Abstract
Genomic alignment of small RNA (smRNA) sequences such as microRNAs poses considerable challenges due to their short length (∼21 nucleotides [nt]) as well as the large size and complexity of plant and animal genomes. While several tools have been developed for high-throughput mapping of longer mRNA-seq reads (>30 nt), there are few that are specifically designed for mapping of smRNA reads including microRNAs. The accuracy of these mappers has not been systematically determined in the case of smRNA-seq. In addition, it is unknown whether these aligners accurately map smRNA reads containing sequence errors and polymorphisms. By using simulated read sets, we determine the alignment sensitivity and accuracy of 16 short-read mappers and quantify their robustness to mismatches, indels, and nontemplated nucleotide additions. These were explored in the context of a plant genome (Oryza sativa, ∼500 Mbp) and a mammalian genome (Homo sapiens, ∼3.1 Gbp). Analysis of simulated and real smRNA-seq data demonstrates that mapper selection impacts differential expression results and interpretation. These results will inform on best practice for smRNA mapping and enable more accurate smRNA detection and quantification of expression and RNA editing.
Keywords: small RNA sequencing, microRNA, next-generation sequencing, short-read aligners, gene expression
INTRODUCTION
Small noncoding RNAs are functional mature transcripts with a length ≤300 nt that do not encode proteins (Mattick and Makunin 2006). This abundant superfamily exists in all domains of life and includes several subclasses that are distinguished by their size, homology, and mechanisms of biogenesis. These include microRNA, siRNA, piRNA, snoRNA, eRNA, and the number of subclasses continues to expand (Morris and Mattick 2014). In plants and animals, microRNAs form complexes with Argonaute family proteins to guide silencing of genes (Farazi et al. 2008). In animals, this occurs by directing post-transcriptional RNA cleavage and translational inhibition (Bartel 2004). In plants and fungi there are descriptions of small RNA-directed silencing via chromatin modification (Bernstein and Allis 2005). MicroRNAs (miRs) are the prototypical small RNA class due to their relatively large gene number in plant and animal genomes, relatively high expression and interspecies sequence conservation (Axtell et al. 2011). The first described roles for microRNAs were in development in Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana (Carrington and Ambros 2003), however, more recent studies demonstrate the importance of microRNAs in signaling, stress response, and disease (Zhang et al. 2006; Mendell and Olson 2012).
Expression analysis is crucial for the understanding of small RNA regulation and is a starting point for initiating reverse genetic functional studies. Quantitative PCR, microarray hybridization, and high-throughput sequencing are commonly used methods for profiling small RNAs (Pritchard et al. 2012). High-throughput small RNA sequencing (smRNA-seq) offers advantages compared to the other methods, specifically by distinguishing very similar smRNA sequences, its unbiased nature allows detection of novel smRNAs and sequence read count measures provide highly accurate quantification of gene expression at high sequencing coverages.
Despite these advantages, there are many bioinformatic challenges in the processing of high-throughput small RNA sequence data. The short sequence length makes these smRNAs difficult to map in large, complex, and repetitive reference genomes. Many small RNAs including biologically important microRNAs are composed of near-identical family members. MicroRNA biogenesis and maturation mechanisms generate fragment lengths of 18–25 nt (Ameres and Zamore 2013) and the current version of miRbase (v21) contains 68 mature human miRs shorter than 18 nt (Kozomara and Griffiths-Jones 2014). Extensive RNA editing introduces sequence differences that make mapping more difficult. Nontemplated extension (NTE) of microRNAs by addition of adenosine and uracil bases to 3′ ends is widespread in animals and plants and may play a role in controlling miR turnover (Ameres and Zamore 2013). Deep sequencing of mouse reveals 3′ NTE to affect ∼20% of miRNA molecules in the hippocampus and ∼15% in reprogrammed stem cells (Zhou et al. 2012). Deamination of adenosine to inosine is common for some microRNAs (termed A-to-I editing) (Kawahara et al. 2007; Ameres and Zamore 2013), which leads to the appearance of single nucleotide mismatches (SNMs) in sequence data. In human, it is estimated that 10%–15% of miRNAs undergo A-to-I RNA editing (Blow et al. 2006). Given these challenges in smRNA-seq read mapping, understanding the limitations of the most commonly used short read mapping software is important to facilitate accurate smRNA-seq bioinformatics analysis.
Previous evaluations have quantified the accuracy and computation speed of aligners for short read genomic DNA and mRNA sequence data (Lindner and Friedel 2012; Shang et al. 2014). Several smRNA-seq pipelines have been evaluated for their sensitivity and accuracy in detecting and quantifying microRNA expression, however many of these pipelines are not open-source, cannot be run on a local computer, can be run only for small subset of species or depend on preexisting alignment software for read mapping (Li et al. 2012; Williamson et al. 2013; Tam et al. 2015). Key information on several of these popular miRNA analysis pipelines is summarized (Table 1).
TABLE 1.
SmRNA/microRNA-seq analysis pipelines in common use

While mapping to known microRNA precursor databases can be a faster and more direct method to quantify microRNA expression, mapping smRNA reads to the entire genome is emerging as a consensus standard procedure (Motameny et al. 2010, Farazi et al. 2012, Stokowy et al. 2014). Genome mapping allows identification of novel smRNAs, and can effectively identify transcripts of RNA classes such as rRNA, tRNA, and mRNA. Differences in smRNA-seq read mapping sensitivity have been observed for different aligners, suggesting that mapper accuracy may be affecting downstream data analysis (Farazi et al. 2012; Williamson et al. 2013). Using spike-ins of several Arabidopsis thaliana microRNA sequences that do not occur in the human genome it was shown that BWA gave truer quantifications of original concentrations than Bowtie, Bowtie2, or Novoalign (Tam et al. 2015).
Despite the importance of accurate smRNA-seq mapping in the confident identification of novel transcripts and expression quantification, rigorous evaluations of short-read mapping software using simulated smRNA-seq data are currently lacking in the scientific literature. In this evaluation, we test the ability of a panel of aligners to accurately map simulated smRNA-seq data sets for human and rice. In addition to studying mapping accuracy of uniquely placed reads, we also investigate the accuracy of multimapping reads which are often ignored in many smRNA-seq studies. These results should provide a framework for best practise in smRNA-seq analysis, enhance detection of smRNA editing and accuracy of differential expression studies.
