Abstract
Background
Some plants develop a breeding system that produces both chasmogamous (CH) and cleistogamous (CL) flowers. However, the underlying molecular mechanism remains elusive.
Results
In the present study, we observed that Viola philippica develops CH flowers with short daylight, whereas an extended photoperiod induces the formation of intermediate CL and CL flowers. In response to long daylight, the respective number and size of petals and stamens was lower and smaller than those of normally developed CH flowers, and a minimum of 14-h light induced complete CL flowers that had no petals but developed two stamens of reduced fertility. The floral ABC model indicates that B-class MADS-box genes largely influence the development of the affected two-whorl floral organs; therefore, we focused on characterizing these genes in V. philippica to understand this particular developmental transition. Three such genes were isolated and respectively designated as VpTM6-1, VpTM6-2, and VpPI. These were differentially expressed during floral development (particularly in petals and stamens) and the highest level of expression was observed in CH flowers; significantly low levels were detected in intermediate CL flowers, and the lowest level in CL flowers. The observed variations in the levels of expression after floral induction and organogenesis apparently occurred in response to variations in photoperiod.
Conclusions
Therefore, inhibition of the development of petals and stamens might be due to the downregulation of B-class MADS-box gene expression by long daylight, thereby inducing the generation of CL flowers. Our work contributes to the understanding of the adaptive evolutionary formation of dimorphic flowers in plants.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0832-2) contains supplementary material, which is available to authorized users.
Keywords: Adaptive evolution, Dimorphic flower, Gene expression, MADS-box gene, Photoperiod, Viola philippica
Background
Flowers are typically composed of four organ types: sepals, petals, stamens, and carpels, which run from the outside of the flower to the center. The ABC model of flower development explains how three major function class genes (A-, B-, and C-class) specify the identity of the four floral organ types. A-class alone controls sepals, A-class in combination with B-class controls petals, B-class in combination with C-class controls stamens, and C-class alone controls carpels [1, 2]. A pair of MADS-box genes, APETALA3 (AP3) and PISTILLATA (PI) in Arabidopsis thaliana, and DEFICIENS (DEF) and GLOBOSA (GLO) in Antirrhinum majus, encodes B-function activity [3–6]. Mutations in either the AP3/DEF or PI/GLO genes results in similar phenotypic variations, wherein petals are transformed into sepals, and stamens into carpels [7–9]. The B-class lineages apparently underwent duplications and subsequent functional divergence in some core eudicots, possibly playing a role in the diversification of floral morphology during evolution [10–16]. For example, in Solanaceae and Leguminosae, the PI lineage duplicated into two GLO-like genes (GLO1 and GLO2), and the AP3 lineage underwent a duplication event at the base of the core eudicots, giving rise to two AP3-like lineages called the euAP3 and paleoAP3 genes [14–20]. The euAP3 genes include AP3/DEF, and the paleoAP3 type genes were named TOMATO MADS BOX GENE 6 (TM6) genes, after the first isolated member from Solanum lycopersicum [21]. Although these paralogous genes are partially redundant, they have largely contributed to the development of stamens and petals. Interestingly, the TM6 lineage was subsequently lost in Arabidopsis and Antirrhinum [22]. In contrast, the AP3 lineage was lost in papaya, which now only contains TM6 [23]. These results indicate that it is possible to retain either or both of the AP3/TM6 paralogous pair and still produce flowers [22].
B-class MADS-box genes are not only involved in the specification of organ identity of petals and stamens, but also in the control of organ maturation. Knocking down AP3/PI at intermediate stages (stages 8–10) in Arabidopsis flowers induces petal-to-sepal transformations that gradually occur in consecutive buds. However, although stamens in these flowers retain their identity, they become increasingly underdeveloped and do not dehisce pollen [24]. Independent petal loss within the buttercup family (Ranunculaceae) is strongly associated with decreased or eliminated expression of a single floral organ identity gene, AP3-3 [25]. In rice, the B-class MADS-box gene, SUPERWOMAN1 (SPW1), specifies the identities of lodicules (equivalent to petals) and stamens. This was supported by the observation in the mutant of this gene, superwoman1-cleistogamy (spw1-cls), of normal stamens, where lodicules were transformed homeotically to lodicule-glume mosaic organs, thereby engendering cleistogamy [26]. The phenotypic effect observed in the downregulation of Medicago truncatula TM6 (MtTM6) activity involved a change in petal shape, as well as some stamens differentiating into filaments and anthers failing to produce pollen grains [14]. Furthermore, severely downregulating either Physalis floridana GLO2 (PFGLO2) or Physalis floridana TM6 (PFTM6) only results in poor pollen maturation [15, 16].
Several plants, including most species in the genus Viola within the family Violaceae are dimorphic cleistogamy plants capable of producing both open potential outcross and closed selfing flowers [27]. Two examples are such as Viola odorata [28, 29] and V. pubescens [30]. The open flowers are also called chasmogamous (CH) flowers, while the closed flowers called cleistogamous (CL) flowers. CL flowers may be energetically less costly to produce, resulting in more resources available for seed production and disruption of locally adapted gene complexes. On the other hand, CH flowers produce genetically variable progeny favored in spatially or temporally heterogeneous habitats [31–33]. Therefore, the evolution of dimorphic flowers at a single event exhibits the highest fitness when each flower type is produced in the environment for which it is best suited [27]. Compared to CH flowers, CL petals are undeveloped and functional stamens become smaller and decreased in number. Moreover, V. odorata CH flowers are formed in response to short daylight, whereas CL flowers emerge in response to long daylight [28, 29]. In V. pubescens, orthologous genes for gibberellins 20 oxidase (VGA20ox) and gibberellins 3 oxidase (VGA3ox) are upregulated in CH flowers compared to that in CL flowers, thereby indicating a role for gibberellins (GA) in the differential production of flower types [34]. However, the molecular mechanisms underlying this developmental transition are largely unknown in Viola. In the present study, we revealed the variations in the development of petals and stamens, which are the major affected floral organs in the formation of dimorphic flowers in V. philippica under different photoperiods. Furthermore, we investigated the potential role of B-class MADS-box genes in the development of the CH-CL breeding system. Three B-class MADS-box genes were cloned and designated as VpTM6-1, VpTM6-2, and VpPI. We determined that the differential expression of these genes during late floral development under the different photoperiods was correlated to the development of CH and CL flowers, thus providing novel insights into the development and evolution of dimorphic flowers in plants.
Results
Different types of floral morphology in V. philippica
V. philippica could develop both CH and CL flowers, although the CL flowers were approximately five to six times smaller than CH flowers (Fig. 1). CH flowers had five large and showy petals, with the lowest protruding slightly at the base into a spur, and five stamens forming a cone surrounding the pistil. Each stamen had four pollen sacs, and the lowest two stamens had noticeable nectar glands attached to them (Fig. 1a, d, g, and j). CL flowers had two stamens without nectar glands, each stamen had two pollen sacs, and the five petals were all undeveloped (Fig. 1b, e, h, and j). In certain conditions, intermediate CL (inCL) flowers developed, and they displayed variable characteristics. Typical observations were: between one and three poorly developed petals, two to five developed stamens, with each stamen having two to four pollen sacs but no nectar glands (Fig. 1c, f, i, and j). In extreme cases, a few poorly developed stamens in inCL flowers had one pollen sac, or even no pollen sac and just a membranous appendage (Fig. 1i and j). In addition, we found that the stamen length, anther length, pistil length, lower petal length, and width of CH flowers were all significantly greater than those of CL and inCL flowers. Conversely, the filament of CL and inCL stamens was distinct compared to that of CH flowers, which was undetectable (Fig. 1d, e, f, and k). No homeosis of floral organs was observed in the CH-CL transition.
Fig. 1.
