Fission yeast Rif1 was heretofore thought to bind telomeres in a Taz1-dependent manner (Kanoh and Ishikawa, 2001), further deepening the mystery of its role in a taz1Δ setting. Here, we address the basis for these observations. We show that when telomeric replication fork progression is challenged, DNA entanglements persist into anaphase, a period in which intense processing activities are observed. We find that Rif1 acts neither to promote telomeric fork stalling nor to prevent fork restart. Rather, it acts via a previously unrecognized role to inhibit the resolution of taz1Δ telomeres during anaphase. Conversely, the timely segregation of centromeric UFBs is promoted by Rif1. Hence, Rif1 acts as a double-edged sword, increasing the efficiency of resolution of WT UFBs while inhibiting resolution of entangled taz1Δ telomeres. We further show that PP1 binding by Rif1 is required for Rif1’s dual roles in facilitating chromosome segregation in the presence of Taz1, while confounding chromosome segregation in the absence of Taz1. Collectively, our data suggest Rif1 as a key universal regulator of the final steps of chromatid segregation.
Results
Telomere Entanglements Persist in Anaphase, when Inefficient Resolution Engenders Problems in the Next Cell Cycle
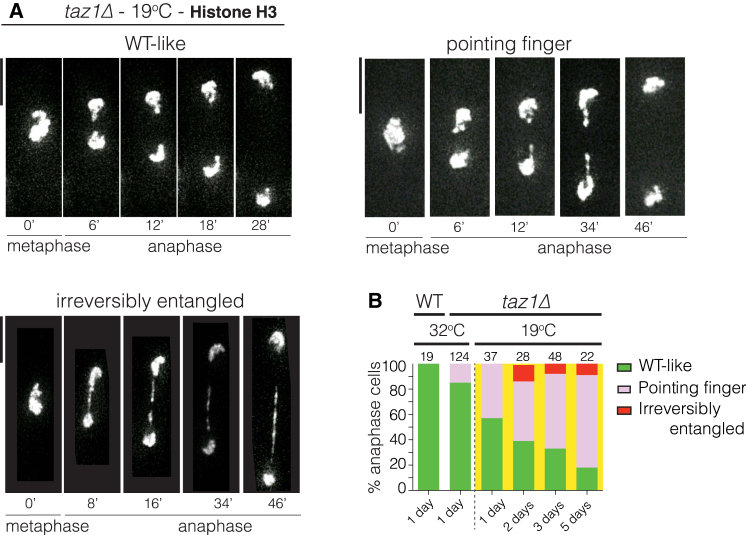
Stalled RFs at taz1Δ telomeres are processed into telomere entanglements (Miller and Cooper, 2003, Miller et al., 2005, Miller et al., 2006), the resolution of which fails at cold temperatures (≤20°C) (Miller and Cooper, 2003). To explore the nature of these entanglements in greater detail, we used live-cell microscopy to analyze taz1Δ cells harboring RFP-tagged histones at 32°C and 19°C. Patterns of chromosome segregation were categorized as wild-type like (WT-like) or aberrant. During WT-like chromosome segregation, sister chromatids equally segregate as compact entities to opposite poles (Figure 1A; Movies S1, S2, and S3). The aberrant category comprises two patterns: one dubbed the “pointing finger,” in which unresolved connections persist between segregating chromosome masses, evinced by protrusions of condensed histone-rich regions. These connections appear to resolve in late anaphase, as they abruptly retract toward the main chromatin masses. The second sub-category, “irreversibly entangled,” refers to cases in which chromatin persists between the main chromosome masses and fails to resolve.
Figure 1.
Telomere Entanglements Fail to Resolve in the Cold
(A) Film stills showing examples of chromosome segregation patterns through mitosis. Chromatin is visualized using Hht1-mRFP (histone H3 tagged at one of the two hht1+ loci). Scale bars in this and all subsequent microscopy images represent 5 μm.
(B) Quantitation of morphologies depicted in (A). Cells grown at 32°C were shifted to 19°C for the stated number of days (x axis labels) and maintained in log phase throughout. Numbers above bars indicate n-values.
At the normal growing temperature (32°C), 15% of taz1Δ cells show the pointing finger pattern, but no reduction of viability (Figure 1B). After shifting cultures from 32°C to 19°C, the proportion of cells displaying aberrant segregation patterns increases and continues to increase progressively with longer incubation at cold temperature (Figure 1B) This progressive increase with successive cell divisions suggests a cumulative accrual of defects due to inapt resolution of telomere entanglements.
taz1Δ Entanglements Occur via Telomeres, Rather than via rDNA, and Are Mainly between Sister Chromatids
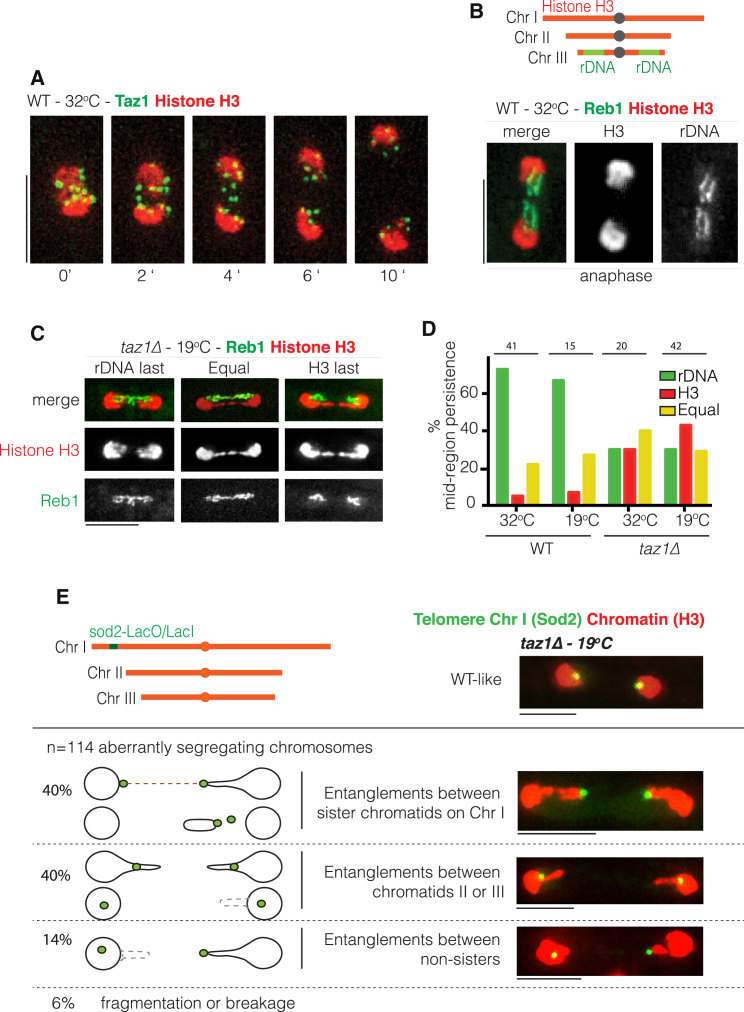
In WT anaphase, we consistently observe four Taz1 foci lagging behind the other eight Taz1 foci that colocalize with the main chromatin masses (Figure 2A). As S. pombe has three chromosomes (and therefore 12 telomeres per nucleus at anaphase onset), we wondered whether the four persisting foci represent sister telomeres of one specific chromosome; in particular, the rDNA repeat regions reside at either end of chr3 (Figure 2B). The rDNA binding factor Reb1 was used to visualize the rDNA (Figure 2B) given that visible histone H3 localization is excluded from the rDNA regions. Reb1-coated chromosome arms remain in the mid-region after the bulk of chromatin has segregated, implying that the four Taz1 foci persisting between chromatin masses in WT anaphase represent the telomeres at either end of chr3 (compare Figure 2A to Figure 2B).