RESULTS
Assessment of microRNA length distribution and sequence modifications
Given the multitude of short read mappers available, we wanted to identify those most suited to small RNA analysis. One of the biggest challenges to microRNA mapping is the sequence length distribution. To explore this, the distribution of read lengths of quality trimmed and adapter clipped smRNA-seq data from previous studies in rice (Oryza sativa) (Barrera-Figueroa et al. 2012; Xu et al. 2014) and human (Homo sapiens) (Mestdagh et al. 2014; Guo et al. 2015) were determined (Table 2). In the data sets analyzed, the proportion of reads shorter than 21 nt is 10%–29%, reads 21–24 nt comprise 19%–60%, and reads >24 nt comprise 11%–71%. Of the reads that mapped to microRNA hairpin containing loci (BWA or Bowtie2 with the default parameters), the proportion of reads shorter than 21 nt is 2%–22%, reads 21–24 nt comprise 43%–76% and reads >24 nt comprise 7%–50%. These results demonstrate that smRNA-seq data contain a considerable number of reads shorter than 21 nt that pose a challenge for accurate genome mapping.
TABLE 2.
Distribution of read lengths and estimates of nontemplated extension (NTE), mismatch (SNM), and indel events in real small Illumina smRNA-seq data

In addition to varying lengths, smRNA-seq possess sequence modifications such as nontemplated extension (NTE) and single nucleotide mismatches (SNMs). The prevalence of these modifications was quantified in reads mapped to hairpin loci (Table 2). NTE modification to 3′ termini accounted for up to 0.33% of mapped smRNA bases in human and up to 0.1% in rice. NTE to 5′ ends accounted for 0.16% bases in human and 0.1% bases in rice. Whether originating from genomic polymorphisms or sequencing errors, SNMs accounted for up to 0.1% of aligned bases in human and up to 0.4% in rice smRNA-seq data, respectively. Indels were less common in the Illumina sequence data, at a rate of 2.3 indels per Mbp in human and 60–140 indels per Mbp in rice. This result indicates that NTE and SNM are prevalent in human and rice smRNA-seq data.
Mapper accuracy with simulated 21-nt hairpin-derived Illumina reads
In order to assess the accuracy of mapping software with microRNA sequence data, we simulated rice and human read sets with Illumina Genome Analyzer IIx error profiles using ART (Huang et al. 2012) using miRbase hairpins as templates and mapped these to the rice and human genome with 16 aligners. Software versions and exact parameters used for these tests are shown in Table 3. A modified F-measure (β = 0.25) that emphasizes precision over recall was used to measure accuracy (Materials and Methods). Hash based aligners underwent prior optimization (using rice 21-nt hairpin-derived reads) as these mappers rely critically on k-mer parameters (Supplemental Table S1). For each mapper that estimates mapping quality (mapQ), the optimum mapQ (based on maximum F-measure) was determined. As expected, as mapQ threshold increased, so did the precision, albeit at the expense of sensitivity (Supplemental Table S2).
TABLE 3.
Alignment software evaluated in the present study

The results testing 16 aligners with a variety of parameter settings are shown in Figure 1. In both rice and human tests, the number of correctly mapped reads identified varied widely from none to 78% (Fig. 1A,C). Bowtie2, BWA, OLego, and Bowtie1 (best strata setting) mapping achieved F0.25 scores greater than 0.95 for rice and human tests (Fig. 1B,D). Overall, the rankings of mappers based on F0.25 scores for rice and human were similar (Spearman ρ = 0.97, P = 0). Subread with default settings did not map 21-nt reads, but the microRNA-specific options suggested by the developers yielded an F0.25 score of 0.92 in rice and human. High-speed aligners STAR and HISAT showed moderate accuracy with F0.25 scores of 0.90 and 0.92, respectively. Stampy with default settings showed poor recall (<15%) but with the “sensitive” parameter, this was improved to ∼60%. Default MicroRazerS, Bowtie1, and Segemehl showed a high recall (>70%) but poor precision (<85%). These results demonstrate a wide range of precision and recall values for the panel of aligners and parameters tested with Illumina-like 21-nt tags.
FIGURE 1.
Alignment of simulated Illumina-like hairpin-derived 21-nt sequences to the genome. For panels A and C, green bars denote correctly mapped reads, yellow bars denote incorrect mapping to non-hairpin genomic locations, red bars denote incorrect mapping to other hairpin loci, and gray bars denote reads unmapped or below map quality threshold. (A,B) Oryza sativa test. (C,D) Homo sapiens test. (B,D) Precision, recall, and F0.25 statistic for each test assessment. Values in parentheses represent optimized mapQ values for filtering uniquely mapped reads.
We investigated the large fraction of reads lacking unambiguous mappings in default BWA and Bowtie2 alignments (Fig. 1A,C). Both aligners mapped >96% of reads, but a large fraction of these reads had a mapQ below the optimized threshold (27%–29% of the whole-read set). The overlap of low mapQ aligned reads between BWA and Bowtie2 in the rice test was 20,876 tags or 26% of the entire rice read set, while this figure was 20% for the human read set. This result shows that up to a quarter of possible hairpin-derived 21-nt reads have ambiguous mapping using these two widely used aligners, suggesting they may also be difficult to map accurately with other aligners. Indeed aligners with substantially higher recall than BWA and Bowtie2 showed the poorest precision.
Mapper accuracy with realistic simulated hairpin-derived reads
Next, we investigated the performance of mappers with realistic read length distribution and sequence variation profiles. We used results from Table 2 to generate synthesized read sets from hairpin loci that have length distribution and variant composition that is within the empirical range. Sequence characteristics used are shown in Figure 2A. These read sets were then aligned to the reference genome using the panel of 16 aligners with mapQ filtering (Supplemental Table S2). After mapping and mapQ filtering, the accuracy results were similar to those for 21-nt reads (Fig. 2B,D). The ranking of mapping procedures based on F0.25 scores is shown in Figure 2C,E, was consistent with 21-nt read analysis shown in Figure 1B,D (Spearman ρ = 0.87, P = 0). Exceptions included STAR aligner that performed better in the realistic read set as compared to the 21-nt read set while default Bowtie2 scored poorer in realistic read set. These results identify mappers that accurately align realistic microRNA sequence reads to the genome.
FIGURE 2.
Alignment of reads with realistic length distribution and error profile. (A) Characteristic length distribution and error profile of sequence sets used. For panels B and D, the color scheme is identical to Figure 1. (B,C) Oryza sativa test. (D,E) Homo sapiens test. (C,E) Precision, recall, and F0.25 statistic for each test assessment.
