Abstract
Staphylococci are commensal bacteria that colonize the epithelial surfaces of humans and many other mammals. These bacteria can also attach to implanted medical devices and develop surface-associated biofilm communities that resist clearance by host defenses and available chemotherapies. These communities are often associated with persistent staphylococcal infections that place a tremendous burden on the healthcare system. Understanding the regulatory program that controls staphylococcal biofilm development, as well as the environmental conditions that modulate this program, has been a focal point of research in recent years. A central regulator controlling biofilm development is a peptide quorum-sensing system, also called the accessory gene regulator or agr system. In the opportunistic pathogen Staphylococcus aureus, the agr system controls production of exo-toxins and exo-enzymes essential for causing infections, and simultaneously, it modulates the ability of this pathogen to attach to surfaces and develop a biofilm, or to disperse from the biofilm state. In this review, we explore advances on the interconnections between the agr quorum-sensing system and biofilm mechanisms, and topics covered include recent findings on how different environmental conditions influence quorum sensing, the impact on biofilm development, and ongoing questions and challenges in the field. As our understanding of the quorum sensing and biofilm interconnection advances, there are growing opportunities to take advantage of this knowledge and develop therapeutic approaches to control staphylococcal infections.
Keywords: biofilm, infection, Methicillin-resistant Staphylococcus aureus (MRSA), quorum sensing, Staphylococcus aureus (S. aureus), biofilms
Introduction
The staphylococci are a large genus of Gram-positive bacteria that live as commensals of mammals (1). These bacteria colonize the skin or mucous membranes and thrive in an asymptomatic relationship with the host. The most notorious species of the genus is Staphylococcus aureus, which persistently colonizes ∼20% of the human adult population, preferably in the anterior nares and secondarily on the skin (2). Despite being a commensal, S. aureus can cause a tremendous range of disease, from simple skin infections to life-threatening ailments, such as sepsis, endocarditis, and osteomyelitis (3). Coupled with this infection diversity, antibiotic resistance levels are rising, with methicillin-resistant S. aureus (MRSA) becoming epidemic in many hospital settings, limiting available treatment options (4, 5). Adding to the challenge is the constant emergence of new strain lineages, such as the USA300 strains that have clonally expanded across the United States (6), making it difficult to keep up with the rapidly evolving nature of this pathogen.
The fact that S. aureus can colonize without harming the host is somewhat surprising considering the depth of secreted virulence factors produced by this pathogen (3, 7, 8). These include an impressive array of pore-forming toxins, degradative enzymes, superantigens, and other immunostimulatory exo-proteins. Indeed, many of these virulence factors are being produced at some level during colonization based on the presence of antibody responses (9). To tailor the virulence factor arsenal, S. aureus relies on intricate layers of regulation that control when and where these weapons are released. One of these regulatory circuits is a cell-to-cell communication system that responds to a peptide signal. When the cell density of S. aureus reaches a critical threshold (e.g. a quorum of cells), the concentration of this signal accumulates and activates a regulatory cascade referred to as “quorum sensing.” This cascade induces the production of exo-enzymes and many of the toxins that make S. aureus a more invasive pathogen (10, 11).
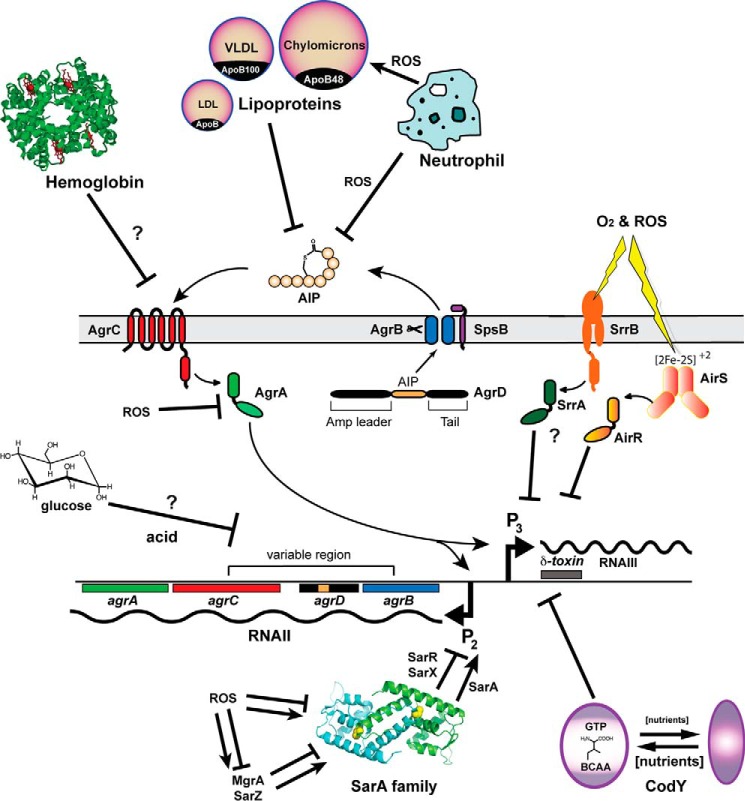
The quorum-sensing system of the staphylococci is called the accessory gene regulator or “agr” system. The central machinery of the system is encoded within the four-gene agrBDCA operon on the chromosome (Fig. 1). The first two genes encode proteins that build the quorum-sensing signal, also called an autoinducing peptide or AIP.2 AgrD is the peptide precursor of the AIP signal, and AgrB is an integral membrane endopeptidase that processes AgrD into the final structure (12, 13), with assistance from the housekeeping type I signal peptidase (14). Outside the cell, the AIP signal accumulates to levels as high as 10 μm (15), and once it overcomes the activation threshold for the AgrC receptor, the signal transduction cascade is activated. The AgrC histidine kinase self-phosphorylates upon activation, and this phosphate is transferred to the response regulator AgrA, which in turn binds four promoters on the chromosome (16). The primary output is the P3 promoter that drives RNAIII expression. The RNAIII transcript is the major effector of the system (10), and higher levels of RNAIII lead to up-regulation of exo-toxins and exo-enzymes. Some of this regulation occurs directly through RNA-protein interactions that promote translation of selected virulence factors (17), and other regulation occurs through RNAIII-dependent inhibition of the translation of the Rot repressor (18, 19). For the other three promoters, AgrA activates the expression of the P2 promoter for the agrBDCA operon (Fig. 1), leading to the autoinduction loop, and AgrA also activates expression of transcripts for the α- and β-phenol-soluble modulins (PSMs) (16).
