Abstract
Rheumatoid arthritis (RA) is a chronic autoimmune inflammatory disease that leads to destructive arthritis. Although the HLA class II locus is the strongest genetic risk factor for rheumatoid arthritis, the relationship between HLA class II alleles and lymphocyte activation remains unclear. We performed immunophenotyping of peripheral blood mononuclear cells on 91 HLA-DRB1-genotyped RA patients and 110 healthy donors. The frequency of memory CXCR4+CD4+ T cells, and not Th1 and Th17 cells, was significantly associated with disease severity by multiple linear regression analysis. RA patients with one or more susceptible HLA-DR haplotypes (shared epitope: SE) displayed a significantly higher frequency of memory CXCR4+CD4+ T cells. Moreover, the frequency of memory CXCR4+CD4+ T cells significantly correlated with the expression level of HLA-DR on B cells, which was elevated in RA patients with SE. In vitro analysis and transcriptomic pathway analysis suggested that the interaction between HLA-DR and T cell receptors is an important regulator of memory CXCR4+CD4+ T cells. Clinically, a higher frequency of memory CXCR4+CD4+ T cells predicted a better response to CTLA4-Ig. Memory CXCR4+CD4+ T cells may serve as a powerful biomarker for unraveling the linkage between HLA-DRB1 genotype and disease activity in RA.
Rheumatoid arthritis (RA) is a chronic autoimmune inflammatory disease that leads to destructive arthritis. Both genetic and environmental factors contribute to RA pathogenesis1. A recent meta-analysis of genome-wide association studies identified as many as 101 RA risk loci2. In particular, the HLA-DRB1 genotype was the first identified and by far the strongest genetic risk factor for RA3,4. The shared epitope (SE), a common amino acid sequence at positions 70–74 of HLA-DRB1, is recognized for its association with anti-cyclic citrullinated peptide antibody (ACPA)-positive RA5. It is thought that citrullinated autoantigen epitopes bind to HLA-DRB1 that contain the SE and are presented to CD4+ T cells, which contribute to autoimmunity6. Moreover, SE is an important risk factor for severe bone destructive disease5,7.
Nevertheless, in spite of tremendous efforts to identify immunological abnormalities in RA, few studies have identified any linkage between SE and adaptive immunity. To understand the immunological role of SE, immune cell populations associated with SE should be identified. The key role of CD4+ T cells in RA pathogenesis is highlighted by the fact that RA genetic risk loci preferentially map to enhancers and promoters active in CD4+ T cell subsets8. Standardized human immunophenotyping has been proposed for classifying CD4+ T cells into conventional Th1, Th2, and Th17 cell types based on their expression of the chemokine receptors CXCR3 and CCR69. Although a number of researchers have examined the frequency of Th1, Th2, Th17, Tfh, and Treg cells in RA, these populations show no clear association with RA disease activity measures, such as Disease Activity Score 28 joints-ESR (DAS28esr) and Health Assessment Questionnaire Disability Index (HAQ)10,11,12,13. Therefore, other markers for CD4+ T cells need to be investigated.
In the RA synovium, there are ectopic lymphoid follicles as well as clonally expanded T cells and antigen-specific B cells that recognize citrullinated autoantigens14,15. These findings strongly suggest that acquired immunity against autoantigens promotes local inflammation in the synovium, such as macrophage activation and inflammatory cytokine production, including TNF-α and IL-6. The chemokine receptor CXCR4 plays a central role in the homing and retention of CD4+ T cells16,17. The CXCR4 ligand CXCL12 (also known as SDF-1) and the recently identified ligand macrophage migration inhibitory factor (MIF) are both produced by synovial fibroblasts and are increased in RA synovium18,19,20. It has also been reported that inflammatory cytokine-activated CD4+ T cells express high levels of CXCR421 and that T-cell-specific CXCR4-deficient mice show a dramatic decrease in the incidence of arthritis22.
Based on these preceding reports, we attempted to identify lymphocyte subsets that are associated with HLA-DRB1 and RA disease activity. We analyzed HLA-DRB1-genotyped RA patients by 24-subset immunophenotyping combined with CXCR4 expression, HLA-DR quantification on antigen-presenting cells, and multiplex serum cytokine analysis.
Results
Study populations
We recruited 91 RA patients and 110 healthy donors (HD) (Table S1). 61 RA patients with at least one HLA-DRB1 SE allele were considered to be SE-positive RA (SE + RA). Among the SE + RA group, 44 patients (72%) had at least one HLA-DRB1 04:05 allele, 14 patients (23%) had at least one 01:01 allele, and 6 patients (10%) had the 04:01 allele. The SE + RA and SE-negative RA (SE-RA) groups showed comparable baseline characteristics, including rheumatoid factor (RF) titer, DAS28esr disease activity score, and HAQ functional disability index. ACPA titer was significantly higher in the SE + RA group compared to the SE-RA group, as reported5.
Memory CD4+ T cells are associated with ACPA and SE positivity in RA
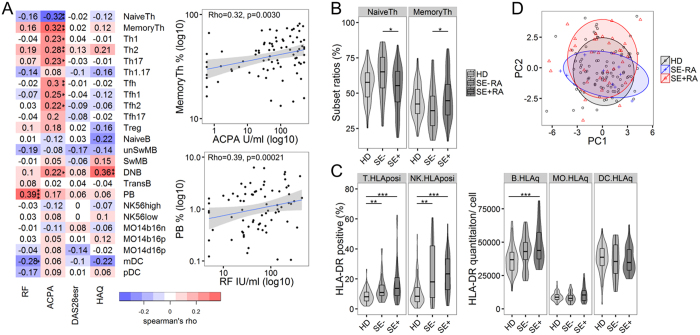
We performed flow cytometric 24-subset immunophenotyping on freshly isolated PBMC in order to assess global immunological changes in RA patients (Table S2, Figure S1). We compared different cell subset frequencies with clinical parameters (RF, ACPA, DAS28esr, and HAQ) in order to identify cell subsets that are associated with clinical sequelae. The percentage of conventional CD4+ T cell subsets, including Th1 and Th17 cells, showed no association with disease activity and HAQ, although the percentage of memory CD4+ T cells (MemoryTh) and ACPA displayed a weak correlation (Fig. 1A). Notably, the MemoryTh ratio was significantly increased in SE + RA (Fig. 1B). These results suggest that memory CD4+ T cells are associated with the production of ACPA and SE positivity. With regard to B cells, the percentages of plasmablasts (PB) correlated with RF titer (Fig. 1A).
Figure 1. Memory CD4+ T cells are associated with ACPA and SE positivity in RA.
(A) Heatmap of correlation matrix between 24 immunophenotyped subsets and four clinical parameters: RF, ACPA, DAS28esr and HAQ in RA patients (n = 91). Representative scatter plots and linear regression line with 95% confidence intervals are shown on the right. See Table S2 for subset definitions. (B,C) Comparison of NaiveTh and MemoryTh subset ratios and HLA-DR expression on T cells, NK cells, B cells, monocytes (MO), and dendritic cells (DC) between healthy donors (HD), shared epitope (SE)-negative RA patients, and SE-positive RA patients (HD; n = 110, SE-RA; n = 30, SE + RA; n = 61). For T cells and NK cells analyses, HLA-DR-positive ratios are used. B cells, MO, and DC analyses are based on flow cytometric HLA-DR quantitative expression per cells. (D) Principal component analysis of 24 immunophenotyped subsets to summarize the differences between HD, SE-RA, and SE + RA. PC1 explained 24% and PC2 explained 14% of the total variance (HD; n = 93, SE-RA; n = 10, SE + RA; n = 35). *p < 0.05 **p < 0.01 ***P < 0.001 Spearman’s tests (A) or Kruskal-Wallis tests with post-hoc Wilcoxon tests with Bonferroni corrections (B,C).