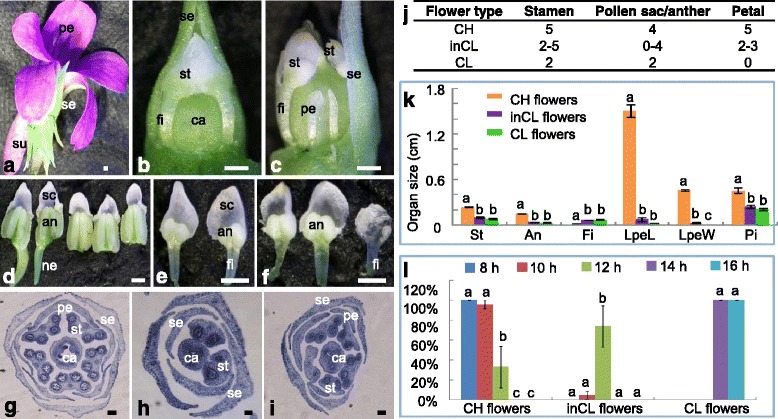
Floral morphological variations under different photoperiods in V. philippica. a CH flower. b CL flower. c inCL flower. d CH flower stamens. Five stamens with invisible filament develop, showing distinct nectar glands. e CL flower stamens. Only two stamens with visible filament develop. f inCL flower stamens. Three to five stamens develop with visible filaments. g-i The cross sections of flowers. g CH flowers. Five stamens and five petals are observed. Each stamen has four pollen sacs. h CL flowers. Two stamens and no petal are detected. Each stamen has two pollen sacs. i inCL flowers. Two to three petals, and two to five stamens develop in these flowers. Each stamen has zero to four pollen sacs. The microsporogenesis in some stamens is inhibited. Bars = 500 um (a–f). Bars = 200 um (g and i), 100 um (h). se, sepal; pe, petal; su, spur; st, stamen; ca, carpel; an, anther; sc, stamen cap; fi, filament; ne, nectar gland. j Variations in the number of floral organs. k Variations in floral organ size. The length of stamens (St), anthers (An), filaments (Fi), lower petals (LpeL), and pistils (Pi), and the width of lower petals (LpeW) are measured. l The CH-CL floral transition under different photoperiods. Standard errors are provided and lower-case letters (a, b, and c) indicate significant differences (P < 0.05)
Photoperiods affect flower development in V. philippica
Under natural conditions, V. philippica produced complete CH flowers in the early spring and a mixture of CH and inCL flowers in late spring and late autumn, whereas complete CL flowers developed in the summer and early autumn, suggesting that dimorphic flower development in V. philippica might be regulated by photoperiods. We thus set five photoperiods (8-, 10-, 12-, 14-, and 16-h daylight) to test this assumption. Results showed that three types of flowers developed under these conditions. Complete CL flowers were formed under long-day lights (14- and 16-h daylight), and CH flowers uniquely developed in 8-h daylight. At 10–12-h daylight, both CH and inCL flowers simultaneously developed and > 90 % of the floral buds were CH flowers under a photoperiod of 10-h daylight. The CH/inCL + CLratio significantly decreased with longer photoperiods (Fig. 1l), indicating that CL flower development was induced by long-day light. Therefore, different photoperiods may affect the development of petals and stamens, thus regulating the formation of different types of flower morphology in V. philippica. To further understand this formation, we next examined the floral organ initiation process.
Organ initiation and development of different flower types in V. philippica
V. philippica is a perennial plant with a (2 + 3) phyllotaxis shoot system, similar to that in V. odorata [28]. Roughly five floral developmental stages were defined based on the position of phyllotaxis using a series of landmark events and observation with a scanning electron microscope. In general, the floral buds closer to the central tender leaf were determined to be relatively young. We comparatively depicted the floral development of CH flowers that developed under short daylights, as well as CL and inCL flowers that developed under long and transitional daylights. The first stage involved the generation of the floral meristem, whereas the second stage showed floral organogenesis. Four whorl floral organ primordia of CH, as well as inCL and CL flowers, were observed in the floral meristem, indicating no obvious differences in the first and second stages in these three types of flowers (Fig. 2a, b, f, g, k, and l). However, at the third stage, after four whorl organ primordia formation, CH flowers had five obvious petals and stamens, whereas CL flowers had two stamens, and other stamens and all petals only consisted of organ primordial structures (Fig. 2c and m). There were two to five stamens, and at least lower petals that were poorly developed in inCL flowers (Fig. 2h). At the fourth stage (Fig. 2d, i, and n), four whorl floral organs of CH flowers continued to develop, and the style was higher than the stamens. The style of the CL and inCL flowers began to bend to the two fully developed stamens. At the fifth stage (Fig. 2e, j, and o), the nectar glands at the base of the two stamens began to appear in the CH flowers. The lower petals, side petals, or other stamens in the inCL flowers were poorly developed to some extent and later became more distinct, whereas five petals and the other three stamens (except for two big stamens) remained in the organ primordial state in the CL flowers (Fig. 2j and o). The filaments of CL and inCL stamens became relatively distinct (Fig. 2j and o). The development of the three stamens and all petals were then completely arrested, and these floral organs remained as primordia in the CL flowers, which developed under long daylight. These comparative observations suggested that early floral development was indistinguishable between CH and CL flowers, whereas the morphological divergence leading to the dimorphic flowers occurred at stage 3. This divergence was mainly attributable to the arrest of petal and stamen development in response to the extension of the photoperiod. Because the growth and differentiation of both petals and stamens were to some extent inhibited in the CH-inCL-CL floral transition in V. philippica with extended photoperiods, we next investigated the role of B-class MADS-box genes in the development of these two-whorl floral organs.
Fig. 2.
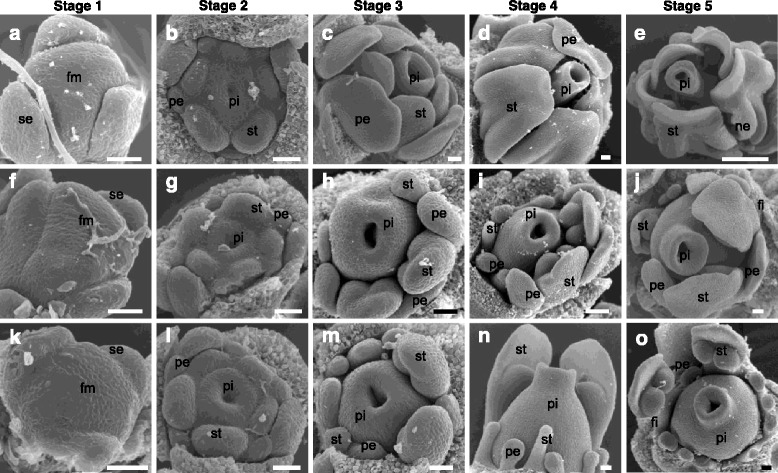
Floral initiation, organogenesis, and development in V. philippica. a–e CH flowers. f–j inCL flowers. k–o CL flowers. Floral induction and development as revealed by scanning electron microscopy (SEM) analysis, and five stages (Stage 1 to 5 as indicated) are roughly defined. se, sepal; pe, petal; st, stamen; ne, nectar; pi, pistil; fi, filament; fm, flower meristem. Bars = 50 um (a–d, f–j, k–o), and 500 um (e)
Isolation and sequencing analysis of B-class MADS-box genes in V. philippica
Using a combination of homologous fragment amplification and rapid amplification of cDNA ends (RACE), we acquired the cDNA of the three genes as putative members of both AP3- and PI-lineages of the MADS-box genes. These two lineage genes usually feature a paleoAP3, euAP3, or PI motif at the end of the C-terminal region [17]. BLAST analysis identified two V. philippica AP3 paralogs that shared the highest sequence identity (66.8 % and 64.6 %) at the amino acid level with putative Petunia TM6 proteins (Additional file 1: Table S1), indicating that these were TM6-like genes, and were thus designated as VpTM6-1 and VpTM6-2. The open reading frame (ORF) of the VpTM6-1 and VpTM6-2 gene was 693 and 687 bp in length, respectively, and encoded predicted polypeptides of 230 and 228 amino acids in length, respectively. Indels and single nucleotide polymorphisms (SNPs), including 41 nucleotide alteration positions were observed between these two cDNA sequences that resulted in 19 amino acid substitutions (Fig. 3a). Furthermore, these two paralogs shared 92.6 % sequence identity at the amino acid level (Additional file 1: Table S1). The four domains (M, I, K and C) of MADS-domain proteins play an essential role in DNA-binding and protein-protein interaction [35, 36]. Distribution of 2 ~ 10 amino acid substitutions, including deletions, was observed in each domain of the hypothetical VpTM6 proteins (Fig. 3a). Most of the amino acid substitutions in VpTM6-2, compared to VpTM6-1, were of different properties, and two of these (K66M, and S63_T64insYV) were predicated to be deleterious in function (Fig. 3a; Additional file 2: Table S2). These divergences thereby suggested functional divergence of the two duplicates. Multiple sequence alignment of these two genes, together with representatives of some previously functionally inferred AP3-like genes, showed that these two AP3 paralogous genes had a paleoAP3 motif instead of an euAP3 motif at the C-terminal end (Additional file 3: Figure S1a), further suggesting that these were putative TM6 orthologs. Phylogenetic analyses using the neighbor-joining (NJ) method further showed that these two AP3 paralogous genes were grouped together with previously reported TM6-like genes, such as the closely homologous genes from tomato, Petunia, and Physalis (Fig. 3b). These analyses clearly confirmed that the isolated two V. philippica AP3 lineage genes were TM6 orthologs. Unfortunately, AP3-lineage genes with the euAP3 motif were not isolated in the present study.
