ABSTRACT
Escherichia coli regulates its metabolism to adapt to changes in the environment, in particular to stressful downshifts in nutrient quality. Such shifts elicit the so-called stringent response, coordinated by the alarmone guanosine tetra- and pentaphosphate [(p)ppGpp]. On sudden amino acid (aa) starvation, RelA [(p)ppGpp synthetase I] activity is stimulated by binding of uncharged tRNAs to a vacant ribosomal site; the (p)ppGpp level increases dramatically and peaks within the time scale of a few minutes. The decrease of the (p)ppGpp level after the peak is mediated by the decreased production of mRNA by (p)ppGpp-associated transcriptional regulation, which reduces the vacant ribosomal A site and thus constitutes negative feedback to the RelA-dependent (p)ppGpp synthesis. Here we showed that on sudden isoleucine starvation, this peak was higher in an E. coli strain that lacks the 10 known mRNase-encoding toxin-antitoxin (TA) modules present in the wild-type (wt) strain. This observation suggested that toxins are part of the negative-feedback mechanism to control the (p)ppGpp level during the early stringent response. We built a ribosome trafficking model to evaluate the fold increase in RelA activity just after the onset of aa starvation. Combining this with a feedback model between the (p)ppGpp level and the mRNA level, we obtained reasonable fits to the experimental data for both strains. The analysis revealed that toxins are activated rapidly, within a minute after the onset of starvation, reducing the mRNA half-life by ∼30%.
IMPORTANCE The early stringent response elicited by amino acid starvation is controlled by a sharp increase of the cellular (p)ppGpp level. Toxin-antitoxin module-encoded mRNases are activated by (p)ppGpp through enhanced degradation of antitoxins. The present work shows that this activation happens over a very short time scale and that the activated mRNases negatively affect the (p)ppGpp level. The proposed mathematical model of (p)ppGpp regulation through the mRNA level highlights the importance of several feedback loops in early (p)ppGpp regulation.
INTRODUCTION
Upon sudden nutritional stress, bacteria cope with the adverse conditions by rapidly producing guanosine tetra- and pentaphosphate [(p)ppGpp], the master regulators of the stringent response (1–4). The “alarmone” (p)ppGpp coordinates multiple physiological processes, such as reduction and adjustment of transcription and activation of biosynthetic pathways (5–9). One of the central topics in stringent response research is to understand how (p)ppGpp triggers these processes at an appropriate time scale.
Numerous measurements revealed that the early (p)ppGpp profile exhibits a sharp peak within a few minutes after the onset of amino acid (aa) starvation (10–18) (e.g., see Fig. 1a). Figure 1b shows the main feedbacks in (p)ppGpp regulation that are relevant to the early stringent response. (p)ppGpp is synthesized by the synthetase RelA and degraded by the hydrolase SpoT. In Escherichia coli, SpoT also has a weak synthetase activity, but RelA is the major producer of (p)ppGpp during aa starvation (7). RelA activity is significantly increased in the presence of uncharged tRNAs and ribosomes (1). The presence of an uncharged tRNA loaded at the ribosomal A site probably allows RelA to rapidly sense aa starvation. In addition, there are positive-feedback mechanisms acting on the level of (p)ppGpp. The hydrolase activity of SpoT may be inhibited by high concentrations of (p)ppGpp (19). This inhibition provides a delay in the normal degradation process and facilitates the accumulation of (p)ppGpp. Furthermore, (p)ppGpp has been shown to increase the activity of RelA in vitro (20). These two factors have positive-feedback effects on the (p)ppGpp concentration (Fig. 1b, gray arrow).
FIG 1.
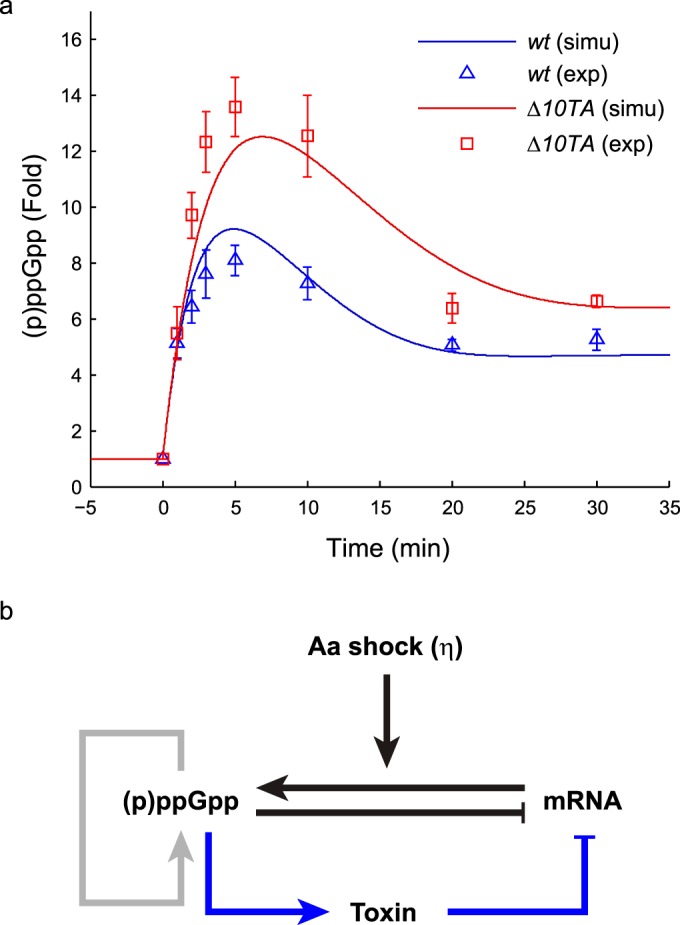
Early stringent response to isoleucine starvation. (a) (p)ppGpp responses of the wild-type MG1655 strain (wt) and a strain with all 10 known toxin-antitoxin loci deleted (Δ10TA) to valine-induced isoleucine starvation at time zero. The levels of (p)ppGpp were normalized to the prestarved level for each strain. The triangles and squares represent average fold increases for three independent measurements, and the error bars represent standard errors. The simulated levels of (p)ppGpp from the model of the early stringent response were plotted as blue and red curves. (b) Schematic illustration of the regulatory network. (p)ppGpp synthesis is triggered by sudden aa starvation imposed on rapidly growing cells. The level of (p)ppGpp is self-amplified by RelA and SpoT, and (p)ppGpp induces transcriptional inhibition. (p)ppGpp also activates Lon protease to release toxins from the toxin-antitoxin complexes to deplete mRNA.
