Abstract
Eukaryotic topoisomerase 2 (Top2) and one of its interacting partners, topoisomerase IIβ binding protein 1 (TopBP1) are two proteins performing essential cellular functions. We mapped the interacting domains of these two proteins using co-immunoprecipitation and pulldown experiments with truncated or mutant Drosophila Top2 with various Ser-to-Ala substitutions. We discovered that the last 20 amino acids of Top2 represent the key region for binding with Mus101 (the Drosophila homolog of TopBP1) and that phosphorylation of Ser-1428 and Ser-1443 is important for Top2 to interact with the N terminus of Mus101, which contains the BRCT1/2 domains. The interaction between Mus101 and the Top2 C-terminal regulatory domain is phosphorylation-dependent because treatment with phosphatase abolishes their association in pulldown assays. The binding affinity of N-terminal Mus101 with a synthetic phosphorylated peptide spanning the last 25 amino acids of Top2 (with Ser(P)-1428 and Ser(P)-1443) was determined by surface plasmon resonance with a Kd of 0.57 μm. In an in vitro decatenation assay, Mus101 can specifically reduce the decatenation activity of Top2, and dephosphorylation of Top2 attenuates this response. Next, we endeavored to establish a cellular system for testing the biological function of Top2-Mus101 interaction. Top2-silenced S2 cells rescued by Top2Δ20, Top2 with 20 amino acids truncated from the C terminus, developed abnormally high chromosome numbers, which implies that Top2-Mus101 interaction is important for maintaining the fidelity of chromosome segregation during mitosis.
Keywords: DNA replication, DNA topoisomerase, Drosophila, protein phosphorylation, RNA interference (RNAi), chromosome missegregation
Introduction
Topoisomerase 2 (Top2)3 is an essential enzyme capable of resolving DNA entanglements generated during replication, transcription, recombination, and chromatin remodeling. During the catalytic cycle, Top2 introduces a transient double strand break in one DNA segment for the passage of a second DNA segment, thereby altering DNA topology (1–3). Although the in vitro Top2 functions have been investigated extensively through biochemical and structural analyses (4–6), one of the emerging questions is how Top2 is regulated in vivo by posttranslational modifications and protein-protein interactions involving the C-terminal domain (CTD) of Top2 (7, 8).
Several proteins, including 14-3-3ϵ, mediator of DNA damage checkpoint 1 (Mdc1), and high-mobility group protein box 1 (HMGB1), have been found to interact with eukaryotic Top2 (9, 10). As demonstrated by in vitro assays, 14-3-3ϵ inhibits the DNA binding of human Top2α (9), whereas HMGB1 enhances the binding and cleavage activities of human Top2α (10). The binding of Mdc1 to human Top2α is phosphorylation-dependent. Via its BRCA1 C terminus (BRCT) domain, Mdc1 interacts with the phosphorylated Ser-1524 of human Top2α, activating the proposed decatenation checkpoint (11).
Topoisomerase IIβ binding protein 1 (TopBP1) is a Top2 binding partner that was also suspected to interact with human Top2β in a phosphorylation-dependent manner. This interaction was first discovered in a yeast two-hybrid system using the C terminus of human Top2β as a bait to screen a HeLa cDNA library (12). However, the interaction between Top2β and TopBP1 was found to be weak in vitro, and the biological role of TopBP1 binding to Top2β has not been elucidated (12). TopBP1 may have multiple roles in DNA replication and damage signaling. Cut, cell untimely torn (Cut5) is a TopBP1 homolog in fission yeast that is essential for DNA replication whose mutation results in abnormal nuclear division. Because mutants of cut5 were identified along with those of top2, Cut5 and Top2 may coordinate closely in the same cellular event (13). TopBP1 in human and chicken and Dpb11 in yeast (TopBP1 homolog in budding yeast) were found enriched on ultrafine anaphase bridges (UFBs). A UFB is a DNA thread connecting sister chromatids that results from unresolved DNA entanglement. The depletion of TopBP1 in chicken and human cell lines causes accumulation of UFBs, hinting that Top2α could function with TopBP1 on UFBs (14, 15). In a recent report, Top2α was found to interact with TopBP1 and recruited to UFBs by TopBP1, suggesting a role of Top2α-TopBP1 in solving DNA topological problems at UFBs for maintaining genome stability (15).
In this study, we examined the interaction between Drosophila Top2 and Mus101, the Drosophila TopBP1 homolog, to gain insights into the regulation of eukaryotic Top2. Using truncated constructs of Top2 and Mus101, we mapped the binding interface of these two proteins. The interaction is phosphorylation-dependent, and a Top2 CTD doubly phosphorylated at Ser-1428 and Ser-1443 is required for the binding. A similar recognition of twin phosphorylated residues by BRCT domains has also been observed in the binding partners of Sld3/Dpb11, Treslin/TopBP1, and Rad9/TopBP1 (16–18). In an in vitro assay, we found that the binding of Mus101 significantly inhibits Top2 decatenation activity, thereby demonstrating the functional effects of this binary protein interaction.
Additionally, we used a plasmid-based shRNA system to address the biological functions of the Top2-Mus101 interaction. Endogenous Top2 in Drosophila Schneider 2 (S2) cells can be almost completely depleted by two sets of shRNA. These cells displayed a G2/M arrest phenotype. Both Top2(S1428A,S1443A) and Top2Δ20, lacking the C-terminal 20 residues, can rescue the G2/M arrest in Top2-depleted S2 cells. However, Top2-depleted S2 cells rescued by Top2Δ20 show an abnormally high number of chromosomes, indicating a significant role of Top2-Mus101 interaction in maintaining the fidelity of chromosome segregation.
Experimental Procedures
Cloning and DNA Constructs
Constructs for Co-immunoprecipitation (Co-IP) Assays
Full-length Drosophila Mus101 was cloned into a modified pMT/V5-His vector (Invitrogen) that contained a FLAG tag as an N-terminal fusion peptide and the hygromycin resistance gene. Each fragment of Mus101, aa 1–350, aa 351–800, aa 801–1150, or aa 1151–1425, was cloned into another modified pMT/V5-His vector, which contained a nuclear localization signal, aa 582–607 from Drosophila RecQ4, as an N-terminal fusion peptide, a FLAG tag as a C-terminal fusion peptide, and the hygromycin resistance gene.
Full-length Drosophila Top2 was cloned into a pMT-puro vector (Addgene, originally from the Sabatini lab) with an N-terminal hemagglutinin (HA) tag. Three truncated Top2 constructs, headless (aa 397–1447), Δ240t (aa 1–1207), and core (397–1207), were individually built in the same way as the full-length Top2 construct, with the addition of a nuclear localization signal, aa 1306–1322 from Drosophila Top2, at their C termini. The CTD was determined previously to be the last 240 amino acids of Top2 (19).
C-terminal Truncated and Mutant Forms of Top2 Constructs for Pulldown Assays
All truncated and mutant Top2 constructs (Top2Δ125t (aa 1–1322), Top2Δ59t (aa 1–1388), Top2Δ20t (aa 1–1427), Top2Δ5t (aa 1–1442), Top2(S1392A,S1396A,S1409A,S1410A,S1428A,S1443A), Top2(S1392A,S1396A,S1409A,S1410A), Top2(S1428A,S1443A), Top2(S1428A), and Top2(S1443A)) were cloned into pMT-puro vectors with N-terminal HA tags.
Mus101 Constructs for Pulldown Assays
Full-length Drosophila Mus101 was cloned into a modified pET41a vector (Novagen) in which a linker containing a PreScission protease cleavage site was added between an N-terminal GST fusion tag and Mus101. In addition, the C terminus of Mus101 was fused with a His6 tag. Mus101[1–350], N-terminal residues 1–350, was made by replacing the full-length Mus101 with Mus101 residues 1–350, preserving the N-terminal GST fusion tag as well as the PreScission protease cleavage site and the C-terminal His6 tag.