FIGURE 1.
Schematic of the agr system in S. aureus including host and environmental factors that have been shown to modulate agr activity. The locus consists of divergent transcripts called RNAII and RNAIII, driven by the P2 and P3 promoters, respectively. The RNAII transcript encodes the core machinery of the system. AgrD is the peptide precursor of the AIP that is processed by and exported through AgrB (and the housekeeping type I signal peptidase SpsB) at the cytoplasmic membrane. Outside the cell, AIP binds to the AgrC receptor, a membrane-bound histidine kinase, which activates AgrC, causing it to auto-phosphorylate, and in turn phosphorylate the AgrA response regulator through a phosphotransfer reaction. Phosphorylated AgrA binds to the P2 and P3 promoters as well as the α-PSM and β-PSM promoters (not shown), resulting in increased transcription. The RNAIII transcript encodes δ-toxin and serves as the primary effector of the agr system by post-transcriptionally regulating the expression of numerous virulence factors. Host (hemoglobin, neutrophil ROS, and lipoproteins) and environmental factors (acid, oxygen and ROS, and nutrients) that can enhance or inhibit agr activity are indicated.
Adding to the complexity of the agr quorum-sensing system is the fact that many species within the Staphylococcus genus contain multiple variants of the system. In a S. aureus clinical isolate, only one copy of the agr system will be present on the chromosome, but the system can be any one of four types, each with the same basic components. The variation is that each system will make a unique AIP signal (e.g. AIP-I, AIP-II, AIP-III, and AIP-IV), and there will be corresponding changes in AgrB and AgrC to build and detect this signal, sometimes called the variable region (Fig. 1). Staphylococcus epidermidis has three different types of the agr system (20), whereas other Staphylococcal species have variable numbers of the system (10). Intraspecies and interspecies signaling with all the AIP types occurs and has been investigated for S. aureus and S. epidermidis (20–22).
The agr system has been the target of multiple comprehensive reviews (10, 11, 23, 24), and for more details not covered herein, the interested reader is referred to these articles. In this minireview, we will focus on the variety of environmental cues that have been demonstrated to impact quorum signaling in the staphylococci. Many of these are cues that are found in the host during colonization or infection, such as the impact of serum proteins and reactive oxygen species on agr function. How some of these agr regulatory changes modulate biofilm development will also be covered. The majority of the studies presented have been performed with S. aureus, and wherever possible, examples with other staphylococci will be included. Finally, some challenges to be overcome and future perspectives are included.
Impact of Environmental Cues on Quorum Sensing
Numerous environmental and metabolic factors such as pH, reactive oxygen species (ROS), and nutrient availability, can modulate agr quorum sensing in S. aureus (all listed in Table 1). The mechanisms by which these factors impact quorum sensing are diverse (Fig. 1), but can be organized roughly into three broad categories: (i) suppression of quorum sensing through the action of ROS, generated by innate immunity cells, upon components of the agr system; (ii) transcriptional up- or down-regulation of agr expression as a consequence of various non-agr regulatory proteins binding within the agr P2-P3 promoter region; and (iii) the up- or down-regulation of agr expression by environmental factors for which the mechanism of action is yet to be determined. It should be noted that there is a burgeoning field of “quorum-quenching” research aimed at identifying small molecule natural products capable of inhibiting quorum sensing, and this area of research has recently been subject of several recent reviews (25–27). This review will focus on the impact of host factors, nutrients, and metabolic state on quorum-sensing function, and the interconnection with biofilm development.
TABLE 1.
List of environmental cues impacting agr function
| Host/Environmental factor | Effect | Mechanism | References |
|---|---|---|---|
| Hemoglobin | Inhibit | Unknown | 60, 61 |
| Serum lipoproteins | Inhibit | Sequestration of all 4 AIP types | 30, 31, 33, 34 |
| SrrAB | Inhibit | SrrA binds agrP2P3 at low O2 concentration | 42, 43 |
| AirRS | Inhibit | Reduced DNA binding activity of AirR at low O2 concentration inhibits agr by unknown mechanism | 41 |
| CodY | Inhibit | CodY binds agrP2P3 under good nutrient availability | 55, 58, 59 |
| SarA | Enhance | Binds to agrP2P3 causing DNA bending that increases transcription | 44–49 |
| SarR, SarX | Inhibit | Compete with AgrA for binding to agrP2P3 | 44–49 |
| MgrA, SarZ, and other SarA family members | Inhibit or enhance | Modulate expression of SarA, SarR, and SarX, as well as expression of one another | 44–49 |
| Glucose | Inhibit | May be due to decrease in pH due to catabolism | 62–65 |
| Acid pH | Inhibit | Unknown | 62 |
| ROS | Inhibit | Met oxidation of AIPs I and IV | 31, 32 |
| ROS | Inhibit | Oxidize lipid component of lipoproteins, resulting in increased AIP sequestration | 30 |
| ROS | Inhibit | Cys oxidation in AgrA inhibits DNA binding | 33 |
| ROS | Inhibit or enhance | Modification of conserved Cys in SarA family of regulators alters DNA binding activity | 50, 51 |
Direct Action of Serum and ROS on agr Components
Studies of global changes in S. aureus gene expression have noted that reduced transcription of agr RNAIII is among the most pronounced effects of growth in fresh serum (28, 29), suggesting that serum contains at least one quorum-sensing inhibitory factor. Multiple lines of evidence (30–33) suggest that ROS generated by innate immunity cells are largely, if not entirely (34), responsible for this quorum-quenching activity.