The decrease of the (p)ppGpp level after a few minutes is conjectured to be caused by the reduced mRNA level, which is known to show a rapid change upon starvation (21, 22). This leads to less RelA synthetase activity through reducing the number of empty A sites, forming a negative-feedback mechanism (Fig. 1b, black arrows). The decrease of mRNAs may be caused by the (p)ppGpp-mediated reduction of transcription (21–23). On longer time scales, effects such as modulation of ribosome copy number (24, 25) and the induction of amino acid synthesis (26) will also affect the (p)ppGpp level, but here we focus on a short time scale.
relBE, one of the 10 known type II toxin-antitoxin (TA) modules encoding mRNases (also called mRNA interferases) in E. coli, is suggested to be activated upon starvation (27). The toxin RelE cleaves mRNAs at the ribosomal A site (28). In nonstressed, rapidly growing cells, RelE activity is neutralized by the antitoxin RelB, which forms a tight complex with RelE (29, 30). During aa starvation, the protein synthesis rate is about 5% of that under nonstarved conditions for the relBE+ strain, while in the relBE mutant strain it is about 10% (27). This indicates that RelE is released to act as a part of the stress response to rapid shutdown of translation. The release is thought to be mediated by (p)ppGpp, as Lon protease, which degrades the antitoxin RelB, is known to exhibit increased activity under nutritional stress (25, 27, 31). Under such conditions, activated Lon is also suggested to degrade the other antitoxins (32, 33). Therefore, it is likely that all 10 corresponding toxins are activated to some degree during aa starvation.
These observations led us to the hypothesis that TA modules may work not only as a downstream component of the stringent response but also as a component to provide negative feedback to the (p)ppGpp level, since activated toxins help to reduce mRNAs that are required to activate RelA (Fig. 1b, blue arrows). Here we experimentally measured the dynamics of the (p)ppGpp level in the wild-type (wt) strain and a strain lacking the 10 TA loci (Δ10TA) (32) under aa starvation conditions to investigate the extent of the feedback as well as changes in the kinetics of the stringent response. We then used quantitative modeling to analyze the observations in a two-step procedure. In the first step, we evaluated the initial starvation signal by analyzing a stochastic model of ribosome translation dynamics, with explicit consideration of binding of uncharged tRNAs to empty ribosomal A sites. In the second step, we used the resulting estimate as an input to a simple feedback model of (p)ppGpp control to study how this initial signal and the regulatory feedbacks mediated the changes in (p)ppGpp and mRNA levels. Fitting the feedback models to the experimental data showed that the feedback shown in Fig. 1b is indeed enough to reproduce the experimental data and revealed that the toxins are likely to be activated immediately after the onset of starvation, within 1 min.
MATERIALS AND METHODS
In vivo (p)ppGpp measurement.
To determine the (p)ppGpp contents of the wt (MG1655) and Δ10TA strains upon valine-induced isoleucine starvation, we modified a previously described method (33). In brief, overnight cultures were diluted 100-fold in 10 ml of MOPS (morpholinepropanesulfonic acid)-glucose minimal medium supplemented with all nucleobases (10 μg ml−1 [each]), as previously described (34), and incubated at 37°C with shaking. The MG1655 and Δ10TA strains were previously reported to have identical growth rates in LB medium (32), and the doubling time in MOPS-glucose minimal medium was just over an hour (∼62 min) for both strains. At an optical density at 600 nm (OD600) of 0.5, cells were diluted 10-fold to an OD600 of 0.05 and were left to grow with shaking at 37°C with H332PO4 (100 µCi ml−1). After 2 to 3 generations (OD600 of 0.2 to 0.3), amino acid starvation was induced by the addition of valine (0.5 mg ml−1). One-hundred-microliter samples were withdrawn before and 1, 2, 3, 5, 10, 20, and 30 min after addition of valine. The reactions were stopped by the addition of 20 μl of ice-cold 2 M formic acid. A 10-μl aliquot of each reaction mixture was loaded on polyethyleneimine (PEI) cellulose thin-layer chromatography (TLC) plates (purchased from GE Healthcare) and separated by chromatography in 1.5 M potassium phosphate at pH 3.4. The TLC plates were revealed by phosphorimaging (GE Healthcare) and analyzed using ImageQuant software (GE Healthcare). The increase in the level of (p)ppGpp was normalized to the basal level (time zero) for each strain.
Ribosome trafficking model. (i) Model description.
We modeled the ribosome traffic upon starvation by extending previous models of translation processes during exponential growth (35–37). A summary of the model is presented in Results. In the model, we considered one mRNA chain modeled as one-dimensional lattice sites, where each site represents one codon. The total length was chosen to be 300 sites, based on the average protein length (38). In every simulation, we randomly generated the mRNA sequence as follows: the first codon was fixed as the start codon, and the remaining codons were randomly generated with probabilities proportional to the usage of the amino acids in E. coli (the usage is available in reference 39). We considered a pool of 15 ribosomes available for translation per mRNA (see Table S1 in the supplemental material). We assumed that each ribosome occupied 11 codons and translated the codon in the middle (35). We did not consider degradation of mRNA, therefore focusing on the steady state of the ribosome traffic.
