Abstract
Triclosan (TCS), an antibacterial compound commonly added to personal care products, could be an endocrine disruptor at low doses. Although TCS has been shown to alter fetal physiology, its effects in the developing fetal brain are unknown. We hypothesize that exposure to TCS during fetal life could affect fetal hypothalamic gene expression. The objective of this study was to use transcriptomics and systems analysis to identify significantly altered biological processes in the late gestation ovine fetal hypothalamus after direct or indirect exposure to low doses of TCS. For direct TCS exposure, chronically catheterized late gestation fetal sheep were infused with vehicle (n = 4) or TCS (250 μg/d; n = 4) iv. For indirect TCS exposure, TCS (100 μg/kg · d; n = 3) or vehicle (n = 3) was infused into the maternal circulation. Fetal hypothalami were collected after 2 days of infusion, and gene expression was measured through microarray. Hierarchical clustering of all samples according to gene expression profiles showed that samples from the TCS-treated animals clustered apart from the controls. Gene set enrichment analysis revealed that fetal hypothalamic genes stimulated by maternal and fetal TCS infusion were significantly enriching for cell cycle, reproductive process, and feeding behavior, whereas the inhibited genes were significantly enriching for chromatin modification and metabolism of steroids, lipoproteins, fatty acids, and glucose (P < .05). In conclusion, short-term infusion of TCS induces vigorous changes in the fetal hypothalamic transcriptomics, which are mainly related to food intake pathways and metabolism. If these changes persist to postnatal life, they could result in adverse consequences in adulthood.
Triclosan (TCS) (5-chloro-2-(2,4-dichlorophenoxy)-phenol) is a synthetic antibacterial compound that is commonly added to personal care products (1). TCS acts as an antibacterial because it interrupts fatty acid synthesis (2) by inhibiting enoyl reductase in bacteria (3). Because of its action on a bacterial enzyme that, except for within the gastrointestinal microbiota (4), is not expressed in the human being, it is thought to be safe for use in humans (5).
TCS specifically has been associated with fetal growth restriction. A recent epidemiologic study showed that maternal urinary TCS concentration was negatively correlated with fetal growth variables in late gestation (6). In rodents, TCS decreases circulating concentrations of thyroid hormones in the pregnant dam and fetus (7, 8), and prolonged exposure to TCS induced fetal growth restriction as well as delayed vaginal opening in the postnatal female offspring (9).
These studies have demonstrated that TCS exposure during fetal life affects fetal and postnatal physiology. However, the effect of TCS on the developing fetal brain remains unknown. We performed the present study to test whether TCS, in doses that are similar to exposure in humans from personal care products (10, 11), has an effect on the fetal hypothalamus. The hypothalamus was selected because it plays a central role in the coordination of fetal autonomic and neuroendocrine function, including control of blood pressure, responses to stress, and control of ingestive behavior postnatally (12). We used the ovine fetus as animal model because the entire gestational equivalent of human brain development occurs in utero (13), and the size of the sheep fetus allows us to analyze molecular responses in defined brain regions.
We have recently used transcriptomics and systems biology analysis to understand the response of the fetal hypothalamus to in utero stressors (14, 15). In the present study, we used this approach to identify the fetal hypothalamic response to TCS using 2 routes of administration (fetal iv or maternal iv). Using this broad transcriptomics approach, we tested the hypothesis that TCS alters fetal hypothalamic gene transcription and that the pattern of changes in the transcriptomics will reveal molecular pathways and potentially neuropeptides or neuronal pathways that are susceptible to TCS in the late gestation fetal hypothalamus.
Materials and Methods
Chemicals
TCS, 99.8% pure, was purchased from TCI America and recrystallized from ethanol:water before administering to sheep. All other chemicals were purchased from laboratory suppliers and were of reagent grade or better.
Animal procedures
All of these experiments were performed in accordance with the Guiding Principles for Research Involving Animals and Human Beings published by the American Physiological Society and were approved by the University of Florida Institutional Animal Care and Use Committee.
To test the fetal response to direct fetal iv infusion of TCS (n = 4) or dimethylsulfoxide:water 50:50 vehicle solution (n = 4), we used the chronically catheterized fetal sheep model of in utero development and physiology. Time-dated pregnant ewes and their fetuses were both surgically prepared with indwelling vascular catheters (femoral arteries and veins, and in the fetuses, an additional catheter in the amniotic fluid). In a second study designed to test the fetal response to maternal iv infusion of TCS, time-dated pregnant ewes (but not their fetuses) were surgically prepared with catheters in femoral arteries and veins or in the external jugular vein to infuse TCS (n = 3) or dimethylsulfoxide vehicle (n = 3).
Food was withheld from the ewes for 24 hours before surgery (124 ± 3-d gestation). Anesthesia was induced by mask with isoflurane, then intubated, and anesthesia was maintained using isoflurane (0.5%–2%) in oxygen. Surgery and postsurgical care were performed as previously described (16). A minimum of 5 days after surgery, we infused TCS at a rate of 0.1 mg/kg · d for 2 days. We estimated fetal body weight to be approximately 2.5 kg; as a result, we infused 0.25 mg/d into each fetus iv in a total volume of 10 mL/d. Body weights of the pregnant ewes in the second study ranged from 63 to 75 kg; therefore, we infused 6.3–7.5 mg/d iv into these ewes. The TCS dose is within the range of likely environmental exposure to TCS from plaque-reducing toothpaste, which contains 0.3% TCS and antibacterial soap, which contains 0.1%–0.2% TCS, as measured in human samples (17).
Sample collection
Blood samples were drawn (5 mL from fetus and ewe in the first study and 5 mL from ewe in the second study) before, and after 1 and 2 days of infusion. At the end of the infusion, we humanely killed the ewes and fetuses with an overdose of sodium pentobarbital. In each study, fetal brains were rapidly removed, dissected into distinct regions, and snap frozen in liquid nitrogen. Tissues collected included the fetal hypothalamus, pituitary, hippocampus, cerebral cortex, medullary brainstem, and cerebellum. Tissues were stored at −80°C until processed for RNA. In the present study, only the RNA isolated from the hypothalamus was analyzed.
RNA extraction and preparation
mRNA was extracted and purified as previously reported (15). RNA integrity numbers of the purified mRNA samples was measured using an Agilent Bioanalyzer, and ranged from 7.4 to 8.7. Deoxyribonuclease treatment, synthesis of cRNA, and labeling were performed as previously described (15). The resulting labeled cRNA was analyzed with the NanoDrop spectrophotometer, and the specific activities and the yields of the cRNAs ranged from 10.2 to 12.8 pmol Cy3/μg RNA and from 3.0 to 9.6 μg, respectively. The labeled cRNA was stored at −80°C until use. Microarray hybridization, washing, and scanning were performed as previously described (14). We used the Agilent-019921 Sheep Gene Expression Microarray 8x15k, G4813A, which we had previously annotated (GEO accession number, GPL14112) (14).
Microarray data preparation
The limma package was employed to import the raw data into R (http://www.r-project.org), perform background correction, and normalize the data using the quantile normalization method (18). Control probes and probes that were less than 10% brighter than the negative control probes were filtered out, and then the intensity of probes with the same probe identity were averaged. In order to eliminate multiple probes and facilitate the biological interpretation of the results, probes that were annotated with the same gene name were collapsed using the MaxMean function of the WGCNA package in R (19). This method assigns the highest intensity value as the expression value of the gene, based on the assumption that the probe with the highest intensity value had the best hybridization to the cDNA. After probe filtering and collapsing, the gene expression list contained 8196 unique official symbols for 8196 genes, out of 15 008 probes in the original platform. Microarray data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE69275.
