Abstract
Impaired estrogen receptor α(ERα) action promotes obesity and metabolic dysfunction in humans and mice; however, the mechanisms underlying these phenotypes remain unknown. Considering that skeletal muscle is a primary tissue responsible for glucose disposal and oxidative metabolism, we established that reduced ERαexpression in muscle is associated with glucose intolerance and adiposity in women and female mice. To test this relationship, we generated muscle-specific ERαknockout (MERKO) mice. Impaired glucose homeostasis and increased adiposity were paralleled by diminished muscle oxidative metabolism and bioactive lipid accumulation in MERKO mice. Aberrant mitochondrial morphology, overproduction of reactive oxygen species, and impairment in basal and stress-induced mitochondrial fission dynamics, driven by imbalanced protein kinase A–regulator of calcineurin 1–calcineurin signaling through dynamin-related protein 1, tracked with reduced oxidative metabolism in MERKO muscle. Although muscle mitochondrial DNA (mtDNA) abundance was similar between the genotypes, ERαdeficiency diminished mtDNA turnover by a balanced reduction in mtDNA replication and degradation. Our findings indicate the retention of dysfunctional mitochondria in MERKO muscle and implicate ERαin the preservation of mitochondrial health and insulin sensitivity as a defense against metabolic disease in women.
INTRODUCTION
Insulin resistance is a central underpinning in the pathogenesis of type 2 diabetes (T2D) and is a defining feature of the metabolic syndrome, a clustering of abnormalities including obesity, glucose intolerance, dyslipidemia, and hypertension (1). Although the incidence of T2D is higher in men than in premenopausal women of a similar age and body mass index (BMI) (2), the mechanisms underlying female protection against metabolic dysfunction and insulin resistance remain unclear (3–5). Unfortunately, after menopause, the incidence of metabolic-related chronic disease increases significantly. Similarly, inactivating mutations in the estrogen receptor α gene (ESR1/Esr1) in both humans and mice recapitulate metabolic dysfunction and heightened disease risk. Although strong relationships between whole-body estrogen action, oxidative function, and insulin sensitivity are now well established (6–8), our understanding of the tissue-specific molecular actions of estrogen and its α form receptor (ERα) in metabolic tissues requires further interrogation.
Skeletal muscle is a primary tissue responsible for oxidative metabolism and insulin-stimulated glucose disposal. Women and female rodents show enhanced insulin sensitivity in skeletal muscle compared with male counterparts when insulin action is normalized to muscle mass (9). In addition, muscle from female human subjects exhibits a higher mitochondrial abundance with enhanced antioxidant scavenging capacity compared with muscle from men and male rodents (10). Under various stress conditions, mitochondria produce excessive amounts of reactive oxygen species (ROS) known to attack lipid bilayers, mutate DNA, and alter the activity of specific enzymes critical for the maintenance of oxidative function (11–14), and female muscle is refractory to these effects of oxidative stress. Secondary to oxidant scavenging, mitochondrial fission-fusion events are engaged to eliminate and replace damaged mitochondrial components as well as stimulate the constant mixing of mitochondria retained in the mitochondrial network (15, 16). In contrast to rapidly dividing cells, which are known to activate apoptosis as a means of quality control, long-lived postmitotic cells, including myocytes, require a more sophisticated mitochondrial surveillance system to preserve cellular health. Thus, effective elimination of damaged mitochondria incapable of coping with changing energy demands is thought to abate inflammation and cellular aging (11, 17).
Here, we tested the impact of skeletal muscle–specific ERα deletion on mitochondrial quality and function in relation to metabolic health. We found that mice with a skeletal muscle–specific ERα knockout (MERKO) displayed obesity and profound skeletal muscle insulin resistance, phenocopying the whole-body ERα homozygous null model previously described by our laboratory (6). Mitochondrial dysfunction in MERKO muscle was distinguished by reduced basal and maximally stimulated oxygen consumption, impaired calcium buffering capacity, and excessive production of ROS. These defects were paralleled by reduced mitochondrial fission and macroautophagy signaling in part as a consequence of imbalanced protein kinaseA (PKA)–regulator of calcineurin 1 (RCAN1)–calcineurin signaling through dynamin-related protein 1 (Drp1). Although mitochondrial DNA (mtDNA) content was similar between the genotypes under basal conditions, a coordinated reduction in mtDNA replication and degradation was observed in ERα-deficient muscle.
Collectively, our data support the notion that ERα promotes mtDNA replication and the turnover of dysfunctional mitochondria in skeletal muscle. In MERKO mice, the retention of damaged mitochondria harboring mutant DNA likely contributes to muscle oxidative stress and inflammation culminating in skeletal muscle insulin resistance. These observations, together with our findings of markedly reduced ERα expression levels in muscle from women who display the metabolic syndrome (MetSyn), underscore the clinical relevance of our work and the central role of muscle ERα in protecting against metabolic disease.
RESULTS
Skeletal muscle ERαexpression correlates with metabolic health in females
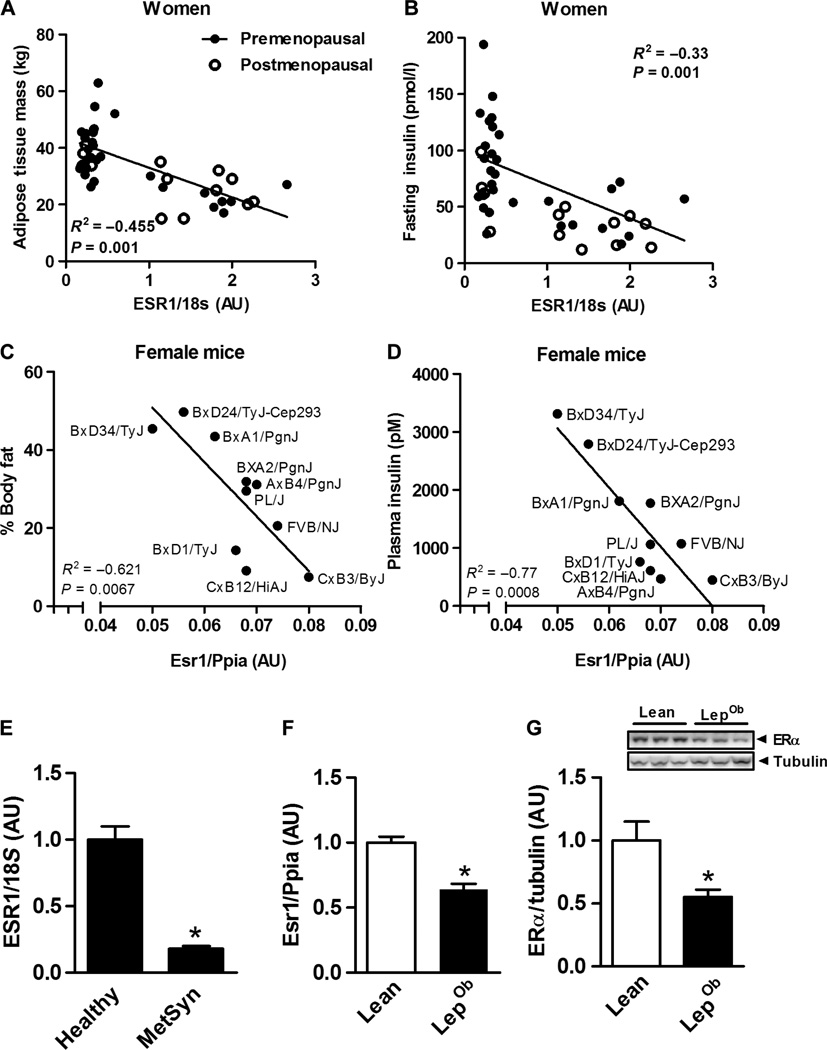
To establish the clinical rationale for our studies in rodents and cells, we first determined the relation between natural variation in muscle ESR1 expression and metabolic phenotypic traits of women. We found that adiposity and fasting insulin are inversely associated with muscle ESR1 expression in women (Fig. 1, A and B), and this relation was phenocopied in a variety of inbred strains of female mice (Fig. 1, C and D). Muscle ESR1 expression was markedly reduced (P = 0.0001) in women who displayed clinical features of MetSyn (Fig. 1E and table S1), and this finding was reproduced in genetically obese mice compared with lean controls (Fig. 1, F and G).
Fig. 1. Skeletal muscle ERαexpression correlates with metabolic health in females.
(A and B) Inverse relationship between Esr1 (natural variation in expression) versus adiposity and fasting insulin in women (n = 42; •, premenopausal n = 29, postmenopausal n = 13). AU, arbitrary units. (C and D) In 10 strains of female mice (•, n = 4 mice per indicated strain; age, 16 weeks), detected by Pearson’s correlation test, *P < 0.05 (Ppia, peptidylprolyl isomerase A housekeeping gene, for comparison]. (E) Muscle ESR1 expression in premenopausal women with (MetSyn) and without (Healthy) the MetSyn (n = 18 to 21 per group). (F and G) Esr1 expression and representative immunoblots of ERα protein in skeletal muscle (quadriceps) from lean and LepOb female mice (n = 6 per genotype; age, 12 weeks). Values are expressed as means ± SEM. All expression values were normalized to 1.0. Mean differences were detected by Student’s t test or analysis of variance (ANOVA); *P < 0.05, between-group comparison.
MERKO mice show impaired glucose homeostasis and insulin action
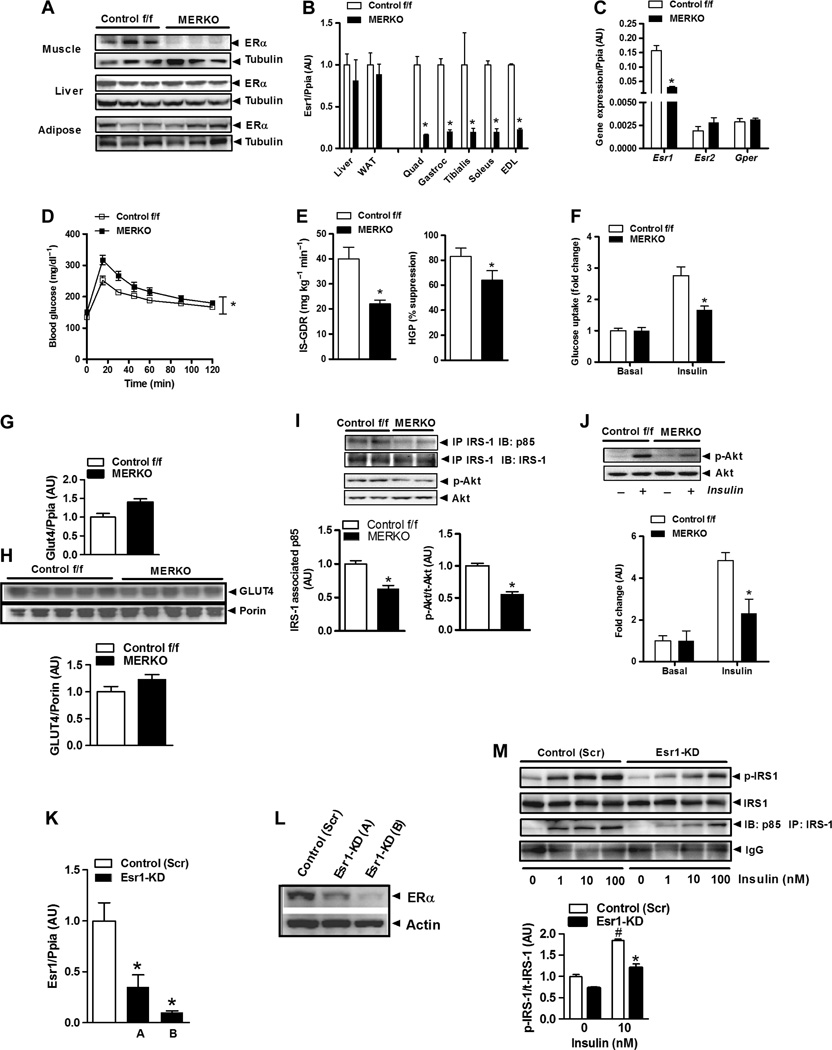
To investigate the impact of muscle Esr1 expression on insulin action, we generated MERKO mice (Fig. 2, A and B). Esr2 (ERβ) is expressed at levels several hundredfold less than Esr1, and no compensatory increase in ERβ mRNA (P = 0.23) or the estrogen-responsive transmembrane G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptor (Gper, GPR30; P = 0.53) was detected in muscle from female MERKO mice (Fig. 2C). Circulating levels of insulin and leptin were elevated (82%, P = 0.01 and 200%, P = 0.001, respectively),whereas circulating adiponectin levels were reduced by 34% (P = 0.004) in 6-hour–fasted MERKO compared with Control f/f mice (table S2). The increased circulating leptin levels in MERKO mice, relative to Control f/f mice, were consistent with a 2.75-fold increase (P = 0.0001; table S2) in gonadal adipose tissue mass and could not be explained by altered food and water consumption (P = 0.095 and P = 0.13, respectively) or ambulatory movement (P = 0.54) compared with Control f/f mice.
Fig. 2. Muscle-specific ERαdeletion impairs glucose tolerance and muscle insulin action.
(A) ERα protein in muscle, liver, and adipose tissue from Control f/f and MERKO female mice (representative immunoblot, n = 3 per genotype). (B) Esr1 expression in muscles [quadriceps, gastrocnemius, tibialis, soleus, and extensor digitorum longus (EDL)] from female MERKO and Control f/f (n = 8 mice per genotype) assessed by quantitative polymerase chain reaction (qPCR). WAT, white adipose tissue. (C) Relative comparison of Esr2 and Gper (ER G protein– coupled receptor) expression in quadriceps from Control f/f and MERKO mice (n = 8 per genotype). P > 0.05. (D) Glucose tolerance [glucose tolerance test (GTT), 1000 mg/kg] in Control f/f and MERKO female mice (n = 6 to 8 per genotype; age, 24 to 26 weeks). *P < 0.05 detected by Student’s t test for AUC. (E) Skeletal muscle and hepatic insulin sensitivity (IS-GDR, insulin-stimulated glucose disposal rate; HGP, hepatic glucose production) assessed by glucose clamp for Control f/f and MERKO mice (n = 6 to 8 mice per genotype; age, 28 weeks). P < 0.05. (F) Ex vivo soleus muscle glucose uptake (fold change from basal; n = 6 mice per genotype). Values for insulin sensitivity are expressed as means ± SEM; *P < 0.05 detected by ANOVA. (G and H) GLUT4 transcript and protein levels in basal 6-hour–fasted quadriceps muscle from Control f/f and MERKO mice (n = 5 to 8 mice per genotype). (I and J) Representative immunoblots and densitometry of insulin signaling (IRS-1–associated p85 and p-Akt) in quadriceps and soleus muscle from glucose clamp and ex vivo glucose uptake studies, respectively, in Control f/f and MERKO mice (n = 6 mice per genotype per condition). P < 0.05. IP, immunoprecipitation; IB, immunoblot. (K to M) Esr1 expression (K), ERα protein levels (L), and insulin signaling (M) (IRS-1pY–and IRS-1–associated p85) in control (Scr) and Esr1-KD C2C12 myotubes (0 to 100 nM insulin; studies performed in triplicate). Densitometric analyses are expressed as means±SEM in arbitrary units normalized to 1.0; *P < 0.05, between-group differences; #P < 0.05, within-group treatment comparison, detected by Student’s t test and ANOVA.
In addition, female MERKO animals showed impaired glucose tolerance (IGT) [area under the glucose curve (AUC) P = 0.025; Fig. 2D] and whole-body insulin resistance [glucose infusion rate (GIR); Fig. 2E and table S2] compared to controls. Marked skeletal muscle insulin resistance in MERKO mice was reflected by a 45% (P = 0.003) reduction in the insulin-stimulated glucose disposal rate (GDR) during glucose clamp studies (Fig. 2E) and a 55% (P = 0.002) reduction in ex vivo insulin-stimulated 2-deoxyglucose glucose uptake into the soleus muscle compared with Control f/f mice (Fig. 2F). MERKO skeletal muscle insulin resistance could not be explained by reduced expression or total protein levels of the insulin-responsive glucose transporter GLUT4 (Fig. 2, G and H) as previously proposed by Barros et al. (18), but rather was a consequence of impaired insulin signal transduction in skeletal muscle (Fig. 2, I and J). Furthermore, in the mouse myoblast C2C12 cell line differentiated to myotubes, Esr1 knockdown (KD) [using lentiviral-mediated Esr1-targeted short hairpin RNA (shRNA)] (Fig. 2, K and L) impaired insulin signal transduction, as evidenced by the observations that insulin-stimulated insulin receptor substrate 1 (IRS-1) tyrosine phosphorylation and IRS-1–p85 association were markedly reduced compared to control scramble shRNA (Scr) (Fig. 2M), thus recapitulating insulin resistance similar to that observed in MERKO mice.
