Abstract
A central topic for conservation science is evaluating how human activities influence global species diversity. Humanity exacerbates extinction rates. But by what mechanisms does humanity drive the emergence of new species? We review human-mediated speciation, compare speciation and known extinctions, and discuss the challenges of using net species diversity as a conservation objective. Humans drive rapid evolution through relocation, domestication, hunting and novel ecosystem creation—and emerging technologies could eventually provide additional mechanisms. The number of species relocated, domesticated and hunted during the Holocene is of comparable magnitude to the number of observed extinctions. While instances of human-mediated speciation are known, the overall effect these mechanisms have upon speciation rates has not yet been quantified. We also explore the importance of anthropogenic influence upon divergence in microorganisms. Even if human activities resulted in no net loss of species diversity by balancing speciation and extinction rates, this would probably be deemed unacceptable. We discuss why, based upon ‘no net loss’ conservation literature—considering phylogenetic diversity and other metrics, risk aversion, taboo trade-offs and spatial heterogeneity. We conclude that evaluating speciation alongside extinction could result in more nuanced understanding of biosphere trends, clarifying what it is we actually value about biodiversity.
Keywords: conservation, diversification, Holocene, no net loss, species
1. Introduction
Understanding and preventing biodiversity loss is of paramount importance to humanity [1–3]. Over the last decade, it has been emphasized that conservation science and practice should consider net, rather than absolute, outcomes of interventions [4–6]. For instance, McDonald-Madden et al. [5] state that for conservation, in general, ‘gains and losses must both be presented as an auditable conservation balance sheet’. There has been a proliferation of policies incorporating the ‘no net loss’ principle, whereby negative impacts on biodiversity associated with human activities are required to be compensated for by conservation actions, theoretically resulting in a neutral net outcome for nature [7]. But globally, it remains common to measure biodiversity declines via the proxy of absolute species losses—more specifically, in terms of the numbers of macroscopic fauna and flora species lost over time.
Both the total number of extant species, and the rate at which those species are disappearing, are highly uncertain [8–10]. Approximately, 1.9 million species have been described [11]. Estimates of the total number of eukaryotic species alive include 5 ± 3 million [10], 8.7 ± 1.3 million [12], less than a million and more than 10 million [11]. Best estimates of extinction rates fall are around 1.0–2.2% of species totals per decade [10,12,13]. But human activities not only drive species extinction. Palkovacs et al. [14] found that human activity is involved in ‘162 of the 198 study systems in which contemporary trait change has been documented in the wild’, and humans have been shown to mediate substantial speciation in plants [15,16]. Such considerations raise the question: in what ways are humans driving speciation alongside extinction, and what is the net anthropogenic contribution towards global species diversity for all taxa? Further, if it transpired that our overall impact on species diversity was neutral, would this be acceptable? Here, we review the literature pertaining to these questions.
Species can be considered ‘separately evolving meta-population lineages’ [17], but there are numerous ways in which different species are delimited. Speciation occurs when a lineage splits into multiple reproductively isolated, genetically distinct sub-populations (cladogenesis), but vagueness in species delimitation means that there is grey area between sub-populations that have developed slightly different traits, and those that are divergent lineages. It is consequently problematic to define exactly when a speciation ‘event’ has occurred; i.e. when one or more new species can be considered to have emerged from those existing [18,19]. In turn, this complicates calculation of speciation rates. Note that speciation, as a term, is not generally applied to ‘anagenesis’ (i.e. where an entire lineage evolves sufficiently over time to effectively become a new species), because the net result is that species richness does not increase.
In this review, we consider anthropogenic activities that result in populations becoming distinct from organisms of the same species, under the criteria currently advocated by evolutionary biologists, e.g. development of new traits, reproductive isolation [17]. We, therefore, include instances of new species emerging, and also anthropogenic mechanisms apparently in the process of driving speciation. The literature on speciation as a general evolutionary process is vast, and has previously been reviewed [19–23], so we emphasize that our focus here is not speciation more broadly. Rather, we build upon the emerging suggestions that human activities could significantly influence speciation on a global scale [16,24], consider anthropogenic speciation mechanisms, and discuss whether these could be significantly influencing global species numbers.
(a). Known species extinctions
Human actions are the main cause of contemporary species extinctions [11]. Although methods for estimating when extinction has occurred are subject to uncertainty, and even charismatic species can mistakenly be classified extinct [25], the number of recorded species extinctions is almost certainly lower than the true number, particularly as some species go extinct before being described [10]. While the number of extinction events approximately over the last 500 years is not yet of the same magnitude seen during the ‘big five’ mass extinction events, the extinction rate is comparable, and could result in a sixth mass extinction event within a few centuries [8,26].
Mammals are perhaps the most well-researched group of living organisms. Incorporating recently extinct species, there are currently 5488 known species of mammal [27]. From the Late Pleistocene (approx. 130 000 years ago) to approximately AD 1000, 177 species of large mammal (more than 10 kg) are known to have become extinct [28]. Estimates for the Holocene (i.e. the last 11 500 years) suggest that 255 mammals became extinct during that period [29]. Similarly, during the Holocene, there have been 523 recorded bird species extinctions [29], of which 129 became extinct since AD 1500 [30]. Although it is not straightforward to determine what caused known extinctions during these time periods, evidence often points to humans [28].
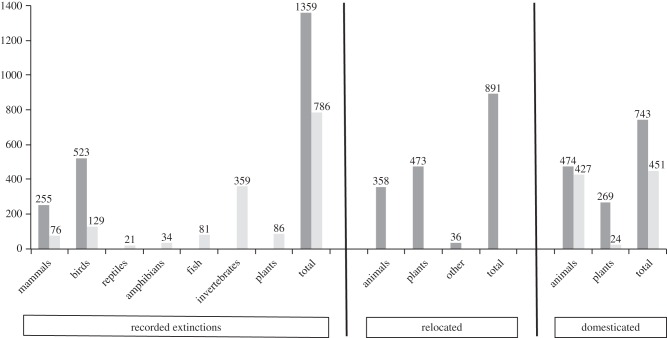
During the more recent time period from AD 1500 to the present day, approximately 784 extinctions have been documented [27,30]. They include: mammals (79), birds (129), reptiles (21), amphibians (34), fish (81), invertebrates (359, of which 291 were molluscs), plants (86) and Protista (1) (figure 1) [27,30,33]. Comparably, Dirzo et al. [9] estimate that 322 species of vertebrate have become extinct since AD 1500, but importantly, highlight that there has been a great loss of invertebrate diversity that is much less studied. For example, Régnier et al. [34] estimate that the actual number of mollusc extinctions is double the IUCN estimate. In addition, it should be noted that the number of species extinctions does not capture the phylogenetic richness of those species, e.g. the magnitude of the loss of evolutionary history associated with ancient or highly diverse lineages.
Figure 1.
Number of recorded animal and plant species extinctions (citations in main text); number of recorded established invasive, i.e. ‘relocated’ species (Global Invasive Species Database [31]); number of domesticated species [32]. Light grey, since AD 1500; dark grey, during the Holocene.
