Abstract
The envelope glycoprotein (Env) gp120/gp41 is required for HIV-1 infection of host cells. Although in general it has been perceived that more Env gives rise to higher infectivity, the precise quantitative dependence of HIV-1 virion infectivity on Env density has remained unknown. Here we have developed a method to examine this dependence. This method involves 1) production of a set of single-cycle HIV-1 virions with varied density of Env on their surface, 2) site-specific labeling of Env-specific antibody Fab with a fluorophore at high efficiency, and 3) optical trapping virometry to measure the number of gp120 molecules on individual HIV-1 virions. The resulting gp120 density per virion is then correlated with the infectivity of the virions measured in cell culture. In the presence of DEAE-dextran, the polycation known to enhance HIV-1 infectivity in cell culture, virion infectivity follows gp120 density as a sigmoidal dependence and reaches an apparent plateau. This quantitative dependence can be described by a Hill equation, with a Hill coefficient of 2.4 ± 0.6. In contrast, in the absence of DEAE-dextran, virion infectivity increases monotonically with gp120 density and no saturation is observed under the experimental conditions. These results provide the first quantitative evidence that Env trimers cooperate on the virion surface to mediate productive infection by HIV-1. Moreover, as a result of the low number of Env trimers on individual virions, the number of additional Env trimers per virion that is required for the optimal infectivity will depend on the inclusion of facilitating agents during infection.
Keywords: glycoprotein, human immunodeficiency virus (HIV), infection, optical tweezers, protein cross-linking, single particle analysis, single-molecule biophysics
Introduction
Like other enveloped viruses, HIV-1 uses the trimer-of-hairpins mechanism to catalyze the fusion between viral and cell membranes, an essential step during the life cycle of all enveloped viruses (1, 2). For HIV-1 virions derived from chronically infected T cell lines, quantitative Western (3) and cryoelectron tomography (4) have revealed that there are on average only 14 Env5 trimers per virion. Recent measurements by Brandenberg et al. (5) using quantitative Western blotting analysis on HIV-1 pseudoparticles derived from transfected 293-T cells also yielded 6 to 20 Env trimers per virion. The number of entry-mediating surface glycoprotein oligomers is lower than for other enveloped viruses (6), and this may have a profound impact on HIV-1 infection, the quantitative aspects of which are being unraveled. For example, recent studies have shown that transmitted founder HIV-1 virions contain more Env than the corresponding chronic virus (7), suggesting that more Env may indeed confer selective advantages in HIV-1 transmission. At the mechanistic level, however, how the Env copy number may enhance virion transmission is not well understood. Although greater amounts of Env have been observed to increase infectivity (8), the dependence of HIV-1 virion infectivity on Env density has not been precisely described or explained.
In this paper, we introduce a quantitative approach to this problem. In our approach, we use a provirus clone that is env− together with a separate wild-type env plasmid (pEnv) to generate HIV-1 virions (9). It is essential that the provirus is env− so that we can titrate pEnv from low to high quantities (8) to generate a set of virions that are expected to carry varied densities of Env on their surface, limited by the pEnv inputs (10). Meanwhile, the resulting virions are only infectious for a single cycle. The infectivity of the virions can thus be correlated with the Env content of these virions that can be directly measured by single-molecule techniques without the complications from multiple rounds of infection.
To determine Env density on individual virions, we have recently developed a technique named optical trapping virometry (OTV) (10). This technique works by optically trapping individual HIV-1 virions in suspension and thereby measuring the number of fluorophores associated with each virion with single-molecule resolution. To use this technique for accurate quantitation of gp120 molecules on individual HIV-1 virions, we have developed a method to site-specifically label a gp120-specific antibody Fab with a fluorophore at >95% efficiency. When the fluorescent Fab against gp120 is incubated with the virus under saturating Fab concentrations, the number of fluorophores associated with each virion thus directly measures the number of gp120 molecules per virion. By applying this method to the set of single-cycle virions and quantitating gp120 density for individual virions in each virion population, we can then quantitatively examine the dependence of HIV-1 infectivity on gp120 density.
Experimental Procedures
Production of HIV-1 Virions
Single-cycle HIV-1 virions were generated and assayed as described recently (9). Briefly, 293T cells were transfected with 1.0 μg of pNL4–3R−E− plasmid, various amounts of pEnv (NL4-3 envelope expression plasmid, the same as pcDNA3.1REC in Ref. 9) and 0.3 μg of pEGFP-Vpr by the use of Mirus LT-1 transfection reagents in a 2-ml culture volume in a 35-mm dish. Culture medium was changed 6 h post-transfection and virions were harvested 24 h post-transfection (9). To determine the infectivity for each population of single-cycle virions, the virion stocks were diluted in complete media either in the presence or absence of 20 μg/ml of DEAE-dextran, and then incubated with TZM-bl indicator cells in a 12-well plate for 2 h at 37 °C, with gentle rocking every 30 min. At the end of 2 h, 1 ml of complete medium was added to each well and the incubation was continued at 37 °C for 48 h with 5% CO2. At the end of 48 h, individual single-cell infectious events were recorded following the procedures as described previously (9). The physical concentrations of the virion particles were determined by p24 ELISA as described (9). The infectivity per virion was calculated by taking the ratio of the infectious events per input volume over the physical particle concentration.
Production, Purification, and Site-specific Labeling of VRC01 Fab
The open reading frames of the light chain and the truncated heavy chain of the VRC01 Fab were synthesized by GenScript USA Inc. and cloned into pCMV/R vector (11) for overexpression of the Fab in the Freestyle 293-F cells. The C-terminal sequence of the truncated heavy chain is as follows: DKTHTCPPDIHHHHHH, where the underlined cysteine residue is Cys free for cross-linking through maleimide chemistry. The production of VRC01 Fab from 293-F cells follows published procedures (12). Briefly, the two plasmids were transfected into 293-F cells at equal masses and the cells were cultured for 6 days at 37 °C with 5% CO2 for protein production. At the end of 6 days post-transfection, the culture supernatant was harvested, filtered through a 0.22-μm filter, and mixed with Ni-NTA-agarose beads for batch binding overnight at 4 °C. On the second morning, the Ni-NTA-agarose beads were packed onto an empty Bio-Rad glass column, and mounted onto Bio-Rad BioLogic DuoFlow 10 system for washing and elution. The column was first washed with buffer A (50 mm Tris, 500 mm NaCl, pH 8.0, at 4 °C) to baseline, and then washed with 4 column volumes of buffer A with 10 mm imidazole, which was followed by gradient elution from 10 to 500 mm increasing concentrations of imidazole in buffer A over 10 column volumes at a flow rate of 1 ml/min. The eluted Fab fragment was then concentrated by centriprep with a molecular mass cut off of 3 kDa and loaded onto an GE Superdex 200 gel filtration column that was equilibrated in buffer B (25 mm HEPES, 0.5 m NaCl, pH 7.0, at 20 °C). The protein was eluted off the column at a flow rate of 0.4 ml/min and its concentration was determined by UV-visible absorbance at 280 nm in 6 m guanidine hydrochloride, with an extinction coefficient of 7.61 × 104 m−1 cm−1 estimated based on the amino acid sequence (13), considering the formation of five disulfide bonds within a single molecule of VRC01 Fab. The eluted protein was then incubated immediately with a 20-fold molar excess of the Alexa 594-maleimide (Invitrogen) solution for cross-linking reaction at 20 °C for a duration of 12 h. At the end of the 12 h, the protein-dye mixture was reloaded onto the Superdex 200 gel filtration column, and eluted at a flow rate of 0.4 ml/min in buffer B. The concentration of the eluted protein was again measured by absorbance at 280 nm (corrected for dye absorbance at this wavelength), and the concentration of the conjugated dye molecule was quantitated simultaneously by absorbance at 590 nm with an extinction coefficient of 9.00 × 104 m−1 cm−1. From 1 liter of 293-F cell culture, we produced ∼100 mg of VRC01 Fab after Ni-NTA column purification. Using 3 mg of this unlabeled VRC01 Fab as the starting material, ∼0.5 mg of site-specifically labeled Fab after labeling and purification through a gel filtration column was produced. All protein purification procedures were conducted at 4 °C unless otherwise noted.
