Abstract
The secondary structure of ribosomal RNA (rRNA) is largely conserved across all kingdoms of life. However, eukaryotes have evolved extra blocks of rRNA sequences, relative to those of prokaryotes, called expansion segments (ES). A thorough characterization of the potential roles of ES remains to be done, possibly because of limitations in the availability of robust systems to study rRNA mutants. We sought to systematically investigate the potential functions, if any, of the ES in 25S rRNA of Saccharomyces cerevisiae by deletion mutagenesis. We deleted 14 of the 16 different eukaryote-specific ES in yeast 25S rRNA individually and assayed their phenotypes. Our results show that all but two of the ES tested are necessary for optimal growth and are required for production of 25S rRNA, suggesting that ES play roles in ribosome biogenesis. Further, we classified expansion segments into groups that participate in early nucleolar, middle, and late nucleoplasmic steps of ribosome biogenesis, by assaying their pre-rRNA processing phenotypes. This study is the first of its kind to systematically identify the functions of eukaryote-specific expansion segments by showing that they play roles in specific steps of ribosome biogenesis. The catalog of phenotypes we identified, combined with previous investigations of the roles ribosomal proteins in large subunit biogenesis, leads us to infer that assembling ribosomes are composed of distinct RNA and protein structural neighborhood clusters that participate in specific steps of ribosome biogenesis.
Keywords: rRNA, eukaryote, ribosome, protein–RNA, ribosome assembly, expansion segments
INTRODUCTION
A universal core secondary structure for ribosomal RNA (rRNA) has been identified across all kingdoms of life (Veldman et al. 1981; Clark et al. 1984; Gutell et al. 1993). However, eukaryotic rRNA has additional blocks of sequences relative to prokaryotic rRNA designated as “expansion segments” (ES) (Ware et al. 1983; Clark et al. 1984; Hassouna et al. 1984). These ES are interspersed throughout the various domains of rRNA secondary structure core at specific, conserved sites (Fig. 1A). They exhibit a striking degree of variability in multiple respects: not all ES are found in all species, they vary in their length and sequence both within and among different species; and they even vary between different rRNA repeats of the same organism (Gonzalez et al. 1985). Hence, ES also have been referred to as variable regions and divergent domains (Gerbi 1996). However, the secondary structures of ES exhibit less variability. In eukaryotes, there are a total of 12 ES in the small subunit rRNA (ESS) and 41 in the large subunit rRNA (ESL) compared to prokaryotic Escherichia coli rRNA. However, only a subset of these ES are present in any given eukaryote species. Though ES have been a continued focus of research, their inherent variability and the limited availability of robust systems to study rRNA mutations have impeded our understanding of their functions.
FIGURE 1.
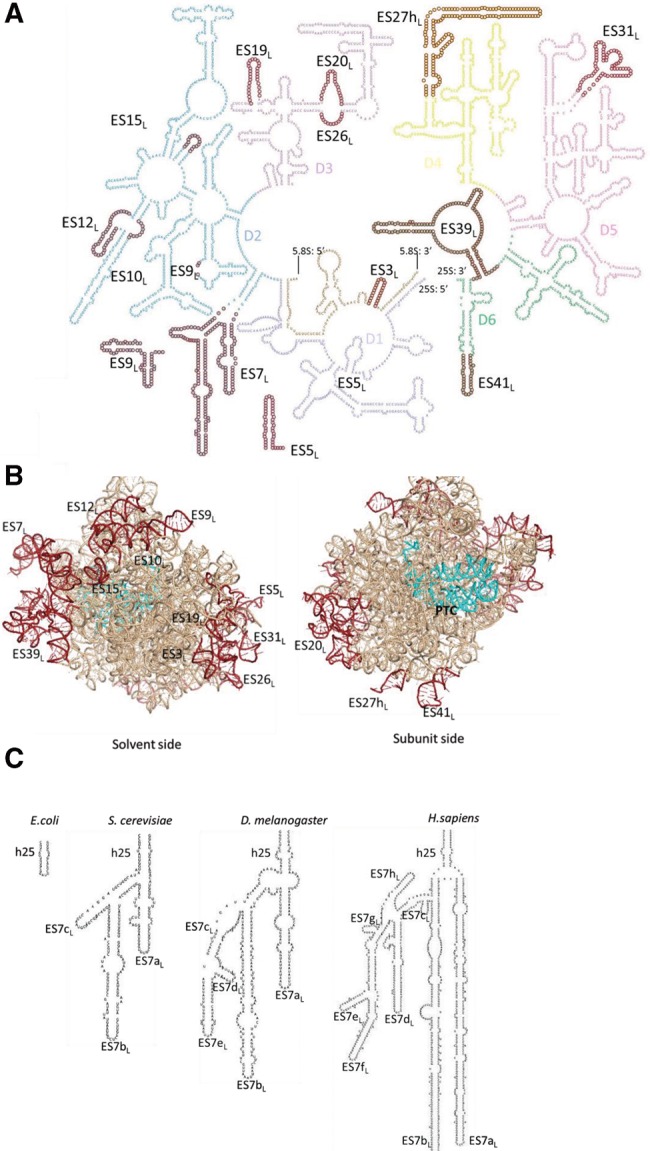
Ribosomal RNA expansion segments in the yeast large ribosomal subunit. (A) Secondary structure of Saccharomyces cerevisiae large subunit ribosomal RNA (rRNA). Different domains are marked by different colors. The ES deletions studied here (14 of the 16 ES in yeast LSU rRNA) are highlighted by maroon circles and labeled. (B) Pymol representation of the large subunit rRNA. ES deletions studied are highlighted in maroon as in A and labeled. The peptidyl transferase center (PTC) is shown in cyan. (C) ES progressively increase in size and complexity in eukaryotes. Helix 25 (h25) is shown as an example to demonstrate the increase in sequence length (not to scale) and the number of helical branches of ES7L, from E. coli through Homo sapiens.
