Significance
Fungal infections pose a serious threat to human health and result in several million deaths annually. To survive in their human host, pathogenic fungi require the disaccharide, trehalose. Significantly, the enzymes that synthesize trehalose are absent in humans, and thus serve as potential targets for novel antifungal intervention. Here, we describe multiple structures of one of the trehalose biosynthetic enzymes, trehalose-6-phosphate phosphatase (Tps2). These structures and germane in vivo and biochemical studies reveal the significance of the Tps2 N-terminal domain in fungal cellular stress responses and the conformational flexibility of the Tps2 C-terminal domain that imposes exquisite substrate specificity and permits efficient catalysis. These structures pave the way for “rational” inhibitor design against Tps2, facilitating antifungal drug design.
Keywords: trehalose-6-phosphate phosphatase, pathogenic fungi, HASDF phosphatase, trehalose-6-phosphate specificity, antifungal inhibitors
Abstract
Trehalose is a disaccharide essential for the survival and virulence of pathogenic fungi. The biosynthesis of trehalose requires trehalose-6-phosphate synthase, Tps1, and trehalose-6-phosphate phosphatase, Tps2. Here, we report the structures of the N-terminal domain of Tps2 (Tps2NTD) from Candida albicans, a transition-state complex of the Tps2 C-terminal trehalose-6-phosphate phosphatase domain (Tps2PD) bound to BeF3 and trehalose, and catalytically dead Tps2PD(D24N) from Cryptococcus neoformans bound to trehalose-6-phosphate (T6P). The Tps2NTD closely resembles the structure of Tps1 but lacks any catalytic activity. The Tps2PD–BeF3–trehalose and Tps2PD(D24N)–T6P complex structures reveal a “closed” conformation that is effected by extensive interactions between each trehalose hydroxyl group and residues of the cap and core domains of the protein, thereby providing exquisite substrate specificity. Disruption of any of the direct substrate–protein residue interactions leads to significant or complete loss of phosphatase activity. Notably, the Tps2PD–BeF3–trehalose complex structure captures an aspartyl-BeF3 covalent adduct, which closely mimics the proposed aspartyl-phosphate intermediate of the phosphatase catalytic cycle. Structures of substrate-free Tps2PD reveal an “open” conformation whereby the cap and core domains separate and visualize the striking conformational changes effected by substrate binding and product release and the role of two hinge regions centered at approximately residues 102–103 and 184–188. Significantly, tps2Δ, tps2NTDΔ, and tps2D705N strains are unable to grow at elevated temperatures. Combined, these studies provide a deeper understanding of the substrate recognition and catalytic mechanism of Tps2 and provide a structural basis for the future design of novel antifungal compounds against a target found in three major fungal pathogens.
Fungal infections, both superficial and invasive, have enormous effects on human health. Superficial infections of skin and nails infect around 1.7 billion people. Invasive fungal infections, primarily opportunistic invasive mycoses, lead to substantial morbidity (more than 2 million cases) and mortality (approximately 50%) (1). Among the opportunistic invasive mycoses, cryptococcosis, candidiasis, and aspergillosis represent by far the most common human infections. The high mortality rate associated with opportunistic invasive mycoses is largely due to the availability of a limited set of fast-acting fungicidal drugs, drug toxicities, drug–drug interactions and rapidly emerging drug resistance (2, 3) combined with serious underlying diseases. Further hampering drug development efforts is the problematic identification of novel antifungal drug targets due to the substantial overlap of essential fungal and mammalian biosynthetic pathways.
One promising novel antifungal target is the trehalose biosynthetic pathway, which does not have a mammalian counterpart. Computer-aided target selection against fungal infections rank the trehalose biosynthetic pathway as a top candidate for antifungal intervention (4). Trehalose, d-glucopyranosyl-(1→1)-d-glucopyranoside, is a nonreducing disaccharide synthesized in bacteria, fungi, lower plants, and invertebrates, but has never been identified in mammals (5). Trehalose levels significantly increase when fungal cells are exposed to multiple external and internal stresses, such as dehydration, heat shock, or oxidative stress (6, 7). Increased production of trehalose has a significant role in protecting proteins and membranes against these environmental stresses (8–10), rendering trehalose essential for fungal cell stress response and survival during infections.
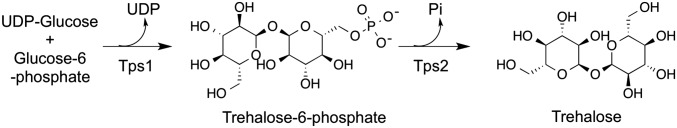
Trehalose is synthesized in most fungi by a two-step enzymatic reaction (Fig. 1). The first step, which is catalyzed by trehalose-6-phosphate synthase (Tps1), is the condensation of UDP-glucose and glucose-6-phosphate (G6P) to form trehalose-6-phosphate (T6P). The second step, which is catalyzed by trehalose-6-phosphate phosphatase (Tps2), is the dephosphorylation of T6P to yield trehalose. Disruption of the tps1 gene in Candida albicans results in impaired hyphae formation and decreased infectivity (11). Disruption of the tps2 gene in C. albicans leads to the accumulation of T6P, which is cytotoxic in high concentrations, decreases yeast growth at higher temperatures, and diminishes infectivity (12, 13). Similarly, the importance of trehalose production and, hence, this biosynthetic pathway, in cell survival has been convincingly demonstrated in other important pathogenic fungi including Cryptococcus (14, 15) and Aspergillus (16, 17). In addition to the protective role of trehalose in multiple stress responses, the trehalose biosynthetic pathway has been recognized as an integral part of basic fungal and plant cellular metabolism and energy homeostasis (18, 19). For instance, overexpression of tps2 in maize decreases T6P concentration and increases maize yield in drought conditions (20). These results further underscore Tps1 and Tps2 as important regulatory proteins for maintaining cellular integrity.
Fig. 1.
Enzyme scheme of trehalose biosynthesis. Trehalose is synthesized by the conversion of glucose-6-phosphate and UDP-glucose to trehalose-6-phosphate (T6P) by Tps1 followed by dephosphorylation of T6P by Tps2.
