ABSTRACT
There are currently 5 million to 10 million human T-lymphotropic virus type 1 (HTLV-1)-infected people, and many of them will develop severe complications resulting from this infection. A vaccine is urgently needed in areas where HTLV-1 is endemic. Many vaccines are best tested in nonhuman primate animal models. As a first step in designing an effective HTLV-1 vaccine, we defined the CD8+ and CD4+ T cell response against simian T-lymphotropic virus type 1 (STLV-1), a virus closely related to HTLV-1, in olive baboons (Papio anubis). Consistent with persistent antigenic exposure, we observed that STLV-1-specific CD8+ T cells displayed an effector memory phenotype and usually expressed CD107a, gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α). To assess the viral targets of the cellular immune response in STLV-1-infected animals, we used intracellular cytokine staining to detect responses against overlapping peptides covering the entire STLV-1 proteome. Our results show that, similarly to humans, the baboon CD8+ T cell response narrowly targeted the Tax protein. Our findings suggest that the STLV-1-infected baboon model may recapitulate some of the important aspects of the human response against HTLV-1 and could be an important tool for the development of immune-based therapy and prophylaxis.
IMPORTANCE HTLV-1 infection can lead to many different and often fatal conditions. A vaccine deployed in areas of high prevalence might reduce the incidence of HTLV-1-induced disease. Unfortunately, there are very few animal models of HTLV-1 infection useful for testing vaccine approaches. Here we describe cellular immune responses in baboons against a closely related virus, STLV-1. We show for the first time that the immune response against STLV-1 in naturally infected baboons is largely directed against the Tax protein. Similar findings in humans and the sequence similarity between the human and baboon viruses suggest that the STLV-1-infected baboon model might be useful for developing a vaccine against HTLV-1.
INTRODUCTION
Human T-lymphotropic virus type 1 (HTLV-1) is a retrovirus that establishes persistent infection in humans (1, 2). It is estimated that at least 5 million to 10 million people worldwide are infected with HTLV-1, with regions of endemicity in Japan, equatorial Africa, the Caribbean, and South America (3, 4). HTLV-1 is the causative agent of adult T cell leukemia/lymphoma (ATL) (5) and HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) (6). Additionally, other diseases have also been associated with chronic HTLV-1 infection, including dermatologic conditions (7), uveitis (8), and myositis (9).
The number of new HTLV-1 infections has been notoriously difficult to estimate due to the lack of reliable HTLV-1 epidemiological data from most regions of the world (4). While it is unclear how many HTLV-1 infections can be prevented on a yearly basis, vaccination of naive individuals and HTLV-1 carriers would likely limit HTLV-1 dissemination and the development of HTLV-1-induced diseases. This vaccine would be most effective when deployed in regions with high numbers of HTLV-1-infected carriers, such as Japan, where up to 1% of the population are HTLV-1 positive (4, 10, 11). Furthermore, the annual incidence of ATL in Japan is approximately 1 in every 1,000 HTLV-1 carriers. Thus, a vaccine that prevents new infections and limits the development of HTLV-1-induced disease would be an important tool to reduce morbidity caused by HTLV-1.
The development of HTLV-1-specific CD8+ T cells may be one of the reasons why most HTLV-1-infected individuals do not develop ATL (12). In particular, Tax-specific CD8+ T cells can be detected in most infected individuals (13–16). In fact, Tax is the dominant target recognized by HTLV-1-specific CD8+ T cells, and it is thought that infected cells expressing Tax are continually destroyed (17–19). In addition, ATL patients who underwent complete remission following hematopoietic stem cell transplantation (HSCT) had expanded Tax-specific CD8+ T cells in vivo (20). These observations suggest that responses against Tax might contribute to better outcomes in HTLV-1-infected patients.
HTLV-1 basic zip factor (HBZ) is also involved in viral chronicity and leukemic transformation (21). Therefore, similarly to a Tax-based vaccine, one could argue that vaccination against HBZ might prevent HTLV-1-induced leukemogenesis (22). Moreover, while Tax transcripts can be detected in only 40% of ATL patients, HBZ is expressed in all ATL patients (23, 24). In fact, Sugata and colleagues generated anti-HBZ-specific CD8+ T cells in vivo in mice as well as in rhesus macaques, using recombinant vaccinia viruses (25). Although this approach is promising, HBZ immunogenicity was poor compared to that of Tax and required multiple boosts. The efficiency of an HBZ-based vaccine will need to be tested against primary human ATL cells.
Previous studies have indicated that HTLV-1 proviral load (PVL) is a major risk factor for HAM/TSP (26, 27). Tax-specific CD8+ T cells have been shown to reduce HTLV-1 PVL and to prevent asymptomatic carriers from developing ATL (28). These findings suggest that a reduction in the HTLV-1 PVL in circulating lymphocytes prevents HTLV-1 carriers from developing ATL and HAM/TSP.
HTLV-1 is classified into six different subtypes, one cosmopolitan subtype (HTLV-1-a) (29), four subtypes restricted to Africa (HTLV-1-b, -d, -e, and -f) (30, 31), and one subtype in descendants of the first settlers of Melanesia and Australia (HTLV-1-c) (31). Simian T-lymphotropic virus type 1 (STLV-1) is closely related to HTLV-1 and infects several nonhuman primate species. Phylogenetic analysis of the conserved tax gene sequences indicates that STLV-1 and HTLV-1 are evolutionarily related (32). Furthermore, STLV-1 Tax and STLV-1 bZIP factor (SBZ) have functions similar to those of their equivalents from HTLV-1 (19, 33). It is well established that Tax interacts with the host transcription factor NF-κB, resulting in the activation of the NF-κB pathway (19). This is critical for transformation, proliferation, and survival of HTLV-1-infected cells, especially in the early phases of infection. Recent evidence showed that hunters in Africa can be infected by HTLV-1 strains that are genetically related to the strains circulating among local nonhuman primates (34). In STLV-1-infected macaques (Macaca tonkeana) and mandrills (Mandrillus sphinx), investigators have described the clonal proliferation of STLV-1-infected cells and preferential infection of CD4+ T cells (35, 36). Interestingly, there is a high frequency of STLV-1 seropositivity in wild and captive baboons (37–41). Phylogenetic analyses of STLV-1 sequences from wild-caught chacma baboons (Papio ursinus) and olive baboons (Papio anubis) suggest cross-species transmissions of STLV-1 in the wild (30, 41, 42). Thus, nonhuman primates can be naturally infected with STLV-1, and some eventually develop STLV-1-associated diseases, including ATL (43, 44).
Olive baboons (Papio anubis) have an immune system similar to that of humans and provide a good animal model of HTLV-1 infection. A recent in vivo study of asymptomatic baboons naturally infected with STLV-1 showed that induction of viral expression with valproate in combination with azidothymidine to prevent viral propagation resulted in a decrease in the PVL. Interestingly, the reduction of the PVL coincided with an accumulation of effector CD8+ T lymphocytes directed against the virus, indicating that these cells could have contributed to the positive outcome (45).
As a prelude to the design of suitable vaccine inserts, we have defined the entire cellular immune response (CD4+ and CD8+ T cells) against STLV-1 in infected baboons. Here we show that cellular responses against STLV-1 are largely restricted to CD8+ T cells. Furthermore, as in HTLV-1-infected humans, Tax is the immunodominant virus-encoded protein target of baboon cellular responses. We have also identified six distinct Tax epitope-rich regions that are targeted by STLV-1-specific CD8+ T cells from assorted baboons. Our results support the use of baboons as models for HTLV-1 vaccine research and further suggest the inclusion of Tax in vaccine compositions.
MATERIALS AND METHODS
Research animals.
The 22 animals used in this study were olive baboons (Papio anubis) housed at the Southwest National Primate Research Center (SNPRC) at the Texas Biomedical Research Institute. They were cared for in accordance with regulations set by the Guide for the Care and Use of Laboratory Animals of the National Research Council (46), as approved by the Texas Biomedical Research Institutional Animal Care and Use Committee.
The study population included 18 STLV-1-infected baboons and 4 uninfected animals used as negative controls (Table 1). We excluded three animals from the study that were serologically reactive to STLV-1 but negative for STLV-1 PVL and CD8 responses. The numbers of male and female baboons were balanced in this study. STLV-1 serology was performed at the SNPRC according to the manufacturer's protocol (47, 48) for the Macaque Tracking multiplexed fluorometric immunoassay (MFIA). This assay is a Luminex bead-based serology test developed by Charles River Labs (CRL) (Wilmington, MA). The performance (specificity and sensitivity) of the MFIA method is comparable to that of serology measurement by enzyme-linked immunosorbent assay (ELISA) (47). In brief, the STLV-1 Luminex multiplex assay used two different bead sets for anti-STLV-1 antibody detection. The first bead set uses HTLV-1 and HTLV-2 whole-virus lysates, whereas the second bead set uses a purified, truncated STLV-1 p21 (12-kDa) protein produced in insect cells. Additionally, beads coated with baculovirus were used as nonspecific assay controls. Data analysis was carried out by using the Charles River MFIA Results Excel Workbook (48) according to the manufacturer's protocol. In summary, an assay score is created based on the reactivity against the HTLV- and STLV-specific beads minus the background. A sample was considered positive when reactivity against both specific beads exceeded the cutoff. The assay result was considered inconclusive if there was reactivity against only one set of beads and negative if no reactivity was detected. Unfortunately, we were unable to estimate the duration of infection, since we did not have longitudinal samples to assess STLV-1 seroconversion.