Recent high-resolution cryo-EM and crystal structures of eukaryotic ribosomes (Spahn et al. 2001; Taylor et al. 2009; Armache et al. 2010; Ben-Shem et al. 2011; Jenner et al. 2012; Anger et al. 2013) have considerably improved our understanding of ES, yielding interesting insights (Fig. 1B). ES are responsible for a majority of the 40% increase in the size of ribosomes from prokaryotes to eukaryotes (Gerbi 1996; Jenner et al. 2012). Their size and the number of helical branches seem to increase progressively in higher eukaryotes (Fig. 1C; Michot and Bachellerie 1987; Michot et al. 1990). Eukaryotic rRNA ES of the large ribosomal subunit are located predominantly in two clusters on the side of the ribosome exposed to the solvent, although they are distributed across the different domains of rRNA secondary structure (Fig. 1A,B). Importantly, ES do not directly interrupt the functional centers of ribosomes (Fig. 1B; Gerbi 1996; Armache et al. 2010; Ben-Shem et al. 2011; Anger et al. 2013). Thus, ribosomes of higher eukaryotes have been described as having a conserved core, with a shell consisting of a flexible outer layer formed by the helical insertions of the ES and eukaryote-specific portions of ribosomal proteins (r-proteins) (Melnikov et al. 2012). In agreement with an earlier study that suggested that ES are on the surface of the ribosome (Han et al. 1994), their clustering on the solvent-exposed surface of the ribosome potentially enables a multitude of molecular interactions by these ES to modulate ribosome assembly and function.
The most interesting question about ES pertains to their function. It has not been determined fully if ES are innocuous, dispensable insertions that arose during the course of evolution, or instead, whether they have specific functions in the cell. Various studies have been carried out to investigate whether ES might have roles in ribosome biogenesis or function. In one example, replacing the ES19L region of yeast 25S rRNA with E. coli or mouse ES sequences was tolerated without apparent loss of function (Musters et al. 1991). Likewise, insertions in the ES5L and ES9S regions of yeast and insertions near the 3′ end of the large subunit rRNA in Tetrahymena were functional (Musters et al. 1989, 1990; Sweeney and Yao 1989). In contrast, a deletion of Tetrahymena ES27L prevented growth (Sweeney et al. 1994). Interestingly, the ES27L deletion could be rescued by replacement with ES27L sequences from another organism, but not with random sequences, leading the authors to hypothesize that these regions share an essential function that is not reflected in their primary sequences. Further evidence that ES might carry out specific functions was revealed by deletion mutagenesis of yeast ES7L and ES27L. The larger deletion mutations of these ES were lethal due to defects in ribosome biogenesis (Jeeninga et al. 1997). For those ES that are essential, several studies supported the notion that their secondary structure is more important for their function than their sequence (Sweeney et al. 1994; Jeeninga et al. 1997).
The high degree of variability observed in expansion segment sequences and their observed location away from the functional centers of ribosomes suggest that they may not directly participate in universally conserved core aspects of translation. However, they could fine-tune translation, potentially in a species-specific manner (Gao et al. 2005; Hashem et al. 2013). For example, some small subunit ES show complementarity with mRNAs, raising the possibility that ES might affect their translation (Gonzalez et al. 1988). Furthermore, it has been suggested that ES could contribute to the functional heterogeneity of Plasmodium ribosomes (Gunderson et al. 1987; Waters et al. 1995). More recent structural studies have identified other potential roles for ES: (i) ES31L and ES41L form additional eukaryote-specific intersubunit bridges in yeast (Spahn et al. 2001); (ii) ES6S, located near the mRNA entry and exit sites, and ES27L, located near the polypeptide exit tunnel, have been suggested to facilitate the access of translation factors to ribosomes (Beckmann et al. 2001; Armache et al. 2010; Anger et al. 2013); (iii) ES24L was proposed to play a role in cotranslational protein localization (Halic et al. 2004). Despite the growing body of evidence indicating the importance of ES in ribosome biogenesis and function, a comprehensive deletion mutagenesis survey that could yield valuable insights into the functions of ES has been lacking.
Here, we systematically investigated the functions of 14 ES of the large subunit rRNA of S. cerevisiae ribosomes, by deleting them individually and studying the mutant phenotypes. We found that 12 of the ES studied are necessary for optimal growth, suggesting that the majority of ES play indispensable functions. The steady-state levels of mature 25S rRNA are severely diminished in all of the mutants that exhibit a growth deficiency, suggestive of improper biogenesis of 25S rRNA. We further categorized ES mutants into early nucleolar, middle, and late nuclear phenotypic groups, based on their pre-rRNA processing phenotype. The observed functional clustering pattern of these rRNA ES functions in the structure of the ribosome resembles those reported for r-proteins (Gamalinda et al. 2014), indicating that distinct neighborhoods in the assembling ribosome might be involved in distinct steps of ribosome biogenesis. We also discuss potential models by which eukaryote-specific protein and RNA elements in the ribosome might have coevolved to perform functions in ribosome biogenesis. Taken together, our data suggest that many ES are indispensable, demonstrate potential functions for ES in ribosome biogenesis, and explain some of their distinct roles in ribosome biogenesis.
RESULTS AND DISCUSSION
ES mutants and the system to assay their phenotype
The presence of around 200 copies of rDNA genes in the yeast genome makes analysis of rDNA mutants challenging. In order to assay the in vivo phenotype of ES deletions, we used a strain carrying a temperature sensitive mutation in a subunit of RNA polymerase I (NOY504), the enzyme that exclusively transcribes rDNA (Nogi et al. 1993). We transformed this strain with a plasmid (pWL160) (Liang and Fournier 1997; Cole and LaRiviere 2008) containing an rDNA unit driven by Pol II from a GAL7 promoter. At the permissive temperature, endogenous chromosomally derived WT rRNA is made as well as rRNA from the plasmid. However, at the restrictive temperature, transcription of chromosomal rDNA is shut off, and the only source of rRNA is the plasmid (pWL160) (Supplemental Fig. 1A). This plasmid contains oligonucleotide tags within ES5L of 25S rRNA and ES3S of 18S rRNA, which permit us to differentiate plasmid-derived rRNA from residual endogenous rRNA.
We introduced 14 ES deletion mutations into plasmid-borne 25S rDNA individually and assayed each for effects on ribosome biogenesis. Although reported nucleotide definitions of ES can vary (Gerbi 1996; Taylor et al. 2009; Armache et al. 2010; Anger et al. 2013), the exact ES deletions examined in this study are indicated in Supplemental Table 1 and highlighted in Figure 1. In cases where an ES deletion involved truncating a helix in the middle, we introduced a tetraloop to maintain stability of the helix and to minimize secondary effects. With the exception of ES4L and ES24L (Peculis and Greer 1998; Cote and Peculis 2001; Halic et al. 2004), we have examined nearly all of the ES of the large subunit rRNA in yeast. Also, we studied a partial deletion of ES27L, as marked in Figure 1, denoted henceforth as ES27hL to represent the deletion of the helix.
