Abstract
One of the current focuses in HIV/AIDS research is to develop a novel therapeutic strategy that can provide a life-long remission of HIV/AIDS without daily drug treatment and ultimately a cure for HIV/AIDS. Hematopoietic stem cell based anti-HIV gene therapy aims to reconstitute patient immune system by transplantation of genetically engineered hematopoietic stem cells with anti-HIV genes. Hematopoietic stem cells can self renew, proliferate and differentiate into mature immune cells. In theory, anti-HIV gene modified hematopoietic stem cells can continuously provide HIV resistant immune cells throughout the life of a patient. Therefore, hematopoietic stem cell based anti-HIV gene therapy has a great potential to provide a life-long remission of HIV/AIDS by a single treatment. Here, we provide a comprehensive review of the recent progress of developing anti-HIV genes, genetic modification of hematopoietic stem progenitor cells, engraftment and reconstitution of anti-HIV gene modified immune cells, HIV inhibition in vitro and in vivo animal models, and human clinical trials.
Keywords: HIV, Hematopoietic stem progenitor cells, anti-HIV genes, RNA interference, genome editing technologies, Zinc Finger nucleases, TALEN, CRISPR/Cas9, humanized mouse models, non-human primate models, clinical trials
Graphical abstract
2 Introduction
The survival and quality of life for HIV (Human Immunodeficiency Virus) infected individuals have significantly improved after the initiation of highly active antiretroviral therapy (HAART) in 1996. The viral load can be suppressed to undetectable levels with the daily drug treatment. However, the viral load rapidly rebounds in a few weeks without medication in majority of patients [1]. Interestingly, 14 HIV patients who had interrupted effective HAART that were initiated very early in the course of their infection showed off-therapy viral load control for several years, suggesting initiating early HAART may open up new therapeutic perspectives for HIV infected patients [2]. However, the majority of patients need to keep HAART for their life. Daily, life-long drug administration creates difficulties with adherence. Additionally, drug associated side effects and the costs of therapy can accumulate greatly over the life of a patient. Chronic HIV infection leads to development of early signs of senile complications with dyslipidemia or cardiovascular diseases in HIV infected patients even under HAART [3,4]. Thus, development of novel therapeutic strategies for a life-long remission of HIV/AIDS (Acquired Immunodeficiency Syndrome) or a HIV cure has become one of the major focuses in current HIV/AIDS research. The first and only case of HIV cure was reported by allogeneic CCR5 deficient bone marrow transplants in 2009. Mr. Timothy Ray Brown, named as the Berlin patient, while being HIV positive, developed Acute Myelogenous Leukemia (AML). Transplantations of naturally HIV resistant CCR5 homozygous Δ32/Δ32 bone marrow stem cells fully reconstituted his immune system with donor derived CCR5 deficient HIV resistant cells. HIV has been undetectable in the Berlin patient for more than eight years [5–7]. The success of “the Berlin patient’s” HIV cure has generated immense interest in promoting HIV cure research. After the first HIV cure case, several efforts have been reported in attempt to achieve a successful second case of HIV cure [8–11]. However, this has not been possible. Some of the limitations include a very low frequency of human leukocyte antigen (HLA) matched natural CCR5 homozygous Δ32/Δ32 allogenic donors, difficult bone marrow transplant procedures and the emergence of naturally resistant HIV strains. A recent clinical case resulted in the rapid emergence of infection with CXCR4 tropic HIV in a patient who had been transplanted with stem cells from a Δ32/Δ32 homozygous CCR5 defective HSPC donor, similar to the Berlin patient, highlighting the fact that viral escape mechanisms may be a major limitation to the CCR5-knockout strategies to control HIV infection [10].
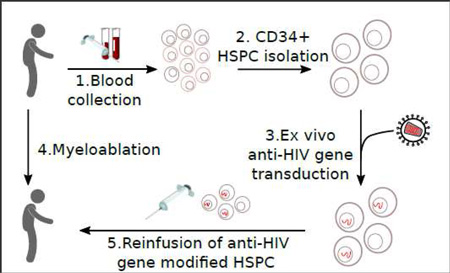
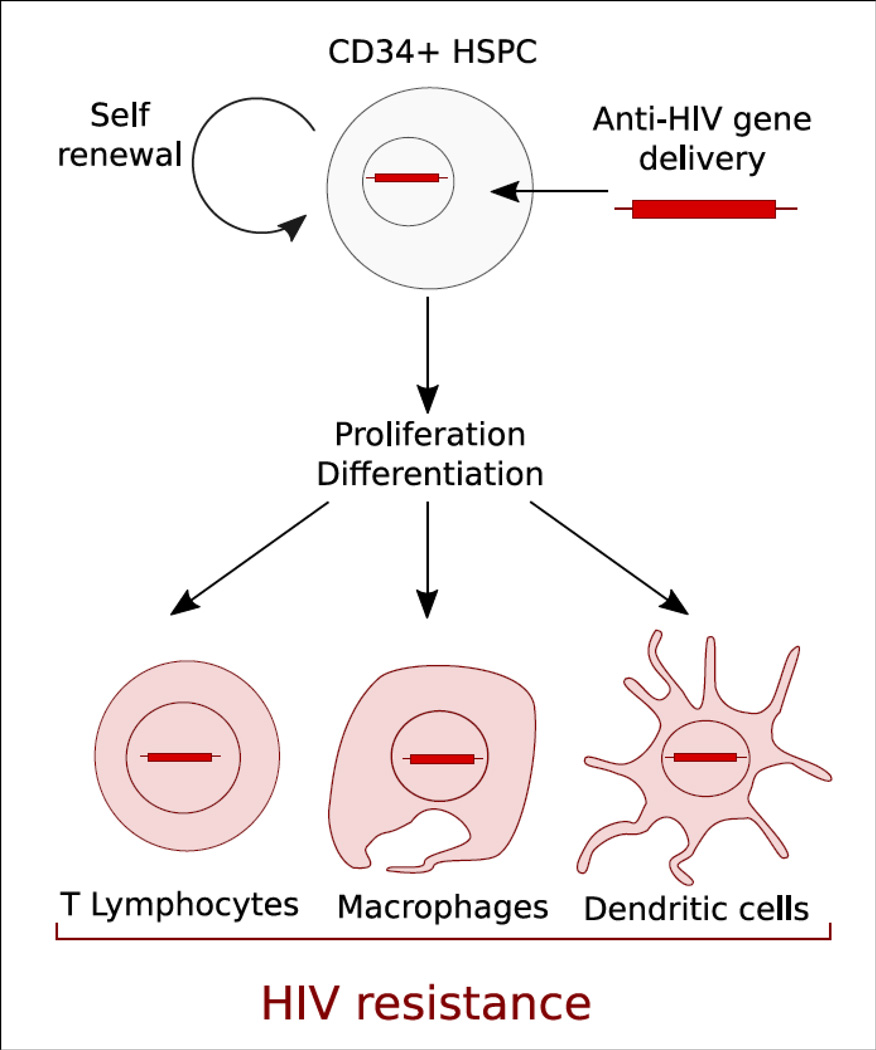
The principal idea of anti-HIV hematopoietic stem/progenitor cell (HSPC) based gene therapy is to genetically engineer patient-derived HSPC and progenies to resist HIV infection (Figure 1). HIV resistant genetically engineered HSPC, in theory, can provide HIV resistant progenies continuously through an entire life of a patient with a single treatment (Figure 2). In this review, we will describe the recent advancements of anti-HIV stem cell based gene therapy strategies, including the anti-HIV gene reagents, anti-HIV gene combinatorial strategies, as well as gene delivery systems for HSPC, in vitro HIV inhibition, anti-HIV gene modified HSPC transplant, hematopoietic reconstitution in vivo in small/large animal models and in human clinical trials (Table 1).
Figure 1.
Genetic modification of CD34+ HSPC to resist HIV infection. Anti-HIV gene modified CD34+ hematopoietic stem/progenitor cells (HSPC) can self renew and proliferate to continuously provide differentiated HIV resistant mature immune cells including T lymphocytes, Macrophages, and Dendritic cells.
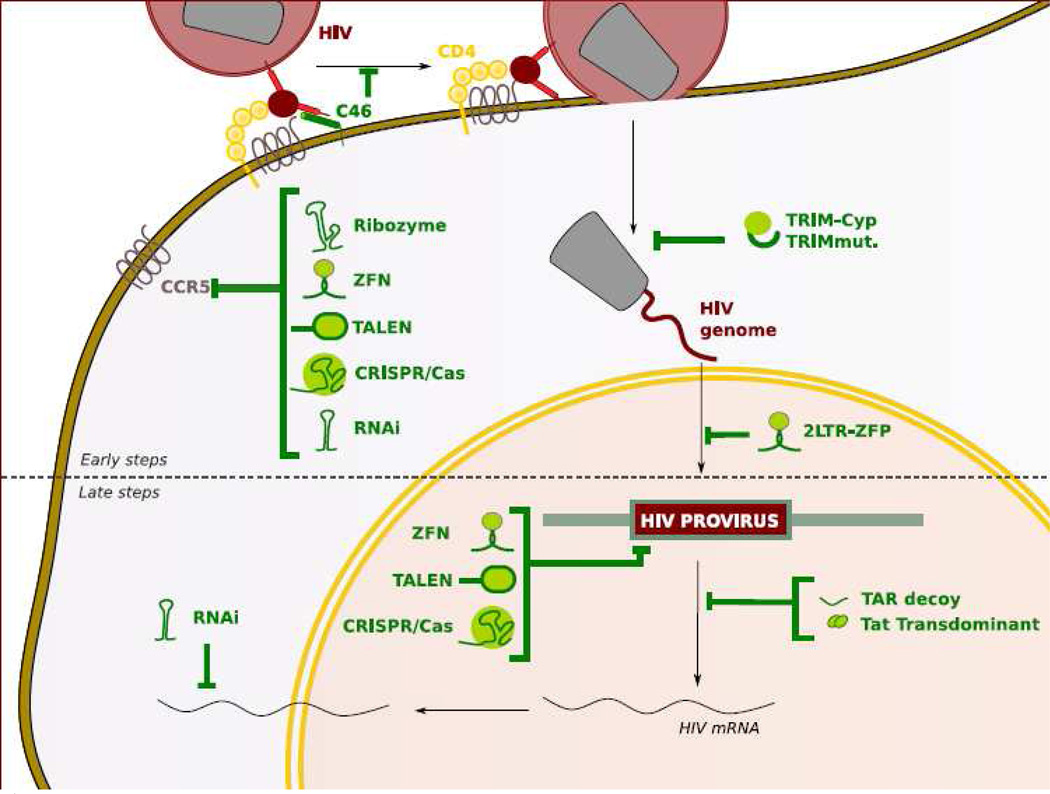
Figure 2.
Anti-HIV genes to inhibit different steps of HIV life cycle. HIV infection can be inhibited by anti-HIV genes in different steps in HIV replication cycle either before (early steps) or after (late steps) HIV integrates into host genome. HIV co-receptor (CCR5 or CXCR4) directed anti HIV genes (ribozyme, ZFN, TALEN, CRISPR/Cas9 and RNAi) and C46 inhibit HIV at the entry steps. TRIMcyp and TRIM5α inhibit HIV after HIV enter into cytoplasm by binding to HIV capsid core structure. 2LTR ZFP inhibit HIV by binding to HIV2LTR DNA. HIV directed ZFN, TALEN, CRISPR/Cas9 introduce indel mutations and excise proviral HIV DNA. RNAi, TAR decoy, tat transdominant can inhibit HIV gene expression at the post transcription step.
Table 1.
A summary of anti HIV HSPC gene therapy research.
