Abstract
Background
Salmon lice, Lepeophtheirus salmonis (Copepoda: Caligidae), are highly important ectoparasites of farmed and wild salmonids, and cause multi-million dollar losses to the salmon aquaculture industry annually. Salmon lice display extensive sexual dimorphism in ontogeny, morphology, physiology, behavior, and more. Therefore, the identification of transcripts with differential expression between males and females (sex-biased transcripts) may help elucidate the relationship between sexual selection and sexually dimorphic characteristics.
Results
Sex-biased transcripts were identified from transcriptome analyses of three L. salmonis populations, including both Atlantic and Pacific subspecies. A total of 35-43 % of all quality-filtered transcripts were sex-biased in L. salmonis, with male-biased transcripts exhibiting higher fold change than female-biased transcripts. For Gene Ontology and functional analyses, a consensus-based approach was used to identify concordantly differentially expressed sex-biased transcripts across the three populations. A total of 127 male-specific transcripts (i.e. those without detectable expression in any female) were identified, and were enriched with reproductive functions (e.g. seminal fluid and male accessory gland proteins). Other sex-biased transcripts involved in morphogenesis, feeding, energy generation, and sensory and immune system development and function were also identified. Interestingly, as observed in model systems, male-biased L. salmonis transcripts were more frequently without annotation compared to female-biased or unbiased transcripts, suggesting higher rates of sequence divergence in male-biased transcripts.
Conclusions
Transcriptome differences between male and female L. salmonis described here provide key insights into the molecular mechanisms controlling sexual dimorphism in L. salmonis. This analysis offers targets for parasite control and provides a foundation for further analyses exploring critical topics such as the interaction between sex and drug resistance, sex-specific factors in host-parasite relationships, and reproductive roles within L. salmonis.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2835-7) contains supplementary material, which is available to authorized users.
Keywords: Copepoda, Evolution, Lepeophtheirus salmonis, Reproduction, Sea lice, Sex-bias, Sexual dimorphism, Transcriptomics
Background
Sexual dimorphism describes the phenotypic differences between sexes of the same species. It is ubiquitous across the animal kingdom and is favored through a combination of sexual selection, intersexual competition for resources, and fundamental differences in reproductive roles [1–3]. Genes overexpressed in one sex relative to the other are known as sex-biased genes, and include genes expressed in both sexes (but higher in one) or genes expressed in only one sex (sex-specific; reviewed in [4, 5]). A large proportion, often greater than 50 %, of genes exhibit sex-biased expression in many species including fruit flies Drosophila spp. [6, 7], the nematode Caenorhabitis elegans [8], parasitic flatworms Schistosoma spp. [9, 10], the water flea Daphnia pulex [11], the African clawed frog Xenopus laevis [12], the songbirds Taeniopygia guttata and Sylvia communis [13], the olive flounder Paralichthys olivaceus [14], the mouse Mus musculus [15], and humans Homo sapiens [16, 17]. This trend is largely driven by expression differences in the gonad. As such, transcriptome profiling is a highly useful approach to understand the mechanisms underlying sexual dimorphism and reproduction.
Crustaceans are one of the most diverse animal taxa, comprising more than 850 families with approximately 67,000 species [18, 19]. They are ecologically important, serving essential roles in the food chain and primary production in marine ecosystems [20]. Furthermore, crustaceans play important roles in aquaculture as both farmed animals (62 species worth over USD 34.8 billion per year; [21, 22]) and as parasites of farmed fish [23]. Most parasitic crustaceans are species within the class Copepoda, which displays a vast array of sexual dimorphism in anatomy, reproductive roles, sensory systems, and host/parasite relationships [23]. One of the most studied parasitic copepod, the salmon louse Lepeophtheirus salmonis, causes more than USD 480 million in losses to the Atlantic Salmon (Salmo salar) aquaculture industry annually [24]. Additionally, drug resistant strains of L. salmonis (and other sea lice species) have emerged globally (reviewed by [25]), threatening the sustainability and productivity of the industry.
Lepeophtheirus salmonis displays sexual dimorphism among several morphological, physiological, and behavioural characters. This phenomenon is observed in the motile parasitic stages of the lice (pre-adult I, II, and adult) although sex-specific differences in cephalothorax size and molt timing are also evident at preceding stages [26]. In addition, males develop faster than females, but they mature at approximately half the size of the adult female [26–28]. Sex differences in the morphology of the genital segment, abdomen, and appendages occur in all motile stages [27]. Distinct sex-associated behavioral characteristics related to reproductive success including frequency of host switching [29–31], mate location [32], blood feeding [33], and mate-guarding [31, 34] have also been reported. Sexually dimorphic physiology is also evident when L. salmonis are exposed to a commonly used antiparasitic compound, emamectin benzoate (EMB). Although EMB-resistance is widespread [25], males consistently show higher tolerance to EMB compared with females, regardless of the overall level of resistance within the population [35–37]. However, the molecular mechanisms underpinning sex-specific anatomy, behavior, and physiology in L. salmonis, and copepods in general, remain poorly understood.
The present study investigates sex-biased gene expression in three populations of L. salmonis using newly-generated transcriptomic data from Pacific Canada L. salmonis as well as a novel analysis of an available published dataset from Atlantic Canada L. salmonis [37]. A consensus-based, meta-analysis approach was used to identify sex-biased transcripts putatively responsible for sexual dimorphism in L. salmonis. Additionally, L. salmonis sequence conservation with related species (UniProt or Conserved Domain Database; e < 10-10) was integrated with sex-biased expression results to investigate sex-specific selective pressure and genomic constraint.
Results
Sex-biased gene expression in L. salmonis
Sex-biased transcripts were identified in three populations of pre-adult II L. salmonis using a 38 K oligonucleotide microarray. Two of the populations were from the Atlantic subspecies L. salmonis salmonis and were collected from separate bay management areas (BMA-2a and BMA-2b) in the Bay of Fundy, New Brunswick [37], and the third was from the Pacific subspecies L. salmonis oncorhynchi [38] collected from the Broughton Archipelago, British Columbia (BC). Eighteen to 21 F1 generation preadult males and females from each population were analyzed in individual microarray hybridizations (total n = 117 individuals and hybridizations). A total of 34.7 – 42.7 % of all unique contigs passing quality control (QC) filters were significantly sex-biased (Benjamini-Hochberg multiple test correction; p < 0.01; fold change (FC) ≥ 1.5) in Atlantic and Pacific L. salmonis (Table 1). Including only the transcripts expressed in both sexes, a Principal Component Analysis (PCA) separated male and female samples along the first principal component (PC1; explaining the most variation) in all three populations, representing 50.2, 39.5 and 53.4 % of the transcriptional variation in BMA-2a, BMA-2b, and Pacific lice, respectively (Fig. 1). No consistent differences were observed between the proportions of transcripts overexpressed in males relative to females in each population (Table 1). Sex-biased transcripts for each population, including p-values, fold changes, annotations, and accession identifiers, can be found in Additional file 1.
Table 1.
Sex-biased contigs in three populations of L. salmonis
| L. salmonis populations | Unique contigs passing QC filter | Proportion sex-biased (%) | Male-biased contigs | Female-biased contigs | Proportion (%) of orphans | ||
|---|---|---|---|---|---|---|---|
| Male-biased | Female-biased | Unbiased | |||||
| Atlantic (BMA-2a) | 11859 | 34.7 | 1955 | 2157 | 45.1 | 37.1 | 33.1 |
| Atlantic (BMA-2b) | 8527 | 40.0 | 1729 | 1682 | 48.9 | 32.8 | 32.4 |
| Pacific | 14923 | 42.7 | 3068 | 3303 | 51.8 | 28.3 | 34.0 |
| Consensus | 7889 | N/A | 368 | 461 | 50.7 | 20.1 | N/A |
Orphans are contigs without annotation (BLASTx; e < 10-10). The numbers listed in this table are representative of unique contigs (i.e. duplicate probes removed). Only unique contigs were considered for each category
Fig. 1.
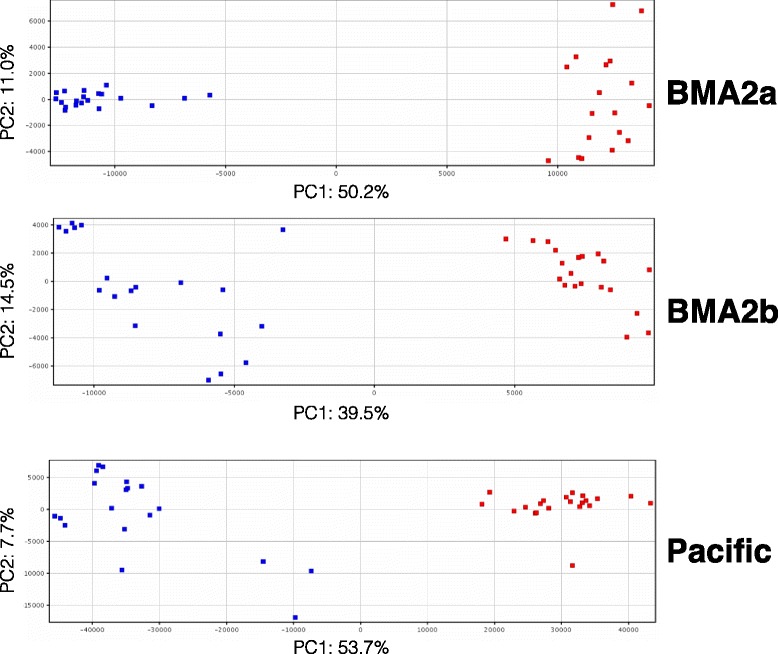
Principal Component Analysis of three populations of L. salmonis. Individual lice are represented in blue (males) and red (females). Sexes are separated on PC1 (x-axis) in all populations by 50.2 %, 39.5 % and 53.4 % for BMA-2a, BMA-2b, and Pacific lice, respectively. Only transcripts expressed by both sexes (i.e. excluding sex-specific probes) were included in PCA analysis
Sequence conservation in sex-biased transcripts
The L. salmonis contigs used for microarray construction [39] were annotated using BLASTx and RPS-BLAST against SwissProt and Conserved Domain Database (CDD; [40]), respectively [39]. Contigs without annotation (e > 10-10) are marked as unknown in the additional files. The proportion of orphans relative to annotated transcripts in male-biased, female-biased, and unbiased categories was assessed for each population. This approach has been used in other model organisms, for example in flies and nematodes [11, 41]. Male-biased transcripts from all three L. salmonis populations had a higher proportion of orphans compared with female-biased and unbiased transcripts (Table 1). Female-biased and unbiased transcripts did not show consistent differences in the proportion of orphans (Table 1). These data suggest lower sequence conservation of male-biased transcripts in L. salmonis.