ERαregulates oxidative metabolism and muscle lipid accumulation in skeletal muscle
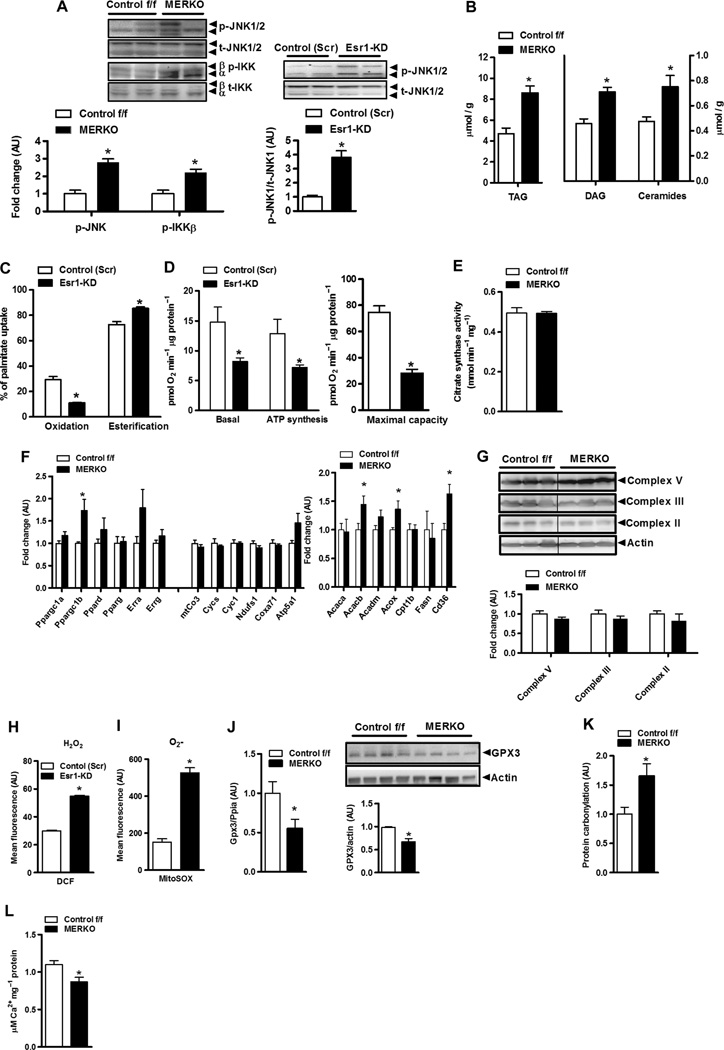
Impaired skeletal muscle insulin action was paralleled by heightened inflammation in MERKO muscle [as shown by increased inflammatory signaling via p-c-Jun N-terminal kinase 1/2 (JNK 1/2) and p–IκB kinase β (IKKβ) relative to controls; P < 0.05, respectively] (Fig. 3A, left panel) and in C2C12 myotubes with Esr1-KD (P < 0.01) (Fig. 3A, right panel). As would be predicted, muscle inflammation paralleled the accumulation of triacylglycerol and bioactive lipid intermediates [diacylglycerol (DAG) and ceramides; Fig. 3B]. To determine whether impaired fatty acid oxidation contributed to the buildup of muscle lipids, we investigated the cellular deposition of [14C]palmitate in C2C12 myotubes with Esr1-KD. The myotube Esr1 deficiency caused a 50% reduction in fatty acid oxidation (P = 0.02) and a concomitant increase in fatty acid storage into complex lipids (P < 0.01) relative to control myotubes (Fig. 3C). This alteration in fatty acid handling was associated with a marked reduction in basal and maximally stimulated cellular oxygen consumption and ATP (adenosine 5′-triphosphate) synthesis (Fig. 3D and fig. S1) as measured by real-time respirometry.
Fig. 3. Esr1 deletion promotes mitochondrial dysfunction, ROS production, and increased muscle inflammation.
(A) Heightened proinflammatory signaling (p-IKKβ and p-JNK 1/2) (fold change over baseline normalized to 1.0) in female MERKO muscle (n = 6 per genotype; left panel) and myotubes (n = 4 per condition; right panel), respectively. P < 0.05. (B) Bioactive lipid intermediates (TAG, triacylglycerol; ceramides) in quadriceps muscle from female Control f/f and MERKO mice (n = 6 muscles pergenotype). P < 0.05. (C and D) Palmitate oxidation and esterification as well as basal and maximal (stimulated using carbonylcyanide p-trifluoromethoxyphenylhydrazone, FCCP) oxygen consumption and ATP synthesis in control (Scr) versus Esr1-KDmyotubes (n = 6). (E) Citrate synthase activity in muscle harvested from Control f/f versus MERKO mice (n = 6 per genotype). (F) Quantitative reverse transcription PCR (RT-PCR) analyses of muscle from Control f/f versus MERKO mice (n = 6 per genotype). (G) Immunoblots and corresponding densitometry of mitochondrial electron transport complexes versus actin loading control in muscle from Control f/f versus MERKO mice (n = 6 per genotype). (H and I) H2O2 and O2− production in control (Scr) versus Esr1- KD C2C12 myocytes. (J) Gpx3 expression and protein in Control f/f versus MERKO muscle (n = 6 per genotype). (K) Oxidative stress–induced protein carbonylation in quadriceps muscle from Control f/f versus MERKO mice (n = 8 per genotype). (L) Calcium buffering capacity/mPTP opening in mitochondria isolated from Control f/f versus MERKO mice (n = 5 per genotype). All values are expressed as means ± SEM. Mean differences detected by Student’s t test and ANOVA where appropriate; *P < 0.05.
Next, we investigated the mechanism underlying diminished oxidative metabolism in ERα-deficient muscle. Considering that obesity and insulin resistance are often associated with a reduction in mitochondrial number (19), we assessed surrogate markers of mitochondrial mass and biogenesis in muscle from female control versus MERKO mice. We observed no difference in maximal activity of muscle citrate synthase (a mitochondrial enzyme that is part of the Krebs cycle) (Fig. 3E), expression of transcriptional regulators of mitochondrial biogenesis (Fig. 3F and tables S3 and S4, microarray analyses), or expression or protein abundance of select components of the mitochondrial electron transport complex (Fig. 3G) between the genotypes. An unanticipated increase in peroxisome proliferator–activated receptor γ coactivator 1β (Ppargc1b), acetyl–coenzyme A (CoA) carboxylase 2 (Acacb), peroxisomal acyl-CoA (Acox), and the fatty acid transporter cluster of differentiation (Cd36) expression was observed in MERKO muscle compared with Control f/f. We found that myocytes that lacked ERα produced markedly elevated amounts of the oxygen radicals H2O2 and O2− (Fig. 3, H and I). In addition, ROS scavenging capacity was diminished because glutathione peroxidase 3 (Gpx3) mRNA and protein levels were significantly reduced in MERKO muscle, relative to control (Fig. 3J), a finding in line with previous reports showing that Gpx3 expression is regulated by ERα (20). Increased oxidative stress promoted protein carbonylation (a kind of protein oxidation) in muscle from MERKO mice compared with Control f/f mice (Fig. 3K). Thus, it is likely that the marked inflammation observed in MERKO muscle was in large part mediated by the cumulative effects of chronic oxidative stress.
An important function of the mitochondria is calcium handling. We found a 21% reduction (P = 0.017) in the maximal calcium concentration that initiated mitochondrial permeability transition pore (mPTP) opening in isolated mitochondria from MERKO versus Control f/f (Fig. 3L and table S3). Recent studies have shown that opening of mPTP links mitochondrial dysfunction to insulin resistance (21). The impairments in calcium handling and ATP production along with the elevation in ROS production by the mitochondria were likely factors underlying decrements in muscle contractile function, because single muscle fibers from MERKO mice fatigued faster than fibers from Control f/f muscle (fig. S2A). Moreover, we observed cytochrome oxidase– negative fiber staining with aging only in MERKO muscle (fig. S2B). Collectively, these data indicate that metabolism requisite to fuel repeated muscular contractions in MERKO mice is limiting and that reduced ERα action accelerates the development of specific muscle phenotypes associated with aging.
In contrast to muscular endurance, peak tension and peak tetanic force were identical between the genotypes, whereas time to peak tension was improved in MERKO muscle compared to Control f/f (fig. S2, C to E). We had initially speculated that ERα deletion might promote muscle atrophy with aging; however, on the contrary, muscle protein levels, including the abundant z-line protein α actinin, were maintained (fig. S2F and table S3). This could be a consequence of a reduction in the expression of atrogenes (genes that influence muscle atrophy; including Foxo1, Fbxo32, andTrim63) in MERKO versus Control f/f mice (fig. S2G). Thus, despite the impairment in muscle oxidative metabolism, muscle size and maximum force were maintained in MERKO, in part, by a compensatory down-regulation of muscle degradation pathways.
ERαregulates mtDNA replication and mitochondrial turnover in muscle
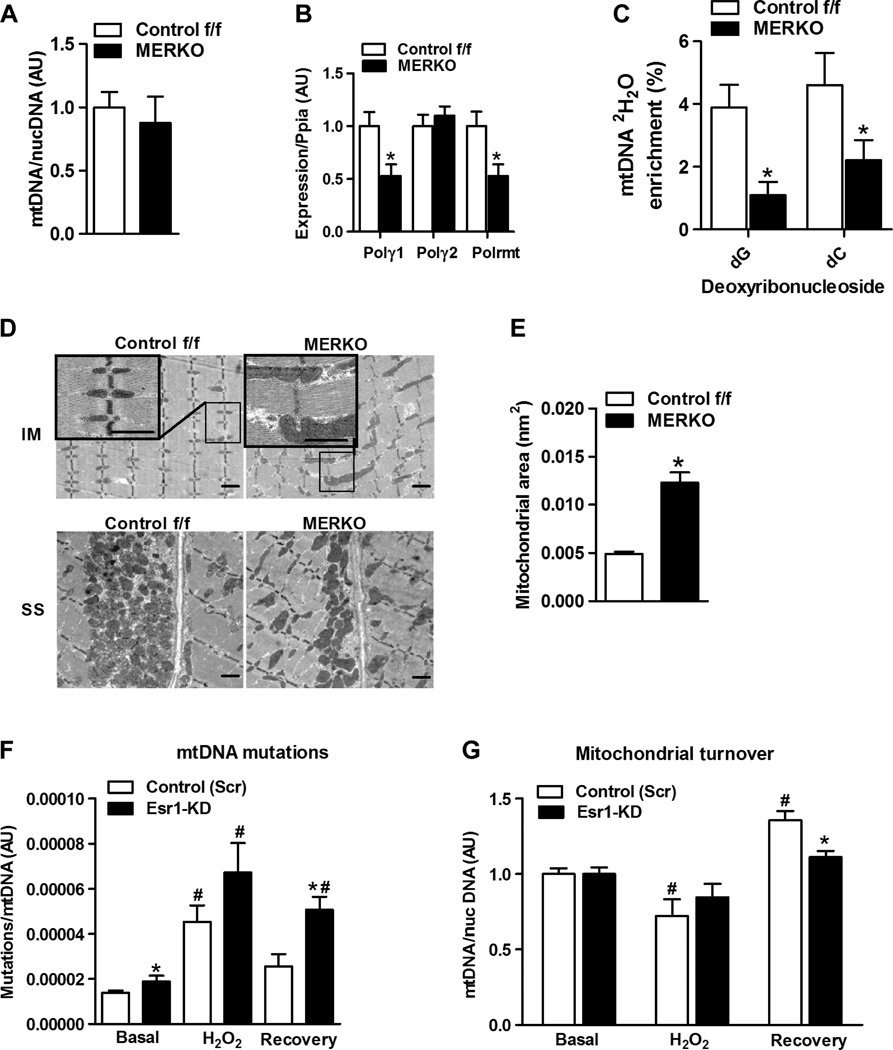
Although we observed no difference in the expression of genes associated with mitochondrial biogenesis or total mtDNA content between the genotypes of female mice (Fig. 4A), we did detect a significant reduction in mtDNA polymerase γ1 (Polγ1) and mtDNA-directed RNA polymerase (Polrmt) expression levels in MERKO versus Control f/f muscle (Fig. 4B). We confirmed regulation of Polγ1 expression by ERα by showing that selective inhibition of ERα using the ERα antagonist ICI 182,780 (fulvestrant) or an ERα DNA binding mutation (ERα DBDΔ) reduced Polγ1 levels in C2C12myotubes and quadriceps muscle, respectively (fig. S3, A and B). To determine whether reduced expression of Polγ1 and Polrmt manifested a functional decrement in mtDNA replication in MERKO muscle, we assessed the incorporation of deuterated water into newly synthesized mtDNA. Consistent with gene expression data, we observed reduced 2H2O labeling of newly synthesized mitochondrial deoxyribonucleosides after heavy water consumption in MERKO versus Control f/f mice (Fig. 4C). In order for the mtDNA pool to be maintained in MERKO muscle to a level similar to that of Control f/f mice, it follows that a reduction in mtDNA replication must be balanced by a reduction in the rate of mtDNA degradation.
Fig. 4. Impaired mtDNA replication and altered mitochondrial morphology in MERKO muscle.
(A) Muscle mtDNA content, expressed as a ratio of mtDNA/nuclear (nuc) DNA (n = 8 per group). (B) Muscle polymerase γ1 (Polγ1), Polγ2, and Polrmt expression. (C) mtDNA replication as measured by 2H2O incorporation into newly synthesized mtDNA (deoxynucleosides, dG and dC) from Control f/f and MERKO mice (n = 6 per genotype). (D) Representative electron micrographs of soleus muscle from female Control f/f and MERKO mice, low- and high-magnification inset (IM, intramyofibrillar; SS, subsarcolemmal; scale bar, 1 µm). (E) Skeletal muscle intermyofibrillar mitochondrial area (n = 3 mice per genotype). (F) mtDNA mutation load was detected by random mutation capture (RMC) at basal (no treatment), after H2O2 treatment, and during recovery from H2O2 treatment. (G) mtDNA copy number at basal, after H2O2 treatment [in the presence of chloramphenicol (Chl) plus cycloheximide (Chx), and during recovery (in the presence of bafilomycin A1 (BafA1)]. All values are expressed as means ± SEM. Mean differences detected by Student’s t test and ANOVA where appropriate; *P < 0.05, between-genotype difference; #P < 0.05, within-genotype between treatment difference.
To examine this notion further, we determined whether mitochondrial morphology and mitochondrial turnover were altered by ERα deficiency. Using transmission electron microscopy, we found that mitochondrial morphology and cellular distribution were altered by ERα inactivation (Fig. 4D). MERKO mitochondria were dysmorphic (enlarged, elongated, and hyperfused), misaligned along the Z-disc, and less densely distributed in the subsarcolemmal compartment compared with mitochondria of Control f/f (Fig. 4D). The mitochondrial area was increased twofold (P = 0.0001) in MERKO muscle compared with Control f/f muscle (Fig. 4E). A similar mitochondrial architecture was observed in muscle from female ERα DBDΔ mice, suggesting that ERα DNA binding is requisite, whereas ERα tethering is insufficient, to alter gene transcription and restore the mitochondrial morphology seen in Control f/f muscle (fig. S3C). Although the cellular architecture of myotubes in culture differs from murine muscle, altered morphological features of MERKO mitochondria (that is, elongation and hyperfusion) were recapitulated in C2C12 Esr1-KD myotubes compared with C2C12 control-Scr myotubes (fig. S3D). An elongated and fused mitochondrial phenotype is often observed as a consequence of mitochondrial stress and is a compensatory response to stabilize the mtDNA and dilute damaged mitochondrial contents across the mitochondrial network (22, 23). Because the lysosome has a cargo-size restriction, mitochondrial elongation is thought to occur as a mechanism to prevent mitophagy (degradation of mitochondria via autophagy). For example, mitochondrial elongation is engaged during starvation when macroautophagy is maximally activated (24–26).
Because muscle mtDNA abundance was identical between the genotypes despite a reduction in the rate of mtDNA replication, a coordinate reduction in mtDNA turnover is required (Fig. 4E) (27). It has been hypothesized that mtDNA replication stress may contribute to increased mtDNA mutation susceptibility because the half-life of mtDNA is increased (27). To test this notion, we determined whether ERα is critical for controlling mtDNA turnover. We quantified mtDNA mutation load and mtDNA abundance as two surrogate markers of mtDNA turnover after H2O2 treatment of control and Esr1-KD myotubes (28). Basal mtDNA mutation load was elevated and failed to return to basal levels in Esr1-KD myotubes during the recovery period when the genotoxic stress was removed (in contrast to control cells) (Fig. 4F). Moreover, although basal mtDNA abundance was similar between the genotypes, mtDNA after H2O2 treatment was reduced from basal only in control cells, suggesting a possible impairment in mtDNA turnover in Esr1-KD cells. During recovery, mtDNA abundance was significantly elevated over basal in control cells, but the recovery response was markedly bluntedforEsr1-KDcells, findings that support our observation of a defect in mtDNA replication in MERKO mice. Thus, our findings suggest that ERα is critical for the elimination of damaged mtDNA as well as for promoting mitochondrial biogenesis during recovery from genotoxic insult (Fig. 4, F and G).
