Abstract
PTEN is a tumor suppressor that functions to negatively regulate the PI3K/AKT pathway as the lipid phosphatase for phosphatidylinositol 3,4,5-triphosphate. Phosphorylation of a cluster of Ser/Thr residues (amino acids 380–385) on the C-terminal tail serves to alter the conformational state of PTEN from an open active state to a closed inhibited state, resulting in a reduction of plasma membrane localization and inhibition of enzyme activity. The relative contribution of each phosphorylation site to PTEN autoinhibition and the structural basis for the conformational closure is still unclear. To further the structural understanding of PTEN regulation by C-terminal tail phosphorylation, we used protein semisynthesis to insert stoichiometric and site-specific phospho-Ser/Thr(s) in the C-terminal tail of PTEN. Additionally, we employed photo-cross-linking to map the intramolecular PTEN interactions of the phospho-tail. Systematic evaluation of the PTEN C-tail phospho-cluster showed autoinhibition, and conformational closure was influenced by the aggregate effect of multiple phospho-sites rather than dominated by a single phosphorylation site. Moreover, photo-cross-linking suggested a direct interaction between the PTEN C-tail and a segment in the N-terminal region of the catalytic domain. Mutagenesis experiments provided additional insights into how the PTEN phospho-tail interacts with both the C2 and catalytic domains.
Keywords: phosphatase, phosphatase and tensin homolog (PTEN), phosphorylation, post-translational modification (PTM), protein conformation, pten
Introduction
PTEN3 is a phosphatidylinositol 3,4,5-triphosphate (PIP3) lipid phosphatase that is frequently inactivated in cancer by mutation, epigenetic silencing, or post-translational modifications (PTMs) (1–8). Loss of PTEN function allows unabated production of PIP3 from PIP2 by PI3Ks and stimulates the AKT protein kinase signaling pathway and cancer cell proliferation (1, 9, 10). Understanding the mechanisms of PTEN regulation is therefore critical to the development of therapeutics that can treat a wide variety of cancers.
PTEN is a ∼45-kDa protein composed of an N-terminal PTP-type phosphatase domain, a midsection phospholipid membrane interacting C2 domain, and a 50-residue regulatory tail (1). A crystal structure of the core PTEN catalytic-C2 region has revealed intimate interactions between the catalytic and C2 domains but omitted the C-terminal tail, which is considered to be flexible and largely unstructured (11). One of the most intensively studied set of PTMs in PTEN is a cluster of four C-terminal phosphorylations at Ser380, Thr382, Thr383, and Ser385 (12–15). These phosphorylation events have been suggested to be catalyzed by protein kinases casein kinase 2 and/or glycogen synthase kinase 3β and demonstrated to drive a conformational change that inhibits membrane interactions and reduces catalytic activity (12–15).
Several reports that shed light on the molecular basis for this phospho-dependent regulatory event suggest that the PTEN tail phosphorylation induces conformational closure involving intramolecular interactions between the tail and the CBR3 loop of the C2 domain of PTEN (14–16). However, important aspects of this structural model remain undetermined. It is not known whether all four tail phosphorylation events are necessary for PTEN conformational closure or whether specific individual sites are critical for this tertiary structural change. Recently, a mutant form of PTEN, ePTEN, was identified through a genetic screen as a form of PTEN that constitutively binds the membrane, despite C-terminal phosphorylation (17). The five mutations in ePTEN, Q17R, R41G, E73D, N262Y, and N329H, are widely spaced throughout the PTEN domains (17). It was hypothesized, but not investigated in detail, that these mutations disrupt the phosphorylation-driven conformational change in PTEN (17). Other studies on PTEN including hydrogen/deuterium exchange experiments have suggested that the catalytic domain and/or the N terminus may also interact with the phosphorylated tail (18).
To help clarify these phospho-PTEN structural issues, we have applied expressed protein ligation (19) to generate several semisynthetic phosphorylated forms of PTEN, and we assessed their activity and tail interactions. We demonstrated that there is an approximately proportional relationship between the number of phosphates and degree of conformational closure. In contrast to expectations, tetraphosphorylated ePTEN (4p-ePTEN) showed a closed conformation similar to that of WT 4p-PTEN. However, mutations of a cluster of Lys and Arg residues in the Cα2 loop relaxed phospho-tail interactions with the PTEN body. The incorporation of a benzolyphenylalanine in the C-tail was employed in photo-cross-linking experiments and provided direct evidence for a tail-catalytic domain interaction. Here, we describe how this biochemical analysis of a new set of semisynthetic PTENs has led to significant insights into the structural basis for the effects of PTEN tail phosphorylation.
Experimental Procedures
Reagents
Soluble diC6-PIP3 was purchased from Avanti Polar Lipids (Alabaster, AL). Anti-phospho-PTEN antibody was from Abcam (ab131107), and anti-total PTEN antibody was from Santa Cruz (sc-7974). Calf intestinal alkaline phosphatase was purchased from New England Biolabs (M0290). MESNA was supplied from Sigma. All canonical Fmoc amino acids were from EMD (Billerica, MA), whereas Fmoc-l-p-benzoylphenylalanine and Fmoc-l-Lys(biotin)-Wang resin were purchased from Novabiochem and Iris Biotech, respectively. All other reagents were of the highest quality and were purchased from either Sigma or Fisher.
Peptide Synthesis
Peptides were synthesized using the PS3 peptide synthesizer or the Prelude X peptide synthesizer from Protein Technologies (Tuscon, AZ) using conventional Fmoc solid phase peptide synthesis strategies. Synthetic peptides were estimated to be more than 90% pure after HPLC, and their correct structures were confirmed by MALDI mass spectrometry. Peptide samples were spotted using the sandwich method with α-cyano-4-hydroxycinnamic acid as the matrix (10 mg/ml α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 5 mm ammonium citrate). Mass spectra were acquired using the Voyager-DE STR biospectrometry work station (Applied Biosystems) by averaging 100 shots using a linear detector in either positive (unphosphorylated peptides) or negative ion (phosphorylated peptides) mode.