2. Human-mediated speciation
The process by which genotypes diverge has been studied extensively, so data on background diversification (the difference between speciation and extinction rates) are available. Plants have median diversification rates of 0.06 new species per species per million years, rates for birds are estimated at 0.15, and mammals at 0.07 [11]. While it has been shown that humans can drive contemporary evolution to a degree that is significantly higher than that from natural causes [24,35], estimates of speciation attributable to human activities do not exist for most organisms.
We posit that human activities can directly or indirectly result in reproductive barriers of various kinds (e.g. geographical, physical) being created between sub-populations of an existing species—or, in different selective pressures being applied to specific members of a species (e.g. by age, size). In both cases, the development of new traits could occur in sub-populations. Given sufficient time this could, at least in some cases, result in cladogenesis. We also assume that there will be some scenarios in which the emergence of new traits, or even full speciation, can be attributed primarily to reproductive isolation or selective pressure caused by human activities, rather than a combination of anthropogenic and non-anthropogenic factors. Demonstrating that a given speciation event is human-mediated requires drawing a direct causal link between anthropogenic impacts on a population, the emergence of new traits in that population, and eventually, genetic divergence. In this section, recognizing that trait change can eventually lead to speciation, and that challenges exist in drawing conclusions as to whether any speciation event is natural or anthropogenic, we review evidence for human-mediated speciation mechanisms.
(a). Relocation
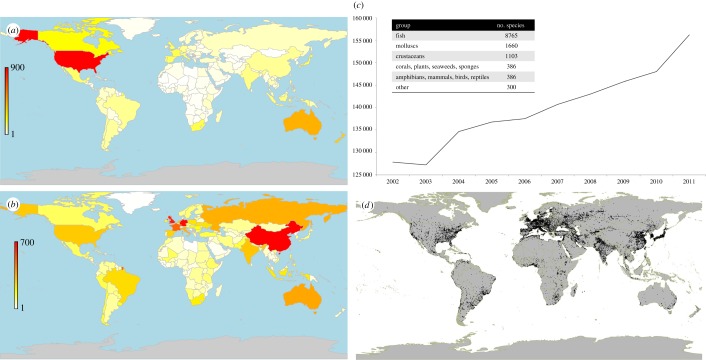
People have transported species to ecosystems in which they are non-native, intentionally or otherwise, for millennia. The establishment of alien invasive species is a threat to global biodiversity, to which many species endangerments are attributed ([36,37]; although see [38]). The Global Invasive Species Database [31], while not comprehensive due to geographical inequalities and biases in detection and survey effort, holds records for 891 distinct invasive species (i.e. established in natural or semi-natural ecosystems or habitat, is an agent of change and threatens native biological diversity), many of which are established in more than one country (figure 2a).
Figure 2.
Example spatial datasets relevant to human-mediated speciation. (a) Relocation—number of invasive alien species recorded, by country (Global Invasive Species Database [31]). Highest number of records = 876 species (USA), median = 26 species (note the database constitutes primarily macroscopic organisms, and inconsistent survey effort across countries). Species counts unique by country only. Grey fill, no data. (b) Domestication—number of domesticated livestock breeds by country, for 38 common species (Domestic Animal Diversity Information System [39]). Highest number of breeds = 709 (China), median = 42. Grey fill, no data. (c) Hunting—total global annual catch of marine species (‘000 tonnes), from 2002 to 2011 [40]. Inset, number of species. (d) Novel ecosystems—areas classified as urban environments (made with www.naturalearthdata.com). (Online version in colour.)
Relocation is also a potential speciation mechanism. Some relocated species undergo rapid evolution [41], which can eventually result in speciation over sufficient timescales [42,43]. For instance, Whitney & Gabler [44] document 38 species that have undergone rapid evolution following introduction, in some cases within 10 years. Buswell et al. [45] found that 70% of introduced plant species studied changed at least one morphological trait (e.g. plant height) during a 150-year period in Australia. Reznick et al. [46] found significant evolution in the life history of guppies Poecilia reticulata (e.g. age of maturation), 11 years after introduction to a new site. The introduction of non-native organisms to provide biological control is another pathway by which relocated species might themselves develop new traits, such as the myxoma virus introduced to Australia to control rabbit populations, and the Entomophaga maimaiga fungus introduced to the USA from Japan [47]. What is not clear is how often rapid evolution in such cases results in actual divergence.
Hybridization (reproduction between members of genetically distinct populations [18]) between native and relocated plant species, or between two different relocated plant species, can result in novel taxa, e.g. Helianthus annuus ssp. texanus (USA), Senecio cambrensis (UK) [48]. Thomas [16] points out that, through relocation and hybridization, more new plant species have appeared in Europe than are documented to have gone extinct over the last three centuries. Invading insects have also been noted to drive evolutionary change in native species via host-race formation, as potentially have invading species of vertebrate [18,42]. Emerging evidence suggests that hybridization may be an important factor in driving speciation for both plant and animal species, and in some cases, comparable with the primary cause (adaptation), although the proportion of hybrids that have resulted in speciation has not yet been quantified [18,49]. Importantly, care should be taken when comparing extinction rates over the distant past and known instances of contemporary species emergence, as past rates might be more difficult to estimate with accuracy than those involving extant species [16].
(b). Domestication
Humans have domesticated 474 animal and 269 plant species approximately over the last 11 000 years (figure 1) [32]. These species encompass a variety of different breeds spread across almost all countries in the world (figure 2b). Any species that has been domesticated is subjected to altered selective pressures, both deliberate and incidental (e.g. [50]).
Domestication has resulted in the documented emergence of novel species: of the world's 40 most important agricultural crop species, six to eight can be considered entirely new [16]. Equally, beyond speciation, it has resulted in very large populations of species representing considerable genetic diversity [51]. Within domesticated species, new traits can emerge: for instance, the domestic dog Canis lupus familiaris is one of the most morphologically diverse vertebrates, represented by 400 breeds [52]. Asian rice was domesticated approximately 8200–13 500 years before the present, and is among the world's most important crops. It could potentially be classified into two distinct sub-species from a single evolutionary origin [53]. Some Triticum (wheat) and Brassica species are entirely new, through hybridization [16]. The full picture is complicated—wheat has also decreased in diversity by some measures since domestication [54], and extensive gene flow between populations of domesticated species likely restricts lineage diversification.
Domestication has, equally, led to increased human interaction with ‘pest’ species, altering selective pressures. For instance, agricultural weeds can evolve resistance to commercial herbicides, sometimes within 10 years of commercial deployment [55]. Although observed resistance represents new trait development and not necessarily speciation, Palumbi [55] discusses ‘Humans as the world's greatest evolutionary force’.
(c). Hunting
Hunting drives new trait development in wild animal populations, influencing broader ecological dynamics [14,56], which could eventually be a precursor to speciation in some cases. Stenseth & Dunlop ([56], see also [57]) compared rate of phenotypic change in 40 populations subject to human harvesting against the rate seen in 20 systems experiencing selection from natural forces only (e.g. Darwin's finches) and 25 systems experiencing other human disturbances (e.g. pollution). They found that recorded rates of change in harvested populations outpaced ‘naturally’ driven changes by 300%. Similarly, Andersen & Brander [58] carry out an evolutionary impact assessment for commercial fisheries, finding that expected rates of evolution attributable to fishing are approximately 0.1–0.6% per year.