ERαdeletion impairs mitochondrial fission-fusion dynamics in skeletal muscle
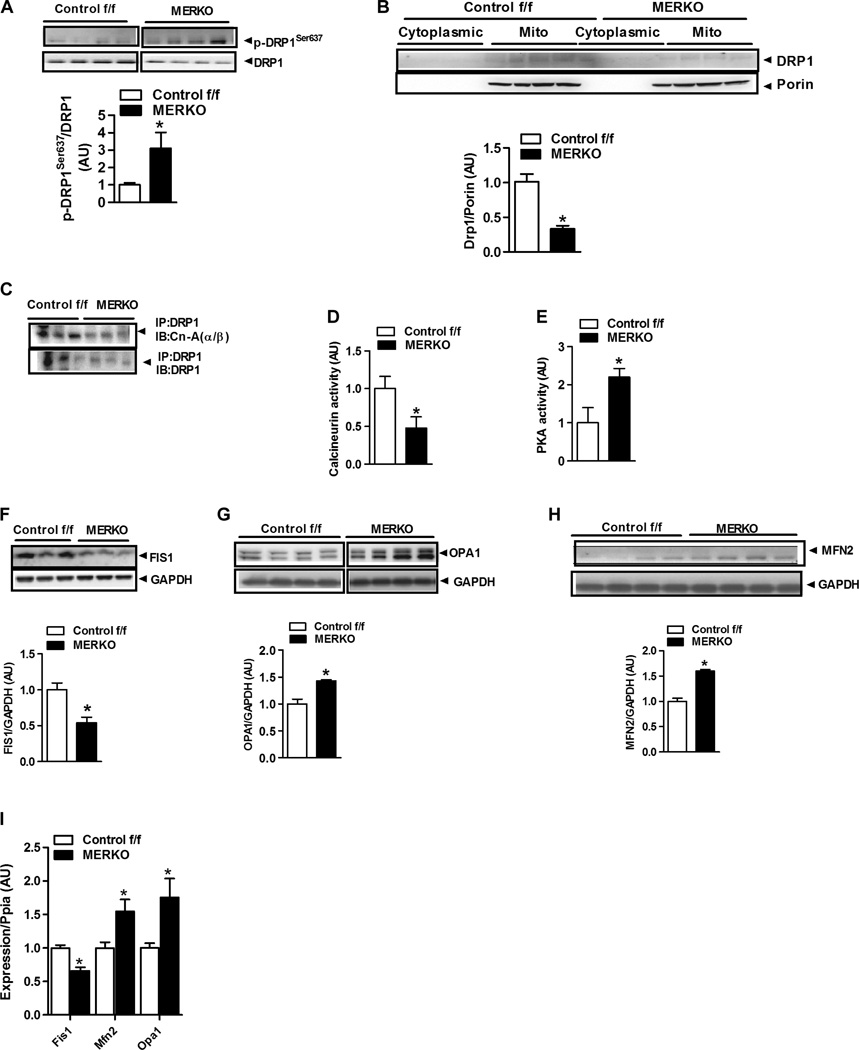
The marked alteration in mitochondrial morphology, reduction in mtDNA replication rate, and impairment in mtDNA turnover in ERα-deficient muscle prompted us to assess effectors of mitochondrial fission-fusion dynamics. On the basis of the mitochondrial hyperfusion phenotype, which showed elongated and highly interconnected mitochondria in muscle of MERKO mice (Fig. 4D), we hypothesized that that there also would be a strong suppression of mitochondrial fission signaling in MERKO muscle. Because mitochondrial fission is responsible for producing smaller organelles and is critical for mitophagy, our findings of altered mitochondrial morphology and impaired mtDNA turnover after genotoxic stress support this hypothesis. To test this notion further, we quantified mitochondrial fission signaling by immunoblotting of MERKO muscle; we observed a threefold increase (P = 0.0001) in DRP1 phosphorylation at the inhibitory residue Ser637 (Fig. 5A) and a reduction inDRP1 protein associated with mitochondria from MERKO muscle compared with Control f/f (Fig. 5B). These data were confirmed in C2C12 myotubes with Esr1-KD (fig. S4). In line with increased DRP1Ser637 phosphorylation, we observed diminished protein interaction between DRP1 and the putative phosphatase calcineurin, which was previously shown to induce mitochondrial fission (Fig. 5C). Moreover, a ~50% reduction in total calcineurin activity was detected in MERKO muscle (Fig. 5D) and in myotubes deficient in ERα (fig. S5, A and B), compared to respective controls.
Fig. 5.
Altered fission-fusion dynamics signaling in muscle from MERKO versus Control f/f mice. (A and B) DRP1Ser637 phosphorylation (A) and DRP1 protein abundance (B) in enriched mitochondrial fractions from female Control f/f and MERKO quadriceps muscle (n = 4; representative immunoblots). Porin, control. (C) Protein-protein association between DRP1 and the phosphatase calcineurin (CnA) (above; n = 3 to 6 per genotype) in muscle from Control f/f and MERKO mice. (D and E) Calcineurin and PKA activity in quadriceps muscle from Control f/f and MERKO mice (n = 6 to 10 per genotype). (F to H) FIS1, OPA1, and MFN2 protein expression in muscle from Control f/f and MERKO mice relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression (control) (n = 6 per genotype). (I) Expression of Fis1, Mfn2, and Opa1 proteins in quadriceps muscle from female Control f/f and MERKO mice relative to Ppia expression (housekeeping gene control) (n = 6 per genotype). All values are expressed as means ± SEM. Mean differences detected by Student’s t test or ANOVA. *P < 0.05, difference between genotypes.
In opposition to the mitochondrial pro-fission enzyme calcineurin, activity and phosphorylation status of PKA—the putative kinase linked with phosphorylation ofDRP1 on the inhibitory Ser637 residue (29)—were elevated (Fig. 5E and fig. S5,C and D) as was the scaffolding protein that associates PKA with the mitochondrion [A kinase anchoring protein 1 (AKAP1)] (table S4 and fig. S4, A and B). Notably, in addition to increased PKA total protein and phospho-PKA in whole-cell muscle lysates, a near twofold increase in PKA was detected in mitochondria-enriched fractions from Esr1-KD myotubes compared with Esr1-replete control cells (fig. S4, C and D).
With respect to additional mitochondria-specific proteins involved in fission-fusion signaling, we observed a 50% (P = 0.005) reduction in the fission-related outer mitochondrial membrane DRP1 anchoring protein, FIS1 (Fig. 5F), in MERKO versus Control f/f. Conversely, the inner and outer mitochondrial membrane fusion proteins, optic atrophy 1 protein (OPA1), and mitofusin 2 (MFN2), respectively, were elevated 50 to 60% in MERKO muscle and Esr1-KD myotubes compared with respective controls (Fig. 5, G and H, and fig. S4, C and F). Alterations in transcript mirrored the observed differences in protein abundance between the genotypes (Fig. 5I). Collectively, these findings strongly suggest that a loss of myocellular ERα dampens mitochondrial fission signaling, promoting unopposed mitochondrial fusion. These signaling data are strongly supported by electron micrographs showing an elongated and hyperfused mitochondrial morphology in muscle from MERKO mice (Fig. 4D).
Mitophagic signaling is impaired in the context of ERαdeficiency
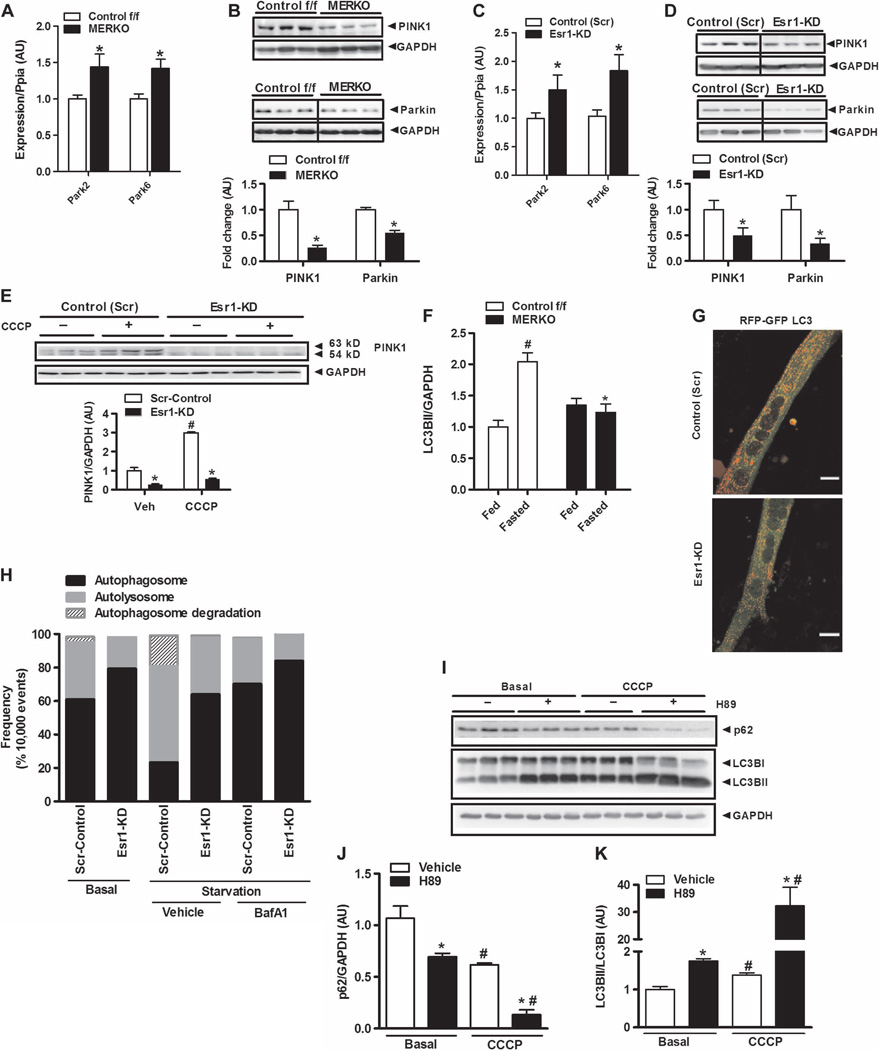
Mitochondria are highly dynamic and constantly undergoing fission-fusion events to repair and selectively retain healthy organelles in the network (30, 31). To investigate whether impaired mitochondrial fission is associated with decrements in mitochondrial quality control in the muscle of MERKO mice, we next assessed mitophagic signaling. The expression levels of Park family members Park2 (Parkin) and Park6 [phosphatase and tensin homolog (PTEN)–induced putative kinase 1 (PINK1)] were significantly elevated in ERα-deficient muscle (Fig. 6A). Despite elevated transcript abundance, PINK1 and Parkin protein levels were reduced by 75% (P = 0.001) and 40% (P = 0.001), respectively, in muscle from MERKO compared with Control f/f mice (Fig. 6B). Similar to findings in skeletal muscle, Park2/6 transcripts were elevated (Fig. 6C), whereas PINK1 and Parkin protein levels in whole-cell lysates were reduced in Esr1-KD versus control (Scr) C2C12 myotubes (Fig. 6D).
Fig. 6. ERαdeficiency impairs mitophagic signaling in muscle.
(A to D) Expression of Park family members (Park2 and Park6) in muscle from female Control f/f and MERKO mice (n = 6 per genotype, A and B) and C2C12 myotubes (n = 6 per group, C and D). (E) CCCP-induced accumulation of full-length (63-kD) PINK1 protein in control (Scr) and Esr1-KD myotubes (n = 6 observations per condition). Veh, vehicle (control). (F) Densitometric analysis of LC3BII protein levels in muscle from fed and fasted (24 hours) female Control f/f versus MERKO mice relative to GAPDH expression (n = 6 mice per condition). (G and H) Confocal microscopy and flow cytometry analyses of dually labeled RFP-GFP-LC3B expressed in control (Scr) and Esr1-KD muscle cells during nutrient deprivation. (G) Reduced red punctae were observed in Esr1-KD muscle cells versus Scr-control, indicating diminished autolysosome formation (scale bar, 1 µm). (H) To more accurately quantify fluorescence signals in muscle cells, flow cytometry analyses were performed in triplicate during nutrient deprivation in the presence and absence of BafA1 (10,000 live events quantified). Bars depicting quantification of dual signal represent autophagosome abundance (closed bars); a single RFP signal represents autolysosome abundance (gray bars); and cells showing loss of signal represent autophagosome degradation (hatched bars). (I to K) The impact of PKA inhibition (H89, 50 µM) on p62 protein levels and LC3B processing in ERα-replete myotubes in the absence or presence of CCCP (20 µM) to stimulate mitophagy (n = 3 per condition). (I) Representative immunoblots and (J and K) densitometric analyses. All values are expressed as means ± SEM. Mean differences detected by Student’s t test or ANOVA. *P < 0.05, difference between genotypes; #P < 0.05, within-group, between-treatment difference.
Because the accumulation of full-length PINK1 protein in the outer mitochondrial membrane is critical for initiation of mitophagy, we performed mitochondrial isolation studies in ERα-replete and ERαdeficient myotubes. We observed a reduced abundance of cytosolic and mitochondria-associated full-length 63-kD PINK1 protein and the 54-kD degradation product in ERα-deficient myotubes compared with control (Scr) in the basal state (Fig. 6E and fig. S4, C and G). To determine whether ERα is critical for surveillance of mitochondrial health, we treated myotubes with the mitochondrial membrane depolarizing agent carbonyl cyanide m-chlorophenylhydrazone (CCCP), a well-described stimulus shown to induce mitophagy. In line with previous reports, CCCP induced an increase in PINK1 protein in ERα-replete control (Scr) myotubes (Fig. 6E, open bars) (32). In contrast to control (Scr) cells, basal PINK1 protein levels were reduced and there was no induction of PINK1 protein above the vehicle control after CCCP treatment in Esr1-KD myotubes (Fig. 6E, closed bars).
Our findings indicate that ERα deficiency causes a reduction in Park family protein levels and implicates increased PINK1/Parkin protein turnover as a potential mechanism for reduced mitophagic signaling in MERKO muscle. Together, these data support the notion that ERα is critical for mitochondrial health surveillance and mitophagic signaling. We recently linked impaired mitophagic signaling to metabolic dysfunction by showing that Parkin inactivation reduces oxidative metabolism and promotes skeletal muscle insulin resistance, similar to phenotypic observations made for female MERKO mice herein (33).
Impaired autophagy in ERα-deficient muscle
Because the turnover of mitochondria is shown to be reliant on intact macroautophagy, we next studied the impact of ERα expression on autophagic signaling under basal and starvation conditions. Macroautopahagy is a cellular housekeeping process that allows for the removal of dysfunctional or unnecessary proteins or organelles. Autophagosomes (spherical structures with double-layer membranes) deliver the unwanted cellular components to the lysosome for degradation. The formation of autophagosomes is incompletely understood but is regulated by a well-conserved family of genes, Atg’s, and microtubule-associated protein light chain 3 (LC3) complexes. LC3 proteins (for our purposes, LC3B) undergo posttranslational modifications during autophagy, including immediate cleavage to LC3BI and subsequent lipidation (phosphatidylethanolamine conjugation of the carboxyl glycine) processing to yield LC3BII, thus allowing for LC3B to become associated with the autophagic vesicle to control autophagosome fusion. In contrast to the marked increase in LC3BII [observed in Control f/f muscle after nutrient deprivation (P = 0.03)], LC3B processing (LC3BI to LC3BII) during starvation was impaired in MERKO muscle; no significant increase in LC3BII protein was detected between the fed to fasted state (P = 0.2; Fig. 6F and fig. S6A). Further, we detected no difference in the expression of the microtubule-associated protein 1B light chain 3 (Maplc3b) LC3B transcript between the MERKO and control genotypes during the basal or fasted conditions (Maplc3b is the gene that encodes the LC3B transcript) (fig. S6B). Although expression of most autophagy-related genes (Atg) is unaltered by nutrient deprivation (34), we observed an induction of Sqstm1 (p62) and Atg7 in Control f/f muscle after fasting, a phenotype that failed to be reproduced in MERKO mice (fig. S6, C and D); Atg7, an E1-like enzyme considered essential in facilitating the association between Atg5 and Atg12 for LC3 activation, was reduced in MERKO muscle even under basal fed conditions (fig. S6D). The increase in basal expression of Atg12, Atg5, and Atg4b might be a compensatory response to the decrement in Atg7 and impaired downstream LC3 processing (fig. S6, E to G) in an attempt to restore autophagic flux.
Our findings in vivo were reproduced in C2C12 cells in culture. A marked accumulation of p62 and reduced LC3BII/LC3BI were observed in Esr1-KD versus control (Scr) myotubes in the basal state and after serum deprivation with and without the lysosomal inhibitor bafilomycin A1 (fig. S6, H and I). Because p62 is an adaptor molecule believed to target ubiquitinated proteins to LC3BII for selective uptake and degradation by the lysosome, increased p62 protein levels, despite its reduced transcript expression, are considered reflective of impaired autophagy (35). Moreover, although we observed no difference in the expression of Maplc3b between the genotypes, LC3BI was consistently elevated in ERα-deficient muscle under basal conditions, suggesting a possible delay in processing of LC3B protein (fig. S6, A, H, and I).
To confirm our signaling findings and provide an additional readout of LC3 processing and autophagolysosome formation, we generated myotubes that expressed LC3B protein with bound tandem red and green fluorescent protein (RFPGFP- LC3B). Because GFP fluorescence is quenched in the acidic pH of the lysosomal compartment, the use of GFP-LC3B is limited to the identification of autophagosomes. However, RFP is pH-resistant and continues to fluoresce at acidic pH; thus, RFP-LC3B can be used to identify both autophagosomes and autolysosomes (36, 37). The apparent reduction in red fluorescence punctae observed in Esr1-KDversus control (Scr) myotubes (Fig. 6G) was fully consistent with reduced autophagic flux, specifically reduced autolysosome formation (36, 37). In addition, flow cytometry studies were performed, providing a more sensitive and quantitative determination of the GFP and RFP signals, including loss of signal determination for assessing starvation-induced LC3Bautophagosome turnover. Consistent with the protein signaling data, functional analyses indicate that ERα inactivation markedly suppressed autolysosome formation and autophagosome turnover in the context of starvation-induced autophagy (Fig. 6H and fig. S7).
One possible explanation for impaired autophagic signaling and delayed autophagosome formation is direct phosphorylation of LC3B by PKA, an event shown to reduce LC3 recruitment to autophagosomes (38). Considering that PKA activity was consistently elevated twofold in both MERKO muscle and C2C12myotubes with Esr1- KD, we next tested the relation between PKA and LC3B maturation in control ERα-replete C2C12 myotubes. Similar to findings previously reported in SH-SY5Y human neuroblastoma cells (38), we found that kinase inhibition by H89 enhanced basal LC3BI-to-LC3BII conversion and p62 degradation. Moreover, these findings were intensified by CCCP treatment, a known inducer of mitophagy (Fig. 6, I to K). Collectively, these data indicate that autophagic signaling is blunted in ERα-deficient muscle and that increased PKA activity might contribute, in part, to the impairment in autophagosome processing during macroautophagy.
