Abstract
Immune checkpoint inhibitor treatment represents a promising approach towards treating cancer and has been shown to be effective in a subset of melanoma, non-small cell lung cancer (NSCLC) and kidney cancers. Recent studies have suggested that the number of non-synonymous mutations (NsM) can be used to select melanoma and NSCLC patients most likely to benefit from checkpoint inhibitor treatment. It is hypothesized that a higher burden of NsMs generates novel epitopes and gene products, detected by the immune system as foreign. We conducted an assessment of NsM across 7,757 tumor samples drawn from 26 cancers sequenced in the Cancer Genome Atlas (TCGA) Project to estimate the subset of cancers (both types and fractions thereof) that fit the profile suggested for melanoma and NSCLC. An additional independent set of 613 tumors drawn from 5 cancers were analyzed for replication. An analysis of the Receiver Operator Characteristic (ROC) curves of published data on checkpoint inhibitor response in melanoma and NSCLC data estimates a cutoff of 192 NsM with 74% sensitivity and 59.3% specificity to discriminate potential clinical benefit. Across the 7,757 samples of TCGA, 16.2% displayed an NsM count that exceeded the threshold of 192. It is notable that more than 30% of bladder, colon, gastric, and endometrial cancers have NsM counts above 192, which was also confirmed in melanoma and NSCLC. Our data could inform the prioritization of tumor types (and subtypes) for possible clinical trials to investigate further indications for effective use of immune checkpoint inhibitors, particularly in adult cancers.
Keywords: Cancer, Immunotherapy, checkpoint inhibitor, TCGA, non-synonymous mutations
INTRODUCTION
The concept of mobilizing the immune system for cancer treatment was postulated more than one hundred years ago, primarily based on anecdotal reports of regression of aggressive cancers in response to co-infection (1). Recently, a new generation of immune therapeutics based on immune checkpoint inhibition, including anti-CTLA-4 (2), anti-PD-1 (3) and anti-PD1-L1 (4), has emerged as a promising development in the treatment of select cancers. In phase 3 clinical trials, immune checkpoint inhibitors have been shown to have improved overall survival in melanoma (5–8), non-small cell lung (NSCLC) (9,10) and kidney cancers (11), compared to the standard treatment. Long-term survival of a subset of patients has been reported and ongoing studies are intended to confirm and possibly extend these exciting observations (12,13). The ability to identify the subset of long-term survivors could have an important impact on treatment options, including first-line as well as salvage therapy. Previously a series of studies have examined somatic alterations to identify possible predictive signatures; these have included studies of gene expression signature associated with immune infiltration (14), neoantigen load (15–17), NRAS mutation status (18) and neoantigen-derived tetrapeptide signature (15). Of these, the neoantigen load is most promising, particularly for treatment of melanoma (15,16) and NSCLC (17).
Non-synonymous somatic mutations can generate neoantigens (19–22), which, in turn, can be recognized by the immune system, triggering an anticancer immune response (23). It has been observed that a higher number of non-synonymous mutations correlates with a response to checkpoint inhibitors in melanoma (15,16) and NSCLC (17). Here, we conducted an analysis of the non-synonymous mutation load across the 7,757 tumor samples drawn from 26 distinct cancers in The Cancer Genome Atlas (TCGA) (24), to infer possible cancers that might be prioritized for subsequent study of immune checkpoint inhibitors.
MATERIAL AND METHODS
TCGA mutation data was analyzed using the Broad Institute GDAC Firehose (25) and downloaded from the TCGA Genome Data Analysis Center (November 2, 2015). Broad Institute GDAC Firehose pipeline uses MuTect for mutation calling and MutSig v2.0 (26) for mutation counts and rates across samples. The following 26 tumor subtypes were analyzed: adrenocortical carcinoma, bladder urothelial carcinoma, brain lower grade glioma, breast invasive carcinoma, cholangiocarcinoma, colon adenocarcinoma, glioblastoma multiforme, head and neck squamous cell carcinoma, kidney chromophobe, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, liver hepatocellular carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, ovarian serous cystadenocarcinoma, pancreatic adenocarcinoma, pheochromocytoma and paraganglioma, prostate adenocarcinoma, rectum adenocarcinoma, skin cutaneous melanoma, stomach adenocarcinoma, testicular germ cell tumors, thyroid carcinoma, uterine carcinosarcoma, uterine corpus endometrial carcinoma, and uveal melanoma. Clinical data for these tumors were downloaded from the TCGA data porta (https://tcga-data.nci.nih.gov) (24) at the same time.