| Anti-HIV gene | Experimental Models | Salient results | References |
|---|---|---|---|
| CCR5 ribozyme | in-vitro | CCR5 reduction and HIV Inhibition in cell line | [21] |
| CCR5 ribozyme, anti-HIV shRNA, TAR decoy | clinical trial phase I/II – NCT01961063 | recruiting | City of Hope Medical Center |
| CCR5 ribozyme, anti-HIV shRNA, TAR decoy | clinical trial phase I/II | Transgene expression circulating cells | [87] |
| CCR5 ribozyme, anti-HIV shRNA, TAR decoy | in-vitro | HIV inhibition in cells line and PBMC | [24] |
| CCR5 ribozyme, anti-HIV shRNA, TAR decoy | in-vitro | HIV inhibition in human primay T cells | [22] |
| CCR5 ribozyme, anti-HIV shRNA, TAR decoy | humanized SCID mouse | HIV inhibition in cells from humanized mice | [86] |
| CCR5 shRNA | in-vitro | CCR5 knock down and HIV inhibition in human primary T cells | [26] |
| CCR5 shRNA | non-human primate | Stable CCR5 knock down in vivo, SIV inhibiton ex vivo | [28] |
| CCR5 shRNA | In-vitro | CCR5 and HIV-1 inhibition in macrophages derived from transduced HSPC | [29] |
| CCR5 shRNA | humanized BLT mouse | CCR5 knock down in lymphoid organs in vivo, HIV inhibitoin ex vivo | [30] |
| CCR5 shRNA/HIV LTRshRNA | humanized BLT mouse | CCR5 knock down and HIV resistance in T-cells | [32] |
| CCR5 shRNA | humanized BLT mouse | HIV resistance in memory T Cells | [31] |
| CCR5 miRNA | humanized NSG mouse | HIV resistance | [99] |
| CCR5 shRNA, C46 | in-vitro | no adverse effect on HSPC differentiation | [33] |
| CCR5 shRNA, C46 | humanized BLT mouse | Viral load reduction and HIV inhibition lymphoid tissue | [60] |
| CCR5 shRNA, C46 | clinical trial phase I/II – NCT01734850 | recruiting | Calimmune, Inc. |
| CCR5 ZFN | In-vitro humanized NOG mouse |
CCR5 kockout and HIV inhibition | [35] |
| CCR5 ZFN | humanized NSG mouse | CCR5 kockout and HIV inhibition | [36] |
| CCR5 ZFN | In-vitro | CCR5 kockout and HIV inhibition | [37] |
| CCR5 ZFN | In-vitro humanized NSG mouse |
CCR5 kockout and HIV inhibition | [39] |
| CCR5 ZFN | clinical trial phase I/II - NCT00842634 |
Safe autologous infusion of modified CD4+ cells | [118] |
| CCR5 ZFN | In-vitro humanized NSG mouse |
CCR5 knock out HSPC multilineage differentiation |
[106] |
| CCR5 ZFN | In-vitro humanized NSG mouse |
CCR5 knock out HIV inhibition |
[108] |
| CCR5 TALEN | In-vitro | CCR5 knock out in cell line | [41] |
| CCR5 TALEN | In-vitro | CCR5 knock out in cell line and primary cells | [40] |
| CCR5 TALEN | In-vitro | CCR5 knock out in iPSC | [48] |
| CCR5 CRISPR/Cas9 | In-vitro | CCR5 knock out in iPSC | [48] |
| CCR5 CRISPR/Cas9 | In-vitro humanized NSG mouse |
CCR5 knock out in HSPC Engraftement of modified HSPC |
[45] |
| CCR5 CRISPR/Cas9 | In-vitro | CCR5 knock out | [44] |
| CCR5 CRISPR/Cas9 | In-vitro | CCR5 knock out | [47] |
| CXCR4 shRNA | In-vitro | CXCR4 knock down and HIV Inhibition | [52] |
| CXCR4 ZFN | In-vitro humanized NSG mouse |
CXCR4 ZFN modified CD4+ T celll enrichment in humanized mice | [53] |
| CXCR4 CRISPR/Cas9 | In-vitro | HindIII restriction site knocked in CXCR4 gene in CD4+ primary cells | [54] |
| CXCR4 CRISPR/Cas9 | In-vitro | CXCR4 knockout and HIV-1 inhibition | [55] |
| CXCR4 ZFN; CCR5 ZFN | humanized NSG mouse | HIV inhibition | [89] |
| C46 | In-vitro | HIV inhibition | [58] |
| C46 | In-vitro | HIV, SIV, and SHIV inhibition | [59] |
| C46 | non-human primate | HIV/SHIV inhibition | [61] |
| C46 | clinical trial phase I/II | Transfert of autologous modified T cells | [62] |
| Human TRIMCyp | In-vitro and PBMC NSG mouse | HIV inhibition and viral load reduction | [65] |
| TRIM5α R322 | In-vitro | HIV inhibition | [67] |
| TRIM5α R332G-R335G | non-human primate | Survival advantage of gene modified cells | [68] |
| hu/maTRIM5α; CCR5 shRNA; TAR decoy | humanized BRG mouse | HIV inhibition | [88] |
| 2LTR-ZFProtein | In-vitro | HIV inhibition | [69] |
| Tre recombinase | In-vitro humanized RAG2 mouse |
Excision of the provirus DNA | [70,71] |
| HIV ZFN | In-vitro | Excision of the provirus from infected primary cells | [72] |
| HIV TALEN | In-vitro | Excision of the provirus from latently infected Jurkat cells | [74] |
| HIV CRISPR/Cas9 | In-vitro | Excision of the provirus from latently infected Jurkat cells | [73] |
| HIV CRISPR/Cas9 | Excision of the provirus and emergece of HIV resistance | [75] | |
| tat-vpr Ribozyme | clinical trial phase I/II | Lower HIV viral load | [117] |
| Tat siRNA | In-vitro | HIV inhibition in macrophages | [79] |
| TAR TALEN | In-vitro | Damage in up to 22% of the provirus integrated in the HeLa/LAV | [80] |
| Tat Nullbasic | In-vitro | HIV inhibition | [81] |
| RRE decoy | clinical trial phase I/II | low levels of gene-containing leukocytes 1 year after gene transfer | [119] |
| Gag-5 shRNA; Pol-1 shRNA; Pol-47shRNA; R/T-5 shRNA | In-vitro | 4shRNA combination prevents HIV escape mutations | [83] |
| CRIPER/Cas9 for latent HIV reactivation | In vitro | Latent HIV reactivation | [75–77] |
| Gag-5 shRNA; Pol-1 shRNA; Pol-47shRNA; R/T-5 shRNA | humanized BRG mouse | HIV inhibition | [84] |
| CD4ζCAR | In-vitro humanized BLT mouse |
HIV inhibition | [112] |
| CCR5Δ32/Δ32 bone marrow transplant | Boston patients | Relapse | [10] |
| CCR5Δ32/Δ32 bone marrow transplant | Berlin patient | An HIV cure | [5,6] |
3 Anti-HIV genes provide resistance to HIV infection
3.1 Targeting the viral entry mechanism
3.1.1 CCR5 inhibition
Development of anti-HIV genes against chemokine receptor CCR5 has become a main focus in anti-HIV HSPC gene therapy research [12,13] CCR5 serves as a major co-receptor for HIV. After HIV binds to the CD4, the primary receptor, subsequent binding to CCR5 is essential for a successful HIV infection. Blocking this early phase of HIV infection can be highly effective in protecting the cells from CCR5 tropic HIV infection before HIV integrates into host genome for establishing stable infection. Importantly, inhibition of CCR5 expression does not cause apparent adverse effects in the hematopoietic and immune system in humans [12], except that individuals with homozygous CCR5 Δ32/Δ32 mutation in CCR5 gene are more susceptible to severe cases of West Nile virus encephalitis [14]. In addition, the CCR5 Δ32/Δ32 mutation in heptatitis C virus infected individuals has been reported to be associated with reduced portal inflammation and milder fibrosis [15–17]. Individuals homozygous for CCR5 Δ32/Δ32 mutation are naturally resistant to HIV transmission, and individuals with heterozygous CCR5 Δ32 mutation show a 2–3 years slower disease progression to AIDS than CCR5 wild type individuals [18]. Ribozyme, RNA interference, Zinc Finger Nuclease (ZFN) and CRISPR/Cas9 genome editing technologies have been developed as CCR5 inhibitors for anti-HIV HSPC gene therapy.
3.1.1.1 Ribozymes mediated CCR5 inhibition
Ribozymes are catalytic RNA molecules with enzymatic functions, capable of cleaving their target RNA [19,20]]. In 2000, Cagnon et al. constructed a hammerhead ribozyme derived from the modified Adenovirus VA1 ribozyme, specifically targeting CCR5 messenger RNA (mRNA). Anti CCR5 ribozyme transduced PM1 cell line were resistant to R5-tropic HIV, but not R4-tropic HIV [21]. In 2005, Li et al. combined this ribozyme with two additional anti-HIV genes, a short hairpin RNA (shRNA) targeting HIV tat and a TAR decoy. HIV Tat is an HIV transcriptional activator. It interacts with the transactivation response element (TAR) in HIV RNA transcripts and promotes the initiation of the viral gene expression and the elongation of HIV transcripts. The tat specific shRNA can induce HIV mRNA degradation through RNA interference [22]. The TAR decoy can bind and sequester HIV tat protein from TAR, thereby inhibiting Tat/TAR interaction [23]. The detail of this combinatorial anti-HIV RNA approach is described in the combinatorial approach section of this review (section 3.6). They demonstrated that transduction of CD34+ HSPC rendered them resistant to HIV [22,24]
3.1.1.2 RNA interference mediated CCR5 knock down
RNA interference (RNAi) is a powerful technology that relies on a small double strand RNA to trigger sequence dependent mRNA degradation through the cellular RNA Induced Silencing Complex (RISC) [25]. RNAi has been utilized to knockdown CCR5 expression. In 2003, Qin and An et al. developed a short hairpin RNA (shRNA) directed to CCR5 that can be efficiently delivered by a lentiviral vector. Their results showed that lentiviral vector transduced primary human CD4+ T cells resulted in a 10-fold CCR5 down-regulation. They also showed that the gene modified CD4+ T cells were 3 to 7 fold more resistant than control cells during a challenge with R5-tropic HIV in vitro [26]. However, for efficient CCR5 knock down with the initially developed shRNA required a strong U6 RNA polymerase promoter to express a large amount of shRNA causing cytotoxicity. Non-toxic shRNA expression required an optimization of shRNA expression level using a transcriptionally weaker H1 RNA polymerase III promoter [27]. In 2007, An et al identified a more potent and non-toxic shRNA directed to CCR5 (sh1005) by an extensive screening of an enzymatically generated CCR5 shRNA library. This sh1005 has been extensively investigated for efficient CCR5 down-regulation in human primary T-cells [28]and CD34+ cell derived macrophages in vitro [29]. sh1005 has also proven to be efficient in modifying human HSPC for the engraftment in humanized BLT mouse model, with results showing multilineage differentiation of sh1005 modified cells and CCR5 down regulation in human CD4+ T cells in lymphoid tissues in vivo [30]. In addition, Shimizu et al. demonstrated that CCR5 down-regulated CD4+ T cells, in particular memory CD4+ T cells, were positively selected after R5-tropic HIV challenge in humanized bone marrow/liver/thymus (BLT) mice [31]. The human version of sh1005 was modified to down-regulate rhesus macaque CCR5 by adjusting a nucleotide mismatch [28]. Modifying CD34+ HSPC with this rhesus macaque adapted sh1005 showed stable engraftment and multilineage hematopoietic cell differentiation, as well as stable CCR5 down regulation in transplanted non-human primates. Importantly, rhesus macaque sh1005 modified CD4+ T cells were resistant to simian immunodeficiency virus (SIV) infection ex vivo [28], details of which have been described in the non-human primate section of this review (section 5.3). More recently, human sh1005 was combined with a second shRNA against HIV-LTR [32] or C46 HIV fusion inhibitor [33] to provide dual anti-HIV genes to inhibit both CCR5 tropic and CXCR4 tropic HIVs.
3.1.1.3 Genome editing technologies for CCR5 knock out
Recently, genome editing strategies have been utilized to knockout CCR5 expression. Zinc Finger Nucleases, TALEN, mega-nuclease, PNAs (Peptide Nucleic Acids), and the more recently discovered CRISPR/CAS9 genome editing technology are capable of introducing insertion and deletion mutations (indels) in CCR5 gene in human cells to confer HIV resistance.
3.1.1.3.1 The Zinc Finger Nucleases (ZFN) mediated CCR5 knock out
The Zinc Finger Nuclease (ZFN) consists of a zinc finger DNA binding domain fused to a DNA cleavage domain from the FokI restriction endonuclease. CCR5 directed ZFN induces nucleotide indels in three steps: First, the CCR5 Zinc Finger domain binds its target site, then FokI cleaves the genomic site, and finally the mutagenic Non Homologous End Joining (NHEJ) system repairs the cleavage with indels. In 2005, Mani et al. published a ZFN targeting CCR5 in-vitro for the first time [34] and since, ZFN have been extensively used to mutate CCR5 genes. In 2008, Perez et al. published a study in which they evaluated the efficiency of a CCR5-targeted ZFN in gene modified autologous CD4+ T cells [35]. In 2010, Holt et al. used this CCR5-ZFN to disrupt up to 17% of CCR5 alleles in human CD34+ HSPC from umbilical cord blood and fetal liver [36]. Transplantation of CCR5-ZFN modified CD34+ HSPC successfully reconstituted humanized mice and provided in vivo HIV resistance to the R5-tropic HIV BaL in humanized mice (see section 5.1 for details). Recently, Saydaminova et al. reported that the same CCR5-ZFN was able to modify human peripheral blood mobilized CD34+ HSPC when delivered and expressed through an adenovirus vector system [37]. Similar to Holt et al., they observed up to 12% CCR5 gene disruption in CCR5-ZFN modified human CD34+ HSPC transplanted NOD/Shi-scid/IL-2Rγnull (NOG) mice. However, ZFN expression negatively affected CD34+ survival rate, engraftment rate and expansion of CD34+ cells by three-fold compared to the un-transduced cells in these transplanted NOG mice [37].
3.1.1.3.2 The ZFN mediated CCR5 knock out and transgene knock in
CCR5 ZFN has a potential to be utilized to knock-in an anti-HIV gene into the CCR5 gene locus by the ZFN induced DNA double strand break (DSB) and cellular Direct Homologous Recombination (DHR) reaction. DHR is a DSB induced cellular mechanism that uses homologous nucleotide sequences as a template to repair the damaged DNA. Therefore, it is possible to take advantage of this system by providing a transgene DNA template containing a sequence homologous to the target as well as a sequence of interest (mutation, reporter gene, therapeutic gene, etc). This “knock-in” approach provides the opportunity to integrate anti-HIV genes into a defined locus (see Heyer, 2010 for detailed review [38]). Wang et al. have successfully inserted an EGFP expression cassette into the CCR5 locus using an Adeno Associated Virus serotype 6 (AAV6) vector in which the genome was replaced by an EGFP expression cassette bordered with CCR5 matching sequence, which could be used as a DNA template for the DHR [39]. This vector was delivered to mobilized-peripheral-blood derived CD34+ cells HSPC before transfection with a mRNA coding a ZFN targeting CCR5. The success of the DHR was highlighted by 2 results: i) the modification in up to 20% of the total alleles, ii) a demonstrable dose-effect relation between the amount of DNA template provided, the allele mutation frequency and the EGFP stable expression. Finally, the results showed that these modified HSPC could be used to engraft humanized mice (see section 5.1 for details). Together, these results suggest that it is possible to insert a transgene into the CCR5 locus and achieve the dual goal of knocking out CCR5 and knocking in genes of interest. Moreover, the EGFP reporter insert can be replaced with a therapeutic anti-HIV gene in the future [38].
3.1.1.3.3 TALEN-mediated CCR5 knock out
Transcription Activator-Like Effector Nucleases (TALENs) consist of DNA binding domains of transcription activator-like effectors from Xanthomonas that have been fused to the non-specific Fok1 nuclease. Like ZFNs, TALENs can be designed to induce sequence specific genome cleavages. TALENs can be more specific than ZFNs due to the longer nucleotide stretch for the TALEN DNA recognition sequence, which ranges anywhere from 33 to 35 nucleotides, as compared to ZFNs which have 3 to 6 nucleotide triplets [38–40]. In 2011, Mussolino et al. developed a TALEN to target CCR5. They transfected HEK 293T cells with a vector expressing CCR5 TALEN and used a ZFN targeting the same CCR5 site as a control. Their results showed that up to 17% of alleles were modified, whereas the control ZFN modified 14% of alleles. They also quantified the off-target effect to only 1% of the CCR2 allele being modified, while ZFN off-target effect on CCR2 reached 11% modification [41]. Mock et al. recently developed a newly designed codon optimized TALEN called CCR5-Uco-TALEN that could knockout the CCR5 gene by more than 50% in primary human T-cells. Importantly, these CCR5 knocked out cells were resistant to R5 tropic HIV infection in vitro [40].
