Significance
This study reports that publicly available gene array expression data together with statistically significant connections’ map successfully predicts licensed drugs able to modify genes of interest. We used this method to predict drugs able to induce A20 [TNFα-induced protein 3 (TNFAIP3)], which is reduced in cystic fibrosis (CF) airway cells, and thus normalize the inflammatory response. Using CF and non-CF airway epithelial cells, ikarugamycin and quercetin had antiinflammatory effects mediated by induction of A20. We confirmed that this was mainly due to A20 induction, because no antiinflammatory effects were seen in bronchial epithelial cells with A20 knockdown. We have identified a process whereby already licensed drugs can be successfully repositioned for chronic inflammatory airway diseases.
Keywords: A20, NF-κB, connectivity mapping, drug repositioning, CF airway inflammation
Abstract
Cystic fibrosis (CF) lung disease is characterized by chronic and exaggerated inflammation in the airways. Despite recent developments to therapeutically overcome the underlying functional defect in the cystic fibrosis transmembrane conductance regulator, there is still an unmet need to also normalize the inflammatory response. The prolonged and heightened inflammatory response in CF is, in part, mediated by a lack of intrinsic down-regulation of the proinflammatory NF-κB pathway. We have previously identified reduced expression of the NF-κB down-regulator A20 in CF as a key target to normalize the inflammatory response. Here, we have used publicly available gene array expression data together with a statistically significant connections’ map (sscMap) to successfully predict drugs already licensed for the use in humans to induce A20 mRNA and protein expression and thereby reduce inflammation. The effect of the predicted drugs on A20 and NF-κB(p65) expression (mRNA) as well as proinflammatory cytokine release (IL-8) in the presence and absence of bacterial LPS was shown in bronchial epithelial cells lines (16HBE14o−, CFBE41o−) and in primary nasal epithelial cells from patients with CF (Phe508del homozygous) and non-CF controls. Additionally, the specificity of the drug action on A20 was confirmed using cell lines with tnfαip3 (A20) knockdown (siRNA). We also show that the A20-inducing effect of ikarugamycin and quercetin is lower in CF-derived airway epithelial cells than in non-CF cells.
The response to pathogens, recognized by pattern recognition receptors including Toll-like receptors (TLRs), triggers an acute innate immune response that is mediated by transcription factors such as nuclear factor kappa-light-chain-enhancer of B cells (NF-κB). NF-κB activation promotes the transcription of inflammatory mediators in a tightly regulated process. However, in individuals with underlying chronic inflammatory diseases, this regulation is compromised, leading to constitutive NF-κB activation and persistent inflammation (1–3).
The development of new first-in-class medicines is costly (approximately $1.2 billion for a single FDA-approved drug) and takes between 10 y and 15 y (4, 5). Many newly developed drugs perform well in the preclinical testing but fail when tested in humans (6). Thus, alternative approaches using predictive models to identify new drugs are needed. Gene expression connectivity mapping (www.broadinstitute.org/cmap/) is an advanced bioinformatics technique to establish the connections among biological states via gene expression profiles/signatures. One major application of connectivity mapping is to identify potential small molecules able to inhibit a disease state or regulate the expression of a small number of genes (7–9). We used an advanced version of connectivity mapping, statistically significant connections’ map (sscMap) (10), which has been successfully applied to phenotypic targeting and predicting effective drugs in cancer (10). However, this has not yet been applied to chronic inflammatory diseases.
Cystic fibrosis (CF) is a chronic multiorgan inflammatory disease, caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene expressed on apical epithelial surfaces. It is the most common lethal genetic disease in Caucasian populations. Lung disease is the primary cause of morbidity and mortality in CF, resulting from dehydration of epithelial surfaces and reduced mucociliary clearance as a consequence of the ionic imbalance created by the CFTR mutation. This leads to a cycle of infection and inflammation associated with a progressive reduction in lung function and eventual respiratory failure. A common feature of CF is the heightened, chronic inflammatory response to Pseudomonas aeruginosa, driven by constitutive NF-κB activation in airway and peripheral blood cells (2, 3, 11). Primary nasal epithelial cells (PNECs) from patients with the common Phe508del/Phe508del mutation and a milder genotype (R117H/Phe508del) show a significant increase in NF-κB(p65), which correlates with disease severity (12).
A20 [TNFα-induced protein 3 (TNFAIP3)] is a central negative regulator of NF-κB activation following stimulation of TLRs and/or TNF receptor and regulates different signaling pathways such as NF-κB and IFN regulatory factor signaling (13). A20 modifies classical immune cells (14, 15) as well as epithelial cells (12), endothelial cells (16), embryonic fibroblasts (17), osteoclasts (18), and pancreatic beta cells (19) and diverse roles have been suggested for A20 in innate immunity, apoptosis, autophagy and antigen processing (13, 15, 16, 20). Within the innate inflammatory immune response, A20 regulates NF-κB signaling at the level of TRAF6 in mouse embryonic fibroblasts and osteoclasts (17, 18). In cultured human airway epithelial cells, A20 is rapidly induced by viral or bacterial compounds (21) and is essential for termination of the TLR4 signal (22). PNECs stimulated with P. aeruginosa LPS show a transient increase in A20, but CF PNECs display lower A20 expression basally and after LPS stimulation (12, 23).
Therefore, A20 induction should have antiinflammatory effects within the tightly regulated NF-κB signaling pathway as shown by the induction of A20 through gibberellin (GA3) in airway epithelial cells. GA3 induced A20, reduced IL-8 secretion, stabilized cytosolic IkBα, and reduced NF-κB (p65) activation (24). Here we set out to identify additional compounds able to induce A20. Thus, we performed a compound search using gene expression connectivity mapping to identify existing drugs that could induce A20 expression.
Results
Connectivity Mapping (sscMap).
The selection of gene array data and creation of the gene signature.
Datasets that passed the selection criteria contained human PNECs and the human bronchial epithelial cell lines CFBE41o−, Calu-3, and IB3-1 analyzed basally and after exposure to P. aeruginosa LPS (Table 1). In total, 76 samples from four different published gene array datasets were used. Linear expression correlation and gene ontology (GO) enrichment analysis for NF-κB pathway genes identified the closest correlates to A20. Table 2 shows the top seven genes that subsequently served as the input to the connectivity mapping process.
Table 1.
Selected GEO gene expression datasets used for the connectivity mapping process
| ID of dataset | Sample type | Platform used | Sample size | Reference |
| GSE2395 | PNECs | HG-U133A and HG-U133B: GPL96/GLP97 | 40 (18 CF {F508del homozygous}, 22 controls), exposed to P. aeruginosa | (56) |
| GSE30439 | CFBE41o− | HG-U133 Plus2: GPL570 | 12 (CFBE41o− and CFBE41o− complemented with wildtype CFTR), ± exposure to PA01 | (57) |
| GSE620 | IB3-1 | HG-U133A: GPL96 | 5 (IB3-1 cells, excluded those with 4-phenylbutyrate treatment) | (58) |
| GSE923 | Calu-3 | HG-U133A: GPL96 | 19 (Calu-3 {F508del/F508del}) infected with P. aeruginosa (mucoid vs. motile strain) | (59) |
Table 2.
Gene expression profile of NF-κB/A20 related genes in CF airway disease
| Gene symbol | Unique ID | Name | Function |
| TNFAIP3 | 202643_s_at | A20 | Ubiquitination, negative regulator of NF-κB |
| ATF3 | 202672_s_at | Activating transcription factor 3 | Binds the CRE, transcriptional repressor (promoters with ATF sites) |
| RAB5C | 201140_s_at | RAB5C | Small GTPase, regulates membrane traffic from plasma membrane to early endosomes, ubiquitously expressed |
| DENND4A | 214787_at | DENN/MADD domain containing 4A | C-myc promoter-binding protein, promotes exchange of GDP to GTP, converting inactive GDP-bound Rab proteins into their active GTP-bound form |
| POM121 | 212178_s_at | POM121 | Nuclear pore protein |
| ICAM1 | 202638_s_at | Also CD54 | Intracellular adhesion molecule 1 |
| PSEN1 | 207782_s_at | Presenilin 1 | Transmembrane protein, proteolytic subunit of γ-secretase, cleaves transmembrane proteins (e.g., β-amyloid) |
Genes behaving in a similar way to the target gene A20 were determined using linear correlation analyses of the selected gene expression datasets.
Prediction of drugs to induce A20 in airway epithelial cells.
This study sought small molecular compounds that may enhance A20 expression and, as a confirming negative control, those compounds that may inhibit A20 expression. Table 3 summarizes the top candidate drugs identified. The column “Z-score” shows the correlation of the drugs with the input gene signature. Positive Z-scores indicate a positive correlation, i.e., the input genes are induced when treated with the particular drug. The significant drugs with the highest positive Z-scores, along with a negative control, were selected for laboratory validation. In addition to P values and Z-scores, stability of the connections was measured by altering the gene signature. The significance of the connections are calculated as the “Perturbation stability.” Drugs with a perturbation stability of 1 represent strong connections that remain significant with perturbation gene signatures. From these predictions, two A20-inducing drugs (ikarugamycin and quercetin) as well as one non–A20-inducing drug (fluvastatin) were chosen for further investigation.
Table 3.
Candidate compounds predicted to induce or reduce A20 expression
| Drug | C-score | P value | Z-score | Drug class |
| Azacyclonol | 0.3496 | 1.00E-05 | 4.8417 | γ-Pipradol, ataractive drug |
| Ikarugamycin | 0.4115 | 6.00E-05 | 4.1275 | Macrolide type |
| Quercetin | 0.33712 | 5.00E-05 | 3.8963 | Flavenoid |
| Karakoline | 0.43145 | 1.00E-05 | 3.8033 | Alkaloid diterpenoid |
| Fluvastatin | −0.35325 | 1.00E-04 | −3.8267 | HMG-CoA reductase inhibitor |
Only drug predictions with a significance value of 1 and a perturbation stability of 1 were regarded as candidate compounds.
Gene Expression of Gene Signature.
Expression of the genes identified as the A20/NF-κB gene signature in CF epithelial cells were analyzed by quantitative real-time (qRT)-PCR (primers are given in Table S1) in 16HBE14o− and CFBE41o− cultured in the presence or absence of LPS for 0–24 h (Fig. 1).
Table S1.