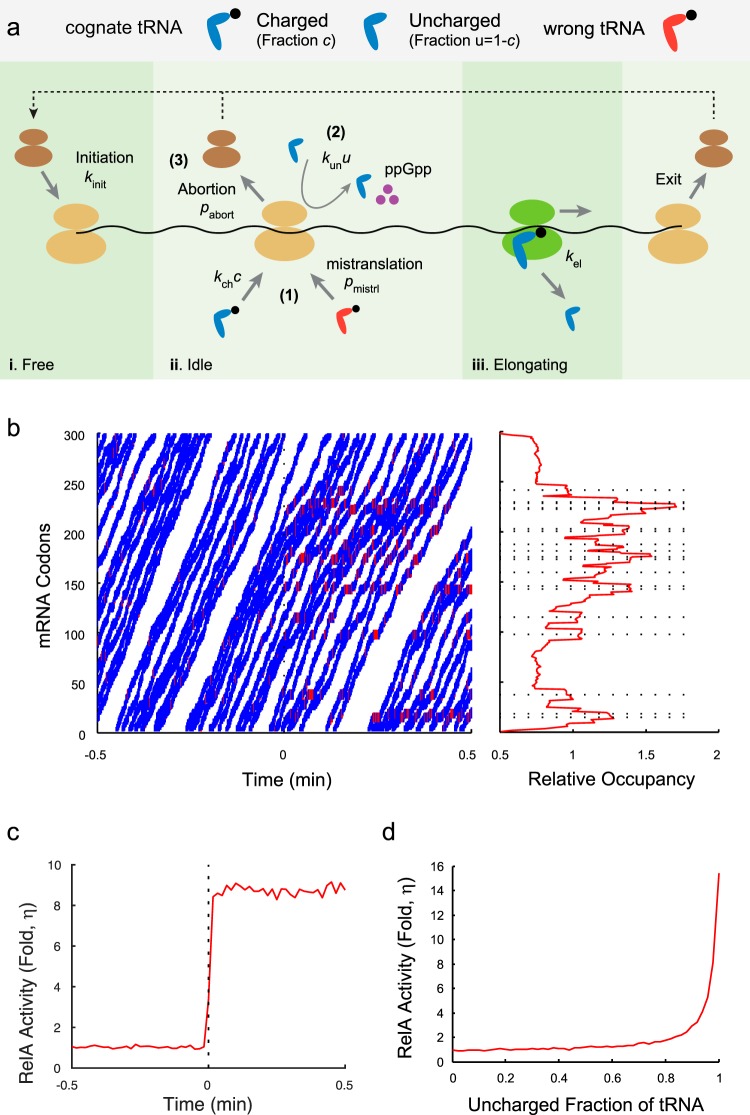
The reactions in the ribosome trafficking model are illustrated in Fig. 2a. We considered that a ribosome takes one of the following three states: (i) the “free” state, where a ribosome is not on the mRNA chain (brown in Fig. 2a); (ii) the “idle” state, where a ribosome is on the mRNA, but the A site is empty (yellow in Fig. 2a); and (iii) the “elongating” state, where a charged tRNA is bound to the ribosomal A site and the ribosome is ready for translocation (green in Fig. 2a). The possible reaction rates are assigned as follows.
FIG 2.
Ribosome trafficking model. (a) Schematic description of the ribosome trafficking model. Free ribosomes may initiate translation at the rate kinit if the start codons of mRNAs are not occupied. Every translating ribosome may bind a charged cognate tRNA at the rate kchc, where c is the charging level of the tRNA, and elongate one codon at the rate kel (1); bind an uncharged cognate tRNA, eject, and produce (p)ppGpp (increase the RelA activity by 1 unit) at the rate kunu, where u = 1 − c gives the uncharged level (2); or abort translation at the rate pabort (3). We also allowed for noncognate tRNA binding to ribosomal A sites at the small rate pmistrl to represent mistranslation. Elongation occurs only if the codon in front is free. Translation is terminated when a ribosome finishes translating all the codons and exits the mRNA. A detailed description of the algorithm is available in Materials and Methods. (b) Ribosome occupancy on mRNA. (Left) Spatiotemporal plot of ribosome traffic on a randomly generated mRNA upon isoleucine starvation at time zero. The lines represent the movement of ribosomes. The blue lines represent normal translation, and the red lines represent (p)ppGpp synthesis. (Right) Relative mean occupancies of mRNA codons by ribosomes in the poststarved steady state. The dashed lines represent the locations of isoleucine codons. (c) Relative RelA activities (η) in the pre- and poststarved states. The averages for 100 independent simulations were plotted as a red curve. The dashed line represents the induction time of starvation. (d) The level of isoleucine starvation was modulated by sampling the uncharged level of tRNAIle (uIle) between 0 and 1. The relative RelA activities (η) in the poststarved steady state were plotted. The figure confirms the previous claim that the stringent response is significant when more than 85% of tRNAs are uncharged (53).
(a) Ribosome in the “free” state (first column in Fig. 2a).
If there are no ribosomes occluding the initiation (i.e., the centers of all translating ribosomes are located at codon 12 or later), a free ribosome may initiate translation by binding to the start codon at the rate kinit and may change its state to “idle.”
(b) Ribosome in the “idle” state (second column in Fig. 2a).
An idle ribosome at the codon encoding amino acid i may experience one of the following reactions:
 |
Reaction 1 represents binding of a charged tRNA to the ribosomal A site, which changes the ribosomal state to “elongating.” A charged cognate tRNA binds at the rate kchci, where ci is the charging level of tRNA for the amino acid i. A noncognate charged tRNA can also bind at the low rate pmistrl, representing the mistranslation process. We used the equation pmistrl = p0Σj≠iξjcj, where ξj is the usage of amino acid j and cj is the charging level for the corresponding tRNA.
Reaction 2 represents the binding of an uncharged cognate tRNA at the rate kun(1 − ci), which triggers the production of (p)ppGpp by 1 unit (in arbitrary units, as we consider only the fold change in levels before and after starvation). We assume that uncharged tRNA is immediately ejected from the A site, with the ribosome remaining in the idle state.
Reaction 3 represents abortion of binding at the rate pabort, where the ribosome dissociates from mRNA and joins the free pool.
(c) Ribosome in the “elongating” state (third column in Fig. 2a).
If the codon in front is not occupied, an elongating ribosome may translocate by one codon, eject the now uncharged tRNA, and change its state to “idle,” with the rate kel. Otherwise, the ribosome will not translocate and will keeps its status.
Finally, when a ribosome finishes translating the last codon, it exits the mRNA chain and changes its state to “free” (fourth column in Fig. 2a).
We implemented the model with the Gillespie algorithm (40), and the parameter values used in the simulation are listed in Table S1 in the supplemental material.
(ii) Data acquisition.