There is evidence that both AgrA and AIP are sensitive to oxidative stress associated with ROS. In the case of AgrA, oxidative stress induces disulfide bond formation (between Cys-199 and Cys-228) that adversely impacts the ability of AgrA to bind to P2-P3 promoter (33). In the case of AIP, ROS can interfere with quorum sensing through two distinct mechanisms, one specific to AIP types I and IV and the other (see next section) applicable to all four AIP types. Exposure, either in vitro or in vivo, of AIPs I or IV to phagocyte-derived ROS can oxidize the side chain of the C-terminal methionine (32), rendering the pheromones unable to activate AgrC. However, the fact that agr expression has been detected, either by quantitative real-time-PCR measurement of RNAIII levels in an agr type I strain (35) or by GFP expression under control of the PSM promoter in an agr type III strain (36), would imply that ROS inhibition of AgrA or AIP for bacteria located within PMN phagosomes is of limited importance. It should be noted that expression of RNAIII, relative to that of GAPDH reference gene, increased in a multiplicity of infection (MOI)-dependent manner (35), suggesting that at higher MOIs the bacteria may overcome the ability of NADPH oxidase to generate sufficient ROS. More importantly, when NADPH oxidase was inhibited by pretreating PMNs with diphenylene iodonium, the increases in expression of RNAIII, relative to expression in post-opsonized S. aureus control bacteria, were 8–37-fold greater than the increases observed for untreated PMNs at similar time points and MOIs (35). This suggests that inhibition of AgrA and/or AIP activities by ROS may make, at least in part, a genuine contribution to the agr-inhibiting activity of serum.
The other serum inhibitory mechanism involves sequestration of AIP by lipoproteins. The details of this mechanism have been unveiled through a series of studies (30, 31, 37). Initially, it was observed that pooled human serum, but not lipoprotein-deficient serum, could inhibit quorum sensing by an agr type I reporter strain during in vitro growth (31). Additional reporter assays narrowed the source of inhibition to LDL and VLDL, and ultimately to the apolipoprotein B (ApoB) component of the lipoproteins. SPR confirmed direct binding of purified ApoB to immobilized AIP-I, indicating that sequestration of AIP-I contributes to the quorum-sensing inhibition by serum. Importantly, soluble AIP-II and AIP-IV were found to inhibit binding of ApoB to immobilized AIP-I, as measured by SPR, suggesting that sequestration of AIP by ApoB may be generally applicable to all four AIP types. A role for ROS generated by NADPH oxidase (Nox2) in the AIP sequestration mechanism was discovered during a follow-up study that investigated the applicability of this mechanism to AIP-III based quorum sensing (30). It was observed that LDL inhibited quorum sensing in an agr type III strain to a lesser extent than purified ApoB, implying that structural differences between free ApoB and ApoB in LDL may impact the ability of ApoB to bind AIP-III. Because oxidation of the lipid component of LDL was known to alter the conformation of ApoB in the LDL, the ability of oxidized LDL (oxLDL) to inhibit AIP-III based quorum sensing was measured and found to be equal to that of purified ApoB. Subsequently, it was demonstrated that reducing the amount of oxLDL in the serum of Nox2 knock-out mice correlated with increased susceptibility to infection by an agr type III strain. The general applicability of AIP sequestration was demonstrated by SPR experiments that showed direct binding of oxLDL to immobilized AIP-II or immobilized AIP-IV. Consistent with the generality of this mechanism, it was recently reported that apolipoprotein B100 in VLDL and apoB48 in chylomicrons also inhibit quorum sensing by AIP sequestration (37). Most recently, it was found that mice with hypolipidemia that reduced ApoB levels in their lungs suffered from increased morbidity and inflammation in an S. aureus pneumonia model (38), highlighting the importance of sequestration of AIP by serum lipoproteins as a mechanism of innate immunity.
In a separate study, James et al. (34) discovered that AgrC point mutations that render AgrC constitutively active overcame quorum-sensing inhibition by serum lipoproteins, leading them to conclude that “sequestration of the AIP is likely to be the only mechanism by which the host innate immune response limits agr expression at the transcriptional level.” Although this finding provides compelling evidence that AIP sequestration may be the dominant inhibitory mechanism, including the fact that oxidation can inactivate AgrA and AIP and that the activities of other regulatory proteins that bind to the P2-P3 promoter region are also sensitive to oxidation (see below), it is probably incorrect to conclude that AIP sequestration is the only innate immunity mechanism for quorum-sensing inhibition. In this regard, it is interesting to note that a study investigating the impact of serum on agr expression that failed to find reduced agr transcription (and, as discussed below, actually found that serum increased agr expression) (39) differed from the aforementioned studies (28, 29) in that the serum was inactivated by heating at 56 °C for 30 min before it was added to S. aureus cultures. Given that biochemical investigation of ApoB thermal unfolding found that ApoB undergoes temperature-induced structural transitions near 56 °C (40), the possibility is raised that Oogai et al. (39) failed to detect reduced agr expression because the ApoB in heat-inactivated serum could no longer bind AIP.
Impact of Oxygen and ROS through Non-agr Regulators
Oxygen availability and ROS generated by innate immunity cells can modulate transcription from the agr P2 and P3 promoters by altering the ability of non-agr regulatory proteins to bind this promoter region. Redox modulation of agr expression is achieved through the combined action of the AirSR (41) and SrrAB (42, 43) two-component systems, both of which reduce agr expression under conditions of low oxygen availability. In the case of AirSR, prolonged exposure to oxygen or oxidative stress destroys the [2Fe-2S] cluster in AirS that is essential for its kinase activity, which in turn alters the phosphorylation, and thus DNA binding activity, of AirR. In the case of SrrAB, the activity of SrrB is sensitive to redox regulation, but the exact mechanism by which kinase activity is modulated is not known. Additional redox regulation of agr expression may be achieved through the collective action of proteins in the SarA protein family of regulators (44–49). All the members of the SarA family share a winged helix structure and possess DNA binding activity (48). Several of the SarA family proteins, SarA, SarR, and SarX, have been shown to bind to the agr P2-P3 promoter region where they either inhibit transcription (SarR and SarX) by competing with AgrA or enhance transcription (SarA) by altering the structure of the agr P2-P3 promoter region. Other members of the family (SarZ and MgrA) either promote or inhibit agr expression by modulating the expression of the SarA family regulators that bind to the promoter region. Collectively, the SarA family of proteins constitutes a complex interconnected regulatory network, the details of which are yet to be fully worked out. Importantly, all the members of the family contain a conserved cysteine that is susceptible to both oxidation and nitrosylation that alters DNA binding activity (50, 51). Thus, the SarA family of proteins provides an additional mechanism by which oxidative or nitrosyl stress, originating either from an innate immune response or from nutritional metabolic conditions, can modulate agr expression. It has further been suggested that this cysteine can be phosphorylated, by kinase Stk1, and that this may represent a mechanism by which cell wall-targeting antibiotics may modulate agr expression (50), because the kinase activity of Stk1 is inhibited by these antibiotics.