Most ESL are necessary for optimal growth
In order to address whether ES are innocuous, dispensable eukaryotic insertions in rRNA, or are essential, we first assayed the viability of the 14 ESL deletion mutants by spotting serial dilutions of cultures onto solid growth media (Fig. 2). At the permissive temperature (left panels), when the chromosome-derived WT rRNA is expressed, all of the mutant strains grew well, as expected. When grown at the restrictive temperature (right panels), 12 of the 14 ES deletion mutants exhibited a growth defect compared to the WT control. The growth phenotype of these 12 ESL deletion mutants closely resembled the no rDNA control. Hence, we refer to them as “lethal” ES mutants or “essential” ES. The slight growth we observe at the restrictive temperature presumably results from the preexisting pool of ribosomes and incomplete shut-off of Pol I. Thus, all of the ESL tested, except ES19L and ES27hL, are necessary for viability, suggesting that most ESL play indispensable functions in the cell.
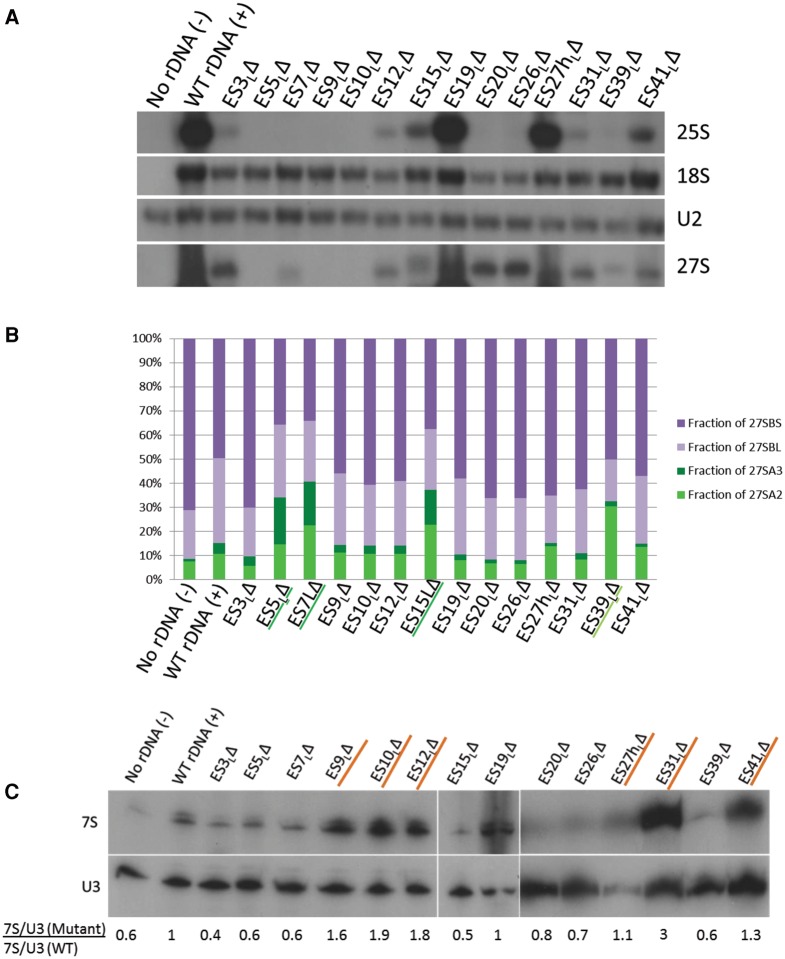
FIGURE 2.
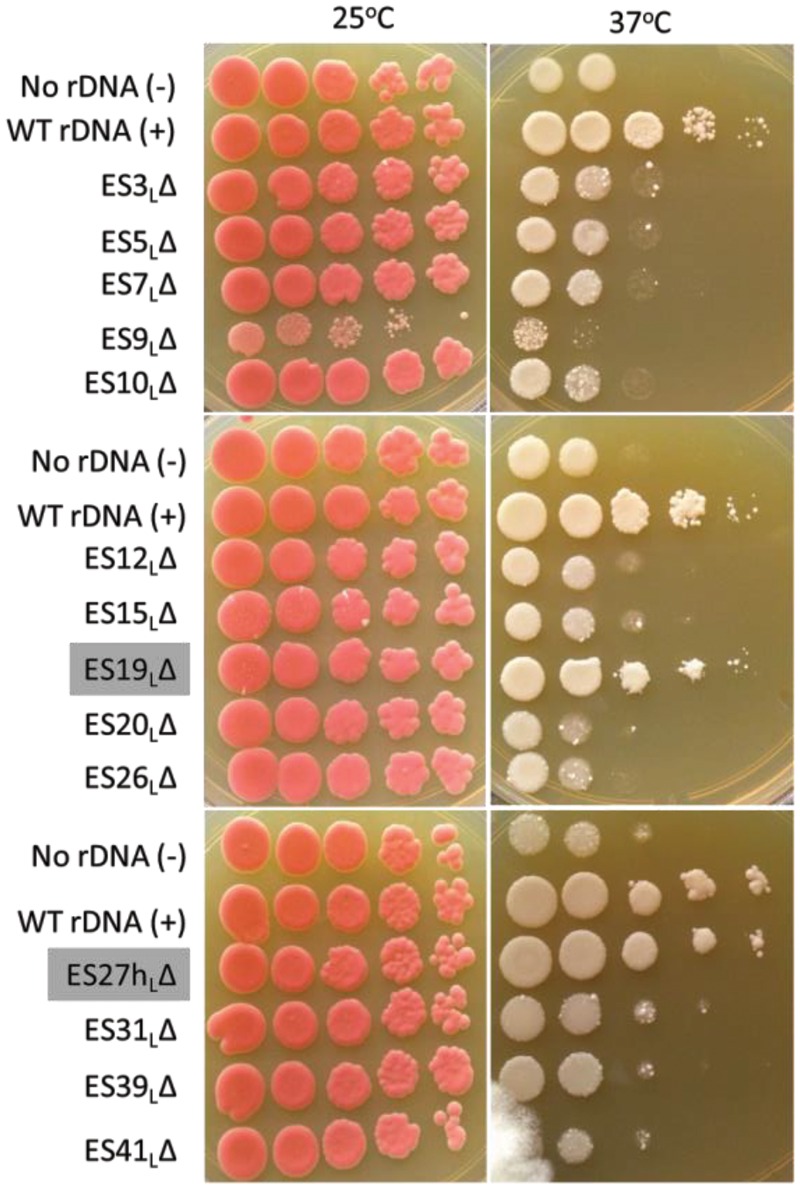
Most large subunit ES are necessary for optimal growth. Serial dilutions of log phase yeast cultures were spotted onto solid medium and grown either at the permissive temperature (25°C, left panels) or at the restrictive temperature (37°C, right panels). Empty vector (“No rDNA”) and wild-type (WT) controls (“WT rDNA”) are indicated. The nonessential ES are highlighted with gray labels. Note that a less dense culture of ES9L was used. Similar results were observed across at least three independent biological replicates.
