Summary
We established the role for epigenetics in regulation of NOTCH signaling in breast cancer. This may constitute a common mechanism of activation of oncogenic signals. Our study provides support for epigenetic-targeting strategies in anticancer approaches.
Abstract
DNA hypomethylation was previously implicated in cancer progression and metastasis. The purpose of this study was to examine whether stilbenoids, resveratrol and pterostilbene thought to exert anticancer effects, target genes with oncogenic function for de novo methylation and silencing, leading to inactivation of related signaling pathways. Following Illumina 450K, genome-wide DNA methylation analysis reveals that stilbenoids alter DNA methylation patterns in breast cancer cells. On average, 75% of differentially methylated genes have increased methylation, and these genes are enriched for oncogenic functions, including NOTCH signaling pathway. MAML2, a coactivator of NOTCH targets, is methylated at the enhancer region and transcriptionally silenced in response to stilbenoids, possibly explaining the downregulation of NOTCH target genes. The increased DNA methylation at MAML2 enhancer coincides with increased occupancy of repressive histone marks and decrease in activating marks. This condensed chromatin structure is associated with binding of DNMT3B and decreased occupancy of OCT1 transcription factor at MAML2 enhancer, suggesting a role of DNMT3B in increasing methylation of MAML2 after stilbenoid treatment. Our results deliver a novel insight into epigenetic regulation of oncogenic signals in cancer and provide support for epigenetic-targeting strategies as an effective anticancer approach.
Introduction
Breast cancer is the most common type of cancer in women and the second most commonly occurring cancer overall worldwide (1,2). Identification of new effective preventive and anticancer strategies is therefore critical. Only 5–10% of breast cancers are hereditary (3,4). The overwhelming majority of cases are sporadic, likely caused by external exposures including estrogens, alcohol use, physical inactivity, and poor diet (3,4). It is estimated that at least 30% of sporadic breast cancer cases are not linked to mutations but have been shown to contain epigenetic alterations, particularly in DNA methylation (5,6).
Epigenetics refers to alterations in gene expression without changes in the underlying DNA sequence and consists of three main components: DNA methylation, histone modifications, and noncoding RNA mechanisms. DNA methylation that occurs predominantly in CpG sequences is considered to be the gatekeeper of gene expression providing stable long-term regulation (7). Simultaneously, DNA methylation has attracted a significant amount of attention for the prevention and treatment of different illnesses with cancer at the forefront, mainly due to the inherent reversibility of epigenetic states (8,9). Hypermethylation of tumor suppressor genes linked to transcriptional silencing and recently reported promoter hypomethylation linked to activation of oncogenes and prometastatic genes have been shown to play a role in cancer initiation, progression and metastasis (8–13).
It was generally assumed that DNA hypomethylation in cancer occurs mainly in repetitive, CpG-sparse regions of the genome (14), in contrast to DNA hypermethylation that targets CpG-rich islands in promoters and first exons (15). However, recent numerous epigenome-wide association studies indicate that hypomethylation also targets promoter regions or enhancers of genes that are involved in functions essential for cancer progression and metastasis (10,13,14). Breast cancer has been associated not only with hypermethylation of tumor suppressor genes (5,6) but also with hypomethylation of oncogenes and pro-metastatic genes. For instance, re-methylation of hypomethylated promoter of urokinase-type plasminogen activator (uPA), a gene inducing metastatic cell behavior, was shown to block breast cancer growth and metastasis (16). Many of the hypomethylated genes in cancer have been shown to fall into oncogenic pathway categories (10). This would suggest that loci-specific DNA hypomethylation in cancer might be associated with activation of oncogenic signals. Interestingly, a number of signaling pathways have been implicated in the development and progression of breast cancer and noteworthy among those is NOTCH signaling (17,18). The NOTCH pathway regulates cell proliferation, survival, differentiation, cell–cell communication, angiogenesis and many other processes essential for tumorigenic potential (19,20).
It is becoming clear that there is a need for novel agents that will also target hypomethylated genes with oncogenic and pro-metastatic function and lead to their methylation and silencing. It would be expected that such compounds remodel the DNA methylation states rather than cause robust on–off changes. They could possibly act through indirect mechanisms resulting in differential changes in the DNA methylation states. Naturally derived compounds that switch cancerous to normal phenotype at minimally toxic doses would be excellent candidates for subtle changes in the DNA methylation profiles. Although limited, there are pieces of evidence demonstrating that bioactive compounds found in food and herbs can modulate gene expression by targeting DNA methylation. Specifically, resveratrol (RSV), a polyphenol from stilbenoid class, reversed hypermethylation and silencing of BRCA1, PTEN, APC and RARbeta2 tumor suppressor genes and inhibited breast cancer growth (5,6,21). Strikingly, RSV-mediated increase in methylation of specific genes has been demonstrated in recent studies in a rat diabetic model where methylation within pro-inflammatory cytokines led to their suppression in response to RSV (22). Similarly, pterostilbene (PTS), which is an analog of RSV, reversed hypomethylation within fasn (fatty acid synthase) gene in obesogenic rats and prevented subsequent gene upregulation (23). Although mechanisms underlying changes in the epigenome mediated by stilbenoids remain to be elucidated, the mode of action targeting both hypermethylated and hypomethylated genes in cancer is promising in cancer therapy and prevention.
To investigate the role of stilbenoids, RSV and PTS, in remodeling the DNA methylation patterns associated with cancer and elaborate on how these epigenetic effects correlate with the anticancer action of the compounds, we used isogenic breast cancer cell lines, lowly invasive MCF10CA1h and highly invasive MCF10CA1a, as a model system. Specifically, we delineated genome-wide DNA methylation landscape in response to RSV and focused on genes that were hypomethylated in breast cancer and whose DNA methylation increased in response to stilbenoids. Among genes with the highest increases in DNA methylation upon RSV exposure, we identified MAML2 (Mastermind (Drosophila)-Like) that is a coactivator of the oncogenic NOTCH signaling pathway (19). At each step, our results are consistent with the hypothesis that stilbenoids target specific genes that are hypomethylated in cancer and encode functional pathways required for cell growth and invasion and that partial reversal of this hypomethylation process by stilbenoids coincides with inhibition of cell growth and invasive properties of breast cancer cells.
Materials and methods
Cell culture and treatment with RSV and PTS
Human breast epithelial MCF10A cell line was purchased from American Type Culture Collection (CRL-10317, USA). Human breast cancer MCF10CA1h and MCF10CA1a cell lines were a gift from Dr Dorothy Teegarden (Purdue University). Please see Supplementary Materials, available at Carcinogenesis Online, for details on cell culture media, culture conditions, cell lines authentication. RSV (Sigma–Aldrich, St Louis, MO) and PTS (Cayman Chem., Ann Arbor, MI) were resuspended in ethanol and 10mM solutions were stored at −20°C. Dilutions of the compounds were freshly prepared prior to adding to the cell medium. Cells were grown in a humidified atmosphere of 5% carbon dioxide at 37°C. Twenty-four hours prior to treatment with RSV or PTS, cells were plated at a density of 2–3×105 per a 10-cm tissue culture dish. Cells were exposed to different RSV or PTS concentrations ranging from 0 to 20 µM for 4 days. Cells were then split 1:50, allowed to attach overnight and exposed to the compounds for additional 4 days (9-day exposure).
Illumina Infinium Human Methylation 450K BeadChip microarray
DNA from control cells (untreated) and treated cells was isolated using standard phenol:chloroform extraction protocol. Genomic DNA was processed for genome-wide DNA methylation analysis using Infinium HumanMethylation 450K BeadChip as described previously (24). Please see Supplementary Materials, available at Carcinogenesis Online, for details. The methylation score for each CpG was represented as a beta value according to the fluorescent intensity ratio with any values between 0 (unmethylated) and 1 (completely methylated). Raw microarray data and processed data are available from Gene Expression Omnibus (GSE80794).
Cell transfection with siRNA
Cells were plated at a density of 4–6×105 per 10-cm tissue culture dish, 24h prior to small interfering RNA (siRNA) treatment. All siRNA sequences were obtained from Dharmacon, including control siRNA (siCtrl), human MAML2 siRNA (siMAML2) and human OCT1 siRNA (siOCT1). The cells were transfected with siRNA using lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) as described previously (10). Concentration of 56nM was used for all siRNAs, which was determined as optimal in our previous studies (10). Please see Supplementary Materials, available at Carcinogenesis Online, for details on siRNA sequences and procedure. The transfection sequence was repeated two or three times depending on the effects on cell viability.
Viability, invasion and anchorage-independent growth assays
Cell viability was determined by trypan blue (Sigma–Aldrich) exclusion test. Cells were harvested after treatment with RSV or PTS on day 4 and day 9. Following 3–5min incubation with trypan blue, the viable and dead cells were counted under the microscope. The results were confirmed by MTT assay (Supplementary Figure S1, available at Carcinogenesis Online). Similarly, the cell viability after treatment with siRNAs was evaluated by trypan blue exclusion test.
Anchorage-independent growth in a 3-D format that resembles an in vivo cellular environment (25) was determined by soft agar assay, whereas the ability of treated cells to invade through extracellular matrix was evaluated by the Cell Invasion Assay Kit (Chemicon Int.) as described previously (10). Please see Supplementary Materials, available at Carcinogenesis Online, for details.