3.1.1.3.4 CRISPR/CAS9 mediated CCR5 knock out
CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats) technology has been utilized to knock-out the CCR5 gene. CRISPR/Cas9 is derived from the type II CRISPR-Cas9 bacterial adaptive immune system [42,43]. The Cas9 protein introduces DSBs into a DNA sequence defined by a guide RNA (gRNA). The customization of the gRNA (typically around 20 nucleotides) enables the Cas9 to cleave a target gene in a site-specific manner, triggering a DNA repair system, such as the NHEJ system and/or the DHR in cases where both chromosomes have already been damaged. Since the discovery of the CRISPR/Cas9 system, it has been extensively used to knockout CCR5. In 2013, Cho et al. were the first to succeed in developing CCR5 directed CRISPR/Cas9. They demonstrated that they could induce double genomic cleavage and mutations in the CCR5 gene by up to 33% after transfection of a Cas9 / gRNA expression cassette in HEK 293T cells [44]. In 2014, Mandal et al. demonstrated that 20–40% CCR5 mutations could be achieved using CRISPR/Cas9 in K562 cells and in human CD34+ HSPC. They also showed that a dual CCR5 gRNA strategy induced 26% of cells with biallelic mutations in CD34+ HSPC [45].
3.1.1.3.5 Caveats and improvement of the genome editing technologies
One major concern of genome editing technologies in their application for anti-HIV HSPC gene therapy is their potential in inducing off-target activities. Depending on the design, ZFNs, TALENs, CRISPR/Cas9 can induce off-target DSB. In general, TALENs are more specific than ZFNs and CRISPR/Cas9 [46]. Cradick et al. showed that their CCR5 directed CRISPR/Cas9 system had a significant off-target occurrence, especially in CCR2 gene locus, due to its sequence similarity to CCR5. They compared the efficiency of CRISPR/Cas9 using five guide RNAs targeting different sites in CCR5 gene and showed that on average, 57% of the CCR5 genes were mutated. However, they also observed that two gRNAs caused mutation in CCR2 at a frequency of 5% and 20% respectively [47]. To improve the specificity of CRISPR/Cas9, recent studies investigated modifications in the gRNA design to minimize off-target effects. In 2014, Ye et al. compared the specificity of the CRISPR/Cas9 system with the more specific TALEN system [48]. They identified a gRNA that did not target CCR2 (8 mismatches) or any other loci due to its uniqueness in the human genome, and compared its specificity to TALENs targeting the same sequence. They demonstrated that 4 times more cells had their genome edited with CRISPR/Cas9 compared to cells edited with TALENs, and that Cas9 introduced mutations in both alleles within 33% of the edited cells (compared to 10% with TALENs) [48]. In the case of CRISPR/Cas9, none of the predicted potential off-targets were mutated. Finally, they showed that monocytes and macrophages derived from these CRISPR/Cas9 modified induced pluripotent stem cells (iPSC) were resistant to R5 tropic HIV infection. The original CRISPR/Cas9 system had a high potential of inducing off-target effects because of its short nucleotide length (20 nucleotides) in gRNAs [47]. Off target effects are also significantly increased by the target DNA / gRNA mismatch tolerance (up to 3 mismatches) of the Cas9 from S.pyogenes [49]. A more specific Cas9 has been isolated from S.aureus. Its higher specificity relies on a longer Protospace Adjacent Motif (PAM) sequence, a short and constant sequence that must be present in 3' end of the targeted DNA for an efficient nuclease activity, as well as a longer size gRNA (up to 23 nucleotide) [50]. Recently, Kleinstiver et al. developed a mutated Cas9 that can significantly reduce off-target indel mutations [51]. Based on the 3D structure of Cas9, they substituted 4 amino acids (N497A ; R661A ; Q695A ; and Q926A) to improve the specificity of Cas9 binding affinity to the gRNA/DNA complex. The mutated Cas9 showed a stringent genome editing activity with undetectable off target effects, providing a hope to develop a highly specific CRISPR/Cas9 system for anti-HIV gene therapy.
3.1.2 CXCR4 inhibition
In anti-HIV gene therapy, while CCR5 has become a primary target to inhibit HIV entry, it is possible that HIV may escape from CCR5 inhibition and a CCR5 directed mono therapy may fail by emergence of X4 tropic HIVs. X4 tropic HIVs uses CXCR4 chemokine receptor for a co-receptor for entry. A recent clinical case resulted in the rapid emergence of CXCR4 tropic HIV in a patient who had been transplanted with stem cells from a Δ32/Δ32 homozygous CCR5 defective HSPC donor, highlighting the fact that viral escape mutations and pre-existing X4 tropic HIVs may be a major limitation to the CCR5 inhibition strategies [10]. Therefore, CXCR4 is another potential target for anti-HIV gene therapy. In 2003, Anderson et al. targeted CXCR4 using shRNA and protected cells against X4 tropic HIV strain, NL4-3 [52]. Genome editing technologies have also been applied to CXCR4 gene to achieve resistance to HIV infection. Yuan et al. used ZFN to knockout CXCR4 in SupT1 CD4+ T cell line as well as in primary T cells thereby successfully inhibiting X4 tropic HIV [53]. In 2015, Schuman et al., as well as Hou et al., showed the CRISPR/Cas9 system could knockout CXCR4 in primary CD4+ lymphocytes [54,55]. Schuman et al. also showed that they can knock-in a transgene in the CRISPR/Cas9 disrupted CXCR4 gene locus by providing a single strand template DNA and therefore inducing DHR. The results showed an overall of 18 to 22% transgene knock-in [54]. While CXCR4 knockdown/out strategy is applicable for protecting mature CD4+ T cells against X4 tropic HIV, CXCR4 knockdown/out on HSPC could have deleterious effects on HSPC growth, migration, and differentiation into mature functional hematopoietic cells, since CXCR4 expression is required for functional CD34+ HSPC migration and quiescence [56,57]. Moreover, CXCR4 double knockout in mice is lethal in utero, or at day 1 post-partum, with embryos presenting bone marrow alteration and severe immune dysfunctions [58–61]. Therefore, CXCR4 knockdown/out strategies should be applied to mature CD4+ T cells, but not to HSPC.
3.1.3 A HIV fusion inhibitor
Another extensively studied an entry step anti-HIV gene is C46. C46 is an HIV fusion inhibitor. It was developed from the C-terminal heptad repeat of HIV glycoprotein (gp) 41 as an anti-HIV gene. It is analogous to the FDA approved soluble peptide drug Enfuvirtide (T20, Fuzeon). C46 has a membrane-anchored domain to stably express on cell surface as an anti-HIV gene product. It interacts with the N-terminal hydrophobic α-helix of HIVgp41, and therefore, interferes with the six-helix bundle formation of HIV and subsequent HIV and cell membrane fusion, all the while rendering cells resistant to HIV [58,59]. C46 is highly potent for inhibition of both R5 tropic and X4 tropic HIV strains in vitro and in vivo humanized mice [59,60]. It has been tested for anti-HIV HSPC based gene therapy experiments in non-human primates [61]. A Phase I clinical trial, where autologous T-cells were transduced with a retroviral vector expressing C46, was infused into patients, with results showing neither gene therapy related adverse effects, nor apparent C46 directed immune reaction [62]. Emergence of resistance mutations associated with C46 were evident in HIV gp120, the N-terminal and C-terminal heptad-repeat of gp41 in vitro. These mutations required passaging HIV infected PM-1 T helper cell line for nearly 200 days in vitro [63]. Therefore, it is possible that HIV may escape from C46, yet, this needs to be examined in vivo animal experiments. To minimize the emergence of HIV escape mutations, a dual anti-HIV combinatorial lentiviral vector for C46 CCR5 shRNA (sh1005) expression has been developed [33,60]. This vector is currently under investigation in a Phase I/II anti-HIV HSPC gene therapy clinical trial (NCT01734850, NCT02390297).
3.2 Host restriction factors derived anti-HIV genes
Research efforts have identified a growing number of host restriction factors that can restrict HIV infection. Host restriction factors serve as endogenous antiviral systems in host cells [64]. They are usually expressed constitutively at low levels and up-regulated upon viral infection through interferon response. They target key components of the virions, and provide a first line of defense against viruses. While HIV has evolved to escape human host restriction factors, some of the human host restriction factors can be genetically modified, and restriction factors from other species may provide strong anti-viral response against HIV [64].
3.2.1 Human TRIMCyp / Mutated human TRIM5α
Rhesus macaque TRIM5α and owl monkey TRIMCyp have been identified as potent HIV restriction factors. TRIM5α is a protein ubiquitously expressed that can bind viral proteins, lead to their ubiquitination, and therefore, can direct them to the proteasome degradation pathway. The human TRIM5α does not inhibit HIV, and only mildly inhibits HIV-2, while rhesus macaque TRIM5α efficiently inhibits HIV, but not SIV. Although these xenogeneic host restriction factors can inhibit HIV, they are not suitable for clinical application because of their potential immunogenicity [64]. Negau. et. al genetically engineered a human version of TRIMCyp (HuTRIMCyp) [65,66]. By replacing the B30.2 domain by CypA, HuTRIMCyp is able to efficiently bind HIV capsid and block HIV infection shortly after cell entry [64–66]. Lentiviral vector delivery and expression of HuTRIMCyp could potently inhibit HIV including primary HIV isolates in primary CD4+ T-cells and macrophages as effectively, and in some cases better than rhesus TRIM5α in vitro. This could potentially be due to the higher capsid binding affinity of the Cyp domain in HuTRIMCyp. In vivo adoptive transfer of HuTRIMCyp transduced human CD4+ T lymphocytes lowered viral load in Rag2−/−cγ−/− immunodeficiency mice. Therefore, among the restriction factors, HuTRIMCyp best fits the criteria to use as an anti-HIV gene therapeutic. Intriguingly, generation of transgenic cats using a lentiviral vector demonstrated uniform rhesus macaque (human was not tested) TRIMCyp transgene expression without apparent health issues. TRIMCyp transgenic cat lymphocytes resisted feline immunodeficiency virus replication. These results further support the potential of TRIMCyp transgene as an anti-HIV gene reagent. Alternatively, human TRIM5α has been engineered to inhibit HIV. Yap et al showed that a single point mutation in the SPRY domain R332P could enable HIV capsid protein binding and efficiently restrict HIV [67]. Recently, Jung et al. demonstrated that lymphocytes expressing an engineered human TRIMα 5332G-R335G efficiently restricted HIV, including an extremely divergent HIV, such as O group strain [68].
3.3 2LTRZFP to inhibit HIV integration
HIV integrates into host DNA genome to stably reside in infected cells as a provirus. Inhibition of HIV infection prior to this step is critical to prevent establishment of chronic HIV infection. Sakkhachornphop et al demonstrated that 2LTRZFP-GFP, an anti-HIV zinc finger protein (ZFP) fused to GFP moiety, targets and binds to the 2-long terminal repeat (2-LTR) in the pre-integration complex, prior to genome integration, with nanomolar affinity [69]. They used a third-generation lentiviral vector and an expression plasmid DNA to deliver the 2LTRZFP-GFP transgene into various cell lines and showed that expression of 2LTRZFP-GFP decreased the genome integration of an HIV based lentiviral vector in HEK 293T cells by 50 percent. They also showed that there was more than 100-fold decrease in HIV capsid p24 protein production in HIV challenged SupT1 cells due to 2LTRZFP-GFP expression, suggesting that the 2LTRZFN-GFP transgene interferes with the integration activity of HIV. In addition, the molecular activity of 2LTRZFP-GFP was evaluated in peripheral blood mononuclear cells, where they also confirmed HIV integration interference by 2LTRZFP-GFP. They confirmed that the 2LTRZFP-GFP has potential for use in HIV gene therapy in the future [69].
3.4 Anti-HIV genes to inhibit later steps of HIV life cycle
CCR5 or CXCR4 gene inhibitors, C46, huTRIMCyp/TRIM5α and 2LTRZFP-GFP are designed to inhibit HIV and to protect cells from stable HIV integration into host cell genome. Although halting HIV infection at early steps before integration is important, complete reliance on early step inhibitors may not be enough to achieve stable control of HIV disease, since early step inhibitors may not be 100% effective as cells that still become infected will produce HIV. Development of anti HIV genes capable of inhibiting HIV at later steps after integration into host cell genome is important to inhibit HIV that are able to get around early inhibition.
3.4.1 Targeting the Provirus
In 2007, Hauber et al. pioneered a new strategy for HIV treatment that required the provirus to be excised from the host DNA [70]. They created a HIV specific Tre recombinase (HIV LTR-specific recombinase) from the Cre (Causes Recombination) recombinase to modify its specificity from loxP site to HIV LTR and recognizes an asymmetric sequence within an HIV LTR. They recently show that the Tre recombinase could excise patient derived HIV provirus from the genome of cell line as well as primary cells [70,71]. Qu et al. developed a ZFN to excise integrated HIV provirus. They infected Jurkat T cells with HIV strain NL4-3 expressing EGFP (NL4-3-EGFP). EGFP positive cells were then sorted and nucleofected with a construct expressing a ZFN targeting the HIV LTR. Three days post nucleofection, they analyzed the genome of the Jurkat cells and detected excised proviruses. They also showed that ZFN could excise provirus from latently infected primary T cells [72]. Ebina et al. have used a CRISPR/Cas9 system to excise latent HIV provirus DNA by developing guide RNAs targeting the HIV LTRs [73]. They demonstrated that transfection of the LTR directed CRISPR/Cas9 system into latently infected Jurkat cells resulted in 30% of the cells losing their provirus. Similarly, Ebina et al. used TALEN excised integrated HIV provirus in latently infected Jurkat cells as well as in latently infected primary T cells. Their results showed 53% of the provirus excised in Jurkat cells. However, when they replicated the experiment with primary PBMC, they did not see any provirus excision or indels in the HIV LTR region [74]. While CRISPR/Cas9 technology has been shown to efficiently inhibit HIV, the NHEJ associated with it enhances the risk of emergence of escape mutants [75]. Alternatively, CRISPR/Cas9 system can be developed to reactivate the provirus in order to purge the HIV reservoir. Zhang et al. combined a transcription activator with a catalytically deficient Cas9 fused with the synergistic activation mediator. Taking advantage of its ability to bind a HIV LTR gRNA, they successfully reactivated the provirus in latently infected cell lines [76]. Similar results were obtained by Saayman et al. and by Limsirichi et al using a deficient Cas9 fused to VP64 transactivation domain [77,78]. While targeting proviruses with anti-HIV genes shows efficient HIV inhibition, it is important to develop a highly efficient gene delivery system to achieve nearly 100% anti-HIV gene transduction in order to achieve an HIV cure.