For the replication set, we downloaded data from 7 studies (27–33) (117 breast, 78 head and neck, 169 lung adenocarcinoma, 118 melanoma, and 131 prostate cancer patients) from TumorPortal (http://www.tumorportal.org/, March 9, 2016) (34). These data had been processed through the same analysis pipeline used for the TCGA set.
A literature search was performed in the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed, October 20, 2015) using combinations of the search terms: cancer, response, mutation, checkpoint inhibitor, CTLA4 and PD1. Studies investigating immune checkpoint response and with published data on NsM count, information on immune checkpoint inhibitors response and number of non-synonymous mutations were extracted for analysis. We found three eligible studies: two from previous melanoma (15,16) and one NSCLC (17) study.
A generalized additive model (GAM) was used to investigate the relationship between the number of NsM and response to immune checkpoint inhibition for the three previous studies (15–17). GAM tries to fit a generalized linear model (such as the linear regression model, and logistic regression model) with its predictors defined by some smooth functions of independent variables. This was chosen because it is flexible to model the effect of the number of NsM on the response, especially when the effect is a nonlinear function of the number of NsM. In addition, receiver operating characteristic (ROC) curves were generated to determine the optimal trade-off between sensitivity and specificity for our analyses. A proposed threshold was determined to be the point closest to a true positive rate (sensitivity) of 1 and a false positive rate (1- specificity) of 0 on the ROC curve of previous clinical studies (15–17), and thus providing the optimal combination of sensitivity and specificity. This point was calculated as the maximum sum of the sensitivity and 1-specificity along the ROC curve. The best NsM threshold was a cut-off for classifying the samples regarding potential clinical benefit for checkpoint inhibitor response.
To compare the number of non-synonymous mutations for TCGA tumor sets with respect to reported staging (stage I and II versus stage III and IV patients) and smoking status (smoking versus non-smoking patients), we used the Kolmogorov–Smirnov test, as it is a nonparametric test to compare the distributions between samples. In order to address the question if stage and smoking could change the number of patients classified with or without potential clinical benefit for checkpoint inhibitor response, we used the Fisher's exact test. Fisher's exact test was also used for comparing previous NSCLC and melanoma studies of checkpoint inhibition response regarding an estimated ROC analysis threshold; it was also employed for the comparison of the TCGA to replication sets. All statistical analyses were done within the R program (v3.0.1).