Inducible Plasmid-based shRNA System for Cellular Assays
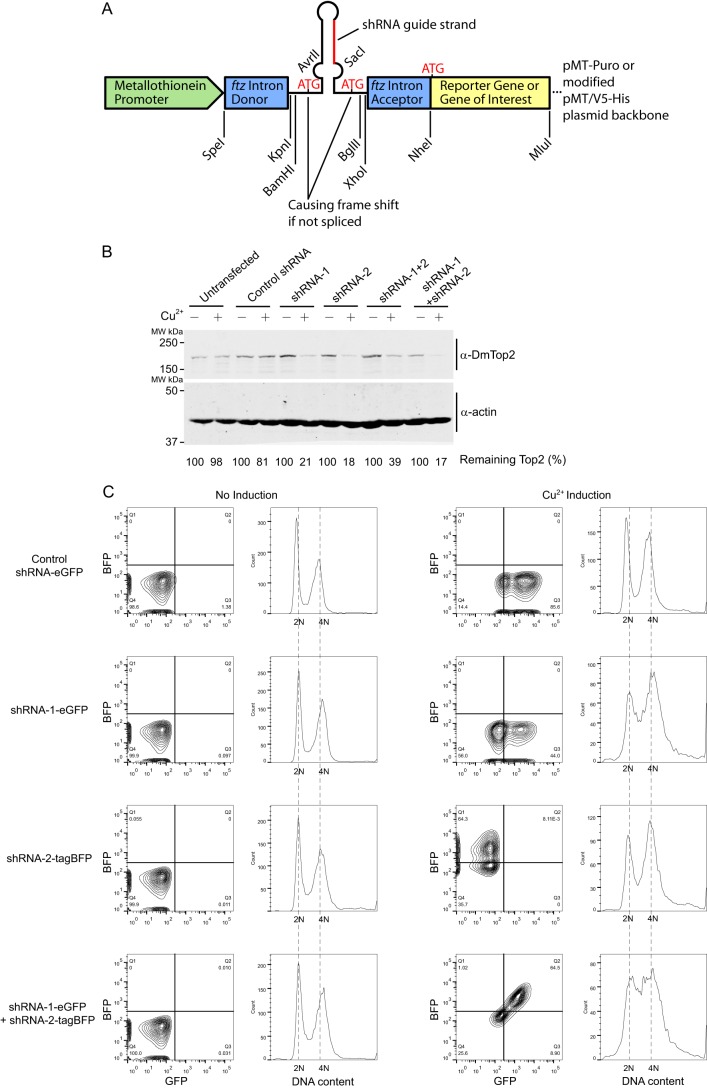
Our inducible plasmid-based shRNA system was adapted from an intron-mediated shmiR expression system by Haley et al. (20), with modifications of restriction enzyme sites that facilitate switching between the constructs of shRNA sequences predicted from the Designer of Small Interfering RNA algorithm (Fig. 6A) (21). The guide strands of the shRNA sequences were as follows: control shRNA, GGTTCTAGTATACTTGGTTGT (scrambled sequence of shRNA-1, not complementary to any genes in the target organism); shRNA-1, TTGTAGTTGGATATCTTCGTG; and shRNA-2, TAAGAGTCCATCTTCTATCAA. The complete cassette, including the ftz intron, pre-miR-1 stem base, shRNA sequence, and reporter gene, was inserted between the SpeI and MluI sites of either a pMT-puro vector (Addgene, originally from the Sabatini laboratory) or a modified pMT/V5-His vector (Invitrogen). To simultaneously express the TOP2 gene (WT or mutants), the reporter gene, eGFP, or tagBFP, was replaced by a sequence of RNAi-resistant Top2(WT), Top2(S1428A,S1443A), or Top2Δ20 with GFP as a C-terminal fusion protein.
FIGURE 6.
Effect of Top2-targeting shRNAs on S2 cells. A, Diagram of the shRNA design. The construct was modified from an intron-mediated shmiR expression system by Haley et al. (20). The exact shRNA targeting sequences are described under “Experimental Procedures.” B, efficiency of an inducible plasmid-based shRNA system on silencing endogenous Top2. Drosophila S2 cells transfected with the indicated shRNA constructs were incubated with or without 500 μm CuSO4 for 4 days. Endogenous Drosophila Top2, DmTop2, was detected by anti-DmTop2 antibodies. shRNA-1 + 2 represents a single construct carrying a tandem-linked shRNA-1 and shRNA-2 cassette, and shRNA-1 + shRNA-2 represents a co-transfection of shRNA-1 and shRNA-2 constructs. The level of remaining Top2 was calculated by dividing the Top2 signal of induced cells by that of uninduced cells. MW, molecular weight. C, cell cycle profiles of cells transfected with inducible shRNA constructs for silencing Top2. Four groups of cells were individually transfected with control shRNA-eGFP, shRNA-1-eGFP, shRNA-2-tagBFP, or shRNA-1-eGFP + shRNA-2-tagBFP. The cells without CuSO4, shown as uninduced controls, expressed an undetectable level of the fluorescent reporter gene (eGFP or tagBFP). Thus, the histograms of GFP- and BFP-negative cells were plotted to represent normal cell cycle profiles. Induced by 500 μm CuSO4, cells with the fluorescent signals of the corresponding reporter genes were gated, and the histograms showed the cell cycle profiles of GFP-positive, BFP-positive, or both positive cells. Cells transfected with shRNA-1-eGFP or shRNA-2-tagBFP showed a slight accumulation at S phase and G2/M. With the transfection of both shRNA-1-eGFP and shRNA-2-tagBFP, marked accumulation of cells at S phase and G2/M was observed.
Transfection of Drosophila S2 Cells
16–24 h before transfection, Drosophila S2 cells were seeded at 1 × 106/ml in 3 ml of Schneider's Drosophila medium (Life Technologies) containing 10% heat-inactivated FBS (HyClone) and 50 μg/ml of gentamicin (Gibco) using 6-well plates at 27 °C. Transfections of plasmids were performed using TransIT-2020 transfection reagent (Mirus). 16–24 h after transfection, the cells were washed and replaced with fresh medium containing 10% FBS. 48 h after transfection, antibiotics, Hygromycin B (300 μg/ml, HyClone), or puromycin (6 μg/ml, Life Technologies) were used for selecting stably transfected cell lines.
Immunoprecipitation Assay with Nuclear Extract of Drosophila S2 Cells
Transfected and stably selected S2 cells were seeded at 2 × 106/ml in 40 ml of Schneider's Drosophila medium containing 10% heat-inactivated FBS and 50 μg/ml of gentamicin (Gibco) using a T-150 flask at 27 °C. After culturing for 4 days, CuSO4 at a final concentration of 500 μm was used to induce the metallothionein promoter (pMT/V5-His and pMT-puro vectors) for 16 h. Cells overexpressing proteins of interest were harvested by centrifuging at 2000 × g for 3 min and resuspended with PBS. All solutions were supplemented with a protease inhibitor mixture that contained 1 mm PMSF (Sigma), 5 μg/ml E-64 (Peptide International), 5 μg/ml leupeptin (Peptide International), 2 μg/ml pepstatin (Peptide International), and 2 μg/ml aprotinin (Peptide International). To obtain the nuclear extract for immunoprecipitation, the cells were lysed with a Dounce homogenizer in a hypotonic buffer (2 mm MgCl2 and 5 mm HEPES buffer (pH 7.4)) and then kept on ice for 10 min. After centrifuging at 16,000 × g for 5 min, the nuclear pellet was washed with a hypotonic buffer before another centrifugation at 16,000 × g for 5 min. The pellet was then resuspended in a nuclear extraction solution (20 mm HEPES buffer (pH 7.4), 400 mm NaCl, 10% glycerol, 0.2% Nonidet P-40 alternative (Calbiochem), and 1 mm EDTA) on ice for 30 min. Nuclear extraction dilution solution (20 mm HEPES buffer (pH 7.4), 10% glycerol, 0.2% Nonidet P-40 alternative, and 1 mm EDTA) was added to bring NaCl down to 250 mm before adding 40 μl of anti-FLAG (Sigma) or anti-HA-agarose beads (Sigma) in the presence of 100 μg/ml ethidium bromide. After incubating at 4 °C for 1 h, antibody-agarose beads were spun down at 2000 × g for 5 min, washed three times with adjusted nuclear extraction solution (250 mm NaCl), resuspended with SDS-PAGE sample buffer, and boiled for 5 min before loading onto SDS-PAGE for Western blotting. Rabbit anti-FLAG antibody (Sigma) and mouse anti-HA antibody (Sigma) were used for detection of FLAG- or HA-tagged proteins.
Purification of Mus101 and Mus101[1–350] for Pulldown Assays
Full-length Mus101 and Mus101[1–350] were purified based on the previously published purification procedure for human TopBP1 (22).