Fig. 3.
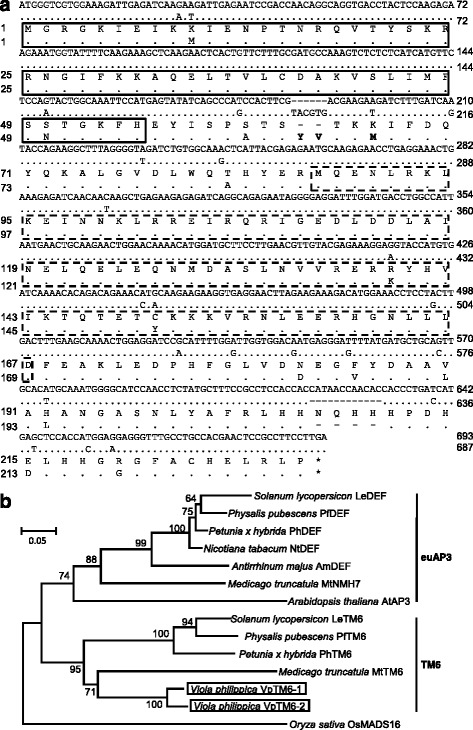
Molecular characterization of AP3-like MADS-box genes in V. philippica. a The ORFs of the VpTM6 genes and its putative peptides. Gaps are introduced to show indels between the two paralogs. The sequences of VpTM6-1 are presented, and the identical sequences in VpTM6-2 are highlighted using dots. The star indicates the stop codon. The M, I, K, and C domains of the hypothetic VpTM6 proteins are indicated. The M domain is highlighted by solid boxes, and the K domain is indicated by dashed boxes. The region between M and K domains is the I domain, while the C domain is behind the K domain. The deleterious amino acid substitutions were highlighted in bold (for details, see Additional file 2: Table S2). b Neighbor-joining (NJ) tree of AP3-like genes. VpTM6-1 and VpTM6-1 are boxed. The accession numbers of other sequences used are LeTM6 (X60759.1), LeAP3 (DQ674532.1), PhTM6 (AF230704.1), PhDEF (X69946.1), MtTM6 (JN412097.1), MtNMH7 (JN412096.1), AtAP3 (NM115294.5), AmDEF (AB516402.1), NtDEF (X96428.1), PfDEF (KC174703.1), and PfTM6 (KC174704.1). OsMADS16 (AF077760.1) is used as outgroup. The species name is listed before the gene name. The numbers next to the nodes are bootstrap values. Multiple sequence alignment for the NJ tree is shown in Additional file 3: (Figure S1a)
BLAST analysis also indicated that we isolated a PI putative ortholog in V. philippica, which we designated as VpPI. The ORF of the VpPI was 636 bp in length, and its cDNA encoded a predicted polypeptide of 211 amino acids in length (Fig. 4a) that was highly similar to Medicago PI (71.3 % amino acid identity) and other PI orthologs from various plant species (Additional file 1: Table S1). Multiple sequence alignment showed that the amino acid sequence of the V. philippica PI-homolog (VpPI) clearly had the PI motif at the C-terminal region (Additional file 3: Figure S1b). Reconstruction of a NJ tree showed VpPI clustering with PI (Fig. 4b). Therefore, VpTM6-1, VpTM6-2, and VpPI belonged to the homologous sequences of the B-class MADS-box genes.
Fig. 4.
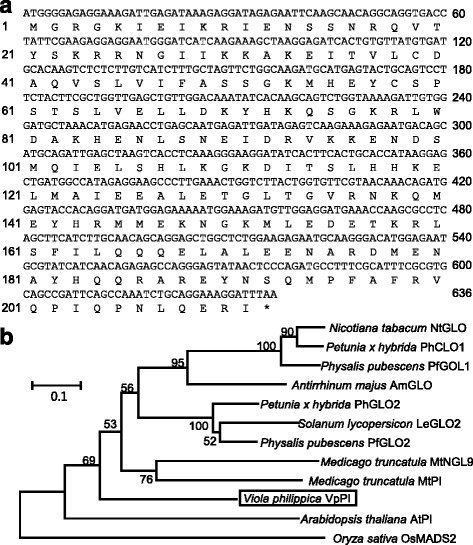
Molecular characterization of PI-like MADS-box genes in V. philippica. a The ORF of VpPI and its putative peptide. The star indicates the stop codon. b MP tree of PI-like genes. VpPI is boxed. The accession numbers of other sequences used are NtGLO (X67959.1), PhGLO1 (M91190.1), PhGLO2 (X69947.1), PfGLO1 (JX467691.1), PfGLO2 (KC174706.1), AmGLO (AB516403.1), LeGLO2 (DQ674531.1), MtNGL9 (FJ403469.1), MtPI (FJ403468.1), and AtPI (NM122031.3). OsMADS2 (L37526.1) is used as outgroup. The species name is listed before the gene name. The numbers next to the nodes are bootstrap values. Multiple sequence alignment for the NJ tree is shown in Additional file 3: (Figure S1b)
Expression of VpTM6 and VpPI during floral induction and organogenesis
Were the poorly developed or undeveloped petals and stamens of inCL and CL flowers in V. philippica related to variations in the expression of B-class MADS-box genes? To answer this question, we first examined the spatial and temporal patterns of expression during floral organogenesis and development of each flower type using in situ hybridization. Since the sequences of VpTM6-1 and VpTM6-2 were highly similar, a probe designed from VpTM6-2 was used in in situ hybridization to detect the expression of these two paralogs (designated as VpTM6). Results showed that VpTM6 and VpPI were continuously expressed from early stage (the second stage) to the later stage (the fifth stage) of floral organogenesis and development. At the floral organogenesis stage, VpTM6 was not only expressed in the primordia that normally developed into petals, stamens and pistils in CH, inCL, and CL flowers, but also in the primordia that either generated poorly developed petals and stamens or were retarded (Additional file 4: Figure S2a–l). In addition, in the later development stage of floral buds, VpTM6 was not only expressed in the developed petals and stamens in CH, inCL, and CL flowers, but also in the poorly developed or undeveloped petals and stamens of inCL and CL flowers. VpPI shared a similar expression domain as VpTM6 during floral organogenesis and development of CH, inCL, and CL flowers (Additional file 4: Figure S2m–x). Sense probes did not show any hybridization signals (Additional file 4: Figure S2y). We expected to observe distinct differences in the levels of expression of these genes between the fully developed and poorly or undeveloped floral organs in correlation to the three different types of flowers. However, no obvious difference in the spatial and temporal patterns of B-class gene expression was observed during floral organogenesis and development under any test conditions. The reason underlying these unexpected observations may be attributable to the decreased suitability of an in situ hybridization technique for quantification. Therefore, we investigated the expression of VpTM6-1, VpTM6-2, and VpPI using qRT-PCR.