Figure 2.
taz1Δ Telomere Entanglements Occur between Both Sister and Non-sister Chromatids and Do Not Involve rDNA
(A) Film stills of a typical WT cell. Telomeres are visualized with Taz1-GFP (endogenously tagged) and chromatin as in Figure 1. Numbers indicate time since metaphase onset.
(B) Above: schematic of S. pombe genome indicating the positions of rDNA on chromosome 3 (chr3). Below: rDNA is viewed via endogenously tagged Reb1 (a transcriptional termination factor specific for rDNA) and chromatin as in Figure 1. In WT anaphase, the rDNA-containing ends of chr3 segregate last (quantified in D).
(C) As in (B). Left: in the absence of taz1+, a subset of cells shows a WT segregation pattern (rDNA last). Middle: example of taz1Δ cell in which non-rDNA and rDNA chromosome ends segregate simultaneously (equal). Right: taz1Δ cell in which the rDNA segregates earlier than other chromosome ends (H3 last).
(D) Quantitation of (C). Cells were grown to log phase at 32°C and then scored after overnight growth at 19°C.
(E) Top left: schematic indicating the position of sod2-lacO/I-GFP near telomere 1L. Below: phenotypes of taz1Δ cells observed via sod2-lacO/I-GFP are indicated. Top right: no entanglements. Chart below: types of aberrant segregation patterns. Top: telomere entanglements involve both chr1 sister chromatids. Panels below: cells with entanglements that do not involve chr1 or involve telomere IL and a different chromosome arm (i.e., between non-sisters). Fragmentation or breakage refers to cells showing multiple sod2-lacO/I-GFP foci or, rarely, both foci at the same pole.
Given that the rDNA-adjacent telomeres tend to segregate last in a WT setting, do the DNA strands persisting between aberrantly segregating chromatin masses in a taz1Δ setting represent rDNA? Whereas rDNA segregates last in ∼70% of WT cells at both 32°C and 19°C, this percentage drops to ∼30% in taz1Δ cells, in which protruding histone-rich regions comprise the majority of persisting mid-region chromatin (Figures 2C and 2D). Hence, the unsegregated regions in taz1Δ anaphase are not confined to the rDNA, but rather comprise the ends of all three chromosomes.
To determine whether taz1Δ entanglements occur between telomeres of sister or non-sister chromatids, we utilized a strain harboring a LacO/LacI-GFP array at the sod2 locus (“sod2-lacO/I-GFP”) ∼80 kb from the left telomere of chr1 (Ding et al., 2004). In taz1Δ cells exhibiting WT-like chromosome segregation, the sod2-lacO/I-GFP foci are always clearly embedded in both of the main chromatin masses at anaphase (Figure 2E, top right). Of those cells demonstrating aberrant chromosome segregation, two GFP foci remain in the mid-region in 40% of cases (Figure 2E), indicating one or more entanglement(s) involving both sisters of chr1. However, in 14% of missegregants, only one sod2-lacO/I-GFP focus persists in the mid-region, whereas the other focus has segregated, indicating an entanglement between telomere IL and a non-sister chromatid. In the remaining ∼40% of defective segregants, both sod2-lacO/I-GFP foci clearly lie within the segregated masses and not within the intervening stretch, suggesting entanglements that did not involve chr1. Hence, stalled taz1Δ RFs preferentially lead to entanglements between sister telomeres, but strand invasion can occur between any combination of telomeres and occasionally link non-sisters. Conceivably, only non-sister entanglements lead to cold sensitivity.
Telomere Entanglements Remain Connected by ssDNA
The existence of unresolved DNA intermediates in the anaphase mid-region of taz1Δ cells prompted us to investigate which proteins co-localize with, and perhaps regulate the resolution of, these DNA stretches. Endogenous tagging of the shelterin component Tpz1, which associates with ss-telomeric overhangs as well as the ds telomere binding proteins, revealed Tpz1 foci between segregating DNA masses (Figure S1A). These foci tend to be interspersed with histone H3 foci in a non-overlapping pattern, suggesting that the intervening DNA comprises interspersed ss and ds regions.
The ssDNA binding complex RPA binds and stabilizes unwound DNA during replication and repair and serves as a platform for repair factors. We investigated RPA localization using an endogenously tagged allele of its Rad11 subunit. Unperturbed WT cells show the expected pattern of RPA localization, with foci appearing only during S phase (Figure 3A). In contrast, taz1Δ cells show one or two pronounced Rad11RPA foci throughout the cell cycle (Carneiro et al., 2010). Intriguingly, the DNA stretching between anaphase taz1Δ chromatin masses is abundantly coated with Rad11RPA at both 32°C and 19°C (Figures 3B and 3C); the stretch can be flanked by one or two particularly large and intense RPA foci, likely representing clumps of entangled ssDNA. Structured illumination microscopy (SIM) reveals that ss-stretches can originate from multiple sites within the same cell, evinced by forked structures (Figures 3C and 3D). Moreover, time-lapse microscopy reveals that the RPA-coated stretches are dynamic, retracting over the course of anaphase (Figure 3E). While the presence of Rad11RPA stretches is observed in taz1Δ cells at both 32°C and 19°C, their frequency in fields of still images of anaphase taz1Δ cells increases when temperature is reduced (from 26% at 32°C to 54% at 19°C).
Figure 3.
RPA Decorates the Strands Persisting between Anaphase taz1Δ Chromatin Masses
(A) Representative images of cells with endogenously tagged Rad11RPA, viewed as in Figure 1. In WT cells, Rad11RPA foci are detectable as foci only during S phase.
(B) taz1Δ cells viewed as in (A). In anaphase, Rad11RPA stretches from one chromatin mass to the other.
(C) Structural illumination microscopy (SIM) images of taz1Δ cells with Rad11RPA tagged as in (A) and histone H3 as in Figure 1.
(D) Magnified and rotated image of the left pole of the cell shown in (C). The forked structure suggests the simultaneous presence of two entanglements.
(E) Film frames of anaphase with tags as in (C). Rad11RPA retracts progressively.
Intrigued by the persistence of ssDNA stretches, we investigated other DNA replication and repair factors that play roles in ssDNA degradation or fill-in. DNA polymerase α (Pol α, encoded by fission yeast swi7+) is a lagging strand DNA polymerase with roles in genome-wide replication and synthesis of the strand complementary to the telomere repeats added by telomerase. In WT cells, Pol α appears in foci only during S phase (Figure S1B). By contrast, Pol α remains associated with telomere entanglements in G2 in taz1Δ cells at 32°C and 19°C. Moreover, these foci persist in anaphase on taz1Δ telomere entanglements at 19°C (Figure S1B).
Rad52, a homologous recombination factor recruited downstream of RPA to promote strand invasion, is required for viability of taz1Δ cells in the cold (Miller and Cooper, 2003). Likewise, Rad52 localizes to the anaphase mid-region in a pattern similar to that of RPARad11 (Figure S1C).