RESULTS
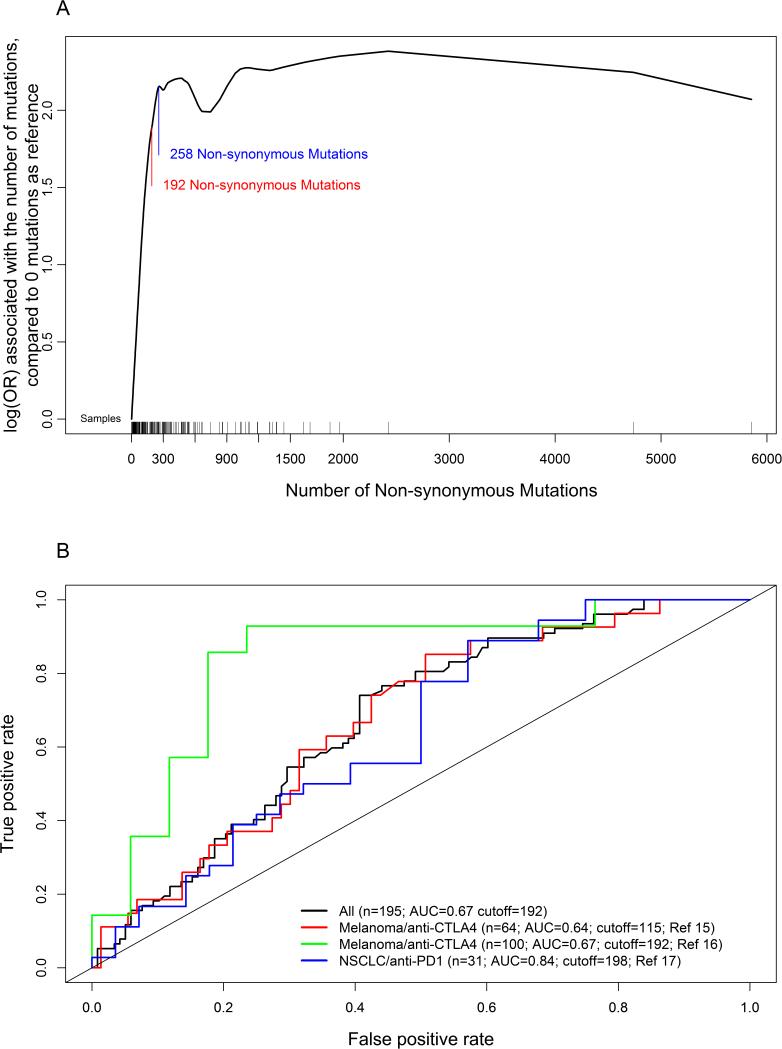
To estimate the fraction of cases in TCGA, by tumor type that could possibly benefit from immune checkpoint inhibition, we analyzed the data with respect to a generalized additive model that relates response for checkpoint inhibitor to the number of NsM on data from 3 previous studies (15–17). Our model suggests that the clinical response increases until 258 NsM at which point there is an observed plateau for the response rate (Figure 1A). The ROC analysis based on these data suggests that 192 NsM is the closer point to the true positive rate (sensitivity) of 1 and false positive rate (1- specificity) of 0, and, therefore, could be a working threshold that optimizes sensitivity (74%) and specificity (59.3%)(Figure 1B). 54.2% (57 out of 105) of the patients from the three studies with ≥192 NsM responded to checkpoint inhibition treatment, while 22.2% with less than 192 NsM (20 out of 90) showed a favorable response (p = 4.8 × 10−6; Supplementary Table 1). Although the data from the NSCLC study indicate a more favorable effect, namely, better area under the curve (AUC) in the ROC analysis (Figure 1B), similar cutoffs were observed for melanoma (cutoff = 192) and NSCLC (cutoff = 198) in tumor specific ROC analyses, suggesting similar parameters influence response to immunotherapy.
Figure 1.
(A) Generalized additive model for the relationship between the number of non-synonymous mutations and response to checkpoint inhibitor using previously published data in melanoma and NSCLC. Y-axis is log(odds ratio) associated with the number of mutations, compared to zero NsM as reference. Blue line: 192 non-synonymous mutations, which is the cutoff based on ROC analysis. Red line: 258 non-synonymous mutations. OR: odds ratio. (B) ROC analysis for the number of non-synonymous mutations using previously published data in melanoma and NSCLC. Black line is the analysis for all combined data; red line is the analysis only for one melanoma study; blue line is the analysis for the other melanoma study; and the green line is the analysis for lung cancer data.