Comparisons between HD and RA revealed immunological abnormalities in all of the major cell populations studied: CD4+ T cells, B cells, NK cells, monocytes, and DC (Figure S2). The increased frequency of HLA-DR-positive T cells and NK cells indicates that these populations are activated in RA patients (Fig. 1C). Furthermore, B cells from SE + RA patients had significantly higher expression of HLA-DR compared to B cells from HD, while monocytes and dendritic cells exhibited similar HLA-DR levels (Fig. 1C). There was no significant difference between SE-HD and SE + HD in HLA-DR expression on B cells (Figure S3). Increased HLA class II and HLA-DR molecules on RA B cells have been reported23,24. The increased HLA-DR expression on SE + RA B cells suggests that B cells may be important for antigen presentation in RA. However, principal component analysis (PCA) of our immunophenotyping data revealed global similarity between HD, SE-RA, and SE + RA patients (Fig. 1D), indicating that conventional immunophenotyping is not specific enough to distinguish between them.
RF and inflammatory cytokines
Correlation analysis of serum cytokine levels showed that RF titer strongly correlated with serum inflammatory cytokines, such as IL-1β, IL-21, IFN-γ, and IL-17A (Figure S4A). Soluble gp130 (sgp130), which neutralizes IL-6 and soluble IL-6 receptor (sIL-6R) complex25, showed a strong negative correlation with RF (Figure S4A). Serum cytokines were not correlated with DAS28esr. Among the cytokines tested, only IL-10 displayed a moderate correlation for HAQ (Rho = 0.44, p = 0.027). The concentrations of multiple inflammatory cytokines were increased in RA, revealing a clear global difference between HD and RA (Figure S4B,C). These data indicate that the inflammatory cytokine environment in RA correlated best with RF, and not ACPA and DAS28esr.
CXCR4 expression is increased on SE + RA memory CD4+ T cells
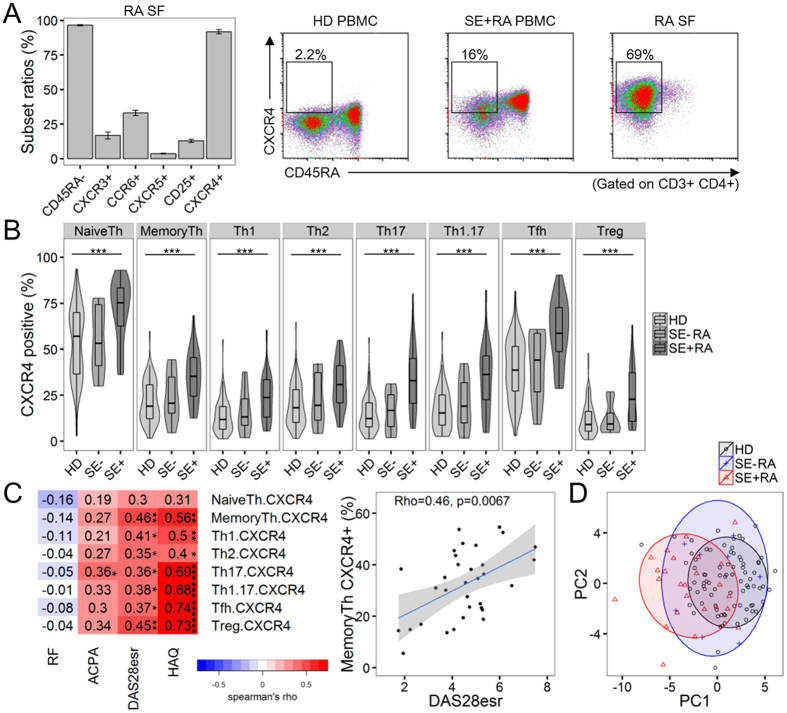
Immunophenotyping of conventional lymphocyte populations and cytokine measurements did not reveal any factors significantly associated with RA disease activity. In accordance with this observation, the expression of Th1, Th17, Tfh, and Treg markers on synovial CD4+ T cells is relatively limited (Fig. 2A). In contrast, most synovial CD4+ T cells expressed CXCR4, as previously reported16. Although only a limited fraction of memory CD4+ T cells expressed CXCR4 in HD, both memory and naive CD4+PBMC T cells showed significantly enhanced CXCR4 expression, especially in SE + RA patients (Fig. 2A,B). There was no significant difference between SE-HD and SE + HD in CXCR4 expression on memory CD4+ T cells (Figure S5). Additionally, enhanced CXCR4 expression was observed in RA patients with high ACPA titers, which also corresponded to SE + RA (Figure S6). CXCR4 expression on various memory CD4+ T cell subsets, such as Tfh or Treg cells, showed moderate to strong correlations with DAS28esr and HAQ (Fig. 2C). In particular, CXCR4 expression on memory CD4+ T cells (MemoryTh), which includes the Th1, Th17, Tfh, and Treg subsets, was associated with DAS28esr in a multiple linear regression model that included CXCR4 expression on various CD4+ T cell subsets (Table S3). Also, CXCR4 expression on memory CD4+ T cells, not the frequency of conventional CD4+ T cell subsets, was associated with DAS28esr by multiple linear regression analysis (Table 1). Similar results were obtained with LASSO regression, a penalized regression model. Although steroid exposure has been reported to be associated with CXCR4 expression26, CXCR4 expression on memory CD4+ T cells was not significantly correlated with treatment drug (PSL or MTX) dose levels in RA. It is possible that treatment drugs might have some effects on CXCR4 expression on CD4+ T cells. The enhanced CXCR4 expression on various helper T cell subsets suggested the presence of generalized immunological modifications on memory CD4+ T cells in RA. When CXCR4 expression on CD4+ T cell subsets was combined with conventional immunophenotyping, we were able to detect a significant difference between HD and SE + RA by PCA, indicating that CXCR4 expression on CD4+ T cells is a key feature discriminating HD and SE + RA (Figs 1D and 2D).
Figure 2. CXCR4 expression on memory CD4+ T cells is correlated with disease activity.
(A) CXCR4 expression on synovial fluid (SF) and PBMC CD4+ T cells. Left: subset ratios of memory (CD45RA-), Th1 (CXCR3), Th17 (CCR6), Tfh (CXCR5), Treg (CD25) and CXCR4 + RA synovial fluid CD4+ T cells (n = 5). Mean ± SEM values. Right: representative plots of CXCR4 expression in PBMC and synovial fluid (SF) samples from healthy donors (HD) and RA patients. The CD45RA−CXCR4+ ratios are shown in the plots. The CXCR4 positivity of the CD45RA- subset (MemoryTh subset) is HD 7.8%, SE + RA 36%, and SE + RA 87% in the representative plots. (B) Comparison of CXCR4 expression on CD4+ T cell subsets among healthy donors (HD), shared epitope (SE)-negative RA patients, and SE-positive RA patients (HD; n = 82, SE-RA; n = 8, SE + RA; n = 27). (C) Correlations between CXCR4 expression on CD4+ T cell subsets and clinical parameters in RA patients (n = 35). A Representative scatter plot is shown on the right, as in Figure 1A. Tfh1, Tfh2, and Tfh17 subpopulations were eliminated from the analysis (B,C). (D) Principal component analysis of immunophenotyped subsets and CXCR4 expression on CD4+ T cell subsets to summarize the differences between HD, SE-RA, and SE + RA. PC1 explained 29% and PC2 explained 16% of the total variance (HD; n = 81, SE-RA; n = 8, SE + RA; n = 25). *p < 0.05 **p < 0.01 ***P < 0.001 Spearman’s test (C) or Kruskal-Wallis tests with post-hoc Wilcoxon tests with Bonferroni corrections (B).