Late floral expression of VpTM6 and VpPI in response to various photoperiods
To reveal the potential role of gene expression variation in response to photoperiods, we chose CH, inCL, and CL flower buds that corresponded to three photoperiods (10-, 12-, and 16-h daylight) for expression study. Two types of flower buds were used: the stage 3 flower buds (Fl1) that started to show distinct differences in morphology among CH, inCL, and CL flowers; and the stage 4 to 5 flower buds, which were designated as Fl2. Mature flowers (Mf) and leaves were also included. Using Fl1 of the CH flowers as control (Fig. 5a–c), we determined that VpTM6-1, VpTM6-2, and VpPI were expressed at extremely low levels in leaves (Fig. 5a–c). In contrast, these genes were mainly expressed in floral tissues, and had a tendency to increase during flower development under all conditions (Fig. 5a–c). Moreover, VpTM6-1 and VpTM6-2 in each floral tissue under 10-h daylight were all higher than in those subjected to 12- and 16-h daylight (Fig. 5a and b), thereby suggesting that the level of expression of VpTM6-1 and VpTM6-2 decreased as the duration of daylight was extended. A similar pattern of expression was observed in VpPI (Fig. 5c). These results indicated that the expression of the isolated MADS-box genes was predominantly expressed in floral organs, and hinted that the expression of these genes during late floral development stages might be regulated by photoperiods.
Fig. 5.
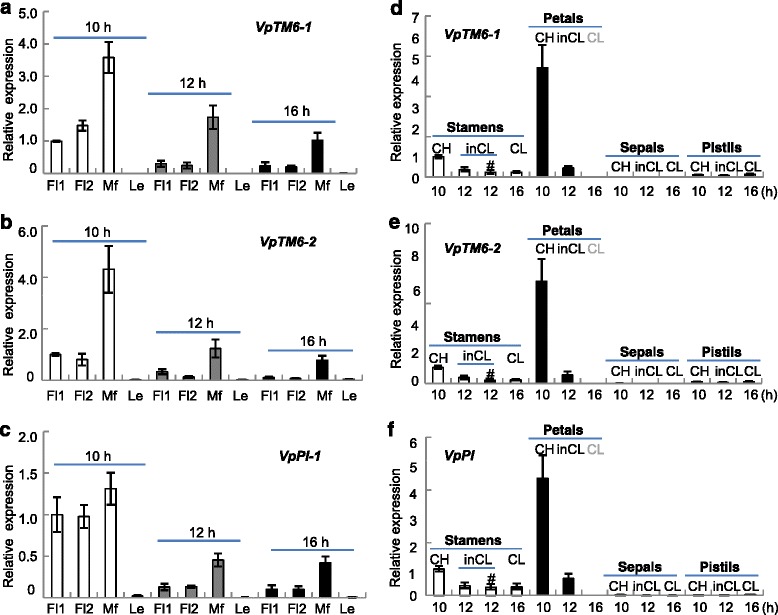
The floral expression of B-class MADS-box genes in response to variations in photoperiod. a–c Relative expression of VpTM6-1, VpTM6-2, and VpPI in flower buds and mature flowers. a VpTM6-1. b VpTM6-2. c VpPI. Gene expression in flower buds around the third stage (Fl1), the fourth to fifth stages (Fl2), mature flowers (Mf) and leaves (Le) under 10- (open columns), 12- (gray columns) and 16-h (black columns) daylight were evaluated using qRT-PCR. The gene expression in Fl1 under 10-h daylight was set to 1. d–f Relative expression of B-class MADS-box genes in floral organs of mature flowers. d VpTM6-1. e VpTM6-2. f VpPI. Gene expression in stamens (open columns), petals (black columns), and pistils (gray columns) from CH, inCL, and CL flowers under the indicated photoperiods were measured using qRT-PCR. No visible expression signal was detected in sepals under various conditions. # indicates poorly developed stamens in inCL flowers. Gray “CL” indicates that the petal was not collected from the CL flowers. The level of gene expression in the stamens of CH flowers subjected to 10-h daylight was set to 1. Three independent biological samples were used in all these analyses. Mean and standard errors are presented
To investigate further, we examined the expression levels of VpTM6-1, VpTM6-2, and VpPI in floral organs of mature flowers (Fig. 5d–f). The floral organs, including sepals, petals, stamens, and pistils, corresponding to different photoperiods, were collected from CH, inCL, and CL flowers. No expression of VpTM6-1 (Fig. 5d), VpTM6-2 (Fig. 5e), and VpPI (Fig. 5f) was detected in the sepals of all flower type, and expressed at a relatively low level in all the pistils. In contrast, these were highly expressed in the petals and stamens of all flowers (Fig. 5d–f). We also observed that the level of expression of VpTM6-1, VpTM6-2, and VpPI in the stamens and petals of the CH flowers were much higher than in the corresponding organs of inCL and CL flowers (Fig. 5d–f). In particular, the level of expression of these genes significantly decreased in poorly developed stamens and petals of inCL flowers (Fig. 5d–f).
These observations indicated that the significant decrease in the expression of B-class MADS-box genes after floral organogenesis was induced by photoperiod extension, playing an essential role in the formation of CL flowers.
Protein-protein interactions between VpTM6s and VpPI
To distinguish the divergence of VpTM6-1 and VpTM6-2, we investigated protein-protein interactions (PPI) among the three B-class MADS-domain proteins using a yeast GAL4 two-hybrid system. Both activating His3 and LacZ reporters respectively enabling cell growth of the transformed yeast and the generation of the blue coloration in the non-lethal ß-galactosidase assay were used to demonstrate PPIs. No toxicity and self-activation of these proteins in yeast were observed, and a dramatic difference in cell growth and ß-galactosidase activity in yeast (Additional file 5: Figure S3) suggested that VpTM6-1 interacted strongly with VpPI in yeast, whereas the interaction between VpTM6-2 and VpPI was very weak, further implying a dramatic difference of transcriptional activity of the two heterodimmers. These results suggest functional divergence of the two VpTM6 paralogs. Nonetheless, no homodimerization of these B-class MADS-domain proteins was observed (Additional file 5: Figure S3).
Discussion
Some plants produce both CH and CL flowers in different ecological conditions during the growth cycle. Extensive studies have focused on how various ecological factors, including water, light intensity, photoperiod, soil fertility, and temperature could affect the CH-CL mixed breeding system [30, 37–39]. The general conclusion is that dimorphic flower development might be the result of adaptive evolution. The present study investigated dimorphic flower development under different photoperiods in V. philippica, including its underlying molecular mechanisms.
Dimorphic flower development and photoperiod regulation
The development of floral organs could be inhibited in nature; however, during the early developmental stage of flowers, all floral organ primordia are generally formed. For example, retardation or arrest of some floral organs after initiation occurs in most unisexual plants. Dioecious plants (such as white campion and Sorrel) and monoecious species (such as cucumber and London plant tree) all go through a hermaphroditic stage early in flower development, followed by differential abortion or arrest of sex organs, which can occur at various stages [40–43]. This phenomenon has also been observed in dimorphic cleistogamy plants. CL flowers of Lamium amplexicaule exhibit the least number of modifications, and no significant morphological differences are observed until pollen meiosis occurs compared to its CH flowers [44]. In the present study, no morphological differences at floral induction and organogenesis were detected in V. philippica. However, at later flower development stages, CH flowers featured normal flower structure, while petal and partial stamen development was retarded in CL flowers. In most cases, CL flowers had no petals and only a few fertile anthers. The arrest of floral organs is ubiquitous in nature for a variety of reasons [27, 28, 30, 40–43]. Similar to observations in temperate plant species [27, 34], the phenology of the two flower types in V. philippica is mainly determined by photoperiod. This phenomenon was also observed in V. odorata, in which CH flowers were formed in response to 11-h or less of daylight, CL flowers developed in response to 14-h or more of daylight, and inCL were produced during transitional periods [28]. Therefore, we confirmed that floral organ morphologic differentiation in V. philippica is determined during organogenesis after floral induction, and is influenced by photoperiod in dimorphic flower plants.