Immobilizing Top2 Proteins on Anti-HA-Agarose Resin for Pulldown Assays
Recombinant Top2 was expressed and purified following a similar protocol as described earlier for the immunoprecipitation assay. To preserve the phosphorylation status of Top2, all solutions were supplemented with 5 mm 2-mercaptoethanol, 5 mm NaF (Sigma), and 1 mm Na3VO4 (Sigma) in addition to a protease inhibitor mixture. The cleared lysate was incubated with a slurry of 50% anti-HA-agarose resin (Sigma) at 4 °C for 2 h in the presence of 100 μg/ml ethidium bromide. The resin was then washed twice with a lysis buffer and three times with a high-salt lysis buffer (20 mm HEPES (pH 7.4), 1 m NaCl, 10% glycerol, 0.1 μm okadaic acid, 5 mm EDTA, and 1% Nonidet P-40) alternative. Truncated or mutant Top2 proteins adsorbed to the resin were stored at −20 °C after equilibrating with a storage solution of a 20 mm HEPES (pH 7.4), 500 mm NaCl, 50% glycerol, 0.1 μm okadaic acid, 5 mm EDTA, and 1% Nonidet P-40 alternative.
For dephosphorylation experiments, immobilized Top2 was washed with an incubation buffer (20 mm HEPES (pH 7.4), 200 mm NaCl, 5% glycerol, 1 mm EDTA, 50 μg/ml bovine serum albumin (Research Organics), 1 mm PMSF, and 5 μg/ml Leupeptin) to eliminate the interference of phosphatase inhibitors. The dephosphorylation of Top2 was accomplished by adding 3 mm MnCl2 and 20 units λ-phosphatase (New England Biolabs) per microliter of resin at 30 °C for 30 min with two controls, one with 3 mm MnCl2 only and the other with 3 mm MnCl2 and λ-phosphatase in the presence of 5 mm NaF, 1 mm Na3VO4, and 0.1 μm okadaic acid. After the treatment, the resin was subjected to pulldown assays or elution of proteins.
Pulldown Assay
The resin containing 2 pmol of immobilized Top2 proteins was washed, resuspended in an incubation buffer, and incubated with Mus101[1–350] or full-length Mus101 at 4 °C for 1 h. After washing the resin with an incubation buffer, the sample was loaded onto 8% SDS-PAGE after boiling with an SDS-PAGE sample buffer. Proteins were detected by Western blotting with rabbit anti-Mus101 antibody for full-length Mus101, mouse anti-His for Mus101[1–350]-His6 (Roche), and mouse anti-HA antibody for HA-tagged Top2 proteins. Signals were detected and quantitated using a LICOR Odyssey scanner.
Decatenation Activity
Purified Drosophila Top2 with or without λ-phosphatase treatment was used for decatenation activity. To purify the Top2 protein after phosphatase treatment, immobilized proteins were treated with or without λ-phosphatase in the presence of 3 mm MnCl2 at 30 °C for 30 min as described above. The resin was washed twice before elution of proteins with 80 μl of 2 μg/μl HA peptide in PBS and then stored at −80 °C.
For the decatenation assays, 0.5 ng of Top2 was added to a 20-μl reaction buffer containing 0.1 μg of kinetoplast DNA (Topogen), 20 mm HEPES (pH 7.4), 100 mm NaCl, 50 mm KCl, 10 mm MgCl2, 0.1 mm EDTA, 50 μg/ml BSA, 5 mm 2-mercaptoethanol, and 1 mm ATP in the presence of 0, 0.625, 1.25, or 2.5 μm Mus101 and incubated at 30 °C for 15 min. The reactions were quenched, and DNA products were analyzed by electrophoresis in 1% agarose gel with ethidium bromide. The levels of of kinetoplast DNA substrate and decatenated product were quantified by ImageJ software. The relative ratio of product DNA was measured by dividing the signals of decatenated kinetoplast DNA (nicked and relaxed) by the total signals.
Peptide Synthesis
Unphosphorylated and doubly phosphorylated C-terminal biotinylated peptides containing the last 25 amino acids of Drosophila Top2, RAVIESDDDDIEIDEDDDDSDFNC-Biotin and RAVIE(pS)DDDDIEIDEDDDDD(pS)DFNC-Biotin, were synthesized by the Peptide Synthesis Core Facility of the Institute of Cellular and Organismic Biology, Academia Sinica, and purified by HPLC. The final products attained >95% purity and were confirmed by electrospray ionization-MS (ESI-MS).
Surface Plasmon Resonance Analysis
Surface plasmon resonance measurements were performed on a BIAcore 2000. A streptavidin sensor chip was derivatized with C-terminal biotinylated unphosphorylated peptide (144 response units) or doubly phosphorylated peptide (145 response units) as described above. Because of the instability of Mus101[1–350], the temperature of the BIAcore machine and samples was maintained at 10 °C. Solutions (100 μl) of Mus101[1–350] at the indicated concentrations in 20 mm HEPES (pH 7.4), 250 mm NaCl, 5% glycerol, 1 mm EDTA, and 0.005% surfactant P20 (GE Healthcare) were flowed across the chip surface at a rate of 20 μl/min. The chip was regenerated by injecting 60 μl of 0.1% SDS (GE Healthcare).
Cell Cycle Analysis by Flow Cytometry
S2 cells were transfected with constructs for expressing inducible shRNA and selected as described above. For testing the effect of shRNA and co-expression of the TOP2 gene, CuSO4 was added to S2 cells seeded at 1 × 106/ml in 3 ml using 6-well plates at 27 °C. After 96 h of culture, 3 × 106/ml S2 cells were collected. The cells were washed with 1 ml of PBS and cross-linked with 1% paraformaldehyde at 4 °C for 1 h. The cross-linked cells were then washed with PBS before permeabilization by 70% ethanol for at least 2 h. After the treatment, the cells were stained with 40 μg/ml propidium iodide (Biolegend) with 40 μg/ml RNase (Affymetrix) at 37 °C for 30 min. Flow cytometry (BD Biosciences FACSCanto II) was then used to select singlet cells from cell aggregates. Based on the reporter genes used, eGFP-positive, tagBFP-positive, or double-positive cells were gated for cell cycle analysis.
Cell Viability Assay
The viability of S2 cells was determined by the membrane permeability of propidium iodide as described previously (23).
Results
The Interaction between Drosophila Top2 and Mus101 Is Mediated through Distinct Domains
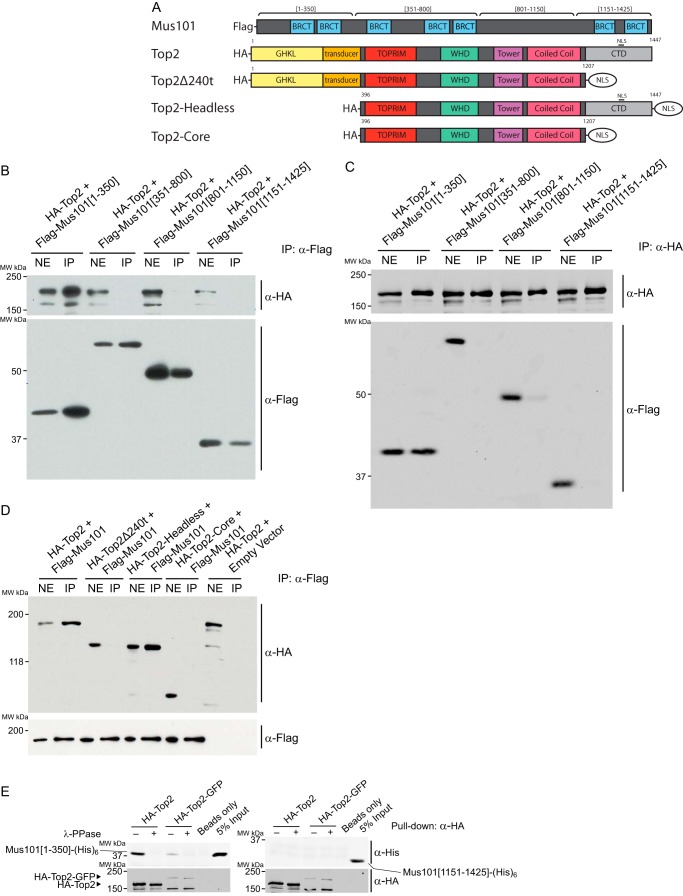
We investigated the possibility that Mus101 can interact with Top2 in the tissue culture cell system. We isolated nuclear extracts from Drosophila S2 cells expressing FLAG-tagged Mus101 and demonstrated its interaction with Top2 by co-IP experiments using anti-FLAG-agarose resin (Fig. 1A). To test whether the interaction is direct or through a mediator molecule, we purified both proteins with distinct tags for pulldown experiments. HA-tagged Top2 was purified from S2 cells in the presence of phosphatase inhibitors to preserve the posttranslational modification of Top2, and His6-tagged Mus101 was purified from Escherichia coli. Mus101 and Top2 can form a binary complex, as demonstrated by the pulldown assays, indicating that their interaction is direct and not through other mediator molecules (Fig. 1, B and C). We used truncated constructs of either Top2 or Mus101 to determine the domains critical for the formation of the binary complex. Using S2 cells, we expressed FLAG-tagged fragments of Mus101, residues 1–350, residues 351–800, residues 801–1150, or residues 1151–1425 (Fig. 2A), in the background of HA-tagged, full-length Top2 to map the binding domain of Mus101. Nuclear extracts prepared from these cells were subjected to a co-IP assay. Mus101[1–350], which contains the N-terminal 350 residues covering the first two BRCT domains (BRCT1/2), bound efficiently with Top2 (Fig. 2B). The reciprocal co-IP experiment also yielded consistent results (Fig. 2C). We additionally confirmed direct interaction by using purified proteins in a pulldown experiment. Under the same binding conditions, Mus101[1–350] could be pulled down by Top2, whereas C-terminal Mus101 (Mus101[1151–1425]) showed no significant interaction with Top2 (Fig. 2E). In previous studies, it was suggested that the C terminus of human TopBP1 is involved in the binding of human Top2α and Top2β (12, 15). However, the Top2 binding region within TopBP1 was not identified. By overexpressing Top2 and preserving phosphorylation with phosphatase inhibitors, we demonstrated that the first 350 amino acids of Mus101 containing BRCT1/2 are responsible for the binding of Top2 in the Drosophila system.