Table 1. Univariate and multivariate relationships between CD4+ T cell subsets and DAS28esr.
| variable | Univariate regression |
Multivariate regression |
||
|---|---|---|---|---|
| β | P value | β | P value | |
| age | 0.19 | 0.08 | Not selected | Not selected |
| sex | −0.033 | 0.76 | Not selected | Not selected |
| disease duration | −0.0087 | 0.94 | −0.24 | 0.14 |
| Shared epitope positivity | 0.028 | 0.79 | Not selected | Not selected |
| RF titer | 0.11 | 0.32 | Not selected | Not selected |
| ACPA titer | 0.25 | 0.022* | 0.17 | 0.32 |
| Th1 | 0.08 | 0.47 | Not selected | Not selected |
| Th2 | 0.2 | 0.07 | Not selected | Not selected |
| Th17 | −0.0072 | 0.94 | −0.24 | 0.15 |
| Tfh | 0.061 | 0.58 | Not selected | Not selected |
| Treg | 0.043 | 0.71 | Not selected | Not selected |
| MemoryTh.CXCR4 | 0.52 | 0.0018** | 0.54 | 0.0023** |
RA n = 32. *p < 0.05, **p < 0.01.
RF, Rheumatoid Factor; ACPA, anti-cyclic citrullinated peptide antibody; DAS28esr, Disease Activity Score 28 joints-ESR.
CXCR4 is widely expressed on multiple immune cells, such as B cells, monocytes, and neutrophils27. In addition to CD4+ T cells, we examined CXCR4 expression on B cell subsets. Of the B cell subsets, only unswitched memory B cells from SE + RA had enhanced CXCR4 expression (Figure S7). Further research is needed to examine the role of SE in CXCR4 expression on innate immune cells. We also analyzed the expression of CCR7, a marker of central memory CD4+ T cells9. There were no significant differences in CCR7 expression on CD4+ T cells between HD, SE-RA, and SE + RA (Figure S8). These results also highlight the significance of enhanced CXCR4 expression on SE + RA CD4+ T cells.
CXCR4 expression is regulated by cytokines and HLA-DR
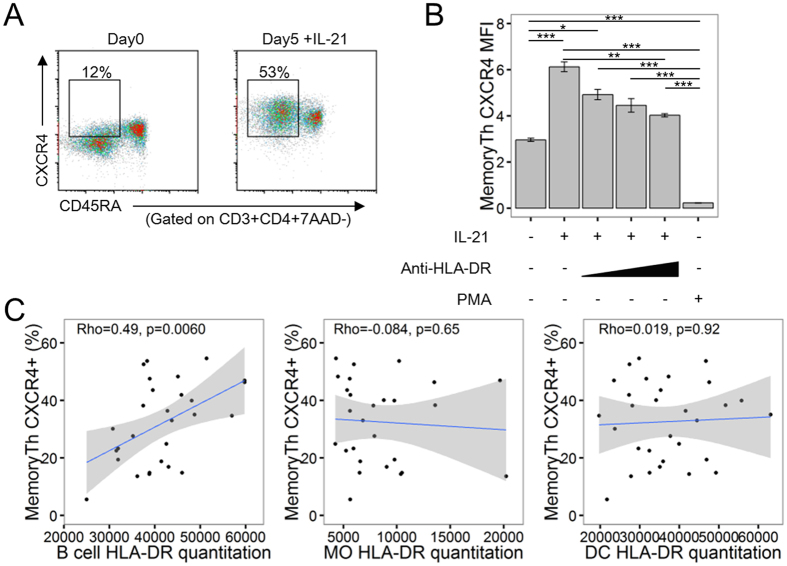
Previous reports have demonstrated that CXCR4 expression on CD4+ T cells is regulated by common gamma chain family cytokines, such as IL-2, IL-4, IL-7, and IL-1528. IL-21 is also a member of the common gamma chain family cytokines and was elevated in the serum of RA patients (Figure S4B). It has been reported that IL-21 is an important cytokine in CD4+ T cell-B cell interactions and is one of the susceptibility genes for RA2,29. Five-day culture of PBMC with IL-21 increased the expression of CXCR4 on memory CD4+ T cells (Fig. 3A,B)30. Furthermore, addition of anti-HLA-DR antibody blocked this increase, suggesting that HLA-DR is directly involved in the elevation of CXCR4 expression (Fig. 3B). Meanwhile, strong TCR signaling induced by PMA decreased CXCR4 expression, consistent with reports that strong TCR stimulation leads to CXCR4 internalization31.
Figure 3. CXCR4 expression on memory CD4+ T cells is regulated by IL-21 and HLA-DR on B cells.
(A,B) HD PBMCs were cultured for 5 days under the indicated conditions. (A) Representative plots of CXCR4 expression from Day 0 and Day 5 PBMC. (B) Day 5 CXCR4 mean fluorescence intensity (MFI) on memory CD4+ T cell subsets in each condition. Mean ± SEM values. Results are representative of two independent experiments. n = 4 per group. (C) Scatter plots of B cell, monocyte (MO), and dendritic cell (DC) HLA-DR quantitative expression value per cells and CXCR4-positive ratios of the MemoryTh subset in RA patients (n = 35). *p < 0.05 **p < 0.01 ***P < 0.001 Unpaired t-tests with Bonferroni corrections.
To elucidate the linkage between antigen-presenting cells and SE + RA memory CD4+ T cells with increased CXCR4 expression, we examined the association between CXCR4 expression on memory CD4+ T cells and HLA-DR expression on antigen-presenting cells. In accordance with the observation that B cells from SE + RA patients had significantly higher HLA-DR expression (Fig. 1C), there was a significant correlation between CXCR4 expression on memory CD4+ T cells and the amount of HLA-DR expressed on B cells (Fig. 3C), while HLA-DR expressed on monocytes and dendritic cells was not significantly correlated. This correlation between CXCR4 expression on memory CD4+ T cells and the amount of HLA-DR expressed on B cells may be specific to RA because it was not observed in HD (Figure S9). These results suggest that the increased CXCR4 expression on memory CD4+ T cells in RA patients is regulated both by inflammatory cytokines and HLA-DR expression on B cells.