TABLE 1.
Animal characteristics
| Animal | STLV-1 serologya | Presence of STLV-1 DNA (CT)b | Frequency of STLV-1-specific CD8+ T cellsc | Gender | Age (yr) | wt (kg) |
|---|---|---|---|---|---|---|
| 11284 | Positive | ++ (33.3) | 0.64 | Female | 21 | 18 |
| 12138 | Positive | ++ (33.2) | 0.43 | Female | 20 | 24 |
| 12427 | Positive | − (ind) | 0.23 | Female | 19 | 22 |
| 12869 | Positive | ++ (33.8) | 3.56 | Female | 18 | 20 |
| 13169 | Positive | − (neg) | 0.20 | Male | 18 | 22 |
| 16666 | Positive | ++++ (27.3) | 2.63 | Male | 13 | 37 |
| 16999 | Positive | +++ (30.4) | 0.85 | Female | 13 | 23 |
| 19522 | Positive | + (36.8) | 0.42 | Male | 10 | 39 |
| 25533 | Positive | ++++ (29.7) | 1.13 | Male | 7 | 26 |
| 26458 | Positive | − (neg) | 0.78 | Male | 9 | 28 |
| 26464 | Positive | +++ (31.9) | 2.86 | Male | 9 | 30 |
| 26498 | Positive | +++ (32.6) | 0.79 | Male | 7 | 30 |
| 27166 | Positive | + (37.8) | 0.45 | Male | 6 | 30 |
| 27181 | Positive | + (36.8) | 0.44 | Female | 8 | 18 |
| 27924 | Positive | +++ (30.8) | 1.13 | Male | 8 | 24 |
| 28328 | Positive | ++ (34.5) | 0.64 | Female | 7 | 22 |
| 30842 | Positive | ++ (33.5) | 2.58 | Female | 3 | 15 |
| 30872 | Positive | + (38.2) | 2.25 | Female | 3 | 13 |
| 15659 | Negative | Not tested | 0.04 | Female | 14 | 16 |
| 30366 | Negative | Not tested | 0.02 | Female | 4 | 16 |
| 30425 | Negative | Not tested | 0.00 | Male | 4 | 20 |
| 30442 | Negative | Not tested | 0.02 | Male | 4 | 23 |
Detection of antibodies against STLV-1 by using the CRL Macaque Tracking Luminex assay.
+ indicates relative PCR levels (number of amplification cycles to reach the detection threshold). ++++, CT of >27 to ≤30; +++, CT of >30 to ≤33; ++, CT of >33 to ≤36; +, CT of >36 to ≤39; − (ind) (CT of >39), indeterminate PCR result; − (neg), no amplification.
Frequency of CD8+ T cells expressing IFN-γ and CD107a upon stimulation with overlapping peptide pools covering the STLV-1 proteome. For any given animal, the frequency of responses to each STLV-1 ORF pool was added in order to compute the frequency of total STLV-1-specific CD8+ T cells.
Blood collection.
We isolated peripheral blood mononuclear cells (PBMCs) from EDTA-treated blood by Ficoll-Paque Plus (GE Healthcare Bio-Sciences, Pittsburgh, PA) density centrifugation. Immunophenotyping studies were performed by polychromatic flow cytometry (as detailed below) using freshly isolated PBMCs.
Detection of STLV-1 proviral DNA.
PCR amplification of STLV-1 sequences was carried out by the California National Primate Research Center Pathogen Detection Laboratory. The procedure was based on a real-time quantitative PCR assay developed for simian betaretrovirus proviral DNA detection (49). In brief, PBMCs were separated by Ficoll density gradient centrifugation and frozen. DNA was extracted from 1.5 × 107 to 4.2 × 107 PBMCs from each animal by using the QIAamp DNA Blood Midi kit (Qiagen, Valencia, CA). The STLV-1 proviral load was estimated by using a semiquantitative TaqMan real-time PCR assay targeting the tax gene, using the following primers and probe: forward primer STLV7333 (5′-CCA GGG TTT GGA CAG AGT CTT C-3′), reverse primer STLV7432 (5′-CCG AAC ATA GTC CCC CAG AGA-3′), and probe STLVJA (6-carboxyfluorescein [FAM]–5′-TCT CCA AAC ACG TAG ACT GGG TAT CCG AAA-3′–6-carboxytetramethylrhodamine [TAMRA]). PCR conditions were 2 min at 50°C, 10 min at 95°C, and 55 cycles of amplification (15 s at 95°C and 1 min at 62°C). All extractions were run in duplicate wells, and the relative STLV-1 PVL was reported as the average reaction threshold cycle (CT). Animal 12427 had discordant amplification replicates (one negative and one positive with a CT of ∼39.5), and this PVL was defined as indeterminate.
Peptide libraries.
Seven hundred sixty-one peptides were required to span the entire STLV-1 proteome. STLV-1 peptides were designed based on the sequence of an olive baboon STLV-1 isolate (GenBank accession number JX987040.1). Individual peptides were 15 amino acid residues long and overlapped by 11 amino acids. These peptides were designed to span the complete STLV-1 proteome, and the proteins were combined into open reading frame (ORF) pools. The peptides were synthesized to >90% purity by GenScript USA Inc. (Piscataway, NJ) and were combined into pools of 10 peptides each for use in intracellular cytokine staining (ICS) and memory phenotyping assays. To determine the sensitivity of our assay and to detect any low-frequency responses to the peptides, PBMCs were first incubated with peptides at concentrations of 0.1 μM, 1.0 μM, 10 μM, and 30 μM. Since our titration experiments showed that a concentration of 10 μM each peptide maximally stimulated T cells, we used this concentration in our experiments (data not shown). The complete list of peptides and pool compositions can be found in Table S1 in the supplemental material.
Intracellular cytokine staining assay.
We performed multiparametric ICS assays by incubating freshly isolated PBMCs with peptide pools spanning the following ORFs: Gag, Pro, polymerase (Pol), Rex, Tax, p30, Env, SBZ, and p13. PBMCs (1.5 × 106 cells) were incubated in a final volume of 200 μl of RPMI 1640 culture medium supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, and l-glutamine for 8 h at 37°C in a 5% CO2 incubator. Incubation was done in the presence of 5 μg/ml of brefeldin A (BioLegend, San Diego, CA), 0.7 μg/ml of Golgi Stop (BD Biosciences, San Jose, CA), and the costimulatory antibodies anti-CD28 (clone L293; BD Biosciences) and anti-CD49d (clone 9F10; BD Biosciences). Anti-CD107a phycoerythrin (PE) (clone H4A3; BioLegend) was also added to the assay mixture. Peptides were added at final concentrations of 10 μM each. PBMCs costimulated in the absence of 15-mer peptides were used as a negative control, and PBMCs stimulated with Lymphocyte Activation Cocktail (LAC; BD Biosciences) were used as a positive control.
We then stained the cells at 4°C with antibodies directed against the surface molecules CD3 (peridinin chlorophyll protein [PerCP]-Cy5.5) (clone SP34-2; BD Biosciences), CD4 (brilliant violet 605 [BV605]) (clone L200; BD Biosciences), CD8 (fluorescein isothiocyanate [FITC]) (clone RPA-T8; BioLegend), CD14 (BV510) (clone M5E2; BioLegend), CD16 (BV510) (clone 3G8; BioLegend), and CD20 (BV510) (clone 2H7; BioLegend). Amine-reactive dye (Invitrogen) was also included in both panels to discriminate between live and dead cells. Cells were then fixed and permeabilized by using Cytofix/Cytoperm (BD Biosciences) prior to staining with antibodies against gamma interferon (IFN-γ) (BV421) (clone 4S.B3; BioLegend), tumor necrosis factor alpha (TNF-α) (allophycocyanin [APC]) (clone MAb11; BD Biosciences), and interleukin-2 (IL-2) (PE-Cy7) (clone MQ1-17H12; BioLegend). For each sample, 250,000 events were acquired by using FACSDiva version 8.0.1 on a special-order research product (SORP) BD LSR II instrument equipped with a 50-mW 405-nm violet laser, a 100-mW 488-nm blue laser, and a 50-mW 640-nm red laser, and data were analyzed with Beckman Coulter Kaluza 1.3 software.
For data analysis, we first created a time gate that included only those events that were recorded within the 5th and 90th percentiles. We then gated on lymphocytes (forward-scatter [FSC]-by-side-scatter [SSC] plot) and excluded aggregates from the analysis by using plots of forward-scatter height (FSC-H) versus forward-scatter width (FSC-W) and side-scatter height by side-scatter width (data not shown). Subsequently, we gated on CD3+ T lymphocytes excluding CD14+, CD16+, CD20+, and dead cells. At this stage, we separated T lymphocyte subsets based on their expression of either CD4 or CD8 (excluding those expressing both markers) and conducted our analyses (of IFN-γ, CD107a, TNF-α, and IL-2 expression) within these two compartments. Therefore, we expressed our results as the median frequency of CD8+ T cells expressing the cytolysis marker CD107a and the cytokine(s). In Fig. 1B, we also analyzed the relative contribution of each protein to the total STLV-1-specific response. In this analysis, we expressed the protein-specific responses as a percentage of the total STLV-1-specific response in a given animal (the sum of frequencies of all STLV-1-specific cells of a given animal equals 100%).
FIG 1.
