Maize homologs of Arabidopsis Oxs2 can interact with the promoter and activate transcription of a gene encoding a putative SAM-dependent methyltransferase, a new factor for enhanced Cd tolerance.
Abstract
Previously the Arabidopsis (Arabidopsis thaliana) zinc finger protein OXIDATIVE STRESS2 (AtOXS2) and four OXS2-like (AtO2L) family members were described to play a role in stress tolerance and stress escape. For stress escape, SOC1 was a target of AtOXS2. However, for stress tolerance, the downstream targets were not identified. We cloned two OXS2 homolog genes from sweet corn, ZmOXS2b and ZmO2L1. Both genes are transiently inducible by Cd treatment. When expressed in Arabidopsis, each enhances tolerance against cadmium. Further analysis showed that ZmOXS2b and ZmO2L1 proteins enhance Cd tolerance in Arabidopsis by activating at least one target gene, that encoding a putative S-adenosyl-l-Met-dependent methyltransferase superfamily protein (AT5G37990), which we named CIMT1. This activation involves the in vivo interaction with a segment of the CIMT1 promoter that contains a BOXS2 motif previously identified as the binding element for AtOXS2. More importantly, CIMT1 is induced by Cd treatment, and overexpression of this gene alone was sufficient to enhance Cd tolerance in Arabidopsis. The connection of ZmOXS2b and ZmO2L1 to Arabidopsis CIMT1 suggests a similar network may exist in maize (Zea mays) and may provide a clue to possibly using a CIMT1 maize homolog to engineer stress tolerance in a major crop.
Cadmium (Cd) is among the most serious heavy metal pollutants (Nriagu and Pacyna, 1988; Patra et al., 2004) and is readily absorbed by leafy vegetables and the grain crop rice. Chronic dietary intake of Cd is associated with various health problems including cancer and cardiovascular diseases (McLaughlin et al., 1999; Waalkes, 2003; Wagner, 1993). An excess amount of Cd in soil reduces the efficiency of plant photosynthesis, absorption of water, and growth (Küpper et al., 2007; Prasad, 1995; Sandalio et al., 2001). Cd inhibition of root elongation leads to lower uptake and transport of nutrients and water from root to shoot (Chen et al., 2003), and in shoot, Cd leads to reduced leaf conductance, CO2 uptake, and stomatal opening (Perfus-Barbeoch et al., 2002).
At the cellular level, Cd affects DNA repair, DNA methylation, gene transcription, and translation (Hartwig and Schwerdtle, 2002; Takiguchi et al., 2003; Waisberg et al., 2003). Cd can also induce concentration-dependent oxidative stress, and in response, plants use enzymatic and nonenzymatic scavenging mechanisms to maintain cellular function (Mittler et al., 2004; Sytar et al., 2012). Chelation, extrusion, and sequestration are among several mechanisms involved in Cd detoxification (Clemens et al., 1999; Kim et al., 2007; Li et al., 1997; Yadav, 2010).
The Arabidopsis (Arabidopsis thaliana) zinc finger protein OXS2 (OXIDATIVE STRESS2) was previously found to induce stress escape through the direct activation of at least one floral integrator gene, SOC1, by binding the BOXS2 cis-element in its promoter (Blanvillain et al., 2011). Because a loss of function in AtOXS2 is more sensitive to stress, AtOXS2 was also proposed to play a role in alleviating stress tolerance. However, overexpression of AtOXS2 alone could not enhance stress tolerance. The lack of an enhanced tolerance phenotype makes it difficult to use AtOXS2 overexpression lines to identify possible downstream stress-responsive genes. In contrast, we found that overexpression in Arabidopsis of the maize (Zea mays) OXS2 homologs ZmOXS2b and ZmO2L1 (OXS2-Like1) was able to enhance Cd tolerance. Therefore, we describe in this study an RNA-seq transcriptome profiling of Cd-treated Arabidopsis wild type and ZmOXS2b or ZmO2L1 transgenic lines and the identification of a group of differentially expressed genes (DEGs). Of eight highly differentially expressed genes tested, DEG23 (AT5G37990) was the only one that enhanced Cd tolerance in Arabidopsis when overexpressed. DEG23 encodes a root specific putative member of the S-adenosyl-l-Met (SAM)-dependent methyltransferases superfamily, which we named CIMT1 (for Cd-inducible methyltransferase 1). Although CIMT1 can be activated by ZmOXS2b or ZmO2L1, it is not activated by AtOXS2. This may help explain why AtOXS2 overexpression does not enhance Cd tolerance. From a crop improvement perspective, the link of ZmOXS2b and ZmO2L1 to CIMT1 suggests a possible connection of ZmOXS2b and ZmO2L1 to CIMT1 homolog(s) in maize that can potentially be used for engineering stress tolerance in a major crop plant.