Generation and Purification of Semisynthetic PTEN
Expressed protein ligation was used to generate different semisynthetic phosphorylated PTENs as previously reported by Bolduc et al. (14). In brief, a C-terminal truncated PTEN (amino acids 1–378) fused to Mycobacterium xenopi GyrA intein, and chitin-binding domain (truncated PTEN-intein-CBD) was subcloned into the pFastBac-1 vector and transformed into DH10Bac competent cells, to produce a bacmid containing the appropriate PTEN sequence. Site-directed mutants were generated by standard QuikChange methods using the primers shown in Table 1. The bacmid containing t-PTEN-intein-CBD was then transfected into Sf-21 insect cells to generate the corresponding baculovirus needed for protein expression. High Five insect cells were infected with the baculovirus at a cell density of 1 million/ml and cultured for 48 h at 27 °C to express truncated PTEN-intein-CBD. The final culture was then harvested by centrifugation, and the cell pellet was lysed with a Dounce homogenizer by 40 strokes in 40 ml of 50 mm HEPES, pH 7.5, 250 mm NaCl, 1 mm EDTA, 10% glycerol, and one tablet of complete EDTA-free protease inhibitor mixture (Roche Diagnostics). The soluble lysate was then incubated with fibrous cellulose (Sigma) for 30 min at 4 °C to remove chitinase. Next, t-PTEN-intein-CBD was immobilized by passing it over chitin resin, followed by rigorous washing with 25 mm HEPES, pH 7.5, 250 mm NaCl, 1 mm EDTA, 0.1% Triton, and 50 mm HEPES, pH 7.5, 250 mm NaCl, 1 mm EDTA. Semisynthetic PTEN was then generated by incubating the t-PTEN-intein-CBD-bound chitin resin for 48–72 h at room temperature with 50 mm HEPES, pH 7.2, 250 mm NaCl, 400 mm MESNA, and 2 mm of the corresponding C-terminal tail peptide. Truncated PTENs were generated by incubating the chitin resin with 50 mm HEPES, pH 7.2, 250 mm NaCl, 400 mm MESNA, and 40 mm cysteine for 24 h at room temperature. Following the ligation reaction, semisynthetic PTEN was eluted from the chitin resin in 50 mm Tris, pH 8.0, 5 mm NaCl, 5 mm DTT, followed by dialysis with the same buffer to remove unreacted peptide and salt from the ligation reaction. The semisynthetic PTEN was then further purified by FPLC anion exchange chromatography (MonoQ; GE Healthcare) using a 240-ml linear gradient (0% to 50% buffer B), whereby buffer A is the dialysis buffer, and buffer B is 50 mm Tris, pH 8.0, 1 m NaCl, 5 mm DTT.
TABLE 1.
Primers for PTEN mutants
| PTEN mutant | Mutation(s) | Primer sequences |
|---|---|---|
| 3R/D-4p-PTEN | R41D | gctatgggatttcctgcagaagcacttgaaggcgtatacag |
| ctgtatacgccttcaagtgcttctgcaggaaatcccatagc | ||
| R47D | aaaccttactacatcatcaatattgttatcgtatacgccttcaagatcttctgcagg | |
| cctgcagaagatcttgaaggcgtatacgataacaatattgatgatgtagtaaggttt | ||
| R74D | cattacaagatatacaatctttgtgctgaagcacattatgacaccgccaa | |
| ttggcggtgtcataatgtgcttcagcacaaagattgtatatcttgtaatg | ||
| Cα2D | K327D, K330D, K332D, and R335D | aatatctagtacttactttaacaaaaaatgatcttgacgatgcaaatgatgacgatgccaacgattacttttctccaaattttaaggtga |
| cagcttcaccttaaaatttggagaaaagtaatcgttggcatcgtcatcatttgcatcgtcaagatcattttttgttaaagtaagtact | ||
| Cα2A | K327A, K330A, K332A, and R335A | cagcttcaccttaaaatttggagaaaagtatgcgttggctgcgtctgcatttgctgcgtcaagatcattttttgttaaagtaagtactag |
| tagtacttactttaacaaaaaatgatcttgacgcagcaaatgcagacgcagccaacgcatacttttctccaaattttaaggtgaagctgt | ||
| ePTEN | Q17R | gagatcgttagcagaaacaaaaggagatatagagaggatggattcgac |
| gtcgaatccatcctctctatatctccttttgtttctgctaacgatctc | ||
| R41G | ctatgggatttcctgcagaaggacttgaaggcgtatac | |
| gtatacgccttcaagtccttctgcaggaaatcccatag | ||
| E73D | acaagatatacaatctttgtgctgatagacattatgacaccgc | |
| gcggtgtcataatgtctatcagcacaaagattgtatatcttgt | ||
| N262Y | caaagtagagttcttccacaaacagtataagatgctaaaaaaggacaaaatgt | |
| acattttgtccttttttagcatcttatactgtttgtggaagaactctactttg | ||
| N329H | caaaaaatgatcttgacaaagcacataaagacaaagccaaccgatac | |
| gtatcggttggctttgtctttatgtgctttgtcaagatcattttttg |
Enzyme Activity Assay
Lipid phosphatase activity of PTEN toward the aqueous soluble substrate diC6-PIP3 was measured by monitoring phosphate release using the Malachite Green assay kit (R&D Biosystems) at 620 nm. Initial velocities were measured for a 25-μl reaction containing 50 mm Tris, pH 8.0, 10 mm BME, 0.05 mg/ml ovalbumin, and varying concentrations of diC6-PIP3 (20–160 μm). The reactions were initiated with 0.5–20 μg of the corresponding PTEN, and reaction rates were measured for 5–10 min at 30 °C. The reactions were then quenched, and inorganic phosphate was measured using the Malachite Green assay kit. Background corrections were performed with quenched enzyme. All measurements were performed in duplicate on at least two separate occasions, and replicates typically agreed within 20%. The kcat/Km measurements derived from these data were obtained using the Michaelis-Menten equation or a linear plot when no evidence of saturation was apparent by visual inspection.
Western Blotting and Alkaline Phosphatase Sensitivity Assay
Semisynthetic phospho-PTEN forms were subjected to alkaline phosphatase treatment over a period of up to 300 min in 50 mm Tris, pH 8.0, 10 mm BME, 1 μm of the corresponding phospho-PTEN, and 1 μm calf intestinal alkaline phosphatase (NEB). The reaction samples were quenched in SDS loading dye at 95 °C, and the fraction of phospho-PTEN remaining was measured by Western blotting analysis using an antibody to the PTEN phospho-C-tail cluster (Abcam) in combination with a HRP-conjugated anti-rabbit secondary antibody. Western positive bands were detected using Amersham Biosciences ECL Western blotting detection reagent with Syngene PXi and quantified by ImageJ image quantification software. Phospho-tail half-lives were calculated using a single exponential decay curve. All measurements were performed at least twice on separate occasions, and half-lives from replicates typically agreed within 30%.
UV-induced C-terminal Tail Cross-linking
An 18-mer tetraphosphorylated C-terminal tail peptide was synthesized with a photoactivatable p-benzoylphenylalanine (Bpa) in the place of Phe392 and an artificial C-terminal lysine that was Nϵ-biotinylated using conventional Fmoc peptide synthesis strategies and ligated to immobilized t-PTEN-intein-CBD using the methods described above. UV-induced cross-linking was performed with 50 μg of PTEN diluted with 50 mm Tris, pH 8.0, 10 mm DTT to ∼150 μg/ml. In a quartz reaction chamber with circulating water at 4 °C and constant stirring, 4p-PTEN containing Bpa was UV-irradiated at 365 nm with a Blak-Ray C50 UV lamp for 3 h. Next, the UV-irradiated 4p-PTEN was treated with bovine alkaline phosphatase (Sigma) at a final concentration of 1 μm for 2 h at room temperature. The alkaline phosphatase-treated cross-linked PTEN samples were subjected to buffer exchange with 50 mm NH4HCO3 followed by reduction with 5 mm tris(2-carboxyethyl)phosphine for 30 min at 60 °C and alkylation with 10 mm 2-chloroacetamide for 30 min at room temperature in the dark. The samples were desalted using Protein Desalting Spin Columns and overnight digestion was conducted using trypsin (2% w/w) for 16 h at 37 °C. The digests were acidified by the addition of trifluoroacetic acid to 0.1% final concentration prior to desalting using C18 solid phase extraction. The eluate was dried in a SpeedVac and reconstituted in avidin column loading buffer for enrichment of the biotinylated peptides. Peptides were eluted from the avidin column with 2 mm biotin buffer (100 mm Na2PO4, pH 7.0, 150 mm NaCl, 2 mm biotin). The peptide concentration was approximated by UV absorption using a NanoDrop (A280 nm), and the samples were reconstituted in a sufficient volume of 0.2% formic acid in water to yield a concentration of 200 fmol/μl prior to LC-MS/MS analysis.
