Abstract
Objective:
To describe the phenotypes in 2 families with vaccinia-related kinase 1 (VRK1) mutations including one novel VRK1 mutation.
Methods:
VRK1 mutations were found by whole exome sequencing in patients presenting with motor neuron disorders.
Results:
We identified pathogenic mutations in the VRK1 gene in the affected members of 2 families. In family 1, compound heterozygous mutations were identified in VRK1, c.356A>G; p.H119R, and c.1072C>T; p.R358*, in 2 siblings with adult onset distal spinal muscular atrophy (SMA). In family 2, a novel VRK1 mutation, c.403G>A; p.G135R and c.583T>G; p.L195V, were identified in a child with motor neuron disease.
Conclusions:
VRK1 mutations can produce adult-onset SMA and motor neuron disease in children without pontocerebellar hypoplasia.
The spinal muscular atrophies (SMA) are an inherited group of conditions characterized by motor neuron loss in the spinal cord and brainstem, causing proximal and distal muscle weakness and atrophy. While 96% of autosomal recessive forms of SMA are associated with mutations in SMN1,1 there are a significant minority of cases with non-SMN1-related phenotypes and diverse genetic causes.2 Similarly, progressive motor neuron disorders involving upper and lower neurons are also clinically and genetically heterogeneous, with overlap among amyotrophic lateral sclerosis (ALS), SMA, and distal hereditary motor neuronopathy and pyramidal tract signs (dHMN + PS).3
The VRK1 gene encodes a ubiquitously expressed serine kinase with a role in embryonic cortical neuronal proliferation and migration as demonstrated in knockout mouse models.4 In cellular models, VRK1 has been shown to be associated with cell cycle regulation, histone modification, DNA repair responses, and disruption of RNA processing,5–7 common pathophysiologic themes underlying motor neuron diseases.
A nonsense mutation, c.1072C>T (p.R358*), in VRK1 has previously been described to cause an infantile onset SMA phenotype associated with pontocerebellar hypoplasia and death in infancy or childhood when present in a homozygous state in a consanguineous family of Ashkenazi Jewish origin.8
Another patient presenting with a complex axonal motor and sensory neuropathy accompanied by microcephaly and cerebral dysgenesis carried the same homozygous c.1072C>T (p.R358*) nonsense mutation.9 In the same study, pathogenic compound heterozygous VRK1 variants, c.G706A (p.V236M) and c.G266A (p.R89Q), have been identified in patients showing a similar distal symmetric polyneuropathy and microcephaly.9
Analysis of a sporadic patient with adult-onset motor neuron disease revealed compound heterozygous variants c.356A>G (p.H119R) and c.961C>T (p.R321C).10
Here we report VRK1 compound heterozygous mutations in 2 families with adult-onset SMA and childhood motor neuron disease.
METHODS
Genetic analysis.
Prior to this study, the affected index patient in family 1 was prescreened for SMN1 mutations. Whole exome sequencing (WES) was performed by Axeq Technologies (Seoul, South Korea) using the Illumina (San Diego, CA) TrueSeq kit to generate sequencing data. Annotated WES data were examined for variants in genes selected for relevance to the phenotype. Preceding Sanger sequencing analysis of SETX, ALS2, SOD1, LMNA, MFN2, and TRPV4 did not identify any pathogenic mutations in the affected individual of family 2; subsequently WES was carried out at the University of Washington Genome Center. Data from VCF files were analyzed using the in-house Seave analysis pipeline (Kinghorn Centre for Clinical Genomics, Sydney, Australia) under homozygous and potentially compound heterozygote models. The effect of amino acid substitutions for sequence variants was assessed using the software SIFT11 and Polyphen2.12
Sanger sequencing was performed to confirm the mutations in the probands and for segregation analysis in family members.
Clinical phenotype: Family 1.
Two affected siblings were from a nonconsanguineous family of Ashkenazi Jewish origin (figure 1). The proband reached normal motor milestones in childhood and had normal growth parameters. In retrospect, she had difficulties performing sporting activities from the age of 15 years. She presented to medical attention at the age of 35 years with progressive lower limb weakness requiring the use of a walking stick. The impairment of motor function was progressive and at age 56 years she required a scooter and long arm crutch to mobilize. There were no significant ocular, bulbar, or sensory symptoms seen over the course of the illness and she had normal intellectual function.
