Abstract
Melioidosis is an emerging, potentially fatal disease caused by Burkholderia pseudomallei, which requires prolonged antibiotic treatment to prevent disease relapse. However, difficulties in laboratory diagnosis of melioidosis may delay treatment and affect disease outcomes. Isolation of B. pseudomallei from clinical specimens has been improved with the use of selective media. However, even with positive cultures, identification of B. pseudomallei can be difficult in clinical microbiology laboratories, especially in non-endemic areas where clinical suspicion is low. Commercial identification systems may fail to distinguish between B. pseudomallei and closely related species such as Burkholderia thailandensis. Genotypic identification of suspected isolates can be achieved by sequencing of gene targets such as groEL which offer higher discriminative power than 16S rRNA. Specific PCR-based identification of B. pseudomallei has also been developed using B. pseudomallei-specific gene targets such as Type III secretion system and Tat-domain protein. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolutionary technique for pathogen identification, has been shown to be potentially useful for rapid identification of B. pseudomallei, although existing databases require optimization by adding reference spectra for B. pseudomallei. Despite these advances in bacterial identification, diagnostic problems encountered in culture-negative cases remain largely unresolved. Although various serological tests have been developed, they are generally unstandardized “in house” assays and have low sensitivities and specificities. Although specific PCR assays have been applied to direct clinical and environmental specimens, the sensitivities for diagnosis remain to be evaluated. Metabolomics is an uprising tool for studying infectious diseases and may offer a novel approach for exploring potential diagnostic biomarkers. The metabolomics profiles of B. pseudomallei culture supernatants can be potentially distinguished from those of related bacterial species including B. thailandensis. Further studies using bacterial cultures and direct patient samples are required to evaluate the potential of metabolomics for improving diagnosis of melioidosis.
Keywords: Melioidosis, Burkholderia pseudomallei, laboratory, diagnosis, metabolomics, biomarkers
Introduction
Melioidosis is a potentially serious disease caused by Burkholderia pseudomallei—a highly pathogenic, Gram-negative β-proteobacterium. B. pseudomallei is a saprophyte found in soil, groundwater, stagnant streams, rice paddies, and ponds.1,2 Although melioidosis is mainly endemic in Southeast Asia and northern Australia, it is also increasingly reported in regions outside the Asia-Pacific region including India,3 Mauritius,4 the Americas,5–7 and Africa.8,9 Melioidosis can present as an acute, subacute, or chronic process. Disease manifestations include subclinical infections, localized abscesses, severe pneumonia, and fulminant sepsis. Case fatality rates ranged from 19 to 36% in endemic areas.10,11 Although the epidemiology and routes of transmission are not yet fully understood, it is believed that melioidosis is acquired through contact with contaminated soil and water by percutaneous inoculation, inhalation of aerosols, and ingestion.12 The incubation period of melioidosis varies widely from two days to 62 years.13 Human cases are often spatially and temporally clustered, following heavy rains and winds with resultant human exposure to soil and water.14,15 B. pseudomallei also causes melioidosis in a wide range of animals in endemic areas.16 In Hong Kong, melioidosis is an endemic disease not only in humans but also in captive marine mammals and birds, including bottlenose dolphins, California sea lions, pilot whales, and zebra doves.17 Treatment of melioidosis can be difficult, as B. pseudomallei is often resistant to multiple antibiotics, and a prolonged course of antibiotics is required to prevent disease relapse.12,18,19 Due to the severity of melioidosis and aerosol transmissibility of the infectious agent, B. pseudomallei has been classified as a category B bioterrorism and Tier 1 select agent by the Center for Disease Control, USA (http://www.bt.cdc.gov/agent/agentlist-category.asp).
Laboratory diagnosis of melioidosis can be difficult. The bacterium is often not readily isolated from clinical specimens and may not be correctly identified even when isolated. Serological tests are neither sensitive nor specific. In the past two decades, laboratory diagnosis of melioidosis has advanced through development of more sensitive tests such as PCR-based diagnostics and rapid specific identification technologies such as gene sequencing and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). In this review, we highlight the major developments in laboratory diagnosis of melioidosis. In addition, metabolomic profiling for identification of potentially novel biomarkers for melioidosis is also discussed.
Culture of B. pseudomallei
Although B. pseudomallei grows on blood agar and MacConkey agar, it is often dismissed as a culture contaminant or misidentified as Pseudomonas species when non-sterile clinical or environmental specimens are cultured. The most widely used selective medium for isolating B. pseudomallei is Ashdown’s medium, which was first described by L. R. Ashdown in 1979.20 Ashdown’s medium contains crystal violet and gentamicin as selecting agents. B. pseudomallei produces characteristic purple, dry, and wrinkled colonies on this medium. Ashdown’s medium should be incubated for at least 96 h because gentamicin may have some inhibitory effects on the growth of B. pseudomallei. The use of an enrichment broth with Ashdown’s medium and colistin for 48 h at 37℃ followed by plating on Ashdown’s medium may further increase the yield but increases time to laboratory diagnosis.21 Other selective media have also been used for clinical isolation of B. pseudomallei. For example, the Burkholderia pseudomallei selective agar (BPSA) and a commercial Burkholderia cepacia medium were both found to have equivalent sensitivity to Ashdown’s medium, but the selectivity of BPSA was lower than that of both Ashdown’s and B. cepacia medium when evaluated with 155 clinical specimens that proved positive for B. pseudomallei.22
Identification of B. pseudomallei
Conventional biochemical tests and commercial kits
Identification of B. pseudomallei poses difficulties in the clinical microbiology laboratory, particularly in localities where B. pseudomallei is rarely found. Even with positive cultures, commercial bacterial identification kits may fail to distinguish between B. pseudomallei and closely related species such as Burkholderia thailandensis (a phenotypically similar but avirulent species) and members of the B. cepacia complex (BCC).23 In particular, differentiation between B. pseudomallei and B. thailandensis would be crucial in guiding clinical management of patients with suspected melioidosis. This is because more than 99% of cases of melioidosis are caused by B. pseudomallei, whereas B. thailandensis causes less than 1% of melioidosis.24 B. pseudomallei can be distinguished from B. thailandensis by arabinose assimilation; B. thailandensis, but not B. pseudomallei, is able to utilize l-arabinose as its sole carbon source.25–28 B. pseudomallei is included in the database of API 20NE and the Vitek 1 and Vitek 2 systems, with variable reported accuracies,23,29–35 and reports of misidentification of B. pseudomallei as other Burkholderia species such as B. cepacia complex.36 In our experience, the accuracy of identification using these commercial systems is around 80%.
Identification by sequencing conserved gene targets
Amplification and sequencing of universal gene targets is a technology that enables timely identification of difficult-to-identify bacteria. Among the various studied gene targets, 16S rRNA gene sequencing is the most widely used for the identification of bacteria in clinical microbiology laboratories.37 The successful use of 16S rRNA gene sequencing for identification of a bacterium to the species level relies on the difference between the 16S rRNA gene sequences of the bacterium and closely related species. In the case of B. pseudomallei, although 16S rRNA gene sequencing is able to distinguish B. pseudomallei from most other Burkholderia species, the difference between the 16S rRNA gene sequences of B. pseudomallei and B. thailandensis is only around 1%. Therefore, 16S rRNA sequencing cannot confidently distinguish between the two species, highlighting the need for alternative gene targets.