Jørgensen et al. [59] state that ‘evolutionary changes’ experienced by commercially exploited fish species are taking place on decadal timescales. This is supported by genetic evidence for phenotypic change in commercially important species, such as European plaice Pleuronectes platessa and Atlantic cod Gadus morhua [60]. Thousands of marine species are currently exploited (figure 2c), some small fraction of which could consequently experience sufficient such changes that speciation occurred. Similar so-called ‘unnatural selection’ has been shown in poaching of elephants Loxodonta africana, trophy hunting for bighorn sheep Ovis canadensis, red deer Cervus elaphus culls and terrestrial snail collecting [61].
Despite the multiple known cases of hunting pressure driving rapid evolution, there are no documented cases of related speciation events yet. Further, some propose that trait changes in hunted species are mainly a result of demographic and environmental factors [62].
(d). Novel ecosystem creation
Most ecosystems are sufficiently altered by human activity to be considered ‘novel’ [63,64], and some are entirely new, e.g. urban environments. Shifting ecosystems between states causes significant biodiversity loss [36], but another effect of creating novel ecosystems is to establish new biological communities. For instance, species respond differently to the conversion of land into urban environments: avoiding, adapting to or even exploiting it [63,65,66]. In turn, new traits can emerge in novel ecosystems. Resident populations of the planktivorous alewife Alosa pseudoharengus, for example, emerged from anadromous ancestors in response to hydropower construction, also altering evolution of prey species Daphnia ambigua [14]. Certain species gain a competitive advantage in novel ecosystems, leading to adaptation—such as fungal diseases emerging faster in agricultural landscapes [67], although it is not always clear whether these species are new.
Anthropogenic change can create new bioclimatic habitats, leading to concurrent changes in species assemblages. For instance, mountaintop plant diversity has been observed to increase under climate change [68], and biodiversity can rise in suburban habitats in comparison to neighbouring ‘natural’ areas [66]. Indeed, regional-scale plant species diversity worldwide is currently increasing as species introductions ‘far outnumber’ extinctions [69].
Novel ecosystems have already been observed to facilitate speciation. The common house mosquito (Culex pipiens) adapted to the environment of the underground railway system in London, UK, establishing a subterranean population. Now named the ‘London Underground mosquito’, Culex pipiens molestus can no longer interbreed with its above-ground counterpart [70]. Forest fragmentation in Mesoamerica appears to have led to Neotropical damselfly Megaloprepus caerulatus diverging into more than one species [71]. Both examples demonstrate that anthropogenic restriction on gene flow between sub-populations can result in speciation.
(e). Future mechanisms
Relocation, domestication, hunting and novel ecosystems are well-established human processes. But emerging technologies could feasibly eventually become mechanisms for driving speciation, if they are not short-lived. Here, we give three examples.
Developments in genetics now enable direct manipulation of genomes, and creation of genetically modified organisms (GMOs). Even in a region like Europe, where the use of GMOs in agriculture is relatively uncommon [72], there are 146 distinct variants of genetically engineered plant are approved or awaiting approval for commercial cultivation [73]. GMOs themselves are not new species, but may have the capacity to generate self-sustaining populations or hybridize with wild species [50,74]. The cultivation of GMOs could eventually, therefore, lead to new self-sustaining lineages.
Technology may soon allow re-creation of extinct species (de-extinction), despite deep practical and moral arguments about doing so [75,76]. At least two approaches to de-extinction (back-breeding, genetic engineering) would not replicate the extinct genome exactly [76], but if successful would result in the emergence of a species that otherwise would not exist. Where to include de-extinction in net extinction rates is questionable if the species only became extinct previously due to human activities.
Thirdly, albeit improbably, humanity could facilitate the movement of organisms to extra-terrestrial bodies. Hundreds of objects have been sent out into the solar system and beyond [77]. Terrestrial bacteria, lichens and even some animals can survive short-term space travel [78–80]. There is, consequently, a non-zero probability of depositing organisms on extra-terrestrial bodies—hence, strict rules concerning sterilization of objects bound for Mars [81]. While the potential extra-terrestrial transfer of organisms has only recently become reality, it will feasibly become common over the timescales projected for humanity to cause major extinction events [26].
(f). Speciation in microorganisms
Global extinction estimates generally focus upon macroscopic organisms [11]. Less is known about the endangerment of microorganisms, and it is uncertain how many free-living microbial lineages are threatened [82,83]. Although extinction of macroscopic species presumably results in co-extinction of parasites and mutualists, few have been documented [84].
Estimates for global prokaryote diversity range from 10 to 50 million species [85,86]. Parasites, including small eukaryotes, constitute 40% of biodiversity in some habitats [83]. So, in principle, establishing global anthropogenic biodiversity impacts requires a better understanding of impacts upon microorganisms. All four mechanisms reviewed above (relocation, domestication, hunting, novel ecosystem creation) likely influence diversification in microorganisms—e.g. water contaminated by effluent in novel urban environments presents opportunities for rapid microbe diversification [87]. But the literature also suggests evidence for diversification occurring in relation to medicine, disease, and the human ‘micro-biome’.
Species causing disease have co-evolved with humans, often from non-human pathogens [88]. Numerous continually adapting species are associated with our activities [89,90], and hybridization between pathogens could result in diversification [49,91]. Further, in fighting disease humans cause pathogens to evolve resistance—hence increasing the ineffectiveness of antibiotics, sometimes within only a year of deployment [55]. Emergence of antibiotic resistance within microbial lineages could be considered one stage in the broader process of speciation [17]. In a recent iteration of a major contemporary database now containing 2107 human pathogens, approximately 40% were human specific, and 175 classified as emerging diseases [92,93].
Other species have co-evolved benignly with humans, and 100 trillion microorganisms inhabit the average person [94], making individual humans a ‘micro-biome’. As a species, human micro-biome diversity is greater than for our closest extant relatives (wild apes) [95]. Despite potential for homogenization through globalization, and loss of ancient human micro-biome assemblages, geographical variation in micro-biome genotypes is large [95,96]. So, the global expansion of humans may itself have led to diversification in these microorganisms. Similar reasoning applies to domesticated species, in which microbial speciation has indeed been observed [97].
In practice, estimating speciation and extinction rates for microorganisms is problematic, and consequently, so is incorporating them into net diversity calculations.
3. Evaluating net outcomes for global species diversity
While human-mediated speciation rates are not quantified for most taxa, they are potentially considerable. Hypothetically then, if humanity drove speciation as fast as extinction with a neutral net outcome for species diversity, would this be acceptable? If species numbers alone reflected our preferences, then species gains should temper concern about extinctions. Yet intuitively, the answer would likely be ‘no’, extinctions cannot acceptably be compensated for in this way. This answer has theoretical support in the literature concerning policies seeking neutral net biodiversity outcomes—the ‘no net loss’ principle [98]. We apply that theory to speciation (figure 3), framing our discussion around challenges for ‘no net loss’ [99].