FIGURE 1.

Interaction between Top2 and Mus101. A, anti-FLAG-agarose beads were incubated with nuclear extract (NE) from Drosophila S2 cells overexpressing FLAG-Mus101. Endogenous Top2 and FLAG-Mus101 were detected in the immunoprecipitation product by rabbit anti-Top2 antibodies and rabbit anti-FLAG antibodies. MW, molecular weight. B and C, direct binding between Top2 and Mus101 was examined with purified proteins by pulldown assays. Mus101-His6 was pulled down by HA-Top2 immobilized on anti-HA-agarose beads. In a reciprocal experiment using nickel-nitrilotriacetic acid (Ni-NTA) resin, HA-Top2 was pulled down by immobilized Mus101-His6. Empty anti-HA-agarose beads and nickel-nitrilotriacetic acid resin were used as controls. HA-Top2 and Mus101-His6 were detected with immunoblot by mouse anti-HA antibodies and rabbit anti-Mus101 antibodies, respectively.
FIGURE 2.
Top2-Mus101 interaction requires the N-terminal domain of Mus101 and the C-terminal regulatory domain of Top2. A, schematic of the expression constructs for Mus101 and the HA-tagged Top2. GHKL, gyrase, Hsp90, histidine kinase, MutL, TOPRIM, topoisomerase-primase; WHD, winged helix domain; NLS, nuclear localization signal. B and C, S2 cells were transfected with ectopic expression vectors for HA-Top2 along with one of the FLAG-tagged Mus101 truncations, aa 1–350, aa 351–800, aa 801–1150, or aa 1151–1425. The nuclear extracts (NE) were prepared from lysed cells and then subjected to immunoprecipitation using anti-FLAG-agarose beads (B) or anti-HA-agarose beads (C). Products of IP were analyzed by immunoblot and detected by rabbit anti-FLAG antibodies for Mus101 fragments and mouse anti-HA antibodies for Top2. Mus101[1–350] is the only fragment that was immunoprecipitated with Top2. MW, molecular weight. D, similar immunoprecipitation experiments were carried out using nuclear extracts from S2 cells expressing FLAG-Mus101 and one of the following Top2 constructs: full-length Top2 (HA-Top2), C terminus-truncated Top2 (HA-Top2Δ240t), N terminus-truncated Top2 (HA-Top2-Headless), or Top2 with truncation at both termini (HA-Top2-Core). HA-Top2 and HA-Top2-headless can immunoprecipitate with FLAG-Mus101. E, 2 pmol of Mus101[1–350]-His6 or Mus101[1151-End]-His6 were tested in a pulldown experiment under the same conditions. HA-Top2 can associate with Mus101[1–350]-His6 but not with Mus101[1151-End]-His6.
To map the interacting domain on Top2, we conducted co-IP experiments using S2 cells expressing FLAG-tagged, full-length Mus101 and HA-tagged Top2 with a truncation of the N terminus (ATPase domain), C terminus (regulatory domain), or both termini (Fig. 2A). The result shows that the interaction is completely abolished with the removal of the C-terminal regulatory domain (Top2Δ240 and -core in Fig. 2D), demonstrating the essential role of the Drosophila Top2 C terminus in binding to Mus101.
The Last 20 Amino Acids of Top2 Are Required for Mus101 Binding
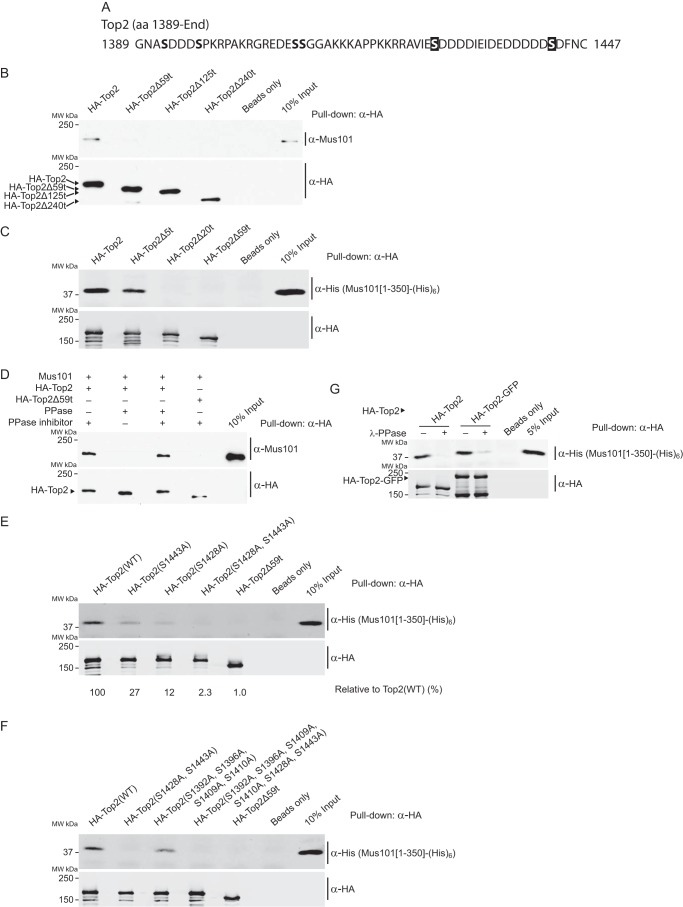
To define the minimal C-terminal elements of Top2 required for the binding of Mus101, we separately purified Top2 constructs with 59 residues, 125 residues, and 240 residues truncated from the C terminus (Top2Δ59t, Top2Δ125t, and Top2Δ240t, respectively) and carried out pulldown experiments with purified Mus101. We found that Mus101 failed to bind any of these truncated Top2 proteins, suggesting that the terminal 59 residues are necessary for Mus101 binding (Fig. 3, A and B). To further refine the mapping of the Top2-interacting motif, we purified truncated Top2 proteins lacking the last five residues and 20 residues (Top2Δ5t and Top2Δ20t, respectively) and tested their binding with the N-terminal 350 aa of Mus101, Mus101[1–350]. Mus101[1–350] did not bind to Top2Δ20t, whereas it bound to Top2Δ5t at a reduced level, which suggests that the last 20 amino acids are required for interacting with Mus101 (Fig. 3C).
FIGURE 3.