The RelA activity was calculated as the number of events where an uncharged tRNA bound to the ribosomal A site in a time window. The number of translating ribosomes was calculated as the number of ribosomes (idle and elongating) on the mRNA chain. The mistranslation fraction was computed by dividing the number of mistranslations by the number of translocations. The trajectories and relative ribosome occupancies in Fig. 2b and in Fig. S2 in the supplemental material were based on a single simulation. The values for other figures were the averages for 100 independent simulations with randomly generated mRNA. We smoothed the data with a window size of 0.05 s for the time series plots. The relative RelA activity (η) was computed by dividing the average RelA activity during the last second of simulation by the prestarved steady-state activity.
For trafficking models without abortion and mistranslation (see Fig. S2 in the supplemental material), we set pabort and pmistrl to zero. For trafficking models for general amino acid starvation (see Fig. S6), we artificially set the usage of one amino acid to be the fraction of starvation and set the charging level for this amino acid to 0.02 after starvation.
Parameter fitting and error of fitting.
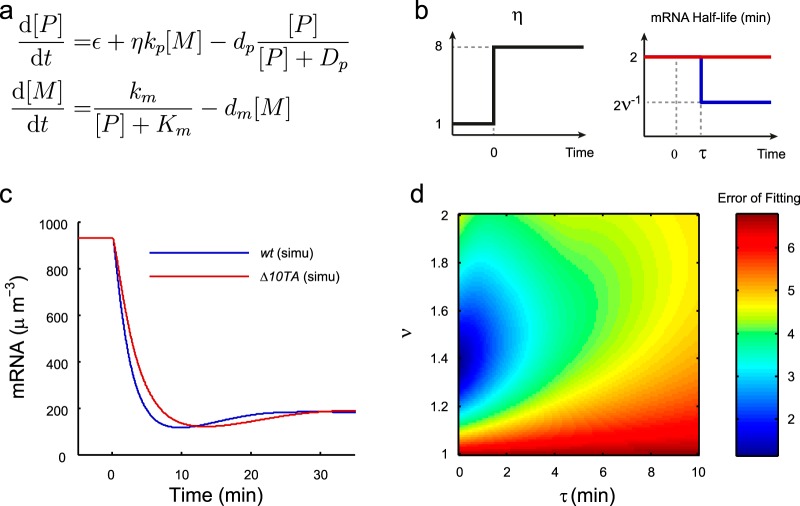
We used a deterministic model of (p)ppGpp-mediated feedback (Fig. 3a) to reproduce the experimental data. The parameters were determined by fitting to the experimental data as follows.
FIG 3.
Model of the early stringent response. (a) Formulation of the model. (b) (Left) Illustration of the relative RelA activity (η) upon isoleucine starvation at time zero. (Right) Illustration of the average mRNA half-life (ln2/dm) upon isoleucine starvation at time zero for both the wt strain (blue) and the Δ10TA strain (red). (c) Simulated concentrations of mRNA for the wt and Δ10TA strains (see Fig. 1a). (d) Effects of the time scale (τ) and strength (ν) of mRNA cleavage activity by toxins on the error of fitting to the wt measurements.
The error of fitting for one strain was defined as follows:
where mexp is the average and σexp is the standard error of the mean for experimentally measured (p)ppGpp levels and msimu is the simulated number of (p)ppGpp molecules. The error of fit was computed by summing over all measured time points except for time zero. In Fig. 1a and in Fig. S4b in the supplemental material, the overall error was defined as the L2 norm of the errors for the two strains. In Fig. 3d and in Fig. S4c, the error was the one for fitting to the wt strain. In Fig. S5c, the error was the one for the Δ10TA strain. The MatLab (Mathworks) function fmincon was used to obtain the parameter values by minimizing the error of fit, and 100 independent minimizations with random initial guesses were carried out to search for globally optimal values.
Global mRNA half-lives.
The measurement of mRNA half-lives was done essentially as described previously (41). Briefly, cells were grown in M9 minimal medium supplemented with 0.2% glucose at 37°C to an OD450 of 0.5. The culture was diluted to an OD450 of 0.1 when the first sample of 200 μl was taken and placed on ice with 10% SDS and 20% trichloroacetic acid (TCA). Immediately after, 1 mg ml−1 rifampin, 50 μg/ml nalidixic acid, and 100 mCi mmol−1 [3H]uridine were added to the culture, and sampling was continued every 30 s for 20 min. Samples were boiled at 95°C for 5 min, applied to nitrate cellulose filters (Whatman), and washed with 10% cold TCA. The filters were transferred to vials, and the incorporated radioactivity was counted in a liquid scintillation counter. When half-lives were measured during aa starvation, the cells were starved with 0.4 mg ml−1 serine hydroxamate (SHX) for 1 h before sampling.
RESULTS
Early stringent response to isoleucine starvation.
“Hyperactivation” of the toxin RelE in the cell was previously shown to be enough to impair the activation of RelA (42). This was suggested to be due to the ribosome-dependent cleavage of mRNA by RelE resulting in inhibition of translation, thus inhibiting RelA from sensing the charging state of the tRNAs (42). More recently, high levels of (p)ppGpp have been suggested to activate Lon protease to degrade all type II antitoxins of E. coli (31). Based on these reports, we wanted to measure the (p)ppGpp profile during the early stringent response in the wt strain (MG1655) and an isogenic strain with all 10 toxin-antitoxin loci deleted (Δ10TA) to quantify the strength of the toxin-mediated negative-feedback loop (Fig. 1a). We induced isoleucine starvation by supplementing the medium with excessive valine (43) at time zero. We quantified the levels of (p)ppGpp 1, 2, 3, 5, 10, 20, and 30 min after the addition, and we normalized the data to the prestarvation level of (p)ppGpp for each strain (see Materials and Methods; see Fig. S1 in the supplemental material). Agreeing with previous reports (10, 12), the (p)ppGpp profiles of both strains exhibited the typical shape of (p)ppGpp accumulation (Fig. 1a). The (p)ppGpp level in the Δ10TA strain reached the maximal value of around a 13-fold increase in 5 min, while the level in the wt strain reached the maximal value of 8-fold within that time frame. The visible difference in 5-min samples suggests that toxins are released rapidly after the starvation signal. Interestingly, the same measurement for the ΔrelBE strain showed a (p)ppGpp profile similar to that of the wt strain, suggesting that this early time difference is not determined solely by the relBE locus (see Fig. S8).
Quantitative analysis by mathematical modeling.