Consensus of sex-biased transcripts in three populations of L. salmonis
To assess the functional impacts of sex-biased expression in L. salmonis, differentially expressed transcripts between sexes from each population were used to generate a consensus list (Fig. 2). A total of 1470 unique transcripts, out of a total of 7889 were shown to be significantly sex-biased in all three populations with 829 of these showing concordant expression profiles (Fig. 2; Additional file 2). Using this consensus list (i.e. requiring concordant differential expression being identified in all three populations), 368 transcripts showed male-bias and 461 were female-biased. As expected from the individual population analyses, consensus male-biased transcripts showed a 2.5-fold higher proportion of orphans compared with those showing female-bias (Table 1). Differences in the degree of sex-biased expression, as measured by fold change (FC) also varied between male- and female-biased transcripts. On average, 84.8 % of the transcripts overexpressed in females had low sex-bias (FC ≥ 1.5 and ≤ 4), whereas transcripts overexpressed in males had equal proportions of high and low sex-bias (Table 2). Interestingly, 127 male-biased transcripts were not expressed above background levels in any of the 58 females assayed and therefore are referred to as male-specific. In contrast, only 20 transcripts were female-specific in the consensus list. Here, fold changes are reported as the range of differential expression between males and females across all populations, unless the transcript was sex-specific, and then it is denoted as such. Fold changes specific to each population for consensus sex-biased transcripts can be found in Additional file 2.
Fig. 2.
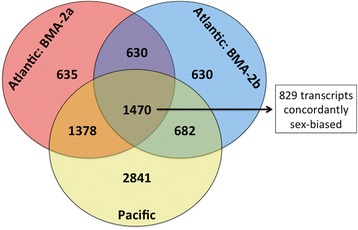
A consensus approach to identify sex-biased transcripts in L. salmonis. Sex-biased transcripts are displayed for each population separately. A consensus on sex-biased expression was achieved by creating a list of transcripts showing concordant differential expression between males and females across all three populations. Sex-biased transcripts for each individual population analysis can be found in Additional file 1 while consensus transcripts are displayed in Additional file 2
Table 2.
Extent of sex-bias in three L. salmonis populations using consensus sex-biased transcripts (Additional file 2)
| Sex-bias | Population | Proportion (%) | |
|---|---|---|---|
| Low fold change (≥1.5 but < 4) | High fold change (≥4 fold) | ||
| MALE | BMA2a | 51.5 | 48.5 |
| BMA2b | 55.3 | 44.7 | |
| Pacific | 37.7 | 62.3 | |
| Mean FC | 49.9 | 50.1 | |
| FEMALE | BMA2a | 85.5 | 14.5 |
| BMA2b | 98.5 | 1.5 | |
| Pacific | 77.3 | 22.7 | |
| Mean FC | 84.8 | 15.2 | |
Mean fold change (FC) was calculated for each transcript using the average fold change across the three populations
Male-biased transcripts in three populations of L. salmonis
The majority of annotated male-biased transcripts had roles in reproduction, for example being accessory gland proteins (Acps) and seminal fluid proteins (SFPs; reviewed by [42]). Transcripts known to regulate proteolysis for reproduction-related functions were highly male-biased in L. salmonis and included 16 proteases and 13 protease inhibitors, 10 of each being male-specific. However, a high degree of variance was observed in the expression of proteolytic transcripts among Pacific males (Fig. 3). To better understand this expression pattern, a transcript similarity assessment using kunitz/BPTI-like toxin (probe ID: C259R052) showed that 110 transcripts were strongly co-expressed (Pearson’s correlation, 0.95 < r < 1.0; Fig. 3). Although Atlantic males showed constitutive expression of these transcripts, Pacific males showed a characteristic “on/off” expression profile, with 10 of 19 individuals showing low, or absence of expression (Fig. 3). This co-expressed transcript list contains numerous representatives from known functional categories of male reproduction including peroxidases, pH regulators, kinases, and transporters, among others (Table 3). As seminal fluid proteins (SFPs) are only expressed in males [43], transcripts exhibiting male-specific expression are putatively assigned as candidate SFPs in L. salmonis. Many of these transcripts also enriched the Swiss-Prot (SP) and Protein Information Resource (PIR) Keyword (SP_PIR_Keyword) secreted (19 transcripts; p < 0.0001, Additional file 3), further supporting the involvement of these transcripts as SFPs or accessory gland proteins (reviewed by [42]).
Fig. 3.
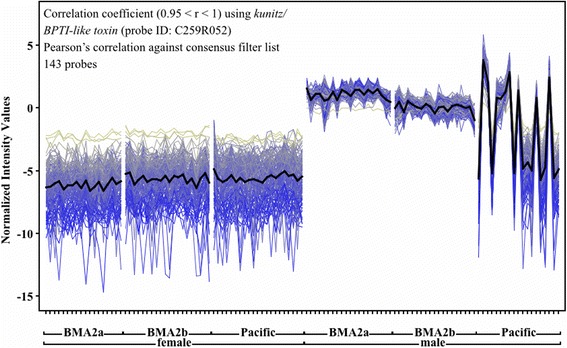
Co-expressed transcripts with putative roles in male reproduction. A total of 110 transcripts were co-expressed with BPTI/kunitz-like toxin (probe ID: C259R052) based on Pearson’s correlation (0.95 < r < 1) against the consensus QC-filtered transcript list (Table 1). Males and females are separated into each population on the x-axis. Normalized relative intensities (y-axis) are represented by log-2 Cy5/Cy3 ratios
Table 3.
Transcripts linked to male reproduction based on expression profiles and annotation
| Functional category | Transcript description | Degree of Male-bias (FC) | Probes | SwissProt Accession | CDD Accession | Ref |
|---|---|---|---|---|---|---|
| Protease inhibitors | Keratin-associated protein 6-2 | 30.8 - 277.1 | C088R043 | O08884 | smart00131 | [95, 96] |
| Kunitz_BPTI | MS | C084R101 | NA | smart00131 | [69, 95–100] | |
| Kunitz_BPTI | MS | C057R056 | NA | smart00131 | ||
| Kunitz_BPTI | MS | C259R083 | NA | pfam00014 | ||
| Kunitz/BPTI-like toxina | MS | C259R052 | B5L5M7 | pfam00014 | ||
| Papilina | MS | C063R028 | Q868Z9 | pfam00014 | [101] | |
| Papilina | MS | C183R014 | ||||
| Papilina | MS | C066R049 | ||||
| Papilina | MS | C142R005 | ||||
| Tissue factor pathway inhibitor 2a | MS | C213R048 | O35536 | pfam00014 | [100, 102] | |
| Antichymotrypsin-2a | 2.9 - 4.2 | C077R006 | P80034 | cd00172 | [59, 95, 96, 101, 103] | |
| SERine Proteinase INhibitors (serpins) | MS | C215R048 | NA | cd00172 | ||
| Serpin-Z10 | 1.6 - 2.4 | C182R015 | Q9SIR9 | cd00172 | ||
| Kinases | Adenylate kinase isoenzyme 1 | 2.1 - 10.4 | C104R155 | P05081 | TIGR01360 | [96, 104] |
| Adenylate kinase isoenzyme 1 | 1.9 – 4.8 | C153R147 | P00571 | TIGR01360 | ||
| Adenylate kinase isoenzyme 1 | 2.1 - 7.1 | C031R080 | ||||
| Casein kinase I isoform alpha | MS | C070R100 | P97633 | cd14016 | [96, 105] | |
| Casein kinase I isoform epsilon | 1.8 - 4.8 | C244R145 | Q9JMK2 | cd00180 | ||
| Hexokinase type 2 | 2.5 - 7.9 | C066R139 | Q9NFT7 | COG5026 | [95, 106] | |
| Probable adenylate kinase isoenzyme F38B2.4 | 1.9 - 5.7 | C212R032 | Q20140 | TIGR01360 | [96, 104] | |
| Probable adenylate kinase isoenzyme F38B2.4 | 2.0 - 5.7 | C028R063 | ||||
| Pyruvate kinase | 2.3 – 5.0 | C020R004 | O62619 | pfam00224 | [95, 96] | |
| Pyruvate kinase | 2.0 – 4.6 | C015R041 | cd00288 | |||
| Pyruvate kinase | 1.9 - 5.0 | C155R159 | ||||
| Proteases | Calpain-A catalytic subunit | MS | C197R005 | Q11002 | smart00720 | [95, 102] |
| Calpain-A catalytic subunit | MS | C018R134 | smart00230 | |||
| Carboxypeptidase Ba | MS | C161R058 | P04069 | cd03860 | [102, 106] | |
| Cytosolic non-specific dipeptidase | MS | C261R120 | Q3ZC84 | COG0624 | [102, 106, 107] | |
| Cytosolic non-specific dipeptidase | 1.9 - 2.9 | C145R086 | Q9D1A2 | COG0624 | ||
| Proprotein convertase subtilisin/kexin type 5a | 2.5 - 4.2 | C118R013 | Q9NJ15 | cd00064 | [96] | |
| Serine protease persephone | MS | C118R020 | Q9VWU1 | smart00020 | [61] | |
| Testisin | 2.2 – 9.8 | C007R130 | Q9JHJ7 | smart00020 | [108, 109] | |
| Tryp_SPc, Trypsin-like serine protease | 5.5 – 322.5 | C158R134 | NA | smart00020 | [95, 96, 103] | |
| Tryp_SPc, Trypsin-like serine protease | MS | C009R051 | ||||
| Tryp_SPc, Trypsin-like serine protease | MS | C008R159 | ||||
| Zinc metalloproteinase nas-15a | 2.1 - 3.7 | C134R018 | P55115 | cd04280 | [95, 103] | |
| Prostasinab | MS | C135R082 | Q16651 | cd00190 | [102, 110] | |
| ZnMc_adamalysin_II_like | MS | C083R024 | NA | cd04269 | [96, 102] | |
| Proclotting enzyme heavy chain | MS | C006R078 | P21902 | smart00020 | [111] | |
| Gamma-glutamyltranspeptidase 1b | 1.7 – 3.4 | C120R152 | P20735 | cl19223 | [112] | |
| pH regulation | Carbonic anhydrase 1 | MS | C183R004 | P83299 | cd00326 | [96, 113–115] |
| Carbonic anhydrase 9 | MS | C196R116 | Q8VHB5 | cd00326 | ||
| Carbonic anhydrase 9 | MS | C131R016 | ||||
| Carbonic anhydrase 9 | MS | C161R087 | ||||
| Structural | Actin | MS | C223R146 | Q92192 | PTZ00004 | [106, 116] |
| Lamin Dm0 | 1.5 - 6.6 | C220R106 | P08928 | pfam00038 | [117] | |
| Outer dense fiber protein 2-like | 2.5 - 5.2 | C121R150 | Q08B20 | pfam02463 | [51, 106] | |
| Kelch-like protein 20 | MS | C022R130 | Q5R7B8 | NA | [96, 118] | |
| Tubulin alpha-2 chain | MS | C160R074 | P06604 | cd02186 | [95, 106] | |
| Transport | Solute carrier family 15 member 1 | 4.1 - 9.3 | C170R033 | P46059 | TIGR00926 | [96, 106] |
| Solute carrier family 2, facilitated glucose transporter member 1 | 1.5 - 2.8 | C192R047 | P11166 | pfam00083 | ||
| Sodium/glucose cotransporter 4 | 1.6 – 3.1 | C170R069 | Q2M3M2 | pfam00474 | ||
| Solute carrier family 22 member 6-B | 2.5 - 4.3 | C072R016 | Q66J52 | TIGR00898 | ||
| Sodium-dependent nutrient amino acid transporter 1 | MS | C167R125 | B4JMC1 | pfam00209 | ||
| Aquaporin-12Ab (LsGlp1_v1; [81]) | 1.7 - 5.3 | C096R035 | Q8IXF9 | NA | [96, 119] | |
| Aquaporin-3 (Lsaqp12L2; [81]) | 3.7 - 10.4 | C030R103 | Q8R2N1 | cd00333 | ||
| Other | Mucin-like glycoprotein | 3.3 - 9.9 | C218R155 | NA | pfam01456 | [102, 120] |
| Major royal jelly protein 3a | MS | C089R070 | Q17060 | NA | [121–124] | |
| Chorion peroxidase heavy chaina | MS | C176R138 | Q9VEG6 | pfam03098 | [125, 126] | |
| Chorion peroxidase heavy chaina | MS | C154R094 | ||||
| Peroxidasea | MS | C026R132 | Q01603 | pfam03098 | ||