Most ESL function in 60S subunit biogenesis
Since ES are eukaryote-specific, we hypothesized that these extra rRNA elements may have evolved to perform eukaryote-specific functions related to the ribosome, including eukaryote-specific features of ribosome biogenesis, nuclear export of nascent ribosomal subunits, and/or eukaryote-specific functions in translation. Since defects in ribosome biogenesis could mask any further defects in translation in these mutants, we decided to begin by assaying for ribosome biogenesis-related functions. To test whether deletions of ES affected synthesis of mature 25S rRNA, we performed a Northern blot of total cellular RNA extracted after shifting to the restrictive temperature (Fig. 3A), and probed for plasmid-derived 25S (first panel, Fig. 3A) and 18S rRNA (second panel, Fig. 3A) using oligonucleotides complementary to the plasmid-specific tag. Strikingly, all the lethal ES deletions resulted in severely diminished, sometimes undetectable levels of mature 25S rRNA, while levels of small subunit 18S rRNA remained relatively stable. This confirms that all of the essential ES play a role in the biogenesis of large ribosomal subunits. Those mutants that can produce WT levels of 25S rRNA also exhibit WT-like growth, suggesting that the observed growth deficiencies in the lethal ES mutants primarily result from defective synthesis of stable 25S rRNA. In principle, another possibility that could explain this result is that: mature 25S rRNA is being made and is subsequently degraded from the translational pool, perhaps by nonfunctional RNA decay (Cole and LaRiviere 2008). However, as shown below, we identified distinct pre-rRNA processing defects in each of our lethal mutants. This suggests that improper biogenesis, perhaps accompanied by turnover during biogenesis as seen before in other ribosomal protein and assembly factor mutants (Sahasranaman et al. 2011; Jakovljevic et al. 2012; Gamalinda et al. 2014), leads to the production of severely decreased levels of stable 25S rRNA in the lethal ES mutants.
FIGURE 3.
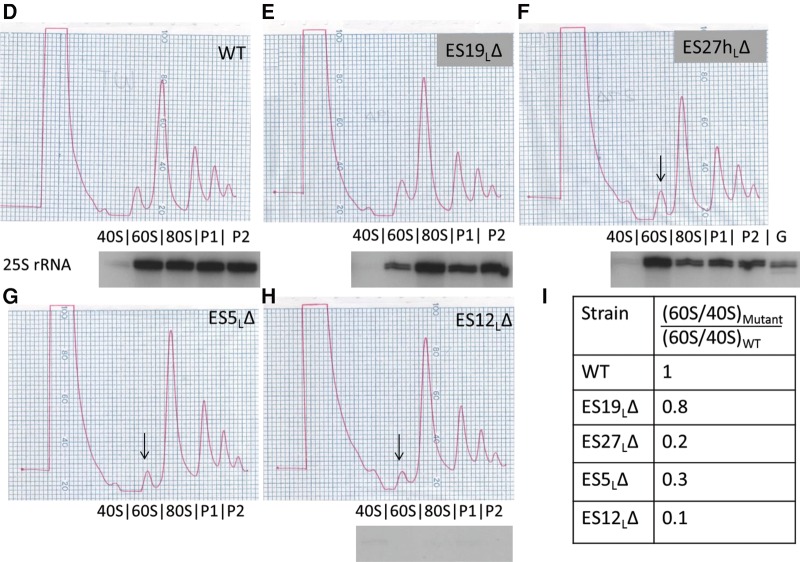
Different ES participate in different steps of large ribosomal subunit assembly. (A) Representative Northern blot of total cellular RNA extracted from ES mutants, probed with various oligonucleotides to detect rRNAs and pre-rRNAs. Similar effects were observed in two different biological replicates. U2 snRNA is the loading control. Only the plasmid-derived 25S, 18S and 27S rRNAs are detected, since plasmid-specific oligonucleotide tags were used to probe for these rRNA species. (B) Relative steady-state levels of various 27S pre-rRNAs (27SA2, 27SA3, 27SBL, 27SBS) in ES mutants, assayed by primer extension. After normalizing each lane with respect to the U2 snRNA loading control, the fraction of each species of pre-rRNA (27SA2, 27SA3, 27SBL, or 27SBS) was calculated as a percentage of total 27S pre-rRNA in that lane (27SA2 + 27SA3 + 27SBL + 27SBS). 27SA processing intermediates are shown in shades of green (27SA2 is light green and 27SA3 is dark green). 27SB processing intermediates are shown in shades of purple (27SBL is light purple and 27SBS is dark purple). An increase in green 27SA pre-rRNA and a concomitant decrease in purple 27SB pre-rRNA are diagnostic of an “early” ES mutant. Note that the levels of pre-rRNA species observed here are a combination of both residual endogenous WT and plasmid-derived mutant pre-rRNA. Shown here are the mean values from three biological replicates. For more details and error bars, see Supplemental Figure 3. (C) Northern blot of total cellular RNA extracted from ES mutants, probed with oligonucleotides that hybridize to 7S pre-rRNA. U3 snoRNA is the loading control. Shown below the blot is the fold increase of 7S pre-rRNA in mutants compared to WT, normalized for loading. A mutant that has a ratio of >1 accumulates 7S pre-rRNA (characteristic of late-acting ES, highlighted in orange), and a mutant that has a ratio of <1 has decreased levels of 7S pre-rRNA (seen in early or middle phenotypes). (D–I) Sucrose gradient centrifugation to assay defects in 60S ribosomal subunit biogenesis in WT cells (D), viable ES mutants (E,F), and representative lethal ES mutants (G,H). Arrows shown in G, H, and F indicate the decreased 60S subunit peak in the lethal ES mutants and in the viable ES27hLΔ mutant, respectively, compared to WT (B). In this assay, defects in 60S subunit biogenesis are discerned exclusively by a decreased 60S peak and a decreased ratio of 60S to 40S subunits, compared to WT. Plasmid-derived 25S rRNA detected by reverse transcription using an oligonucleotide complementary to the plasmid-specific tag is shown underneath the polysome curves to indicate those fractions that contain plasmid-derived mature 25S rRNA. As shown in F, a sequencing lane with G was used to map the 5′ end of 25S rRNA. The primer extension assay could not be performed on the ES5LΔ mutant in G, since this deletion encompasses the plasmid-specific tag, thereby eliminating the binding site of the reverse transcription primer. (G) Ratios of area under the curve of 60S/40S subunits in mutant compared to WT, quantified from D–H. All the ES mutants shown, except ES19LΔ, have a multifold decrease in 60S/40S ratio compared to WT, indicative of defects in 60S subunit biogenesis.