In our previous study, we have amplified and sequenced the groEL genes of seven strains of B. thailandensis and six strains of B. pseudomallei. We observed that the groEL gene nucleotide sequences of the B. pseudomallei strains showed <97.6% nucleotide identity with those of B. thailandensis.38 This means that the groEL gene sequences offer a higher discriminatory power between B. pseudomallei and B. thailandensis than 16S rRNA gene sequences. In the circumstances that suspected B. pseudomallei colonies are isolated in clinical specimens and arabinose assimilation is not available for differentiation from B. thailandensis, groEL would be a better gene target for identification of B. pseudomallei. We have also described the use of groEL gene sequencing for diagnosing a case of seronegative melioidosis in an 84-year-old patient with acute bacteremic pneumonia.39 An aerobic Gram-negative bacterium was isolated from the blood and sputum of the patient, with phenotypic characteristics and antibiotic susceptibility patterns suggestive of B. pseudomallei. However, the Vitek 1 system (GNI+) could not identify the isolate and sera for antibody against B. pseudomallei were negative.39 The diagnosis was confirmed by groEL gene sequencing which showed unambiguously that the isolate was B. pseudomallei.39
Identification by PCR using B. pseudomallei specific primers
Despite the recognition of the genus Pseudomonas as a heterogeneous group since the 1970s, the rectifying re-classification of the Pseudomonas RNA homology group II under the new genus Burkholderia and the transfer of Pseudomonas pseudomallei to Burkholderia pseudomallei only occurred in 1992.40 As a result, the early success in specific PCR-based identification of B. pseudomallei was, at least, partly fortuitous. By sequencing and comparing the sequences of the 23S rRNA gene of B. pseudomallei (then classified under the genus Pseudomonas) and “closely-related” species including Pseudomonas aeruginosa and Pseudomonas putida, a B. pseudomallei-specific 18-bp rDNA probe was the first to be identified and applied in direct PCR identification.41 Of note, this rDNA-targeting nucleotide probe also detected the glanders pathogen Burkholderia mallei. This observation heralded the discovery of B. mallei as a subclade of B. pseudomallei by multilocus sequence typing almost a decade later.17
The challenge remains to identify specific primers that can differentiate B. pseudomallei from closely related species like B. thailandensis. Early assays continued to be designed based on certain “first principles”: the first multiplex PCR assay capable of differentiating B. pseudomallei and B. thailandensis was based on small stable differences in their 16S rRNA genes.42 A subsequent PCR assay was devised to target amplicon size differences due to a 15-bp deletion in the variable domain of the B. thailandensis flagellin gene.43 While such assays do not target the phenotypic differences or virulence determinants of the organisms, they are nonetheless effective provided that they are extensively validated.44
When the complete genome sequences of B. pseudomallei45 and other closely related species46,47 were published in the 2000s, PCR assays based on observed interspecific differences in individual genetic loci continued to be developed,48,49 combined,50 or variably adopted into quantitative PCR assays,51–57 which have been exhaustively reviewed by Lowe et al.58 These included assays based on Type III secretion system genes and single nucleotide polymorphisms in conserved regions such as the BurkDiff assay.48,57,59 On the genomics front, a comparative proteomics study successfully identified two specific protein markers useful for discrimination between B. pseudomallei and B. thailandensis,60 leading to a large-scale in silico proteomic analysis of more than 40 Burkholderia genomes resulting in the systematic identification of 12 promising targets.61 Apart from use in organism identification, targets identified by comparative genomic and proteomic approaches are likely to be important determinants of virulence, adaptability, and evolutionary biology of B. pseudomallei.62
In our laboratory, PCR identification of B. pseudomallei isolates is performed by a multiplex PCR assay derived from the pan-genomic study mentioned earlier.61 A number of design features were integrated to make the assay both versatile and robust. First, the endpoint PCR was designed with flexible adoption to quantitative PCR in mind: the primers were situated on highly conserved regions of the putative Tat-domain protein gene and the B. pseudomallei-specific amplicon was just 110 bp in size, suitable for detection by both endpoint and real-time PCR. Second, the multiplex assay was tailored to the clinical microbiology laboratory as it offered discrimination of B. pseudomallei from avirulent B. thailandensis and the common opportunistic pathogen BCC species. Furthermore, specificity validation of the assay was performed computationally using the NCBI genome database (http://www.ncbi.nlm.nih.gov/genome/), empirically using genetically similar organisms including then unsequenced species such as Burkholderia gladioli and additionally with spiked sputum and environmental soil samples.61 Recent improvements of our endpoint PCR protocol have decreased the cycling time to less than 30 min (unpublished data), making rapid specific identification of B. pseudomallei possible without quantitative or real-time PCR equipment.63
MALDI-TOF MS
MALDI-TOF MS has recently emerged as a revolutionary technique for pathogen identification, yielding rapid, accurate, and highly reproducible results at a lower price than any other methods routinely used in clinical laboratories. The methodology is easy, with only minimal quantity of bacteria required and results available within minutes. As a result, this technique is increasingly being integrated into many clinical laboratories. It has been shown to be useful for the identification of various non-fermenting gram-negative bacilli including some Burkholderia species. In particular, several studies have addressed the potential of MALDI-TOF MS for B. pseudomallei, all using the Bruker MALDI Biotyper system.64–67
In our previous study which included 52 B. pseudomallei strains and three B. thailandensis strains, MALDI-TOF MS was found to be potentially useful for the identification of B. pseudomallei and B. thailandensis using the direct transfer method and MALDI Biotyper 3.0 equipped with Reference Library v3.1.2.0 (Bruker Daltonik).64 The Biotyper library contained 41 Burkholderia main spectra from 26 species including one from B. thailandensis but not B. pseudomallei. The B. pseudomallei test strains were only identified correctly to the species level (score of top match ≥2.0 and score of second match lower by ≥10%) when 21 B. pseudomallei strains were added to the database. The three B. thailandensis strains were misidentified as B. pseudomallei. Nevertheless, addition of one of the B. thailandensis strains in the Bruker database enabled the correct identification of the other two B. thailandensis isolates. Therefore, the misidentification of B. thailandensis is likely due to the inadequate number of spectra to cover intraspecies variability. In another study using the Biotyper library expanded with two B. pseudomallei strains, two new suspected B. pseudomallei isolates from patients with septicemia were successfully identified and the procedure reduced the time to definitive diagnosis by more than 24 h.67 Similarly, a study which included 10 B. pseudomallei strains also showed that B. pseudomallei and B. thailandensis could be identified if a dedicated subset of the reference spectra library, including three B. pseudomallei strains, was used.66 A recent report has also stressed the importance of using Bruker’s Security-Relevant (SR) library and the inclusion of these potentially hazardous agents in clinical laboratories.65 Although the Biotyper reference library does not contain select agents such as B. pseudomallei, Bruker’s SR library does, which can be obtained by users and searched simultaneously with the Biotyper reference library. The authors also attempted to test the SR library for identifying two B. pseudomallei isolates. The two isolates were identified as B. mallei or B. pseudomallei (both score >2), suggesting that the SR library cannot distinguish B. pseudomallei from B. mallei.
Laboratory exposures to B. pseudomallei have always been a concern. A recent case of accidental laboratory exposure to B. pseudomallei was reported in the United States, following genus-level identification by MALDI-TOF MS.68 The patient had recently traveled to Thailand before presenting with urinary tract infection. The possibility of B. pseudomallei from the urine culture was only suspected after MALDI-TOF MS identification as B. thailandensis with score 1.864. The risk of laboratory exposure to B. pseudomallei can be minimized by a high index of suspicion and inactivation with ethanol and/or protein extraction for any suspicious bacterial isolates in Biosafety Level II cabinets before processing for MALDI-TOF MS.
In conclusion, MALDI-TOF MS is potentially useful for rapid identification of B. pseudomallei in clinical laboratories. However, existing databases do not contain enough spectra for this bacterium, which is uncommonly encountered outside endemic areas. Optimization of the databases by adding more reference spectra for B. pseudomallei and related species is critical to enable accurate identification, especially in countries where melioidosis is prevalent, and should be adopted in standard MALDI-TOF MS libraries used in clinical laboratories.