Virus Neutralization Assay
NL4-3 virus neutralization by VRC01 Fab was carried out as described (10). Briefly, the HIV-1 NL4-3 virions were prepared by transfecting 293T cells with 1.0 μg of pNL4–3R−E− plasmid and 2 μg of pEnv (encoding full-length NL4-3 Env, including the cytoplasmic tail) and 0.3 μg of pEGFP-Vpr per 2-ml culture. HIV-1 virions were incubated with Fab at varied concentrations for 1 h, at 20 °C, and then diluted 10-fold with complete medium for inoculation of TZM-bl cells at 37 °C for 2 h. After the addition of fresh medium to each well in a 12-well plate, the cells were cultured at 37 °C with 5% CO2 for another 48 h, and then blue cells were counted (14). The number of blue cells generated from virus infection without Fab treatment was normalized to 100%. The number of blue cells generated from virus treated with either unlabeled or Alexa 594-labeled VRC01 Fab was expressed as a percentage of the blue cell counts in the absence of Fab and plotted as a function of Fab concentration. The neutralization data were fitted to a sigmoid function (15) with top and bottom plateaus constrained to 100 and 0% to determine IC50 values and Hill slopes.
Gp120 Shedding Assay
The gp120 shedding assay was performed as described previously (10). Briefly, we transfected 293T cells with pEnv, the plasmid encoding NL4-3 Env. This generates uncleaved Env precursors and Env complexes, including gp120 subunits, expressed on the cell surface. The day before the transfection, 4 × 105 of 293T cells per well were seeded in a 12-well plate, and incubated at 37 °C with 5% CO2 in complete medium. The next day the cells had reached ∼50% confluence and were transfected with 0.2 μg of pEnv by the use of the TransIT LT-1 transfection reagent (Mirus Bio, Madison, WI). 24 h after the transfection, cell culture medium was removed and 100 μl of VRC01 IgG at varied concentrations (0.4, 2, 10, and 50 μg/ml), diluted in Dulbecco's phosphate-buffered saline, was added to the cells. The cells were incubated with the antibodies for 1 h at 37 °C, gentle rocking was performed every 20 min. Then the supernatants were collected and placed on ice, and centrifuged at 1,000 × g for 5 min at 4 °C to remove cells and cell debris. These supernatants were mixed with equal volumes of 2× Laemmli sample buffer containing β-mercaptoethanol, immediately vortexed vigorously, and then incubated at 95 °C for 5 min. These samples were loaded onto a SDS-PAGE. Proteins were blotted by electrotransfer onto supported nitrocellulose membranes. To detect gp120, the membranes were incubated first with sheep anti-gp120 polyclonal serum (catalog number 288, NIH AIDS Reagent Reference program) and then with donkey anti-sheep IgG alkaline phosphatase-conjugated secondary antibody (catalog number A5187, Sigma). The enzyme reaction was then developed, and the resulting bands scanned and quantitated with ImageJ as described (10). In similar gp120 shedding assays, the presence of gp160 in the sample supernatant after 293T cell transfection has been described in the literature by several different groups, including ours (9, 10, 16, 17). Gp160 proteins are present in the supernatant in the absence of any ligand addition, suggesting that they stem from gp160-bearing microvesicles secreted by the cells; gp120 in the supernatant could originate from proteolytically processed Env expressed on the cell surface or incorporated into microvesicles.
Optical Tweezers and Two-photon Fluorescence (TPF) Experiments
The trapping of individual virions in suspension was done as described previously (10) with a few modifications. Throughout, a home-made optical tweezer instrument using a tapered amplifier diode laser at 830 nm (SYS-420-830-1000, Sacher LaserTechnik LLC, Germany) was used for optical trapping and simultaneous TPF measurements of individual HIV-1 virions (18). Briefly, the live virus stock thawed from −80 °C was first incubated with 20 μg/ml of Alexa 594-labeled VRC01 Fab (Alexa 594-Fab) for 1 h at 20 °C in the dark, and then diluted in PBS to a concentration of 0.6–1.6 × 108 virions/ml and injected into a microfluidic chamber for optical trapping. The diameter of individual virions was measured as described previously (10). An electron-multiplying charge-coupled device camera (Evolve, Photometrics) was used for all fluorescence detection with single-molecule sensitivity. The TPF from EGFP was monitored by the use of an emission filter (HQ525/50m, Chroma) and the TPF of Alexa 594 was monitored with another emission filter (ET645/75, Chroma). A laser power of 130.8 milliwatts at the focus was used throughout for optical trapping and simultaneous TPF excitation so that fluorescence intensity can be directly compared across experiments. Individual HIV-1 virions were identified based on an EGFP-positive signal and a measured diameter that lies between 96 and 216 nm as described previously (10). The initial Alexa 594 TPF prior to photobleaching was averaged and recorded per virion before sequential measurement of EGFP TPF. The residual leakage of the EGFP channel fluorescence into the Alexa 594 channel was experimentally measured and quantified by linear regression as described previously (10) and this information was used to correct for Alexa 594 TPF throughout. The TPF was collected as described by Nis-Element Software (Nikon), with an EM gain of 200 and exposure time of either 100 or 400 ms unless otherwise noted. All trapping and TPF experiments were conducted at a constant temperature of 20.0 ± 0.2 °C. The step-finding algorithm that we developed recently (19) was used to identify individual photobleaching steps from real time TPF trajectories. Throughout this work, all errors are standard deviations unless otherwise noted.
Results
Purification and Site-specific Labeling of VRC01 Fab
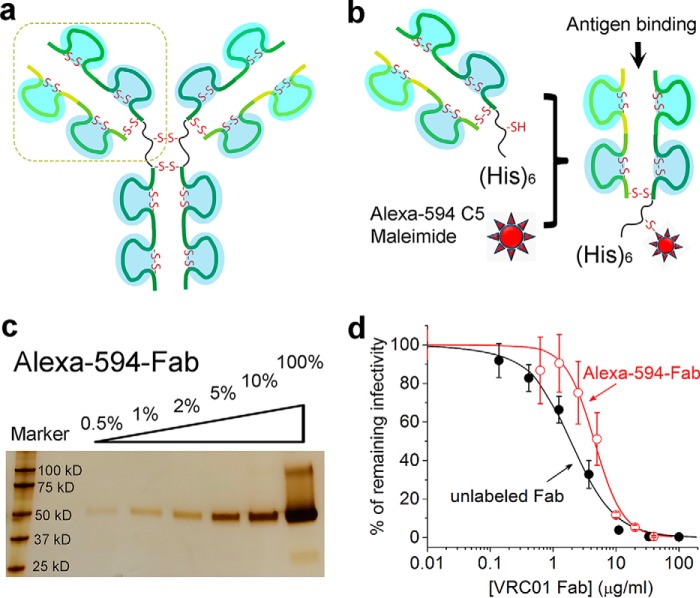
To make site specifically labeled Fab without interfering with its epitope binding is challenging because typical IgG molecules contain a number of cysteine residues that form disulfide bonds (Fig. 1a). Analogously to Galimidi et al. (20), we have developed a potentially generic approach to site specifically label the Fab fragment of an antibody without compromising its antigen binding affinity. This strategy is schematically shown in Fig. 1b, where a Fab fragment carries a single free cysteine for labeling through maleimide chemistry. The Fab is expressed in the FreeStyle 293-F cells (Invitrogen) upon transfection with two plasmids: one encodes the light chain, and the other encodes a truncated heavy chain of the IgG antibody. This truncation is located between the two cysteine residues in the original hinge region of the full-length IgG (denoted by the dashed line in Fig. 1a). We further added a hexahistidine tag at the C terminus of the truncated heavy chain for ease of protein purification by Ni-NTA technology (Fig. 1b). The cysteine close to the C terminus of the truncated heavy chain is free for labeling in the Fab because of the absence of Fc which holds the original two heavy chains together. We have developed a protocol for purification and labeling of a Fab fragment from the monoclonal antibody VRC01, which binds to the CD4-binding site (CD4bs) on gp120 with high affinity and potently neutralizes 91% of all major circulating HIV-1 isolates (12). This binding does not induce gp120 shedding (16) (Fig. 2), which is in contrast to soluble CD4 that induces shedding of gp120 under identical experimental conditions as we published previously (10). Thus VRC01 is well suited for quantitation of gp120 molecules on a virion. We purified this protein first through a Ni-NTA affinity column and a GE S200 gel filtration column, and then labeled it with Alexa 594-maleimide by incubation at room temperature for 12 h. The labeled Fab was further purified through an S200 gel-filtration column to remove excess free dye molecules. As shown in Fig. 1c for the final labeled protein on a non-reducing polyacrylamide gel, we obtained a Fab fragment that was more than 95% pure based on quantitation of band intensity. The labeling efficiency was 97% as determined by UV-visible absorbance spectrum. Moreover, we determined the mass for both unlabeled and labeled VRC01 Fab by positive ion electrospray mass spectrometry. The measured mass for unlabeled VRC01 Fab was 52334.6 Da. The mass for labeled VRC01 Fab was determined to be 53222.0 Da. The difference in mass between the two proteins is 887.4 Da, which very well matches the expected addition of a single Alexa 594-C5 maleimide group (885.0 Da) onto the Fab molecule.