Various ESL function in different steps of ribosome assembly
Ribosome biogenesis proceeds in a series of pre-rRNA processing steps (Supplemental Fig. 2), which have been used to demarcate the progress of ribosome assembly in biogenesis mutants. Thus, we examined which pre-rRNA processing steps are affected in each of the ES mutants by Northern blotting and primer extension. We defined “early”-acting ES as those, when deleted, result in an increase in levels of 27SA2 and 27SA3 pre-rRNA accompanied by a relative decrease in 27SB pre-rRNA. These early processing steps occur in the nucleolus. We defined “late”-acting ES as those, when deleted, result in an increase of 7S pre-rRNA, the processing of which begins in the nucleoplasm. ES mutants that result in an increase of 27S pre-rRNA that cannot be explained by an increase in 27SA pre-rRNA (while not resulting in an increase in 7S pre-rRNA) were categorized as “middle”-acting, presumably resulting in inefficient processing of 27SB pre-rRNA.
Compared to the WT rDNA control, we observed that the ES39ΔL mutant showed increased levels of 27SA2 pre-rRNA relative to later 27S pre-rRNA processing intermediates. In addition, ES5ΔL, ES7ΔL, and ES15ΔL showed a relative increase of the 27SA3 pre-rRNA species accompanied by a concomitant decrease in 27SBS pre-rRNA. Hence, we categorize ES39ΔL, ES5ΔL, ES7ΔL, and ES15ΔL (indicated by text underlined in green in Fig. 3B) as early-acting ES, with ES39ΔL affecting 27SA2 processing and the remainder affecting 27SA3 pre-rRNA processing.
Since the 27SB pre-rRNA intermediate is relatively long-lived (Kos and Tollervey 2010), and the primer extension assay described above measures the combined levels of endogenous WT rRNA and rRNA derived from the plasmid, we were not able to unequivocally determine increases in 27SB pre-rRNA levels, characteristic of the “middle” steps. However, for several mutants, most noticeably ES3ΔL, ES20ΔL, and ES26ΔL, we were able to infer plasmid-derived 27S (A + B) pre-rRNA accumulation (Fig. 3A, fourth panel) using the plasmid-specific oligonucleotide complementary to 25S rRNA. There are two considerations worth noting here. First, the relative lifetimes of various pre-rRNA intermediates are different (Kos and Tollervey 2010); hence the effects on the long-lived 27SB pre-rRNA may be overrepresented in the steady-state Northern blot assay shown in Figure 3A. Second, it has been shown that preribosomes become increasingly stable as assembly proceeds (Gamalinda et al. 2014). Since this steady-state measurement is indicative of a combination of accumulation and turnover, there may be a bias toward later, more stable particles containing 27SB pre-rRNA. Based on these considerations, we conclude that ES3L, ES20L, and ES26L fall into the “middle”-acting category, which includes those that help in processing of 27SB pre-rRNA.
To identify “late”-acting ES mutants, we assayed for effects on 7S pre-rRNA processing by Northern blotting of RNAs, using U3 snoRNA as a loading control (Fig. 3C). After normalizing for loading, we calculated the ratio of levels of 7S pre-rRNA in the mutants to 7S pre-rRNA in the WT control in the same blot with the same exposure. Using this assay, we identified ES9L, ES10L, ES12L, ES31L, and ES41L among the essential ES with a late-acting function, evident by 7S pre-rRNA accumulation when they are deleted. Of these, ES31ΔL seemed to possess the highest accumulation of 7S pre-rRNA. Those ES mutants that result in an accumulation of 7S pre-rRNA are underlined in orange in Figure 3C. In addition, we observed that 7S pre-rRNA levels decreased in the early- and middle-acting ES, consistent with inefficient processing of upstream 27S pre-rRNAs.
Viable ES mutants can synthesize mature 25S rRNA
To further understand the effect of nonlethal ES mutations, we performed sucrose gradient fractionation to separate free ribosomal subunits and polyribosomes in the viable ES mutants (Fig. 3E,F) as well as in representative lethal ES mutants (Fig. 3G,H). These mutant strains, along with a WT control (Fig. 3D), were grown at the permissive temperature and then shifted to the restrictive temperature for 6 h prior to preparing extracts for gradient analysis. If any of these mutants had defects in 60S subunit biogenesis, we expect that levels of free 60S subunits will be decreased compared to 40S subunits (reflected as a decrease in 60S/40S subunit ratio, in comparison with the WT strain, Fig. 3I). Among the viable ES mutants, ES19ΔL resembled the WT control. However, the nonlethal ES27hΔL exhibited a decreased 60S peak and a decrease in the 60S/40S ratio, indicating that this part of ES27L may possibly play a role that is dispensable in 60S subunit biogenesis. This could potentially reflect a slight inefficiency in the 7S pre-rRNA processing step observed in this mutant (Fig. 3C). Processing of 7S pre-rRNA has been shown to be dispensable for the normal function of the ribosome (Rodriguez-Galan et al. 2015), hence it is not surprising that this mutant yields a slight 7S pre-rRNA processing defect while it exhibits no growth defect. Consistent with their inability to make mature 25S rRNA, the lethal mutants ES5ΔL and ES12ΔL showed a decreased 60S peak and exhibited a multifold decrease in the 60S/40S ratio, compared to the WT control. We surmise that other lethal ESL deletion mutants would exhibit a similar effect in this assay. Note that, under the conditions of this assay, synthesis of new WT rRNA from the chromosome is largely blocked. However, a preexisting pool of WT rRNA remains in these cells. Thus, we do not expect to observe other characteristic indicators of a 60S subunit biogenesis defect such as a diminished 80S peak or halfmer polysomes.
To further confirm that the observed effects arose from the plasmid-derived rRNA, we performed a primer extension assay using the plasmid-specific 25S rRNA tag. We detected the 5′ end of 25S rRNA in the 60S, 80S, and polyribosome fractions P1 and P2 of the WT, ES19ΔL and ES27hΔL extracts (but not in the free 40S subunit fraction, as expected). This result is consistent with our observation that the viable mutants can synthesize plasmid-derived mature 25S rRNA at near WT levels. Conversely, in the lethal mutant ES12ΔL, we were not able to detect plasmid-derived 25S rRNA in any fraction, indicating severe defects in large subunit biogenesis.