LC-MS/MS
Chromatographic separation was performed using a Dionex Ultimate 3000 RSLCnano system (Thermo Scientific) with a 75 μm x 15 cm Acclaim PepMap100 separating column (Thermo Scientific) protected by a 2 cm guard column (Thermo Scientific). The mobile phase flow rate was 300 nL/min and consisted of 0.1% formic acid in water (A) and 0.1% formic acid, 95% acetonitrile (B). MS analysis was performed using an LTQ Orbitrap Velos Pro mass spectrometer (Thermo Scientific). The spray voltage was set at 2.2 kV. Orbitrap spectra were collected from 400–1800 m/z at a resolution of 30,000 followed by data-dependent HCD MS/MS (at a resolution of 7500, collision energy 35%, activation time 0.1 ms) of the 10 most abundant ions using an isolation width of 2.0 Da. Charge state screening was enabled to reject the generation of MS/MS spectra for unassigned and singly charged precursor ions. A dynamic exclusion time of 40 s was used to discriminate against previously selected ions.
Data Analysis
Mass spectrometry data from the cross-linked PTEN samples were analyzed using Crossfinder version 1.0 (20, 21) with the default parameters. Prior to analysis with Crossfinder, MS/MS data were converted to mzXML files using ProteoWizard.
Results
Individual Tail Phosphorylation Effects on PTEN Conformation
To assess the specific effects of the individual Ser/Thr phosphorylations among Ser380, Thr382, Thr383, and Ser385, we employed expressed protein ligation to generate individual semisynthetic phospho-forms of PTEN. In this way, amino acids 1–378 were produced as an intein fusion protein using a baculovirus expression system and then reacted with MESNA to generate the PTEN recombinant fragment C-terminal thioester (Fig. 1, A and B). In addition, we synthesized a series of eight new 25-mer N-Cys containing mono-, di-, and tri-phosphorylated C-terminal peptides, along with the unphosphorylated and tetraphosphorylated peptides prepared previously. These peptides were then chemoselectively ligated individually to the recombinant PTEN thioester with high efficiency (>80%) and then the semisynthetic full-length PTEN proteins purified using anion exchange chromatography to >90% purity (Fig. 1C). As shown previously (14), the introduction of Cys379 (in place of Tyr379) and the conditions of ligation are well tolerated by PTEN regarding its catalytic activity.
FIGURE 1.
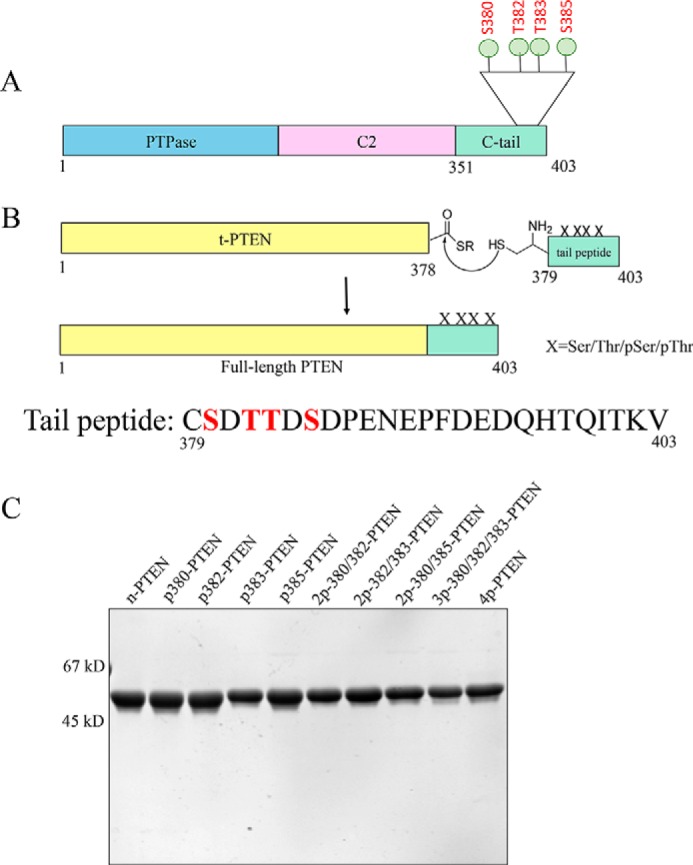
Generation of unmodified, mono-, di-, tri-, and tetraphosphorylated semisynthetic PTEN proteins. A, PTEN is composed of a protein-tyrosine phosphatase (PTPase) domain, a C2 domain, and a regulatory C-terminal tail. The cluster of phosphorylation (Ser380, Thr382, Thr383, and Ser385) is highlighted. B, C-terminal t-PTEN (amino acids 1–378) with a thioester at the C terminus is generated from intein fusion, treated with MESNA, and then ligated to the synthetic peptide containing different combinations of Ser(P) and Thr(P) (mono: p380, p382, p383, and p385; di: 2p-380/382, 2p-380/385, and 2p-382/383; tri: 3p-380/382/383; and tetra: 4p-380/382/383/385). X, Ser/Thr/Ser(P)/Thr(P). C, Coomassie-stained 10% SDS-PAGE gel of the set of differentially phosphorylated semisynthetic PTEN proteins. Ligation of t-PTEN-thioester and the specific peptide proceeds at a constant rate for 48 h, and the full-length PTEN is further purified by FPLC-anion exchange chromatography using MonoQ column. The final protein is >90% pure. First lane, n-PTEN; second lane, p380-PTEN; third lane, p382-PTEN; fourth lane, p383-PTEN; fifth lane, p385-PTEN; sixth lane, 2p-380/382-PTEN; seventh lane, 2p-382/383-PTEN; eighth lane, 2p-380/385-PTEN; ninth lane, 3p-380/382/383-PTEN; tenth lane, 4p-PTEN.
We then measured the PIP3 phosphatase activity of these semisynthetic phospho-PTENs using soluble diC6-PIP3 substrate and monitoring inorganic phosphate release using malachite green. As shown in Fig. 2 and Table 2, the monophosphorylated forms p380-PTEN, p382-PTEN, p383-PTEN, and p385-PTEN all showed a modest reduction in catalytic efficiency, with each showing a kcat/Km ∼3-fold below that of unphosphorylated PTEN (WT n-PTEN). The three di-phosphorylated PTENs 2p-380/382-PTEN, 2p-382/383-PTEN, and 2p-380/385-PTEN each hydrolyze diC6-PIP3 ∼6-fold below that of WT n-PTEN. The triphosphorylated PTEN form 3p-380/382/383-PTEN showed approximately a 12-fold reduction in catalytic efficiency compared with WT n-PTEN, and its catalytic efficiency was very similar to that of tetraphosphorylated (WT 4p-PTEN). These data suggest that no single phospho-modification of the Ser/Thr C-terminal cluster is dominant and that each is partially additive in antagonizing catalysis.