Drp1 is critical for insulin action and oxidative metabolism
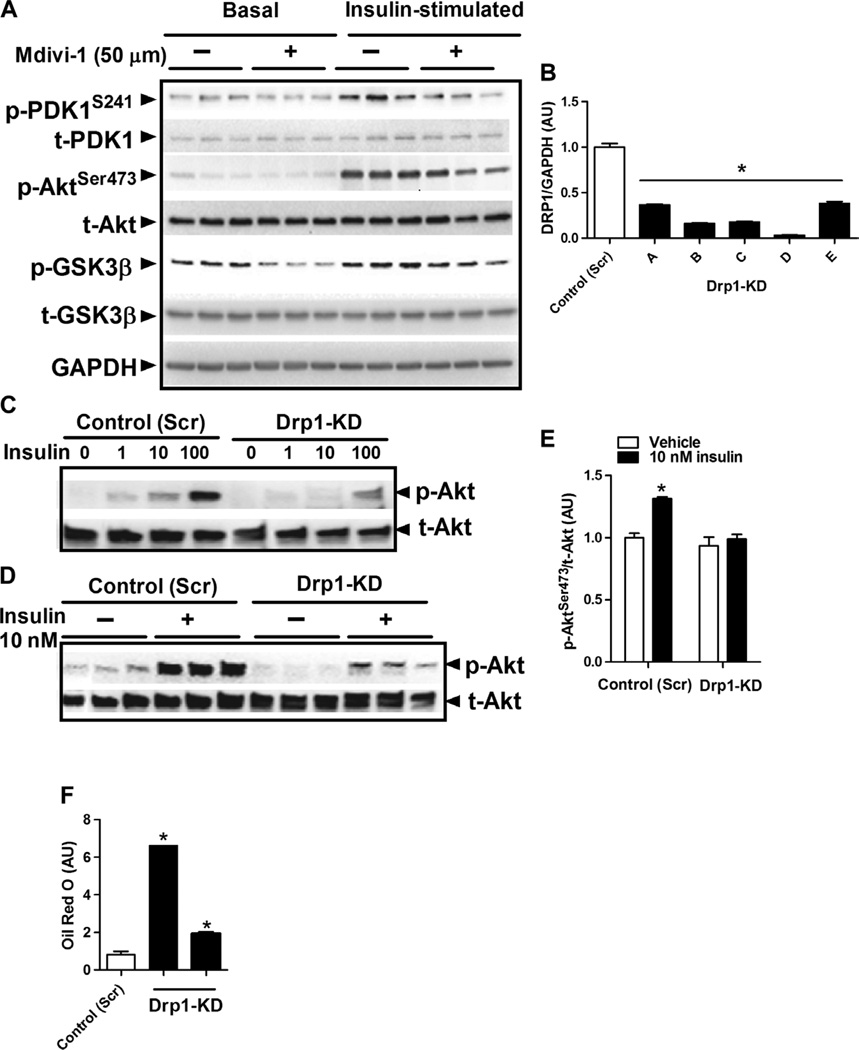
To test directly whether impairment of mitochondrial fission could induce similar phenotypic outcomes as those observed in Esr1-KD cells, we performed chemical inhibition and gene deletion of Drp1. First, we treated murine myotubes with the Drp1 inhibitor Mdivi-1 and found that insulin-stimulated phosphorylation of pyruvate dehydrogenase kinase (PDK), Akt, and glycogen synthase kinase 3β (GSK3β) was reduced compared to vehicle-treated basal control myotubes (Fig. 7A). Next, we used shRNA to knock down Drp1 in myotubes (Fig. 7B). Similar to findings for studies of Drp1 inhibition, Drp1-KD markedly impaired insulin action in myotubes (Fig. 7, C to E), and similar to MERKO muscle, lipid levels were elevated in Drp1-KD myotubes (Fig. 7F). In aggregate, these data suggest that Drp1 action is critical for the maintenance of insulin sensitivity and potentially oxidative metabolism, and thus, impaired fission dynamics might in part underlie the pathogenic phenotypes observed in MERKO muscle.
Fig. 7. Impaired mitochondrial fission signaling through DRP1 impairs oxidative metabolism and induces insulin resistance.
(A) Insulin signaling (p-PDK1S241, p-AktSer473, and p-GSK3β) in C2C12 cells treated with the Drp1 selective inhibitor Mdivi-1 (50 µM) versus vehicle control (n = 6 per condition). (B) DRP1 protein levels in C2C12 myotubes relative to GAPDH expression, with Drp1-KD achieved by lentiviral delivery of shRNA (five clones tested, A to E). (C to E) Drp1-KD impaired insulin signaling in C2C12 myotubes (1, 10, and 100 nM insulin treatment). (F) Drp1-KD promoted accumulation of lipid in myotubes as determined by Oil Red O staining (n = 6 observations per genotype) (using two of the five shRNA lentiviral clones, C and D). Values are expressed as means ± SEM. Mean differences detected by Student’s t test or ANOVA. *P < 0.05, difference between genotypes.
The oxidative stress–responsive Rcan1 links impaired fission with insulin resistance
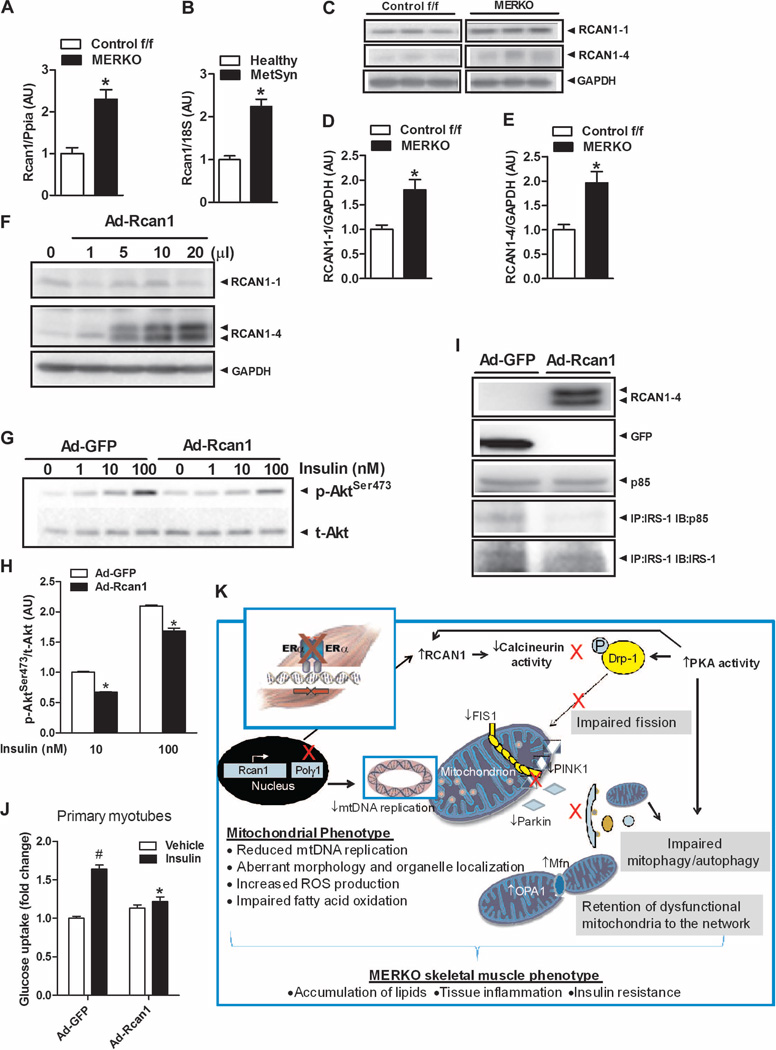
Because ERα is a hormone nuclear receptor involved in gene activation and repression, we screened microarrays to identify genes differentially expressed in skeletal muscle between MERKO and Control f/f. Rcan1 (also calledDscr1 andMcip1)—a known inhibitor of calcineurin (39–41) whose encoding gene is responsive to oxidative stress (39, 40)—was upregulated 6.8-fold (P = 0.01) by ERα deletion (table S4); we confirmed this finding by qPCR (Fig. 8A). Rcan1 expression was also increased in muscle from premenopausal women with MetSyn (Fig. 8B). Similar to RCAN1 mRNA transcript abundance in muscle, the RCAN1-1 and RCAN1-4 protein levels also were significantly elevated (Fig. 8, C to E), and these findings were reproduced in C2C12 myotubes with Esr1-KD (fig. S5, C, E, and F). Although increased Rcan1 mRNA transcript could account for the elevated RCAN1 protein levels in MERKO muscle, PKA is also known to phosphorylate and stabilize RCAN1 protein, thus increasing its half-life (42). When Rcan1 was overexpressed in C2C12 or primary myotubes, insulin action was impaired (Fig. 8, F to J). Thus, together, our findings suggest a conserved mechanism by which ERα regulates a PKA-Rcan1-calcineurin signaling axis to control mitochondrial fission and insulin action (Fig. 8K).
Fig. 8. Rcan1 is up-regulatedin ERα-deficient muscle and impairs insulin action.
(A and B) Rcan1 gene expression is elevated in female MERKO muscle (n = 8 per genotype) and in human muscle from women with MetSyn (n = 8 per group) compared with respective controls. (C to E) Rcan1-1 and Rcan1-4 protein levels are elevated in MERKO muscle versus Control f/f (n = 8 per genotype). (F to J) Rcan1 overexpression by adenovirus (Ad) infection of C2C12 or primary myotubes impairs insulin action as shown by (G) reduced insulin–stimulated p-AktSer473 (1, 10, and 100 nM insulin), (I) IRS-1–p85 association (50 nM insulin), and (J) 2-deoxyglucose uptake (n = 3 per condition in triplicate, 50 nM insulin). All values are expressed as means ± SEM. Mean differences detected by Student’s t test and ANOVA. *P < 0.05, difference between genotypes; #P<0.05 within-group, between-treatment difference. (K) Schematic overview of the MERKO phenotype. Skeletal muscle–specific ERα deletion reduced mtDNA replication and impaired muscle oxidative metabolism, despite maintenance of mtDNA copy number. Elevated Rcan1 and PKA reduced calcineurin activity levels, promoting elongated, hyperfused mitochondria in female MERKO muscle. The morphological changes coupled with an imbalanced PKA-calcineurin axis blunted mitochondrial fission signaling through DRP1 and impaired macroautophagy, processes critical for mitochondrial turnover, mitophagy. Unopposed fusion of the outer and inner mitochondrial membranes was permitted by increased expression of mitochondria-specific fusion proteins Mfn2 and OPA1. The retention of damaged mitochondria to the network was paralleled by increased ROS production, inflammation, and insulin resistance in skeletal muscle from female MERKO mice.
DISCUSSION
Reduced estrogen action is clinically tied to obesity, metabolic dysfunction, and increased risk of chronic disease. However, the molecular actions of ERα and the tissues responsible for conferring these clinical outcomes remain inadequately understood. In the current investigation, we studied the importance of skeletal muscle ERα action and the impact of skeletal muscle–specific receptor inactivation on the manifestation of metabolic dysfunction. We focused our efforts on skeletal muscle because it is a predominant tissue responsible for fatty acid metabolism and whole-body insulin-stimulated glucose disposal.
First, we established the clinical rationale for our studies by showing that ERα expression was reduced in muscle from women displaying clinical features of the MetSyn. Moreover, skeletal muscle ESR1/Esr1 was highly predictive of fasting insulin concentration and adiposity in women and female mice. To interrogate this relationship further, we performed a selective deletion of Esr1 from skeletal muscle in female mice and myotubes in culture. ERα deletion caused marked insulin resistance in skeletal muscle and cultured myotubes compared to control ERα-replete muscle and cells. Studies in myotubes indicated that the defects in insulin action are a direct effect of Esr1 deficiency within that cell type and that muscle insulin resistance in MERKO mice occurs independently of, but is likely exacerbated by, secondary phenotypes in adipose tissue and liver. Targeted deletion of ERα from skeletal muscle fully recapitulated the insulin resistance and obesity observed in whole-body Esr1−/− mice, and the metabolic dysfunction appears more severe than other mouse harboring cell-specific deletions of Esr1 (43–46).
Paralleling insulin resistance in ERα-deficient myocytes, there was a marked reduction in mitochondrial functionality including diminished rates of oxygen consumption, reduced fatty acid oxidation, impaired calcium buffering capacity/heightened susceptibility for mPTP opening, and increased ROS production. It is reasonable to conclude that these combinatory defects in mitochondrial function contributed to tissue inflammation and impaired insulin action in skeletal muscle, because each of these features of mitochondrial dysfunction has independently been shown to promote inflammatory signaling and insulin resistance (21, 47, 48).
Underlying these alterations in mitochondrial function in ERαdeficient muscle were defects in mtDNA replication and organelle quality control including impairments in mitochondrial health surveillance, fission-fusion dynamics, and autophagy. In addition to reductions in PINK1 and Parkin signaling, critical for the initiation of mitophagy and subsequent elimination of damaged mitochondrial contents, aberrant signaling through the PKA-Rcan1-calcineurin- Drp1 axis was linked with imbalanced fission-fusion dynamics and delayed autophagic flux. Whether a primary defect in mitochondrial replication was the initiating factor responsible for driving a compensatory reduction in mitophagy to sustain mtDNA abundance in MERKO muscle remains to be determined. However, it appears that total mtDNA content was preserved in MERKO muscle at the expense of organelle health.
As previously described, a marked alteration in mitochondrial morphology characterized by enlarged, tubulated, and, in many cases, fused mitochondria extending across multiple z-lines of myofibrils is entirely consistent with heightened mitochondrial stress (22). Similar morphological features of mitochondria were observed previously when comparing electron micrographs of muscle from obese versus lean human subjects and young versus aged mice (19, 49). A mitochondrial hyperfusion phenotype is thought to be a protective mechanism to dilute damaged organelle contents, for example, DNA and enzymes, across the mitochondrial network (25, 50). Although the mechanisms underlying mitochondrial targeting for turnover remain incompletely understood, it is believed that in addition to PINK1 and Parkin signaling, organelle size alone may serve as an important factor excluding a mitochondrion from autophagosome incorporation and lysosomal degradation (25, 51). Thus, the morphology of MERKO mitochondria alone supports our hypothesis that ERα deletion induces mitochondrial stress and impaired organelle turnover.
It is widely accepted that overproduction of ROS, decreased ROS scavenging capacity, and impaired DNA and protein repair mechanisms contribute to mtDNA damage and cellular aging (52). Considering that MERKO muscle exhibited decrements in all three processes, it follows that ERα inactivation is likely to accelerate muscle aging. Indeed, Esr1-KD myotubes showed increased susceptibility of mtDNA to mutations induced experimentally by exogenous H2O2. Cells are typically equipped with a sensitive mitochondrial surveillance system to combat the damaging effects of excessive mitochondrial ROS production by elimination of compromised mitochondrial components via mitophagy. We show that this surveillance system, specifically PINK1 and Parkin responsiveness to CCCP and H2O2, was markedly depressed in the absence of ERα. Muscle from MERKO mice phenocopied features of loss-of-function mutations in PINK1 and Parkin in model organisms including impaired mitochondrial function, retention of enlarged organelles, sensitivity to oxidative stress, and derangements in metabolism contributing to reduced longevity (53, 54). Polymorphisms in Park family members have now been linked with T2D susceptibility and aging-related disease in humans (55). More recently, we confirmed this clinical relationship in mice by showing that muscle Parkin inactivation impairs mitochondrial function and insulin action (33).
Mitochondrial fission is thought to be a central process required for the autophagic degradation of damaged mitochondrial components (that is, mitophagy). In mammals, mitochondrial fission is primarily regulated by the dynamin-related GTPase (guanosine triphosphatase) DRP1. Upon stimulation, DRP1 translocates to the mitochondria and via self-assembly acts to remodel the outer mitochondrial membrane, making the organelle physically available for digestion by the lysosome, relocalization by the cytoskeleton, or fusion with other mitochondria for retention to the network (16, 56, 57). Similar to findings in ERα-deficient muscle, loss-of-function Drp1 mutant cells are fission-incompetent and exhibit a fused and extensively tubulated mitochondrial morphology that is associated with reduced ATP synthesis, increased ROS production (58), and mtDNA replication stress–linked DNA damage (59). Our studies in Drp1-KD muscle cells recapitulate these published findings and strongly implicate a link between impaired fission signaling and the development of insulin resistance. Indeed, recent findings by Drummond et al. show reduced Drp1 and Parkin expression levels in muscle paralleled by hyperinsulinemia in inactive elderly women compared with younger female controls (60).
In further support of fission incompetency in the context of ERα inactivation, calcineurin, the fission-promoting Ser/Thr phosphatase (61, 62) shown to dephosphorylate and activate DRP1, was reduced in expression (regulatory subunit CnB), overall activity, and protein association with Drp1 in MERKO muscle compared with Control f/f. Impaired calcineurin action was paralleled by a marked increase in transcript and protein expression of the calcineurin inhibitor RCAN1 in both MERKO muscle and Esr1-KDmyotubes. Notably, RCAN1 expression was also significantly increased in muscle from women displaying clinical features of the MetSyn. PKA-mediated phosphorylation of RCAN1 enhances its inhibitory effect on calcineurin, which, given the increased phosphorylation and total activity of PKA seen in parallel with a marked phosphorylation of DRP1Ser637 in MERKO muscle, is suggestive of a primary role of PKA-Rcan1–mediated repression of fission in Esr1-deficient muscle by calcineurin-DRP1 inactivation. Our findings support that ERα exerts a strong regulatory function over the PKA-calcineurin-Rcan1-DRP1 axis to control mitochondrial dynamics and maintain a healthy organelle network.