PCR primer sequences
| Gene | Accession number | Forward (left) primer | Reverse (right) primer |
| ATF3 | NM_001674.3 | caagtgcatctttgcctcaa | cacccgaggtacagacact |
| RAB5C | NM_201434.2 | ctgaatgacccgactggaat | aaaggtgcaggtggaatgac |
| DENND4A | NM_001144823.1 | gtggcaaagacgaacactgt | acaggcagagggtatttgct |
| POM121 | NM_172020.4 | atcacctctgccagtccatc | atcacctctgccagtccatc |
| ICAM1 | AY225514.1 | ccataaggtcttgcctccaa | cattggagtctgctgggaat |
| PSEN1 | NM_000021.3 | ctggggaggacaaaggtgat | cctcgtccctcaaatctggt |
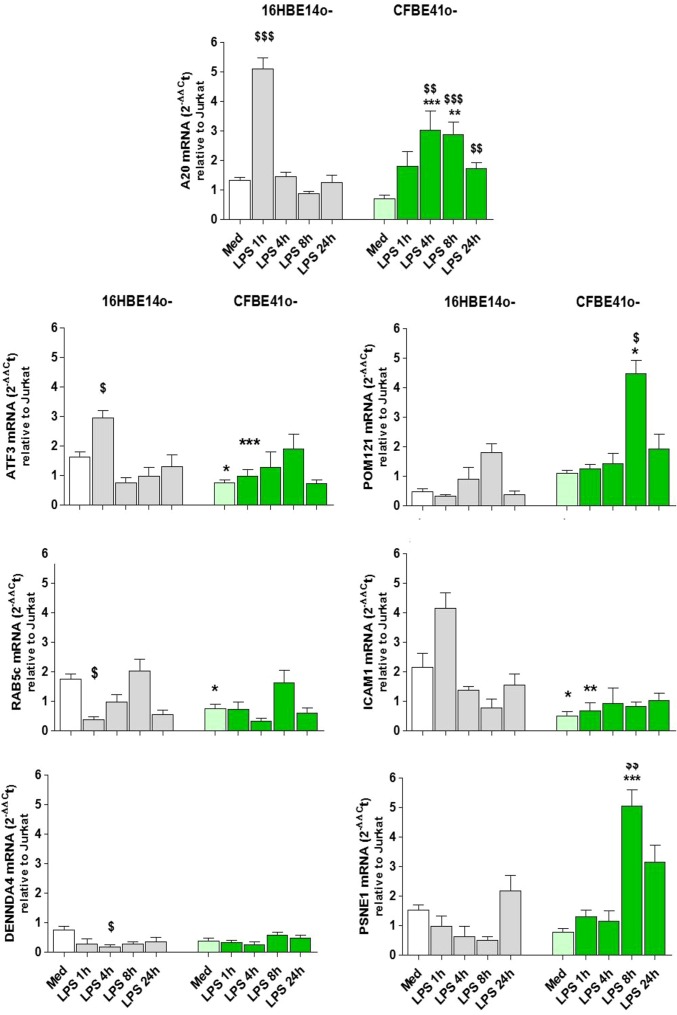
Fig. 1.
Gene expression profile of the gene signature genes associated with A20. The 16HBE14o− (grey) and CFBE41o− (green) were stimulated (LPS, 10 μg/mL, 0–24 h), and mRNA levels of A20, ATF3, Rab5c, DENNDA4, POM121, ICAM-1, and PSNE1 were determined as described; $, significant difference compared with medium control; *, significant differences between genotypes.
Basal expression.
CFBE41o− show significantly lower mRNA expression for A20, ATF3, Rab5c, and ICAM1 compared with 16HBE14o− (all P < 0.05, n = 5). Expression of DENNDA4 and PSNE1 was also lower in CFBE41o−, but this did not reach significance.
LPS-induced expression.
In 16HBE14o−, A20 mRNA is rapidly induced, with expression peaking 1 h after LPS exposure (P < 0.001 compared with medium, n = 5), whereas CFBE41o− show significantly lower (at 1 h, P < 0.001 vs. 16HBE14o−, n = 5) and delayed (maximal induction at 4–8 h, P < 0.01 and 0.001 vs. medium, n = 5) induction upon LPS stimulation. After LPS, ATF3 and ICAM1 expression was significantly lower in CFBE41o− compared with 16HBE14o− (P < 0.01, n = 5). Pom121 and PSNE1 expression increased in CFBE41o− compared with medium (8 h, P < 0.05 and P < 0.01) and in CFBE41o− compared with 16HBE14o− (8 h, P < 0.05 and P < 0.001). Non-CF 16HBE14o− showed a significant reduction in DENNDA4 and Rab5c expression compared with medium (1 h, P < 0.05; and 4 h, P < 0.05, respectively), but there was no significant change in the expression of these genes in CFBE41o−.
Effect of A20-Inducing Drugs on Cell Lines.
Lactate dehydrogenase release in drug-exposed 16HBE14o− and CFBE41o−.
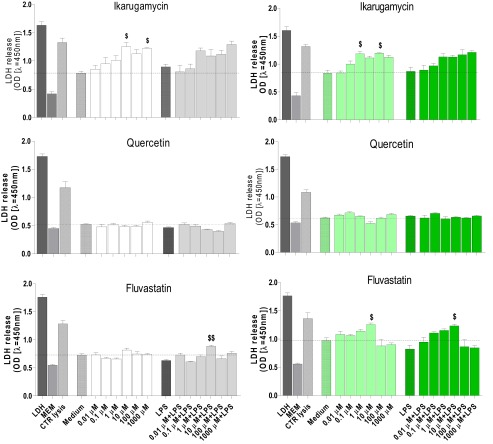
Lactate dehydrogenase (LDH) release was measured after exposure to the drugs alone (0.01–1,000 µM) and with LPS stimulation (Fig. S1). Quercetin did not cause any LDH release. Overall, fluvastatin was almost without effect on LDH; the exceptions were a slight but statistically significant increase at 10 µM alone (CFBE41o−) and in the presence of LPS (in CFBE41o− and 16HBE14o−). Ikarugamycin (1, 100 µM) caused a significantly higher LDH release in both cell types. In LPS-stimulated cells, 0.1 μM ikarugamycin and higher concentrations showed a higher LDH release compared with LPS alone, but this did not reach statistical significance (Fig. S1).
Fig. S1.
Effect of the predicted A20-inducing drugs on LDH release. Cell lines 16HBE14o− (grey bars) and CFBE41o− (green bars) were preincubated for 1 h with ikarugamycin, quercetin, or fluvastatin (0.01–1,000 μM) and then stimulated with LPS (10 μg/mL) for 24 h. LDH release was determined in the cell-free supernatant as described in Materials and Methods.
LPS-stimulated IL-8 release in drug-pretreated 16HBE14o− and CFBE41o−.
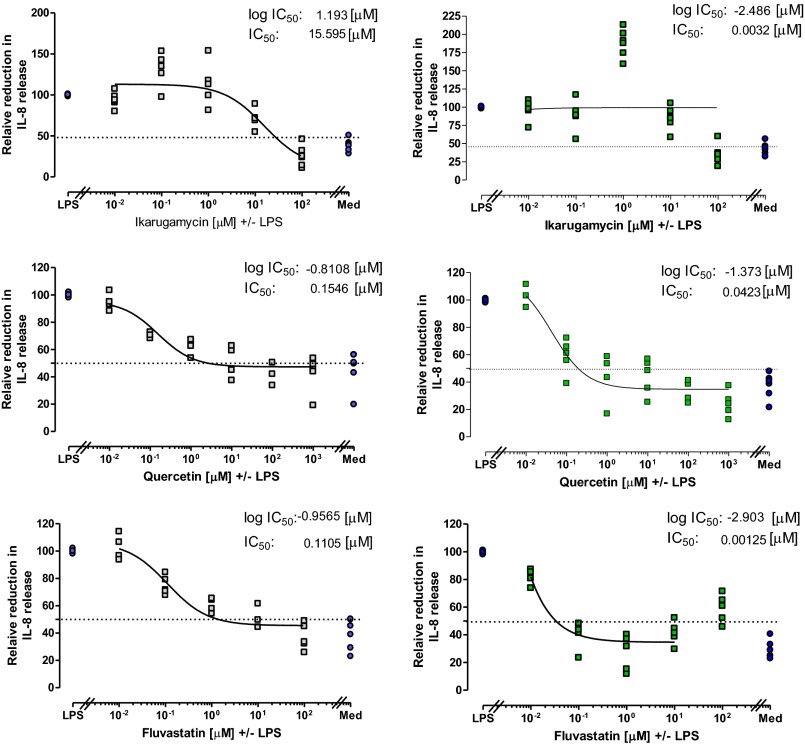
To assess the antiinflammatory potential of the selected drugs, cells were pretreated with the drug for 1 h, stimulated with LPS, the IL-8 release was measured, and the relative IC50 was calculated (Table S2). In 16HBE14o−, all drugs reduced IL-8 release by at least 50% with an IC50 of 15.6 μM for ikarugamycin, 0.09 μM for quercetin, and 0.11 μM for fluvastatin. In CFBE41o−, only quercitin (IC50 0.03 μM) and fluvastatin (IC50 0.001 μM) pretreatment were able to reduce the IL-8 release by 50%. In contrast, pretreatment of CFBE41o− with 1 μM ikarugamycin caused a significant increase in IL-8 release compared with LPS alone (LPS 269.9 ± 47.9 pg/mL vs. 590.7 ± 82.6 pg/mL, P < 0.05, n = 5). Therefore, a meaningful calculation of the relative IC50 for IL-8 release in ikarugamycin treated CFBE41o− cells was not possible.
Table S2.
Relative IC50 and concentrations brought forward for further investigations in PNECs
| Drug | 16HBE14o− | CFBE41o− | No effect concentration, μM | Significant reduction in IL-8, μM | ||
| Log IC50 | Rel. IC50, μM | Log IC50 | Rel. IC50, μM | |||
| Ikarugamycin | 1.193 | 15.59 | NP | NP | 0.01 | 1 |
| Quercetin | −1.049 | 0.09 | −1.522 | 0.03 | 0.1 | 100 |
| Fluvastatin | −0.956 | 0.11 | −2.903 | 0.001 | 0.1 | 1 |
Cell lines (16HBE14o− and CFBE41o−) were preincubated with ikarugamycin, quercetin, or fluvastatin (0.01–100 μM), stimulated (LPS, 10 μg/mL, 24 h) and IL-8 release determined. The relative IC50 was calculated as the logarithmic inhibitory drug concentration versus the relative IL-8 release achieving 50% of inhibition and back-calculated to give the IC50 (in micromolars).