Aligner accuracy subject to varying read length
Read lengths vary considerably in smRNA-seq data but it is unknown which aligners are most accurate over a broad range of read lengths. In order to assess this, we generated synthetic sequence reads (16–25 nt) derived from known hairpin loci from human and rice that perfectly match the reference genome. In order to focus on unique alignments, our read set excluded sequences that appeared more than once. Alignment rates were lower for shorter 18-nt reads as compared to 21- and 24-nt reads (Fig. 3A,E). Results from other read lengths are presented in Supplemental Figure S1. At 21-nt length, default Bowtie1 and Segemehl produced high false mapping rates of ∼15% in rice and ∼8% in human; these reads were assigned to incorrect hairpin loci as well as non-hairpin regions (Fig. 3B,F). BBMap, Mosaik, and Stampy mapped a relatively small proportion of 21-nt reads with these settings (<85%) but improved with 24-nt reads (Fig. 3C,G). Default Subread did not map 18-nt reads with the parameters tested. Interestingly, at 18-nt HISAT, Segemehl and Stampy (sens) correctly mapped in excess of 70% of reads in rice but in human, none were mapped. This is likely due to internal parameters of these aligners that are dependent on genome size for judging the accuracy of each read mapping position. This trend for better recall in rice mapping as compared to human is also apparent at read lengths of 16 and 17 nt (Supplemental Fig. S1). Bowtie2, SOAP2, GEM, STAR, and Bowtie1 (best strata setting) scored high F-measures (>0.95) in both human and rice tests (Fig. 3D,H). These findings demonstrate differences in mapper accuracy using perfectly matched reads. These differences are more pronounced at shorter read lengths and in larger reference genomes.
FIGURE 3.
Alignment of perfectly matching hairpin-derived sequence reads of varying length. For panels A–C, E–F, the color scheme is identical to Figure 1. (A–D) Oryza sativa reads. (E–H) Homo sapiens reads. (A,E) 18-nt reads. (B,F) 21-nt reads. (C,G) 24-nt reads. (D,H) Average precision, recall, and F0.25 measure for read sets 16–25 nt.
Aligner accuracy with nontemplated terminal extension
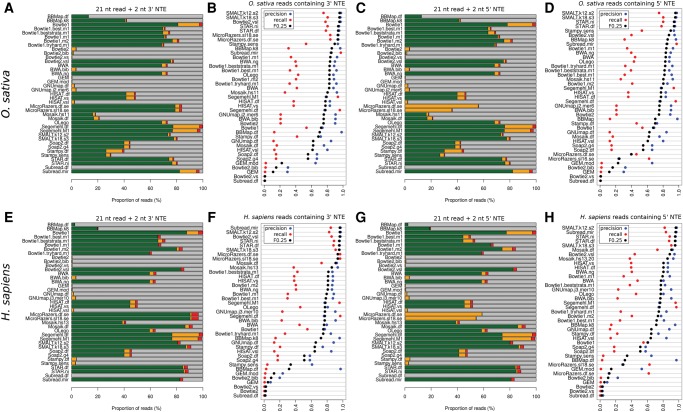
We sought to identify alignment software that could correctly map reads with nontemplated extensions (NTE). We added up to 4 nt to the 3′ or 5′ end of the 21-nt simulated read set, mapped them to the genome and analyzed the mapping positions for all aligners to generate F0.25 scores (Fig. 4). Remaining data used to generate F0.25 scores are shown in Supplemental Figure S2. The addition of 1 nt to the 3′ end reduced the sensitivity of GEM, Bowtie2 (vs, bib, and df parameters), SOAP2, and HISAT aligners by at least 40%. Bowtie2 (vsl), Subread (mir), SMALT, STAR, and MicroRazerS recorded the highest F0.25 accuracy scores for 3′ NTE reads (Fig. 4A,B,E,F). NTE to the 5′ end yielded results that were largely consistent with 3′ NTE, except for MicroRazerS which was able to correctly map most 3′ NTE reads but not with 5′ NTE (Fig. 4C,D,G,H). These results demonstrate that commonly used mappers show differing ability to correctly align reads containing NTEs.
FIGURE 4.
Alignment of 21-nt hairpin-derived reads with nontemplated extension. For panels A,C,E,G the color scheme is identical to Figure 1. (A–D) Oryza sativa reads. (E–H) Homo sapiens reads. (A,E) 21-nt reads with 2nt NTE to the 3′ end. (B,F) Precision, recall, and F0.25 accuracy metric for 21-nt reads with 1–4 nt added to the 3′ end. (C,G) 21-nt reads with 2-nt NTE to the 5′ end. (D,H) Precision, recall, and F0.25 metric for 21-nt reads with 1–4 nt added to the 5′ end.
Aligner accuracy with reads containing mismatches
MicroRNAs are subject to A-to-I editing, genomic variation, and sequencing errors that manifest as single nucleotide mismatches (SNMs). We investigated the ability of aligners to correctly map reads containing SNMs. In order to do this, we added up to two SNMs to the 21-nt reads then analyzed mapping positions (Fig. 5). GEM, HISAT, Bowtie2, SOAP, and Subread mapping rates were reduced by over 50% with the incorporation of one SNM (Fig. 5A,D). BWA, Bowtie2, OLego, and Bowtie1 (best strata) were the most accurate mappers with reads containing two SNMs (Fig. 5B,E) and gave high F0.25 scores in human and rice (Fig. 5C,F). This analysis demonstrates that commonly used mappers have differing ability to map SNM containing reads, and that mapping of these reads is less accurate in larger genomes.
FIGURE 5.
Alignment of 21-nt hairpin-derived reads containing single nucleotide mismatches (SNM). For panels A,B,D,E the color scheme is identical to Figure 1. (A–C) Oryza sativa reads. (D–F) Homo sapiens reads. (A,D) 21-nt reads containing one SNM. (B,E) 21-nt reads containing two SNMs. (C,F) Precision, recall, and F0.25 scores for 21-nt reads containing one to two SNMs.
Aligner accuracy with reads containing indels
To determine aligner robustness to indels, we introduced up to two single nucleotide insertions or deletions to the 21-nt read set, as well as single indels up to 2 nt in length, followed by mapping position analysis (Fig. 6). Single nucleotide insertions and deletions had a drastic effect on alignment rates and accuracy with recall reduced by >50% (Fig. 6A,C,E,G). In rice, BWA and OLego were the most accurate in mapping insertion-containing reads (Fig. 6A,B), while Mosaik (hs11) and SMALT (k12,s2) were most accurate in mapping deletion-containing reads (Fig. 6C,D). Erroneous mapping of indel containing reads was more prevalent in the larger human genome (Fig. 6E,G) with most mappers yielding poor F0.25 scores (<0.5) for insertions and deletion tests (Fig. 6F,H). Remaining data used to generate F0.25 scores are shown in Supplemental Figure S3. These data indicate microRNA sized reads containing indels are subject to of spurious alignment.