Checkpoint Arrest Is Provoked in Response to Inappropriate Resolution of Telomere Entanglements
When taz1Δ cells are grown at 32°C, their telomeres co-localize with components of the DNA damage response pathway, yet they avoid cell-cycle arrest (Carneiro et al., 2010). This avoidance was shown to stem from local histone modification state; methylation of histone H4 lysine 20 is required for binding by the checkpoint mediator Crb253BP1, but H4-K20 methylation is excluded from telomeric chromatin (Carneiro et al., 2010). In contrast, taz1Δ cells do activate the checkpoint at 19°C, as indicated by Rad3ATR/Chk1-dependent cell elongation (Miller and Cooper, 2003). The accrual of taz1Δ chromosomal defects over time at 19°C suggests an explanation for this. Whereas stalled taz1Δ RFs, which occur at 32°C as well as at 19°C, fail to trigger checkpoint activation, damage arising from transmission of inappropriately resolved telomere entanglements into the next cell cycle may trigger checkpoint activation. To evaluate this possibility, we monitored several hallmarks of cell-cycle progression and/or arrest. Both WT and taz1Δ cells respond within 1 hr of cold treatment by accumulating transiently in the mono-nucleate state, indicating a brief metaphase arrest. In WT cultures, binucleate cells, representing progression through mitosis, start to arise at 3 hr; soon thereafter, RPA appears in such binucleates, indicating S phase initiation (note that fission yeast begins S phase before septation is complete; Figures S2A and S2B). In taz1Δ cultures, this timing is altered. taz1Δ cell length increases gradually (Figure S2C). Initiation of anaphase following cold induction is delayed compared to WT (Figures S2A and S2B), indicating a brief metaphase arrest, as reported previously (Miller and Cooper, 2003). Consistent with the idea that G2/M DNA damage checkpoint instigation follows progression to the following cell cycle, Crb253BP1 foci start to appear in taz1Δ cells 3 hr after the temperature shift and increase in number thereafter (Figure S2D). Moreover, taz1Δ cell elongation at ≤20°C requires Crb253BP1 (Figure S2E). Hence, Crb253BP1 foci fail to form in the first S/G2 phase following temperature shift. Instead, they form later, in response to damage incurred during M phase and carried into subsequent cell cycles.
Rif1 Is Specifically Involved in Telomere Entanglement
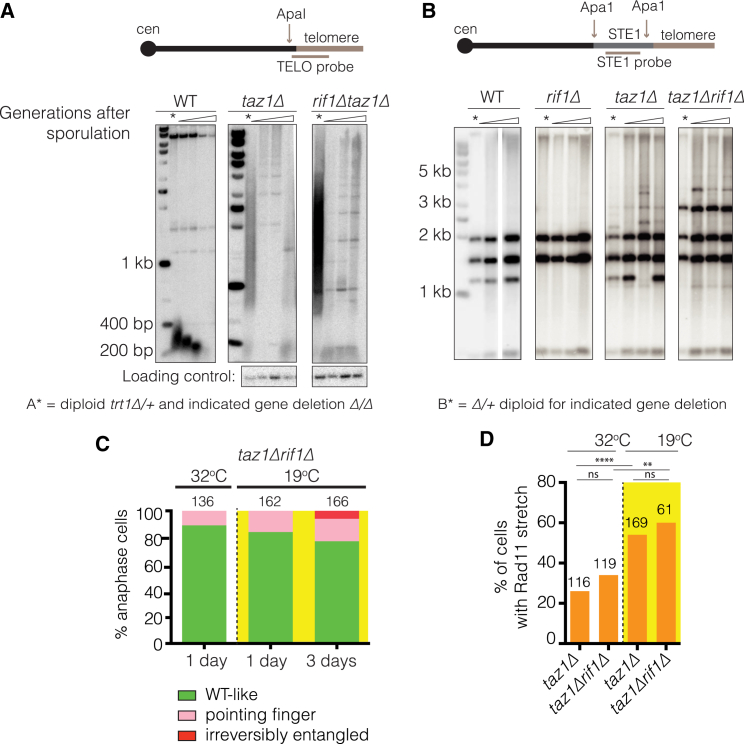
Rif1 has been implicated as causative of telomere entanglements through the observation that rif1+ deletion rescues taz1Δ cold sensitivity (Miller et al., 2005). Rif1’s involvement could stem from a role in initiating fork stalling events, in processing stalled forks, or in downstream processes that promote the formation—or inhibit the resolution—of entanglements (Germe et al., 2009, Rog et al., 2009). To tease apart these possibilities, we addressed each stage of the genesis of entanglements. We analyzed telomere replication intermediates using 2D gel electrophoresis. As previously reported, taz1Δ telomeres generate a distinct “plume” pattern reflecting the inability of forks to reach the end of the elongated taz1Δ telomeres (Miller et al., 2006) (Figure S3A). This taz1Δ plume pattern is unaffected by rif1+ deletion. Moreover, the fork stalling seen at an internally inserted telomere repeat stretch in a taz1Δ setting is unaffected by rif1+ deletion (Figure S3B). Therefore, Rif1’s role in telomere entanglement does not reflect a role in the initial genesis of stalled RFs.
The inability to fully replicate taz1Δ telomeres renders their maintenance fully dependent on telomerase; hence, in the absence of the telomerase catalytic subunit (Trt1), telomeres are rapidly lost despite their initial over-elongation (Miller et al., 2006). Consistent with the observation that Rif1’s presence has no impact on fork stalling, trt1+ deletion in a taz1Δrif1Δ background yields the same rapid loss of telomeres as seen in the taz1Δ background (Figure 4A). This result also rules out a role for Rif1 in converting stalled RFs to irreversibly stalled RFs and contrasts with the role of Rqh1 in that conversion (Rog et al., 2009). Accordingly, rif1+ deletion fails to rescue taz1Δ sub-telomeric hyper-recombination or the propensity of taz1Δ telomeres to survive telomerase loss via hyper-recombination (Figure 4B), both outcomes of stalled taz1Δ RFs. We conclude that Rif1 specifically acts in the generation or resolution of telomere entanglements downstream of irreversibly stalled taz1Δ RFs.
Figure 4.
Rif1 Is Involved in neither taz1Δ Telomeric Fork Stalling nor Processing of Stalled Forks
(A and B) Top: diagrams of restriction sites and probes used. The TELO probe contains telomere repeats and 32 bp of sub-telomeric sequence. (A) Diploids heterozygous for trt1+(trt1+/trt1Δ), but homozygous for taz1Δ and/or rif1Δ, as indicated, were induced to sporulate. Progeny haploids were restreaked to single colonies every 3 days. At each restreak, samples of genomic DNA were extracted, and telomere length was assessed by Southern blot analysis. A probe to a PCR-amplified fragment of SPAC4A8.02c was used as an internal loading control. Asterisks above blots indicate parental diploids, with the following lanes taken from progressive restreaks. (B) Sub-telomeric recombination is monitored via stability of the restriction site pattern over time. Single colonies were successively restreaked at 32°C. Every 2 days, a single colony was transferred to a new plate and subjected to genomic DNA isolation and Southern blot analysis. The loss of Rif1 does not affect restriction pattern stability in WT or taz1Δ settings.
(C) Effect of rif1 deletion on taz1Δ chromosome segregation morphologies. Quantitation as in Figure 1B.
(D) Frequencies of visible anaphase Rad11RPA-GFP stretches. Log phase cultures were shifted from 32°C to 19°C for 3 days. Such stretches were never detected in WT anaphase cells. ns, not significant, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
See also Figures S2 and S3.
To explore Rif1’s role downstream of the stalled RFs, we analyzed chromosome segregation dynamics. A clear reduction in chromosome segregation defects is observed when comparing taz1Δrif1Δ and taz1Δ cells grown in the cold (compare Figure 1B with Figure 4C). However, the occurrence of Rad11RPA -bound stretches between sister chromatids during anaphase is not affected by the loss of Rif1 (Figure 4D). These observations suggest that Rif1 is involved not in forming entanglements, but rather in inhibiting their proper resolution.