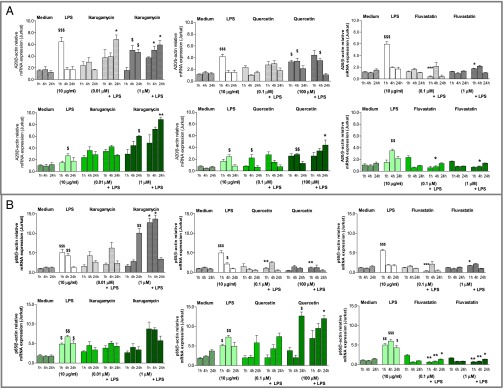
A20 mRNA induction in drug-treated 16HBE14o− and CFBE41o−.
To elucidate if ikarugamycin and quercetin facilitate their antiinflammatory action through the induction of A20 as predicted, A20 mRNA was determined by qRT-PCR. Fluvastatin was included as a negative control. Using the LDH and IL-8 release data, two drug concentrations were selected for further investigations (Table S2). In 16HBE14o−, LPS stimulation caused a significant induction of A20 1 h after stimulation (Fig. 2). Ikarugamycin (0.01 µM) alone did not cause a significant induction of A20, but additional LPS stimulation caused a significant A20 induction at 24 h (P < 0.05 vs. LPS at 24 h). One micromolar of ikarugamycin significantly induced A20 at 4 h on its own, but the higher A20 expression in the presence of LPS (1−24 h) did not reach statistical significance. Quercetin pretreatment did not induce significant levels of A20 mRNA at 0.1 μM, alone or in the presence of LPS. However, at 100 μM quercetin, A20 mRNA was significantly induced alone at 1 h and at 4 h (P < 0.05 vs. medium 1 h and 4 h) and in the presence of LPS at 4 h (P < 0.05 vs. LPS at 4 h). Fluvastatin alone did not induce A20 mRNA at any time or concentration (Fig. 2), and, in the presence of LPS, fluvastatin pretreatment caused a significant reduction in A20 mRNA at both concentrations tested (P < 0.001 for 0.1 μM+LPS at 1 h vs. LPS at 1 h; P < 0.05 for 1 μM+LPS at 1 h vs. LPS at 1 h). In CFBE41o−, LPS significantly induced A20 at 4 h (P < 0.05), but induction levels were lower than in 16HBE14o− (Fig. 2). Ikarugamycin (1 µM) induced significant levels of A20 at 4 h and at 24 h (both P < 0.05); this was further increased in the presence of LPS at 4 h (P < 0.05 vs. LPS at 4 h) and at 24 h (P < 0.01 vs. LPS at 24 h). Quercetin treatment alone significantly induced A20 at 4 h at both concentrations (P < 0.05 0.1 μM vs. medium at 4 h and P < 0.01 100 μM vs. medium at 4 h). In the presence of LPS, only 100 μM quercetin caused a significant induction of A20 at 24 h (P < 0.05). Fluvastatin alone did not induce A20 mRNA at any time or concentration (Fig. 2), and, in the presence of LPS, fluvastatin caused a significant reduction in A20 mRNA at both concentrations tested (P < 0.05 for 0.1 μM+LPS at 4 h vs. LPS 4 h; P < 0.05 for 1 μM+LPS at 4 h vs. LPS at 4 h).
Fig. 2.
A20 (A) and p65 (B) mRNA expression of ikarugamycin-, quercetin-, and fluvastatin-treated cells. Cell lines 16HBE14o− (grey) and CFBE41o− (green) were preincubated with ikarugamycin (0.01, 1 μM, Left), quercetin (0.1, 100 μM, Center), or fluvastatin (0.1, 1 μM, Right) stimulated (LPS, 10 μg/mL, 0–24 h), A20 and p65 mRNA determined (qRT-PCR) and expressed as A20/β-actin or p65//β-actin relative to the internal control. $, significant difference compared to medium control; *, significant differences between genotypes.
Effect of selected components on NF-κB (p65) mRNA in 16HBE14o− and CFBE41o−.
Next we investigated if A20 induction altered NF-κB(p65) mRNA levels (Fig. 2). LPS stimulation caused a significant induction of p65 1 h after stimulation in 16HBE14o− (P < 0.01 – P < 0.001 vs. medium). Ikarugamycin (0.01 µM) was without significant effect on p65 mRNA. At 1 μM, ikarugamycin alone induced p65 at 24 h (P < 0.01 vs. medium at 24 h) and, in the presence of LPS, at 1 h and at 4 h (both P < 0.05 vs. LPS). Quercetin alone showed no effect on p65 mRNA levels, but, when stimulated with LPS, both concentrations of quercetin (0.1, 100 μM) significantly reduced p65 mRNA levels at 1 h (P < 0.01 vs. LPS at 1 h). Similar to quercetin, fluvastatin alone showed no effect on p65 mRNA levels, but, after LPS stimulation, fluvastatin (0.1, 100 μM) significantly reduced p65 mRNA levels at 1 h (P < 0.001 and P < 0.05 vs. LPS at 1 h). In CFBE41o−, LPS significantly induced p65 at 1 h, 4 h, and 24 h (P < 0.05–0.001 vs. medium), and, overall, CFBE41o− exhibited higher expression levels of p65 at 4 h and at 24 h than 16HBE14o− (Fig. 2). Ikarugamycin was without significant effect on p65 mRNA expression at any concentration or time point, although overall expression levels appeared higher at 1 μM, when stimulated with LPS (Fig. 2). In CFBE41o−, quercetin (0.1 μM) did not affect p65 mRNA levels. At 100 μM, quercetin significantly induced p65 (P < 0.05: quercetin alone 100 μM at 24 h vs. medium at 24 h and quercetin 100 μM+LPS 24 h vs. LPS at 24 h). Fluvastatin caused a significant reduction in p65 mRNA at both concentrations and all time points after LPS stimulation (P < 0.05 and P < 0.01 for 0.1 μM+LPS vs. LPS; P < 0.05 and P < 0.01 for 1 μM+LPS vs. LPS) (Fig. 2).
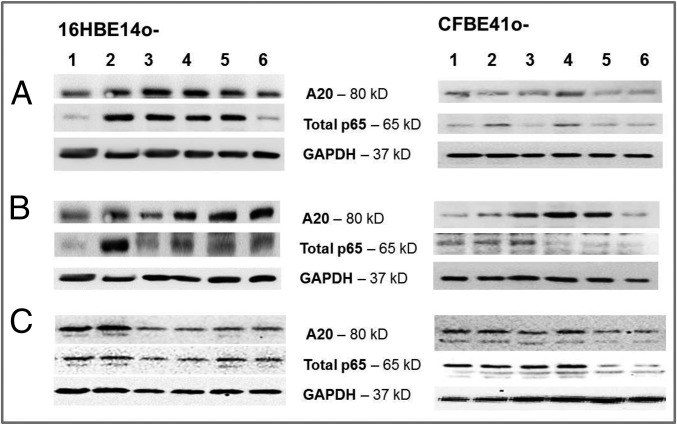
Effect of predicted drugs on A20 and p65 protein expression.
We then determined the effect of the drugs on cytosolic A20 and p65 protein by western blotting using the same selected concentrations as before. Ikarugamycin (0.01 µM) induced A20 protein in both 16HBE14o− and CFBE41o−, with less A20 protein induction at 1 μM. Ikarugamycin also induced cytosolic p65 in both cell types (Fig. 3A). Quercetin treatment caused a strong induction of A20 protein at both concentrations (0.1, 100 μM) in 16HBE14o− and, to a lower degree, in CFBE41o−. Quercetin (100 µM) reduced cytosolic p65 in 16HBE14o− and in CFBE41o− (Fig. 3B). Fluvastatin did not induce A20 protein at either concentration (0.1, 1 μM) in both 16HBE14o− and CFBE41o− cells. Fluvastatin pretreatment reduced cytosolic p65 protein in 16HBE14o− cells, although this was only apparent at the higher concentration in CFBE41o− (Fig. 3C).
Fig. 3.
Effect of ikarugamycin, quercetin, and fluvastatin on A20 and p65 protein expression. The 16HBE14o− and CFBE41o− were preincubated with (A) ikarugamycin (0.01, 1 μM), (B) quercetin (0.1, 100 μM), or (C) fluvastatin (0.1, 1 μM) and stimulated (LPS, 10 μg/mL, 0–24 h). Cytosolic A20 and p65 protein were determined by western blotting: 1, Ctr; 2, LPS; 3, drug at lower concentration; 4, drug at lower concentration + LPS; 5, drug at higher concentration; and 6, drug at higher concentration + LPS.
Specificity of the drug effect on A20 mRNA expression using A20 siRNA.
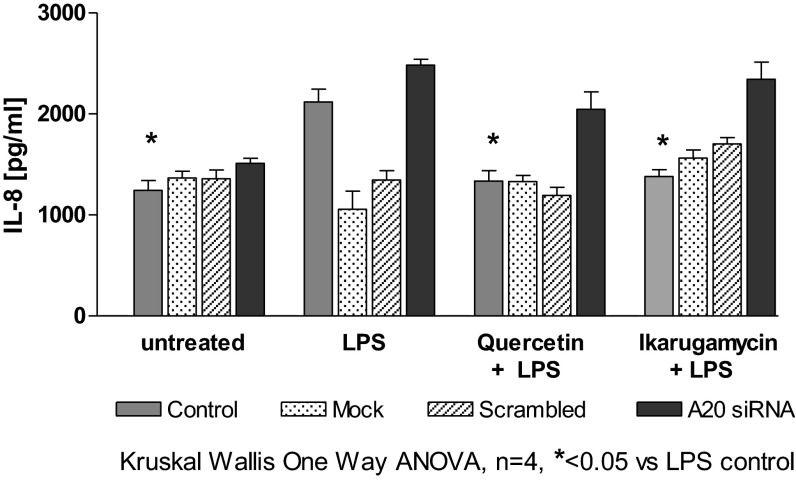
To confirm that the effect of the selected drugs is facilitated through A20 induction, we used siRNA to knock down A20 expression in 16HBE14o− cells as previously described (25). Cells were pretreated with quercetin or ikarugamycin before LPS, and IL-8 was determined. Results (Fig. 4) showed that, in 16HBE14o−, LPS significantly induced IL-8 (P < 0.05 compared with untreated control), but, when A20 was knocked down, IL-8 increased further (although not significantly different from LPS alone). When cells were pretreated with quercetin (100 μM) or ikarugamycin (1 μM), the LPS-induced IL-8 release was significantly reduced (P < 0.05). However, when A20 was knocked down, IL-8 levels were not different from LPS control (Fig. 4).