The last 20 amino acids containing two potential phosphorylated serines (Ser-1428 and Ser-1443) are the minimal requirement for the binding of Mus101. A, sequence of the last 59 amino acids of Top2. B, in a pulldown experiment, purified full-length Mus101 was incubated with immobilized full-length Top2 (HA-Top2), truncated Top2 lacking the C-terminal 59, 125, or 240 amino acid residues (HA-Top2Δ59t, HA-Top2Δ125t, and HA-Top2Δ240t) on anti-HA-agarose beads. Only HA-Top2 is able to pull down Mus101. MW, molecular weight. C, after incubation with truncated Top2 lacking the C-terminal 5, 20, or 59 amino acid residues (HA-Top2Δ5t, HA-Top2Δ20t, and HA-Top2Δ59t) on anti-HA-agarose beads, Mus101[1–350]-His6 was pulled down by HA-Top2 and HA-Top2Δ5t but not HA-Top2Δ20t. D, binding of Mus101-His6 to HA-Top2 with or without treatment with λ-phosphatase or treatment with inhibited λ-phosphatase was compared by pulldown assays. HA-Top2Δ59t served as a negative control. Dephosphorylated HA-Top2 failed to pull down Mus101-His6. E, Top2 mutants with alanine substituted for serine at residue 1428, residue 1443, and both (HA-Top2(S1428A), HA-Top2(S1443A), and HA-Top2(S1428A,S1443A), respectively), were tested in pulldown experiments, and the quantified signals were normalized to the positive control of wild-type Top2(WT). Mus101[1–350]-His6 can pull down HA-Top2(S1428A) (27% of Top2(WT)) and HA-Top2(S1443A) (12% of Top2(WT)) but not HA-Top2(S1428A,S1443A) (less than 3% of Top2(WT)). F, HA-Top2(S1392A,S1396A,S1409A,S1410A,S1428A,S1443A), HA-Top2(S1392A,S1396A,S1409A,S1410A), and HA-Top2(S1428A,S1443A) were tested in pulldown experiments. Substitutions of Ser-1392, Ser-1396, Ser-1410, and Ser-1409 for Ala did not abrogate the binding of Mus101. G, Mus101[1–350]-His6 can pull down both HA-Top2 and HA-Top2-GFP at a similar level, indicating that C-terminal GFP fusion does not interfere with its phosphorylation-dependent binding with Mus101[1–350]-His6.
Phosphorylation of Both Ser-1428 and Ser-1443 Is Required for Efficient Binding to Mus101
Although BRCT domains are characterized as binding motifs for phosphorylated protein modules, the conserved BRCT1/2 domains of TopBP1 and its homologs have been found to interact with several binding partners in their doubly phosphorylated forms (16–18). Because the N-terminal 350 aa of Mus101, which contains BRCT1/2 domains, is required for Top2 binding, we set out to test whether Top2 interacts with Mus101 in a phosphorylation-dependent manner and, furthermore, whether a doubly phosphorylated form is required. Dephosphorylation of Top2 by λ-phosphatase could abolish its binding with Mus101 in a pulldown experiment whereas the negative controls could not, suggesting that phosphorylation is required for Mus101 binding (Fig. 3D). Taken together with our mapping results, there are only two potential phosphorylation sites in the terminal 20 amino acids, Ser-1428 and Ser-1443. We generated site-specific mutants changing one or both of these residues from Ser to Ala and tested the Mus101 binding ability of these Top2 mutants (Fig. 3A). Although the single Ser substitution mutants, Top2(S1428A) and Top2(S1443A), diminished Mus101 binding, only the double substitutions, Top2(S1428A,S1443A), completely abolished the binding (Fig. 3E). To confirm that these are the only critical Ser residues that are involved in phosphorylation-dependent interaction, we also examined Top2 with four additional Ser substitutions Top2(S1392A,S1396A,S1409A,S1410A) that are upstream of Ser-1428 and Ser-1443, and none of these Ser/Ala substitutions affected their binding to Mus101[1–350] (Fig. 3F). Thus, among the last 59 amino acids, Ser-1428 and Ser-1443 are apparently the only two phosphorylated sites required for the interaction.
The Phosphorylated Top2 Peptide Interacts with Mus101[1–350]
We confirmed the sufficiency of the Mus101 binding motif at the C terminus of Top2 by synthesizing phosphorylated and unphosphorylated peptides spanning the last 25 amino acids of Top2 with a C-terminal biotinylation. After peptide immobilization on streptavidin-agarose beads, Mus101[1–350] was pulled down by phosphorylated peptide but not by unphosphorylated peptide (Fig. 4A). The binding affinity between the phosphorylated Top2 peptide and Mus101[1–350] was determined by surface plasmon resonance with a Kd of 0.57 ± 0.04 μm, whereas the binding affinity between unphosphorylated Top2 peptide and Mus101[1–350] was not detectable (Fig. 4B). Our pulldown and surface plasmon resonance experiments strongly support the requirement of phosphorylation in Top2-Mus101 binding and demonstrate that the last 25 amino acids of Top2 itself are sufficient for high-affinity Mus101 binding.
FIGURE 4.
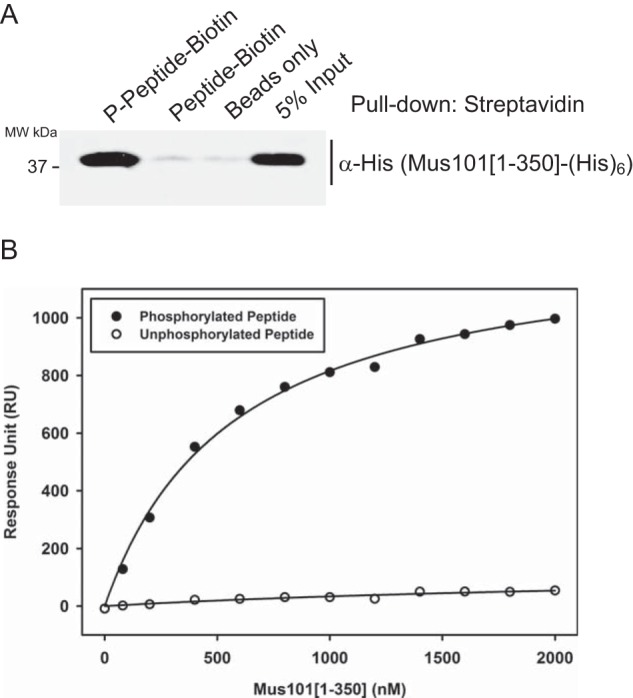
Phosphorylated Top2 peptide interacts with Mus101[1–350]-His6. C-terminal biotinylated synthetic peptides, phosphorylated (Ser(P)-1428 and Ser(P)-1443) or unphosphorylated, containing the last 25 amino acids of Top2, were examined with a pulldown assay and a surface plasmon resonance experiment. A, Mus101[1–350]-His6 was incubated with immobilized peptides on streptavidin-agarose beads in the pulldown experiment, and only phosphorylated peptide was found to have binding activity. MW, molecular weight. B, 145 response units (RU) of phosphorylated or unphosphorylated peptides were immobilized on separate channels of a streptavidin chip for a surface plasmon resonance experiment. Different concentrations of Mus101[1–350]-His6 (from 0–2 μm) were tested, and sensorgrams were recorded. Saturated response units of each condition were plotted against the concentrations of Mus101[1–350]-His6. The Kd of Mus101[1–350]-His6 to phosphorylated Top2 peptide was determined to be 0.57 ± 0.04 μm.
Mus101 Binding Inhibits Top2 Decatenation Activity
Although not required for its catalytic function, the C-terminal domain of Top2 can play a role in regulating its biochemical and biological functions. Phosphorylation of C-terminal residues or its binding to proteins, including HMBG1 and 14-3-3ϵ, can both modulate Top2 activities (9, 10, 24–28). Because we showed here a robust phosphorylation-dependent interaction between Top2 and Mus101, we then considered whether such binding could also affect a key biochemical activity, such as DNA decatenation by Top2. We used purified, λ-phosphatase-treated Top2 to abolish the interaction with Mus101. In the presence of Mus101, the decatenation activity of phosphorylated Top2 decreased with an increasing amount of Mus101, whereas the activity of dephosphorylated Top2 was unaffected until the Mus101 concentration exceeded 1.5 μm, suggesting that the binding of Mus101 inhibits Top2 decatenation activity and that the sensitivity of Top2 to Mus101 inhibition is also phosphorylation-dependent (Fig. 5, A and B).
FIGURE 5.
