Abstract

Carbon–carbon bond formation is the key reaction for organic synthesis to construct the carbon framework of organic molecules. The review gives a selection of biocatalytic C–C-bond-forming reactions which have been investigated during the last 5 years and which have already been proven to be applicable for organic synthesis. In most cases, the reactions lead to products functionalized at the site of C–C-bond formation (e.g., α-hydroxy ketones, aminoalcohols, diols, 1,4-diketones, etc.) or allow to decorate aromatic and heteroaromatic molecules. Furthermore, examples for cyclization of (non)natural precursors leading to saturated carbocycles are given as well as the stereoselective cyclopropanation of olefins affording cyclopropanes. Although many tools are already available, recent research also makes it clear that nature provides an even broader set of enzymes to perform specific C–C coupling reactions. The possibilities are without limit; however, a big library of variants for different types of reactions is required to have the specific enzyme for a desired specific (stereoselective) reaction at hand.
Keywords: C−C coupling, C−C-bond formation, biocatalysis, multifunctional products, lyases, transferases, oxidases
1. Introduction
The creation of carbon–carbon (C–C) bonds is the key reaction for organic synthesis to set up the carbon backbone of every organic molecule by connecting smaller substructures in order to gain more complex molecules.1 Thereby the generation of multifunctional products and asymmetry is of inestimable value but at the same time challenging. Consequently, sophisticated methods for asymmetric synthesis as well as protection/deprotection strategies have to be considered. In this context, biocatalytic C–C couplings represent a concise (asymmetric) alternative for the preparation of multifunctional products.2 Biocatalysis is continuously gaining impact for the synthesis of organic molecules, due to the development and identification of improved enzymes enabled by recently advanced methods of molecular and computational biology.3 Recent biocatalytic developments of selected highly active biocatalysts applicable for organic synthesis with tight control of chemo-, regio-, and stereoselectivity are the focus of this Perspective. Enzymes discussed involve aldolases, thiamine-diphosphate (ThDP)-dependent carboligases, Pictet–Spenglerases, oxidases, prenyltransferases, squalene/hopene cyclases or engineered hemoproteins for cyclopropanation. The Perspective has been structured according to the functional group arising from the C–C-bond-forming step or—if not applicable—according to the product formed (Scheme 1). Each functional group or type of product is discussed individually, thereby focusing on enzymes and methodologies which were applied for organic synthesis to transform especially non-natural substrates leading to (multifunctional) targets with high optical purity. In a final chapter, selected examples of promising, novel identified C–C-coupling enzymes will be presented which might get more into focus in the next years. Polyketide synthases,4 methyltransferases, or hydroxy nitrilases are out of the scope of this review.2e
Scheme 1. Main Functional Groups and Type of Products Obtained by Biocatalytic C–C-Bond Formation at the Site of C–C-Bond Formation Discussed in This Perspective.
The multifunctional products are formed by reaction of an acceptor (acceptor part marked in blue) and a donor (red). Additionally saturated carbocycles and cyclopropanes are reviewed subsequently.
2. C–C-Bond Formation Leading to 1,2-Diols Employing Aldolases
2.1. Overview
The most common C–C-bond-forming reaction in organic chemistry is most likely the aldol addition.5 In nature, this reaction is catalyzed by aldolases which perform the reversible and stereoselective addition of a donor to an acceptor.6 Coupling for instance an α-hydroxy carbonyl compound as donor with an aldehyde as acceptor will lead to a 1,2-diol moiety at the site of C–C-bond formation (Scheme 2). The reaction allows the simultaneous formation of two chiral centers.
Scheme 2. General Biocatalytic Route toward the Chiral 1,2-Diol Functionality.

DHAP: dihydroxyacetone phosphate; DHA: dihydroxyacetone; HA: hydroxyacetone; HB: hydroxybutantone; GO: glycolaldehyde. FSA: d-fructose-6-phosphate aldolase; FruA: fructose-1,6-bisphosphate aldolase; RhuA: rhamnose-1-phosphate aldolase; FucA: fuculose-1-phosphate aldolase; TagA: tagatose-1,6-bisphosphate aldolase.
These 1,2-diol-forming aldolases are mainly exploited for the synthesis of carbohydrates and analogues and can be generally divided into (i) dihydroxyacetone phosphate-dependent (DHAP) aldolases and (ii) DHAP-independent aldolases. Important representatives within the DHAP-family are the commercially available fructose-1,6-bisphosphate aldolase (FruA) from rabbit muscle (RAMA), fuculose-1-phosphate aldolase (FucA) from E. coli, tagatose-1,6-bisphosphate aldolase (TagA) and rhamnose-1-phosphate aldolases from E. coli (RhuAEc) or thermophilic Thermotoga maritima (Rhu1PATm).7 The latter has just recently been characterized, and its synthetic potential is currently under investigation.8
Aside from DHAP-dependent aldolases, d-fructose-6-phosphate aldolase from E. coli (FSA) represents an exception. Its unique feature to accept nonphosphorylated donor substrates such as dihydroxyacetone (DHA, also weakly accepted by RhuAEc9), hydroxyacetone (HA), hydroxybutantone (HB), or glycolaldehyde (GO) instead of (expensive) phosphate analogues as well as its unbiased stereoselectivity [syn-configured adducts (3S,4R)] makes this enzyme an attractive target for biocatalytic synthesis. The versatility of the wild-type FSA was already demonstrated in multiple syntheses yielding carbohydrates and analogues,10 whereas most recent investigations focused on the immobilization and scale-up techniques, respectively.11 Detailed knowledge of the enzyme structure,12 the catalytically relevant active-site residues and the catalytic mechanism of FSA has enabled protein engineering to further extend its substrate scope.13 Several robust variants with different substrate preferences have been evolved (e.g., FSA-A129S, FSA-A129S/A165G, FSA-L107Y/A129G or the triple variant FSA-L107Y/A129G/A165G). Moreover, for the first time, a novel variant which exhibits an outstanding donor tolerance toward sterically demanding nucleophiles has just been reported.14 The variant FSA-L107/L163A is able to accept HA-derivatives as donor, including linear, branched and constrained C3 to C7 1-hydroxyalkan-2-ones as well as several DHA-derived ethers (Scheme 3).
Scheme 3. Variant FSA-L107A/L163A Is Able To Accept a Broad Set of Donors Including Long-Chained, Branched or Constrained Alkyl Residues as well as Ether Residues.
Transformations of 3-hydroxybutanal with this unique double variant revealed that it retained the native d-threo diastereoselectivity, thus providing the corresponding C2-substituted sugar analogues with perfect stereoselectivities and high isolated yields (25–89%).
Other enzymes showing promiscuous activity for aldol formation and Michael addition such as lipases15 and the investigated engineered 4-oxalocrotonate tautomerase16 possessing a N-terminal proline as reactive moiety have been reviewed recently.2c
2.2. Carboyhydrates and O-Phosphorylated Sugars and Derivatives
A series of aldose carbohydrates with up to four chiral centers was constructed by sequential de novo connection of simple achiral aldehyde precursors in one pot. Reacting 2 equivalents of glycolaldehyde and 1 equivalent of an acceptor aldehyde (R1CHO) resulted in the desired aldose derivatives (Scheme 4).17
Scheme 4. Biocatalytic of d- and l-Hexose Derivatives from Simple Achiral Precursors by Engineered E. coli-FSA Variants.
Stereochemical control was assured by engineered variants of FSA, which afforded d-aldoses with excellent diastereoselectivities, high conversions (50–98%), and isolated yields. Two double variants (FSA-A129T/S166G and FSA-A129T/A165G) and one triple variant (FSA-A129T/A165G/S166G) emerged as best candidates for the preparation of several deoxy- and O-substituted-d-hexoses. To prepare the corresponding l-aldoses, two FSA variants were operated in a stepwise fashion, whereby for installing the inverted C5 center of the l-aldose, for example, the variant FSA-A129G was employed in the second step (conversions 41–81%).
Although FSA variants proved to work well, also the original wild-type enzyme is still extensively used, which is mainly due to its superior preference for hydroxyacetone compared to most FSA variants. For instance, WT-FSA was selected as catalyst for the preparation of several O-phosphorylated d-ketoses: The cross-aldol addition of glycolaldehyde phosphate and hydroxyacetone furnished 1-deoxy-d-xylulose-5-phosphate, an interesting chemotherapeutic target, with >97% conversion, high isolated yield (85%), and excellent stereoselectivity (only d-threo-configured products). Using the same methodology but d-ribose-5-phophate with hydroxyacetone or 1-hydroxy-2-butanone as substrates, several C8 and C9 d-ketoses were made available as well (77–79% yield, Scheme 5).18 Moreover, therapeutically relevant O-phosphorylated sugars like d-arabinose-5-phophate, d-fructose-6-phosphate, or 1-deoxy-d-fructose-6-phosphate were prepared at 100 mg scale via WT-FSA-coupled cascades.19
Scheme 5. Wild-Type FSA Used for the Aldol Reaction of Glycolaldehyde Phosphate or d-Ribose-5-phosphate with Hydroxyacetone or 1-Hydroxy-2-butanone.
More recently, a combination of a ThDP-dependent carboligase and an aldolase was embedded in a four-step chemoenzymatic route to access 5- or 6-C-aryl carbohydrates with up to good overall yields (11–82%) (Scheme 6).20 In the first step, an enzymatic benzoin reaction of an aromatic aldehyde with dimethoxyacetaldehyde was performed using benzaldehyde lyase from Pseudomonas fluorescens (BAL). After reduction of the resulting 2R-hydroxyketone and removal of the protection group, an aldol addition of (di)hydroxy acetone or glycolaldehyde was carried out by selected FSA variants or RhuA. Although the FSA-catalyzed aldol reactions were stereoselective yielding 6-C-aryl-l-sorbose and 5-C-aryl-l-xylose derivatives, epimeric products with respect to the C4-carbon of the corresponding 6-C-arylated l-fructoses and l-tagatoses were obtained with RhuA.
Scheme 6. Four-Step Two-Enzyme Chemoenzymatic Synthesis of 5- or 6-C-Aryl Carbohydrates Using a Combination of the ThDP-Dependent BAL with the Aldolases FSA or RhuA.
The stereogenic center marked with an asterisk refers to the position where epimerization occurred.
2.3. Imino- and Nitrocyclitols
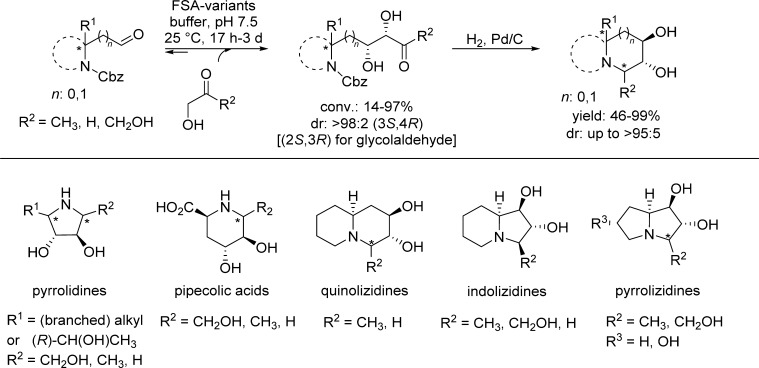
Sugar-related imino- and nitrocyclitols display various biological activities including inhibition of glycosidases, glycotransferases, or hexoaminidases.21 Nitrocyclitols in particular are synthetic precursors of aminocyclitols22 which have important antibacterial, antiviral, or antifungal activities. Moreover, natural nitrosugars have been described.23 The application of WT-FSA for the preparation of several small pyrrolidine and piperidine-based iminocyclitols from azido- and Cbz-aminoaldehydes has already been reported.24 Recently, a structure-guided redesign of several FSA variants expanded the acceptor scope toward bulky α-substituted N-Cbz-aminoaldehydes. Sufficient space in the catalyst’s active site was conferred by altering two essential residues, S166G and R134X (X = P, S, V) followed by combining them with well-established variants from earlier studies. This finally allowed accessing a broad set of “constrained iminocyclitols” like pyrrolizidines, indolizidines, or quinolizidines by FSA with up to >90% de (Scheme 7).25
Scheme 7. Chemoenzymatic Route Toward Iminocyclitols Combining a FSA-Catalyzed Aldol Addition of Chiral Bulky N-Cbz-Aminoaldehydes and a Reductive Amination (H2, Pd/C).
Stereogenic centers marked with an asterisk refer to the positions where epimerization and/or racemization occurred.
While aldol additions with dihydroxyacetone or hydroxyacteone resulted in excellent stereoselectivities [>98:2, (3S,4R)], partial racemization of some N-Cbz-aminoaldehydes during aldol additions with glycolaldehyde was observed depending on the FSA variant used. The reductive amination step was highly stereoselective for the aldol adducts derived from (R)-configured acceptors, furnishing a broad set of linear and branched alkyl-substituted pyrrolidines, quinolizidines, indolizidines, as well as a pipecolic acid derivative as a single stereoisomer. The pyrrolizidine series as well as iminocyclitols resulting from the corresponding (S)-N-Cbz-aminoaldehydes were obtained as a mixture of epimers. Besides FSA-based catalysis, more conformationally constrained iminocyclitols, including indolizidines and quinolizidines were recently prepared via chemoenzymatic methodologies involving the DHAP-dependent aldolases FucA or RhuAEc.26 Furthermore, several benzopyrrolizidine- and cyclohexapyrrolizidine-type cyclitols were readily obtained from a three-step chemoenzymatic synthesis using FucA-F131A variant and an acid phosphate.27 Due to their strong α-glycosidase inhibitory properties,28 pharmaceutically relevant pyrrolidine- and piperidine-based iminocyclitols such as 1,4-dideoxy-1,4-imino-d-arabintol (DAB), its corresponding enantiomer LAB, pipecolic acids, or the nutritionally relevant d-fagomine represent attractive target products of multistep chemoenzymatic syntheses involving the powerful DHAP-independent FSA.29
By coupling l- or d-threonine aldolases with DHAP/DHA aldolases and a reductive amination step, polyhydroxypipecolic acid analogues were obtained as a single diastereoisomer and with excellent recoveries (>99% yield).30 Moreover, the suitability of selected FSA variants to operate in microreactors as “free enzyme” was demonstrated for the synthesis of d-fagomine precursors.31 The same precursor was prepared with high isolated yields (79%) in a three-enzyme cascade, employing FSA-A129S, horse-liver dehydrogenase (HLADH) for the oxidation of the achiral starting material as well as the cofactor recycling enzyme NADH-oxidase (NOX).32
Several stereoselective cascades for the synthesis of nitrocyclitols have been developed. Most of them required at least two enzymes, namely, a DHAP-dependent aldolase and a phytase or phosphatase for phosphate hydrolysis (Scheme 8).33 The sequence consists of the enzyme-catalyzed aldol reaction followed by a spontaneous, stereoselective nitro-Henry reaction of the aldol adduct, furnishing the final nitrocyclitol. Current examples on aldolase-catalyzed nitrocyclitol synthesis mainly focused on optimizations with respect to isolated yield, reaction steps and the employed aldolase. In this context, the need of chemically synthesized DHAP was recently eliminated by performing an ATP-dependent in situ preparation of DHAP from DHA, catalyzed by dihydroxyacetone kinase (DHAK) and an acetate kinase (AK) for ATP-regeneration (Scheme 8, B).34
Scheme 8. Stereoselective Chemoenzymatic Routes Toward Nitrocyclitols: (A) Wildtype-FSA or FSA-A129S-Catalyzed Aldol Reaction Using DHA or HA as Donor; (B) DHAP-Dependent FruA- or RhuA-Catalyzed Aldol Reaction in Combination with the ATP-Dependent Dihydroxyacetone Kinase (DHAK) for the In Situ Preparation of DHAP.
Additionally, an ATP-regeneration system is required.
Choosing either FucA or RhuA, nitrocyclitols with different stereoselectivities at the C2- and C3-carbon were obtained. While FucA provided the expected (2R,3S)-configuration in most cases, RhuA was much less reliable for the expected (2R,3R)-configuration, hence leading to product mixtures. The Henry reaction, however, was highly stereoselective at the C1- and C6-carbon resulting in a trans relative configuration for R′ and NO2.
In another example, a concise one-pot-one-enzyme chemoenzymatic process using wild-type FSA or the variant FSA-A129S was established. Depending on the donor (DHA or HA), several new (deoxy C7) nitrocyclitols were prepared starting from nitrobutanal-related aldehydes (Scheme 8A).35
Despite being independent of DHAP, the main benefits of this route were improved isolated yields (up to 71% compared to FruA33a,33b) and the FSA-based high stereocontrol at the C2- and C3-carbon, respectively. However, rather low diastereoselectivites were obtained, especially if C1 was a methyl group (derived from HA), leading to mixtures of four isomers, which were separated by flash chromatography.