Transcriptome analysis of CXCR4-expressing memory CD4+ T cells
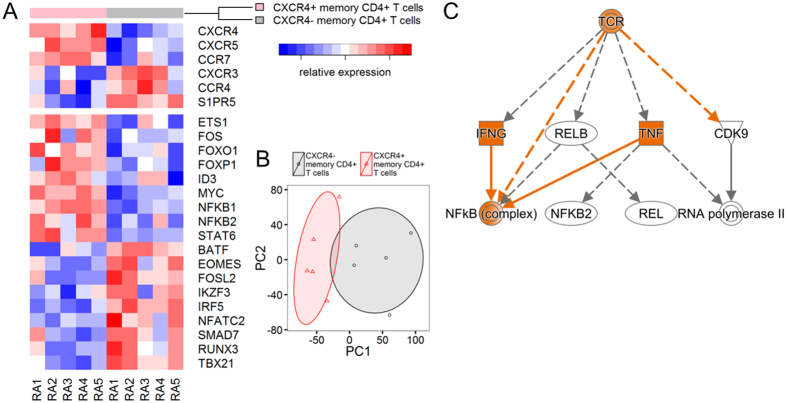
We compared the transcriptome of CXCR4+ memory CD4+ T cells of RA patients with CXCR4− memory CD4+ T cells by RNA-seq. Chemokine receptors, such as CXCR4, CXCR5, and CCR7, and transcription factors, such as MYC, NFKB1, and NFKB2, were differentially upregulated in the CXCR4+ population (Fig. 4A). Comparison of the entire transcriptome of CXCR4+ and CXCR4− memory CD4+ T cells revealed clear global differences (Fig. 4B), suggesting that CXCR4-expressing memory CD4+ T cells are a distinct subset in RA. Pathway analysis revealed that MYC-related protein synthesis and the mTOR pathway were activated in the CXCR4+ population (Figure S10), consistent with a previous report that showed that MYC controls metabolic reprogramming upon T cell activation32. Moreover, T cell receptor (TCR) signaling was the most significant upstream regulator of transcriptomic changes in the CXCR4+population (Fig. 4C and Figure S10D), in agreement with our observation that HLA-DR and CXCR4 expression on memory CD4+ T cells are associated (Fig. 3).
Figure 4. Transcriptome differences in CXCR4+ memory CD4+ T cells and CXCR4− memory CD4+ T cells from RA patients.
(A) Heatmap of selected receptors and transcription factors differentially expressed (FDR < 0.05) in CXCR4+ and CXCR4- memory CD4+ T cells from five RA patients with active disease (RA1-5). Red color represents the upregulation and blue color represents the downregulation of respective genes. (n = 5). (B) Principal component analysis of whole transcriptome to summarize differences between CXCR4+ and CXCR4− memory CD4+ T cells. PC1 explained 24% and PC2 explained 12% of the total variance. (n = 5). (C) The IPA mechanistic network identified the TCR as the most significant upstream regulator of CXCR4+ memory CD4+ T cells upregulated genes (p = 1.67E-08, Z-score = 1.256). Orange color represents more confident predicted activation.
CXCR4 expression predicts a better response to CTLA4-Ig in RA patients
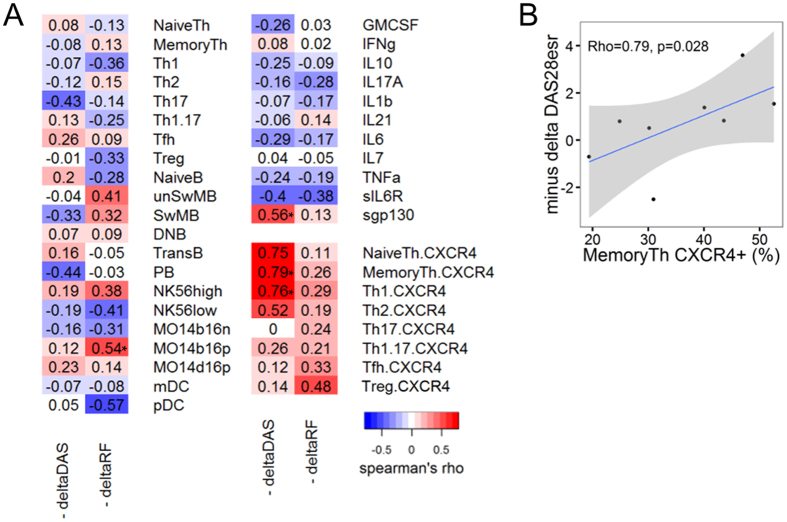
We next investigated the relationship between the therapeutic efficacy of biologics and immunological parameters examined above. Immunophenotyping and serum cytokine analysis were performed when CTLA4-Ig treatment was initiated and after 6 months of treatment in 20 RA patients. We also examined the clinical efficacy of CTLA4-Ig in RA patients, and found that it partially reverses immunological abnormalities in RA, specifically that CTLA4-Ig decreased the ratio of inflammatory CD14brightCD16+ monocytes and increased the ratio of mDCs (Figures S2 and S11)33,34,35. Decreased HLA-DR expression on T cells and decreased inflammatory cytokines were observed in parallel with a decrease in DAS28esr. CTLA4-Ig administration significantly reduced CXCR4 expression on naive CD4+ T cells (Figure S11D). Importantly, although CTLA4-Ig treatment did not decrease HLA-DR expression on B cells or CXCR4 expression on memory CD4+ T cells, higher CXCR4 expression on memory CD4+ T cells was predictive of a better response to CTLA4-Ig treatment (Fig. 5 and Figure S11). This result also confirms the importance of CXCR4 expression on memory CD4+ T cells in RA and supports its role as an immunological biomarker.
Figure 5. Increased CXCR4 expression on memory CD4+ T cells predicts a better response to CTLA4-Ig.
(A) Correlations between baseline immunophenotyping subsets, cytokines, soluble cytokine receptors, and CXCR4 expression on CD4+ T cell subsets and clinical responses to CTLA4-Ig treatment in RA patients (n = 8–20). Tfh1, Tfh2, and Tfh17 subpopulations were eliminated from the analysis. (B) Representative scatter plot between baseline MemoryTh CXCR4 positivity (%) and DAS28esr change from baseline following CTLA4-Ig treatment (Minus delta DAS28esr; n = 8). *p < 0.05 Spearman’s test.