Fig. 4.
The antiinflammatory effect of ikarugamycin and quercetin is mediated by A20 induction. The 16HBE14o− with and without knockdown of A20 was preincubated with ikarugamycin (1 μM) or quercetin (100 μM) and stimulated, and IL-8 was analyzed. Drug treatment caused a significant reduction in IL-8 (P < 0.05) compared with LPS stimulation alone. A20 knockdown resulted in a lack of the antiinflammatory effect of the drug tested.
Effect of A20-Inducing Drugs on PNECs.
Effect on IL-8 release.
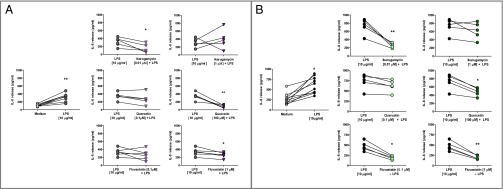
LPS significantly induced IL-8 release from PNECs from non-CF and CF patients (non-CF: P < 0.01, CF: P < 0.05, Wilcoxon paired test; Fig. 5). IL-8 release from CF PNECs was significantly higher than from non-CF PNECs (600.6 ± 62.8 pg/mL vs. 315.8 ± 36.1 pg/mL, P < 0.01, Mann−Whitney test). In non-CF PNECs (Fig. 5A), pretreatment with ikarugamycin at 0.01 μM, but not at 1 μM, significantly reduced LPS-induced IL-8 release (P < 0.05). In quercetin- and fluvastatin-treated non-CF PNECs, only the higher concentrations tested (quercetin: 100 μM and fluvastatin: 1 μM) significantly reduced IL-8 release (P < 0.01 and P < 0.05, respectively; Fig. 5A). PNECs from patients with CF showed similar results with a significant IL-8 reduction at the lower concentration of ikarugamycin (0.01 μM, P < 0.01) and the higher concentration of quercetin (100 μM, P < 0.05). In CF PNECs, fluvastatin treatment significantly reduced IL-8 release at both concentrations tested (0.1 μM, P < 0.05; 1 μM, P < 0.01; Fig. 5B).
Fig. 5.
Effect of ikarugamycin, quercetin, and fluvastatin on IL-8 release from PNECs from (A) healthy controls and (B) patients with CF. The release of IL-8 (picograms per milliliter) was determined using a commercially available IL-8 ELISA kit. Statistical analysis was performed using Wilcoxon paired ranked t test.
A20 induction in PNECs.
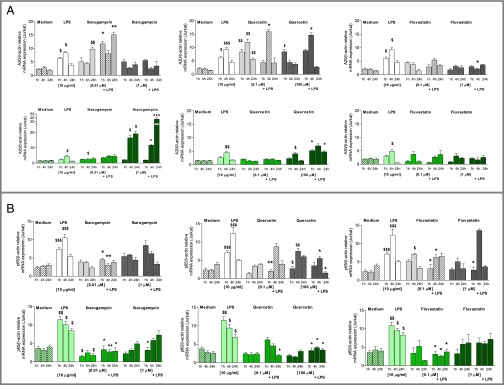
In PNECs from non-CF control subjects (Fig. 6), LPS stimulation resulted in a rapid and significant up-regulation of A20 mRNA within 1 h and a peak expression at 4 h (P < 0.05, LPS vs. medium at 1 h and 4 h). Ikarugamycin alone at 0.01 μM increased A20 mRNA, which reached significance at 24 h (P < 0.01), and, in the presence of LPS, this increase was significantly higher at 1 h and 24 h than LPS alone (P < 0.05 and P < 0.01 vs. LPS). Ikarugamycin (1 μM) had no effect on A20 mRNA induction, either alone or in the presence of LPS. Quercetin (0.1 μM) caused a significant induction of A20 compared with medium control at 1 h, 4 h, and 24 h (all P < 0.01), with expression levels similar to those induced by LPS (Fig. 6), and this was maintained in the presence of LPS, with a peak A20 induction at 4 h. The higher concentration of quercetin (100 μM) significantly induced A20 mRNA at 1 h (P < 0.05), and subsequent stimulation with LPS resulted in significantly increased A20 mRNA levels at 4 h (P < 0.05 vs. LPS 4 h). Fluvastatin treatment did not induce A20 mRNA expression, but 1 μM reduced A20 mRNA at 4 h LPS (Fig. 6).
Fig. 6.
Effect of ikarugamycin, quercetin, and fluvastatin on A20 (A) and p65 (B) mRNA expression in PNECs from healthy controls (grey) and patients with CF (green). Cells were preincubated with ikarugamycin (Right), quercetin (Center), or fluvastatin (Left) at the indicated concentrations (0.01–100 μM) and then stimulated (LPS, 10 μg/mL, 0–24 h). A20 and p65 mRNA was determined by qRT-PCR and expressed as A20/β-actin or p65//β-actin relative to internal control. $, significant difference compared to medium control; *, significant differences between genotypes.
In CF PNECs, LPS-induced A20 mRNA expression was lower than in non-CF PNECs, but, in CF PNECs, LPS induced significant levels of A20 mRNA 4 h after LPS (P < 0.05 vs. medium 4 h). Ikarugamycin at 0.01 μM (alone and in the presence of LPS) had no effect on A20 mRNA levels (Fig. 6), but 1 μM ikarugamycin significantly induced A20 mRNA at 4 h and 24 h alone (P < 0.05 vs. medium control) and in the presence of LPS (P < 0.05 and P < 0.001 vs. LPS). Similarly, quercetin treatment with the lower concentration (0.01 μM) alone and in the presence of LPS had no effect on A20 mRNA levels (Fig. 6), whereas 100 μM quercetin significantly induced A20 mRNA alone (4 h, P < 0.05 vs. medium control) and, additionally, above LPS induction (1 h and 24 h, both P < 0.05 vs. LPS). Similar to non-CF PNECs, fluvastatin treatment of CF PNECs had no significant effect on A20 mRNA expression levels.
NF-κB (p65) induction in PNECs.
PNECs from non-CF control subjects respond to LPS exposure with a significant increase in NF-κB (p65) at 1 h and 4 h (P < 0.001 vs. medium control). Thereafter, p65 mRNA expression returns to its corresponding medium control value (Fig. 6). PNECs from patients with CF, however, show the expected high levels of p65 throughout the 24 h studied (P < 0.01 at 1 h, P < 0.05 at 4 h and 24 h vs. medium control) (Fig. 6).
In non-CF PNECs, ikarugamycin treatment alone did not change p65 mRNA levels (vs. medium control), but, in the presence of LPS, p65 mRNA was significantly reduced (P < 0.05 at 1 h, P < 0.01 at 4 h vs. LPS). Overall, the higher concentration of ikarugamycin induced p65 mRNA levels with a significant increase at 4 h (P < 0.05 vs. medium control). However, in the presence of LPS, p65 levels remained not significantly different from those after LPS exposure at 1 h and 24 h but were significantly lower compared with LPS alone at 4 h (P < 0.05) (Fig. 6). The lower concentration of 0.01 μM quercetin alone did not modify basal p65 mRNA. After subsequent LPS challenge, p65 mRNA significantly decreased at 1 h (P < 0.01 vs. LPS) but then increased in a similar manner to LPS alone. However, 100 μM quercetin alone significantly reduced p65 induction at 1 h and 4 h (P < 0.05 and p0.01 vs. medium control). In the presence of LPS, this reduction of p65 mRNA reached statistical significance at 4 h and 24 h (both P < 0.05 vs. LPS) (Fig. 6). Fluvastatin (0.1 μM) induced p65 at 4 h (P < 0.05 vs. medium 4 h), but p65 levels remain significantly lower when LPS is added (P < 0.05, vs. LPS at 1 h and 4 h). The higher concentration of fluvastatin (1 μM) did not change p65 levels alone, but, after addition of LPS, p65 mRNA was initially reduced (1 h P < 0.05 vs. LPS) but then induced similarly to LPS alone (Fig. 6).
When PNECs from patients with CF were pretreated with ikarugamycin, p65 levels dropped significantly at 0.01 μM ikarugamycin in the absence or presence of LPS (P < 0.05–0.01). At the higher concentration of 1 μM, p65 levels appeared lower, but this only reached statistical significance at 1 h (P < 0.05 vs. LPS) (Fig. 6). Pretreatment of CF PNECs with quercetin did not affect p65 levels alone at either concentration tested, but, at 0.1 μM, quercetin in the presence of LPS significantly reduced p65 at 24 h (P < 0.05 vs. LPS). Treatment with 100 μM quercetin significantly reduced p65 mRNA in the presence of LPS at all time points (all P < 0.05 vs. LPS) (Fig. 6). Fluvastatin alone had no significant effect on p65 mRNA levels, but significantly reduced LPS induced p65 at 1 h, at 4 h, and at 24 h (P < 0.05 vs. medium at 1 h, at 4 h, or at 24 h), whereas the higher concentration of fluvastatin (1 μM) showed no significant effect on basal or LPS-induced p65 mRNA (Fig. 6).
Discussion
Airways infection and the subsequent inflammation are deleterious for patients suffering from CF. Current drugs targeting the mutated CFTR (potentiators/correctors) improve expression and function of CFTR on epithelial surfaces, and patients showed improved lung function and reduced frequency of pulmonary exacerbations, hospitalization, and use of i.v. antibiotics, but augmented CFTR function failed to reduce inflammatory markers in sputum (e.g., IL-1, -6, -8) (26), and heterogeneous responses to the treatment have been reported (27), suggesting that CFTR correction/potentiation may not directly improve the underlying compromised immune response. The negative NF-κB regulator A20 (TNFAIP3) is reduced in CF airway epithelial cells, basally and after LPS stimulation (23), and is associated with markers of inflammation and decreased lung function (12). A20 silencing increased TRAF6 and NF-κB activity (18), and A20 overexpression had protective effects in airway inflammation in asthmatic mice (28), suggesting that A20 augmentation normalizes the inflammatory response in the airways.