We aimed to quantitatively understand the obtained (p)ppGpp profiles (Fig. 1a). The first signal of aa starvation is the activation of RelA. This is promoted by the binding of uncharged tRNAs to the ribosomal A sites, which results in (p)ppGpp production per mRNA. This leads the above-mentioned feedback loops to take effect and alter the (p)ppGpp levels further. For a quantitative analysis, we needed to separate the initial direct signal of the aa starvation from the effect of the feedbacks. We therefore quantified the early stringent response in two steps. In the first step, we analyzed a ribosome trafficking model (Fig. 2a) (see Materials and Methods for details) where the binding of charged and uncharged tRNAs to the translating ribosomes was explicitly considered. From the model, we evaluated the initial direct signal of aa starvation (η), defined as the fold change in the (p)ppGpp production rate per mRNA just after aa starvation. No regulatory feedbacks or effects of toxins were considered at this step. In the next step, we used this η value as an input to the (p)ppGpp regulatory feedback model in which we would study the effects of feedbacks.
Ribosome trafficking model to quantify the starvation signal.
The ribosome trafficking model simulated the movements of multiple ribosomes on a typical mRNA of 300 codons (38). We modeled the charging levels for the tRNA cognate for each amino acid explicitly, and the codons of the mRNA chain were randomly chosen, with probabilities proportional to the usage of their cognate amino acids (with the usage of the isoleucine codons being around 6% [39]). We excluded mRNA degradation and cleavage because these processes took place on a longer time scale (see below). Consequently, we cannot distinguish between the wt and Δ10TA strains here. Each ribosome stochastically initiates translation at the rate kinit when there is no occlusion by the other ribosomes. The kinetics of the translating ribosome was modeled with several possible reaction steps to address starvation-induced (p)ppGpp production. When translating a codon, a ribosome can be bound by a charged cognate tRNA with the rate kchc, with c being the charging level of the corresponding tRNA, or it can be bound by an uncharged cognate tRNA with the rate kunu, with u = 1 − c denoting the fraction of the uncharged cognate tRNA. If an uncharged tRNA is bound, (p)ppGpp molecules are produced and the tRNA is ejected, while if a charged tRNA is bound, the ribosome moves to the next codon at the rate kel as long as the codon in front is not occupied by another ribosome. When a ribosome reaches the end of the mRNA, it is released to join the free ribosomes and ready to initiate another round of translation. We also accounted for mistranslation (44) and the abortion of the translating ribosome mediated by transfer-messenger RNA (tmRNA) (45), but with realistic parameters they did not have a significant effect on the result (see below). Parameters were chosen to fit previously published data (see Table S1 in the supplemental material).
Figure 2b (left panel) presents a typical spatiotemporal plot of ribosome traffic with isoleucine starvation at time zero. The charging levels (c) of all tRNAs in the prestarved state were set to be 0.8 (23, 46), and we reduced the charging level of tRNAIle (cIle) to 0.02 (see Table S1 in the supplemental material) from time zero and kept the levels of other tRNAs unchanged to simulate isoleucine starvation. The immediate reduction in the charging level can be justified by the rapid turnover of tRNA (47). We saw that ribosomes elongated rapidly along the mRNA chain in the prestarved state (trajectories shown in blue), with few events of (p)ppGpp synthesis (shown in red). Upon starvation, (p)ppGpp synthesis events occurred more frequently. The average translation elongation time for ribosomes, however, decreased 20% due to frequent stalling, a number similar to the previously reported 15% reduction (21). To visualize the ribosome movement further, we plotted the relative occupancy of each mRNA codon by ribosomes in the poststarved state (Fig. 2b, right panel). Ribosomes were shown to occupy isoleucine codons (dashed lines) more often than the unstarved ones did, and ribosome jamming was visualized by the stair-like occupancy curve upstream of the starved codons. No visible difference was observed between the occupancies at unstarved codons close to the 5′ end and those close to the 3′ end, meaning that translation abortion was rare.
To quantify the starvation signal, we computed the average statistics for 100 independent simulations. The relative RelA activity (η), defined as the ratio of the frequencies of (p)ppGpp synthesis events in the poststarved state and the prestarved one, increased about 8-fold almost immediately upon starvation (Fig. 2c). This fast convergence pinpointed that depletion of the amino acid pool can rapidly and accurately be sensed through the binding of uncharged tRNAs and suggested that the initial increase of (p)ppGpp synthesis upon aa starvation can be decoupled from slower biological processes, including the mRNA cleavage and regulatory feedbacks (Fig. 1b).
We fitted the parameters so that the fraction of mistranslated isoleucine codons in the prestarved state was 0.5% (44). We found that mistranslation did not have a significant impact on the relative RelA activity, as a trafficking model without this process generated a 10-fold increase in RelA activity after the onset of starvation (see Fig. S2 and S3d in the supplemental material).
The fold change of (p)ppGpp synthesis upon starvation (η) was sensitive to the charging level of isoleucyl-tRNA at starvation, as a cIle value of 0.04 gave a 5-fold increase and a cIle value of 0 gave a 15-fold increase (Fig. 2d). We adopted a cIle value of 0.02 as a typical value, based on measurements showing the charging level of tRNALeu being between 1% and 3% upon leucine starvation (23) and on the assumption of a similar value for isoleucine starvation due to the similarity in the amino acid structures. This gave an η value of 8, and we used this value as an immediate input to the control of (p)ppGpp on aa starvation as we analyzed a model of (p)ppGpp regulatory feedbacks (see the next section).
A feedback model of the early stringent response suggests a rapid activation of toxins.
In the second step of our quantitative analysis, we looked at how the initial aa starvation signal and the regulatory feedbacks mediated the early dynamics of (p)ppGpp levels by constructing a simple feedback model. The model equations were formulated as shown in Fig. 3a, where we considered the concentrations of (p)ppGpp ([P]) and mRNA ([M]). We assumed that the toxins were not active in the wt before aa starvation and therefore assumed no difference in model parameters before the starvation. We modulated the mRNA degradation rate in the wt strain after aa starvation (Fig. 3b, right panel, and see below), as mRNAs should have shorter half-lives in the wt strain with the toxins' mRNase activity.