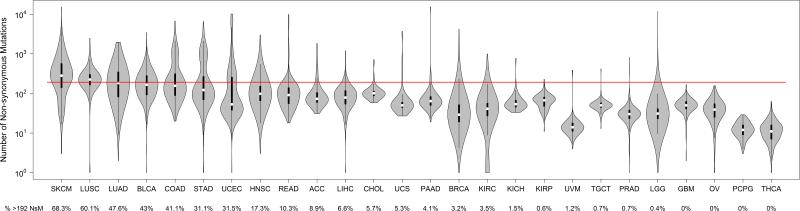
To investigate the landscape of NsM across 26 cancers reported in TCGA, we applied 192 NsM as a threshold for the 7,757 TCGA samples from 26 different tumor types (Figure 2). Across the TCGA set, 16.2% displayed an NsM count that exceeded 192 and thus, could be in a subset more likely to respond to immune checkpoint inhibitors. For specific cancer types, we confirmed that the majority of melanoma (68.3%) and lung tumors (squamous cell = 60.1%; adenocarcinoma = 47.6%) have NsM greater than 192. We also noted that four additional cancers have at least 30% of tumors with an NsM count greater than 192, namely bladder (43%), colon (41.1%), gastric (31.1%) and endometrial (31.5%). We suggest these cancers could be prioritized for immune checkpoint inhibition studies because a larger fraction harbors a sufficiently high burden of NsM. Not surprisingly, these tumors are known to be associated with strong environmental factors, and exhibit higher loads of somatic mutations (35).
Figure 2.
Violin plot for the number of non-synonymous mutations (log10) across 26 tumor types in TCGA data. Red line is the cutoff for 192 non-synonymous mutations. ACC: adrenocortical carcinoma; BLCA: bladder urothelial carcinoma; LLG: brain lower grade glioma; BRCA: breast invasive carcinoma, cholangiocarcinoma; COAD: colon adenocarcinoma; GBM: glioblastoma multiforme; HNSC: head and neck squamous cell carcinoma; KICH: kidney chromophobe; KIRC: kidney renal clear cell carcinoma; KIRP: kidney renal papillary cell carcinoma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; OV: ovarian serous cystadenocarcinoma; PAAD: pancreatic adenocarcinoma; PCPG: pheochromocytoma and paraganglioma; PRAD: prostate adenocarcinoma; READ: rectum adenocarcinoma; SKCM: skin cutaneous melanoma; STAD: stomach adenocarcinoma; TGCT: testicular germ cell tumors; THCA: thyroid carcinoma; UCS: uterine carcinosarcoma; UCEC: uterine corpus endometrial carcinoma; and UVM: uveal melanoma.
We replicated these results using independent non-TCGA data from 7 studies (27–33) processed though the same Broad Firehose pipeline applied to the TCGA data. There were no significant differences observed for melanoma, lung adenocarcinoma, head and neck, breast and prostate cancer between TCGA and the TumorPortal set, which served as an independent replication set, specifically regarding the 192 NsM threshold (Supplementary Figure 1 and Supplementary Table 2).
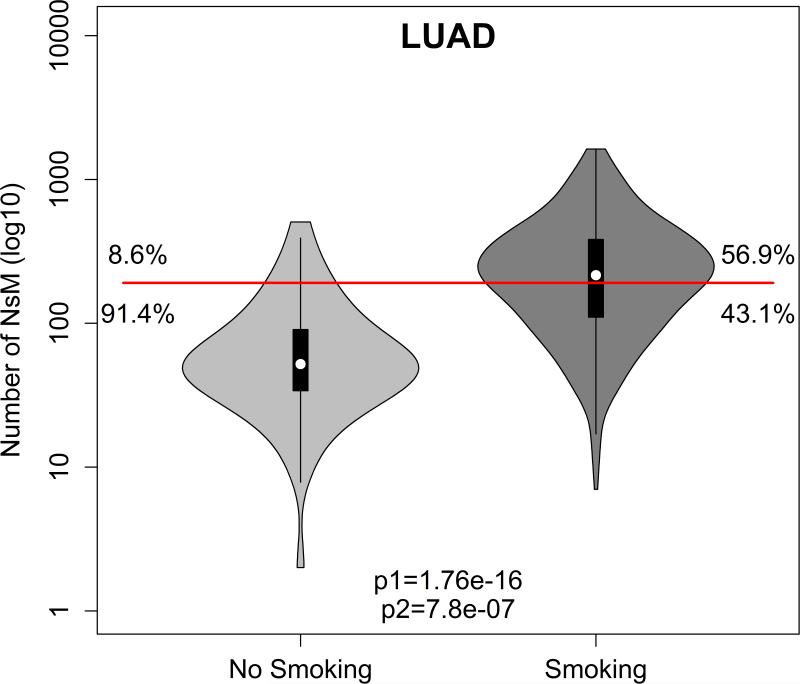
We further evaluated the role of one of the strongest environmental exposures, smoking, in 8 TCGA tumor subtypes in which tobacco use has been implicated as a major risk factor: bladder urothelial carcinoma, head and neck squamous cell carcinoma, kidney chromophobe, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, lung adenocarcinoma, lung squamous cell carcinoma and pancreatic adenocarcinoma. In a preliminary analysis, limited by small numbers for the non-smoking subset, both head and neck (p = 3.5 × 10−4) and lung adenocarcinoma (p = 1.7 × 10−16) smoking patients have more mutations than non-smoking patients (Supplementary Figure 2), but lung adenocarcinoma was the one subtype in which smoking correlated with a higher NsM (p = 7.8 × 10−7; OR = 6.62; Figure 3).