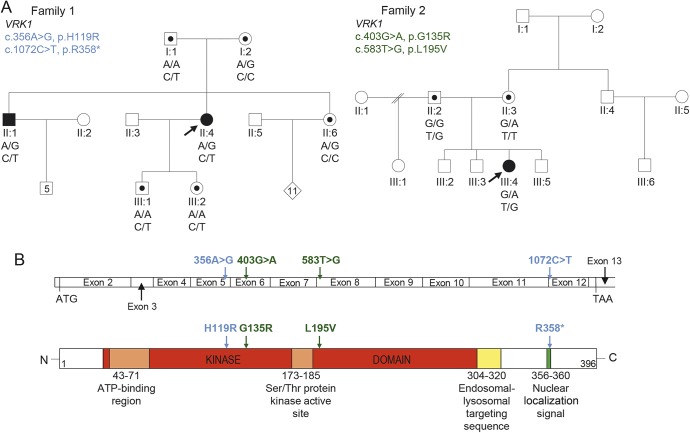
Figure 1. Compound heterozygous VRK1 mutations in adult-onset distal spinal muscular atrophy and childhood amyotrophic lateral sclerosis.
(A) Segregation of compound heterozygous sequence variants in family 1 and 2. Solid symbols indicate affected individuals. Symbols with dots illustrate carriers. Arrows indicate proband. Genotypes are demonstrated below tested individuals. (B) Schematic graph of the VRK1 coding region and the corresponding VRK1 protein shows the position of mutations identified in family 1 (blue) and 2 (green).
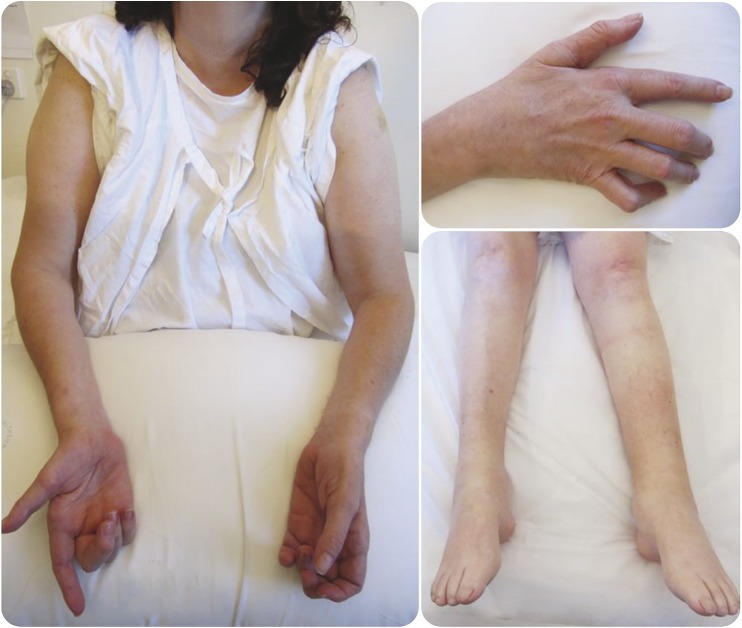
Clinical examination was consistent with a profound symmetric distal muscle wasting affecting the upper and lower limbs symmetrically (figure 2). There was a predominantly distal pattern of weakness of greater severity in the lower than upper limbs, with relative preservation of proximal strength. Reflexes were absent in the lower limbs but symmetrically brisk in the upper limbs. There was no evidence of cerebellar ataxia.
Figure 2. Clinical images of the spinal muscular atrophy proband.
Clinical images demonstrate symmetrical, distal wasting in legs and arms.