3.5 Anti-HIV genes to inhibit HIV gene expression
3.5.1 HIV tat and transactivation response element (TAR) inhibitors
HIV Tat is an HIV transcriptional activator. Tat interacts with the transactivation response element (TAR) in HIV RNA transcripts and promotes the initiation of the viral gene expression and the elongation of HIV transcripts. Lee et al. developed a small interfering RNA directed to tat to inhibit HIV transcription and demonstrated HIV inhibition in tat shRNA transduced macrophages [79]. More recently, Strong et al. used a TALEN that specifically disrupted the highly conserved sequence in the HIV TAR domain. They measured TAR-TALEN efficiency by transfecting an expressing plasmid into HeLa-tat-III/LTR/d1EGFP, a cell line that stably expressed the EGFP under the control of HIV LTR. Their results showed a reduction of 55 to 60% of EGFP cells. They also showed that they could damage up to 22% of the provirus integrated in the HeLa/LAV cell line [80]. A TAR decoy was also developed by Li et al to inhibit Tat/TAR interaction [22]. Finally, Meredith et al used a Tat negative transdominant named Nullbasic to successfully inhibit HIV. Nullbasic is a Tat mutant in which all the arginine-rich domains have been replaced by glycine or alanine. Nullbasic inhibits unspliced as well as single spliced viral mRNA, in addition to HIV Rev [81].
3.6 Anti-HIV gene combinations
The success of HAART has highlighted the necessity to develop strategies that combine multiple anti-HIV genes to effectively suppress HIV and prevent resistant HIV escape mutants. The multi-target approach might also be a key element to a successful gene therapy strategy as in a recent clinical case, it was reported that though a CCR5 Δ32/Δ32 HSPC donor transplanted patient was resistant to CCR5 tropic HIV, it led to the rapid emergence of CXCR4 tropic HIV [10]. This case highlighted the importance of anti-HIV gene combinatorial strategies in HSPC gene therapy. Dr. Berkhout's group has extensively studied the emergence of escape mutant in cells engineered to express various shRNA targeting HIV mRNA [82–85]. Ter Brake et al. tested 4 different shRNA targeting gag, pol, or tat/rev and their results show that the combination of the 4 shRNA provided potent HIV inhibition. Moreover, in a mix population of engineered and non-engineered cells, HIV escape mutants could be detected if one shRNA was used to protect cells, however no escape mutants could be detected if three (or more) different shRNAs were combined [83]. Anderson et al. developed a triple combination of anti-HIV gene HSPC gene therapy strategy using three RNA based anti-HIV genes. Not only did they use an shRNA targeting tat and rev mRNA, but also in order to tackle tat more efficiently, they co-express a transactivation response (TAR) decoy that sequesters Tat from binding to TAR [22,86]. They combined this two anti-HIV gene with a ribozyme targeting CCR5 to create a single lentiviral vector (called Triple-R). Triple-R was then used to transduce CD34+ cells, which were then immediately injected into the thymic graft, so as to reconstitute humanized SCID mice [86]]. This triple anti-HIV gene therapy strategy has also been evaluated in a Phase I/II clinical trial [87]]. In 2012, Walker et al. combined an shRNA targeting CCR5, a human-rhesus macaque chimeric TRIM5α, and a TAR decoy into a lentiviral vector [88]. This vector was used to transduce CD34+ HSPC, which were then engrafted into humanized mice (see section 5.1). Ringpis et al. co-expressed sh1005 and an shRNA targeting the HIV LTR (sh516) from a lentiviral vector [32]]. A combination of the two shRNAs inhibited HIV replication by a factor of 6.9 times less expression. They also showed that vector-transduced PBMC express marker genes similarly to control vector transduced PBMC and do not alter viability. Importantly, they showed that mobilized peripheral blood CD34+ HSPC transduced with this vector maintained shRNA expression in vitro without altering their differentiation potential. Wolstein et al. published a combination of sh1005 and C46 in a lentiviral vector. Vector transduced MOLT4/CCR5 and primary PBMC were resistant to both X4 and R5 tropic HIVs. (NL4-3 and BaL, respectively) [33]. The same vector combining sh1005 and C46 was then used to modify CD34+ HSPC and successfully reconstitute humanized BLT mice that supported multilineage human hematopoietic reconstitution. HIV viral load was inhibited after HIV challenge in the reconstituted BLT mice [60]]. In order to block the entry of both X4 and R5 tropic HIV, Didigu et al. simultaneously knocked-out genes for both CCR5 and CXCR4 co-receptors using ZFNs delivered by Adenovirus vector in SupT1 cell lines as well as in primary human CD4+ T cells [89]]. While an efficient disruption of both genes occurred in 9% of the SupT1 cells, the disruption of CCR5 or CXCR4 was observed in 20% of the primary CD4+ T cells. They also demonstrated that ZFN treatment of SupT1 cells, as well as, primary CD4+ T lymphocytes resulted in these cells having a survival advantage after HIV challenge in vitro.
4 Anti HIV gene delivery system for anti-HIV HSPC gene therapy
Successful treatment of HIV/AIDS by anti-HIV HSPC gene therapy requires an efficient and safe gene delivery system for anti-HIV HSPC. Various gene delivery vector systems have been developed for anti HIV gene delivery [90]. Among them, lentiviral vectors (LV) are versatile in delivering single or multiple anti-HIV genes from a single vector. Lentiviral vectors (LV) are versatile in delivering single or multiple anti-HIV genes from a single vector. Because LV integrates into host cell genome, stable anti-HIV transgene expression can be achieved from a genome integrated LV. LV can infect dividing cells as well as non-dividing cells including CD34+ HSPC [91]]. LV pseudotyped with vesicular stomatitis virus glycoprotein G (VSV-G) can transduce many cell types [92], including CD34+ HSPC [90,91] with high titer vector stocks [94]. VSV-G pseudotyped LVs have been mainly used for ex vivo HSPC gene delivery. LV has been used in clinical trials for several diseases [69,82–85], including X-linked adrenoleuko-dystrophy [[95], metachromatic leukodystrophy and Wiskott-Aldrich syndrome [96,97]), and X-linked severe combined immunodeficiency, and the Fanconi anemia [98]. Lentiviral vector delivery of β-globulin transgene through HSPC has provided a therapeutic effect to a β-thalassemia patient. However, vector integration analysis indicated an unexpected clonal expansion of treated cells in this case. In the field of the HIV research, LV have been extensively utilized for the delivery of anti-HIV genes that require stable expression for efficient HIV inhibition, such as CCR5 shRNA [28,79,99], LTR RNA [[28,32]], tat/rev shRNA, TRIM5α mutant, CCR5 ribozyme, TAR decoy [[22,79,86], C46 [[60]], or combinations of these genes. Anti-HIV genes transduced through HSPC have been stably expressed in vitro [[22,33,100]], in vivo animal models [[28,32,99]]. Although lentiviral vectors have been the first choice for anti-HIV HSPC gene delivery to CD34+ HSPC for a long term stable expression of anti-HIV genes. Recently developed genome editing technologies (ZFN, TALENs, CRISPR/Cas9) utilize a variety of gene delivery systems. These genome editing technologies only require transient transgene expression to induce double strand DNA breaks and NHEJ mediated DNA repair, subsequently introducing indels and gene knockout. It is also important to optimize the duration of the genome editing transgene expression for maximum specificity and limiting off target effects and toxicity. Therefore, the delivery of genome editing strategies for anti-HIV HSPC gene therapy has been achieved by transfection, nanoparticles, adenoviral vector, adeno-associated based vector or non-integrating LV vector systems. Ye at al. have successfully transfected TALENs and CRISPR/CAS system to thrombopoietin (TPO), stem cell factor (SCF), and FMS-like tyrosine kinase 3 ligand (Flt-3L) stimulated iPSC using electroporation. Their results show a successful disruption of both CCR5 alleles in 33% of the colonies [48]]. In addition, as previously mentioned, Wang et al have successfully transfected CD34+ cells with a mRNA coding a ZFN targeting CCR5 [[39]. Recently, several nanoparticle-based gene delivery systems have been developed for anti HIV gene therapies. Several nanoparticles systems have been used for anti–HIV gene delivery in CD34+ HSPC. Several research teams have engineered “vaults” as delivery systems. Vaults are naturally occuring ovoid ribonucleoprotein nanocapsules present ubiquitously in eukaryotic cells. Their size usually varies between 40–70 nM and weight 10–15MDa. Interestingly, they are stable and non-immunogenic [101,102]. Modified vaults have been used to deliver cDNA [103] or protein [102] into cells. Nanoparticles have been also developed as an anti-HIV gene delivery system. Lipid nanoparticles are biomimetic 50–200nM nano vectors that can deliver lipophilic complex [103] or RNAi [83]. Polymer nanocapsules build a polymer cage that can integrate a protein cross-linker degradable by hydrolyzation or by specific enzyme [104]. This approach has recently been used to deliver silencing RNA (siRNA) targeting CCR5 to mobilized CD34+ cells. Depending on the cross-linker ratio, the authors reported up 99% transduction efficiency in cytokine mobilized human CD34+ cells cultured in SCF, TPO, and Flt-3 medium. They also successfully used these nanocapsules to transfer a gRNA that is able to excise HIV provirus from CEM-T-cell, the Cas9 cDNA being delivered through polyethyleneimine/dioleoylphosphatidylethanolamine (PEI-PE) transfection [104]. Adenovirus (AdV) has several advantages over other means of transduction: they can carry therapeutic gene DNA as large as 35kb (compared to 10kb for lentiviral system), they can be grown at high titer, and they can infect non-dividing cells. Moreover, they can be engineered to become helper-dependent ('gutless' AdVs) and, therefore, are safe for in-patient use.
Adenovirus also exist in several serotype that can be use to infect different cell types. However, AdV are not efficient in infecting HSPC. In order to optimize this system for HSPC, Shayakhmetov et al. created a chimeric AdV5 based vector with AdV35 fibers that can infect TPO, SCF and IL-3 stimulated enriched bone marrow CD34+ cells with an efficiency of 54% [[105]]. Chimeric AdV5/35 have been used to successfully deliver a ZFN targeting CCR5 to HSPC. In 2013, Li et al. showed that pretreatment with PKC increased by more than 25% CCR5 disruption in HSPC isolated from granulocyte-CSF–mobilized peripheral blood [106]]. They also showed that HSPC modified with their protocol could engraft on humanized mice and support multilineage differentiation. Recently, Saydaminova et al. also added a miR-183-5p and miR-218-5p based regulation to their AdV5/35 for their CCR5-ZFN transgene and efficiently transduced HSPC [37] as described in the Zinc Finger Nuclease section (3.1.1.3) of this review. Adeno-Associated Virus (AAV) vector with capsid from serotype 6 (AAV6) and 9 (AAV9) have been used to CD34+ HSPC transduction [39,107]. Wang et al. used AAV6 vectors to transduce knock-in template carrying EGFP expression cassette into HSPC. They inserted GFP into the CCR5 locus at a mean frequency of 17% in mobilized peripheral blood CD34+ cells cultured in medium supplemented with SCF, TPO, IL-6, and Flt-3. They obtained similar results (19%) with fetal liver CD34+ [39]. Smith et al. showed that AAV are naturally present in HSPC from blood donors (AAVHSPC). Most of these AAVHSPC are related to AAV9 and belong the Clade F. They also showed that these AAV have potential as a base for vectors targeting CD34+ cells and transgene expression [107]
5 In vivo investigation of anti-HIV gene HSPC therapy
Investigations of anti-HIV gene HSPC therapy in animal model have provided critical information for the engraftment of anti-HIV gene modified HSPC, reconstitution of anti-HIV hematopoietic system and HIV inhibition in vivo. Humanized mouse models are ideal to test anti-HIV gene therapy strategies using human HSPC to validate the use of therapeutic vectors prior to large animals and human clinical trials. In these xenotransplantation mouse models, newly generated anti-HIV genes and delivery vectors can be rigorously investigated on their efficiency and safety of vector-mediated HSPC transduction, simultaneous expression of anti-HIV gene products, extent of multilineage hematopoietic differentiation of transduced cells, selective protection against HIV infection, and ability to stably control HIV replication in vivo in small animal model settings.