To find agents to induce A20 in CF, we used sscMap, which has been widely used in drug development for uncovering potential new indications for existing drugs as well as predicting side effects (29). Using disease-specific publicly available gene array data [Gene Expression Omnibus (GEO) datasets], we used connectivity mapping to, firstly, identify the target gene (A20) related gene signature and to, secondly, predict already licensed drugs to induce A20 expression. We included a total of 76 gene array data from PNECs and cell lines commonly used in CF research (Table 1). Gene array databases were first selected in August 2013, but a recent (January 2016) search revealed no further significant published gene array data on CF (primary nasal) epithelial cells.
The applied linear regression model (Pearson’s correlation coefficient) is an established robust method to identify the correlates of a known gene expression estimating the strength of a linear relationship between two random normally distributed variables (30). The application of the linear regression model with GO selection revealed a gene signature of six genes in additional to the seed gene A20 (Table 2), and we confirmed that the expression of these genes is similarly reduced in CF epithelial cells, basally and after LPS stimulation (Fig. 1). The identified A20 correlates were ATF3, a transcriptional repressor that binds to cAMP response elements (CRE); RAB5C, a small ubiquitously expressed GTPase; DENND4A, which encodes the C-Myc Promoter Binding Protein (MBP-1); POM121, a nuclear transmembrane protein and essential component of the nuclear pore complex; ICAM1, a cell surface glycoprotein typically expressed on endothelial and immune cells, especially during inflammation; and PSEN 1 (Presenilin 1), a catalytic component of γ-secretase and a downstream regulatory element antagonist modulator (DREAM) binding protein. Further descriptions of these genes and their involvements in inflammation can be found in Supporting Information. These genes, as a combined gene signature, were then input into the sscMap process comparing the gene expression of the gene signature with the gene expression in the reference database (www.broadinstitute.org), which was obtained from systematic microarray gene expression profiling.
The sscMap predicted a short list of drugs that should modify the expression profile of the gene signature genes, including A20. Those drugs included azacyclonol, ikarugamycin, quercetin, and karakoline (Table 3). Azacyclonol is a drug used in psychotic individuals (25). We excluded azacyclonol, as its use requires special permission through relevant government authorities. Interestingly, the antihistamine terfenadine is metabolized to azacyclonol and terfenadine (31). Karakoline is a highly toxic plant diterpenoid (32), and the pharmacological effects of preparations of Aconitum roots are attributed to diterpenoid alkaloids (33). The antiinflammatory activity of gibberellin (GA3), also a plant-derived diterpenoid, is mediated through A20 induction (24). We therefore selected ikarugamycin and quercetin for further studies.
Ikarugamycin is a macrolide antibiotic with cytostatic effects against Gram-positive bacteria. We show that ikarugamycin exhibits antiinflammatory properties in LPS-stimulated airway cells. In 16HBE14o−, ikarugamycin showed a dose-dependent reduction of LPS-induced IL-8 release (Fig. S2), through induction of A20 and reduction of p65 (Fig. 2). The 16HBE14o− and CFBE41o− did not show reduced cell viability at concentrations lower than 1 μM; higher concentrations increased LDH release, suggesting a cytotoxic effect (Fig. S1). CFBE41o− appear more sensitive to ikarugamycin treatment (Fig. S2), which made it difficult to calculate a meaningful relative IC50 value (Table S2), although p65 protein expression was not increased (Fig. 2). In HL-60 cells, ikarugamycin reduced cell viability and increased DNA fragmentation starting at 0.1 µM (IC50 of 0.22 μM), whereas MCF-7 cells and peripheral blood mononuclear cells showed higher resistance. Furthermore, ikarugamycin treatment of HL-60 cells caused a significant caspase activation, increase in intracellular calcium, and p38 MAP kinase activation (34). However, investigating the proapoptotic mechanisms in bronchial epithelial cells was beyond the scope of this study. Nonetheless, our ikarugamycin data at near-cytotoxic levels add valuable information: Firstly, sscMap correctly predicted that ikarugamycin would induce A20 mRNA, but sscMap does not predict the physiological effect of the gene induction. CF cells overall show a limited ability to induce A20; however, our results show that—given the right stimulus—CF cells are indeed able to induce A20 mRNA, and the high induction of A20 at near-cytotoxic levels may be able to counteract the proapoptotic stimulation of ikarugamycin.
Fig. S2.
Effect of the predicted A20-inducing drugs on IL-8 release. Cell lines 16HBE14o− (grey) and CFBE41o− (green) were preincubated for 1 h with ikarugamycin (Top), quercetin (Middle), or fluvastatin (Bottom) at (0.01–100 μM) and then stimulated with LPS (10 μg/mL) for 24 h. IL-8 release was determined in the cell-free supernatant as described in Materials and Methods.
Quercetin, a flavonoid, is known for its antiinflammatory effects. In vivo studies have shown antioxidant, antiinflammatory, antitumor and even antiinfectious properties of quercetin, which are promoted through its effects on signaling pathways such as NF-κB (35). In lung epithelial cells, quercetin inhibited IL-1 and TNF-α–induced IκBα degradation and NF-κB activity through modification of the MAPK pathway (AP-1) (36). The sscMap correctly predicted that quercetin can induce A20 mRNA, adding a previously unidentified mechanism for the antiinflammatory effects of quercetin. It also significantly reduced LPS-induced IL-8 release in both cell types, with a relative IC50 of 0.15 and 0.04 μM in 16HBE14o− and CFBE41o−, respectively. Quercetin at concentrations up to 1,000 μM did not show any cytotoxicity, although, in neuronal cell cultures, quercetin higher than 100 μM was cytotoxic (37). Within the in vivo antioxidant network, quercetin has been described to be oxidized and to yield an ortho-quinone, which, in absence of reducing glutathione, can oxidize protein thiols, and thereby impair enzyme activities (38). We have not investigated the antioxidant status of our cell culture, but we took precautions to minimize oxidation when preparing our quercetin dilutions.
To further investigate the A20-dependent mechanism of the antiinflammatory action of quercetin and ikarugamycin, we used A20 knockdown in 16HBE14o−. As previously described for the A20-inducing antiinflammatory compound gibberellin (24), we were able to confirm that the antiinflammatory effect of the predicted drugs was indeed mainly mediated by the induction of A20.
We also tested fluvastatin, which was predicted not to affect or reduce A20 gene expression (negative Z-score). Although fluvastatin exerted antiinflammatory effects (IL-8) in both cell lines, our data show that this was not mediated by the induction of A20 (mRNA), clearly confirming the sscMap prediction. In asthma, fluvastatin inhibits eosinophil adhesion to ICAM-1 (39) and fibroblast proliferation (40). Using similar concentrations, we did not observe any reduced proliferation. Fluvastatin at a concentration range similar to those we used reduced basal and LPS-induced IL-8 release from LPS-stimulated whole blood cells, with CF cells appearing more sensitive to fluvastatin than control cells (IC50: 19.1 μM in non-CF cells, 4.6 μM in CF blood cells) (41). In isolated LPS-stimulated peripheral blood monocytes from patients with chronic kidney disease, fluvastatin had a significant antiinflammatory effect (IL-8, IL-6) at a concentration range of 0.0001–1 μM (42). Patients with heart transplants and control subjects receiving 40 mg fluvastatin per day for 4 wk showed a significant reduction in total cholesterol levels and a maximum blood fluvastatin concentration of 2.11 μM and 3.77 μM. These studies suggest that we have covered a physiologically relevant range of fluvastatin. However, fluvastatin metabolism may be affected by concomitant therapies, especially substances competing with cytochrome enzymes, and, in such cases, fluvastatin levels may need to be monitored (43). Any reactions with other therapies (as they would appear in patients with CF) were not investigated in our work, as they would have been beyond the scope of the study.
Similar to ikarugamycin, fluvastatin has been described to have proapoptotic effects, e.g., in human lymphoma cells or human smooth muscle cells and in rat neonatal cardiac myocytes or rat vascular smooth muscle cells (44, 45), mediated through activation of caspase-3, reactive oxygen species, and activation of p38 MAPK (44, 46). However, statins, through their inhibition of 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA), but not through induction of A20, may still have therapeutic potential in airway and systemic inflammation in CF (41).
Overall, our study shows that connectivity mapping (sscMap) can predict A20-inducing drugs. Pretreatment of cells with both ikarugamycin and quercetin reduces LPS-induced IL-8 secretion by induction of A20. In non-CF PNECs, both drugs up-regulated A20 and reduced IL-8 and p65 mRNA at lower concentrations than in cell lines. CF PNECs, however, have a reduced and delayed A20-inducing response to LPS and to the tested drugs, and a significant A20 induction appears at the higher concentration of the drugs tested, which might be near to the cytotoxic effect. A20 reduces apoptosis (47, 48), and mutational loss of A20 resulted in rapid apoptosis and inflammation in hematopoietic cells (49). We did not determine markers of apoptosis in our study, but the huge increase in A20 mRNA may indicate a possible counteraction to proapoptotic changes in response to ikarugamycin treatment. Of particular interest, this may indicate a higher susceptibility/sensitivity of CF cells to proapoptotic stimuli.
Our study has several limitations. Firstly, for the sscMap process, a huge number of gene array samples are required, and, although the database search gave a high number of initial results, upon detailed inspection, several gene array studies could not be included. Connectivity mapping uses gene array data run on Affimetrix platforms, and we selected those performed on these platforms. Several published gene array studies were performed in cell lines. However, the majority of samples selected were PNECs (n = 40), but we also included data from cell lines. Furthermore, every published dataset has been performed using a specific experimental design with respect to treatments and time points. We selected experiments that used either no stimulation or exposure to P. aeruginosa LPS or to P. aeruginosa itself. A sample size of 50–100 individual samples is a statistically acceptable sample size for sscMap to produce an unbiased result, and we used 76 individual gene array samples.
Secondly, the reference database was generated using different cell lines: MCF7, HL60, PC3, and SKMEL5. Although all of human origin, none of these cell lines are airway-derived. We therefore confirmed the effect of the predicted drugs in airway-relevant and disease-specific cell lines, determined an effective drug concentration in our disease model, and confirmed their effect in primary cells. Additionally, factors such as the interaction between various signaling pathways and the interplay between genes can affect the functions of the predicted and validated drugs when used in humans.