DNA extraction and pyrosequencing
DNA bisulfite conversion was performed as previously described (10,26). Specific bisulfite-converted DNA sequences were amplified with HotStar Taq DNA polymerase (Qiagen) using biotinylated primers listed in Supplementary Table S1A, available at Carcinogenesis Online. The biotinylated DNA strands were pyrosequenced in the PyroMarkTMQ24 instrument (Biotage, Qiagen) as previously described (27). Data were analyzed using PyroMarkTMQ24 software.
RNA extraction and QPCR
Total RNA was isolated using TRIzol (Roche Diagnostics) according to the manufacturer’s protocol and as described previously (10). Please see Supplementary Materials, available at Carcinogenesis Online, for details. Primers used in QPCR are listed in Supplementary Table S1B, available at Carcinogenesis Online. Quantification was performed using a standard curve and analyzed by the Roche LightCycler 480 software.
Western blot analyses
Total protein extract was obtained as described previously (10). Please see Supplementary Materials, available at Carcinogenesis Online, for details. The proteins were immunoblotted with anti-MAML2 (Abcam, ab57824) or anti-OCT1 (Millipore, MAB5434) antibody at 1:1000 dilution, followed by a secondary anti-mouse (Millipore, 12–349) IgG antibody at 1:2000 dilution. The membranes were blotted with an anti-β-actin antibody as loading control (Millipore, MABT523).
ChIP and qChIP
Chromatin immunoprecipitation (ChIP) was performed as previously described (28,29). Please see Supplementary Materials, available at Carcinogenesis Online, for details. The following primary antibodies were used: anti-acetyl-Histone H3Lys27 rabbit antibody (H3K27ac, Millipore, 07-360), anti-acetyl-Histone H3Lys9 rabbit antibody (H3K9ac, Millipore, 07-352), anti-trimethyl-Histone H3Lys27 rabbit antibody (H3K27me3, Millipore, 07-449), anti-DNMT3B rat antibody (Millipore, MABE305) or anti-OCT1 mouse antibody (Millipore, MAB5434). ChIP DNA was used as a template for QPCR (quantitative ChIP [qChIP]). Twenty-five nanogram input, antibody-bound and IgG-bound DNA was used as starting material in all conditions. Levels of H3K27ac, H3K9ac, H3K27me3, DNMT3B, and OCT1 binding were expressed as (Bound-IgG)/Input. Primers used for ChIP validation are depicted in Supplementary Table S1C, available at Carcinogenesis Online.
Statistical analysis
Raw methylation data from Human Methylation 450K microarrays were pre-processed using GenomeStudio and IMA (Illumina Methylation Analyzer for 450K, R/Bioconductor) including quality control, background correction, normalization, probe scaling and adjustment for batch effect. Differential methylation analysis between sample groups was conducted using linear models (R Bioconductor package limma). Specifically, limma uses an empirical Bayes moderated t-test, computed for each probe, which is similar to a t-test, except that standard errors have been moderated using information from the full set of probes (30). A methylation difference greater than 0.05 with a moderated t-test P ˂ 0.05 was considered statistically significant.
Statistical analysis of pyrosequencing, QPCR, qChIP, invasion, viability, soft agar assays was performed using the unpaired t-test with two-tailed distribution. Each value represents the mean ± SD of three independent experiments. The results were considered statistically significant when P < 0.05.
Results
Cancer-specific effects of RSV and PTS in inhibition of cell growth and invasiveness
Our hypothesis was that both RSV and PTS inhibit cancer cell growth and invasiveness, at least partially, through epigenetic regulation of gene expression. High cytotoxic doses of an agent with potential epigenetic activity can impede epigenetic reprogramming (31). Hence, minimally cytotoxic doses are recommended for studying epigenetic effects as was shown for epigenetic drugs, 5-aza-cytidine (5-aza-CR) and 5-aza-deoxycytidine (5-aza-CdR) (31). We therefore examined the effect of the compounds on cell growth at different concentrations using trypan blue exclusion test to quantify viable/death cell ratio. The results of cell viability were further validated by MTT assay. The treatments of MCF10A human immortalized mammary epithelial cells (used here as a normal cell model) and MCF10CA1h and MCF10CA1a human breast cancer cells were conducted for 4 or 9 days in order to establish time-dependent effects. MCF10CA1h and MCF10CA1a cells are derived from xenografts of MCF10A-ras cells that were generated by transfecting MCF10A mammary epithelial cells with constitutively active T24 Harvey-ras oncogene. MCF10CA1h and MCF10CA1a cells form well-differentiated and poorly differentiated malignant tumors in the xenograft models, respectively. Thus, MCF10CA1h cells have low invasive properties, contrary to MCF10CA1a cells that have characteristics of highly invasive phenotype. The three cell lines used in the present study (mammary epithelial cells, cancer lowly invasive and cancer highly invasive) are isogenic and thereby constitute an excellent model to study differential epigenetic effects during breast cancer progression.
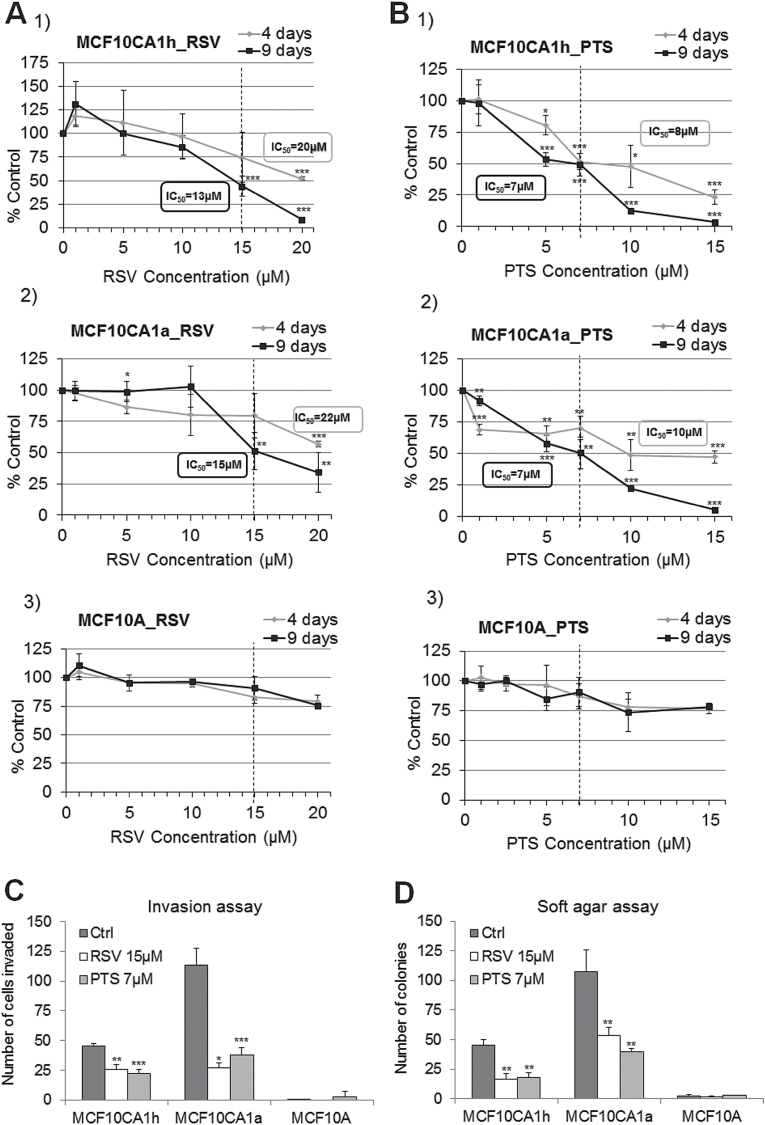
Treatment of both cancer cell lines with RSV or PTS resulted in a significant inhibition of cell growth as compared with control cells (treated with vehicle-ethanol) (Figure 1A and B, Supplementary Figure S1, available at Carcinogenesis Online). These effects were dose- and time dependent in both cell lines treated with RSV or PTS. The compounds caused 50% inhibition of cell viability (IC50) at doses equal to 15 µM for RSV and 7 µM for PTS (Figure 1A and B). Highly invasive cells were slightly more resistant to the treatments than lowly invasive cells. To compare the effects of RSV or PTS treatments in cancer cells with that in mammary epithelial cells, we challenged MCF10A cells with different doses of the compounds for 4 and 9 days. A concentration range 0–20 µM exerted only modest effects on viability of mammary epithelial cells (Figure 1A and B). Because the established 50% inhibition of cell viability concentrations were minimally toxic in both cancer cell lines (<10% of dead cells) and did not significantly affect mammary epithelial cells (>90% of cells viable), hence these doses were chosen for further experiments.
Figure 1.