Figure 3.
Violin plot for the number of non-synonymous mutations for smoking (dark gray) and (light gray) non-smoking patients for lung adenocarcinoma (LUAD). p1 is Bonferroni corrected p-value for Kolmogorov-Smirnov test, while p2 is for Bonferroni corrected p-value for Fisher's exact test.
Although clinical trials with checkpoint inhibitors have been conducted in patients with advanced disease, often stage IV, immunotherapy could have an important role in adjuvant therapy. The assessment of NsM count could provide an opportunity to investigate stage-related immunotherapy response rates. First, we compared the number of non-synonymous mutations between stage I and II patients to stage III and IV patients (Supplementary Figure 3). After correction for multiple comparisons, only thyroid cancer stages III and IV have more non-synonymous mutations than stages I and II (p = 4.1 × 10−8), although this difference is inconsequential since thyroid tumors do not show NsM loads near the 192 threshold for immunotherapy response. Second, we evaluated if stage could influence the number of patients above or below our 192 NsM threshold. Again, there was no difference in response rate based on NsM when we compared stage I and II to stage III and IV. It is important, however, to note that even early stage TCGA tumor specimens display a bias toward larger tumor size since the inclusion criteria for TCGA samples is skewed toward larger samples due to material requirements for genomic characterization.
DISCUSSION
Data from previous studies of melanoma and NSCLC response to immune checkpoint inhibitors has suggested a foundation for developing a strategy to consider stratification of cancers based on the NsM load, primarily because of the observed correlation between a higher somatic burden and clinical response (15,16) (17). We applied an empirically derived cutoff of 192 NsM count based on three previous studies to 7,757 TCGA cancer patients across 26 different tumor subtypes, and replicated in 613 cancer patients across 5 tumor subtypes. In addition to melanoma and lung cancer which have data from clinical trials indicating a potential benefit of immune checkpoint inhibitors, our analysis suggests bladder, colon, gastric, and endometrial cancers could be high priorities for future immune checkpoint inhibition studies based on a higher fraction of tumors (30%) with greater than 192 NsM. A subset of cancers with a very low fraction (<5%) of tumors with an NsM above 192 might not be the highest priority for new studies; these include cancers of the thyroid, ovary, prostate, kidney, testicle, breast as well as glioblastomas, pheochromocytomas and paragangliomas. Some of these cancers have displayed few drivers and lower overall burden of mutations (36).
In a comparison of the three published studies, using an ROC assessment for NsM against the checkpoint response rate, we observed similar cutoffs when comparing tumor types, but interestingly, the NSCLC study treated with anti-PD-1 had better AUC, which suggests perhaps better accuracy for NsM to predict immunotherapy response. PD-1 blockade occurs on T cells which can attach to tumor cells, while CTLA-4 blockade occurs on the dendritic cell which presents antigens to the T cell (37). Despite the fact that the NSCLC study ROC analysis used a small sample size, it is plausible that NsM may be a better proxy for events related to blockade closer to the mechanisms of T cell tumor attack.