FIGURE 2.
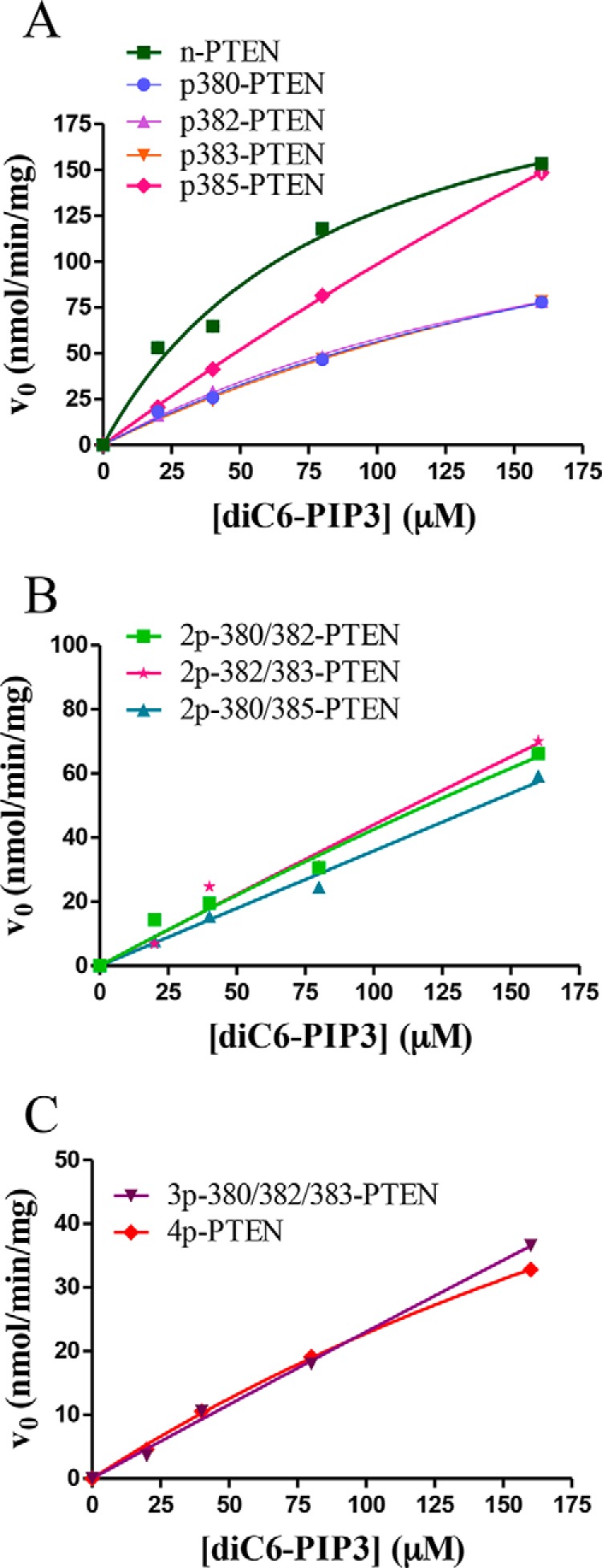
Catalytic activity of the set of differentially phosphorylated semisynthetic PTENs toward a range of diC6-PIP3 substrate concentrations. A, n-PTEN and monophosphorylated PTEN kinetics. B, diphosphorylated PTEN kinetics. C, tri- and tetraphosphorylated PTEN kinetics (n = 2).
TABLE 2.
Catalytic efficiency and tail phosphate reactivity with alkaline phosphatase for different PTENs
The errors are reported as ±S.E. ND, not determined.
| Semisynthetic PTENs | PTEN kcat/Km | Phospho-tail half-life after alkaline phosphatase treatment t½ |
|---|---|---|
| ×102m−1 s−1 | min | |
| WT n-PTEN | 22 ± 5 | ND |
| p380-PTEN | 6 ± 1 | 3 ± 2 |
| p382-PTEN | 7 ± 1 | ND |
| p383-PTEN | 6 ± 4 | ND |
| p385-PTEN | 9 ± 3 | ND |
| 2p-380/382-PTEN | 3.1 ± 0.1 | 6 ± 1 |
| 2p-382/383-PTEN | 3.4 ± 0.2 | ND |
| 2p-380/385-PTEN | 2.9 ± 0.2 | 20 ± 5 |
| 3p-380/382/383-PTEN | 1.8 ± 0.1 | 96 ± 13 |
| WT 4p-PTEN | 1.7 ± 0.1 | 104 ± 17 |
| WT t-PTEN | 18 ± 5 | ND |
| Cα2D-tPTEN | 12 ± 3 | ND |
| Cα2D-4p-PTEN | 3.3 ± 0.1 | 16 ± 5 |
| Cα2A-tPTEN | 15 ± 2 | ND |
| Cα2A-4p-PTEN | 3.1 ± 0.2 | 15 ± 2 |
| n-ePTEN | 49 ± 23 | ND |
| 4p-ePTEN | 8 ± 1 | 141 ± 26 |
| 3R/D-4p-PTEN | ND | 69 ± 13 |
To assess the effects of the particular phosphorylation events on PTEN conformation, the mono-, di-, and tri-phosphorylated semisynthetic PTENs were treated with alkaline phosphatase to gauge tail accessibility. As reported previously, natively folded 4p-PTEN versus denatured 4p-PTEN shows resistance to tail phosphate removal catalyzed by the nonspecific hydrolase alkaline phosphatase (14). The resistance of natively folded 4p-PTEN to alkaline phosphatase is understood to be caused by the masking of the phospho-tail through its intramolecular interactions with the PTEN body. We first determined which of the mono-,di-, and tri-phosphorylated semisynthetic PTENs are recognized by a commercially available phospho-PTEN Ab used in Western blotting. We found that all forms of p380-containing semisynthetic PTEN gave a strong Western blot signal with the anti-phospho-PTEN Ab, but those PTEN forms lacking p380 were not reliably detected (Fig. 3A). Thus, we proceeded to analyze the kinetics of alkaline phosphatase-catalyzed dephosphorylation of p380-PTEN, 2p-380/382-PTEN, 2p-380/385-PTEN, and 3p-380/382/385-PTEN benchmarked to WT 4p-PTEN standard (Fig. 3, B and C, and Table 2). These measurements showed that p380-PTEN was the most rapidly dephosphorylated by alkaline phosphatase (t½ = 3 min), 30-fold faster than WT 4p-PTEN (t½ = 104 min), and was followed closely by 2p-380/382-PTEN (t½ = 6 min), which was ∼20-fold faster than WT 4p-PTEN. 2p-380/385-PTEN (t½ = 20 min) was dephosphorylated by alkaline phosphatase ∼5-fold faster than WT 4p-PTEN. The sensitivity of 3p-380/382/385-PTEN (t½ = 96 min) to alkaline phosphatase was nearly identical to that of WT 4p-PTEN. Taken together, the pattern of alkaline phosphatase sensitivity correlates fairly well with the levels of semisynthetic PTEN catalytic activity, indicating that the individual phosphorylation events show partial additivity in driving the tail interaction with the PTEN body.
FIGURE 3.