Cancer-specific effects of RSV and PTS: inhibition of cell growth and invasive capacities. (A,B) Effect on cell growth after 4-day- and 9-day treatment with RSV (A) and PTS (B) at 5–20 µM concentrations in MCF10CA1h (1) and MCF10CA1a (2) breast cancer cells and in MCF10A (3) immortalized mammary epithelial cells; (C,D) effect on cell invasion (C) and anchorage-independent growth (D) as measured by Boyden chamber invasion assay and soft agar, respectively, upon 9-day exposure to 15 µM RSV and 7 µM PTS. All results represent mean ± SD of three independent experiments; ***P < 0.001, **P < 0.01, *P < 0.05.
Stilbenoids at 50% inhibition of cell viability concentrations significantly reduced invasive properties of breast cancer cells (Figure 1C) and their ability to anchorage-independent growth, a measure of oncogenesis (Figure 1D). The invasion and soft agar assays confirm high invasive potential of MCF10CA1a cells. The number of invaded cells and colonies formed in soft agar was nearly 2.5-fold higher for MCF10CA1a than for MCF10CA1h cells (Figure 1C and D). No such effects were seen in MCF10A cells that do not invade through the extracellular matrix and do not form colonies in soft agar (Figure 1C and D).
The landscape of DNA methylation in MCF10CA1h and MCF10CA1a breast cancer cells upon RSV exposure
Our previous work shows that RSV activates several methylation-silenced tumor suppressor genes in breast cancer (5,6). Previous data generated in epigenome-wide association studies provide increasing evidence that changes in DNA methylation are involved in cancer initiation and development (10,32). We reasoned therefore that reversing aberrant DNA methylation changes in cancer cells could be one of the mechanisms of the anticancer action of stilbenoids. Although it was demonstrated that stilbenoids like RSV can modulate gene expression of candidate tumor suppressors by modifying DNA methylation (5,6,21), their effects, particularly with respect to genes with oncogenic functions, have yet to be addressed. This is of high interest as genes with oncogenic and pro-metastatic functions were demonstrated to be commonly hypomethylated and activated in tumors versus normal tissue and drive cell transformation (10,13,32–34). In order to elaborate on the effects of stilbenoids on DNA methylation patterns and elucidate a possible molecular mechanism, we examined, using Illumina 450K array, genome-wide patterns of DNA methylation in MCF10CA1h and MCF10CA1a cells exposed to 15 µM RSV for 9 days (Figure 2). Please see Supplementary Materials, available at Carcinogenesis Online, for details on the Illumina 450K microarray platform.
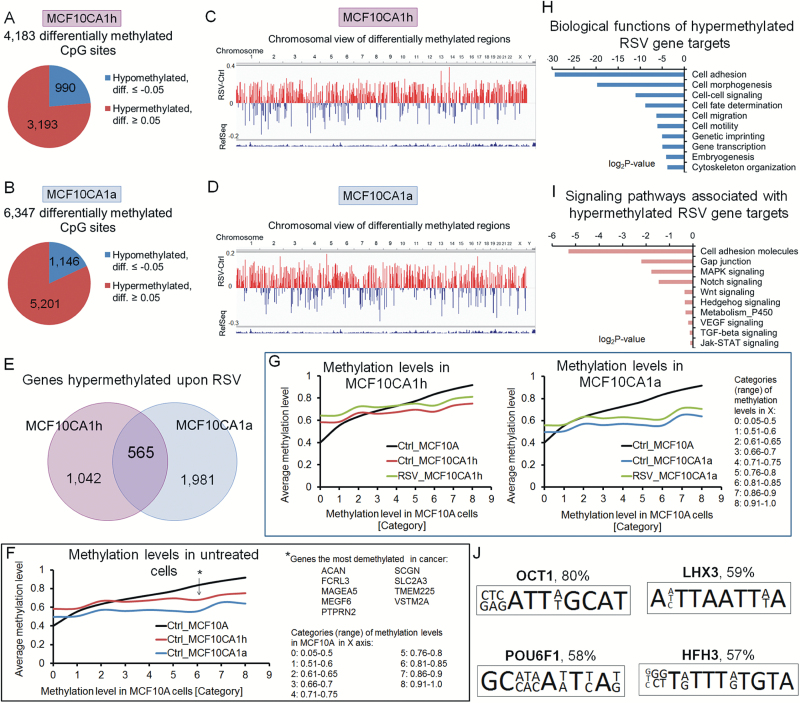
Figure 2.
RSV changes the landscape of DNA methylation in breast cancer cells and increases DNA methylation in genes that are hypomethylated in breast cancer cells. (A,B) Pie charts of differentially methylated CpG sites with the difference of at least 0.05 between RSV-treated and control MCF10CA1h (A) and MCF10CA1a (B) breast cancer cells (P ˂ 0.05, limma t-test). The landscape of DNA methylation was delineated using Illumina 450K microarray platform in breast cancer cells treated with RSV at 15 µM for 9 days (n = 3 per group). Diff. refers to differential methylation (delta beta, RSV minus control). (C,D) Chromosomal views of differentially methylated CpG sites. Each CpG site with significant difference of at least 0.05 between RSV and control cells was plotted into a bar track of a chromosomal view using IGV visualization tool. Each bar corresponds to a single, differentially methylated CpG site, with blue indicating hypomethylation and red indicating hypermethylation in RSV versus control (RSV minus Ctrl). (E) Venn diagram showing overlap between the hypermethylated genes in lowly invasive MCF10CA1h and highly invasive MCF10CA1a breast cancer cells in response to RSV. (F,G) Basal levels of methylation of ‘hypermethylated RSV targets’ as determined by the genome-wide microarray data in untreated MCF10A (normal), MCF10CA1h (cancer lowly invasive) and MCF10CA1a (cancer highly invasive) cells (F), as well as in MCF10CA1h and MCF10CA1a breast cancer cells exposed to 15 µM RSV for 9 days (G). The basal level of methylation of these genes in breast cancer cells is compared with the methylation levels in MCF10A cells. (H,I) Functional analyses using GO, KEGG and DAVID knowledgebase indicate biological functions (H) and pathways (I) associated with genes corresponding to CpG sites hypermethylated in response to RSV in both MCF10CA1h and MCF10CA1a. (J) Transcription factors and their binding motifs enriched for RSV hypermethylated genes in breast cancer. Putative transcription factor binding sites were predicted using TransFac. Percentage of RSV hypermethylated genes containing the motif is indicated.
We identified 4183 differentially methylated CpG sites in MCF10CA1h and 6347 differentially methylated CpG sites in MCF10CA1a cells upon 9-day treatment with 15 µM RSV as compared with control cells (differential methylation ≤−0.05 or ≥0.05, nominal P < 0.05, limma t-test) (Figure 2A and B). Chromosomal views of these differences were plotted using the Integrative Genomics Viewer (IGV) visualization tool (Figure 2C and D). Each vertical bar corresponds to a single differentially methylated CpG site, with blue indicating hypomethylation and red indicating hypermethylation in RSV versus control.
Most genes with tumor suppressor role in cancer have hypermethylated promoters and hypomethylated bodies compared with normal cells, which reflects their transcriptional silencing. Activated oncogenes would demonstrate the opposite patterns of DNA methylation. Taking into account the anticancer effects of RSV, we would expect that RSV treatment will decrease DNA methylation within promoters of potential tumor suppressor genes but increase DNA methylation within regulatory regions of potential oncogenes. Certain regions in the body of genes, such as CpG islands and enhancers, can play a regulatory role in gene transcription, thereby reflecting DNA methylation patterns in gene promoters. Approximately 36% of CpG sites whose methylation is changed upon treatment with RSV are located outside of gene promoters in gene bodies. Interestingly, 74% of those sites in gene bodies are found in regulatory regions, including CpG islands and enhancers. Examples of such sites in MCF10CA1h and MCF10CA1a breast cancer cells are depicted in Supplementary Figure S2 (right panel, available at Carcinogenesis Online), along with maps of the genes (representative pictures for MCF10CA1a in the left panel). We selected two genes with potential oncogenic role, MAML2 and GLI2, and two tumor suppressor genes, SEMA3A and HOXA9. An overview of DNA methylation level at CpG sites covered on Illumina 450K array for MAML2 shows hypomethylation in the promoter and hypermethylation within the gene body in cancer cells (MCF10CA1a) versus ‘normal’ breast cell model (MCF10A) (Supplementary Figure S2A, available at Carcinogenesis Online). Non-regulatory region in the body is hypermethylated (red stars), whereas hypomethylated fragments in the body correspond to enhancer regions (gray shaded areas with green stars). Methylation patterns of those enhancers reflect the patterns in the promoter that would indicate their regulatory influences. The profile of DNA methylation throughout MAML2 promoter and gene body in cancer cells is characteristic of active genes. Similar patterns are observed in GLI2 (Supplementary Figure S2B, available at Carcinogenesis Online). CpG island located in the gene body is heavily hypomethylated in cancer cells versus normal (MCF10A), which reflects patterns in the promoter and would be consistent with active gene transcription. Upon treatment of breast cancer cells with RSV, methylation levels of CpG sites within enhancers of MAML2 and GLI2 rise toward the levels present in normal cells (Supplementary Figure S2A and B, available at Carcinogenesis Online). Contrary to active oncogenes, tumor suppressor gene SEMA3A has hypomethylated gene body with no assigned regulatory role, whereas 5ʹ-UTR and enhancer located in the gene body are hypermethylated in cancer versus normal similarly to the promoter (Supplementary Figure S2C, available at Carcinogenesis Online). Another interesting example is tumor suppressor gene HOXA9, where CpG island that encompasses the promoter and gene body is heavily hypermethylated (Supplementary Figure S2D, available at Carcinogenesis Online). These patterns for SEMA3A and HOXA9 tumor suppressor genes would reflect transcriptional silencing in cancer versus normal. RSV exposure leads to decrease in methylation of CpG sites in SEMA3A enhancer and in the CpG island of GLI2, bringing the levels closer to the levels in normal cells.