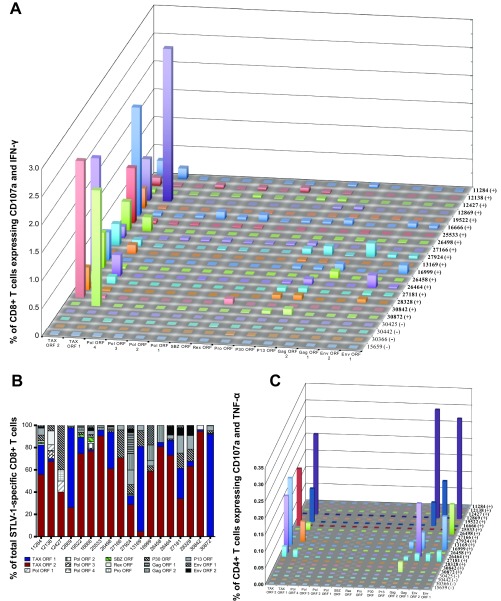
STLV-1-specific CD8+ T cells preferentially target Tax. To determine the targets of the immune responses against STLV-1, we stimulated PBMCs with pools of 10 15-mer peptides overlapping by 11 amino acids, spanning the STLV-1 proteins Env, Gag, p13, p30, Pro, Rex, Pol, SBZ, and Tax, and measured T cell responses by ICS. (A) CD8+ T cell responses against peptides covering the entire STLV-1 proteome. (B) Relative recognition of each STLV-1 protein by STLV-1-specific cells shows that Tax is the preferential target of CD8+ T cells in almost every infected animal. (C) CD4+ T cell responses against peptides covering the STLV-1 proteome. Infected animals are denoted by (+), and uninfected animals are indicated by (−).
We noticed that the hallmark of an antigen/peptide-specific response in our assay was the expression of CD107a in combination with one of the cytokines in CD8+ T cells and double positivity for IFN-γ and either TNF-α or IL-2 in CD4+ T cells (data not shown).
As controls, we stimulated STLV-1-negative animals with the same peptides as those used on the STLV-1-infected animals. Additionally, we included a sample without stimulation for assessment of background response levels. In order for a response to be considered positive, the following criterion had to be fulfilled: the peptide-specific response (percentage of CD8+ T cells that are CD107a+ and cytokine positive) minus the response to costimulation in the absence of peptide must be greater than the response of STLV-1-naive animals to the same peptide. The same formula was also applied to CD4+ T cell populations. In uninfected animals, the average response to any given peptide stimulus was below 0.01%. Experiments were done based on animals and peptide availability. Experiments with Env, Gag, and Tax were conducted three times for 10 infected animals (animals 11284, 12138, 12427, 12869, 19522, 16666, 25533, 26498, 27166, and 27924) and for the 4 naive animals (animals 30425, 30442, 30366, and 15659). We expanded this initial measurement to include responses to p13 and p30 (shown as averages of data from at least two experiments) and to Pol, Pro, Rex, and SBZ (one experiment). Finally, we tested an additional set of 8 infected animals (animals 13169, 16999, 26458, 26464, 27181, 28328, 30842, and 30872) once for the entire STLV-1 proteome and twice for Tax. The frequency of responding cells was expressed as a median value when three or more independent measurements were performed and as averages otherwise. The total STLV-1-specific CD8+ T cell response was expressed as a sum of the background-subtracted median responses to the ORF pools.
Memory phenotype staining assay.
We stimulated freshly isolated PBMCs (described above) with Tax peptide pools and assessed cytokine production and the expression of the degranulation marker CD107a. We stained the cells at room temperature with antibodies directed against the surface molecules CD3 (PerCP-Cy5.5) (clone SP34-2; BD Biosciences), CD8 (PE-Cy7) (clone RPA-T8; BioLegend), CD4 (BV711) (clone L200; BD Biosciences), CCR7 (FITC) (clone 150503; BD Biosciences), CD14 (BV510) (clone M5E2; BioLegend), CD16 (BV510) (clone 3G8; BioLegend), and CD20 (BV510) (clone 2H7; BioLegend). The fixable viability dye eFluor780 (eBioscience) was also included to discriminate between live and dead cells. Cells were then fixed (as described above) prior to staining with antibodies against IFN-γ (BV421) (clone 4S.B3; BioLegend), TNF-α (BV421) (clone MAb11; BioLegend), and IL-2 (BV421) (clone MQ1-17H12; BioLegend). We gated on the CD107a+ responding cells (i.e., expressing IFN-γ, TNF-α, and/or IL-2) to define the memory phenotype of the responding T lymphocytes. The memory phenotype of Tax-specific CD8+ T cells from STLV-1-infected baboons was defined based on the expression of CD45RA and the chemokine receptor CCR7. Effector memory T cells (TEM cells) were defined as CD45RA− CCR7−, T effector memory RA+ cells (TEMRA cells) were defined as CCR7− CD45RA+, central memory T cells (TCM cells) were defined as CD45RA− CCR7+, and naive cells were defined as CCR7+ CD45RA+.
RESULTS
Proviral loads in naturally infected baboons.
To characterize STLV-1 infection in olive baboons, we measured the relative STLV-1 PVLs using a semiquantitative real-time PCR for 18 infected baboons (9 males and 9 females) (Table 1). This procedure was done at the Pathogen Detection Laboratory (California National Primate Research Center) and was based on a real-time quantitative PCR assay developed for simian betaretrovirus proviral DNA detection (49). Because standards for STLV-1 were not available, we expressed the relative levels of STLV-1 PVLs as the number of amplification cycles to reach a detection threshold (CT). We observed a wide range of PVL values among the animals. For instance, animal 16666 (CT of 27.3) had detectable amplification more than 10 PCR cycles earlier than did animal 30872 (CT of 38.2). Despite measuring PVLs in most baboons with STLV-1-specific serological responses, we were unable to detect viral nucleic acid in two seropositive animals (animals 13169 and 26458), and one sample was indeterminate (animal 12427). Finally, three serologically reactive animals (animals 13326, 26138, and 29033) did not have measurable PVL or T cell responses and were excluded from the study.
CD8+ T cells target Tax in STLV-1-infected baboons.
Understanding immune-mediated control of STLV-1 in infected baboons could lead to insights into how to design an effective vaccine. Thus, our first objective was to determine the viral targets of the cellular immune response against STLV-1. We evaluated T cell responses to 761 overlapping peptides covering the entire STLV-1 proteome in 18 STLV-1-infected baboons and 4 uninfected controls by ICS (Table 1). Cellular responses to one or more STLV-1 peptide pools were detected in all baboons enrolled in the study (frequencies of STLV-1-specific CD8+ T cells were 0.2 to 3.56% of CD8+ T cells). Of note, none of the seronegative baboons had measurable responses (Table 1).
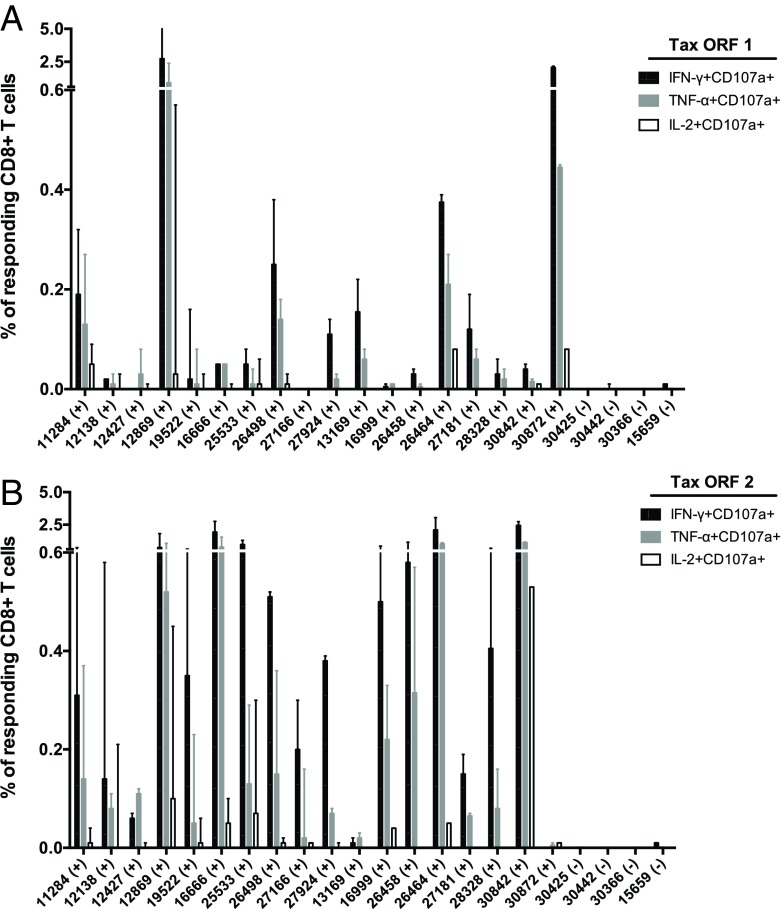
We conducted ICS assays to characterize the frequency of STLV-1-specific CD8+ and CD4+ T lymphocytes (Fig. 1). To map responses against STLV-1, we created peptide pools spanning the open reading frames (ORFs) of the proteins Gag, Pol, Env, Pro, Tax, Rex, p13, p30, and SBZ. These “ORF pools” contained up to 60 peptides. If a protein could not be covered in a single ORF pool, we created subsets of pools (e.g., Tax ORF1 and Tax ORF2). Interestingly, we observed that STLV-1-specific CD8+ T cells largely targeted epitopes in Tax and to a lesser extent the regulatory proteins p13, p30, Rex, and SBZ (Fig. 1A and B). A dominant response to Tax was observed in the 18 STLV-1-positive monkeys (Fig. 1B). Indeed, CD8+ T cell responses against p13, p30, and SBZ made up only a small fraction of the total response against the virus (Fig. 1B). For eight STLV-1-infected animals, we observed low frequencies of CD4+ T cells recognizing Env, Gag, and Tax (Fig. 1C). Tax-specific CD8+ T cells preferentially responded to Tax ORF2 and expressed IFN-γ, TNF-α, and IL-2 (Fig. 2). Tax-specific CD8+ T cell responses more frequently expressed CD107a+ and IFN-γ+ than other measured parameters (Fig. 2). Despite the overwhelming dominance of Tax, most ORF peptide pools elicited responses from one or more animals. The only exception was Pol ORF4, with only marginal responses being detected (Fig. 1).