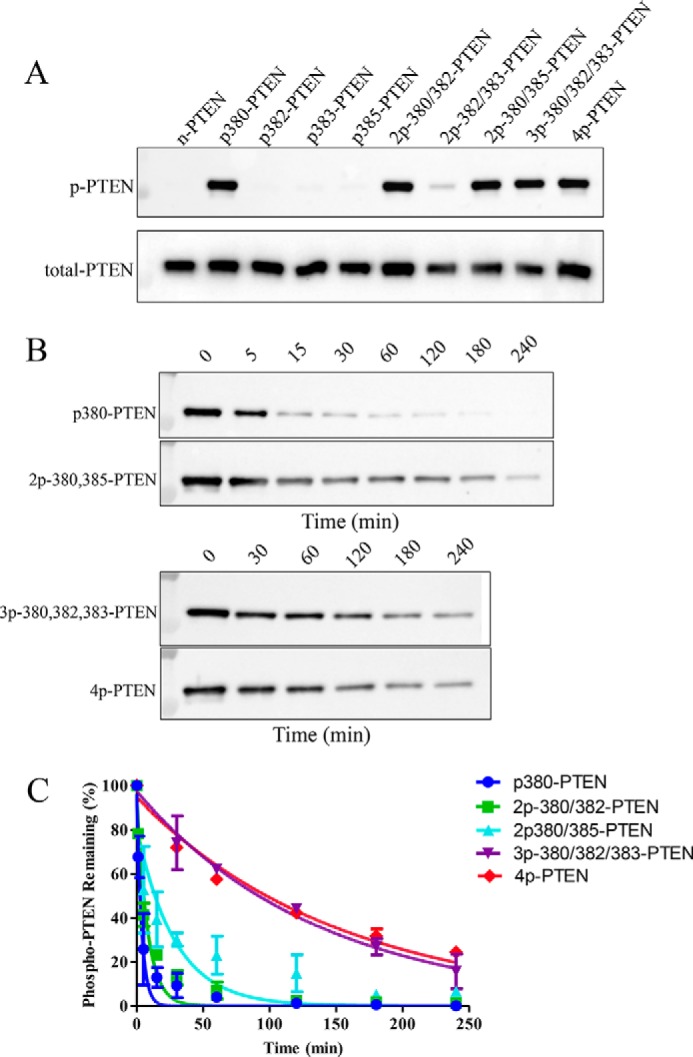
Alkaline phosphatase sensitivity of the set of differentially phosphorylated semisynthetic PTENs. A, Western blot of the set of differentially phosphorylated semisynthetic PTENs using an anti-phospho-PTEN (Ser380, Thr382, and Thr383) antibody. First lane, n-PTEN; second lane, p380-PTEN; third lane, p382-PTEN; fourth lane, p383-PTEN; fifth lane, p385-PTEN; sixth lane, 2p-380/382-PTEN; seventh lane, 2p-382/383-PTEN; eighth lane, 2p380/385-PTEN; ninth lane, 3p-380/382/383-PTEN; tenth lane, 4p-PTEN. B, time course of the rate of dephosphorylation of the p380-containing semisynthetic PTENs after alkaline phosphatase treatment analyzed by Western blotting. C, quantification of the time courses of the Western blot data in Fig. 3B (n = 2). Error bars indicate ± standard error.
Analysis of Mutant Semisynthetic 4p-PTENs
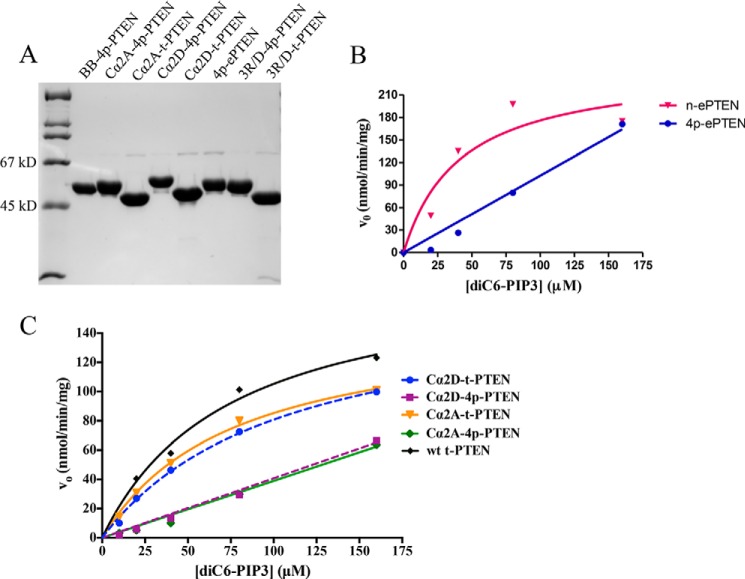
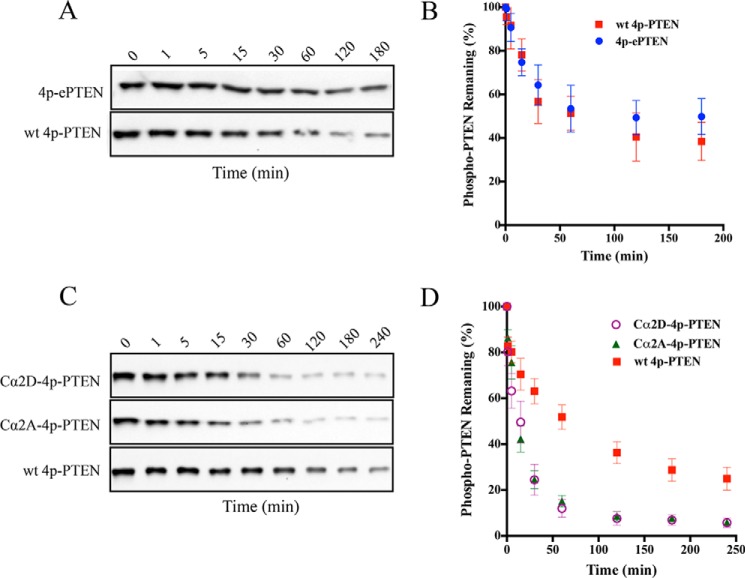
We next combined site-directed mutagenesis along with expressed protein ligation to prepare three semisynthetic 4p-PTEN proteins to investigate the biochemical properties of the ePTEN mutant residues and those of the PTEN Cα2 loop. As previously mentioned, cellular expression of ePTEN indicates that it localizes to the plasma membrane, suggesting that tail phosphorylation may be unable to drive a closed PTEN conformation (17). The Cα2 loop of PTEN has been implicated in membrane binding and in hydrogen/deuterium exchange experiments to participate in PTEN phospho-tail-mediated conformational change (11, 18). The 4p-ePTEN mutant containing Q17R, R41G, E73D, N262Y, and N329H was prepared by semisynthesis as described above, as were two 4p-PTEN Cα2 mutants, Cα2D (K327D, K330D, K332D, and R335D) and Cα2A (K327A, K330A, K332A, and R335A) to >90% purity (Fig. 4). Interestingly both n-ePTEN and 4p-ePTEN had 2- and 5-fold higher diC6-PIP3 phosphatase catalytic efficiencies compared with their WT counterparts (Fig. 4 and Table 2). However, the diC6-PIP3 phosphatase catalytic efficiency of 4p-ePTEN was a marked 6-fold lower than that of n-ePTEN (Fig. 4B and Table 2), suggesting that tail phosphorylation could drive ePTEN into a closed state. Furthermore, 4p-ePTEN alkaline phosphatase sensitivity showed a slightly (∼40%) longer half-life of phospho-tail hydrolysis relative to that of WT 4p-PTEN (Fig. 5, A and B, and Table 2). These results suggest that the point mutants of ePTEN do not substantially weaken phospho-tail-PTEN body interactions, and there is an alternative, unidentified explanation for ePTEN membrane localization.