Floral expression of VpTM6 and VpPI in response to variations in photoperiod
Genome-wide gene expression patterns from cross-species microarray analysis of Cardamine kokaiensis of Brassicaceae has suggested that a substantial amount of genes, including petal and stamen developmental genes, are expressed differentially between CH and CL flowers [45]. However, the key regulatory gene in controlling the CH-CL transition has not been identified. In the present study, the floral organs most affected in the CH-CL transition as the photoperiod was extended were the petals and stamens. The floral ABC model indicates that B-class MADS-box genes determine the development of the two types of floral organs [1, 7, 46]. Modification of these genes either in terms of sequence or expression in various plants leads to the morphological diversification of related floral organs [13, 14, 25, 47–52]. To understand the molecular switches related to this CH-CL transition, we investigated these genes in V. philippica. Three B-class MADS-box genes, VpTM6-1, VpTM6-2, and VpPI were isolated. The coding sequence variation of these B-class gene homologs does not immediately suggest any functional novelties relative to those in non-dimorphic flower model plants, such as Arabidopsis and Petunia. Nonetheless, the expression of these genes was significantly altered in CH, CL, and inCL flower development. In line with no distinguishable differences during early flower development and no observed floral homeosis, these genes shared a relatively similar expression pattern during floral organogenesis of CH, CL, and inCL flowers. However, in the later flower development stage, where distinct floral morphology (particularly the size/number of petals and stamens) were observed in CL and inCL flowers, the expression of VpTM6-1, VpTM6-2, and VpPI in flowers (particularly in petals and stamens) significantly decreased in CL and inCL flowers, especially compared to CH flowers. The variations in expression pattern were concomitantly consistent with the floral organ divergence that generates CH, inCL, and CL flowers in extended photoperiods. The downregulation of these B-class MADS-box genes occurred during the late developmental stages was not possible to alter the organ identity of petals and stamens, but might result in organ size reduction and developmental abortion. This was further supported by the observation that the extent of reduction in organ size/number was not strictly equivalent to the decrease extent of B-class gene expression in these organs (Additional file 6: Table S3), indicating that the drop in expression should not be a result of the lower proportion of petal/stamen tissues in CL and inCL flowers to those in CH flowers. Nonetheless, the CH-CL transition, as an indirect result of the downregulation in the expression of these B-class genes, cannot be excluded. Given the postdevelopmental role of B-class MADS-box genes [24, 50–52], we therefore concluded that long daylight inhibited the development of petals and stamens by directly or indirectly inhibiting the expression of VpTM6-1, VpTM6-2, and VpPI genes to produce CL flowers.
B-class MADS-box genes, encoding bifunctional transcription factors, activate or repress a substantial number of downstream targets [24], thus acting as a context-dependent transcriptional switch that directs flower development: either floral organ identity, or postdevelopment. Since floral homeosis did not occur in the CH-CL transition, possible postdevelopmental mechanisms of B-class genes in Viola were further discussed. In Arabidopsis, AP3 and PI positively regulate the expression of SPOROCYTELESS/NOZZLE (SPL/NZZ) and NAP (for NAC-LIKE, ACTIVATED BY AP3/PI, also known as NO APICAL MERISTEM) [24, 53]. SPL/NZZ is necessary for the formation of anther walls, and is required during the late stages of stamen development for microsporogenesis and consequent pollen formation [54–57]. On the other hand, NAP is involved in the transition between cell division and cell expansion phases during the growth of petals and stamens [53]. AP3/PI also suppresses certain downstream genes, such as GNC and GNC-LIKE (GNL), that encode two GATA-type zinc finger proteins, which in turn regulate sugar response and nitrate metabolism genes, thereby providing a link between organ development and nutrient sensing [58]. Short photoperiods induce the expression of VGA20ox and VGA3ox, which increases gibberellins that are also involved in CH-CL flower development in V. pubescens [34]. Gibberellins regulate the expression of specific floral genes in Arabidopsis such as the B-class MADS-box genes [59, 60]. Therefore, upon perceiving regulatory signals such as photoperiod (or gibberellins), we assumed that the differential expression of these Viola B-class MADS-box genes might influence dimorphic flower development by regulating homologous genes SPL/NZZ, NAP, and GNC, which in turn affects the growth and development of petals and stamens. However, further functional studies involving genetic manipulation of V. philippica should be conducted to verify this hypothesis.
The novel role of B-class MADS-box genes in dimorphic flower development
B-class MADS-box genes include both AP3 and PI lineages that evolved to play a primary role in petal and stamen development and in the establishment of stamen functionality [12–16, 18, 61]. Coding sequence variations that lead to different PPIs associated with these proteins play an essential role in this functional evolutionary process [15, 16], whereas altering its expression pattern facilitates the acquisition of additional functions, thereby leading to new floral morphologies [17–19, 25, 50–52]. The transcriptional regulation of these genes roughly includes the initiation of expression during early floral stages and the maintenance of expression through the majority of floral organ development [6, 9, 46–48, 62, 63]. When expression is affected during floral initiation, homeotic alterations usually occur, resulting in multiple corolla or calyces [7–9, 15, 64, 65]. However, when the expression is altered during late developmental stages, the maturation or function of the organs is affected [13–16, 24, 40–43]. Compared to previously identified B-class MADS-box genes [3–6], VpTM6-1, VpTM6-2, and VpPI in V. philippica shared a similar expression pattern during flower development, indicating that these genes may play a conserved role in establishing floral organ identity. However, we found that differential expression of these genes during late developmental stages of CH and CL flowers is regulated by variations in photoperiod, thereby suggesting a novel role for these B-class MADS-box genes in dimorphic flower development.
Floral morphology, such as petal size/shape instead of change of organ identity, could be regulated by artificial control of B-class gene expression, such as in M. truncatula [14] and tomato [13], whereas severely downregulating either PFGLO2 or PFTM6 only results in the production of immature pollen in P. floridana [15, 16]. Interestingly, regulating gene expression to generate distinct floral morphology also occurs in nature, such as in the decrease in B-class MADS-box genes to generate CL flowers in V. philippica. The molecular mechanism underlying this regulation of B-class MADS-box genes in response to photoperiods in petals and stamens requires further investigation. Currently, at least two levels of divergence have been observed. VpTM6-1, VpTM6-2, and VpPI are expressed at higher levels in petals than in stamens in CH flowers. However, this decrease in the level of expression was more extensive in petals than in stamens, correlating to the absence of petals and the reduction to two fertile stamens in CL flowers. Moreover, VpPI interacted strongly with VpTM6-1, but did weakly with VpTM6-2, which was indicative of VpTM6-1 and VpTM6-2 divergence in dimerization activity and transcriptional activity. Such differences in heterodimerization capabilities have been observed in species that harbor duplicated genes such as Petunia and Physalis [12, 15, 16]. Since a single amino acid change in the I doamin is sufficient to alter PI-like dimerization activity during maize domestication [66], that this type of divergence might result from extensive seuquence divergence of the two VpTM6 proteins, particularly the two deleterious alterations (K66M, and S63_T64insYV) in the I domain, should also be considered in understanding the functional divergence of B-class MADS-box genes in V. philippica.
Conclusions
The comprehensive investigation of floral MADS-box genes could facilitate better understanding of CH and CL flower development in V. philippica. Nevertheless, to our knowledge, we were the first time to reveal that the differential floral expression of B-class MADS-box genes after floral induction and organogenesis in response to variations in photoperiod is associated with the development of the CH-CL breeding system in Viola. Our findings present new insight into the development and evolution of dimorphic flowers.