FIG 2.
Cytokine expression by Tax-specific CD8+ T cells. We tested the ability of Tax-specific CD8+ T cells to express IFN-γ, TNF-α, and IL-2 using ICS assays. We stimulated PMBCs with 15-mer peptide pools spanning Tax amino acids 1 to 171 (Tax ORF1) (A) and amino acids 161 to 353 (Tax ORF2) (B). Infected animals are denoted by (+), and uninfected animals are indicated by (−). Results are expressed as the median of the responses, and error bars indicate the range.
CD8+ T cells from several STLV-1-infected baboons target Tax pool 7.
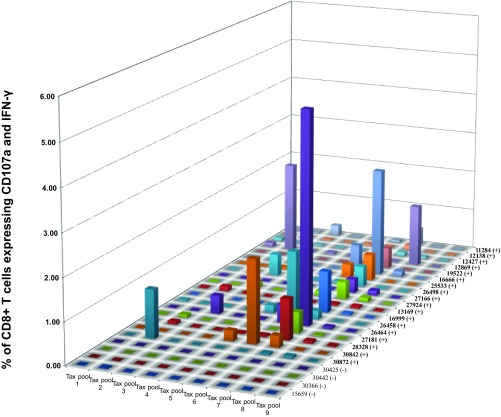
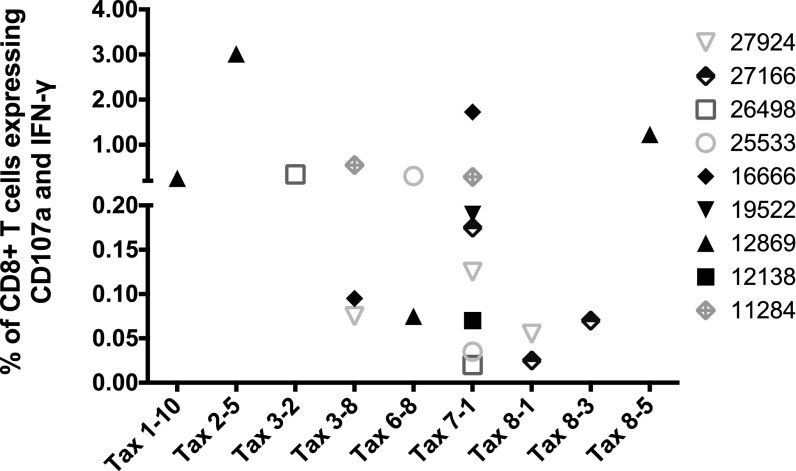
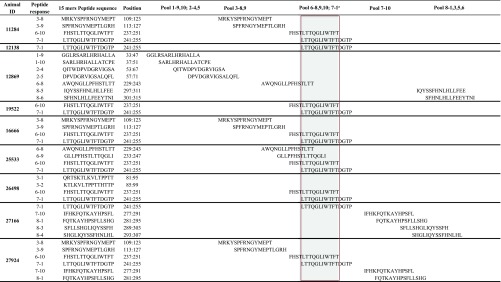
To identify the most immunogenic regions of Tax, we then fine-mapped the Tax-specific CD8+ T cell response. We first used pools consisting of only 10 15-mer peptides (Tax pools 1 to 9). We then further defined the targets of the CD8+ T cell responses using individual peptides. Pool 7 was preferentially recognized: 13 of the 18 animals mounted robust responses against this pool (Fig. 3), which spans Tax amino acids 241 to 291. While CD8+ T cell responses targeting Tax pool 7 accounted for the highest Tax-specific responses in 12/18 of the STLV-1-infected baboons, other animals (6/18) mounted their highest-frequency responses against other Tax pools (Tax pools 2, 3, 5, 6, and 8) (Fig. 3). We then selected 9 animals based on availability, and with diverse response characteristics (differing in STLV-1 response magnitudes, peptide pool reactivities, and PVLs), in order to define individual peptides containing putative immunodominant CD8+ T cell epitopes. After breaking down these pools into their individual peptides, we detected responses against peptides in pools 1, 2, 3, 6, 7, and 8 (Fig. 4 and Table 2). The most immunogenic peptide in pool 7 (7-1 [Tax241LTTQGLIWTFTDGTP255]) elicited responses in 8 of 9 tested infected animals (Fig. 4). Our results suggest that Tax241–255 contains a dominant epitope recognized by STLV-1-specific CD8+ T cells in infected baboons. Other peptides that harbor putative immunodominant epitopes are also listed in Table 2.
FIG 3.
The majority of STLV-1-infected animals target Tax amino acids 241 to 291, covered by Tax pool 7. PBMCs from the indicated animals were isolated and tested in ICS assays against small Tax pools of 10 peptides. Tax pool 7 (Tax241–291) was recognized by CD8+ T cells from most STLV-1-infected animals.
FIG 4.
Identification of STLV-1-derived peptides containing CD8+ T cell epitopes. CD8+ T cell responses against synthetic 15-mer peptides overlapping by 11 amino acids and covering the entire sequence of the Tax protein were tested in ICS assays. Results are expressed as the averages of data from two independent experiments.
TABLE 2.
Amino acid sequences of the Tax regions targeted by CD8+ T cells in nine olive baboons (Papio anubis)
a The shaded area indicates amino acid sequences common among immunogenic peptides in pools 6 and 7.
Tax-specific CD8+ T cells express effector memory markers.
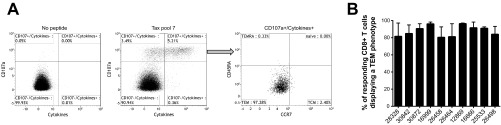
Next, we sought to characterize the memory phenotype of Tax-specific CD8+ T cells in STLV-1-infected monkeys. In this study, we defined the memory phenotype of Tax-specific CD8+ T cells from STLV-1-infected baboons based on the expression of CD45RA and the chemokine receptor CCR7 (50, 51). To determine the memory phenotype of Tax-specific CD8+ T cells, we stimulated PBMCs from 10 animals with their best-responding Tax peptide pool (usually pool 6 or 7) and assessed the expression of CD107a and cytokines. We gated on responding CD8+ T cells (CD107a+ cells expressing at least one of the cytokines IFN-γ, TNF-α, or IL-2) to define their memory phenotype (Fig. 5A). Indeed, the majority of Tax-specific cells displayed a TEM phenotype in all 10 tested animals (Fig. 5B). Previously, other groups reported the accumulation of effector memory lymphocytes in STLV-1-infected baboons after the animals were treated with valproate to induce viral expression and azidothymidine to prevent viral propagation (45). Thus, in STLV-1-infected baboons, it is possible that continuous exposure to viral antigen results in the generation of TEM cells.
FIG 5.
Tax-specific CD8+ T cells display an effector memory (TEM) phenotype. (A) PBMCs from animal 26464 were stimulated with Tax pool 7 to determine the memory phenotype of Tax-specific CD8+ T cells. We gated on the responding cells (CD107+ cells expressing IFN-γ+, TNF-α+, and/or IL-2+) to define the Tax-specific cells. The memory phenotype of the responding cells was then defined based on the expression of CD45RA and the chemokine receptor CCR7. TEM cells were defined as CD45RA− CCR7−, T effector memory RA+ cells (TEMRA) were defined as CCR7− CD45RA+, central memory T cells (TCM) were defined as CD45RA− CCR7+, and naive cells were defined as CCR7+ CD45RA+. (B) TEM phenotype of Tax-specific CD8+ T cells from 10 animals. The results represent the averages of data from two independent experiments. Error bars indicate the range.
DISCUSSION
A vaccine could be an important tool to reduce the burden of HTLV-1-induced diseases. Unfortunately, there are very few animal models useful for testing HTLV-1 vaccine approaches (52). Olive baboons (Papio anubis) are a nonhuman primate species that can be naturally infected with STLV-1, a virus closely related to HTLV-1. Sometimes, these STLV-1-infected baboons develop diseases that parallel clinical outcomes of HTLV-1 infection (40–42, 44, 52, 53). In fact, STLV-1-infected animals in the Southwest National Primate Research Center baboon colony develop lymphosarcoma at a higher rate than do uninfected animals (54–56). Thus, it is possible that understanding immune-mediated control of STLV-1 replication could help inform the development of HTLV-1 vaccines. With this goal in mind, we sought to characterize the cellular immune responses of STLV-1-infected baboons. In the present study, we have identified the immunogenic regions of the STLV-1 proteome in naturally infected baboons. Only a few reports have described STLV-1 in naturally infected monkeys (35, 45), but none have characterized the specificities of the T cell responses. Here we provided the first characterization of the preferential CD8+ and CD4+ T cell targets in the STLV-1 proteome. We also show that, similarly to HTLV-1-infected humans (13), the cellular responses in STLV-1-infected baboons preferentially target the Tax protein.