FIGURE 6.
Alignment of 21-nt hairpin-derived reads with indels. For panels A,C,E,G the color scheme is identical to Figure 1. (A–D) Oryza sativa reads. (E–H) Homo sapiens reads. (A,E) 21-nt reads with 1-nt insertion. (B,F) Precision, recall, and F0.25 scores for 21-nt reads with insertions. (C,G) 21-nt reads with 1-nt deletion. (D,H) Precision, recall, and F0.25 scores for 21-nt reads with deletions.
Non-hairpin genes are not a major source of error for microRNA quantification
False mapping of “uniquely” mapped reads to the genome remains a problem for short read sequencing (Menzel et al. 2013). To evaluate whether this is likely to impact smRNA-seq studies, we quantified the proportion of short reads from protein-coding genes that were erroneously mapped to hairpin regions using a range of different read lengths. Protein-coding cDNA sequences that overlap hairpin regions were excluded. After mapping, we determined that at a read length of 21 nt, fewer than 0.1% of mRNA derived tags mapped to hairpin regions (Fig. 7A,C). We noted that STAR with default settings showed up to 0.16% of rice 24-nt reads were falsely mapped to hairpin loci (Supplemental Fig. S4). We determined the F0.25 statistic for these mRNA derived tags 18–25 nt (Fig. 7B,D) and found mapper rankings correlated significantly (Spearman ρ = 0.84, P = 0) with tests using hairpin-derived tags of the same length (Fig. 3D,H). This result suggests that many of the aligners evaluated show falsely mapped mRNA tags do not represent a major source of error with respect to microRNA quantification.
FIGURE 7.
Alignment of short simulated Illumina-like mRNA-derived sequences to the genome. For panels A and C, 21-nt read results are shown. Green bars denote correctly mapped reads, yellow bars denote incorrect mapping to protein-coding locations, purple bars represent incorrect mapping to non-mRNA and non-hairpin loci, and gray bars denote reads unmapped or below map quality threshold. Red bars denote incorrect mapping to hairpin loci; these are plotted on enlarged axes for visibility. Precision, recall, and F0.25 statistic are summarized for read lengths 16–25 nt for O. sativa (B) and H. sapiens (D).
Summary of simulated miRNA read-mapping results
The F0.25 scores determined above were used to rank aligners from 1 (most accurate) to 39 (least accurate) for each relevant test (Figs. 1–5). An overall score was determined by calculating the sum of ranks for all tests excluding indel tests (Fig. 8). Bowtie2 (vsl), Bowtie1 (best strata), and BWA (ng) scored favorably, while default Subread, Stampy, BBMap, Bowtie1, and Segemehl scored poorly.
FIGURE 8.
Summary of unique mapping accuracy. Each aligner is assigned a rank based upon its F0.25 score for each test (1; most accurate, 35; least accurate). These include ART-simulated 21-nt reads (ART), simulated realistic reads (SIM), perfectly matching reads (PERF). Read sets containing 3′ NTE, 5′ NTE, single-nucleotide mismatches (SNM), insertions (INS), and deletions (DEL). Overall rank is based on the sum of ranks for each test excluding insertions and deletions. For GNUmap and Mosaik, the optimized hash sizes for rice and human alignments are summarized on one line.
Mapper selection impacts differentially expressed miRNA detection
In order to determine whether mapper selection is likely to influence downstream results in smRNA profiling experiments, we simulated a microRNA-seq profiling experiment (results include fold changes and sequence files) using Polyester (Frazee et al. 2015). Hairpin-derived reads were mapped with highly ranked Bowtie1 (best strata), Bowtie2 (vsl), BWA, middle-ranked MicroRazerS (sl18.se.pa), and lowly ranked default Bowtie1.
Multidimensional scaling analysis showed Polyester simulated profiles were distinct to any aligner-processed data. Bowtie1 generated profiles were distinct as compared to other aligners such as BWA, Bowtie1 (best strata), Bowtie2 (vsl), and MicroRazerS that were clustered (Fig. 9A,C). This is shown quantitatively using Pearson correlation analysis (Fig 9B,E). Interestingly, while default Bowtie1 yielded the highest Pearson correlation with the ground truth profile, this mapper also gave the highest number of false positive and false negative differentially expressed genes (DEGs, FDR≤0.05) as determined with edgeR (Fig. 9C,F). The resulting F1 and F0.25 measures were lower for default Bowtie1 as compared to Bowtie1 (best strata), Bowtie2 (vsl), BWA, and MicroRazerS. These results demonstrate that mapper selection impacts miRNA expression profiles.
FIGURE 9.
Assessment of aligner accuracy using simulated small RNA-seq experiments. MicroRNA expression fold changes and Illumina-like read sets were generated by Polyester (three control and three treatment samples) and were aligned to the reference genome, followed by read counting and differential analysis. (A–C) Oryza sativa test. (D–F) Homo sapiens test. (A,D) MDS plots for simulated small RNA-seq data sets, only control data sets are shown. (B,E) Pairwise correlation scatterplots for one sample (c1). (C,F) Occurrence of true positives (TP), false positives (FP), false negatives (FN), alongside F1 and F0.25 statistics for lists of differentially expressed genes after differential analysis with edgeR (FDR ≤ 0.05).
Mapper selection impacts gene detection in real smRNA-seq data
Next, we sought to determine the effect of mapper selection by analyzing a publicly available smRNA-seq data set (Guo et al. 2015) with the same panel of five aligners used in Figure 9. Default Bowtie1 yielded the highest proportion of mapped reads and the largest proportion mapped to non-genic regions (Table 4), with MicroRazerS showing the next highest nongenic mapping rate. Default Bowtie1 gave the highest number of genes detected above the threshold (1242 genes above 10 tags per sample on average) as compared to Bowtie2 (vsl) that detected 426 genes. The higher number of detected genes by default Bowtie1 was not contributed by higher detection of microRNA genes, rather protein-coding genes and other non-microRNA classes were over-represented (Fig. 10A). Venn diagram showed that these protein-coding genes were largely unique to default Bowtie1, whereas most microRNA genes were common to all mappers (Fig. 10B). Multidimensional scaling analysis showed signatures in default Bowtie1 and MicroRazerS mapped data were distinct from BWA, Bowtie2 (vsl), and Bowtie1 (best strata) (Fig. 10C). Pearson correlation analysis quantified that MicroRazerS and default Bowtie1 results were dissimilar to Bowtie1 (best strata) and Bowtie2 (vsl) (Fig. 10D). Mismatches in mapQ20 alignments were highest for MicroRazerS and lowest for Bowtie2 (vsl). Mismatch rate was lowest for Bowtie2 (vsl) over the entire read length (Fig. 10E) and for all mismatch types (Fig. 10F). These results are consistent with simulated data and show that mapper selection impacts the number and type of transcripts detected in smRNA-seq data. Moreover accurate mappers yield highly correlated results in contrast to poorly ranked mappers that show a lesser degree of correlation.