Impact of Nutrients through Non-agr Regulators
Nutrient and growth phase regulation of agr expression appears to occur primarily through the pleotropic CodY repressor (52–58). CodY provides a mechanism for Gram-positive bacteria to adapt to conditions of starvation (54). Under conditions of good nutrient availability, branched chain amino acids and GTP are bound by CodY, which promotes binding of CodY to DNA, where it inhibits transcription by competing with RNA polymerase or preventing transcriptional elongation. When nutrients are depleted, branched chain amino acids and GTP are not available for binding to CodY, and in turn, CodY no longer binds DNA and repression of transcription is relieved (54). In the case of the agr locus, deletion of CodY has been shown to result in enhanced transcription (58, 59), and overexpression of CodY has been shown to result in decreased expression, as would be expected from a CodY-regulated locus. Consistent with these findings, a CodY binding sequence was identified within the agr P2-P3 promoter region (55). Taken together, CodY is likely important for growth phase regulation of agr expression.
Impact of Miscellaneous Factors by Unknown Mechanisms
There are a number of environmental factors whose presence reduces transcription of the agr locus through unknown or not well understood mechanisms. Among these factors are iron (39), hemoglobin (60, 61), and glucose (62–65). In the aforementioned study that failed to detect repression of agr in the presence of serum (39), the addition of heat-inactivated serum actually resulted in increased agr transcription. Interestingly, inclusion of FeCl3 in the medium eliminated the increase in transcription, suggesting that (in the absence of AIP sequestration by serum lipoproteins) the normally low iron concentration of serum may actually be permissive toward agr transcription and that the addition of iron somehow suppresses agr transcription. If this inference is true, the mechanism by which iron would inhibit agr transcription is unknown. In the case of hemoglobin (60, 61), it was found that the addition of hemoglobin, globin chains, or even peptides derived from globin chains could reduce agr transcription. The underlying mechanism by which globin-derived peptides inhibit agr expression is unknown. In the case of glucose, it has long been known that the addition of glucose suppresses agr transcription (62), but interestingly, inhibition was not relieved (63) by deletion of catabolite control protein A (CcpA), which is consistent with the lack of a catabolite-responsive element at the agr locus. It appears that the decrease in pH that is associated with the catabolism of glucose may be responsible for glucose-associated inhibition, because inhibition could be reduced by buffering the growth media or by growing the bacteria in a fermenter in which pH and glucose concentration could be held constant (62). Moreover, it was found that lowering the pH of the growth media in the absence of glucose could mimic the inhibition observed in the presence of glucose (65). Exactly how reduced pH inhibits agr transcription remains unclear. Perhaps even more mysterious was the finding that there was no transcription of agr in the ccpA knock-out mutant (63), implying the possible existence of an agr transcriptional repressor whose own expression is repressed by CcpA. Whether such a repressor, if it exists, is among the known repressors of agr transcription, or is a novel protein, is unknown. Given that serum contains thousands of proteins (66) and small molecule metabolites (67) that can interact with S. aureus, it seems highly likely that other factors capable of modulating agr expression will be found.
Quorum Sensing and Biofilm Interconnections
The interconnection between the agr quorum-sensing system and staphylococcal biofilm development has been a focal point of many studies. Through in vitro studies, there was some early surprise in the field that inactivation of the agr system either had little impact (68), or in fact enhanced adherence and biofilm development in S. aureus and S. epidermidis (69, 70). The prevailing view in the broader bacterial biofilm field is that quorum sensing is required to build a biofilm (71), making these observations somewhat of an anomaly. These early staphylococcal studies were followed by a time course analysis by Yarwood et al. (72) that demonstrated that pockets of an S. aureus biofilm would activate the agr system, and these regions of activated cells would leave the biofilm to seed a new site. When the level of the AIP signal was controlled, the entire biofilm could be dispersed and resensitized to antibiotics (64). This mechanism was conserved across strains and even functions in the emerging S. aureus USA300 isolates (64, 73). Similar observations of quorum-sensing control over dispersal of biofilm cells was also demonstrated in S. epidermidis (74), indicating that this is a conserved mechanism across the staphylococci.
Unraveling the agr-mediated biofilm dispersal mechanism has drawn considerable attention (75, 76). The agr regulatory system controls a myriad of secreted toxins and enzymes, and identifying the exact agents responsible for dispersal has represented a challenge. Early studies suggested that surfactant properties of δ-toxin could be responsible (69), considering that many bacterially produced surfactants have known anti-biofilm effects (77). As it became evident that δ-toxin was part of a larger family of PSM peptides (78), further studies demonstrated that multiple PSMs are important in modulating S. aureus biofilm structure (79). These studies were extended in S. epidermidis to demonstrate that the agr system and PSMs are necessary to disperse from a biofilm and spread to new sites during infection (74). In contrast, other studies have pointed to the secreted proteases as being important mediators of the biofilm inhibitory properties of quorum sensing (64). This observation stems from the fact that most clinical isolates of S. aureus make biofilms that are protease-labile (45, 64, 73, 80), and S. aureus secretes many extracellular proteases that are known to self-cleave surface adhesins (81, 82). Biochemical and genetic studies have narrowed the proteases responsible to the staphopains (83), which are two cysteine proteases secreted by S. aureus. However, the target(s) of these enzymes have not yet been identified, although it seems likely to be one of the many surface proteins that have been linked to biofilm formation (84). Taken together, it is known that both the PSMs and secreted proteases are tightly regulated by the agr system (16, 18), and thus it seems probable that coordinated effort of these factors is responsible for quorum sensing-mediated biofilm dispersal.