The synthesis rate of (p)ppGpp was formulated in two terms: the first term accounted for basal production, including SpoT-mediated synthesis, and the second one accounted for the RelA-mediated production proportional to the mRNA level. The starvation dependence of this term was represented in the parameter η. η is equal to 1 before starvation and increases to another constant at starvation to represent the fold change of the (p)ppGpp synthesis rate per mRNA after starvation. For isoleucine starvation, we chose the poststarvation value of η to be 8, based on the presented ribosome trafficking model analysis (Fig. 3b, left panel).
This value of η suggests that the starvation signal alone was not enough to induce a 13-fold increase in the (p)ppGpp profile for the Δ10TA strain (Fig. 1a). We therefore needed to include some positive feedbacks on the (p)ppGpp concentration to get the peak high enough.
The degradation of (p)ppGpp was modeled with saturation in the Michaelis-Menten form to capture the starvation-dependent hydrolysis activity (19). It turned out that this saturated degradation could provide enough positive feedback to accumulate (p)ppGpp more than the fold increase of η to fit the data, so for simplicity, we focused on this result in the main text. A version of the model with additional positive feedback for (p)ppGpp synthesis (20) is presented in Fig. S4 in the supplemental material, which also gave a reasonable fit.
Note that we also analyzed a model without any positive feedback, but with a high value for η, because it is possible to produce a strong starvation signal by modulating tRNA charging levels (Fig. 2d) at starvation. However, a good fit to the experimental curves required an unrealistically long mRNA half-life (see Fig. S5 in the supplemental material). Thus, we concluded that the model with a lower η value and with some positive feedbacks is more realistic.
For the mRNA level, we modeled the transcription rate with [P]-dependent repression of the Michaelis-Menten form. We assumed a constant degradation of mRNA, with a half-life of 2 min (dm = ln2/mRNA half-life) (48, 49). For the wt strain, we considered the additional toxin-mediated mRNA cleavage by modulating the half-lives of mRNA. By changing the half-lives of mRNA from 2 min to 2ν−1 min at time τ min after starvation (Fig. 3b, right panel), we accounted for both the time scale of toxin activation (τ) and the strength of the mRNA cleavage activity (ν).
We obtained the parameter values from the literature and by fitting to the experimental curves (see Materials and Methods and see Table S2 in the supplemental material). We obtained reasonable fits to the data for both strains (Fig. 1a). Our model illustrated that saturated degradation of the (p)ppGpp level can provide a delay to the peak to fit the 5-min time scale of the (p)ppGpp peak. Without the saturated degradation, even with a sufficiently strong starvation signal, a good fit required a long mRNA half-life during the rise of the peak but a short one during the fall of the peak even for the Δ10TA strain, a phenomenon that is unlikely to occur (data not shown).
The best fit to the wt strain gave the estimate that the toxins were released shortly (τ of ∼10 s) after isoleucine starvation, and the mRNA half-lives decreased by around 30% (ν ≈ 1.4). This gave a faster drop in the mRNA level in the wt strain than in the Δ10TA strain (Fig. 3c), reducing the (p)ppGpp peak height (Fig. 1a). To verify this further, we sampled the values of τ and ν over a wide range and computed the distance between the simulated curves and the experimental (p)ppGpp profile (or the error of fit) (see Materials and Methods). Figure 3d shows that a good fit required the effective half-life of mRNA to be reduced 20% to 40% and for toxins to be released within 1 min after starvation. Therefore, our analysis suggests that a rapid and mild mRNA cleavage activity is induced from the toxin-antitoxin complexes in the cytoplasm.
Our model could generate predictions consistent with experimental measurements. For example, the model predicted that the wt strain would show a 4-fold reduction in the transcription rate {computed by the term km/(Km + [P])} within 10 min after the onset of isoleucine starvation (Fig. 3). Earlier literature reported a similarly rapid transcription inhibition (21, 22, 50). O'Farrell (50) also reported that the transcription rate of the wt strain dropped to 30% to 35% of the unstarved level under isoleucine starvation conditions, and this number was consistent with our model's prediction. Furthermore, if we assumed that the translation rate (aa incorporation rate) could be estimated by the product of the mRNA level and the translation elongation rate, we could predict translation rates from our model. We showed earlier that the ribosome trafficking model predicted that the translation elongation rate dropped 20% after aa starvation. Combined with the mRNA levels, we predicted that the translation rate dropped to 15% of the prestarved level. This number is in accordance with the previously reported values of 10% to 13% (50).
DISCUSSION
In this study, we combined experimental measurements and modeling to study the early dynamics of (p)ppGpp levels after isoleucine starvation. Our models demonstrate that the feedbacks through the mRNA level are enough to reproduce the (p)ppGpp peak. We also showed that the TA systems not only modulate physiological activities in the poststarved steady state (27) but also serve as a negative feedback on (p)ppGpp levels during the early stringent response. Our analysis predicts that activation of toxins occurs rapidly (within a minute) after the onset of starvation and, on average, induces an ∼30% reduction of the mRNA half-life.
Although our modeling framework was developed for valine-induced isoleucine starvation, it can be applied to a wide range of experimental setups by explicitly setting the charging levels of tRNAs and the fraction of starved codons. The ribosome trafficking model predicts a sublinear increase of relative RelA activity (η) on the fraction of starved amino acids compared to total amino acid usage (e.g., 2% for histidine starvation and 16% for isoleucine and leucine starvation) (see Fig. S6a in the supplemental material). The early (p)ppGpp response curve in different situations can be obtained by applying the obtained fold increase of RelA activity (η) to the proposed model, which naturally predicts increasing (p)ppGpp peak heights with starvation of more frequently used amino acids (Fig. 4). Following the procedure described in Results, one may also utilize our modeling framework to estimate the mRNA levels and translation rates shortly after aa starvation. For example, our model predicts a 3-fold reduction in the mRNA level and a 70% drop in the translation rate within 10 min after the onset of histidine starvation, while the numbers are 10-fold and 95% for isoleucine/leucine starvation (see Fig. S6d and e).
FIG 4.