Discussion
In spite of the importance of the HLA-DRB1 genotype for the development of ACPA-positive RA and bone destruction5,7, little is known about its association with lymphocyte subsets. In this study, we identified that the HLA-DRB1 disease-susceptible haplotype, SE, is related to PBMC memory CD4+ T cell subset and CXCR4 expression on memory CD4+ T cells (Figs 1B and 2B). Increased CXCR4 expression on memory CD4+ T cells from SE + RA patients may explain the linkage between HLA-DRB1 genotype and destructive arthritis (Fig. 2C and Table 1). The moderate correlation between CXCR4 expression on memory CD4+ T cells and DAS28esr/ HAQ suggests that synovial migration or retention of memory CD4+ T cells by increased CXCR4 expression is associated with sustained autoimmunity and local inflammation that eventually lead to functional disability due to bone destruction. Furthermore, the quantitative increase of HLA-DR expression on SE + RA B cells and its correlation to CXCR4 expression levels on memory CD4+ T cells suggest the importance of B cells as antigen-presenting cells in RA (Figs 1C and 3C). In addition, there were no significant differences between SE-HD and SE + HD in HLA-DR expression on B cells or CXCR4 expression on memory CD4+ T cells (Figures S3 and S5), which suggests that the relationship between SE and enhanced HLA-DR expression on B cells or enhanced CXCR4 expression on memory CD4+ T cells could be specific to RA. It has been reported that increased HLA-DR molecules on antigen-presenting cells, especially B cells in RA, can efficiently present low-affinity peptides to T cells24. Some factors other than HLA-DRB1 SE alleles may contribute to the increased HLA-DR expression on SE + RA B cells. Although stimulation related response expression quantitative trait locus (reQTL) effect36 of HLA-DRB1 SE alleles is one candidate mechanism under inflammation in RA, further examination is required to address this important point. Since TCR signaling is the most significant upstream regulator of transcriptomic changes in CXCR4-expressing memory CD4+ T cells (Fig. 4C), the quantitative increase in HLA-DR expression on B cells could contribute to the increased CXCR4 expression observed on memory CD4+ T cells in RA, and the increased amount of serum IL-21 might also play a role (Fig. 3B and Figure S4B). Furthermore, CXCR4 expression on the most expanded memory CD4+ T cell clones was up-regulated compared to non-expanded clones in the RA synovium37. However, it is possible that increased CXCR4 expression on memory CD4+ T cells reflects non-antigen-specific T cell activation because increased CXCR4 expression was observed in various CD4+ T cell subsets, and strong TCR signaling decreased CXCR4 expression in vitro (Fig. 3B). Inflammatory cytokines, especially TNF-α and IL-6, can also activate CD4+ T cells and contribute to increased CXCR4 expression21. Regardless, inflammatory cytokine concentrations were not associated with HLA-DRB1 genotype or RA disease activity (Figure S4). Our data strongly suggest the close link between HLA-DR, CXCR4-expressing memory CD4+ T cells, and disease activity in RA.
In spite of the higher expression of CXCR4 on naive CD4+ T cells compared to memory CD4+ T cells, CXCR4 expression on naive CD4+ T cells was not associated with RA disease activity (Fig. 2 and Table S3). Although CXCR4 is a receptor for HIV-1 entry into CD4+ cells, memory CD4+ T cells are preferentially infected and harbor more integrated proviral DNA than naive CD4+ T cells38. It has been reported that memory CD4+ T cells possess higher cortical actin and chemotactic actin activity than naive CD4+ T cells, thus showing higher susceptibility to CXCL12 and HIV-139. This implies that there is a functional difference between CXCR4 on memory CD4+ T cells and CXCR4 on naive CD4+ T cells, which may be related to the preferential association between CXCR4-expressing memory CD4+ T cells and RA disease activity.
ACPA had a stronger correlation to HLA-DRB1 SE and clinical resistance to therapy than RF40. Our data suggest different mechanisms for the generation of these two autoantibodies. We showed that RF is significantly correlated with the frequency of plasmablasts and inflammatory cytokines (Fig. 1A and Figure S4A), which suggests that the inflammatory environment and B cell activation may induce the production of RF. Meanwhile, ACPA was correlated with the frequency of memory CD4+ T cells, suggesting a role for acquired immunity to citrullinated peptides in the pathogenesis of ACPA, especially in SE + RA patients.
This study identified a previously unrecognized predictor for the efficacy of CTLA4-Ig. Increased CXCR4 expression on memory CD4+ T cells predicted a better response to CTLA4-Ig (Fig. 5). We did not observe any direct effects of CTLA4-Ig on CXCR4 expression on memory CD4+ T cells or HLA-DR expression on B cells (Figure S11). However, because CTLA4-Ig significantly suppressed CXCR4 expression on naive CD4+ T cells (Figure S11D), it is possible that CTLA4-Ig may suppress CXCR4 expression on memory CD4+ T cells after a longer period of treatment. Clinically, a better response to CTLA4-Ig has been reported for RA patients with high ACPA titers41. CTLA4-Ig blocks CD28 costimulation, thereby interfering with the interaction between CD4+ T cells and antigen-presenting cells, such as B cells. Increased CXCR4 expression on memory CD4+ T cells, which also corresponds to high ACPA titers (Figure S6), could be related to stronger interactions between CD4+ T cells and B cells. Another possibility is that higher CXCR4 expression may be associated with enhanced migration of memory CD4+ T cells to the joints. Monocytes/macrophages could produce inflammatory cytokines upon interactions with the migrated CD4+ T cells, and this pathogenic process may be inhibited by CTLA4-Ig. Further research is needed to elucidate the mechanism of the improved CTLA4-Ig responses in patients with higher CXCR4 expression.
In summary, we revealed genetic-immunological interactions that impact RA pathogenesis. Our data show that increased CXCR4 expression on memory CD4+ T cells correlates with disease activity and response to CTLA4-Ig treatment. Therefore, an immunological abnormality that induces enhanced HLA-DR expression on B cells and CXCR4 expression on RA memory CD4+ T cells may impact RA pathology and could potentially be a therapeutic target.
Methods
RA patients and healthy donors
We recruited 91 RA patients from April 2013 to March 2015 who fulfilled the 1987 revised American College of Rheumatology Criteria or the 2010 American College of Rheumatology/European League Against Rheumatism classification criteria. Patients with active infection or malignancy were excluded. The following clinical data were collected (Table S1): age, sex, disease duration, methotrexate (MTX) usage, conventional and biological disease-modifying antirheumatic drugs (DMARDs) usage, rheumatoid factor (RF) titer, anti-cyclic citrullinated peptide antibody (ACPA) titer, Disease Activity Score 28 joints-ESR (DAS28esr) and Health Assessment Questionnaire Disability Index (HAQ)10,11. RF titers were measured with latex coagulating nephelometry (cut off value is 15 IU/ml). ACPA titers were measured by chemiluminescence enzyme immunoassay (cut of value is 4.5 U/ml, Medical and Biological Laboratories, Japan). 22 patients were recruited at the initiation of CTLA4-Ig treatment, and 20 patients that continued treatment were reanalyzed after 6 months. Two patients discontinued CTLA4-Ig treatment due to ineffectiveness and pneumonia. Synovial fluid samples were derived from therapeutic arthrocentesis. 110 healthy donors (HD) were screened with a questionnaire to disqualify HD with diseases requiring treatment. All donors provided written informed consent, and the use of human peripheral blood and synovial fluid samples was approved by the Ethical Committee of the University of Tokyo Hospital (No. 10154 and G3582). The methods were carried out in accordance with the approved guidelines.
Human PBMC isolation and flow cytometry
Preliminary immunophenotyping analyses with standard cryopreserved samples exhibited substantial differences from fresh samples42. Therefore, all collected samples were freshly analyzed by flow cytometry. Human peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Paque Plus density gradient centrifugation (GE healthcare). Erythrocytes were lysed with ammonium chloride potassium buffer, and non-specific binding was blocked with Fc-gamma receptor antibodies. Staining was performed with predefined cell and antibody concentrations. The following antibodies were used: Human Fc Receptor Binding Inhibitor Purified (eBioscience), CD3-PE-Cy7 (UCHT1, BioLegend), CD3-PerCP-Cy5.5 (UCHT1, BioLegend), CD4-PerCP-Cy5.5 (OKT4, BioLegend), CD4-V500 (RPA-T4, BD Biosciences), CD11c-Brilliant Violet 421 (B-ly6, BD Biosciences), CD14-FITC (M5E2, BioLegend), CD16-PerCP-Cy5.5 (3G8, BioLegend), CD19-APC-Cy7 (HIB19, BioLegend), CD19-V500 (HIB19, BD Biosciences), CD24-PE (ML5, BD Biosciences), CD25-Brilliant Violet 421 (BC96, BioLegend), CD25-PE-Cy7 (BC96, eBioscience), CD27-FITC (O323, eBioscience), CD38-PE-Cy7 (HIT2, BioLegend), CD45RA-APC-Cy7 (HI100, BioLegend), CD56 -APC-Cy7 (HCD56, BioLegend), CD123-APC (AC145, Miltenyi), CD127-PE-Cy7 (eBioRDR5, eBioscience), CXCR3-Brilliant Violet 421 (1C6, BD Biosciences), CXCR4-APC (12G5, BD Biosciences), CXCR5-Alexa Fluor 488 (RF8B2, BD Biosciences), CCR6-PE (11A9, BD Biosciences), CCR7-PerCP-Cy5.5 (G043H7, Biolegend), HLA-DR-PE (L243, eBioscience), IgD-Brilliant Violet 421 (IA6-2, BD Biosciences). Flow cytometric analysis was performed on an 8-color MoFlo XDP (Beckman Coulter).