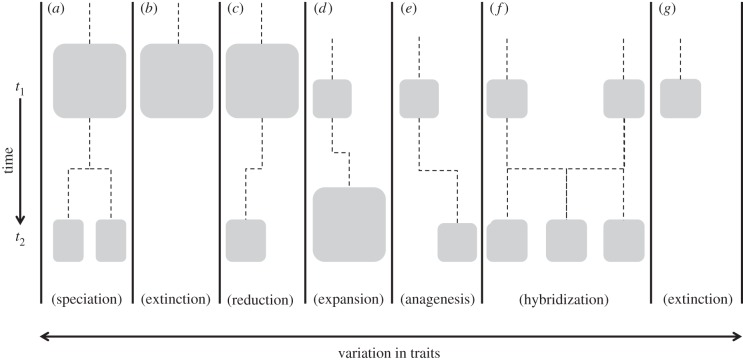
Figure 3.
Schematic illustrating trait change through time for a hypothetical set of species. Each species is represented at some time = t1 by a block, the size of the block denotes abundance. Different outcomes occur for each species over time (evaluated at some later time = t2). (a) Speciation results in two distinct species. (b) Abundant species goes extinct. (c) Species abundance significantly reduced, although sub-population with certain traits remains. (d) Species abundance increases, adopting a more varied set of traits. (e) Species adopts a sufficiently new set of traits to become a new species (anagenesis). (f) Two species hybridize, the hybrid eventually becomes a distinct species. (g) Another species extinction. Challenges: though species richness remains constant (eight species), overall phylogenetic diversity and abundance are substantially reduced at t2; if trends are human-mediated, the counterfactual ‘natural’ scenario at t2 is unknown; composition and distribution of extant species changes from t1 to t2; and extinctions might be intrinsically unacceptable.
(a). Species diversity as a metric
That species gains might not be considered fungible with losses shines a spotlight on one weakness of ‘species’ as a fundamental unit for conservation. Although extinctions are a widely used indicator of biodiversity trends, they inadequately capture why biodiversity decline is important. Also relevant are changes in abundance [9], range reductions, trophic downgrading [100] and loss of dynamics, e.g. migration [101]. Equally, replacing lost species from very different phylogenies with a comparable number diverging from extant relatives would result in loss of evolutionary distinctness [102]. Full compensation in species numbers alongside a loss of phylogenetic diversity would never represent true ‘no net loss’ of biodiversity per se. So, as species diversity alone is an insufficient unit for capturing conservation importance, neither is ‘no net loss of species diversity’ an adequate objective. Species is an especially problematic metric for microorganisms [103,104].
Not to suggest that species-based metrics serve no purpose for conservation, indeed, our reasoning applies to other measures too—for instance, abundance. Losses in wild fauna abundance during the Anthropocene [9] are not satisfactorily mitigated by concurrent increases in abundance of relatively homogeneous domesticated species (e.g. the 22 bn poultry or 1.5 bn cattle worldwide [39]). As changes in abundance occurred in similar species with useful traits, the broader loss, e.g. of phylogenetic diversity is again not reflected. Achieving neutral net outcomes for biodiversity in relation to any single metric cannot be considered acceptable [99].
(b). Counterfactuals and timescales
Human activities can also suppress speciation—for instance, by limiting species population sizes and ranges, thus reducing establishment of geographical isolates [105,106]. Alongside reducing isolates, large-scale losses of global species abundance [9] would correspond to losses in trait diversity within species, and losses of entire sub-species—reducing natural speciation, perhaps far outweighing any anthropogenic contribution towards increasing rates. Equally, regarding mechanisms for increased speciation we have explored here, relocation could, alongside loss of environmental heterogeneity, lead to a degree of interspecific hybridization that reduces speciation rates [107]. That speciation could be both increased and decreased by human activity demonstrates the complexity of calculating human-driven speciation rates, which is partly a problem of establishing a robust counterfactual of ‘natural’ diversification. Developing counterfactuals is a broader problem for ‘no net loss’ conservation [4,108].
Timescales are crucial in calculating net biodiversity outcomes [99]. As discussed, a human-driven mass extinction event could happen within 200 years [26]. Considering mammal diversification rates [11] and known species [27], one might simplistically expect a background global diversification approximately (5488 × e0.07) – 5488 = 398 new species of mammal in the first million years. But it is non-trivial to estimate diversification over a timescale as short as 200 years (and to isolate human-driven speciation rates) for reasons including the difficulty in defining when a new species has actually emerged.
Extinction events might intuitively be expected to progress more rapidly than speciation events. But recent research has suggested that speciation might occur quickly and often—just that new species rarely persist, making persistence over time an important consideration [109]. In turn, this would make it more problematic to evaluate whether short-lived speciation events are artificial or whether they would have occurred naturally anyway. Finally, any estimate of human-driven speciation rates is limited by uncertainty concerning technological and social change over the timescales needed for even rapid evolution to occur.
(c). Spatial heterogeneity
Species extinctions exhibit substantial regional variation (e.g. [28]). In turn, associated losses of ecosystem function have likely varied in magnitude and timing, in different parts of the world. Such spatial and temporal heterogeneity makes it problematic to propose an ecologically defensible calculation of net outcomes [99].
The challenge also extends to speciation. If the speciation mechanisms discussed in this article significantly increased diversification rates, then the rate would be influenced by, for example, the number of invasive species or domesticated breeds in any given region. There could be considerable heterogeneity in the distribution of invasive species and livestock diversity (figure 2), in turn, suggesting heterogeneous influences upon speciation rates under these assumptions. Similarly, the intensity of novel ecosystem creation varies spatially (e.g. [40]) (figure 2d). ‘No net loss’ type calculations would thus likely be different in different regions—perhaps with a net gain in some areas, and net loss in others—even if no net loss were approximately achieved overall. Such regional inequality in loss–gain trades is likely to be deemed unacceptable by conservationists [110].
(d). Uneven trades
Even if all species were of objectively equal value, the human mind fundamentally weights losses more highly than gains [111,112]. So, a promised species gain would have to be perceived as having greater value than an existing species lost, to ensure a neutral net outcome was experienced. In addition, there is uncertainty—known extant species are valued for utility and existence, whereas unknown novel species cannot be. Uncertainty has yet to be satisfactorily factored into net biodiversity calculations [99]. Further, the prospect of ‘artificially’ gaining novel species through human activities is unlikely to elicit the feeling that they would confer benefits to offset losses of extant ‘natural’ species. Indeed, many people might find the prospect of an artificially biodiverse world just as daunting as an artificially impoverished one. If we presume the natural to have intrinsically greater value than the artificial, then assessment of net biodiversity outcomes is further complicated.
Finally, framing human-driven losses and gains of species diversity as trades may be unethical, depending upon the nature of the trades. Daw et al. [113] outline taboo trade-offs, referring to trades in ecosystem services between ‘sacred’ and ‘secular’ values. In the context of this article, natural wild species might have ‘sacred’ value—whereas economic gain, livestock and other species which human civilization has benefitted (e.g. Rattus norvegicus) might be seen as having more ‘secular’ or even negative value. If so, comparing the loss of diverse wild species with gains in multiple closely related artificial species would be morally incommensurable [113].