Unsupervised data analysis
An exploratory microarray data analysis was performed initially, to determine the relatedness between microarray samples according gene expression profiles. Microarray samples were: TCS infused into the fetus (fetal TCS [TCSF], n = 4), TCS infused into the ewe (maternal TCS [TCSM], n = 3), saline infused into the fetus (ControlF, n = 4), and saline infused into the ewe (ControlM, n = 3). Correlations between samples were evaluated by application of hierarchical clustering and principal component (PC) analysis methods. For hierarchical clustering, we used the open software Cluster 3.0 (20). Data was adjusted by centering the gene expressions by the median. Gene expression and microarray samples were clustered according to centroid linkage, after similarity measurement by centered correlation. The PCs of the data were calculated with the function prcomp from the stats package for R (21). The first and second PCs were plotted in a two-dimensional plot using the plot function from the graphics package for R. The 14 data points were colored according their group.
Gene set enrichment analysis (GSEA)
This analytical method considers first the gene expression profiles from 2 classes to rank genes according to the strength of the correlation with a class. Then, given a priori defined sets of genes, the method will determine whether genes (belonging to the same set) are randomly distributed throughout the ranked list or mainly found at the top or bottom, ie, significantly overexpressed in one or other class (22). For our study, we defined the gene sets as genes sharing the same biological process. A significant biological process was defined by a nominal P < .05, after 1000 permutations. GSEA (http://software.broadinstitute.org/gsea) is an open software developed at the Broad Institute of Massachusetts Institute of Technology and Harvard, which determines whether an a priori defined set of genes shows statistically differences between 2 biological states (22, 23). One limitation of this tool is that relies on biological information from previous experiments; for this reason, the ability of GSEA to identify processes relevant to fetal hypothalamic function might be underpowered.
Transcription factor analysis
WebGestalt was used for detection of transcription factor binding sites that are statistically significantly overrepresented among the differentially regulated genes. This analysis is based on an ORACLE relational database GeneKeyDB, which uses a strong gene and protein centric viewpoint (24).
Statistical analysis
The GSEA method finds significant biological processes in the data independently of statistical analysis. However, in order to corroborate the results we applied a moderated t test to find differentially expressed genes (DEGs) in the data to then perform ontology analysis of them. The limma package was used for the moderated t test, applying the empirical Bayes method proposed by Smyth (25), which produces more stable estimates when the number of replicates is small. Genes were considered differentially expressed with P < .05. Gene ontology analysis for the significant up or down-regulated genes was performed with the DAVID database (26), searching for enriched biological processes (P < .05) in the data.
Quantitative real-time (qRT)-PCR validation
The RNA samples extracted from the fetal hypothalami of the 4 groups were converted to cDNA with a High Capacity cDNA Archive kit using the methodology recommended by the kit manufacturer (Applied Biosystems). The newly synthesized cDNA was stored at −20°C until qRT-PCR was performed. The following genes were selected for further validation by qRT-PCR: agouti-related protein (AGRP), agouti-signaling protein (ASIP), proopiomelanocortin (POMC), and fatty acid synthase (FASN).
Relative expression of selected genes was determined using primers (Sigma-Aldrich) and SYBR Green PCR Master Mix (Applied Biosystems). Primers were designed with Primer Express software (Applied Biosystems) from the corresponding ovine mRNA. Primers sequences and accession numbers are reported in Supplemental Table 1. All primer pairs had efficiencies greater than 95%. The abundance of β-actin mRNA was determined in each sample, using primers and VIC TaqMan probes designed from the ovine β-actin sequence and TaqMan qRT-PCR master mix (Applied Biosystems). All samples were run in triplicate for each gene and for β-actin. There were no differences in β-actin expression among the groups. Relative mRNA expression of each gene was calculated by determining change in threshold cycle (ΔCt) between the mean Ct for each gene and the mean Ct for β-actin mRNA from the same sample. The effect of TCSM or TCSF infusion on each gene was analyzed by ANOVA using the ΔCt values. Data were graphed as the mean fold change in expression relative to the respective control group; fold change in each sample was calculated as 2−ΔΔCt, where ΔΔCt is the difference between ΔCt in each sample and the mean ΔCt in the control group. For all statistical analyses, the criterion for achieving statistical significance was P < .05.
Results
Unsupervised data analysis
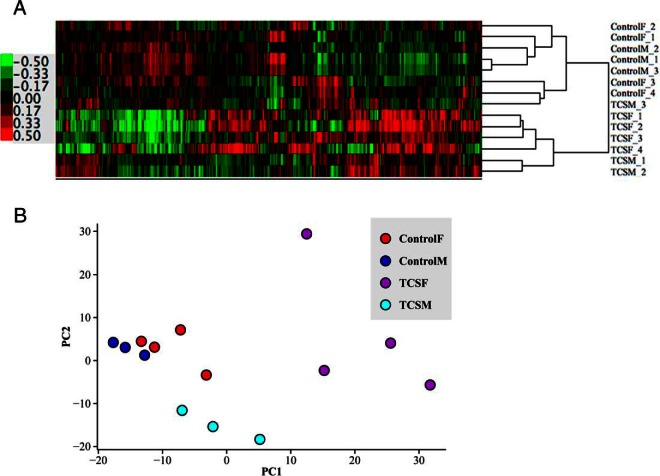
The heat map resulting from the hierarchical clustering method and the PCs plot are shown in Figure 1. The heat map (Figure 1A) demonstrates that all samples belonging to TCSF are clustered together, and they are similar to TCSM, except for 1 sample (TCSM_3) that clusters with ControlF_4 sample. The PC analysis (Figure 1B) corroborates the relatedness of the samples. Visually, control samples are closer to TCSM samples (although they are not mixed), whereas TCSF segregated apart from the other groups. These analyses suggest that TCS infusion induces a strong effect in fetal hypothalamic gene expression, and this effect is more pronounced if the TCS is infused directly into the fetus.
Figure 1. Unsupervised analysis of the microarray samples.
The heat map (A) and the principal component (PC) plot (B) show that samples belonging to the same group tend to cluster together.
Gene set enrichment analysis
The GSEA analysis was performed selecting the biological processes (from gene ontology) as the a priori defined gene sets. Comparisons were done between TCSM and ControlM samples and between TCSF and ControlF samples. Each gene set has a leading-edge subset, that is, the core of genes that accounts for the enrichment signal. If enriched biological processes were redundant, a leading edge analysis was performed. This analysis determines common genes in the leading-edge subsets between similar biological processes. For each group, the enriched biological processes and only the genes belonging to the leading edges are shown in Table 1.
Table 1.