5.1 In vivo investigations of anti-HIV HSPC gene therapy
strategies in humanized mouse models Anti-HIV HSPC based gene therapy strategies have been examined in vivo in several different humanized mouse models. Humanized mice have been extensively developed whereby some elements of the human immune system have been reconstituted in severe combined immune deficiency (SCID) mice, such as human CD34+ cell injected Rag2−/− γc−/−, or NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mouse models. Anti-HIV gene modified CD34+ cells were transplanted into irradiated neonatal mice, successfully engrafted in mice and reconstituted anti-HIV gene expressing multilineage cells in these mice. In 2010, Holt et al. published a study in which they optimized the delivery of CCR5-specific ZFNs to human umbilical cord blood derived CD34+ HSPCs and transplanted the modified cells into NSG mice, as NSG mice support both human hematopoiesis and HIV infection [36]. They observed that transplantation of ZFN-modified human CD34+ HSPC led to multilineage hematopoietic differentiated cells with CCR5 disruption of 5–7% in humanized mice. Moreover, upon secondary transplantation, all animals had engrafted and human cells in the blood of the secondary transplanted mice had levels of CCR5 disruption that exceeded the level in the original donor marrow (12–20%). Furthermore, infection of the mice with a CCR5-tropic HIV Bal led to rapid selection for CCR5 negative human cells, a significant reduction in viral load and protection of human T-cell populations from HIV infection [36]. Using the same CCR5-ZFN as Holt and coworkers, in 2015, Saydaminova et al. modified PBMC derived CD34+ HSPCs and were able to achieve up to 12% CCR5 gene disruption in gene modified cells in bone marrow transplanted NOD/Shi-scid/IL-2Rγnull (NOG) mice. However, ZFN expression negatively affected transduced CD34+ cell survival rate, engraftment rate and expansion of transduced CD34+ cells as compared to the untransduced HSPC. They speculated that this negative effect could be related to ZFN expression over extended periods of time, thereby causing potential cytotoxicity [37]]. More recently, the same group has been conducting preclinical studies to evaluate the safety and efficacy of an anti-HIV gene therapy based on CCR5-targeted ZFNs to disrupt the CCR5 gene in mobilized peripheral blood derived HSPCs in a humanized mouse model. They found that intravenous injection of mobilized blood HSPC into adult NSG mice after myeloablation using irradiation gave engraftment level approaching 100% and lasted for more than 6 months. They also determined that pretreating the HSPC with protein kinase C activators increased CCR5 distribution rates to >25%, while still supporting the engraftment of adult NSG mice [108]. Also based on the results by Holt and coworkers, Li et al. published a study in 2013 in which they evaluated disruption of CCR5 gene expression in HSPCs isolated from granulocyte colony-stimulating factor (CSF) mobilized adult blood using a recombinant adenoviral vector encoding a CCR5-specific pair ZFN [106]. They demonstrated that CCR5-ZFN RNA and protein expression from the adenoviral vector is enhanced by pretreatment of HSPC with protein kinase C (PKC) activators, thereby resulting in >99% gene-modified cells and >25% CCR5 gene disruption, due to the activation of the MEK/ERK signaling pathway. Furthermore, by using an optimized dose of PKC activator and adenoviral vector they were able to generate CCR5-modified HSPCs, which successfully engrafted in a humanized NSG mouse model, although at a low level, to support multilineage differentiation in vivo. Also, HIV challenge of these mice successfully reconstituted with CCR5-ZFN–treated HSPC resulted in greater level of CCR5 disruption in both CD4+ T-cells and CD8+ T-cells [106]. Li et al had previously reported the use of a combinatorial vector co-expressing a tat/rev siRNA, a nucleolar TAR RNA decoy, and a CCR5-targeting ribozyme from independent RNA Polymerase III promoters to ensure persistent antiviral transgene expression in all hematopoietic cells [22]]. They conducted a pilot clinical trial in four AIDS patients undergoing therapy and HSPC transplantation for lymphoma where a portion of the HSPCs was modified with this combinatorial vector. They observed rapid engraftment of gene-modified cells, persistent multilineage transgene expression with no serious adverse events, demonstrating that the treatment was safe feasible and long lasting [[87]]. However, the frequency of gene marking in peripheral blood mononuclear cells ranged from 0.02 to 0.32% and was extremely low to clinically assess antiviral efficacy.
To overcome this hurdle, Chung et al developed an O(6)-methylguanine-DNA-methyltransferase- P(140)K mutant (MGMT-P140K) expressing combinatorial lentiviral vector using the minichromosome maintenance complex component 7 (MCM7) platform that allowed simultaneous expression of three antiviral RNAs in a micro RNA cassette from a single Pol II U1 promoter, eliminating the need for multiple promoters and thereby reducing chances of vector toxicity [1]. HSPCs were transduced with an MGMT-P140K expressing vector and used to transplant immunodeficient NSG mice and the mice were treated with bis-chloroethylnitrosourea (BCNU) and O(6)-benzylguanine (O6-BG). They demonstrated that though engraftment of the bone marrow and spleen with human (CD45+) cells was significantly reduced in treated animal relative to the control mice, the frequency of transduced CD45+ cells in the bone marrow and spleen was enriched 10- to 15-fold in the treated populations as compared to the control mice. These data demonstrates that selective enrichment of HIV-target cells occurred in vivo [1]. In a recent study by Myburgh et al., they reported a novel microRNA (miRNA)-based gene knockdown strategy that used a triple-hairpin cassette targeting CCR5 resulting in >90% CCR5 knockdown in HeLa cell line [99]. Furthermore, they assessed whether genetically modified CD34+ HSPC with this lentiviral vector construct results in down-regulation of CCR5 in progeny cells in humanized NSG mice and whether it could protect against HIV challenge ex vivo as well as in vivo. Humanized NSG mice were transplanted with transduced CD34+ human HSPCs and were called FACS-sorted R5 knockdown mice. The FACS-sorted R5 knockdown mice had markedly lower viral loads than the FACS-sorted negative mice and control mice over 28 weeks. Moreover, CD4+ T cells from the FACS-sorted negative mice decreased steadily upon infection (day 0, 55%; day 134, 20%) whereas, the FACS-sorted R5 knockdown mice showed an increase in CD4+ T cells (day 0, 33%; day 196, 65%). Therefore, transplantation of transduced CD34+ HSPC produced mice with high levels of genetically modified CCR5 knockdown CD4+ T cells in vivo, thereby resulting in long-term inhibition of HIV replication in vivo and persistence of HIV target cells in the mice [99]]. Wang et al. transduced fetal liver CD34+ cells with CCR5-GFP AAV6 vector followed by transfection with CCR5 ZFN mRNA [39]. They then engrafted the modified CD34+ cells into neonatal NSG mice and monitored them over 16 weeks. Analysis of peripheral blood bone marrow and spleen at various time points revealed robust development of human CD45+ cells at higher levels compared to the mice that received untreated control HSPCs. Moreover, they reported that the treated HSPCs were capable of multilineage differentiation. They also examined the levels of genome editing in the human cells that developed in the mice over time. Using flow cytometry for cells from the CCR5-GFP mice, Wang et al. also examined the levels of genome editing in cells in both the circulation and tissues. They found significant levels of genome editing in CD45+ population as well as several lineages. Using In-out PCR, they also confirmed the site-specific GFP insertion at the CCR5 locus in the blood and tissues of individual mice from the CCR5-GFP-ZFN group. Furthermore, Wang et al. evaluated the ability of the genome-edited HSPCs to persist after secondary transplantations using bone marrow harvested from the CCR5-GFP-ZFN mice. The bone marrow of the secondary transplant NSG recipients was then analyzed at 20 weeks post secondary transplant and revealed that the HSPCs had frequencies of genome editing that were at similar or higher levels when compared to the bone marrow samples from the primary donors [[39]]. In 2008, ter Brake et al. demonstrated that multiple shRNA could be efficiently expressed in a single lentiviral vector, if expression is driven be a different promoter for each shRNA. They combined three anti-HIV shRNAs, each specifically targeting a highly conserved region of [[83]]. In addition to being able to assess the potency of anti-HIV genes in humanized mouse models, these models are also valuable and are sensitive preclinical tools to evaluate the hematopoietic safety of gene therapy strategies. In 2013, Centlivre et al. evaluated the preclinical safety of four anti-HIV shRNA candidates targeting the viral capside, integrase, protease and tat/rev open-reading frames among all HIV subtypes in human HSPCs [84]. They reported that out of the four shRNA candidates, a single shRNA targeting Gag gene exhibited a negative impact on the development of human hematopoietic cells. They demonstrated the safety of the shRNAs targeting the Polymerase and Tat/Rev genes, as they did not affect the in vivo development of the human immune system. Furthermore, when these three shRNAs were combined into a single lentiviral vector (R3A), simultaneous expression of these three shRNAs did not affect the in vivo multilineage differentiation of HSPCs. Finally, they also showed that the shRNA transduced CD4+ T cells were resistant to HIV infection. Their results suggested that combinatorial shRNA treatment not only exhibits additive HIV inhibition but could also prevent the selection of escape virus variants [84]]. In a study published in 2012 by Walker et al., they carried out an in vivo evaluation of a combination anti-HIV lentiviral vector in humanized NOD-RAG1−/− IL2rγ−/− knockout (NRG) mice [88]. This combination vector contained a human/rhesus macaque TRIM5 isoform, a CCR5 shRNA, and a TAR decoy. Upon transplantation with anti-HIV lentiviral vector-transduced human cord blood derived CD34+ HSPCs, Walker et al. observed 17.5% engraftment of modified cells as well as multilineage hematopoiesis in the peripheral blood and in various lymphoid organs of engrafted humanized NRG mice. Furthermore, Anti-HIV vector-transduced CD34+ HSPC displayed normal development of various immune cells, including T cells, B cells, and macrophages. The anti-HIV vector-transduced cells also displayed knockdown of cell surface CCR5 (7.4%) due to the expression of the CCR5 shRNA, whereas in the untransduced cells, the CCR5 expression was 46%. After in vivo challenge with either an R5-tropic or X4-tropic strains of HIV, maintenance of human CD4+ cell levels and a selective survival advantage of anti-HIV gene-modified cells were observed in engrafted mice [[88]. In a proof-of-concept study by Kalomoiris et al., the in vitro safety and improved efficacy of HSPC gene therapy was demonstrated by using a triple combination, pre-selective, anti-HIV vector that incorporated a selectable marker, human CD25, which is expressed on the surface of transduced cells [109]. This was done to avoid rejection of transduced cells and allow for the purification of vector-transduced cells prior to transplantation. Human CD25 is a low affinity IL-2 receptor alpha subunit, and is not expressed on the surface of HSPCs or within HSPCs, with no direct intracellular signaling. Upon expressing a mutated/truncated form of human CD25 on the surface of HSPCs and purification of the transduced cells, safety of the enriched population of anti-HIV vector-transduced HSPCs was observed along with potent HIV inhibition. They were able to obtain a highly enriched (94.2%) population of HIV resistant HSPC for transplantation [109]. In a follow-up study, the same group evaluated the in vivo safety and efficacy of the tCD25 pre-selective combination anti-HIV lentiviral vector in umbilical cord derived human HSPCs and their immune cell subsets in NRG mice. In vivo safety, engraftment, stable chimerism, and multilineage hematopoiesis was successfully observed when the tCD25 pre-selective combination anti-HIV lentiviral vector transduced HSPCs were transplanted into NRG mice [110].
5.2 In vivo investigations of anti-HIV HSPC gene therapy strategies in humanized marrow/liver/thymus (BLT) transplanted mouse model
Humanized BLT mouse model allows us to investigate genetically engineered human HSPC reconstitution in vivo in bone marrow, the human thymus-like organoid (thy/liv), and secondary lymphoid organs, including the gut, the major site of HIV replication [111]. Human T-lineage lymphocyte reconstitution is robust in BLT mice because of the co-transplantation of human fetal thymus and liver that creates thymus like organoid (thy/liv) under the kidney capsule and CD34+ HSPC. The human immune cells within this BLT mouse model are susceptible to infection by HIV, making this mouse model an ideal basis to investigate the potential for genetically engineered HSPC to resist HIV infection. We are the first group to use BLT mice to test HSPC-based gene therapy strategies using an shRNA against CCR5 [30]. Shimizu et al. utilized the humanized BLT murine model to perform in vivo experiments in which they transduced fetal liver derived CD34+ HSPCs with a lentiviral vector carrying shRNA targeting CCR5 coreceptor (sh1005). They successfully showed that effective down-regulation of HIV co-receptor CCR5 protected mouse-derived human splenocytes, ex vivo [30], and CD4+ T cells, in vivo, [31] from CCR5 tropic HIV infection. Conversion of HIV tropism to CXCR4 was not evident in their experimental setting. In 2012, Ringpis et al. published a study in which they co-expressed sh1005 and a shRNA targeting the HIV LTR (sh516) from a single lentiviral vector [32]. They transduced CD34+ HSPC with the dual shRNA vector and transplanted humanized BLT mice with the modified cells. Twelve weeks post engraftment, they evaluated the reconstitution in the humanized BLT mice and observed that the modified CD45+ cells represented 20 to 80% of peripheral blood cells. Moreover, they observed modified CD45+ cells in bone marrow, spleen and GALT as well, suggesting that dual sh1005/sh516-transduced HSPCs could successfully engraft and differentiate into human T-cells in peripheral blood and systemic lymphoid organs. They also showed up to 7.5% CCR5 down-regulation in various tissues. Moreover, they demonstrated that the dual shRNA modified HSPCs were resistant to both R5 tropic and X4 tropic HIV infection [32]. Burke et al. recently published a study in which they assessed the efficacy of the LVsh5/C46 dual vector in vivo. They transduced human-fetal liver derived CD34+ HSPCs with the dual lentiviral vector, and transplanted the modified cells into NSG mice to generate humanized BLT mice [60]. They observed that both LVsh5/C46-transduced CD34+ HSPC and non-modified CD34+ HSPC both displayed robust engraftment of human CD45+ cells in vivo, with modified cells showing 78% engraftment, with control cells showing 87% engraftment. They also reported that not only do LVsh5/C46-modified CD34+ HSPCs efficiently engraft in vivo, but they also undergo multilineage hematopoietic development, including CD4+ T-cell differentiation in peripheral blood as various lymphoid tissues. Moreover, these genetically modified cells could persist in the mice for over six months. Finally, they demonstrated that LVsh5/C46 vector-transduced human CD34+ HSPC can efficiently down-regulate CCR5 expression in human CD4+ T lymphocytes in multiple lymphoid tissues and engineer cellular resistance to both R5-tropic and X4-tropic HIV infection [60]. Chimeric antigen receptors (CARs) redirect T cells toward malignancies and have become a much talked about method of treatment, representing a broad-based approach of engineering. One prototype CAR for treating HIV infection is the CD4ζ chimeric antigen receptor (CD4ζ CAR). The CD4ζ CAR molecule is a hybrid molecule consisting of the extracellular and transmembrane domains of the human CD4 molecule fused to the signaling domain of the CD3 complex ζ-chain. Thus, when CD4 recognizes and engages HIV gp120 envelope protein on HIV infected cells, the CAR-modified cell is triggered and activated via ζ-chain signaling. CD4ζ CAR-modified T cells were reported to inhibit viral replication and kill HIV-infected cells in vitro. Recently, Zhen et al. explored the ability of CD4ζ CAR to allow gene-modified HSPCs to produce multilineage immune cells that target HIV infection in vivo using the humanized BLT mouse model [112]. Furthermore, to protect the gene modified cells from HIV infection, they combined CD4ζ CAR with efficient anti-HIV reagents (CCR5shRNA/sh516/CD4ζ CAR; Triple CAR). They demonstrated that the cells carrying the Triple CAR could elicit HIV-specific T-cell responses following antigen encounter. Moreover, the genetic modification of HSPCs with a lentiviral vector containing the Triple CAR can result in the multilineage reconstitution of HIV-specific cells that are protected from infection, and can lower viral loads in vivo following HIV challenge [112].