TABLE 4.
Alignment metrics for a real human smRNA-seq data set

FIGURE 10.
Effect of different mapping algorithms on detection of microRNA genes and genes of other classes. Human smRNA data (from GEO accession GSE60036; Guo et al. 2015) were mapped with Default Bowtie1 (bt1), Bowtie1 best strata (bt1bs), Bowtie2 very sensitive local (bt2vsl), MicroRazerS (mr), and BWA. (A) Number of genes of varying Ensembl biotypes detected above the expression threshold (10 reads per sample on average). (B) Overlap between lists of detected genes based on different alignment algorithms. Only miRNA and protein-coding genes are shown. (C) Multidimensional scaling plot for three selected data sets: SRR1535280, SRR1535283, and SRR1535293. (D) One data set (SRR1535280) was selected to show the Pearson correlation in tag counts for different aligners. Pearson correlation coefficient shown in upper diagonal and scatterplot of tag counts in lower diagonal. (E) Profile of sequence mismatches across read length for all data sets. (F) Base mismatch profile for all data sets.
Mapper selection impacts differential gene expression results of smRNA-seq data
Given that gene detection was significantly altered by mapper selection using real data, we investigated whether this impacts differential expression results. The gene count matrices generated by the five mappers underwent edgeR differential analysis to compare Actinomycin D (4 h) and untreated samples in biological triplicate. Default Bowtie1 identified the largest number of significant DEGs, followed by MicroRazerS (Fig. 11A). Venn diagram showed that most protein-coding DEGs identified by default Bowtie1 and MicroRazerS were unique to each aligner, while only four microRNA genes were common to all mappers (Fig. 11B).
FIGURE 11.
Effect of mapping algorithms on differential expression of microRNA genes and genes of other classes. Comparison of the results of differential smRNA analysis with different aligners. Human smRNA data (from GEO accession GSE60036; Guo et al. 2015) were mapped with Default Bowtie1 (bt1), Bowtie1 best strata (bt1bs), Bowtie2 very sensitive local (bt2vsl), MicroRazerS (mr), and BWA. (A) Number of significant DEGs (FDR < 0.05) from each Ensembl gene biotype class determined by edgeR with data produced by these four mapping procedures. (B) Overlap between lists of differentially expressed genes based on different alignment algorithms. Only miRNA and protein-coding genes are shown. The five mappers were used to determine expression of miR-125b-1 (C) and miR-33b (D) before and after Actinomycin D treatment. Error bars denote standard deviation. (E) P-value curve of edgeR results for each of the five mappers evaluated. (F) Tagwise dispersion distribution determined by edgeR analysis for each of the five mappers evaluated. The common dispersion values are shown (inset).
Given the overlap between DEG lists was below expectations, we investigated whether microRNA genes validated by qRT-PCR were found to be statistically significant in sequence data aligned with different mappers. The differential expression trend of miR-125b-1 and miR-33b identified by all five aligners here was consistent with that reported previously (Guo et al. 2015). However, statistical significance of miR-125b-1 was variable, with FDR ≤ 0.05 for all aligners except default Bowtie1 (Fig. 11C). Statistical significance of miR-33b also varied, with FDR ≤ 0.05 for all aligners except default Bowtie1 and MicroRazerS (Fig. 11D). Bowtie2 (vsl) provided the most statistically significant differences for both miR-125b-1 and miR-33b, with nominal P-values an order of magnitude smaller than the other aligners evaluated. To investigate this in more detail, nominal and FDR adjusted edgeR P-values obtained from count matrices derived from each mapper were plotted, showing that FDR P-value correction of default Bowtie1 P-values was only slightly more strict than that of other aligners (Fig. 11E). As fold changes did not vary dramatically between mappers, we investigated whether there were differences in dispersions. Cumulative distribution analysis shows overall tagwise dispersions (gene-wise measures of variance) were smallest for Bowtie2 (vsl) and largest for default Bowtie1 and MicroRazerS (Fig. 11F). In line with this, the common dispersion (experiment-wise measure of variance) was smallest for Bowtie2 (vsl) and largest for default Bowtie1 and MicroRazerS (Fig. 11F inset). These data show that poor mapper selection results in higher dispersions which represent a source of error that impacts on significance values. Lowly ranked mappers uniquely identified a higher number of protein-coding DEGs that may be false positives.
Aligner accuracy with multiply mapped reads
In some cases, rather than discarding reads with multiple mapping locations, researchers may want to know all mapping locations of these tags. We generated a read set consisting of hairpin-derived sequences 18–24 nt in length expected to map to multiple hairpin loci. These tags were mapped to the genome using BLAT (Kent 2002) with sensitive parameters to establish ground truth. Parameters of the 16 aligners in our panel were adjusted to report multiple alignments (Supplemental Table S3). No mapQ filtering was undertaken. The precision and recall values were determined for each read with 100 or fewer BLAT hits. The average values of precision and recall were used to calculate the F1-measure (Fig. 12).
FIGURE 12.
Alignment of 18–24 nt hairpin-derived multimapping reads. Precision and recall were calculated for each read based upon ground truth estimated from BLAT mapping positions. The mean values for precision and recall are reported for each test. (A) Oryza sativa reads. (B) Homo sapiens reads.
Bowtie1 (best strata) performed well overall (F1 = 0.908 for rice and 0.808 for human), although with other parameter settings, Bowtie1 scored poorly for precision. MicroRazerS scored highly in the human read set when compared to rice, while Segemehl scored higher for the rice read set than human. Bowtie2 scored only moderately well (F1 ∼0.7 to ∼0.8) compared to other aligners, which was largely due to lower precision. These results identify mappers suitable for multimapping read analysis.