The majority of differentially methylated CpG sites (76–82%) showed elevated levels of methylation after RSV treatment (Figure 2A–E). Interestingly, in approximately 70%, hypermethylated CpG sites were located in gene enhancers and/or CpG islands/shores/shelves, regions important for gene transcription. Validation by pyrosequencing of eight hypermethylated CpG sites linked to eight genes, selected based on large difference and statistical significance, confirmed the distinct DNA methylation pattern in RSV-treated cancer cells versus control cells (Supplementary Figure S3, available at Carcinogenesis Online). One of the highest differences was detected for MOBP in MCF10CA1a cells (25% increase) and PRKG1 in MCF10CA1h (20% increase; Supplementary Figure S3, available at Carcinogenesis Online). Interestingly, both genes are involved in regulation of cellular signal transduction (35,36).
Hypermethylated CpG sites corresponded to 1607 genes in MCF10CA1h and to 2546 genes in MCF10CA1a cells (Figure 2E, list of genes in Supplementary Table S2, available at Carcinogenesis Online). Among these genes, we determined 565 genes that are hypermethylated in response to RSV in both breast cancer cell lines (Figure 2E, Supplementary Table S2, available at Carcinogenesis Online). This overlap was significant as measured by Fisher’s exact test (P = 2.2E-16). For convenience, we call this set of genes ‘hypermethylated RSV targets’. This group potentially includes genes hypomethylated in cancer compared with normal tissue and RSV would act to increase their methylation to levels present in mammary epithelial cells. Interestingly, we found that 85% of hypermethylated RSV targets, whose basal methylation level in mammary epithelial MCF10A cells is higher than 0.5, are hypomethylated in breast cancer cells (Figure 2F). The extent of hypomethylation is more robust in highly invasive MCF10CA1a cancer cells compared with lowly invasive MCF10CA1h cells (Figure 2F–G). Among the most hypomethylated genes in cancer, we identified several with known oncogenic functions, including MAGEA5 (37), FCRL3 (38) and SLC2A3 (39) (Figure 2F). Exposure to RSV increases DNA methylation within hypomethylated genes to a similar extent in cancer cells with high or low invasive properties (Figure 2G). For part of those genes, methylation levels come back to the levels observed in mammary epithelial MCF10A cells (normal cell model; Figure 2G). As we previously demonstrated, genes hypomethylated in cancer are those with oncogenic and pro-metastatic functions and hypomethylation is one of the mechanisms of their upregulation (10,33,40). Indeed, the bioinformatics analysis using GO, KEGG and DAVID knowledgebase indicate that hypermethylated RSV targets are associated with functions that are essential for cancer formation and progression (Figure 2H). Furthermore, these genes are enriched in pathways, including MAPK, NOTCH, WNT and VEGF, which were reported before to drive cellular transformation, angiogenesis and metastasis (Figure 2I). It may suggest that these hypermethylated RSV targets are important for cancer development and progression. We therefore hypothesized that RSV leads to increased methylation within regulatory regions of these genes and causes their transcriptional silencing that may be one of the mechanisms of RSV-mediated anticancer effects. The analysis of transcription factor binding sites within regions hypermethylated by RSV further supports our hypothesis. These regions are highly enriched for motifs for OCT1 transcription factor that was shown to be associated with cancer promotion (41–43). OCT1 has putative binding sites within 80% of these hypermethylated regions (TransFac prediction; Figure 2J).
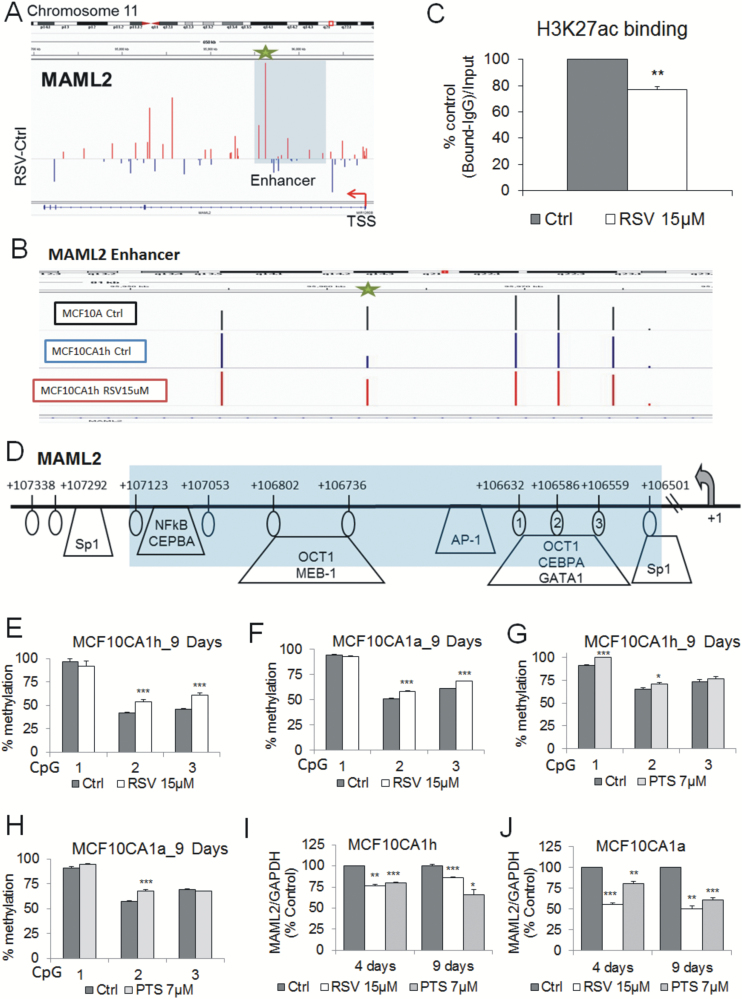
One of the top canonical pathways associated with genes hypermethylated in response to RSV is NOTCH signaling (Figure 2I) that was shown to have oncogenic functions in breast cancer (17,18). One of the most robustly hypermethylated genes in RSV-treated cancer cells was MAML2 that acts as a positive regulator of NOTCH pathway (19). The difference in DNA methylation within MAML2 in RSV treated versus control cells was determined as 0.37 in MCF10CA1h and 0.1 in MCF10CA1a cells based on the array data. These elevated levels of methylation were identified in a fragment corresponding to the predictive enhancer within MAML2 gene body (detailed map of the region in array track, Figure 3A). Methylation levels achieved after RSV treatment in cancer cells closely reflected patterns in MCF10A cells (normal cell model) as shown on the array track in Figure 3B (representative picture for MCF10CA1h cells, please see Supplementary Figure S4A, available at Carcinogenesis Online, for pyrosequencing results). It has been demonstrated that active enhancers are enriched with H3K27ac (44,45). We confirmed by ChIP assay that this histone mark is present at MAML2 in cancer cells and its occupancy is significantly reduced by RSV (Figure 3C). These data strongly support the role of this region as a gene enhancer and the role of RSV in decreasing the activity of this enhancer. The detailed map in Figure 3D shows the exact position of CpG sites in the region targeted by RSV. Thus, we elaborated in the present report on epigenetic mechanisms of RSV in regulating MAML2 expression in breast carcinogenesis and consequences of this regulation for NOTCH activity.
Figure 3.
Hypermethylation and silencing of MAML2 in response to RSV or PTS in breast cancer cells. (A) Magnified display from chromosome 11 demonstrates hypermethylation within MAML2 enhancer as determined by Illumina 450K microarray (representative picture for MCF10CA1h cells, red indicating hypermethylation upon treatment with 15 µM RSV on day 9). The region within MAML2 enhancer containing the most significantly methylated probes is shaded in light blue. CpG site with the highest difference in methylation is marked with green star. (B) Illumina 450K microarray tracks demonstrating methylation levels at the most differentially methylated CpG site (green star) within MAML2 enhancer in mammary epithelial MCF10A cells (‘normal’ cell model) and in MCF10CA1h breast cancer cells untreated or treated with 15 µM RSV for 9 days. (C) Occupancy of histone H3 acetylation at lysine 27 (H3K27ac) within the MAML2 tested region in MCF10CA1a cells after 9-day treatment with 15 µM RSV as assessed by qChIP and expressed as a percentage of the binding level in control cells. H3K27ac marks active enhancers. (D) A map of the MAML2 enhancer where the light blue-shaded region represents the entire fragment tested by pyrosequencing and qChIP. The CpG sites, whose elevated methylation was validated by pyrosequencing, are circled and numbered. Putative transcription factor binding sites are indicated as predicted using TransFac. (E–H) Average methylation status of CpG sites in the MAML2 enhancer as determined by pyrosequencing in MCF10CA1h (E,G) and MCF10CA1a (F,H) breast cancer cells exposed for 9 days to 15 µM RSV (E,F) or 7 µM PTS (G,H). Numbers in X axis correspond to CpG sites within the tested region. (I,J) The effects of 4 day- and 9 day-treatment with 15µM RSV or 7 µM PTS on expression of MAML2 in MCF10CA1h (I) or MCF10CA1a (J) breast cancer cells as determined by QPCR. All results represent mean ± SD of three independent experiments; ***P < 0.001, **P < 0.01, *P < 0.05.