The location of ES in specific neighborhoods of the 60S subunit correlates with their ribosome assembly phenotype
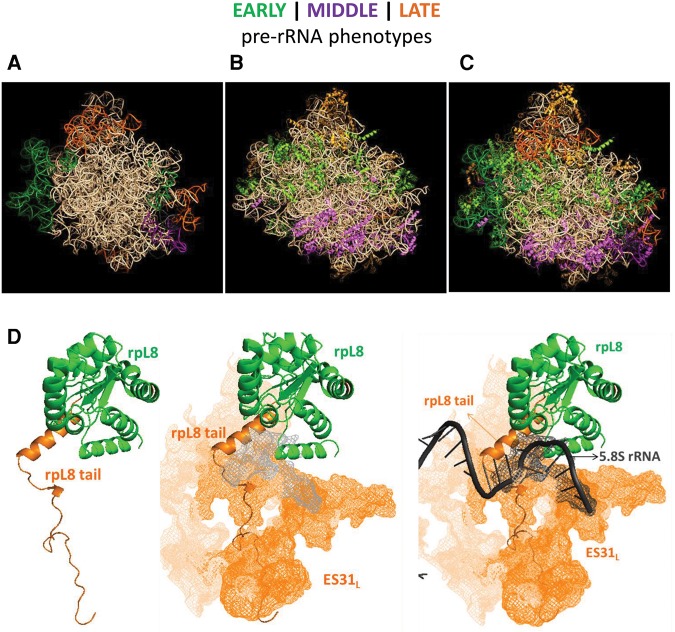
Putting the results together, we have discussed additional information about each ESL individually in Supplemental Data. We have highlighted relevant previous information about that particular ESL and interpreted the results of our ESL mutations in light of the contacts they make with r-proteins in mature subunits. Having developed a catalog of ES phenotypes, we sought to understand whether the reported phenomenon of clustering of r-proteins according to their function in successive steps of ribosome biogenesis (Gamalinda et al. 2014) extends to ES. To visualize their locations, we color-coded early, middle, and late-acting ES and r-proteins in shades of green, purple, or orange, respectively (Fig. 4). Recognizing that ES are localized only to the periphery of the solvent interface of the large subunit, as opposed to r-proteins that are distributed more evenly, we observed that the early-acting ES localize to the equatorial belt, the middle-acting ES localize near the bottom third of the subunit, and the late-acting ES localize near the central protuberance (Fig. 4A). This pattern of ES clustering according to their function in successive steps of ribosome biogenesis is reminiscent of the functional clustering of r-proteins reported previously (Fig. 4B,C).
FIGURE 4.
A model for ES functions and neighborhood-specific effects in ribosome biogenesis. (A) Structural model of the large ribosomal subunit showing ES color-coded according to the pre-rRNA processing steps in which they function. Green-colored ES are necessary for 27SA processing (early steps) and localize to the “equatorial belt.” Purple-colored ES participate in 27S processing, but not specifically 27SA processing (middle steps). These ES localize near the bottom third of the solvent side of the ribosome, near the polypeptide exit tunnel. Orange-colored ES participate in 7S pre-rRNA processing (late steps) and localize near the central protuberance. (B) The same color scheme (early, green; middle, purple; late, orange) is used to represent previously known r-protein depletion phenotypes. (C) Superimposition of A on B. See Results and Discussion for details of the model. (D) A case study of eukaryote-specific r-protein-rRNA elements. The left panel shows the body of r-protein L8 (in green, consistent with its “early” role in ribosome assembly) and its eukaryote-specific N-terminal extension (in orange, consistent with its “late” role in ribosome assembly). The center panel shows the contact established between the “late”-acting ES31L (shown as an orange-colored mesh) and the extension of L8. The right panel shows the 3′ end of 5.8S rRNA (dark gray) to denote the terminal site of pre-rRNA processing of ITS2 by the exosome. ES31L is in close contact with the 3′ end of 5.8S rRNA.
In order to explore this further, we have represented each of the ES studied, along with the r-proteins that make contact with that particular ES, in mature yeast ribosomes (Supplemental Fig. 4A–C; Supplemental Table 1; Ben-Shem et al. 2011). It is apparent that in the majority of cases, the phenotype caused by deletion of a particular ES corresponds to the phenotype caused by depletion of the r-proteins that it contacts.
This functional clustering of both rRNA and r-proteins on the solvent interface has interesting implications. First, it adds credence to the possibility that as the ribosome is constructed, there may be neighborhood-specific effects on pre-rRNA processing, folding, and stability. A hierarchy of r-protein assembly exists where r-proteins that participate in specific steps cluster to specific neighborhoods. Our results suggest that this hierarchy might extend to RNA–protein neighborhoods, not simply r-proteins, in the assembling ribosome (Shajani et al. 2011; Woodson 2011; Gamalinda et al. 2014).
A case study of ES31L
Perhaps the only glaring exception to the neighborhood-specific separation of ribosome assembly functions of ES arises from the phenotype of the deletion of ES31L. Although the location of ES31L in the “equatorial belt” predicts that it might have an early-acting phenotype, in fact, we observed a predominantly late phenotype for ES31ΔL. To better understand this apparent discrepancy, we looked at this area of the ribosome in more detail to build a model for the function of ES31L based on its contact with r-protein L8 (Fig. 4D). Depletion of L8 causes an early phenotype (Jakovljevic et al. 2012), consistent with its location in the equatorial belt. On the other hand, deletion of the eukaryote-specific N-terminal extension of L8 that intimately contacts ES31L, causes a later, 7S pre-rRNA accumulation phenotype (Tutuncuoglu et al. 2016), which is similar to the phenotype that ES31ΔL exhibits. Thus, what initially appeared to be an exception to the neighborhood-specific phenotype rule, turned out to be an informative sign about how eukaryote-specific RNA–protein elements that contact each other, exhibit similar functions in ribosome assembly. Strikingly, the 3′ end of 5.8S rRNA, where processing of 7S pre-rRNA terminates during late steps of 60S subunit biogenesis, is encompassed by ES31L. This indicates that potential proximity effects may be at play, resulting in a late processing phenotype in this mutant.