In overview, our studies highlight the critical importance of skeletal muscle ERα for the maintenance of mitochondrial health and metabolic homeostasis. This research provides clinically relevant evidence that natural variation in expression of muscle ERα is predictive of adiposity, and importantly, we showed that muscle ESR1 expression is diminished in women displaying clinical features of the MetSyn. Our findings have clear implications for understanding the pathobiology of metabolic dysfunction in women, and support that strategies to maintain or elevate ERα action in muscle may be of therapeutic benefit in ameliorating complications associated with metabolic-related disease.
MATERIALS AND METHODS
Study design
The objective of this research was to understand the role of muscle ERα in regulating metabolic homeostasis and insulin sensitivity. First, to establish a clinical rationale for our studies in genetically engineered mice, we determined the relationship between muscle ERα expression and indices of metabolic function using historical deidentified data and sample from a human study previously conducted by Pederson and colleagues from the University of Copenhagen (63, 64) and mouse data and samples from previous studies performed on mice of the University of California, Los Angeles, Hybrid Mouse Diversity Panel (UCLA HMDP) (65). Each subject provided written informed consent before the original study participation, and the study procedures were approved by the Scientific-Ethical Committee of the Frederiksberg and Copenhagen counties and are in accordance with the principles of the Declaration of Helsinki. All procedures in rodents were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the Animal Subjects Committee of UCLA. Because muscle ERα expression was inversely correlated with adiposity and fasting insulin levels, we next determined whether muscle ERα levels were reduced in women with MetSyn compared with lean healthy controls. Indeed, we observed a marked reduction in muscle ERα levels in women with MetSyn.
To determine whether the metabolic dysfunction in MetSyn subjects might be causally related to the reduction in muscle ERα expression levels, next we generated MERKO mice using the standard Lox-Cre approach. We performed an exhaustive phenotypic evaluation of at least 10 cohorts of MERKO and Control f/f mice using a variety of in vivo and ex vivo approaches. The number of animals used for each study was determined by Power calculations using a desired P value of 0.05 (animal numbers for each study are indicated in the figure legends), and all studies performed to assess glucose homeostasis and insulin action were blinded for animal genotype. In addition to studies in rodents, we generated primary myotubes from muscles obtained from Control f/f and MERKO mice as well as C2C12 cells with shRNA-mediated KD of ERα using a lentiviral-mediated approach. Nearly all in vitro studies were performed in triplicate with a minimum of three independent replicates as indicated in the figure legends.
Animals
The strategy for the generation of the floxed exon 3 ERα (ERαf/f) was described in (66). Floxed mice were crossed with a muscle-specific Cre transgenic mouse (MCK-Cre) line and bred to obtain the following genotypes of females: MCK-Cre-ERαf/f (MERKO) and ERαf/f CRE-negative control mice (Control f/f). Animals were studied at various ages as indicated. Primers used for genotyping are in table S5. Tissues from 10 strains of female inbred mice (16 weeks of age)—(i) FVB/NJ, (ii) PL/J, (iii) BXA2/PgnJ, (iv) AxB4/PgnJ, (v) CxB3/ByJ, (vi) BxD34/TyJ, (vii) BxD24/TyJ-Cep293, (viii) BxD24/TyJ-Cep293, (ix) CxB12/HiAJ, and (x) BxD1/TyJ—were obtained from the Lusis Laboratory HMDP UCLA. Lean wild-type and obese (LepOb) female mice were obtained from Jackson Laboratories at 8 weeks of age and studied at 12 weeks of age. Skeletal muscle from ERα DBDΔ mice was obtained from K. Korach as previously described (67).
Control f/f and MERKO mice were maintained on a normal chow standard diet and divided into two general groups: basal or insulin-stimulated. Within these categories, mice were further divided into groups depending on the duration of fasting (6 to 24 hours) or whether they were used for in vivo (glucose clamps) or ex vivo (soleus muscle 2-deoxyglucose uptake assays) assessment of insulin sensitivity. Animals used for the deuterium studies to assess mtDNA replication were weight-matched and administered an intraperitoneal injection of 2H2O (600µl; Cambridge Isotope Laboratories) followed by ad libitum drinking of heavy water [4% (v/v) deuteriumoxide] for 30 days before euthanasia and hindlimb tissue harvest for mtDNA isolation and subsequent mass spectrometry analyses (as described below).
Human subjects
We recruited potential female volunteers by advertising on the Copenhagen hospital staff Web sites and in local newspapers. The potential volunteers visited the laboratory for an initial screening in the morning after an overnight fast. A health examination was conducted by a physician followed by a 2-hour 75-g oral glucose tolerance test (OGTT) and a maximum working rate (Pmax) test on a cycle ergometer (Ergometric 839E, Monark). Volunteers included in the study were either lean healthy individuals (n = 18; 9 premenopausal and 9 postmenopausal) or individuals classified as MetSyn showing IGT and/or insulin resistance (n = 25; 21 premenopausal and 4 postmenopausal) and in addition met the following criteria: overweight (BMI, >25 kg/m2), aged 30 to 60 years, and a low VO2max for their age and sex (68). Exclusion criteria were diabetes, impaired fasting glucose, blood pressure >140/90 mmHg, smoking, and any diagnosed systemic disease. All of the premenopausal women were in the mid to late follicular phase of their menstrual cycle (days 10 to 14), and oral contraceptives were included as an exclusion criteria. Subject characteristics are presented in table S1 and were, in part, previously described (64). The test day started at 8:00 a.m., after an overnight fast. Subjects were asked to refrain from drinking coffee, tea, soft drinks, or alcohol 24 hours before, and from performing any rigorous physical exercise 72 hours before the test day. Weight and height were measured to the nearest 0.1 kg and 0.5 cm, respectively. Blood pressure was measured twice in the supine position, with 10min of rest preceding each measurement, using an automatic sphygmomanometer (Greenlight 300, AC Cossor & Son Accoson Works) placed on the left arm.
Volunteers were then subjected to a 3-hour 75-g OGTT. A catheter was placed in an antecubital vein. Within a 5-min period, subjects drank 75 g of glucose (anhydrous glucose, Unikem) diluted in water (5 dl). Eight blood samples of 7 ml (the baseline sample contained 16ml) were taken for the determination of blood glucose and plasma concentrations of insulin. The blood samples were collected before glucose ingestion at 0min and at 10, 20, 30, 60, 90, 120, and 180 min after glucose ingestion. Blood samples were immediately centrifuged for 15 min at 2400g and 4°C. Plasma was removed and pipetted into 3 × 1.5–ml Eppendorf tubes (Brand), and stored at −80°C for subsequent analysis. Blood glucose and circulating hormone levels including insulin, estradiol, follicle-stimulating hormone, and luteinizing hormone were measured by the Department of Clinical Biochemistry, Rigshospitalet.
At 11:30 a.m., the volunteers received a light lunch and then were scanned in a dual-energy x-ray absorptiometry scanner (GE Medical Systems Lunar, Prodigy Advance) to estimate whole body fat percentage using software version 8.8. After a ~10-min rest on a hospital bed in the supine position, a muscle biopsy was obtained from the vastus lateralis using the Bergstrom percutaneous needle biopsy technique with suction. Biopsy samples (100 to 200 mg) were immediately frozen in liquid nitrogen and stored at −80°C until analyzed.
Animal characteristics
Blood was drawn from 6-hour–fasted, 20-week-old female mice and analyzed for circulating factors: glucose (HemoCue), insulin, leptin, and adiponectin (Millipore; table S2). After a 2-week recovery, GTTs (1 g/kg dextrose) were performed on 6-hour–fasted female mice as previously described (69, 70). Feeding and movement were recorded over 48 hours after animals were acclimated to metabolic chambers (Oxymax, Columbus Instruments).
Hyperinsulinemic-euglycemic clamp studies
Two weeks after the GTT, dual catheters were surgically placed in the right jugular vein, and glucose clamp studies were performed on female mice 3 days after surgery as previously described (69, 70). All animals were fasted for 6 hours before the clamp and studied in the conscious state. Basal glucose turnover was determined after a 90-min constant infusion of (5.0 µCi/hour, 0.12 ml/hour) d-[3-3H]glucose (PerkinElmer). After the basal period, glucose (50% dextrose, Abbott Laboratories) and insulin (12 mU kg−1 min−1, Novo Nordisk Pharmaceutical Industries) plus tracer (5.0 µCi/hour) infusions were initiated simultaneously, and glucose levels clamped at euglycemia (~120 mg/dl) using a variable GIR. At steady state, the total GDR, measured by tracer dilution technique, is equal to the sum of the rate of endogenous or hepatic glucose production and the exogenous (cold) GIR (69–71). The insulin-stimulated component of the total GDR is equal to the total GDR minus the basal glucose turnover rate.
Immunoblot analysis
Mouse tissue samples were pulverized in liquid nitrogen and homogenized in radioimmunoprecipitation assay (RIPA) lysis buffer containing freshly added protease (cOmplete EDTA-free, Roche) and phosphatase inhibitors (Sigma). All lysates were clarified, centrifuged, and resolved by SDS–polyacrylamide gel electrophoresis. Samples were transferred to polyvinylidene difluoride membranes and subsequently probed with the following antibodies for protein and phospho-protein detection: ERα and tubulin (Santa Cruz Biotechnology);GLUT4, Mfn2, and RCAN1-1/1-4 (Sigma-Aldrich);GAPDH and α-actinin (Millipore); pan-actin, AKAP, t-Akt/p-AktSer473, IKKβ/p-IKKα/βSer180/181, JNK/ p-JNKSer183/Tyr185, Parkin (4211 and 2132), LC3B, tDRP1/p-DRP1Ser616/637, and t-PKA/p-PKA (Cell Signaling); FIS1 (GeneTex); OPA1 (BD Biosciences); GPX3 and calcineurin pan and α/β (R&D Systems);MitoProfile OXPHOS (MS604/ab110413; CI-NDUFB8 20 kD, CII-SDHB 30 kD, CIII-UQCR2 Core protein 2 48 kD, CIV-MTCO1 40 kD, and CVATP5A 55 kD) and Porin (MitoSciences, Abcam); PINK1 (Cayman Chemical); p62 (Progen Biotechnik GmbH); and GFP (Abcam). To analyze tyrosine phosphorylation of IRS-1 (Santa Cruz Biotechnology), lysates were immunoprecipitated overnight with IRS-1 or IRS-2 antibody (Santa Cruz Biotechnology) and immobilized on protein A (Upstate Millipore) or G agarose beads (Santa Cruz Biotechnology). Phosphorylation was detected by immunoblotting with anti-pY20 antibody (Cell Signaling). Additionally, immunoprecipitates were also immunoblotted for PI 3-kinase (phosphatidylinositol 3-kinase) p85 protein (Santa Cruz Biotechnology). Densitometric analysis was performed using Bio-Rad Quantity One image software.
Muscle lipids and lipid intermediates
Lipids were extracted from the quadriceps muscle by the Folch method (72). Triacylglycerol was saponified in an ethanol/KOH solution at 60°C, and glycerol content was determined fluorometrically. DAG and ceramide were extracted and quantified as previously described (72–74). Lipid accumulation in Drp1KD myotubes was assessed by Oil Red O staining. Differentiated cells were fixed for 1 hour in 10% formalin. After washing with 60% isopropanol, cells were stained with Oil Red O for 10 min. Cells were rinsed with water and then imaged. Quantification was performed by eluting Oil Red O with 100% isopropanol for 10 min and by measuring optical density at 500 nm for a 0.5-s reading.
Quantitative reverse transcription PCR
Tissues were first homogenized using TRIzol reagent, and RNA was isolated and further cleaned using RNeasy columns (Qiagen) with DNase digestion. For RNA isolation from cells, the RNeasy Plus Kit was used as per the manufacturer’s instructions. Complementary DNA synthesis was performed using 1 to 3 µg of RNA with SuperScript II Reverse Transcriptase (Invitrogen). PCRs were prepared using iQSYBR Green Supermix (Bio-Rad). All PCRs were performed in a Bio-Rad MyiQ Real-Time Detection System. Quantification of a given gene, expressed as relative mRNA level compared with control, was calculated after normalization to a standard housekeeping gene (β-actin, Ppia). We performed separate control experiments to ensure that the efficiencies of target and reference amplification are equal, as described in the User Bulletin 2 from Applied Biosystems. Primer pairs were designed using Primer Express 2.0 software (Applied Biosystems) or previously published sequences (70, 75, 76). Primer sets were selected spanning at least one exon-exon junction when possible and were checked for specificity using BLAST (Basic Local Alignment Search Tool; National Center for Biotechnology Information). The specificity of the PCR amplification was confirmed by melting curve analysis ensuring that a single product with its characteristic melting temperature was obtained. To determine mtDNA abundance, total DNA was isolated from cells or tissues using the DNeasy Kit (Qiagen), and the mtCO3 oligos and SDHA (succinate dehydrogenase complex subunit A) were used for quantification of mitochondrial and nuclear genomes. Primer sequences for the specific target genes analyzed are in table S5.
Illumina microarray analyses
Microarray analysis was performed using the Illumina Bead Chip System (MouseRef-8 v2.0) by the Southern California Genome Consortium on RNA isolated from quadriceps as described above. Each of the eight separate arrays on the chip (four Control f/f and four MERKO) was performed on pooled RNA (from three mice) that was subjected to purity and integrity analysis using an Agilent Bioanalyzer. Array internal control and basic analysis was performed using GenomeStudio (v2010.3). Findings for differential gene expression MERKO versus Control f/f are presented at 5% false discovery rate.
Cell culture, transfections, and treatments
C2C12 cells were maintained in high-glucose Dulbecco’smodifiedEagle’s medium (DMEM) 10% fetal bovine serum with penicillin/streptomycin. To obtain C2C12myotubes, cells were allowed to reach confluence and the medium was switched for high-glucoseDMEM2%horse serum with penicillin/streptomycin for 5 to 7 days. Bafilomycin A1 (25 nM) and cycloheximide (Sigma; 20 µM) were used to block lysosomal degradation and protein synthesis, respectively. Chloramphenicol (Sigma; 100 µg/ml) was used to block mitochondrial protein synthesis, and carbonyl cyanide m-chlorophenylhydrazone (CCCP; 20 µM) is a mitochondrial uncoupler used to induce mitophagy. H89 kinase inhibitor (Tocris; 50 µM) was used to test the impact of PKA inhibition on LC3B processing.
For the induction of mtDNA mutations with H2O2, C2C12 cells were treated with 0.5mMH2O2 (Sigma) for 24 hours and left to recover in growth medium for an additional 24 hours. Mitochondria were isolated using a dounce homogenizer and the Mitochondria Isolation Kit for Cultured Cells according to the manufacturers’ instructions (Pierce/Thermo Scientific), and mtDNA was extracted using DNeasy extraction kit (Qiagen). Afterward, mtDNA was digested with endonuclease Taq I for 6 hours with the addition of 100 U of enzyme every hour. Then, two sets of primers were used in real-time qPCR as described for RMC assay (77). The first set flanks one Taq I site that is amplified only when there is a pointmutation in any of the bases of the Taq I target site. The second set amplifies an adjacent region not containing a Taq I site, which quantifies the total amount of mtDNA in the sample (table S5). Point mutation frequency was calculated by the amount of mutant molecules normalized to the amount of total mtDNA molecules. For tandem Lc3b imaging, C2C12 myocytes were transfected with tandem mRfp-Gfp-Lc3 plasmid (Addgene plasmid #21074) onto glass coverslips using Lipofectamine Plus reagent (Invitrogen).
Lentiviral-induced ERαKD in C2C12 myocytes
To achieve Esr1-KD, lentiviral particles (Sigma) carrying shRNA targeted to ERα were used to transduce C2C12 myoblasts. After selecting positive transformants with puromycin (5 µg/ml), the selected clones were expanded and analyzed for KD efficiency as measured by qRTPCR and immunoblotting. The resulting cultures were then used for subsequent assays in the undifferentiated and differentiated states.
Mitochondria isolation from C2C12 myotubes
Mitochondria were isolated from myotubes as instructed (Mitochondria Isolation Kit for Culture Cells, Thermo Scientific, 89874). Briefly, 2 × 107 cell pellet was resuspended with mitochondria isolation reagent A and vortexed at medium speed for 5 s and then incubated on ice for 2 min. After incubation, 10 µl of reagent B was added and vortexed at maximum speed for 5 s followed by an additional incubation on ice for5min. After adding 800 µl of reagent C, tubes were inverted and then centrifuged (700g for 10min at 4°C). In a new tube, the supernatant was centrifuged again (12,000g for 15min at 4°C). The mitochondrial pellet was spun (12,000g for 5 min) and cleaned again by adding 500 µl of reagent C before mitochondrial lysis using RIPA buffer.
Insulin signal transduction in C2C12 myotubes
C2C12 myotube insulin signaling was performed in 12-well culture plates. Briefly, cells were serum-starved for 2 hours in low-glucose DMEM containing 0.1% bovine serum albumin (BSA), then placed in phosphate-buffered saline (PBS) containing 0.1% BSA for 30 min, and allowed to equilibrate. Cells were then stimulated with or without insulin (dose response 1, 10, and 100 nM) in the same medium for 15 min. Insulin signaling was terminated by four sequential washes with ice-cold PBS before lysis in 0.3 M NaOH.
Fatty acid oxidation and esterification in myocytes
Myotube fatty acid oxidation and esterification were performed in six-well culture plates, as adapted from the method previously described (78, 79). Briefly, cells were incubated in low-glucose DMEM containing 1% fatty acid–free BSA and 0.5mMpalmitate for 1 hour. After this, the medium was replaced with the addition of [14C]palmitate (1 µCi/ml) and allowed to incubate for a further 2 hours. Complete oxidation of palmitate was measured by benzethonium hydroxide captured 14CO2, liberated from acid-treated culture medium. Cellular aqueous and esterified lipids were isolated from cell monolayers by sequential inorganic and organic phase separations, respectively. Calculations were normalized to the specific activity of [14C]palmitate in the medium.