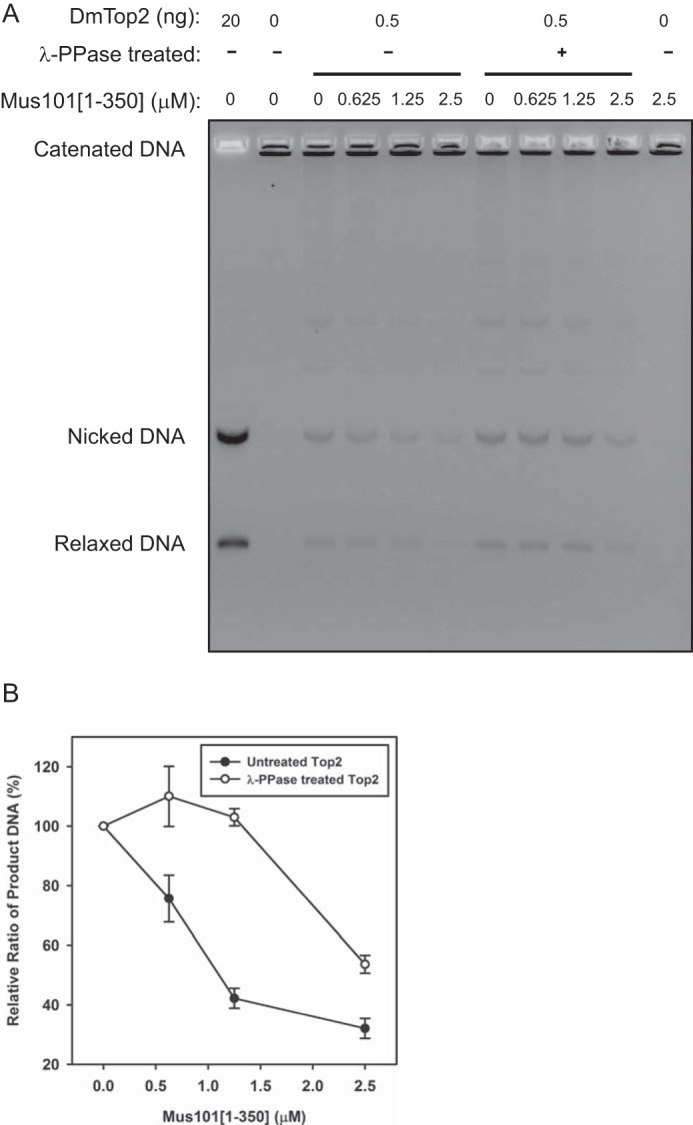
Mus101 inhibits Top2-mediated kinetoplast DNA decatenation. A, 0.5 μg of untreated or λ-phosphatase (λ-PPase)-treated Top2 was incubated with 0.1 μg of kinetoplast DNA in the presence of Mus101[1–350]-His6 (at 0, 0625, 1.25, or 2.5 μm) at 30 °C for 15 min. The samples were analyzed by electrophoresis in 1% agarose gel with EtBr. B, triplicate results were quantified, and relative ratios of DNA products were plotted against Mus101[1–350]-His6 concentrations. Top2 (untreated) decatenation activity reduces significantly with the increasing amount of Mus101.
Top2-silenced S2 Cells Accumulate Products of Incomplete DNA Segregation
Because of the essential functions of Drosophila Top2 and Mus101, it is difficult to decipher the biological function of the Top2-Mus101 interaction by using loss-of-function genetic analysis. Instead, we proceeded by knocking down the endogenous enzyme and substituting it with an ectopically expressed mutant Top2. We adapted an intron-mediated shmiR expression system to generate an inducible plasmid-based shRNA system to simultaneously silence endogenous Top2 and express the WT or mutant Top2 (Fig. 6A) (20). Using the Designer of Small Interfering RNA algorithm, we chose two sets of shRNA sequences for the TOP2 gene, with one targeting the protein-coding region and the other targeting the 3′ UTR (21). In case of insufficient knockdown efficiency by a single shRNA, we employed two approaches to combine the effect of two shRNAs either from two separate plasmids or with tandem-linked shRNAs on a single plasmid (20). As described by Haley et al. (20), the shRNA sequence is embedded in an ftz intron upstream of a GFP reporter gene, which allowed us to track the induction of the shRNA at the single cell level by fluorescence-activated cell sorting. By examining the protein level of endogenous Top2, all four constructs of the shRNA expression system successfully knocked down the endogenous Top2 (Fig. 6B). Because Top2 is essential for cell growth, depleting Top2 using shRNA should show defects in the cell cycle. We used flow cytometry to monitor the cell cycle profile in S2 cells transfected with shRNAs. S2 cells transfected with shRNA-1 or shRNA-2 accumulated at S and G2/M phases (Fig. 6C). However, in addition to an even higher level of accumulation at S and G2/M phases, we observed severe cell cycle arrest and cell death in cells transfected with both shRNA-1 and shRNA-2, indicating that the combination of two sets of shRNAs achieved a better depletion of Top2 (Figs. 6C and 7).
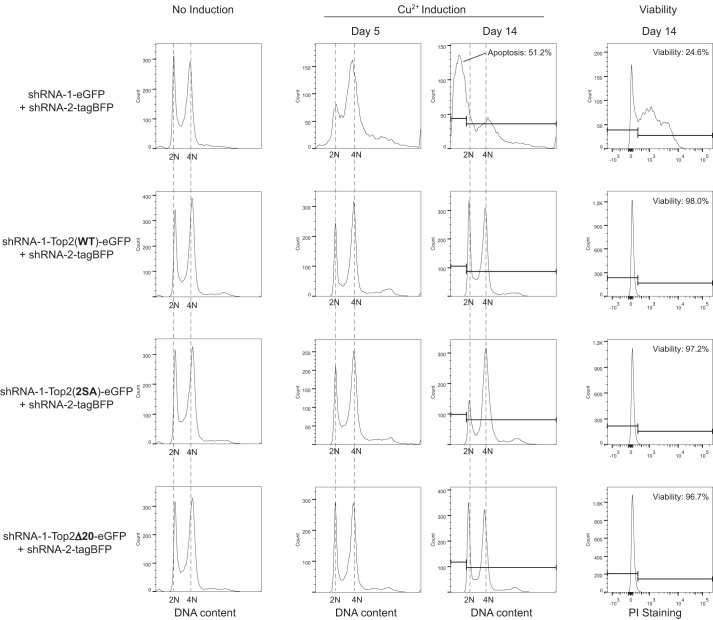
FIGURE 7.
Cell cycle defect and loss of viability caused by Top2 depletion were rescued by Top2(WT), Top2(2SA), and Top2Δ20. Four groups of cells were transfected with the constructs indicated on the left. After 100 μm CuSO4 induction, the cell cycle profiles were recorded on days 5 and 14. The cells without CuSO4 induction are shown as controls for normal cell cycle distribution. After 14 days of induction, cells expressing shRNA-1-eGFP and shRNA-2-tagBFP accumulated at sub-G1 on day 14, and 75.4% of cells were non-viable (propidium iodide (PI)-permeable). Top2-depleted cells rescued by Top2(WT), Top2(2SA), or Top2Δ20 appeared to be viable and normal in cell cycle distribution on day 14.
For simultaneous expression of shRNA and a TOP2-rescuing gene, we replaced one of the GFP reporter genes with an RNAi-resistant Top2(WT), Top2(S1428A,S1443A) (hereafter Top2(2SA)), or Top2Δ20 with GFP fused at the C terminus. We also confirmed that fusion of GFP does not affect the binding of Mus101 (Fig. 3G). In addition to S and G2/M accumulation, the viability of S2 cells was severely affected after continuously knocking down endogenous Top for 14 days (Fig. 7). More than 50% of Top2-silenced cells were at a sub-G1 population, representing cells with DNA fragmentation, likely caused by cell apoptosis. Probed by the permeability of propidium iodide, Top2-silenced cells were found to be less than 25% viable (Fig. 7). Defects in the cell cycle and viability caused by depletion of endogenous Top2 were rescued by the expression of Top2(WT), Top2(2SA), or Top2Δ20 (Fig. 7).
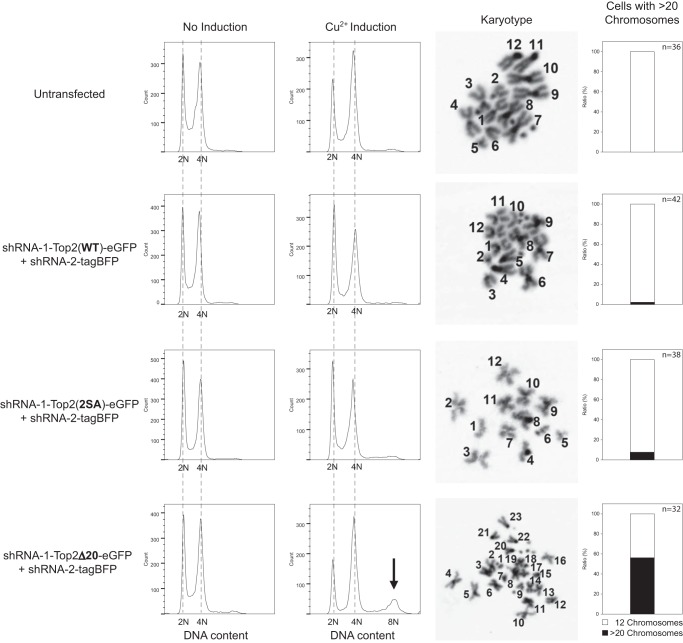
No cell cycle defects were observed after replacing endogenous Top2 with Top2(2SA) or Top2Δ20 for 14 days. However, after 29 days of shRNA and transgene expression, Top2-silenced cells rescued by Top2Δ20 showed an increasing population with higher DNA content (Fig. 8). To ascertain whether the additional DNA content was from increasing the copy number of chromosomes or from the alteration in chromosome size through genome rearrangement, we analyzed the karyotypes of untransfected S2 cells and Top2-silenced S2 cells rescued by wild-type or mutant Top2. Although 12 long chromosomes were normally seen in untransfected cells, high copy numbers (>20) of chromosomes were observed in the Top2-silenced S2 cells rescued by Top2Δ20, indicating incomplete segregation during mitosis (Fig. 8). Our data suggest that abolishing the Top2-Mus101 interaction by completely removing the Mus101 binding interface in Top2 CTD impairs the fidelity of chromosome segregation, with daughter cells continuously undergoing nondisjunction, thereby leading to increasing the DNA content.