| Energy | Fructose-bisphosphate aldolase | 2.0 – 3.4 | C230R040 | O52402 | PRK09197 | [59, 80, 104] |
| Fructose-bisphosphate aldolase | 1.8 – 3.0 | C085R145 | P14540 | PRK09197 | ||
| Fructose-bisphosphate aldolase | 1.9 – 3.3 | C069R104 | P14540 | cd00946 | ||
| Glycogen phosphorylase | 1.7 – 2.5 | C107R029 | Q9XTL9 | cd04300 | ||
| Glycogen phosphorylase | 1.6 – 3.2 | C085R148 | Q9XTL9 | cd04300 | ||
| Glycogen phosphorylase, brain form | 1.6 – 3.5 | C171R004 | Q3B7M9 | cd04300 | ||
| Male Fertility | Protein ref(2)Pb | 1.6 - 3.9 | C036R126 | Q24629 | cd14320 | [127] |
References are provided based on the identification of similarly annotated sequences or proteins involved in male reproduction (e.g. spermatogenesis, seminal fluid proteins, testis expression) in related organisms. The degree of male-bias is indicated by a range of fold change (FC) across populations while MS indicates the listed probes showed male-specific expression. Transcripts denoted with a have a signal peptide for secretion (SwissProt) while those with b were annotated using e < 10-5 due to the absence of homology at higher stringency. Transcripts without annotation are represented by NA (no annotation). Each transcript has a unique contig ID which can be found in Additional file 2 using the probes listed here
Several other male-biased transcripts had putative roles in morphogenesis and the nervous system. Male-biased transcripts were enriched for cellular component assembly involved in morphogenesis (4 transcripts, p = 0.03), ossification (here probably calcification; 4 transcripts, p = 0.02), and Z disc (5 transcripts, p < 0.0001; Additional file 3). Additionally, male-biased transcripts were enriched for potassium ion binding (4 transcripts, p = 0.02), calcium ion binding (12 transcripts, p = 0.03), ion channel activity (5 transcripts, p = 0.04), and solute:cation symporter activity (5 transcripts, p = 0.01; Additional file 3), showing differences in sensory-system related functions. Other sex-biased transcripts involved in the nervous system, including their sex-biased expression profiles, are reported in Fig. 4. As salmon lice display sexually dimorphic patterns of mobility (i.e. mate location [32] and frequency of host switching [31]) and responses to neurotoxic drugs [37, 44], these transcripts will serve as important markers to better understand sex-related differences in the L. salmonis sensory system.
Fig. 4.
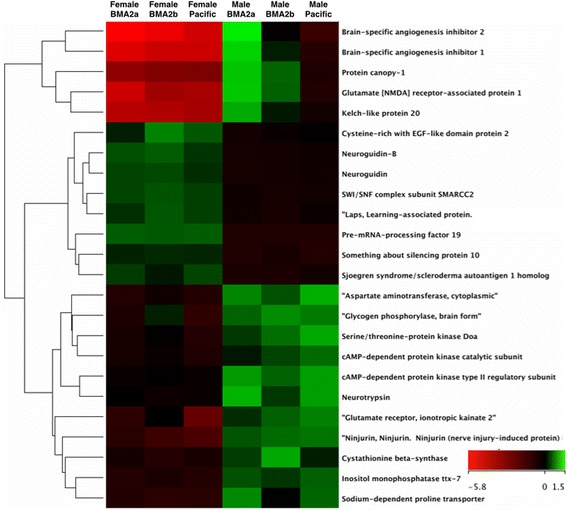
Candidate sex-biased transcripts involved with sensory system related functions in L. salmonis. Transcripts with high expression relative to Cy3 reference pool are green while low expressing transcripts relative to the Cy3 reference pool are red
Female-biased transcripts in three populations of L. salmonis
Female-biased transcripts were enriched for basic molecular processes including RNA processing (60 transcripts, p < 0.0001), ribosome biogenesis (38 transcripts, p < 0.0001), and transcription (41 transcripts, p < 0.0001) (Additional file 3). Within these categories, some multi-subunit protein complexes were completely female-biased including chaperonin-containing T-complex (CCT-complex; 5 transcripts, p = 0.0002), Nup107-160 complex (4 transcripts, p = 0.02), spliceosome (11 transcripts, p = 0.02), histone deacetylase complex (4 transcripts, p = 0.04) and ribonucleoprotein complex (30 transcripts, p = 0.0009). Protein complex formation was also enriched in the female-biased list, for example the GO category macromolecular complex subunit organization (28 transcripts, p < 0.0001).
Several transcripts related to cell division and organization were overexpressed in females and some significantly enriched GO categories included cell cycle (35 transcripts, p < 0.0001), DNA replication (17 transcripts, p < 0.0001), and cell proliferation (13 transcripts, p = 0.004) (Additional file 3). Furthermore, GO enrichment of ATP binding (60 transcripts, p < 0.0001), ATP-dependent helicase activity (10 transcripts, p = 0.001), and ATPase activity (15 transcripts, p = 0.005; Additional file 3) indicated female-biased energy generation.
As observed with male-biased transcripts, female-biased transcripts were also enriched for reproductive functions. The GO category in utero embryonic development (8 transcripts, p = 0.005; Additional file 3) was significantly enriched despite the absence of mating across all experiments (female lice are not sexually mature at the pre-adult stage; [34]). These included nuclear autoantigenic sperm protein (FC = 2.7 – 10.0), pre-mRNA processing factor 19 (FC = 1.6 – 2.6), and protein arginine N-methyltransferase 1, among others (FC = 1.7 – 2.5; Additional file 3). Female-biased transcripts were also enriched for nuclear hormone receptor binding (4 transcripts, p = 0.02) with an additional female-specific transcript containing the c4 zinc finger common to nuclear hormone receptors (Zn_C4; CDD: smart00390; e = 2.14-15). Several other female-biased transcripts not included in GO categories also have potential roles in reproduction including piwi-like protein 1 (FC = 2.6 – 8.3) and peroxiredoxin 1 (FC = 1.6 – 3.1). Lastly, transcripts involved in sex determination were female-biased in all populations, including prohibitin-2 ([45]; FC = 1.5 – 9.6) and pre-mRNA-splicing regulator female-lethal(2)D ([46]; FC = 1.7 – 2.4).
Some transcripts related to morphology, feeding, and detoxification were also female-biased across all populations. Transcripts such as serine proteinase stubble catalytic chain (female-specific), la protein homolog (FC = 1.5 – 2.5), and digestive organ expansion factor homolog (FC = 1.7 – 2.9), are involved in development, while trypsin-1 (FC = 3.1 – 12.0) and quinone oxidoreductase (FC = 2.1 – 116.5) have putative roles in feeding and detoxification, respectively. Lastly, immune-related transcripts, like rhotekin-2 (female-specific), ras-related protein Rab-32 (female-specific), and complement component 1 Q subcomponent-binding protein, mitochondrial (FC = 1.6 – 4.0), were female-biased in all L. salmonis populations. Female-biased transcripts therefore represent several candidates responsible for controlling sexual dimorphism at the molecular level in L. salmonis. Based on these findings, immunity, energy expenditure, and organogenesis are examples of previously unknown sexual dimorphism in salmon lice.
Annotation of L. salmonis sex-biased orphan contigs
Sex-biased orphan contigs were compared to known sequences using UniProt (e ≤ 10-5) to augment novel transcript discovery in the non-model L. salmonis. Although this threshold is less conservative than that used for the original microarray annotation [39], it has been frequently used in other sea lice transcriptomic studies [47–49], being generally acceptable for gene annotation [50]. This method was used for novel transcript discovery only and these newly annotated transcripts were not included in GO analyses. A total of 16 female-biased and 12 male-biased transcripts were subsequently annotated (Additional file 2) using this method. This additional annotation did not substantially change observations made on sequence divergence differences in male and female-biased transcripts, and many of the newly annotated female-biased transcripts had similar functions to those identified in the enrichment analysis described above, including transcription, translation, and cell cycle (Additional file 3).
Potential links to reproduction were observed among several of the newly annotated male-biased orphans, including prostasin (male-specific), gamma-glutamyltranspeptidase (FC = 1.7 – 3.4), and protein ref(2)P (FC = 1.6 – 2.3) (Table 3). A male-specific transcript, c-factor, was also identified. However, the function of this transcript in L. salmonis remains unknown. Additionally, male-biased transcripts involved in neuromuscular development and function were discovered, including excitatory amino acid transporter 3 (SLC1A1; FC = 2.3 - 3), twitchin (FC = 1.6 – 5.7), and sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (FC = 1.5 – 7.6). Probe identifiers, e-values, bitscores, SwissProt accessions, and descriptions for newly annotated transcripts can be found in Additional file 2.
Discussion
The economically and ecologically important parasite, salmon lice L. salmonis (Copepoda: Caligidae) displays sexual dimorphism in ontogeny, morphology, physiology, and behavior [26–35]. However, little is known about the molecular mechanisms that control these traits and the possible interactions they may have with chemical response/resistance, host-parasite interactions, and overall population dynamics. Here, sex-biased transcripts were identified within three populations of L. salmonis (including two subspecies; [38]) from the Atlantic and Pacific coasts of Canada. Our observation revealed a large proportion (34.7 – 42.7 %) of sex-biased expression (Table 1) consistent with findings in other arthropods [6, 11], nematodes [8], amphibians [12], fish [51], birds [13], and mammals [17]. Principal component analysis supported this finding as males and females were separated on PC1 in all populations. These data suggest large differences in baseline gene expression between male and female L. salmonis that could potentially impact parasite control strategies. For example, routine lice counting and staging is the primary management strategy used to inform treatment regimes in aquaculture. However, pre-adult male and female L. salmonis are often grouped into a single category for these counts [52, 53]. The basal differences in gene expression reported here and the interactive effects of sex with chemical treatment and resistance described elsewhere [37], suggests a more informative strategy would be to separately count male and female preadult L. salmonis in both farm and laboratory settings whenever possible.