3. C–C-Bond Formation Leading to 1,2-Aminoalcohols: (α-Quaternary)-β-Hydroxy-α-Amino Acids
From a chemical point of view, the stereoselective formation of the 1,2-aminoalcohol scaffold exhibiting additionally a quaternary stereogenic center is considered as a major challenge. Nevertheless, chiral vicinal aminoalcohols are of utmost importance for biotechnological and biomedical purposes. In this context, (α-quaternary)-β-hydroxy-α-amino acids are key products and precursors for multiple fields of application, including immunology, neurology, cancer therapy, infectious diseases, or biomimetic studies.36
Biocatalytically, the 1,2-aminoalcohol motif can be accessed via numerous (multi)enzymatic reactions,37 but in terms of C–C-bond formation, an aldolase-catalyzed addition of an amino donor (shown in red) to a desired aldehyde acceptor (shown in blue) allows the stereoselective, direct implementation of a vicinal aminoalcohol-functionality into a β-hydroxy-α-amino acid final product (Scheme 9).7a
Scheme 9. General Biocatalytic Route toward the Chiral 1,2-Aminoalcohol Functionality at the Site of C–C-Bond Formation.

Gly: glycine; d-Ala: d-alanine; d-Ser: d-serine; d-Cys: d-cysteine; SHMT: serine hydroxymethyltransferase; LTA: l-threonine aldolase; DTA: d-threonine aldolase.
The two aldolases, serine hydroxymethyltransferase (SHMT) and l- or d-specific threonine aldolase (LTA and DTA), are pyridoxal-5′-phosphate (PLP)-dependent enzymes which, based on their donor preference, catalyze the enantioselective and diastereoselective formation of either β-hydroxy-α-amino acids (donor = Gly) or α,α-disubstituted-β-hydroxy-α-amino acids (donor = d-Ala, d-Ser, d-Cys).2e,2f,38 The latter acids bear a quaternary stereogenic center and were recently made available upon finding the first natural aldolases which tolerate amino acid donors other than glycine.36a,39 In general, the stereoselectivity at Cα is enzyme-dependent and was found to be perfect for all so-far identified aldolases (ee >99%); however, low or no stereocontrol at the Cβ resulted mostly in syn/anti mixtures with poor diastereoselectivities. To address this issue, ongoing investigations are majorly focused on enzyme engineering as well as the development of appropriate screening techniques for the identification of improved enzymes.40
Following up on previous studies which aimed for a broader donor tolerance, a single variant of the SHMT from Streptococcus thermophilus (SHMTSth) has been developed recently via rational design with high affinity for d-Ser and d-Ala.41 The SHMTSthY55T-variant catalyzed aldol additions of d-Ser to a broad set of aldehydes with low to almost perfect diastereoselectivities (Scheme 10).
Scheme 10. Chiral α,α-Disubstituted-β-Hydroxy-α-Amino Acids Obtained with SHMTSth or LTA-Av.
Selected examples using either d-Ala or d-Ser are shown. Substrate loadings for SHMTSth: acceptor (2.5 mM), donor (10 mM), 24 h. For LTA-Av: acceptor (50 mM), donor (500 mM), 24 h. n.d.: not determined. n.c.: not converted.
The resulting α,α-disubstituted-β-hydroxy-α-amino acids were always anti with respect to the α-amino and β-hydroxy group, whereas more syn-product (and lower de) was obtained if d-Ala or Gly were employed instead.
At the same time, five novel l- and d-specific threonine aldolases were identified which accept d-Ala and d-Ser. From the investigated LTAs and DTAs in this study, the l-specific TA originating from Aeromonas veronii (LTA-Av) was superior with respect to conversions and furthermore favored the anti-diastereoisomer with moderate de values in most cases.42 The DTAs generally gave lower conversions and preferred the syn-diastereoisomers. The l-selective enzyme LTA-Av was employed for stereocomplementary aldol additions to linear, branched and (hetero)aromatic substrates similar as the SHMT from Streptococcus thermophilus (Scheme 10).
4. C–C-Bond Formation Leading to α-Hydroxyketones
4.1. Overview
α-Hydroxyketones represent an ubiquitous structural motif with a range of applications relevant for the fine chemical industry as well as for the pharmaceutical sector. They widely occur in antidepressants, antifungal agents, antitumor antibiotics (Olivomycin A) or inhibitors of farsenyl transferase (Kurasoin A and B) or amyloid-β-protein (treatment of Alzheimer’s disease).43 Additionally, they are important synthetic intermediates for the preparation of diols or aminoalcohols.
ThDP-dependent enzymes are recognized as a powerful tool for the preparation of enantiopure α-hydroxyketones via C–C-bond formation.44 The key step in the catalytic process involves the reaction of a donor substrate with the thiazolium ring of the cofactor ThDP (Vitamin B1) to produce a highly reactive intermediate via polarity reversal (Umpolung reaction). This intermediate, often referred to as “activated aldehyde”, is described by the mesomeric formulas of a 2α-carbanion and an enamine (Scheme 11). The activated aldehyde-intermediate subsequently attacks a more electrophilic “acceptor” species. If this acceptor is an aldehyde, α-hydroxyketones with a sec-alcohols moiety (sec-α-hydroxyketones) will be formed, whereas ketones as acceptor yield α-hydroxyketones with a tert-alcohol functionality (tert-α-hydroxyketones).
Scheme 11. Acyloin Derivatives Bearing a sec-Alcohol (sec-α-Hydroxyketones).
Phenylacetylcarbinol derivatives (PACs), 2-hydroxypropiophenone analogues (2-HPPs), benzoins, aliphatic acyloins or δ-hydroxy-γ-keto acids are obtained depending on the donor preference of the employed carboligase.
4.2. sec-α-Hydroxyketones/sec-Acyloins
Depending on the donor/acceptor and the employed enzyme various products may be obtained such as phenylacetylcarbinol derivatives (PACs), 2-hydroxypropiophenone analogues (2-HPPs), δ-hydroxy-γ-keto acids, benzoins, or aliphatic acyloins. The latter can be formed by cross-carboligations but mostly arise from homocoupling reactions in which the enzyme uses a single substrate as donor (shown in red) as well as acceptor (blue) (Scheme 11).
The synthetic utility of (R)-phenylacetylcarbinol was demonstrated in recently published two-step one-pot cascades for the preparation of the (1R)-isomers of nor(pseudo)ephedrine [N(P)E] (Scheme 12).45 Excellent stereoselectivity for the corresponding phenylpropanolamines (ee 98−99%, de 97–99%) was assured by coupling the (R)-selective acetohydroxyacid synthase (AHAS-I) from E. coli either with a (R)- or (S)-selective ω-transaminase (ω-TA) from various species. The synthesis of (1R,2R)-norpseudoephedrine went to completion and space-time yields of up to 26 g L–1 d–1 were reached.46 In an analogous reaction, the (R)-selective pyruvate decarboxylase (PDC) from S. cerevisiae was used instead of AHAS-I.47 Due to a lack of a highly (S)-selective carboligase, the corresponding (1S)-isomers were accessed employing enantiocomplementary alcohol dehydrogenases (ADHs) instead.
Scheme 12. Two-Step Cascade Involving (R)-Selective Carboligases and Enantiocomplementary ω-TAs for the Synthesis of the (1R)-Isomers of Nor(pseudo)ephedrine [N(P)E].
More asymmetric cross carboligations of pyruvate with a broad set of aromatic acceptor aldehydes were recently performed with the cyclohexane-1,2-dione hydrolase (CDH)48 from Azoracus sp. CDH turned out to be a powerful tool for the transformation of several electron-rich and electron-deficient benzaldehydes into highly enantioenriched (R)-PACs (ee 92–99%) with up to quantitative conversions.49 Notably, the enzyme also converted sterically demanding substrates, such as t-Bu-or i-Pr-substituted benzaldehydes or 2-naphthaldehyde with >99% conversion and excellent ees (Scheme 13).
Scheme 13. Chiral sec-α-Hydroxyketones Obtained with CDH or ApPDC Variants.
Selected examples are shown. Substrate loadings for CDH: acceptor (10 mM), pyruvate (25 mM), 48 h; ApPDC variants: acceptor (18 mM), 24–48 h.
Semipreparative scale biotransformation of selected bulky-substrates afforded enantiopure products with high isolated yields (87–90%).
CDH was also found to catalyze the formation of (S)-acetoin, a food additive found in many dietary products such as butter, yoghurt, apples, among others, by either homocoupling of pyruvate or acetaldehyde or cross-coupling reactions of pyruvate and acetaldehyde. The aliphatic acetoin is thereby obtained with moderate to good conversions and ees ranging from 87 to 95% depending on the reaction temperature. Mechanistic studies utilizing 13C-labeled substrates further revealed that CDH formed (S)-acetoin solely from pyruvate in an acetolactate-independent pathway.50 This is in contrast to other studied ThDP-dependent enzymes which showed an α-hydroxy-β-keto acid intermediate prior to the formation of almost racemic acetoin.51 An alternative reaction pathway of the CDH is believed to explain the obtained outstanding enantioselectivities.
Another extensively investigated nondecarboxylative ThDP-dependent carboligase is the (R)-selective benzaldehyde lyase (BAL) from Pseudomonas fluorescens (see also Scheme 6).52 BAL-catalyzed carboligations were used for a kinetic resolution transforming preferentially one enantiomer of a racemic α-chiral aldehyde to setup two chiral centers in one reaction (Scheme 14).53 In the case in which benzaldehyde served as donor, perfect enantioselectivities were obtained.
Scheme 14. BAL-Catalyzed Diastereoselective Condensation of Benzaldehyde and Racemic (±)-2-Methyl-Alkyl-Aldehydes Affording Enantiopure 2-HPP Derivatives.
n.r.: not reported.
The diastereoselectivity increased with increasing chain length of the coupled acceptor aldehyde. The authors explained this finding by envisaging that the less sterically demanding (±)-2-methyl butanal could be arranged in a favored and a disfavored manner in the active-site, whereas the sterically more demanding (±)-2-methylpentanal would dock exclusively in its favored position. In general, the syn-products were favored, while the diastereoselectivities of the corresponding PAC-byproducts (formed when benzaldehyde is the acceptor) were poor in all cases. Notably, if enantiopure (S)-2-methylbutanal was employed instead of the racemic precursor, the corresponding PAC byproduct was formed with perfect diastereoselectivity.
Furthermore, the versatility of BAL was demonstrated by embedding the biocatalyst in a one-pot two-step cascade (Scheme 15).54 In situ oxidation of (biobased) aliphatic alcohols to the corresponding aldehydes by Hansenula sp. oxidase and a catalase for disproportionation of hydrogenperoxide followed by the BAL-catalyzed C–C-bond formation yielded optically pure 2-hydroxypropiophenone and analogues. Short-chain aliphatic alcohols (methanol to butanol) were readily accepted in the cascade, thereby achieving excellent enantioselectivities (98–99%) and moderate to excellent conversions (15–99%) of the final products. Reactions with longer-chain, branched, or allylic alcohols however were not successful, except yielding quantitative amounts of (R)-benzoin, as a result of the homocoupling of benzaldehyde as an undesired side-reaction. Testing other oxidases as well as investigating an organocatalytic approach with TEMPO for the oxidation step failed to afford more complex aldehydes. In spite of the low compatibility of BAL with oxidizing agents, BAL tolerated several organic cosolvents (e.g., DMSO, MTBE, butanol, 2-MeTHF, etc.) or deep-eutectic solvents.55 For instance, (R)-benzoin was produced in buffer/2-MeTHF mixtures (5% v/v) employing BAL (half-life 178 ± 8 h) with a productivity of 10 g L–1 h–1.56
Scheme 15. Cascade To Transform (Biobased) Aliphatic Alcohols and Benzaldehyde to Optically Pure 2-Hydroxypropiophenone and Analogues Employing an Alcohol Oxidase and the Benzaldehyde Lyase (BAL).
Hydrogen peroxide is disproportionated by a catalase.
Benzoin served as precursor for the preparation of benzil, a commonly used photoinitiator in polymer synthesis. Whereas numerous biocatalytic approaches for the direct preparation of (R)-benzoin and analogues have been established in the past,57 enzymes catalyzing the formation of the respective (S)-enantiomers have been identified only very recently. By tuning the substrate binding-site of a (S)-selective variant of pyruvate decarboxylase from Acetobacter pasteurianus (ApPDC) by rational design, the direct enzymatic homocoupling of commercially available benzaldehyde derivatives to yield optically active (S)-benzoins was achieved.58 An amino acid sequence-based approach as well as analyzing the active-site characteristics of both enzymes (BAL and ApPDC) allowed the creation of a “chimeric enzyme” by combining the (S)-pocket of ApPDC-E469G and the large donor binding site of the (R)-selective BAL. Consequently, meta- and para-substituted (S)-benzoins with good conversions (up to 97%) and high ee values (85−99%) were prepared. While a triple variant (ApPDC-E469G/T384G/I468A) provided products with high conversions but imperfect ee, the variant differing at four amino acid positions (ApPDC-E469G/T348G/I468A/W543F) displayed superior enantioselectivities and moderate conversions (Scheme 16). Moreover, selected substrates were transformed on a preparative scale yielding product concentrations of up to 4 g L–1.
Scheme 16. Direct Asymmetric Synthesis of Meta- and Para-Substituted (S)-Benzoins Catalyzed by Variants of the ThDP-Dependent Pyruvate Decarboxylase from Acetobacter pasteurianus (ApPDC).
In contrast to the aforementioned carboligases, the ThDP-dependent enzyme MenD accepts α-ketoglutarate (α-KG) as donor, thereby accessing another type of sec-α-hydroxyketones as products, namely, δ-hydroxy-γ-keto acids (Scheme 17). MenD from E. coli (EcMenD) is involved in the menaquinone biosynthesis, where it catalyzes the 1,4-addition of α-ketoglutarate (α-KG) to its natural substrate isochorismate.59 Apart from its physiological reaction, MenD was also found to be an excellent catalyst for the (R)-selective 1,2-addition of α-KG to aromatic aldehydes to afford aromatic δ-hydroxy-γ-keto acids with excellent ee values and conversions (>99%), respectively.60 Besides exploiting the excellent (R)-selectivity of MenD, the enzyme was recently subjected to extensive engineering work in order to invert its stereoselectivity. Consequently, the (S)-pocket concept61 led to the design of several powerful double variants, being able to preferentially convert meta-substituted benzaldehydes to the corresponding (S)-δ-hydroxy-γ-keto acids.62 Even more recently, MenD originating from Bacillus subtilis (BsMenD) was similarly engineered toward (S)-selectivity and several benzaldehydes were coupled to α-KG with moderate to high conversions and improved enantioselectivities (compared to EcMenD variants, Scheme 17).63
Scheme 17. Chiral δ-Hydroxy-γ-Keto Acids Obtained with EcMenD or BsMenD Variants.
Selected examples are shown. Substrate loadings: acceptor (20 mM), α-KG (50 mM), 22 h.
4.3. tert-α-Hydroxyketones, α-Hydroxy-1,3-Diketones, and α-Alkyl-α,β-Dihydroxyketones
The preparation of optically pure tertiary alcohols is generally considered as a challenge in organic synthesis. This is mainly attributed to the enhanced steric requirement at the prochiral quaternary carbon atom which often leads to low asymmetric induction.
Additionally to chemical methodologies,64 different biocatalytic routes have been successfully established to yield enantioenriched tertiary alcohols. Besides enzymatic kinetic resolution, reduction of ketones or hydroxylation,65 the direct C–C-bond formation represents an attractive option. In this context, several ThDP-dependent enzymes were recently found to catalyze the formation of tert-α-hydroxyketones accepting ketones as acceptor.
The frequent occurrence of the tert-α-hydroxyketone motif in numerous natural products, biologically active compounds or components relevant for surface protections in the coating industry66 promoted the investigation of such ThDP-dependent enzymes. Depending on the biocatalyst and substrates, two major pathways can be distinguished: (i) asymmetric ketone-donor cross-couplings, whereby pyruvate, butan-2,3-dione, acetoin or methylacetoin are ligated to (activated) ketones or (ii) asymmetric ketone–ketone homocouplings of 1,2-diketones yielding α-hydroxy-1,3-diketones (Scheme 18).
Scheme 18. Biocatalytic Routes toward tert-α-Hydroxyketones Using ThDP-Enzymes: (A) Asymmetric Cross-Coupling of Ketones with a Set of Specific Donors Yielding General tert-α-Hydroxyketones; (B) Asymmetric Homo-Coupling of 1,2-Diketones Yielding α-Hydroxy-α,β-diketones (and a Prochiral Byproduct).

YerE: carboligase from Yersinia pseudotuberculosis; AAS: acetylacetoin synthase; CDH: cyclohexane-1,2-dione hydrolase; Ao:DCPIP OR: acetoin:dichlorophenolindophenol oxidoreductase.