Our subjects had variable levels of PBMC-associated STLV-1 and virus-specific T cell responses (Table 1). Perhaps surprisingly, there was no statistically significant correlation of PVLs with the magnitude of STLV-1-specific CD8+ T cell responses (r = −0.4860 and P = 0.0580 for PVL [CT] versus frequency of STLV-1-specific CD8+ T cells [percent] as determined by Spearman's rank-order correlation). Despite the lack of statistical significance, there appears to be a trend associating the frequency of T cell responses with the PVL. For instance, the highest frequencies of STLV-1-specific cellular responses were detected in animals with relatively high PVLs (animals 16666, 26464, 12869, 30842, and 30872). While T cell responses were readily detectable in all animals with a measurable PVL, we detected only modest CD8+ and CD4+ T cell responses in animals with the lowest PVL burden. The exception was animal 26458, with high-frequency CD8+ T cell responses against Tax ORF2 (Fig. 1) despite an undetectable PVL, at least with the PCR primers used. The magnitude of the HTLV-1-specific CD8+ T cell response is often associated with PVLs in infected humans (12). However, these results are difficult to interpret in terms of infection or disease outcomes. For instance, higher frequencies of HTLV-1-specific CD8+ T cells are associated with a low PVL and a reduced risk of the inflammatory disease HAM/TSP (57). Nonetheless, other groups reported that higher levels of HTLV-1-specific cellular responses were associated with higher PVLs (58, 59), suggesting that the T cell response was merely fluctuating with virus levels rather than being a determinant of PVLs. Thus, parameters associated with T cells can be a cause and/or consequence of HTLV-1 control. Of note, deciphering the antiviral effects of T cells requires comprehensive longitudinal investigations that are outside the scope of the present study. While we observed an inverse relationship between the frequency of STLV-1-specific CD8+ T cells and STLV-1 PVLs, more work is needed to understand the predictive value of this association in terms of disease outcomes.
We also found that cellular immune responses against STLV-1 comprise mainly CD8+ T cells targeting discrete regions of the Tax protein. We found that the majority of STLV-1-specific CD8+ T cells produce IFN-γ, TNF-α, and, less frequently, IL-2 (Fig. 2). Furthermore, we observed that the majority of the IFN-γ-producing CD8+ T cells also expressed the degranulation marker CD107a. This finding suggests that these antigen-specific CD8+ T cells are cytotoxic and therefore capable of killing target cells.
There are, however, a number of caveats to this kind of ICS analysis. A potential limitation of response mapping by ICS or enzyme-linked immunosorbent spot (ELISPOT) assays, especially with single cytokines (e.g., IFN-γ), is that the cytokine-responding cells may not be representative of the entire virus-specific response. Therefore, to avoid potential underrepresentation of the responding cells, our analysis included cytokines (IFN-γ, TNF-α, and IL-2) and the degranulation marker CD107a (60). Our ICS assay is limited by the ability of CD8+ T cells to be stimulated by their cognate peptides and express the markers that we can detect. Ideally, one would validate the sensitivity of this assay by comparing the frequency of measured responses by ICS to the detection of antigen-specific cells by major histocompatibility complex (MHC) class I tetramer staining, which is independent of the ability of the cells to secrete cytokines. In most situations, the frequencies measured by ICS and tetramer staining are similar, if the ICS assay is sufficiently sensitive. This validation would provide further confirmation that ICS is able to optimally detect antigen-specific cells. Unfortunately, there are no baboon MHC class I tetramers currently available. While it remains possible that we have underestimated low-frequency responses, we have considerable experience with this ICS assay and have extensively characterized it in the context of a simian immunodeficiency virus (SIV)-infected Indian rhesus macaque model (61–63). Furthermore, we detected high-frequency responses against the Tax protein, a finding is similar to those in previous reports of HTLV-1-specific T cell responses in humans (13, 20, 64).
To develop a CD8+ T cell-based vaccine against a pathogen, it will be important to define the preferred targets of these effector cells. During human immunodeficiency virus (HIV) or SIV infection, for example, the CD8+ T cell response selects for escape mutants in almost all of the SIV proteins (61, 65). Furthermore, SIV- and HIV-specific CD8+ T cells target the entire virus. Here we demonstrate that the anti-STLV-1 CD8+ T cell response is instead highly focused on Tax in during the chronic phase. Additionally, the targets of STLV-1-specific CD8+ T cells in baboons closely resemble their HTLV-1-directed counterparts in humans. Specifically, peptide sequences recognized by CD8+ T cells in STLV-1-infected baboons aligned with known HTLV-1 CD8+ T cell epitopes restricted by HLA-A*02, HLA-A*24, HLA-B*15, and HLA-B*52 (16, 66, 67). These results suggest that not only do both human and baboon preferentially recognize Tax, they also share similarity in the recognized epitopes within Tax. These similarities thus suggest that STLV-1-infected baboons will be a suitable model for HTLV-1 vaccine research.
Analysis of freshly isolated PBMCs indicates that most HTLV-1-infected cells express low to undetectable levels of viral antigens in vivo (68, 69). As a result, a strategy to develop a vaccine will likely need to overcome this low level of viral expression, and an immunodominant protein might be the ideal immunogen for this task. Here we show that high-frequency CD8+ T cell responses against STLV-1 principally target Tax in STLV-1-infected baboons. We have also identified six distinct Tax regions that are targeted by STLV-1-specific CD8+ T cells. While other HTLV-1 antigens, like HBZ, could be used as a vaccine target, their immunogenicity is usually weak and requires several boosts (25). Furthermore, Suemori and colleagues demonstrated that HBZ-specific CD8+ T cells are scarcely detectable in HTLV-1 infections using HLA-A*0201/HBZ26–34 tetramer analysis (70). In addition, Rowan and colleagues concluded that HBZ epitope presentation by primary cells is significantly less efficient than that of Tax (71). These results are consistent with the results of our present study. Of 18 naturally STLV-1-infected animals, we observed SBZ-specific CD8+ T cells at >0.1% frequencies only in animal 16666 (Fig. 1A). Even in this animal, SBZ marginally contributed to the total magnitude of the STLV-1-specific CD8+ T cell response, with only 3.75% of the STLV-1-specific cells targeting this protein (Fig. 1B). It is possible, however, that the use of immunodominant viral proteins such as Tax would overcome the limitations imposed by the low level of HTLV-1 protein expression and could be useful for the development of an effective HTLV-1 vaccine.
An important theoretical consideration for the development of a Tax-specific ATL vaccine is that ATL cells often lack the ability to express Tax (72). Previously reported data suggest that Tax is required to maintain persistent infection in the early phases of the long progression to ATL, but this protein is dispensable in established ATL (73, 74). Immortalization of infected T cells is a major mechanism by which HTLV-1 and STLV-1 achieve persistence. Once infection is established, the genetic changes in the host genome are responsible for genome instability, and most viral proteins are no longer needed to maintain the growth-immortalized state (74). Tax is therefore a critical protein important to establish and maintain immortalization prior to more serious progression. In this scenario, a vaccine strategy that targets an HTLV-1 protein that is required for initiating and maintaining the growth-immortalized state makes sense to prevent HTLV-1-induced leukemogenesis. Accordingly, there is evidence for the presence of sufficient levels of Tax expression for the CD8+ T cell response to be effective in vivo (20, 75). In fact, tax/rex mRNA is the most abundant transcript of HTLV-1-infected cells (76). Recently, Suehiro et al. and Melief et al. developed a therapeutic vaccine consisting of dendritic cells (DCs) pulsed with Tax peptides designed to augment the HTLV-1 Tax-specific CD8+ T cell response in ATL patients (75, 77). The outcome of this pilot study indicated that the Tax peptide-pulsed DC vaccine is safe and a promising immunotherapy for ATL. Additionally, Tax-specific CD8+ T cells inhibited the growth of HTLV-1-transformed tumor cells in rats inoculated with a rat HTLV-1-infected T cell line (78). Furthermore, immunization with a mutant Tax DNA induced tumor-suppressive responses in this rat model (78). Thus, strategies aiming to augment the HTLV-1 Tax-specific CD8+ T cell response may be a promising immunotherapy for ATL, in particular for individuals in early infection stages.
T cell-mediated control of HTLV-1 infection is thought to depend on the frequency and function of virus-specific CD8+ T cells (12, 79, 80). The accumulation of cells with an effector memory phenotype was previously correlated with lower STLV-1 PVLs in baboons (45). Here we observed that over 80% of Tax-specific CD8+ T cells differentiated into a TEM phenotype in all 10 tested animals (Fig. 5). TEM cells, in contrast to other memory T cell phenotypes, proliferate less but are capable of migrating to peripheral tissues and exert immediate antiviral activity (79). However, we did not observe a correlation between the TEM cell frequency and PVL (r = −0.4545 and P = 0.1912 for PVL [CT] versus Tax-specific TEM cell frequency [percent] as determined by Spearman's rank-order correlation). Of note, the baboons in our cohort did not display signs of disease, so these infections could be considered examples of controlled or early-stage STLV-1 infections. Thus, it remains possible that we did not see an association because we did not have sufficient TEM and STLV-1 infection variability or did not observe the outcomes longitudinally. Future research will address the association of T cell phenotypes with the control of STLV-1.