A potential model for ES function: ES may have coevolved as scaffolds for eukaryote-specific protein elements in the assembling ribosome
In this study, we observed that there is functional similarity in ribosome biogenesis between ES rRNA and neighboring r-proteins. Furthermore, it has been previously pointed out that eukaryotic RNA elements and eukaryotic r-proteins and their extensions localize together on the solvent-exposed side of the ribosome and make numerous contacts with each other (Ben-Shem et al. 2011). Several eukaryote-specific assembly factors such as Rlp7, Arx1, Rrp5, and Nop7 bind to ES (Granneman et al. 2011; Bradatsch et al. 2012; Babiano et al. 2013; Dembowski et al. 2013; Lebaron et al. 2013). Combined with our observation about similar functions in ribosome assembly for eukaryote-specific ES31ΔL and the eukaryote-specific extension of L8, these lines of evidence raise the possibility that eukaryote-specific protein–RNA elements may have coevolved, and perform similar functions in the cell. It is tempting to suggest that ES may have coevolved with the r-proteins in order to provide a scaffold for binding of many eukaryote-specific protein elements, including assembly factors, r-proteins and their extensions, and translation factors. Once extant technical challenges are overcome, mutations in eukaryote-specific regions of proteins in tandem with eukaryote-specific RNA elements may lead to interesting insights about the function of these eukaryotic elements.
In addition, using this study as a starting point and adopting an evolutionary standpoint, it would be interesting to replace yeast ES with those of other eukaryotes and assay for rescue of those phenotypes with corresponding eukaryotic proteins, to understand how these ES have continued to evolve. Also, whether the precise functions of specific ES in biogenesis and translation are conserved in other eukaryotes remains to be investigated. Further, having established that these ES participate in ribosome biogenesis, it is tempting to ask if rRNA ES might play more direct eukaryote-specific roles in pre-rRNA processing or folding, in addition to serving as eukaryote-specific scaffolds.
In summary, we have shown that most eukaryote-specific rRNA ES of the large ribosomal subunit are indispensable and participate in various steps of ribosome biogenesis. While complementing a vast body of literature about the functions of r-proteins, this study settles the long-standing debate about the dispensability of ES and identifies a catalog of specific functions of these eukaryote-specific insertions in ribosome biogenesis. These results provide a framework for further investigation of eukaryote-specific features of ribosome assembly and function and pave the way for many further studies of the evolution, structure, biogenesis and function of the ribosome.
MATERIALS AND METHODS
Construction and growth of yeast strains
As the parent strain, we used NOY504 (JWY10423, MATa rrn4::LEU2 ade2-101 ura3-1 trp1-1 leu2-3,112 his3-11 canl-100), which lacks the Rpa12 subunit of RNA polymerase 1, rendering it temperature sensitive for growth (a gift from the Nomura laboratory) (Nogi et al. 1993). This strain was transformed with the plasmid pWL160 (a gift from the M. Fournier laboratory) encoding WT RNA as the positive control, or the pRS314 vector as the “no rDNA” negative control.
Handling of the NOY504-based strains was done essentially as described previously (Henry et al. 1994). Briefly, small cultures of the controls and the mutant yeast strains were grown to log phase at 25°C in galactose minimal medium. Cultures were then diluted, and endogenous ribosomal RNA was depleted by shifting cultures to 37°C for 6 h. The mid log-phase cells were then harvested by centrifugation at 5000 rpm for 5 min at 4°C for further experiments.
Mutagenesis
The size and location of ES deletions were devised from definitions of ES insertion regions from Gerbi (1996) and secondary structures derived from comparative analysis (Gutell et al. 1993). Mutagenesis of the ES regions (Supplemental Table 1) was carried out in the pWL160 plasmid using the Quikchange Lightning kit from Agilent Technologies. Mutagenesis was verified by DNA sequencing. Sequences for oligonucleotides used in this study are available upon request.
Growth assays of ES mutants
Small 5-mL cultures of the control and mutant strains were grown in C-Trp+Gal media overnight. Tenfold serial dilutions of yeast strains bearing either the controls (the positive control is JWY10430: NOY504 with pWL160, and the negative control is JWY10429: NOY504 with pRS314) or one of the 14 mutant plasmids from Supplemental Table 1 (JWY10431-JWY10444) were spotted onto YEPD solid medium starting with an OD of 1 and grown either at 25°C and photographed after 3 d, or grown at 37°C and photographed after 5 d.
Sucrose gradient analysis
Gradient fractionation of preribosomes, ribosomes, and polyribosomes, and subsequent analysis, were carried out as described (Talkish et al. 2014). The image was digitized using the PlotDigitizer software. The area under the curve for each peak was calculated using the trapezoidal rule, and subsequently, 60S/40S ratios were determined using Excel.
RNA analysis
RNA extraction and subsequent Northern blotting assays were performed as described previously (Horsey et al. 2004). Assaying steady state pre-rRNA processing by primer extension was carried out using the Transcriptor reverse transcriptase (Roche) as described (Talkish et al. 2014).
Illustrations
The PDB file of the crystal structure of yeast ribosomes (PDB ID: 4V88) (Ben-Shem et al. 2011) was manipulated using PyMol software to produce the structural images (The PyMOL molecular graphics system, version 1.8, Schrodinger LLC). RiboVision software (Bernier et al. 2014) was used for generation of some of the secondary structures shown here.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health to J.L.W. (GM028301) and the David Scaife Family Charitable Foundation (141RA01). We thank the members of the Woolford laboratory for critical reading of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.056705.116.