DISCUSSION
Read mapping is an early step in smRNA-seq data analysis, therefore the choice of mapper may influence downstream results including differential expression or novel smRNA identification. The use of simulated read sets allows empirical evaluation of mapper sensitivity and specificity given that the origin of the reads is known a priori. This strategy is the standard for determining of read mapper accuracy (Holtgrewe et al. 2011). Prior work in the evaluation of smRNA detection and quantification was performed using real sequence data, where it is impossible to determine absolute accuracy due to lack of ground truth (Li et al. 2012). A recent study using simulated reads examined a small number of microRNA pipelines and did not test general-purpose aligners (Williamson et al. 2013). Our work is the first evaluation of smRNA aligner accuracy using simulated reads incorporating sequence variations that are relevant to microRNA biology in plants and animals. Our evaluation shows a considerable difference between alignment softwares in terms of accuracy in the case where short hairpin-derived reads are mapped to a genome.
The optimal selection of a mapper for smRNA-seq analysis will depend on the distribution of lengths, inclusion/exclusion of multimapping reads, polymorphism types and error profiles in smRNA-seq data. In our analysis of public smRNA-seq data, we find that size distribution can vary considerably between experiments. This variability may be technical and due to the manual excision of a narrow DNA band from polyacrylamide gels, a step commonly used to exclude adapter-only fragments. Differences observed in the proportion of sequence variants are also likely to have some technical basis, with varying sequencing chemistry versions, base calling software versions, cluster densities, library preparation kit versions among other factors playing some role in addition to biological variability. Overall, 3′ NTE are the most common sequence variation type in human smRNA-seq data followed by SNM. In rice, SNM are the most common at up to 0.7% of bases.
Regarding uniquely mapped read analysis, Subread, BBMap, and Stampy with default settings yield mapping rates <50% for 21-nt reads and as such, show poor sensitivity compared to other aligners and are therefore not recommended for smRNA-seq mapping. Bowtie1 with default settings demonstrated poor accuracy in most tests and provided the third lowest accuracy overall. A recent report has highlighted considerable strand-bias in Bowtie1 when using the default settings and cautioned that this is likely to have negatively impacted previously published analyses (Axtell 2014). In contrast, Bowtie1 using the “best strata” setting was highly accurate in our tests (including multimapped reads), and is used in microRNA prediction pipelines such as miRDeep2 and microRNA target prediction (StarBase) (Friedländer et al. 2008; Hackenberg et al. 2011). Despite being considered a specialist smRNA aligner, MicroRazerS scored only moderately in most tests compared to general-purpose short read aligners but was highly ranked for accuracy with human multimapping reads. SOAP2 and GEM were accurate with perfectly matching reads but gave poor alignment rates and accuracy with 3′ and 5′ NTE. These observations suggest these aligners may not correctly identify NTE in real smRNA-seq data. SOAP2 was accurate with perfectly matching reads including multimapped reads, but performed poorly with SNM-containing reads and so may prevent identification of A-to-I editing. GNUMAP scored moderately in most tests, including good results for multi-mapping reads, but required hash size optimization for each reference genome.
STAR is one of the fastest aligners for RNA-seq data (Dobin et al. 2013), and among the top-ranked aligners for accurately mapping NTE reads but showed poorer handling of SNM. Using STAR with the no-intron (ni) option produced marginally better results across most tests compared to the default settings. Subread (mir settings) does not map reads <19 nt with the parameters used, but at longer read lengths, performs well in mapping NTE containing reads. BWA is commonly used in smRNA-seq analysis and showed good accuracy with SNM containing reads but only moderate sensitivity with perfectly matching reads <18 nt; running BWA disallowing gap openings yielded marginally better results in most tests compared to the default settings. Results from OLego were similar to BWA in most tests. SMALT aligner (k12.s2 setting) was very accurate with NTE-containing reads but did not score highly with perfectly matching reads. Bowtie2 (vs) was accurate with perfect reads but intolerant of NTE and SNM. Bowtie2 (vsl) performed well throughout most tests, especially 3′ NTE reads.
Overall Bowtie2 with the “very sensitive local” option was ranked the most accurate mapper in our tests, and is our recommendation for analysis of uniquely placed reads. Bowtie1 with “best strata” setting could be appropriate when SNMs are more frequent than NTEs. Whereas Bowtie1 does not give an estimation of mapping quality (i.e., either 0 or 255), Bowtie2 does provide this feature and is useful for moderating the effects of ambiguously mapped reads. When specifically looking for accuracy in aligning multimapped reads, Bowtie1 “best strata” was ranked highly in both rice and human tests and is our recommendation. In large genomes such as human and rice, indel-containing smRNA-seq reads are not accurately mapped with any software, and as such it may be prudent to disallow internal gaps in read alignments.
Our comparison of selected mappers with real human smRNA-seq data demonstrates that the choice of mapper is likely to have an impact on the detection of RNA editing, with sequence mismatch rates being strongly affected by the choice of mapper, consistent with previous RNA editing studies (de Hoon et al. 2010). Importantly, differential smRNA expression results from simulated and real data were strongly affected by the choice of alignment software, with default Bowtie1 performing worst in the simulated differential miRNA-seq test, and in real smRNA-seq identifying a relatively large number of protein-coding DEGs not identified by other mappers. Bowtie2 “very sensitive local” detects fewer genes and DEGs as a likely consequence of its higher mapping stringency. Importantly, we also show that Bowtie2 with very sensitive local settings provides better accuracy with simulated differential miRNA-seq data and in real data analysis generates smaller P-values for true positives compared to other aligners and this is not solely due to multiple testing correction but smaller measures of variation (dispersion).
MATERIALS AND METHODS
Quantification of sequence modifications in smRNA-seq data
The distribution of smRNA-lengths as well as the abundance of modifications (NTEs, SNMs, and indels) were determined from real smRNA-seq data. Rice and human smRNA-seq data were downloaded from GEO (www.ncbi.nlm.nih.gov/geo/), with the following accession numbers: GSE26357, GSE62200, GSE49816, and GSE60036 (Barrera-Figueroa et al. 2012; Mestdagh et al. 2014; Xu et al. 2014; Guo et al. 2015). SRA toolkit (v2.1.7) was used to convert to fastq format and after removing bases with base call quality less than 20 and adapter clipping (FastxToolkit v0.0.14), the reads were mapped to the respective genomes with BWA (v0.7.10) and Bowtie2 (v2.1.0) with default settings. Oryza sativa and Homo sapiens reference genomes were downloaded from Ensembl (Oryza_sativa.IRGSP-1.0.26.dna.genome.fa.gz and Homo_sapiens.GRCh38.dna.primary_assembly.fa.gz). Reads with mapQ<20 were omitted from downstream analysis. A custom UNIX shell script was used to determine read length distribution and abundance of NTEs, internal SNM and indels in sequence tags located within known microRNA hairpin regions obtained from miRbase (Kozomara and Griffiths-Jones 2014). All custom scripts have been deposited to SourceForge (https://sourceforge.net/projects/microrna-alignment-evaluation/).