There were no consistent differences between the proportion of male-biased relative to female-biased transcripts in L. salmonis (Table 1), despite observations in other crustaceans like Caligus rogercresseyi, another species of parasitic copepod, and Daphnia pulex which suggested a slight over-representation of male-biased transcripts [11, 54]. However, we did find that a larger proportion of male-biased transcripts had higher fold change than those with female-bias. In this study, sex-biased expression was assessed across multiple populations of L. salmonis reared under similar conditions, thus offering a more comprehensive overview of gene expression differences than is obtained in single cohort transcriptome analyses. However the sex-biased transcripts identified here were limited to ~22,000 unique contig sequences (i.e. transcripts) on the microarray and it was not possible to examine some of the more complex facets of sex-biased expression (i.e. sex-specific alternative transcripts [55]). Additionally, studies using whole-body homogenates offer general patterns of sex-bias but lack the resolution to detect sex-biased expression in individual tissues [7] and control for differences in tissue allometry [56]. Tissue-specific extractions will be an important tool for future studies examining sex-biased gene expression in L. salmonis. Nonetheless, characterization of sex-biased transcripts reported here provides numerous molecular targets putatively underlying sexual dimorphism in L. salmonis, offering important insight for pest management and future drug development for this non-model organism.
Discovery of sex-biased transcripts related to reproduction in L. salmonis
Male-biased transcripts displayed greater sex-bias compared with female-biased transcripts in L. salmonis, similar to findings in D. melanogaster [6]. Among the highly male-biased transcripts, 127 were not expressed above background detection in any of the 58 females assayed in this study. These transcripts represent putative seminal fluid proteins (SFPs) based on their expression profiles (SFPs are only expressed in males; [43]) and annotations (Table 3 and references therein). SFPs are transferred to females during mating, impacting a variety of physiological processes such as sperm storage, egg production, feeding, behavior, and receptivity to re-mating (reviewed in [42]). Although genes involved in reproduction tend to evolve rapidly [57], many of the functional constituents of seminal fluid are conserved from arthropods to mammals [43, 58]. For example, genes involved in proteolysis are essential for sperm transfer, storage, and activity, among other functions [59, 60]. A total of 10 proteases and 10 protease-inhibitors were identified as male-specific in this study with several others showing male-bias (Table 3). All protease inhibitors putatively involved in reproduction contained at least one kunitz or serpin domain (CDD accessions: pfam00014 and cd00172, respectively), which are the predominant classes of protease inhibitors in seminal fluid [59]. In turn, proteases such as serine protease persephone and calpain-A catalytic subunit also had relevant annotations to SFPs based on their ability to modulate toll signaling in D. melanogaster [61, 62]. The transfer of immunomodulatory and antimicrobial SFPs may aid females in defending against infections that are introduced during mating [60, 63]. Many of the L. salmonis proteases assigned as SFPs also contained trypsin-like domains (CDD: cd00190 and smart00020) including prostasin, proclotting enzyme heavy chain, and three additional transcripts without SwissProt annotation (i.e. CDD only). These transcripts represent important targets for understanding the proteolytic events controlling reproduction and mating in L. salmonis.
Female-biased transcripts also had putative reproductive functions, including five with nuclear hormone receptor activity, one of which was female-specific. Recently, a sex-linked SNP in the L. salmonis prohibitin-2 gene with a pattern of female-biased expression was identified in three strains of L. salmonis from Scotland [45]. Similarly, a transcript annotated as prohibitin-2 was female-biased in all three populations of L. salmonis assayed in this study. The prohibitin-2 gene is likely involved in sex determination [45] along with pre-mRNA-splicing regulator female-lethal(2)D [46], which was also female-biased in L. salmonis.
Overall, functional enrichment for reproduction was less clear in female-biased transcripts compared with male-biased transcripts. Many of the putative female-biased transcripts potentially involved with reproduction had GO annotation with many other functional categories and only a small number were female-specific. For example, nuclear autoantigenic sperm protein (NASP) and histone deacetylase 1 (HD1), were female-biased in L. salmonis and have reproductive roles in similar species [54, 64, 65]. However, these transcripts were in the enriched GO category in utero embryonic development (p = 0.005), which is taxonomically constrained [66] and inconsistent with the virgin status of the lice used here [34]. Furthermore, the functions of NASP and HD1 differ in other species such as C. elegans in which they are important for female development and male-specific gene repression [67]. These discrepancies make it difficult to identify putative female-biased reproduction genes in the present work. In an earlier study by Eichner and colleagues [47], L. salmonis genes involved in reproduction were overexpressed in adult females compared to preadult II females. As the present study used preadult II females, it may have missed the identification of some female reproductive genes that are induced later in development than the stages evaluated here. Future studies assessing the expression and localization of these transcripts will offer insight on their exact functions. Much work remains to identify female-biased reproductive genes in L. salmonis, including the extensive changes likely to occur in female mating-responsive genes post transfer of male spermatophores (e.g. see [68, 71, 72]).
In general, sex-biased genes evolve faster than unbiased genes, with those exhibiting male-bias showing the highest rates of evolution (reviewed by [4]). This trend is heavily influenced by sex-biased transcripts involved in reproduction as these typically exhibit higher than normal rates of positive selection (reviewed by [4, 57]). Only male-biased transcripts showed a consistently higher number of orphans in each L. salmonis population (45.1 – 51.8 %) when compared to female-biased (28.3 – 37.1 %) and unbiased (32.4 – 34.0 %) transcripts. This trend was largely driven by the putative SFPs in this study (i.e. male-specific transcripts), of which 65.4 % were orphans. As higher rates of nucleotide substitutions are often caused by a relaxed functional constraint or greater positive selection on certain transcripts (reviewed in [4, 57]), higher-resolution analyses including non-synonymous/synonymous mutation ratios (dN/dS; [70, 71] and codon-usage bias [72, 73] will be important in clarifying the effects of selection on these L. salmonis genes. These analyses will yield important information regarding evolutionary processes affecting reproduction, population dynamics, and drug resistance. In particular, it will be important to identify whether genes involved in drug resistance are similar to those involved in sex-biased expression as this will provide insight on molecular mechanisms behind the higher rate of drug resistance in male L. salmonis. Specifically this will inform on whether the increased resistance is due to inherent physiological factors that differ between the sexes, or to the evolutionary rate of resistance mechanisms.
Sex-biased transcription related to sexually dimorphic phenotypes of L. salmonis
L. salmonis exhibits extensive sexual dimorphism in morphology at the pre-adult and adult stages [27]. Several sex-biased transcripts discovered here have related functions including the male-biased transcripts enriching the GO categories cellular components involved in morphogenesis (4 transcripts, p = 0.03) and Z-disc (sarcomere; 5 transcripts, p < 0.0001). Additionally, serine proteinase stubble catalytic chain was female-specific in L. salmonis and is required for proper formation of appendages in Drosophila spp. [74]. Therefore, these transcripts represent targets for understanding sexually dimorphic morphology including features that are important in mating (described in [27, 31, 34]). The sex-biased orphans described here are also ideal candidates for understanding mate guarding, a common mating behavior in crustaceans, which is known to be sexually antagonistic [75]. Therefore, transcripts involved in mate guarding are unlikely to be annotated due to rapid sexual selection [75] and taxonomic constraint [50].
Other molecular links to sexual dimorphism in L. salmonis were observed in transcripts such as longitudinal lacking protein, trypsin-1, and digestive organ expansion factor, which were female-biased in this study and have roles in salivary gland, trachea, and digestive organ development and function [76, 77]. Based on female L. salmonis having a greater requirement for blood in the meal [33], these transcripts will serve as important markers for understanding sexually dimorphic feeding patterns and related host-parasite interactions.
Male-biased L. salmonis transcripts also enriched several GO categories related to the sensory system, which is generally known to be more refined in male copepods [23]. These included potassium ion binding (4 transcripts, p = 0.02), solute:cation symporter activity (5 transcripts, p = 0.01), and calcium ion binding (12 transcripts, p = 0.03), among others (Additional file 3). Males are known to transfer between salmonid hosts more frequently than females [30, 31] with mate location primarily being the responsibility of the male [32]. Thus, transcripts involved in the sensory system and muscle development (e.g. Z disc; Additional file 3) represent putative targets for understanding neuromuscular differences related to increased mobility in males. Furthermore, chemical cues are essential components of host and mate location in L. salmonis and serve to optimize the probability of mating through balancing the proportion of males relative to females on each host [29]. Chemosensory signaling and behavioral responses to non-host semiochemical treatments known to interfere with host recognition are sexually dimorphic in L. salmonis [32, 78]. Based on the high evolutionary rates of genes involved in chemoreception [79], sex-biased orphans should be considered candidate targets for understanding the chemical ecology of L. salmonis.
The identification of sex-biased transcripts in L. salmonis also provided preliminary evidence for sexually dimorphic characteristics previously unknown in salmon lice. For example, immune-related transcripts such as rhotekin-2 and ras-related protein Rab-32 were female-specific in this study while other related transcripts were female-biased (Additional file 2). Gene Ontology analysis also indicated a higher energy expenditure in female L. salmonis based on the female-biased expression of 15 transcripts enriching ATPase activity coupled and 60 transcripts enriching ATP binding (Additional file 3). This is a similar finding to the GO analysis of sex-biased expression in D. pulex [11], and may serve as an explanation for the lower frequency of inter-host transfer in female L. salmonis [30, 31]. Although male-biased transcripts such as glycogen phosphorylase, fructose-bisphosphate aldolase, and hexokinase-2 enriched the GO category glucose metabolic process (7 transcripts; p =0.02), these are likely acting as an energy source for spermatogenesis [80]. These data indicate an increased basal energy demand in female L. salmonis that is important for understanding observed sex differences in drug tolerance and resistance [35, 37]. Collectively, the candidate sex-biased transcripts described here represent putative markers controlling energy expenditure, morphology, and the immune and sensory systems of L. salmonis.
Strengths and limitations of using a consensus-based approach to identify sex-bias
Recently, seven aquaporin paralogs were characterized in L. salmonis, each with different expression profiles across stage and sex [81]. Two of these aquaporins (LsGlp1_v1 (KR005660.1) and Lsaqp12L2 (KR005666.1)) showed male-specific and male-biased expression, respectively. In the present work, two male-biased contigs annotated as aquaporin 3 (BT121448.1) and aquaporin-12A (BT121051.1; see Table 3) showed more than 99 % sequence similarity to LsGlp1_v1 and Lsaqp12L2, respectively (Additional file 4). Therefore, aquaporin 3 and LsGlp1_v1 appear to be the same transcript based on sequence alignment, as are aquaporin-12A and Lsaqp12L2. Here, aquaporin 3 was only expressed above background fluorescence in one of the 58 females assayed, supporting the expression profile previously reported [81]. Additionally, aquaporin-3 was shown to be highly male-biased in the closely related C. rogercresseyi [54], suggesting this gene is important for a male-specific function in salmon lice. This type of transcriptomic consensus will be important for functional categorizations in future sea lice studies.