FIGURE 1.
Specific labeling of VRC01 Fab by Alexa 594-maleimide. a, schematic of a full-length IgG. b, site-specific labeling strategy for VRC01 Fab. c, silver staining to assess the purity of Alexa 594-labeled VRC01 Fab using a 4–15% gradient polyacrylamide gel. The 100% corresponds to 3 μg of purified protein. d, neutralization of NL4-3 virions by purified VRC01 Fab.
FIGURE 2.
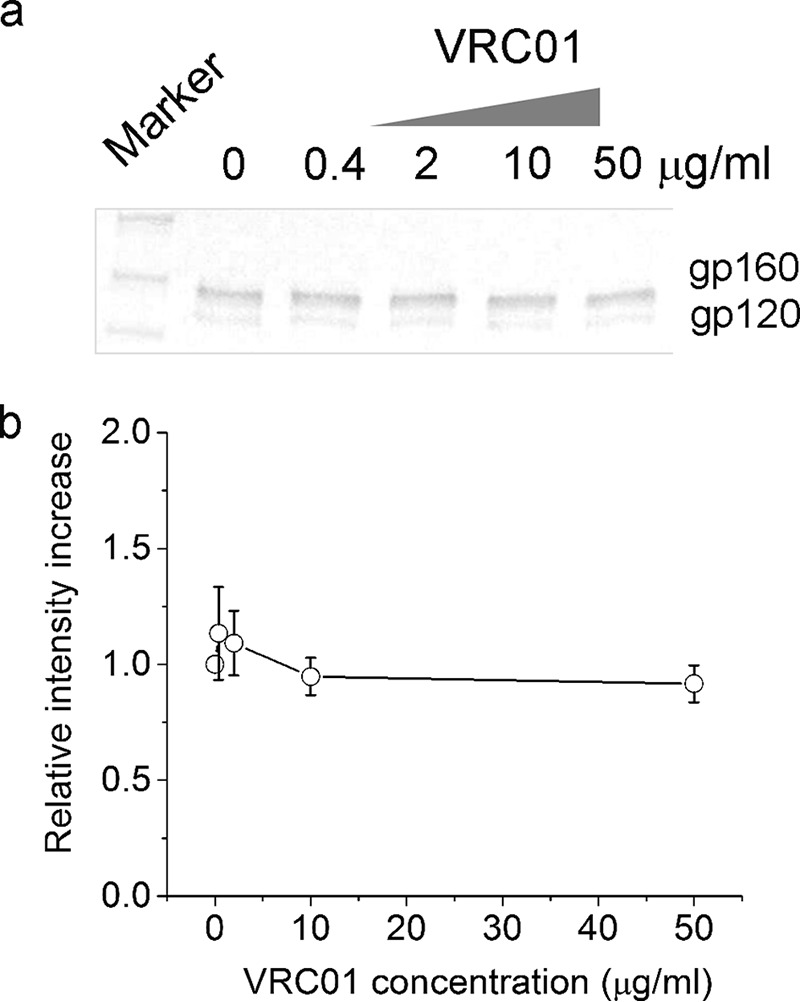
VRC01 antibody induces negligible shedding of NL4-3 gp120. a, Western blotting analysis of gp120 and gp160 proteins in the culture supernatant upon incubation with VRC01 IgG at various concentrations. b, quantitation of the gp120 band intensity from panel a as a function of VRC01 IgG concentration. The positive control for this assay, demonstrating gp120 shedding in response to addition of various concentrations of CD4 under identical cell and incubation conditions, was published previously (10).
Furthermore, the labeled VRC01 Fab is able to neutralize NL4-3 virus. As shown in Fig. 1d, the efficiency of neutralization was not significantly compromised by the Alexa 594 label. The IC50 value for the labeled Fab derived from fitting to a sigmoid function (red curve) was 4.5 ± 1.1 μg/ml, which should be compared with 1.9 ± 0.5 μg/ml for the unlabeled Fab (black curve). 95% infectivity was neutralized by the labeled VRC01 Fab at 20 μg/ml. The neutralizing capacity of the labeled Fab is expected, as the site of labeling is distant from the antigen binding site (Fig. 1b).
Recently, the dissociation constant, Kd, of VRC01 Fab toward the BG505 stabilized, soluble trimers has been determined by surface plasmon resonance (21), which is less than 1 nm at 25 °C in the presence of 150 mm NaCl. This affinity is likely to be higher for NL4-3 functional trimers, which would suggest a >99.9% occupancy (>400-fold excess of the Kd) on unconstrained CD4bs at a Fab concentration of 20 μg/ml. However, the stoichiometry of Fab binding to the CD4bs on trimers on the virion surface is a more complex issue, as this stoichiometry could be influenced by spontaneous shedding of gp120 before the assay of detection, accessibility, and other factors. Cryo-electron microscopy revealed that three VRC01 Fabs can bind to a single Env trimer (22). Recently, the stoichiometry of CD4bs-specific Fabs has been studied with stabilized, soluble BG505 Env trimers as immobilized antigens by surface plasmon resonance (15, 21). The apparent maximum binding observed from these experiments for these Fabs ranged from 1.3 to 3.0 paratopes per trimer; for the VRC01 Fab, as estimated by surface plasmon resonance, it was 1.6 (15). This number is likely to be higher for native Env trimers on the virion surface that do not have the stabilizing mutations. On the other hand, NL4-3 Env may also differ from the BG505 Env used in those studies. The similar potency and efficacy (>95%) of the Fab with and without Alexa 594, as shown in Fig. 1d, indicate largely unrestrained binding of both forms to functional trimers on NL4-3 virions. Because the virus neutralization assay (Fig. 1d) was done with the very same virus preparations (NL4-3) under the same incubation conditions as the measurement of gp120 numbers by OTV (“Experimental Procedures”), Alexa 594-Fab can probe at least a minimal set of gp120 molecules that are required for infection, out of the total gp120 molecules present on individual NL4–3 virions. As a result, the number of Alexa 594 fluorophores that we can quantitate using single-molecule fluorescence will thus estimate the minimum number of gp120 molecules on individual virions that are required for infection. A potential caveat apart from the stoichiometry is that the binding of VRC01 to gp120 may not discriminate between functional trimeric and nonfunctional forms of Env (15) present on the pseudovirions (23). Thus, if there are gp120s in nonfunctional forms of Env present on these virions and these gp120s are also bound by the labeled Fab under the same conditions, our result may overestimate the number of gp120 molecules that participate in viral infection. In conclusion, all uncertainties about the quantitation of gp120 present in functional Env trimers cannot be eliminated, and whether the potential errors might cancel each other remains to be investigated.