4. Conclusion and future directions
We have examined mechanisms by which human activities could be driving rapid evolution, consequently, increasing speciation rates. Regardless, there is an ongoing biodiversity challenge to be met. But by considering net human influence on biodiversity, conservation scientists will achieve a more complete understanding of how we are changing the biosphere. We recognize that a key limitation overall is the blurred line as to when an actual speciation event has occurred [17], and that the definition of ‘species’ remains vague.
Although we cannot currently quantify human-mediated speciation rates, numerous studies have found human activities to materially influence species' evolution. Given the range of species affected, this influence may be significant, and deserves further investigation. Under each mechanism we have discussed here, existing datasets could support exploration for many taxa (figure 2), which we suggest is an important avenue for exploration. Microorganisms in particular deserve more attention from conservation biologists. Further, emerging technologies could eventually lead to human-mediated speciation—but not in the near future, if ever. While less pressing from that perspective, it is important to better understand the timescales over which these technologies might develop.
Conceptual barriers prevent neutral net outcomes for species diversity seeming acceptable—barriers that are technical, social and ethical. The range and strength of such barriers requires further interdisciplinary exploration between biologists and social scientists, establishing what (if not absolute species diversity) society truly wants to conserve about biodiversity, thereby improving conservation science and practice.
In conclusion, it is not currently possible to quantify exactly how many speciation events have been caused through human activities, or how significant this process is. Yet it is clearly a phenomenon worthy of further attention from conservation science, given examples of human-influenced speciation events do exist, as do multiple anthropogenic mechanisms for driving rapid evolution. Consideration of speciation alongside extinction may well prove important in developing a better understanding of our impact upon global biodiversity. Merely considering the issue leads to deeper questions: how we use species as a fundamental metric, what species we value and why.
Acknowledgements
We thank the Invasive Species Specialist Group for extracting data from the Global Invasive Species Database. K. Marske and A. Stensgaard at the Center for Macroecology, Evolution and Climate (CMEC) commented upon the manuscript. We acknowledge useful suggestions provided by two anonymous reviewers.
Authors' contributions
J.W.B. developed the concept, performed the review, retrieved the relevant datasets and drafted the manuscript; M.M. drafted the manuscript and provided critical revisions. Both authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
J.W.B. is funded by a Marie Skłodowska-Curie Fellowship at the University of Copenhagen, and acknowledges the Danish National Research Foundation for funding CMEC (grant no. DNRF96). M.M. is funded by ARC Future Fellowship FT140100516.
References
- 1.Cardinale BJ, et al. 2012. Biodiversity loss and its impact on humanity. Nature 486, 59–67. ( 10.1038/nature11148) [DOI] [PubMed] [Google Scholar]
- 2.Hooper DU, et al. 2012. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature 486, 105–108. ( 10.1038/nature11118) [DOI] [PubMed] [Google Scholar]
- 3.Rockström J, et al. 2009. A safe operating space for humanity. Nature 461, 472–475. ( 10.1038/461472a) [DOI] [PubMed] [Google Scholar]
- 4.Bull JW, Gordon A, Law EA, Suttle KB, Milner-Gulland EJ. 2014. Importance of baseline specification in evaluating conservation interventions and achieving no net loss of biodiversity. Conserv. Biol. 28, 799–809. ( 10.1111/cobi.12243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald-Madden E, et al. 2009. ‘True’ conservation progress. Science 323, 43–44. ( 10.1126/science.1164342) [DOI] [PubMed] [Google Scholar]
- 6.Ferraro PJ, Pattanayak SK. 2006. Money for nothing? A call for empirical evaluation of biodiversity conservation investments. PLoS Biol. 4, 482–488. ( 10.1371/journal.pbio.0040105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madsen B, Carroll N, Kandy D, Bennett G. 2011. State of biodiversity markets report: offset and compensation programs worldwide. Washington, DC: Forest Trends. [Google Scholar]
- 8.Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM. 2015. Accelerated modern human-induced species losses: entering the sixth mass extinction. Sci. Adv. 1, e1400253 ( 10.1126/sciadv.1400253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dirzo R, Young HS, Galetti M, Ceballos G, Isaac NJB, Collen B. 2014. Defaunation in the Anthropocene. Science 345, 401–406. ( 10.1126/science.1251817) [DOI] [PubMed] [Google Scholar]
- 10.Costello MJ, May RM, Stork NE. 2013. Can we name Earth's species before they go extinct? Science 339, 413–416. ( 10.1126/science.1230318) [DOI] [PubMed] [Google Scholar]
- 11.Pimm SL, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO. 2014. The biodiversity of species and their rates of extinction, distribution, and protection. Science 344, 1246752-1. ( 10.1126/science.1246752) [DOI] [PubMed] [Google Scholar]
- 12.Mora C, Rollo A, Tittensor DP. 2013. Comment on ‘Can we name Earth's species before they go extinct?’ Science 341, 237-c. ( 10.1126/science.1237254) [DOI] [PubMed] [Google Scholar]
- 13.Stork NE. 2009. Re-assessing current extinction rates. Biodiv. Conserv. 19, 357–371. ( 10.1007/s10531-009-9761-9) [DOI] [Google Scholar]
- 14.Palkovacs EP, Kinnison MT, Correa C, Dalton CM, Hendry AP. 2012. Fates beyond traits: ecological consequences of human-induced trait change. Evol. Appl. 5, 183–191. ( 10.1111/j.1752-4571.2011.00212.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas CD. 2013. The Anthropocene could raise biological diversity. Nature 502, 7 ( 10.1038/502007a) [DOI] [PubMed] [Google Scholar]
- 16.Thomas CD. 2015. Rapid acceleration of plant speciation during the Anthropocene. Ttends Ecol. Evol. 30, 448–455. ( 10.1016/j.tree.2015.05.009) [DOI] [PubMed] [Google Scholar]
- 17.De Quieroz K. 2007. Species concepts and species delimitation. Syst. Biol. 56, 879–886. ( 10.1080/10635150701701083) [DOI] [PubMed] [Google Scholar]
- 18.Abbott R, et al. 2013. Hybridization and speciation. J. Evol. Biol. 26, 229–246. ( 10.1111/j.1420-9101.2012.02599.x) [DOI] [PubMed] [Google Scholar]
- 19.Fitzpatrick BM, Fordyce JA, Gavrilets S. 2009. Pattern, process and geographic modes of speciation. J. Evol. Biol. 22, 2342–2347. ( 10.1111/j.1420-9101.2009.01833.x) [DOI] [PubMed] [Google Scholar]
- 20.Sobel JM, Chen GF, Watt LR, Schemske DW. 2010. The biology of speciation. Evolution 64, 295–315. ( 10.1111/j.1558-5646.2009.00877.x) [DOI] [PubMed] [Google Scholar]
- 21.Wolf JBW, Lindell J, Backström N. 2010. Speciation genetics: current status and evolving approaches. Phil. Trans. R. Soc. B 365, 1717–1733. ( 10.1098/rstb.2010.0023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulter D. 2009. Evidence for ecological speciation and its alternative. Science 323, 737–741. ( 10.1126/science.1160006) [DOI] [PubMed] [Google Scholar]
- 23.Nosil P, Harmon LJ, Seehausen O. 2009. Ecological explanations for (incomplete) speciation. Trends Ecol. Evol. 24, 145–156. ( 10.1016/j.tree.2008.10.011) [DOI] [PubMed] [Google Scholar]
- 24.Stockwell CA, Hendry AP, Kinnison MT. 2003. Contemporary evolution meets conservation biology. Trends Ecol. Evol. 18, 94–101. ( 10.1016/S0169-5347(02)00044-7) [DOI] [Google Scholar]
- 25.Fitzpatrick JW, et al. 2005. Ivory-billed woodpecker (Campephilus principalis) persists in continental North America. Science 308, 1460–1462. ( 10.1126/science.1114103) [DOI] [PubMed] [Google Scholar]
- 26.Barnosky AD, et al. 2011. Has the Earth's sixth mass extinction already arrived? Nature 471, 51–57. ( 10.1038/nature09678) [DOI] [PubMed] [Google Scholar]
- 27.IUCN (International Union for the Conservation of Nature). 2015. IUCN Red List of Threatened Species. See http://www.iucnredlist.org (accessed 30 January 2015).