Within the 1407 transcripts shown to be sex-biased in Atlantic and Pacific populations, only 829 showed concordant expression profiles. The majority of transcripts in the discordant list (Additional file 2) were involved in cuticle formation and molting and, therefore, transcripts that potentially oscillate in expression levels at different molt intervals [47, 82] were eliminated from this interpretation. However, the consensus identification of sex-biased transcripts in L. salmonis did prove to be over-conservative in some cases, causing particular transcripts with putative sex-bias to be overlooked. For instance, trypsin-4 (probe: C054R168), annotated from Anopheles gambiae and involved in host seeking behavior and blood feeding [83], was female-specific in BMA2a and in Pacific L. salmonis. However, this probe did not pass quality filters in the BMA2b population and was eliminated from the consensus list. Female L. salmonis are known to feed more heavily on blood than males [33], with certain trypsins known to be involved in digestion and immune evasion on salmonid hosts [84–86]. Additionally, a similar transcript annotated to trypsin-1 was female-biased in this study (Additional file 2). Therefore, monitoring individual population analyses from this work is also important for identifying potential sex-biased markers in L. salmonis. Nonetheless, the consensus set of sex-biased transcripts identified here supports the characterization of L. salmonis transcript as markers for reproduction, morphogenesis, behavior, and other sexually dimorphic traits for targeted approaches (i.e. knock-out/knock-down, recombinant production, in vitro characterization, etc.) in future studies. This improved understanding of sex-biased gene expression in L. salmonis will inform future studies examining host-parasite interactions, drug resistance, reproduction, and novel drug discovery.
Conclusions
A consensus-based, meta-analysis approach was used to analyze the L. salmonis transcriptome, clearly identifying sex-biased transcripts associated with sexually dimorphic traits. Specifically, male-biased transcripts showed higher degrees of sex-bias and lower sequence similarity compared with female-biased transcripts. The enrichment of male-biased transcripts associated with reproduction was likely responsible for these trends. Our results provided insights into known and novel forms of sexual dimorphism in L. salmonis including immunity, energy expenditure, morphology, feeding, and mobility. These sexual dimorphisms will be important to consider for industry-relevant applications in areas such as parasiticidal drug response, reproductive roles, and host-parasite relationships. The current work shows that sex-biased gene expression is abundant in the pre-adult L. salmonis transcriptome and is likely to control several aspects of sexual dimorphism in this species.
Methods
Lepeophtheirus salmonis populations and collections
Adult L. salmonis were collected from Atlantic salmon aquaculture farms on the Atlantic and Pacific coasts of Canada. Two populations of Atlantic L. salmonis were collected in the spring of 2013 from Bay Management Area 2a (BMA-2a; Back Bay) and 2b (BMA-2b; Grand Manan) in the Bay of Fundy, New Brunswick (NB), as described in full previously [35, 37]. A third population representing Pacific L. salmonis was collected from the Broughton Archipelago, British Columbia (BC) in 2010. Atlantic and Pacific L. salmonis are considered allopatric subspecies [38]. For all collections, egg strings were removed from adult females and larvae reared to the infective copepodid stage in static seawater hatch systems as previously described [35]. Copepodids (F1 generation) were then used to infect Atlantic salmon (Salmo salar) and allowed to develop to the pre-adult II stage. Pre-adult (F1) lice from all populations were used in 24 h in vitro EMB bioassays (described below) before collection and storage at -80 °C for RNA extraction.
Atlantic and Pacific L. salmonis Microarray Datasets
Two microarray datasets were used to compare sex differences in L. salmonis from the Atlantic (2013 collection) and Pacific (2010 collection) coasts of Canada. The Atlantic dataset was accessed from NCBI through Gene Expression Omnibus (GEO) accession GSE56024 [37]. In this study a total of 77 pre-adult Atlantic L. salmonis, 38 females and 39 males from BMA-2a and BMA-2b were exposed to four concentrations of EMB (0.1, 25, 300, and 1000 ppb) and a seawater control, as previously reported. The bioassay protocol was identical for both populations. This study compared the effects of EMB on L. salmonis including the interactions between population (BMA-2a is more EMB-resistant than BMA-2b; [37, 87]) and sex. However, baseline differences between males and females were not reported. The Pacific dataset was provided by the same laboratory group (B. Koop and S. Jones, unpublished data), which exposed 39 pre-adult Pacific L. salmonis, 21 females and 19 males, to low doses of EMB (0.01, and 0.1 ppb) or a seawater control. Lower doses of EMB were selected based on the high EMB-sensitivity of this population [88]. For all F1 generation cultures, lice were maintained in filtered sea water at 10 ± 2 °C and 32 ± 2 ppt. The Pacific dataset has been uploaded to GEO under the accession GSE73734.
A 38 K oligonucleotide microarray (eArray, Agilent) designed with expressed sequence tags (ESTs) from Atlantic and Pacific L. salmonis [39] was used to analyze all lice in this study. Annotation of each contig was completed using BLASTx and RPS-BLAST against SwissProt and Conserved Domain Database (e < 10-10), respectively. A total of 18 – 21 hybridizations for each sex and population combination (117 total hybridizations) were completed. Hybridizations were completed using methods for sample preparation, microarray hybridization, and scanning as previously reported [37, 89]. Briefly, all slides were scanned using a Perkin Elmer ScanArray® at 5 μm resolution and optimized PMT intensities (1-2 % of array spots saturated). Filtering and quantification was completed using Imagene 8.1 (Biodiscovery) before completing statistical analyses in GeneSpring GX v12.6 (Agilent). A quality control (QC) filtered probe list was created for each population (Table 1) with probes included for statistical analysis only if at least 65 % of the samples in any one condition had raw fluorescent intensities ≥ 500 and showed no poor quality spots.
Sex-biased Gene Expression in L. salmonis
Microarray data was used to characterize baseline expression differences between male and female L. salmonis. Sex-biased probes were identified using a two-way ANOVA with sex and EMB as explanatory variables (Benjamini-Hochberg multiple test correction; p < 0.01; fold change (FC) ≥ 1.5). The effects of EMB on Atlantic L. salmonis transcriptomes was minor [37] and a significant transcriptomic effect was not detected in Pacific L. salmonis used here (i.e. no probes showed differential expression by EMB). The effects of EMB exposures were also controlled for in the statistical model by only using transcripts affected by a main effect of sex and through consensus-based analyses (described below). All probes with a main effect of sex for each individual population are compiled into Additional file 1.
Sex-biased probes from each population were used to create a consensus list for functional analyses. This list was limited to probes exhibiting significant and concordant sex-bias in all three populations of L. salmonis described here (Fig. 2). Sex-biased probes from individual population analysis can be found in Additional file 1, while sex-biased probes identified using the consensus sex-biased method can be found in Additional file 2. As duplicate probes represent unique contigs (i.e. transcripts) on the array [39], contig IDs are also included in the additional files. The variation between duplicate probes of consensus sex-biased transcripts is quantified in Additional file 2. Only unique transcripts are used to calculate the proportion of sex-biased expression in Atlantic and Pacific L. salmonis.
Differences in the degree of sex-bias were assessed by binning transcripts based on their degree of differential expression between male and female L. salmonis [6]. Transcripts with low sex-bias were those overexpressed by a fold change (FC) of ≥ 1.5 but < 4, while highly sex-biased transcripts had a FC ≥ 4. A mean FC value representing all three populations was also included (Additional file 2). Any transcript that did not pass the background QC filter in 100 % of the individuals within one sex was considered to be sex-specific in this study. However, based on lower limits of detection for microarrays, these transcripts may not be biologically sex-specific. A transcript similarity assessment was also completed using all QC-filtered probes against kunitz/BPTI-like toxin (probe ID: C259R052). Transcripts with similar expression patterns to kunitz/BPTI-like toxin were determined using a Pearson’s correlation (0.95 < r < 1.0). These transcripts are described in Additional file 2.
Functional enrichment of the consensus sex-biased transcript list was done using Gene Ontology (GO), InterPro, and SwissProt (SP) and Protein Information Resource (PIR) Keywords (SP_PIR_Keywords) with DAVID bioinformatics [90–92] using a modified Fisher’s exact test (p < 0.05; genes/enrichment category ≥ 4). Unique SwissProt accession ID’s were compared against a QC filter background list designed to include transcripts passing QC filters in all populations (Table 1). To reduce redundancy of Gene Ontology categories, GO Trimming was used with an 80 % soft trim threshold [93]. All GO analyses can be found in Additional file 3.
Sequence conservation in sex-biased and unbiased transcripts
To investigate rates of sequence divergence in sex-biased transcripts, unique contigs passing QC filter for each population were binned into one of three categories: Male-biased (overexpressed by males), female-biased (overexpressed by females) or unbiased (no expression difference between sexes). All contigs on the microarray were annotated using UniProt [94] and Conserved Domain Database (NCBI, [40]) with the best match being the alignment with the lowest Expect value followed by the highest bitscore [39]. Transcripts with no significant match (e > 1.0-10) were considered orphans (labeled as “unknown” in additional files). To assess the degree of sequence conservation between male-biased, female-biased and unbiased transcripts, the proportion of orphans relative to annotated transcripts in each of these categories was assessed based on similar methodologies [11, 41]. Sequence conservation in male-biased and female-biased transcripts from the consensus list was also analyzed (Table 1).
Novel transcript discovery using sex-biased orphans
Based on the robust approach using a consensus sex-biased transcript list, orphans showing concordant sex-bias across all populations were re-annotated using a less conservative threshold of e > 1.0-5. This annotation threshold is common amongst other sea lice transcriptomic studies and is generally considered an acceptable cut-off for annotation [50]. This annotation was used for exploratory transcript prediction but not for the main analysis or the Gene Ontology enrichment analysis. Sex-biased orphan annotation results can also be found in Additional file 2.
Abbreviations
BMA, Bay Management Area; CDD, Conserved Domain Database; EMB, emamectin benzoate; FC, fold change; GEO, Gene Expression Omnibus; GO, Gene Ontology; PCA, principal component analysis; QC, quality filtered; SFP, seminal fluid proteins; SP_PIR_Keywords, SwissProt and Protein Information Resource Keywords.
Acknowledgements
This work was supported by Elanco Fish Health Research Chair; NSERC Discovery (610108); ACOA-AIF TREAT2 (199308); and Innovation PEI – Development and Commercialization grant DCFG (210205-70). JDP was supported by NSERC PGSD3 (290948462). Thanks to Drs. Shona Whyte, Spencer Greenwood, Laura Braden, and John Burka for comments on early drafts of the manuscript. Also, thanks to the aquatics staff at AVC for their assistance in fish husbandry and animal care.
Funding
This work was supported by Elanco Fish Health Research Chair; NSERC Discovery (610108); ACOA-AIF TREAT2 (199308); and Innovation PEI – Development and Commercialization grant DCFG (210205-70). JDP was supported by NSERC PGSD3 (290948462).
Availability of data and materials
The datasets supporting the results of this article are available in the Gene Expression Omnibus repository, GSE73734 and GSE56024.