The first reported enzyme which was exploited for the preparation of a broad range of cyclic and open-chain tert-α-hydroxyketones (Scheme 18A) with good conversions (20–99%) and high ee values (up to 96%) is the ThDP-dependent flavoenzyme YerE from Yersinia pseudotuberculosis.67 Although YerE relies on pyruvate as donor substrate, several ketones, including 1,2-diketones and even β-ketoesters were transformed on a preparative scale with reasonable isolated yields (9–39%). At about the same time, crude extracts of Bacillus licheniformis were reported to catalyze a similar biotransformation of 1,2-diketones into α-hydroxy-1,3-diketones (up to 62% yield and 91% ee) via homocoupling (Scheme 18B).68 Further investigations have led to the identification of the ThDP-dependent acetylacetoin synthase (AAS) as the involved biocatalyst. Subsequently, the broad applicability of crude and partially purified AAS was demonstrated in multiple reactions, including coupling of the AAS to the NADH-dependent acetylacetoin reductase (AAR) to access syn-α-alkyl-α,β-dihydroxyketones with high enantioselectivities (>95%) (Scheme 19).69 Interestingly, the corresponding prochiral diketone byproducts were not reduced by the strictly (S)-stereospecific AAR and could be separated from the final products by flash chromatography. The AAS-AAR-combination was used by the same group for the synthesis of the chiral green tea flavor compound 3-hydroxy-3-methylnonane-2,4-dione.70 Furthermore, the stability of AAS was very recently demonstrated by operating the enzyme in flow-mode fixed-bed microreactors. Reasonable volumetric activities (3.5 U g–1) as well as the long-term stability of the immobilized biocatalyst (up to 15 days) have turned the enzyme into an attractive target for future applications.71
Scheme 19. Cascade for the Synthesis of Enantioenriched syn-α-alkyl-α,β-Dihydroxyketones Using Crude Extracts of Bacillus licheniformis Containing the Acetylacetoin Synthase (AAS) and Acetylacetoin Reductase (AAR).
Another enzyme named acetoin:dichlorophenolindophenol oxidoreductase (Ao:DCPIP OR) was also described to catalyze a ThDP-dependent oxidative cleavage of acetoin.72 In 2015, Ao:DCPIP OR and AAS, both from Bacillus licheniformis, were heterologously expressed in E. coli, purified and compared for their ability to catalyze the aforementioned cross- and homoligations of several 1,2-diketones. Interestingly, both enzymes showed remarkable similarities in terms of product composition and enantioselectivities; however, it remains to be clarified whether AAS and Ao:DCPIP OR are actually the same enzyme.73
The so-far hardly suppressed side-reaction in the homocoupling of 1,2-diketones (see Scheme 18B) could be successfully avoided by employing methylacetoin (3-hydroxy-3-methyl-butan-2-one) as alternative donor for Ao:DCPIP OR-catalyzed carboligations.
Apart from AAS and YerE, a double variant of the already above-discussed CDH was recently designed and applied for the preparation α-hydroxyketones with a tert-alcohol function. While the WT-CDH solely performs hydrolysis of its model substrate cyclohexane-2,3-dione to 6-oxohexanoic acid,48c the hydrolytic activity was successfully knocked down by mutations at residues crucial for the original cleavage activity (H28, N484).74 The double CDH-variant H28A/N484A was then able to perform a C–C-bond formation between pyruvate and cyclohexane-2,3-dione, yielding the corresponding cyclic tert-α-hydroxyketone with 88% ee (Scheme 20).75
Scheme 20. Reversing the C–C-Cleavage Activity of CDH to C–C-Ligation by Inserting Two Mutations at H28 and N484.
CDH-variant H28A/N484A preferred pyruvate as donor substrate (butan-2,3-dione was only weakly accepted) and furthermore displayed a similar acceptor tolerance as YerE and Ao:DCPIP OR. When comparing YerE, Ao:DCPIP OR, and the CDH variant in terms of catalytic performance under optimized conditions for a selected set of substrates, it turned out that the CDH variant provided products with higher enantioselectivities while the obtained conversions were generally lower than with the other two enzymes (Scheme 21).
Scheme 21. Chiral tert-α-Hydroxyketones Obtained with YerE, CDH-Variant H28A/N484A or Ao:DCPIP OR.
Selected examples are shown. Substrate concentrations for YerE: acceptor (20 mM), pyruvate (50 mM); CDH-variant: acceptor (20 mM), pyruvate (50 mM); Ao:DCPIP OR: acceptor (10 mM), methylacetoin (10 mM). n.d. not determined.
Interestingly, YerE and the CDH variant formed the (S)-enantiomer using 3,4-hexandione as substrate (Scheme 21, bottom row – middle), while the (R)-enantiomer was obtained with Ao:DCPIP OR. Notably, the latter enzyme also showed excellent activity toward 2-oxopropanamide which has never been tested with any other ThDP-dependent enzyme before. Despite still requiring optimizations to achieve generally higher stereoselectivities, the biocatalytic toolbox for the preparation of tertiary alcohols via direct C–C-bond formation evolves as an alternative to chemical routes.
5. 1,4-Dicarbonyls: 1,4-Diketones and γ-Keto Acids
While ThDP-dependent carboligases are mostly known for their ability to catalyze a 1,2-addition of a ThDP-bound “activated aldehyde” to the carbonyl moiety of an aldehyde or ketone, resulting in the formation of an α-hydroxyketone (vide supra), selected members of this enzyme family were also found to perform 1,4-additions.44a,44b This biocatalytic version of a Stetter reaction is feasible due to the electron-poor C=C-double bond of the Michael acceptor (shown in blue), hence allowing a donor (red) to attack the β-carbon of the substrate (Scheme 22).
Scheme 22. General Biocatalytic Stetter Reaction Toward Chiral 1,4-Dicarbonyls.

α-KG: α-ketoglutarate; PigD: ThDP-dependent carboligase from Serratia macescens; MenD: 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase from E. coli; SeAAS: ThDP-dependent carboligase from Saccharopolyspora erythraea; HapD: ThDP-dependent carboligase from Hahella chejuensis..
The enzyme PigD from Serratia marcescens, the first identified “stetterase”, adds (decarboxylated) pyruvate (Pyr) to α,β-unsaturated ketones, therefore yielding enantioenriched 1,4-diketones.76 Prompted by this discovery as well as the importance of the 1,4-diketone products as building blocks for heterocycle synthesis (Paal-Knorr), sequence-based homology searches recently led to the identification of two new members of the “stetterase-family”, namely, SeAAS from Saccharopolyspora erythraea and HapD from Hahella chejuensis.77 Both enzymes show similar enantioselectivities as PigD (Scheme 23); conversions were sometimes lower and also the soluble expression of these new enzymes remains to be improved.
Scheme 23. 1,4-Dicarbonyls Obtained with PigD, SeAAS, HapD, or MenD.
Selected examples are shown. Substrate concentrations for PigD, SeAAS, HapD: acceptor (20 mM), pyruvate (25 mM), reaction volume (1.5 mL), 24 h. Substrate concentrations for MenD: acceptor (15 mM), α-KG (30 mM), 24 h.
Along with the classical “stetterases”, the nonsequence related ThDP-dependent MenD from E. coli, catalyzes a Stetter-type addition of α-ketoglutarate (α-KG) to its natural substrate isochorismate. First attempts to improve this 1,4-addition activity toward β-substituted α,β-unsaturated acids were limited to (2S,3R)-2,3-dihydroxy-2,3-dihydrobenzoate. A focus has subsequently been set toward the 1,2-additions of α-KG to various aromatic aldehydes, yielding δ-hydroxy-γ-keto acids (vide supra).59b Investigations on an extended substrate scope revealed that MenD indeed performs Stetter-type reactions, thereby converting α-KG and acrylates and acrylonitrile into γ-keto acids with moderate to good conversions (Scheme 23, bottom row).78 Although the acceptor scope is still very limited, these results are a good starting point for further optimizations.
6. C–C Formation at Aromatic and Heteroaromatic Carbons
6.1. Overview
The C–C-bond formation reactions in this chapter include reactions on indole, aniline and phenol moieties catalyzed by several different enzyme classes such as lyases (chapter 6.2–6.4), transferases (6.5), and oxidoreductases (6.6–6.9). For instance, the Pictet–Spengler reaction79 is catalyzed by strictosidine synthases80 condensing tryptamine and secologanin affording (S)-strictosidine (chapter 6.2) and by norcoclaurine synthases catalyzing the formation of (S)-norcoclaurine from dopamine and 4-hydroxyphenylacetaldehyde (chapter 6.3). Tyrosine phenol lyase catalyzes the formation of tyrosine derivatives from pyruvate, phenols, and ammonia (chapter 6.4). Regarding the group of transferases, prenyltransferases from the dimethylallyltryptophane synthase (DMATS) superfamily are reviewed catalyzing the transfer of the isoprene unit from dimethylallyl diphosphate (DMAPP) onto aromatic ring systems such as indole or phenol moieties in a regioselective fashion (chapter 6.5). SAM-dependent transferases for C–C-bond formation or hydroxynitrile lyases were less investigated during the previous years and have been reviewed elsewhere.2e
Finally, oxidoreductases perform C–C formation at the expense of molecular oxygen. Examples of oxidoreductases are laccases (chapter 6.6–6.8) and the berberine bridge enzyme which perform inter- and intramolecular oxidative bond formations preferentially on phenolic substrates (chapter 6.9).
In general, different reaction types can be distinguished (Scheme 24): (i) intermolecular reaction by attaching (functional) moieties onto the ring scaffold with (Scheme 24A) and without cyclization (Scheme 24B) as well as (ii) intramolecular cyclization reaction (Scheme 24C).
Scheme 24. General Overview on C–C-Bond Formations Involving Aromatic Moieties: (A) Intermolecular C–C Bond Formations without Ring Formation; (B) Intermolecular C–C Bond Formations Involving a Cyclization Step; (C) Intramolecular Cyclization By the Berberine Bridge Enzyme (BBE).

6.2. Biocatalytic Pictet–Spengler Reaction Leading to Tetrahydro-β-carbolines
Tetrahydro-β-carbolines are indole alkaloids exhibiting strong biological activity; for example, some of them are known as potent inhibitors of monoamine oxidase and thus find application in the treatment of depression and anxiety disorders.81 Strictosidine synthase (STR)79,80 catalyzes the Pictet–Spengler condensation of the natural substrates tryptamine and the aldehyde secologanin to form (S)-strictosidine (Scheme 25). It is reported to be mainly restricted to close derivatives of secologanin as aldehyde substrate but accepts various substituted tryptamine derivatives.82 Nevertheless, spectroscopic data indicated that the strictosidine synthase from Ophiorrhiza pumila might transform other aldehydes at 1 mM substrate concentration.83
Scheme 25. Strictosidine and Derivatives Obtained via Strictosidine-Synthase-Catalyzed Cyclization: (A) Natural Reaction of Tryptamine and Secologanin and Recent Examples of Derivatives of the Aromatic Core; (B) Cascade Transforming Prochiral Ketones to Optically Pure Amine Intermediates Which Were Condensed with Secologanin to Various Indole Substituted 3-Methyl Strictosidine Derivatives.
In recent studies, preparative transformations have been reported by applying flow chemistry condensing tryptamine or 7-aza-tryptamine with secologanin giving strictosidine (1.0 g, 73% yield) and 12-aza-strictosidine (0.7 g, 50% yield) using immobilized enzyme (Scheme 25A).84 Additionally 1H-indole-ethanamine was accepted at 10 mM substrate concentration to give access to strictosidine analogues harboring the piperazino[1,2-a]indole scaffold (Scheme 25A).85
Very recently, a two-step biocatalytic cascade consisting of the simultaneous amination of ketones employing ω-transaminases and the Pictet–Spengler condensation with secologanin by STRs gave access to (S)-strictosidine derivatives with an additional stereogenic center at C3 of the tetrahydro-β-carboline core (Scheme 25B). By using stereocomplementary transaminases from Arthrobacter sp. and Silibacter pomeroyi, both epimers were obtained with high diastereoseletivities.86 Protein engineering of the STR from Rauvolfia serpentina by a small focused library of circular permutated variants demonstrated that the enzyme tolerates termini relocation without loss of its overall structure and yielded several active variants, however with lower activity toward the enzyme’s natural substrates.87
6.3. Biocatalytic Pictet–Spengler Reaction Leading to 1,2,3,4-Tetrahydroisoquinolines
The 1,2,3,4-tetrahydroisoquinoline structural motif can be found in several antitumor antibiotics.88 Norcoclaurine, a 1-benzyltetrahydroisoquinoline alkaloid, was found to act as β2-adrenergic receptor agonist used in the treatment of asthma and possesses also antiplatelet and antithrombotic activity.89 The formation of (S)-norcoclaurine, the precursor of all benzylisoquinoline alkaloids, is catalyzed by norcoclaurine synthases (NCS) by stereoselective C–C coupling of dopamine and 4-hydroxyphenylacetaldehyde (Scheme 26).
Scheme 26. Norcoclaurins Synthase (NCS)-Catalyzed Pictet–Spengler Reaction and Its Scope.
Several studies on the substrate scope of NCSs from Thalictrum flavum (TfNCS) and Coptis japonica (CjNCS2) revealed a broad aldehyde substrate tolerance as both enzymes transform several phenylacetaldehyde derivatives substituted with various electron-withdrawing or -donating groups, heteroatoms, and aliphatic side chains.90 Notably, CjNCS2 also accepted linear aliphatic aldehydes as well as α-substituted alkyl groups, the latter however with lower activity90c while TfNCS accepted naphtha-1-ylacetaldehyde.90b In contrast to the aldehyde substrate, only small variations on the dopamine substrate are allowed, as the meta-hydroxy moiety of dopamine is crucial for turnover. Thus, metaraminol and 3-hydroxy-phenethylamine were accepted as non-natural amine substrates.90a The exceptional tolerance toward aldehyde substrates is consistent with docking studies performed with the structure of TfNCS, which state that the most favorable binding modes for reaction intermediates expose only the R-group of the aldehyde to the solvent. The poor acceptance of α-substituted aldehydes by TfNCS can be rationalized by steric hindrance and was improved by the variant L76A, which improved the activity toward (S)- and (R)-citronellal.91 To prove the applicability of NCSs, optically active 1-substituted 1,2,3,4-tetrahydroisoquinolines were synthesized on preparative scale (10 mg/mL) affording the products in good to excellent conversions (86–99%) and high optical purity (95–98% ee).90c By simultaneously combining the norcoclaurine synthase with a transaminase, selected (S)-benzylisoquinoline alkaloids were prepared via a cascade (Scheme 27).92 Starting from the amine, a fraction of the amine was deaminated using a transaminase and pyruvate as formal oxidant giving the second substrate for the norcoclaurine synthase, the aldehyde. Conversions up to 86% were obtained.
Scheme 27. Two-Enzyme Cascade Using Only Amine as Substrate To Prepare Benzylisoquinoline Alkaloids.
6.4. p-Vinylphenols Using a Lyase in the C–C Forming Step
para-Vinylphenols are widely used as building blocks of polymers, which can be used for advanced materials as for example chemical or biological sensors93 or flame retardants.94
A biocatalytic cascade reaction transforming para-substituted phenol derivatives into the corresponding para-vinylphenols was set up employing three simultaneous enzymatic steps (Scheme 28).95 In the first step a tyrosine phenol lyase (TPL)96 from Citrobacter freudii catalyzed the C–C coupling between phenol derivatives and pyruvate in the presence of ammonia yielding tyrosine derivatives. The obtained intermediate is deaminated by tyrosine ammonia lyase (TAL) from Rhodobacter spharoides to give a coumaric acid derivative which is finally decarboxylated by ferulic acid decarboxylase (FAD) from Enterobater sp. A variety of 2- or 3-substituted phenols as well as 2,3-disubstituted phenols were quantitatively transformed giving high isolated yields on a preparative scale. Tyrosine phenol lyase has also recently been used in a cascade starting from substituted benzenes, which were enzymatically hydroxylated to the corresponding phenol derivative and then coupled to pyruvate yielding tyrosine derivatives.97 By combining tyrosine phenol lyase with a decarboxylase tyramine was obtained.98
Scheme 28. Para-Selective Vinylation of Phenols via a Three-Step Cascade Employing a Tyrosine Phenol Lyase in the C–C-Forming Step.
TAL: tyrosine ammonia lyase; FAD: ferulic acid decarboxylase.