RESULTS
OXS2 Homologs Induced by Cd in Maize
An initial BLAST analysis with Arabidopsis OXS2 protein sequence found two homologous proteins in the maize genome. Based on the degree of homology to AtOXS2 (Supplemental Fig. S1A), and consistent with rice OXS2 proteins, we named the two maize homologs ZmOXS2b (ACN25172, 45.4% identity with AtOXS2) and ZmO2L1 (OXS2-Like1, NP_001145979, 42.1% identity with AtOXS2) (http://www.ebi.ac.uk/Tools/psa). In July 2014, a new maize homolog sequence (XP_008665236) was deposited into the database, and we refer to it as ZmOXS2a since it is most closely related to rice OXS2a (Supplemental Fig. S1A). However, since this work started before the discovery of ZmOXS2a, it is not included in this study.
We cloned the protein coding sequences of ZmOXS2b and ZmO2L1 from the genome of the South China sweet corn cultivar FengTian 1 and found a few coding sequence differences from those of the NCBI (Supplemental Table S1). Similar to the AtOXS2 family proteins, both ZmOXS2 homologs have two ANKYRIN repeats and two zinc finger domains (Supplemental Fig. S1B).
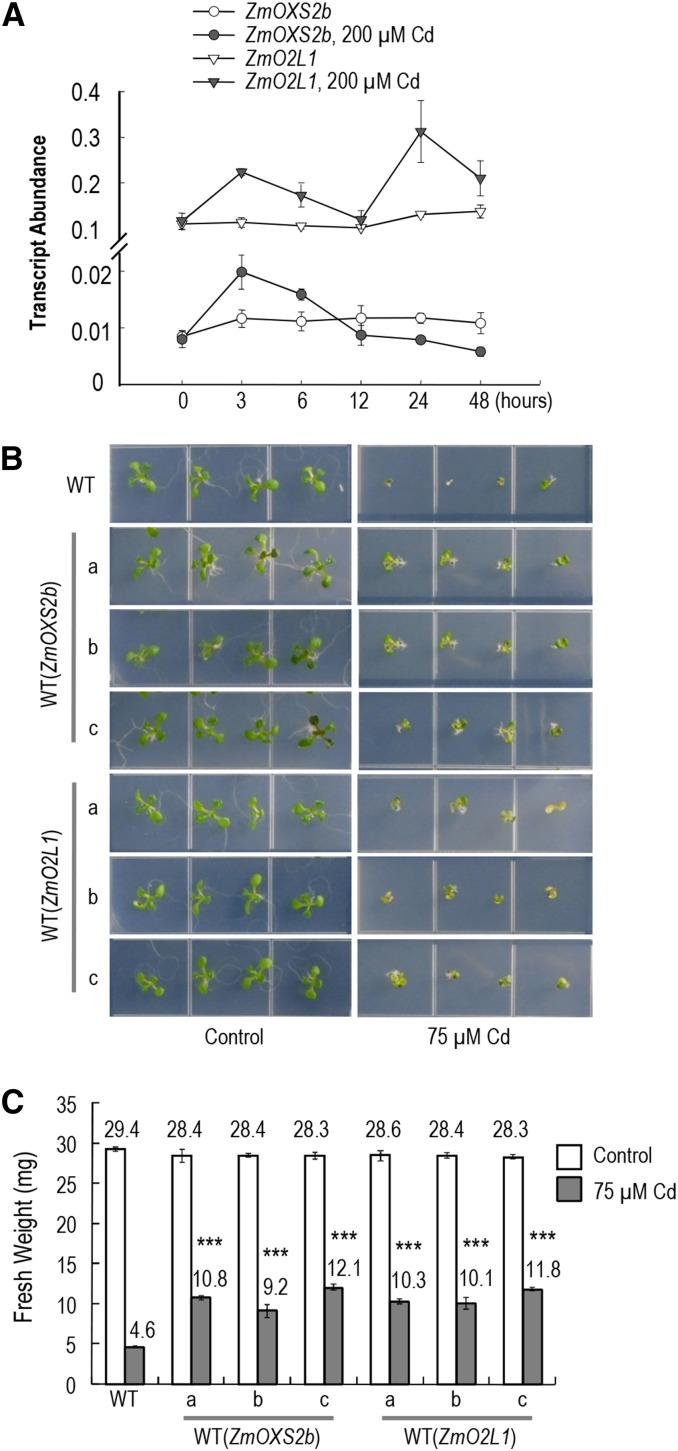
To test whether expression of ZmOXS2b and ZmO2L1 responds to stress, maize seedlings were grown in nutrient solution without or with 200 µM CdCl2. Leaves were collected at different time points after Cd treatment. Quantitative reverse transcription PCR (qRT-PCR) data show that the mRNA abundance for both genes was steady in the absence of Cd and with ZmO2L1 expressing at a higher level (Fig. 1A). In the presence of Cd, expression increased by 2-fold 3 h after treatment, but returned to basal level at the 12-h time point. At the 24- and 48-h time points, ZmO2L1 expression again climbed and dipped, while that of ZmOXS2b gradually dropped to its lowest level. Although the mRNA abundance after extended Cd treatment is difficult to interpret, it is clear that both genes respond to Cd within 3 h with accumulation of mRNA.
Figure 1.