Challenges in the Field and Future Perspectives
Staphylococcal quorum-sensing responses and the interconnections with biofilm development have drawn considerable interest from scientists and likely will continue to do so going forward, given that these two social behaviors are almost universally linked among bacteria (71). Although extensive knowledge regarding the dynamics of quorum sensing during in vitro culture has been gained, how quorum sensing functions during the natural commensal state of staphylococci remains almost completely unknown. Considering that the predominant state of these bacteria is not as a pathogen, it seems likely that the agr system may have a role in colonization. Consistent with this possibility, the agr system was recently found necessary for effective colonization of the skin by S. epidermidis (20). In the case of S. aureus, how quorum-sensing kinetics are controlled in ways that best suit the commensal lifestyle is unclear, and gaining insight into this dynamic may be particularly challenging given the profusion of inflammatory and tissue-damaging secreted virulence factors made by this pathogen. Although colonization studies (7, 85) demonstrate that agr is repressed in this state, and as outlined above, many studies indicate that numerous host factors (e.g. serum components, ROS, low pH) are capable of repressing agr function, the reference state for these studies was growth under idealized laboratory culture conditions. Therefore the possibility remains that the reduced level of agr function within the host is adequate for the needs of the S. aureus cells during colonization. Although this possibility has not been examined during colonization, it has been addressed in various infection studies, and indeed, time course imaging studies have demonstrated that agr does activate in waves above baseline within an abscess (86). Also, following neutrophil influx to an infection, the first responders to S. aureus, the neutrophils, phagocytose the invading pathogen, and the agr system activates within the confined space of the phagosome (35, 36). Adding another layer of complexity, S. aureus can form intricate interactions with host proteins such as fibrinogen and agglutinate as an immune evasion strategy (87, 88), and hyperactivation of the agr system has been observed within these clumps (89), potentially due to the increased local concentration of the AIP signal. Thus, there is a tremendous amount of varying information concerning host impact on quorum-sensing function and staphylococcal lifestyle, and the context and questions being asked are important to understanding of this complex behavior.
Deciphering the role of quorum sensing during chronic biofilm infections is also complicated by seemingly contradictory observations between infection models and in vitro biofilm experiments. In osteomyelitis, agr mutants display a decreased ability to establish infection (90), and similar observations were made with agr mutants using a model of infective endocarditis (91). Time course imaging studies within an endocarditis vegetation show that the agr system does indeed activate during the infection (92). Taken together, these animal studies suggest that agr function is necessary for developing a chronic biofilm infection. These findings seem in direct contrast to the in vitro biofilm studies (outlined above) that demonstrate that biofilms only form on abiotic materials when agr is inhibited, or disperse when agr is activated. However, all of these in vitro biofilm studies were done in the absence of host defenses, and many of the critical toxins, superantigens, and exo-enzymes needed to subvert these defenses are in fact regulated by the agr system (10). Thus, there are conflicting forces at work within a biofilm infection, with agr-regulated secreted virulence factors being necessary for staphylococcal survival in the host, while some of these same factors can inhibit biofilm structure. Going forward, the function and dynamics of quorum sensing during a biofilm infection are in clear need of further investigation, and the preliminary observations suggest that the paradigms from in vitro studies may not hold once in the host. Future progress in understanding the role of agr quorum sensing in chronic biofilm infection models and colonization studies will benefit from improved methodologies for quantifying agr quorum-sensing kinetics in vivo (86, 93).
How does all of this knowledge help develop therapies for treating staphylococcal infections? With the rising resistance levels in S. aureus, there is a pressing need for innovative approaches to treat this pathogen to maintain antibiotic stewardship (94). The agr system represents a compelling target for therapeutic development based on its critical nature during infection (25–27), but complexities in the literature and the early biofilm studies have led some to question this approach (95). However, it is increasingly clear that in most types of infections, acute or chronic, the presence of a functional agr system is important and that agr activity during infection can be detected. It seems possible that the ability of S. aureus to agglutinate with fibrinogen and other host components can protect agr signaling and maintain a functional quorum-sensing system during infection (87–89), suggesting that the system evolved to stay active in the host, likely to help staphylococci fight the immune system. The anomaly to this condition is the presence of a foreign body where staphylococcal biofilms can form and the presence of agr-negative strains is frequent (70), implying that quorum-sensing inhibition could actually enhance the biofilm infection (95). Whether this is always true remains to be determined, but it is well known that clearance of a foreign body infection requires a more involved treatment approach than other types of infections (96, 97). Considering that the majority of S. aureus infections are treatable skin infections, and not on implanted materials, it seems prudent to continue developing alternative strategies to prevent or clear these infections and preserve the most critical antibiotics for life-threatening ailments (94).
Tremendous progress in our understanding of staphylococcal quorum sensing and biofilm development has been made in recent years, but important questions remain. Going forward, studies will need to address how quorum sensing functions in the colonization state and during chronic infections, and the development of improved real-time animal imaging approaches may be necessary to advance the field. How other commensal flora impact staphylococcal quorum sensing and biofilm development has also received little attention, expect for some initial examples (98), despite the tremendous interest in microbiome studies (99). More careful analysis of the contribution of host components to these various mechanisms will also be critical. Addressing these shortcomings in the field, as well as limiting the current biases from in vitro studies, will be important in future studies to take the field further and to use this information for improving treatment options for antibiotic-resistant staphylococcal infections.
This work was supported by National Institutes of Health Grant AI083211 (to A. R. H.) and U.S. Department of Veterans Affairs Grant I01 BX002711 (to A. R. H.). This is the fourth article in the Thematic Minireview series “Biofilms.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- AIP
- autoinducing peptide
- PSM
- phenol-soluble modulin
- ROS
- reactive oxygen species
- MOI
- multiplicity of infection
- PMN
- polymorphonuclear neutrophil
- ApoB
- apolipoprotein B
- oxLDL
- oxidized LDL
- CcpA
- catabolite control protein A.