The aim of our study was to investigate the potential of sscMap to predict A20-inducing drugs from a list of drugs already licensed for the use in humans to make them available for drug repositioning. As a proof of concept, we have focused on the LPS-induced expression of A20, p65, and cytokines IL-6 and IL-8. However, in addition to its direct regulation of TLR-induced NF-κB activation, A20 is also involved in the negative regulation of the NLRP3 inflammasome via TLR3/4-(TRIF)-RIPK3 (50) and may also inhibit inflammation-induced regulated necrosis (necroptosis) via RIPK3 (51), adding further levels of action and complexity to the antiinflammatory action of A20. Therefore, future work analyzing further NF-κB driven cytokines such as TNF-α and IL-1β would also indicate if the predicted drugs are able to modify A20 action on the inflammasome.
Summary
There is a need for a need for alternative antiinflammatory drugs for patients with CF, as restoring CFTR function with potentiators and correctors does not directly affect the inherent innate immune defect. The exaggerated inflammatory response is, in part, due to the lack of the NF-κB regulator A20, and pharmacological induction of A20 is antiinflammatory. We have shown here that sscMap is a potent tool to predict effective drugs that can modify A20 without totally inhibiting NF-κB. This is particularly important in the clinical setting, as pharmacological suppression of inflammation may increase the incidence of infective exacerbations (52).
Our study also suggests that pharmacological induction of A20 may be less efficient in CF airway cells, but, given the appropriate stimulation, A20 induction is indeed possible. Tiruppathi et al. (53) recently showed that A20 induction may be regulated not only via NF-κB but also through the opposing effects of the repressor DREAM and transcription factor USF1 (upstream stimulatory factor 1).
In addition, A20-inducing drugs have to be carefully adjusted as, in addition to the A20 induction, e.g., ikarugamycin can be proapoptotic. A20 inhibits TNF-induced proapoptotic signaling by inhibiting both the activation of caspase 8 and the activation of c-Jun (54). However, neither the drug-induced proapoptotic mechanisms nor the A20-induced antiapoptotic mechanisms have been investigated in this study; our observation of cytotoxicity despite high antiapoptotic A20 mRNA levels may suggest an overriding mechanism. Therefore, although sscMap successfully predicts drugs to modify A20, the effect of the candidate drugs must to be confirmed in a suitable model system to optimize treatments.
Materials and Methods
Selection of Gene Array Data.
A search of PubMed GEO datasets (www.ncbi.nlm.nih.gov/gds/) was performed in August 2013 using the search terms “cystic fibrosis,” “epithelial cells,” “airways,” and “primary cells.” Datasets that passed the search criteria and were compatible to Affymetrix Human Genome U133A Array were selected.
Connectivity Mapping (sscMap).
Gene expression profiles were generated using Affimetrix Gene chip Microarray, and the relative expression of treatment vs. control was sorted in descending order, giving rise to ∼22,000 rank-ordered genes and their expression.
Determination of the Gene Signature.
A gene signature (a set of genes that behaves in the same way or uniquely under a biological state) was created. A20 (TNFAIP3) was used as the known seed gene, and a linear regression model was applied to create the gene signature from all selected GEO data. A20 correlates were identified by calculating the Pearson correlation coefficient between the expression of A20 and other genes using the formula (where x̄ and ȳ are the sample means of the two arrays of values and r was calculated in Excel)
The correlation coefficient r demonstrates the association between A20 and other genes, either in a positive or negative direction. The significance of the observed correlations was measured by calculating the corresponding P values and applying a stringent P value threshold 1/N, where N is the number of genes analyzed. The significant r values were selected as correlates of A20. GO enrichment analysis (geneontology.org/) was then applied to further filter the A20 correlates, identifying those related to NF-κB. The subsequent gene signature (including A20) was used as the input query to evaluate the connection between them and the reference profiles (GEO, accession no. GSE5258) (8). Based on the principles of Lamb’s connectivity mapping, we used a simpler and more robust method (9, 10), called sscMap, to determine the connections between the gene signature and the reference profile. The similarity between the gene signature and each gene expression reference profile was assessed via a connection score. Connection scores are a function of expression profile and the query gene signature, which is expected to reflect the underlying connection between them. The sscMap applies a robust and improved scoring system based on the following formulae:
and
[where gi represents the ith gene in the signature, s(gi) is its signed rank in the signature, and R(gi) is this gene's signed rank in the reference profile]. To calculate the P value, after calculating the connection strength between a gene signature and the reference profile, a large number of random gene signatures are created, the same number of connection scores are calculated, and the proportion of scores higher than the observed score in absolute values is the P value. In addition to controlling false positives, sscMap is extended to measure the stability of the connections discovered by gene signature perturbation. To implement this, one gene was left out from the gene signature to derive the perturbation gene signature, and the changes in the significant connections were observed. The connections that stay stable over the changes were given the perturbation stability score, defined as the fraction of times a drug remained significant under the perturbation process (55).
Cell Culture.
The bronchial epithelial cell lines 16HBE14o− (control) and CFBE41o− (CF, F508del/F508del), obtained from D. Gruenert (University of California, San Francisco), were cultured as described (32). PNECs from CF patients (all F508del/F508del, n = 5) and healthy volunteers [n = 5, informed consent given; research ethics approval from the Office for Research Ethics Committees Northern Ireland (ORECI) 07/NIR02/23] were cultured as previously described (46). Control participants did not have any acute airways disease at the time of sampling, or a history of any chronic airways inflammation.
Cell Culture Stimulations.
Cells were exposed to the selected drugs (ikarugamycin, quercetin, fluvastatin (all Sigma-Aldrich, SML0188, Q0125, SML0038, 0.01–1,000 µM) at 0–1,000 μM for 1 h before LPS stimulation (P. aeruginosa LPS, Sigma-Aldrich, L9143, 10 µg/mL, up to 24 h). Stock solutions of the drugs were kept at −20 °C for up to 3 mo. To minimize oxidation or degradation of the compounds, the working dilutions were freshly prepared.
Determination of LPS-Induced Cytokine Release (IL-6 and IL-8).
IL-6 and IL-8 in cell-free culture supernatants were measured by a commercially available ELISA (PeproTech EC Ltd.) according to the manufacturer’s instructions.
LDH Cytotoxicity Assay.
LDH release into cell culture supernatants was determined using an LDH-Cytotoxicity Assay Kit (BioVision Ltd.) according to the manufacturer’s instructions.
Quantitative Real-Time PCR.
Total RNA was extracted (GenElute, RTN350; Sigma-Aldrich) and quantified. Equal amounts of RNA (250 ng) were reverse transcribed into cDNA (High-Capacity cDNA Reverse Transcription Kit; Applied Biosystems), and quantitative RT-PCR was performed (LightCycler thermal cycler system; Roche). Expression of A20, p65, and β-actin was assessed using primer sequences previously described (23) and given in Table S1. Relative expression to β-actin was calculated as ΔΔCt. Jurkat cell cDNA acted as an internal calibrator for all experiments and was used to determine differences in basal gene expression.
Western Blotting.
Cytosolic protein expression was determined by western blotting after extraction in RIPA buffer containing protease inhibitors (cOmplete, Mini; Roche). Lysates were separated by SDS/PAGE and PVDF membranes incubated with 1 µg/mL primary antibody A20 (ab74037; Abcam) or p65 (C-20; Santa Cruz Biotechnology), washed, incubated with appropriate horseradish peroxidase-conjugated antibody, and visualized on a BioRadChemi Doc XRS system (BioRad). Anti−GAPDH-HRP (ab-9484; Abcam) was used as a loading control.
Transfections.
The 16HBE14o− were seeded at 4 × 104 cells per well and allowed to attach overnight. Custom FlexiTube siRNA (QIAGEN) was designed against TNFAIP3, and both cell lines were transfected with 50 nM siRNA and Lipofectamine Transfection Reagent (Invitrogen) over 72 h. All experiments included mock transfection and scrambled controls. Gene silencing was assessed by quantitative PCR (qPCR) as described above, with knockdown of 74% ± 7.2 (n = 5) achieved.
Statistical Analysis.
All data are presented as the means ± SEM. Differences between groups were analyzed using the Kruskal−Wallis nonparametric ANOVA with Dunn’s posttest (*P < 0.05, **P < 0.01, and ***P < 0.001). In Figs. 1, 2, and 6 the symbol “$” denotes a significant difference compared with medium control, while an asterisk (*) denotes significant differences between groups (CF vs. non-CF, comparing the same time points or LPS vs. treatment+LPS). The logarithmic inhibitor concentration versus the relative IL-8 response achieving a 50% inhibition was calculated as the relative IC50. GraphPad Prism was used to plot graphs and to analyze the data.
SI Materials and Methods
Primers were designed using gene accession numbers and Primer3 open-source PCR primer design software and obtained from Invitrogen Ltd. Primer sequences are given in Table S1.
SI Results
The following six genes were identified as the A20-associated genes in CF by Pearson’s correlation that made up the gene signature: ATF3, RAB5C, DENND4A, POM121, ICAM1, and PSEN 1.
Activating Transcription Factor 3 (ATF3) is a transcriptional repressor that binds to CRE. It is stress-inducible with proapoptotic and immunomodulatory function. Induced via various TLR signaling pathways (e.g., TLRs), ATF3 negatively regulates genes associated with inflammation, including IL1β, IL6, IL12, TNFα, and CCL4 (MIP1β), and protects against infection by inhibiting the transcription of inflammatory mediators such as IL-6 in mice (60). In epithelial cells, ATF3 protein inhibits LPS-dependent CXCL1 production and thereby the recruitment of neutrophils to the site of inflammation (61). A dysregulation of ATF3 has been suggested to underlie various inflammatory diseases (62), and therefore ATF3 has been identified as a key molecular target against bacterial sepsis.
RAB5C is a small, ubiquitously expressed GTPase with a major role in the regulation of membrane traffic from plasma membrane to early endosomes (63). We have previously shown reduced endosome and endolysosome formation in CF epithelial cells inflammation but had not investigated RAB5C (64). However, considering its role in endolysosome formation, reduced RAB5C expression in CF airway epithelium, as shown in our mRNA data, may significantly contribute to the prolonged inflammatory response.