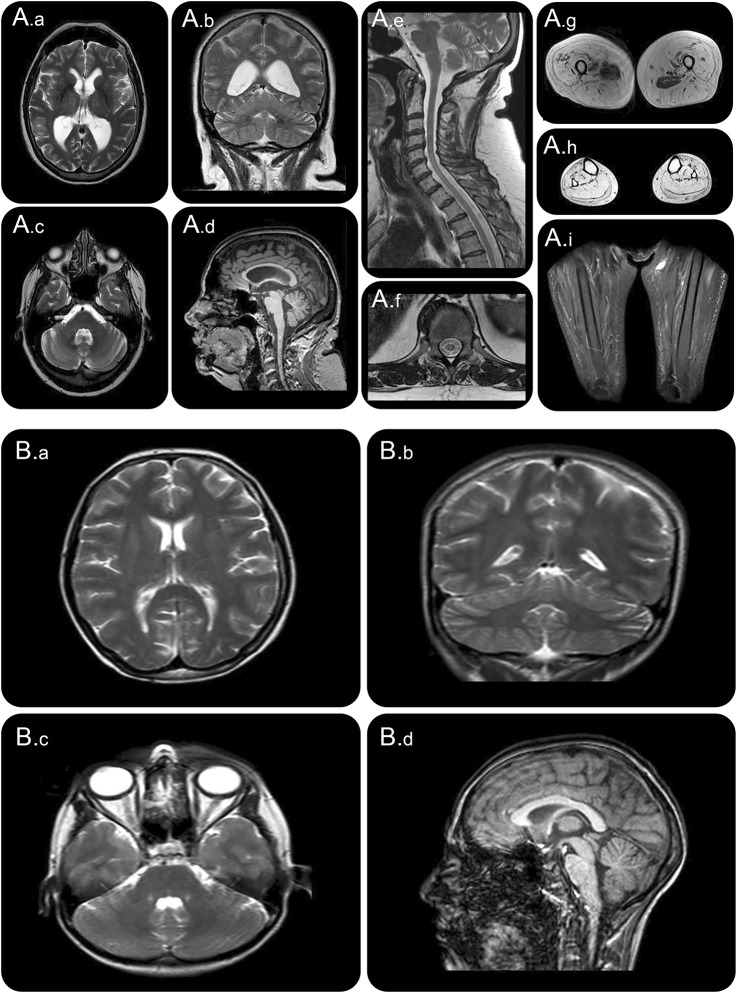
Neurophysiology showed preservation of sensory responses but severe and progressive reduction in compound motor action potential amplitudes with relative preservation of conduction velocities. EMG identified evidence of chronic and active denervation consistent with motor neuron pathology. Motor evoked potentials from the upper limbs were prolonged with a central component suggesting upper motor neuron involvement in the disease, which is consistent with MRI findings of atrophy of the spinal cord (figure 3A). A brain MRI showed nonspecific mild to moderate generalized atrophy. Pontocerebellar hypoplasia or other developmental anomalies as reported in other patients previously described with VRK1 mutations were absent (figure 3). Respiratory muscle weakness resulted in reduced mean inspiratory and expiratory pressures and a nonprogressive elevation of blood gas CO2. Other respiratory parameters had been stable for 5 years with no requirement for assisted ventilation. Serum creatine kinase was not elevated and an MRI of the lower limbs showed profound proximal and distal muscle atrophy with fat replacement. There was relative preservation of the iliopsoas and adductor compartments bilaterally (figure 3).
Figure 3. MRI of the spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS) proband.
(A) MRI of proband in family 1 (SMA). (A.a–A.c) T2 images and (A.d) sagittal T1 image demonstrate mild nonspecific generalized atrophy and absence of pontocerebellar hypoplasia. (A.e) T2 image sagittal and (A.f) transverse spinal image demonstrate cord atrophy. (A.g–A.i) T2 images of upper thigh, calf, and STIR sequence of thigh demonstrate diffuse fatty replacement of muscle and relative sparing of adductor compartments. (B) MRI of proband in family 2 (ALS). (B.a–B.c) T2 images and (B.d) sagittal T1 image demonstrate absence of pontocerebellar hypoplasia.
The proband has a similarly affected older brother with a slightly later onset of progressive motor weakness in the late teenage years, progression to requiring a walking stick in his early 30s, and the requirement for an articulated walking device and wheelchair in his late 40s. Neurophysiology showed a chronic active denervating process.
Clinical phenotype: Family 2.
The patient was a 3-year-old girl with short stature and microcephaly who presented with initial distal lower limb muscle weakness and amyotrophy combined with pathologically brisk deep tendon reflexes and normal sensation. Weakness progressed in a caudalocephalic direction, with loss of ambulation by 10 years of age and complete dependence for activities of daily living at 18 years. Severe thoracolumbar scoliosis required surgical rod insertion at age 14 years. Respiratory dysfunction developed with dyspnea and weak cough accompanied by reduction of forced vital capacity to below 40% predicted. Speech became progressively hoarse and soft. Ocular signs of brainstem or cerebellar dysfunction were absent. Skeletal and endocrine assessments for short stature did not identify any specific etiology.