- 28.Sandom CJ, Faurby S, Sandel B, Svenning J-C. 2014. Global Late Quaternary megafauna extinctions linked to humans, not climate change. Proc. R. Soc. B 281, 20133254 ( 10.1098/rspb.2013.3254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turvey ST (ed.). 2009. Holocene extinctions. Oxford, UK: Oxford University Press. [Google Scholar]
- 30.Baillie JE, Hilton-Taylor C, Stuart SN (eds). 2004. A global species assessment. Gland, Switzerland: The World Conservation Union; See www.iucnredlist.org [Google Scholar]
- 31.Invasive Species Specialist Group. 2015. Global invasive species database. See http://www.issg.org/database/welcome/ (accessed 30 January 2015).
- 32.Duarte CM, Marbá N, Holmer M. 2007. Rapid domestication of marine species. Science 316, 382–383. ( 10.1126/science.1138042) [DOI] [PubMed] [Google Scholar]
- 33.Monastersky R. 2014. Life under threat. Nature 516, 160. [DOI] [PubMed] [Google Scholar]
- 34.Régnier C, Fontaine B, Bouchet P. 2009. Not knowing, not recording, not listing: numerous unnoticed mollusk extinctions. Conserv. Biol. 23, 1214–1221. ( 10.1111/j.1523-1739.2009.01245.x) [DOI] [PubMed] [Google Scholar]
- 35.Hendry AP, Farrugia TJ, Kinnison MT. 2008. Human influences on rates of phenotypic change in wild animal populations. Mol. Ecol. 17, 20–29. ( 10.1111/j.1365-294X.2007.03428.x) [DOI] [PubMed] [Google Scholar]
- 36.Butchart SHM, et al. 2010. Global biodiversity: indicators of recent declines. Science 328, 1164–1168. ( 10.1126/science.1187512) [DOI] [PubMed] [Google Scholar]
- 37.Molnar JL, Gamboa RL, Revenga C, Spalding MD. 2008. Assessing the global threat of invasive species to marine biodiversity. Front. Ecol. Environ. 6, 485–492. ( 10.1890/070064) [DOI] [Google Scholar]
- 38.Gurevitch J, Padilla DK. 2004. Are invasive species a major cause of extinctions? Trends Ecol. Evol. 19, 470–474. ( 10.1016/j.tree.2004.07.005) [DOI] [PubMed] [Google Scholar]
- 39.DAD-IS (Domestic Animal Diversity Information System). 2015. Hosted by the Food and Agriculture Organisation of the United Nations (FAO). See http://dad.fao.org (accessed 30 January 2015).
- 40.FAO (Food and Agricultural Organisation of the United Nations). 2011. FAO yearbook: fishery and aquaculture statistics. Rome, Italy: Fisheries and Aquaculture Department of the FAO. [Google Scholar]
- 41.Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ. 2008. Adaptive evolution in invasive species. Trends Ecol. Evol. 13, 288–294. ( 10.1016/j.tplants.2008.03.004) [DOI] [PubMed] [Google Scholar]
- 42.Lee CE. 2002. Evolutionary genetics of invasive species. Trends Ecol. Evol. 17, 386–391. ( 10.1016/S0169-5347(02)02554-5) [DOI] [Google Scholar]
- 43.Mooney HA, Cleland EE. 2001. The evolutionary impact of invasive species. Proc. Natl Acad. Sci. USA 98, 5446–5451. ( 10.1073/pnas.091093398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitney KD, Gabler CA. 2008. Rapid evolution in introduced species, ‘invasive traits’ and recipient communities: challenges for predicting invasive potential. Div. Distr. 14, 569–580. ( 10.1111/j.1472-4642.2008.00473.x) [DOI] [Google Scholar]
- 45.Buswell JM, Moles AT, Hartley S. 2011. Is rapid evolution common in introduced plant species? J. Ecol. 99, 214–224. ( 10.1111/j.1365-2745.2010.01759.x) [DOI] [Google Scholar]
- 46.Reznick DA, Bryga H, Endler JA. 1990. Experimentally induced life-history evolution in a natural population. Nature 346, 357–359. ( 10.1038/346357a0) [DOI] [Google Scholar]
- 47.Roderick GK, Navajas M. 2003. Genes in new environments: genetics and evolution in biological control. Nat. Rev. 4, 889–899. ( 10.1038/nrg1201) [DOI] [PubMed] [Google Scholar]
- 48.Abbott RJ. 1992. Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol. Evol. 7, 401–405. ( 10.1016/0169-5347(92)90020-C) [DOI] [PubMed] [Google Scholar]
- 49.Mallet J. 2007. Hybrid speciation. Nat. Rev. 446, 279–283. ( 10.1038/nature05706) [DOI] [PubMed] [Google Scholar]
- 50.Thrall PH, et al. 2011. Evolution in agriculture: the application of evolutionary approaches to the management of biotic interactions in agro-ecosystems. Evol. Appl. 4, 200–215. ( 10.1111/j.1752-4571.2010.00179.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruford MW, Bradley DG, Luikart G. 2003. DNA markers reveal the complexity of livestock domestication. Nature 4, 900–910. ( 10.1038/nrg1203) [DOI] [PubMed] [Google Scholar]
- 52.Streitberger K, Schweizer M, Kropatsch R, Dekomien G, Distl O, Fischer MS, Epplen JT, Hertwig ST. 2012. Rapid genetic diversification within dog breeds as evidenced by a case study on Schnauzers. Anim. Genet. 43, 577–586. ( 10.1111/j.1365-2052.2011.02300.x) [DOI] [PubMed] [Google Scholar]