References
- 1.Becker K., Heilmann C., and Peters G. (2014) Coagulase-negative staphylococci. Clin. Microbiol. Rev. 27, 870–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wertheim H. F., Melles D. C., Vos M. C., van Leeuwen W., van Belkum A., Verbrugh H. A., and Nouwen J. L. (2005) The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5, 751–762 [DOI] [PubMed] [Google Scholar]
- 3.Lowy F. D. (1998) Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 4.DeLeo F. R., Otto M., Kreiswirth B. N., and Chambers H. F. (2010) Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375, 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLeo F. R., and Chambers H. F. (2009) Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J. Clin. Invest. 119, 2464–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy A. D., Otto M., Braughton K. R., Whitney A. R., Chen L., Mathema B., Mediavilla J. R., Byrne K. A., Parkins L. D., Tenover F. C., Kreiswirth B. N., Musser J. M., and DeLeo F. R. (2008) Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc. Natl. Acad. Sci. U.S.A. 105, 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiedrowski M. R., Paharik A. E., Ackermann L. W., Shelton A. U., Singh S. B., Starner T. D., and Horswill A. R. (2016) Development of an in vitro colonization model to investigate Staphylococcus aureus interactions with airway epithelia. Cell. Microbiol. 18, 720–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon R. J., and Lowy F. D. (2008) Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46, Suppl. 5, S350–S359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtfreter S., Nguyen T. T., Wertheim H., Steil L., Kusch H., Truong Q. P., Engelmann S., Hecker M., Völker U., van Belkum A., and Bröker B. M. (2009) Human immune proteome in experimental colonization with Staphylococcus aureus. Clin. Vaccine Immunol. 16, 1607–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thoendel M., Kavanaugh J. S., Flack C. E., and Horswill A. R. (2011) Peptide signaling in the staphylococci. Chem. Rev. 111, 117–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novick R. P., and Geisinger E. (2008) Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564 [DOI] [PubMed] [Google Scholar]
- 12.Thoendel M., and Horswill A. R. (2013) Random mutagenesis and topology analysis of the autoinducing peptide biosynthesis proteins in Staphylococcus aureus. Mol. Microbiol. 87, 318–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thoendel M., and Horswill A. R. (2009) Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J. Biol. Chem. 284, 21828–21838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kavanaugh J. S., Thoendel M., and Horswill A. R. (2007) A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol. Microbiol. 65, 780–798 [DOI] [PubMed] [Google Scholar]
- 15.Junio H. A., Todd D. A., Ettefagh K. A., Ehrmann B. M., Kavanaugh J. S., Horswill A. R., and Cech N. B. (2013) Quantitative analysis of autoinducing peptide I (AIP-I) from Staphylococcus aureus cultures using ultrahigh performance liquid chromatography-high resolving power mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 930, 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Queck S. Y., Jameson-Lee M., Villaruz A. E., Bach T. H., Khan B. A., Sturdevant D. E., Ricklefs S. M., Li M., and Otto M. (2008) RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morfeldt E., Taylor D., von Gabain A., and Arvidson S. (1995) Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14, 4569–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mootz J. M., Benson M. A., Heim C. E., Crosby H. A., Kavanaugh J. S., Dunman P. M., Kielian T., Torres V. J., and Horswill A. R. (2015) Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol. Microbiol. 96, 388–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geisinger E., Adhikari R. P., Jin R., Ross H. F., and Novick R. P. (2006) Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 61, 1038–1048 [DOI] [PubMed] [Google Scholar]
- 20.Olson M. E., Todd D. A., Schaeffer C. R., Paharik A. E., Van Dyke M. J., Büttner H., Dunman P. M., Rohde H., Cech N. B., Fey P. D., and Horswill A. R. (2014) Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J. Bacteriol. 196, 3482–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto M., Echner H., Voelter W., and Götz F. (2001) Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69, 1957–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji G., Beavis R., and Novick R. P. (1997) Bacterial interference caused by autoinducing peptide variants. Science 276, 2027–2030 [DOI] [PubMed] [Google Scholar]
- 23.Yarwood J. M., and Schlievert P. M. (2003) Quorum sensing in Staphylococcus infections. J. Clin. Invest. 112, 1620–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le K. Y., and Otto M. (2015) Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 6, 1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cech N. B., and Horswill A. R. (2013) Small-molecule quorum quenchers to prevent Staphylococcus aureus infection. Future Microbiol. 8, 1511–1514 [DOI] [PubMed] [Google Scholar]
- 26.Gray B., Hall P., and Gresham H. (2013) Targeting agr- and agr-like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. Sensors (Basel) 13, 5130–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon C. P., Williams P., and Chan W. C. (2013) Attenuating Staphylococcus aureus virulence gene regulation: a medicinal chemistry perspective. J. Med. Chem. 56, 1389–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yarwood J. M., McCormick J. K., Paustian M. L., Kapur V., and Schlievert P. M. (2002) Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184, 1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malachowa N., Whitney A. R., Kobayashi S. D., Sturdevant D. E., Kennedy A. D., Braughton K. R., Shabb D. W., Diep B. A., Chambers H. F., Otto M., and DeLeo F. R. (2011) Global changes in Staphylococcus aureus gene expression in human blood. PLoS ONE 6, e18617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall P. R., Elmore B. O., Spang C. H., Alexander S. M., Manifold-Wheeler B. C., Castleman M. J., Daly S. M., Peterson M. M., Sully E. K., Femling J. K., Otto M., Horswill A. R., Timmins G. S., and Gresham H. D. (2013) Nox2 modification of LDL is essential for optimal apolipoprotein B-mediated control of agr type III Staphylococcus aureus quorum-sensing. PLoS Pathog. 