Immunophenotyping
Subset definitions are summarized in Table S2. We classified CD4+ T cells, B cells, natural killer (NK) cells, monocytes, and dendritic cells (DC) based on Human Immunology Project classification9 and also added modifications for subsets already reported to be important in RA. First, for the classification of CD4+ T cells, the follicular helper T cell (Tfh) subset and its subclassifications were added because Tfh, a specialized CD4+ T cell subset that helps B cell differentiation and antibody production, could potentially be pathogenic in RA (Figure S1)43,44,45,46. Second, for the classification of monocytes, HLA-DR positivity was added to rigorously exclude granulocytes, and three precise subset classifications were employed based on previous reports showing the expansion and high inflammatory cytokine production of CD14brightCD16+ monocytes in RA33,47,48. The ratio of each subset is represented as the percentage of the parent population (Table S2). CXCR4-positive ratios were determined based on isotype controls (APC Mouse IgG2a Kappa Isotype Control, MOPC-173, Biolegend). As CXCR4-positive ratios on CD4+ T cells were correlated with age49, we adjusted CXCR4-positive ratios of each CD4+ T cell subset by linear regression for age when comparing HD and RA.
HLA-DRB1 typing and quantitative HLA-DR expression analysis
The HLA-DRB1 alleles of RA patients were genotyped by the polymerase chain reaction-sequence-based typing (PCR-SBT) method (SRL, Japan). HD were genotyped by Infinium OmniExpressExome (Illumina). Quality control (QC) of genotyping data was performed by PLINK 1.9050. PCA of genotype data was performed by EIGENSTRAT51. Pre-phasing was performed by SHAPEIT and imputation was performed by IMPUTE2 with 1000 Genome Project Phase1 version 3 reference data52,53. Four-digit HLA alleles were imputated with SNP2HLA software54,55. The following alleles were considered to carry shared epitopes; 01:01, 01:02, 04:01, 04:04, 04:05, 04:08, 04:10, 10:01, 14:02, 14:0656. Using a monoclonal antibody to a common epitope of HLA-DR, cell surface HLA-DR quantitation analysis was performed with QuantiBrite PE (BD Biosciences), as instructed. The HLA-DR-PE antibody concentration we used was confirmed to saturate cell-surface HLA-DR24.
Multiplex cytokine analysis
Serum samples were stored at −30 °C until assays were performed. To reduce false amplification by heterophilic antibodies, HeteroBlock (Omega Biologicals) was added to all serum samples to achieve a final concentration of 150 μg/ml57. Serum concentrations of GM-CSF, IFN-γ, IL-10, IL-17A, IL-1b, IL-21, IL-4, IL-6, TNF-α, IL-7, sIL-6R and spg130 were measured using the Milliplex MAP kit (the Human Soluble Cytokine Receptor Magnetic Bead Panel and the Human High Sensitivity T Cell Magnetic Panel, Merck Millipore) and the BioPlex 3D system (Bio-Rad), according to the manufacturer’s instructions. To avoid inter-assay variation, all samples were analyzed at one time with the same kit lot number. Because IL-4 concentrations were above the detection limit only in 3.8% (3/78) of the samples, IL-4 was excluded from further analysis. Other cytokines or soluble cytokine receptors were detected in 88–100% of the samples.
PBMC culture
PBMC samples (1 × 106 cells/mL) were cultured in RPMI 1640 medium supplemented with 10% FBS (Equitech-Bio), 2 mM γ-glutamine (Sigma), 100 U/ml penicillin (Sigma), and 100 μg/ml Streptomycin (Sigma) in a humidified incubator at 37 °C, 5% CO2, for 120 hours. Recombinant human IL-21 (Peprotech) was added at 10 ng/ml. Phorbol myristate acetate (PMA, Sigma) was added at 25 ng/ml. To block the association between HLA-DR and T cell receptors, anti-HLA DR monoclonal antibodies (L243, Biolegend) were added to final concentrations of 0.1 μg/mL, 1 μg/mL, and 10 μg/mL. Viable 7-Amino-Actinomycin D (7-AAD, Biolegend)-negative CD3+ CD4+ CD45RA- cells were analyzed for CXCR4 expression with flow cytometry.
RNA-seq
CD3+CD4+CD45RA-CXCR4+ cells (CXCR4+ memory CD4+ T cells) and CD3+CD4+CD45RA−CXCR4− cells (CXCR4- memory CD4+ T cells) were sorted from five RA patients with active disease (DAS28esr > 3.2) by MofloXDP. Upper and lower 33rd percentiles of CXCR4 expression were considered to be CXCR4+ and CXCR4- populations of memory CD4+ T cells, respectively, in this RNA-seq analysis. 4.0–10 × 104 cells were sorted and stored at −80 oC until use. Library preparation was performed using a TruSeq Stranded mRNA Sample Prep Kit (Illumina). For PCR enrichment of the library, an additional 20 cycles were added to the manufacturer’s instructions because of the limited RNA input. The quality of the libraries was validated on an Agilent Technologies 2100 Bioanalyzer, and paired-end sequencing was performed by MiSeq (Illumina). 1.0–2.1 × 106 reads were obtained for each sample. Illumina Truseq adaptors were trimmed by Cutadapt58 and low-quality ends (phred score < 20) were also trimmed by FASTX-Toolkit (http://hannonlab.cshl.edu/fastx-toolkit). Mapping was performed with STAR59 with UCSC hg19 reference sequence (http://genome.ucsc.edu/). The uniquely mapped rate was 93.08 to 94.63%. Assignment of reads to genes was performed by HTSeq60 based on the UCSC gene model. Differential expression analysis was performed with a generalized linear model based edgeR61. 922 genes were upregulated in CXCR4high memory CD4+ T cells and 997 genes were downregulated with FDR < 0.05. Log2 counts-per-million (logCPM) values for each gene were used for heatmap and principal component analysis. Network analysis of upregulated and downregulated genes was performed with Ingenuity Pathways Analysis (IPA, Ingenuity).