All of HIV challenge experiments were performed in already reconstituted humanized mice with anti-HIV gene modified cells.
5.3 In vivo investigations of anti-HIV HSPC gene therapy strategies in non human primates
To translate an HIV/AIDS gene therapy to the clinic, it is important to determine if hematopoietic repopulating stem cells transduced with proposed anti-HIV vectors can efficiently expand and also engraft in an animal model that closely mimic human transplantation in a large animal model setting, such as non-human primates, in order to translate the findings rapidly to a clinical setting. In 2007, An et al. published a study in which they examined the rhesus macaque adapted CCR5-shRNA (sh1005) in non-human primates by lentiviral vector transduction of mobilized peripheral blood CD34+ HSPC and transplant. An et al. carried out a single mismatched nucleotide mutation in the human sh1005 sequence to match the rhesus macaques CCR5 target sequence, and showed that there was stable engraftment of the modified HSPCs, multilineage hematopoietic differentiation, as well as stable down-regulation of rhesus macaque CCR5 expression. Furthermore, rhesus macaque sh1005 modified CD4+ T cells were resistant to simian immunodeficiency virus (SIV) infection ex vivo [28]. Kim et al. investigated properties of a large number of heterogeneous clones following the four rhesus macaques that underwent a sh1005-modified CD34+ cell transplant, and evaluated how long-term and stable hematopoietic reconstitution was achieved through the combined contributions of thousands of clones [113]. Even though there are variations in HSPC utilization over months or years, total gene marking remained relatively constant for over ten years in these non-human primates. Also, normal repopulation of myeloid and lymphoid cell lineages was maintained by a few stable clones that had a relatively balanced multilineage output. The results suggest tremendous diversity, as well as intricate control of HSPC repopulation. These sh1005 transduced macaque hematopoietic cells were examined for clonal tracking analysis using lentiviral vector integration sites [113]. Their results demonstrate that vector integration sites were random for more than twelve years, indicating polyclonal multilineage differentiation of vector transduced HSPC in vivo. In 2009, Trobridge et al. evaluated HSPC gene transfer of an anti-HIV lentiviral vector carrying an HIV fusion inhibitor (C46) in the pigtailed macaque (Macaca nemestrina) model [61]. The lentiviral vectors inhibited both HIV and simian immunodeficiency virus (SIV)/HIV (SHIV) chimera virus infection, and also expressed MGMT-P140K transgene to select gene-modified cells. Following transplantation and MGMT-mediated selection of transduced CD34+ cells derived from bone marrow, they demonstrated transgene expression in over 7% of stem-cell derived lymphocytes, which enabled them to demonstrate protection from SHIV in lymphocytes derived from gene-modified macaque, long-term repopulating cells that expressed C46. On secondary transplantation, they observed a significant 4-fold increase of gene-modified cells after challenge of lymphocytes from one macaque that received HSPCs transduced with an anti-HIV vector (p<0.02). All vectors that were used were HIV-based and therefore, could efficiently transduce human cells. Moreover, the transgenes that Trobridge et al. used were also found in SHIV, therefore their findings could be rapidly translated to the clinical setting [61]. Furthermore, in 2013, Younan et al. incorporated the fusion inhibitor C46 and a chemoresistant transgene MGMT-P140K into HSPCs, and these were then transplanted into pigtail macaques followed by treatment with chemotherapeutic agents to select gene-modified cells and then onto Simian-Human Immunodeficiency Virus (SHIV) challenge [114]. The control macaques did not receive the fusion-inhibitor expression cassette, whereas the C46 macaques did. Following SHIV challenge, C46 macaques, but not control macaques, showed a positive selection of >90% gene-modified CD4+ T cells in peripheral blood, gastrointestinal tract, and lymph nodes. C46 macaques also maintained high frequencies of gene-modified CD4+ T cells, an increase in non-modified CD4+T cells, enhanced cytotoxic T lymphocyte function, and various antibody responses. This was the first study in a nonhuman primate model to demonstrate the protective effect of transplanted gene modified, infection-resistant HSPCs against SHIV challenge. Although a limited number of animals were used in this study, the results indicated that C46 infection-resistant, HSPC-derived CD4+ T cells, and other potential target cells, are resistant to infection in vivo and may lead to an enhanced SHIV-specific immune response resulting in reduced plasma viremia and decreased viral pathogenesis [114]. In another study by Younan et al. published in 2015, they studied the effect of ART on HSPC transplantation. Following infection by SHIV and ART, pigtail macaques underwent autologous HSPC transplantation with lentiviral vector transduced CD34+ cells expressing the C46 anti-HIV fusion protein. They showed that SHIV+, ART-treated animals had very low gene marking levels after HSPC transplantation. Pre-transduction CD34+ cells contained detectable levels of all three ART drugs, likely contributing to the low gene transfer efficiency. Moreover, T-cell subset analysis demonstrated a high percentage of CCR5-expressing CD4+ T-cells after the transplantation, suggesting that an extended ART interruption time may be required for more efficient lentiviral transduction. These results would help establish protocols for lentiviral vector-mediated gene modification strategies in clinical settings where the patient population may already be on ART [115].
5.4 Anti HIV HSPC based gene therapy clinical trials
Over the past years, five anti-HIV HSPC based gene therapy strategies have been investigated in clinical trials [87,116–119]]. In 1999, Kohn et al. reported a clinical trial evaluating the safety and feasibility of the approach of transplanting modified HSPCs. They used CD34+ cells derived from the bone marrow from four HIV infected pediatric subjects, and performed in vitro transduction with a retroviral vector carrying a rev-responsive element (RRE) decoy gene. They then reinfused the cells into these subjects with no evidence of adverse effects. Unfortunately, the levels of gene-marking in peripheral blood samples after one year were extremely low (<0.01%) [119]]. In 2000, Bauer et al. reported a Phase I trial testing three classes of anti-HIV genes:(1) dominant-negative mutants of the HIV regulatory genes tat and rev, (2) a small synthetic portion of the rev-responsive element (RRE) to serve as a decoy to sequester HIV Rev protein, and (3) hammerhead ribozymes, to cleave HIV mRNA [116]. Bone marrow CD34+ HSPCs were harvested from four HIV infected pediatric patients, and transduced with a Moloney murine leukemia virus (MMLV) based retroviral vector carrying an RRE decoy gene. The cells were re-infused into the subjects, without complications, showing that gene transfer in pediatric AIDS patients is safe and feasible. Cells expressing these genes showed significant inhibition of HIV replication (>95–99.5%) compared to control cells. However, the level of gene-marking in peripheral blood leukocytes was low (<0.01%) and only seen in the first months following cell infusion, thereby, resulting in poor sustained gene expression. It is necessary to achieve a higher level of gene transfer in order to produce high percentages of T-lymphocytes and immune cells that are resistant to HIV infection [116]. Mitsuyasu et al. reported a Phase II trial for testing autologous CD34+ HSPC transduced with anti-HIV ribozyme (OZ1) targeting Tat/Vpr in a MMLV based retroviral vector (NCT00074997) [117]. This study, the first randomized, double-blind, placebo-controlled, Phase II cell-delivered gene transfer clinical trial, was conducted in 74 HIV infected adults who received a tat/vpr specific anti-HIV ribozyme (OZ1), or placebo delivered in autologous CD34+ hematopoietic progenitor cells. These participants were then followed for 100 weeks. The outcomes of this trial were to assess if OZ1-transduced CD34+ HSPCs would engraft, divide and differentiate in vivo to produce a pool of mature myeloid and lymphoid cells that are protected from productive HIV replication. There were no OZ1-related adverse events. Throughout the 100 weeks, CD4+ lymphocyte counts were higher in the OZ1 group, and HIV viral loads were consistently lower in the OZ1 group as compared to the control group. This study provided the first indication that cell-delivered gene transfer is safe and biologically active in HIV patients. However, the mature T-lymphocytes had a shortened lifespan and insufficient homing to the bone marrow, engraftment (gene marking <0.38%) and differentiation through the myeloid and lymphoid compartments of transduced CD34+ cells [117]. DiGuisto et al. reported a trial in which HIV+ patients undergoing autologous transplantation for lymphoma were treated with gene-modified peripheral blood derived (CD34+) HSPCs expressing three RNA-based anti-HIV moieties (Tat/Rev shRNA, TAR decoy and CCR5 ribozyme). The CD34+ HSPCs were transduced using a self-inactivating lentiviral vector (SIN) carrying the three anti-HIV moieties. All four infused patients engrafted by day 11 post-infusion, and showed no unexpected infusion related toxicities. However, in this study as well, persistently low gene marking (<0.2%) in multiple cell lineages was observed at low levels for up to 24 months. Moreover, there was no evidence of any immediate therapeutic benefit of these genetically modified HSPCs [87]. However, this trial warranted the need to develop improved transduction processes and revise transplant procedures to preferentially infuse only transduced cells are likely to lead to higher levels of engrafted genetically modified cells. Currently, in a trial sponsored by CalImmune, Inc., Mitsuyasu and coworkers are testing the CCR5-sh1005 lentiviral vector which has been further modified to express the C46 fusion inhibitor that blocks the fusion between HIV envelope and the host cell membrane. This combination of the CCR5-sh1005 and C46 fusion inhibitor into a single lentiviral vector (named as Cal-1) has been tested in various animal models and is currently being assessed in a Phase I/II trial in patients with chronic HIV infection (NCT01734850, NCT02390297) [62]. This is an early phase research study where they will evaluate whether the Cal-1 vector is safe and if it can protect the immune system from HIV infection. Long Term Follow-up of this phase includes detection of delayed adverse events in recipients of CD4+ T lymphocytes and/or CD34+ HSPC transduced with Cal-1. The preclinical study successfully demonstrated that the Cal-1 vector was able to protect gene-modified CD34+ HSPCs from both CXCR4- and CCR5- tropic HIV strains, while being non-toxic, non-inflammatory and with no adverse effects on HSPC differentiation [33]. Recently, Tang and coworkers completed a clinical trial of CCR5-targeted ZFNs modified HSPC transplant sponsored by Sangamo Biosciences (NCT01044654). These CCR5-targeted ZFNs have been evaluated in a clinical trial with gene modified autologous CD4+ T cells [35]. A Phase I/II trial assessed the efficacy of targeting autologous T cells genetically modified at the CCR5 gene by ZFN (NCT01252641, NCT00842634). They enrolled twelve patients in an open-label, nonrandomized, uncontrolled study of a single dose of ZFN-modified autologous CD4 T cells. The primary outcome was safety as assessed by appearance of treatment-related adverse events. Secondary outcomes included assessing the increases in the CD4 T-cell count, persistence of the modified cells, homing to gut mucosa, and effects on viral load. Although there was one serious adverse event, it was attributed to a transfusion reaction. The median CD4 T-cell count increased upon infusion of ZFN-modified CD4+ T cells. Due to the increase in the number of gene-modified T-cells, the blood level of HIV DNA decreased in most patients and HIV RNA became undetectable in one of four patients. Therefore, CCR5-modified autologous CD4 T-cell infusions are safe within the limits of this study [118]. The ZFN mediated disruption of CCR5 in HSPC may provide us with a more durable anti-viral effect as it may give rise to CCR5−/− cells in both the lymphoid and myeloid compartments that HIV infects.
6 Conclusions
Anti-HIV HSPC based gene therapy has emerged as a potentially powerful approach to develop as an HIV cure strategy since the first case of HIV cure achieved by HIV resistant bone marrow transplants. Numerous anti-HIV genes capable of inhibiting different steps of HIV infection have been developed with state-of-the-art technologies including RNAi, ZFN, TALEN, and CRISPR/Cas9. Some of these anti-HIV genes have shown promising results in vitro and in vivo animal experiments to protect HSPC and their progenies from HIV infection. Over the past years, five anti-HIV HSPC based gene therapy strategies have been investigated in clinical trials ([69,83–85,101]). These clinical trials have demonstrated that safety and feasibility of anti-HIV gene modification of HSPC and engraftment in patients. However, due to the low engraftment rate of genetically modified HSPC, cells expressing anti-HIV reagents were not reconstituted at sufficient levels to achieve therapeutic benefit ([69,83,84,101]). Therefore, for successful application of the anti-HIV HSPC gene therapy to achieve HIV cure, the engraftment of anti-HIV gene modified HSPC needs to be improved. Second, a safe myeloablation procedure needs to be established when applying anti-HIV gene modified HSPC to a broader patient population. The use of intensive myeloablation procedures, which were justified with leukemia in the first case of HIV cure, will be difficult to routinely apply. Two anti-HIV HSPC based gene therapy clinical trials are investigating optimized myeloablation procedures for (sh1005-C46 (NCT01734850, NCT02390297) and CCR5 ZFN(NCT01044654). These clinical trials will provide important results for the successful translations of anti-HIV HSPC based gene therapy into clinical use for the future.
Acknowledgments
We thank Jennifer Lam, Wendy Hsuan Wen Hung, Jenny J Hong, Kevin M Kim, and Yujin Jung for critical reading and edits of our manuscript. This research was supported by the NIAID 1R01AI100652, 1 U19 AI117941-01, the UCLA AIDS Institute and the UCLA Center for AIDS Research NIH/NIAID AI028697, and Charity Treks. We thank the UCLA CFAR Gene and Cellular Therapy Core Facility for providing fetal tissues and CD34+ HSPC, the CFAR Humanized Mouse Core Facility for the animal housing.