Methods
Plant materials and growth conditions
Seeds of V. philippica were collected and stored at the Northwest Normal University (Lanzhou, Gansu, China). They are available upon request. The seeds were sterilized by immersion in 30 % sodium hypochlorite solution for 20 min, and then rinsed three times with distilled water before sowing in triangular flasks containing MS (Murashige and Skoog) culture medium. After the fourth true leaves developed, the seedlings were transplanted to individual 100-mm (0.5 L) diameter plastic pots containing peat and vermiculite (v/v = 2:1). The plants were grown at 22-28 °C under 8-, 10-, 12-, 14-, and 16-h daylight, respectively, in a growth chamber. The temperature was kept by air-conditioners. The humidity was kept around 48 %, and light intensity was 116 μmol m−2s−1. The plants under different daylight were grown in parallel on the different shelves. The daylight length was controlled by time controller in each cultivated shelf. To avoid the reciprocal influence, each shelf was enclosed by black cloth that was manually removed every day10:00 am for 8 h for ventilation.
Flower morphology observation
The morphology of flower buds was observed under a stereomicroscope (Olympus SZ61, Tokyo, Japan). The ratio of CH, inCL, and CL flowers was evaluated under 8-, 10-, 12-, 14-, and 16-h daylight, respectively. One hundred plants were observed in each photoperiod, and about 200 flower buds were counted in each case. To measure floral organ size, 10 mature flowers of each type (CH, inCL, and CL) were analyzed. The images were captured using a camera linked to a stereomicroscope (Olympus SZ61, Tokyo, Japan).
Scanning electron microscopy and histological analyses
Flower buds at different stages were fixed in a 3:1 (v/v) ethanol:glacial acetic acid solution and kept at 4 °C, dehydrated through an increasing ethanol gradient of up to 100 % ethanol, and dried in a critical point drier. Samples were imaged using a scanning electron microscope (Hitachi S-450, Tokyo, Japan). For histological analyses, the whole green flower buds were dyed for 30 h using Love’s hematoxylin to dark red color. Then, the dyed flower buds were washed in running water for 5 h to remove excess dye until these became blue. Finally, the blue flower buds were dehydrated through an increasing ethanol gradient, cleared using xylene, and embedded in Paraplast (Sigma P3683, St. Louis, Missouri, USA). Cross sections (7-μm thickness) of the flower buds were mounted on the slides with water at 40 °C. The wax was cleared from the slides by washing with 100 % xylene. The images of the cleared slides were finally photographed under a Leica microscope (DMI4000B, Wetzlar, Germany).
Sequence isolation
The cDNA fragments of the targeted genes were isolated using degenerate primers (Additional file 7: Table S4) that were designed based on the conserved regions of the APETALA3 (AP3) and PISTILLATA (PI) orthologs from various plant species in GenBank (http://www.ncbi.nlm.nih.gov/). The full-length cDNA was assembled using 3′/5′-rapid amplification of cDNA ends (RACE). Universal 3′ and 5′ PCR primers were supplied by the SMARTer™ cDNA Library Construction Kit (Clontech, Mountain View, California, USA). After the RACE experiments, the full-length cDNA was amplified by routine RT-PCR using gene-specific primers (Additional file 7: Table S4). The cycling program consisted of an initial denaturation at 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 55 °C (degenerate primers) or 68 °C (gene-specific primers) for 30 s, 72 °C for 30 s, and a final extension of 72 °C for 10 min.
Sequence divergence evaluation and phylogenetic analysis
Neutral or deleterious amino acid mutations were predicted by PROVEAN, the protein variation effect analyzer with default parameter setting (http://provean.jcvi.org/index.php). The amino acid sequences of these genes were aligned using CLUSTALW1.81 under default settings with manual adjustments. Gaps were introduced for proper alignment. The neighbor-joining phylogeny trees using protein sequences were constructed using MEGA5 [67]. Bootstrap values were based on 1,000 replicates.
qRT-PCR
The plant growth cycle of V. philippica was too long, few floral buds developed under 8-h daylight. Therefore, stages 3, 4, and 5 floral buds, young leaves, and floral organs of mature flowers were respectively collected under 10-, 12- and 16-h light period in the glasshouse. Total RNA was extracted using TRIzol reagent (TIANGEN, Beijing, China). For qRT-PCR analysis, a PrimeScript RT Reagent Kit (TaKaRa, Dalian, China), SYBR Premix EX Taq II (TaKaRa, Dalian, China), and gene-specific primers (Additional file 7: Table S4) were used. An 18S ribosomal RNA gene was used as internal reference (AB354544.1). The primers were designed using DNAMAN, and evaluating of their specificity and primer efficiency indicated that they were suitable for comparative qRT-PCR analysis (Additional file 8: Figure S4). The amplification conditions were 95 °C for 30 s for one cycle, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. Three independent biological samples were used. Expression levels were calculated according to Livak and Schmittgen [68].
RNA in situ hybridization
Floral buds at various developmental stages were fixed in 4 % (wt/vol = 4 g/100 mL) paraformaldehyde and embedded in Paraplast (Sigma P3683, St. Louis, Missouri, USA). When ATG was set as 1, around 356-bp fragment of VpTM6-2 (positions 331–687) and VpPI (positions 236–591) was used as template for both sense and antisense probe synthesis using the DIG RNA labeling kit (Roche, Mannheim, Germany) and T7 RNA polymerase (Roche, Mannheim, Germany). Hybridization was performed as described by Javelle [69], with the alteration that we washed the slides at a temperature of 50 °C. Sections of floral tissues of CH, inCL and CL flowers were incubated in the same hybridization solution for each probe. Images were captured with a Leica microscope (DMI4000B, Wetzlar, Germany).
Yeast two-hybrid analysis
The ORFs of VpTM6-1, VpTM6-1, and VpPI were cloned into the pGADT7 or PGBKT7 vector (Clontech, Mountain View, California, USA). The co-transformed yeast cells of BD and AD fusion plasmids were selected by growth on SD plates lacking leucine (Leu) and tryptophan (Trp). Interactions were analyzed on the SD plates lacking Leu, Trp, adenine (Ade), and histidine (His), and further confirmed by the non-lethal ß-galactosidase activity assay in the yeast strain AH109. Before checking PPIs, the toxicity and self-activation capability of these B-class MADS-domain proteins were checked. These experiments were performed according to Gong and He [70].
Sequencing and primer information
Sequencing of all constructs and primer synthesis were performed by Huada (Beijing, China). All primers used in this study are presented in Additional file 7: (Table S4).
Abbreviations
AP3, APETALA3; cDNA, complementary DNA; CH, chasmogamous; CL, cleistogamous. GLO, GLOBOSA; NJ, neighbor-joining; ORF, open reading frame; PI, PISTILLATA; PPI, protein-protein interaction; qRT-PCR, quantitative RT-PCR; RACE, rapid amplification of cDNA ends; SNP, single nucleotide polymorphism; TM6, TOMATO MADS BOX GENE 6
Acknowledgements
The assistance of Dr. PC Gong (State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences) in yeast two-hybrid analysis and PROVEAN prediction is acknowledged.
Funding
This work was supported by the NSFC grants (31560066 and 31260054), the Young Teachers Improving Program from the Northwest Normal University (NWNU-LKQN-13-20), and the CAS/SAFEA International Partner Program for Creative Research Teams of “Systematic and Evolution Botany”.
Availability of data and materials
All relevant supporting data can be found within the additional files accompanying this article. Sequence data described in this article can be found in GenBank (http://www.ncbi.nlm.nih.gov) under the accessions of KU318322 (VpTM6-1), KU318323 (VpTM6-2), and KU318324 (VpPI).
Authors’ contributions
QXL, KS and CYH conceived and designed the work. KS identified plant species. QXL performed all experiments. JZ participated in the in situ hybridizations. QDH and JW involved in RNA extraction and expression study. QXL, KS and CYH analyzed the data. QXL, CYH and KS wrote the manuscript. All authors have read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
Protein sequence variation of AP3- (a) and PI-lineage (b) genes. (PDF 169 kb)
Amino acid divergence between the two VpTM6 paralogs. (PDF 177 kb)
Multiple sequence alignment of B-class MADS-box genes. (PDF 312 kb)
Gene expression during floral organogenesis and development. (PDF 590 kb)
Protein-protein interactions in yeast. (PDF 180 kb)
The reduction extent of organ size and gene expression in CH-CL transition. (PDF 336 kb)
The primers used in the present study. (PDF 252 kb)
Primer evaluation for qRT-PCR. (PDF 342 kb)
Contributor Information
Qiaoxia Li, Email: liqiaoxia8024@163.com.