C. albicans Tps2 is an 888-aa residue protein that can be divided into two structural domains. The N-terminal domain (Tps2NTD) is 534-aa residues and shares significant sequence similarity to C. albicans Tps1 and Escherichia coli OtsA (the bacterial Tps1 homolog) but is missing key catalytic residues and, hence, is posited to be a “pseudo-Tps1.” Such a large, two domain-containing trehalose-6-phosphatase is unlike the bacterial Tps2 proteins, which are much smaller. For example, the E. coli Tps2 protein (OtsB) is only 266 aa residues and does not have a comparable Tps2NTD. Thus, the structure and particularly the function of the Tps2NTD remain enigmatic. The C-terminal domain of the C. albicans Tps2 protein (Tps2PD) encompasses amino acid residues 535 through 888 and contains the putative phosphatase domain. Indeed, sequence comparisons reveal that Tps2PD is a member of the haloacid dehydrogenase superfamily (HADSF) phosphatases (21, 22), enzymes that recognize a broad spectrum of substrates. To date only structures of substrate-free Tps2 from Brugia malayi (22, 23) and the Tps2-related protein from Thermoplasma acidophilum (24) have been determined, thus leaving the mechanisms of T6P binding specificity and catalysis by Tps2 enzymes unknown.
Our understanding of the substrate specificity and catalytic mechanism of any Tps2 has been hindered significantly by the lack of germane structural data, which consequently has delayed structure-guided design of novel inhibitors. Here, we report, to our knowledge, the first structure of any fungal pathogen Tps2 protein and, more important, the first transition-state structure of the C. albicans Tps2PD in complex with trehalose and BeF3. We also report here the structure of the “closed” state of the Cryptococcus neoformans Tps2PD, which has had the conserved nucleophilic aspartate mutated to asparagine, in complex with T6P and the substrate-free “open” state of the Aspergillus fumigatus Tps2PD. Our structural and biochemical analyses reveal the basis of substrate specificity and conformational flexibility used by the enzyme for catalysis, and provide key insight into the catalytic mechanism of trehalose-6-phosphate phosphatases, thus paving the way for the initiation of structure-guided inhibitor design.
Results and Discussion
Structure of the C. albicans Tps2NTD.
The Tps2NTD structure (residues 1–534) was determined to 2.56 Å resolution by molecular replacement using the structure of E. coli OtsA, a Tps1 homolog (PDB ID code 1UQU) (25), as a search model. Selected crystallographic data and refinement statistics are listed in SI Appendix, Table S2. Electron density was poor for the N-terminal domain, with 58 residues (residues 1–17, 45–61, and 91–114 from chain B) missing from the structure, likely contributing to a slightly higher Rfree. These disordered regions are consistent with secondary structure prediction by PSIPRED (26). However, deletion of these disordered residues renders Tps2NTD insoluble and necessitated their presence during protein purification and for crystallization.
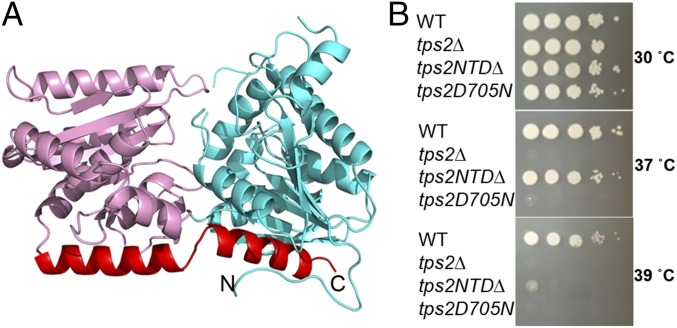
The overall structure of the Tps2NTD is similar to that of other enzymes belonging to the GT-B class of glycosyltransferases (27) and contains N- and C-terminal Rossmann-fold domains and a C-terminal helix that crosses and interacts with each Rossmann-fold domain (Fig. 2A). As anticipated, a DALI search (28) revealed that the structure most similar to the Tps2NTD is that of E. coli OtsA with a rmsd of 2.2 Å for 433 corresponding residues. Structural superposition of the Tps2NTD and OtsA reveals the highly conserved Rossmann fold of the C-terminal domain and C-terminal helix that spans both domains of the Tps2NTD. Compared with E. coli OtsA, the most notable difference between the structures is the presence of an antiparallel β-strand in OtsA that is substituted by α2 in Tps2NTD (SI Appendix, Fig. S1).
Fig. 2.
Structure of the C. albicans Tps2NTD and functional importance of Tps2. (A) Cartoon diagram of the Tps2NTD. The N-terminal and C-terminal Rossmann-fold domains are depicted as ribbons and colored cyan and pink, whereas the C-terminal helix that interacts with both domains is colored red. (B) The phosphatase domain of Tps2 is sufficient to rescue the temperature-sensitive phenotype of tps2∆ in C. neoformans at 37 °C but not at 39 °C, whereas altering a single amino acid residue (D705N), predicted to eliminate Tps2 activity, failed to restore growth at elevated temperatures. The strains were incubated for 3 d.
Despite the overall structural and sequence similarity of Tps2NTD and OtsA, alignment of their sequences reveals that OtsA residues G22, involved in UDP binding, and R9, Y76, and R300, involved in G6P binding (29), are replaced respectively with a tyrosine, serine, tryptophan, and a deletion in Tps2NTD (SI Appendix, Fig. S2). This finding suggested strongly that the Tps2NTD could not synthesize T6P. Indeed, Tps2NTD has no detectable glycosyltransferase activity. No T6P phosphatase activity was detected as well. At present it is unclear whether Tps2NTD has any other enzymatic activity, and its role in C. albicans and other pathogenic fungi thus remains enigmatic. Despite the lack of a trehalose biosynthesis function of the Tps2NTD, deletion of this domain in C. neoformans resulted in a temperature-sensitive phenotype at 39 °C (Fig. 2B), suggesting Tps2NTD is functionally essential for cell survival at elevated temperature.