Atlantic data set described and uploaded to GEO in [37].
Reviewer access link for Pacific lice (GSE73734): http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73734.
Authors’ contributions
JDP: Meta-analyses of sex-biased transcripts from all data sets, result interpretation, wrote manuscript. BJGS: Experimental design, microarray experiments, transcriptome analysis, and writing. SRMJ: Experimental design, Pacific lice collection, writing. BFK: Experimental design, analysis, writing. MDF: Experimental design, Atlantic lice collection, writing. All authors contributed to this work and have read and approved the most recent version of the manuscript.
Authors’ information
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
For Atlantic lice experiments, please see [37]. The Pacific lice experiment was approved under the Fisheries and Oceans Canada Pacific Region Animal Care Committee protocol number 09-001.
Additional files
Sex-biased and sex-specific transcripts in individual populations. (XLSX 1955 kb)
Consensus sex-biased and sex-specific transcripts, co-expressed “kunitz cluster”, and newly annotated sex-biased orphans. (XLSX 340 kb)
ene Ontology, SP-PIR_Keywords, and InterPro. (XLSX 93 kb)
Sequence alignment of L. salmonis aquaporins. (TXT 16 kb)
Contributor Information
Jordan D. Poley, Email: jpoley@upei.ca
Ben J. G. Sutherland, Email: ben.sutherland.1@ulaval.ca
Simon R. M. Jones, Email: simon.jones@dfo-mpo.gc.ca
Ben F. Koop, Email: bkoop@uvic.ca
Mark D. Fast, Email: mfast@upei.ca
References
- 1.Darwin C. The Descent of Man VOL I. 1871.
- 2.Hedrick AV, Temeles EJ. The evolution of sexual dimorphism in animals: Hypotheses and tests. Trends Ecol Evol. 1989;4:136–138. doi: 10.1016/0169-5347(89)90212-7. [DOI] [PubMed] [Google Scholar]
- 3.Slatkin M. Ecological causes of sexual dimorphism. Soc Study Evol. 1984;38:622–630. doi: 10.1111/j.1558-5646.1984.tb00327.x. [DOI] [PubMed] [Google Scholar]
- 4.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- 5.Parsch J, Ellegren H. The evolutionary causes and consequences of sex-biased gene expression. Nat Rev Genet. 2013;14:83–87. doi: 10.1038/nrg3376. [DOI] [PubMed] [Google Scholar]
- 6.Ranz JM, Castíuo-davís CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 7.Catalán A, Hutter S, Parsch J. Population and sex differences in Drosophila melanogaster brain gene expression. BMC Genomics. 2012;13:654. doi: 10.1186/1471-2164-13-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 9.Fitzpatrick JM, Johansen MV, Johnston DA, Dunne DW, Hoffmann KF. Gender-associated gene expression in two related strains of Schistosoma japonicum. Mol Biochem Parasitol. 2004;136:191–209. doi: 10.1016/j.molbiopara.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Fitzpatrick JM, Johnston DA, Williams GW, Williams DJ, Freeman TC, Dunne DW, Hoffmann KF. An oligonucleotide microarray for transcriptome analysis of Schistosoma mansoni and its application/use to investigate gender-associated gene expression. Mol Biochem Parasitol. 2005;141:1–13. doi: 10.1016/j.molbiopara.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Eads BD, Colbourne JK, Bohuski E, Andrews J. Profiling sex-biased gene expression during parthenogenetic reproduction in Daphnia pulex. BMC Genomics. 2007;8:464. doi: 10.1186/1471-2164-8-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malone JH, Hawkins DL, Michalak P. Sex-biased gene expression in a ZW sex determination system. J Mol Evol. 2006;63:427–436. doi: 10.1007/s00239-005-0263-4. [DOI] [PubMed] [Google Scholar]
- 13.Naurin S, Hansson B, Hasselquist D, Kim Y-H, Bensch S. The sex-biased brain: sexual dimorphism in gene expression in two species of songbirds. BMC Genomics. 2011;12:37. doi: 10.1186/1471-2164-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan Z, You F, Wang L, Weng S, Wu Z, Hu J, Zou Y, Tan X, Zhang P. Gonadal transcriptome analysis of male and female olive flounder (Paralichthys olivaceus) Biomed Res Int. 2014;2014:1–10. doi: 10.1155/2014/291067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Pääbo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Am Assoc Adv Sci. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 17.Rinn JL, Snyder M. Sexual dimorphism in mammalian gene expression. Trends Genet. 2005;21:298–305. doi: 10.1016/j.tig.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Ahyong ST, Lowry JK, Alonso M, Bamber RN, Boxshall GA, Castro P, Gerken S, Karaman GS, Goy JW, Jones DS, Meland K, Rogers DC, Svavarsson J. Animal Biodiversity: An outline of higher-level classification and survey of taxonomic richness. Volume 1817. Auckland, New Zealand: Magnolia Press; 2011.
- 19.Martin JW, Davis GE. An Updated Classification of the Recent Crustacea. 2001.
- 20.Carpenter SR, Kitchell JF, Hodgson JR. Fish predation and herbivory can regulate lake ecosystems. Bioscience. 1985;35:634–639. doi: 10.2307/1309989. [DOI] [Google Scholar]
- 21.FAO Fisheries and Aquaculture Department. World Review of Fisheries and Aquaculture: Part 1. 2012.
- 22.FAO Fisheries and Aquaculture Department: Global Aquaculture Production Statistics for the year 2011. 2013. ftp://ftp.fao.org/FI/news/GlobalAquacultureProductionStatistics2011.pdf.
- 23.Boxshall G. Crustacean parasites. In: Marine Parasitology. Edited by Rohde K.; 2005.
- 24.Costello MJ. The global economic cost of sea lice to the salmonid farming industry. J Fish Dis. 2009;32:115–118. doi: 10.1111/j.1365-2761.2008.01011.x. [DOI] [PubMed] [Google Scholar]
- 25.Aaen SM, Helgesen KO, Bakke MJ, Kaur K, Horsberg TE. Drug resistance in sea lice: a threat to salmonid aquaculture. Trends Parasitol. 2015;31:72–81. doi: 10.1016/j.pt.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Eichner C, Hamre LA, Nilsen F. Instar growth and molt increments in Lepeophtheirus salmonis (Copepoda: Caligidae) chalimus larvae. Parasitol Int. 2015;64:86–96. doi: 10.1016/j.parint.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Johnson SC, Albright LJ. The developmental stages of Lepeophtheirus salmonis (Kroyer, 1837) (Copepoda : Caligidae) Can J Zool. 1991;69:929–950. doi: 10.1139/z91-138. [DOI] [Google Scholar]
- 28.Hamre LA, Eichner C, Caipang CMA, Dalvin ST, Bron JE, Nilsen F, Boxshall G, Skern-Mauritzen R. The Salmon Louse Lepeophtheirus salmonis (Copepoda: Caligidae) Life Cycle Has Only Two Chalimus Stages. PLoS One. 2013;8:e73539. doi: 10.1371/journal.pone.0073539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephenson JF. The chemical cues of male sea lice Lepeophtheirus salmonis encourage others to move between host Atlantic salmon Salmo salar. J Fish Biol. 2012;81:1118–1123. doi: 10.1111/j.1095-8649.2012.03347.x. [DOI] [PubMed] [Google Scholar]
- 30.Wotton JH, Purcell S, Covello J, Koop B, Fast M. Comparison of host selection and gene expression of adult Lepeophtheirus salmonis and Salmo salar during a cohabitation of initially infected and uninfected fish. J Aquac Res Dev. 2014;03:226–33. doi: 10.4172/2155-9546.1000226. [DOI] [Google Scholar]
- 31.Hull MQ, Pike AW, Mordue AJ, Rae GH. Patterns of pair formation and mating in an ectoparasitic caligid copepod Lepeophtheirus salmonis (Kroyer 1837): implications for its sensory and mating biology. Philos Trans R Soc B Biol Sci. 1998;353:753–764. doi: 10.1098/rstb.1998.0241. [DOI] [Google Scholar]
- 32.Ingvarsdóttir A, Birkett MA, Duce I, Mordue W, Pickett JA, Wadhams LJ, Mordue AJ. Role of semiochemicals in mate location by parasitic sea louse, Lepeophtheirus salmonis. J Chem Ecol. 2002;28:2107–2117. doi: 10.1023/A:1020762314603. [DOI] [PubMed] [Google Scholar]
- 33.Brandal P, Egidius E, Romslo I. Host blood: a major food component for the parasitic copepod, Lepeophtheirus salmonis Kroyer, 1838 (Crustacea caligidae) Nor J Zool. 1976;24:341–343. [Google Scholar]
- 34.Ritchie G, Luntz AJM, Pikeb AW, Raec GH. Observations on mating and reproductive behaviour of Lepeophtheirus salmonis, Kroyer (Copepoda : Caligidae) J Exp Mar Bio Ecol. 1996;201:285–298. doi: 10.1016/0022-0981(96)00008-1. [DOI] [Google Scholar]
- 35.Igboeli OO, Burka JF, Fast MD. Sea lice population and sex differences in P-glycoprotein expression and emamectin benzoate resistance on salmon farms in the Bay of Fundy, New Brunswick, Canada. Pest Manag Sci. 2013;70:905–914. doi: 10.1002/ps.3620. [DOI] [PubMed] [Google Scholar]
- 36.Whyte SK, Westcott JD, Elmoslemany A, Hammell KL, Revie CW. A fixed-dose approach to conducting emamectin benzoate tolerance assessments on field-collected sea lice, Lepeophtheirus salmonis. J Fish Dis. 2013;36:283–292. doi: 10.1111/jfd.12055. [DOI] [PubMed] [Google Scholar]
- 37.Sutherland BJG, Poley JD, Igboeli OO, Jantzen JR, Fast MD, Koop BF, Jones SRM. Transcriptomic responses to emamectin benzoate in Pacific and Atlantic Canada salmon lice Lepeophtheirus salmonis with differing levels of drug resistance. Evol Appl. 2015;8:133–148. doi: 10.1111/eva.12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skern-mauritzen R, Torrissen O, Glover KA. Pacific and Atlantic Lepeophtheirus salmonis (Krøyer, 1838) are allopatric subspecies : Lepeophtheirus salmonis salmonis and L. salmonis oncorhynchi subspecies novo. BMC Genet. 2014;15:1–9. doi: 10.1186/1471-2156-15-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yasuike M, Leong J, Jantzen SG, von Schalburg KR, Nilsen F, Jones SRM, Koop BF. Genomic resources for sea lice: Analysis of ESTs and mitochondrial genomes. Mar Biotechnol. 2012;14:155–166. doi: 10.1007/s10126-011-9398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cutter AD, Ward S. Sexual and temporal dynamics of molecular evolution in C. elegans development. Mol Biol Evol. 2005;22:178–188. doi: 10.1093/molbev/msh267. [DOI] [PubMed] [Google Scholar]
- 42.Avila FW, Sirot LK, LaFlamme BA, Rubinstein DC, Wolfner MF. Insect seminal fluid proteins: Identification and function. Annu Rev Entemology. 2011;56:21–40. doi: 10.1146/annurev-ento-120709-144823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman T. Seminal fluid-mediated fitness traits in Drosophila. Heredity. 2001;87:511–521. doi: 10.1046/j.1365-2540.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- 44.Poley JD, Igboeli OO, Fast MD. Towards a consensus: Multiple experiments provide evidence for constitutive expression differences among sexes and populations of sea lice (Lepeophtheirus salmonis) related to emamectin benzoate resistance. Aquaculture. 2015;448:445–50. doi: 10.1016/j.aquaculture.2015.06.026. [DOI] [Google Scholar]