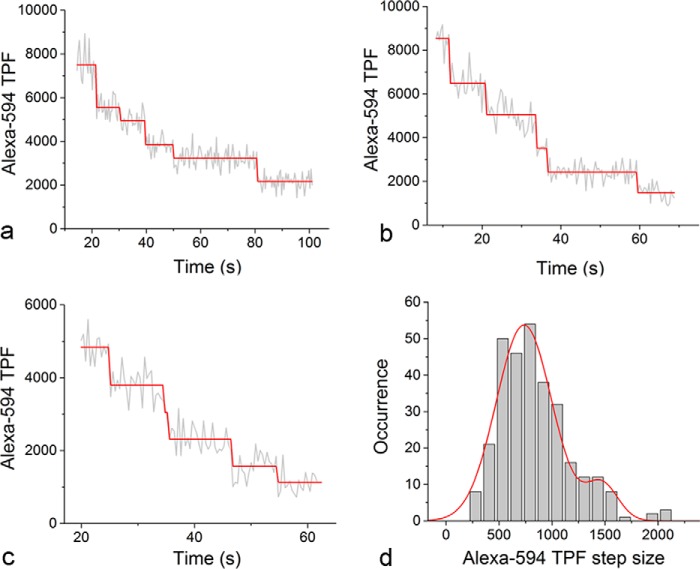
TPF Intensity of a Single Alexa 594 Fluorophore on HIV-1 Virions
To quantitate the number of Alexa 594 fluorophores on individual virions, we took the ratio comparison approach using a single molecule as a fluorescence standard, which has been well established in quantitative fluorescence microscopy (24, 25). To determine the TPF intensity of a single Alexa 594 fluorophore, we have taken advantage of the spontaneous photobleaching of Alexa 594. When individual virions bound by Alexa 594-Fab are optically trapped, the TPF of Alexa 594 will bleach to extinction (10), and the time course of this process allows us to visualize the stochastic photobleaching of individual molecules over time. As shown in Fig. 3, a–c, these real-time trajectories from individual virions show clear steps of TPF photobleaching. We have used a step-detection algorithm that we developed previously (19) to identify steps from these real-time fluorescence traces (red lines in Fig. 3, a–c). The histogram of the individual photobleaching step sizes for Alexa 594 molecules (n = 303) could be well described by the sum of two Gaussians (red curve in Fig. 3d), one centered at 736 ± 263 analog-to-digital units (mean ± S.D.) and the other centered at 1472 ± 151 analog-to-digital units. Under a statistical significance level of 5%, Pearson's χ-square test (26) selected the double-Gaussian distribution (p value = 0.112) and rejected the single-Gaussian distribution (p value = 2.33 × 10−15) as the model to describe the data. The difference in peak values is exactly 2-fold. As we have observed previously, the secondary peak may result from the photobleaching of two Alexa 594 molecules that occurred almost simultaneously, which could not be resolved by either the finite camera exposure time or the step-finding algorithm (10, 27). The TPF intensity of a single Alexa 594 fluorophore can then be used to calculate the total number of Alexa 594 molecules bound to the virion based on a ratio comparison (24, 25) with the initial TPF intensity of Alexa 594 associated with individual virions.
FIGURE 3.
TPF intensity of single Alexa-594 molecules on HIV-1 virions. a–c, representative TPF time trajectories from Alexa 594 molecules associated with individual HIV-1 virions (gray traces). The step-finding algorithm that we developed previously (19) was used to identify steps from these real time trajectories, which are shown as red lines. d, the histogram of the individual photobleaching step sizes for Alexa 594 molecules identified from the step-finding algorithm (n = 303), which could be described by the sum of two Gaussians (red curve).
Determination of the Number of Gp120 Molecules on Individual Virions
To quantitate the dependence of HIV-1 virion infectivity as a function of gp120 density on individual HIV-1 virions, we produced a set of HIV-1 virions that carry different numbers of Env molecules by varying the envelope plasmid input (pEnv, 0–4 μg) during virion production from 293T cells, as we described recently (10). To quantitate gp120 density on individual virions, we incubated virions with 20 μg/ml of Alexa 594-labeled VRC01 Fab, prepared as described above, and then measured the initial Alexa 594 TPF intensity for individual virions as we trapped them individually. Based on the TPF intensity of a single Alexa 594 fluorophore that we have measured directly from these trapped virions (Fig. 3), we then calculated the number of gp120 molecules from the initial Alexa 594 TPF intensity. We have measured NL4-3 virions prepared with 0, 0.01, 0.1, 0.2, 1, 2, and 4 μg of pEnv. Fig. 4 shows a compendium of histograms for the number of gp120 molecules per virion. Throughout, most of the distributions were very broad, consistent with what we have measured previously using b12 IgG labeled with Alexa 594 through NHS-ester chemistry (10). The mean, standard deviation, and median numbers of gp120 molecules per virion are listed in Table 1 for each of these virus populations. Because we can clearly distinguish between virions with a single molecule of Alexa 594 fluorophore versus those without (10), we have also included the fraction of virions without any gp120 in Table 1 for each population of the virus.
FIGURE 4.
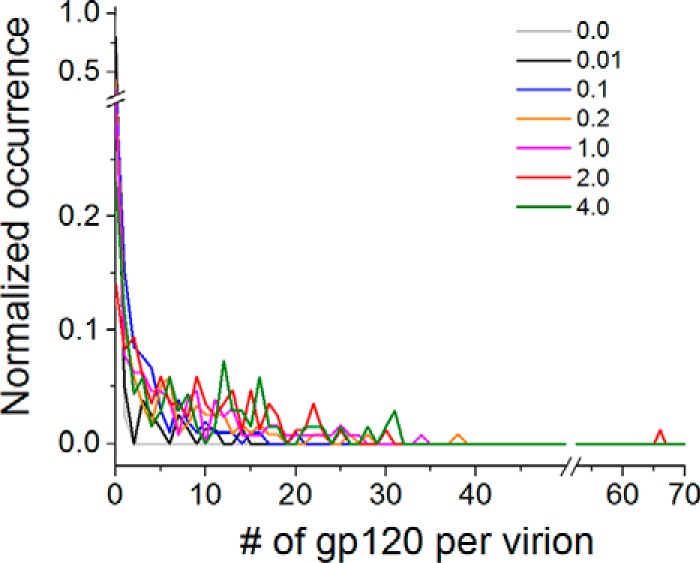
Histograms for the number (#) of gp120 molecules per virion determined from OTV. The histograms, with gray, black, blue, orange, purple, red, and green curves are for virions from 0 (n = 80), 0.01 (n = 79), 0.1 (n = 105), 0.2 (n = 119), 1.0 (n = 129), 2.0 (n = 85) and 4.0 μg of pEnv (n = 69), respectively.
TABLE 1.