C. albicans Tps2 is one component of the trehalose biosynthetic complex, which also consists of Tps1 and the trehalose synthase regulatory protein (Tps3). The formation of a multiprotein complex containing Tps1, Tps2, and a Tps3 homolog has been demonstrated in Saccharomyces cerevisiae (30), and such complex formation appears necessary for proper function in fungi because OtsA, the E. coli Tps1, which does not form a complex with OtsB, the E. coli Tps2, is unable to complement a S. cerevisiae Tps1 deletion strain (31). The presence of the Tps2NTD, which is structurally similar to OtsA, suggests its potential role in hetero–protein complex formation as the key residues in the conserved dimer interface of E. coli OtsA (29) are also present in the C. albicans Tps1, Tps2NTD, and Tps3NTD (SI Appendix, Fig. S2). Although direct interactions among the C. albicans Tps2NTD, Tps3NTD, and Tps1 have not yet been reported, the presence of this conserved interface suggests the potential for the Tps2NTD and Tps3NTD to bind to Tps1 and, thus, play a role in the formation of the trehalose biosynthetic complex in Candida and probably in Aspergillus and Cryptococcal species. One of the most likely roles for the formation of at least a Tps1-Tps2 heterodimer, but certainly not the only possibility, is substrate sequestration of T6P and its delivery to the latter enzyme. Beyond its function as an intermediate in the biosynthesis of trehalose, T6P has been demonstrated to be an important regulatory molecule in both yeast and plant (18, 32). Intriguingly, the Km of Tps2PD for T6P is 0.87 mM, a surprisingly high concentration in comparison with the reported cytosolic concentration (100 μM) of T6P (12, 33, 34). This difference in Km and cytosolic T6P concentration requires a mechanism for controlling the free T6P concentration in vivo. The formation of a trehalose biosynthetic complex, composed of at least Tps1 and Tps2, would reduce cytosolic T6P concentration and increase T6P concentrations in proximity to Tps2 by trapping T6P in the complex (34), thereby allowing the efficient dephosphorylation of this signaling molecule and cytotoxin, leading to the production of trehalose.
Structure of the C. albicans Tps2PD.
To determine the structure of the C. albicans Tps2PD, a series of truncated proteins that encompassed the putative phosphatase domain were created and purified. Crystals of a Tps2PD construct comprising residues 1–299 of that domain were obtained in the presence of beryllium fluoride (BeF3) and trehalose and diffracted to 2.0 Å resolution. This Tps2PD truncate is catalytically active and displays a kcat of 36.8 ± 1.5 s−1 and Km of 867.4 ± 69.7 μM with specificity for T6P alone (SI Appendix, Fig. S3 and Table S1). These values are similar to those reported for the Tps2 from B. malayi (23). The structure was determined by using single-wavelength anomalous dispersion (SAD) methods and selenomethionine (SeMet)-substituted Tps2PD. The model was later refined by using a higher resolution native dataset (SI Appendix, Table S2). There are two molecules in the asymmetric unit, which adopt nearly identical conformations (rmsd = 0.25 Å). The Tps2PD structure and size exclusion chromatography indicate that Tps2PD is monomeric in solution. All subsequent results and discussion focus on molecule A.
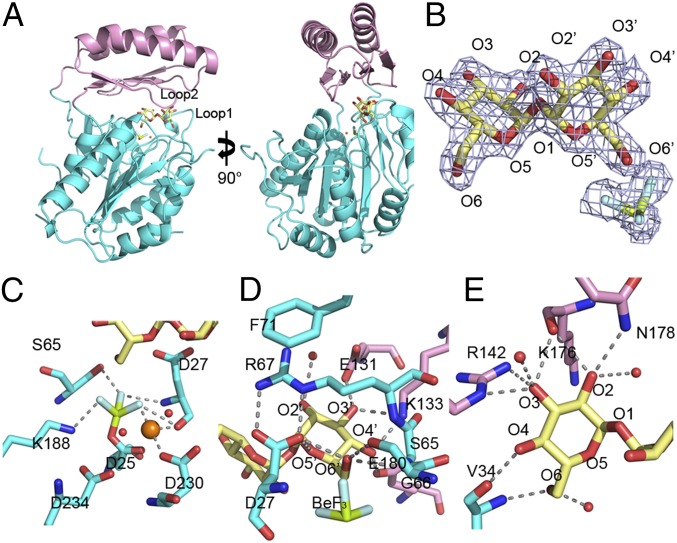
The Tps2PD structure consists of a core domain (residues 1–105 and 186–299), which is a modified Rossmann fold, and a cap domain (residues 106–185), which is inserted between the two halves of the core domain (Fig. 3A and SI Appendix, Fig. S4). Residues 206–214 were not seen in electron density maps and, thus, are not included in the structure. The Rossmann fold of the core domain is formed by six parallel β-strands flanked by seven α-helices. An additional β-strand, β5, is antiparallel with respect to the core β-sheet. A loop that is C-terminal to β5 links the core and cap domains. The cap domain is composed of four antiparallel β-strands and two α-helices. The cap domain reconnects to the core domain by a loop C-terminal of α7. The insertion position and topology of the cap domain is typically found in subfamily 2B of HADSF phosphatases (35). Unambiguous electron density for trehalose, BeF3, which is covalently linked to the conserved nucleophilic aspartate residue, D25, and a Mg2+ ion is observed (Fig. 3B) and demarcates the active site of this phosphatase. Thus, to our knowledge the structure reveals the first transition-state structure of any T6P-specific phosphatase.
Fig. 3.