FIGURE 4.
SDS-PAGE analysis and catalytic activity of a series of mutant semisynthetic PTENs toward a range of diC6-PIP3 substrate concentrations. A, Coomassie-stained 10% SDS-PAGE gel of semisynthetic PTEN mutants. First lane, molecular mass markers; second lane, BB-4p-PTEN; third lane, Cα2A-4p-PTEN; fourth lane, Cα2A-t-PTEN; fifth lane, Cα2D-4p-PTEN; sixth lane, Cα2D-t-PTEN; seventh lane, 4p-ePTEN; eighth lane, 3R/D-4p-PTEN; ninth lane, 3R/D-t-PTEN. B, catalytic activity of ePTEN forms with diC6-PIP3 (n = 2): Q17R, R41G, E73D, N262Y, and N329H. C, catalytic activity of Cα2 loop mutant PTEN forms with diC6-PIP3. Cα2D, K327D, K330D, K332D, and R335D; Cα2A, K327A, K330A, K332A, and R335A (n = 2).
FIGURE 5.
Alkaline phosphatase sensitivity of different semisynthetic phospho-PTEN mutants. A, Western blot of time course from the alkaline phosphatase-treated 4p-ePTEN (Q17R, R41G, E73D, N262Y, and N329H) and WT 4p-PTEN. B, quantification of the alkaline phosphatase assay for 4p-ePTEN and WT 4pPTEN (n = 3). C, Western blot of time course from the alkaline phosphatase treated Cα2D-4p-PTEN (K327D, K330D, K332D, and R335D), Cα2A-4p-PTEN (K327A, K330A, K332A, R335A), and WT 4p-PTEN. D, quantification of the alkaline phosphatase assay for 4p-Cα2 loop mutants and WT 4p-PTEN (n = 6). Error bars indicate ± standard error.
In contrast, analysis of the 4p-PTEN Cα2 mutants demonstrated that this region participates significantly in phosphorylation-mediated conformational change. Both the Cα2D and Cα2A 4p-PTEN mutants showed increased catalytic efficiencies of diC6-PIP3 hydrolysis relative to that of 4p-PTEN (Fig. 4C and Table 2). Although still lower than the truncated PTEN Cα2 mutants that lack phospho-tails, the lipid phosphatase activities of these 4p-PTEN Cα2 mutants suggest that replacement of these Lys and Arg residues weakened phospho-tail interactions in phosphorylated PTEN. Furthermore, both Cα2D and Cα2A 4p-PTEN exhibited a marked 6-fold greater sensitivity to alkaline phosphatase-mediated tail dephosphorylation compared with WT 4p-PTEN. The fact that both the Asp and Ala replacements of the Lys/Arg residues in the Cα2 4p-PTEN mutants display similar behaviors is consistent with the possibility that one or more of the Lys/Arg side chains are making important electrostatic interactions with residues in the anionic PTEN tail.
Photo-cross-linking of Benzoylphenylalanine Containing 4p-PTEN
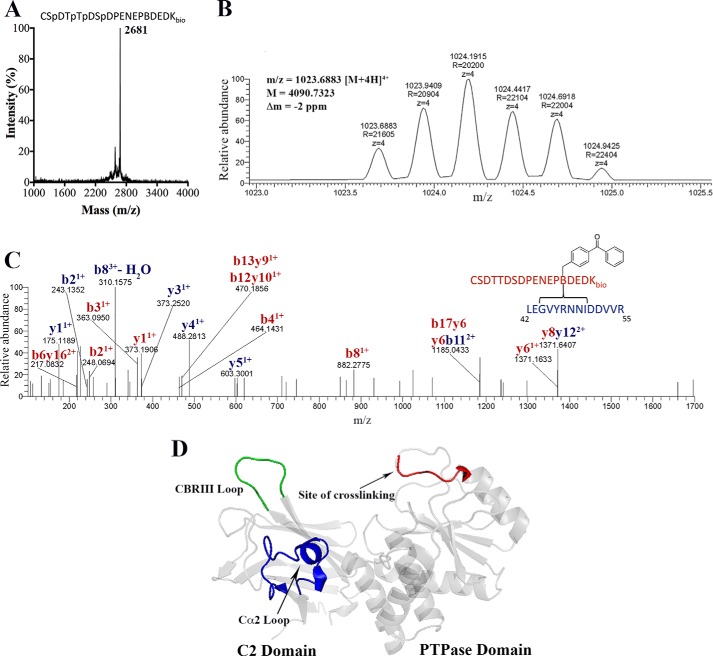
To further investigate the structural basis of phospho-tail-body interactions within 4p-PTEN, we pursued a photo-cross-linking strategy. In this regard, Phe392 was replaced with the photo-activatable Bpa and a C-terminal biotin-modified Lys. In this semisynthetic strategy, a 18-mer N-Cys peptide (Fig. 6A) was used rather than the full-length 25-mer tail for synthetic ease and because prior studies have shown the 4p-17-mer tail is sufficient to promote high affinity interaction with the core of PTEN (14). This Bpa/biotin-modified semisynthetic 4p-PTEN (BB-4p-PTEN) was efficiently produced and appeared greater than 90% pure after anion exchange chromatography (Fig. 4A). Irradiation of BB-4p-PTEN was followed by alkaline phosphatase treatment (to remove the mass spectrometry electrospray ionization suppressive effects of the phosphates) and trypsinization and avidin treatment to enrich for biotin-containing peptides. These peptides were then subjected to LC-MS/MS, and then the data were analyzed using Crossfinder (20, 21). This led to the identification of a cross-linked peptide that contained a C-terminal peptide (379CSDTTDSDPENEP(B)DEDKbio396) attached to an N-terminal peptide (42LEGVYRNNIDDVVR55) (Fig. 6, B and C). The assignment of this cross-linked peptide was based on a combination of its intact molecular mass (4090.7 Da), as well as its fragmentation pattern using tandem mass spectrometry, although the site of conjugation to the N-terminal peptide (between Gly45 and Ile50) could not be precisely defined (Fig. 6C). This cross-linked peptide was not detected in the absence of UV irradiation. These data thus suggest that the C-tail can access a conformation that places the C-terminal peptide in proximity to the N-terminal catalytic domain segment containing amino acids 42–55.
FIGURE 6.
BB-4p-PTEN, photo-cross-linking, and mass spectrometry analysis. A, MALDI-TOF MS spectrum of 18-mer synthetic peptide containing tetraphosphorylation (Ser380, Thr382, Thr383, and Ser385), Bpa at position 392, and a C-terminal biotinylated lysine used for expressed protein ligation. B, isotopic distribution of a 4+ charged cross-linked peptide with the reported mass, m/z, and Δm from Crossfinder. C, high resolution tandem-MS spectrum of the cross-linked peptide. D, PTEN crystal structure (Protein Data Bank code 1D5R) highlighting the site of cross-linking and other important areas shown to be involved in phospho-C-tail binding.