Statistics for the number of gp120 molecules per virion probed by OTV
The concentration of plasmid DNA was determined in 10 mm sodium phosphate buffer with 100 mm NaCl, pH 7.0, at 20 °C and the use of an extinction coefficient of 20 mg·ml−1 cm−1 at 260 nm. Varied amounts of plasmids were obtained by serial dilution of the stock solutions, yielding typical coefficients of variance for the final concentrations of less than 5%. For each population of virions, virus was freshly thawed from the −80 °C stock and incubated with 20 μg/ml of VRC01 Alexa 594-Fab for 1 h at 20 °C. The mixture was then diluted in PBS buffer and injected into a microfluidic chamber for optical trapping and TPF measurement of Alexa 594 molecules. The number of gp120 molecules per virion was determined by normalizing the initial TPF of each individual virions against the mean TPF intensity of a single Alexa 594 fluorophore, as we measured and reported in Fig. 3. Because both gp120 density and virus infectivity were measured for each population of viruses, the errors in preparing and quantifying the pEnv did not enter into the quantitative analysis and would have zero influence on our correlation results. The standard deviations for the percentages of particles without gp120 were estimated based on a set of bootstrap analysis of the single-particle data collected for each population of viruses as follows: for a total of N particles from a virus population at a given pEnv amount, we randomly sampled this pool for N times with allowance for resampling. This procedure was repeated 1,000 times to yield the standard deviations for the fractions of particles without gp120.
| pEnv input (μg) and No. of particles measured | Mean | Standard deviation | Median | Percentage of particles without gp120 |
|---|---|---|---|---|
| % | ||||
| 0 (n = 80) | 0.01 | 0.2 | 0 | 98 ± 2 |
| 0.01 (n = 79) | 1.0 | 2.7 | 0 | 80 ± 5 |
| 0.1 (n = 105) | 2.5 | 3.9 | 1 | 44 ± 5 |
| 0.2 (n = 119) | 4.5 | 6.8 | 1 | 44 ± 5 |
| 1 (n = 129) | 5.9 | 7.5 | 3 | 34 ± 4 |
| 2 (n = 85) | 8.7 | 9.6 | 6 | 14 ± 4 |
| 4 (n = 69) | 8.1 | 8.7 | 6 | 23 ± 5 |
As we see from Table 1, at pEnv = 0, when the HIV-1 particles have no gp120 on their surface, 2 of 80 particles showed an apparent Alexa 594 TPF that corresponds to one Alexa 594 molecule associated per virion. This residual background (2.5%) indicates that the Alexa 594-Fab is indeed highly specific toward gp120 under current conditions. This low percentage of nonspecific binding is essential for our approach to quantitate the number of gp120 molecules on individual virions.
As we increased pEnv, the mean number of gp120 molecules per virion increased, and reached a maximum at pEnv = 2.0 μg. Meanwhile, the fraction of virion particles without gp120 also decreased. To analyze HIV-1 virion infectivity as a function of gp120 density on individual HIV-1 virions, we measured the infectivity for these seven populations of virions using the standard TZM-bl assay both in the presence and absence of DEAE-dextran, and then examined the correlation by plotting the virion infectivity as a function of average gp120 number per virion that we determined from OTV.
As shown in Fig. 5a, the infectivity of virions in the presence of 20 μg/ml of DEAE-dextran increased with the average number of gp120 molecules per virion in a sigmoidal fashion, which displayed an apparent plateau when there were on average 8–10 gp120 molecules per virion. Notably, this dependence is very well described by the Hill equation (28). The resulting fitted curve is overlaid on the experimental data, which yields a Hill coefficient of 2.4 ± 0.6, a maximum infectivity (%) of 0.29 ± 0.02, and an average number of gp120 molecules of 2.4 ± 0.3 per virion at 50% of the maximum infectivity. All the errors are standard errors from the nonlinear least squares fitting with a 95% confidence interval. In contrast, the infectivity of virions in the absence of DEAE-dextran monotonically increases within the experimental conditions tested (Fig. 5b). Although the lack of an apparent plateau allows for alternative explanations of these data, the slightly concave trend suggests that cooperativity may occur under these conditions as well. To examine this quantitative dependence in the absence of DEAE-dextran, we also fitted these experimental data to the Hill equation. The resulting fitted curve is overlaid on the experimental data, which yields a Hill coefficient of 2.0 ± 7.6, a maximum infectivity (%) of 0.3 ± 18.3, and an average number of gp120 molecules of 26 ± 950 per virion to reach 50% of the maximum infectivity. These large errors for the fitting parameters are due to the lack of maximum infectivity in the data, and speak to the fact that the dependence of virion infectivity in the absence of DEAE-dextran is very different from that in its presence. However, we note that both the values for the Hill coefficient and the maximum infectivity are very close to the corresponding values in the presence of DEAE-dextran: 2.0 compared with 2.4 and 0.3% compared with 0.29%. The major difference is in the average number of gp120 molecules required to reach 50% of maximum infectivity: 26 compared with 2.4 in the presence of DEAE-dextran. Within the same framework of the Hill equation, this comparison suggests that the cooperativity among gp120 molecules is likely to occur regardless of the inclusion of DEAE-dextran. Rather, the effect of DEAE-dextran is to produce a plateau at a significantly lower number of gp120 molecules.
FIGURE 5.
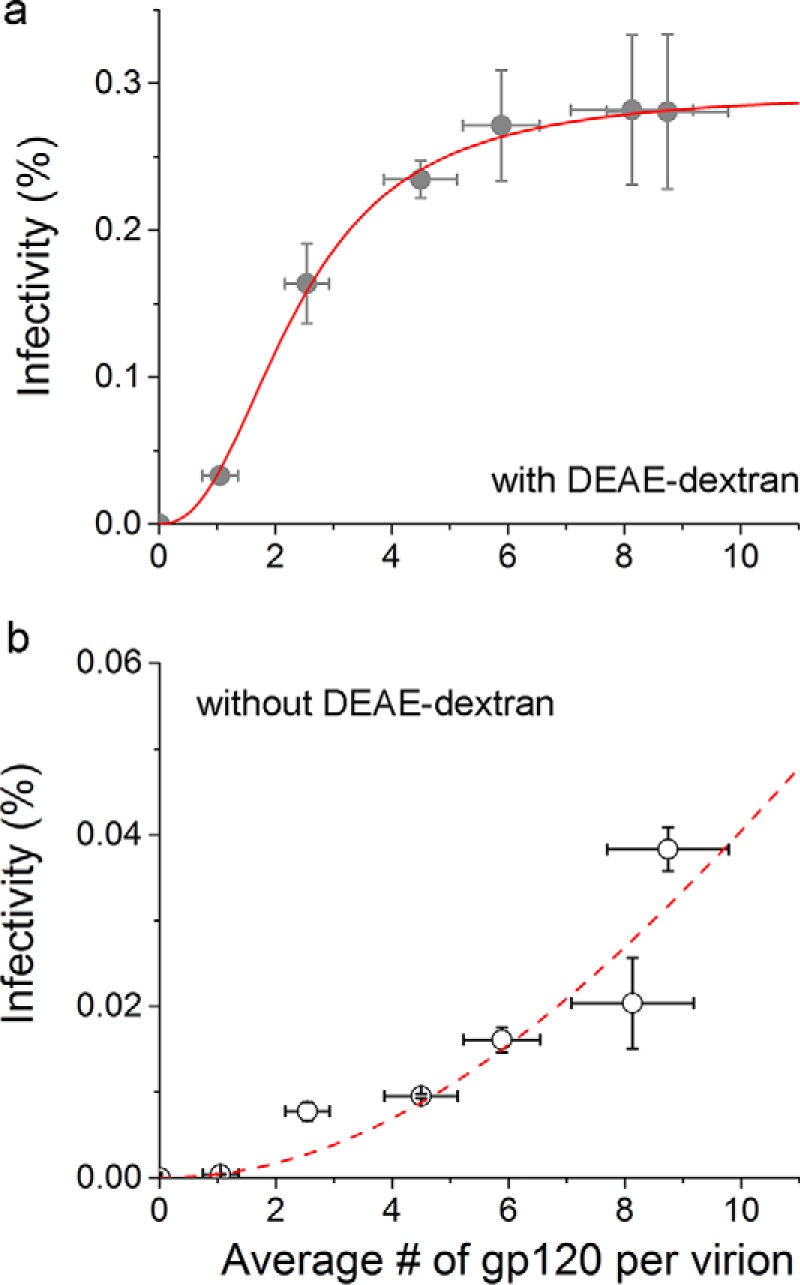
Relationship between HIV-1 virion infectivity and the average number of gp120 molecules per virion in the presence (a) or absence (b) of DEAE-dextran. The symbols are experimental data, where the error bars for infectivity are standard deviations derived from three independent repeats of the same experiment, and the error bars for the average number of gp120 molecules per virion are mean ± S.E. derived from single-particle data. The adjusted R-square values for the fittings are 0.9975 and 0.8293, respectively. The dashed curve in panel b shows a sigmoid function with large uncertainties in the fitted parameters.