Mammalian Rif1 has been implicated in the DNA damage response as a 53BP1-dependent inhibitor of 5′ end resection (Chapman et al., 2013, Di Virgilio et al., 2013, Escribano-Díaz et al., 2013, Zimmermann et al., 2013). The observations that fission yeast rif1+ deletion yields neither DNA damage sensitivity (Davé et al., 2014, Hayano et al., 2012) nor altered levels of 5′ resection in WT or taz1Δ settings (Miller et al., 2005) weigh against the notion that SpRif1 generates taz1Δ cold sensitivity by inhibiting 5′ resection. Moreover, it is unlikely that SpRif1 is involved in channeling repair to NHEJ as is mammalian Rif1 (Chapman et al., 2013, Di Virgilio et al., 2013, Zimmermann et al., 2013, Escribano-Díaz et al., 2013), since rif1+ deletion has no impact on the NHEJ-dependent telomere fusions seen in G1 arrested taz1Δ cells (Miller et al., 2005). To explore this further, we investigated the role of the fission yeast 53BP1 ortholog, Crb2, in taz1Δ telomere entanglement. crb2+ deletion fails to suppress taz1Δ cold sensitivity (Figure S2F), again indicating that Rif1 acts independently of Crb2 to exacerbate taz1Δ entanglements.
Rif1 Is Present in the Anaphase Mid-region
If Rif1 inhibits the resolution of telomere entanglements at anaphase, then we might expect it to localize to the anaphase mid-region. To investigate this possibility, we turned to live microscopy. C-terminally GFP-tagged Rif1 acts as a rif1 null, rescuing both taz1Δ cold sensitivity and the lethality of the DDK mutant hsk1-89 (Figures S4A and S4B). Moreover, whereas C-terminally Myc-GFP-tagged Rif1 is functional, expression levels yield insufficient fluorophore signal for detailed analysis. We therefore placed N-terminally tagged Rif1 under control of the inducible nmt41 promoter. Growth of nmt41-GFP-rif1 cells under partially repressed conditions allows visualization of GFP-Rif1 without affecting viability in either WT or taz1Δ backgrounds (Figure S4C). GFP-Rif1 appears as a nuclear haze with distinct foci co-localizing with Taz1, as previously shown (Kanoh and Ishikawa, 2001) (Figure S4D). Surprisingly, Rif1 shows two distinct categories of localization in WT mitotic cells. First, Rif1 localizes to segregating Taz1 foci, which appears at all telomeres (Figure S4E). Second, Rif1 forms a stretch of foci between segregated anaphase chromatin masses (Figure 5A). Higher resolution live imaging suggests two Rif1 “sliding arms” in the mid-region; these arms shorten on spindle elongation (Figure 5C).
Figure 5.
Rif Localizes to Anaphase Mid-region
(A) Frames from films of WT cells at 32°C or taz1Δ cells grown to log phase at 32°C and then shifted to 19°C for 24 hr. Rif1 is N-terminally GFP-tagged under nmt41 promoter control and histone H3 tagged as in Figure 1. GFP-Rif1 lingers in the mid-region as cells complete anaphase. Compare with Figure S4E.
(B) Quantitation of GFP-Rif1 stretches during WT-like or aberrant chromosome segregation in taz1Δ cells.
(C) SIM of GFP-Rif1 in anaphase. For each panel, the image is rotated to the orientation that yields the clearest signal. The pattern suggests interlocking helical strands (diagram below).
(D) Cells with either an extra copy of the gene encoding α-tubulin (atb2+, under nmt1 control) or Rif1 tagged as in (A) were grown at 32°C and mixed in one microscope dish. Anaphase, as detected via the presence of a mitotic spindle or two segregating nuclei harboring GFP-Rif1, was filmed. At time point 0’, MBC was added and cells were imaged every 2 min to follow MBC-induced spindle depolymerization. Dissipation of the Atb2 signal (i.e., the spindle) is clearly seen; nonetheless, GFP-Rif1 in adjacent cells (subject to identical MBC treatment in the same dish) clearly persists in the mid-region (red arrow).
See also Figure S4.
Rif1’s localization to the anaphase mid-region appears at a time and place not occupied by Taz1 (compare Figure 5A with Figure 2A). Accordingly, this category of Rif1 foci is seen in both taz1+ and taz1Δ cells. However, these Rif1 foci are more often visible between aberrantly segregating than properly segregating chromosomes in taz1Δ cells (Figure 5B). Enhanced visibility reflects either longer duration or higher frequency of Rif1 mid-region localization in mis-segregants, consistent with a role in inhibiting resolution of entanglements.
How is Rif1 recruited to the anaphase mid-region? A previous study suggested that human Rif1 binds anaphase microtubules (Xu and Blackburn, 2004), whereas a more recent study localized Rif1 to UFBs (Hengeveld et al., 2015). To determine whether spindle microtubules are required for Rif1 localization, we assessed whether spindle depolymerization would affect Rif1 localization. While treatment of live cells with methyl 2-benzimidazole carbamate (MBC) efficiently depolymerizes spindle microtubules (Figure 5D), Rif1 foci remain unaltered by MBC treatment, indicating spindle-independent localization. Moreover, overexposure of the histone H3 signal reveals a thin stretch between anaphase chromatids, either co-localizing with or adjacent to Rif1 (Figure S4F).
Rif1 Promotes Timely Resolution of Centromere-Proximal UFBs
Are our observations that Rif1 inhibits resolution of taz1Δ telomeric entanglements relevant to other types of entangled structures? To expand our analysis beyond telomeric DNA, we turned to a non-telomeric region. A Tomato-tagged Tet repressor (TetR) bound to a repetitive tetO array integrated within the centromere of chr2 (“cenII-tetO/R-Tomato”) blocks efficient RF progression and appears in UFBs (Sofueva et al., 2011). cenII-tetO/R-Tomato forms a single bright focus in each G2 cell, whereas unbound TetI generates a general nuclear haze in cells lacking the tetO array (Figure 6A). In WT cells at 32°C, the tetO/R-Tomato focus separates at metaphase to two foci that segregate neatly to opposite poles during anaphase. Intriguingly, however, in cultures grown at 19°C for 3 days, 8% of WT cells exhibit a tetO/R-Tomato stretch between the separating chromatid masses. Remarkably, rif1+ deletion greatly exacerbated the occurrence of these UFBs, most pronouncedly at 19°C (Figures 6B and 6C). Hence, like UFBs comprising telomeric entanglements, non-telomeric UFBs arise more frequently and/or persist longer at cold temperature. However, in direct opposition to its role in telomeric entanglements, Rif1 plays a key cold-specific role in limiting the appearance of tetO/R-containing UFBs.
Figure 6.
Rif1 Promotes Efficient UFB Resolution in WT Cells at ≤20°C and Requires PP1 Interaction
(A) Schematic indicates the location of the tetO array bound by TetR-tomato. Below: G2 cells expressing TetR-tomato with or without cenII-tetO array.
(B) Differential interference contrast (top) and fluorescence (bottom) images of TetR-tomato in anaphase cells without (−) or with (+) cenII-tetO. Interpretive diagrams are shown below.
(C) Quantitation of UFB frequencies. Cells with cenII-tetO/R-tomato were grown to log phase at 32°C and then shifted to 19°C for 3 days. ∗∗∗∗p < 0.0001.
(D) 5-fold serial dilutions of log phase cells (32°C) were stamped and incubated at 32°C (2 days) or 19°C (5 days). rif1-PP1 generates variable suppression of taz1Δ cold sensitivity.
(E) Quantitation of cenII-tetO/R-tomato-UFBs at 19°C. ns, not significant, ∗p < 0.05, ∗∗∗∗p < 0.0001.
See also Figure S5.