ZmOXS2 family members in stress response. A, ZmOXS2b and ZmO2L1 transcript abundance in maize (relative to EF1-α control) determined by qRT-PCR. Fifteen-day-old maize seedlings exposed to 0 or 200 µm CdCl2. Error bars indicate ±sd from three independent experiments. B, Arabidopsis plants grown on 0.5× MS plates horizontally without or with 75 μm CdCl2 for 11 d. Three independent homozygous lines transgenic for each gene shown. Representative data from three reproducible experiments with T3 seedlings. C, Average fresh weight of 20 T4 seedlings (11 d old) grown on 0.5× MS without or with 75 μm CdCl2 (measured in batches of five seedlings). Error bars indicate ±sd from three independent experiments. P value of Student’s t test: transgenic plants compared with wild-type plants. ***P < 0.001
ZmOXS2b and ZmO2L1 Enhance Cd Tolerance in Arabidopsis
ZmOXS2b or ZmO2L1 was fused behind the Cauliflower mosaic virus 35S RNA promoter in a binary vector to yield p35S::ZmOXS2b or p35S::ZmO2L1, respectively, for transformation into Arabidopsis (cv Columbia). In addition, p35S::ZmOXS2b was also transformed into mutant oxs2-1 and p35S::ZmO2L1 into mutant o2l1-1. Approximately 10 independent transgenic lines were obtained for each transformation. Unlike overexpression of AtOXS2 that caused early flowering (Blanvillain et al., 2011), we did not observe early flowering with the four classes of transgenic lines: WT(ZmOXS2b), WT(ZmO2L1), oxs2-1(ZmOXS2b), and o2l1-1 (ZmO2L1). Three independent lines from each class were randomly selected and homozygous plants obtained for further analysis. RT-PCR verified the expression of ZmOXS2b and ZmO2L1 and the transgenic plants were compared to the wild type control against several types of stresses: Cd (75 µm), diamide (1 and 2 mm), NaCl (100 and 150 mm), mannitol (100 and 200 mm), abscisic acid (1.5 and 3 µm), heat (37°C, 3 h), and cold (4°C, 3 h). In the absence of stress, or for the tested stress conditions other than Cd, the phenotype of the transgenic plants was indistinguishable from that of the untransformed control. In Cd (75 µm), the root length, shoot growth, and biomass of WT(ZmOXS2b) and WT(ZmO2L1) plants were greater than the untransformed controls (Fig. 1, B and C; Supplemental Fig. S2A). Likewise, the root length and shoot growth of oxs2-1(ZmOXS2b) and o2l1-1(ZmO2L1) plants were also greater than oxs2-1 and the wild type (Supplemental Fig. S2, B and C).