Abbreviations
- AAV
adeno associated virus
- Adv
adenovirus
- AIDS
acquired immunodeficiency syndrome
- AML
acute myelogenous leukemia
- BLT
bone marrow/liver/thymus
- BCNU
bis-chloroethylnitrosourea
- CARs
chimeric antigen receptors
- cART
combined antiretroviral therapy
- CypA
cyclophilin A
- Cre
causes recombination
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- DHR
Direct Homologous Recombination
- DSB
Double Strand Breaks
- EGFP
enhanced green fluorescent protein
- Flt-3L
FMS-like tyrosine kinase 3 ligand
- gRNA
guide RNA
- HuTRIMCyp
human version of TRIMCyp
- HAART
Highly Active Antiretroviral Therapy
- HIV
human immunodeficiency virus
- HLA
human leukocyte antigen
- HSPC
hematopoietic stem progenitor cell
- HIC
HIV controllers
- Indels
insertion and deletions
- iPSC
induced pluripotent stem cells
- LTR
long terminal repeat
- LV
Lentiviral vector
- MCM7
minichromosome maintenance complex component 7
- mRNA
messengerRNA
- miRNA
microRNA
- MMLV
moloney murine leukemia virus
- MGMT-P(140)K
O(6)-methylguanine-DNA-methyltransferase- P(140)K mutant
- NRG
NOD-RAG1−/−IL2rγ−/−mice
- NSG mice
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice
- NOG mice
NOD/Shi-scid/IL-2Rγnull mice
- NHEJ
non homologous ends joining
- O6-BG
O(6)-benzylguanine
- PAM
protospace adjacent motif
- PEI/PE
Polyethyleneimine/ dioleoylphosphatidylethanolamine
- PKC
protein kinase C
- PNA
peptide nucleic acids
- PPIase
peptidyl-prolyl isomerase
- RISC
RNA Induced Silencing Complex
- RNAi
RNA interference
- SCF
stem cell factor
- shRNA
short hairpin RNA
- SCID
severe combined immune deficiency
- siRNA
small interfering RNA
- TALEN
transcription Activator-Like Effector Nucleases
- TAR
transactivation response element
- TPO
thrombopoietin
- VSV-G
vesicular stomatitis virus glycoprotein G
- ZFP
zinc finger protein
- ZFN
zinc finger nuclease
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Olivier Pernet, Email: opernet@ucla.edu.
Swati Seth Yadav, Email: swatisyadav@ucla.edu.
Dong Sung An, Email: an@ucla.edu.
References
- 1.Chung J, Scherer LJ, Gu A, Gardner AM, Torres-Coronado M, Epps EW, et al. Optimized Lentiviral Vectors for HIV Gene Therapy: Multiplexed Expression of Small RNAs and Inclusion of MGMTP140K Drug Resistance Gene. Mol. Ther. 2014;22:952–963. doi: 10.1038/mt.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sáez-Cirión A, Pancino G. HIV controllers: a genetically determined or inducible phenotype? Immunol. Rev. 2013;254:281–294. doi: 10.1111/imr.12076. [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG. HIV Infection, Inflammation, Immunosenescence, and Aging. Annu. Rev. Med. 2011;62:141. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramana KV, Sabitha V, Rao R. A Study of Alternate Biomarkers in HIV Disease and Evaluating their Efficacy in Predicting T CD4+ Cell Counts and Disease Progression in Resource Poor Settings in Highly Active Antiretroviral Therapy (HAART) Era. J. Clin. Diagn. Res. JCDR. 2013;7:1332–1335. doi: 10.7860/JCDR/2013/5306.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allers K, Schneider T. CCR5Δ32 mutation and HIV infection: basis for curative HIV therapy. Curr. Opin. Virol. 2015;14:24–29. doi: 10.1016/j.coviro.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Hütter G, Nowak D, Mossner M, Ganepola S, Müßig A, Allers K, et al. Long-Term Control of HIV by CCR5 Delta32/Delta32 Stem-Cell Transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 7.Brown TR. I Am the Berlin Patient: A Personal Reflection. AIDS Res. Hum. Retroviruses. 2015;31:2–3. doi: 10.1089/aid.2014.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hütter G, Blüthgen C, Neumann M, Reinwald M, Nowak D, Klüter H. Coregulation of HIV-1 dependency factors in individuals heterozygous to the CCR5-delta32 deletion. AIDS Res. Ther. 2013;10:26. doi: 10.1186/1742-6405-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Persaud D, Palumbo PE, Ziemniak C, Hughes MD, Alvero CG, Luzuriaga K, et al. Dynamics of the resting CD4+ T-cell latent HIV reservoir in infants initiating HAART less than 6 months of age. AIDS Lond. Engl. 2012;26:1483–1490. doi: 10.1097/QAD.0b013e3283553638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kordelas L, Verheyen J, Esser S. Shift of HIV Tropism in Stem-Cell Transplantation with CCR5 Delta32 Mutation. N. Engl. J. Med. 2014;371:880–882. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- 11.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee T-H, et al. Antiretroviral-Free HIV-1 Remission and Viral Rebound Following Allogeneic Stem Cell Transplantation: A Report of Two Cases. Ann. Intern. Med. 2014;161:319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiem H-P, Jerome KR, Deeks SG, McCune JM. Hematopoietic-Stem-Cell-Based Gene Therapy for HIV Disease. Cell Stem Cell. 2012;10:137–147. doi: 10.1016/j.stem.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hütter G, Bodor J, Ledger S, Boyd M, Millington M, Tsie M, et al. CCR5 Targeted Cell Therapy for HIV and Prevention of Viral Escape. Viruses. 2015;7:4186–4203. doi: 10.3390/v7082816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass WG, McDermott DH, Lim JK, Lekhong S, Yu SF, Frank WA, et al. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J. Exp. Med. 2006;203:35–40. doi: 10.1084/jem.20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellier S, Frodsham AJ, Hennig BJW, Klenerman P, Knapp S, Ramaley P, et al. Association of genetic variants of the chemokine receptor CCR5 and its ligands, RANTES and MCP-2, with outcome of HCV infection. Hepatology. 2003;38:1468–1476. doi: 10.1016/j.hep.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 16.Promrat K, McDermott DH, Gonzalez CM, Kleiner DE, Koziol DE, Lessie M, et al. Associations of chemokine system polymorphisms with clinical outcomes and treatment responses of chronic hepatitis C. Gastroenterology. 2003;124:352–360. doi: 10.1053/gast.2003.50061. [DOI] [PubMed] [Google Scholar]
- 17.Wald O, Pappo O, Ari ZB, Azzaria E, Wiess ID, Gafnovitch I, et al. The CCR5Δ32 allele is associated with reduced liver inflammation in hepatitis C virus infection. Eur. J. Immunogenet. 2004;31:249–252. doi: 10.1111/j.1365-2370.2004.00482.x. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nat. Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 19.Sarver N, Cantin EM, Chang PS, Zaia JA, Ladne PA, Stephens DA, et al. Ribozymes as potential anti-HIV-1 therapeutic agents. Science. 1990;247:1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- 20.Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nat. Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cagnon L, Rossi JJ. Downregulation of the CCR5 β-Chemokine Receptor and Inhibition of HIV-1 Infection by Stable VA1-Ribozyme Chimeric Transcripts. Antisense Nucleic Acid Drug Dev. 2000;10:251–261. doi: 10.1089/108729000421439. [DOI] [PubMed] [Google Scholar]
- 22.Li M-J, Kim J, Li S, Zaia J, Yee J-K, Anderson J, et al. Long-Term Inhibition of HIV-1 Infection in Primary Hematopoietic Cells by Lentiviral Vector Delivery of a Triple Combination of Anti-HIV shRNA, Anti-CCR5 Ribozyme, and a Nucleolar-Localizing TAR Decoy. Mol. Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 23.Michienzi A, Li S, Zaia JA, Rossi JJ. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc. Natl. Acad. Sci. 2002;99:14047–14052. doi: 10.1073/pnas.212229599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M-J, Bauer G, Michienzi A, Yee J-K, Lee N-S, Kim J, et al. Inhibition of HIV-1 Infection by Lentiviral Vectors Expressing pol III-promoted anti-HIV RNAs. Mol. Ther. 2003;8:196–206. doi: 10.1016/s1525-0016(03)00165-5. [DOI] [PubMed] [Google Scholar]
- 25.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 26.Qin X-F, An DS, Chen ISY, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An DS, Qin FX-F, Auyeung VC, Mao SH, Kung SKP, Baltimore D, et al. Optimization and Functional Effects of Stable Short Hairpin RNA Expression in Primary Human Lymphocytes via Lentiviral Vectors. Mol. Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An DS, Donahue RE, Kamata M, Poon B, Metzger M, Mao S-H, et al. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc. Natl. Acad. Sci. 2007;104:13110–13115. doi: 10.1073/pnas.0705474104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang M, Kamata M, Chen KN, Pariente N, An DS, Chen ISY. Inhibition of HIV-1 infection by a unique short hairpin RNA to chemokine receptor 5 delivered into macrophages through hematopoietic progenitor cell transduction. J. Gene Med. 2010;12:255–265. doi: 10.1002/jgm.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu S, Hong P, Arumugam B, Pokomo L, Boyer J, Koizumi N, et al. A highly efficient short hairpin RNA potently down-regulates CCR5 expression in systemic lymphoid organs in the hu-BLT mouse model. Blood. 2010;115:1534–1544. doi: 10.1182/blood-2009-04-215855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimizu S, Ringpis G-E, Marsden MD, Cortado RV, Wilhalme HM, Elashoff D, et al. RNAi-Mediated CCR5 Knockdown Provides HIV-1 Resistance to Memory T Cells in Humanized BLT Mice. Mol. Ther. — Nucleic Acids. 2015;4:e227. doi: 10.1038/mtna.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ringpis G-EE, Shimizu S, Arokium H, Camba-Colón J, Carroll MV, Cortado R, et al. Engineering HIV-1-Resistant T-Cells from Short-Hairpin RNA-Expressing Hematopoietic Stem/Progenitor Cells in Humanized BLT Mice. PLOS ONE. 2012;7:e53492. doi: 10.1371/journal.pone.0053492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolstein O, Boyd M, Millington M, Impey H, Boyer J, Howe A, et al. Preclinical safety and efficacy of an anti-HIV-1 lentiviral vector containing a short hairpin RNA to CCR5 and the C46 fusion inhibitor. Mol. Ther. — Methods Clin. Dev. 2014;1:11. doi: 10.1038/mtm.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mani M, Kandavelou K, Dy FJ, Durai S, Chandrasegaran S. Design, engineering, and characterization of zinc finger nucleases. Biochem. Biophys. Res. Commun. 2005;335:447–457. doi: 10.1016/j.bbrc.2005.07.089. [DOI] [PubMed] [Google Scholar]
- 35.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saydaminova K, Ye X, Wang H, Richter M, Ho M, Chen H, et al. Efficient genome editing in hematopoietic stem cells with helper-dependent Ad5/35 vectors expressing site-specific endonucleases under microRNA regulation. Mol. Ther. Methods Clin. Dev. 2015;1:14057. doi: 10.1038/mtm.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heyer W-D, Ehmsen KT, Liu J. Regulation of Homologous Recombination in Eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Exline CM, DeClercq JJ, Llewellyn GN, Hayward SB, Li PW-L, et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015 doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mock U, Machowicz R, Hauber I, Horn S, Abramowski P, Berdien B, et al. mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res. 2015;43:5560–5571. doi: 10.1093/nar/gkv469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 45.Mandal PK, Ferreira LMR, Collins R, Meissner TB, Boutwell CL, Friesen M, et al. Efficient Ablation of Genes in Human Hematopoietic Stem and Effector Cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 50.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson J, Banerjea A, Planelles V, Akkina R. Potent Suppression of HIV Type 1 Infection By a Short Hairpin Anti-CXCR4 siRNA. AIDS Res. Hum. Retroviruses. 2003;19:699–706. doi: 10.1089/088922203322280928. [DOI] [PubMed] [Google Scholar]
- 53.Yuan J, Wang J, Crain K, Fearns C, Kim KA, Hua KL, et al. Zinc-finger Nuclease Editing of Human cxcr4 Promotes HIV-1 CD4+ T Cell Resistance and Enrichment. Mol. Ther. 2012;20:849–859. doi: 10.1038/mt.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou P, Chen S, Wang S, Yu X, Chen Y, Jiang M, et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci. Rep. 2015;5:15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tu TC, Nagano M, Yamashita T, Hamada H, Ohneda K, Kimura K, et al. A Chemokine Receptor, CXCR4, Which Is Regulated by Hypoxia-Inducible Factor 2α, Is Crucial for Functional EPC Migration to Ischemic Tissue and Wound Repair. Stem Cells Dev. 2015 doi: 10.1089/scd.2015.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lapidot T. Mechanism of human stem cell migration and repopulation of NOD/SCID and B2mnull NOD/SCID mice. The role of SDF-1/CXCR4 interactions. Ann. N. Y. Acad. Sci. 2001;938:83–95. doi: 10.1111/j.1749-6632.2001.tb03577.x. [DOI] [PubMed] [Google Scholar]
- 58.Hildinger M, Dittmar MT, Schult-Dietrich P, Fehse B, Schnierle BS, Thaler S, et al. Membrane-Anchored Peptide Inhibits Human Immunodeficiency Virus Entry. J. Virol. 2001;75:3038–3042. doi: 10.1128/JVI.75.6.3038-3042.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zahn RC, Hermann FG, Kim E-Y, Rett MD, Wolinsky SM, Johnson RP, et al. Efficient Entry Inhibition of Human and Non-Human Primate Immunodeficiency Virus by Cell Surface-Expressed gp41-Derived Peptides. Gene Ther. 2008;15:1210–1222. doi: 10.1038/gt.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burke BP, Levin BR, Zhang J, Sahakyan A, Boyer J, Carroll MV, et al. Engineering Cellular Resistance to HIV-1 Infection In Vivo Using a Dual Therapeutic Lentiviral Vector. Mol. Ther. — Nucleic Acids. 2015;4:e236. doi: 10.1038/mtna.2015.10. [DOI] [PubMed] [Google Scholar]