Cellular oxygen consumption in myocytes
Real-time measurement of oxygen consumption and proton production in cells was carried out using the XF24 Extracellular Flux Analyzer (Seahorse Bioscience). To directly assess mitochondrial metabolism, measurements of oxygen consumption were made continuously while cells were sequentially treated with oligomycin (ATP synthase inhibitor, which allows determination of the levels of ATP synthesis), FCCP (an uncoupler, which allows determination of maximal mitochondrial respiratory capacity), and rotenone/myxothiazol (inhibitors of complex I/III of the electron transport chain, which allow determination of the nonmitochondrial oxygen consumption).
ROS production
C2C12 myoblasts were washed and incubated in low-glucose DMEM at 37°C, 5% CO2 in the dark with 25 µMcarboxy-H2DCF-DA (Molecular Probes), washed with PBS and incubated for 15min with 5 µMmitoSOX (Molecular Probes), washed and quickly trypsinized, pelleted, and retained on ice. Cells were resuspended in FACS (fluorescence-activated cell sorting) buffer (PBS 3%BSA) with DAPI (4′,6-diamidino-2-phenylindole) (25 µg/ml) and analyzed immediately by flow cytometry on an LSRII (Becton Dickinson) with FlowJo software (Tree Star Inc.). Unstained and single stains were used for establishing compensation and gates, and only live cells (DAPI-negative) were analyzed by the software.
Ex vivo skeletal muscle glucose uptake
Whole-muscle ex vivo glucose uptake was assessed using 2-deoxyglucose, with minor changes to that described previously (80). Briefly, soleus muscles were carefully excised from anesthetized animals and immediately incubated for 30 min in complete Krebs-Henseleit buffer with or without insulin (60 µU/ml) at 35°C. Muscles were then transferred to the same buffer containing [3H]2-deoxyglucose (3 µCi/ml) and [14C]mannitol (0.053 µCi/ml), and incubated for exactly 20min before being blotted and snap-frozen. Muscles were homogenized in lysis buffer and counted for radioactivity or subjected to Western blotting. Glucose uptake was standardized to the nonspecific uptake of mannitol and estimated as micromole of glucose uptake per gramof tissue.
Citrate synthase activity
Citrate synthase enzyme activity assays were performed on soleus and gastrocnemius muscle using standard methods, as previously described (81).
Mitochondrial calcium retention capacity
Preparation of mitochondria was adapted from quick isolation method, previously described (82). Gastrocnemius muscles were placed in isolation buffer A (70 mM sucrose, 210 mM mannitol, 1m MEDTA, 50 mM tris, pH 7.4). The tissue was finely minced with scissors and digested with bacterial proteinase type XXIV (0.2 mg/ml) (Sigma) for 5 min on ice, washed, and quickly homogenized in a Potter-Elvehjem tissue grinder in the same buffer supplemented with 1% albumin. The homogenate was centrifuged at 1300g for 3min, and the supernatant was centrifuged at 10,000g for 10 min. The mitochondrial pellet was resuspended in isolation buffer B (150 mM sucrose, 50 mM KCl, 2 mM KH2PO4, succinic acid, and 20 mM tris-HCl, pH 7.4). The initiation of mPTP opening was assessed after in vitro Ca2+ overload as previously described (82, 83). Free Ca2+ concentration outside the mitochondria was recorded with 0.5 µM calcium green-5N (Molecular Probes) (excitation, 500 nm; emission, 530 nm). Isolated mitochondria (500 µg of protein) were suspended in 2 ml of buffer B. Samples were preincubated in continuous stirring for 90 s in the spectrofluorometer cell, and CaCl2 pulses (200 pmol of CaCl2) were applied every 60 s. The calcium retention capacity was defined as the amount of Ca2+ required to trigger Ca2+ release and is expressed as micromole of CaCl2 per milligram of mitochondrial protein.
Muscular force and endurance analyses
Mice were anesthetized (sodium pentobarbital, 60 mg/kg), and the soleus muscle was dissected with both tendons intact. The muscles were mounted in a chamber continuously perfused with an oxygenated (95% O2, 5% CO2) Tyrode’s solution (121 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 0.5m M MgCl2, 0.4 mM NaH2PO4, 24 mM NaHCO3, 5.5mM glucose, 0.1 mM EGTA) at room temperature to ensure no unstirred layers around the muscle, sufficient oxygenation to the core of the muscle, and stable pH (7.4). One tendon was tied with silk thread to a force transducer and the other tendon was tied to an adjustable tube at the opposite end of the chamber, allowing muscle length to be altered incrementally to determine optimal muscle length (Lo) and the length at which maximal tetanic force is produced. Muscles were stimulated at supramaximal voltage using platinum electrodes placed on either side of the muscle. After 15 min of equilibration, the muscle was tested for maximal isometric tetanic force and fatigue.
Maximal isometric tetanic force was elicited with a 500-ms train duration and 0.2-ms pulse duration at 80 Hz. Fatigue was measured using isometric tetanic contractions of 500-ms train duration and 0.2-ms pulse duration at 50 Hz. Stimulation frequency was increased every 2 min in a progressive manner (from a contraction every 8 to 4 to 3 to 2 to 1 s) and terminated when the developed force had fallen to ~50%of the initial maximal developed force. Muscle fatigue was determined by the duration of time to reach 60% of the maximal developed force. After testing, the cross-sectional area was calculated as previously described (84), and the maximal isometric tetanic force (Po) was expressed as N/cm2.
Electron microscopy
Muscles were harvested and immediately immersed in 2% glutaraldehyde in PBS for 2 hours at room temperature and then at 4°C overnight. Fixed tissues were washed and postfixed in a solution of 1% OsO4 for 2 hours. After further washes in buffer, tissues were dehydrated through serial immersions in graded ethanol solutions (50 to 100%), passed through propylene oxide, and infiltrated in mixtures of Epon 812 and propylene oxide 1:1 and then 2:1 for 2 hours each and then in pure Epon 812 overnight. Embedding was then performed in pure Epon 812, and curing was done at 60°C for 48 hours. Muscle longitudinal sections of 60-nm thickness were cut using an ultramicrotome (RMC MTX). The sections were double-stained in aqueous solutions of 8% uranyl acetate for 25 min at 60°C and lead citrate for 3 min at room temperature. Thin sections were subsequently examined with a 100CX JEOL electron microscope at the UCLA Brain Research Institute. The mitochondrial area was assessed for 300 to 350 organelles per mouse (n = 3) per genotype using ImageJ.
Confocal microscopy and flow cytometry
To detect autophagy, both C2C12 control scramble (Scr) and Esr1-KD myocytes were incubated with Rfp-Gfp-Lc3b (tandem Lc3) adenovirus generated using the ViraPower Adenoviral Expression System (Invitrogen) according to the manufacturer’s instructions. Briefly, the coding sequence for tandem Lc3 with flanking BP sites was amplified from the Addgene plasmid #21074 using High-Fidelity Pfx50 DNA polymerase (Invitrogen). Entry clones for tandem Lc3 were obtained by recombination of this purified DNA fragment with pDONR221 vector using Gateway BP Clonase II. Tandem Lc3 insert was transferred from the entry clones into the pAd/CMV/V5-DEST vector (Invitrogen) using the Gateway LR Clonase II enzyme mix. Recombinant adenoviral purified plasmid was used to generate adenoviral particles according to vector manufacturer’s instructions (Invitrogen). Adenoviral titer was determined using the Adeno-X Rapid Titer Kit (Clontech). Autophagy was induced with nutrient starvation in Hanks’ balanced salt solution (Gibco) for 4 or 8 hours followed with or without autophagy inhibition by 25 nM bafilomycin A1 (Sigma) for another 4 hours. After treatment, cells were washed twice with PBS, fixed in 4% paraformaldehyde for 15 min at room temperature, and resuspended in FACS buffer for analysis on an LSRII (Becton Dickinson) using FlowJo software (Tree Star Inc.). Untreated cells, cells infected with adenovirus carrying EGFP-LC3, and cells transfected with mRFP-LC3 plasmid were used for establishing compensation and gates.
For tandem LC3B imaging by confocal microscopy, confluent C2C12 myocytes were transfected with tandem Rfp-Gfp-LC3 plasmid (Addgene plasmid #21074) onto glass coverslips using Lipofectamine Plus reagent (Invitrogen). Cells then were induced to differentiate to myotubes for 5 days. For imaging, cells were nutrient-starved with Hanks’ balanced salt solution for 4 hours and then fixed for 15 min in 4% paraformaldehyde-PBS at room temperature, washed with PBS twice, and sealed before being visualized on a Leica TCS-SP2 AOBS confocal microscope.
Newly synthesized mtDNA labeling with 2H2O
A highly enriched population of mitochondria were isolated from hindlimb muscles obtained from female Control f/f and MERKO mice after 30 days of heavy water (Cambridge Isotope Laboratories; deuterium oxide) consumption with modifications to previously described techniques (85, 86). mtDNA was isolated, assessed for purity (Control f/f 91.5 ± 2.62% versus MERKO 92 ± 1.54%; P = 0.756), and digested with a cocktail of nuclease enzymes using a commercial kit following the manufacturers’ protocol (DNA Degradase Plus, Zymo Research; 2.5 µl of 10× DNA Degradase Reaction buffer, 1 µl of DNA Degradase Plus, and water to make a total reaction volume of 25 µl). After incubation (37°C, >1 hour), aqueous formic acid was added (25 µl; 0.1%, v/v) to yield a final concentration of 40 ng of digested DNA/µl. The DNA hydrolysis samples were injected onto a reversed-phase column (Hypercarb 2.1 × 100 mm, 3-µm particle size, Thermo Scientific) equilibrated with buffer A (0.1% aqueous formic acid) and eluted (200 µl/min) with an increasing concentration of buffer B (methanol: min/%B; 0/5, 2/5, 12/50, 14/50, 15/5, 20/5). The effluent from the column was directed to an electrospray ion source (Agilent Jet Stream) connected to a triple quadrupole mass spectrometer (Agilent 6460 QQQ) operating in the positive ion neutral loss mode of 116, 117, 118, and 119, which corresponds to the loss of a neutral fragment of unlabeled deoxyribose (dR), one deuterium–labeled dR (dR+1), two deuterium–labeled dR (dR+2), and three deuterium–labeled dR (dR+3). The intensity of precursor ion transitions from either dR loss was recorded. The precursor ion or the nucleosides of interest were the following: deoxycytosine (dC) mass/ charge ratio (m/z) 228.1, thymidine (dT) m/z 243.1, deoxyguanosine (dG) m/z 268.1, and deoxyadenosine (dA) m/z 252.1. The measured peak area for the nucleosides of a specific dR loss was measured and analyzed (Agilent MassHunter Quantitative Analysis, version B.04.00). The measured percentage of labeled nucleosides was calculated by the ratio of the labeled to the total peak area counts for each nucleoside.
Statistics
Values presented are expressed as means ± SEM. Statistical analyses were performed using Student’s t test when comparing two groups of samples or one-way ANOVA with Tukey’s post hoc comparison for identification of significance within and between groups using SPSS (Graduate Pack) or GraphPad Prism 5 (GraphPad Software). Significance was set a priori at P < 0.05.
Supplementary Material
Acknowledgments
We thank J. Olefsky and H. Cohen from the University of California, San Diego/UCLA Diabetes Research Center for their continued support of our work; D. Chan, M. Guo, D. Shackelford, and A. van der Bliek for their helpful discussions regarding mitochondrial fusion, fission, and mitophagy; C. Pan and C. Rau for assistance with bioinformatics; D. Henstridge for assistance with the isolation of primary muscle cells; S. Kohan from the UCLA Brain Research Institute for assistance with electron microscopy analyses; R. Boyadjian for assistance with measuring circulating factors; W. Foster for assistance with the PKA activity assay; K. Wroblewski for assistance with metabolic chamber analyses; and L. Nielsen for assistance with qRT-PCR analyses on human muscle.
Funding: These studies were supported in part by research grants from the NIH [DK060484, DK073227, and Nuclear Receptor Signaling Atlas (NURSA) Data Source Project grant DK097748], UCLA Department of Medicine, and UCLA Iris Cantor Women’s Health Center–UCLA Clinical and Translational Science Institute (UL1TR000124) to A.L.H. and Z.Z. V.R. was supported by the Spanish Ministry of Health, and B.G.D. was supported by the Australian National Health and Medical Research Council and the UCLA Jonsson Comprehensive Cancer Center. During the study period, the Centre of Inflammation and Metabolism was supported by a grant from the Danish National Research Foundation (DNRF55) to B.K.P. A.J.L. is supported by NIH (grants HL28481 and HL30568), and B.W.P. is supported by NIH R00 grant HL123021. The Centre for Physical Activity Research is supported by a grant from TrygFonden. A.J.L. is supported by NIH grants HL28481 and HL30568, and L.V. and K.R. are supported in part by NIH grant HL28481. The Seahorse XF24 instrument was acquired by a shared instrument grant from the NIH National Center for Research Resources (S10RR026744). P.T. is an investigator of the Howard Hughes Medical Institute and is supported by NIH (DK063491).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/8/334/334ra54/DC1
Table S1. Basal metabolic characteristics in female subjects.
Table S2. Animal characteristics after a 6-hour fast in the basal or clamped state.
Table S3. Annotation of functionally enriched terms from Illumina microarray analyses on quadriceps muscle from female Control f/f versus MERKO mice.
Table S4. Microarray analyses on quadriceps muscle from Control f/f versus MERKO mice.
Table S5. Primer sequences for qPCR analyses on tissues and for genotyping of Control f/f and MERKO mice.
Fig. S1. Real-time respirometry in C2C12 myotubes.
Fig. S2. Muscle peak tension and time to fatigue.
Fig. S3. Polγ1 expression is regulated by ERα in muscle.
Fig. S4. Altered mitochondrial fission-fusion signaling in ERα-deficient C2C12 myotubes.
Fig. S5. Calcineurin-PKA-Rcan1 axis in C2C12 myotubes with Esr1-KD.
Fig. S6. Expression of autophagy-related genes in MERKO muscle.
Fig. S7. Autophagic flux studies in C2C12 myocytes with Esr1-KD.