Direct detection by specific primer PCR amplification
The sensitive and specific detection of B. pseudomallei from clinical and environmental specimens by specific primer PCR is dependent upon both assay- and sample-related factors. Having been involved in the design, development, and validation of a multiplex endpoint PCR assay for the specific detection of B. pseudomallei from cultured and uncultured clinical specimens and the subsequent adoption of the modified assay in the environmental survey of a long-term, large-scale epidemiological study of melioidosis, we illustrate the considerations involved in the design of a practical assay.
Direct detection of B. pseudomallei from clinical samples
Since the early days of B. pseudomallei detection using specific primer PCR, blood and sputum have been the clinical specimens used in assay evaluation.41,69 Arguably, contamination by the oropharyngeal flora or co-isolation of other lower respiratory tract colonizers in patients with cystic fibrosis or bronchiectasis render sputum samples a formidable challenge to the empirical specificity of the assay. Yet, studies have shown that sputum represents a good sample for PCR detection, as it often contains high bacterial load of B. pseudomallei.69 An assay for the direct detection of B. pseudomallei from an uncultured sputum sample must not erroneously identify any of the normal flora and, more importantly, the commonly misidentified colonizer species B. cepacia complex as B. pseudomallei, or vice versa, from clinical management35,36,70,71 and laboratory safety perspectives.72
While some assays were designed to differentiate B. pseudomallei and other Burkholderia species,73,74 other PCR assays from the pregenomic era require caution with interpretation as they were known to have suboptimal sensitivity, affected by “anomalous sequence variation” or may even misidentify B. cepacia complex as B. pseudomallei.75 Therefore, in the development of a direct B. pseudomallei PCR assay for sputum specimens, both in silico and experimental validation remain essential. Theoretically, there is always the risk of false-positives due to non-specific PCR amplification of host DNA; nonetheless, to our knowledge, this has not been reported in the literature and any concerns regarding this possibility can be addressed using the NCBI Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/).
Direct detection from environmental samples
Natural soil harbors a diverse bacterial and fungal community, making non-selective culture and subsequent isolation of individual species impractical. Another factor hindering the successful development of a direct PCR assay for B. pseudomallei is the presence of potent DNA polymerase inhibitors in soil. There are at least three ways in which this may be overcome: selective enrichment culture to increase the B. pseudomallei and, accordingly, template DNA concentration, DNA purification with effective inhibitor removal and/or adopting an inhibitor-resistant PCR polymerase.
Selective enrichment culture, with or without subsequent DNA purification, increases the total number of DNA templates for PCR amplification to occur. The standard medium for enrichment is the Ashdown’s medium as described earlier.76 Nonetheless, this standard medium also supports the growth of other soil inhabitants including the B. pseudomallei-like organism B. thailandensis.77–79 As a result, a direct soil PCR assay must be able to distinguish among B. pseudomallei, B. thailandensis, and the BCC species.61 The ability to discriminate B. pseudomallei from B. mallei in such an assay is non-essential because B. mallei does not persist in the environment80 and is inhibited by the Ashdown’s medium.79
Humic acid and other high molecular weight compounds in soil are potent PCR inhibitors that are known to co-purify with DNA.81 Despite extensive attempts, DNA purification from soil sample is either difficult to scale up82,83 or highly dependent upon specific soil types.83,84 From our experience, selective culture enrichment, DNA extract purification, and template dilution can be combined and optimized for a particular soil type to achieve sensitivities superior to traditional culture and isolation.85 The use of mutant DNA polymerases to overcome inhibitors inherent to crude soil samples remains an area of active research86,87 and will likely enhance the efficiency of direct detection of B. pseudomallei from soil and other complex environmental specimens in the near future.
Serological diagnosis
The isolation of B. pseudomallei from clinical specimens is still considered to be the “gold standard” for melioidosis diagnosis. However, culture has a low diagnostic sensitivity in patients with melioidosis.88 Even among culture positive patients, isolation of the agent takes time and expertise, resulting in delayed institution of correct treatment. Therefore, serological tests are often performed as a preliminary test in endemic areas to expedite the diagnosis. There are many antibody detection formats currently in use.89–91 These tests are generally unstandardized “in house” assays. The performance characteristics of these serological tests are ambiguous as most studies involve small groups of patients and investigators compare their assays against an imperfect gold standard (bacterial culture). Even so, serological tests generally have lower sensitivity than culture. Interpretation of a positive qualitative antibody test is also difficult in endemic areas with high background seroprevalence rates, which affects test specificity for diagnosis of melioidosis disease states. The situation is further complicated by an incomplete understanding of the time frame of the melioidosis antibody response upon exposure or re-exposure. The relative importance of IgM and IgG detection in melioidosis diagnosis is also unclear. Therefore, serological tests are, at best, adjuncts to culture-based diagnosis and cannot be recommended for routine diagnosis in endemic areas at this stage.92 Novel ELISA assays, immunochromatographic tests (ICTs), and antigen detection tests should make use of a “pooled gold standard” of culture, molecular, and multiple serological tests for better delineation of assay performance. The important serodiagnostic assay platforms for melioidosis are described below.
Indirect hemagglutination assay (IHA)
The IHA is the earliest described serological test for melioidosis and is still routinely performed in many melioidosis endemic areas. Sheep erythrocytes are sensitized with crude antigen derived from local clinical B. pseudomallei strains.89,90 The sensitized erythrocytes are then added to serial dilutions of heat-inactivated patient sera. The IHA titer is the highest dilution of patient serum that causes distinct agglutination of erythrocytes. Raised IHA titers appear to mostly reflect an IgM response based on antibody fractionation studies; however, the precise antigenic targets are still unknown and are likely to be highly variable between laboratories using different strains.91,93 The test is frequently performed using single patient sera and cut-off values are assigned based on background seropositivity in the population; interpretation can be difficult in rural endemic settings with a high seroprevalence rate. Test sensitivity varies with disease status: less than 60% of patients with acute culture positive melioidosis are IHA positive at presentation; however, the sensitivity appears to improve in patients with chronic disease.93 False positive IHA titers may occur in patients with systemic P. aeruginosa infections.90 Acutely bacteremic patients appear to be more likely to have negative IHA, limiting the usefulness of this assay for diagnosis of severe melioidosis.93,94 This paradoxical seronegativity may reflect defective humoral immune responses in these patients although negative titers may simply reflect the early presentation. Time to IHA seroconversion is unpredictable and may even fail to occur in 30% of patients.93 Consequences of persistent IHA seropositivity or seroreversion on disease outcomes are unclear.
Enzyme-linked immunosorbent assay (ELISA)
IgM and IgG ELISAs have been described for the serodiagnosis of melioidosis. Such tests are rapid and avoid the observer bias of IHAs. Bacterial lipopolysaccharide (LPS) is a commonly used antigen. Whole cells, exopolysaccharide (EPS), and antibody affinity-purified EPS are also used in some centers.95 There is little evidence that any particular antigen type offers superior diagnostic sensitivity for melioidosis although LPS-based assays may offer improved specificity. Heterogeneity in LPS among clinical strains of B. pseudomallei is well recognized96,97; the effect of this variability on the performance of LPS-based ELISA is unknown. The use of purified recombinant antigen is an attractive option, offering standardization and reproducibility. However, a trial of outer membrane protein (Omp3, Omp85), type VI secretion system protein (TssD-5), and serine protease MprA (smBpF4) based IgG ELISA demonstrated only modest sensitivity when used singly or in combinations (62% compared to culture for TssD-5-based ELISA).98 There is some data to support the use of recombinant flagellin protein derived from B. pseudomallei or non-pathogenic B. thailandensis; however, comparisons against LPS-based ELISA and IHA are still pending.99 We have previously reported the cloning of the B. pseudomallei groEL and maIE gene, which encode immunogenic proteins; a clinical trial of an ELISA using these recombinant proteins is pending.100,101 The performance parameters of the ELISA assay are further affected by the optical density (OD) cutoff used—resetting cutoffs at a lower OD value based on unbiased receiver operating characteristic curves using Bayesian latent-class models improved sensitivity to 80% with no compromise in specificity.102 Further studies comparing the relative importance of IgM and IgG responses in melioidosis diagnosis are required.