6.5. Prenylated Indoles and Phenols
As review articles on prenyltransferases (PTs) were published recently,99 only a brief overview is given on selected reactions with regard to substrate and alkyl donor. For instance, the tyrosine O-prenyltransferases SirD (Leptospheria maculans) and TyrPT (Aspergillus niger) were able to catalyze the C7-prenylation of l-tryptophan derivatives (Scheme 29A).100 This is the natural reaction carried out by the tryptophan C7-prenyltransferase 7-DMATS from A. fumigatus, which accepted also tyrosine and derivatives as substrates by catalyzing the O-prenylation of the phenolic OH of tyrosine and its derivatives.101 In contrast, C3-prenylation of tyrosine can be observed by the tryptophan C4-prenyltransferase FgaPT2 (A. fumigatus). The enzyme was subsequently significantly improved by the single point mutation K174F, exerting >200% relative activity compared to the wild type (Scheme 29B).102
Scheme 29. Selected Recent Examples of Prenylation: (A) Tyrosine O-Prenyltransferases SirD (Leptospheria maculans) and TyrPT (Aspergillus niger) Catalyzed C7-Prenylation of l-Tryptophan Derivatives; (B) C3-Prenylation of Tyrosine by the Tryptophan C4-Prenyltransferase FgaPT2 (A. fumigatus); (C) C5-Benzylation of l-Tryptophan by FgaPT2; (D) C4-Prenylation of 1-Naphthol by the Prenyltransferase BAE61387 from A. oryzae; (E) Prenyltransferase from A. fumigatus Active on Nonaromatic Carbon Atoms.

In contrast to the rather high flexibility of prenyltransferases toward their aromatic prenyl acceptors, a strict specificity regarding their prenyl donor has been assumed. Nevertheless, previous studies already revealed that some prenyltransferases from the dimethylallyltryptophane synthase superfamily also accept unnatural alkyl donors with similar structures as the natural prenyl donor dimethylallyl diphosphate (DMAPP). Recently, however, five different PTs were found to be able to catalyze the benzylation of their preferred substrates using benzyl diphosphate as donor substrate. The best results were achieved by the l-tryptophan prenyltransferase FgaPT2, giving conversions of up to 83% (400 μg/mL enzyme, substr. conc.: 1 mM tryptophan, 2 mM benzyl diphosphate, 16 h) for the C5-benzylation of l-tryptophan (Scheme 29C). Furthermore, also several tryptophan derivatives were accepted and benzylated either at the C5 or C6 position.103 The prenyltransferase BAE61387 from A. oryzae catalyzes the prenylation of hydroxynaphthalenes and was shown to accept not only prenyl but also geranyl and farnesyl as alkyl donors.104 It selectively prenylated C4 of 1-naphthol (Scheme 29D) and 1,7-dihydroxynaphtahalene as well as C3 of 2,7-dihydroxynaphthalene, thus, the para- and ortho-positon of the hydroxyl group of the same benzene ring, respectively.
Additionally, a prenyltransferase active on nonaromatic carbon atoms has been identified in A. fumigatus. Subsequently, it also produced a novel α-prenylindolylbutenone from indolylbutenone as prenyl acceptor (Scheme 29E) [conversion of 46% in the case of (E)-4-(1H-indol-3-yl)but-3-en-2one].105
6.6. Oxidative C–C Coupling To Form Carbazoles by a Laccase
Carbazoles exhibit not only a large variety of biological activities such as antimicrobial, anticancer, or anti-inflammatory properties,106 but they are also interesting building blocks for luminescent polymers or photovoltaic devices.107 A symmetric carbazole derivative was the product of a biotransformation of a meta, para-disubstituted arylamine using the laccase CotA from Bacillus subtilis (Scheme 30).108 This intramolecular oxidative coupling led to the formation of a carbazole framework rather than phenazine or phenoxazinone frameworks, which were reported for related substrates. After the biotransformation of 2,4-diamindiphenylamine with CotA, the insoluble product was isolated as dark purple solid.
Scheme 30. Formation of a Carbazole Derivative Using the Laccase CotA from Bacillus subtilis.
6.7. Oxidative C–C Coupling To Form Dimerized Phenols by a Laccase
Biphenols such as the well-known bisphenol A or 4,4′-biphenol are known for their cytotoxic and estrogenic activities, while 2,2′-biphenol possesses anti-inflammatory properties.109 Owing to their biological activities, biphenols are for example used as building blocks for herbicides.110 The commercially available laccase from Myceliophthora thermophila (Novozym 51003) catalyzed the selective oxidative C–C homocoupling reaction of ortho- and para-diphenolic as well as monophenolic compounds. Especially coupling reactions of 2,6-disubstituted phenols showed to be highly selective and yielded symmetrical products (Scheme 31). The applicability was demonstrated on a multigram scale transforming 2,6-diisopropyl phenol (10 g) with 0.01 mol % biocatalyst loading, which afforded the product in 70% isolated yield. The oxidation product can easily be reduced to the antibacterial agent dipropophol.111
Scheme 31. Dimerization of Phenols Using the Commercial Laccase from Myceliophthora thermophila.
Novozym 51003.
6.8. Oxidative C–C Coupling To Form Benzo[b]furans by a Laccase
Benzo[b]furan derivatives are target molecules for the pharmaceutical industry due to their wide range of biological activities, as they act as antifungal,112 antimicrobial113 and antitumor agents.114
Catechols were transformed with 1,3-dicarbonyls to benzo[b]furans by a Michael addition reaction with in situ generation of o-quinone employing a commercially available laccase from Myceliophthora thermophila (Suberase) (Scheme 32).115 Besides linear 1,3-dicarbonyls also cyclohexane-1,3-dione derivatives were accepted which were substituted by methyl-, dimethyl- or phenyl-groups at the C5 position (meta to both carboxy moieties). This approach eliminated the use of Lewis acid and lipase, which were used in previous methods,116 and afforded benzo[b]furan products with potent cytostatic effects against several cancer cell lines.
Scheme 32. Formation of Benzo[B]furans by Oxidative Coupling of Catechols with 1,3-Dicarbonyls Using Laccases.
6.9. Oxidative C–C Coupling To Yield Berbines by the Berberine Bridge Enzyme
Berbines are plant alkaloids from the protoberberine group of benzylisoquinoline alkaloids and have a long history of medicinal use. They show diverse biological activities such as antimicrobial117 or anti-inflammatory118 effects. Furthermore, it has been demonstrated that berbines possess central nervous system activities and might act as an herbal antidepressant119 as well as protectant against Alzheimer’s disease.120 (S)-Scoulerine, the branch point intermediate for the pathway leading to berberine, is synthesized from (S)-reticuline by intramolecular, oxidative C–C-bond formation of the so-called berberine-bridge by the flavin-dependent berberine bridge enzyme (BBE).
Various racemic non-natural substrates were subjected to kinetic resolution via enantioselective oxidative ring closure by BBE from California poppy (Eschscholzia californica) yielding the (S)-berbine derivatives and the unreacted (R)-substrates both in excellent optical purity and in good to excellent isolated yields (Scheme 33A).121 The maximal conversion levels of 50% were reached with perfect enantioselecitvity of the enzyme (E > 200). The applicability of the enzyme was demonstrated employing BBE in preparative scale reactions of 500 mg substrate, which yielded the optically pure products.
Scheme 33. (A) Substrate Scope of the Berberine Bridge Enzyme (BBE) Catalyzing C–C-Bond Formation at the Expense of Molecular Oxygen in a Kinetic Resolution; (B) Deracemization To Transform Both Substrate Enantiomers of Benzylisoquinolines to Optically Pure Berbine Derivatives.
To overcome the limitation of a kinetic resolution of the BBE-catalyzed C–C coupling, a deracemization process was developed employing simultaneous kinetic resolution and stereoinversion (Scheme 33B). A variant of the monoamine oxidase from Aspergillus niger (MAO-N variant D11) was utilized to oxidize the (R)-amine substrate not transformed by BBE to the corresponding achiral iminium intermediate. The latter was nonstereoselectively reduced with morpholine·BH3, which did not inhibit the BBE unlike other reducing agents. The redox cascade of MAO/BBE/borane could be performed stepwise or in a concurrent fashion and yielded the optically pure (S)-product with conversions of up to 98%. The applicability of this deracemization process was demonstrated by preparative scale biotransformations (150–165 mg substrate).122
7. Saturated Carbo- and Heterocycles Obtained by Squalene-Hopene Cyclase
Terpenes and the related functionalized terpenoids are composed of differing numbers of isoprene units and represent a highly abundant class of natural products acting as flavors, hormones, or pigments. The large diversity of these compounds can be attributed to terpene cyclases, which catalyze the cyclization reaction of terpene backbones to cyclic terpenoids. The one-step poly cyclization reaction of squalene to the pentacyclic hopene, for example, is catalyzed by squalene-hopene cyclase (SHC), whereby their mode of action resembles a Brønsted acid catalyst (Scheme 34). It has already been demonstrated that the substrate scope of SHCs is not limited to its natural substrate squalene, as several truncated analogues were accepted as already summarized in a review.123
Scheme 34. Cyclization Reactions of Squalene by Squalene-Hopene Cyclase (SHC) to the Pentacyclic Terpenes Hopene and Hopanol.
Recently, also functionalized isoprene backbones were identified as substrates for several (engineered) SHCs (Table 1). The SHCs AasSHC (Alicyclobacillus acidocaldarius) and ZmoSHC1 (Zymomonas mobilis) also accepted substrates which are shortened in their polyisoprene backbone (C16–C13) and functionalized by a hydroxy, carboxy, or keto group. These substrates were converted to bi- or triheterocyclic compounds such as cyclic ethers, lactones, and enol ethers (entries 1–5).124 Mutagenesis studies on AasSHC and a SHC from Zymomonas mobilis (ZMO-1548 gene product) further expanded the substrate scope toward citronellal and geraniol (entries 6–8). Introduction of the single amino acid exchange F486C in ZMO-1548 as well as other SHCs from Z. mobilis, A. pasteurianus, B. japonicum, and A. acidocaldarius led to citronellal cyclase activity while squalene cyclization was reduced (entry 8). The most remarkable results with regard to stereopreference and conversion of rac-citronellal were obtained with ZMO-1548 with the highest ratio of isopurgenol to neo-isopurgenol formation and ZMO-0872 (Z. mobilis) and a total conversion of 75% respectively.125 Mutations introduced on several sites in the AacSHC allowed cyclization of geraniol, 6,7-epoxygeraniol and citronellal, giving rise to cyclogeraniol and isopurgenol products. Notably, a variant was generated (I261A), which was highly selective for (S)-citronellal and led to the formation of (−)-iso-isopurgenol with a selectivity of 99%.126
Table 1. Non-Natural Substrates and Products of Squalene-Hopene Cyclases.
Isolated yield.
8. Cyclopropanation of Olefins and Olefination of Aldehydes
Since the cyclopropanation of olefins does not occur in nature but has been described for transition-state metal catalysis,127 P450 enzymes were engineered to perform this reaction stereoselectively128 using diazoacetate as non-natural reagent (Scheme 35A). While hemin on its own produced mainly the racemic E-product for the cyclopropanation of styrene derivatives, a variant (P411BM3-CIS) showed preference for the Z-product (Scheme 35A). P411 enzymes are obtained by an exchange of the proximal cysteine-ligand of the iron to a serine to facilitate reduction to catalytic active Fe2+.129 Ees up to 75% were reached for the E-product and up to 87% ee for the Z-isomer using evolved enzymes. It is worth mentioning that the introduction of a single additional mutation (I263A) in P411BM3-CIS reversed the diastereoselectivity again to the initial E-preference. Related transformations were achieved employing engineered myoglobin.130 Incorporating a threonine (T268) to alanine mutation into a panel of P450 scaffolds, enantioselective catalysts for all possible diastereomers in the model reaction of styrene with ethyl diazoacetate were identified.131
Scheme 35. P450- and P411-Catalyzed Cyclopropanation of Styrene Derivatives (A) and Acrylamides (B) as well as Cyclopropanation of Styrenes Using a Prolyl Oligopeptidase Scaffold Containing a Di-Rhodium Catalyst (C); (D) Myoglobin-Heme-Catalyzed Olefination of Aldehydes.
One variant (P450BM3-HStar T268A-C400H-L437W-V78M-L181 V), possessing the axial cysteine changed to a histidine thereby increasing the reduction potential even further, was used to catalyze the cyclopropanation of acrylamides (Scheme 35B).132
Similar transformations were catalyzed by an artificial metalloenzyme formed by the incorporation of a dirhodium catalyst into the engineered prolyl oligopeptidase scaffold Pfu POP-ZA4-HFF-1 from Pyrococcus furiosus (Scheme 35C). The construct accepted a range of styrene and donor–acceptor carbene precursors yielding 14–73% of the desired cyclopropanation products with up to 92% ee.133
Following the concept of engineering heme-containing proteins to catalyze non-natural reactions, the variant Mb(F43 V,V68F) of sperm whale myoglobin was found to catalyze the olefination of benzaldehyde derivatives with α-diazo esters in the presence of AsPh3 (Scheme 35D). The catalyst exhibited excellent diastereoselectivity for the E-products (up to >99% de) as well as high chemoselectivity toward olefination over carbine dimerization. In general, electron-deficient benzaldehydes were less reactive than their electron-rich counterparts and conversions never exceeded 50% in spite of the high TONs (up to 3400) that were observed.134
9. C–C-Bond-Forming Enzymes To Be Exploited in the Future
While the previous chapters presented enzymatic reactions which have already been exploited for organic synthesis transforming non-natural substrates, the focus in this section is on promising enzymes which were either just recently discovered or their non-natural substrate scope needs to be tested or further engineering is required prior to application. Promiscuous activity of enzymes e.g. lipases for C–C-bond formation, is summarized in recent reviews.2c,135 Additionally the design of artificial metal enzymes like for Diels–Alder reactions or Friedel–Crafts alkylations may gain impact.135,136
9.1. Cyclases Transforming Squalene
Although several studies have focused on the squalene-hopene cyclase and its substrate promiscuity, enzymes cyclizing squalene to other cyclic terpenes have been identified but barely investigated with regard to non-natural substrate scope and applicability in biocatalytic processes (Scheme 36). For instance, squalene is also the natural substrate of the migrated hopene synthases CPH and CPFa from Colysis pothifolia, which catalyze the cyclization to hop-17(21)-ene and fern-9(11)-ene, respectively. The activity of these two enzymes could be interconverted by introducing one single amino acid exchange in each case (CPH Q276 V and CPFa V281Q), thereby identifying the amino acid residues responsible for controlling the number of 1,2-hydride and methyl shifts in the mechanism.137 Furthermore, the tetraprenyl-β-curcumene cyclase from Bacillus megaterium (BmeTC) is capable of converting squalene into onoceranoxide via a bicyclic intermediate. Furthermore, a two-step reaction of squalene with the AasSHC variant D377C and BmeTC afforded (+)-ambrein, the main constituent of ambergris, which is used in perfume production, however in low yields (3.4%).138 Finally, it has been shown that the natural product of SHC, hopene, can be further utilized by tetrahymanol synthase (THS) from Methylomicrobium alcaliophilum, yielding the sterol surrogate tetrahymanol.139
Scheme 36. Cyclization Products and Follow Up Products Obtained by Squalene Cyclizing Enzymes.
9.2. C–C Ring Closure Reactions in Terpenoid and Alkaloid Biosyntheses
In this subsection, the major focus was placed on C–C-bond formation observed in ring-closing reactions in the biosynthesis of terpenoids and alkaloids.
The biosynthetic gene cluster from Solanum lycopersicon, which catalyzes the synthesis of the diterpenoid lycosantalonol, was reconstructed in E. coli. Cyclization of (Z,Z,Z)-nerylneryl diphosphate by the terpene synthase SlTPS21 generates lycosantalene, a tricyclene core ring structure with a neryl side chain, of which the cis double bond is in vivo transformed to an α-hydroxy keto moiety yielding lycosantalonol (Scheme 37).140
Scheme 37. Cyclization of (Z,Z,Z)-Nerylneryl Diphosphate by the Terpene Synthase SlTPS21 Generates Lycosantalene.
Two terpene synthases which were active on farnesyl diphosphate were identified in Valeriana officinalis: VoTPS7 catalyzes the cyclization to germacrene C, whereas VoTPS1 synthesizes valerena-1,10-diene, which was verified by recombinant overexpression in E. coli and yeast (Scheme 38).141 Both intermediates are further converted in the biosynthesis to valerone and valerenic acid, two bioactive terpenoids found in the herbal preparations of Valeriana officinalis.
Scheme 38. Cyclization Farnesyl Diphosphate Leading Either to Germacrene C or Valerena-1,10-diene.
The conversion of (−)-yatein to (−)-deoxypodophyllotoxin is catalyzed by a 2-oxoglutarate/Fe(II)-dependent dioxygenase (2-ODD) from Podophyllum hexandrum (mayapple), closing the core cyclohexane ring of the aryltetralin scaffold (Scheme 39).142 Ten enzymes—six involved in the conversion of coniferyl alcohol to (−)-podophyllooxin—were recombinantly coexpressed in Nicotiana banthamiana (tobacco), enabling the synthesis of (−)-4′-desmethylepipodophyllotoxin, the immediate precursor of etoposide, which is used in chemotherapy regimens.
Scheme 39. 2-Oxoglutarate/Fe(II)-Dependent Dioxygenase (2-ODD) Closes the Six-Membered Ring Transforming (−)-Yatein to (−)-Deoxypodophyllotoxin.