In our assessment of the TCGA melanoma set, 68.3% of patients could have potential clinical benefit, a rate in agreement with a 1 year overall survival rate from an anti-PD-1 phase III clinical trial (65.5 to 78.9%) in melanoma without BRAF mutation (38). For anti-CTLA-4, plus dacarbazine, the range of 1 year survival is between 41.2 to 53.7% for melanoma patients (8), which is lower than our analysis predicts, however, anti-CTLA-4 appears to be less efficacious than anti-PD-1. Early reports from a phase I clinical trial testing a combination of anti-PD-1 and anti-CTLA-4 has shown an 82% 1 year overall survival (39).
The evaluation of lung cancer in TCGA also revealed that 60.1% of squamous cell patients have an NsM score above the threshold of 192 and could be more likely to respond to treatment with checkpoint inhibitors; interestingly the fraction for adenocarcinoma is 47.6%. In the anti-PD-1 phase III squamous cell lung cancer clinical trial, the 1 year overall survival rate range was 34 to 50% (9); the rate for the non-squamous cell lung cancer phase III was 45 to 56% 1-year overall survival rate (10). Our analysis may be overestimating benefit for squamous cell patients due to the high mutation rate induced by smoking. On the other hand, smoking adenocarcinoma patients show a higher mutation burden, which increases the fraction of cases with an NsM above the 192 threshold. These results correlate with data from lung cancer clinical trials where smoking patients do show significantly higher response rates to checkpoint inhibitors than non-smoking patients (40).
Surprisingly, we did not observe an effect by the stage of the disease; for instance, there was no significant difference in the load of non-synonymous mutations in stage III and IV compared to stage I and II. This result could be due to tumor selection bias because TCGA general guidelines for collecting samples required larger tumors capable of yielding enough genetic material for analysis. Alternatively, anti-CTLA-4 is active for adjuvant therapy in melanoma stage III patients which suggests that activity is not restricted to stage IV patients. Although there is not a high level of evidence for checkpoint inhibitors action on stages I and II, often early stage tumors have not accumulated as many non-synonymous mutations. Further studies are needed to determine if immune checkpoint inhibition therapy is indicated for earlier stage lung cancer and melanoma.
Currently, only data from clinical trials conducted in melanoma and NSCLC are available to model a threshold for stratification of therapy. Our determination of an NsM of 192 is limited by the sample sizes and studies available. More precise estimates should emerge from ongoing studies, which could, in turn, inform our understanding of what may emerge as a more complex stratification model. In our model, clear cell renal cancer could have a lower response rate to immunotherapy based on NsM (3.5% of tumors with more than 192 NsM), but a recent clinical trial showed survival improvement from 19.6 to 25 months for anti-PD-1 treatment on the second or third line of treatment, compared to Everolimus (11). On the other hand, 43% of colon cancer and 10.3% of rectal cancers have more than the 192 NsM. To the best of our knowledge, there are no published phase III clinical trials from colorectal cancer, however, some studies suggest that the subset of colorectal cancers with mismatch repair-deficient would have better immune checkpoint inhibition response (41,42). Furthermore, other biomarkers could emerge that might improve the algorithms for choosing immune checkpoint inhibition, either as a first-line or salvage therapy. In turn, differences in the response rate for immune checkpoint inhibition therapy could lead to cancer-specific thresholds, and perhaps stage variables may also be informative.
In conclusion, we have reported on available somatic tumor data to develop a stratification model in which response rate to immune checkpoint inhibitors correlates with NsM burden. Although further validation is needed, we suggest that information on NsM could be useful in selecting tumor types that are most likely to respond to immune checkpoint inhibitors for future clinical trials. Clearly, more data are needed to move beyond a linear threshold for NsM and establish a model with greater accuracy for developing robust approaches towards precision medicine for select cancers (43).
Supplementary Material
Acknowledgments
Financial support: This study was supported by the Intramural Research Program of the National Institutes of Health.
Footnotes
Conflicts of interest: The authors declare that they have no competing interests.