FIGURE 8.
Long-term expression of Top2Δ20 in Top2-depleted cells triggers the accumulation of a population with doubled DNA content. On day 29 of CuSO4 induction, the cell cycle profiles of S2 cells transfected with the indicated constructs were recorded. Top2-depleted cells rescued by Top2Δ20 showed a peak representing high DNA content, whereas cells rescued by Top2(WT) or Top2(2SA) showed no significant difference from untransfected cells. The arrow marks the cell population with polyploidy. Among 32 Top2-depleted cells rescued by Top2Δ20, observed by karyotype analysis, 18 cells contained more than 20 chromosomes, whereas only 14 cells appeared to have 12 long chromosomes as in untransfected cells. The elevated population of cells with more than 20 chromosomes observed in karyotype analysis was also concordant with the appearance of a population of cells with higher DNA content from flow cytometry.
Discussion
In this study, we demonstrated that phosphorylated Ser-1428 and Ser-1443 of Drosophila Top2 are required for interaction with the N terminus of Mus101. The binding of the N-terminal domain of Mus101 to the Top2 CTD was found to inhibit the decatenation activity of Top2 in vitro. In the cellular assay, we were able to achieve almost complete depletion of endogenous Top2 by transfecting cells with two sets of shRNA, causing strong accumulation at S and G2/M phases followed by cell death. Furthermore, Top2-silenced cells rescued by Top2Δ20 had an increasing population with higher DNA content, suggesting that the Top2-Mus101 interaction is important for faithful DNA segregation.
Drosophila Top2 Binds the N terminus of Mus101
Our finding regarding the Top2-Mus101 function in DNA segregation supports a recent discovery that human Top2α-TopBP1 interaction is responsible for recruiting Top2α to UFBs during mitosis (15). Based on our mapping results from co-IP and pulldown experiments, it is evident that doubly phosphorylated Drosophila Top2 interacts with the N-terminal 350 residues of Mus101. This conclusion differs from earlier work suggesting that human Top2α and Top2β bind to the C terminus of TopBP1 (12). In the previous study, the C-terminal domain of TopBP1 containing BRCT7/8 was deduced to interact with Top2α because Top2α failed to co-immunoprecipitate with a truncated TopBP1 with BRCT7/8 deleted or co-localize at UFBs with this mutant TopBP1. However, removing the C-terminal domain of TopBP1 is known to result in several cellular consequences, including mislocalization of TopBP1 and abolition of the ATR-TopBP1 interaction (29, 30). One therefore cannot rule out the possibility that loss of the TopBP1-Top2α interaction at UFBs may be an indirect effect of interrupting a protein interaction network involving TopBP1. Our results showed that the C terminus of Mus101 (Mus101[1151–1425]) does not interact with Top2 by co-immunoprecipitation and with pulldown assays using purified components. Although we have direct biochemical evidence to demonstrate the interacting domains for TopBp1-Top2, different Top2 interacting domains may be utilized during different cellular processes. We also cannot rule out the possibility that regulation of the Top2-Mus101 interaction may vary among different species. Further investigation is needed to evaluate how Top2α interacts with TopBP1.
Regulation of the Top2-Mus101 Interaction
BRCT1/2 of Mus101 and its homologs are important for the initiation of replication as well as DNA damage repair signaling. On one hand, for assembling the preinitiation complex during DNA replication, yeast Sld3 and human Treslin, the functional homolog of yeast Sld3, must be doubly phosphorylated by CDK on [S/TP] motifs for the binding of BRCT1/2 of yeast Dpb11 and human TopBP1, respectively (17, 31, 32). On the other hand, Rad9, a member of the 9-1-1 clamp complex involved in the DNA damage response, interacts with BRCT1/2 of TopBP1 when Rad9 is phosphorylated by casein kinase 2 on the [S/T]XX[D/E] motifs (18). In this study, we found that BRCT1/2 of Mus101 is likely involved in a third event, DNA segregation. BRCT1/2 of Mus101 seems to be versatile enough to accommodate the binding of different sequence motifs phosphorylated by kinases from various signaling pathways. According to previous studies, Drosophila Top2 has been shown to be phosphorylated by casein kinase 2 (25). After examining the C-terminal sequence of Drosophila Top2, casein kinase 2 may be responsible for phosphorylating Ser-1428 but less likely for Ser-1443, which is not in a conserved [S/T]XX[D/E] motif. We suspect that there may be other kinases required for the phosphorylation. Two likely candidate kinases, Aurora B kinase and Polo-like kinase 1 (Plk1), were recently discovered to phosphorylate human Top2α. They are both crucial regulators in mitosis (33–35). Given the DNA segregation defect we observed after abolishing the Top2-Mus101 interaction, one of these two kinases may modulate the interaction by phosphorylating the C-terminal domain of Top2.
Top2Δ20 Abolishes the Top2-Mus101 Interaction
Although we demonstrated that two serine phosphorylation sites in the last 20 amino acids of Drosophila Top2 are required for the binding of Mus101 by in vitro assays, our cellular experiments showed that mitotic defects were only observed in Top2-deficient cells rescued by Top2Δ20 but not by Top2(2SA). It is possible that the remaining aspartate- and glutamate-rich sequence without phosphorylation may still serve as a weak binding platform for Mus101. Top2(2SA) driven by a strong metallothionein promotor might be abundant enough to achieve low-affinity binding with Mus101 and mitigate mitotic defects. The segregation defects in the Top2Δ20 mutant after prolonged culturing are possibly because of the fact that the removal of both phosphorylation sites and the flanking amino acids completely abrogates binding with Mus101 even when overexpressed.
Maintaining DNA Segregation Fidelity
Although previous studies have shown that SUMOylation of the CTD of eukaryotic Top2 is required for its localization at the centromere for DNA segregation (36, 37), we found that phosphorylation of two specific serines in Drosophila Top2 may have a role in DNA segregation. How the C-terminal domain of Top2 and its phosphorylation can promote fidelity in chromosome segregation remains to be elucidated. It is plausible that they play a key role in coordinating the state of chromosome condensation during metaphase with the mitotic apparatus for disjoining the replicated chromosomes.
After chromosomes are condensed by the condensin complex and eukaryotic Top2, sister chromatids are aligned in the mitotic plate between spindles and pulled by kinetochore-attached microtubules. To prevent aberrant segregation, the spindle assembly checkpoint monitors the proper attachment of microtubules on kinetochores and the tension build-up between sister kinetochores. Cohesin also contributes to the balancing of the pulling force by encircling sister chromatids with its dimerized ring (38). For smooth segregation without physical obstructions in anaphase, Plk1 and Aurora B kinase facilitate and regulate the release of cohesion, thus allowing chromosome segregation in a timely manner (35, 39). Eukaryotic Top2 could be phosphorylated by either Plk1 or Aurora B kinase and interact with Mus101. The inhibition of strand passage activity of phosphorylated Top2 through the binding of Mus101 is analogous to the inhibition by SUMOylation at Lys-660 to prevent recatenation of segregated chromosomes (40). It is plausible that the inhibition of decatenation activity of Top2 regulates the proper timing of chromosome disjunction only when it is appropriate. Through the phosphorylation by Plk1 or Aurora B kinase, the level of DNA-DNA catenation and cohesin-DNA interaction can be simultaneously adjusted to promote proper chromosome segregation in anaphase.
Author Contributions
Y. T. S. C., J. W., P. M., and T. S. H. designed the study. Y. T. S. C. and J. W. carried out the research. Y. T. S. C., J. W., P. M., and T. S. H. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank I-Jay Chen for initial cloning of the Mus101 expression constructs.