Phosphatase Binding by Rif1 Is Required for Rif1’s Anaphase Activity
The SILK/RvXF motif at the N-terminal domain of Rif1 has been identified as the PP1 binding site (Sreesankar et al., 2012). Hence, while telomeres normally replicate late in S phase, the Rif1-PP1 mutant allele, which compromises PP1 binding, confers early telomere replication (Mattarocci et al., 2014, Davé et al., 2014, Hiraga et al., 2014). To determine whether phosphatase regulation mediates Rif1’s roles in telomere length regulation or anaphase resolution of mid-region DNA structures, we assessed the effects of rif1-PP1 on these parameters. Most cells harboring Rif1-PP1 show telomere elongation reminiscent of that conferred by rif1Δ (Figure S5A, left). This telomere elongation phenotype is incompletely penetrant with independent rif1-PP1 isolates showing variable telomere length (Figure S5A, right), suggesting that the extent of PP1 exclusion varies between clones. Nonetheless, the clear occurrence of rif1Δ telomere length in rif1-PP1 cells indicates that telomere length regulation by Rif1 is mediated, at least in part, by PP1 control. Consistently, cells harboring the Rif1-7A allele, which confers constitutive binding of PP1 to Rif1, show WT telomere length (Figure S5A).
To address the role of Rif1-associated PP1 in telomeric entanglement, we asked whether rif1-PP1 rescues taz1Δ cold sensitivity as does rif1Δ. As was the case for telomere length regulation, clonal variation was observed, with approximately half of the rif1-PP1 clones rescuing taz1Δ cold sensitivity (Figure 6D). This phenotypic mosaicism varies over successive generations; on restreaking for single colonies, a rif1-PP1 clone that rescues cold sensitivity can give rise to clones that fail to rescue and vice versa (data not shown). Such reversibility indicates that the mosaicism is an inherent property of the rif1-PP1 mutations rather than the emergence of suppressor mutations. Furthermore, deletion of the gene encoding the Dis2 PP1 rescues taz1Δ cold sensitivity to an extent comparable with rif1Δ and rif1-PP1 (Figure S5B).
If Rif1’s role in inhibiting the resolution of telomere entanglements stems from the same activity that confers Rif1’s role in stimulating non-telomeric UFB resolution, then Rif1-PP1 should phenocopy rif1Δ in exacerbating the presence of UFBs. Remarkably, introduction of rif1-PP1 into strains harboring cenII-tetO/R-Tomato generates a clonally variable increase in visible anaphase bridge formation (Figure 6E). Collectively, we conclude that Rif1-mediated recruitment of phosphatase activity to UFBs and telomere entanglements dictates the success of their resolution.
The Activities of Rif1 in S Phase Are Separable from Rif1’s Anaphase Role in Regulating UFB Resolution
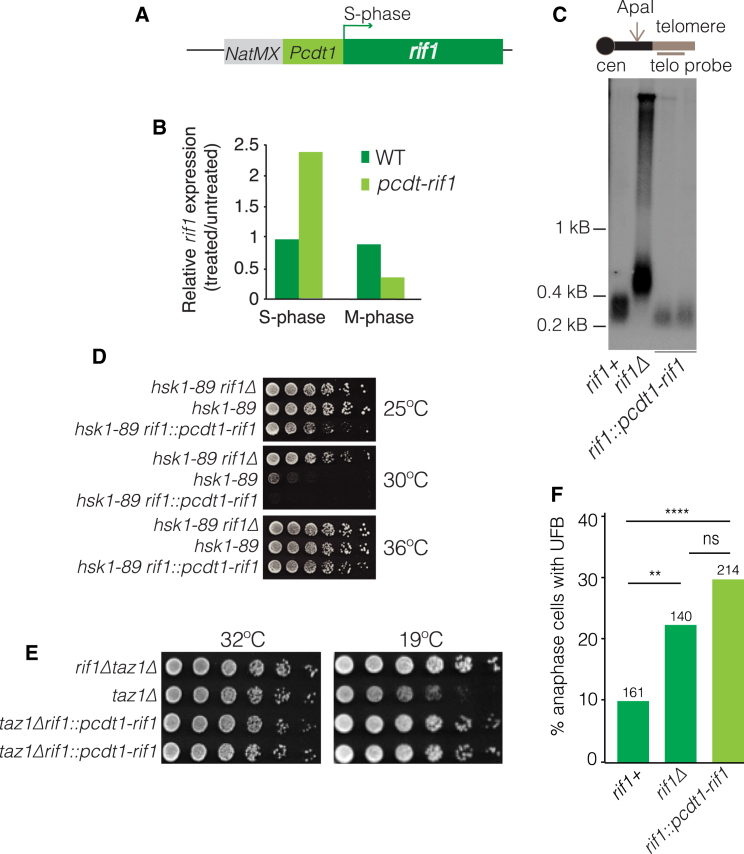
Our observations that rif1+ deletion has no impact on taz1Δ telomeric fork progression, hyper-recombination, or immediate telomere loss on trt1+ deletion, along with the co-localization of Rif1 with DNA stretches in the anaphase mid-region, strongly suggest that Rif1 acts at anaphase to regulate entanglement resolution and that this activity is not a by-product of Rif1’s roles in controlling S-phase DNA transactions (e.g., replication origin firing or DSB resection). To further explore this idea, we sought to construct a Rif1 allele that would separate S- and M-phase functions. To achieve this, we placed Rif1 under control of the S-phase-specific promoter of cdt1+; the cdt1+ promoter, along with a selectable marker at its 5′ end, was inserted directly upstream of endogenous rif1+ (see Figure 7A for details). While WT rif1+ is expressed to similar levels during S- or M-phase arrest, cells carrying pcdt1-rif1 yield higher rif1+ expression levels than WT cells during S-phase arrest and lower levels than WT during an M-phase arrest (Figure 7B).
Figure 7.
Rif1’s Anaphase and S-Phase Functions Are Separate
(A) Schematic of approach to induce S-phase-specific rif1+ expression. The cdt1+ promoter region (640 bp, including the binding site for the S-phase transcription factor Cdc10 and cdt1 transcription start site and 5′ UTR) was inserted immediately upstream of the endogenous rif1 ORF. NatMx provides a selectable marker.
(B) Relative rif1 transcript levels were determined by qRT-PCR and normalized to levels of act1. (S phase) Cells were grown in media containing 15 mM hydroxyurea (Sigma) for 4 hr at 32°C. (M phase) Cells were treated with 50 μg/ml TBZ (thiabendazole, a microtubule-depolymerizing agent) for 2 hr at 25°C. Arrest was verified via septation index.
(C) pcdt1-rif1 confers WT telomere length, confirming S-phase function. Southern blot as in Figure 4. Two individual clones of the pcdt1-rif1 strain were analyzed.
(D) pcdt1-rif1 confers WT Rif1 function with respect to Hsk1-mediated replication control. 5-fold serial dilutions of log phase cells at 25°C (permissive for hsk1-89) incubated at 25°C, 30°C, or 36°C. While rif1Δ restores growth of hsk1-89 cells at a non-permissive (30°C) temperature, pcdt1-rif1 behaves as rif1+ in failing to do so, confirming Rif1 functionality in S phase. Replicates grown at 36°C serve as loading control.
(E) Dilution assay as in (D). pcdt1-rif1 behaves as rif1Δ by rescuing taz1Δ cold sensitivity.