The halogenated polycycles merochlorin A and B are produced by the marine bacterium Streptomyces sp. strain CNH-189 and feature unique ring systems–a bicycle[3.2.1]octadione and a 6–5–5-fused tricycle (Scheme 40). The final step in the biosynthesis is catalyzed by the vanadium-dependent haloperoxidase Mcl24 and involves a site-selective naphthol chlorination, followed by oxidative dearomatization and terpene cyclization.143 This reaction sequence has already inspired the development of a chemical counterpart, where premerochlorin is treated with N-chlorosuccinimide in the presence of two equivalents of triethylamine, affording a product mixture of merochlorin derivatives in 30% yield. The selectivity profile of the chemical and the enzymatic oxidative cyclization however is inverted, as Mcl24 chlorinates the naphthol prior to cyclization whereas the chemical synthesis used prefers the reversed order.144
Scheme 40. Vanadium-Dependent Haloperoxidase Mcl24 Enables a Site-Selective Naphthol Chlorination, Followed by Oxidative Dearomatization and Terpene Cyclization.
The biosynthesis of ergot alkaloids involves the common intermediate chanoclavine-I, which is produced from tryptophane and dimethylallylpyrophosphate over three steps involving four enzymes. Deletion experiments in Aspergillus fumigatus and Claviceps purpurea indicated that the last step might be catalyzed by two enzymes—EasE and EasC—involving a cyclization step of N-methyl-4-(dimethylallyl)tryptophan (Scheme 41). The mechanism of this reaction remains unknown; however, similarities of EasC to peroxisomal catalases and EasE to berberine bridge enzymes have been recognized. Chanoclavine-I was produced in Saccharomyces cerevisiae by expression of genes from Aspergillus japonicus (dmaW, easE, and easC) and Aspergillus fumigatus (easF); however, only low titers of 0.75 mg L–1 were observed.145 Additionally, the biosynthetic pathway for cycloclavine synthesis—a complex ergot alkaloid containing a cyclopropyl moiety—was reconstructed in S. cerevisiae by expressing four additional genes from A. japonicus (easD, easA, easG, and easH), which enabled synthesis of the desired product in final titers of 529 mg L–1 by fed-batch fermentation.146
Scheme 41. Transformation of N-Methyl-4-(dimethylallyl)tryptophan to Chanoclavine I Presumably Catalyzed by the Two Enzymes EasE and EasC.
9.3. Redox-Enzymes and a Pd-Suzukiase Leading to Biaryls
Axially chiral biaryls are found as motif in natural products and are also of interest for synthetic chemistry, as for example, BINAP and BINOL, which serve as chiral auxiliaries or ligands of chiral catalysts. In nature, the generation of these compounds usually involves oxidative coupling of two phenol moieties by, for example, laccases, peroxidases, or cytochrome P450 enzymes (Scheme 42),147 but also, coupling of pyrroles by flavin-dependent enzymes has been observed.148 While P450 enzymes are able to produce optically active products, laccases and peroxidases catalyze the formation of radicals but not the actual C–C coupling, which results in racemic products. However, regio- and stereoselectivity for laccase reactions can be induced by the presence of a dirigent protein (DIR),149 which binds specifically the radical intermediates but is not catalytically active.
Scheme 42. Asymmetric C–C Coupling of Phenols Leading to Biaryls or Other Dimers Employing P450 Enzymes or Laccases in the Presence of a Dirigent Protein.
Recently, the dirigent protein GHDIR4 was discovered in Gossypium hirsutum var. marie-galante and purified after expression in transgenic plant cells. The recombinant protein was successfully employed to the bimolecular radical coupling of hemigossypol by laccase (Trametes versicolor) obtaining (P)-(+)-gossypol in >80% ee.150 Dirigent proteins not only control the formation of biaryls but also the coupling reaction of coniferyl alcohol to pinoresinol such as the proteins identified in Arabidopsis thaliana (AtDIR5, AtDIR6) and Schizandra chinensis (ScDIR). The proteins were heterologously expressed in insect and plant cells, respectively. Both AtDIR5 and AtDIR6 afforded (−)-pinoresinol (70% ee and 36% ee respectively) upon incubation with laccase (T. versicolor) and the natural substrate, coniferyl alcohol, while ScDIR led to the formation of the antipode (+)-pinoresionol (60% ee). By generating a variant of ScDIR harboring a short region of AtDIR6, the stereoselectivity of the enzyme was reversed, allowing first insights in regio-controlling coupling selectivity.151
In addition to the dirigent proteins, a P450 enzyme catalyzing the direct regio- and stereoselective oxidative coupling of demethylsiderin to (P)-orlandin, a precursor of (P)-(+)-kotanin, was identified in Aspergillus niger. The genetic cluster for the biosynthesis of kotanin consisting of a polyketide synthase (KtnS), an O-methyltransferase (KtnB) and a cytochrome P450 monooxygenase (KtnC) were identified.152 Similar gene clusters were furthermore found in other bicoumarin-producing fungi including Aspergillus flavus, A. alliaceus, and A. oryzae but also in Emericella desertorum. The P450 enzyme of the latter (DesC) as well as KtnC were heterologously expressed in S. cerevisiae and employed to phenol coupling reactions in vivo by feeding 7-demethylsiderin, the precursor of (P)-orlandin and (M)-desertorin A; however, only partial conversions of the substrate was observed (3–7% and 35–55% at 0.5 and 0.05 mM substrate concentration, respectively). Although both enzymes catalyze the homocoupling of 7-demethylsiderin, they differ in their regioselectivity, as KtnC catalyzes the symmetric dimerization to P-orlandin while DesC affords the asymmetric dimer M-desertorin A. Other monomeric coumarins were not accepted by either of the enzymes, indicating a stringent substrate specificity of both “bicoumarin synthases”.153 Gene clusters responsible for the biosynthesis of axially chiral biaryl compounds are not only present in fungi and plants but also in bacteria, since three of these clusters were identified in Streptomyces species. The P450 enzyme JulI was heterologously expressed in E. coli and successfully employed to C–C coupling reactions of julichrome Q6. Genome analysis of different Streptomycis strains indicated the presence of similar gene clusters.154
A different approach made use of an artificial Pd-dependent metal enzyme. Anchoring a biotinylated monophosphine palladium complex within a streptavidin scaffold afforded a stereoselective catalyst applicable for C–C-bond formation according to the well know Suzuki–Miyaura cross-coupling reaction.155 Consequently, the artificial enzyme was named “Suzukiase”. Using an evolved protein, a variety of atropisomeric biaryls were obtained with up to 90% ee (Scheme 43). TONs were in general in the range of 80 up to 160. While the presence of a phenolic moiety is required for the P450 or laccase-catalyzed reactions, the Suzuki–Miyaura needs one substrate activated by boronic acid and one by halogen.
Scheme 43. Stereoselective Aryl Coupling Using a Pd-Dependent Artificial “Suzukiase”.
9.4. Pyruvate Aldolases for the Preparation of 4-Hydroxy-2-oxoacids
The versatility of DHA(P)-dependent aldolases such as FSA, FruA, or RhuA has been proven detailed in a previous chapter. A second class of aldolases, namely, the pyruvate aldolases are currently subject to extensive engineering studies. Naturally, these enzymes catalyze the retro aldol cleavage of a 4-hydroxy-2-oxoacid skeleton into pyruvate and an aldehyde (Scheme 44). The synthetic direction of this reaction is particularly interesting since the enzymes are independent of phosphorylated donors and the formed chiral 4-hydroxy-oxoacid products occur in a variety of natural products with antimicrobial properties.156 In this context, the 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolases157 from E. coli and a closely related isoform from Thermatoga maritima as well as the BphI aldolase from Burkholderia xenovorans(158) are the most important representatives among these catabolic enzymes.
Scheme 44. Pyruvate-Aldolase-Catalyzed C–C-Bond Cleavage/Formation of 4-Hydroxy-Oxoacids.
KDPG aldolases from E. coli and Thermatoga maritima and the BphI aldolase from Burkholderia xenovorans are in the focus of current engineering research in order to widen their substrate specificity.
Recently, important progress has been made in terms of active-site remodeling of the EcKDPG in order to gain highly efficient mutants with enhanced substrate specificity.159 By combining the two mutations S184L and T161S, both crucial to substrate recognition and stereocontrol, respectively, a powerful double mutant (EcKDPG-T161S/S184L) evolved which is able to cleave the non-natural substrate (4S)-2-keto-4-hydroxy-4-(2′-pyridyl)butyrate (S-KHPB) with 450-fold improved kcat/KM-values compared to the wild type. This corresponds to TON of 240 s–1 for S-KHPB which is approximately 3 times higher than that of the WT-enzyme for its natural substrate KDPG (83 s–1). Similarly, the kcat/KM value for cleaving 2-keto-4-hydroxyoctonoate (KHO) by a TmKDPG-A30 V single mutant was improved by 25-fold (compared to the WT) by directed evolution approaches.160 While the native BphI-aldolase exhibits strict (S)-selectivity over the stereogenic center at C4, which is created in the natural product (4S)-hydroxy-2-oxopentanoate [(S)-HOPA], the stereoselectivity was successfully inverted to (R) by rational design.161 Several active-site residues, namely, L89, L87, and Y290 are key to substrate recognition and also determine the stereoselectivty of the enzyme.162 By combining variants (Y290F with either L87N or L87W), two BphI double variants with exclusive (R)-stereoselectivity were created for the first time. Although the catalytic efficiencies of BphI-L87N/Y290F and BphI-L87W/Y290F were ∼40-fold lower for (R)-HOPA compared to the cleavage efficiency of WT-BphI for (S)-HOPA, the novel enzymes are able to synthesize (4R)-hydroxy-oxoacids with up to eight carbons in length and good stereoselectivity, as confirmed by polarimetric analysis.161
10. Conclusion and Outlook
Over the last five years, tremendous advances have been achieved in the field of biocatalytic C–C-bond formation applicable for organic synthesis. This is especially true for enzymatic reactions (i) which have previously been considered as difficult or exotic due to the requirement of special activation of the substrate (e.g., by phosphorylation), (ii) and/or due to limited substrate scope or (iii) which were not known, yet. Identification of novel enzymes and enzyme engineering163 helped to overcome many limitations. Although there is a huge potential in using the biocatalytic toolbox to cut short and simplify organic synthetic routes, its broad applicability is only recognized slowly by synthetic chemists. Thus, the area of enzymatic C–C-bond formation is underused, probably as a result of a lack of awareness of how they might be used to construct organic molecules.164 This might be due to the fact that almost every selective transformation requires a specific enzyme, depending on the substrate. Due to the high chemo- and stereoselectivity of enzymes, a universal catalyst (e.g., for all aldol reactions) is therefore elusive. Thus, a specific C–C-bond formation with desired perfect stereoselectivity calls very often for a unique enzyme.
C–C-bond formations are ubiquitously found in nature for the stereoselective assembling of small building blocks to bigger molecules. Deduced from the huge number of different natural products, many different C–C-bond formation reactions are realized in nature, for which the enzyme responsible was not identified yet. These enzymes “just” need to be identified and applied to opening novel synthetic routes. Enzyme engineering might be requested to allow higher substrate loads and gain better activity. It can be expected that the set of stable enzymes performing unique C–C-bond formations not achievable with established metal-/organo-catalysts will expand significantly in the future. This will enable efficient short synthetic routes to the target molecules via novel (stereoselective) transformations.
N.G.S. has been supported by the Austrian BMWFJ, BMVIT, SFG, Standortagentur Tirol and ZIT through the Austrian FFG-COMET-Funding Program. E.E. was supported by the Austrian Science Fund (FWF, project W9, DK Molecular Enzymology). COST Action CM1303 “Systems Biocatalysis” is acknowledged.
The authors declare no competing financial interest.
References