Differentially Regulated Genes Identified from RNA-Seq Analysis
The Cd tolerance phenotype associated with constitutive expression of ZmOXS2b or ZmO2L1 is likely due to a gene expression change caused by the putative transcription factors. As AtOXS2 is known to activate promoters of some flowering genes (Blanvillain et al., 2011), it would seem plausible that ZmOXS2b and ZmO2L1 may recognize heterologous promoters from a similar protein-structure/DNA sequence interaction. To examine if the gene expression pattern has changed, an RNA-seq analysis was conducted on Cd-treated plants comparing WT(ZmOXS2b) and WT(ZmO2L1) against the wild-type control.
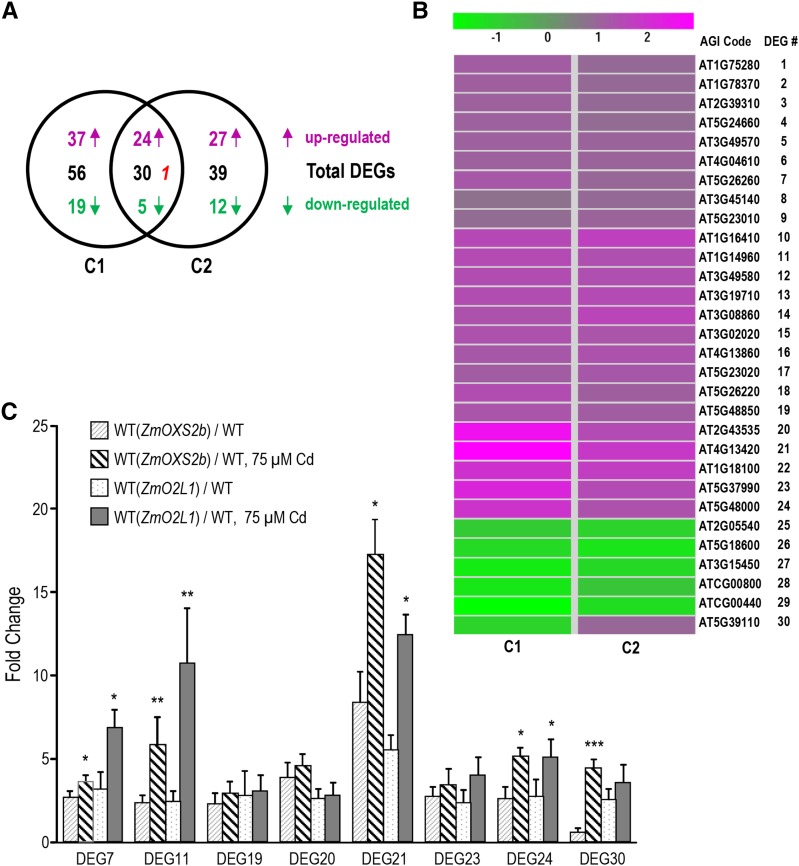
The expression profile of WT(ZmOXS2b) compared to the wild-type control is hereafter referred to as comparison 1 or C1; likewise, WT(ZmO2L1) compared to the wild type is C2. DEGs with statistically significant change (up-regulated by at least 1.67-fold or down-regulated by at least 0.59-fold, NOISeq Q value over 0.8; Tarazona et al., 2011) in either C1 or C2 were selected (Supplemental Table S2), consisting of 86 DEGs in C1 and 69 DEGs in C2. However, since 30 of them are common to both C1 and C2 (Fig. 2A), the total number of