- 53.Molina J, et al. 2011. Molecular evidence for a single evolutionary origin of domesticated rice. Proc. Natl Acad. Sci. USA 108, 8351–8356. ( 10.1073/pnas.1104686108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haudry A, et al. 2007. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24, 1506–1517. ( 10.1093/molbev/msm077) [DOI] [PubMed] [Google Scholar]
- 55.Palumbi SR. 2001. Humans as the world's greatest evolutionary force. Science 293, 1786–1790. ( 10.1126/science.293.5536.1786) [DOI] [PubMed] [Google Scholar]
- 56.Stenseth NC, Dunlop ES. 2009. Unnatural selection. Nature 457, 803–804. ( 10.1038/457803a) [DOI] [PubMed] [Google Scholar]
- 57.Darimont CT, Carlson SM, Kinnison MT, Paquet PC, Reimchen TE, Wilmers CC. 2009. Human predators outpace other agents of trait change in the wild. Proc. Natl Acad. Sci. USA 106, 952–954. ( 10.1073/pnas.0809235106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersen KH, Brander K. 2009. Expected rate of fisheries-induced evolution is slow. Proc. Natl Acad. Sci. USA 106, 11 657–11 660. ( 10.1073/pnas.0901690106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jørgensen C, et al. 2007. Managing evolving fish stocks. Science 318, 1247–1248. ( 10.1126/science.1148089) [DOI] [PubMed] [Google Scholar]
- 60.Kuparinen A, Merila J. 2007. Detecting and managing fisheries-induced evolution. Trends Ecol. Evol. 22, 652–659. ( 10.1016/j.tree.2007.08.011) [DOI] [PubMed] [Google Scholar]
- 61.Allendorf FW, Hard JJ. 2009. Human-induced evolution caused by unnatural selection through harvest of wild animals. Proc. Natl Acad. Sci. USA 106, 9987–9994. ( 10.1073/pnas.0901069106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Traill LW, Schindler S, Coulson T. 2014. Demography, not inheritance, drives phenotypic change in hunted bighorn sheep. Proc. Natl Acad. Sci. USA 111, 13 223–13 228. ( 10.1073/pnas.1407508111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seastedt TR, Hobbs RJ, Suding KN. 2008. Management of novel ecosystems: are novel approaches required? Front. Ecol. Environ. 6, 547–553. ( 10.1890/070046) [DOI] [Google Scholar]
- 64.Chapin FS III, Starfield AM. 1997. Time lags and novel ecosystems in response to transient climate change in Arctic Alaska. Clim. Change 35, 449–461. ( 10.1023/A:1005337705025) [DOI] [Google Scholar]
- 65.Hunter P. 2007. The human impact on biological diversity. EMBO Rep. 8, 316–318. ( 10.1038/sj.embor.7400951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKinney ML. 2002. Urbanization, biodiversity, and conservation. BioScience 52, 883–890. ( 10.1641/0006-3568(2002)052%5B0883:UBAC%5D2.0.CO;2) [DOI] [Google Scholar]
- 67.Gladieux P, Guerin F, Giraud T, Caffier V, Lemaire C, Parisi L, Didelot F, Le Cam B. 2011. Emergence of novel fungal pathogens by ecological speciation: importance of the reduced viability of immigrants. Mol. Ecol. 20, 4521–4532. ( 10.1111/j.1365-294X.2011.05288.x) [DOI] [PubMed] [Google Scholar]
- 68.Jackson ST, Sax DS. 2009. Balancing biodiversity in a changing environment: extinction debt, immigration credit and species turnover. Trends Ecol. Evol. 25, 153–160. ( 10.1016/j.tree.2009.10.001) [DOI] [PubMed] [Google Scholar]
- 69.Mascaro J, Hughes RF, Schnitzer SA. 2012. Novel forests maintain ecosystem processes after the decline of native tree species. Ecol. Monograph 82, 221–228. ( 10.1890/11-1014.1) [DOI] [Google Scholar]
- 70.Byrne K, Nichols RA. 1999. Culex pipiens in London underground tunnels: differentiation between surface and subterranean populations. Heredity 82, 7–15. ( 10.1038/sj.hdy.6884120) [DOI] [PubMed] [Google Scholar]
- 71.Feindt W, Fincke O, Hadrys H. 2014. Still a one species genus? Strong genetic diversification in the world's largest living odonate, the Neotropical damselfly Megaloprepus caerulatus. Conserv. Genet. 15, 469–481. ( 10.1007/s10592-013-0554-z) [DOI] [Google Scholar]
- 72.Uzogara SG. 2000. The impact of genetic modification of human foods in the 21st century: a review. Biotech. Adv. 18, 179–206. ( 10.1016/S0734-9750(00)00033-1) [DOI] [PubMed] [Google Scholar]
- 73.GMO Database. 2015. GMO Compass. See http://www.gmo-compass.org/eng/gmo/db/ (accessed 30 January 2015).
- 74.Wolfenbarger LL, Phifer PR. 2000. The ecological risks and benefits of genetically engineered plants. Science 290, 2088–2093. ( 10.1126/science.290.5499.2088) [DOI] [PubMed] [Google Scholar]
- 75.Jørgensen D. 2013. Reintroduction and de-extinction. BioScience 63, 719–720. ( 10.1093/bioscience/63.9.719) [DOI] [Google Scholar]
- 76.Sherkow JS, Greely HT. 2013. What if extinction is not forever? Science 340, 32–33. ( 10.1126/science.1236965) [DOI] [PubMed] [Google Scholar]
- 77.Johnston WR. 2014. Deep space probes and other manmade objects beyond near Earth space. Johnston's Archive. See http://www.johnstonsarchive.net/astro/awrjp493.html (accessed 30 January 2015).