ICTs
ICTs in the form of commercial point-of-care test strips and cassettes have been developed for detecting melioidosis IgG and IgM. Limited evaluations103,104 suggest that these tests enjoy comparable diagnostic sensitivity to other serological methods such as IHA, ELISA, and indirect immunofluorescence assay (IFA). However, these tests are not yet widely available.
Other antibody detection methods
Complement fixation test for melioidosis is cumbersome, time consuming, and rarely performed. Indirect IFA is used in some centers to detect serum antibodies against whole cell B. pseudomallei antigen coated on slides.105,106 There are a few evaluations supporting routine IFA IgM and/or IgG for melioidosis diagnosis, but implementation requires a fluorescence microscope, which may be difficult in some endemic areas. The IFA is a part of the battery of diagnostic tests against which emerging diagnostic methods can be compared.
Antigen detection tests
Detection of specific B. pseudomallei antigens in clinical specimens should provide excellent positive predictive value for melioidosis. Direct IFA and antibody sandwich ELISAs, which make use of rabbit or mouse monoclonal antiboides raised against B. pseudomallei crude whole cell extract have been described.107,108 The sensitivity is significantly lower than culture but may be useful for rapid screening of clinical specimens from severely ill patients that are likely to contain a high bacterial load. Recently, a specific lateral flow immunoassay detecting the bacterial capsular polysaccharide using high affinity monoclonal antibodies has been described.109 Further clinical evaluations of this promising assay are proceeding and are likely to be valuable in endemic settings.
Metabolomic profiling for identification of novel biomarkers
Metabolomics is an important tool in microbiology and infectious diseases research, providing a revolutionary method to study both the pathogen and the pathogen-specific host response. It involves the systematic study of the small-molecule metabolite profiles of a cell, tissue, or organism, which are the end products of cellular processes. Using statistical analyses, the metabolic profiles from different cells or systems can be compared, which can be used to differentiate between different biological systems and identify potential novel biomarkers specific to these systems. In particular, liquid chromatography–mass spectrometry has been increasingly utilized in both untargeted profiling and targeted quantitation approaches to determine the changes of proteins, lipids, and metabolites in biochemical pathways. Metabolomics has also been recently applied to characterize infectious diseases or pathogens.110–114 Using this approach, metabolomic data obtained from urine samples have been used to distinguish healthy subjects from patients with infections such as pneumococcal disease and urinary tract infections.115–117 Another study using nuclear magnetic resonance spectroscopy-based metabolomics showed that the metabolic profile of sera from tuberculosis patients can be distinguished from those from healthy controls.118
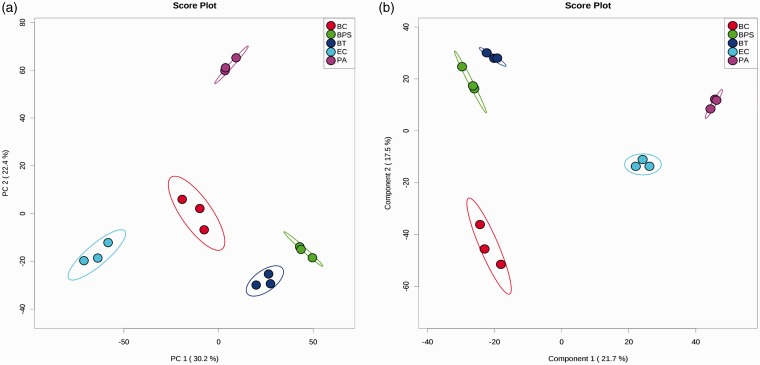
Although no studies have reported the use of metabolomics for B. pseudomallei, the technique is potentially useful to explore specific biomarkers for identification and diagnosis. In our pilot study using culture supernatants from B. pseudomallei and related species for metabolomics profiling by ultra-high performance liquid chromatography-electrospray ionization-quadruple time-of-flight mass spectrometry (UHPLC-ESI-Q-TOF-MS), we showed that B. pseudomallei can be potentially distinguished from B. thailandensis, B. cepacia complex, P. aeruginosa as well as Escherichia coli by principal component analysis (PCA) and partial-least squares discrimination analysis (PLS-DA) (Figure 1). Although the B. pseudomallei strains were most closely related to B. thailandensis strains, the two groups are still clearly separated from each other. Differentiation between B. pseudomallei and B. thailandensis infections is crucial in guiding clinical management of patients, since more than 99% of cases of melioidosis are caused by B. pseudomallei strains, while B. thailandensis is much less virulent. Our preliminary data suggested that the metabolomes of B. pseudomallei cultures are significantly different from those of related Burkholderia species as well as common Gram-negative bacteria. Although a more comprehensive study using more isolates from each bacterial species is required to draw conclusions, it is likely that B. pseudomallei produces specific metabolites that are not found in other bacteria. These specific metabolites may represent potential biomarkers for bacterial identification. Further studies on both bacterial cultures and direct patient samples are required to evaluate the potential of metabolomics in the discovery of novel biomarkers which may help improve clinical diagnosis of melioidosis and expand our knowledge on disease pathogenesis.
Figure 1.
(a) PCA score plot and (b) PLS-DA score plot generated using MetaboAnalyst 3.0 (www.metaboanalyst.ca)119 in positive mode. Filtered culture supernatants were subject to UHPLC-ESI-Q-TOF-MS using Agilent 1290 UHPLC (Agilent Technologies, Santa Clara, CA USA). PLS-DA models were validated using R2 and Q2 based on leave one out cross-validation (LOOCV). Five-component model was selected as optimized model with R2 = 1.00 and Q2 = 0.87. The significance of the model was demonstrated by permutation test with 2000 testing iterations using separation distance and P value < 0.001 was obtained. BC: B. cepacia complex; BPS: B. pseudomallei; BT: B. thailandensis; PA: P. aeruginosa; EC: E. coli. Three strains from each bacterial species were used for culture in RPMI 1640 medium ((#22400-089, Gibco, Carlsbad, CA, USA). (A color version of this figure is available in the online journal.)
Concluding remarks
Despite its medical importance, melioidosis remains a relatively under-studied disease. Although accurate diagnosis of melioidosis is important to guide antibiotic regimen and prevent relapse, laboratory diagnosis may not be straightforward, especially in culture negative cases. Recent advances in molecular diagnostics have dramatically improved the accurate identification of B. pseudomallei isolated from clinical specimens. Since PCR amplification and sequencing of genes such as groEL may be expensive and time consuming, PCR using specific primers represents a more convenient alternative for species identification from positive cultures. As MALDI-TOF MS becomes increasingly available in clinical microbiology laboratories, the technique is expected to further accelerate the routine identification of suspicious isolates. However, as B. pseudomallei is not included in the reference spectra of the Biotyper library, expansion of databases with reference strains is critical in achieving accurate identification by individual laboratories. As existing serological assays do not always offer satisfactory sensitivities and specificities for diagnosis of culture negative melioidosis, exploration of novel biomarkers, which can be detected in body fluids of patients, using metabolomic profiling may improve diagnosis of this emerging disease.
ACKNOWLEDGEMENTS
This work is partly supported by the HKSAR Research Fund for the Control of Infectious Diseases (Commissioned Study HK-09-01-11) of the Food and Health Bureau; University Development Fund and Strategic Research Fund, The University of Hong Kong; and donation from Ms. Eunice Lam on emerging infectious disease research.