This work was supported by National Institute of Health Grants GM29006 (to T. S. H.) and GM45190 (to P. M.), Taiwan's Ministry of Science and Technology Grant 334122 (to T. S. H.), and the Howard Hughes Medical Institute (to P. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Top2
- topoisomerase 2
- CTD
- C-terminal domain
- BRCT
- BRCA1 C terminus
- UFB
- ultrafine anaphase bridge
- IP
- immunoprecipitation
- aa
- amino acids
- eGFP
- enhanced GFP
- BFP
- blue fluorescent protein.
References
- 1. Liu L. F., Rowe T. C., Yang L., Tewey K. M., and Chen G. L. (1983) Cleavage of DNA by mammalian DNA topoisomerase II. J. Biol. Chem. 258, 15365–15370 [PubMed] [Google Scholar]
- 2. Morrison A., and Cozzarelli N. R. (1979) Site-specific cleavage of DNA by E. coli DNA gyrase. Cell 17, 175–184 [DOI] [PubMed] [Google Scholar]
- 3. Sander M., and Hsieh T. (1983) Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem. 258, 8421–8428 [PubMed] [Google Scholar]
- 4. Baird C. L., Harkins T. T., Morris S. K., and Lindsley J. E. (1999) Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl. Acad. Sci. U.S.A. 96, 13685–13690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harkins T. T., Lewis T. J., and Lindsley J. E. (1998) Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II: 2: kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry 37, 7299–7312 [DOI] [PubMed] [Google Scholar]
- 6. Schmidt B. H., Osheroff N., and Berger J. M. (2012) Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity. Nat. Struct. Mol. Biol. 19, 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nitiss J. L. (2009) DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9, 327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen S. H., Chan N. L., and Hsieh T. S. (2013) New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 82, 139–170 [DOI] [PubMed] [Google Scholar]
- 9. Kurz E. U., Leader K. B., Kroll D. J., Clark M., and Gieseler F. (2000) Modulation of human DNA topoisomerase IIα function by interaction with 14-3-3ϵ. J. Biol. Chem. 275, 13948–13954 [DOI] [PubMed] [Google Scholar]
- 10. Stros M., Bacíková A., Polanská E., Stokrová J., and Strauss F. (2007) HMGB1 interacts with human topoisomerase IIα and stimulates its catalytic activity. Nucleic Acids Res. 35, 5001–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J., Gong Z., and Chen J. (2011) MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J. Cell Biol. 193, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamane K., Kawabata M., and Tsuruo T. (1997) A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur. J. Biochem. 250, 794–799 [DOI] [PubMed] [Google Scholar]
- 13. Saka Y., Fantes P., Sutani T., McInerny C., Creanor J., and Yanagida M. (1994) Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 13, 5319–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Germann S. M., Schramke V., Pedersen R. T., Gallina I., Eckert-Boulet N., Oestergaard V. H., and Lisby M. (2014) TopBP1/Dpb11 binds DNA anaphase bridges to prevent genome instability. J. Cell Biol. 204, 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Broderick R., Nieminuszczy J., Blackford A. N., Winczura A., and Niedzwiedz W. (2015) TOPBP1 recruits TOP2A to ultra-fine anaphase bridges to aid in their resolution. Nat. Commun. 6, 6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moyer S. E., Lewis P. W., and Botchan M. R. (2006) Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U.S.A. 103, 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boos D., Sanchez-Pulido L., Rappas M., Pearl L. H., Oliver A. W., Ponting C. P., and Diffley J. F. (2011) Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr. Biol. 21, 1152–1157 [DOI] [PubMed] [Google Scholar]
- 18. Takeishi Y., Ohashi E., Ogawa K., Masai H., Obuse C., and Tsurimoto T. (2010) Casein kinase 2-dependent phosphorylation of human Rad9 mediates the interaction between human Rad9-Hus1-Rad1 complex and TopBP1. Genes Cells 15, 761–771 [DOI] [PubMed] [Google Scholar]
- 19. Crenshaw D. G., and Hsieh T. (1993) Function of the hydrophilic carboxyl terminus of type II DNA topoisomerase from Drosophila melanogaster: I: in vitro studies. J. Biol. Chem. 268, 21328–21334 [PubMed] [Google Scholar]
- 20. Haley B., Foys B., and Levine M. (2010) Vectors and parameters that enhance the efficacy of RNAi-mediated gene disruption in transgenic Drosophila. Proc. Natl. Acad. Sci. U.S.A. 107, 11435–11440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vert J. P., Foveau N., Lajaunie C., and Vandenbrouck Y. (2006) An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics 7, 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rappas M., Oliver A. W., and Pearl L. H. (2011) Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res. 39, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Darzynkiewicz Z., Bruno S., Del Bino G., Gorczyca W., Hotz M. A., Lassota P., and Traganos F. (1992) Features of apoptotic cells measured by flow cytometry. Cytometry 13, 795–808 [DOI] [PubMed] [Google Scholar]
- 24. Sander M., Nolan J. M., and Hsieh T. (1984) A protein kinase activity tightly associated with Drosophila type II DNA topoisomerase. Proc. Natl. Acad. Sci. U.S.A. 81, 6938–6942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ackerman P., Glover C. V., and Osheroff N. (1985) Phosphorylation of DNA topoisomerase II by casein kinase II: modulation of eukaryotic topoisomerase II activity in vitro. Proc. Natl. Acad. Sci. U.S.A. 82, 3164–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chikamori K., Grabowski D. R., Kinter M., Willard B. B., Yadav S., Aebersold R. H., Bukowski R. M., Hickson I. D., Andersen A. H., Ganapathi R., and Ganapathi M. K. (2003) Phosphorylation of serine 1106 in the catalytic domain of topoisomerase II α regulates enzymatic activity and drug sensitivity. J. Biol. Chem. 278, 12696–12702 [DOI] [PubMed] [Google Scholar]
- 27. DeVore R. F., Corbett A. H., and Osheroff N. (1992) Phosphorylation of topoisomerase II by casein kinase II and protein kinase C: effects on enzyme-mediated DNA cleavage/religation and sensitivity to the antineoplastic drugs etoposide and 4′-(9-acridinylamino)methane-sulfon-m-anisidide. Cancer Res. 52, 2156–2161 [PubMed] [Google Scholar]
- 28. Qi X., Hou S., Lepp A., Li R., Basir Z., Lou Z., and Chen G. (2011) Phosphorylation and stabilization of topoisomerase IIα Protein by p38γ mitogen-activated protein kinase sensitize breast cancer cells to its poisons. J. Biol. Chem. 286, 35883–35890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu S., Shiotani B., Lahiri M., Maréchal A., Tse A., Leung C. C., Glover J. N., Yang X. H., and Zou L. (2011) ATR autophosphorylation as a molecular switch for checkpoint activation. Mol. Cell 43, 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bai L., Michael W. M., and Yan S. (2014) Importin β-dependent nuclear import of TopBP1 in ATR-Chk1 checkpoint in Xenopus egg extracts. Cell. Signal. 26, 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zegerman P., and Diffley J. F. (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445, 281–285 [DOI] [PubMed] [Google Scholar]
- 32. Kumagai A., Shevchenko A., Shevchenko A., and Dunphy W. G. (2011) Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J. Cell Biol. 193, 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morrison C., Henzing A. J., Jensen O. N., Osheroff N., Dodson H., Kandels-Lewis S. E., Adams R. R., and Earnshaw W. C. (2002) Proteomic analysis of human metaphase chromosomes reveals topoisomerase II α as an Aurora B substrate. Nucleic Acids Res. 30, 5318–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H., Wang Y., and Liu X. (2008) Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIα in cell cycle progression. J. Biol. Chem. 283, 6209–6221 [DOI] [PubMed] [Google Scholar]
- 35. Losada A., Hirano M., and Hirano T. (2002) Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 16, 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Azuma Y., Arnaoutov A., Anan T., and Dasso M. (2005) PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 24, 2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Agostinho M., Santos V., Ferreira F., Costa R., Cardoso J., Pinheiro I., Rino J., Jaffray E., Hay R. T., and Ferreira J. (2008) Conjugation of human topoisomerase 2 α with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 68, 2409–2418 [DOI] [PubMed] [Google Scholar]
- 38. Bloom K. S. (2014) Centromeric heterochromatin: the primordial segregation machine. Annu. Rev. Genet. 48, 457–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Onn I., Heidinger-Pauli J. M., Guacci V., Unal E., and Koshland D. E. (2008) Sister chromatid cohesion: a simple concept with a complex reality. Annu. Rev. Cell Dev. Biol. 24, 105–129 [DOI] [PubMed] [Google Scholar]
- 40. Ryu H., Furuta M., Kirkpatrick D., Gygi S. P., and Azuma Y. (2010) PIASy-dependent SUMOylation regulates DNA topoisomerase IIα activity. J. Cell Biol. 191, 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]