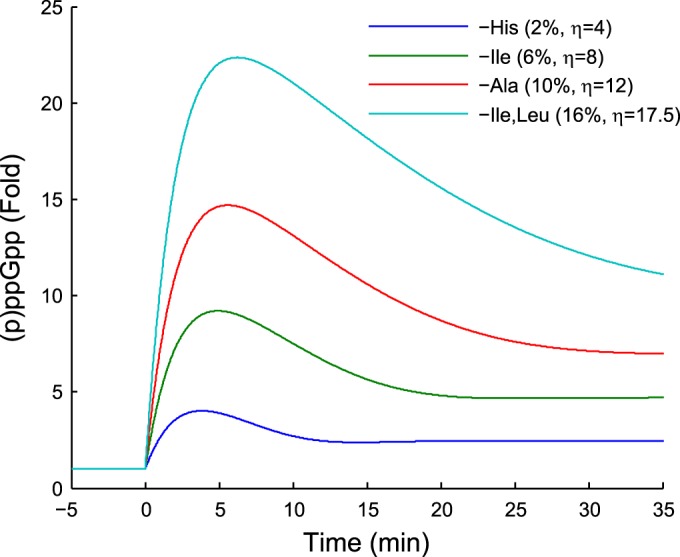
Predicted ppGpp levels upon general amino acid starvation. The charging level of the starved codons was set to be 0.02 during starvation. The percentages in the legend represent the usage of the starved amino acids, and the values for η indicate the strengths of the corresponding starvation signals. The prediction assumes that the toxins are released immediately upon starvation and that the reduction of mRNA half-lives by toxins is not modulated by the number of starved codons.
Our findings on the rapid activation of the toxins by (p)ppGpp support the idea that the TA systems may serve as an immediate protection mechanism against stress. Meanwhile, it is probably uneconomical for cells to enter a complete shutdown of translation, regardless of the extent of adversity. This suggests that rather weak mRNA cleavage activity from the toxins probably facilitates the entry of cells into a temporary state (for aa starvation, the “hunger state,” as suggested previously [18]) to evaluate the extent of austerity, ensuring that the cell may recover without a high cost in metabolism if the stress is modest or transient.
Since we focused only on the early behavior of aa starvation, our model might be inconsistent with experimental data on long-term behavior. Our model predicts that the Δ10TA strain has a lower transcription rate [due to higher (p)ppGpp levels] and longer mRNA half-lives (due to the absence of toxins) than those of the wt strain, resulting in the two strains having similar mRNA levels and translation rates (Fig. 3c). This prediction does not match a previous report where RelE alone was shown to reduce translation rates 2-fold in the long term (27). This inconsistency can be explained by long-term behaviors of the transcription rates and mRNA half-lives which were not considered in the current model. First, the model parameters related to transcription rates were fitted to the early stringent response, where the downregulation of growth-related genes is dominant (26, 51). Consequently, the model prediction primarily reflected the differences between the two strains in the expression of these genes, and the (p)ppGpp-dependent upregulation of stress response genes in the poststarved steady state was less covered. If we also account for the stress response genes, then the transcription rate may exhibit a smaller difference between the two strains than that seen with model prediction. Second, our model did not consider many factors modulating the global mRNA stability in the poststarved state. In reality, a change in mRNA species (26) and protection of mRNA from degradation by stalled ribosomes (41, 52) may increase the stability. To check this, we measured the average mRNA half-lives in the pre- and poststarved (60 min after aa starvation) states (see Materials and Methods). We found that mRNA half-lives in the Δ10TA strain indeed increased 2-fold, from 1.8 min before starvation to 4.4 min in the poststarved steady state. Meanwhile, the wt strain showed less stabilization (from 1.5 min to 2.4 min) (see Fig. S7 in the supplemental material). As a result, the mRNA decay rate in the wt strain was 2-fold higher than that in the Δ10TA strain in the poststarved state. In summary, the possibility that the two strains have similar transcription rates but a 2-fold difference in mRNA decay rates indicates a 2-fold difference in the cellular mRNA levels in the long term, explaining the translation rate differences in the two strains. It would be interesting to extend our present model to include these effects so that it also becomes applicable to long-term behavior after aa starvation.
Supplementary Material
ACKNOWLEDGMENT
We thank Steen Pedersen for fruitful discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00062-16.
REFERENCES
- 1.Haseltine WA, Block R. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci U S A 70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallant J, Palmer L, Pao CC. 1977. Anomalous synthesis of ppGpp in growing cells. Cell 11:181–185. doi: 10.1016/0092-8674(77)90329-4. [DOI] [PubMed] [Google Scholar]
- 3.Battesti A, Bouveret E. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol 62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 4.Vinella D, Albrecht C, Cashel M, D'Ari R. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol 56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- 5.Gallant JA. 1979. Stringent control in E. coli. Annu Rev Genet 13:393–415. doi: 10.1146/annurev.ge.13.120179.002141. [DOI] [PubMed] [Google Scholar]
- 6.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 7.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 8.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. 2013. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott M, Klumpp S, Mateescu EM, Hwa T. 2014. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol Syst Biol 10:747. doi: 10.15252/msb.20145379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryals J, Little R, Bremer H. 1982. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol 151:1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cashel M. 1969. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem 244:3133–3141. [PubMed] [Google Scholar]
- 12.Lagosky PA, Chang FN. 1980. Influence of amino acid starvation on guanosine 5′-diphosphate 3′-diphosphate basal-level synthesis in Escherichia coli. J Bacteriol 144:499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazzarini RA, Cashel M, Gallant J. 1971. On the regulation of guanosine tetraphosphate levels in stringent and relaxed strains of Escherichia coli. J Biol Chem 246:4381–4385. [PubMed] [Google Scholar]
- 14.Hughes J, Mellows G. 1978. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Escherichia coli by pseudomonic acid. Biochem J 176:305–318. doi: 10.1042/bj1760305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metzger S, Schreiber G, Aizenman E, Cashel M, Glaser GJ. 1989. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem 264:21146–21152. [PubMed] [Google Scholar]
- 16.Lund E, Kjeldgaard NO. 1972. Metabolism of guanosine tetraphosphate in Escherichia coli. Eur J Biochem 28:316–326. doi: 10.1111/j.1432-1033.1972.tb01916.x. [DOI] [PubMed] [Google Scholar]
- 17.Fiil NP, Willumsen BM, Friesen JD, von Meyenburg K. 1977. Interaction of alleles of the relA, relC and spoT genes in Escherichia coli: analysis of the interconversion of GTP, ppGpp and pppGpp. Mol Gen Genet 150:87–101. doi: 10.1007/BF02425329. [DOI] [PubMed] [Google Scholar]