Neurophysiology studies at age 11 years demonstrated severe reduction in compound muscle action potential amplitudes combined with chronic and active denervation, consistent with motor neuropathy/neuronopathy. Sensory responses were initially preserved but subsequently were reduced on repeat testing at age 19 years. Muscle histopathology showed marked atrophy of entire fascicles, confirming a significant neurogenic process.
Brain and spine MRI at 7 and 16 years were normal. Brain MRI at 20 years demonstrated normal structure (figure 3B). The clinical possibility of juvenile ALS, with the presence of upper and lower motor neuron signs in more than 2 spinal regions and clinical progression, prompted Sanger sequencing as described above.
Standard protocol approvals, registrations, and patient consents.
Participating individuals were enrolled through the Neurogenetics Clinic Concord Hospital and the Paediatric Neuromuscular clinic at Sydney Children's Hospital Randwick. Genomic DNA was isolated from peripheral blood or saliva. These procedures were performed with informed consent according to protocols approved by the Human Ethics Committees of Sydney Local Health District, Concord Hospital (HREC/11/CRGH/105), and South Eastern Sydney Local Health District (HREC/13/POWH/203), Australia.
RESULTS
Testing identified compound heterozygous VRK1 mutations NM_003384.2 (VRK1):c.(356A>G); (1072C>T) (p.[H119R];[R358*]) in the individuals with SMA in family 1 and NM_003384.2 (VRK1):c.(403 G>A); (583T>G) (p.[G135R];[L195V]) in the patient with juvenile ALS in family 2. No pathogenic variants were identified in other genes associated with SMA or ALS in either patient or genes associated with short stature.
DISCUSSION
The present cases with adult-onset distal SMA and childhood motor neuron disease broaden the clinical spectrum of patients with VRK1 mutations. The identification of VRK1 mutations among these various motor phenotypes, with clinical overlap among juvenile ALS, dHMN + PS, and SMA, may serve to further unite concepts of pathogenesis among motor neuron disorders.
The results for family 1 are consistent with an autosomal recessive mode of inheritance with segregation of both VRK1 variants with the disease (figure 1A). The c.1072C>T mutation is well-characterized and described as pathogenic in the Ashkenazi Jewish population when in a homozygous state.8 The c.356A>G variant (ESP frequency <0.01%) encodes a histidine residue that is conserved across multiple vertebrate species and is in the protein kinase domain of the VRK1 gene (figure 1B). It was assessed with in silico tools (Polyphen2 score 1 and 0.999, PROVEAN13 score −3.804) and is classified as damaging. The c.356A>G missense substitution was reported in one recent case of a 32-year-old man with a 5-year history of ALS without cerebellar hypoplasia.9 This sequence variant is most likely to be pathogenic as it is present in both the affected members of family 1 and a reported sporadic ALS case.10
In family 2, both substituted amino acids are highly conserved across multiple species and the frequency of the novel c.583T>G mutation in public databases was 2 in 121382. Online database tools supported pathogenicity for c.403 G>A and c.583T>G mutations (PolyPhen2 scores 1 and 0.997, probably damaging; SIFT score 0 and 0, deleterious; and CADD score 22.6 and 19.05, respectively). The mutations were Sanger sequenced for confirmation and each parent was found to carry one of the variants, supporting autosomal recessive inheritance (figure 1).
Three-dimensional structural protein analysis of the p.Gly135Arg VRK1 variant with the HOPE14 Protein structure analysis suite database suggested a difference in charge, size, and hydrophobicity resulting in disturbances in local structure and protein misfolding. Similarly, the p.Leu195Val VRK1 variant altered the amino acid size, causing an empty space in the core of the protein.
Other phenotypes have recently been associated with VRK1 mutations. Gonzaga-Jauregui et al.9 described childhood-onset distal sensory motor axonal neuropathy with microcephaly with a simplified gyral pattern and in a second case there was a similar motor and sensory axonal neuropathy with an underdeveloped cerebellar vermis.
The latter case and this report suggest that VRK1 is not only important in development of motor system architecture, but are also important in the longer term in maintenance of motor neurons. VRK1 mutations should therefore be considered in the differential diagnosis of patients presenting with adult-onset SMA and childhood motor neuron disease.