Structure of the C. albicans Tps2PD–BeF3–trehalose transition-state complex. (A) Two views of the structure of the Tps2PD–BeF3–trehalose transition-state complex. The core domain and cap domains are shown as cartoons and colored cyan and pink. The trehalose and BeF3 are shown as sticks and the Mg2+ as an orange sphere. (B) A 2Fo-Fc electron density map of the bound trehalose and covalently bound BeF3, shown as light blue mesh and contoured at 1.5 σ. Trehalose is shown as pale yellow sticks. The oxygen atoms are labeled where primed oxygens are found on the glucose ring close to the catalytic residue D25. The beryllium and fluoride are colored olive and light cyan, respectively. (C) View of core-domain residues involved in transition-state stabilization, as depicted by the D25-BeF3 covalent link and Mg2+ ion coordination. The cap-domain residues are shown as atom-colored cyan sticks, and the magnesium ion and waters are depicted as spheres and colored orange and red, respectively. Selected hydrogen bonds are shown by dashed lines. (D) View of residues and solvent involved in binding the catalytic-residue proximal glucose moiety of trehalose. Residues from the cap domain are shown as atom-colored pink sticks and from the core domain as atom-colored cyan sticks. Hydrogen bonds are shown by dashed lines. (E) View of residues and solvent involved in binding the catalytic-residue distal glucose moiety of trehalose. Hydrogen bonds are shown by dashed lines.
Substrate Binding Pocket of the C. albicans Tps2PD.
Similar to other HADSF phosphatases, the C. albicans Tps2PD reveals four conserved motifs for nucleophilic catalysis (21, 22, 35). Residue D25 (motif I) is covalently bonded to the BeF3 and, therefore, presents a visualization of the aspartyl-phosphate intermediate that is formed during the nucleophilic attack of its carboxylate side chain (Fig. 3C and SI Appendix, Fig. S5 A and B). Residues S65 (motif II) and K188 (motif III) make hydrogen bonds and electrostatic interactions with the BeF3, contributing to the stabilization of the aspartyl-phosphate intermediate. A Mg2+ ion also interacts with the BeF3 potentially to neutralize the charge of the covalent intermediate and facilitate the subsequent removal of the phosphate group from D25. The Mg2+ ion is positioned in the active site through interactions with the backbone carbonyl group of residue D27, a motif I residue and the second aspartate of the characteristic DXD motif of the HADSF phosphatases, and the side chain of D230 (motif IV). Two water molecules are also seen in the active site, both of which interact with the Mg2+ ion. These water molecules also interact with the side chains of residues D230 and D234 (motif IV). These interactions result in the octahedral coordination of the Mg2+ ion. In the current structure, these water molecules also make protein-water–mediated hydrogen bonds to the trehalose, likely increasing the specificity and binding affinity of Tps2PD for T6P.
The two glucose moieties of trehalose engage in multiple interactions with residues of the extensive substrate-binding pocket of Tps2PD, both directly and indirectly through water-mediated interactions (Fig. 3 D and E and SI Appendix, Fig. S5 C and D). To adopt the correct conformation for nucleophilic attack by residue D25, the trehalose O6′ atom, from which the phosphate group of T6P is cleaved, makes an extensive hydrogen bond network with Tps2PD core residues S65, G66, and R67. These interactions orient the substrate into an ideal position for nucleophilic attack by D25. Additional interactions with this glucose molecule include hydrogen bonds from the side chain of cap residue K133 to both the O3′ and O4′ hydroxyl groups. O3′ and O4′ also interact with the side chains of cap residues E131 and E180, respectively. The O2′ hydroxyl group is positioned in the active site by its water-mediated interactions with the side chains of residues E131 and R67. The ring oxygen of this glucose moiety, O5′, interacts with carboxylate group of residue D27. These interactions ensure the proper chemical environment and orientation of the O6′-phosphate bond for nucleophilic attack by the D25 carboxylate side chain (Fig. 3 C and D). Additionally, the proper alignment of the substrate is secured by cation-π stacking between the side chains of residues R67 and F71 whereby the guanidinium side chain of R67 is locked into place over the glucose ring by two electrostatic interactions with the carboxyl group of D27 and the phenylalanine side chain (Fig. 3D).
The other glucose moiety of trehalose also engages in extensive interactions with the Tsp2PD catalytic pocket (Fig. 3E). Its O2 hydroxyl group makes hydrogen bonds to the backbone carbonyl moiety of residue K176, the N178 side chain, and a water molecule. This water is involved in a hydrogen-bonding network that links this glucose to residue D231 (motif IV) and a water molecule that coordinates the Mg2+ ion. O3 makes two hydrogen bonds to the side chain of residue R142 and also interacts with the K176 backbone. Both H140 and A177 coordinate one water molecule to bind to O3 as well. The O4 hydroxyl group forms a hydrogen bond with the V34 backbone carbonyl group, whereas O6 forms hydrogen bonds to both the V34 backbone amide group and a water molecule. This water molecule interacts with P32 and D27 (motif I), again demonstrating the strong link between key catalytic residues and both glucose moieties of trehalose. Only the glucosyl ring oxygen atom O5 is not engaged in hydrogen bonding to either Tps2PD or solvent.
Thus, residues belonging to both the cap and core domains play key roles in substrate binding and specificity. However, the key catalytic residues, including the nucleophile D25 and those residues responsible for the direct or indirect stabilization of the transition state, appear to be found primarily in the core domain. Two additional contributors to trehalose binding are loop 1 (residues 24–37) and loop 2 (residues 140–146). As noted above, both loops host residues, P32 and V34 from loop 1 and H140 and R142 from loop 2, that make hydrogen bonds to the trehalose. Substrate binding brings the two loops into proximity, which effectively shields the active site from the bulk solvent (Fig. 3A). Interestingly only one weak interaction, a D36–R143 electrostatic contact, is observed between the loops. A limited number of “pocket-closing” interactions might facilitate active site reopening and substrate departure.
Tps2PD Active Site Mutations.