Statistics
Categorical data were tested with Fisher’s exact test. Normality was tested with the Kolmogorov–Smirnov test. Differences between groups of normally distributed continuous data were tested with unpaired t-tests with Bonferroni corrections. Differences between groups of non-normally distributed continuous data were tested for significance as follows: nonparametric Mann–Whitney U test to compare two groups and the Kruskal–Wallis test with post-hoc Wilcoxon test with Bonferroni corrections to compare three groups. Correlations were evaluated by nonparametric Spearman’s rank correlation coefficients. Missing values were excluded pairwise in the correlation analysis. Samples with missing values were excluded from principal component analysis. CD4+ T cell subsets were examined for an association between CXCR4 expression and DAS28esr by univariate and multivariate linear regression analyses. Age, sex, disease duration, shared epitope positivity, RF titer, and ACPA titer were added as covariates. In the multivariate regression model selection, an exhaustive search was performed to maximize adjusted R-squared. Least absolute shrinkage and selection operator (LASSO) regression was also performed62. P values less than 0.05 were considered significant. All statistical analyses were performed with R version 3.1.2 (R Foundation for Statistical Computing).
Additional Information
How to cite this article: Nagafuchi, Y. et al. Immunophenotyping of rheumatoid arthritis reveals a linkage between HLA-DRB1 genotype, CXCR4 expression on memory CD4+ T cells, and disease activity. Sci. Rep. 6, 29338; doi: 10.1038/srep29338 (2016).
Supplementary Material
Acknowledgments
We thank the patients and healthy volunteers who participated in this research. We thank K. Watada, J. Takezawa, A. Inoue, and M. Anraku for their excellent technical assistance. We thank A. Yamaguchi at Shimokitazawa Hospital (Tokyo, Japan) for providing synovial fluid samples. We thank K. Kubo, H. Harada, M. Shibuya, Y. Iwasaki, K. Michishita, T. Eri, Y. Seri, and M. Ohshima for their help in collecting patients’ data. This study was supported by the Ministry of Health, Labour and Welfare, Ministry of Education, Culture, Sports, Science and Technology KAKENHI Grant-in-Aid for Scientific Research (S) (23229007), Grant-in-Aid for Scientific Research (C) (26461462), Grant-in-Aid for challenging Exploratory Research (25670459) and the Japan Society for the Promotion of Science. This work was supported in part by grants from the Cell Science Research Foundation and Bristol-Myers Squibb (BMS).
Footnotes
Keishi Fujio received financial support or fees from Astellas, BMS, Daiichi-Sankyo, MitsubishiTanabe, Pfizer, Santen, Takeda, Chugai, Eisai, Taisho Toyama, UCB, and Janssen. Kazuhiko Yamamoto received financial support or fees from AbbVie, Astellas, BMS, Daiichi-Sankyo, MitsubishiTanabe, Pfizer, Sanofi, Santen, Takeda, Teijin, Boehringer Ingelheim, Chugai, Eisai, Ono, Taisho Toyama, UCB, ImmunoFuture, Asahi Kasei, and Janssen. All other authors declare no competing financial interests.
Author Contributions Y.N., H.S., S.S., K.F. and K.Y. conceived, designed and analyzed the experiments, and contributed to writing the manuscript. Y.N. carried out all experiments with the exceptions noted below. Y.N., S.N., R.K., Y.T., H.T., K.S. and N.H. collected clinical data and performed the flow cytometric immunophenotyping experiments. S.T. and H.K. collected clinical data. Y.N., K.I., A.S. and Y.K. performed RNA-seq. Y.O. performed HLA-DRB1 typing of healthy donors.
References
- McInnes I. B. & Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med 365, 2205–2219, 10.7748/phc2011.11.21.9.29.c8797 (2011). [DOI] [PubMed] [Google Scholar]
- Okada Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381, 10.1038/nature12873 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen P. K., Silver J. & Winchester R. J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 30, 1205–1213 (1987). [DOI] [PubMed] [Google Scholar]
- Diogo D., Okada Y. & Plenge R. M. Genome-wide association studies to advance our understanding of critical cell types and pathways in rheumatoid arthritis: recent findings and challenges. Curr Opin Rheumatol 26, 85–92, 10.1097/BOR.0000000000000012 (2014). [DOI] [PubMed] [Google Scholar]
- Huizinga T. W. et al. Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA-DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis Rheum 52, 3433–3438, 10.1002/art.21385 (2005). [DOI] [PubMed] [Google Scholar]
- Scally S. W. et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J Exp Med 210, 2569–2582, 10.1084/jem.20131241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J. D. et al. Impact of shared epitope genotype and ethnicity on erosive disease: a meta-analysis of 3,240 rheumatoid arthritis patients. Arthritis Rheum 50, 400–412, 10.1002/art.20006 (2004). [DOI] [PubMed] [Google Scholar]
- Farh K. K. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343, 10.1038/nature13835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker H. T., McCoy J. P. & Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol 12, 191–200, 10.1038/nri3158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevoo M. L. et al. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum 38, 44–48 (1995). [DOI] [PubMed] [Google Scholar]
- Fries J. F., Spitz P., Kraines R. G. & Holman H. R. Measurement of patient outcome in arthritis. Arthritis Rheum 23, 137–145 (1980). [DOI] [PubMed] [Google Scholar]
- Arroyo-Villa I. et al. Constitutively altered frequencies of circulating follicullar helper T cell counterparts and their subsets in rheumatoid arthritis. Arthritis Res Ther 16, 500, 10.1186/s13075-014-0500-6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi J. et al. Peripheral blood CD4(+)CD25(+)CD127(low) regulatory T cells are significantly increased by tocilizumab treatment in patients with rheumatoid arthritis: increase in regulatory T cells correlates with clinical response. Arthritis Res Ther 17, 10, 10.1186/s13075-015-0526-4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarenbeek P. L. et al. Inflamed target tissue provides a specific niche for highly expanded T-cell clones in early human autoimmune disease. Ann Rheum Dis 71, 1088–1093, 10.1136/annrheumdis-2011-200612 (2012). [DOI] [PubMed] [Google Scholar]
- Amara K. et al. Monoclonal IgG antibodies generated from joint-derived B cells of RA patients have a strong bias toward citrullinated autoantigen recognition. J Exp Med 210, 445–455, 10.1084/jem.20121486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nanki T. et al. Stromal cell-derived factor-1-CXC chemokine receptor 4 interactions play a central role in CD4+ T cell accumulation in rheumatoid arthritis synovium. J Immunol 165, 6590–6598 (2000). [DOI] [PubMed] [Google Scholar]
- Buckley C. D. et al. Persistent induction of the chemokine receptor CXCR4 by TGF-beta 1 on synovial T cells contributes to their accumulation within the rheumatoid synovium. J Immunol 165, 3423–3429 (2000). [DOI] [PubMed] [Google Scholar]
- Kanbe K., Takagishi K. & Chen Q. Stimulation of matrix metalloprotease 3 release from human chondrocytes by the interaction of stromal cell-derived factor 1 and CXC chemokine receptor 4. Arthritis Rheum 46, 130–137 (2002). [DOI] [PubMed] [Google Scholar]
- Bernhagen J. et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med 13, 587–596, 10.1038/nm1567 (2007). [DOI] [PubMed] [Google Scholar]
- Leech M. et al. Macrophage migration inhibitory factor in rheumatoid arthritis: evidence of proinflammatory function and regulation by glucocorticoids. Arthritis Rheum 42, 1601–1608, (1999). [DOI] [PubMed] [Google Scholar]