Author contributions: These authors were involved in data collection and analysis: V.R. (performed in vitro insulin action, mitochondrial real-time respirometry, immunoblotting qPCR, RMC assays, and confocal microscopy studies), B.G.D. (cloning and fatty acid oxidation and insulin action assays), Z.Z. (studies of Drp1 action and mitochondrial isolations, immunoblotting, and qPCR), J.P. (performed genotyping, animal husbandry, immunoblotting, and qPCR), N.Y.K. (performed animal husbandry, genotyping, and immunoblotting), T.S. (performed animal husbandry and qPCR), P.D. (performed animal husbandry, genotyping, plasma analyses, and qPCR), K.W. and J.W. (performed muscle histological analyses and assisted with the RMC assays), T.Q.d.A.V. (designed and synthesized adenovirus), A.H.F. and S.B. (performed flow cytometry analyses of autophagy), T.L. (performed assessment of deuterium labeling), J.P.W. (performed mitochondrial proteomics analyses), S.W.B. (performed metabolic chamber analyses), B.W.P. (performed all studies associated with UCLA HMDP), L.V. (performed mitochondrial real-time respirometry), H.S. and J.C.B. (performed mitochondrial calcium homeostasis mPTP analyses), M.J.W. (performed bioactive lipid analyses in muscle), S.S. (muscle function testing), T.A. and M.K. (human study design and data collection), and A.L.H. (performed all in vivo studies in mice). These authors directed the research: A.L.H. (orchestrated the entire project and directed in vivo and ex vivo studies as well as performed data integration), E.S. and L.T. (directed mitochondrial calcium handling studies), P.T. (directed studies of indirect calorimetry and movement), K.R. (directed the real-time respirometry studies), B.K.P. (directed all human studies), and A.J.L. (directed studies associated with UCLA HMDP and gene arrays performed on f/f and MERKO mouse muscle). These authors contributed reagents, samples, or instrumentation: S.C.H. and K.S.K. (provided the ERα floxed and ERα DBD mutant mouse lines), P.T. (provided metabolic chambers), S.S. (provided instrumentation and reagents to assess muscle function), L.T. and E.S. (provided instrumentation and reagents to assess calcium fluxes in isolated mitochondria), K.R. (provided Seahorse Biosciences XF24 instrument), B.K.P. (provided reagents and tissues for human muscle analyses), A.J.L. (provided data and muscle samples from the HMDP studies), C.R. (provided mass spectroscopy instrumentation and reagents for the deuterium studies), and J.P.W. (provided mass spectroscopy instrumentation and reagents for the mitochondrial proteomics analyses). These authors contributed to drafting of the original manuscript: V.R., B.G.D., Z.Z., and A.L.H. All authors contributed to the final drafting of the manuscript.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.DeFronzo RA, Bonadonna RC, Ferrannini E. Pathogenesis of NIDDM: A balanced overview. Diabetes Care. 1992;15:318–368. doi: 10.2337/diacare.15.3.318. [DOI] [PubMed] [Google Scholar]
- 2.Park Y-W, Zhu S, Palaniappan L, Heshka S, Carnethon MR, Heymsfield SB. The metabolic syndrome: Prevalence and associated risk factor findings in the US population from the Third National Health and Nutrition Examination Survey, 1988–1994. Arch. Intern. Med. 2003;163:427–436. doi: 10.1001/archinte.163.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hevener A, Reichart D, Janez A, Olefsky J. Female rats do not exhibit free fatty acid–induced insulin resistance. Diabetes. 2002;51:1907–1912. doi: 10.2337/diabetes.51.6.1907. [DOI] [PubMed] [Google Scholar]
- 4.Gómez-Pérez Y, Amengual-Cladera E, Català-Niell A, Thomàs-Moyà E, Gianotti M, Proenza AM, Lladó I. Gender dimorphism in high-fat-diet-induced insulin resistance in skeletal muscle of aged rats. Cell. Physiol. Biochem. 2008;22:539–548. doi: 10.1159/000185538. [DOI] [PubMed] [Google Scholar]
- 5.Frias JP, Macaraeg GB, Ofrecio J, Yu JG, Olefsky JM, Kruszynska YT. Decreased susceptibility to fatty acid–induced peripheral tissue insulin resistance in women. Diabetes. 2001;50:1344–1350. doi: 10.2337/diabetes.50.6.1344. [DOI] [PubMed] [Google Scholar]
- 6.Ribas V, Nguyen MTA, Henstridge DC, Nguyen A-K, Beaven SW, Watt MJ, Hevener AL. Impaired oxidative metabolism and inflammation are associated with insulin resistance in ERα-deficient mice. Am. J. Physiol. Endocrinol. Metab. 2010;298:E304–E319. doi: 10.1152/ajpendo.00504.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Eon TM, Souza SC, Aronovitz M, Obin MS, Fried SK, Greenberg AS. Estrogen regulation of adiposity and fuel partitioning. Evidence of genomic and non-genomic regulation of lipogenic and oxidative pathways. J. Biol. Chem. 2005;280:35983–35991. doi: 10.1074/jbc.M507339200. [DOI] [PubMed] [Google Scholar]
- 8.Campbell SE, Mehan KA, Tunstall RJ, Febbraio MA, Cameron-Smith D. 17β-Estradiol upregulates the expression of peroxisome proliferator-activated receptor α and lipid oxidative genes in skeletal muscle. J. Mol. Endocrinol. 2003;31:37–45. doi: 10.1677/jme.0.0310037. [DOI] [PubMed] [Google Scholar]
- 9.Yki-Järvinen H. Sex and insulin sensitivity. Metabolism. 1984;33:1011–1015. doi: 10.1016/0026-0495(84)90229-4. [DOI] [PubMed] [Google Scholar]
- 10.Borrás C, Sastre J, García-Sala D, Lloret A, Pallardó FV, Viña J. Mitochondria from females exhibit higher antioxidant gene expression and lower oxidative damage than males. Free Radic. Biol. Med. 2003;34:546–552. doi: 10.1016/s0891-5849(02)01356-4. [DOI] [PubMed] [Google Scholar]
- 11.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, Akira S, Yamamoto A, Komuro I, Otsu K. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salminen A, Kaarniranta K, Kauppinen A. Inflammaging: Disturbed interplay between autophagy and inflammasomes. Aging. 2012;4:166–175. doi: 10.18632/aging.100444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 14.Tschopp J. Mitochondria: Sovereign of inflammation? Eur. J. Immunol. 2011;41:1196–1202. doi: 10.1002/eji.201141436. [DOI] [PubMed] [Google Scholar]
- 15.Chan DC. Mitochondria: Dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 17.Mai S, Muster B, Bereiter-Hahn J, Jendrach M. Autophagy proteins LC3B, ATG5 and ATG12 participate in quality control after mitochondrial damage and influence lifespan. Autophagy. 2012;8:47–62. doi: 10.4161/auto.8.1.18174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barros RPA, Machado UF, Warner M, Gustafsson J-A. Muscle GLUT4 regulation by estrogen receptors ERβ and ERα. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1605–1608. doi: 10.1073/pnas.0510391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes. 2005;54:8–14. doi: 10.2337/diabetes.54.1.8. [DOI] [PubMed] [Google Scholar]
- 20.Baltgalvis KA, Greising SM, Warren GL, Lowe DA. Estrogen regulates estrogen receptors and antioxidant gene expression in mouse skeletal muscle. PLOS One. 2010;5:e10164. doi: 10.1371/journal.pone.0010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taddeo EP, Laker RC, Breen DS, Akhtar YN, Kenwood BM, Liao JA, Zhang M, Fazakerley DJ, Tomsig JL, Harris TE, Keller SR, Chow JD, Lynch KR, Chokki M, Molkentin JD, Turner N, James DE, Yan Z, Hoehn KL. Opening of the mitochondrial permeability transition pore links mitochondrial dysfunction to insulin resistance in skeletal muscle. Mol. Metab. 2014;3:124–134. doi: 10.1016/j.molmet.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Bliek AM. Fussy mitochondria fuse in response to stress. EMBO J. 2009;28:1533–1534. doi: 10.1038/emboj.2009.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rambold AS, Kostelecky B, Lippincott-Schwartz J. Fuse or die: Shaping mitochondrial fate during starvation. Commun. Integr. Biol. 2011;4:752–754. doi: 10.4161/cib.17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rambold AS, Kostelecky B, Lippincott-Schwartz J. Together we are stronger: Fusion protects mitochondria from autophagosomal degradation. Autophagy. 2011;7:1568–1569. doi: 10.4161/auto.7.12.17992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam ZY, Gruber J, Halliwell B, Gunawan R. Mathematical modeling of the role of mitochondrial fusion and fission in mitochondrial DNA maintenance. PLOS One. 2013;8:e76230. doi: 10.1371/journal.pone.0076230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat. Genet. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- 29.Gomes LC, Scorrano L. Mitochondrial elongation during autophagy: A stereotypical response to survive in difficult times. Autophagy. 2011;7:1251–1253. doi: 10.4161/auto.7.10.16771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer CS, Osellame LD, Stojanovski D, Ryan MT. The regulation of mitochondrial morphology: Intricate mechanisms and dynamic machinery. Cell. Signal. 2011;23:1534–1545. doi: 10.1016/j.cellsig.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 31.Kowald A, Kirkwood TBL. Evolution of the mitochondrial fusion–fission cycle and its role in aging. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10237–10242. doi: 10.1073/pnas.1101604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narendra DP, Jin SM, Tanaka A, Suen D-F, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLOS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drew BG, Ribas V, Le JA, Henstridge DC, Phun J, Zhou Z, Soleymani T, Daraei P, Sitz D, Vergnes L, Wanagat J, Reue K, Febbraio MA, Hevener AL. HSP72 is a mitochondrial stress sensor critical for Parkin action, oxidative metabolism, and insulin sensitivity in skeletal muscle. Diabetes. 2014;63:1488–1505. doi: 10.2337/db13-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin L-S, Choi A, Chu CT, Chung J, Clark RSB, Clarke PGH, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Dennis PB, Dennis PA, Demarchi F, Deretic V, Devenish RJ, Di Sano F, Dice JF, Distelhorst CW, Dinesh-Kumar SP, Eissa NT, DiFiglia M, Djavaheri-Mergny M, Dorsey FC, Dröge W, Dron M, Dunn WA, Jr, Duszenko M, Elazar Z, Esclatine A, Eskelinen E-L, Fésüs L, Finley KD, Fuentes JM, Fueyo-Margareto J, Fujisaki K, Galliot B, Gao F-B, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, González-Estévez C, Gorski SM, Gottlieb RA, Häussinger D, He Y-W, Heidenreich K, Hill JA, Høyer-Hansen M, Hu X, Huang W-P, Iwasaki A, Jäättelä M, Jackson WT, Jiang X, Jin SV, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JAKW, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovács AL, Kroemer G, Kuan C-Y, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei H-Y, Levine B, Lieberman AP, Lim K-L, Lin F-C, Liou W, Liu LF, Lopez-Berestein G, López-Otín C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Meléndez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Münz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nürnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri IS, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny AA, Silva-Zacarin ECM, Simon H-U, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Tallóczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcátegui NL, van der Klei IJ, Vaquero EC, Vellai T, Vogel MW, Wang H-G, Webster P, Xi Z, Xiao G, Yahalom J, Yang J-M, Yap GS, Yin X-M, Yoshimori T, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 37.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi RK, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NYO, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai X-Y, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ETW, Juhász G, Bartholomew CR, Bassham DC, Bast RC, Jr, Batoko H, Bay B-H, Beau I, Béchet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin J-P, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahová M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac V, Carneiro LAM, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Chae H-J, Chai C-Y, Chan DC, Chan EY, Chang RC-C, Che C-M, Chen C-C, Chen G-C, Chen G-Q, Chen M, Chen Q, Chen SS-L, Chen W, Chen X, Chen X, Chen X, Chen Y-G, Chen Y, Chen Y, Chen Y-J, Chen Z, Cheng A, Cheng CHK, Cheng Y, Cheong H, Cheong J-H, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang H-L, Chiarelli R, Chiba T, Chin L-S, Chiou S-H, Chisari FV, Cho CH, Cho D-H, Choi AMK, Choi D, Choi KS, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Chuang T-H, Chueh S-H, Chun T, Chwae Y-J, Chye M-L, Ciarcia R, Ciriolo MR, Clague MJ, Clark RSB, Clarke PGH, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronjé MJ, Cuervo AM, Cullen JJ, Czaja MJ, D’Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, de Groot JF, de Haan CAM, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LMD, Demarchi F, Deng YZ, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Ding W-X, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff KE, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Eissa NT, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Eng KE, Engelbrecht A-M, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Eskelinen E-L, Espert L, Espina V, Fan H, Fan J, Fan Q-W, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Farre J-C, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fésüs L, Feuer R, Figueiredo-Pereira ME, Fimia GM, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Gao F-B, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Ginsberg HN, Golab J, Goligorsky MS, Golstein P, Gomez-Manzano C, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, González-Estévez C, González-Polo RA, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Granato GE, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Guan J-L, Guan K-L, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson ÅB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Harper JW, Harris AL, Harris J, Harris SD, Hashimoto M, Haspel JA, Hayashi S, Hazelhurst LA, He Y, He Y-W, Hébert M-J, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Höhfeld J, Holyoake TL, Hong M-H, Hood DA, Hotamisligil GS, Houwerzijl EJ, Høyer-Hansen M, Hu B, Hu CA, Hu H-M, Hua Y, Huang C, Huang J, Huang S, Huang W-P, Huber TB, Huh W-K, Hung T-H, Hupp TR, Hur GM, Hurley JB, Hussain SNA, Hussey PJ, Hwang JJ, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Isobe K, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jäättelä M, Jackson GR, Jackson WT, Janji B, Jendrach M, Jeon J-H, Jeung E-B, Jiang H, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jiang X, Jiménez A, Jin M, Jin SV, Joe CO, Johansen T, Johnson DE, Johnson GVW, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhász G, Juillerat-Jeanneret L, Jung CH, Jung Y-K, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kågedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Kang KB, Kang KI, Kang R, Kang Y-A, Kanki T, Kanneganti T-D, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Ke P-Y, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JAKW, Kiger AA, Kihara A, Kim DR, Kim D-H, Kim D-H, Kim E-K, Kim H-R, Kim J-S, Kim JH, Kim JC, Kim JK, Kim PK, Kim SW, Kim Y-S, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BCB, Koch JC, Koga H, Koh J-Y, Koh YH, Koike M, Komatsu M, Kominami E, Kong HJ, Kong W-J, Korolchuk VI, Kotake Y, Koukourakis MI, Kouri Flores JB, Kovács AL, Kraft C, Krainc D, Krämer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Krüger R, Krut O, Ktistakis NT, Kuan C-Y, Kucharczyk R, Kumar A, Kumar R, Kumar S, Kundu M, Kung H-J, Kurz T, Kwon HJ, La Spada AR, Lafont F, Lamark T, Landry J, Lane JD, Lapaquette P, Laporte JF, László L, Lavandero S, Lavoie JN, Layfield R, Lazo PA, Le W, Le Cam L, Ledbetter DJ, Lee AJX, Lee B-W, Lee GM, Lee J, Lee J, Lee M, Lee M-S, Lee SH, Leeuwenburgh C, Legembre P, Legouis R, Lehmann M, Lei H-Y, Lei Q-Y, Leib DA, Leiro J, Lemasters JJ, Lemoine A, Lesniak MS, Lev D, Levenson VV, Levine B, Levy E, Li F, Li J-L, Li L, Li S, Li W, Li X-J, Li Y-B, Li Y-P, Liang C, Liang Q, Liao Y-F, Liberski PP, Lieberman A, Lim HJ, Lim K-L, Lim K, Lin C-F, Lin F-C, Lin J, Lin JD, Lin K, Lin W-W, Lin W-C, Lin Y-L, Linden R, Lingor P, Lippincott-Schwartz J, Lisanti MP, Liton PB, Liu B, Liu C-F, Liu K, Liu L, Liu QA, Liu W, Liu Y-C, Liu Y, Lockshin RA, Lok C-N, Lonial S, Loos B, Lopez-Berestein G, López-Otín C, Lossi L, Lotze MT, Low P, Lu B, Lu Z, Luciano F, Lukacs NW, Lund AH, Lynch-Day MA, Ma Y, Macian F, MacKeigan JP, Macleod KF, Madeo F, Maiuri L, Maiuri MC, Malagoli D, Malicdan MCV, Malorni W, Man N, Mandelkow E-M, Manon S, Manov I, Mao K, Mao X, Mao Z, Marambaud P, Marazziti D, Marcel YL, Marchbank K, Marchetti P, Marciniak SJ, Marcondes M, Mardi M, Marfe G, Mariño G, Markaki M, Marten MR, Martin SJ, Martinand-Mari C, Martinet W, Martinez-Vicente M, Masini M, Matarrese P, Matsuo S, Matteoni R, Mayer A, Mazure NM, McConkey DJ, McConnell MJ, McDermott C, McDonald C, McInerney GM, McKenna SL, McLaughlin B, McLean PJ, McMaster CR, McQuibban GA, Meijer AJ, Meisler MH, Meléndez A, Melia TJ, Melino G, Mena MA, Menendez JA, Menna-Barreto RFS, Menon MB, Menzies FM, Mercer CA, Merighi A, Merry DE, Meschini S, Meyer CG, Meyer TF, Miao C-Y, Miao J-Y, Michels PAM, Michiels C, Mijaljica D, Milojkovic A, Minucci S, Miracco C, Miranti CK, Mitroulis I, Miyazawa K, Mizushima N, Mograbi B, Mohseni S, Molero X, Mollereau B, Mollinedo F, Momoi T, Monastyrska I, Monick MM, Monteiro MJ, Moore MN, Mora R, Moreau K, Moreira PI, Moriyasu Y, Moscat J, Mostowy S, Mottram JC, Motyl