Tps2PD dephosphorylates T6P with a kcat of 36.8 ± 1.5 s−1 and Km of 867.4 ± 67.9 μM, values that are similar to those reported for B. malayi Tps2 (23) (kcat = 24 ± 2 s−1, Km = 360 ± 60 μM). To assess the impact of the structurally identified catalytic and substrate-binding residues on Tps2PD phosphatase activity, a series of mutations were introduced into C. albicans Tps2PD. Mutation of the Tps2PD nucleophile aspartate residue, D25, to asparagine completely abolishes Tps2PD enzymatic activity, highlighting the critical importance of the nucleophile in phosphatase activity (SI Appendix, Table S1). Alanine substitutions of conserved residues S65 (motif II) and trehalose-binding residues E131, K133, and E180 showed significantly impaired activity with decreases in (kcat/Km) of 9.5- to 840-fold compared with WT. As observed for B. malayi Tps2, the decreased activity is attributed to lower kcat rather than higher Km. Interestingly, mutation of either R67 or E230 to alanine abolished activity. Together, the high-resolution structure of the Tps2PD transition state and Tps2PD phosphatase activity assays reveal those residues critical for T6P binding and Tps2PD catalytic activity.
Closed-Conformation Structure of the C. neoformans Tps2PD(D24N)–T6P Complex.
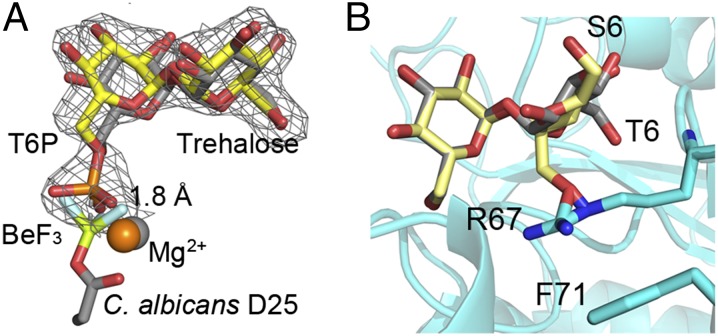
The C. neoformans Tps2PD (residues 1–306 of that domain)–T6P complex structure was determined to 2.15 Å resolution by molecular replacement using the C. albicans Tps2PD structure as a search model. Selected crystallographic data and refinement statistics are listed in SI Appendix, Table S3. To capture T6P in the binding pocket, the Tps2PD nucleophile aspartate, D24, was mutated to asparagine. Despite multiple attempts, we were unable to obtain crystals of the C. albicans Tps2PD(D25N)–T6P complex. As seen for the C. albicans Tps2PD–BeF3–trehalose complex, the C. neoformans Tps2PD(D24N)–T6P complex adopts a closed conformation (SI Appendix, Fig. S6) with the two proteins, taking essentially identical structures (rmsd = 1.0 Å). Furthermore, clear electron density for T6P was detected in the binding pocket, providing, to our knowledge, the first view of a Tps2PD–substrate complex (Fig. 4A). In addition to the overall structural similarity, the high sequence identity (46%), the complete conservation of all active site residues (SI Appendix, Fig. S4), and conserved active site interactions (SI Appendix, Fig. S7) indicate that the C. neoformans Tps2PD uses the same nucleophilic catalytic mechanism and substrate binding mechanism as the C. albicans Tps2PD.
Fig. 4.
Superpositions reveal key steps in catalysis and substrate specificity of Tps2. (A) View of T6P in the C. neoformans Tps2PD(D24N) closed conformation structure after superposition of the protein onto the Tps2 from the C. albicans Tps2–BeF3–trehalose transition-state complex. Note the significant overlap of the T6P and trehalose sugars. T6P from the C. neoformans Tps2PD structure is shown as atom-colored yellow sticks. A 2Fo-Fc electron density map of T6P bound to the C. neoformans Tps2PD is shown as gray mesh and contoured at 1.5 σ. Trehalose and the D25-BeF3 covalent adduct are shown as atom-colored gray sticks. Mg2+ ions from the C. neoformans and C. albicans Tps2PD structure are shown as orange and gray spheres, respectively. (B) Superposition of the glucose moieties of trehalose and sucrose in the C. albicans Tps2PD–BeF3–trehalose–Mg2+ transition-state complex structure. Trehalose and sucrose are shown as atom-colored gray and yellow sticks, respectively. The 6 position of the phosphorylated glucose and fructose moieties of T6P and S6P is labeled as T6 and S6. The side chains of residues R67 and F71 are shown as cyan- and atom-colored sticks.
Superposition of the C. albicans Tps2PD–BeF3–trehalose and C. neoformans Tps2PD(D24N)–T6P complexes reveals insight into the reaction coordinate of these T6P phosphatases (Fig. 4A). In the C. albicans Tps2PD transition-state structure, the BeF3 phosphate mimic is covalently bonded to the carboxylate side chain of residue D25, whereas in the C. neoformans Tps2PD(D24N)–T6P complex structure, the phosphorus atom of the T6P hydrogen bonds with the side chain of N24 and is 1.8 Å removed from the Be atom. The two structures thus provide a visualization of phosphoryl group transfer from T6P to aspartate during the nucleophilic attack, which appears to require the slight repositioning of the Mg2+ ion by ∼0.5 Å. This nucleophile mutation (D-N) was also introduced into C. neoformans (annotated as tps2D705N) and showed a temperature-sensitive growth phenotype at 37 °C (Fig. 2B) that was comparable to a tps2Δ deletion mutant. Interestingly, the tps2D705N cells had a pronounced defect in cell division and failed to produce large capsules under capsule-inducing conditions (SI Appendix, Fig. S8). These findings highlight that Tps2 phosphatase activity is important for virulence traits critical for Cryptococcus to adapt to the host environment as well as for proper growth and underscores the significance of T6P concentration regulation. It also suggests that blocking the activity of the enzyme has more severe consequences compared with complete loss of the protein. Temperature in humans remains a constant host factor that critically affects fungal pathogen survival; therefore, a Tps2-specific inhibitor would be predicted to eliminate C. neoformans infections.
Substrate Specificity of Tps2PD.