Authors’ contributions
All authors participated in the critical review and editing of the manuscript; SKPL, SS, CCH, WNC, and PCYW carried out literature search and writing of the manuscript. WNC and KCL performed experiments and data analysis.
References
- 1.Kanaphun P, Thirawattanasuk N, Suputtamongkol Y, Naigowit P, Dance DA, Smith MD, White NJ. Serology and carriage of Pseudomonas pseudomallei: a prospective study in 1000 hospitalized children in northeast Thailand. J Infect Dis 1993; 167: 230–3. [DOI] [PubMed] [Google Scholar]
- 2.Rattanavong S, Wuthiekanun V, Langla S, Amornchai P, Sirisouk J, Phetsouvanh R, Moore CE, Peacock SJ, Buisson Y, Newton PN. Randomized soil survey of the distribution of Burkholderia pseudomallei in rice fields in Laos. Appl Environ Microbiol 2011; 77: 532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prakash A, Thavaselvam D, Kumar A, Kumar A, Arora S, Tiwari S, Barua A, Sathyaseelan K. Isolation, identification and characterization of Burkholderia pseudomallei from soil of coastal region of India. Springerplus 2014; 3: 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Issack MI, Bundhun CD, Gokhool H. Melioidosis in Mauritius. Emerg Infect Dis 2005; 11: 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inglis TJJ, Rolim DB, Sousa ADQ. Melioidosis in the Americas. Am J Trop Med Hyg 2006; 75: 947–54. [PubMed] [Google Scholar]
- 6.O’Sullivan BP, Torres B, Conidi G, Smole S, Gauthier C, Stauffer KE, Glass MB, Gee JE, Blaney D, Smith TL. Burkholderia pseudomallei infection in a child with cystic fibrosis: acquisition in the Western Hemisphere. Chest 2011; 140: 239–42. [DOI] [PubMed] [Google Scholar]
- 7.Brilhante RSN, Bandeira TJPG, Cordeiro RA, Grangeiro TB, Lima RAC, Ribeiro JF, Castelo-Branco DSCM, Rodrigues JLN, Coelho ICB, Magalhaes FG, Rocha MFG, Sidrim JJC. Clinical-epidemiological features of 13 cases of melioidosis in Brazil. J Clin Microbiol 2012; 50: 3349–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katangwe T, Purcell J, Bar-Zeev N, Denis B, Montgomery J, Alaerts M, Heyderman RS, Dance DAB, Kennedy N, Feasey N, Moxon CA. Human melioidosis, Malawi, 2011. Emerg Infect Dis 2013; 19: 981–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morosini MI, Quereda C, Gil H, Anda P, Nunez-Murga M, Canton R, Lopez-Velez R. Melioidosis in traveler from Africa to Spain. Emerg Infect Dis 2013; 19: 1656–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Currie BJ, Fisher DA, Howard DM, Burrow JN, Lo D, Selva-Nayagam S, Anstey NM, Huffam SE, Snelling PL, Marks PJ, Stephens DP, Lum GD, Jacups SP, Krause VL. Endemic melioidosis in tropical northern Australia: a 10-year prospective study and review of the literature. Clin Infect Dis 2000; 31: 981–6. [DOI] [PubMed] [Google Scholar]
- 11.Bhengsri S, Baggett HC, Jorakate P, Kaewpan A, Prapasiri P, Naorat S, Thamthitiwat S, Tanwisaid K, Chantra S, Salika P, Dejsirilert S, Peruski LF, Maloney SA. Incidence of bacteremic melioidosis in eastern and northeastern Thailand. Am J Trop Med Hyg 2011; 85: 117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev 2005; 18: 383–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ngauy V, Lemeshev Y, Sadkowski L, Crawford G. Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II. J Clin Microbiol 2005; 43: 970–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng AC, Jacups SP, Gal D, Mayo M, Currie BJ. Extreme weather events and environmental contamination are associated with case-clusters of melioidosis in the Northern Territory of Australia. Int J Epidemiol 2006; 35: 323–9. [DOI] [PubMed] [Google Scholar]
- 15.Currie BJ, Mayo M, Anstey NM, Donohoe P, Haase A, Kemp DJ. A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am J Trop Med Hyg 2001; 65: 177–9. [DOI] [PubMed] [Google Scholar]
- 16.Choy JL, Mayo M, Janmaat A, Currie BJ. Animal melioidosis in Australia. Acta Trop 2000; 74: 153–8. [DOI] [PubMed] [Google Scholar]
- 17.Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Spratt BG. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol 2003; 41: 2068–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng AC. Melioidosis: advances in diagnosis and treatment. Curr Opin Infect Dis 2010; 23: 554–9. [DOI] [PubMed] [Google Scholar]
- 19.Wuthiekanun V, Peacock SJ. Management of melioidosis. Expert Rev Anti-infect Ther 2006; 4: 445–55. [DOI] [PubMed] [Google Scholar]
- 20.Ashdown LR. An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathology 1979; 11: 293–7. [DOI] [PubMed] [Google Scholar]
- 21.Cheng AC, Wuthiekanun V, Limmathurosakul D, Wongsuvan G, Day NPJ, Peacock SJ. Role of selective and nonselective media for isolation of Burkholderia pseudomallei from throat swabs of patients with melioidosis. J Clin Microbiol 2006; 44: 2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peacock SJ, Chieng G, Cheng AC, Dance DAB, Amornchai P, Wongsuvan G, Teerawattanasook N, Chierakul W, Day NPJ, Wuthiekanun V. Comparison of Ashdown's medium, Burkholderia cepacia medium, and Burkholderia pseudomallei selective agar for clinical isolation of Burkholderia pseudomallei. J Clin Microbiol 2005; 43: 5359–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inglis TJJ, Merritt A, Chidlow G, Aravena-Roman M, Harnett G. Comparison of diagnostic laboratory methods for identification of Burkholderia pseudomallei. J Clin Microbiol 2005; 43: 2201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woods DE. Species versus biotype status. J Clin Microbiol 1999; 37: 3786–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wuthiekanun V, Smith MD, Dance DA, Walsh AL, Pitt TL, White NJ. Biochemical characteristics of clinical and environmental isolates of Burkholderia pseudomallei. J Med Microbiol 1996; 45: 408–12. [DOI] [PubMed] [Google Scholar]
- 26.Moore RA, Reckseidler-Zenteno S, Kim H, Nierman W, Yu Y, Tuanyok A, Warawa J, DeShazer D, Woods DE. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect Immun 2004; 72: 4172–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith MD, Angus BJ, Wuthiekanun V, White NJ. Arabinose assimilation defines a nonvirulent biotype of Burkholderia pseudomallei. Infect Immun 1997; 65: 4319–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brett PJ, DeShazer D, Woods DE. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol 1998; 48: 317–20. [DOI] [PubMed] [Google Scholar]
- 29.Amornchai P, Chierakul W, Wuthiekanun V, Mahakhunkijcharoen Y, Phetsouvanh R, Currie BJ, Newton PN, van Vinh Chau N, Wongratanacheewin S, Day NP, Peacock SJ. Accuracy of Burkholderia pseudomallei identification using the API 20NE system and a latex agglutination test. J Clin Microbiol 2007; 45: 3774–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dance DA, Wuthiekanun V, Naigowit P, White NJ. Identification of Pseudomonas pseudomallei in clinical practice: use of simple screening tests and API 20NE. J Clin Pathol 1989; 42: 645–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deepak RN, Crawley B, Phang E. Burkholderia pseudomallei identification: a comparison between the API 20NE and VITEK2GN systems. Trans R Soc Trop Med Hyg 2008; 102: S42–4. [DOI] [PubMed] [Google Scholar]