Simulation and mapping of Illumina-like 21-nt miRNA sequences
Simulated 21-nt microRNA reads with realistic error profiles were generated with ART (Huang et al. 2012) using miRbase hairpin sequences as templates. The read sets comprised 80,400 rice reads and 128,980 human reads that equates to 100-fold coverage over hairpin containing genomic loci. These read sets and those described in the following sections are available in our SourceForge repository. Sixteen short read mappers were selected, and used to identify unique genome alignments. The software versions, command lines used and references for all aligners including those optimized are shown (Table 2). Hash- or seed-based mappers BBMap, GNUMAP, MicroRazerS, Mosaik, and SMALT were initially optimized for k-mer size based upon a modified F-measure (see below) for 21-nt rice reads with a minimum mapping quality of 20 (Supplemental Table S1). For most other aligners, we tested other mapping parameters including those suggested by the software authors or those described in previous reports.
Evaluating mapping accuracy
Bedtools intersect was used to determine whether reads aligned to miRbase hairpin regions (Quinlan and Hall 2010). Reads were classified as “correctly mapped” if they were mapped to a location of the original hairpin; “incorrect miR” if they were mapped to a different hairpin location; “incorrect other” if they mapped to a non-hairpin location; “unmapped” if below the mapQ threshold or not mapped at all. Precision is defined as the proportion of mapped reads that are placed correctly. Recall is defined as the proportion of reads that align with mapQ value over the specified threshold. For each mapper, an optimum mapQ value was determined based upon the modified F-measure. MapQ thresholds tested were: 0, 1, 2, 3, 4, 5, 10, 15, 20, 25, and 30 (Supplemental Table S2). The F-measure is a summary statistic of precision and recall. For analysis of uniquely mapped reads, we utilize the F0.25 measure (β = 0.25) that weights precision approximately four times more than recall.
Evaluating mapping accuracy with synthetic read sets of variable length or containing sequence variations
Synthetic sequence reads were generated from hairpin regions at each nucleotide (nt) position with lengths 16–25 nt using a custom script. Any reads with identical duplicate sequences were removed. The rice read sets contained 28,056–30,846 sequence tags and the human read sets contained 28,226–42,071 sequence tags. Synthetic 21-nt reads were mutated using the msbar utility from the EMBOSS package (Rice et al. 2000). Up to two single nucleotide mismatches (SNM), single nucleotide insertions, and single nucleotide deletions were incorporated. Single indels up to 2 nt in length were also incorporated using msbar. To simulate nontemplated extension (NTE), up to four random nonreference bases were added to the 3′ or 5′ ends using a custom script.
Evaluating mapping accuracy with short protein-coding tags
We again utilized ART with the default settings to generate short tags derived from protein-coding cDNA sequences at lengths from 16 to 25 nt. Any protein-coding gene that overlaps a miRbase21 hairpin region was excluded from the template sequence set. The read sets comprised (1,933,571–3,034,322) rice reads and (1,102,243–1,734,494) human reads that equates to twofold coverage over protein-coding loci.
Effect of aligner accuracy on differential expression analysis
We utilized Polyester software (Frazee et al. 2015) to simulate fold changes and fastq read sets for miRbase hairpin loci with two sample groups each with three replicates. Fold change values used were 0.5, 1, and 2, with proportions 0.05, 0.9, and 0.05, respectively. Fastq files were generated with an average of 100 reads per transcript, a length of 21 nt and built-in “illumina 4” error profile. Reads were mapped to the respective genome using Bowtie1 (default), Bowtie1 (best strata), Bowtie2 (vsl), BWA (df), and MicroRazers (sl18.se.pa). Summarization of read alignments was performed by feature Counts (Liao et al. 2014) with miRbase v21 hairpin annotation and Ensembl protein-coding gene annotation (Homo_sapiens.GRCh38.78.gtf). For Bowtie2 (vsl) and BWA (df), the optimized mapQ threshold identified previously was used. Genes with fewer than 10 reads per sample on average were excluded from downstream analysis. Multidimensional scaling analysis was performed using the cmdscale function in R. Correlation analysis was performed by the Pearson method to compare the same data sets processed by different aligners. Differential analysis of sample group replicates was performed by edgeR (Robinson et al. 2010). P-values ≤ 0.05 after Benjamini–Hochberg false-discovery rate (FDR) adjustment were considered significant.
Effect of aligner accuracy on expression quantification and differential expression calling
Small RNA-seq data from a recently submitted data set (Guo et al. 2015) (GEO accession GSE60036) underwent conversion to fastq format, quality trimming and adapter clipping as above, with a minimum read length of 16 nt. Reads were mapped to the human genome using Bowtie1 (default), Bowtie1 (best strata), Bowtie2 (vsl), BWA (df), and MicroRazers (sl18.se.pa). Mismatch profiles were generated by RSeqQC (Wang et al. 2012).
Evaluating accuracy of multiply mapped reads
Synthetic sequence reads were generated at each nucleotide (nt) position of hairpin loci at lengths 18–24 nt as above, however reads with identical duplicate sequences were retained and reads with unique sequences were removed. This generated a single read set for rice (20,147 tags) and human (20,774) with a range of lengths (18–24 nt). Modifications to the alignment command lines are given in Supplemental Table S3. In order to establish “ground truth” mapping of this read set, we used BLAT (Kent 2002) with the following parameters “-minScore=10 -tileSize=8 -fine -stepSize=4” and extracted exact BLAT hits. Tags with >100 BLAT hits were excluded. There were 18,371 and 19,600 tags from rice and human, respectively, with 100 or fewer BLAT hits. For evaluating high-throughput aligners, a custom script was used to determine true positives, false positives, and false negatives for each sequence read with reference to the BLAT result. Precision and recall was then calculated for each read and the average values were reported for each alignment software. The F1 measure was used to rank overall precision and recall for multimapping tests.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Lazarus for bioinformatics support. The authors acknowledge funding support from the Juvenile Diabetes Research Foundation International (JDRF), the National Health and Medical Research Council (NHMRC), and the National Heart Foundation of Australia (NHF). A.E-O. is a Senior Research Fellow supported by the NHMRC. This work was supported in part by the Victorian Government's Operational Infrastructure Support Program.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.055509.115.