- 61.Trobridge GD, Wu RA, Beard BC, Chiu SY, Muñoz NM, von Laer D, et al. Protection of Stem Cell-Derived Lymphocytes in a Primate AIDS Gene Therapy Model after In Vivo Selection. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0007693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Lunzen J, Glaunsinger T, Stahmer I, von Baehr V, Baum C, Schilz A, et al. Transfer of Autologous Gene-modified T Cells in HIV-infected Patients with Advanced Immunodeficiency and Drug-resistant Virus. Mol. Ther. 2007;15:1024–1033. doi: 10.1038/mt.sj.6300124. [DOI] [PubMed] [Google Scholar]
- 63.Hermann FG, Egerer L, Brauer F, Gerum C, Schwalbe H, Dietrich U, et al. Mutations in gp120 Contribute to the Resistance of Human Immunodeficiency Virus Type 1 to Membrane-Anchored C-Peptide maC46. J. Virol. 2009;83:4844–4853. doi: 10.1128/JVI.00666-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan E, Towers GJ, Qasim W. Gene Therapy Strategies to Exploit TRIM Derived Restriction Factors against HIV-1. Viruses. 2014;6:243–263. doi: 10.3390/v6010243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neagu MR, Ziegler P, Pertel T, Strambio-De-Castillia C, Grütter C, Martinetti G, et al. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J. Clin. Invest. 2009;119:3035–3047. doi: 10.1172/JCI39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luban J. Cyclophilin A, TRIM5, and Resistance to Human Immunodeficiency Virus Type 1 Infection. J. Virol. 2007;81:1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. CB. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 68.Jung U, Urak K, Veillette M, Nepveu-Traversy M-É, Pham QT, Hamel S, et al. Preclinical Assessment of Mutant Human TRIM5α as an Anti-HIV-1 Transgene. Hum. Gene Ther. 2015;26:664–679. doi: 10.1089/hum.2015.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakkhachornphop S, Barbas CF, Keawvichit R, Wongworapat K, Tayapiwatana C. Zinc Finger Protein Designed to Target 2-Long Terminal Repeat Junctions Interferes with Human Immunodeficiency Virus Integration. Hum. Gene Ther. 2012;23:932–942. doi: 10.1089/hum.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 Proviral DNA Excision Using an Evolved Recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 71.Karpinski J, Hauber I, Chemnitz J, Schäfer C, Paszkowski-Rogacz M, Chakraborty D, et al. Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity. Nat. Biotechnol. 2016;34:401–409. doi: 10.1038/nbt.3467. [DOI] [PubMed] [Google Scholar]
- 72.Qu X, Wang P, Ding D, Li L, Wang H, Ma L, et al. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 2013;41:7771–7782. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ebina H, Kanemura Y, Misawa N, Sakuma T, Kobayashi T, Yamamoto T, et al. A High Excision Potential of TALENs for Integrated DNA of HIV-Based Lentiviral Vector. PLOS ONE. 2015;10:e0120047. doi: 10.1371/journal.pone.0120047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang G, Zhao N, Berkhout B, Das AT. CRISPR-Cas9 Can Inhibit HIV-1 Replication but NHEJ Repair Facilitates Virus Escape. Mol. Ther. 2016;24:522–526. doi: 10.1038/mt.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Yin C, Zhang T, Li F, Yang W, Kaminski R, et al. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci. Rep. 2015;5 doi: 10.1038/srep16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saayman SM, Lazar DC, Scott TA, Hart JR, Takahashi M, Burnett JC, et al. Potent and Targeted Activation of Latent HIV-1 Using the CRISPR/dCas9 Activator Complex. Mol. Ther. 2016;24:488–498. doi: 10.1038/mt.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Limsirichai P, Gaj T, Schaffer DV. CRISPR-mediated Activation of Latent HIV-1 Expression. Mol. Ther. 2016;24:499–507. doi: 10.1038/mt.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee M-TM, Coburn GA, McClure MO, Cullen BR. Inhibition of Human Immunodeficiency Virus Type 1 Replication in Primary Macrophages by Using Tat- or CCR5-Specific Small Interfering RNAs Expressed from a Lentivirus Vector. J. Virol. 2003;77:11964. doi: 10.1128/JVI.77.22.11964-11972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strong CL, Guerra HP, Mathew KR, Roy N, Simpson LR, Schiller MR. Damaging the Integrated HIV Proviral DNA with TALENs. PLOS ONE. 2015;10:e0125652. doi: 10.1371/journal.pone.0125652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meredith LW, Sivakumaran H, Major L, Suhrbier A, Harrich D. Potent Inhibition of HIV-1 Replication by a Tat Mutant. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0007769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA Interference: A Multiple shRNA Approach. Mol. Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 83.ter ter Brake O, ’t Hooft K, Liu YP, Centlivre M, Jasmijn von Eije K, Berkhout B. Lentiviral Vector Design for Multiple shRNA Expression and Durable HIV-1 Inhibition. Mol. Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 84.Centlivre M, Legrand N, Klamer S, Liu YP, von Eije KJ, Bohne M, et al. Preclinical In Vivo Evaluation of the Safety of a Multi-shRNA-based Gene Therapy Against HIV-1. Mol. Ther. — Nucleic Acids. 2013;2:e120. doi: 10.1038/mtna.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Herrera-Carrillo E, Berkhout B. The impact of HIV-1 genetic diversity on the efficacy of a combinatorial RNAi-based gene therapy. Gene Ther. 2015;22:485–495. doi: 10.1038/gt.2015.11. [DOI] [PubMed] [Google Scholar]
- 86.Anderson J, Li M-J, Palmer B, Remling L, Li S, Yam P, et al. Safety and Efficacy of a Lentiviral Vector Containing Three Anti-HIV Genes—CCR5 Ribozyme, Tat-rev siRNA, and TAR Decoy—in SCID-hu Mouse–Derived T Cells. Mol. Ther. 2007;15:1182–1188. doi: 10.1038/sj.mt.6300157. [DOI] [PubMed] [Google Scholar]
- 87.DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walker JE, Chen RX, McGee J, Nacey C, Pollard RB, Abedi M, et al. Generation of an HIV-1-Resistant Immune System with CD34+ Hematopoietic Stem Cells Transduced with a Triple-Combination Anti-HIV Lentiviral Vector. J. Virol. 2012;86:5719–5729. doi: 10.1128/JVI.06300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Didigu CA, Wilen CB, Wang J, Duong J, Secreto AJ, Danet-Desnoyers GA, et al. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood. 2014;123:61. doi: 10.1182/blood-2013-08-521229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lanao JM, Briones E, Colino CI. Recent advances in delivery systems for anti-HIV1 therapy. J. Drug Target. 2007;15:21–36. doi: 10.1080/10611860600942178. [DOI] [PubMed] [Google Scholar]
- 91.Miyoshi H, Smith KA, Mosier DE, Verma IM, Torbett BE. Transduction of Human CD34+ Cells That Mediate Long-Term Engraftment of NOD/SCID Mice by HIV Vectors. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]
- 92.Reiser J, Harmison G, Kluepfel-Stahl S, Brady RO, Karlsson S, Schubert M. Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park SW, Pyo C-W, Choi S-Y. High-efficiency lentiviral transduction of primary human CD34+ hematopoietic cells with low-dose viral inocula. Biotechnol. Lett. 2014;37:281–288. doi: 10.1007/s10529-014-1678-z. [DOI] [PubMed] [Google Scholar]
- 94.Mochizuki H, Schwartz JP, Tanaka K, Brady RO, Reiser J. High-Titer Human Immunodeficiency Virus Type 1-Based Vector Systems for Gene Delivery into Nondividing Cells. J. Virol. 1998;72:8873. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 96.Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, et al. Lentivirus-based Gene Therapy of Hematopoietic Stem Cells in Wiskott-Aldrich Syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 98.Tolar J, Becker PS, Clapp DW, Hanenberg H, de Heredia CD, Kiem H-P, et al. Gene Therapy for Fanconi Anemia: One Step Closer to the Clinic. Hum. Gene Ther. 2012;23:141–144. doi: 10.1089/hum.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Myburgh R, Ivic S, Pepper MS, Gers-Huber G, Li D, Audigé A, et al. Lentivector Knockdown of CCR5 in Hematopoietic Stem and Progenitor Cells Confers Functional and Persistent HIV-1 Resistance in Humanized Mice. J. Virol. 2015;89:6761–6772. doi: 10.1128/JVI.00277-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shimizu S, Yadav SS, An DS. Stable Delivery of CCR5-Directed shRNA into Human Primary Peripheral Blood Mononuclear Cells and Hematopoietic Stem/Progenitor Cells via a Lentiviral Vector. In: Shum K, Rossi J, editors. SiRNA Deliv. Methods. New York, NY: Springer New York; 2016. [accessed November 12, 2015]. pp. 235–248. http://link.springer.com/10.1007/978-1-4939-3112-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kong LB, Siva AC, Rome LH, Stewart PL. Structure of the vault, a ubiquitous celular component. Structure. 1999;7:371–379. doi: 10.1016/s0969-2126(99)80050-1. [DOI] [PubMed] [Google Scholar]
- 102.Champion CI, Kickhoefer VA, Liu G, Moniz RJ, Freed AS, Bergmann LL, et al. A Vault Nanoparticle Vaccine Induces Protective Mucosal Immunity. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0005409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kar UK, Srivastava MK, Andersson Å, Baratelli F, Huang M, Kickhoefer VA, et al. Novel CCL21-Vault Nanocapsule Intratumoral Delivery Inhibits Lung Cancer Growth. PLOS ONE. 2011;6:e18758. doi: 10.1371/journal.pone.0018758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shin J-Y, Suh D, Kim JM, Choi H-G, Kim JA, Ko JJ, et al. Low molecular weight polyethylenimine for efficient transfection of human hematopoietic and umbilical cord blood-derived CD34+ cells. Biochim. Biophys. Acta BBA - Gen. Subj. 2005;1725:377–384. doi: 10.1016/j.bbagen.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 105.Shayakhmetov DM, Papayannopoulou T, Stamatoyannopoulos G, Lieber A. Efficient Gene Transfer into Human CD34+ Cells by a Retargeted Adenovirus Vector. J. Virol. 2000;74:2567–2583. doi: 10.1128/jvi.74.6.2567-2583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li L, Krymskaya L, Wang J, Henley J, Rao A, Cao L-F, et al. Genomic Editing of the HIV-1 Coreceptor CCR5 in Adult Hematopoietic Stem and Progenitor Cells Using Zinc Finger Nucleases. Mol. Ther. 2013;21:1259–1269. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smith LJ, Ul-Hasan T, Carvaines SK, Van Vliet K, Yang E, Wong KK, et al. Gene Transfer Properties and Structural Modeling of Human Stem Cell-derived AAV. Mol. Ther. 2014;22:1625–1634. doi: 10.1038/mt.2014.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hofer U, Henley JE, Exline CM, Mulhern O, Lopez E, Cannon PM. Pre-clinical Modeling of CCR5 Knockout in Human Hematopoietic Stem Cells by Zinc Finger Nucleases Using Humanized Mice. J. Infect. Dis. 2013;208:S160–S164. doi: 10.1093/infdis/jit382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kalomoiris S, Lawson J, Chen RX, Bauer G, Nolta JA, Anderson JS. CD25 Preselective Anti-HIV Vectors for Improved HIV Gene Therapy. Hum. Gene Ther. Methods. 2012;23:366–375. doi: 10.1089/hgtb.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barclay SL, Yang Y, Zhang S, Fong R, Barraza A, Nolta JA, et al. Safety and Efficacy of a tCD25 Preselective Combination Anti-HIV Lentiviral Vector in Human Hematopoietic Stem and Progenitor Cells. STEM CELLS. 2015;33:870–879. doi: 10.1002/stem.1919. [DOI] [PubMed] [Google Scholar]
- 111.Melkus MW, Estes JD, Padgett-Thomas A, Gatlin J, Denton PW, Othieno FA, et al. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat. Med. 2006;12:1316–1322. doi: 10.1038/nm1431. [DOI] [PubMed] [Google Scholar]
- 112.Zhen A, Kamata M, Rezek V, Rick J, Levin B, Kasparian S, et al. HIV-specific Immunity Derived From Chimeric Antigen Receptor-engineered Stem Cells. Mol. Ther. 2015;23:1358–1367. doi: 10.1038/mt.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim S, Kim N, Presson AP, Metzger ME, Bonifacino AC, Sehl M, et al. Dynamics of HSPC Repopulation in Nonhuman Primates Revealed by a Decade-Long Clonal-Tracking Study. Cell Stem Cell. 2014;14:473–485. doi: 10.1016/j.stem.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Younan PM, Polacino P, Kowalski JP, Peterson CW, Maurice NJ, Williams NP, et al. Positive selection of mC46-expressing CD4+ T cells and maintenance of virus specific immunity in a primate AIDS model. Blood. 2013;122:179–187. doi: 10.1182/blood-2013-01-482224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Younan PM, Peterson CW, Polacino P, Kowalski JP, Obenza W, Miller HW, et al. Lentivirus-mediated Gene Transfer in Hematopoietic Stem Cells Is Impaired in SHIV-infected, ART-treated Nonhuman Primates. Mol. Ther. J. Am. Soc. Gene Ther. 2015;23:943–951. doi: 10.1038/mt.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bauer G, Selander D, Engel B, Carbonaro D, Csik S, Rawlings S, et al. Gene Therapy for Pediatric AIDS. Ann. N. Y. Acad. Sci. 2000;918:318–329. doi: 10.1111/j.1749-6632.2000.tb05501.x. [DOI] [PubMed] [Google Scholar]
- 117.Mitsuyasu RT, Merigan TC, Carr A, Zack JA, Winters MA, Workman C, et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat. Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene Editing of CCR5 in Autologous CD4 T Cells of Persons Infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kohn DB, Bauer G, Rice CR, Rothschild JC, Carbonaro DA, Valdez P, et al. A Clinical Trial of Retroviral-Mediated Transfer of arev-Responsive Element Decoy Gene Into CD34+ Cells From the Bone Marrow of Human Immunodeficiency Virus-1–Infected Children. Blood. 1999;94:368–371. [PubMed] [Google Scholar]