DENND4A encodes a C-Myc Promoter Binding Protein (MBP-1), where it acts as a general transcriptional repressor (65) and down-regulates gene expression of the promotor C-Myc (66).
POM121 encodes a transmembrane protein localized to the inner nuclear membrane. It is an essential component of the nuclear pore complexes (NPCs), which span the nuclear membrane, allowing transport between the cell and its environment, and POM121 is required to target proteins to the NPCs (67). POM121 is the only gene from the gene signature whose expression was found not to be reduced in CFBE41o−. However, within the scope of this study, we did not investigate the expression of POM121 further. Furthermore, it may also suggest increased nuclear pore complex activity and nuclear−cytoplasmatic shuttling of proteins during inflammation (68).
Intracellular adhesion molecule 1 (ICAM1) is a cell surface glycoprotein that is typically expressed on endothelial and immune cells, especially during inflammation. ICAM-1 binds to integrins CD11a/b/CD18 and enables leukocyte attachment to the endothelium and trans-endothelial migration into the peripheral tissue. Similarly, in CF airways, ICAM-1 (together with other adhesion molecules) facilitates the adherence of neutrophils to the epithelium, whereas blocking ICAM-1 on CF bronchial epithelial cells significantly inhibits neutrophil adherence (69). However, using the linear correlation on the gene expression data of CF cell lines and CF PNECs gene arrays predicted reduced basal ICAM-1 expression, which is supported by our mRNA data. The CF cell lines IB3-1 and CFTE29o− cells express higher levels of ICAM protein than non-CF control cell lines (70), although no data on CFBE41o− are published.
Presenilins (PSEN), are catalytic components of γ-secretase, cleaving transmembrane proteins (e.g., Notch pathway, processing of β-amyloid), but PSNE1 has also been identified as a DREAM binding protein. This is particularly interesting as DREAM acts as a repressor for A20 (53). Mice with homozygous disruptions of PSEN1 die in late embryogenesis (71, 72), but these studies have also revealed PSEN1 as a proteolytic enzymes involved in the pathobiology of pulmonary fibrosis (73). Furthermore, conditional double knockout of PSEN1 and PSEN2 in the forebrain of mice show a robust inflammatory response that gradually expands to systemic tissues (74), further supporting a role for PSEN1 in regulating inflammation.
These genes as a combined gene signature were then input into the connectivity mapping process, which compared the gene expression of the gene signature with the gene expression in the reference database.
Acknowledgments
The authors thank all of the CF patients and volunteers who took part in this study. We thank Declan McGuigan and Fiona Manderson Koivula (University of Ulster) for technical assistance with the transfection experiments. B.M. and H.W. were, in part, supported through a summer studentship obtained from the CF Trust UK. S.-D.Z. was supported by BBSRC/MRC/EPSRC cofunded Grant BB/I009051/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520289113/-/DCSupplemental.
References
- 1.Knorre A, Wagner M, Schaefer HE, Colledge WH, Pahl HL. DeltaF508-CFTR causes constitutive NF-kappaB activation through an ER-overload response in cystic fibrosis lungs. Biol Chem. 2002;383(2):271–282. doi: 10.1515/BC.2002.029. [DOI] [PubMed] [Google Scholar]
- 2.Cohen TS, Prince A. Cystic fibrosis: A mucosal immunodeficiency syndrome. Nat Med. 2012;18(4):509–519. doi: 10.1038/nm.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell TS, Stecenko AA, Christman JW. Dysregulated NF-κB activation in cystic fibrosis: Evidence for a primary inflammatory disorder. Am J Physiol Lung Cell Mol Physiol. 2001;281(1):L69–L70. doi: 10.1152/ajplung.2001.281.1.L69. [DOI] [PubMed] [Google Scholar]
- 4.Paul SM, et al. How to improve R&D productivity: The pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9(3):203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 5.DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: New estimates of drug development costs. J Health Econ. 2003;22(2):151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 6.Mak IWY, Evaniew N, Ghert M. Lost in translation: Animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6(2):114–118. [PMC free article] [PubMed] [Google Scholar]
- 7.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 8.Zhang SD, Gant TW. A simple and robust method for connecting small-molecule drugs using gene-expression signatures. BMC Bioinformatics. 2008;9:258. doi: 10.1186/1471-2105-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang SD, Gant TW. sscMap: An extensible Java application for connecting small-molecule drugs using gene-expression signatures. BMC Bioinformatics. 2009;10:236. doi: 10.1186/1471-2105-10-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsey JM, et al. Entinostat prevents leukemia maintenance in a collaborating oncogene-dependent model of cytogenetically normal acute myeloid leukemia. Stem Cells. 2013;31(7):1434–1445. doi: 10.1002/stem.1398. [DOI] [PubMed] [Google Scholar]
- 11.Zaman MM, et al. Interleukin 8 secretion from monocytes of subjects heterozygous for the deltaF508 cystic fibrosis transmembrane conductance regulator gene mutation is altered. Clin Diagn Lab Immunol. 2004;11(5):819–824. doi: 10.1128/CDLI.11.5.819-824.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly C, Williams MT, Elborn JS, Ennis M, Schock BC. Expression of the inflammatory regulator A20 correlates with lung function in patients with cystic fibrosis. J Cyst Fibros. 2013;12(4):411–415. doi: 10.1016/j.jcf.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Catrysse L, Vereecke L, Beyaert R, van Loo G. A20 in inflammation and autoimmunity. Trends Immunol. 2014;35(1):22–31. doi: 10.1016/j.it.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Li M, et al. A20 overexpression alleviates pristine-induced lupus nephritis by inhibiting the NF-κB and NLRP3 inflammasome activation in macrophages of mice. Int J Clin Exp Med. 2015;8(10):17430–17440. [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuzawa Y, et al. TNFAIP3 promotes survival of CD4 T cells by restricting MTOR and promoting autophagy. Autophagy. 2015;11(7):1052–1062. doi: 10.1080/15548627.2015.1055439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu G, Zhang Y, Guo L. Ubiquitin E3 ligase A20 is required in degradation of microbial superantigens in vascular endothelial cells. Cell Biochem Biophys. 2013;66(3):649–655. doi: 10.1007/s12013-012-9509-0. [DOI] [PubMed] [Google Scholar]
- 17.Shembade N, Ma A, Harhaj EW. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327(5969):1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mabilleau G, Chappard D, Sabokbar A. Role of the A20-TRAF6 axis in lipopolysaccharide-mediated osteoclastogenesis. J Biol Chem. 2011;286(5):3242–3249. doi: 10.1074/jbc.M110.150300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catrysse L, et al. A20 deficiency sensitizes pancreatic beta cells to cytokine-induced apoptosis in vitro but does not influence type 1 diabetes development in vivo. Cell Death Dis. 2015;6:e1918. doi: 10.1038/cddis.2015.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An YF, et al. Ubiquitin E3 ligase A20 facilitates processing microbial product in nasal epithelial cells. J Biol Chem. 2012;287(42):35318–35323. doi: 10.1074/jbc.M112.392639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onose A, et al. An inhibitory effect of A20 on NF-κB activation in airway epithelium upon influenza virus infection. Eur J Pharmacol. 2006;541(3):198–204. doi: 10.1016/j.ejphar.2006.03.073. [DOI] [PubMed] [Google Scholar]
- 22.Gon Y, et al. A20 inhibits toll-like receptor 2- and 4-mediated interleukin-8 synthesis in airway epithelial cells. Am J Respir Cell Mol Biol. 2004;31(3):330–336. doi: 10.1165/rcmb.2003-0438OC. [DOI] [PubMed] [Google Scholar]
- 23.Kelly C, et al. Expression of the nuclear factor-κB inhibitor A20 is altered in the cystic fibrosis epithelium. Eur Respir J. 2013;41(6):1315–1323. doi: 10.1183/09031936.00032412. [DOI] [PubMed] [Google Scholar]
- 24.Reihill JA, et al. Induction of the inflammatory regulator A20 by gibberellic acid in airway epithelial cells. Br J Pharmacol. 2016;173(4):778–789. doi: 10.1111/bph.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odland TM. Azacyclonol (frenquel) hydrochloride in the treatment of chronic schizophrenia; a double-blind, controlled study. J Am Med Assoc. 1957;165(4):333–335. doi: 10.1001/jama.1957.02980220017005. [DOI] [PubMed] [Google Scholar]
- 26.Rowe SM, et al. GOAL Investigators of the Cystic Fibrosis Foundation Therapeutics Development Network Clinical mechanism of the cystic fibrosis transmembrane conductance regulator potentiator ivacaftor in G551D-mediated cystic fibrosis. Am J Respir Crit Care Med. 2014;190(2):175–184. doi: 10.1164/rccm.201404-0703OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones AM, Barry PJ. Lumacaftor/ivacaftor for patients homozygous for Phe508del-CFTR: Should we curb our enthusiasm? Thorax. 2015;70(7):615–616. doi: 10.1136/thoraxjnl-2015-207369. [DOI] [PubMed] [Google Scholar]
- 28.Kang NI, et al. A20 attenuates allergic airway inflammation in mice. J Immunol. 2009;183(2):1488–1495. doi: 10.4049/jimmunol.0900163. [DOI] [PubMed] [Google Scholar]
- 29.Qu XA, Rajpal DK. Applications of Connectivity Map in drug discovery and development. Drug Discov Today. 2012;17(23-24):1289–1298. doi: 10.1016/j.drudis.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 30.Casson RJ, Farmer LDM. Understanding and checking the assumptions of linear regression: A primer for medical researchers. Clin Experiment Ophthalmol. 2014;42(6):590–596. doi: 10.1111/ceo.12358. [DOI] [PubMed] [Google Scholar]
- 31.Ling KH, et al. Metabolism of terfenadine associated with CYP3A(4) activity in human hepatic microsomes. Drug Metab Dispos. 1995;23(6):631–636. [PubMed] [Google Scholar]
- 32.Díaz JG, Ruiz JG, Herz W. Alkaloids from Delphinium pentagynum. Phytochemistry. 2004;65(14):2123–2127. doi: 10.1016/j.phytochem.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 33.Ameri A. The effects of Aconitum alkaloids on the central nervous system. Prog Neurobiol. 1998;56(2):211–235. doi: 10.1016/s0301-0082(98)00037-9. [DOI] [PubMed] [Google Scholar]