- Bryant J., Ahern D. J. & Brennan F. M. CXCR4 and vascular cell adhesion molecule 1 are key chemokine/adhesion receptors in the migration of cytokine-activated T cells. Arthritis Rheum 64, 2137–2146, 10.1002/art.34394 (2012). [DOI] [PubMed] [Google Scholar]
- Chung S. H. et al. CXC chemokine receptor 4 expressed in T cells plays an important role in the development of collagen-induced arthritis. Arthritis Res Ther 12, R188, 10.1186/ar3158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliaou J. F. et al. Increase of class II HLA molecules on the membrane of B lymphocytes from patients with rheumatoid arthritis. Autoimmunity 1, 217–222 (1988). [DOI] [PubMed] [Google Scholar]
- Kerlan-Candon S. et al. Specific overexpression of rheumatoid arthritis-associated HLA-DR alleles and presentation of low-affinity peptides. Arthritis Rheum 44, 1281–1292, (2001). [DOI] [PubMed] [Google Scholar]
- Calabrese L. H. & Rose-John S. IL-6 biology: implications for clinical targeting in rheumatic disease. Nat Rev Rheumatol 10, 720–727, 10.1038/nrrheum.2014.127 (2014). [DOI] [PubMed] [Google Scholar]
- Besedovsky L. et al. Cortisol increases CXCR4 expression but does not affect CD62L and CCR7 levels on specific T cell subsets in humans. Am J Physiol Endocrinol Metab 306, E1322–1329, 10.1152/ajpendo.00678.2013 (2014). [DOI] [PubMed] [Google Scholar]
- Wang A. et al. Dysregulated expression of CXCR4/CXCL12 in subsets of patients with systemic lupus erythematosus. Arthritis Rheum 62, 3436–3446, 10.1002/art.27685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan P. et al. Cytokines and cell surface molecules independently induce CXCR4 expression on CD4+CCR7+ human memory T cells. J Immunol 165, 716–724 (2000). [DOI] [PubMed] [Google Scholar]
- Spolski R. & Leonard W. J. Interleukin-21: a double-edged sword with therapeutic potential. Nat Rev Drug Discov 13, 379–395, 10.1038/nrd4296 (2014). [DOI] [PubMed] [Google Scholar]
- Liao W., Lin J. X. & Leonard W. J. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol 23, 598–604, 10.1016/j.coi.2011.08.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo M. et al. Activation of blood T lymphocytes down-regulates CXCR4 expression and interferes with propagation of X4 HIV strains. Eur J Immunol 28, 3192–3204, (1998). [DOI] [PubMed] [Google Scholar]
- Wang R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882, 10.1016/j.immuni.2011.09.021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossol M., Kraus S., Pierer M., Baerwald C. & Wagner U. The CD14(bright) CD16+ monocyte subset is expanded in rheumatoid arthritis and promotes expansion of the Th17 cell population. Arthritis Rheum 64, 671–677, 10.1002/art.33418 (2012). [DOI] [PubMed] [Google Scholar]
- Jongbloed S. L. et al. Enumeration and phenotypical analysis of distinct dendritic cell subsets in psoriatic arthritis and rheumatoid arthritis. Arthritis Res Ther 8, R15, ar1864 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richez C. et al. Myeloid dendritic cells correlate with clinical response whereas plasmacytoid dendritic cells impact autoantibody development in rheumatoid arthritis patients treated with infliximab. Arthritis Res Ther 11, R100, 10.1186/ar2746 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. N. et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 343, 1246980, 10.1126/science.1246980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki K. et al. Quantitative and qualitative characterization of expanded CD4(+) T cell clones in rheumatoid arthritis patients. Sci Rep 5, 12937, 10.1038/srep12937 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomont N. et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15, 893–900, 10.1038/nm.1972 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. A dichotomy in cortical actin and chemotactic actin activity between human memory and naive T cells contributes to their differential susceptibility to HIV-1 infection. J Biol Chem 287, 35455–35469, 10.1074/jbc.M112.362400 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Cruyssen B. et al. Anti-citrullinated protein/peptide antibodies (ACPA) in rheumatoid arthritis: specificity and relation with rheumatoid factor. Autoimmun Rev 4, 468–474, 10.1016/j.autrev.2005.04.018 (2005). [DOI] [PubMed] [Google Scholar]
- Sokolove J. et al. Impact of baseline anti-cyclic citrullinated peptide-2 antibody concentration on efficacy outcomes following treatment with subcutaneous abatacept or adalimumab: 2-year results from the AMPLE trial. Ann Rheum Dis, 10.1136/annrheumdis-2015-207942 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama W. M., Thompson M. L. & Ehrhardt R. O. Cryopreservation and thawing of cells. Curr Protoc Immunol Appendix 3, 3G, 10.1002/0471142735.ima03gs99 (2012). [DOI] [PubMed] [Google Scholar]
- Morita R. et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 34, 108–121, 10.1016/j.immuni.2010.12.012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. et al. Increased frequency of circulating follicular helper T cells in patients with rheumatoid arthritis. J Clin Endocrinol Metab 2012, 827480, 10.1155/2012/827480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. et al. A regulatory effect of IL-21 on T follicular helper-like cell and B cell in rheumatoid arthritis. Arthritis Res Ther 14, R255, 10.1186/ar4100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata S. et al. Activation of Syk in peripheral blood B cells in patients with rheumatoid arthritis: a potential target for abatacept therapy. Arthritis Rheumatol 67, 63–73, 10.1002/art.38895 (2015). [DOI] [PubMed] [Google Scholar]
- Autissier P., Soulas C., Burdo T. H. & Williams K. C. Evaluation of a 12-color flow cytometry panel to study lymphocyte, monocyte, and dendritic cell subsets in humans. Cytometry A 77, 410–419, 10.1002/cyto.a.20859 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B. R. et al. Functional phenotype of synovial monocytes modulating inflammatory T-cell responses in rheumatoid arthritis (RA). PloS one 9, e109775, 10.1371/journal.pone.0109775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cané S., Ponnappan S. & Ponnappan U. Altered regulation of CXCR4 expression during aging contributes to increased CXCL12-dependent chemotactic migration of CD4(+) T cells. Aging Cell 11, 651–658, 10.1111/j.1474-9726.2012.00830.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575, 10.1086/519795 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909, 10.1038/ng1847 (2006). [DOI] [PubMed] [Google Scholar]
- Delaneau O., Marchini J. & Zagury J. F. A linear complexity phasing method for thousands of genomes. Nat Methods 9, 179–181, 10.1038/nmeth.1785 (2012). [DOI] [PubMed] [Google Scholar]
- Howie B., Marchini J. & Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 1, 457–470, 10.1534/g3.111.001198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8, e64683, 10.1371/journal.pone.0064683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y. et al. Construction of a population-specific HLA imputation reference panel and its application to Graves’ disease risk in Japanese. Nat Genet 47, 798–802, 10.1038/ng.3310 (2015). [DOI] [PubMed] [Google Scholar]
- Fisher B. A. et al. Smoking, the HLA-DRB1 shared epitope and ACPA fine-specificity in Koreans with rheumatoid arthritis: evidence for more than one pathogenic pathway linking smoking to disease. Ann Rheum Dis 73, 741–747, 10.1136/annrheumdis-2012-202535 (2014). [DOI] [PubMed] [Google Scholar]
- Todd D. J. et al. Erroneous augmentation of multiplex assay measurements in patients with rheumatoid arthritis due to heterophilic binding by serum rheumatoid factor. Arthritis Rheum 63, 894–903, 10.1002/art.30213 (2011). [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T. & Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Series B. 58, 267–288 (1996). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.