REFERENCES
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. 2013. Structures of the human and Drosophila 80S ribosome. Nature 497: 80–85. [DOI] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. 2010. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc Natl Acad Sci 107: 19748–19753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, Badis G, Saveanu C, Namane A, Doyen A, Diaz-Quintana A, Jacquier A, Fromont-Racine M, de la Cruz J. 2013. Yeast ribosomal protein L7 and its homologue Rlp7 are simultaneously present at distinct sites on pre-60S ribosomal particles. Nucleic Acids Res 41: 9461–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. 2001. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107: 361–372. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334: 1524–1529. [DOI] [PubMed] [Google Scholar]
- Bernier CR, Petrov AS, Waterbury CC, Jett J, Li F, Freil LE, Xiong X, Wang L, Migliozzi BL, Hershkovits E, et al. 2014. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss 169: 195–207. [DOI] [PubMed] [Google Scholar]
- Bradatsch B, Leidig C, Granneman S, Gnadig M, Tollervey D, Bottcher B, Beckmann R, Hurt E. 2012. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol 19: 1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CG, Tague BW, Ware VC, Gerbi SA. 1984. Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res 12: 6197–6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ. 2008. Chapter 12. Analysis of nonfunctional ribosomal RNA decay in Saccharomyces cerevisiae. Methods Enzymol 449: 239–259. [DOI] [PubMed] [Google Scholar]
- Cote CA, Peculis BA. 2001. Role of the ITS2-proximal stem and evidence for indirect recognition of processing sites in pre-rRNA processing in yeast. Nucleic Acids Res 29: 2106–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembowski JA, Ramesh M, McManus CJ, Woolford JL Jr. 2013. Identification of the binding site of Rlp7 on assembling 60S ribosomal subunits in Saccharomyces cerevisiae. RNA 19: 1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, Lin L, Woolford JL Jr. 2014. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev 28: 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Ayub MJ, Levin MJ, Frank J. 2005. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc Natl Acad Sci 102: 10206–10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi S. 1996. Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In Ribosomal RNA structure, evolution, processing, and function in protein biosynthesis (ed. Zimmermann RA, Dahlberg AE), pp. 71–87. Telford-CRC Press, Boca Raton, FL. [Google Scholar]
- Gonzalez IL, Gorski JL, Campen TJ, Dorney DJ, Erickson JM, Sylvester JE, Schmickel RD. 1985. Variation among human 28S ribosomal RNA genes. Proc Natl Acad Sci 82: 7666–7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez IL, Sylvester JE, Schmickel RD. 1988. Human 28S ribosomal RNA sequence heterogeneity. Nucleic Acids Res 16: 10213–10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Tollervey D. 2011. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J 30: 4006–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson JH, Sogin ML, Wollett G, Hollingdale M, de la Cruz VF, Waters AP, McCutchan TF. 1987. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science 238: 933–937. [DOI] [PubMed] [Google Scholar]
- Gutell RR, Gray MW, Schnare MN. 1993. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res 21: 3055–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R. 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427: 808–814. [DOI] [PubMed] [Google Scholar]
- Han H, Schepartz A, Pellegrini M, Dervan PB. 1994. Mapping RNA regions in eukaryotic ribosomes that are accessible to methidiumpropyl-EDTA•Fe(II) and EDTA•Fe(II). Biochemistry 33: 9831–9844. [DOI] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CU, Pestova TV, Frank J. 2013. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 153: 1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassouna N, Michot B, Bachellerie JP. 1984. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res 12: 3563–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J 13: 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsey EW, Jakovljevic J, Miles TD, Harnpicharnchai P, Woolford JL Jr. 2004. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA 10: 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, Milkereit P, Woolford JL Jr. 2012. Ribosomal proteins L7 and L8 function in concert with six A3 assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA 18: 1805–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeninga RE, Van Delft Y, de Graaff-Vincent M, Dirks-Mulder A, Venema J, Raue HA. 1997. Variable regions V13 and V3 of Saccharomyces cerevisiae contain structural features essential for normal biogenesis and stability of 5.8S and 25S rRNA. RNA 3: 476–488. [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Melnikov S, Garreau de Loubresse N, Ben-Shem A, Iskakova M, Urzhumtsev A, Meskauskas A, Dinman J, Yusupova G, Yusupov M. 2012. Crystal structure of the 80S yeast ribosome. Curr Opin Struct Biol 22: 759–767. [DOI] [PubMed] [Google Scholar]
- Kos M, Tollervey D. 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron S, Segerstolpe A, French SL, Dudnakova T, de Lima Alves F, Granneman S, Rappsilber J, Beyer AL, Wieslander L, Tollervey D. 2013. Rrp5 binding at multiple sites coordinates pre-rRNA processing and assembly. Mol Cell 52: 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WQ, Fournier MJ. 1997. Synthesis of functional eukaryotic ribosomal RNAs in trans: development of a novel in vivo rDNA system for dissecting ribosome biogenesis. Proc Natl Acad Sci 94: 2864–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol 19: 560–567. [DOI] [PubMed] [Google Scholar]
- Michot B, Bachellerie JP. 1987. Comparisons of large subunit rRNAs reveal some eukaryote-specific elements of secondary structure. Biochimie 69: 11–23. [DOI] [PubMed] [Google Scholar]
- Michot B, Qu LH, Bachellerie JP. 1990. Evolution of large-subunit rRNA structure. The diversification of divergent D3 domain among major phylogenetic groups. Eur J Biochem 188: 219–229. [DOI] [PubMed] [Google Scholar]
- Musters W, Venema J, van der Linden G, van Heerikhuizen H, Klootwijk J, Planta RJ. 1989. A system for the analysis of yeast ribosomal DNA mutations. Mol Cell Biol 9: 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W, Boon K, van der Sande CA, van Heerikhuizen H, Planta RJ. 1990. Functional analysis of transcribed spacers of yeast ribosomal DNA. EMBO J 9: 3989–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W, Concalves PM, Boon K, Raue HA, van Heerikhuizen H, Planta RJ. 1991. The conserved GTPase center and variable region V9 from Saccharomyces cerevisiae 26S rRNA can be replaced by their equivalents from other prokaryotes or eukaryotes without detectable loss of ribosomal function. Proc Natl Acad Sci 88: 1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y, Yano R, Dodd J, Carles C, Nomura M. 1993. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol Cell Biol 13: 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis BA, Greer CL. 1998. The structure of the ITS2-proximal stem is required for pre-rRNA processing in yeast. RNA 4: 1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Galan O, Garcia-Gomez JJ, Kressler D, de la Cruz J. 2015. Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae. RNA Biol 12: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL Jr. 2011. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J 30: 4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. 2011. Assembly of bacterial ribosomes. Annu Rev Biochem 80: 501–526. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell 107: 373–386. [DOI] [PubMed] [Google Scholar]
- Sweeney R, Yao MC. 1989. Identifying functional regions of rRNA by insertion mutagenesis and complete gene replacement in Tetrahymena thermophila. EMBO J 8: 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney R, Chen L, Yao MC. 1994. An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol Cell Biol 14: 4203–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, Campbell IW, Sahasranaman A, Jakovljevic J, Woolford JL Jr. 2014. Ribosome assembly factors Pwp1 and Nop12 are important for folding of 5.8S rRNA during ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 34: 1863–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, Harvey SC, Frank J. 2009. Comprehensive molecular structure of the eukaryotic ribosome. Structure 17: 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutuncuoglu B, Jakovljevic J, Wu S, Gao N, Woolford JL Jr. 2016. The N-terminal extension of yeast ribosomal protein L8 is involved in two major remodeling events during late nuclear stages of 60S ribosomal subunit assembly. RNA 10.1261/rna.055798.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman GM, Klootwijk J, de Regt VC, Planta RJ, Branlant C, Krol A, Ebel JP. 1981. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res 9: 6935–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware VC, Tague BW, Clark CG, Gourse RL, Brand RC, Gerbi SA. 1983. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res 11: 7795–7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AP, White W, McCutchan TF. 1995. The structure of the large subunit rRNA expressed in blood stages of Plasmodium falciparum. Mol Biochem Parasitol 72: 227–237. [DOI] [PubMed] [Google Scholar]
- Woodson SA. 2011. RNA folding pathways and the self-assembly of ribosomes. Acc Chem Res 44: 1312–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.