MAML2 is epigenetically silenced in response to RSV and PTS, which is associated with decreased NOTCH signaling activity
Hypermethylation within MAML2 enhancer was quantitatively validated by pyrosequencing of the region encompassing three CpG sites that are circled and numbered in MAML2 enhancer map depicted in Figure 3D. Exposure of lowly and highly invasive breast cancer cells to 15 μM RSV resulted in significant increase in methylation at CpG#2 and CpG#3 on day 9 (Figure 3E and F). PTS at 7 μM caused increase in methylation at CpG#2 and/or CpG#1 in both cancer cells (Figure 3G and H). To determine whether these changes in DNA methylation are biologically relevant, we tested MAML2 expression upon treatment with the compounds. Using QPCR, we detected downregulation of MAML2 in both cancer cell lines treated with RSV or PTS for 4 or 9 days (Figure 3I and J). The effects in invasive MCF10CA1a cells were stronger, especially upon RSV treatment.
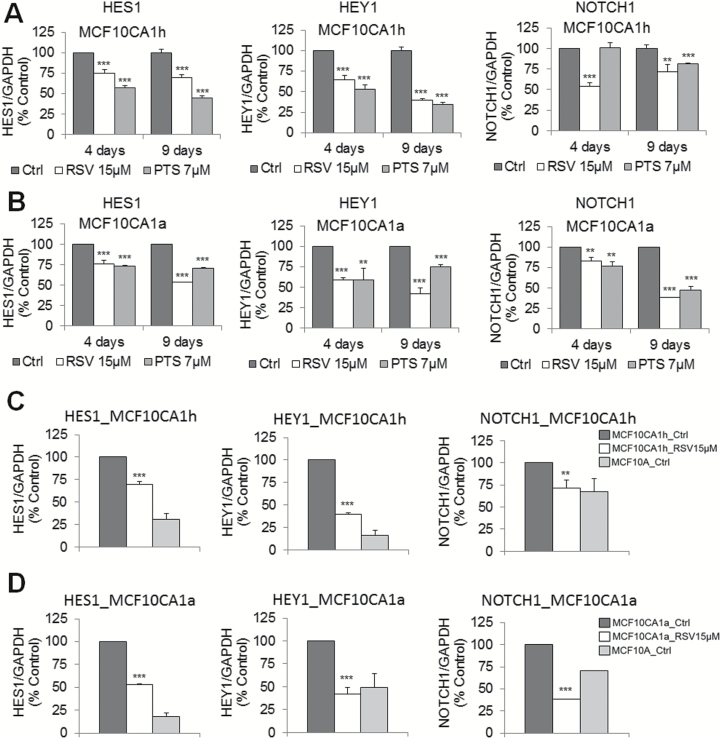
If changes in methylation and expression of MAML2 in response to RSV or PTS are functionally relevant, attenuation of NOTCH signaling should be observed. Indeed, we found that all three NOTCH target genes that we tested are downregulated upon treatment of breast cancer cells with stilbenoids (Figure 4). RSV and PTS caused 20–65% decrease in expression of the tested genes in MCF10CA1h cells at both time points, 4 and 9 days (Figure 4A). Similar effects were observed in highly invasive MCF10CA1a cells (Figure 4B). As shown in Figure 4C and 4D (representative results for RSV), the levels of expression of NOTCH target genes achieved upon treatments closely reflect the levels observed in mammary epithelial MCF10A cells (normal cell model). Expression comes back to the levels close to normal for all the genes and reaches normal levels for NOTCH1 in lowly invasive MCF10CA1h cells and for HEY1 and NOTCH1 in highly invasive MCF10CA1a cells (Figure 4C and D).
Figure 4.
Epigenetic silencing of MAML2 in response to RSV or PTS is associated with attenuation of the activity of the NOTCH signaling pathway. (A,B) The effects of 4-day and 9-day treatment with 15 µM RSV or 7 µM PTS on expression of NOTCH target genes, HES1, HEY1 and NOTCH1, in MCF10CA1h (A) and MCF10CA1a (B) breast cancer cells as determined by QPCR. Expression was expressed as a percentage of mRNA levels in untreated cells (% control). (C,D) Levels of expression of NOTCH target genes in untreated mammary epithelial MCF10A cells (‘normal’ cell model) and in MCF10CA1h (C) and MCF10CA1a (D) breast cancer cells untreated or upon 9-day treatment with 15 µM RSV. Expression was assessed by QPCR and expressed as a percentage of mRNA levels in untreated cancer cells (% control). All results represent mean ± SD of three independent experiments; ***P < 0.001, **P < 0.01, *P < 0.05.
Interestingly, in MCF10A-ras breast cancer cells that are characterized by low invasive properties (Supplementary Figure S4B, available at Carcinogenesis Online) and low ability to anchorage-independent growth (Supplementary Figure S4C, available at Carcinogenesis Online), MAML2 is expressed at a similar level as in mammary epithelial MCF10A cells (normal cell model) (Supplementary Figure S4D, available at Carcinogenesis Online). This corresponds to similar high DNA methylation levels of MAML2 enhancer in MCF10A-ras and MCF10A, as compared with highly invasive MCF10CA1a cells (Supplementary Figure S4E, available at Carcinogenesis Online), where higher MAML2 expression is observed (Supplementary Figure S4D, available at Carcinogenesis Online). Furthermore, all three NOTCH target genes are expressed at normal levels in MCF10A-ras breast cancer cells (Supplementary Figure S4F, available at Carcinogenesis Online), which indicate low activity of NOTCH signaling. Thus, we suggest that low MAML2 expression in cancer cells is associated with reduced NOTCH activity and low invasive potential of cancer cells.
Depletion of MAML2 with siRNA mimics inhibitory effects of stilbenoids on NOTCH signaling
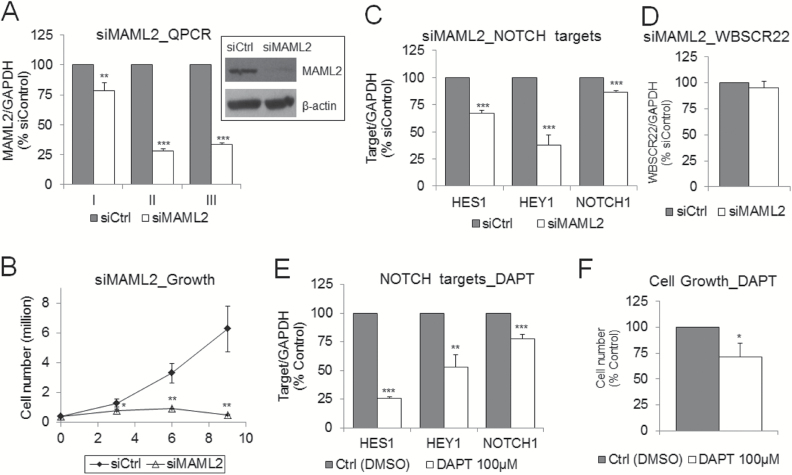
If NOTCH inhibition upon treatment with stilbenoids is associated with stilbenoid-mediated MAML2 silencing, we should observe similar effects on NOTCH pathway upon MAML2 depletion. Indeed, MAML2 depletion in invasive MCF10CA1a cells, as confirmed by QPCR and western blot (Figure 5A), resulted in inhibition of cancer cell growth (Figure 5B) and suppression of NOTCH target genes (Figure 5C) as compared with cells treated with scrambled siRNA (siCtrl). MAML2 depletion did not affect the expression of WBSCR22 (MERM1), which is a non-NOTCH target gene involved in invasion and metastasis (46) (Figure 5D). Lack of the effect on WBSCR22 strengthens evidence that the observed decrease in cancer cell growth upon siMAML2 is a result of NOTCH signaling inhibition. Importantly, each of the four sets of siRNA present in SmartPool mixture downregulated MAML2 expression and inhibited cancer cell growth when used separately (Supplementary Figure S5A–C, available at Carcinogenesis Online). Stronger downregulation of MAML2 upon siRNA2 or siRNA3 corresponded to stronger inhibition of cell growth (Supplementary Figure S5A and B, available at Carcinogenesis Online). These data demonstrate that the results we present are not an idiosyncrasy of the siRNA sequence used and provide high confidence that we are measuring effects of MAML2 depletion. We further show that treatment of MCF10CA1a cells with NOTCH signaling inhibitor, DAPT, causes similar downregulation of NOTCH target genes as siMAML2 (Figure 5E) and reduces cancer cell growth (Figure 5F). DAPT inhibits γ-secretase and therefore blocks cleavage of NOTCH receptor and production of the Notch intracellular domain that translocates into the nucleus and transactivates the target gene. Similar effects observed upon siMAML2 and DAPT treatments provide strong support for the oncogenic role of NOTCH signaling in breast cancer and for MAML2 in promoting the activity of NOTCH pathway.