(F) Frequencies of cenII-tetO/R-tomato-UFBs are quantified as in Figure 6C. pcdt1-rif1 phenocopies rif1Δ in elevating UFB levels, tying UFB resolution to Rif1 function outside of S phase. ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
Functional expression of Rif1 during S/G2 was further tested by analyzing telomere length. Telomeres show WT length in pcdt1-rif1 cells, indicating WT Rif1 function in S/G2 (Figure 7C). Moreover, pcdt1-rif1 fails to phenocopy rif1Δ in rescuing the DDK mutant hsk1-89, again indicating WT Rif1 activity during S phase (Figure 7D). Strikingly, however, pcdt1-rif1 fully mimics the rif1+ null for rescue of taz1Δ cold sensitivity (Figure 7E). Moreover, pcdt1-rif1 fully mimics the opposing effect of rif1+ null in exacerbating the presence of non-telomeric UFBs harboring cenII-tetO/R-Tomato (Figure 7F) These data demonstrate that Rif1’s activity at anaphase is indeed separable from its S-phase activities and that its anaphase activity is specifically responsible both for inhibiting resolution of telomere entanglements and stimulating resolution of non-telomeric UFBs.
Discussion
This work sheds light on the complexity of resolving different types of DNA entanglements during anaphase, a period in which active repair and DNA synthesis processes remain underway, as evinced by the appearance of RPA, DNA polymerase α, and associated factors on telomeric entanglements in the anaphase mid-region. We have found a key role for Rif1 in regulating the cell’s final attempt to separate persisting associations between segregating chromosomes. Remarkably, Rif1 can either help or hinder, depending on context. Rif1 helps this process by promoting timely resolution of the “WT” UFBs represented by cenII-tetO/R and hinders by inhibiting the resolution, and perhaps further interweaving, telomeric entanglements formed in taz1Δ cells. A unifying model for these seemingly paradoxical activities of Rif1 is presented below.
Rif1 in the taz1Δ Context
Loss of taz1+ results in a host of dysfunctional telomere phenotypes, but taz1Δ cells are surprisingly viable, unless they are grown at cold temperatures (Miller and Cooper, 2003). Genetic and physical analyses have demonstrated that taz1Δ telomere entanglements originate from stalled RFs processed by Rqh1 to form DNA structures refractory to resumption of fork progression (Rog et al., 2009). Although resection at stalled taz1Δ RFs may play a role in entanglement (O. Rog and J.P.C., unpublished data), resection alone does not generate entanglements, as rap1Δ telomeres suffer excessive resection with little entanglement (Miller et al., 2005). It is more likely that aberrant strand invasion events generate these entanglements.
Our microscopy-based analysis has uncovered a variety of proteins associated with the DNA stretches that span the segregating chromatin masses. Alternating patterns suggest that the DNA stretches comprise ssDNA regions (devoid of nucleosomes but coated with RPA) interspersed with dsDNA-wrapped nucleosomes. The RPA patterns are dynamic, with RPA-coated filaments retracting into one of the daughter cells during anaphase. We speculate that such retraction represents resolution of entanglement.
The observation that taz1Δ cells lose viability progressively over time in the cold suggests that resolution can be imperfect, leading to aberrant structures in the next cell cycle. Indeed, taz1Δ cells grown at cold temperatures accumulate high levels of DSBs (Miller and Cooper, 2003), likely a readout of inappropriate resolution or breakage of entangled chromosomes upon cell division. Consistently, Crb253BP1 foci only begin to appear after 3-hr growth of taz1Δ cells at 19°C; hence, Crb2 is recruited to DSBs, the genesis of which follows failed telomere entanglement resolution. This phenomenon resembles the progressive DSB accumulation reported to originate from UFBs generated by late replication intermediates; these UFBs trigger DSBs in daughter nuclei in the ensuing G1 phase (Chan et al., 2009).
The basis for the cold specificity of failed entanglement resolution is not yet known. Possibilities include the enhanced propensity for partially unwound DNA structures to reanneal in suboptimal conformations in the cold or a cold-sensitive activity of some DNA processing protein that limits entanglement. Whatever the basis for cold specificity, it serves as a useful property in linking roles of Rif1 that are specific to ≤20°C. These roles include not only the inhibition of taz1Δ entanglement resolution but also promotion of the robust resolution of non-telomeric UFBs. Indeed, the shared cold specificity of Rif1’s involvement in taz1Δ entanglements and cenII-tetO/R UFBs provides compelling evidence for a common etiology.
Rif1: A Double-Edged Sword
While UFBs have been loosely defined as thin DNA strands not detectable by conventional DNA dyes, the term is applied to essentially distinct structures (see Introduction). Hence, the recognition of particular UFB types, and/or the processes that channel each type into an appropriate resolution pathway, may be prone to inaccuracy. Here, we have found that Rif1 may be responsible for such inaccuracy when UFBs harbor entangled telomeres. A plausible scenario is that Rif1 promotes the same anaphase activity at non-telomeric UFBs, entangled sister telomeres, and entangled non-sister telomeres; while this activity is beneficial for resolution of sister telomere entanglements, it is deleterious for non-sister telomeric entanglements. Indeed, while telomere entanglements mainly occur between sister chromatids, they connect non-sisters sufficiently frequently to account for the loss of viability we have observed at ≤20°C. One highly speculative but concrete idea for the basis for such opposing activities involves topoisomerase II (Top2). Given our previous work implicating Top2 in the resolution of telomere entanglements (Germe et al., 2009), Rif1 may promote Top2 activity in the specific orientation that removes entanglements between segments along a single chromosome. However, Top2 activity in this specific orientation could be envisioned to further entwine those telomeric entanglements that involve inter-chromosomal interactions.
Where Does Rif1 Bind?
The dynamic localization pattern of Rif1 places it precisely where resolution-promoting factors act during anaphase. We find that Rif1 localizes to the anaphase mid-region in a Taz1-independent fashion and lingers long after the bulk chromatin masses have segregated, coating thin stretches whose co-localization with histones is detectable only in single z stack microscopic sections. The first precedent for such Rif1 localization was reported in human cells, in which it was suggested to coincide with microtubules (Xu and Blackburn, 2004). Very recently, human Rif1 was reported to localize to UFBs in a microtubule-independent manner (Hengeveld et al., 2015). Likewise, our data show microtubule independent localization of Rif1 to the anaphase mid-region. Rif1 chromatin immunoprecipitation (ChIP)-chip assays have demonstrated both Taz1-dependent and Taz1-independent Rif1 binding sites, as well as differences in binding site profiles through the cell cycle (Hayano et al., 2012). Intriguingly, many of the Taz1-independent Rif1 binding sites correspond to sequences with the propensity to form G-quartet structures and indeed, a binding preference for the G-quartet configuration was demonstrated. It is conceivable that such secondary structures might form in the ss telomeric DNA regions intervening between anaphase chromosome masses, and might contribute to Rif1 recruitment. However, as the Rif1-regulated UFBs measured in the cenII-tetO/R assay lack G-quartet potential, the anaphase Rif1 recruitment mode cannot be primarily through G-quartet binding. We hypothesize that Rif1’s recruitment to anaphase DNA structures occurs via non-sequence specific binding to ss/ds junctions, as Rif1 shows a binding preference for such regions (Xu et al., 2010), and entangled telomeres and non-telomeric UFBs are both replete with such ss/ds junctions.
Independent Roles for Rif1 in S Phase and Anaphase
A plethora of circumstantial evidence, along with construction of a key separation-of-function rif1 allele, indicates that the function of Rif1 at anaphase is separable from its S/G2 phase functions. While Rif1 regulates origin firing and replication timing (Kanoh et al., 2015, Yamazaki et al., 2013), both taz1Δ and taz1Δ rif1Δ cells exhibit deregulated telomere replication timing (Dehé et al., 2012, Hayano et al., 2012), ruling out the possibility that altered replication timing underlies the effect of Rif1 on taz1Δ telomere entanglements. Moreover, Rif1 is clearly uninvolved in replication fork stalling at taz1Δ telomeres, sub-telomeric hyper-recombination, and the immediate telomere loss upon trt1+ deletion seen in taz1Δ cells. These observations, along with the clear Taz1-independent localization of Rif1 to the anaphase mid-region, most parsimoniously place Rif1 as a regulator of entanglement resolution at anaphase.