Enriched Biological Processes and Core Genes on Fetal Hypothalamic Samples Up- or Down-Regulated by Maternal or Fetal Infusion of TCS
| Biological Process | Core Genes | P Value |
|---|---|---|
| Up-regulated by TCSM infusion | ||
| Mitotic cell cycle | KIF15 | AURKA | KIF2C | SUGT1 | FBXO5 | NDC80 | DLGAP5 | BIRC5 | CENPE | ESPL1 | MPHOSPH6 | SMAD3 | UBE2C | TPX2 | KIF11 | CCNA2 | BUB1 | SKP2 | MAD2L2 | TTK | NCAPH | KNTC1 | SMC4 | CDC123 | PLK1 | CLIP1 | KPNA2 | <.001 |
| Cell-cell signaling | CGA | GHRH | TSHB | TFAP2C | POMC | NPY | ADM | CALCA | GNRH1 | CCL5 | TSHR | PMP22 | AREG | EDN3 | EFNA4 | TBX3 | GCH1 | WISP1 | CXCL5 | SST | KCNN3 | PTHLH | GHRL | SYN2 | MERTK | PRG3 | NQO1 | GRM5 | SH2D1A | <.001 |
| Female pregnancy | ADM | TRO | SFRP4 | TAC3 | FSHB | OVGP1 | SPRR2E | PTHLH | GHRL | OXTR | FLT1 | <.001 |
| Cell adhesion | SAA1 | CYTIP | IL8 | EMCN | TGM2 | IL12A | ARHGDIB | SPN | .003 |
| G protein-coupled receptor | AGRP | GHRH | TSHB | NPY | CALCA | TAC3 | TSHR | PTAFR | GCG | GLP1R | CD3E | SST | IL8 | PTHLH | GHRL | CXCR5 | SSTR5 | CRHR1 | ADRB1 | GRM5 | RGS14 | CXCR6 | CCR3 | CCRL2 | GHRHR | PTGER1 | UCN | CXCR2 | RGS1 | PIK3CG | APOC3 | HCRTR1 | ANG | OPRM1 | GPRC5C | CXCL12 | CXCR4 | CCL2 | HRH2 | APOA1 | AGTR1 | .003 |
| Reproductive process | ADM | TRO | SFRP4 | TAC3 | ACE2 | FSHB | OVGP1 | TOP2A | SMAD3 | TBX3 | SPRR2E | IL8 | PTHLH | GHRL | OXTR | CRHR1 | IDE | FLT1 | CXCR6 | MKKS | .003 |
| Behavior | AGRP | SAA1 | CCL28 | NPY | CCL5 | PTAFR | GCG | CXCL5 | GHRL | IL8 | AGTR2 | BRS3 | PMCH | CCR3 | CCRL2 | CXCR2 | SPN | HCRTR1 | OPRM1 | CXCL12 | CXCR4 | LECT2 | CCL2 | .005 |
| Down-regulated by TCSM infusion | ||
| Chromatin/histone modification | PRMT7 | KDM4A | PHB | KAT5 | SIRT2 | UBE2N | WHSC1L1 | NSD1 | RBM14 | .03 |
| Protein processing | SCLY | OAZ1 | ASRGL1 | NFS1 | ADI1 | WARS | GCLM | SLC7A5 | HGD | HPRT1 | BBOX1 | GLUD1 | ALDH6A1 | SLC7A4 | SMS | ATF4 | MAT2B | PEPD | BCKDHA | DHPS | FPGS | ALDH5A1 | CCBL1 | QDPR | PAH | BCAT1 | DARS | GOT1 | PLOD1 | .03 |
| Lipoprotein metabolic process | APOA2 | SCARF1 | FNTB | PIGO | DPM1 | LDLR | PPT1 | .04 |
| Steroid metabolic Process | SCARB1 | HSD17B11 | STARD3 | STUB1 | CLN6 | PPARD | NPC1 | APOL2 | HSD17B6 | IL4 | | .05 |
| Fatty acid metabolic process | ACADM | ADIPOR1 | ALDH5A1 | CD74 | CPT1A | CROT | ECHS1 | FADS2 | FASN | HACL1 | HADHB | PPARD | PTGES3 | .05 |
| Up-regulated by TCSF infusion | ||
| Cell-cell signaling | GHRH | CXCL10 | GNRH1 | SH2D1A | S100A9 | MME | PRG3 | CALCA | CCL4 | CCL15 | CCL5 | IL18 | GCHFR | CHRNB1 | TFAP2C | GRM4 | TNFRSF11A | SST | WISP1 | SYPL1 | GCH1 | AREG | C1QA | SNAI2 | POMC | NPY | MGST2 | SYN2 | ASIP | KLF10 | GRM5 | CXCL13 | BMP2 | INHBA | .005 |
| Inflammatory response | S100A12 | CXCL10 | S100A9 | ANXA1 | CCL4 | CCL5 | HDAC4 | IL1RAP | AIF1 | CYBB | .006 |
| Cytokine production | TLR1 | PRG3 | CALCA | IL6 | IL18 | SRGN | SMAD3 | TLR3 | INHBA | EBI3 | TLR6 | HIF1A | CEBPG | APOA2 | SMAD4 | INHA | ATP6AP2 | SFTPD | MALT1 | |
| Regulation of secretion | GHRH | ANG | SRGN | INHBA | DNAJC1 | APOA2 | MYO6 | INHA | .01 |
| Cellular homeostasis | F2RL1 | CALCA | CD55 | CCL15 | CCL5 | MT2A | ATP1A1 | SAA1 | CP | GPR98 | .03 |
| Behavior | CXCL10 | AGRP | CCL15 | CCL5 | SAA1 | PMCH | NPY | BRS3 | ACSL4 | CCL2 | PLAUR | CCRL2 | CXCL13 | .04 |
| Down-regulated by TCSF infusion | ||
| Chromatin/histone modification | HDAC6 | CREBBP | UBE2N | NSD1 | SET | EHMT1 | RBM14 | KAT5 | KDM4A | SIRT2 | PPARGC1A | .01 |
| Lipoprotein metabolic process | SCARF1 | FNTB | PPT1 | PIGO | LDLR | .03 |
| CNS development | EIF2B1 | TNC | NNAT | PROP1 | UBE3A | CTNS | SMARCA1 | EIF2B2 | ROBO2 | S100B | BTD | ARNT2 | PITPNM1 | DRP2 | SLIT1 | BPTF | PAX6 | GPR56 | RNF103 | SLIT3 | ALDH5A1 | NCOA6 | NPAS2 | SHH | PPT1 | NKX2–2 | .03 |
| Gamete generation | FSHB | CDYL | PARN | PICK1 | USP9 × | TOB2 | MAST2 | HMGCR | TESK1 | SERPINA5 | RUVBL1 | DEDD | BRD2 | .05 |
GLP1R, glucagon-like peptide receptor.
Statistical analysis
The total number of DEG was higher for the comparison between TCSF and ControlF samples than for TCSM and ControlM samples, as expected after the unsupervised analysis results (2740 and 1461 DEGs, respectively) (Supplemental Table 2, A–D). There were significant DEG overlaps between TCSM and TCSF infusion for both up-regulated and down-regulated genes (P < .00001 by Pearson's χ2 test) (Figure 2). However, there were a much larger number of genes stimulated or inhibited only by TCSM or TCSF infusion, as shown in Figure 2.
Figure 2. Venn diagrams of DEGs.
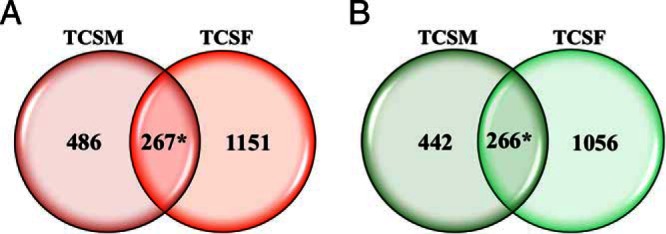
The Venn diagrams show significant overlap between DEG up-regulated (A) or down-regulated (B) by TCSM and TCSF infusion (*, P < .00001 by Pearson's χ2 test).
Ontology analysis
An extended number of biological processes were significantly enriched (P < .05) with the up or down-regulated genes by TCSM or TCSF infusion (Supplemental Table 3, A–D, respectively), which could be summarized in the following biological processes.