9, e1003166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson M. M., Mack J. L., Hall P. R., Alsup A. A., Alexander S. M., Sully E. K., Sawires Y. S., Cheung A. L., Otto M., and Gresham H. D. (2008) Apolipoprotein B is an innate barrier against invasive Staphylococcus aureus infection. Cell Host Microbe 4, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothfork J. M., Timmins G. S., Harris M. N., Chen X., Lusis A. J., Otto M., Cheung A. L., and Gresham H. D. (2004) Inactivation of a bacterial virulence pheromone by phagocyte-derived oxidants: new role for the NADPH oxidase in host defense. Proc. Natl. Acad. Sci. U.S.A. 101, 13867–13872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun F., Liang H., Kong X., Xie S., Cho H., Deng X., Ji Q., Zhang H., Alvarez S., Hicks L. M., Bae T., Luo C., Jiang H., and He C. (2012) Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. Proc. Natl. Acad. Sci. U.S.A. 109, 9095–9100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James E. H., Edwards A. M., and Wigneshweraraj S. (2013) Transcriptional downregulation of agr expression in Staphylococcus aureus during growth in human serum can be overcome by constitutively active mutant forms of the sensor kinase AgrC. FEMS Microbiol. Lett. 349, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang Y. Y., Schwartz J., Thoendel M., Ackermann L. W., Horswill A. R., and Nauseef W. M. (2010) agr-dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J. Innate Immun. 2, 546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surewaard B. G., Nijland R., Spaan A. N., Kruijtzer J. A., de Haas C. J, and van Strijp J. A. (2012) Inactivation of staphylococcal phenol soluble modulins by serum lipoprotein particles. PLoS Pathog. 8, e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elmore B. O., Triplett K. D., and Hall P. R. (2015) Apolipoprotein B48, the structural component of chylomicrons, is sufficient to antagonize Staphylococcus aureus quorum-sensing. PLoS ONE 10, e0125027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manifold-Wheeler B. C., Elmore B. O., Triplett K. D., Castleman M. J., Otto M., and Hall P. R. (2016) Serum lipoproteins are critical for pulmonary innate defense against Staphylococcus aureus quorum sensing. J. Immunol. 196, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oogai Y., Matsuo M., Hashimoto M., Kato F., Sugai M., and Komatsuzawa H. (2011) Expression of virulence factors by Staphylococcus aureus grown in serum. Appl. Environ. Microbiol. 77, 8097–8105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh M. T., and Atkinson D. (1990) Calorimetric and spectroscopic investigation of the unfolding of human apolipoprotein B. J. Lipid Res. 31, 1051–1062 [PubMed] [Google Scholar]
- 41.Sun F., Ji Q., Jones M. B., Deng X., Liang H., Frank B., Telser J., Peterson S. N., Bae T., and He C. (2012) AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J. Am. Chem. Soc. 134, 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilde A. D., Snyder D. J., Putnam N. E., Valentino M. D., Hammer N. D., Lonergan Z. R., Hinger S. A., Aysanoa E. E., Blanchard C., Dunman P. M., Wasserman G. A., Chen J., Shopsin B., Gilmore M. S., Skaar E. P., and Cassat J. E. (2015) Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog. 11, e1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yarwood J. M., McCormick J. K., and Schlievert P. M. (2001) Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183, 1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyes D., Andrey D. O., Monod A., Kelley W. L., Zhang G., and Cheung A. L. (2011) Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J. Bacteriol. 193, 6020–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lauderdale K. J., Boles B. R., Cheung A. L., and Horswill A. R. (2009) Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77, 1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheung A. L., Nishina K. A., Trotonda M. P., and Tamber S. (2008) The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40, 355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manna A. C., and Cheung A. L. (2006) Transcriptional regulation of the agr locus and the identification of DNA binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol. Microbiol. 60, 1289–1301 [DOI] [PubMed] [Google Scholar]
- 48.Cheung A. L., and Zhang G. (2002) Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7, d1825–1842 [DOI] [PubMed] [Google Scholar]
- 49.Chien Y., and Cheung A. L. (1998) Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 273, 2645–2652 [DOI] [PubMed] [Google Scholar]
- 50.Sun F., Ding Y., Ji Q., Liang Z., Deng X., Wong C. C., Yi C., Zhang L., Xie S., Alvarez S., Hicks L. M., Luo C., Jiang H., Lan L., and He C. (2012) Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 109, 15461–15466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poor C. B., Chen P. R., Duguid E., Rice P. A., and He C. (2009) Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J. Biol. Chem. 284, 23517–23524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montgomery C. P., Boyle-Vavra S., Roux A., Ebine K., Sonenshein A. L., and Daum R. S. (2012) CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect. Immun. 80, 2382–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivera F. E., Miller H. K., Kolar S. L., Stevens S. M. Jr., and Shaw L. N. (2012) The impact of CodY on virulence determinant production in community-associated methicillin-resistant Staphylococcus aureus. Proteomics 12, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stenz L., Francois P., Whiteson K., Wolz C., Linder P., and Schrenzel J. (2011) The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol. Med. Microbiol. 62, 123–139 [DOI] [PubMed] [Google Scholar]
- 55.Majerczyk C. D., Dunman P. M., Luong T. T., Lee C. Y., Sadykov M. R., Somerville G. A., Bodi K., and Sonenshein A. L. (2010) Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192, 2861–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brinsmade S. R., Kleijn R. J., Sauer U., and Sonenshein A. L. (2010) Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J. Bacteriol. 192, 6357–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pohl K., Francois P., Stenz L., Schlink F., Geiger T., Herbert S., Goerke C., Schrenzel J., and Wolz C. (2009) CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191, 2953–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Majerczyk C. D., Sadykov M. R., Luong T. T., Lee C., Somerville G. A., and Sonenshein A. L. (2008) Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190, 2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roux A., Todd D. A., Velázquez J. V., Cech N. B., and Sonenshein A. L. (2014) CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J. Bacteriol. 196, 1184–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pynnonen M., Stephenson R. E., Schwartz K., Hernandez M., and Boles B. R. (2011) Hemoglobin promotes Staphylococcus aureus nasal colonization. PLoS Pathog. 7, e1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schlievert P. M., Case L. C., Nemeth K. A., Davis C. C., Sun Y., Qin W., Wang F., Brosnahan A. J., Mleziva J. A., Peterson M. L., and Jones B. E. (2007) Alpha and beta chains of hemoglobin inhibit production of Staphylococcus aureus exotoxins. Biochemistry 46, 14349–14358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Regassa L. B., Novick R. P., and Betley M. J. (1992) Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 60, 3381–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seidl K., Stucki M., Ruegg M., Goerke C., Wolz C., Harris L., Berger-Bächi B., and Bischoff M. (2006) Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50, 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boles B. R., and Horswill A. R. (2008) agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4, e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weinrick B., Dunman P. M., McAleese F., Murphy E., Projan S. J., Fang Y., and Novick R. P. (2004) Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186, 8407–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adkins J. N., Varnum S. M., Auberry K. J., Moore R. J., Angell N. H., Smith R. D., Springer D. L., and Pounds J. G. (2002) Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics 1, 947–955 [DOI] [PubMed] [Google Scholar]