REFERENCES
- 1.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–9. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–6. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 3.Iwai Y, Terawaki S, Honjo T. PD-1 blockade inhibits hematogenous spread of poorly immunogenic tumor cells by enhanced recruitment of effector T cells. Int Immunol. 2005;17:133–44. doi: 10.1093/intimm/dxh194. [DOI] [PubMed] [Google Scholar]
- 4.Dong H, Chen L. B7-H1 pathway and its role in the evasion of tumor immunity. J Mol Med (Berl) 2003;81:281–7. doi: 10.1007/s00109-003-0430-2. [DOI] [PubMed] [Google Scholar]
- 5.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber JS, D'Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16:375–84. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 8.Robert C, Thomas L, Bondarenko I, O'Day S, Weber J, Garbe C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–26. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 9.Brahmer J, Reckamp KL, Baas P, Crinó L, Eberhardt WEE, Poddubskaya E, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. N Engl J Med. 2015;373:123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non–Small-Cell Lung Cancer. N Engl J Med. 2015;373:1627–39. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med. 2015;373:1803–13. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, et al. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J Clin Oncol. 2015;33:1889–94. doi: 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maio M, Grob J-J, Aamdal S, Bondarenko I, Robert C, Thomas L, et al. Five-Year Survival Rates for Treatment-Naive Patients With Advanced Melanoma Who Received Ipilimumab Plus Dacarbazine in a Phase III Trial. J Clin Oncol. 2015;33:1191–6. doi: 10.1200/JCO.2014.56.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and Genetic Properties of Tumors Associated with Local Immune Cytolytic Activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson DB, Lovly CM, Flavin M, Panageas KS, Ayers GD, Zhao Z, et al. Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Cancer Immunol Res. 2015;3:288–95. doi: 10.1158/2326-6066.CIR-14-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–81. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–5. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castle JC, Kreiter S, Diekmann J, Löwer M, van de Roemer N, de Graaf J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–91. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 23.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Center BITGDA Analysis-ready standardized TCGA data from Broad GDAC Firehose stddata__2015_06_01 run. Broad Inst MIT Harvard. 2015 [Google Scholar]
- 26.Lawrence MS, Stojanov P, Polak P, Kryukov G V, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancerassociated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat J-P, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodis E, Watson IR, Kryukov G V, Arold ST, Imielinski M, Theurillat J-P, et al. A Landscape of Driver Mutations in Melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012 doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the Hallmarks of Lung Adenocarcinoma with Massively Parallel Sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–9. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postow MA, Callahan MK, Wolchok JD. Immune Checkpoint Blockade in Cancer Therapy. J Clin Oncol. 2015;33:1974–82. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, et al. Nivolumab in Previously Untreated Melanoma without BRAF Mutation. N Engl J Med. 2015;372:320–30. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 39.Sznol M, Kluger HM, Callahan MK, Postow MA, Gordon RA, Segal NH, et al. Survival, response duration, and activity by BRAF mutation (MT) status of nivolumab (NIVO, anti-PD-1, BMS-936558, ONO-4538) and ipilimumab (IPI) concurrent therapy in advanced melanoma (MEL). J Clin Oncol. 2014;32(suppl):5s. abstr LBA9003^ 2014. [Google Scholar]
- 40.Gettinger SN, Horn L, Gandhi L, Spigel DR, Antonia SJ, Rizvi NA, et al. Overall Survival and Long-Term Safety of Nivolumab (Anti-Programmed Death 1 Antibody, BMS-936558, ONO-4538) in Patients With Previously Treated Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2015;33:2004–12. doi: 10.1200/JCO.2014.58.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipson EJ, Sharfman WH, Drake CG, Wollner I, Taube JM, Anders RA, et al. Durable Cancer Regression Off-Treatment and Effective Reinduction Therapy with an Anti-PD-1 Antibody. Clin Cancer Res. 2013;19:462–8. doi: 10.1158/1078-0432.CCR-12-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins FS, Varmus H. A New Initiative on Precision Medicine. N Engl J Med. 2015;372:793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.