For TCSM, the more representative up-regulated biological processes were: mitosis, cell cycle, cell adhesion, female pregnancy, response to hormone stimulus, developmental maturation, calcium ion homeostasis, feeding behavior, and regulation of inflammatory response and cytokine production (Supplemental Table 3A). Representative down-regulated biological processes were: nitrogen compound biosynthesis process, cellular homeostasis, adaptive and humoral immune response, response to hypoxia, leukocyte migration, generation of precursor metabolites and energy, protein maturation, response to steroid hormone stimulus, proteolysis, and monosaccharide metabolic process (Supplemental Table 3B).
For TCSF, representative up-regulated biological processes were: regulation of apoptosis, cell-cell signaling, female pregnancy, inflammatory response, regulation of cytokine production, homeostatic process, response to insulin stimulus, and regulation of secretion (Supplemental Table 3C). Representative down-regulated biological processes were: chromatin/histone modification, actin filament-based process, in utero development, regulation of neuron differentiation, protein catabolic process, glucose catabolic process, cholesterol metabolic process, and fatty acid metabolic process (Supplemental Table 3D).
The ontology analysis performed with the DEG, selected after the statistical analysis, shows that most of the enriched biological processes correspond with the findings of the GSEA, corroborating these results. Also, the ontology analysis refined the results, because it demonstrated a wider range of significant biological processes.
Transcription factor analysis
Results of transcription factor analysis are reported in Table 2. Analysis transcription factor binding sites revealed that SP1, ELK1, LEF1, ERR1, estrogen receptor (ER)α, glucocorticoid receptor (GR), thyroid hormone receptor (T3R), and isoforms of peroxisome proliferator-activated receptor (PPAR) are significantly associated with genes up- and down-regulated by either maternal or fetal infusion of TCS. Genes up-regulated by TCSF infusion but not altered by maternal infusions are associated with nuclear transcription factor Y α (NFY) and GA-binding protein α-chain (GABP). Conversely, genes down-regulated by TCSF infusion but not altered by maternal infusions are associated with paired box gene 4 (PAX4) and sterol regulatory element-binding protein (SREBP). Other transcription factors were associated with responses in more than 1 group (eg, c-Myc, which is associated with both up- and down-regulated genes after fetal infusion and down-regulated genes after maternal infusion).
Table 2.
Transcription Factor-Binding Sites Significantly Overrepresented Among Genes Up- or Down-Regulated by Maternal or Fetal Infusion of TCS
| Transcription Factor | Site | Genes/Total | Adjusted Probability | |
|---|---|---|---|---|
| Up-regulated by TCSF infusion | ||||
| 1 | SP1 | hsa_GGGCGGR_V$SP1_Q6 | 265/2891 | 9.87 e-57 |
| 2 | ELK1 | hsa_SCGGAAGY_V$ELK1_O2 | 160/1176 | 8.13 e-55 |
| 3 | NRF1 | hsa_RCGCANGCGY_V$NRF1_Q6 | 121/894 | 1.03 e-40 |
| 4 | FOX04 | hsa_TTGTTT_V$FOX04_O1 | 157/2037 | 5.28 e-24 |
| 5 | NFY | hsa_GATTGGY_V$NFY_Q6_O1 | 101/1141 | 3.56 e-19 |
| 6 | GABP | hsa_MGGAAGTG_V$GABP_B | 76/744 | 9.21 e-18 |
| 7 | unknown | hsa_TGCGCANK_unknown | 63/537 | 1.2 e-17 |
| 8 | NFAT | hsa_TGGAAA_V$NFAT_Q4_O1 | 133/1871 | 2.5 e-17 |
| 9 | LEF1 | hsa_CTTTGT_V$LEF1_Q2 | 136/1939 | 2.64 e-17 |
| 10 | MYC | hsa_CACGTG_V$MYC_Q2 | 86/1015 | 2.55 e-15 |
| T3R | hsa_V$T3R_Q6 | 17/248 | 0.0048 | |
| ERR1 | hsa_TGACCTTY_V$ERR1_Q2 | 73/1023 | 8.99 e-10 | |
| ERR1 | hsa_V$ERR1_Q2 | 20/257 | 0.0007 | |
| ER | hsa_V$ER_Q6_O2 | 16/251 | 0.011 | |
| ER | hsa_V$ER_Q6_O1 | 16/264 | 0.0159 | |
| GR | hsa_V$GR_O1 | 19/202 | 9.76 e-5 | |
| GR | hsa_V$GR_Q6 | 17/267 | 0.009 | |
| GRE | hsa_V$GRE_C | 9/124 | 0.0255 | |
| LXR | hsa_V$LXR_Q3 | 8/74 | 0.005 | |
| PPARG | hsa_V$PPARG_O1 | 8/46 | 0.0003 | |
| PPAR | hsa_V$PPAR_DR1_Q2 | 17/257 | 0.0066 | |
| Down-regulated by TCSF infusion | ||||
| 1 | SP1 | hsa_GGGCGGR_V$SP1_Q6 | 319/2891 | 1.04 e-94 |
| 2 | MAZ | hsa_GGGAGGRR_V$MAZ_Q6 | 212/2250 | 9.23 e-49 |
| 3 | MYC | hsa_CACGTG_V$MYC_Q2 | 135/1015 | 1.92 e-46 |
| 4 | LEF1 | hsa_CTTTGT_V$LEF1_Q2 | 185/1939 | 4.6 e-43 |
| 5 | E12 | hsa_CAGCTG_V$E12_Q6 | 201/2450 | 2.06 e-37 |
| 6 | ELK1 | hsa_SCGGAAGY_V$ELK1_O2 | 130/1176 | 3.5 e-36 |
| 7 | FOX04 | hsa_TTGTTT_V$FOX04_O1 | 167/2037 | 1.11 e-30 |
| 8 | NRF1 | hsa_RCGCANGCGY_V$NRF1_Q6 | 103/894 | 5.75 e-30 |
| 9 | PAX4 | hsa_GGGTGGRR_V$PAX4_O3 | 125/1278 | 1.17 e-29 |
| 10 | SREBP1 | hsa_TCANNTGAY_V$SREBP1_O1 | 72/466 | 1.19 e-28 |
| ERR1 | hsa_TGACCTY_V$ERR1_Q2 | 106/1023 | 4.24 e-27 | |