- 45.Carmichael SN, Bekaert M, Taggart JB, Christie HRL, Bassett DI, Bron JE, Skuce PJ, Gharbi K, Skern-Mauritzen R, Sturm A. Identification of a sex-linked SNP marker in the salmon louse (Lepeophtheirus salmonis) using RAD sequencing. PLoS One. 2013;8:e77832. doi: 10.1371/journal.pone.0077832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Granadino B, Campuzano S, Sanchez L. The Drosophila melanogaster fl(2)d gene is needed the female-specific splicing of sex-lethal RNA. EMBO J. 1990;9:2597–2602. doi: 10.1002/j.1460-2075.1990.tb07441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eichner C, Frost P, Dysvik B, Jonassen I, Kristiansen B, Nilsen F. Salmon louse (Lepeophtheirus salmonis) transcriptomes during post molting maturation and egg production, revealed using EST-sequencing and microarray analysis. BMC Genomics. 2008;15:1–15. doi: 10.1186/1471-2164-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carmichael SN, Bron JE, Taggart JB, Ireland JH, Bekaert M, Burgess ST, Skuce PJ, Nisbet AJ, Gharbi K, Sturm A. Salmon lice (Lepeophtheirus salmonis) showing varying emamectin benzoate susceptibilities differ in neuronal acetylcholine receptor and GABA-gated chloride channel mRNA expression. BMC Genomics. 2013;14:408. doi: 10.1186/1471-2164-14-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valenzuela-Muñoz V, Sturm A, Gallardo-Escárate C. Transcriptomic insights on the ABC transporter gene family in the salmon louse Caligus rogercresseyi. Parasit Vectors. 2015;8:1–14. doi: 10.1186/s13071-015-0801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Small CM, Carney GE, Mo Q, Vannucci M, Jones AG. A microarray analysis of sex- and gonad-biased gene expression in the zebrafish: evidence for masculinization of the transcriptome. BMC Genomics. 2009;10:579. doi: 10.1186/1471-2164-10-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lees F, Baillie M, Gettinby G, Revie CW. The efficacy of emamectin benzoate against infestations of Lepeophtheirus salmonis on farmed Atlantic salmon (Salmo salar L) in Scotland, 2002-2006. PLoS One. 2008;3:e1549. doi: 10.1371/journal.pone.0001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whyte SK, Westcott JD, Jimenez D, Revie CW, Hammell KL. Assessment of sea lice (Lepeophtheirus salmonis) management in New Brunswick, canada using deltamethrin (Alphamax®) through clinical field treatment and laboratory bioassay responses. Aquaculture. 2014;422–423:54–62. doi: 10.1016/j.aquaculture.2013.11.027. [DOI] [Google Scholar]
- 54.Farlora R, Araya-Garay J, Gallardo-Escárate C. Discovery of sex-related genes through high-throughput transcriptome sequencing from the salmon louse Caligus rogercresseyi. Mar Genomics. 2014;15:85–93. doi: 10.1016/j.margen.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 55.McIntyre LM, Bono LM, Genissel A, Westerman R, Junk D, Telonis-Scott M, Harshman L, Wayne ML, Kopp A, Nuzhdin SV. Sex-specific expression of alternative transcripts in Drosophila. Genome Biol. 2006;7:R79. doi: 10.1186/gb-2006-7-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 57.Swanson WJ, Vacquier VD. Reproductive Protein Evolution. Annu Rev Ecol Syst. 2002;33:161–179. doi: 10.1146/annurev.ecolsys.33.010802.150439. [DOI] [Google Scholar]
- 58.Mueller JL, Ripoll DR, Aquadro CF, Wolfner MF. Comparative structural modeling and inference of conserved protein classes in Drosophila seminal fluid. Proc Natl Acad Sci U S A. 2004;101:13542–13547. doi: 10.1073/pnas.0405579101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laflamme BA, Wolfner MF. Identification and function of proteolysis regulators in seminal fluid. Mol Reprod Dev. 2013;80:80–101. doi: 10.1002/mrd.22130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGraw LA, Gibson G, Clark AG, Wolfner MF. Genes regulated by mating, sperm, or seminal fluid proteins in mated female Drosophila mealnogaster. Curr Biol. 2004;14:1509–1514. doi: 10.1016/j.cub.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 61.Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart J-M. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. 2002;297:114–116. doi: 10.1126/science.1072391. [DOI] [PubMed] [Google Scholar]
- 62.Fontenele M, Lim B, Oliveira D, Buffolo M, Perlman DH, Schupbach T, Araujo H. Calpain A modulates toll responses by limited Cactus/IkB proteolysis. Mol Biol Cell. 2013;24:2966–2980. doi: 10.1091/mbc.E13-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng J, Zipperlen P, Kubli E. Drosophila sex-peptide stimulates female innate immune system after mating via the toll and Imd pathways. Curr Biol. 2005;15:1690–1694. doi: 10.1016/j.cub.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 64.Karoonuthaisiri N, Sittikankeaw K, Preechaphol R, Kalachikov S, Wongsurawat T, Uawisetwathana U, Russo JJ, Ju J, Klinbunga S, Kirtikara K. ReproArray(GTS): a cDNA microarray for identification of reproduction-related genes in the giant tiger shrimp Penaeus monodon and characterization of a novel nuclear autoantigenic sperm protein (NASP) gene. Comp Biochem Physiol, Part D: Genomics Proteomics. 2009;4:90–99. doi: 10.1016/j.cbd.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 65.Wang M, Shi J-L, Cheng G-Y, Hu Y-Q, Xu C. The antibody against a nuclear autoantigenic sperm protein can result in reproductive failure. Asian J Androl. 2009;11:183–192. doi: 10.1038/aja.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deegan-Clark JI, Dimmer EC, Mungall CJ. Formalization of taxon-based constraints to detect inconsistencies in annotation and ontology development. BMC Bioinformatics. 2010;11:530. doi: 10.1186/1471-2105-11-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grote P, Conradt B. The PLZF-like Protein TRA-4 Cooperates with the Gli-like Transcription Factor TRA-1 to Promote Female Development in C. elegans. Dev Cell. 2006;11:561–573. doi: 10.1016/j.devcel.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 68.Mack PD, Kapelnikov A, Heifetz Y, Bender M. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:10358–10363. doi: 10.1073/pnas.0604046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rogers DW, Whitten MMA, Thailayil J, Soichot J, Levashina EA, Catteruccia F. Molecular and cellular components of the mating machinery in Anopheles gambiae females. Proc Natl Acad Sci U S A. 2008;105:19390–19395. doi: 10.1073/pnas.0809723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Z, Hambuch TM, Parsch J. Molecular evolution of sex-biased genes in Drosophila. Mol Biol Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]
- 71.Muse SV. Estimating synonymous and nonsynonymous substitution rates. Mol Biol Evol. 1996;13:105–114. doi: 10.1093/oxfordjournals.molbev.a025549. [DOI] [PubMed] [Google Scholar]
- 72.Powell JR, Moriyama EN. Evolution of codon usage bias in Drosophila. Proc Natl Acad Sci U S A. 1997;94:7784–7790. doi: 10.1073/pnas.94.15.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- 74.Appel LF, Prout M, Abu-Shumays R, Hammonds A, Garbe JC, Fristrom D, Fristrom J. The Drosophila Stubble-stubbloid gene encodes an apparent transmembrane serine protease required for epithelial morphogenesis. Proc Natl Acad Sci U S A. 1993;90:4937–4941. doi: 10.1073/pnas.90.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jormalainen V. Precopulatory mate guarding in crustaceans: male competitive strategy and intersexual conflict. Q Rev Biol. 1998;73:275–304. doi: 10.1086/420306. [DOI] [Google Scholar]
- 76.Kerman BE, Cheshire AM, Myat MM, Andrew DJ. Ribbon modulates apical membrane during tube elongation through crumbs and moesin. Dev Biol. 2008;320:278–288. doi: 10.1016/j.ydbio.2008.05.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tao T, Shi H, Huang D, Peng J. Def functions as a cell autonomous factor in organogenesis of digestive organs in zebrafish. PLoS One. 2013;8:e58858. doi: 10.1371/journal.pone.0058858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hastie LC, Wallace C, Birkett MA, Douglas A, Jones O, AJ M (L), Ritchie G, Pickett JA, Webster JL, Bowman AS. Prevalence and infection intensity of sea lice (Lepeophtheirus salmonis) on Atlantic salmon (Salmo salar) host is reduced by the non-host compound 2-aminoacetophenone. Aquaculture. 2013;410-411:179–183. doi: 10.1016/j.aquaculture.2013.06.035. [DOI] [Google Scholar]
- 79.Sánchez-Gracia A, Vieira FG, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity. 2009;103:208–216. doi: 10.1038/hdy.2009.55. [DOI] [PubMed] [Google Scholar]
- 80.Cheah Y, Yang W. Functions of essential nutrition for high quality spermatogenesis. Adv Biosci Biotechnol. 2011;02:182–197. doi: 10.4236/abb.2011.24029. [DOI] [Google Scholar]
- 81.Stavang JA, Chauvigné F, Kongshaug H, Cerdà J, Nilsen F, Finn RN. Phylogenomic and functional analyses of salmon lice aquaporins uncover the molecular diversity of the superfamily in Arthropoda. BMC Genomics. 2015;16:618. doi: 10.1186/s12864-015-1814-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turek M, Bringmann H. Gene expression changes of Caenorhabditis elegans larvae during molting and sleep-like lethargus. PLoS One. 2014;9:e113269. doi: 10.1371/journal.pone.0113269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muller H, Catteruccia F, Vizioli J, Torre D, Crisanti A. Constitutive and blood meal-induced trypsin genes in Anopheles gambiae. Exp Parasitol. 1995;81:371–385. doi: 10.1006/expr.1995.1128. [DOI] [PubMed] [Google Scholar]
- 84.Fast AMD, Burka JF, Johnson SC, Ross NW. Enzymes released from Lepeophtheirus salmonis in response to mucus from different salmonids. Am Soc Parasitol. 2003;89:7–13. doi: 10.1645/0022-3395(2003)089[0007:ERFLSI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 85.Fast MD, Johnson SC, Eddy TD, Pinto D, Ross NW. Lepeophtheirus salmonis secretory/excretory products and their effects on Atlantic salmon immune gene regulation. Parasite Immunol. 2007;29:179–189. doi: 10.1111/j.1365-3024.2007.00932.x. [DOI] [PubMed] [Google Scholar]