To further probe the importance of this interaction, we generated semisynthetic 4p-PTEN containing three point mutations in the vicinity of the three-dimensional surface of the cross-linked N-terminal segment, R41D, R47D, and R74D (3R/D). Even unligated 3R/D PTEN mutant showed significantly impaired catalytic activity (Fig. 7A), as expected from prior studies that have investigated the phosphatase effects of mutating Arg47 (22, 23). Thus, it was not feasible to use diC6-PIP3 hydrolytic activity to gain insight into the phosphorylation effects of 3R/D-4p-PTEN. To assess the conformation of the 3R/D-4p-PTEN, we relied on alkaline phosphatase sensitivity. In these experiments, we found that 3R/D-4p-PTEN (t½ = 63 min) showed approximately a 2-fold rate increase in alkaline phosphatase-mediated dephosphorylation compared with 4p-PTEN (Fig. 7, B and C, and Table 2). These data support at least a modest contribution of the N-terminal catalytic segment in stabilizing the closed conformation of 4p-PTEN.
FIGURE 7.
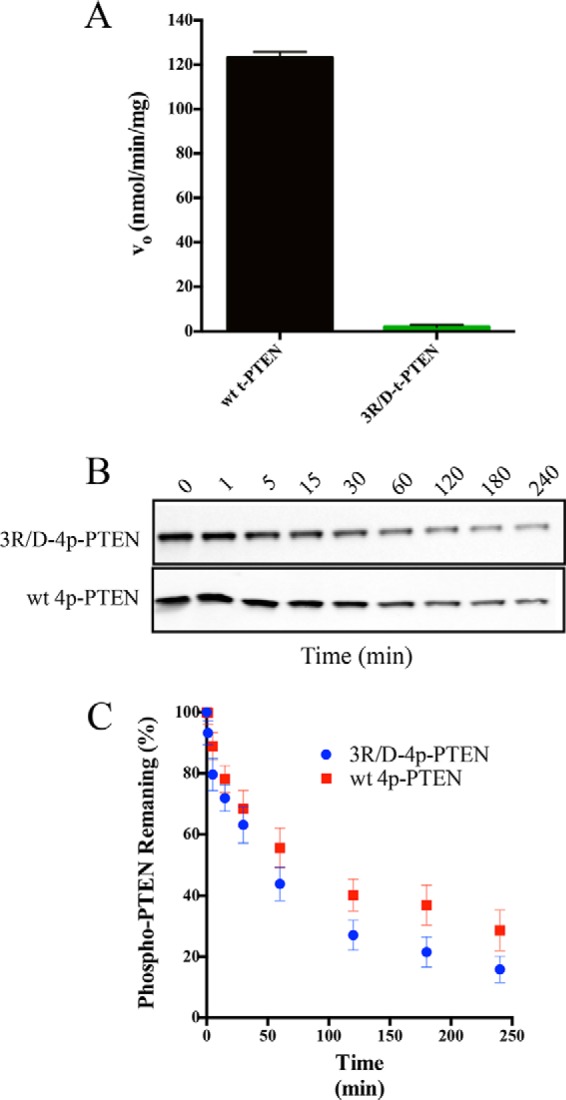
Conformational analysis of the 3R/D PTEN mutant. A, catalytic activity of WT t-PTEN (123 ± 4 nmol/min/mg) compared with 3R/D t-PTEN (<4 nmol/min/mg) with 160 μm diC6-PIP3 (n = 2). B, Western blot of time course of dephosphorylation of alkaline phosphatase-treated 3R/D-4p-PTEN (R41D, R47D, and R74D) and WT 4p-PTEN. C, quantification of the alkaline phosphatase assay for 3R/D-4p-PTEN and WT 4p-PTEN (n = 6). Error bars indicate ± standard error.
Discussion
The application of protein semisynthesis, mutagenesis, and photo-cross-linking has been integrated to refine our understanding of how phosphorylation of a cluster of four C-terminal Ser/Thr residues drives a conformational change in PTEN. Prior to this work, it was unclear whether any single phosphorylation site in PTEN would be predominantly or wholly responsible for inducing the PTEN conformational change, and the influence of the three other sites would be negligible. Our data shown here support the idea that each of the four sites can contribute incrementally to stabilizing the closed PTEN conformation, and at least three sites are needed to match the full effects of tetraphosphorylated PTEN. These results imply that the surface of interaction between the phospho-tail and the PTEN body may be rather broad to encompass multiple contact sites with these phosphates. These findings also suggest the concept that a dynamic stepwise degree of PTEN conformational closure may occur by modifying only a subset of the tail Ser/Thr residues, which in turn could give rise to a sliding scale of cellular signaling effects. Our results also highlight that the antibody used here cannot readily distinguish between monophosphorylated p380 and the multiply modified forms that contain p380. This suggests the need for caution in interpreting Western blots and immunocytochemistry data that rely on this reagent for signaling analysis.
Several models have been proposed regarding where the phospho-tail in PTEN contacts the core of the protein (14–16). In the current study, we found additional evidence for key interactions between the PTEN C2 domain and the phospho-tail. Prior studies have shown that the CBR3 loop of the PTEN C2 domain is involved in the phospho-tail interactions (14), with an 8-fold enhanced sensitivity to alkaline phosphatase-catalyzed tail dephosphorylation compared with WT 4p-PTEN. However, although the penta-Lys (260, 263, 266, 267, and 269) to penta-Asp CBR3 loop 4p-PTEN mutant appears to have a relatively open structure, mutation of the penta-Lys motif to penta-Ala is quite similar to WT 4p-PTEN (14). Thus, it appears that none of the CBR3 loop Lys residues are making key electrostatic interactions with the phospho-tail, and only when replaced with the negatively charged Asp residues do they prevent conformational closure, arguing for a more indirect role in stabilizing the closed conformation. In contrast, as shown here the Cα2 Lys/Arg side chains cannot be substituted functionally by Ala, suggesting that these basic residues are more directly involved in tail phospho-interactions.
Some prior experiments including hydrogen/deuterium exchange, mutagenesis, and protease sensitivity have suggested that the catalytic domain of PTEN might also somehow participate in phospho-tail interactions (14, 15, 18). Through the use of photo-cross-linking, we find that at least part of the phospho-tail binds to an N-terminal segment in the PTEN catalytic domain. Mutagenesis of this segment is consistent with this region playing a partial role in mediating the conformational closure of phosphorylated PTEN. If we consider the energetic sum of the alkaline phosphatase sensitivity parameters of the N-terminal segment (2-fold), the Cα2 loop (6-fold), and the CBR3 loop (8-fold) mutants reported here and previously, we can more than account for the 25-fold protection measured when comparing folded and unfolded 4p-PTEN. Thus, it is plausible that these three PTEN surfaces are fully responsible for locking the phospho-tail to the PTEN body (14). It is perhaps noteworthy that this N-terminal segment as well as the CBR3 loop and the Cα2 loop all have been implicated in PTEN membrane recruitment (1, 18, 22, 23). It is therefore reasonable to postulate that the PTEN phosphate tail acts as a molecular mimic of the phospholipid membrane. These results underscore the very large interaction surface that may govern the 4p-PTEN conformational closure.