REFERENCES
- Ameres SL, Zamore PD. 2013. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 14: 475–488. [DOI] [PubMed] [Google Scholar]
- Axtell MJ. 2013. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19: 740–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. 2014. Butter: high-precision genomic alignment of small RNA-seq data. bioRxiv 10.1101/007427. [DOI] [Google Scholar]
- Axtell MJ, Westholm JO, Lai EC. 2011. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol 12: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera-Figueroa BE, Gao L, Wu Z, Zhou X, Zhu J, Jin H, Liu R, Zhu JK. 2012. High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol 12: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. 2005. RNA meets chromatin. Genes Dev 19: 1635–1655. [DOI] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. 2006. RNA editing of human microRNAs. Genome Biol 7: R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. 2003. Role of microRNAs in plant and animal development. Science 301: 336–338. [DOI] [PubMed] [Google Scholar]
- Clement NL, Snell Q, Clement MJ, Hollenhorst PC, Purwar J, Graves BJ, Cairns BR, Johnson WE. 2010. The GNUMAP algorithm: unbiased probabilistic mapping of oligonucleotides from next-generation sequencing. Bioinformatics 26: 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Taft RJ, Hashimoto T, Kanamori-Katayama M, Kawaji H, Kawano M, Kishima M, Lassmann T, Faulkner GJ, Mattick JS, Daub CO, Carninci P, Kawai J, Suzuki H, Hayashizaki Y. 2010. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res 20: 257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emde AK, Grunert M, Weese D, Reinert K, Sperling SR. 2010. MicroRazerS: rapid alignment of small RNA reads. Bioinformatics 26: 123–124. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. 2008. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 135: 1201–1214. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Brown M, Morozov P, Ten Hoeve JJ, Ben-Dov IZ, Hovestadt V, Hafner M, Renwick N, Mihailović A, Wessels LF, et al. 2012. Bioinformatic analysis of barcoded cDNA libraries for small RNA profiling by next-generation sequencing. Methods 58: 171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazee AC, Jaffe AE, Langmead B, Leek JT. 2015. Polyester: simulating RNA-seq datasets with differential transcript expression. Bioinformatics 31: 2778–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40: 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Liu J, Elfenbein SJ, Ma Y, Zhong M, Qiu C, Ding Y, Lu J. 2015. Characterization of the mammalian miRNA turnover landscape. Nucleic Acids Res 43: 2326–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Markmann K, Pedersen CN, Stougaard J, Andersen SU. 2012. shortran: a pipeline for small RNA-seq data analysis. Bioinformatics 28: 2698–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg M, Rodríguez-Ezpeleta N, Aransay AM. 2011. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res 39: 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermüller J. 2009. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol 5: e1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtgrewe M, Emde AK, Weese D, Reinert K. 2011. A novel and well-defined benchmarking method for second generation read mapping. BMC Bioinformatics 12: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PJ, Liu YC, Lee CC, Lin WC, Gan RR, Lyu PC, Tang P. 2010. DSAP: deep-sequencing small RNA analysis pipeline. Nucleic Acids Res 38: W385–W391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Li L, Myers JR, Marth GT. 2012. ART: a next-generation sequencing read simulator. Bioinformatics 28: 593–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. 2007. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WP, Stromberg MP, Ward A, Stewart C, Garrison EP, Marth GT. 2014. MOSAIK: a hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS One 9: e90581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Liu F, Vongsangnak W, Jing Q, Shen B. 2012. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res 40: 4298–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Lindner R, Friedel CC. 2012. A comprehensive evaluation of alignment algorithms in the context of RNA-seq. PLoS One 7: e52403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21: 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco-Sola S, Sammeth M, Guigó R, Ribeca P. 2012. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods 9: 1185–1188. [DOI] [PubMed] [Google Scholar]
- Mathelier A, Carbone A. 2010. MIReNA: finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics 26: 2226–2234. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. 2006. Non-coding RNA. Hum Mol Genet 15: R17–R29. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Olson EN. 2012. MicroRNAs in stress signaling and human disease. Cell 148: 1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel P, Frellsen J, Plass M, Rasmussen SH, Krogh A. 2013. On the accuracy of short read mapping. Methods Mol Biol 1038: 39–59. [DOI] [PubMed] [Google Scholar]
- Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D'Andrade P, DeMayo M, Dennis L, et al. 2014. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 11: 809–815. [DOI] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. 2014. The rise of regulatory RNA. Nat Rev Genet 15: 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motameny S, Wolters S, Nürnberg P, Schumacher B. 2010. Next generation sequencing of miRNAs—strategies, resources and methods. Genes (Basel) 1: 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Cheng HH, Tewari M. 2012. MicroRNA profiling: approaches and considerations. Nat Rev Genet 13: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen R, Gan I, Modai S, Sukacheov A, Dror G, Halperin E, Shomron N. 2010. miRNAkey: a software for microRNA deep sequencing analysis. Bioinformatics 26: 2615–2616. [DOI] [PubMed] [Google Scholar]
- Shang J, Zhu F, Vongsangnak W, Tang Y, Zhang W, Shen B. 2014. Evaluation and comparison of multiple aligners for next-generation sequencing data analysis. Biomed Res Int 2014: 309650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V. 2012. The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28: 2059–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokowy T, Eszlinger M, Świerniak M, Fujarewicz K, Jarząb B, Paschke R, Krohn K. 2014. Analysis options for high-throughput sequencing in miRNA expression profiling. BMC Res Notes 7: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Tsao MS, McPherson JD. 2015. Optimization of miRNA-seq data preprocessing. Brief Bioinform 16: 950–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. 2009. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics 10: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W. 2012. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28: 2184–2185. [DOI] [PubMed] [Google Scholar]
- Williamson V, Kim A, Xie B, McMichael GO, Gao Y, Vladimirov V. 2013. Detecting miRNAs in deep-sequencing data: a software performance comparison and evaluation. Brief Bioinform 14: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Anczuków O, Krainer AR, Zhang MQ, Zhang C. 2013a. OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic Acids Res 41: 5149–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Liu Q, Wang X, Zheng J, Wang T, You M, Sheng Sun Z, Shi Q. 2013b. mirTools 2.0 for non-coding RNA discovery, profiling, and functional annotation based on high-throughput sequencing. RNA Biol 10: 1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Bai H, Liu C, Chen E, Chen Q, Zhuang J, Shen B. 2014. Genome-wide analysis of microRNAs and their target genes related to leaf senescence of rice. PLoS One 9: e114313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. 2006. Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289: 3–16. [DOI] [PubMed] [Google Scholar]
- Zhou H, Arcila ML, Li Z, Lee EJ, Henzler C, Liu J, Rana TM, Kosik KS. 2012. Deep annotation of mouse iso-miR and iso-moR variation. Nucleic Acids Res 40: 5864–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.