- 86.Kvamme BO, Skern R, Frost P, Nilsen F. Molecular characterisation of five trypsin-like peptidase transcripts from the salmon louse (Lepeophtheirus salmonis) intestine. Int J Parasitol. 2004;34:823–832. doi: 10.1016/j.ijpara.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 87.Jones PG, Hammell KL, Gettinby G, Revie CW. Detection of emamectin benzoate tolerance emergence in different life stages of sea lice, Lepeophtheirus salmonis, on farmed Atlantic salmon, Salmo salar L. J Fish Dis. 2013;36:209–220. doi: 10.1111/jfd.12022. [DOI] [PubMed] [Google Scholar]
- 88.Saksida SM, Morrison D, McKenzie P, Milligan B, Downey E, Boyce B, Eaves A. Use of Atlantic salmon, Salmo salar L., farm treatment data and bioassays to assess for resistance of sea lice, Lepeophtheirus salmonis, to emamectin benzoate (SLICE®) in British Columbia, Canada. J Fish Dis. 2013;36:515–520. doi: 10.1111/jfd.12018. [DOI] [PubMed] [Google Scholar]
- 89.Sutherland BJG, Jantzen SG, Yasuike M, Sanderson DS, Koop BF, Jones SRM. Transcriptomics of coping strategies in free-swimming Lepeophtheirus salmonis (Copepoda) larvae responding to abiotic stress. Mol Ecol. 2012;21:6000–6014. doi: 10.1111/mec.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang D, Sherman B, Lempick R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 92.Huang D, Sherman B, Lempick R. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jantzen SG, Sutherland BJ, Minkley DR, Koop BF. GO Trimming: Systematically reducing redundancy in large Gene Ontology datasets. BMC Res Notes. 2011;4:267. doi: 10.1186/1756-0500-4-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Consortium U. UniProt: a hub for protein information. Nucleic Acids Res. 2014;43:204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sonenshine DE, Bissinger BW, Egekwu N, Donohue KV, Khalil SM, Roe RM. First transcriptome of the testis-vas deferens-male accessory gland and proteome of the spermatophore from Dermacentor variabilis (acari: Ixodidae) PLoS One. 2011;6:e24711. doi: 10.1371/journal.pone.0024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dorus S, Wasbrough E, Busby J, Wilkins E, Karr T. Sperm proteomics reveals intensified sexual selection on mouse sperm membrane and acrosome genes. Mol Biol Evol. 2010;27:1235–1246. doi: 10.1093/molbev/msq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Findlay GD, Yi X, Maccoss MJ, Swanson WJ. Proteomics Reveals Novel Drosophila Seminal Fluid Proteins Transferred at Mating. Plos Biol. 2008;6:1417–1426. doi: 10.1371/journal.pbio.0060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Costa AFDV, Gasser RB, Dias SRC, Rabelo EML. Male-enriched transcription of genes encoding ASPs and Kunitz-type protease inhibitors in Ancylostoma species. Mol Cell Probes. 2009;23:298–303. doi: 10.1016/j.mcp.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 99.Veselsky L, Jonakova V, Cechova D. A Kunitz Type of Proteinase Inhibitor Isolated from Boar Seminal Vesicle Fluid. Andrologia. 1985;17:352–358. doi: 10.1111/j.1439-0272.1985.tb01020.x. [DOI] [PubMed] [Google Scholar]
- 100.Clauss A, Persson M, Lilja H, Lundwall Å. Three genes expressing Kunitz domains in the epididymis are related to genes of WFDC-type protease inhibitors and semen coagulum proteins in spite of lacking similarity between their protein products. BMC Biochem. 2011;12:55. doi: 10.1186/1471-2091-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.South A, Sirot LK, Lewis SM. Identification of predicted seminal fluid proteins in Tribolium castaneum. Insect Mol Biol. 2011;20:447–456. doi: 10.1111/j.1365-2583.2011.01083.x. [DOI] [PubMed] [Google Scholar]
- 102.Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7:R40. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ram KR, Wolfner MF. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr Comp Biol. 2007;47:427–445. doi: 10.1093/icb/icm046. [DOI] [PubMed] [Google Scholar]
- 104.Konno A, Shiba K, Cai C, Inaba K. Branchial cilia and sperm flagella recruit distinct axonemal components. PLoS One. 2015;10:e0126005. doi: 10.1371/journal.pone.0126005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Muhlrad PJ, Ward S. Spermiogenesis initiation in Caenorhabditis elegans involves a casein kinase 1 encoded by the spe-6 gene. Genetics. 2002;161:143–155. doi: 10.1093/genetics/161.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baer B, Heazlewood JL, Taylor NL, Eubel H, Millar AH. The seminal fluid proteome of the honeybee Apis mellifera. Proteomics. 2009;9:2085–2097. doi: 10.1002/pmic.200800708. [DOI] [PubMed] [Google Scholar]
- 107.Braswell WE, Andrés JA, Maroja LS, Harrison RG, Howard DJ, Swanson WJ. Identification and comparative analysis of accessory gland proteins in Orthoptera. Genome. 2006;49:1069–1080. doi: 10.1139/g06-061. [DOI] [PubMed] [Google Scholar]
- 108.Netzel-Arnett S, Bugge TH, Hess RA, Carnes K, Stringer BW, Scarman AL, Hooper JD, Tonks ID, Kay GF, Antalis TM. The glycosylphosphatidylinositol-anchored serine protease PRSS21 (testisin) imparts murine epididymal sperm cell maturation and fertilizing ability. Biol Reprod. 2009;81:921–932. doi: 10.1095/biolreprod.109.076273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scarman AL, Hooper JD, Boucaut KJ, Sit ML, Webb GC, Normyle JF, Antalis TM. Organization and chromosomal localization of the murine Testisin gene encoding a serine protease temporally expressed during spermatogenesis. Eur J Biochem. 2001;268:1250–1258. doi: 10.1046/j.1432-1327.2001.01986.x. [DOI] [PubMed] [Google Scholar]
- 110.Yu JX, Chao L, Chao J. Prostasin is a novel human serine proteinase from seminal fluid. Purification, tissue distribution, and localization in prostate gland. J Biol Chem. 1994;269:18843–18848. [PubMed] [Google Scholar]
- 111.Kelleher ES, Watts TD, LaFlamme BA, Haynes PA, Markow TA. Proteomic analysis of Drosophila mojavensis male accessory glands suggests novel classes of seminal fluid proteins. Insect Biochem Mol Biol. 2009;39:366–371. doi: 10.1016/j.ibmb.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 112.Walker MJ, Rylett CM, Keen JN, Audsley N, Sajid M, Shirras AD, Isaac RE. Proteomic identification of Drosophila melanogaster male accessory gland proteins, including a pro-cathepsin and a soluble gamma-glutamyl transpeptidase. Proteome Sci. 2006;4:9. doi: 10.1186/1477-5956-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mawson CA, Fischer MI. Zinc and carbonic anhydrase in human semen. Biochem J. 1953;55:696–700. doi: 10.1042/bj0550696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Inaba K, Dréanno C, Cosson J. Control of flatfish sperm motility by CO2 and carbonic anhydrase. Cell Motil Cytoskeleton. 2003;55:174–187. doi: 10.1002/cm.10119. [DOI] [PubMed] [Google Scholar]
- 115.Holm L, Ridderstråle Y, Knutsson P. Localisation of carbonic anhydrase in the sperm storing regions of the domestic hen oviduct. Cells Tissues Organs. 1996;156:253–260. doi: 10.1159/000147853. [DOI] [PubMed] [Google Scholar]
- 116.Reinhardt K, Wong CH, Georgiou AS. Detection of seminal fluid proteins in the bed bug, Cimex lectularius, using two-dimensional gel electrophoresis and mass spectrometry. Parasitology. 2009;136:283–292. doi: 10.1017/S0031182008005362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen H, Chen X, Zheng Y. The nuclear lamina regulates germline stem cell niche organization via modulation of EGFR signalling. Cell Stem Cell. 2013;13:73–86. doi: 10.1016/j.stem.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yan W, Ma L, Burns KH, Matzuk MM. Haploinsufficiency of kelch-like protein homolog 10 causes infertility in male mice. Proc Natl Acad Sci U S A. 2004;101:7793–7798. doi: 10.1073/pnas.0308025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang CG, Lamitina T, Agre P, Strange K. Functional analysis of the aquaporin gene family in Caenorhabditis elegans. Am J Physiol Cell Physiol. 2007;292:C1867–C1873. doi: 10.1152/ajpcell.00514.2006. [DOI] [PubMed] [Google Scholar]
- 120.Milardi D, Grande G, Vincenzoni F, Messana I, Pontecorvi A, De Marinis L, Castagnola M, Marana R. Proteomic approach in the identification of fertility pattern in seminal plasma of fertile men. Fertil Steril. 2012;97:67–73. doi: 10.1016/j.fertnstert.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 121.Drapeau MD, Albert S, Kucharski R, Prusko C, Maleszka R. Evolution of the yellow/major royal jelly protein family and the emergence of social behavior in honey bees. Genome Res. 2006;16:1385–1394. doi: 10.1101/gr.5012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schmitzová J, Klaudiny J, Albert Š, Schröder W, Schreckengost W, Hanes J, Júdová J, Šimúth J. A family of major royal jelly proteins of the honeybee Apis mellifera L. Cell Mol Life Sci. 1998;54:1020–1030. doi: 10.1007/s000180050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Albert Š, Klaudiny J, Šimúth J. Molecular characterization of MRJP3, highly polymorphic protein of honeybee (Apis mellifera) royal jelly. Insect Biochem Mol Biol. 1999;29:427–434. doi: 10.1016/S0965-1748(99)00019-3. [DOI] [PubMed] [Google Scholar]
- 124.Albert S, Bhattacharya D, Klaudiny J, Schmitzová J, Simúth J. The family of major royal jelly proteins and its evolution. J Mol Evol. 1999;49:290–297. doi: 10.1007/PL00006551. [DOI] [PubMed] [Google Scholar]
- 125.Tootle TL, Spradling AC. Drosophila Pxt: a cyclooxygenase-like facilitator of follicle maturation. Development. 2008;135:839–847. doi: 10.1242/dev.017590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Konstandi OA, Papassideri IS, Stravopodis DJ, Kenoutis CA, Hasan Z, Katsorchis T, Wever R, Margaritis LH. The enzymatic component of Drosophila melanogaster chorion is the Pxd peroxidase. Insect Biochem Mol Biol. 2005;35:1043–1057. doi: 10.1016/j.ibmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 127.Dezelee S, Bras F, Contamine D, Lopez-Ferber M, Segretain D, Teninges D. Molecular analysis of ref(2)P, a Drosophila gene implicated in sigma rhabdovirus multiplication and necessary for male fertility. EMBO J. 1989;8:3437–3446. doi: 10.1002/j.1460-2075.1989.tb08508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the results of this article are available in the Gene Expression Omnibus repository, GSE73734 and GSE56024.
Atlantic data set described and uploaded to GEO in [37].
Reviewer access link for Pacific lice (GSE73734): http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73734.