- 32.Glass MB, Popovic T. Preliminary evaluation of the API 20NE and RapID NF plus systems for rapid identification of Burkholderia pseudomallei and B. mallei. J Clin Microbiol 2005; 43: 479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe P, Engler C, Norton R. Comparison of automated and nonautomated systems for identification of Burkholderia pseudomallei. J Clin Microbiol 2002; 40: 4625–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowe P, Haswell H, Lewis K. Use of various common isolation media to evaluate the new VITEK 2 colorimetric GN Card for identification of Burkholderia pseudomallei. J Clin Microbiol 2006; 44: 854–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inglis TJ, Chiang D, Lee GS, Chor-Kiang L. Potential misidentification of Burkholderia pseudomallei by API 20NE. Pathology 1998; 30: 62–4. [DOI] [PubMed] [Google Scholar]
- 36.Zong Z, Wang X, Deng Y, Zhou T. Misidentification of Burkholderia pseudomallei as Burkholderia cepacia by the VITEK 2 system. J Med Microbiol 2012; 61: 1483–4. [DOI] [PubMed] [Google Scholar]
- 37.Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect 2008; 14: 908–34. [DOI] [PubMed] [Google Scholar]
- 38.Woo PC, Woo GK, Lau SK, Wong SS, Yuen Ky. Single gene target bacterial identification. groEL gene sequencing for discriminating clinical isolates of Burkholderia pseudomallei and Burkholderia thailandensis. Diagn Microbiol Infect Dis 2002; 44: 143–9. [DOI] [PubMed] [Google Scholar]
- 39.Woo PCY, Lau SKP, Woo GKS, Fung AMY, Ngan AHY, Hui W-T, Yuen K-Y. Seronegative bacteremic melioidosis caused by Burkholderia pseudomallei with ambiguous biochemical profile: clinical importance of accurate identification by 16S rRNA gene and groEL gene sequencing. J Clin Microbiol 2003; 41: 3973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol 1992; 36: 1251–75. [DOI] [PubMed] [Google Scholar]
- 41.Lew AE, Desmarchelier PM. Detection of Pseudomonas pseudomallei by PCR and hybridization. J Clin Microbiol 1994; 32: 1326–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dharakul T, Tassaneetrithep B, Trakulsomboon S, Songsivilai S. Phylogenetic analysis of Ara+ and Ara- Burkholderia pseudomallei isolates and development of a multiplex PCR procedure for rapid discrimination between the two biotypes. J Clin Microbiol 1999; 37: 1906–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wajanarogana S, Sonthayanon P, Wuthiekanun V, Panyim S, Simpson AJ, Tungpradabkul S. Stable marker on flagellin gene sequences related to arabinose non-assimilating pathogenic Burkholderia pseudomallei. Microbiol Immunol 1999; 43: 995–1001. [DOI] [PubMed] [Google Scholar]
- 44.Kunakorn M, Raksakait K, Sethaudom C, Sermswan RW, Dharakul T. Comparison of three PCR primer sets for diagnosis of septicemic melioidosis. Acta Trop 2000; 74: 247–51. [DOI] [PubMed] [Google Scholar]
- 45.Holden MTG, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PCF, Parkhill J. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA 2004; 101: 14240–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, Feldblyum T, Ulrich RL, Ronning CM, Brinkac LM, Daugherty SC, Davidsen TD, Deboy RT, Dimitrov G, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Khouri H, Kolonay JF, Madupu R, Mohammoud Y, Nelson WC, Radune D, Romero CM, Sarria S, Selengut J, Shamblin C, Sullivan SA, White O, Yu Y, Zafar N, Zhou L, Fraser CM. Structural flexibility in the Burkholderia mallei genome. Proc Natl Acad Sci USA 2004; 101: 14246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EP, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 2009; 191: 261–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thibault FM, Valade E, Vidal DR. Identification and discrimination of Burkholderia pseudomallei, B. mallei, and B. thailandensis by real-time PCR targeting type III secretion system genes. J Clin Microbiol 2004; 42: 5871–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee MA, Wang D, Yap EH. Detection and differentiation of Burkholderia pseudomallei, Burkholderia mallei and Burkholderia thailandensis by multiplex PCR. FEMS Immunol Med Microbiol 2005; 43: 413–7. [DOI] [PubMed] [Google Scholar]
- 50.Koh SF, Tay ST, Sermswan R, Wongratanacheewin S, Chua KH, Puthucheary SD. Development of a multiplex PCR assay for rapid identification of Burkholderia pseudomallei, Burkholderia thailandensis, Burkholderia mallei and Burkholderia cepacia complex. J Microbiol Methods 2012; 90: 305–8. [DOI] [PubMed] [Google Scholar]
- 51.Tomaso H, Scholz HC, Al Dahouk S, Pitt TL, Treu TM, Neubauer H. Development of 5' nuclease real-time PCR assays for the rapid identification of the burkholderia mallei//Burkholderia pseudomallei complex. Diagn Mol Pathol 2004; 13: 247–53. [DOI] [PubMed] [Google Scholar]
- 52.Tomaso H, Pitt TL, Landt O, Al Dahouk S, Scholz HC, Reisinger EC, Sprague LD, Rathmann I, Neubauer H. Rapid presumptive identification of Burkholderia pseudomallei with real-time PCR assays using fluorescent hybridization probes. Mol Cell Probes 2005; 19: 9–20. [DOI] [PubMed] [Google Scholar]
- 53.Tomaso H, Scholz HC, Al Dahouk S, Eickhoff M, Treu TM, Wernery R, Wernery U, Neubauer H. Development of a 5'-nuclease real-time PCR assay targeting fliP for the rapid identification of Burkholderia mallei in clinical samples. Clin Chem 2006; 52: 307–10. [DOI] [PubMed] [Google Scholar]
- 54.U’Ren JM, Van Ert MN, Schupp JM, Easterday WR, Simonson TS, Okinaka RT, Pearson T, Keim P. Use of a real-time PCR TaqMan assay for rapid identification and differentiation of Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol 2005; 43: 5771–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Supaprom C, Wang D, Leelayuwat C, Thaewpia W, Susaengrat W, Koh V, Ooi EE, Lertmemongkolchai G, Liu Y. Development of real-time PCR assays and evaluation of their potential use for rapid detection of Burkholderia pseudomallei in clinical blood specimens. J Clin Microbiol 2007; 45: 2894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chantratita N, Wuthiekanun V, Limmathurotsakul D, Thanwisai A, Chantratita W, Day NP, Peacock SJ. Prospective clinical evaluation of the accuracy of 16S rRNA real-time PCR assay for the diagnosis of melioidosis. Am J Trop Med Hyg 2007; 77: 814–7. [PubMed] [Google Scholar]
- 57.Novak RT, Glass MB, Gee JE, Gal D, Mayo MJ, Currie BJ, Wilkins PP. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J Clin Microbiol 2006; 44: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowe W, March JK, Bunnell AJ, O’Neill KL, Robison RA. PCR-based methodologies used to detect and differentiate the Burkholderia pseudomallei complex: B. pseudomallei, B. mallei, and B. thailandensis. Curr Issues Mol Biol 2013; 16: 23–54. [PubMed] [Google Scholar]
- 59.Bowers JR, Engelthaler DM, Ginther JL, Pearson T, Peacock SJ, Tuanyok A, Wagner DM, Currie BJ, Keim PS. BurkDiff: a real-time PCR allelic discrimination assay for Burkholderia pseudomallei and B. mallei. PLoS One 2010; 5: e15413–e15413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wongtrakoongate P, Mongkoldhumrongkul N, Chaijan S, Kamchonwongpaisan S, Tungpradabkul S. Comparative proteomic profiles and the potential markers between Burkholderia pseudomallei and Burkholderia thailandensis. Mol Cell Probes 2007; 21: 81–91. [DOI] [PubMed] [Google Scholar]