Previous biochemical characterization of Mycobacterium smegmatis (36) and B. malayi Tps2 revealed a strong preference for T6P as a substrate. C. albicans Tps2PD shows similar substrate specificity for T6P and, indeed, does not dephosphorylate sucrose-6-phosphate (S6P), glucose-6-phosphate (G6P), or paranitrophenylphosphate (pNPP), the “universal” phosphatase substrate (SI Appendix, Fig. S3). The structures of the Tps2PD–transition state and Tps2PD(D24N)–T6P complexes reveal the underlying molecular basis of the specificity of this phosphatase for T6P and, particularly, its discrimination against S6P. Superposition of the glucose moiety of T6P, which does not contain the phosphate that is cleaved, and the glucose moiety of sucrose places the fructose ring of the latter sugar proximal to the nucleophilic aspartate (Fig. 4B). However, the structural differences between the fructose of S6P and the glucose of T6P would result not only in the loss of several hydrogen bonds but more critically the steric clash of the fructose ring with the guanidinium side chain of residue R67. Residue R67 is unable to avoid this clash because of its stacking interaction with residue F71. It should be emphasized that this clash would be even more significant if the six position of the fructose sugar were phosphorylated. Furthermore, this phosphate would be positioned incorrectly for catalysis.
The strict conformational requirements of the Tps2PD binding pocket eliminate other potential sugar phosphate substrates for Tps2PD. However, Tps2PD demonstrates no activity against G6P, which can be positioned into the active site without steric clash because it can occupy the identical position of the glucose-6-phosphate moiety of T6P. However, the catalytic pocket is composed of two glucose-binding sites and the nonphosphorylated glucose moiety of T6P is critical for substrate affinity by interacting extensively with the protein (Fig. 3). Loss of these interactions would decrease Tps2PD binding affinity for the G6P significantly and require extremely high G6P concentrations for enzymatic activity, which are not physiological relevant. Importantly, the glucose moiety of T6P binds to residues of loop 1 and loop 2 and contributes to the conformational change necessary for the enzyme to take its closed, catalytically competent conformation.
Tps2PD Undergoes a Large Conformational Change Upon Substrate Binding.
Superpositions of the individual core and cap domains of B. malayi Tps2PD (residues 206–492, excluding the N-terminal domain) onto the corresponding core and cap domains of the C. albicans Tps2PD reveal rmsd of 3.9 Å and 1.9 Å, respectively (SI Appendix, Fig. S9). Superposition of the entire B. malayi Tps2PD onto the C. albicans Tps2PD reveals an rmsd of 5.6 Å for 277 corresponding Cα atoms. This large rmsd is due to a substrate binding-induced conformational change within the protein. Indeed, such a conformational transition from a substrate-free “open” conformation to a substrate-bound “closed” conformation is critical for the exclusion of bulk solvent and catalysis. However, sequence alignment between C. albicans Tps2PD and B. malayi Tps2 reveals only 24% sequence identity, and more importantly, no conservation of the residues involved in substrate recognition, suggesting a significant evolutionary difference between the two species. Therefore, we sought to solve the structure of a substrate-free Tps2PD from a more closely related fungal species to verify the observed Tps2PD conformational flexibility. Thus, we determined the substrate-free structure of Tps2PD from the human pathogenic mold A. fumigatus, which shares 58% sequence identity to the C. albicans Tps2PD including all residues involved in T6P binding and catalysis (SI Appendix, Fig. S4). We obtained three different crystal forms of the A. fumigatus Tps2PD (forms 1, 2, and 3). The structure of form 1 was solved by SAD using a SeMet-substituted protein crystal, whereas the structures of forms 2 and 3 were determined by molecular replacement using the structure of crystal form 1 as the search model.
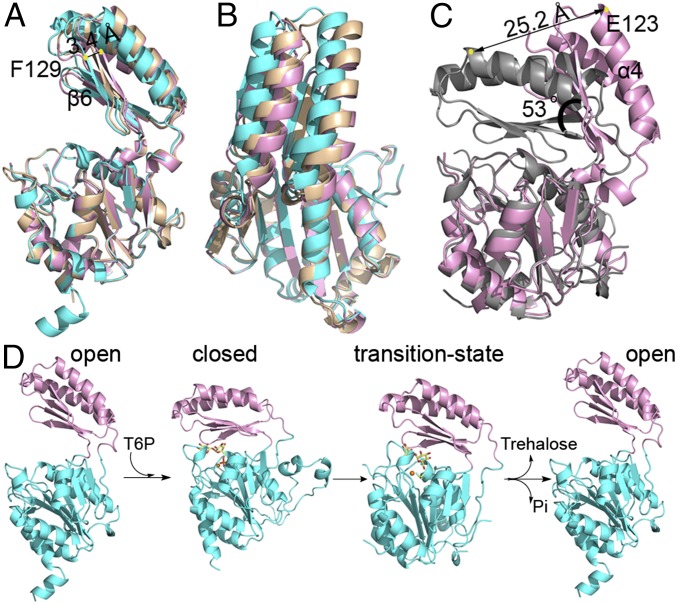
As expected, the A. fumigatus Tps2PD structures exhibit the conserved overall fold of a HADSF phosphatase with the core Rossmann-fold domain and cap domain. However, quite different from the transition-state-Tps2PD and T6P-bound Tps2PD(D24N) structures, all three A. fumigatus structures assume conformations in which the core and cap domains have opened significantly but to different degrees (Fig. 5 A and B). Superposition of the three A. fumigatus core domains (residues 1–106 and 184–270) reveals strong structural conservation (rmsd of 0.3 Å). However, a clear difference in the relative positions of their cap domains is observed, and this superposition reveals β6 of the cap domain, as denoted by F129, is 3.4 Å closer to the core in form 1 than in form 2, hence underscoring a significant conformational flexibility of Tps2PD in the absence of substrate (Fig. 5A). Superposition of the C. albicans Tps2PD transition-state structure with the open conformation that is observed for A. fumigatus Tps2PD form 2 visualizes the large conformational change between the two states (Fig. 5C). Despite the strong structural similarity of the core and cap domains (rmsd of 0.88 Å and 0.48 Å, respectively) when the core domains of the transition-state structure and form 2 substrate-free structure are overlaid, the tip of cap domain, as denoted by E123 of helix 4, is translocated 25.2 Å. Further analysis of this movement using DynDom (37) reveals a 53° rotation upon substrate binding, which is centered about C. albicans residues 102–103 and 184–188 that are located on the linker between the core and cap domain and form a hinge (Fig. 5C). Thus, the structures of the highly related fungal Tps2PDs support the idea that type 2B HADSF phosphatases adopt a large conformational change upon substrate binding and dephosphorylation (Fig. 5D), although, interestingly, the extent of the conformational change appears to vary among different phosphatases. For example, a 28° rotation is observed for substrate binding by the sucrose-6-phosphate phosphatase (38).