In sum, here we show that anti-STLV-1 cellular immunity in baboons resembles HTLV-1-specific T cell responses in humans in their preferential targeting of Tax. There are reasons to believe that a Tax-specific CD8+ T cell response may be particularly protective. First, Tax is the immunodominant target of CD8+ T cell responses in controlled HTLV-1 and STLV-1 infections. Second, HLA-A*02, which is associated with protection in southern Japan, binds several Tax epitopes (67). Third, the selective pressure exerted on Tax may be higher in asymptomatic carriers than in those who have developed HAM/TSP (81). Fourth, Tax-specific CD8+ T cells can suppress tumor growth in vivo in a rat model of ATL (78). The Tax protein is therefore predicted to be a useful immunogen for HTLV-1 vaccine development.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Luis Giavedoni, Director, Immunology Core Laboratories, SNPRC, for STLV-1 serological testing of the animals used in this study and the veterinary staff for their research support. We thank JoAnn Yee from the California National Primate Research Center, University of California, Davis, for performing the STLV-1 PVL assay. We also thank Delia Gutman, Leydi Guzman, and the members of the Watkins laboratory for technical and administrative assistance.
Funding Statement
This work is supported by the Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, and HTLV-1 Program Project internal funds. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00281-16.
REFERENCES
- 1.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci U S A 77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallo RC. 2005. The discovery of the first human retrovirus: HTLV-1 and HTLV-2. Retrovirology 2:17. doi: 10.1186/1742-4690-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Proietti FA, Carneiro-Proietti AB, Catalan-Soares BC, Murphy EL. 2005. Global epidemiology of HTLV-I infection and associated diseases. Oncogene 24:6058–6068. doi: 10.1038/sj.onc.1208968. [DOI] [PubMed] [Google Scholar]
- 4.Gessain A, Cassar O. 2012. Epidemiological aspects and world distribution of HTLV-1 infection. Front Microbiol 3:388. doi: 10.3389/fmicb.2012.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida M, Seiki M, Yamaguchi K, Takatsuki K. 1984. Monoclonal integration of human T-cell leukemia provirus in all primary tumors of adult T-cell leukemia suggests causative role of human T-cell leukemia virus in the disease. Proc Natl Acad Sci U S A 81:2534–2537. doi: 10.1073/pnas.81.8.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gessain A, Barin F, Vernant JC, Gout O, Maurs L, Calender A, de The G. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii:407–410. [DOI] [PubMed] [Google Scholar]
- 7.LaGrenade L, Hanchard B, Fletcher V, Cranston B, Blattner W. 1990. Infective dermatitis of Jamaican children: a marker for HTLV-I infection. Lancet 336:1345–1347. doi: 10.1016/0140-6736(90)92896-P. [DOI] [PubMed] [Google Scholar]
- 8.Mochizuki M, Watanabe T, Yamaguchi K, Takatsuki K, Yoshimura K, Shirao M, Nakashima S, Mori S, Araki S, Miyata N. 1992. HTLV-I uveitis: a distinct clinical entity caused by HTLV-I. Jpn J Cancer Res 83:236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuura E, Umehara F, Nose H, Higuchi I, Matsuoka E, Izumi K, Kubota R, Saito M, Izumo S, Arimura K, Osame M. 2008. Inclusion body myositis associated with human T-lymphotropic virus-type I infection: eleven patients from an endemic area in Japan. J Neuropathol Exp Neurol 67:41–49. doi: 10.1097/nen.0b013e31815f38b7. [DOI] [PubMed] [Google Scholar]
- 10.Satake M, Yamada Y, Atogami S, Yamaguchi K. 2015. The incidence of adult T-cell leukemia/lymphoma among human T-lymphotropic virus type 1 carriers in Japan. Leuk Lymphoma 56:1806–1812. doi: 10.3109/10428194.2014.964700. [DOI] [PubMed] [Google Scholar]
- 11.Satake M, Yamaguchi K, Tadokoro K. 2012. Current prevalence of HTLV-1 in Japan as determined by screening of blood donors. J Med Virol 84:327–335. doi: 10.1002/jmv.23181. [DOI] [PubMed] [Google Scholar]
- 12.Bangham CR. 2009. CTL quality and the control of human retroviral infections. Eur J Immunol 39:1700–1712. doi: 10.1002/eji.200939451. [DOI] [PubMed] [Google Scholar]
- 13.Goon PK, Biancardi A, Fast N, Igakura T, Hanon E, Mosley AJ, Asquith B, Gould KG, Marshall S, Taylor GP, Bangham CR. 2004. Human T cell lymphotropic virus (HTLV) type-1-specific CD8+ T cells: frequency and immunodominance hierarchy. J Infect Dis 189:2294–2298. doi: 10.1086/420832. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson S, Shida H, McFarlin DE, Fauci AS, Koenig S. 1990. Circulating CD8+ cytotoxic T lymphocytes specific for HTLV-I pX in patients with HTLV-I associated neurological disease. Nature 348:245–248. doi: 10.1038/348245a0. [DOI] [PubMed] [Google Scholar]
- 15.Kannagi M, Harada S, Maruyama I, Inoko H, Igarashi H, Kuwashima G, Sato S, Morita M, Kidokoro M, Sugimoto M, Funahashi S, Osame M, Shida H. 1991. Predominant recognition of human T cell leukemia virus type I (HTLV-I) pX gene products by human CD8+ cytotoxic T cells directed against HTLV-I-infected cells. Int Immunol 3:761–767. doi: 10.1093/intimm/3.8.761. [DOI] [PubMed] [Google Scholar]
- 16.Parker CE, Nightingale S, Taylor GP, Weber J, Bangham CR. 1994. Circulating anti-Tax cytotoxic T lymphocytes from human T-cell leukemia virus type I-infected people, with and without tropical spastic paraparesis, recognize multiple epitopes simultaneously. J Virol 68:2860–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanon E, Hall S, Taylor GP, Saito M, Davis R, Tanaka Y, Usuku K, Osame M, Weber JN, Bangham CR. 2000. Abundant Tax protein expression in CD4+ T cells infected with human T-cell lymphotropic virus type I (HTLV-I) is prevented by cytotoxic T lymphocytes. Blood 95:1386–1392. [PubMed] [Google Scholar]
- 18.Parker CE, Daenke S, Nightingale S, Bangham CR. 1992. Activated, HTLV-1-specific cytotoxic T-lymphocytes are found in healthy seropositives as well as in patients with tropical spastic paraparesis. Virology 188:628–636. doi: 10.1016/0042-6822(92)90517-S. [DOI] [PubMed] [Google Scholar]
- 19.Currer R, Van Duyne R, Jaworski E, Guendel I, Sampey G, Das R, Narayanan A, Kashanchi F. 2012. HTLV tax: a fascinating multifunctional co-regulator of viral and cellular pathways. Front Microbiol 3:406. doi: 10.3389/fmicb.2012.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harashima N, Kurihara K, Utsunomiya A, Tanosaki R, Hanabuchi S, Masuda M, Ohashi T, Fukui F, Hasegawa A, Masuda T, Takaue Y, Okamura J, Kannagi M. 2004. Graft-versus-Tax response in adult T-cell leukemia patients after hematopoietic stem cell transplantation. Cancer Res 64:391–399. doi: 10.1158/0008-5472.CAN-03-1452. [DOI] [PubMed] [Google Scholar]
- 21.Mesnard JM, Barbeau B, Cesaire R, Peloponese JM. 2015. Roles of HTLV-1 basic zip factor (HBZ) in viral chronicity and leukemic transformation. Potential new therapeutic approaches to prevent and treat HTLV-1-related diseases. Viruses 7:6490–6505. doi: 10.3390/v7122952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahieux R. 2015. A vaccine against HTLV-1 HBZ makes sense. Blood 126:1052–1053. doi: 10.1182/blood-2015-06-652040. [DOI] [PubMed] [Google Scholar]
- 23.Matsuoka M, Jeang KT. 2007. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer 7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- 24.Satou Y, Yasunaga J, Yoshida M, Matsuoka M. 2006. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci U S A 103:720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugata K, Yasunaga JI, Mitobe Y, Miura M, Miyazato P, Kohara M, Matsuoka M. 10 June 2015. Protective effect of cytotoxic T lymphocytes targeting HTLV-1 bZIP factor. Blood doi: 10.1182/blood-2015-04-641118. [DOI] [PubMed] [Google Scholar]
- 26.Nagai M, Usuku K, Matsumoto W, Kodama D, Takenouchi N, Moritoyo T, Hashiguchi S, Ichinose M, Bangham CR, Izumo S, Osame M. 1998. Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J Neurovirol 4:586–593. doi: 10.3109/13550289809114225. [DOI] [PubMed] [Google Scholar]
- 27.Taylor GP, Tosswill JH, Matutes E, Daenke S, Hall S, Bain BJ, Davis R, Thomas D, Rossor M, Bangham CR, Weber JN. 1999. Prospective study of HTLV-I infection in an initially asymptomatic cohort. J Acquir Immune Defic Syndr 22:92–100. doi: 10.1097/00042560-199909010-00012. [DOI] [PubMed] [Google Scholar]