Figure 5.
Functional role of MAML2 in regulation of the NOTCH signaling pathway. (A) MAML2 expression quantified by QPCR after first (I), second (II) and third (III) transfection and by western blot after second transfection with scrambled siRNA (siCtrl) or siRNA directed to MAML2 (siMAML2). (B) Effect on cell growth after first (day 3), second (day 6) and third (day 9) transfection with siCtrl or siMAML2. (C,D) Effect of MAML2 depletion (second transfection) on expression of NOTCH target genes, HES1, HEY1 and NOTCH1 (C), and on a non-NOTCH target gene, WBSCR22 (D), in MCF10CA1a breast cancer cells as determined by QPCR. (E) Effect of 72-h exposure to NOTCH inhibitor, DAPT, at 100 µM concentration, on expression of NOTCH target genes, HES1, HEY1 and NOTCH1, as determined by QPCR and on cell growth in MCF10CA1a breast cancer cells. All results represent mean ± SD of three independent experiments; ***P < 0.001, **P < 0.01, *P < 0.05.
Chromatin condensation and transcription factor occupancy at MAML2 enhancer upon treatment with RSV
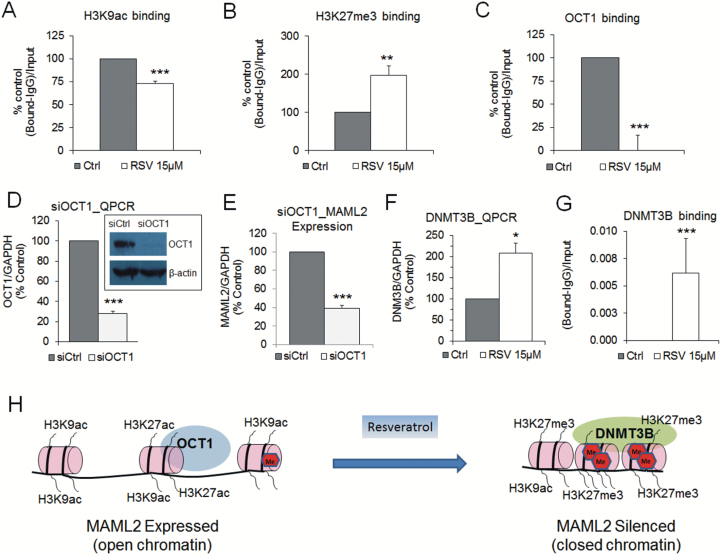
Stilbenoids, especially RSV, lead to more profound downregulation of MAML2 in highly invasive cancer cells than in lowly invasive cells, despite similar increase in DNA methylation at MAML2 enhancer (Figure 3). We reasoned that other epigenetic marks including histone modifications may be involved in stilbenoid-mediated regulation of MAML2 transcription. Using ChIP, we assessed the occupancy of H3K9acetylation (activating histone mark) and H3K27tri-methylation (repressive histone mark) at MAML2. We found 30% decrease in H3K9ac (Figure 6A) and 2-fold increase in H3K27me3 occupancy (Figure 6B) at the same region within MAML2 enhancer where higher DNA methylation levels were detected in response to RSV. These data indicate epigenetic reprogramming at the tested MAML2 region that results in marks characteristic of condensed chromatin structure and is consistent with MAML2 transcriptional silencing.
Figure 6.
Changes in histone modifications and in occupancy of DNMT3B and transcription factor OCT1 within MAML2 enhancer in breast cancer cells in response to RSV; functional role of OCT1 in regulation of MAML2 expression. (A–C) Occupancy of histone H3 acetylation at lysine 9 (H3K9ac, activating mark) (A), histone H3 trimethylation at lysine 27 (H3K27me3, repressive mark) (B) and transcription factor OCT1 (C) within the MAML2 enhancer in MCF10CA1a cells in response to 9-day treatment with 15 µM RSV as assessed by qChIP and expressed as a percentage of the binding level in control cells. (D) OCT1 expression quantified by QPCR and western blot after first transfection with scrambled siRNA (siCtrl) or siRNA directed to OCT1 (siOCT1). (E) Effect of OCT1 depletion (first transfection) on expression of MAML2 in MCF10CA1a breast cancer cells as determined by QPCR. (F) Effect of 9-day treatment with 15 µM RSV on DNMT3B expression in MCF10CA1a breast cancer cells as determined by QPCR. (G) Binding of DNMT3B to the MAML2 tested region in MCF10CA1a cells in response to 9-day treatment with 15 µM RSV as assessed by qChIP. (H) The scheme displaying changes in DNA methylation, histone marks and occupancy of DNMT3B and OCT1 at MAML2 enhancer in response to RSV, as suggested by our study. All results represent mean ± SD of three independent experiments; ***P < 0.001, **P < 0.01, *P < 0.05.
Gene silencing could be a consequence of blocked access of transcription factors to the recognized DNA sequence because of changes in the chromatin state. We computed putative transcription factor binding elements within MAML2 enhancer using TransFac. Three transcription factors, OCT1, CEBPA and GATA1, were found to have predictive binding sites at the region encompassing three CpG sites assessed in pyrosequencing (Figure 3D). As shown in Figure 2J, motifs for OCT1 were found to be the most common within regions hypermethylated by RSV. We therefore explored OCT1 binding to MAML2 and tested how this binding is affected by stilbenoid-mediated epigenetic changes within MAML2 enhancer. Using ChIP, we confirmed OCT1 binding to MAML2 enhancer (Figure 6C, control cells). The binding dramatically decreases upon treatment of MCF10CA1a with 15 μM RSV (Figure 6C). The lack of OCT1 binding may be directly linked to MAML2 transcriptional silencing. To test this hypothesis, we depleted OCT1 in MCF10CA1a cells using SmartPool siRNA mixture and four siRNA sequences separately, as confirmed by QPCR and western blot (Figure 6D, Supplementary Figure S5D–F, available at Carcinogenesis Online) and tested the effect of OCT1 absence on MAML2 expression (Figure 6D and E). Indeed, OCT1 depletion led to substantial MAML2 downregulation (Figure 6E) and produced similar effects on cancer cell growth as siMAML2 (Supplementary Figure S5D, available at Carcinogenesis Online). These findings suggest the role of OCT1 in regulation of MAML2 transcription.
DNMT3B occupancy at MAML2 enhancer is increased upon treatment with RSV
Stilbenoids lead to increase in DNA methylation within MAML2 enhancer that coincides with MAML2 transcriptional silencing. DNA methylation reaction is catalyzed by DNA methyltransferases, including maintenance DNMT1, and de novo DNMT3A and 3B. While RSV and PTS caused attenuation or no change of DNMT1 and DNMT3A, we detected upregulation of DNMT3B in breast cancer cells (Figure 6F, Supplementary Figure S6, available at Carcinogenesis Online). A question arose whether DNMT3B is involved in stilbenoid-mediated increased methylation within MAML2. We therefore tested DNMT3B binding using ChIP. Treatment with 15 μM RSV led to DNMT3B enrichment at MAML2 enhancer in MCF10CA1a cells, whereas no binding was detected in control untreated cells (Figure 6G). The fragment bound by DNMT3B overlapped with the fragment whose increased methylation was detected by microarray and pyrosequencing. It suggests the role of DNMT3B in increased methylation of MAML2 upon RSV exposure.
Discussion
Attention in the field of epigenetic cancer research has been focused on the increase in DNA methylation of tumor suppressor genes as an early event in cancer development (47). It is well accepted that promoter hypermethylation leads to silencing of tumor suppressor genes and that reversal of repression of these genes results in anticancer effects (5,6). Hence, the main focus in epigenetic pharmacology was on developing strategies that will inhibit DNA methylation and remove aberrant methylation marks from tumor suppressor genes. However, it is becoming clear that promoter hypomethylation is as prevalent in cancer as hypermethylation (10). Recent genome-wide DNA methylation studies in cancer report a similar number of hypomethylated and hypermethylated gene promoters (10,13,48). Interestingly, the hypomethylated genes share common functions relevant to cancer promotion. Our recent studies show that differentially methylated genes that distinguish noninvasive cancer cells from highly invasive cells include predominantly genes that are hypomethylated in invasive cells and may account for their highly invasive potential (24,49). These pieces of evidence indicate a need for developing therapeutic strategies that would be aimed at reversal of both hypermethylation of tumor suppressor genes and hypomethylation of genes with cancer-promoting functions (e.g. oncogenes and pro-metastatic genes). Such compounds would mediate subtle changes in the DNA methylation patterns rather than dramatic off–on changes and lead to reprogramming of multiple functional gene networks. Excellent candidates for such action are naturally derived compounds, such as RSV and PTS, which bring about profound phenotypic changes at minimally toxic doses. Indeed, the global picture emerging from our genome-wide DNA methylation study is that most of the changes in DNA methylation in response to RSV are moderate indicating reprogramming of the patterns (Figure 2).