T, Moussa CE-H, Müller S, Muller S, Münger K, Münz C, Murphy LO, Murphy ME, Musarò A, Mysorekar I, Nagata E, Nagata K, Nahimana A, Nair U, Nakagawa T, Nakahira K, Nakano H, Nakatogawa H, Nanjundan M, Naqvi NI, Narendra DP, Narita M, Navarro M, Nawrocki ST, Nazarko TY, Nemchenko A, Netea MG, Neufeld TP, Ney PA, Nezis IP, Nguyen HP, Nie D, Nishino I, Nislow C, Nixon RA, Noda T, Noegel AA, Nogalska A, Noguchi S, Notterpek L, Novak I, Nozaki T, Nukina N, Nürnberger T, Nyfeler B, Obara K, Oberley TD, Oddo S, Ogawa M, Ohashi T, Okamoto K, Oleinick NL, Oliver FJ, Olsen LJ, Olsson S, Opota O, Osborne TF, Ostrander GK, Otsu K, Ou J-J, Ouimet M, Overholtzer M, Ozpolat B, Paganetti P, Pagnini U, Pallet N, Palmer GE, Palumbo C, Pan T, Panaretakis T, Pandey UB, Papackova Z, Papassideri I, Paris I, Park J, Park OK, Parys JB, Parzych KR, Patschan S, Patterson C, Pattingre S, Pawelek JM, Peng J, Perlmutter DH, Perrotta I, Perry G, Pervaiz S, Peter M, Peters GJ, Petersen M, Petrovski G, Phang JM, Piacentini M, Pierre P, Pierrefite-Carle V, Pierron G, Pinkas-Kramarski R, Piras A, Piri N, Platanias LC, Pöggeler S, Poirot M, Poletti A, Poüs C, Pozuelo-Rubio M, Prætorius-Ibba M, Prasad A, Prescott M, Priault M, Produit-Zengaffinen N, Progulske-Fox A, Proikas-Cezanne T, Przedborski S, Przyklenk K, Puertollano R, Puyal J, Qian S-B, Qin L, Qin Z-H, Quaggin SE, Raben N, Rabinowich H, Rabkin SW, Rahman I, Rami A, Ramm G, Randall G, Randow F, Rao VA, Rathmell JC, Ravikumar B, Ray SK, Reed BH, Reed JC, Reggiori F, Régnier-Vigouroux A, Reichert AS, Reiners JJ, Jr, Reiter RJ, Ren J, Revuelta JL, Rhodes CJ, Ritis K, Rizzo E, Robbins J, Roberge M, Roca H, Roccheri MC, Rocchi S, Rodemann HP, Rodríguez de Córdoba S, Rohrer B, Roninson IB, Rosen K, Rost-Roszkowska MM, Rouis M, Rouschop KMA, Rovetta F, Rubin BP, Rubinsztein DC, Ruckdeschel K, Rucker EB, III, Rudich A, Rudolf E, Ruiz-Opazo N, Russo R, Rusten TE, Ryan KM, Ryter SW, Sabatini DM, Sadoshima J, Saha T, Saitoh T, Sakagami H, Sakai Y, Salekdeh GH, Salomoni P, Salvaterra PM, Salvesen G, Salvioli R, Sanchez AMJ, Sánchez-Alcázar JA, Sánchez-Prieto R, Sandri M, Sankar U, Sansanwal P, Santambrogio L, Saran S, Sarkar S, Sarwal M, Sasakawa C, Sasnauskiene A, Sass M, Sato K, Sato M, Schapira AHV, Scharl M, Schätzl HM, Scheper W, Schiaffino S, Schneider C, Schneider ME, Schneider-Stock R, Schoenlein PV, Schorderet DF, Schüller C, Schwartz GK, Scorrano L, Sealy L, Seglen PO, Segura-Aguilar J, Seiliez I, Seleverstov O, Sell C, Seo JB, Separovic D, Setaluri V, Setoguchi T, Settembre C, Shacka JJ, Shanmugam M, Shapiro IM, Shaulian E, Shaw RJ, Shelhamer JH, Shen H-M, Shen W-C, Sheng Z-H, Shi Y, Shibuya K, Shidoji Y, Shieh J-J, Shih C-M, Shimada Y, Shimizu S, Shintani T, Shirihai OS, Shore GC, Sibirny AA, Sidhu SB, Sikorska B, Silva-Zacarin ECM, Simmons A, Simon AK, Simon H-U, Simone C, Simonsen A, Sinclair DA, Singh R, Sinha D, Sinicrope FA, Sirko A, Siu PM, Sivridis E, Skop V, Skulachev VP, Slack RS, Smaili SS, Smith DR, Soengas MS, Soldati T, Song X, Sood AK, Soong TW, Sotgia F, Spector SA, Spies CD, Springer W, Srinivasula SM, Stefanis L, Steffan JS, Stendel R, Stenmark H, Stephanou A, Stern ST, Sternberg C, Stork B, Strålfors P, Subauste CS, Sui X, Sulzer D, Sun J, Sun S-Y, Sun Z-J, Sung JJY, Suzuki K, Suzuki T, Swanson MS, Swanton C, Sweeney ST, Sy L-K, Szabadkai G, Tabas I, Taegtmeyer H, Tafani M, Takács-Vellai K, Takano Y, Takegawa K, Takemura G, Takeshita F, Talbot NJ, Tan KSW, Tanaka K, Tang D, Tang D, Tanida I, Tannous BA, Tavernarakis N, Taylor GS, Taylor GA, Taylor JP, Terada LS, Terman A, Tettamanti G, Thevissen K, Thompson CB, Thorburn A, Thumm M, Tian F, Tian Y, Tocchini-Valentini G, Tolkovsky AM, Tomino Y, Tönges L, Tooze SA, Tournier C, Tower J, Towns R, Trajkovic V, Travassos LH, Tsai T-F, Tschan MP, Tsubata T, Tsung A, Turk B, Turner LS, Tyagi SC, Uchiyama Y, Ueno T, Umekawa M, Umemiya-Shirafuji R, Unni VK, Vaccaro MI, Valente EM, Van den Berghe G, van der Klei IJ, van Doorn WG, van Dyk LF, van Egmond M, van Grunsven LA, Vandenabeele P, Vandenberghe WP, Vanhorebeek I, Vaquero EC, Velasco G, Vellai T, Vicencio JM, Vierstra RD, Vila M, Vindis C, Viola G, Viscomi MT, Voitsekhovskaja OV, von Haefen C, Votruba M, Wada K, Wade-Martins R, Walker CL, Walsh CM, Walter J, Wan X-B, Wang A, Wang C, Wang D, Wang F, Wang F, Wang G, Wang H, Wang H-G, Wang H-D, Wang J, Wang K, Wang M, Wang RC, Wang X, Wang XJ, Wang Y-J, Wang Y, Wang Z-B, Wang ZC, Wang Z, Wansink DG, Ward DM, Watada H, Waters SL, Webster P, Wei L, Weihl CC, Weiss WA, Welford SM, Wen L-P, Whitehouse CA, Whitton JL, Whitworth AJ, Wileman T, Wiley JW, Wilkinson S, Willbold D, Williams RL, Williamson PR, Wouters BG, Wu C, Wu D-C, Wu WKK, Wyttenbach A, Xavier RJ, Xi Z, Xia P, Xiao G, Xie Z, Xie Z, Xu D, Xu J, Xu L, Xu X, Yamamoto A, Yamamoto A, Yamashina S, Yamashita M, Yan X, Yanagida M, Yang D-S, Yang E, Yang J-M, Yang SY, Yang W, Yang WY, Yang Z, Yao M-C, Yao T-P, Yeganeh B, Yen W-L, Yin J-J, Yin X-M, Yoo O-J, Yoon G, Yoon S-Y, Yorimitsu T, Yoshikawa Y, Yoshimori T, Yoshimoto K, You HJ, Youle RJ, Younes A, Yu L, Yu L, Yu S-W, Yu WH, Yuan Z-M, Yue Z, Yun C-H, Yuzaki M, Zabirnyk O, Silva-Zacarin E, Zacks D, Zacksenhaus E, Zaffaroni N, Zakeri Z, Zeh HJ, III, Zeitlin SO, Zhang H, Zhang H-L, Zhang J, Zhang J-P, Zhang L, Zhang L, Zhang M-Y, Zhang XD, Zhao M, Zhao Y-F, Zhao Y, Zhao ZJ, Zheng X, Zhivotovsky B, Zhong Q, Zhou C-Z, Zhu C, Zhu W-G, Zhu X-F, Zhu X, Zhu Y, Zoladek T, Zong W-X, Zorzano A, Zschocke J, Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherra SJ, III, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, Chu CT. Regulation of the autophagy protein LC3 by phosphorylation. J. Cell Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peiris H, Dubach D, Jessup CF, Unterweger P, Raghupathi R, Muyderman H, Zanin MP, Mackenzie K, Pritchard MA, Keating DJ. RCAN1 regulates mitochondrial function and increases susceptibility to oxidative stress in mammalian cells. Oxid. Med. Cell. Longev. 2014;2014:520316. doi: 10.1155/2014/520316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ermak G, Harris CD, Davies KJA. The DSCR1 (Adapt78) isoform 1 protein calcipressin 1 inhibits calcineurin and protects against acute calcium-mediated stress damage, including transient oxidative stress. FASEB J. 2002;16:814–824. doi: 10.1096/fj.01-0846com. [DOI] [PubMed] [Google Scholar]
- 41.Rothermel B, Vega RB, Yang J, Wu H, Bassel-Duby R, Williams RS. A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J. Biol. Chem. 2000;275:8719–8725. doi: 10.1074/jbc.275.12.8719. [DOI] [PubMed] [Google Scholar]
- 42.Kim SS, Oh Y, Chung KC, Seo SR. Protein kinase A phosphorylates Down syndrome critical region 1 (RCAN1) Biochem. Biophys. Res. Commun. 2012;418:657–661. doi: 10.1016/j.bbrc.2012.01.071. [DOI] [PubMed] [Google Scholar]
- 43.Davis KE, Neinast MD, Sun K, Skiles WM, Bills JD, Zehr JA, Zeve D, Hahner LD, Cox DW, Gent LM, Xu Y, Wang ZV, Khan SA, Clegg DJ. The sexually dimorphic role of adipose and adipocyte estrogen receptors in modulating adipose tissue expansion, inflammation, and fibrosis. Mol. Metab. 2013;2:227–242. doi: 10.1016/j.molmet.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y, Nedungadi TP, Zhu L, Sobhani N, Irani BG, Davis KE, Zhang X, Zou F, Gent LM, Hahner LD, Khan SA, Elias CF, Elmquist JK, Clegg DJ. Distinct hypothalamic neurons mediate estrogenic effects on energy homeostasis and reproduction. Cell Metab. 2011;14:453–465. doi: 10.1016/j.cmet.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kilic G, Alvarez-Mercado AI, Zarrouki B, Opland D, Liew CW, Alonso LC, Myers MG, Jr, Jonas J-C, Poitout V, Kulkarni RN, Mauvais-Jarvis F. The islet estrogen receptor-α is induced by hyperglycemia and protects against oxidative stress-induced insulin-deficient diabetes. PLOS One. 2014;9:e87941. doi: 10.1371/journal.pone.0087941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu L, Brown WC, Cai Q, Krust A, Chambon P, McGuinness OP, Stafford JM. Estrogen treatment after ovariectomy protects against fatty liver and may improve pathway-selective insulin resistance. Diabetes. 2013;62:424–434. doi: 10.2337/db11-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin SD, McGee SL. The role of mitochondria in the aetiology of insulin resistance and type 2 diabetes. Biochim. Biophys. Acta. 2014;1840:1303–1312. doi: 10.1016/j.bbagen.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 48.Kim J, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ. Res. 2008;102:401–414. doi: 10.1161/CIRCRESAHA.107.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leduc-Gaudet J-P, Picard M, St-Jean Pelletier F, Sgarioto N, Auger M-J, Vallée J, Robitaille R, St-Pierre DH, Gouspillou G. Mitochondrial morphology is altered in atrophied skeletal muscle of aged mice. Oncotarget. 2015;6:17923–17937. doi: 10.18632/oncotarget.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nunnari J, Suomalainen A. Mitochondria: In sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallace DC. Mitochondrial DNA mutations in diseases of energy metabolism. J. Bioenerg. Biomembr. 1994;26:241–250. doi: 10.1007/BF00763096. [DOI] [PubMed] [Google Scholar]
- 53.Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheele C, Nielsen AR, Walden TB, Sewell DA, Fischer CP, Brogan RJ, Petrovic N, Larsson O, Tesch PA, Wennmalm K, Hutchinson DS, Cannon B, Wahlestedt C, Pedersen BK, Timmons JA. Altered regulation of the PINK1 locus: A link between type 2 diabetes and neurodegeneration? FASEB J. 2007;21:3653–3665. doi: 10.1096/fj.07-8520com. [DOI] [PubMed] [Google Scholar]
- 56.Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 57.Santel A, Frank S. Shaping mitochondria: The complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life. 2008;60:448–455. doi: 10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- 58.Smirnova E, Shurland D-L, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qian W, Choi S, Gibson GA, Watkins SC, Bakkenist CJ, Van Houten B. Mitochondrial hyperfusion induced by loss of the fission protein Drp1 causes ATM-dependent G2/M arrest and aneuploidy through DNA replication stress. J. Cell Sci. 2012;125:5745–5757. doi: 10.1242/jcs.109769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drummond MJ, Addison O, Brunker L, Hopkins PN, McClain DA, LaStayo PC, Marcus RL. Downregulation of E3 ubiquitin ligases and mitophagy-related genes in skeletal muscle of physically inactive, frail older women: A cross-sectional comparison. J. Gerontol. A Biol. Sci. Med. Sci. 2014;69:1040–1048. doi: 10.1093/gerona/glu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elgass K, Pakay J, Ryan MT, Palmer CS. Recent advances into the understanding of mitochondrial fission. Biochim. Biophys. Acta. 2013;1833:150–161. doi: 10.1016/j.bbamcr.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 62.Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nielsen S, Scheele C, Yfanti C, Åkerström T, Nielsen AR, Pedersen BK, Laye M. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J. Physiol. 2010;588:4029–4037. doi: 10.1113/jphysiol.2010.189860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen S, Hvid T, Kelly M, Lindegaard B, Dethlefsen C, Winding K, Mathur N, Scheele C, Pedersen BK, Laye MJ. Muscle specific miRNAs are induced by testosterone and independently upregulated by age. Front. Physiol. 2014;4:394. doi: 10.3389/fphys.2013.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parks BW, Sallam T, Mehrabian M, Psychogios N, Hui ST, Norheim F, Castellani LW, Rau CD, Pan C, Phun J, Zhou Z, Yang W-P, Neuhaus I, Gargalovic PS, Kirchgessner TG, Graham M, Lee R, Tontonoz P, Gerszten RE, Hevener AL, Lusis AJ. Genetic architecture of insulin resistance in the mouse. Cell Metab. 2015;21:334–346. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hewitt SC, Kissling GE, Fieselman KE, Jayes FL, Gerrish KE, Korach KS. Biological and biochemical consequences of global deletion of exon 3 from the ERα gene. FASEB J. 2010;24:4660–4667. doi: 10.1096/fj.10-163428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahlbory-Dieker DL, Stride BD, Leder G, Schkoldow J, Trölenberg S, Seidel H, Otto C, Sommer A, Parker MG, Schütz G, Wintermantel TM. DNA binding by estrogen receptor-α is essential for the transcriptional response to estrogen in the liver and the uterus. Mol. Endocrinol. 2009;23:1544–1555. doi: 10.1210/me.2009-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aåstrand P-O. Human physical fitness with special reference to sex and age. Physiol. Rev. 1956;36:307–335. doi: 10.1152/physrev.1956.36.3.307. [DOI] [PubMed] [Google Scholar]
- 69.Hevener AL, He W, Barak Y, Le J, Bandyopadhyay G, Olson P, Wilkes J, Evans RM, Olefsky J. Muscle-specific Pparg deletion causes insulin resistance. Nat. Med. 2003;9:1491–1497. doi: 10.1038/nm956. [DOI] [PubMed] [Google Scholar]
- 70.Hevener AL, Olefsky JM, Reichart D, Nguyen MTA, Bandyopadyhay G, Leung H-Y, Watt MJ, Benner C, Febbraio MA, Nguyen A-K, Folian B, Subramaniam S, Gonzalez FJ, Glass CK, Ricote M. Macrophage PPARγ is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J. Clin. Invest. 2007;117:1658–1669. doi: 10.1172/JCI31561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steele R. Influences of glucose loading and of injected insulin on hepatic glucose output. Ann. N.Y. Acad. Sci. 1959;82:420–430. doi: 10.1111/j.1749-6632.1959.tb44923.x. [DOI] [PubMed] [Google Scholar]
- 72.Frayn KN, Maycock PF. Skeletal muscle triacylglycerol in the rat: Methods for sampling and measurement, and studies of biological variability. J. Lipid Res. 1980;21:139–144. [PubMed] [Google Scholar]
- 73.Preiss J, Loomis CR, Bishop WR, Stein R, Niedel JE, Bell RM. Quantitative measurement of sn-1,2-diacylglycerols present in platelets, hepatocytes, and ras- and sis-transformed normal rat kidney cells. J. Biol. Chem. 1986;261:8597–8600. [PubMed] [Google Scholar]
- 74.Allred JB, Guy DG. Determination of coenzyme A and acetyl CoA in tissue extracts. Anal. Biochem. 1969;29:293–299. doi: 10.1016/0003-2697(69)90312-1. [DOI] [PubMed] [Google Scholar]
- 75.Nguyen MTA, Favelyukis S, Nguyen A-K, Reichart D, Scott PA, Jenn A, Liu-Bryan R, Glass CK, Neels JG, Olefsky JM. A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J. Biol. Chem. 2007;282:35279–35292. doi: 10.1074/jbc.M706762200. [DOI] [PubMed] [Google Scholar]
- 76.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 77.Vermulst M, Bielas JH, Loeb LA. Quantification of random mutations in the mitochondrial genome. Methods. 2008;46:263–268. doi: 10.1016/j.ymeth.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bruce CR, Hoy AJ, Turner N, Watt MJ, Allen TL, Carpenter K, Cooney GJ, Febbraio MA, Kraegen EW. Overexpression of carnitine palmitoyltransferase-1 in skeletal muscle is sufficient to enhance fatty acid oxidation and improve high-fat diet-induced insulin resistance. Diabetes. 2009;58:550–558. doi: 10.2337/db08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen MB, McAinch AJ, Macaulay SL, Castelli LA, O’Brien PE, Dixon JB, Cameron-Smith D, Kemp BE, Steinberg GR. Impaired activation of AMP-kinase and fatty acid oxidation by globular adiponectin in cultured human skeletal muscle of obese type 2 diabetics. J. Clin. Endocrinol. Metab. 2005;90:3665–3672. doi: 10.1210/jc.2004-1980. [DOI] [PubMed] [Google Scholar]
- 80.McCurdy CE, Cartee GD. Akt2 is essential for the full effect of calorie restriction on insulin-stimulated glucose uptake in skeletal muscle. Diabetes. 2005;54:1349–1356. doi: 10.2337/diabetes.54.5.1349. [DOI] [PubMed] [Google Scholar]
- 81.Vercauteren K, Pasko RA, Gleyzer N, Marino VM, Scarpulla RC. PGC-1-related coactivator: Immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol. Cell. Biol. 2006;26:7409–7419. doi: 10.1128/MCB.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bopassa JC, Eghbali M, Toro L, Stefani E. A novel estrogen receptor GPER inhibits mitochondria permeability transition pore opening and protects the heart against ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol. 2010;298:H16–H23. doi: 10.1152/ajpheart.00588.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gomez L, Paillard M, Thibault H, Derumeaux G, Ovize M. Inhibition of GSK3β by post-conditioning is required to prevent opening of the mitochondrial permeability transition pore during reperfusion. Circulation. 2008;117:2761–2768. doi: 10.1161/CIRCULATIONAHA.107.755066. [DOI] [PubMed] [Google Scholar]
- 84.Close RI. The relations between sarcomere length and characteristics of isometric twitch contractions of frog sartorius muscle. J. Physiol. 1972;220:745–762. doi: 10.1113/jphysiol.1972.sp009733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bandsma RHJ, Rake J-P, Visser G, Neese RA, Hellerstein MK, van Duyvenvoorde W, Princen HMG, Stellaard F, Smit GPA, Kuipers F. Increased lipogenesis and resistance of lipoproteins to oxidative modification in two patients with glycogen storage disease type 1a. J. Pediatr. 2002;140:256–260. doi: 10.1067/mpd.2002.121382. [DOI] [PubMed] [Google Scholar]
- 86.Zimmerman EG, Akins DR, Planz JV, Schurr MJ. A rapid procedure for isolating mitochondrial DNA. Gene Anal. Tech. 1988;5:102–104. doi: 10.1016/0735-0651(88)90004-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.