Qingdi Huo, Email: huoqingdi@hotmail.com.
Juan Wang, Email: damoshuihen@163.com.
Jing Zhao, Email: zhj1201@126.com.
Kun Sun, Email: kunsun@163.com.
Chaoying He, Email: chaoying@ibcas.ac.cn.
References
- 1.Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–7. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 2.Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–9. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 3.Sommer H, Beltran JP, Huijser P, Pape H, Lonnig WE, Saedler H, Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 1990;9:605–13. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–97. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- 5.Trobner W, Ramirez L, Motte P, Hue I, Huijser P, Lonnig WE, Saedler H, Sommer H, Schwarz-Sommer Z. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992;13:4693–704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene. PISTILLATA. Genes Dev. 1994;8:1548–60. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- 7.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill JP, Lord EM. Floral development in Arabidopsis thaliana: a comparison of the wild type and the homeotic pistillata mutant. Can J Bot. 1989;67:2922–36. doi: 10.1139/b89-375. [DOI] [Google Scholar]
- 9.Jack T, Fox GL, Meyerowitz EM. Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell. 1994;76:703–16. doi: 10.1016/0092-8674(94)90509-6. [DOI] [PubMed] [Google Scholar]
- 10.van der Krol AR, Brunelle A, Tsuchimoto S, Chua NH. Functional analysis of Petunia floral homeotic MADS box gene pMADS1. Genes Dev. 1993;7:1214–28. doi: 10.1101/gad.7.7a.1214. [DOI] [PubMed] [Google Scholar]
- 11.Tsuchimoto S, Mayama T, van der Krol A, Ohtsubo E. The whorl-specific action of a Petunia class B floral homeotic gene. Genes Cells. 2000;5:89–99. doi: 10.1046/j.1365-2443.2000.00308.x. [DOI] [PubMed] [Google Scholar]
- 12.Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T. The duplicated B–class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrid flower development. Plant Cell. 2004;16:741–54. doi: 10.1105/tpc.019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Martino G, Pan I, Emmanuel E, Levy A, Irish VF. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell. 2006;18:1833–45. doi: 10.1105/tpc.106.042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roque E, Serwatowska J, Rochina MC, Wen JQ, Mysore KS, Yenush L, Beltrán JP, Caňas LA. Functional specialization of duplicated AP3-like genes in Medicago truncatula. Plant J. 2013;73:663–75. doi: 10.1111/tpj.12068. [DOI] [PubMed] [Google Scholar]
- 15.Zhang JS, Li ZC, Zhao J, Zhang SH, Quan H, Zhao M, He CY. Deciphering the Physalis floridana double-layered-lantern1 mutant provides insights into functional divergence of the GLOBOSA duplicates within the Solanaceae. Plant Physiol. 2014;164:748–64. doi: 10.1104/pp.113.233072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang SH, Zhang JS, Zhao J, He CY. Distinct subfunctionalization and neofunctionalization of the B-class MADS-box genes in Physalis floridana. Planta. 2015;241:387–402. doi: 10.1007/s00425-014-2190-3. [DOI] [PubMed] [Google Scholar]
- 17.Kramer EM, Dorit RL, Irish VF. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149:765–83. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandenbussche M, Theissen G, Van de Peer Y, Gerats T. Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frame shift mutations. Nucleic Acids Res. 2003;31:4401–9. doi: 10.1093/nar/gkg642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramer EM, Su HJ, Wu JM, Hu JM. A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the APETALA3 gene lineage. BMC Evol Biol. 2006;6:30. doi: 10.1186/1471-2148-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benlloch R, Roque E, Ferrándiz C, Cosson V, Caballero T, Penmetsa RV, Beltrán JP, Cañas LA, Ratet P, Madueño F. Analysis of B function in legumes: PISTILLATA proteins do not require the PI motif for floral organ development in Medicago truncatula. Plant J. 2009;60:102–11. doi: 10.1111/j.1365-313X.2009.03939.x. [DOI] [PubMed] [Google Scholar]
- 21.Pnueli L, Abu-Abeid M, Zamir D, Nacken W, Schwarz-Sommer Z, Lifschitz E. The MADS box gene family in tomato: expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1991;1:255–66. doi: 10.1111/j.1365-313X.1991.00255.x. [DOI] [PubMed] [Google Scholar]
- 22.Causier B, Castillo R, Xue Y, Schwarz-Sommer Z, Davies B. Tracing the evolution of the floral homeotic B- and C-function genes through genome synteny. Mol Biol Evol. 2010;27:2651–64. doi: 10.1093/molbev/msq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ackerman CM, Yu Q, Kim S, Paull RE, Moore PH, Ming R. B-class MADS-box genes in trioecious papaya: two paleoAP3 paralogs, CpTM6-1 and CpTM6-2, and a PI ortholog CpPI. Planta. 2008;227:741–53. doi: 10.1007/s00425-007-0653-5. [DOI] [PubMed] [Google Scholar]
- 24.Wuest SE, O’Maoileidigha DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci U S A. 2012;109:13452–7. doi: 10.1073/pnas.1207075109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R, Guo C, Zhang WG, Wang PP, Li L, Duan XS, Zhao L, Shan HY, Hodges SA, Kramer EM, Ren Y, Kong HZ. Disruption of the petal identity gene APETALA3-3 is highly correlated with loss of petals within the buttercup family (Ranunculaceae). Proc Natl Acad Sci U S A. 2013;110:5074–9. [DOI] [PMC free article] [PubMed]
- 26.Yoshida H, Itoh JI, Ohmori S, Miyoshi K, Horigome A, Uchida E, Kimizu M, Matsumura Y, Kusaba M, Satoh H, Nagato Y. Superwoman1-cleistogamy, a hopeful allele for gene containment in GM rice. Plant Biotech J. 2007;5:835–46. doi: 10.1111/j.1467-7652.2007.00291.x. [DOI] [PubMed] [Google Scholar]
- 27.Culley TM, Klooster MR. The cleistogamous breeding system: A review of its frequency, evolution, and ecology in angiosperms. Bot Rev. 2007;73:1–30. doi: 10.1663/0006-8101(2007)73[1:TCBSAR]2.0.CO;2. [DOI] [Google Scholar]
- 28.Mayers AM, Lord EM. Comparative flower development in the cleistogamous species Viola odorata. I. A growth rate study. Am J Bot. 1983;70:1548–55. doi: 10.2307/2443353. [DOI] [Google Scholar]
- 29.Mayers AM, Lord EM. Comparative flower development in the cleistogamous species Viola odorata. II.An organographic study. Am J Bot. 1983;70:1556–63. doi: 10.2307/2443354. [DOI] [Google Scholar]
- 30.Culley TM. Reproductive biology and delayed selfing in Viola pubescens (Violaceae), an understory herb with chasmogamous and cleistogamous flowers. Int J Plant Sci. 2002;163:113–22. doi: 10.1086/324180. [DOI] [Google Scholar]
- 31.Mitchell-Olds T, Waller DM. Relative performance of selfed and outcrossed progeny in Impatiens capensis. Evolution. 1985;39:533–44. doi: 10.2307/2408651. [DOI] [PubMed] [Google Scholar]
- 32.Antlfinger AE. Field germination and seedling growth of CH and CL progeny of Impatiens capensis (Balsaminaceae) Am J Bot. 1986;73:1267–73. doi: 10.2307/2444061. [DOI] [Google Scholar]
- 33.Holsinger KE. Dispersal and plant mating systems: the evolution of self-fertilization in subdivided populations. Evolution. 1986;40:405–13. doi: 10.2307/2408818. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Ballard HE, McNally RR, Wyatt SE. Gibberellins are involved but not sufficient to trigger a shift between chasmogamous-cleistogamous flower types in Viola pubescens. J Torrey Bot Soc. 2013;140:1–8. doi: 10.3159/TORREY-D-12-00044.1. [DOI] [Google Scholar]