| ERR1 | hsa_V$ERR1_Q2 | 32/257 | 9.22 e-11 | |
| ER | hsa_V$ER_Q6_O1 | 29/264 | 9.70 e-9 | |
| ER | hsa_V$ER_Q6 | 26/273 | 7.32 e-7 | |
| ER | hsa_V$GR_Q6_O2 | 22/251 | 1.53 e-5 | |
| GR | hsa_V$GR_Q6_O1 | 22/276 | 5.88 e-5 | |
| GR | hsa_V$GR_Q6 | 20/267 | 0.0003 | |
| GR | hsa_V$GR_O1 | 14/202 | 0.0038 | |
| GRE | hsa_V$GRE_C | 9/124 | 0.0138 | |
| PPAR | hsa_V$PPAR_DR1_Q2 | 24/257 | 2.53 e-6 | |
| PPARA | hsa_V$PPARA_O2 | 10/127 | 0.0057 | |
| PPARG | hsa_V$PPARG_O1 | 5/46 | 0.0138 | |
| T3R | hsa_V$T3R_Q6 | 19/248 | 0.0003 | |
| LXR | hsa_V$LXR_DR4_Q3 | 8/91 | 0.0069 | |
| LXR | hsa_V$LXR_Q3 | 7/74 | 0.0076 | |
| Up-regulated by TCSM infusion | ||||
| 1 | SP1 | hsa_GGGCGGR_V$SP1_Q6 | 152/2891 | 2.11 e-35 |
| 2 | MAZ | hsa_GGGAGGRR_V$MAZ_Q6 | 115/2250 | 5.15 e-25 |
| 3 | E12 | hsa_CAGGTG_V$E12_Q6 | 105/2450 | 4.56 e-17 |
| 4 | NRF1 | hsa_RCGCANGCGY_V$NRF1_Q6 | 56/894 | 1.88 e-15 |
| 5 | FOX04 | hsa_TTGTTT_V$FOX04_O1 | 90/2037 | 1.88 e-15 |
| 6 | ELK1 | hsa_SCGGAAGY_V$ELK1_O2 | 65/1176 | 1.88 e-15 |
| 7 | E2F1 | hsa_V$ERF1_Q6 | 29/228 | 1.88 e-15 |
| 8 | LEF1 | hsa_CTTTGT_V$LEF1_Q2 | 86/1939 | 5.45 e-15 |
| 9 | E2F1 | hsa_V$ERF1_Q3 | 29/240 | 5.45 e-15 |
| 10 | ETS2 | hsa_RYTTCCTG_V$ETS2_B | 58/1074 | 2.08 e-13 |
| ERR1 | hsa_TGACCTY_V$ERR1_Q2 | 29/1023 | 0.009 | |
| GR | hsa_V$GR_Q6 | 13/267 | 0.00019 | |
| GRE | hsa_V$GRE_C | 7/124 | 0.0111 | |
| GR | hsa_V$GR_O1 | 9/202 | 0.0149 | |
| GR | hsa_V$GR_Q6_O1 | 10/276 | 0.0308 | |
| PPAR | hsa_V$PPAR_DR1_Q2 | 12/257 | 0.0040 | |
| PPARA | hsa_V$PPARA_O2 | 7/127 | 0.0121 | |
| ER | None | |||
| PPARG | None | |||
| T3R | None | |||
| LXR | None | |||
| Down-regulated by TCSM infusion | ||||
| 1 | SP1 | hsa_GGGCGGR_V$SP1_Q6 | 162/2891 | 1.17 e-43 |
| 2 | E12 | hsa_CAGGTG_V$E12_Q6 | 117/2450 | 2.64 e-24 |
| 3 | ERR1 | hsa_TGACCTY_V$ERR1_Q2 | 72/1023 | 9.61 e-24 |
| 4 | NFAT | hsa_TGGAAA_V$NFAT_Q4_O1 | 95/1871 | 2.1 e-21 |
| 5 | MYC | hsa_CACGTG_V$MYC_Q2 | 68/1015 | 2.35 e-21 |
| 6 | LEF1 | hsa_CTTTGT_V$LEF1_Q2 | 94/1939 | 6.14 e-20 |
| 7 | unknown | hsa_AACTTT_unknown | 90/1859 | 4.81 e-19 |
| 8 | NFY | hsa_GATTGGY_V$NFY_Q6_O1 | 65/1141 | 6.46 e-17 |
| 9 | ELK1 | hsa_SCGGAAGY_V$ELK1_O2 | 66/1176 | 6.52 e-17 |
| 10 | AP4 | hsa_CAGCTG_V$AP4_Q5 | 75/1502 | 1.55 e-16 |
| ERR1 | hsa_V$ERR1_Q2 | 21/257 | 2.16 e-8 | |
| ER | hsa_V$ER_Q6_O1 | 14/264 | 0.0004 | |
| ER | hsa_V$ER_Q6 | 13/273 | 0.0015 | |
| ER | hsa_V$GR_Q6_O2 | 12/251 | 0.0019 | |
| GR | hsa_V$GR_Q6 | 11/267 | 0.0078 | |
| GR | hsa_V$GR_O1 | 8/202 | 0.0249 | |
| GRE | hsa_V$GRE_C | 6/124 | 0.023 | |
| T3R | hsa_V$T3R_Q6 | 10/248 | 0.0121 | |
| PPAR | hsa_V$PPAR_DR1_Q2 | 13/257 | 0.001 | |
| PPARA | hsa_V$PPARA_O2 | 7/127 | 0.0084 | |
| LXR | hsa_V$LXR_DR4_Q3 | 10/91 | 1.38 e-5 | |
| LXR | hsa_V$LXR_Q3 | 7/74 | 0.0007 | |
Main effects of both TCSM and TCSF infusions
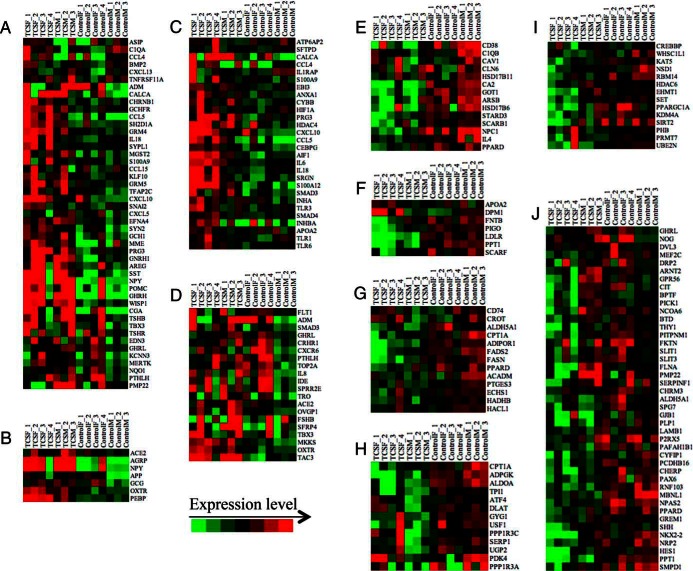
The GSEA performed with all the genes, and the ontology analysis performed only with the DEG, show that some biological processes are equally affected by both TCSM and TCSF infusion, whereas others appear to be affected only if the TCS is infused to the ewe or to the fetus. To explore this concept, we elaborated heat maps with the genes of selected biological processes (displayed in Figure 3). For the main biological processes induced by TCS infusion, the heat maps show that genes related to cell signaling (Figure 3A) and feeding behavior (Figure 3B) are stimulated by TCS infusion in both fetus and mother. Genes involved in inflammatory response (Figure 3C) are induced by TCSF infusion rather than by TCSM infusion; on the other hand, genes related with the reproductive process (Figure 3D) are induced in TCSM infusion. For selected biological processes down-regulated by TCS infusion, the heat maps suggest that steroid metabolic process (Figure 3E), lipoprotein metabolic process (Figure 3F), and fatty acid metabolic process (Figure 3G) are inhibited by both TCSM and TCSF infusion. Glucose metabolic process (Figure 3H) is inhibited by TCSM infusion rather than fetal infusion, whereas chromatin modification (Figure 3I) and central nervous system (CNS) development (Figure 3J) are inhibited by TCSF infusion.
Figure 3. Heat maps of core genes involved in selected biological processes (BPs) affected by TCSM or TCSF infusion.
Heat maps from A–D represent BP increased by TCS infusion, whereas heat maps from E–J belong to BP decreased by TCS. These BP are (A) cell signaling, (B) feeding behavior, (C) inflammatory response, (D) reproduction, (E) steroid metabolic process, (F) lipoprotein metabolic process, (G) fatty acid metabolic process, (H) glucose metabolic process, (I) chromatin modification, and (J) CNS development.