We and others previously observed that RSV modulates DNA methylation within promoters of tumor suppressor genes leading to reactivation of those genes in breast cancer (5,6,21). However, the effects on oncogenes and pro-metastatic genes are still largely unknown. One would expect that anticancer action of RSV will be associated with downregulation of oncogenes and pro-metastatic genes, contrary to the effects on tumor suppressor genes observed so far. In the present study, we tested the possibility that stilbenoids remodel the DNA methylation landscape in breast cancer and target genes with oncogenic function for methylation and silencing. This would account, at least partially, for the anticancer action of these compounds. Treatment with stilbenoids resulted in a dose- and time-dependent inhibition in cell proliferative and invasive properties in breast cancer cells (Figure 1). These effects were cancer specific, as only marginal changes in cell viability were detected in mammary epithelial MCF10A cells within a dose range of 0–20 µM (Figure 1). One possible explanation for this remarkable observation is that RSV and PTS affect genes that are dysregulated in cancer but not in normal cells. For instance, the compounds lead to silencing of genes that are activated in cancer. Since these genes are already silenced in normal cells, the compounds would have no impact on these genes.
We propose a model whereby stilbenoids reduces tumorigenic features and metastatic potential of cancer cells by increased DNA methylation and silencing of genes and signaling pathways that promote cancer. Genome-wide analysis of the DNA methylation landscape in breast cancer cells revealed remodeling of the patterns in response to RSV (Figure 2). Indeed, the majority of differentially methylated CpG sites were hypermethylated in response to RSV versus control (Figure 2A–D, Supplementary Figure S3, available at Carcinogenesis Online). These hypermethylated CpG sites were predominantly located in genes involved in critical oncogenic signaling pathways and functions essential for cancer development and progression (Figure 2H and I). The increase in DNA methylation could be potentially linked to silencing of these genes and mediate anticancer effects of stilbenoids. This is further supported by our next observation. When we compared the basal methylation levels of hypermethylated RSV targets between cancer cells and MCF10A cells (our normal cell model), we found that most of the genes targeted by RSV are highly methylated in normal cells and lose methyl marks in cancer cells (Figure 2F). Thus, their methylation levels are coming back toward normal levels upon RSV treatment (Figure 2G). Genes that are the most demethylated in cancer cells and targeted by RSV for increase in methylation are listed in the right panel of Figure 2F. Functional analysis of these genes indicates that they are involved in tumor progression (MAGEA5, FCRL3) (37,38), angiogenesis (MEGF6) (50), increase in migration of metastatic cancer cells (PTPRN2, SCGN) (51,52) and stimulation of cell proliferation (SCGN, SLC2A3) (39).
One of the pathways associated with genes hypermethylated in response to RSV (Figure 2E) is NOTCH signaling with reported oncogenic role in breast cancer (17,18). Thus, targeting this pathway may be an important therapeutic strategy in breast cancer. Our genome-wide study in RSV-treated breast cancer cells indicated increase in methylation within the enhancer region of MAML2 (Figure 3) that is a coactivator of NOTCH target genes (19,20). Higher DNA methylation levels at MAML2 enhancer were linked to decrease in occupancy of H3K27ac (Figure 3C) that is a histone modification specific to active enhancers (45,46). Furthermore, we detected decrease in activating and increase in repressive histone marks at this region (Figure 6A and B). Cumulatively, these changes in DNA methylation and histone modifications at MAML2 enhancer in response to stilbenoids suggest condensed chromatin structure and could account for observed decrease in MAML2 expression (Figure 3I and J). Interestingly, both RSV and PTS were demonstrated to exert similar mode of action on the expression of several genes in rat diabetic models where DNA methylation and silencing were detected in response to the compounds (22,23).
Epigenetic silencing of MAML2 in response to RSV should have consequences for the activity of NOTCH signaling (19,20). We found that stilbenoids downregulate all three targets of the pathway suggesting negative regulation of signal transduction (Figure 4). MAML2 knockdown exerts similar effects (Figure 5). It indicates the functional link between MAML2 and NOTCH and suggests that the effects of stilbenoids on NOTCH signaling are at least partially linked to stilbenoid-mediated MAML2 epigenetic silencing. Our results are consistent with a recent report where NOTCH pathway was shown to be attenuated by RSV in cervical cancer (53). With the present findings, we deliver insights into the mechanisms of regulation of the pathway by stilbenoids and their relevance in breast carcinogenesis. Epigenetic targeting of MAML2 and subsequently NOTCH pathway may have important in vivo implications. Publicly available genome-wide data show hypomethylation of MAML2 enhancer in breast tumors compared with normal breast tissue (TCGA, Supplementary Figure S7A, available at Carcinogenesis Online) and overexpression of MAML2 in breast tumors and other cancers (Oncomine, Supplementary Figure S7B, available at Carcinogenesis Online). In addition, we observe robust upregulation of Maml2 and Notch target genes in cancer versus normal in N-methyl nitrosourea-induced mammary carcinogenesis in Sprague–Dawley rats (Supplementary Figure S7C, available at Carcinogenesis Online). These pieces of evidence give strong support for future investigations of in vivo effects of stilbenoids on epigenetic regulation of oncogenic NOTCH pathway and tumor formation.
What mechanism is responsible for stilbenoid-mediated increase in DNA methylation at MAML2 enhancer? The classic explanation would involve DNA methyltransferases. We found that de novo methyltransferase DNMT3B binds to MAML2 enhancer upon treatment with stilbenoids (Figure 6G). This coincides with decrease in OCT1 occupancy (Figure 6C). Our findings indicate that OCT1 is a candidate transcription factor that regulates MAML2 transcription. It binds to MAML2 enhancer in untreated cancer cells (Figure 6C) and its depletion with siRNA leads to MAML2 suppression and mimics the effects of siMAML2 on cancer cell growth (Figure 6D and E, Supplementary Figure S5D, available at Carcinogenesis Online). Importantly, regions hypermethylated in response to RSV based on the array data are highly enriched for OCT1 motifs (TransFac). These RSV-targeted regions are located in genes that are associated with oncogenic signaling and other cancer-promoting functions (Figure 2H–I), including genes that are strongly demethylated in cancer cells compared with mammary epithelial MCF10A cells (normal cell model) (Figure 2F and G). Their oncogenic potential is consistent with OCT1 function as an enhancer of tumor malignancy (41–43). It raises an interesting question whether OCT1 dictates specificity of stilbenoids in targeting this set of genes for hypermethylation. This intriguing hypothesis remains to be tested in future projects.
Our findings support a model of stilbenoids action proposed in Figure 6H. In breast cancer cells, MAML2 is expressed that is associated with low DNA methylation at MAML2 enhancer and enrichment with H3K27ac and H3K9ac (open chromatin structure). Hence, OCT1 binds to its recognized sequence within this region, the gene is transcribed and NOTCH signaling is active. In the presence of RSV, chromatin structure becomes condensed gaining DNA methylation and H3K27me3. OCT1 binding is impaired and DNMT3B binding is facilitated that may underlie DNA methylation and transcriptional silencing (Figure 6H). Interestingly, RSV brings methylation levels within MAML2 very close to the levels observed in normal cells (Figure 3B, Supplementary Figure S4A, available at Carcinogenesis Online). Changes in methylation coexist with changes in histone modifications as presented in Figures 3 and 6. This suggests DNA methylation and histone modifications can cooperate together to affect gene expression and thus the anticancer effect of RSV. This might be a general pattern for genes targeted by RSV because methylation levels for some RSV targets increase but do not reach normal levels (Figure 2G).
In the present study, we determine for the first time genome-wide landscapes of DNA methylation in response to RSV. Our findings reveal that stilbenoids target for methylation genes with oncogenic and pro-metastatic function, many of which are regulators of signaling pathways critical in cancer such as NOTCH among others. Stilbenoids lead to epigenetic silencing of MAML2, the activator of NOTCH pathway, which is associated with increased occupancy of DNMT3B and decreased binding of OCT1 transcription factor at MAML2 enhancer. Suppression of MAML2 results in inactivation of NOTCH signaling. We deliver here scientific evidence for stilbenoids as agents that could combat epigenetic activation of genes involved in breast cancer development and progression and hence could be effective in anticancer strategies.
Supplementary material
Supplementary Materials, Supplementary Figures S1–S7 and Supplementary Tables S1 and S2 can be found at http://carcin.oxfordjournals.org/
Funding
Indiana Clinical and Translational Sciences Institute funded, in part by grant number UL1TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award; the Women’s Global Health Institute (WGHI MEER); USDA National Institute of Food and Agriculture (Hatch project 1005656); and the Purdue University Center for Cancer Research Undergraduate Research Program granted to B.S. J.M.F. and K.J.F. were funded by Breast Cancer Now and acknowledge funding support from Cancer Research UK (A13086) and the National Institute for Health Research (NIHR) Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London.