- 78.Jönsson KI, Rabbow E, Schill RO, Harms-Ringdahl M, Rettberg P. 2008. Tardigrades survive exposure to space in low Earth orbit. Curr. Biol. 18, R729–R731. ( 10.1016/j.cub.2008.06.048) [DOI] [PubMed] [Google Scholar]
- 79.Sancho LG, de la Torre R, Horneck G, Ascaso C, de los Rios A, Pintado A, Wierzchos J, Schuster M. 2007. Lichens survive in space: results from the 2005 LICHENS experiment. Astrobiology 7, 443–454. ( 10.1089/ast.2006.0046) [DOI] [PubMed] [Google Scholar]
- 80.Rothschild LJ, Mancinelli RL. 2001. Life in extreme environments. Nature 409, 1092–1101. ( 10.1038/35059215) [DOI] [PubMed] [Google Scholar]
- 81.Glavin DP, Dworkin JP, Lupisella M, Kminek G, Rummel JD. 2004. Biological contamination studies of lunar landing sites: implications for future planetary protection and life detection on the Moon and Mars. Int. J. Astrobiol. 3, 265–271. ( 10.1017/S1473550404001958) [DOI] [Google Scholar]
- 82.Weinbauer MG, Rassoulzadegan F. 2007. Extinction of microbes: evidence and potential consequences. Endanger. Species Res. 3, 205–215. ( 10.3354/esr003205) [DOI] [Google Scholar]
- 83.Dobson A, Lafferty KD, Kuris AM, Hechinger RF, Jetz W. 2008. Homage to Linnaeus: how many parasites? How many hosts? Proc. Natl Acad. Sci. USA 105, 11 482–11 489. ( 10.1073/pnas.0803232105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dunn RR, Harris NC, Colwell RK, Koh LP, Sodhi NS. 2009. The sixth mass extinction: are most endangered species parasites and mutualists? Proc. R. Soc. B 276, 3037–3045. ( 10.1098/rspb.2009.0413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cohan FM, Koeppel AF. 2008. The origins of ecological diversity in prokaryotes. Curr. Biol. 18, R1024–R1034. ( 10.1016/j.cub.2008.09.014) [DOI] [PubMed] [Google Scholar]
- 86.Curtis TP, Sloan WT, Scannell JW. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl Acad. Sci. USA 99, 10 494–10 499. ( 10.1073/pnas.142680199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cho JC, Kim SJ. 2000. Increase in bacterial community diversity in subsurface aquifers receiving livestock wastewater input. Appl. Environ. Microbiol. 66, 956–965. ( 10.1128/AEM.66.3.956-965.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wolfe ND, Dunavan CP, Diamond J. 2007. Origins of major human infectious diseases. Nat. Rev. 447, 279–283. ( 10.1038/nature05775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nesse RM, Stearns SC. 2008. The great opportunity: evolutionary applications to medicine and public health. Evol. Appl. 1, 28–48. ( 10.1111/j.1752-4571.2007.00006.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451, 990–993. ( 10.1038/nature06536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.King KC, Stelkens RB, Webster JP, Smith DF, Brockhurst MA. 2015. Hybridization in parasites: consequences for adaptive evolution, pathogenesis, and public health in a changing world. PLoS Pathogens 11, e1005098 ( 10.1371/journal.ppat.1005098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wardeh M, Risley C, McIntyre MK, Setzkorn C, Baylis M. 2015. Database of host–pathogen and related species interactions, and their global distribution. Sci. Data 2, 150049 ( 10.1038/sdata.2015.49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cleaveland S, Laurenson MK, Taylor LH. 2001. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Phil. Trans. R. Soc. Lond B 356, 991–999. ( 10.1098/rstb.2001.0889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host–bacterial mutualism in the human intestine. Science 307, 1915–1920. ( 10.1126/science.1104816) [DOI] [PubMed] [Google Scholar]
- 95.Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, Lonsdorf EV, Pusey AE, Peeters M, Hahn BH, Ochman H. 2014. Rapid changes in the gut microbiome during human evolution. Proc. Natl Acad. Sci. USA 111, 16 431–16 435. ( 10.1073/pnas.1419136111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yatsunenko T, et al. 2012. Human gut microbiome viewed across age and geography. Nature 486, 222–228. ( 10.1038/nature11053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cabañes FJ, Theelen B, Castellá G, Boekhout T. 2007. Two new lipid-dependent Malassezia species from domestic animals. FEMS Yeast Res. 7, 1064–1076. ( 10.1111/j.1567-1364.2007.00217.x) [DOI] [PubMed] [Google Scholar]
- 98.Maron M, et al. In Press Taming a wicked problem: resolving controversies in biodiversity offsetting. BioScience. See http://bioscience.oxfordjournals.org/content/early/2016/04/08/biosci.biw038.abstract . (http://bioscience.oxfordjournals.org/content/early/2016/04/08/biosci.biw038.abstract 10.1093/biosci/biw038. ( doi:10.1093/biosci/biw038 ) [DOI] [Google Scholar]
- 99.Bull JW, Suttle KB, Gordon A, Singh NJ, Milner-Gulland EJ. 2013. Biodiversity offsets in theory and practice. Oryx 47, 369–380. ( 10.1017/S003060531200172X) [DOI] [Google Scholar]
- 100.Estes JA, et al. 2011. Trophic downgrading of planet Earth. Science 333, 301–306. ( 10.1126/science.1205106) [DOI] [PubMed] [Google Scholar]
- 101.Harris G, Thirgood S, Hopcraft JGC, Cromsigt JPGM, Berger J. 2009. Global decline in aggregated migrations of large terrestrial mammals. Endanger. Species. Res. 7, 55–76. ( 10.3354/esr00173) [DOI] [Google Scholar]
- 102.Jetz W, Thomas GH, Joy JB, Redding DW, Hartmann K, Mooers AO. 2014. Global distribution and conservation of evolutionary distinctness in birds. Curr. Biol. 24, 919–930. ( 10.1016/j.cub.2014.03.011) [DOI] [PubMed] [Google Scholar]
- 103.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl Acad. Sci. USA 102, 2567–2572. ( 10.1073/pnas.0409727102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lawrence JG. 2002. Gene transfer in bacteria: speciation without species? Theor. Pop. Biol. 61, 449–460. ( 10.1006/tpbi.2002.1587) [DOI] [PubMed] [Google Scholar]
- 105.Rosenzweig ML. 2001. Loss of speciation rate will impoverish future diversity. Proc. Natl Acad. Sci. USA 98, 5404–5410. ( 10.1073/pnas.101092798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rosenzweig ML. 2003. Reconciliation ecology and the future of species diversity. Oryx 37, 194–205. ( 10.1017/S0030605303000371) [DOI] [Google Scholar]
- 107.Seehausen O, Takimoto G, Roy D, Jokela J. 2008. Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol. Ecol. 17, 30–44. ( 10.1111/j.1365-294X.2007.03529.x) [DOI] [PubMed] [Google Scholar]
- 108.Maron M, Bull JW, Evans MC, Gordon A. 2015. Locking in loss: baselines of decline in Australian biodiversity offset policies. Biol. Conserv. 192, 504–512. ( 10.1016/j.biocon.2015.05.017) [DOI] [Google Scholar]
- 109.Rosenblum EB, Sarver BAJ, Brown JW, Des Roches S, Hardwick KM, Hether TD, Eastman JM, Pennell MW, Harmon LJ. 2012. Goldilocks meets Santa Rosalia: an ephemeral speciation model explains patterns of diversification across time scales. Evol. Biol. 39, 255–261. ( 10.1007/s11692-012-9171-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bull JW, Hardy MJ, Moilanen A, Gordon A. 2015. Categories of flexibility in biodiversity offsetting, and the implications of out-of-kind ecological compensation. Biol. Conserv. 192, 522–532. ( 10.1016/j.biocon.2015.08.003) [DOI] [Google Scholar]
- 111.Kahneman D, Tversky A. 1984. Choices, values, and frames. Am. Psychol. 39, 341–350. ( 10.1037/0003-066X.39.4.341) [DOI] [Google Scholar]
- 112.Burgman M. 2005. Risks and decisions for conservation and environmental management. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 113.Daw TM, et al. 2015. Evaluating taboo trade-offs in ecosystems services and human well-being. Proc. Natl Acad. Sci. USA 112, 6949–6954. ( 10.1073/pnas.1414900112) [DOI] [PMC free article] [PubMed] [Google Scholar]