It is worth noting that the infectivity in the presence of DEAE-dextran was above 0 at an average of one gp120 molecule per virion. This does not suggest that a single molecule of gp120 is sufficient to support HIV-1 infection; rather, this average of one gp120 per virion results from averaging over all virion particles that are present in this population. Under this condition, where the pEnv input is 0.01 μg, which is limiting, the cells have produced a significant portion of HIV-1 virions without any gp120 (see the black distribution in histograms in Fig. 4, and also Table 1 for fraction of particles that have no gp120). As a result, it lowers the mean number of gp120 molecules per virion. The infectivity that we measured results from a subset of those virions that carry gp120 on their surface. Because we have measured the gp120 number for each single particle, this allows us to calculate the average gp120 number for those virions that have at least one gp120 on them (Env+ virions). For this subset, the mean gp120 per virion when infectivity is detectable is actually 5.2; the data do not exclude that the required number is closer to ∼9, where the inflection for the plateaus is, but they indicate it is not much higher than that. The dependence of virion infectivity on the number of gp120 molecules per Env+ virion, i.e. virions with at least one gp120, is shown in Fig. 6, a and b, for data in the presence and absence of DEAE-dextran, respectively.
FIGURE 6.
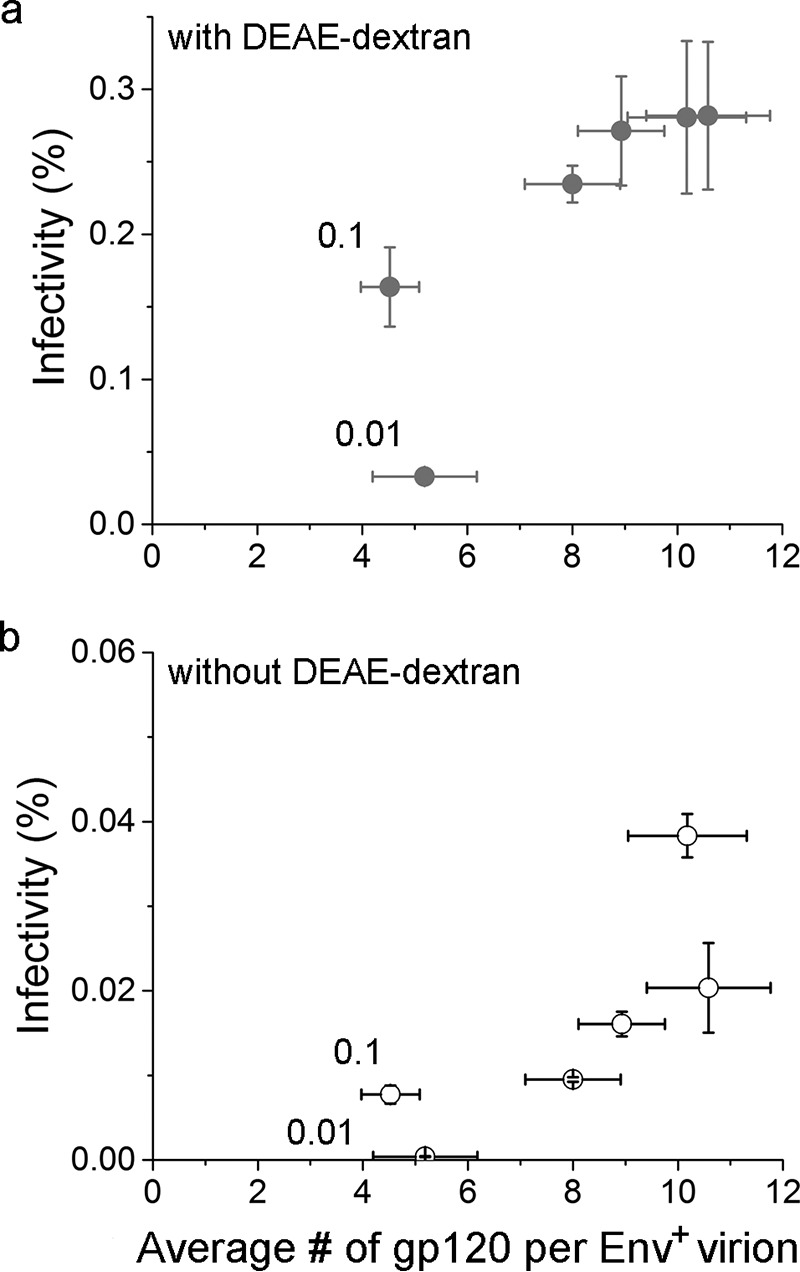
Relationship between HIV-1 virion infectivity and the average number of gp120 molecules per Env+ virion in the presence (a) or absence (b) of DEAE-dextran. The error bars for infectivity are standard deviations derived from three independent repeats of the same experiment, and the error bars for the average number of gp120 molecules per Env+ virion are mean ± S.E. derived from single-particle data. The data points for virions prepared with pEnv = 0.01 and 0.1 μg are labeled.
It is worth noting that, although the virions produced with 0.01 and 0.1 μg of pEnv have on average an identical number of gp120 molecules per Env+ virion within error, the apparent infectivity for the 0.1 virus group is higher because the fraction of virions that carry gp120 in this group is almost 3-fold higher than that for the 0.01 virus group (Table 1). This interpretation applies to the data both in the presence and absence of DEAE-dextran. Furthermore, it is worth noting that within the framework of classical cooperativity (29), all species of virions need to be included when applying the Hill equation, also those that do not carry gp120. The vertical dependence observed in Fig. 6 for Env+ virions is consistent with cooperativity and further suggests that an absolute minimum number of gp120 molecules per virion is required for infection.
Discussion
In this paper, we have prepared a site-specifically labeled VRC01 Fab fragment and purified this labeled protein to homogeneity. Because this Fab is labeled with a single fluorophore through maleimide chemistry and we have achieved 97% labeling efficiency, it allows us to quantitate the number of gp120 molecules on individual virions with single-molecule resolution using the OTV technique that we recently developed without the compounding effects of 1) NHS-ester chemistry for labeling of fluorophores, which produces multiple fluorophores per antibody; and 2) the potential cross-linking between the CD4bs' of two trimers by a single IgG.
To examine the relationship between virion infectivity and the density of gp120 molecules on individual virions, we have used a provirus clone that is env− together with a separate wt Env expression plasmid (pEnv) to generate HIV-1 virions (9). This design allows us to titrate pEnv from low to high quantities (8) to generate a set of virions that are expected to carry varied densities of Env on their surface, determined by the pEnv inputs. Because the resulting virions are only infectious for a single cycle, the infectivity of the virions can thus be compared with the gp120 density on these virions directly measured by OTV. This approach allows one to quantitatively determine the relationship between the two, without the complications from multiple rounds of infection.
As shown in Fig. 5 both in the presence and absence of DEAE-dextran, the viral infectivity increased with the gp120 density per virion. In the presence of DEAE-dextran, the infectivity increased with gp120 density in a sigmoidal fashion that can be well described by a Hill equation. This result suggests that HIV-1 gp120 displays a positive cooperativity and the 97% infectivity is only achieved when on average nine gp120 molecules are present on a virion (pEnv = 2.0 μg). In contrast, the data in the absence of DEAE-dextran showed no apparent plateau. Although the data also showed a trend of sigmoidal dependence, evidenced by the fitting to a Hill equation, this fitting predicts that the infectivity will not reach 90% of the projected plateau until there are on average 78 gp120 molecules per virion.