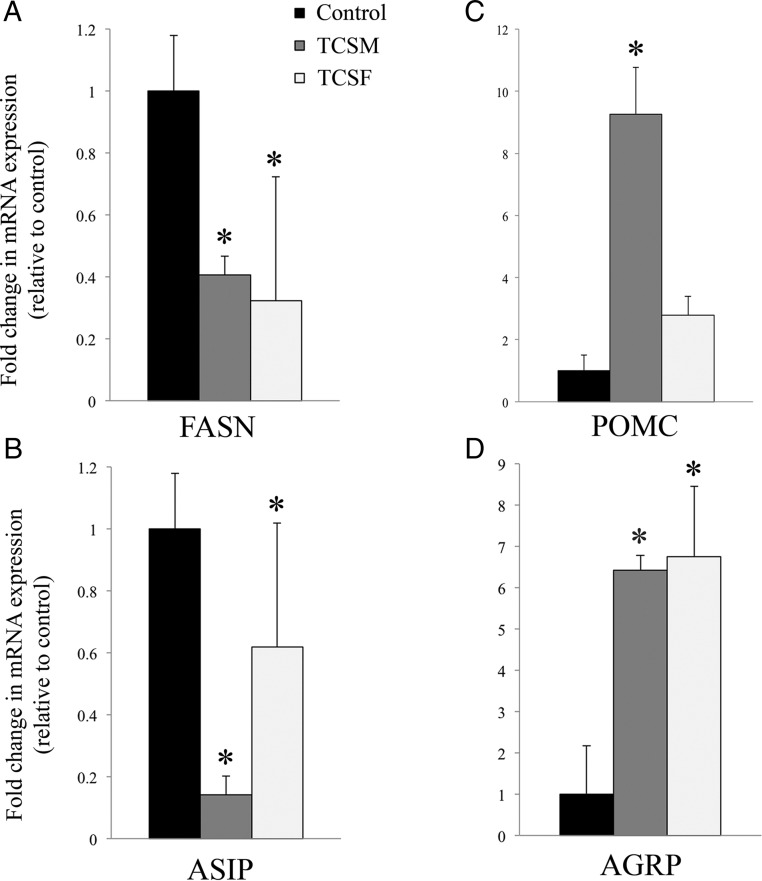
Quantitative PCR confirmation of altered genes related to fatty acid metabolism and appetite
Differential expression of several key regulator genes was confirmed by qRT-PCR. Expressions of FASN and ASIP were significantly decreased by either fetal or maternal infusion of TCS (Figure 4, A and B, respectively), whereas expressions of POMC and AGRP were significantly increased by either maternal or fetal infusion of TCS (Figure 4, C and D).
Figure 4. mRNA expression measured by qRT-PCR in fetal hypothalami after TCSM or TCSF infusion.
mRNA expression for each group is shown as fold change with respect to the controls for the down-regulated genes: (A) FASN and (B) ASIP; and up-regulated genes: (C) POMC and (D) AGRP. *, statistically significant difference (P < .05) in mRNA expression between TCS and control group.
Discussion
The results of this study reveal that 2 days of low-dose TCS infusion, designed to mimic exposure from the use of personal care products containing TCS, induces strong changes in gene expression in the fetal hypothalamus, effects that are even more profound after direct fetal exposure. Conceptually, the changes in hypothalamic gene expression may reflect the neuroendocrine response to the disturbances in fetal physiology after exposure to TCS.
TCS is a known inhibitor of the enoyl reductase activity of the type II FASN enzyme complex in bacteria: it is this action as enzyme inhibitor that is the basis of its antibacterial action (3). TCS also inhibits the type I FASN in mammals (encoded by the FASN gene), although at much higher concentrations (27). Interestingly, the major transcriptomics responses to both maternal and TCSF infusion are related to fatty acid and lipoprotein metabolism and regulation of food intake and energy balance. TCS exposure decreased the expression of genes related to the metabolism of lipoproteins (Figure 3D), fatty acids (Figure 3G), and glucose (Figure 3H), including a reduction in genes associated with fatty acid metabolism, including a reduction of FASN, as determined by the array and confirmed by qRT-PCR (Figure 4A). Although we do not know whether the effect of TCS on fetal hypothalamic FASN is direct, several of the genes affected by TCS are known to be related to each other mechanistically. FASN gene transcription is increased by an additive effect of insulin and agouti protein, the product of the ASIP gene, because FASN promoter has an agouti-response region distinct from the insulin-response element (28); ASIP was down-regulated by (maternal or fetal) infusion of TCS (Figure 4B).
TCS also altered the expression of genes important for regulation of food intake in postnatal animals. In the present study, we found that both AGRP mRNA and POMC abundance were significantly increased (Figure 4, C and D). The AGRP, encoded by the AGRP gene, is known to be an important neuropeptide with respect to hypothalamic control systems (29). AGRP is expressed by the arcuate neurons (30), which coexpress NPY. These neurons are found in the arcuate nucleus as an adjacent but distinct population from those that produce POMC, the precursor of the melanocortin α melanocyte stimulating hormone (31). Both ASIP and AGRP are inverse agonists at the melanocortin 4 receptor in vitro (32). The neuropeptides NPY and AGRP increase feed intake (orexigenic), whereas α melanocyte stimulating hormone (cleaved from POMC) is anorectic (reviewed in Ref. 33). Although AGRP and POMC are expressed in discrete populations of neurons in the adult, and have opposing effects on food consumption, in the developing brain, these neurons develop from the same lineage. The development of hypothalamic neurons is not completely understood, but in rodents neurogenin 3 (NEUROG3) is known to be expressed in the progenitor neurons, and appears to lead to greater POMC expression and reduced NPY and AGRP expression (34), although there are some POMC neurons that do not arise from neurogenin positive neurons. Neurogenin is on our array and was not differentially expressed with TCS exposure, consistent with its expression in the period of development preceding neuropeptide expression. In rodents, maturation of connections of mature neurons expressing NPY/AGRP or POMC occurs postnatally (35), but these components of the neuronal network that regulate appetite are expressed in the arcuate nucleus of the ovine fetal hypothalamus from at least 110 days of gestation (36). Expression of the orexigenic genes NPY and AGRP in the arcuate of the late gestation fetal sheep are increased by maternal undernutrition (37), whereas glucose infusion increased fetal POMC but not NPY or AGRP expression (38). Increased expression of mRNA for both the orexigenic and anorexic neuropeptides after TCS infusion suggests that neuronal maturation in the ovine fetal hypothalamus may be altered. Expression of other hypothalamic neurohormones or neuropetides, were also increased by TCSM infusion, including GHRH, GNRH, IGF-2, prolactin-releasing hormone, and somatostatin (SST), suggesting that multiple neuronal populations are affected by TCS exposure. Because expression of genes associated with mitosis, cell cycle, and developmental maturation were up-regulated, this suggests that TCS may stimulate expansion of, and neuropeptide expression in, a variety of hypothalamic neurons important for growth, energy, and food intake postnatally.
Maternal TCS administration increased expression of ghrelin (GHRL) and glucagon-like peptide receptor, which would be expected to have opposing effects on activity of neurons in the orexigenic-anoretic pathways. In postnatal animals, GHRL increases activity of NPY neurons, resulting in increased food intake (reviewed in Ref. 39), whereas glucagon-like peptide-1 inhibits these responses in fasting rats (40). The expression of the neuropeptides is also influenced by leptin (LEP) and insulin. In the postnatal animal, insulin inhibits the expression of NPY/AGRP neurons and increases the expression of the POMC neurons, whereas LEP has the opposite effect (reviewed in Ref. 41). In keeping with the observation that there was not differential expression of POMC and AGRP or NPY in these fetuses, expression of insulin and LEP signaling molecules did not appear to be differentially expressed after either maternal or fetal infusion of TCS; expression of the insulin receptor and insulin substrate 1, and of LEP and its receptor, were not significantly changed, nor were expression of suppressor of cytokine signaling 3, signal transducer and activator of transcription 3, or forkhead box O1. Downstream signaling molecules (40). The postnatal implications of the increases in expression of both sides of this balance, that is, both anorexigenic and orexigenic neuropeptides, and the neuropeptides that stimulate their release in fetal life are not clear. The adjusted array data suggests that the AGRP expression is more markedly increased (8.3-fold) as compared with POMC expression (2-fold), suggesting that the stimulation of orexigenic pathways may be greater. It is unknown, however, to what extent maturation of these pathways may be disrupted by TCS, altering the neonates ability to match energy needs and food intake in postnatal life.