- 61.Ho CC, Lau CC, Martelli P, Chan SY, Tse CW, Wu AK, Yuen KY, Lau SK, Woo PC. Novel pan-genomic analysis approach in target selection for multiplex PCR identification and detection of Burkholderia pseudomallei, Burkholderia thailandensis, and Burkholderia cepacia complex species: a proof-of-concept study. J Clin Microbiol 2011; 49: 814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nandi T, Ong C, Singh AP, Boddey J, Atkins T, Sarkar-Tyson M, Essex-Lopresti AE, Chua HH, Pearson T, Kreisberg JF, Nilsson C, Ariyaratne P, Ronning C, Losada L, Ruan Y, Sung WK, Woods D, Titball RW, Beacham I, Peak I, Keim P, Nierman WC, Tan P. A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence. PLoS Pathogens 2010; 6: e1000845–e1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foster A, Laurin N. Development of a fast PCR protocol enabling rapid generation of AmpFlSTR(R) Identifiler(R) profiles for genotyping of human DNA. Invest Genet 2012; 3: 6–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lau SK, Tang BS, Curreem SO, Chan TM, Martelli P, Tse CW, Wu AK, Yuen KY, Woo PC. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of Burkholderia pseudomallei: importance of expanding databases with pathogens endemic to different localities. J Clin Microbiol 2012; 50: 3142–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cunningham SA, Patel R. Importance of using Bruker’s security-relevant library for Biotyper identification of Burkholderia pseudomallei, Brucella species, and Francisella tularensis. J Clin Microbiol 2013; 51: 1639–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karger A, Stock R, Ziller M, Elschner MC, Bettin B, Melzer F, Maier T, Kostrzewa M, Scholz HC, Neubauer H, Tomaso H. Rapid identification of Burkholderia mallei and Burkholderia pseudomallei by intact cell matrix-assisted laser desorption/ionisation mass spectrometric typing. BMC Microbiol 2012; 12: 229–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Inglis TJJ, Healy PE, Fremlin LJ, Golledge CL. Use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis for rapid confirmation of Burkholderia pseudomallei in septicemic melioidosis. Am J Trop Med Hyg 2012; 86: 1039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dingle TC, Butler-Wu SM, Abbott AN. Accidental exposure to Burkholderia pseudomallei in the laboratory in the era of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2014; 52: 3490–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gal D, Mayo M, Spencer E, Cheng AC, Currie BJ. Short report: application of a polymerase chain reaction to detect Burkholderia pseudomallei in clinical specimens from patients with suspected melioidosis. Am J Trop Med Hyg 2005; 73: 1162–4. [PubMed] [Google Scholar]
- 70.Weissert C, Dollenmaier G, Rafeiner P, Riehm J, Schultze D. Burkholderia pseudomallei misidentified by automated system. Emerg Infect Dis 2009; 15: 1799–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Carroll MR, Kidd TJ, Coulter C, Smith HV, Rose BR, Harbour C, Bell SC. Burkholderia pseudomallei: another emerging pathogen in cystic fibrosis. Thorax 2003; 58: 1087–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schlech WF, Turchik JB, Westlake RE, Klein GC, Band JD, Weaver RE. Laboratory-acquired infection with Pseudomonas pseudomallei (melioidosis). N Engl J Med 1981; 305: 1133–5. [DOI] [PubMed] [Google Scholar]
- 73.Bauernfeind A, Schneider I, Jungwirth R, Roller C. Discrimination of Burkholderia gladioli from other Burkholderia species detectable in cystic fibrosis patients by PCR. J Clin Microbiol 1998; 36: 2748–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suppiah J, Thimma JS, Cheah SH, Vadivelu J. Development and evaluation of polymerase chain reaction assay to detect Burkholderia genus and to differentiate the species in clinical specimens. FEMS Microbiol Lett 2010; 306: 9–14. [DOI] [PubMed] [Google Scholar]
- 75.Merritt A, Inglis TJ, Chidlow G, Harnett G. PCR-based identification of Burkholderia pseudomallei. Rev Inst Med Trop Sao Paulo 2006; 48: 239–44. [DOI] [PubMed] [Google Scholar]
- 76.Ashdown LR, Clarke SG. Evaluation of culture techniques for isolation of Pseudomonas pseudomallei from soil. Appl Environ Microbiol 1992; 58: 4011–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ashdown LR. Rapid differentiation of Pseudomonas pseudomallei from Pseudomonas cepacia. Lett Appl Microbiol 1992; 14: 203–5. [DOI] [PubMed] [Google Scholar]
- 78.Francis A, Aiyar S, Yean CY, Naing L, Ravichandran M. An improved selective and differential medium for the isolation of Burkholderia pseudomallei from clinical specimens. Diagn Microbiol Infect Dis 2006; 55: 95–9. [DOI] [PubMed] [Google Scholar]
- 79.Glass MB, Beesley CA, Wilkins PP, Hoffmaster AR. Comparison of four selective media for the isolation of Burkholderia mallei and Burkholderia pseudomallei. Am J Trop Med Hyg 2009; 80: 1023–8. [PubMed] [Google Scholar]
- 80.Inglis TJ, Sagripanti JL. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl Environ Microbiol 2006; 72: 6865–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilson IG. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 1997; 63: 3741–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moreira D. Efficient removal of PCR inhibitors using agarose-embedded DNA preparations. Nucleic Acids Res 1998; 26: 3309–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miller DN, Bryant JE, Madsen EL, Ghiorse WC. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol 1999; 65: 4715–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dineen SM, Aranda Rt, Anders DL, Robertson JM. An evaluation of commercial DNA extraction kits for the isolation of bacterial spore DNA from soil. J Appl Microbiol 2010; 109: 1886–96. [DOI] [PubMed] [Google Scholar]
- 85.Lau SKP, Chan SY, Curreem SOT, Hui SW, Lau CCY, Lee P, Ho CC, Martell P, Woo PCY. Burkholderia pseudomallei in soil samples from an oceanarium in Hong Kong detected using a sensitive PCR assay. Emerg Microbes Infect 2014; 3: e69–e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baar C, d’Abbadie M, Vaisman A, Arana ME, Hofreiter M, Woodgate R, Kunkel TA, Holliger P. Molecular breeding of polymerases for resistance to environmental inhibitors. Nucleic Acids Res 2011; 39: e51–e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kermekchiev MB, Kirilova LI, Vail EE, Barnes WM. Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples. Nucleic Acids Res 2009; 37: e40–e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Limmathurotsakul D, Jamsen K, Arayawichanont A, Simpson JA, White LJ, Lee SJ, Wuthiekanun V, Chantratita N, Cheng A, Day NPJ, Verzilli C, Peacock SJ. Defining the true sensitivity of culture for the diagnosis of melioidosis using Bayesian latent class models. PLoS One 2010;5:e12485. [DOI] [PMC free article] [PubMed]
- 89.Ileri SZ. The indirect haemagglutination test in the diagnosis of melioidosis in goats. Br Vet J 1965; 121: 164–70. [DOI] [PubMed] [Google Scholar]
- 90.Alexander AD, Huxsoll DL, Warner AR, Jr., Shepler V, Dorsey A. Serological diagnosis of human melioidosis with indirect hemagglutination and complement fixation tests. Appl Microbiol 1970; 20: 825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ashdown LR. Indirect haemagglutination test for melioidosis. Med J Aust 1987; 147: 364–5. [DOI] [PubMed] [Google Scholar]