The crucial clincher in our conception of Rif1 having separable roles in S/G2 and anaphase comes from the cdt1-rif1 allele, which places Rif1 expression under the control of an S-phase-specific promoter. This allele retains the essentiality of DDK kinase activity and confers WT telomere length, indicating robust Rif1 expression during S/G2. Nonetheless, cdt1-rif1 clearly phenocopies the null in rescuing the cold sensitivity of taz1Δ cells and exacerbating the appearance of non-telomeric UFBs in the cold. Hence, S/G2 expression of Rif1 is irrelevant for entanglement resolution. Not only do these results separate S/G2 and anaphase Rif1 functions, but also they further highlight the connection between Rif1’s opposing roles in resolving taz1Δ versus cenII-tetO/R UFBs, as both are products of Rif1 activity outside of S/G2.
Collectively, three hot-off-the-press reports reinforce the relevance of this study to our understanding human chromosome segregation and cancer. First, overexpression of hTRF2 leads to stalled telomeric RFs (Nera et al., 2015) reminiscent of those seen in taz1Δ fission yeast. Accordingly, telomere-containing UFBs are seen on hTRF2 overexpression, as are chromosome fusions presumably resulting from botched attempts of repair damage carried over from defective anaphases; this suite of phenotypes closely resembles those seen in taz1Δ cells grown in the cold. Given that hTRF2 is often overexpressed in cancer cells, such entanglement-mediated chromosome fusions are likely causative agents in tumorigenesis. The recent demonstration that hRIF1 localizes to UFBs further highlights the parallels with our system, suggesting that the anaphase role of fission yeast Rif1 is likely to be conserved. Finally, anaphase bridges resulting from dominant-negative inhibition of human TRF2 were shown to trigger chromothripsis when nuclear membrane rupture allows the cytoplasmic nuclease TREX1 to access the bridging DNA (Maciejowski et al., 2015). While the NHEJ-mediated telomere bridges induced by TRF2 inhibition are distinct from the entanglements studied here, the downstream DSBs in ensuing cell cycles could lead to NHEJ. This distinction highlights the need to understand the full range and regulation of anaphase chromosome separation pathways along with their downstream consequences.
Rif1 as a PP1 Platform Differently Utilized through Time and Space
While the full list of players and mechanistic details of Rif1 action at anaphase remain open questions, these anaphase activities share a dependence on PP1 binding with Rif1’s role in coordinating the timing and initiation of DNA replication. The rif1-PP1 allele shows variable penetrance with respect to telomere length, attenuation of taz1Δ cold sensitivity, and exacerbation of cenII-tetO/R-mediated UFBs alike. This variable penetrance may stem from the existence of two PP1 proteins in fission yeast, Sds21 and Dis2; while the rif1-PP1 mutation vastly reduces the recruitment of Sds21 to telomeres, it may be more permissive to residual recruitment of Dis2 (Davé et al., 2014). This exchangeable interaction of the two PP1s could result in stochastically variable phosphatase activity.
The PP1 linkage suggests a unifying model for the apparently pleiotropic roles of Rif1. Binding to chromatin, whether via interactions with Taz1, G-quadruplex structures, ss/dsDNA junctions, or as-yet-undefined means, recruits PP1 activity to the relevant place and time. Once there, Rif1 will oppose local phosphorylation of PP1 substrates, creating the seemingly diverse set of chromatin-related functions attributed to Rif1. At replication origins, localized dephosphorylation will inhibit MCM-mediated origin firing; at sites of DNA damage, desphosphorylation may inhibit local resection activities; and at replicating telomeres, dephosphorylation clearly inhibits the extent of telomerase-mediated telomere extension. Likewise, dephosphorylation of kinase substrates localizing to the anaphase mid-region is likely to regulate resolution activities and may also control the rate of spindle elongation (data not shown). A candidate mid-region kinase ripe for opposition by Rif1-associated PP1 is Aurora B, a key component of the chromosomal passenger complex that relocates from kinetochores to the anaphase mid-region. Hence, Rif1 may be an adaptable chromatin-phosphatase interface, exploited to provide a brake on diverse phospho-proteins that act on partially unwound DNA.
Experimental Procedures
Construction of S-rif1
To generate the rif1::pcdt1-rif1 strain, four fragments (one comprising the 200-bp sequence immediately upstream of rif1+, one a 1.2-kb natMX6 cassette [Sato et al., 2005], one comprising 640 bp of the cdt1+ promoter sequence [Hofmann and Beach, 1994], and one comprising the first 200 bp of the rif1+ ORF) were PCR amplified. The four fragments were then assembled into the pUC19 cloning vector using the Gibson Assembly Kit using the manufacturer’s protocol (NEB). Correct orientation of all four fragments was confirmed by sequencing. The resultant natMX6-pcdt1-rif1 construct was PCR amplified and transformed into WT cells; successful integration was confirmed by PCR. The pcdt1-rif1 taz1Δ strain was made in two ways: first, by crossing the pcdt1-rif1 strain with a taz1Δ strain (Figure 7E) and, second, by transforming the pcdt1-rif1 construct into taz1Δ cells as, well as into a heterozygous diploid (rif1+/pcdt-rif1 taz1+/Δ). Regardless of which of these strategies were used, all pcdt1-rif1 taz1Δ strains showed rescue of taz1Δ cold sensitivity.
Author Contributions
S.Z. and J.P.C. designed and interpreted this study with contributions from N.S. and R.K.N. S.Z. performed the experiments in Figures 1−5 and S1−S5 with help from N.S. N.S. performed the work on non-telomeric UFBs, with help from S.Z. and R.K.N. R.K.N. performed the experiments on S-Rif1. S.Z. and J.P.C. wrote the paper, and all authors edited it. J.P.C. obtained funding for this study.
Acknowledgments
We thank all members of the J.P.C. lab for useful discussion and experimental help, and special thanks for advice go to our former lab members Jessica Greenwood and Ofer Rog. We thank Alexander Kelly for valuable discussion and Alessandro Bianchi and Miguel Godinho Ferreira for kind gifts of reagents and advice. This work was supported by Cancer Research UK and the National Cancer Institute.
Published: June 16, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, one table, and four movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.05.077.
Supplemental Information
Film (stills in Figure 1A), showing a taz1Δ cell, harboring fluorescent histone H3 (see Figure 1A) and grown at 19°C, with a “wt-like” chromosome segregation pattern.
Film of a taz1Δ cell tagged as in Figure 1A exhibiting the “pointing finger” segregation phenotype (see Figure 1A and text).
Film of a taz1Δ cell tagged as in Figure 1A demonstrating the “irreversibly entangled” segregation pattern (see Figure 1A and text).
Film (stills in Figure 3E) of taz1Δ cell carrying mRFP-tagged histone H3 and GFP-tagged RPARad11. This cell, grown at 19°C, shows a strand of RPA between segregating chromosomes retracting into one of the daughter nuclei.