- 34.Popescu R, et al. Ikarugamycin induces DNA damage, intracellular calcium increase, p38 MAP kinase activation and apoptosis in HL-60 human promyelocytic leukemia cells. Mutat Res. 2011;709-710:60–66. doi: 10.1016/j.mrfmmm.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Askari G, Ghiasvand R, Feizi A, Ghanadian SM, Karimian J. The effect of quercetin supplementation on selected markers of inflammation and oxidative stress. J Res Med Sci. 2012;17(7):637–641. [PMC free article] [PubMed] [Google Scholar]
- 36.Ying B, et al. Quercetin inhibits IL-1 beta-induced ICAM-1 expression in pulmonary epithelial cell line A549 through the MAPK pathways. Mol Biol Rep. 2009;36(7):1825–1832. doi: 10.1007/s11033-008-9386-1. [DOI] [PubMed] [Google Scholar]
- 37.Dajas F. Life or death: Neuroprotective and anticancer effects of quercetin. J Ethnopharmacol. 2012;143(2):383–396. doi: 10.1016/j.jep.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Boots AW, Kubben N, Haenen GR, Bast A. Oxidized quercetin reacts with thiols rather than with ascorbate: Implication for quercetin supplementation. Biochem Biophys Res Commun. 2003;308(3):560–565. doi: 10.1016/s0006-291x(03)01438-4. [DOI] [PubMed] [Google Scholar]
- 39.Robinson AJ, et al. Fluvastatin and lovastatin inhibit granulocyte macrophage-colony stimulating factor-stimulated human eosinophil adhesion to inter-cellular adhesion molecule-1 under flow conditions. Clin Exp Allergy. 2009;39(12):1866–1874. doi: 10.1111/j.1365-2222.2009.03334.x. [DOI] [PubMed] [Google Scholar]
- 40.Folli C, et al. Effect of statins on fibroblasts from human nasal polyps and turbinates. Eur Ann Allergy Clin Immunol. 2008;40(3):84–89. [PubMed] [Google Scholar]
- 41.Jouneau S, et al. Anti-inflammatory effect of fluvastatin on IL-8 production induced by Pseudomonas aeruginosa and Aspergillus fumigatus antigens in cystic fibrosis. PLoS One. 2011;6(8):e22655. doi: 10.1371/journal.pone.0022655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mantuano E, et al. Simvastatin and fluvastatin reduce interleukin-6 and interleukin-8 lipopolysaccharide (LPS) stimulated production by isolated human monocytes from chronic kidney disease patients. Biomed Pharmacother. 2007;61(6):360–365. doi: 10.1016/j.biopha.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Park JW, et al. Pharmacokinetics and pharmacodynamics of fluvastatin in heart transplant recipients taking cyclosporine A. J Cardiovasc Pharmacol Ther. 2001;6(4):351–361. doi: 10.1177/107424840100600404. [DOI] [PubMed] [Google Scholar]
- 44.Qi XF, et al. HMG-CoA reductase inhibitors induce apoptosis of lymphoma cells by promoting ROS generation and regulating Akt, Erk and p38 signals via suppression of mevalonate pathway. Cell Death Dis. 2013;4:e518. doi: 10.1038/cddis.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi M, et al. Fluvastatin enhances apoptosis in cytokine-stimulated vascular smooth muscle cells. J Cardiovasc Pharmacol. 2002;39(2):310–317. doi: 10.1097/00005344-200202000-00018. [DOI] [PubMed] [Google Scholar]
- 46.de Courcey F, et al. Development of primary human nasal epithelial cell cultures for the study of cystic fibrosis pathophysiology. Am J Physiol Cell Physiol. 2012;303(11):C1173–C1179. doi: 10.1152/ajpcell.00384.2011. [DOI] [PubMed] [Google Scholar]
- 47.Gray ST, Arvelo MB, Hasenkamp W, Bach FH, Ferran C. A20 inhibits cytokine-induced apoptosis and nuclear factor κB–dependent gene activation in islets. J Exp Med. 1999;190(8):1135–1146. doi: 10.1084/jem.190.8.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He KL, Ting AT. A20 inhibits tumor necrosis factor (TNF) alpha-induced apoptosis by disrupting recruitment of TRADD and RIP to the TNF receptor 1 complex in Jurkat T cells. Mol Cell Biol. 2002;22(17):6034–6045. doi: 10.1128/MCB.22.17.6034-6045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagamachi A, et al. Acquired deficiency of A20 results in rapid apoptosis, systemic inflammation, and abnormal hematopoietic stem cell function. PLoS One. 2014;9(1):e87425. doi: 10.1371/journal.pone.0087425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duong BH, et al. A20 restricts ubiquitination of pro-interleukin-1β protein complexes and suppresses NLRP3 inflammasome activity. Immunity. 2015;42(1):55–67. doi: 10.1016/j.immuni.2014.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onizawa M, et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat Immunol. 2015;16(6):618–627. doi: 10.1038/ni.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konstan MW, et al. Investigators and Coordinators of BI Trial 543.45 A randomized double blind, placebo controlled phase 2 trial of BIIL 284 BS (an LTB4 receptor antagonist) for the treatment of lung disease in children and adults with cystic fibrosis. J Cyst Fibros. 2014;13(2):148–155. doi: 10.1016/j.jcf.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tiruppathi C, et al. The transcription factor DREAM represses the deubiquitinase A20 and mediates inflammation. Nat Immunol. 2014;15(3):239–247. doi: 10.1038/ni.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lademann U, Kallunki T, Jäättelä M. A20 zinc finger protein inhibits TNF-induced apoptosis and stress response early in the signaling cascades and independently of binding to TRAF2 or 14-3-3 proteins. Cell Death Differ. 2001;8(3):265–272. doi: 10.1038/sj.cdd.4400805. [DOI] [PubMed] [Google Scholar]
- 55.McArt DG, Zhang SD. Identification of candidate small-molecule therapeutics to cancer by gene-signature perturbation in connectivity mapping. PLoS One. 2011;6(1):e16382. doi: 10.1371/journal.pone.0016382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wright JM, et al. Respiratory epithelial gene expression in patients with mild and severe cystic fibrosis lung disease. Am J Respir Cell Mol Biol. 2006;35(3):327–336. doi: 10.1165/rcmb.2005-0359OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hampton TH, et al. Does the F508-CFTR mutation induce a proinflammatory response in human airway epithelial cells? Am J Physiol Lung Cell Mol Physiol. 2012;303(6):L509–L518. doi: 10.1152/ajplung.00226.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wright JM, Zeitlin PL, Cebotaru L, Guggino SE, Guggino WB. Gene expression profile analysis of 4-phenylbutyrate treatment of IB3-1 bronchial epithelial cell line demonstrates a major influence on heat-shock proteins. Physiol Genomics. 2004;16(2):204–211. doi: 10.1152/physiolgenomics.00160.2003. [DOI] [PubMed] [Google Scholar]
- 59.Cobb LM, Mychaleckyj JC, Wozniak DJ, López-Boado YS. Pseudomonas aeruginosa flagellin and alginate elicit very distinct gene expression patterns in airway epithelial cells: Implications for cystic fibrosis disease. J Immunol. 2004;173(9):5659–5670. doi: 10.4049/jimmunol.173.9.5659. [DOI] [PubMed] [Google Scholar]
- 60.Lai P-F, et al. ATF3 protects against LPS-induced inflammation in mice via inhibiting HMGB1 expression. J Evidence-Based Complementary Altern Med. 2013;2013:716481. doi: 10.1155/2013/716481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boespflug ND, et al. ATF3 is a novel regulator of mouse neutrophil migration. Blood. 2014;123(13):2084–2093. doi: 10.1182/blood-2013-06-510909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hai T, Wolford CC, Chang YS. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: Is modulation of inflammation a unifying component? Gene Expr. 2010;15(1):1–11. doi: 10.3727/105221610x12819686555015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeigerer A, et al. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 2012;485(7399):465–470. doi: 10.1038/nature11133. [DOI] [PubMed] [Google Scholar]
- 64.Kelly C, et al. Toll-like receptor 4 is not targeted to the lysosome in cystic fibrosis airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2013;304(5):L371–L382. doi: 10.1152/ajplung.00372.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tvrdík D, et al. Downregulation of myc promoter-binding protein 1 (MBP-1) in growth-arrested malignant B cells. Folia Biol (Praha) 2007;53(6):207–215. [PubMed] [Google Scholar]
- 66.Sedoris KC, Thomas SD, Miller DM. c-myc promoter binding protein regulates the cellular response to an altered glucose concentration. Biochemistry. 2007;46(29):8659–8668. doi: 10.1021/bi7003558. [DOI] [PubMed] [Google Scholar]
- 67.Funakoshi T, et al. Two distinct human POM121 genes: Requirement for the formation of nuclear pore complexes. FEBS Lett. 2007;581(25):4910–4916. doi: 10.1016/j.febslet.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 68.Depping R, Jelkmann W, Kosyna FK. Nuclear-cytoplasmatic shuttling of proteins in control of cellular oxygen sensing. J Mol Med (Berl) 2015;93(6):599–608. doi: 10.1007/s00109-015-1276-0. [DOI] [PubMed] [Google Scholar]
- 69.Tabary O, et al. Adherence of airway neutrophils and inflammatory response are increased in CF airway epithelial cell-neutrophil interactions. Am J Physiol Lung Cell Mol Physiol. 2006;290(3):L588–L596. doi: 10.1152/ajplung.00013.2005. [DOI] [PubMed] [Google Scholar]
- 70.Greene CM, et al. TLR-induced inflammation in cystic fibrosis and non-cystic fibrosis airway epithelial cells. J Immunol. 2005;174(3):1638–1646. doi: 10.4049/jimmunol.174.3.1638. [DOI] [PubMed] [Google Scholar]
- 71.Shen J, et al. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell. 1997;89(4):629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 72.Wong PC, et al. Presenilin 1 is required for Notch1 and DII1 expression in the paraxial mesoderm. Nature. 1997;387(6630):288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- 73.Herreman A, et al. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc Natl Acad Sci USA. 1999;96(21):11872–11877. doi: 10.1073/pnas.96.21.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang X, et al. Increased inflammatory response both in brain and in periphery in presenilin 1 and presenilin 2 conditional double knock-out mice. J Alzheimers Dis. 2009;18(3):515–523. doi: 10.3233/JAD-2009-1164. [DOI] [PubMed] [Google Scholar]