Transcription factor analysis revealed a substantial overlap between groups of up- and down-regulated genes. This result may reflect an indirect action of TCS on the fetal hypothalamus, perhaps with changes in gene expression after TCS-mediated alterations in maternal-fetal metabolism. One insight into the action of TCS might be the involvement of SREBP (which may reflect changes in local sterol biosynthesis), and GABP (which may reflect changes in mitochondrial function) in the down- and up-regulation of genes after TCSF infusion.
A well-known action of TCS postnatally is disruption of the hypothalamus-pituitary-thyroid hormone axis. Exposure to TCS, either during pregnancy or lactation, has been related to maternal and neonatal hypothyroidism (7–9). These studies established the association of the resulting hypothyroxinemia to increased hepatic catabolism. However, a study by Paul et al (8) suggests that other modes of actions could contribute to the observed hypothyroxinemia. TCS inhibits sulfotransferase family 1E member 1 (supra vita) and sulfotransferase family 1B member 1 enzymes that catalyzes the sulfonation of iodothyronines, such as the prohormone T4 and the active hormone T3, for their inactivation and excretion (42). In the present study, we found an important down-regulation of the hypothalamic TRH receptor (TRHR) when TCS was administered to the pregnant ewe (>2-fold, measured by microarray). TRHRs are located in the lateral hypothalamus and in the ventromedial hypothalamus, areas that are important in food intake and energy balance. Central TRH administration is known to decrease food intake, although the site and mechanism of this action is not well characterized (43). TRH neurons are located in the paraventricular nucleus, but they could project to the lateral hypothalamus to inhibit the secretion of the orexigenic peptides such as melanin-concentrating hormone (44). NPY neurons in the arcuate nucleus innervate and inhibit TRH neurons in the paraventricular nucleus (45). NPY also reduces pro-TRH processing (46). Although a direct link has not been established so far, an increase of the orexigenic NPY may down-regulate TRHRs. Accordingly, food restriction in rats reduced hypothalamic mRNA levels of both TRHRα and TRHRβ and TRH but increased the levels of NPY (47). Thus, the strong down-regulation of TRHR in hypothalamus after TCS treatment supports the concept that TCS interrupts fetal energy balance.
Thyroid hormone signaling is essential for brain development, because thyroid hormone acts on regulatory genes that shape the brain during fetal life (48). This could explain inhibition of genes related to CNS development after TCSF infusion (Figure 3J). Although it is interesting that genes containing transcription factor binding site for T3R were found in all 4 experimental groups (Table 2), it is not possible to know whether there is a cause-and-effect relationship between tissue T3 concentrations and transcriptomics response without direct measurements of T3 concentrations.
Conclusion
This study demonstrated that a short-term infusion of TCS, either in the pregnant ewe or in the ovine fetus, induces vigorous changes in the fetal hypothalamic transcriptomics, which are primarily related to pathways important for lipid metabolism, food intake, and energy balance postnatally. Although we are mindful of the fact that these are short-term infusions, the pattern of gene expression suggests to us the possibility that exposure to TCS during fetal life might predispose the newborn lamb to disturbances of the food intake and energy homeostasis, the thyroid hormone system, and the reproductive function, with possible consequences in the pattern of growth.
Acknowledgments
We thank Ms Xiaoying (Lisa) Fang and Ms Kristina Steinfeldt for their expert technical assistance. We also thank the Genomics Division of the University of Florida's Interdisciplinary Center for Biotech Research for the use of the Agilent Bioanalyzer and Agilent scanner.
This work was supported by the National Institutes of Health Grant ES019651 and by an American Recovery and Reinvestment Act of 2009 Supplement to Grant HD057561 (to C.E.W.), and by the National Institutes of Health Grant T32DK076541 (to E.I.C. [trainee] and C.E.W. [PI]). This work was also supported by a grant from the Opportunity Fund of the University of Florida.
Disclosure Summary: The authors have nothing to disclose.
For News & Views see page 2575
- AGRP
- agouti-related protein
- ASIP
- agouti-signaling protein
- CNS
- central nervous system
- ΔCt
- change in threshold cycle
- ControlF
- group in which saline is infused into the fetus
- ControlM
- group in which saline is infused into the mother
- DEG
- differentially expressed gene
- ELK1
- ETS domain containing protein
- ER
- estrogen receptor
- ERR1
- estrogen-related receptor α
- FASN
- fatty acid synthase
- GABP
- GA-binding protein α-chain
- GHRL
- ghrelin
- GR
- glucocorticoid receptor
- GHRH
- growth hormone releasing hormone
- GNRH
- gonadotropin releasing hormone
- GSEA
- gene set enrichment analysis
- LEF1
- lymphoid enhancer binding factor 1
- LEP
- leptin
- NFY
- nuclear transcription factor Y α
- PAX4
- paired box gene 4
- PC
- principal component
- POMC
- proopiomelanocortin
- PPAR
- peroxisome proliferator-activated receptor
- qRT
- quantitative real time
- SP1
- specificity protein 1
- SREBP
- sterol regulatory element-binding protein
- SST
- somatostatin
- TCS
- triclosan
- TCSF
- fetal TCS
- TCSM
- maternal TCS
- T3R
- thyroid hormone receptor
- TRHR
- TRH receptor.