- 28.Akimoto M, Kozako T, Sawada T, Matsushita K, Ozaki A, Hamada H, Kawada H, Yoshimitsu M, Tokunaga M, Haraguchi K, Uozumi K, Arima N, Tei C. 2007. Anti-HTLV-1 tax antibody and tax-specific cytotoxic T lymphocyte are associated with a reduction in HTLV-1 proviral load in asymptomatic carriers. J Med Virol 79:977–986. doi: 10.1002/jmv.20807. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita M, Ido E, Miura T, Hayami M. 1996. Molecular epidemiology of HTLV-I in the world. J Acquir Immune Defic Syndr Hum Retrovirol 13(Suppl 1):S124–S131. [DOI] [PubMed] [Google Scholar]
- 30.Mahieux R, Ibrahim F, Mauclere P, Herve V, Michel P, Tekaia F, Chappey C, Garin B, Van Der Ryst E, Guillemain B, Ledru E, Delaporte E, de The G, Gessain A. 1997. Molecular epidemiology of 58 new African human T-cell leukemia virus type 1 (HTLV-1) strains: identification of a new and distinct HTLV-1 molecular subtype in Central Africa and in Pygmies. J Virol 71:1317–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verdonck K, Gonzalez E, Van Dooren S, Vandamme AM, Vanham G, Gotuzzo E. 2007. Human T-lymphotropic virus 1: recent knowledge about an ancient infection. Lancet Infect Dis 7:266–281. doi: 10.1016/S1473-3099(07)70081-6. [DOI] [PubMed] [Google Scholar]
- 32.Giri A, Slattery JP, Heneine W, Gessain A, Rivadeneira E, Desrosiers RC, Rosen L, Anthony R, Pamungkas J, Iskandriati D, Richards AL, Herve V, McClure H, O'Brien SJ, Franchini G. 1997. The tax gene sequences form two divergent monophyletic lineages corresponding to types I and II of simian and human T-cell leukemia/lymphotropic viruses. Virology 231:96–104. doi: 10.1006/viro.1997.8511. [DOI] [PubMed] [Google Scholar]
- 33.Miura M, Yasunaga J, Tanabe J, Sugata K, Zhao T, Ma G, Miyazato P, Ohshima K, Kaneko A, Watanabe A, Saito A, Akari H, Matsuoka M. 2013. Characterization of simian T-cell leukemia virus type 1 in naturally infected Japanese macaques as a model of HTLV-1 infection. Retrovirology 10:118. doi: 10.1186/1742-4690-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazanji M, Mouinga-Ondeme A, Lekana-Douki-Etenna S, Caron M, Makuwa M, Mahieux R, Gessain A. 2015. Origin of HTLV-1 in hunters of nonhuman primates in Central Africa. J Infect Dis 211:361–365. doi: 10.1093/infdis/jiu464. [DOI] [PubMed] [Google Scholar]
- 35.Gabet AS, Gessain A, Wattel E. 2003. High simian T-cell leukemia virus type 1 proviral loads combined with genetic stability as a result of cell-associated provirus replication in naturally infected, asymptomatic monkeys. Int J Cancer 107:74–83. doi: 10.1002/ijc.11329. [DOI] [PubMed] [Google Scholar]
- 36.Souquiere S, Mouinga-Ondeme A, Makuwa M, Hermine O, Kazanji M. 2009. Dynamic interaction between STLV-1 proviral load and T-cell response during chronic infection and after immunosuppression in non-human primates. PLoS One 4:e6050. doi: 10.1371/journal.pone.0006050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.d'Offay JM, Eberle R, Sucol Y, Schoelkopf L, White MA, Valentine BD, White GL, Lerche NW. 2007. Transmission dynamics of simian T-lymphotropic virus type 1 (STLV1) in a baboon breeding colony: predominance of female-to-female transmission. Comp Med 57:105–114. [PubMed] [Google Scholar]
- 38.Ishikawa K, Fukasawa M, Tsujimoto H, Else JG, Isahakia M, Ubhi NK, Ishida T, Takenaka O, Kawamoto Y, Shotake T, Ohsawa H, Ivanoff B, Cooper RW, Frost E, Grant FC, Spriatna Y, Sutarman K, Abe K, Yamamoto K, Hayami M. 1987. Serological survey and virus isolation of simian T-cell leukemia/T-lymphotropic virus type I (STLV-I) in non-human primates in their native countries. Int J Cancer 40:233–239. doi: 10.1002/ijc.2910400219. [DOI] [PubMed] [Google Scholar]
- 39.Saksena NK, Herve V, Durand JP, Leguenno B, Diop OM, Digouette JP, Mathiot C, Muller MC, Love JL, Dube S, Sherman MP, Benz PM, Erensoy S, Galat-Luong A, Galat G, Paul B, Dube DK, Sinoussi FB, Poiesz BJ. 1994. Seroepidemiologic, molecular, and phylogenetic analyses of simian T-cell leukemia viruses (STLV-I) from various naturally infected monkey species from central and western Africa. Virology 198:297–310. doi: 10.1006/viro.1994.1033. [DOI] [PubMed] [Google Scholar]
- 40.Takemura T, Yamashita M, Shimada MK, Ohkura S, Shotake T, Ikeda M, Miura T, Hayami M. 2002. High prevalence of simian T-lymphotropic virus type L in wild Ethiopian baboons. J Virol 76:1642–1648. doi: 10.1128/JVI.76.4.1642-1648.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahieux R, Chappey C, Meertens L, Mauclere P, Lewis J, Gessain A. 2000. Molecular characterization and phylogenetic analyses of a new simian T cell lymphotropic virus type 1 in a wild-caught African baboon (Papio anubis) with an indeterminate STLV type 2-like serology. AIDS Res Hum Retroviruses 16:2043–2048. doi: 10.1089/088922200750054774. [DOI] [PubMed] [Google Scholar]
- 42.Mahieux R, Pecon-Slattery J, Chen GM, Gessain A. 1998. Evolutionary inferences of novel simian T lymphotropic virus type 1 from wild-caught chacma (Papio ursinus) and olive baboons (Papio anubis). Virology 251:71–84. doi: 10.1006/viro.1998.9377. [DOI] [PubMed] [Google Scholar]
- 43.Allan JS, Leland M, Broussard S, Mone J, Hubbard G. 2001. Simian T-cell lymphotropic viruses (STLVs) and lymphomas in African nonhuman primates. Cancer Invest 19:383–395. doi: 10.1081/CNV-100103133. [DOI] [PubMed] [Google Scholar]
- 44.d'Offay JM, Eberle R, Wolf RF, Kosanke SD, Doocy KR, Ayalew S, Mansfeild KG, White GL. 2013. Simian T-lymphotropic virus-associated lymphoma in 2 naturally infected baboons: T-cell clonal expansion and immune response during tumor development. Comp Med 63:288–294. [PMC free article] [PubMed] [Google Scholar]
- 45.Afonso PV, Mekaouche M, Mortreux F, Toulza F, Moriceau A, Wattel E, Gessain A, Bangham CR, Dubreuil G, Plumelle Y, Hermine O, Estaquier J, Mahieux R. 2010. Highly active antiretroviral treatment against STLV-1 infection combining reverse transcriptase and HDAC inhibitors. Blood 116:3802–3808. doi: 10.1182/blood-2010-02-270751. [DOI] [PubMed] [Google Scholar]
- 46.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 47.Liao Q, Guo H, Tang M, Touzjian N, Lerche NW, Lu Y, Yee JL. 2011. Simultaneous detection of antibodies to five simian viruses in nonhuman primates using recombinant viral protein based multiplex microbead immunoassays. J Virol Methods 178:143–152. doi: 10.1016/j.jviromet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wunderlich ML, Dodge ME, Dhawan RK, Shek WR. 12 December 2011. Multiplexed fluorometric immunoassay testing methodology and troubleshooting. J Vis Exp doi: 10.3791/3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White JA, Todd PA, Rosenthal AN, Yee JL, Grant R, Lerche NW. 2009. Development of a generic real-time PCR assay for simultaneous detection of proviral DNA of simian betaretrovirus serotypes 1, 2, 3, 4 and 5 and secondary uniplex assays for specific serotype identification. J Virol Methods 162:148–154. doi: 10.1016/j.jviromet.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson-Nauroth JM, Graber J, Yao K, Jacobson S, Calabresi PA. 2006. Memory lineage relationships in HTLV-1-specific CD8+ cytotoxic T cells. J Neuroimmunol 176:115–124. doi: 10.1016/j.jneuroim.2006.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lugli E, Dominguez MH, Gattinoni L, Chattopadhyay PK, Bolton DL, Song K, Klatt NR, Brenchley JM, Vaccari M, Gostick E, Price DA, Waldmann TA, Restifo NP, Franchini G, Roederer M. 2013. Superior T memory stem cell persistence supports long-lived T cell memory. J Clin Invest 123:594–599. doi: 10.1172/JCI66327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lairmore MD, Silverman L, Ratner L. 2005. Animal models for human T-lymphotropic virus type 1 (HTLV-1) infection and transformation. Oncogene 24:6005–6015. doi: 10.1038/sj.onc.1208974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voevodin A, Samilchuk E, Schatzl H, Boeri E, Franchini G. 1996. Interspecies transmission of macaque simian T-cell leukemia/lymphoma virus type 1 in baboons resulted in an outbreak of malignant lymphoma. J Virol 70:1633–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hubbard GB, Mone JP, Allan JS, Davis KJ III, Leland MM, Banks PM, Smir B. 1993. Spontaneously generated non-Hodgkin's lymphoma in twenty-seven simian T-cell leukemia virus type 1 antibody-positive baboons (Papio species). Lab Anim Sci 43:301–309. [PubMed] [Google Scholar]