Fig. 5.
Structures Tps2PD in the open conformation and the catalytic cycle of Tps2. (A) Side view of A. fumigatus Tps2PD structures in the open conformation from three different crystal forms. The Tps2PD structures are depicted as cartoon and displayed after the superposition of their respective core domains. Crystal forms 1, 2, and 3 are colored in cyan, pink, and wheat, respectively. (B) “Top” view of the superposition of the three A. fumigatus Tps2PD structures. (C) Structural superposition of the core domains of A. fumigatus Tps2PD crystal form 2 and the C. albicans Tps2PD transition-state complex. The yellow dots depict the locations of the Cα atom of residue E123 in the closed and open conformations. (D) The Tps2PD catalytic cycle requires conformational changes: Open/apo enzyme ⇒ closed/substrate bound ⇒ transition state/aspartylphosphate formation ⇒ open/product release effected by substrate binding, catalysis, and product release.
In summary, we report here the Tps2NTD structure from C. albicans and suggest its potential function in the formation of a trehalose biosynthetic complex. The formation of this complex would regulate the concentration of free T6P and increase trehalose production efficiency. Further biochemical and cellular experiments are required to understand the stereochemistry and function of this complex. We also report here the structures of Tps2PD in multiple conformational states. Combined with biochemical and cellular analyses, we demonstrate the critical roles of key residues in substrate specificity and provide insight into the dephosphorylation mechanism of this T6P phosphatase. The conformational flexibility displayed by Tps2PD structures deepens our understanding of Tps2 and other general HADSF phosphatases. Significantly, the highly specific active site of Tps2PD distinguishes this enzyme from other mammalian phosphatases and, thus, may alleviate potential off-target toxicity of Tps2PD-specific inhibitors. Further, sequence alignment of the C. albicans Tps2PD with those of C. neoformans and A. fumigatus reveals 46% and 58% sequence identity, respectively. Residues involved in nucleophilic catalysis and critically, substrate binding, are conserved among these pathogenic fungi species. The high conservation of these residues in the pathogenic fungal Tps2PD suggests that this enzyme is a viable broad-spectrum antifungal drug target, paving the way for antifungal drug design targeting the trehalose biosynthetic pathway.
Methods
All proteins were expressed in bacterial systems and purified by Ni2+-NTA affinity column chromatography and size exclusion chromatography. Crystals were obtained by hanging-drop vapor diffusion, and data were collected at the Advanced Light Source or Advanced Photon Source synchrotrons. Detailed protocols can be found in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Nam Tonthat and Ivan Birukou for assistance with data collection and processing; Dr. Ziqiang Guan at Duke University for mass spectrometry sample analysis; Dr. Jane S. Richardson for her input; and the Advanced Photon Source for access to beamlines ID-22 and BM-22 and the Advanced Light Source for access to beamlines 5.0.1 and 5.0.2. The work was funded by National Institutes of Health Grants 1P01AI104533 (to R.G.B., R.E.L., and J.R.P.), and R01AI073896, and R01AI093257 (to J.R.P.); and the American Lebanese Syrian Associated Charities, St. Jude Children’s Research Hospital (R.E.L.). E.J.W. was funded by National Institutes of Health Grant 5T32AI052080-12.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.S.G. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5DXF, 5DXI, 5DX9, 5DXL, 5DXN, and 5DXO).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601774113/-/DCSupplemental.
References
- 1.Brown GD, et al. Hidden killers: Human fungal infections. Sci Transl Med. 2012;4(165):165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Ghannoum MA, Rice LB. Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12(4):501–517. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowen LE. The evolution of fungal drug resistance: Modulating the trajectory from genotype to phenotype. Nat Rev Microbiol. 2008;6(3):187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- 4.Spaltmann F, Blunck M, Ziegelbauer K. Computer-aided target selection-prioritizing targets for antifungal drug discovery. Drug Discov Today. 1999;4(1):17–26. doi: 10.1016/s1359-6446(98)01278-1. [DOI] [PubMed] [Google Scholar]
- 5.Gancedo C, Flores CL. The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res. 2004;4(4-5):351–359. doi: 10.1016/S1567-1356(03)00222-8. [DOI] [PubMed] [Google Scholar]
- 6.Benaroudj N, Lee DH, Goldberg AL. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J Biol Chem. 2001;276(26):24261–24267. doi: 10.1074/jbc.M101487200. [DOI] [PubMed] [Google Scholar]
- 7.Hottiger T, Boller T, Wiemken A. Rapid changes of heat and desiccation tolerance correlated with changes of trehalose content in Saccharomyces cerevisiae cells subjected to temperature shifts. FEBS Lett. 1987;220(1):113–115. doi: 10.1016/0014-5793(87)80886-4. [DOI] [PubMed] [Google Scholar]
- 8.Crowe JH, Crowe LM, Chapman D. Preservation of membranes in anhydrobiotic organisms: The role of trehalose. Science. 1984;223(4637):701–703. doi: 10.1126/science.223.4637.701. [DOI] [PubMed] [Google Scholar]
- 9.Crowe JH, Hoekstra FA, Crowe LM. Anhydrobiosis. Annu Rev Physiol. 1992;54:579–599. doi: 10.1146/annurev.ph.54.030192.003051. [DOI] [PubMed] [Google Scholar]
- 10.Hottiger T, Devirgilio C, Hall MN, Boller T, Wiemken A. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur J Biochem. 1994;219(1–2):187–193. doi: 10.1111/j.1432-1033.1994.tb19929.x. [DOI] [PubMed] [Google Scholar]