Could the appearance of cooperativity under current experimental conditions result from virion aggregation? We repudiate this possibility for the following reasons. First, as we showed previously, the individual TZM-bl cell infectious events in the cell culture show very good linear correlation with the concentrations of virions (9), which argues against virion aggregation. Second, under current infection assay conditions, i.e. with a virion particle concentration well below 4 × 107/ml, the concentration-dependent aggregation of virions is negligible as we showed recently (10). Third, the concentration of DEAE-dextran that we used is in huge molar excess over that of HIV-1 virions, a condition that does not favor aggregation of virions. Indeed, we have conducted single-virion tracking experiments using fluorescently labeled NL4-3 pseudovirions in the presence of DEAE-dextran. The intensity of individual virions that we measured provides evidence against virion aggregation under current infection conditions.
The above data have important implications for HIV-1 infectivity. First, they suggest that at the single-particle level, HIV-1 infectivity increases with the number of Env trimers on its surface once they have surpassed a lower threshold, and reaches a full infectivity when the number of gp120 molecules exceeds a certain upper threshold (30–33). Virion-cell binding may well depend on the gp120 density per virion and thus contribute to the observed cooperativity phenomena. Second, DEAE-dextran has a marked impact on this threshold number of gp120 molecules required for the full infectivity. A study by Platt et al. (34) revealed that DEAE-dextran enhances virion infectivity by stabilizing adsorption of virions on the host-cell surface. Our current results favor this interpretation. Our data indicate that DEAE-dextran is playing a role of compensating for a paucity of Env to facilitate entry and infection by HIV-1 virions. The presence of DEAE-dextran could therefore potentially mask the effects of differences in gp120 density among virions. Conversely, in the absence of DEAE-dextran, significantly more Env molecules are needed to reach a theoretical full infectivity. In conclusion, infectivity of a virion may not be an all-or-nothing quantity but a spectrum of propensities with a lower and an upper threshold, as has been hypothesized (5, 30–33, 35).
The low density of HIV Env on virions may have a profound impact on HIV transmission and host immune responses, the quantitative aspects of which are just beginning to be understood (6, 20). Transmission of HIV-1 from an infected person to a new host usually occurs through a mucosal port of entry. In that environment, the conditions may lack any enhancing factors such as the DEAE-dextran, regularly included in in vitro experiments based on the TZM-bl cell line, for example, with the aim of determining the potency of neutralizing antibodies. As shown here, DEAE-dextran lowers the critical amounts of Env on virions required for optimal infectivity. Indeed, under in vivo conditions, in the absence of such enhancing factors, the quantitative requirements on Env may be greater. For example, transmitted or founder strains of HIV-1 have been shown to have higher Env densities on virions than viruses isolated during chronic infection (7). These variables are important to vaccine research, because the ratio of how many functional trimers a virion has to how many it requires for infection is a critical parameter in quantitative modeling of virion neutralization and probably influences the sensitivity of virions to neutralization (35).
The current study raises questions of what the mechanistic bases for a lower and higher Env-trimer threshold would be. The findings relate these questions to the mechanism of enhancement of infection by DEAE-dextran. Hypothetically, a lower threshold would be set by the small number of trimers that can minimally support the membrane fusion between the viral and the cell membrane, which allows the packaged viral genome to be released into the cytosol. In their physical form, this small number of trimers may cluster together on the virion surface (17) and may eventually form a so-called entry claw (36) that mediates membrane fusion and fusion pore dilation (37). Additional trimers may increase the propensity of virions to attach to the cell surface incrementally up to a limit beyond which more trimers cease to enhance the attachment process: the upper threshold, which could be influenced by both viral and host-cell factors. As observed from current studies, the inclusion of DEAE-dextran clearly decreases this upper threshold for NL4-3 virions in TZM-bl cells; the lower threshold, which may be less variable, can thus be closer to or further from the upper one depending on the infection conditions. As suggested by Platt et al. (34), the enhancing effect of DEAE-dextran could operate at or after the attachment stage, possibly preventing virion dissociation, thereby increasing the still small fraction of virions that get a chance to infect. Moreover, in the cell line used here, productive infection may involve endocytosis of the receptor-virion complex and trafficking to an endosomal compartment where fusion and cytoplasmic entry of the viral core occurs (38, 39). Whether DEAE-dextran merely enhances the chances of endocytosis rather than dissociation, or also promotes Env-receptor interactions inside the endosome, and how such effects modulate the apparent lower and higher thresholds of Env requirements, remain to be elucidated. In light of these potential mechanisms of DEAE-dextran, it is worth considering charged molecules present on the host-cell surface that may modulate HIV-1 attachment. It has been suggested that HIV-1 attachment to negatively charged heparan sulfate proteoglycans on the cell surface operates most strongly for tissue culture-adapted strains that have accumulated several positively charged amino acid residues in the V3 loops of their gp120 molecules. Specifically, it was shown in CHO cells that the enhancing effect of hexadimethrine bromide (Polybrene) on infection is negatively correlated with the positive charge in V3 loop (40), the greater the number of positive charges, the weaker enhancement by Polybrene. In this context our findings may be somewhat surprising, in that the TZM-bl cells, which are derived from HeLa cells, have preponderant heparan sulfate proteoglycans (41) and the V3 loop of NL4-3 Env is rich in basic residues. This comparison suggests that both the cell type and the precise composition of the polycations modulate these effects quantitatively, which might explain the strong enhancement of NL4-3 infection by DEAE-dextran in the current experiments.
In conclusion, the current findings strongly favor a propensity view of Env function and viral entry: the quantitative aspects of Env mediation are likely to vary with HIV-1 strain, cell type, and experimental conditions such as the presence of polycations or spinoculation (42). The approach that we have developed here to examine the impact of Env density on HIV virion infectivity can be readily adopted for the study of other HIV strains and target cell types.
Author Contributions
W. C. directed the project; M. C. D. and J. H. K. conducted OTV experiments; J. H. K. cloned, purified, and labeled VRC01 Fab; H. S. prepared HIV-1 virions; M. C. D., J. H. K., and W. C. performed the analysis; W. C. and P. J. K. wrote the paper.
Acknowledgments
We thank Dr. John R. Mascola and Dr. Stephen D. Schmidt for kindly providing the monoclonal antibody production and purification protocol. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH): pNL4-3 from Dr. Malcolm Martin; pNL4-3.Luc.R-E- from Dr. Nathaniel Landau; pEGFP-Vpr from Warner C. Greene; and TZM-bl cells from Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc.
This work was supported, in whole or in part, by National Institutes of Health Director's New Innovator Award 1DP2OD008693-01 (to W. C.), National Science Foundation CAREER Award CHE1149670 (to W. C.), and Research Grant 5-FY10-490 from the March of Dimes Foundation (to W. C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no competing financial interests.
- Env
- envelope glycoprotein
- gp
- glycoprotein
- OTV
- optical trapping virometry
- TPF
- two-photon fluorescence
- EGFP
- enhanced green fluorescent protein
- Ni-NTA
- nickel-nitrilotriacetic acid.