- 67.Psychogios N., Hau D. D., Peng J., Guo A. C., Mandal R., Bouatra S., Sinelnikov I., Krishnamurthy R., Eisner R., Gautam B., Young N., Xia J., Knox C., Dong E., Huang P., et al. (2011) The human serum metabolome. PLoS ONE 6, e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beenken K. E., Blevins J. S., and Smeltzer M. S. (2003) Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71, 4206–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vuong C., Saenz H. L., Götz F., and Otto M. (2000) Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182, 1688–1693 [DOI] [PubMed] [Google Scholar]
- 70.Vuong C., Kocianova S., Yao Y., Carmody A. B., and Otto M. (2004) Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190, 1498–1505 [DOI] [PubMed] [Google Scholar]
- 71.Parsek M. R., and Greenberg E. P. (2005) Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13, 27–33 [DOI] [PubMed] [Google Scholar]
- 72.Yarwood J. M., Bartels D. J., Volper E. M., and Greenberg E. P. (2004) Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lauderdale K. J., Malone C. L., Boles B. R., Morcuende J., and Horswill A. R. (2010) Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J. Orthop. Res. 28, 55–61 [DOI] [PubMed] [Google Scholar]
- 74.Wang R., Khan B. A., Cheung G. Y., Bach T. H., Jameson-Lee M., Kong K. F., Queck S. Y., and Otto M. (2011) Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest. 121, 238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boles B. R., and Horswill A. R. (2011) Staphylococcal biofilm disassembly. Trends Microbiol. 19, 449–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lister J. L., and Horswill A. R. (2014) Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 4, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banat I. M., De Rienzo M. A., and Quinn G. A. (2014) Microbial biofilms: biosurfactants as antibiofilm agents. Appl. Microbiol. Biotechnol. 98, 9915–9929 [DOI] [PubMed] [Google Scholar]
- 78.Peschel A., and Otto M. (2013) Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 11, 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Periasamy S., Joo H. S., Duong A. C., Bach T. H., Tan V. Y., Chatterjee S. S., Cheung G. Y., and Otto M. (2012) How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1281–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O'Neill E., Pozzi C., Houston P., Smyth D., Humphreys H., Robinson D. A., and O'Gara J. P. (2007) Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 45, 1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karlsson A., Saravia-Otten P., Tegmark K., Morfeldt E., and Arvidson S. (2001) Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69, 4742–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McGavin M. J., Zahradka C., Rice K., and Scott J. E. (1997) Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect. Immun. 65, 2621–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mootz J. M., Malone C. L., Shaw L. N., and Horswill A. R. (2013) Staphopains modulate Staphylococcus aureus biofilm integrity. Infect. Immun. 81, 3227–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Speziale P., Pietrocola G., Foster T. J., and Geoghegan J. A. (2014) Protein-based biofilm matrices in Staphylococci. Front. Cell. Infect. Microbiol. 4, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burian M., Rautenberg M., Kohler T., Fritz M., Krismer B., Unger C., Hoffmann W. H., Peschel A., Wolz C., and Goerke C. (2010) Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J. Infect. Dis. 201, 1414–1421 [DOI] [PubMed] [Google Scholar]
- 86.Wright J. S. 3rd, Jin R., and Novick R. P. (2005) Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. U.S.A. 102, 1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thammavongsa V., Kim H. K., Missiakas D., and Schneewind O. (2015) Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 13, 529–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walker J. N., Crosby H. A., Spaulding A. R., Salgado-Pabón W., Malone C. L., Rosenthal C. B., Schlievert P. M., Boyd J. M., and Horswill A. R. (2013) The Staphylococcus aureus ArlRS two-component system is a novel regulator of agglutination and pathogenesis. PLoS Pathog. 9, e1003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rothfork J. M., Dessus-Babus S., Van Wamel W. J., Cheung A. L., and Gresham H. D. (2003) Fibrinogen depletion attenuates Staphylococcus aureus infection by preventing density-dependent virulence gene up-regulation. J. Immunol. 171, 5389–5395 [DOI] [PubMed] [Google Scholar]
- 90.Gillaspy A. F., Hickmon S. G., Skinner R. A., Thomas J. R., Nelson C. L., and Smeltzer M. S. (1995) Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63, 3373–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheung A. L., Eberhardt K. J., Chung E., Yeaman M. R., Sullam P. M., Ramos M., and Bayer A. S. (1994) Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Invest. 94, 1815–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiong Y. Q., Van Wamel W., Nast C. C., Yeaman M. R., Cheung A. L., and Bayer A. S. (2002) Activation and transcriptional interaction between agr RNAII and RNAIII in Staphylococcus aureus in vitro and in an experimental endocarditis model. J. Infect. Dis. 186, 668–677 [DOI] [PubMed] [Google Scholar]
- 93.Quave C. L., and Horswill A. R. (2014) Flipping the switch: tools for detecting small molecule inhibitors of staphylococcal virulence. Front. Microbiol. 5, 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spellberg B., Bartlett J. G., and Gilbert D. N. (2013) The future of antibiotics and resistance. N. Engl. J. Med. 368, 299–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Otto M. (2004) Quorum-sensing control in Staphylococci – a target for antimicrobial drug therapy? FEMS Microbiol. Lett. 241, 135–141 [DOI] [PubMed] [Google Scholar]
- 96.Kiedrowski M. R., and Horswill A. R. (2011) New approaches for treating staphylococcal biofilm infections. Ann. N.Y. Acad. Sci. 1241, 104–121 [DOI] [PubMed] [Google Scholar]
- 97.Zimmerli W., Trampuz A., and Ochsner P. E. (2004) Prosthetic-joint infections. N. Engl. J. Med. 351, 1645–1654 [DOI] [PubMed] [Google Scholar]
- 98.Wollenberg M. S., Claesen J., Escapa I. F., Aldridge K. L., Fischbach M. A., and Lemon K. P. (2014) Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. MBio 5, e01286–01214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cho I., and Blaser M. J. (2012) The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]