- a Van de Vyver S.; Roman-Leshkov Y. Angew. Chem., Int. Ed. 2015, 54, 12554–12561. 10.1002/anie.201503701. [DOI] [PubMed] [Google Scholar]; b Liu Q.; Jackstell R.; Beller M. Angew. Chem., Int. Ed. 2013, 52, 13871–13873. 10.1002/anie.201307865. [DOI] [PubMed] [Google Scholar]; c Tsubogo T.; Ishiwata T.; Kobayashi S. Angew. Chem., Int. Ed. 2013, 52, 6590–6604. 10.1002/anie.201210066. [DOI] [PubMed] [Google Scholar]; d Schonherr H.; Cernak T. Angew. Chem., Int. Ed. 2013, 52, 12256–12267. 10.1002/anie.201303207. [DOI] [PubMed] [Google Scholar]; e Cho S. H.; Kim J. Y.; Kwak J.; Chang S. Chem. Soc. Rev. 2011, 40, 5068–5083. 10.1039/c1cs15082k. [DOI] [PubMed] [Google Scholar]; f Monnier F.; Taillefer M. Angew. Chem., Int. Ed. 2009, 48, 6954–6971. 10.1002/anie.200804497. [DOI] [PubMed] [Google Scholar]; g Alexakis A.; Backvall J. E.; Krause N.; Pamies O.; Dieguez M. Chem. Rev. 2008, 108, 2796–2823. 10.1021/cr0683515. [DOI] [PubMed] [Google Scholar]; h Trost B. M.; Toste F. D.; Pinkerton A. B. Chem. Rev. 2001, 101, 2067–2096. 10.1021/cr000666b. [DOI] [PubMed] [Google Scholar]
- a Faber K.; Fessner W. D.; Turner N. J.. Biocatalysis in Organic Synthesis. Science of Synthesis, Vol. 1–3; Georg Thieme: Stuttgart, 2015. [Google Scholar]; b Lechner H.; Pressnitz D.; Kroutil W. Biotechnol. Adv. 2015, 33, 457–480. 10.1016/j.biotechadv.2015.01.012. [DOI] [PubMed] [Google Scholar]; c Miao Y. F.; Rahimi M.; Geertsema E. M.; Poelarends G. J. Curr. Opin. Chem. Biol. 2015, 25, 115–123. 10.1016/j.cbpa.2014.12.020. [DOI] [PubMed] [Google Scholar]; d Wessjohann L. A.; Keim J.; Weigel B.; Dippe M. Curr. Opin. Chem. Biol. 2013, 17, 229–235. 10.1016/j.cbpa.2013.02.016. [DOI] [PubMed] [Google Scholar]; e Fesko K.; Gruber-Khadjawi M. ChemCatChem 2013, 5, 1248–1272. 10.1002/cctc.201200709. [DOI] [Google Scholar]; f Müller M. Adv. Synth. Catal. 2012, 354, 3161–3174. 10.1002/adsc.201100655. [DOI] [Google Scholar]; g Faber K.Biotransformations in Organic Chemistry, 6th revised ed.; Springer: Heidelberg, 2011. [Google Scholar]; h Resch V.; Schrittwieser J. H.; Siirola E.; Kroutil W. Curr. Opin. Biotechnol. 2011, 22, 793–799. 10.1016/j.copbio.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; i Brovetto M.; Gamenara D.; Mendez P. S.; Seoane G. A. Chem. Rev. 2011, 111, 4346–4403. 10.1021/cr100299p. [DOI] [PubMed] [Google Scholar]
- a Bornscheuer U. T. Angew. Chem., Int. Ed. 2016, 55, 4372–4373. 10.1002/anie.201510042. [DOI] [PubMed] [Google Scholar]; b Kries H.; O’Connor S. E. Curr. Opin. Chem. Biol. 2016, 31, 22–30. 10.1016/j.cbpa.2015.12.006. [DOI] [PubMed] [Google Scholar]; c Choi J. M.; Han S. S.; Kim H. S. Biotechnol. Adv. 2015, 33, 1443–1454. 10.1016/j.biotechadv.2015.02.014. [DOI] [PubMed] [Google Scholar]; d Prier C. K.; Arnold F. H. J. Am. Chem. Soc. 2015, 137, 13992–14006. 10.1021/jacs.5b09348. [DOI] [PubMed] [Google Scholar]; e Nestl B. M.; Hammer S. C.; Nebel B. A.; Hauer B. Angew. Chem., Int. Ed. 2014, 53, 3070–3095. 10.1002/anie.201302195. [DOI] [PubMed] [Google Scholar]
- a Abe I. Curr. Opin. Chem. Biol. 2012, 16, 179–185. 10.1016/j.cbpa.2011.12.016. [DOI] [PubMed] [Google Scholar]; b Wong F. T.; Khosla C. Curr. Opin. Chem. Biol. 2012, 16, 117–123. 10.1016/j.cbpa.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Beutner G. L.; Denmark S. E. Angew. Chem., Int. Ed. 2013, 52, 9086–9096. 10.1002/anie.201302084. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Machajewski T. D.; Wong C. H. Angew. Chem., Int. Ed. 2000, 39, 1352–1374. . [DOI] [PubMed] [Google Scholar]
- Chen L. R.; Dumas D. P.; Wong C. H. J. Am. Chem. Soc. 1992, 114, 741–748. 10.1021/ja00028a050. [DOI] [Google Scholar]
- a Clapes P.; Garrabou X. Adv. Synth. Catal. 2011, 353, 2263–2283. 10.1002/adsc.201100236. [DOI] [Google Scholar]; b Clapes P.; Fessner W. D.; Sprenger G. A.; Samland A. K. Curr. Opin. Chem. Biol. 2010, 14, 154–167. 10.1016/j.cbpa.2009.11.029. [DOI] [PubMed] [Google Scholar]
- a Oroz-Guinea I.; Hernández K.; Camps Bres F.; Guérard-Hélaine C.; Lemaire M.; Clapés P.; García-Junceda E. Adv. Synth. Catal. 2015, 357, 1951–1960. 10.1002/adsc.201500187. [DOI] [Google Scholar]; b Oroz-Guinea I.; Sánchez-Moreno I.; Mena M.; García-Junceda E. Appl. Microbiol. Biotechnol. 2015, 99, 3057–3068. 10.1007/s00253-014-6123-7. [DOI] [PubMed] [Google Scholar]; c Babich L.; Hartog A. F.; van Hemert L. J. C.; Rutjes F. P. J. T.; Wever R. ChemSusChem 2012, 5, 2348–2353. 10.1002/cssc.201200468. [DOI] [PubMed] [Google Scholar]; d Babich L.; van Hemert L. J. C.; Bury A.; Hartog A. F.; Falcicchio P.; van der Oost J.; van Herk T.; Wever R.; Rutjes F. P. J. T. Green Chem. 2011, 13, 2895–2900. 10.1039/c1gc15429j. [DOI] [Google Scholar]
- Garrabou X.; Joglar J.; Parella T.; Crehuet R.; Bujons J.; Clapés P. Adv. Synth. Catal. 2011, 353, 89–99. 10.1002/adsc.201000719. [DOI] [Google Scholar]
- a Garrabou X.; Castillo J. A.; Guerard-Helaine C.; Parella T.; Joglar J.; Lemaire M.; Clapes P. Angew. Chem., Int. Ed. 2009, 48, 5521–5525. 10.1002/anie.200902065. [DOI] [PubMed] [Google Scholar]; b Concia A. L.; Lozano C.; Castillo J. A.; Parella T.; Joglar J.; Clapés P. Chem. - Eur. J. 2009, 15, 3808–3816. 10.1002/chem.200802532. [DOI] [PubMed] [Google Scholar]
- Mahdi R.; Guérard-Hélaine C.; Laroche C.; Michaud P.; Prévot V.; Forano C.; Lemaire M. J. Mol. Catal. B: Enzym. 2015, 122, 204–211. 10.1016/j.molcatb.2015.07.014. [DOI] [Google Scholar]
- Thorell S.; Schürmann M.; Sprenger G. A.; Schneider G. J. Mol. Biol. 2002, 319, 161–171. 10.1016/S0022-2836(02)00258-9. [DOI] [PubMed] [Google Scholar]
- a Szekrenyi A.; Soler A.; Garrabou X.; Guérard-Hélaine C.; Parella T.; Joglar J.; Lemaire M.; Bujons J.; Clapés P. Chem. - Eur. J. 2014, 20, 12572–12583. 10.1002/chem.201403281. [DOI] [PubMed] [Google Scholar]; b Gutierrez M.; Parella T.; Joglar J.; Bujons J.; Clapes P. Chem. Commun. 2011, 47, 5762–5764. 10.1039/c1cc11069a. [DOI] [PubMed] [Google Scholar]; c Castillo J. A.; Guérard-Hélaine C.; Gutiérrez M.; Garrabou X.; Sancelme M.; Schürmann M.; Inoue T.; Hélaine V.; Charmantray F.; Gefflaut T.; Hecquet L.; Joglar J.; Clapés P.; Sprenger G. A.; Lemaire M. Adv. Synth. Catal. 2010, 352, 1039–1046. 10.1002/adsc.200900772. [DOI] [Google Scholar]
- Güclü D.; Szekrenyi A.; Garrabou X.; Kickstein M.; Junker S.; Clapés P.; Fessner W.-D. ACS Catal. 2016, 6, 1848–1852. 10.1021/acscatal.5b02805. [DOI] [Google Scholar]
- Xie Z. B.; Wang N.; Zhou L. H.; Wan F.; He T.; Le Z. G.; Yu X. Q. ChemCatChem 2013, 5, 1935–1940. 10.1002/cctc.201200890. [DOI] [Google Scholar]
- a Zandvoort E.; Geertsema E. M.; Quax W. J.; Poelarends G. J. ChemBioChem 2012, 13, 1274–1277. 10.1002/cbic.201200225. [DOI] [PubMed] [Google Scholar]; b Narancic T.; Radivojevic J.; Jovanovic P.; Francuski D.; Bigovic M.; Maslak V.; Savic V.; Vasiljevic B.; O’Connor K. E.; Nikodinovic-Runic J. Bioresour. Technol. 2013, 142, 462–468. 10.1016/j.biortech.2013.05.074. [DOI] [PubMed] [Google Scholar]
- Szekrenyi A.; Garrabou X.; Parella T.; Joglar J.; Bujons J.; Clapés P. Nat. Chem. 2015, 7, 724–729. 10.1038/nchem.2321. [DOI] [PubMed] [Google Scholar]
- Guerard-Helaine C.; Debacker M.; Clapes P.; Szekrenyi A.; Helaine V.; Lemaire M. Green Chem. 2014, 16, 1109–1113. 10.1039/C3GC42140F. [DOI] [Google Scholar]
- Sánchez-Moreno I.; Hélaine V.; Poupard N.; Charmantray F.; Légeret B.; Hecquet L.; García-Junceda E.; Wohlgemuth R.; Guérard-Hélaine C.; Lemaire M. Adv. Synth. Catal. 2012, 354, 1725–1730. 10.1002/adsc.201200150. [DOI] [Google Scholar]
- Hernández K.; Parella T.; Joglar J.; Bujons J.; Pohl M.; Clapés P. Chem. - Eur. J. 2015, 21, 3335–3346. 10.1002/chem.201406156. [DOI] [PubMed] [Google Scholar]
- Stuetz A. E.Iminosugars as Glycosidase Inhibitors: Nojirimycin and Beyond; Wiley-VCH: Weinheim, 1999. [Google Scholar]
- a Duchek J.; Adams D. R.; Hudlicky T. Chem. Rev. 2011, 111, 4223–4258. 10.1021/cr1004138. [DOI] [PubMed] [Google Scholar]; b Grondal C.; Jeanty M.; Enders D. Nat. Chem. 2010, 2, 167–178. 10.1038/nchem.539. [DOI] [PubMed] [Google Scholar]; c Chen X.; Fan Y.; Zheng Y.; Shen Y. Chem. Rev. 2003, 103, 1955–1977. 10.1021/cr0102260. [DOI] [PubMed] [Google Scholar]
- a Timmons S. C.; Thorson J. S. Curr. Opin. Chem. Biol. 2008, 12, 297–305. 10.1016/j.cbpa.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hu Y.; Al-Mestarihi A.; Grimes C. L.; Kahne D.; Bachmann B. O. J. Am. Chem. Soc. 2008, 130, 15756–15757. 10.1021/ja8051415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M.; Hong Z.; Liang P.-H.; Dean S. M.; Whalen L. J.; Greenberg W. A.; Wong C.-H. J. Am. Chem. Soc. 2007, 129, 14811–14817. 10.1021/ja073911i. [DOI] [PubMed] [Google Scholar]
- Soler A.; Gutiérrez M. L.; Bujons J.; Parella T.; Minguillon C.; Joglar J.; Clapés P. Adv. Synth. Catal. 2015, 357, 1787–1807. 10.1002/adsc.201500073. [DOI] [Google Scholar]
- a Concia A. L.; Gómez L.; Parella T.; Joglar J.; Clapés P. J. Org. Chem. 2014, 79, 5386–5389. 10.1021/jo500991p. [DOI] [PubMed] [Google Scholar]; b Gomez L.; Garrabou X.; Joglar J.; Bujons J.; Parella T.; Vilaplana C.; Cardona P. J.; Clapes P. Org. Biomol. Chem. 2012, 10, 6309–6321. 10.1039/c2ob25943e. [DOI] [PubMed] [Google Scholar]
- Laborda P.; Sayago F. J.; Cativiela C.; Parella T.; Joglar J.; Clapés P. Org. Lett. 2014, 16, 1422–1425. 10.1021/ol5002158. [DOI] [PubMed] [Google Scholar]
- a Gómez L.; Molinar-Toribio E.; Calvo-Torras M. Á.; Adelantado C.; Juan M. E.; Planas J. M.; Cañas X.; Lozano C.; Pumarola S.; Clapés P.; Torres J. L. Br. J. Nutr. 2012, 107, 1739–1746. 10.1017/S0007114511005009. [DOI] [PubMed] [Google Scholar]; b Malle B. M.; Lundt I.; Wrodnigg T. M. Org. Biomol. Chem. 2008, 6, 1779–1786. 10.1039/b719631h. [DOI] [PubMed] [Google Scholar]; c From Synthesis of Iminosugars to Therapeutic Applications; Compain P. M., Martin O. R., Eds.; John Wiley & Sons: Chicester, 2007. [Google Scholar]
- Concia A. L.; Gomez L.; Bujons J.; Parella T.; Vilaplana C.; Cardona P. J.; Joglar J.; Clapes P. Org. Biomol. Chem. 2013, 11, 2005–2021. 10.1039/c3ob27343a. [DOI] [PubMed] [Google Scholar]
- Soler A.; Garrabou X.; Hernandez K.; Gutierrez M. L.; Busto E.; Bujons J.; Parella T.; Joglar J.; Clapes P. Adv. Synth. Catal. 2014, 356, 3007–3024. 10.1002/adsc.201400453. [DOI] [Google Scholar]
- Sudar M.; Findrik Z.; Vasić-Rački Đ.; Clapés P.; Lozano C. Enzyme Microb. Technol. 2013, 53, 38–45. 10.1016/j.enzmictec.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Sudar M.; Findrik Z.; Vasic-Racki D.; Soler A.; Clapes P. RSC Adv. 2015, 5, 69819–69828. 10.1039/C5RA14414K. [DOI] [Google Scholar]
- a El Blidi L.; Assaf Z.; Camps Bres F.; Veschambre H.; Théry V.; Bolte J.; Lemaire M. ChemCatChem 2009, 1, 463–471. 10.1002/cctc.200900151. [DOI] [Google Scholar]; b El Blidi L.; Ahbala M.; Bolte J.; Lemaire M. Tetrahedron: Asymmetry 2006, 17, 2684–2688. 10.1016/j.tetasy.2006.09.010. [DOI] [Google Scholar]; c El Blidi L.; Crestia D.; Gallienne E.; Demuynck C.; Bolte J.; Lemaire M. Tetrahedron: Asymmetry 2004, 15, 2951–2954. 10.1016/j.tetasy.2004.06.029. [DOI] [Google Scholar]
- Camps Bres F.; Guérard-Hélaine C.; Hélaine V.; Fernandes C.; Sánchez-Moreno I.; Traïkia M.; García-Junceda E.; Lemaire M. J. Mol. Catal. B: Enzym. 2015, 114, 50–57. 10.1016/j.molcatb.2014.10.016. [DOI] [Google Scholar]
- Camps Bres F.; Guérard-Hélaine C.; Fernandes C.; Castillo J. A.; Lemaire M. Tetrahedron: Asymmetry 2013, 24, 1075–1081. 10.1016/j.tetasy.2013.07.016. [DOI] [Google Scholar]
- a Dückers N.; Baer K.; Simon S.; Gröger H.; Hummel W. Appl. Microbiol. Biotechnol. 2010, 88, 409–424. 10.1007/s00253-010-2751-8. [DOI] [PubMed] [Google Scholar]; b Vogt H.; Brase S. Org. Biomol. Chem. 2007, 5, 406–430. 10.1039/B611091F. [DOI] [PubMed] [Google Scholar]; c Zhang W. H.; Otting G.; Jackson C. J. Curr. Opin. Struct. Biol. 2013, 23, 581–587. 10.1016/j.sbi.2013.06.009. [DOI] [PubMed] [Google Scholar]; d Tanaka M. Chem. Pharm. Bull. 2007, 55, 349–358. 10.1248/cpb.55.349. [DOI] [PubMed] [Google Scholar]; e Miller M. J.; Mattingly P. G.; Morrison M. A.; Kerwin J. F. J. Am. Chem. Soc. 1980, 102, 7026–7032. 10.1021/ja00543a021. [DOI] [Google Scholar]
- Sehl T.; Maugeri Z.; Rother D. J. Mol. Catal. B: Enzym. 2015, 114, 65–71. 10.1016/j.molcatb.2014.12.005. [DOI] [Google Scholar]
- Gutierrez M. L.; Garrabou X.; Agosta E.; Servi S.; Parella T.; Joglar J.; Clapés P. Chem. - Eur. J. 2008, 14, 4647–4656. 10.1002/chem.200800031. [DOI] [PubMed] [Google Scholar]
- a Baer K.; Dückers N.; Hummel W.; Gröger H. ChemCatChem 2010, 2, 939–942. 10.1002/cctc.201000007. [DOI] [PubMed] [Google Scholar]; b Fesko K.; Uhl M.; Steinreiber J.; Gruber K.; Griengl H. Angew. Chem., Int. Ed. 2010, 49, 121–124. 10.1002/anie.200904395. [DOI] [PubMed] [Google Scholar]; c Nozaki H.; Kuroda S.; Watanabe K.; Yokozeki K. J. Mol. Catal. B: Enzym. 2009, 59, 237–242. 10.1016/j.molcatb.2008.06.007. [DOI] [Google Scholar]
- Fesko K. Appl. Microbiol. Biotechnol. 2016, 100, 2579–2590. 10.1007/s00253-015-7218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez K.; Zelen I.; Petrillo G.; Usón I.; Wandtke C. M.; Bujons J.; Joglar J.; Parella T.; Clapés P. Angew. Chem., Int. Ed. 2015, 54, 3013–3017. 10.1002/anie.201411484. [DOI] [PubMed] [Google Scholar]
- Fesko K.; Strohmeier G.; Breinbauer R. Appl. Microbiol. Biotechnol. 2015, 99, 9651–9661. 10.1007/s00253-015-6803-y. [DOI] [PubMed] [Google Scholar]