References
- 1. White J. M., Delos S. E., Brecher M., and Schornberg K. (2008) Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43, 189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kielian M., and Rey F. A. (2006) Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chertova E., Bess J. W. Jr., Crise B. J., Sowder I. R. II, Schaden T. M., Hilburn J. M., Hoxie J. A., Benveniste R. E., Lifson J. D., Henderson L. E., and Arthur L. O. (2002) Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76, 5315–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu P., Liu J., Bess J. Jr., Chertova E., Lifson J. D., Grisé H., Ofek G. A., Taylor K. A., and Roux K. H. (2006) Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 [DOI] [PubMed] [Google Scholar]
- 5. Brandenberg O. F., Magnus C., Rusert P., Regoes R. R., and Trkola A. (2015) Different infectivity of HIV-1 strains is linked to number of envelope trimers required for entry. PLoS Pathog. 11, e1004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klein J. S., and Bjorkman P. J. (2010) Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 6, e1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parrish N. F., Gao F., Li H., Giorgi E. E., Barbian H. J., Parrish E. H., Zajic L., Iyer S. S., Decker J. M., Kumar A., Hora B., Berg A., Cai F., Hopper J., Denny T. N., et al. (2013) Phenotypic properties of transmitted founder HIV-1. Proc. Natl. Acad. Sci. U.S.A. 110, 6626–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yuste E., Johnson W., Pavlakis G. N., and Desrosiers R. C. (2005) Virion envelope content, infectivity, and neutralization sensitivity of simian immunodeficiency virus. J. Virol. 79, 12455–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim J. H., Song H., Austin J. L., and Cheng W. (2013) Optimized infectivity of the cell-free single-cycle human immunodeficiency viruses type 1 (HIV-1) and its restriction by host cells. PLOS One 8, e67170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pang Y., Song H., Kim J. H., Hou X., and Cheng W. (2014) Optical trapping of individual human immunodeficiency viruses in culture fluid reveals heterogeneity with single-molecule resolution. Nat. Nanotechnol. 9, 624–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barouch D. H., Yang Z. Y., Kong W. P., Korioth-Schmitz B., Sumida S. M., Truitt D. M., Kishko M. G., Arthur J. C., Miura A., Mascola J. R., Letvin N. L., and Nabel G. J. (2005) A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J. Virol. 79, 8828–8834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu X., Yang Z. Y., Li Y., Hogerkorp C. M., Schief W. R., Seaman M. S., Zhou T., Schmidt S. D., Wu L., Xu L., Longo N. S., McKee K., O'Dell S., Louder M. K., Wycuff D. L., et al. (2010) Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gill S. C., and von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino-acid sequence data. Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 14. Sanders R. W., Derking R., Cupo A., Julien J. P., Yasmeen A., de Val N., Kim H. J., Blattner C., de la Peña A. T., Korzun J., Golabek M., de Los Reyes K., Ketas T. J., van Gils M. J., et al. (2013) A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yasmeen A., Ringe R., Derking R., Cupo A., Julien J. P., Burton D. R., Ward A. B., Wilson I. A., Sanders R. W., Moore J. P., and Klasse P. J. (2014) Differential binding of neutralizing and non-neutralizing antibodies to native-like soluble HIV-1 Env trimers, uncleaved Env proteins, and monomeric subunits. Retrovirology 11, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y., O'Dell S., Walker L. M., Wu X., Guenaga J., Feng Y., Schmidt S. D., McKee K., Louder M. K., Ledgerwood J. E., Graham B. S., Haynes B. F., Burton D. R., Wyatt R. T., and Mascola J. R. (2011) Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J. Virol. 85, 8954–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chojnacki J., Staudt T., Glass B., Bingen P., Engelhardt J., Anders M., Schneider J., Müller B., Hell S. W., and Kräusslich H. G. (2012) Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338, 524–528 [DOI] [PubMed] [Google Scholar]
- 18. Cheng W., Hou X., and Ye F. (2010) Use of tapered amplifier diode laser for biological-friendly high-resolution optical trapping. Opt. Lett. 35, 2988–2990 [DOI] [PubMed] [Google Scholar]
- 19. Arunajadai S. G., and Cheng W. (2013) Step detection in single-molecule real time trajectories embedded in correlated noise. PLoS ONE 8, e59279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Galimidi R. P., Klein J. S., Politzer M. S., Bai S., Seaman M. S., Nussenzweig M. C., West A. P. Jr., and Bjorkman P. J. (2015) Intra-spike crosslinking overcomes antibody evasion by HIV-1. Cell 160, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Behrens A. J., Vasiljevic S., Pritchard L. K., Harvey D. J., Andev R. S., Krumm S. A., Struwe W. B., Cupo A., Kumar A., Zitzmann N., Seabright G. E., Kramer H. B., Spencer D. I., Royle L., Lee J. H., et al. (2016) Composition and antigenic effects of individual glycan sites of a trimeric HIV-1 envelope glycoprotein. Cell Rep. 14, 2695–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tran E. E., Borgnia M. J., Kuybeda O., Schauder D. M., Bartesaghi A., Frank G. A., Sapiro G., Milne J. L., and Subramaniam S. (2012) Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8, e1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moore P. L., Crooks E. T., Porter L., Zhu P., Cayanan C. S., Grise H., Corcoran P., Zwick M. B., Franti M., Morris L., Roux K. H., Burton D. R., and Binley J. M. (2006) Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80, 2515–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coffman V. C., and Wu J. Q. (2012) Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem. Sci. 37, 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawrimore J., Bloom K. S., and Salmon E. D. (2011) Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 195, 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Evans R. D. (1969) The Atomic Nucleus, Chapter 27, pp. 774–784, McGraw-Hill, Bombay - New Delhi, India [Google Scholar]
- 27. Hou X., and Cheng W. (2011) Single-molecule detection using continuous wave excitation of two-photon fluorescence. Opt. Lett. 36, 3185–3187 [DOI] [PubMed] [Google Scholar]
- 28. Hill A. V. (1913) The combinations of haemoglobin with oxygen and with carbon monoxide. I. Biochem. J. 7, 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wyman J., and Gill S. J. (1990) Binding and Linkage: Functional Chemistry of Biological Macromolecules, University Science Books, Mill Valley, CA [Google Scholar]
- 30. Klasse P. J. (2012) The molecular basis of HIV entry. Cell. Microbiol. 14, 1183–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brandenberg O. F., Magnus C., Regoes R. R., and Trkola A. (2015) The HIV-1 entry process: a stoichiometric view. Trends Microbiol. 23, 763–774 [DOI] [PubMed] [Google Scholar]
- 32. Klasse P. J. (2007) Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology 369, 245–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Magnus C., Rusert P., Bonhoeffer S., Trkola A., and Regoes R. R. (2009) Estimating the stoichiometry of human immunodeficiency virus entry. J. Virol. 83, 1523–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Platt E. J., Kozak S. L., Durnin J. P., Hope T. J., and Kabat D. (2010) Rapid dissociation of HIV-1 from cultured cells severely limits infectivity assays, causes the inactivation ascribed to entry inhibitors, and masks the inherently high level of infectivity of virions. J. Virol. 84, 3106–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klasse P. J., and Moore J. P. (1996) Quantitative model of antibody- and soluble CD4-mediated neutralization of primary isolates and T-cell line-adapted strains of human immunodeficiency virus type 1. J. Virol. 70, 3668–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sougrat R., Bartesaghi A., Lifson J. D., Bennett A. E., Bess J. W., Zabransky D. J., and Subramaniam S. (2007) Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 3, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blumenthal R., Durell S., and Viard M. (2012) HIV entry and envelope glycoprotein-mediated fusion. J. Biol. Chem. 287, 40841–40849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Daecke J., Fackler O. T., Dittmar M. T., and Kräusslich H. G. (2005) Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J. Virol. 79, 1581–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miyauchi K., Kim Y., Latinovic O., Morozov V., and Melikyan G. B. (2009) HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137, 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y. J., Hatziioannou T., Zang T., Braaten D., Luban J., Goff S. P., and Bieniasz P. D. (2002) Envelope-dependent, cyclophilin-independent effects of glycosaminoglycans on human immunodeficiency virus type 1 attachment and infection. J. Virol. 76, 6332–6343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mondor I., Ugolini S., and Sattentau Q. J. (1998) Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J. Virol. 72, 3623–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Doherty U., Swiggard W. J., and Malim M. H. (2000) Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74, 10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]