- 18.Traxler MF, Zacharia VM, Marquardt S, Summers SM, Nguyen H-T, Stark SE, Conway T. 2011. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol Microbiol 79:830–845. doi: 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray DK, Bremer H. 1996. Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J Mol Biol 259:41–57. doi: 10.1006/jmbi.1996.0300. [DOI] [PubMed] [Google Scholar]
- 20.Shyp V, Tankov S, Ermakov A, Kudrin P, English BP, Ehrenberg M, Tenson T, Elf J, Hauryliuk V. 2012. Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep 13:835–839. doi: 10.1038/embor.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sørensen MA, Jensen KF, Pedersen S. 1994. High concentrations of ppGpp decrease the RNA chain growth rate: implications for protein synthesis and translational fidelity during amino acid starvation in Escherichia coli. J Mol Biol 236:441–454. doi: 10.1006/jmbi.1994.1156. [DOI] [PubMed] [Google Scholar]
- 22.Svitil AL, Cashel M, Zyskind JW. 1993. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem 268:2307–2311. [PubMed] [Google Scholar]
- 23.Sørensen MA. 2001. Charging levels of four tRNA species in Escherichia coli Rel+ and Rel− strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J Mol Biol 307:785–798. doi: 10.1006/jmbi.2001.4525. [DOI] [PubMed] [Google Scholar]
- 24.Bremer H, Dennis P. 2008. Feedback control of ribosome function in Escherichia coli. Biochimie 90:493–499. doi: 10.1016/j.biochi.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. 2001. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 26.Durfee T, Hansen A-M, Zhi H, Blattner FR, Jin DJ. 2008. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol 190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A 98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131–140. doi: 10.1016/S0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 29.Galvani C, Terry J, Ishiguro EE. 2001. Purification of the RelB and RelE proteins of Escherichia coli: RelE binds to RelB and to ribosomes. J Bacteriol 183:2700–2703. doi: 10.1128/JB.183.8.2700-2703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen K, Christensen SK, Gerdes K. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol 45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 31.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 32.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Germain E, Roghanian M, Gerdes K, Maisonneuve E. 2015. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci U S A 112:5171–5176. doi: 10.1073/pnas.1423536112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Cashel M. 1994. Detection of (p)ppGpp accumulation patterns in Escherichia coli mutants. Methods Mol Genet 3:341–356. [Google Scholar]
- 35.Mitarai N, Sneppen K, Pedersen S. 2008. Ribosome collisions and translation efficiency: optimization by codon usage and mRNA destabilization. J Mol Biol 382:236–245. doi: 10.1016/j.jmb.2008.06.068. [DOI] [PubMed] [Google Scholar]
- 36.Ciandrini L, Stansfield I, Romano MC. 2010. Role of the particle's stepping cycle in an asymmetric exclusion process: a model of mRNA translation. Phys Rev E Stat Nonlin Soft Matter Phys 81:051904. doi: 10.1103/PhysRevE.81.051904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitarai N, Pedersen S. 2013. Control of ribosome traffic by position-dependent choice of synonymous codons. Phys Biol 10:056011. doi: 10.1088/1478-3975/10/5/056011. [DOI] [PubMed] [Google Scholar]
- 38.Sundararaj S, Guo A, Habibi-Nazhad B, Rouani M, Stothard P, Ellison M, Wishart DS. 2004. The CyberCell Database (CCDB): a comprehensive, self-updating, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic Acids Res 32:D293–D295. doi: 10.1093/nar/gkh108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pramanik J, Keasling JD. 1997. Stoichiometric model of Escherichia coli metabolism: incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol Bioeng 56:398–421. doi:. [DOI] [PubMed] [Google Scholar]
- 40.Gillespie DT. 1977. Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81:2340–2361. doi: 10.1021/j100540a008. [DOI] [Google Scholar]
- 41.Pato ML, Bennett PM, von Meyenburg K. 1973. Messenger ribonucleic acid synthesis and degradation in Escherichia coli during inhibition of translation. J Bacteriol 116:710–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christensen SK, Gerdes K. 2004. Delayed-relaxed response explained by hyperactivation of RelE. Mol Microbiol 53:587–597. doi: 10.1111/j.1365-2958.2004.04127.x. [DOI] [PubMed] [Google Scholar]
- 43.Umbarger HE, Brown B. 1955. Isoleucine and valine metabolism in Escherichia coli. V. Antagonism between isoleucine and valine. J Bacteriol 70:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker J. 1989. Errors and alternatives in reading the universal genetic code. Microbiol Mol Biol Rev 53:273–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keiler KC, Waller PRH, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 46.Dittmar KA, Sørensen MA, Elf J, Ehrenberg M, Pan T. 2005. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep 6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gouy M, Grantham R. 1980. Polypeptide elongation and tRNA cycling in Escherichia coli: a dynamic approach. FEBS Lett 115:151–155. doi: 10.1016/0014-5793(80)81155-0. [DOI] [PubMed] [Google Scholar]
- 48.Pedersen S, Reeh S, Friesen JD. 1978. Functional mRNA half lives in E. coli. Mol Gen Genet 166:329–336. [DOI] [PubMed] [Google Scholar]
- 49.Pedersen M, Nissen S, Mitarai N, Svenningsen SL, Sneppen K, Pedersen S. 2011. The functional half-life of an mRNA depends on the ribosome spacing in an early coding region. J Mol Biol 407:35–44. doi: 10.1016/j.jmb.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 50.O'Farrell PH. 1978. The suppression of defective translation by ppGpp and its role in the stringent response. Cell 14:545–557. doi: 10.1016/0092-8674(78)90241-6. [DOI] [PubMed] [Google Scholar]
- 51.Bremer H, Dennis PP. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p 1553–1569. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 52.Reimers JM, Schmidt KH, Longacre A, Reschke DK, Wright BE. 2004. Increased transcription rates correlate with increased reversion rates in leuB and argH Escherichia coli auxotrophs. Microbiology 150:1457–1466. doi: 10.1099/mic.0.26954-0. [DOI] [PubMed] [Google Scholar]
- 53.Goldman E, Jakubowski H. 1990. Uncharged tRNA, protein synthesis, and the bacterial stringent response. Mol Microbiol 4:2035–2040. doi: 10.1111/j.1365-2958.1990.tb00563.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.