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- RSV
resveratrol
- PTS
pterostilbene
- siRNA
small interfering RNA
- ChIP
chromatin immunoprecipitation
- qChIP
quantitative chromatin immunoprecipitation
References
- 1. Siegel R.L., et al. (2015) Cancer statistics, 2015. CA. Cancer J. Clin., 65, 5–29. [DOI] [PubMed] [Google Scholar]
- 2. Ly D., et al. (2013) An international comparison of male and female breast cancer incidence rates. Int. J. Cancer, 132, 1918–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lacey J.V., Jr., et al. (2009) Breast cancer epidemiology according to recognized breast cancer risk factors in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial Cohort. BMC Cancer, 9, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Danaei G., et al. (2005) Causes of cancer in the world: comparative risk assessment of nine behavioural and environmental risk factors. Lancet, 366, 1784–1793. [DOI] [PubMed] [Google Scholar]
- 5. Stefanska B., et al. (2010) Hypomethylation and induction of retinoic acid receptor beta 2 by concurrent action of adenosine analogues and natural compounds in breast cancer cells. Eur. J. Pharmacol., 638, 47–53. [DOI] [PubMed] [Google Scholar]
- 6. Stefanska B., et al. (2012) Comparative effects of retinoic acid, vitamin D and resveratrol alone and in combination with adenosine analogues on methylation and expression of phosphatase and tensin homologue tumour suppressor gene in breast cancer cells. Br. J. Nutr., 107, 781–790. [DOI] [PubMed] [Google Scholar]
- 7. Cedar H., et al. (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet., 10, 295–304. [DOI] [PubMed] [Google Scholar]
- 8. Baylin S.B., et al. (2001) Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet., 10, 687–692. [DOI] [PubMed] [Google Scholar]
- 9. Nilsson E., et al. (2014) Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes, 63, 2962–2976. [DOI] [PubMed] [Google Scholar]
- 10. Stefanska B., et al. (2011) Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res., 71, 5891–5903. [DOI] [PubMed] [Google Scholar]
- 11. Jones P.A. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet., 13, 484–492. [DOI] [PubMed] [Google Scholar]
- 12. Szyf M., et al. (2004) DNA methylation and breast cancer. Biochem. Pharmacol., 68, 1187–1197. [DOI] [PubMed] [Google Scholar]
- 13. Mayol G., et al. (2012) DNA hypomethylation affects cancer-related biological functions and genes relevant in neuroblastoma pathogenesis. PLoS One, 7, e48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hatada I., et al. (2006) Genome-wide profiling of promoter methylation in human. Oncogene, 25, 3059–3064. [DOI] [PubMed] [Google Scholar]
- 15. Baylin S.B., et al. (1998) Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res., 72, 141–196. [PubMed] [Google Scholar]
- 16. Pakneshan P., et al. (2004) Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J. Biol. Chem., 279, 31735–31744. [DOI] [PubMed] [Google Scholar]
- 17. Suman S., et al. (2013) Silencing NOTCH signaling causes growth arrest in both breast cancer stem cells and breast cancer cells. Br. J. Cancer, 109, 2587–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bolós V., et al. (2013) Notch activation stimulates migration of breast cancer cells and promotes tumor growth. Breast Cancer Res., 15, R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin S.E., et al. (2002) Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J. Biol. Chem., 277, 50612–50620. [DOI] [PubMed] [Google Scholar]
- 20. Grabher C., et al. (2006) Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer, 6, 347–359. [DOI] [PubMed] [Google Scholar]
- 21. Papoutsis A.J., et al. (2012) BRCA-1 promoter hypermethylation and silencing induced by the aromatic hydrocarbon receptor-ligand TCDD are prevented by resveratrol in MCF-7 cells. J. Nutr. Biochem., 23, 1324–1332. [DOI] [PubMed] [Google Scholar]
- 22. Lou X.D., et al. (2014) Effects of resveratrol on the expression and DNA methylation of cytokine genes in diabetic rat aortas. Arch. Immunol. Ther. Exp. (Warsz)., 62, 329–340. [DOI] [PubMed] [Google Scholar]
- 23. Gracia A., et al. (2014) Fatty acid synthase methylation levels in adipose tissue: effects of an obesogenic diet and phenol compounds. Genes Nutr., 9, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheishvili D., et al. (2015) A common promoter hypomethylation signature in invasive breast, liver and prostate cancer cell lines reveals novel targets involved in cancer invasiveness. Oncotarget, 6, 33253–33268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Larco J.E., et al. (1978) Growth factors from murine sarcoma virus-transformed cells. Proc. Natl Acad. Sci. USA, 75, 4001–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Colella S., et al. (2003) Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques, 35, 146–150. [DOI] [PubMed] [Google Scholar]
- 27. Tost J., et al. (2007) DNA methylation analysis by pyrosequencing. Nat. Protoc., 2, 2265–2275. [DOI] [PubMed] [Google Scholar]
- 28. Brown S.E., et al. (2008) DNA demethylation induced by the methyl-CpG-binding domain protein MBD3. Gene, 420, 99–106. [DOI] [PubMed] [Google Scholar]
- 29. Peng G.H., et al. (2013) Double chromatin immunoprecipitation: analysis of target co-occupancy of retinal transcription factors. Methods Mol. Biol., 935, 311–328. [DOI] [PubMed] [Google Scholar]
- 30. Wilhelm-Benartzi C.S., et al. (2013) Review of processing and analysis methods for DNA methylation array data. Br. J. Cancer, 109, 1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsai H.C., et al. (2012) Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell, 21, 430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stefanska B., et al. (2013) Discovery and validation of DNA hypomethylation biomarkers for liver cancer using HRM-specific probes. PLoS One, 8, e68439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stefanska B., et al. (2013) Transcription onset of genes critical in liver carcinogenesis is epigenetically regulated by methylated DNA binding protein MBD2. Carcinogenesis, 34, 2738–2749. [DOI] [PubMed] [Google Scholar]
- 34. Shao C., et al. (2011) Integrated, genome-wide screening for hypomethylated oncogenes in salivary gland adenoid cystic carcinoma. Clin. Cancer Res., 17, 4320–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma A.K., et al. (2008) Probing the interaction between the coiled coil leucine zipper of cGMP-dependent protein kinase Ialpha and the C terminus of the myosin binding subunit of the myosin light chain phosphatase. J. Biol. Chem., 283, 32860–32869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Primiani C.T., et al. (2014) Coordinated gene expression of neuroinflammatory and cell signaling markers in dorsolateral prefrontal cortex during human brain development and aging. PLoS One, 9, e110972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cappell K.M., et al. (2012) Multiple cancer testis antigens function to support tumor cell mitotic fidelity. Mol. Cell. Biol., 32, 4131–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wysocka M., et al. (2014) CD164 and FCRL3 are highly expressed on CD4+CD26- T cells in Sézary syndrome patients. J. Invest. Dermatol., 134, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Starska K., et al. (2015) Gene and protein expression of glucose transporter 1 and glucose transporter 3 in human laryngeal cancer-the relationship with regulatory hypoxia-inducible factor-1α expression, tumor invasiveness, and patient prognosis. Tumour Biol., 36, 2309–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stefanska B., et al. (2014) Genome-wide study of hypomethylated and induced genes in liver cancer patients unravels novel anticancer targets. Clin Cancer Res. [DOI] [PubMed] [Google Scholar]
- 41. Hwang-Verslues W.W., et al. (2013) Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc. Natl. Acad. Sci. USA, 110, 12331–12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Y., et al. (2015) Octamer transcription factor 1 mediates epithelial-mesenchymal transition in colorectal cancer. Tumour Biol, 36, 9941–9946. [DOI] [PubMed] [Google Scholar]
- 43. Xiao S., et al. (2014) High expression of octamer transcription factor 1 in cervical cancer. Oncol. Lett., 7, 1889–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Consortium E.P. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hon G.C., et al. (2009) Predictive chromatin signatures in the mammalian genome. Hum. Mol. Genet., 18(R2), R195–R201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakazawa Y., et al. (2011) The novel metastasis promoter Merm1/Wbscr22 enhances tumor cell survival in the vasculature by suppressing Zac1/p53-dependent apoptosis. Cancer Res., 71, 1146–1155. [DOI] [PubMed] [Google Scholar]
- 47. Fernandez A.F., et al. (2012) A DNA methylation fingerprint of 1628 human samples. Genome Res., 22, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brennan K., et al. (2012) Is there a link between genome-wide hypomethylation in blood and cancer risk? Cancer Prev. Res. (Phila)., 5, 1345–1357. [DOI] [PubMed] [Google Scholar]
- 49. Shukeir N., et al. (2015) Pharmacological methyl group donors block skeletal metastasis in vitro and in vivo. Br J Pharmacol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mas V.R., et al. (2007) Angiogenesis soluble factors as hepatocellular carcinoma noninvasive markers for monitoring hepatitis C virus cirrhotic patients awaiting liver transplantation. Transplantation, 84, 1262–1271. [DOI] [PubMed] [Google Scholar]
- 51. Sengelaub C.A., et al. (2016) PTPRN2 and PLCβ1 promote metastatic breast cancer cell migration through PI(4,5)P2-dependent actin remodeling. EMBO J., 35, 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ilhan A., et al. (2011) Expression of secretagogin in clear-cell renal cell carcinomas is associated with a high metastasis rate. Hum. Pathol., 42, 641–648. [DOI] [PubMed] [Google Scholar]
- 53. Zhang P., et al. (2014) Biological significance and therapeutic implication of resveratrol-inhibited Wnt, Notch and STAT3 signaling in cervical cancer cells. Genes Cancer, 5, 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.