- 11.Zaragoza O, Blazquez MA, Gancedo C. Disruption of the Candida albicans TPS1 gene encoding trehalose-6-phosphate synthase impairs formation of hyphae and decreases infectivity. J Bacteriol. 1998;180(15):3809–3815. doi: 10.1128/jb.180.15.3809-3815.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Dijck P, De Rop L, Szlufcik K, Van Ael E, Thevelein JM. Disruption of the Candida albicans TPS2 gene encoding trehalose-6-phosphate phosphatase decreases infectivity without affecting hypha formation. Infect Immun. 2002;70(4):1772–1782. doi: 10.1128/IAI.70.4.1772-1782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Esparza M, et al. Role of trehalose-6P phosphatase (TPS2) in stress tolerance and resistance to macrophage killing in Candida albicans. Int J Med Microbiol. 2009;299(6):453–464. doi: 10.1016/j.ijmm.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Ngamskulrungroj P, et al. The trehalose synthesis pathway is an integral part of the virulence composite for Cryptococcus gattii. Infect Immun. 2009;77(10):4584–4596. doi: 10.1128/IAI.00565-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petzold EW, et al. Characterization and regulation of the trehalose synthesis pathway and its importance in the pathogenicity of Cryptococcus neoformans. Infect Immun. 2006;74(10):5877–5887. doi: 10.1128/IAI.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puttikamonkul S, et al. Trehalose 6-phosphate phosphatase is required for cell wall integrity and fungal virulence but not trehalose biosynthesis in the human fungal pathogen Aspergillus fumigatus. Mol Microbiol. 2010;77(4):891–911. doi: 10.1111/j.1365-2958.2010.07254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Bader N, et al. Role of trehalose biosynthesis in Aspergillus fumigatus development, stress response, and virulence. Infect Immun. 2010;78(7):3007–3018. doi: 10.1128/IAI.00813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M. Trehalose metabolism in plants. Plant J. 2014;79(4):544–567. doi: 10.1111/tpj.12509. [DOI] [PubMed] [Google Scholar]
- 19.van Heerden JH, et al. Lost in transition: Start-up of glycolysis yields subpopulations of nongrowing cells. Science. 2014;343(6174):1245114. doi: 10.1126/science.1245114. [DOI] [PubMed] [Google Scholar]
- 20.Nuccio ML, et al. Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat Biotechnol. 2015;33(8):862–869. doi: 10.1038/nbt.3277. [DOI] [PubMed] [Google Scholar]
- 21.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361(5):1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 22.Allen KN, Dunaway-Mariano D. Markers of fitness in a successful enzyme superfamily. Curr Opin Struct Biol. 2009;19(6):658–665. doi: 10.1016/j.sbi.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farelli JD, et al. Structure of the trehalose-6-phosphate phosphatase from Brugia malayi reveals key design principles for anthelmintic drugs. PLoS Pathog. 2014;10(7):e1004245. doi: 10.1371/journal.ppat.1004245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao KN, et al. Crystal structure of trehalose-6-phosphate phosphatase-related protein: Biochemical and biological implications. Protein Sci. 2006;15(7):1735–1744. doi: 10.1110/ps.062096606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson RP, Tarling CA, Roberts S, Withers SG, Davies GJ. The donor subsite of trehalose-6-phosphate synthase: Binary complexes with UDP-glucose and UDP-2-deoxy-2-fluoro-glucose at 2 A resolution. J Biol Chem. 2004;279(3):1950–1955. doi: 10.1074/jbc.M307643200. [DOI] [PubMed] [Google Scholar]
- 26.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 27.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibson RP, Turkenburg JP, Charnock SJ, Lloyd R, Davies GJ. Insights into trehalose synthesis provided by the structure of the retaining glucosyltransferase OtsA. Chem Biol. 2002;9(12):1337–1346. doi: 10.1016/s1074-5521(02)00292-2. [DOI] [PubMed] [Google Scholar]
- 30.Bell W, et al. Composition and functional analysis of the Saccharomyces cerevisiae trehalose synthase complex. J Biol Chem. 1998;273(50):33311–33319. doi: 10.1074/jbc.273.50.33311. [DOI] [PubMed] [Google Scholar]
- 31.Bonini BM, et al. Expression of Escherichia coli otsA in a Saccharomyces cerevisiae tps1 mutant restores trehalose 6-phosphate levels and partly restores growth and fermentation with glucose and control of glucose influx into glycolysis. Biochem J. 2000;350(Pt 1):261–268. [PMC free article] [PubMed] [Google Scholar]
- 32.Thevelein JM, Hohmann S. Trehalose synthase: Guard to the gate of glycolysis in yeast? Trends Biochem Sci. 1995;20(1):3–10. doi: 10.1016/s0968-0004(00)88938-0. [DOI] [PubMed] [Google Scholar]
- 33.Lunn JE, et al. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J. 2006;397(1):139–148. doi: 10.1042/BJ20060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Vaeck C, Wera S, van Dijck P, Thevelein JM. Analysis and modification of trehalose 6-phosphate levels in the yeast Saccharomyces cerevisiae with the use of Bacillus subtilis phosphotrehalase. Biochem J. 2001;353(Pt 1):157–162. [PMC free article] [PubMed] [Google Scholar]
- 35.Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: Evolution of a catalytic scaffold. Trends Biochem Sci. 2004;29(9):495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Klutts S, et al. Purification, cloning, expression, and properties of mycobacterial trehalose-phosphate phosphatase. J Biol Chem. 2003;278(4):2093–2100. doi: 10.1074/jbc.M209937200. [DOI] [PubMed] [Google Scholar]
- 37.Poornam GP, Matsumoto A, Ishida H, Hayward S. A method for the analysis of domain movements in large biomolecular complexes. Proteins. 2009;76(1):201–212. doi: 10.1002/prot.22339. [DOI] [PubMed] [Google Scholar]
- 38.Fieulaine S, Lunn JE, Borel F, Ferrer J-L. The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell. Plant Cell. 2005;17(7):2049–2058. doi: 10.1105/tpc.105.031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.