GLOSSARY
- ALS
amyotrophic lateral sclerosis
- dHMN + PS
distal hereditary motor neuronopathy and pyramidal tract signs
- SMA
spinal muscular atrophies
- WES
whole exome sequencing
AUTHOR CONTRIBUTIONS
Marion Stoll: organized and performed genetic testing on proband and family members (family 1), performed bioinformatics analysis and interpretation of WES data, writing of the manuscript. Hooi Ling Teoh: collected data of the proband (family 2), writing of the manuscript. James H.F. Lee: clinically examined and interviewed the proband (family 1), writing of the manuscript. Stephen Reddel: clinically examined and performed neurophysiology on proband (family 1), critical revision of manuscript. Ying Zhu: supervised the creation of the bioinformatic pipeline for genomic analysis, assisted with bioinformatics analysis and interpretation of whole exome data, critical revision of manuscript. Michael Buckley: organized and performed genetic testing on proband and family members (family 2), interpretation of variant results, critical revision of manuscript. Hugo Sampaio: clinically examined and interviewed the proband (family 2), critical revision of manuscript. Tony Roscioli: obtained research funding for genomic sequencing, developed genomic consent forms for patient enrolment, supervised the creation of the bioinformatic pipeline for genomic analysis, performed bioinformatics analysis and interpretation of WES data, critical revision of manuscript. Michelle Farrar: provided clinical and scientific direction for the manuscript, drafted, reviewed and approved the final manuscript. Garth Nicholson: provided clinical and scientific direction for the manuscript, planned, drafted, reviewed, and finalized the manuscript, clinically examined, characterized, and interviewed the proband (family 1).
STUDY FUNDING
Supported by Motor Neurone Disease Research Institute of Australia. No industry, governmental, or institutional funding.
DISCLOSURE
M. Stoll reports no disclosures relevant to the manuscript. H. Teoh received scholarship support from the Thyne Reid Foundation. J. Lee, S. Reddel, Y. Zhu, M. Buckley, H. Sampaio, T. Roscioli, M. Farrar, and G. Nicholson report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Lefebvre S, Burglen L, Reboullet S, et al. . Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995;80:155–165. [DOI] [PubMed] [Google Scholar]
- 2.Farrar MA, Kiernan MC. The genetics of spinal muscular atrophy: progress and challenges. Neurotherapeutics 2015;12:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Jonghe P, Auer-Grumbach M, Irobi J, et al. . Autosomal dominant juvenile amyotrophic lateral sclerosis and distal hereditary motor neuronopathy with pyramidal tract signs: synonyms for the same disorder? Brain 2002;125:1320–1325. [DOI] [PubMed] [Google Scholar]
- 4.Vinograd-Byk H, Sapir T, Cantarero L, et al. . The spinal muscular atrophy with pontocerebellar hypoplasia gene VRK1 regulates neuronal migration through an amyloid-beta precursor protein-dependent mechanism. J Neurosci 2015;35:936–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzano M, Sanz-Garcia M, Monsalve DM, Moura DS, Lazo PA. VRK1 chromatin kinase phosphorylates H2AX and is required for foci formation induced by DNA damage. Epigenetics 2015;10:373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vega FM, Sevilla A, Lazo PA. p53 Stabilization and accumulation induced by human vaccinia-related kinase 1. Mol Cell Biol 2004;24:10366–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valbuena AL-SI, Lazo PA. Human VRK1 is an early response gene and its loss causes a block in cell cycle progression. PLoS One 2008;3:e1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renbaum P, Kellerman E, Jaron R, et al. . Spinal muscular atrophy with pontocerebellar hypoplasia is caused by a mutation in the VRK1 gene. Am J Hum Genet 2009;85:281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzaga-Jauregui C, Lotze T, Jamal L, et al. . Mutations in VRK1 associated with complex motor and sensory axonal neuropathy plus microcephaly. JAMA Neurol 2013;70:1491–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen TP, Biliciler S, Wiszniewski W, Sheikh K. Expanding phenotype of VRK1 mutations in motor neuron disease. J Clin Neuromuscul Dis 2015;17:69–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073–1081. [DOI] [PubMed] [Google Scholar]
- 12.Adzhubei IA, Schmidt S, Peshkin L, et al. . A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015;31:2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases: an e-Science approach with life scientist friendly interfaces. BMC Bioinformatics 2010;11:548. [DOI] [PMC free article] [PubMed] [Google Scholar]