- 35.Immink RG, Kaufmann K, Angenent GC. The “ABC” of MADS domain protein behaviour and interactions. Semin Cell Dev Biol. 2010;21:87–93. doi: 10.1016/j.semcdb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Theissen G. Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol. 2001;4:75–85. doi: 10.1016/S1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- 37.Clavijo ERD, Jimenez MJ. Cleistogamy and chasmogamy in Ceratocapnos heterocarpa (Fumariaceae) Int J Plant Sci. 1993;154:325–33. doi: 10.1086/297113. [DOI] [Google Scholar]
- 38.Corff JL. Effects of light and nutrient availability on chasmogamy and cleistogamy in an understory tropical herb, Calathea micans (Marantaceae) Am J Bot. 1993;80:1392–9. doi: 10.2307/2445667. [DOI] [Google Scholar]
- 39.Sigrist MR, Sazima M. Ruellia brevifolia (Pohl) Ezcurra (Acanthaceae): flowering phenology, pollination biology and reproduction. Braz J Bot. 2002;25:35–42. doi: 10.1590/S0100-84042002000100006. [DOI] [Google Scholar]
- 40.Hardenack S, Ye D, Saedler H, Grant S. Comparison of MADS box gene expression in developing male and female flowers of the dioecious plant white campion. Plant Cell. 1994;6:1775–87. doi: 10.1105/tpc.6.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ainsworth C, Crossley S, Buchanan-Wollaston V, Thangavelu M, Parke J. Male and female flowers of the dioecious plant Sorrel show different patterns of MADS box gene expression. Plant Cell. 1995;7:1583–98. doi: 10.1105/tpc.7.10.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kater MM, Franken J, Carney KJ, Colombo L, Angenent GC. Sex determination in the monoecious species cucumber is confined to specific floral whorls. Plant Cell. 2001;13:481–93. doi: 10.1105/tpc.13.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z, Liu G, Zhang J, Lu S, Yi S, Bao M. Cloning and characterization of paleoAP3-like MADS-box gene in London plane tree. Biol Plantarum. 2012;56:585–9. doi: 10.1007/s10535-012-0112-4. [DOI] [Google Scholar]
- 44.Lord EM. Floral morphogenesis in Lamium amphlexicaule L. (Labiatae) with a model for the evolution of the cleistogamous flower. Botanical Gazette. 1982;143:63–72. doi: 10.1086/337271. [DOI] [Google Scholar]
- 45.Morinaga SI, Nagano AJ, Miyazaki S, Kubo M, Demura T, Fukuda H, Sakai S, Hasebe M. Ecogenomics of cleistogamous and chasmogamous flowering: genome-wide gene expression patterns from cross-species microarray analysis in Cardamine kokaiensis (Brassicaceae) J Ecol. 2008;96:1086–97. doi: 10.1111/j.1365-2745.2008.01392.x. [DOI] [Google Scholar]
- 46.Krizek BA, Meyerowitz EM. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development. 1996;122:11–22. doi: 10.1242/dev.122.1.11. [DOI] [PubMed] [Google Scholar]
- 47.Hill TA, Day CD, Zondlo SC. Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development. 1998;125:1711–21. doi: 10.1242/dev.125.9.1711. [DOI] [PubMed] [Google Scholar]
- 48.Honma T, Goto K. The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development. 2000;127:2021–30. doi: 10.1242/dev.127.10.2021. [DOI] [PubMed] [Google Scholar]
- 49.Stellari GM, Jaramillo MA, Kramer EM. Evolution of the APETALA3 and PISTILLATA lineages of MADS-box containing genes in basal angiosperms. Mol Biol Evol. 2004;21:506–19. doi: 10.1093/molbev/msh044. [DOI] [PubMed] [Google Scholar]
- 50.Kramer M, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago PM. Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell. 2007;19:750–66. doi: 10.1105/tpc.107.050385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma B, Guo C, Kong H, Kramer EM. Petal-specific subfunctionalization of an APETALA3 paralog in the Ranunculales and its implications for petal evolution. New Phytol. 2011;191:870–83. doi: 10.1111/j.1469-8137.2011.03744.x. [DOI] [PubMed] [Google Scholar]
- 52.Sharma B, Kramer EM. Sub- and neo-functionalization of APETALA3 paralogs have contributed to the evolution of novel floral organ identity in Aquilegia (columbine, Ranunculaceae) New Phytol. 2013;197:949–57. doi: 10.1111/nph.12078. [DOI] [PubMed] [Google Scholar]
- 53.Sablowski RWM, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell. 1998;92:93–103. doi: 10.1016/S0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 54.Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1999;96:11664–9. doi: 10.1073/pnas.96.20.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang WC, Ye D, Xu J, Sundaresan V. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Genes Dev. 1999;13:2108–17. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann J, Meyerowitz EM. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature. 2004;15:356–60. doi: 10.1038/nature02733. [DOI] [PubMed] [Google Scholar]
- 57.Liu XD, Huang J, Parameswaran S, Ito T, Seubert B, Auer M, Rymaszewski A, Jia GX. The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol. 2009;151:1401–11. doi: 10.1104/pp.109.145896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mara CD, Irish VF. Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol. 2008;147:707–18. doi: 10.1104/pp.107.115634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blazquez MA, Weigel D. Integration of floral inductive signals in Arabidopsis. Nature. 2000;404:889–92. doi: 10.1038/35009125. [DOI] [PubMed] [Google Scholar]
- 60.Yu H, Ito T, Zhao Y, Peng J, Kumar P, Meyerowitz EM. Floral homeotic genes are targets of gibberellin signaling in flower development. Proc Natl Acad Sci U S A. 2004;101:7827–32. doi: 10.1073/pnas.0402377101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rijpkema AS, Royaert S, Zethof J, van der Weerden G, Gerats T, Vandenbussche M. Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell. 2006;18:1819–32. doi: 10.1105/tpc.106.042937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lonnig WE, Saedler H, Sommer H. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and auto regulation of its persistent expression throughout flower development. EMBO J. 1992;11:251–63. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tilly JJ, Allen DW, Jack T. The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development. 1998;125:1647–57. doi: 10.1242/dev.125.9.1647. [DOI] [PubMed] [Google Scholar]
- 64.Kanno A, Saeki H, Kameya T, Saedler H, Theißen G. Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana) Plant Mol Biol. 2003;52:831–41. doi: 10.1023/A:1025070827979. [DOI] [PubMed] [Google Scholar]
- 65.Hofer KA, Ruonala R, Albert VA. The double-corolla phenotype in the Hawaiian lobelioid genus Clermontia involves ectopic expression of PISTILLATA B-function MADS box gene homologs. EvoDevo. 2012;3:26. doi: 10.1186/2041-9139-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bartlett M, Thompson B, Brabazon H, Del Gizzi R, Zhang T, Whipple C. Evolutionary dynamics of floral homeotic transcription factor protein-protein interactions. Mol Biol Evol. 2016;33:1486–501. doi: 10.1093/molbev/msw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–△△CT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Javelle M, Marco CF, Timmermans M. In situ hybridization for the precise localization of transcripts in plants. J Vis Exp. 2011;57:3328. doi: 10.3791/3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gong PC, Zhao M, He CY. Slow co-evolution of the MAGO and Y14 protein families is required for the maintenance of their obligate heterodimerization mode. PLoS One. 2014;1:e84842. doi: 10.1371/journal.pone.0084842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant supporting data can be found within the additional files accompanying this article. Sequence data described in this article can be found in GenBank (http://www.ncbi.nlm.nih.gov) under the accessions of KU318322 (VpTM6-1), KU318323 (VpTM6-2), and KU318324 (VpPI).