- Hoyos P.; Sinisterra J.-V.; Molinari F.; Alcántara A. R.; Domínguez de María P. Acc. Chem. Res. 2010, 43, 288–299. 10.1021/ar900196n. [DOI] [PubMed] [Google Scholar]
- a Müller M.; Sprenger G. A.; Pohl M. Curr. Opin. Chem. Biol. 2013, 17, 261–270. 10.1016/j.cbpa.2013.02.017. [DOI] [PubMed] [Google Scholar]; b Hailes H. C.; Rother D.; Müller M.; Westphal R.; Ward J. M.; Pleiss J.; Vogel C.; Pohl M. FEBS J. 2013, 280, 6374–6394. 10.1111/febs.12496. [DOI] [PubMed] [Google Scholar]; c Müller M.; Gocke D.; Pohl M. FEBS J. 2009, 276, 2894–2904. 10.1111/j.1742-4658.2009.07017.x. [DOI] [PubMed] [Google Scholar]; d Pohl M.; Sprenger G. A.; Muller M. Curr. Opin. Biotechnol. 2004, 15, 335–342. 10.1016/j.copbio.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Sehl T.; Hailes H. C.; Ward J. M.; Wardenga R.; von Lieres E.; Offermann H.; Westphal R.; Pohl M.; Rother D. Angew. Chem., Int. Ed. 2013, 52, 6772–6775. 10.1002/anie.201300718. [DOI] [PubMed] [Google Scholar]
- Sehl T.; Hailes H. C.; Ward J. M.; Menyes U.; Pohl M.; Rother D. Green Chem. 2014, 16, 3341–3348. 10.1039/C4GC00100A. [DOI] [Google Scholar]
- Wu X.; Fei M.; Chen Y.; Wang Z.; Chen Y. Appl. Microbiol. Biotechnol. 2014, 98, 7399–7408. 10.1007/s00253-014-5797-1. [DOI] [PubMed] [Google Scholar]
- a Harder J. Arch. Microbiol. 1997, 168, 199–204. 10.1007/s002030050488. [DOI] [Google Scholar]; b Fraas S.; Steinbach A. K.; Tabbert A.; Harder J.; Ermler U.; Tittmann K.; Meyer A.; Kroneck P. M. H. J. Mol. Catal. B: Enzym. 2009, 61, 47–49. 10.1016/j.molcatb.2009.03.021. [DOI] [Google Scholar]; c Steinbach A. K.; Fraas S.; Harder J.; Tabbert A.; Brinkmann H.; Meyer A.; Ermler U.; Kroneck P. M. H. J. Bacteriol. 2011, 193, 6760–6769. 10.1128/JB.05348-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschonsky S.; Waltzer S.; Fraas S.; Wacker T.; Andrade S. L. A.; Kroneck P. M. H.; Müller M. ChemBioChem 2014, 15, 389–392. 10.1002/cbic.201300673. [DOI] [PubMed] [Google Scholar]
- Loschonsky S.; Waltzer S.; Brecht V.; Müller M. ChemCatChem 2014, 6, 969–972. 10.1002/cctc.201300904. [DOI] [Google Scholar]
- a Beigi M.; Loschonsky S.; Lehwald P.; Brecht V.; Andrade S. L. A.; Leeper F. J.; Hummel W.; Muller M. Org. Biomol. Chem. 2013, 11, 252–256. 10.1039/C2OB26981C. [DOI] [PubMed] [Google Scholar]; b Bornemann S.; Crout D. H. G.; Dalton H.; Hutchinson D. W.; Dean G.; Thomson N.; Turner M. M. J. Chem. Soc., Perkin Trans. 1 1993, 309–311. 10.1039/p19930000309. [DOI] [Google Scholar]
- a Mosbacher T. G.; Mueller M.; Schulz G. E. FEBS J. 2005, 272, 6067–6076. 10.1111/j.1742-4658.2005.04998.x. [DOI] [PubMed] [Google Scholar]; b González B.; Vicuña R. J. Bacteriol. 1989, 171, 2401–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C. R.; Perez-Sanchez M.; Dominguez de Maria P. Org. Biomol. Chem. 2013, 11, 2000–2004. 10.1039/c2ob27344f. [DOI] [PubMed] [Google Scholar]
- Pérez-Sánchez M.; Müller C. R.; Domínguez de María P. ChemCatChem 2013, 5, 2512–2516. 10.1002/cctc.201300093. [DOI] [Google Scholar]
- Maugeri Z.; Domínguez de María P. J. Mol. Catal. B: Enzym. 2014, 107, 120–123. 10.1016/j.molcatb.2014.06.003. [DOI] [Google Scholar]
- Shanmuganathan S.; Natalia D.; van den Wittenboer A.; Kohlmann C.; Greiner L.; Dominguez de Maria P. Green Chem. 2010, 12, 2240–2245. 10.1039/c0gc00590h. [DOI] [Google Scholar]
- a Demir A. S.; Şeşenoglu Ö.; Eren E.; Hosrik B.; Pohl M.; Janzen E.; Kolter D.; Feldmann R.; Dünkelmann P.; Müller M. Adv. Synth. Catal. 2002, 344, 96–103. . [DOI] [Google Scholar]; b Dünkelmann P.; Kolter-Jung D.; Nitsche A.; Demir A. S.; Siegert P.; Lingen B.; Baumann M.; Pohl M.; Müller M. J. Am. Chem. Soc. 2002, 124, 12084–12085. 10.1021/ja0271476. [DOI] [PubMed] [Google Scholar]
- Westphal R.; Vogel C.; Schmitz C.; Pleiss J.; Müller M.; Pohl M.; Rother D. Angew. Chem., Int. Ed. 2014, 53, 9376–9379. 10.1002/anie.201405069. [DOI] [PubMed] [Google Scholar]
- a Jiang M.; Cao Y.; Guo Z. F.; Chen M.; Chen X.; Guo Z. Biochemistry 2007, 46, 10979–10989. 10.1021/bi700810x. [DOI] [PubMed] [Google Scholar]; b Kurutsch A.; Richter M.; Brecht V.; Sprenger G. A.; Müller M. J. Mol. Catal. B: Enzym. 2009, 61, 56–66. 10.1016/j.molcatb.2009.03.011. [DOI] [Google Scholar]
- Beigi M.; Waltzer S.; Fries A.; Eggeling L.; Sprenger G. A.; Müller M. Org. Lett. 2013, 15, 452–455. 10.1021/ol3031186. [DOI] [PubMed] [Google Scholar]
- Rother D.; Kolter G.; Gerhards T.; Berthold C. L.; Gauchenova E.; Knoll M.; Pleiss J.; Müller M.; Schneider G.; Pohl M. ChemCatChem 2011, 3, 1587–1596. 10.1002/cctc.201100054. [DOI] [Google Scholar]
- a Westphal R.; Waltzer S.; Mackfeld U.; Widmann M.; Pleiss J.; Beigi M.; Muller M.; Rother D.; Pohl M. Chem. Commun. 2013, 49, 2061–2063. 10.1039/c3cc38607d. [DOI] [PubMed] [Google Scholar]; b Westphal R.; Hahn D.; Mackfeld U.; Waltzer S.; Beigi M.; Widmann M.; Vogel C.; Pleiss J.; Müller M.; Rother D.; Pohl M. ChemCatChem 2013, 5, 3587–3594. 10.1002/cctc.201300318. [DOI] [PubMed] [Google Scholar]
- Westphal R.; Jansen S.; Vogel C.; Pleiss J.; Müller M.; Rother D.; Pohl M. ChemCatChem 2014, 6, 1082–1088. 10.1002/cctc.201300690. [DOI] [Google Scholar]
- a He H.; Qi C.; Hu X.; Guan Y.; Jiang H. Green Chem. 2014, 16, 3729–3733. 10.1039/C4GC00522H. [DOI] [Google Scholar]; b Gormisky P. E.; White M. C. J. Am. Chem. Soc. 2013, 135, 14052–14055. 10.1021/ja407388y. [DOI] [PubMed] [Google Scholar]; c Rose C. A.; Gundala S.; Fagan C. L.; Franz J. F.; Connon S. J.; Zeitler K. Chem. Sci. 2012, 3, 735–740. 10.1039/C2SC00622G. [DOI] [Google Scholar]; d Stymiest J. L.; Bagutski V.; French R. M.; Aggarwal V. K. Nature 2008, 456, 778–782. 10.1038/nature07592. [DOI] [PubMed] [Google Scholar]; e Enders D.; Niemeier O.; Balensiefer T. Angew. Chem., Int. Ed. 2006, 45, 1463–1467. 10.1002/anie.200503885. [DOI] [PubMed] [Google Scholar]
- Müller M. ChemBioEng Rev. 2014, 1, 14–26. 10.1002/cben.201300005. [DOI] [Google Scholar]
- Liang Y.-F.; Jiao N. Angew. Chem., Int. Ed. 2014, 53, 548–552. 10.1002/anie.201308698. [DOI] [PubMed] [Google Scholar]
- Lehwald P.; Richter M.; Röhr C.; Liu H.-w.; Müller M. Angew. Chem., Int. Ed. 2010, 49, 2389–2392. 10.1002/anie.200906181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannini P. P.; Pedrini P.; Venturi V.; Fantin G.; Medici A. J. Mol. Catal. B: Enzym. 2010, 64, 113–117. 10.1016/j.molcatb.2010.03.001. [DOI] [Google Scholar]
- a Giovannini P. P.; Fantin G.; Massi A.; Venturi V.; Pedrini P. Org. Biomol. Chem. 2011, 9, 8038–8045. 10.1039/c1ob05928a. [DOI] [PubMed] [Google Scholar]; b Giovannini P. P.; Mantovani M.; Grandini A.; Medici A.; Pedrini P. J. Mol. Catal. B: Enzym. 2011, 69, 15–20. 10.1016/j.molcatb.2010.12.004. [DOI] [Google Scholar]
- Bortolini O.; Giovannini P. P.; Maietti S.; Massi A.; Pedrini P.; Sacchetti G.; Venturi V. J. Mol. Catal. B: Enzym. 2013, 85–86, 93–98. 10.1016/j.molcatb.2012.08.015. [DOI] [Google Scholar]
- Giovannini P. P.; Bortolini O.; Cavazzini A.; Greco R.; Fantin G.; Massi A. Green Chem. 2014, 16, 3904–3915. 10.1039/C4GC00838C. [DOI] [Google Scholar]
- Xiao Z.; Xu P. Crit. Rev. Microbiol. 2007, 33, 127–140. 10.1080/10408410701364604. [DOI] [PubMed] [Google Scholar]
- Bernacchia G.; Bortolini O.; De Bastiani M.; Lerin L. A.; Loschonsky S.; Massi A.; Müller M.; Giovannini P. P. Angew. Chem., Int. Ed. 2015, 54, 7171–7175. 10.1002/anie.201502102. [DOI] [PubMed] [Google Scholar]
- Steinbach A.; Fraas S.; Harder J.; Warkentin E.; Kroneck P. M. H.; Ermler U. FEBS J. 2012, 279, 1209–1219. 10.1111/j.1742-4658.2012.08513.x. [DOI] [PubMed] [Google Scholar]
- Loschonsky S.; Wacker T.; Waltzer S.; Giovannini P. P.; McLeish M. J.; Andrade S. L. A.; Müller M. Angew. Chem., Int. Ed. 2014, 53, 14402–14406. 10.1002/anie.201408287. [DOI] [PubMed] [Google Scholar]
- Dresen C.; Richter M.; Pohl M.; Lüdeke S.; Müller M. Angew. Chem., Int. Ed. 2010, 49, 6600–6603. 10.1002/anie.201000632. [DOI] [PubMed] [Google Scholar]
- Kasparyan E.; Richter M.; Dresen C.; Walter L.; Fuchs G.; Leeper F.; Wacker T.; Andrade S. A.; Kolter G.; Pohl M.; Müller M. Appl. Microbiol. Biotechnol. 2014, 98, 9681–9690. 10.1007/s00253-014-5850-0. [DOI] [PubMed] [Google Scholar]
- Beigi M.; Waltzer S.; Zarei M.; Müller M. J. Biotechnol. 2014, 191, 64–68. 10.1016/j.jbiotec.2014.07.451. [DOI] [PubMed] [Google Scholar]
- Stockigt J.; Antonchick A. P.; Wu F. R.; Waldmann H. Angew. Chem., Int. Ed. 2011, 50, 8538–8564. 10.1002/anie.201008071. [DOI] [PubMed] [Google Scholar]
- Zhu H. J.; Kercmar P.; Wu F. R.; Rajendran C.; Sun L. L.; Wang M. T.; Stockigt J. Curr. Med. Chem. 2015, 22, 1880–1888. 10.2174/0929867322666150408110919. [DOI] [PubMed] [Google Scholar]
- Herraiz T. J. Agric. Food Chem. 2000, 48, 4900–4904. 10.1021/jf000508l. [DOI] [PubMed] [Google Scholar]
- a Chen S.; Galan M. C.; Coltharp C.; O’Connor S. E. Chem. Biol. 2006, 13, 1137–1141. 10.1016/j.chembiol.2006.10.009. [DOI] [PubMed] [Google Scholar]; b Stockigt J.; Barleben L.; Panjikar S.; Loris E. A. Plant Physiol. Biochem. 2008, 46, 340–355. 10.1016/j.plaphy.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Bernhardt P.; Usera A. R.; O’Connor S. E. Tetrahedron Lett. 2010, 51, 4400–4402. 10.1016/j.tetlet.2010.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H. B.; Zhu H. J.; Zhang L. A.; Yang L. Q.; Yu Y. P.; Stockigt J. Chem. - Asian J. 2010, 5, 2400–2404. 10.1002/asia.201000520. [DOI] [PubMed] [Google Scholar]
- Wu F. R.; Zhu H. J.; Sun L. L.; Rajendran C.; Wang M. T.; Ren X.; Panjikar S.; Cherkasov A.; Zou H. B.; Stockigt J. J. Am. Chem. Soc. 2012, 134, 1498–1500. 10.1021/ja211524d. [DOI] [PubMed] [Google Scholar]
- Fischereder E.-M.; Pressnitz D.; Kroutil W. ACS Catal. 2016, 6, 23–30. 10.1021/acscatal.5b01839. [DOI] [Google Scholar]
- Fischereder E.; Pressnitz D.; Kroutil W.; Lutz S. Bioorg. Med. Chem. 2014, 22, 5633–5637. 10.1016/j.bmc.2014.06.023. [DOI] [PubMed] [Google Scholar]
- Scott J. D.; Williams R. M. Chem. Rev. 2002, 102, 1669–1730. 10.1021/cr010212u. [DOI] [PubMed] [Google Scholar]
- a Bai G.; Yang Y.; Shi Q.; Liu Z.; Zhang Q.; Zhu Y.-y. Acta Pharmacol. Sin. 2008, 29, 1187–1194. 10.1111/j.1745-7254.2008.00859.x. [DOI] [PubMed] [Google Scholar]; b Pyo M. K.; Lee D.-H.; Kim D.-H.; Lee J.-H.; Moon J.-C.; Chang K. C.; Yun-Choi H. S. Bioorg. Med. Chem. Lett. 2008, 18, 4110–4114. 10.1016/j.bmcl.2008.05.094. [DOI] [PubMed] [Google Scholar]
- a Pesnot T.; Gershater M. C.; Ward J. M.; Hailes H. C. Adv. Synth. Catal. 2012, 354, 2997–3008. 10.1002/adsc.201200641. [DOI] [Google Scholar]; b Ruff B. M.; Bräse S.; O’Connor S. E. Tetrahedron Lett. 2012, 53, 1071–1074. 10.1016/j.tetlet.2011.12.089. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Nishihachijo M.; Hirai Y.; Kawano S.; Nishiyama A.; Minami H.; Katayama T.; Yasohara Y.; Sato F.; Kumagai H. Biosci., Biotechnol., Biochem. 2014, 78, 701–707. 10.1080/09168451.2014.890039. [DOI] [PubMed] [Google Scholar]; d Maresh J. J.; Crowe S. O.; Ralko A. A.; Aparece M. D.; Murphy C. M.; Krzeszowiec M.; Mullowney M. W. Tetrahedron Lett. 2014, 55, 5047–5051. 10.1016/j.tetlet.2014.07.043. [DOI] [Google Scholar]; e Bonamore A.; Rovardi I.; Gasparrini F.; Baiocco P.; Barba M.; Molinaro C.; Botta B.; Boffi A.; Macone A. Green Chem. 2010, 12, 1623–1627. 10.1039/c0gc00036a. [DOI] [Google Scholar]
- Lichman B. R.; Gershater M. C.; Lamming E. D.; Pesnot T.; Sula A.; Keep N. H.; Hailes H. C.; Ward J. M. FEBS J. 2015, 282, 1137–1151. 10.1111/febs.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichman B. R.; Lamming E. D.; Pesnot T.; Smith J. M.; Hailes H. C.; Ward J. M. Green Chem. 2015, 17, 852–855. 10.1039/C4GC02325K. [DOI] [Google Scholar]
- Roberts M. E.; Mannsfeld S. C. B.; Queraltó N.; Reese C.; Locklin J.; Knoll W.; Bao Z. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 12134–12139. 10.1073/pnas.0802105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.; Chang J. Y. J. Mater. Chem. 2010, 20, 749–754. 10.1039/B920203J. [DOI] [Google Scholar]
- Busto E.; Simon R. C.; Kroutil W. Angew. Chem., Int. Ed. 2015, 54, 10899–10902. 10.1002/anie.201505696. [DOI] [PubMed] [Google Scholar]
- a Seisser B.; Zinkl R.; Gruber K.; Kaufmann F.; Hafner A.; Kroutil W. Adv. Synth. Catal. 2010, 352, 731–736. 10.1002/adsc.200900826. [DOI] [Google Scholar]; b Yu Y.; Zhou Q.; Wang L.; Liu X. H.; Zhang W.; Hu M. R.; Dong J. S.; Li J. S.; Lv X. X.; Ouyang H. L.; Li H.; Gao F.; Gong W. M.; Lu Y.; Wang J. Y. Chem. Sci. 2015, 6, 3881–3885. 10.1039/C5SC01126D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennig A.; Busto E.; Kroutil W.; Faber K. ACS Catal. 2015, 5, 7503–7506. 10.1021/acscatal.5b02129. [DOI] [Google Scholar]
- Zhang H.; Lu Y.; Wu S.; Wei Y.; Liu Q.; Liu L.; Jiao Q. J. Mol. Catal. B: Enzym. 2016, 124, 38–44. 10.1016/j.molcatb.2015.11.024. [DOI] [Google Scholar]
- a Fan A.; Winkelblech J.; Li S.-M. Appl. Microbiol. Biotechnol. 2015, 99, 7399–7415. 10.1007/s00253-015-6813-9. [DOI] [PubMed] [Google Scholar]; b Winkelblech J.; Fan A.; Li S.-M. Appl. Microbiol. Biotechnol. 2015, 99, 7379–7397. 10.1007/s00253-015-6811-y. [DOI] [PubMed] [Google Scholar]