The observation that 4p-ePTEN behaves conformationally similarly to WT 4p-PTEN was unexpected, given the dramatic cellular phenotype associated with ePTEN (17). We speculate that these ePTEN point mutations may strengthen a specific protein-protein or protein lipid interaction between PTEN and a molecule not yet identified. Future work will be needed to address these possibilities that could uncover new targets to enhance cellular PTEN functions.
This study also illustrates the power of expressed protein ligation in elucidating how a complex series of PTMs can alter the structure of a protein (24). Because mass spectrometry has identified hundreds of thousands of PTMs in proteins, it has become daunting to cope with illuminating the structural and functional effects of these PTMs at an individual protein level. The incorporation of four phosphorylations, a benzoylphenylalanine, and a biotin tag site-specifically into a single folded protein of this size would have been difficult to achieve without expressed protein ligation. Expressed protein ligation when coupled with other biochemical, biophysical, and analytical approaches thus offers atomic precision in working through the 21st century challenges of protein science.
Author Contributions
Z. C., D. R. D., and P. A. C. designed the experiments, and Z. C., D. R. D., D. H., and D. M. B. performed the biochemical experiments in this manuscript. S. N. T. designed and performed the protein mass spectrometry experiments and analyzed the data along with D. R. D. and P. A. C. All authors contributed to the analysis of the data. The manuscript was drafted by Z. C., D. R. D., and P. A. C., and all authors contributed to writing and editing the manuscript.
Acknowledgments
We thank Felix Mueller-Planitz, Chan-Hyun Na, Peter Devreotes, Miho Iijima, Mario Amzel, and Sandra Gabelli for helpful discussions. We thank Jun Liu's lab for assistance with photo-cross-linking.
This work was supported by National Institutes of Health Grant CA74305. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PTEN
- phosphatase and tensin homolog
- Bpa
- benzoylphenylalanine
- BB-4p-PTEN
- Bpa/biotin-modified semisynthetic 4p-PTEN
- diC6-PIP3
- 1,2-dihexanoyl-sn-glycero-3-(phosphoinositol-3′,4′,5′-trisphosphate)
- PIP3
- phosphatidylinositol 3,4,5-triphosphate
- t-PTEN
- truncated PTEN
- n-PTEN
- unphosphorylated PTEN
- 4p-PTEN
- tetraphosphorylated PTEN
- PTM
- post-translational modification
- MESNA
- sodium 2-mercaptoethanesulfonate
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl.
References
- 1. Worby C. A., and Dixon J. E. (2014) PTEN. Annu. Rev. Biochem. 83, 641–669 [DOI] [PubMed] [Google Scholar]
- 2. Maehama T., and Dixon J. E. (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273, 13375–13378 [DOI] [PubMed] [Google Scholar]
- 3. Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S. I., Puc J., Miliaresis C., Rodgers L., McCombie R., Bigner S. H., Giovanella B. C., Ittmann M., Tycko B., Hibshoosh H., Wigler M. H., and Parsons R. (1997) PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947 [DOI] [PubMed] [Google Scholar]
- 4. Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., and Patel T. (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salvesen H. B., MacDonald N., Ryan A., Jacobs I. J., Lynch E. D., Akslen L. A., and Das S. (2001) PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. Int. J. Cancer 91, 22–26 [DOI] [PubMed] [Google Scholar]
- 6. Wang X., and Jiang X. (2008) Post-translational regulation of PTEN. Oncogene 27, 5454–5463 [DOI] [PubMed] [Google Scholar]
- 7. Wang X., and Jiang X. (2008) PTEN: a default gate-keeping tumor suppressor with a versatile tail. Cell Res. 18, 807–816 [DOI] [PubMed] [Google Scholar]
- 8. Vazquez F., Ramaswamy S., Nakamura N., and Sellers W. R. (2000) Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell Biol. 20, 5010–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun H., Lesche R., Li D. M., Liliental J., Zhang H., Gao J., Gavrilova N., Mueller B., Liu X., and Wu H. (1999) PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 96, 6199–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Myers M. P., Pass I., Batty I. H., Van der Kaay J., Stolarov J. P., Hemmings B. A., Wigler M. H., Downes C. P., and Tonks N. K. (1998) The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl. Acad. Sci. U.S.A. 95, 13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee J. O., Yang H., Georgescu M. M., Di Cristofano A., Maehama T., Shi Y., Dixon J. E., Pandolfi P., and Pavletich N. P. (1999) Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 99, 323–334 [DOI] [PubMed] [Google Scholar]
- 12. Torres J., and Pulido R. (2001) The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus: implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 276, 993–998 [DOI] [PubMed] [Google Scholar]
- 13. Al-Khouri A. M., Ma Y., Togo S. H., Williams S., and Mustelin T. (2005) Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3β. J. Biol. Chem. 280, 35195–35202 [DOI] [PubMed] [Google Scholar]
- 14. Bolduc D., Rahdar M., Tu-Sekine B., Sivakumaren S. C., Raben D., Amzel L. M., Devreotes P., Gabelli S. B., and Cole P. (2013) Phosphorylation-mediated PTEN conformational closure and deactivation revealed with protein semisynthesis. Elife 2, e00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rahdar M., Inoue T., Meyer T., Zhang J., Vazquez F., and Devreotes P. N. (2009) A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc. Natl. Acad. Sci. U.S.A. 106, 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Odriozola L., Singh G., Hoang T., and Chan A. M. (2007) Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J. Biol. Chem. 282, 23306–23315 [DOI] [PubMed] [Google Scholar]
- 17. Nguyen H. N., Yang J. M., Afkari Y., Park B. H., Sesaki H., Devreotes P. N., and Iijima M. (2014) Engineering ePTEN, an enhanced PTEN with increased tumor suppressor activities. Proc. Natl. Acad. Sci. U.S.A. 111, E2684–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Masson G. R., Perisic O., Burke J. E., and Williams R. L. (2016) The intrinsically disordered tails of PTEN and PTEN-L have distinct roles in regulating substrate specificity and membrane activity. Biochem. J. 473, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muir T. W., Sondhi D., and Cole P. A. (1998) Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. U.S.A. 95, 6705–6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mueller-Planitz F. (2015) Crossfinder-assisted mapping of protein crosslinks formed by site-specifically incorporated crosslinkers. Bioinformatics 31, 2043–2045 [DOI] [PubMed] [Google Scholar]
- 21. Forne I., Ludwigsen J., Imhof A., Becker P. B., and Mueller-Planitz F. (2012) Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive crosslinkers and mass spectrometry. Mol. Cell. Proteomics 11, M111.012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Q., Wei Y., Mottamal M., Roberts M. F., and Krilov G. (2010) Understanding the stereospecific interactions of 3-deoxyphosphatidylinositol derivatives with the PTEN phosphatase domain. J. Mol. Graph. Model. 29, 102–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei Y., Stec B., Redfield A. G., Weerapana E., and Roberts M. F. (2015) Phospholipid-binding sites of phosphatase and tensin homolog (PTEN): exploring the mechanism of phosphatidylinositol 4,5-bisphosphate activation. J. Biol. Chem. 290, 1592–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Z., and Cole P. A. (2015) Synthetic approaches to protein phosphorylation. Curr. Opin. Chem Biol. 28, 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]