References
- Barefield C., Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 2012;40:7358–7367. doi: 10.1093/nar/gks407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C., Körner R., Hofmann K., Nigg E.A. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- Carneiro T., Khair L., Reis C.C., Borges V., Moser B.A., Nakamura T.M., Ferreira M.G. Telomeres avoid end detection by severing the checkpoint signal transduction pathway. Nature. 2010;467:228–232. doi: 10.1038/nature09353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.L., Hickson I.D. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin. Cell Dev. Biol. 2011;22:906–912. doi: 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Chan K.L., North P.S., Hickson I.D. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.L., Palmai-Pallag T., Ying S., Hickson I.D. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Chapman J.R., Barral P., Vannier J.B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D., Boulton S.J. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornacchia D., Dileep V., Quivy J.P., Foti R., Tili F., Santarella-Mellwig R., Antony C., Almouzni G., Gilbert D.M., Buonomo S.B. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J. 2012;31:3678–3690. doi: 10.1038/emboj.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davé A., Cooley C., Garg M., Bianchi A. Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep. 2014;7:53–61. doi: 10.1016/j.celrep.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehé P.M., Rog O., Ferreira M.G., Greenwood J., Cooper J.P. Taz1 enforces cell-cycle regulation of telomere synthesis. Mol. Cell. 2012;46:797–808. doi: 10.1016/j.molcel.2012.04.022. [DOI] [PubMed] [Google Scholar]
- Di Virgilio M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A.D., Feldhahn N., Resch W., Oliveira T.Y., Chait B.T. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D.Q., Yamamoto A., Haraguchi T., Hiraoka Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 2004;6:329–341. doi: 10.1016/s1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Germann S.M., Schramke V., Pedersen R.T., Gallina I., Eckert-Boulet N., Oestergaard V.H., Lisby M. TopBP1/Dpb11 binds DNA anaphase bridges to prevent genome instability. J. Cell Biol. 2014;204:45–59. doi: 10.1083/jcb.201305157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germe T., Miller K., Cooper J.P. A non-canonical function of topoisomerase II in disentangling dysfunctional telomeres. EMBO J. 2009;28:2803–2811. doi: 10.1038/emboj.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J., Polakova S., Zhang L., Tolić-Nørrelykke I.M., Cimini D. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 2011;21:374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C.F., Sussel L., Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- Hayano M., Kanoh Y., Matsumoto S., Renard-Guillet C., Shirahige K., Masai H. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012;26:137–150. doi: 10.1101/gad.178491.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengeveld R.C., de Boer H.R., Schoonen P.M., de Vries E.G., Lens S.M., van Vugt M.A. Rif1 is required for resolution of ultrafine DNA bridges in anaphase to ensure genomic stability. Dev. Cell. 2015;34:466–474. doi: 10.1016/j.devcel.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Hiraga S., Alvino G.M., Chang F., Lian H.Y., Sridhar A., Kubota T., Brewer B.J., Weinreich M., Raghuraman M.K., Donaldson A.D. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 2014;28:372–383. doi: 10.1101/gad.231258.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J.F., Beach D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J. 1994;13:425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J., Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- Kanoh Y., Matsumoto S., Fukatsu R., Kakusho N., Kono N., Renard-Guillet C., Masuda K., Iida K., Nagasawa K., Shirahige K., Masai H. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol. 2015;22:889–897. doi: 10.1038/nsmb.3102. [DOI] [PubMed] [Google Scholar]
- Lian H.Y., Robertson E.D., Hiraga S., Alvino G.M., Collingwood D., McCune H.J., Sridhar A., Brewer B.J., Raghuraman M.K., Donaldson A.D. The effect of Ku on telomere replication time is mediated by telomere length but is independent of histone tail acetylation. Mol. Biol. Cell. 2011;22:1753–1765. doi: 10.1091/mbc.E10-06-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grøfte M., Chan K.L., Hickson I.D., Bartek J., Lukas J. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- Maciejowski J., Li Y., Bosco N., Campbell P.J., de Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattarocci S., Shyian M., Lemmens L., Damay P., Altintas D.M., Shi T., Bartholomew C.R., Thomä N.H., Hardy C.F., Shore D. Rif1 controls DNA replication timing in yeast through the PP1 phosphatase Glc7. Cell Rep. 2014;7:62–69. doi: 10.1016/j.celrep.2014.03.010. [DOI] [PubMed] [Google Scholar]
- Miller K.M., Cooper J.P. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol. Cell. 2003;11:303–313. doi: 10.1016/s1097-2765(03)00041-8. [DOI] [PubMed] [Google Scholar]
- Miller K.M., Ferreira M.G., Cooper J.P. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005;24:3128–3135. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.M., Rog O., Cooper J.P. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- Muñoz P., Blanco R., de Carcer G., Schoeftner S., Benetti R., Flores J.M., Malumbres M., Blasco M.A. TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol. Cell. Biol. 2009;29:1608–1625. doi: 10.1128/MCB.01339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nera B., Huang H.S., Lai T., Xu L. Elevated levels of TRF2 induce telomeric ultrafine anaphase bridges and rapid telomere deletions. Nat. Commun. 2015;6:10132. doi: 10.1038/ncomms10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi T., Muramatsu Y., Yoshida H., Seimiya H. TRF1 ensures the centromeric function of Aurora-B and proper chromosome segregation. Mol. Cell. Biol. 2014;34:2464–2478. doi: 10.1128/MCB.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O., Miller K.M., Ferreira M.G., Cooper J.P. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol. Cell. 2009;33:559–569. doi: 10.1016/j.molcel.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Sato M., Dhut S., Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T., Bunker R.D., Mattarocci S., Ribeyre C., Faty M., Gut H., Scrima A., Rass U., Rubin S.M., Shore D., Thomä N.H. Rif1 and Rif2 shape telomere function and architecture through multivalent Rap1 interactions. Cell. 2013;153:1340–1353. doi: 10.1016/j.cell.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Silverman J., Takai H., Buonomo S.B., Eisenhaber F., de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 2004;18:2108–2119. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S., Osman F., Lorenz A., Steinacher R., Castagnetti S., Ledesma J., Whitby M.C. Ultrafine anaphase bridges, broken DNA and illegitimate recombination induced by a replication fork barrier. Nucleic Acids Res. 2011;39:6568–6584. doi: 10.1093/nar/gkr340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreesankar E., Senthilkumar R., Bharathi V., Mishra R.K., Mishra K. Functional diversification of yeast telomere associated protein, Rif1, in higher eukaryotes. BMC Genomics. 2012;13:255. doi: 10.1186/1471-2164-13-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Blackburn E.H. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J. Cell Biol. 2004;167:819–830. doi: 10.1083/jcb.200408181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Muniandy P., Leo E., Yin J., Thangavel S., Shen X., Ii M., Agama K., Guo R., Fox D., 3rd Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J. 2010;29:3140–3155. doi: 10.1038/emboj.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Rushton M.D., Maringele L. A novel checkpoint and RPA inhibitory pathway regulated by Rif1. PLoS Genet. 2011;7:e1002417. doi: 10.1371/journal.pgen.1002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Hayano M., Masai H. Replication timing regulation of eukaryotic replicons: Rif1 as a global regulator of replication timing. Trends Genet. 2013;29:449–460. doi: 10.1016/j.tig.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Zimmermann M., Lottersberger F., Buonomo S.B., Sfeir A., de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Film (stills in Figure 1A), showing a taz1Δ cell, harboring fluorescent histone H3 (see Figure 1A) and grown at 19°C, with a “wt-like” chromosome segregation pattern.
Film of a taz1Δ cell tagged as in Figure 1A exhibiting the “pointing finger” segregation phenotype (see Figure 1A and text).
Film of a taz1Δ cell tagged as in Figure 1A demonstrating the “irreversibly entangled” segregation pattern (see Figure 1A and text).
Film (stills in Figure 3E) of taz1Δ cell carrying mRFP-tagged histone H3 and GFP-tagged RPARad11. This cell, grown at 19°C, shows a strand of RPA between segregating chromosomes retracting into one of the daughter nuclei.