- Fan A.; Chen H.; Wu R.; Xu H.; Li S.-M. Appl. Microbiol. Biotechnol. 2014, 98, 10119–10129. 10.1007/s00253-014-5872-7. [DOI] [PubMed] [Google Scholar]
- Fan A.; Li S.-M. Tetrahedron Lett. 2014, 55, 5199–5202. 10.1016/j.tetlet.2014.07.080. [DOI] [Google Scholar]
- Fan A.; Zocher G.; Stec E.; Stehle T.; Li S.-M. J. Biol. Chem. 2015, 290, 1364–1373. 10.1074/jbc.M114.623413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebhold M.; Li S.-M. Org. Lett. 2013, 15, 5834–5837. 10.1021/ol4029012. [DOI] [PubMed] [Google Scholar]
- Pockrandt D.; Sack C.; Kosiol T.; Li S.-M. Appl. Microbiol. Biotechnol. 2014, 98, 4987–4994. 10.1007/s00253-014-5509-x. [DOI] [PubMed] [Google Scholar]
- Chen J.; Morita H.; Wakimoto T.; Mori T.; Noguchi H.; Abe I. Org. Lett. 2012, 14, 3080–3083. 10.1021/ol301129x. [DOI] [PubMed] [Google Scholar]
- Knölker H.-J.; Reddy K. R. Chem. Rev. 2002, 102, 4303–4428. 10.1021/cr020059j. [DOI] [PubMed] [Google Scholar]
- Grazulevicius J. V.; Strohriegl P.; Pielichowski J.; Pielichowski K. Prog. Polym. Sci. 2003, 28, 1297–1353. 10.1016/S0079-6700(03)00036-4. [DOI] [Google Scholar]
- Sousa A. C.; Piedade M. F. M. M.; Martins L. O.; Robalo M. P. Green Chem. 2015, 17, 1429–1433. 10.1039/C4GC02511C. [DOI] [Google Scholar]
- Murakami Y.; Ishii H.; Hoshina S.; Takada N.; Ueki A.; Tanaka S.; Kadoma Y.; Ito S.; Machino M.; Fujisawa S. Anticancer Res. 2009, 29, 2403–2410. [PubMed] [Google Scholar]
- Francke R.; Reingruber R.; Schollmeyer D.; Waldvogel S. R. J. Agric. Food Chem. 2013, 61, 4709–4714. 10.1021/jf401081g. [DOI] [PubMed] [Google Scholar]
- Engelmann C.; Illner S.; Kragl U. Process Biochem. 2015, 50, 1591–1599. 10.1016/j.procbio.2015.05.011. [DOI] [Google Scholar]
- Masubuchi M.; Kawasaki K.-i.; Ebiike H.; Ikeda Y.; Tsujii S.; Sogabe S.; Fujii T.; Sakata K.; Shiratori Y.; Aoki Y.; Ohtsuka T.; Shimma N. Bioorg. Med. Chem. Lett. 2001, 11, 1833–1837. 10.1016/S0960-894X(01)00319-5. [DOI] [PubMed] [Google Scholar]
- Wahab Khan M.; Jahangir Alam M.; Rashid M. A.; Chowdhury R. Bioorg. Med. Chem. 2005, 13, 4796–4805. 10.1016/j.bmc.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Lee S. K.; Cui B.; Mehta R. R.; Kinghorn A. D.; Pezzuto J. M. Chem.-Biol. Interact. 1998, 115, 215–228. 10.1016/S0009-2797(98)00073-8. [DOI] [PubMed] [Google Scholar]
- Wellington K. W.; Qwebani-Ogunleye T.; Kolesnikova N. I.; Brady D.; de Koning C. B. Arch. Pharm. 2013, 346, 266–277. 10.1002/ardp.201200413. [DOI] [PubMed] [Google Scholar]
- a Witayakran S.; Ragauskas A. J. Eur. J. Org. Chem. 2009, 2009, 358–363. 10.1002/ejoc.200800791. [DOI] [Google Scholar]; b Witayakran S.; Gelbaum L.; Ragauskas A. J. Tetrahedron 2007, 63, 10958–10962. 10.1016/j.tet.2007.08.066. [DOI] [Google Scholar]
- Yan D.; Jin C.; Xiao X.-H.; Dong X.-P. J. Biochem. Biophys. Methods 2008, 70, 845–849. 10.1016/j.jbbm.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Jang S.; Kim B. H.; Lee W.-Y.; An S. J.; Choi H. G.; Jeon B. H.; Chung H.-T.; Rho J.-R.; Kim Y.-J.; Chai K.-Y. Arch. Pharmacal Res. 2004, 27, 923–929. 10.1007/BF02975845. [DOI] [PubMed] [Google Scholar]
- Kulkarni S. K.; Dhir A. Eur. J. Pharmacol. 2008, 589, 163–172. 10.1016/j.ejphar.2008.05.043. [DOI] [PubMed] [Google Scholar]
- Huang L.; Shi A.; He F.; Li X. Bioorg. Med. Chem. 2010, 18, 1244–1251. 10.1016/j.bmc.2009.12.035. [DOI] [PubMed] [Google Scholar]
- a Resch V.; Schrittwieser J. H.; Wallner S.; Macheroux P.; Kroutil W. Adv. Synth. Catal. 2011, 353, 2377–2383. 10.1002/adsc.201100233. [DOI] [Google Scholar]; b Schrittwieser J. H.; Resch V.; Sattler J. H.; Lienhart W.-D.; Durchschein K.; Winkler A.; Gruber K.; Macheroux P.; Kroutil W. Angew. Chem., Int. Ed. 2011, 50, 1068–1071. 10.1002/anie.201006268. [DOI] [PubMed] [Google Scholar]; c Schrittwieser J. H.; Resch V.; Wallner S.; Lienhart W.-D.; Sattler J. H.; Resch J.; Macheroux P.; Kroutil W. J. Org. Chem. 2011, 76, 6703–6714. 10.1021/jo201056f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrittwieser J. H.; Groenendaal B.; Resch V.; Ghislieri D.; Wallner S.; Fischereder E.-M.; Fuchs E.; Grischek B.; Sattler J. H.; Macheroux P.; Turner N. J.; Kroutil W. Angew. Chem., Int. Ed. 2014, 53, 3731–3734. 10.1002/anie.201400027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer S. C.; Syrén P.-O.; Seitz M.; Nestl B. M.; Hauer B. Curr. Opin. Chem. Biol. 2013, 17, 293–300. 10.1016/j.cbpa.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Seitz M.; Syrén P.-O.; Steiner L.; Klebensberger J.; Nestl B. M.; Hauer B. ChemBioChem 2013, 14, 436–439. 10.1002/cbic.201300018. [DOI] [PubMed] [Google Scholar]
- Siedenburg G.; Breuer M.; Jendrossek D. Appl. Microbiol. Biotechnol. 2013, 97, 1571–1580. 10.1007/s00253-012-4008-1. [DOI] [PubMed] [Google Scholar]
- Hammer S. C.; Marjanovic A.; Dominicus J. M.; Nestl B. M.; Hauer B. Nat. Chem. Biol. 2015, 11, 121–126. 10.1038/nchembio.1719. [DOI] [PubMed] [Google Scholar]
- Davies H. M. L.; Manning J. R. Nature 2008, 451, 417–424. 10.1038/nature06485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho P. S.; Brustad E. M.; Kannan A.; Arnold F. H. Science 2013, 339, 307–310. 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]
- Coelho P. S.; Wang Z. J.; Ener M. E.; Baril S. A.; Kannan A.; Arnold F. H.; Brustad E. M. Nat. Chem. Biol. 2013, 9, 485–U433. 10.1038/nchembio.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeaux M.; Tyagi V.; Fasan R. Angew. Chem., Int. Ed. 2015, 54, 1744–1748. 10.1002/anie.201409928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gober J. G.; Rydeen A. E.; Gibson-O’Grady E. J.; Leuthaeuser J. B.; Fetrow J. S.; Brustad E. M. ChemBioChem 2016, 17, 394–397. 10.1002/cbic.201500624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. J.; Renata H.; Peck N. E.; Farwell C. C.; Coelho P. S.; Arnold F. H. Angew. Chem., Int. Ed. 2014, 53, 6810–6813. 10.1002/anie.201402809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P.; Yang H.; Ellis-Guardiola K.; Lewis J. C. Nat. Commun. 2015, 6, 7789. 10.1038/ncomms8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi V.; Fasan R. Angew. Chem., Int. Ed. 2016, 55, 2512–2516. 10.1002/anie.201508817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Iglesias M.; Gotor-Fernandez V. Chem. Rec. 2015, 15, 743–759. 10.1002/tcr.201500008. [DOI] [PubMed] [Google Scholar]
- a Ghattas W.; Cotchico-Alonso L.; Marechal J. D.; Urvoas A.; Rousseau M.; Mahy J. P.; Ricoux R. ChemBioChem 2016, 17, 433–440. 10.1002/cbic.201500445. [DOI] [PubMed] [Google Scholar]; b Bos J.; Fusetti F.; Driessen A. J. M.; Roelfes G. Angew. Chem., Int. Ed. 2012, 51, 7472–7475. 10.1002/anie.201202070. [DOI] [PubMed] [Google Scholar]; c Bos J.; Browne W. R.; Driessen A. J. M.; Roelfes G. J. Am. Chem. Soc. 2015, 137, 9796–9799. 10.1021/jacs.5b05790. [DOI] [PubMed] [Google Scholar]
- Shinozaki J.; Hiruta M.; Okada T.; Masuda K. ChemBioChem 2016, 17, 65–70. 10.1002/cbic.201500511. [DOI] [PubMed] [Google Scholar]
- Ueda D.; Hoshino T.; Sato T. J. Am. Chem. Soc. 2013, 135, 18335–18338. 10.1021/ja4107226. [DOI] [PubMed] [Google Scholar]
- Banta A. B.; Wei J. H.; Welander P. V. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 13478–13483. 10.1073/pnas.1511482112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J.; Matsuba Y.; Hong Y. J.; Jackson A. J.; Tantillo D. J.; Pichersky E.; Peters R. J. J. Am. Chem. Soc. 2014, 136, 16951–16953. 10.1021/ja508477e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo Y.-S.; Nybo S. E.; Chittiboyina A. G.; Weerasooriya A. D.; Wang Y.-H.; Góngora-Castillo E.; Vaillancourt B.; Buell C. R.; DellaPenna D.; Celiz M. D.; Jones A. D.; Wurtele E. S.; Ransom N.; Dudareva N.; Shaaban K. A.; Tibrewal N.; Chandra S.; Smillie T.; Khan I. A.; Coates R. M.; Watt D. S.; Chappell J. J. Biol. Chem. 2013, 288, 3163–3173. 10.1074/jbc.M112.415836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau W.; Sattely E. S. Science 2015, 349, 1224–1228. 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel R.; Kaysser L.; Villaume M. T.; Diethelm S.; Carbullido M. K.; Baran P. S.; Moore B. S. Angew. Chem., Int. Ed. 2014, 53, 11019–11022. 10.1002/anie.201405694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethelm S.; Teufel R.; Kaysser L.; Moore B. S. Angew. Chem., Int. Ed. 2014, 53, 11023–11026. 10.1002/anie.201405696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C. A. F.; Folly C.; Hatsch A.; Molt A.; Schröder H.; O’Connor S. E.; Naesby M. Microb. Cell Fact. 2014, 13, 95. 10.1186/s12934-014-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczyk D.; Caputi L.; Hatsch A.; Nielsen C. A. F.; Diefenbacher M.; Klein J.; Molt A.; Schröder H.; Cheng J. Z.; Naesby M.; O’Connor S. E. Angew. Chem. Int. Ed. 2015, 54, 5117–5121. 10.1002/ange.201410002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldemir H.; Richarz R.; Gulder T. A. M. Angew. Chem., Int. Ed. 2014, 53, 8286–8293. 10.1002/anie.201401075. [DOI] [PubMed] [Google Scholar]
- Yamanaka K.; Ryan K. S.; Gulder T. A. M.; Hughes C. C.; Moore B. S. J. Am. Chem. Soc. 2012, 134, 12434–12437. 10.1021/ja305670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickel B.; Schaller A. Appl. Microbiol. Biotechnol. 2013, 97, 8427–8438. 10.1007/s00253-013-5167-4. [DOI] [PubMed] [Google Scholar]
- Effenberger I.; Zhang B.; Li L.; Wang Q.; Liu Y.; Klaiber I.; Pfannstiel J.; Wang Q.; Schaller A. Angew. Chem., Int. Ed. 2015, 54, 14660–14663. 10.1002/anie.201507543. [DOI] [PubMed] [Google Scholar]
- Kim K.-W.; Moinuddin S. G. A.; Atwell K. M.; Costa M. A.; Davin L. B.; Lewis N. G. J. Biol. Chem. 2012, 287, 33957–33972. 10.1074/jbc.M112.387423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil Girol C.; Fisch K. M.; Heinekamp T.; Günther S.; Hüttel W.; Piel J.; Brakhage A. A.; Müller M. Angew. Chem., Int. Ed. 2012, 51, 9788–9791. 10.1002/anie.201203603. [DOI] [PubMed] [Google Scholar]
- Mazzaferro L. S.; Hüttel W.; Fries A.; Müller M. J. Am. Chem. Soc. 2015, 137, 12289–12295. 10.1021/jacs.5b06776. [DOI] [PubMed] [Google Scholar]
- Präg A.; Grüning B. A.; Häckh M.; Lüdeke S.; Wilde M.; Luzhetskyy A.; Richter M.; Luzhetska M.; Günther S.; Müller M. J. Am. Chem. Soc. 2014, 136, 6195–6198. 10.1021/ja501630w. [DOI] [PubMed] [Google Scholar]
- Chatterjee A.; Mallin H.; Klehr J.; Vallapurackal J.; Finke A. D.; Vera L.; Marsh M.; Ward T. R. Chem. Sci. 2016, 7, 673–677. 10.1039/C5SC03116H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Ling H.; Wang G.; Tian Y.; Liu G.; Tan H. Biochem. Biophys. Res. Commun. 2007, 361, 196–201. 10.1016/j.bbrc.2007.07.016. [DOI] [PubMed] [Google Scholar]; b Henderson D. P.; Shelton M. C.; Cotterill I. C.; Toone E. J. J. Org. Chem. 1997, 62, 7910–7911. 10.1021/jo971549s. [DOI] [PubMed] [Google Scholar]
- Shelton M. C.; Cotterill I. C.; Novak S. T. A.; Poonawala R. M.; Sudarshan S.; Toone E. J. J. Am. Chem. Soc. 1996, 118, 2117–2125. 10.1021/ja952596+. [DOI] [Google Scholar]
- a Carere J.; Baker P.; Seah S. Y. K. Biochemistry 2011, 50, 8407–8416. 10.1021/bi200960j. [DOI] [PubMed] [Google Scholar]; b Baker P.; Pan D.; Carere J.; Rossi A.; Wang W.; Seah S. Y. K. Biochemistry 2009, 48, 6551–6558. 10.1021/bi9006644. [DOI] [PubMed] [Google Scholar]
- a Cheriyan M.; Toone E. J.; Fierke C. A. Biochemistry 2012, 51, 1658–1668. 10.1021/bi201899b. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Cheriyan M.; Toone E. J.; Fierke C. A. Protein Sci. 2007, 16, 2368–2377. 10.1110/ps.073042907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheriyan M.; Walters M. J.; Kang B. D.; Anzaldi L. L.; Toone E. J.; Fierke C. A. Bioorg. Med. Chem. 2011, 19, 6447–6453. 10.1016/j.bmc.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P.; Seah S. Y. K. J. Am. Chem. Soc. 2012, 134, 507–513. 10.1021/ja208754r. [DOI] [PubMed] [Google Scholar]
- Baker P.; Carere J.; Seah S. Y. K. Biochemistry 2011, 50, 3559–3569. 10.1021/bi101947g. [DOI] [PubMed] [Google Scholar]
- a Porter J. L.; Rusli R. A.; Ollis D. L. ChemBioChem 2016, 17, 197–203. 10.1002/cbic.201500280. [DOI] [PubMed] [Google Scholar]; b Denard C. A.; Ren H. Q.; Zhao H. M. Curr. Opin. Chem. Biol. 2015, 25, 55–64. 10.1016/j.cbpa.2014.12.036. [DOI] [PubMed] [Google Scholar]; c Renata H.; Wang Z. J.; Arnold F. H. Angew. Chem., Int. Ed. 2015, 54, 3351–3367. 10.1002/anie.201409470. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Ilie A.; Reetz M. T. Isr. J. Chem. 2015, 55, 51–60. 10.1002/ijch.201400087. [DOI] [Google Scholar]; e Bornscheuer U. T.; Huisman G. W.; Kazlauskas R. J.; Lutz S.; Moore J. C.; Robins K. Nature 2012, 485, 185–194. 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]; f Acevedo-Rocha C. G.; Hoebenreich S.; Reetz M. T. Methods Mol. Biol. 2014, 1179, 103–128. 10.1007/978-1-4939-1053-3_7. [DOI] [PubMed] [Google Scholar]
- Turner N. J.; O’Reilly E. Nat. Chem. Biol. 2013, 9, 285–288. 10.1038/nchembio.1235. [DOI] [PubMed] [Google Scholar]