References
- 1. Dann AB, Hontela A. Triclosan: environmental exposure, toxicity and mechanisms of action. J Appl Toxicol. 2011;31:285–311. [DOI] [PubMed] [Google Scholar]
- 2. Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem. 1999;274:11110–11114. [DOI] [PubMed] [Google Scholar]
- 3. McMurry LM, Oethinger M, Levy SB. Triclosan targets lipid synthesis. Nature. 1998;394(6693):531–532. [DOI] [PubMed] [Google Scholar]
- 4. Angelakis E, Armougom F, Carriere F, et al. A metagenomic investigation of the duodenal microbiota reveals links with obesity. PLoS One. 2015;10:e0137784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhargava HN, Leonard PA. Triclosan: applications and safety. Am J Infect Control. 1996;24:209–218. [DOI] [PubMed] [Google Scholar]
- 6. Philippat C, Botton J, Calafat AM, Ye X, Charles MA, Slama R. Prenatal exposure to phenols and growth in boys. Epidemiology. 2014;25:625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paul KB, Hedge JM, Devito MJ, Crofton KM. Developmental triclosan exposure decreases maternal and neonatal thyroxine in rats. Environ Toxicol Chem. 2010;29:2840–2844. [DOI] [PubMed] [Google Scholar]
- 8. Paul KB, Hedge JM, Bansal R, et al. Developmental triclosan exposure decreases maternal, fetal, and early neonatal thyroxine: a dynamic and kinetic evaluation of a putative mode-of-action. Toxicology. 2012;300:31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodríguez PE, Sanchez MS. Maternal exposure to triclosan impairs thyroid homeostasis and female pubertal development in Wistar rat offspring. J Toxicol Environ Health A. 2010;73:1678–1688. [DOI] [PubMed] [Google Scholar]
- 10. Arbuckle TE, Marro L, Davis K, et al. Exposure to free and conjugated forms of bisphenol A and triclosan among pregnant women in the MIREC cohort. Environ Health Perspect. 2015;123:277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Urinary concentrations of triclosan in the U.S. population: 2003–2004. Environ Health Perspect. 2008;116:303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McDonald TJ, Nathanielsz PW. Bilateral destruction of the fetal paraventricular nuclei prolongs gestation in sheep. Am J Obstet Gynecol. 1991;165:764–770. [DOI] [PubMed] [Google Scholar]
- 13. Ramadoss J, Lunde ER, Chen WJ, West JR, Cudd TA. Temporal vulnerability of fetal cerebellar Purkinje cells to chronic binge alcohol exposure: ovine model. Alcohol Clin Exp Res. 2007;31:1738–1745. [DOI] [PubMed] [Google Scholar]
- 14. Rabaglino MB, Richards E, Denslow N, Keller-Wood M, Wood CE. Genomics of estradiol-3-sulfate action in the ovine fetal hypothalamus. Physiol Genomics. 2012;44:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wood CE, Rabaglino MB, Chang EI, Denslow N, Keller-Wood M, Richards E. Genomics of the fetal hypothalamic cellular response to transient hypoxia: endocrine, immune, and metabolic responses. Physiol Genomics. 2013;45:521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winikor J, Schlaerth C, Rabaglino MB, Cousins R, Sutherland M, Wood CE. Complex actions of estradiol-3-sulfate in late gestation fetal brain. Reprod Sci. 2011;18:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ye X, Wong LY, Jia LT, Needham LL, Calafat AM. Stability of the conjugated species of environmental phenols and parabens in human serum. Environ Int. 2009;35:1160–1163. [DOI] [PubMed] [Google Scholar]
- 18. Smyth GK. Limma: linear models for microarray data. In: Gentleman VC, Dudoit S, Irizarry R, Huber W, eds. Bioinformatics and Computational Biology Solutions using R and Bioconductor, R. New York, NY: Springer; 2005:397–420. [Google Scholar]
- 19. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. [DOI] [PubMed] [Google Scholar]
- 21. Venables WN, Ripley BD. Modern Applied Statistics with S. New York, NY: Springer-Verlag; 2002. [Google Scholar]
- 22. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. [DOI] [PubMed] [Google Scholar]
- 24. Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 26. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 27. Liu B, Wang Y, Fillgrove KL, Anderson VE. Triclosan inhibits enoyl-reductase of type I fatty acid synthase in vitro and is cytotoxic to MCF-7 and SKBr-3 breast cancer cells. Cancer Chemother Pharmacol. 2002;49:187–193. [DOI] [PubMed] [Google Scholar]
- 28. Claycombe KJ, Wang Y, Jones BH, et al. Transcriptional regulation of the adipocyte fatty acid synthase gene by agouti: interaction with insulin. Physiol Genomics. 2000;3:157–162. [DOI] [PubMed] [Google Scholar]
- 29. Dinulescu DM, Cone RD. Agouti and agouti-related protein: analogies and contrasts. J Biol Chem. 2000;275:6695–6698. [DOI] [PubMed] [Google Scholar]
- 30. Broberger C, Johansen J, Johansson C, Schalling M, Hökfelt T. The neuropeptide Y/agouti gene-related protein (AGRP) brain circuitry in normal, anorectic, and monosodium glutamate-treated mice. Proc Natl Acad Sci USA. 1998;95:15043–15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hahn TM, Breininger JF, Baskin DG, Schwartz MW. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat Neurosci. 1998;1:271–272. [DOI] [PubMed] [Google Scholar]
- 32. Chai BX, Neubig RR, Millhauser GL, et al. Inverse agonist activity of agouti and agouti-related protein. Peptides. 2003;24:603–609. [DOI] [PubMed] [Google Scholar]
- 33. Morton GJ, Schwartz MW. The NPY/AgRP neuron and energy homeostasis. Int J Obes Relat Metab Disord. 2001;25(suppl 5):S56–S62. [DOI] [PubMed] [Google Scholar]
- 34. Pelling M, Anthwal N, McNay D, et al. Differential requirements for neurogenin 3 in the development of POMC and NPY neurons in the hypothalamus. Dev Biol. 2011;349:406–416. [DOI] [PubMed] [Google Scholar]
- 35. Bouret SG, Draper SJ, Simerly RB. Formation of projection pathways from the arcuate nucleus of the hypothalamus to hypothalamic regions implicated in the neural control of feeding behavior in mice. J Neurosci. 2004;24:2797–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mühlhäusler BS, McMillen IC, Rouzaud G, et al. Appetite regulatory neuropeptides are expressed in the sheep hypothalamus before birth. J Neuroendocrinol. 2004;16:502–507. [DOI] [PubMed] [Google Scholar]
- 37. Adam CL, Williams PA, Milne JS, Aitken RP, Wallace JM. Orexigenic gene expression in late gestation ovine fetal hypothalamus is sensitive to maternal undernutrition and realimentation. J Neuroendocrinol. 2015;27:765–771. [DOI] [PubMed] [Google Scholar]
- 38. Mühlhäusler BS, Adam CL, Marrocco EM, et al. Impact of glucose infusion on the structural and functional characteristics of adipose tissue and on hypothalamic gene expression for appetite regulatory neuropeptides in the sheep fetus during late gestation. J Physiol. 2005;565:185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ferrini F, Salio C, Lossi L, Merighi A. Ghrelin in central neurons. Curr Neuropharmacol. 2009;7:37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seo S, Ju S, Chung H, Lee D, Park S. Acute effects of glucagon-like peptide-1 on hypothalamic neuropeptide and AMP activated kinase expression in fasted rats. Endocr J. 2008;55:867–874. [DOI] [PubMed] [Google Scholar]
- 41. Varela L, Horvath TL. Leptin and insulin pathways in POMC and AgRP neurons that modulate energy balance and glucose homeostasis. EMBO Rep. 2012;13:1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kester MH, van Dijk CH, Tibboel D, et al. Sulfation of thyroid hormone by estrogen sulfotransferase. J Clin Endocrinol Metab. 1999;84:2577–2580. [DOI] [PubMed] [Google Scholar]
- 43. Nillni EA. Regulation of the hypothalamic thyrotropin releasing hormone (TRH) neuron by neuronal and peripheral inputs. Front Neuroendocrinol. 2010;31:134–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwartz MW, Woods SC, Porte D, Jr, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404:661–671. [DOI] [PubMed] [Google Scholar]
- 45. Fekete C, Sarkar S, Rand WM, et al. Neuropeptide Y1 and Y5 receptors mediate the effects of neuropeptide Y on the hypothalamic-pituitary-thyroid axis. Endocrinology. 2002;143:4513–4519. [DOI] [PubMed] [Google Scholar]
- 46. Cyr NE, Toorie AM, Steger JS, et al. Mechanisms by which the orexigen NPY regulates anorexigenic α-MSH and TRH. Am J Physiol Endocrinol Metab. 2013;304:E640–E650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lindblom J, Haitina T, Fredriksson R, Schiöth HB. Differential regulation of nuclear receptors, neuropeptides and peptide hormones in the hypothalamus and pituitary of food restricted rats. Brain Res Mol Brain Res. 2005;133:37–46. [DOI] [PubMed] [Google Scholar]
- 48. Rovet JF. The role of thyroid hormones for brain development and cognitive function. Endocr Dev. 2014;26:26–43. [DOI] [PubMed] [Google Scholar]