- 55.Dick EJ Jr, Owston MA, David JM, Sharp RM, Rouse S, Hubbard GB. 2014. Mortality in captive baboons (Papio spp.): a-23-year study. J Med Primatol 43:169–196. doi: 10.1111/jmp.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dick EJ., Jr 2014. Pulmonary lymphosarcoma (STLV-1) in a baboon. Abstr 23rd Annu Southcentral Div Conf Slide Semin Charles Louis Davis Found, Galveston, TX. [Google Scholar]
- 57.Vine AM, Heaps AG, Kaftantzi L, Mosley A, Asquith B, Witkover A, Thompson G, Saito M, Goon PK, Carr L, Martinez-Murillo F, Taylor GP, Bangham CR. 2004. The role of CTLs in persistent viral infection: cytolytic gene expression in CD8+ lymphocytes distinguishes between individuals with a high or low proviral load of human T cell lymphotropic virus type 1. J Immunol 173:5121–5129. doi: 10.4049/jimmunol.173.8.5121. [DOI] [PubMed] [Google Scholar]
- 58.Kubota R, Nagai M, Kawanishi T, Osame M, Jacobson S. 2000. Increased HTLV type 1 tax specific CD8+ cells in HTLV type 1-asociated myelopathy/tropical spastic paraparesis: correlation with HTLV type 1 proviral load. AIDS Res Hum Retroviruses 16:1705–1709. doi: 10.1089/08892220050193182. [DOI] [PubMed] [Google Scholar]
- 59.Wodarz D, Hall SE, Usuku K, Osame M, Ogg GS, McMichael AJ, Nowak MA, Bangham CR. 2001. Cytotoxic T-cell abundance and virus load in human immunodeficiency virus type 1 and human T-cell leukaemia virus type 1. Proc Biol Sci 268:1215–1221. doi: 10.1098/rspb.2001.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. 2003. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods 281:65–78. doi: 10.1016/S0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 61.Martins MA, Tully DC, Cruz MA, Power KA, Veloso de Santana MG, Bean DJ, Ogilvie CB, Gadgil R, Lima NS, Magnani DM, Ejima K, Allison DB, Piatak M Jr, Altman JD, Parks CL, Rakasz EG, Capuano S III, Galler R, Bonaldo MC, Lifson JD, Allen TM, Watkins DI. 2015. Vaccine-induced simian immunodeficiency virus-specific CD8+ T-cell responses focused on a single Nef epitope select for escape variants shortly after infection. J Virol 89:10802–10820. doi: 10.1128/JVI.01440-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, Piaskowski SM, Duan L, Seese A, Gladden AD, Weisgrau KL, Furlott JR, Kim YI, Veloso de Santana MG, Rakasz E, Capuano S III, Wilson NA, Bonaldo MC, Galler R, Allison DB, Piatak M Jr, Haase AT, Lifson JD, Allen TM, Watkins DI. 2012. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature 491:129–133. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sacha JB, Buechler MB, Newman LP, Reed J, Wallace LT, Loffredo JT, Wilson NA, Watkins DI. 2010. Simian immunodeficiency virus-specific CD8+ T cells recognize Vpr- and Rev-derived epitopes early after infection. J Virol 84:10907–10912. doi: 10.1128/JVI.01357-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka Y, Nakasone H, Yamazaki R, Wada H, Ishihara Y, Kawamura K, Sakamoto K, Ashizawa M, Machishima T, Sato M, Terasako K, Kimura S, Kikuchi M, Okuda S, Kako S, Kanda J, Tanihara A, Nishida J, Kanda Y. 2012. Long-term persistence of limited HTLV-I Tax-specific cytotoxic T cell clones in a patient with adult T cell leukemia/lymphoma after allogeneic stem cell transplantation. J Clin Immunol 32:1340–1352. doi: 10.1007/s10875-012-9729-5. [DOI] [PubMed] [Google Scholar]
- 65.Goulder PJ, Watkins DI. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol 4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 66.Pique C, Connan F, Levilain JP, Choppin J, Dokhelar MC. 1996. Among all human T-cell leukemia virus type 1 proteins, Tax, polymerase, and envelope proteins are predicted as preferential targets for the HLA-A2-restricted cytotoxic T-cell response. J Virol 70:4919–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yashiki S, Fujiyoshi T, Arima N, Osame M, Yoshinaga M, Nagata Y, Tara M, Nomura K, Utsunomiya A, Hanada S, Tajima K, Sonoda S. 2001. HLA-A*26, HLA-B*4002, HLA-B*4006, and HLA-B*4801 alleles predispose to adult T cell leukemia: the limited recognition of HTLV type 1 tax peptide anchor motifs and epitopes to generate anti-HTLV type 1 tax CD8(+) cytotoxic T lymphocytes. AIDS Res Hum Retroviruses 17:1047–1061. doi: 10.1089/088922201300343735. [DOI] [PubMed] [Google Scholar]
- 68.Gessain A, Louie A, Gout O, Gallo RC, Franchini G. 1991. Human T-cell leukemia-lymphoma virus type I (HTLV-I) expression in fresh peripheral blood mononuclear cells from patients with tropical spastic paraparesis/HTLV-I-associated myelopathy. J Virol 65:1628–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richardson JH, Hollsberg P, Windhagen A, Child LA, Hafler DA, Lever AM. 1997. Variable immortalizing potential and frequent virus latency in blood-derived T-cell clones infected with human T-cell leukemia virus type I. Blood 89:3303–3314. [PubMed] [Google Scholar]
- 70.Suemori K, Fujiwara H, Ochi T, Ogawa T, Matsuoka M, Matsumoto T, Mesnard JM, Yasukawa M. 2009. HBZ is an immunogenic protein, but not a target antigen for human T-cell leukemia virus type 1-specific cytotoxic T lymphocytes. J Gen Virol 90:1806–1811. doi: 10.1099/vir.0.010199-0. [DOI] [PubMed] [Google Scholar]
- 71.Rowan AG, Suemori K, Fujiwara H, Yasukawa M, Tanaka Y, Taylor GP, Bangham CR. 2014. Cytotoxic T lymphocyte lysis of HTLV-1 infected cells is limited by weak HBZ protein expression, but non-specifically enhanced on induction of Tax expression. Retrovirology 11:116. doi: 10.1186/s12977-014-0116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeda S, Maeda M, Morikawa S, Taniguchi Y, Yasunaga J, Nosaka K, Tanaka Y, Matsuoka M. 2004. Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int J Cancer 109:559–567. doi: 10.1002/ijc.20007. [DOI] [PubMed] [Google Scholar]
- 73.Azran I, Schavinsky-Khrapunsky Y, Aboud M. 2004. Role of Tax protein in human T-cell leukemia virus type-I leukemogenicity. Retrovirology 1:20. doi: 10.1186/1742-4690-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mortreux F, Gabet AS, Wattel E. 2003. Molecular and cellular aspects of HTLV-1 associated leukemogenesis in vivo. Leukemia 17:26–38. doi: 10.1038/sj.leu.2402777. [DOI] [PubMed] [Google Scholar]
- 75.Suehiro Y, Hasegawa A, Iino T, Sasada A, Watanabe N, Matsuoka M, Takamori A, Tanosaki R, Utsunomiya A, Choi I, Fukuda T, Miura O, Takaishi S, Teshima T, Akashi K, Kannagi M, Uike N, Okamura J. 2015. Clinical outcomes of a novel therapeutic vaccine with Tax peptide-pulsed dendritic cells for adult T cell leukaemia/lymphoma in a pilot study. Br J Haematol 169:356–367. doi: 10.1111/bjh.13302. [DOI] [PubMed] [Google Scholar]
- 76.Li M, Kesic M, Yin H, Yu L, Green PL. 2009. Kinetic analysis of human T-cell leukemia virus type 1 gene expression in cell culture and infected animals. J Virol 83:3788–3797. doi: 10.1128/JVI.02315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Melief CJ, van Hall T, Arens R, Ossendorp F, van der Burg SH. 2015. Therapeutic cancer vaccines. J Clin Invest 125:3401–3412. doi: 10.1172/JCI80009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ohashi T, Hanabuchi S, Kato H, Tateno H, Takemura F, Tsukahara T, Koya Y, Hasegawa A, Masuda T, Kannagi M. 2000. Prevention of adult T-cell leukemia-like lymphoproliferative disease in rats by adoptively transferred T cells from a donor immunized with human T-cell leukemia virus type 1 Tax-coding DNA vaccine. J Virol 74:9610–9616. doi: 10.1128/JVI.74.20.9610-9616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sallusto F, Geginat J, Lanzavecchia A. 2004. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol 22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 80.Bangham CR, Meekings K, Toulza F, Nejmeddine M, Majorovits E, Asquith B, Taylor GP. 2009. The immune control of HTLV-1 infection: selection forces and dynamics. Front Biosci (Landmark Ed) 14:2889–2903. [DOI] [PubMed] [Google Scholar]
- 81.Kubota R, Hanada K, Furukawa Y, Arimura K, Osame M, Gojobori T, Izumo S. 2007. Genetic stability of human T lymphotropic virus type I despite antiviral pressures by CTLs. J Immunol 178:5966–5972. doi: 10.4049/jimmunol.178.9.5966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.