- 92.Kelly H, Bennett N, Murray S, O’Grady KA. The blue book: Guidelines for the control of infectious diseases, Melbourne: Communicable Diseases Section, Victorian Government Department of Human Services, 2005. [Google Scholar]
- 93.Cheng AC, O’Brien M, Freeman K, Lum G, Currie BJ. Indirect hemagglutination assay in patients with melioidosis in northern Australia. Am J Trop Med Hyg 2006; 74: 330–4. [PubMed] [Google Scholar]
- 94.Harris PN, Ketheesan N, Owens L, Norton RE. Clinical features that affect indirect-hemagglutination-assay responses to Burkholderia pseudomallei. Clin Vaccine Immunol 2009; 16: 924–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chantratita N, Wuthiekanun V, Thanwisai A, Limmathurotsakul D, Cheng AC, Chierakul W, Day NP, Peacock SJ. Accuracy of enzyme-linked immunosorbent assay using crude and purified antigens for serodiagnosis of melioidosis. Clin Vaccine Immunol 2007; 14: 110–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Anuntagool N, Aramsri P, Panichakul, Wuthiekanun VR, Kinoshita R, White NJ, Sirisinha S. Antigenic heterogeneity of lipopolysaccharide among Burkholderia pseudomallei clinical isolates. Southeast Asian J Trop Med Public Health 2000; 31: 146–52. [PubMed] [Google Scholar]
- 97.Anuntagool N, Wuthiekanun V, White NJ, Currie BJ, Sermswan RW, Wongratanacheewin S, Taweechaisupapong S, Chaiyaroj SC, Sirisinha S. Lipopolysaccharide heterogeneity among Burkholderia pseudomallei from different geographic and clinical origins. Am J Trop Med Hyg 2006; 74: 348–52. [PubMed] [Google Scholar]
- 98.Hara Y, Chin CY, Mohamed R, Puthucheary SD, Nathan S. Multiple-antigen ELISA for melioidosis—a novel approach to the improved serodiagnosis of melioidosis. BMC Infect Dis 2013; 13: 165–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wajanarogana S, Nimnuch P, Thongmee A, Kritsiriwuthinan K. Potential of recombinant flagellin fragment from Burkholderia thailandensis as an antigen for melioidosis antibody detection by indirect ELISA. Mol Cell Probes 2013; 27: 98–102. [DOI] [PubMed] [Google Scholar]
- 100.Woo PC, Leung PK, Tsoi HW, Yuen KY. Cloning and characterisation of malE in Burkholderia pseudomallei. J Med Microbiol 2001; 50: 330–8. [DOI] [PubMed] [Google Scholar]
- 101.Woo PC, Leung PK, Wong SS, Ho PL, Yuen KY. groEL encodes a highly antigenic protein in Burkholderia pseudomallei. Clin Diagn Lab Immunol 2001; 8: 832–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Limmathurotsakul D, Chantratita N, Teerawattanasook N, Piriyagitpaiboon K, Thanwisai A, Wuthiekanun V, Day NP, Cooper B, Peacock SJ. Enzyme-linked immunosorbent assay for the diagnosis of melioidosis: better than we thought. Clin Infect Dis 2011; 52: 1024–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cuzzubbo AJ, Chenthamarakshan V, Vadivelu J, Puthucheary SD, Rowland D, Devine PL. Evaluation of a new commercially available immunoglobulin M and immunoglobulin G immunochromatographic test for diagnosis of melioidosis infection. J Clin Microbiol 2000; 38: 1670–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O’Brien M, Freeman K, Lum G, Cheng AC, Jacups SP, Currie BJ. Further evaluation of a rapid diagnostic test for melioidosis in an area of endemicity. J Clin Microbiol 2004; 42: 2239–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mathai E, Jesudason MV, Anbarasu A. Indirect immunofluorescent antibody test for the rapid diagnosis of melioidosis. Indian J Med Res 2003; 118: 68–70. [PubMed] [Google Scholar]
- 106.Khupulsup K, Petchclai B. Application of indirect hemagglutination test and indirect fluorescent antibody test for IgM antibody for diagnosis of melioidosis in Thailand. Am J Trop Med Hyg 1986; 35: 366–9. [DOI] [PubMed] [Google Scholar]
- 107.Anuntagool A, Intachote P, Naigowit P, Sirisinha S. Rapid antigen detection assay for identification of Burkholderia (Pseudomonas) pseudomallei infection. J Clin Microbiol 1996; 34: 975–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Walsh AL, Smith MD, Wuthiekanun V, Suputtamongkol Y, Desakorn V, Chaowagul W, White NJ. Immunofluorescence microscopy for the rapid diagnosis of melioidosis. J Clin Pathol 1994; 47: 377–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Houghton RL, Reed DE, Hubbard MA, Dillon MJ, Chen H, Currie BJ, Mayo M, Sarovich DS, Theobald V, Limmathurotsakul D, Wongsuvan G, Chantratita N, Peacock SJ, Hoffmaster AR, Duval B, Brett PJ, Burtnick MN, Aucoin DP. Development of a prototype lateral flow immunoassay (LFI) for the rapid diagnosis of melioidosis. PLoS Negl Trop Dis 2014;8:e2727. [DOI] [PMC free article] [PubMed]
- 110.Olivier LD., I An overview of tuberculosis treatments and diagnostics. What role could metabolomics play? J Cell Tissue Res 2011; 11: 2655–71. [Google Scholar]
- 111.Meissner-Roloff RJKG, Warren RM, Loots DT. A metabolomics investigation of a hyper- and hypo-virulent phenotype of Beijing lineage M. tuberculosis. Metabolomics 2012; 8: 1194–203. [Google Scholar]
- 112.Tam EW, Chen JH, Lau EC, Ngan AH, Fung KS, Lee KC, Lam CW, Yuen KY, Lau SK, Woo PC. Misidentification of Aspergillus nomius and Aspergillus tamarii as Aspergillus flavus: characterization by internal transcribed spacer, beta-Tubulin, and calmodulin gene sequencing, metabolic fingerprinting, and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2014; 52: 1153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.To KK, Fung AM, Teng JL, Curreem SO, Lee KC, Yuen KY, Lam CW, Lau SK, Woo PC. Characterization of a Tsukamurella pseudo-outbreak by phenotypic tests, 16S rRNA sequencing, pulsed-field gel electrophoresis, and metabolic footprinting. J Clin Microbiol 2013; 51: 334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Woo PC, Lam CW, Tam EW, Leung CK, Wong SS, Lau SK, Yuen KY. First discovery of two polyketide synthase genes for mitorubrinic acid and mitorubrinol yellow pigment biosynthesis and implications in virulence of Penicillium marneffei. PLoS Negl Trop Dis 2012; 6: e1871–e1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Van QN, Klose JR, Lucas DA, Prieto DA, Luke B, Collins J, Burt SK, Chmurny GN, Issaq HJ, Conrads TP, Veenstra TD, Keay SK. The use of urine proteomic and metabonomic patterns for the diagnosis of interstitial cystitis and bacterial cystitis. Dis Markers 2003; 19: 169–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lv H, Hung CS, Chaturvedi KS, Hooton TM, Henderson JP. Development of an integrated metabolomic profiling approach for infectious diseases research. Analyst 2011; 136: 4752–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mahadevan S, Shah SL, Marrie TJ, Slupsky CM. Analysis of metabolomic data using support vector machines. Anal Chem 2008; 80: 7562–70. [DOI] [PubMed] [Google Scholar]
- 118.Zhou A, Ni J, Xu Z, Wang Y, Lu S, Sha W, Karakousis PC, Yao YF. Application of (1)h NMR spectroscopy-based metabolomics to sera of tuberculosis patients. J Proteome Res 2013; 12: 4642–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kuhl C, Tautenhahn R, Bottcher C, Larson TR, Neumann S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal Chem 2012; 84: 283–9. [DOI] [PMC free article] [PubMed] [Google Scholar]