DEGs is 125. Gene Ontology analysis shows that many of these DEGs are involved in oxidoreductase activity, arsenate reductase activity, and iron ion binding (Supplemental Table S3). In C1, 61 DEGs are up-regulated and 25 down-regulated in WT(ZmOXS2b). Thirty-eight of these 86 DEGs are listed as involved in stress response, defense response, or metal ion transport (Supplemental Table S2). In C2, 52 DEGs are up-regulated and 17 down-regulated in WT(ZmO2L1). Twenty-eight of these 69 DEGs in C2 are listed as involved in stress responses, defense response, or metal ion transport (Supplemental Table S2). Given that 30 DEGs are common to C1 and C2, we narrowed the focus to this smaller group. The 30 members were named DEG1 to DEG30 from top to bottom in the clustering analysis map (Fig. 2B). They behave similarly in both transgenic lines except that DEG30 (AT5G39110) was down-regulated in WT(ZmOXS2b) but up-regulated in WT(ZmO2L1) (Fig. 2B). Among these 30 DEGs, 14 are annotated to be stress related or metal ion transport genes (Supplemental Table S2).
Figure 2.
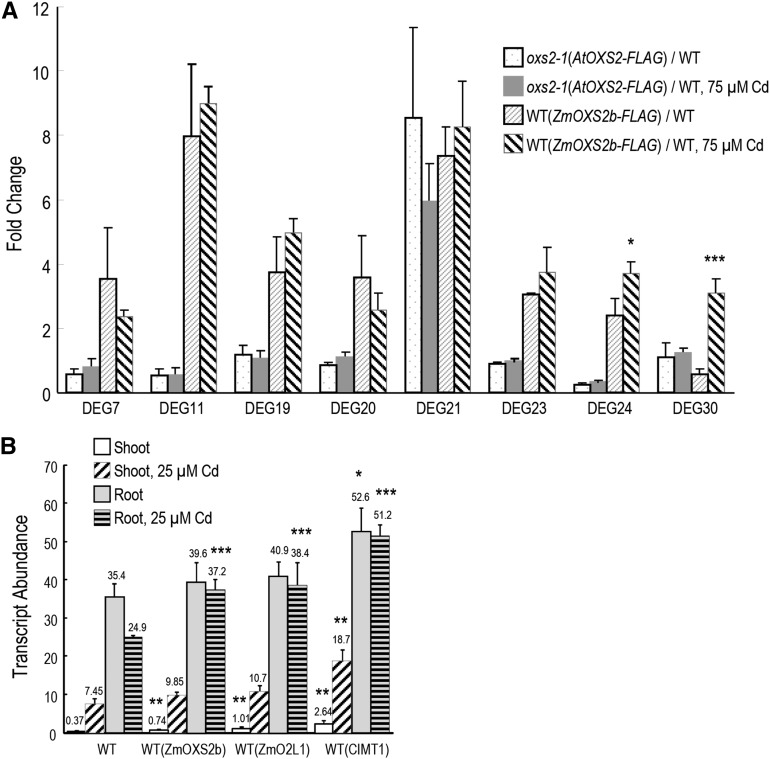
RNA-seq analysis and qRT-PCR verification. A, Venn diagram of DEGs. C1 = WT(ZmOXS2b) versus the wild type; C2 = WT(ZmO2L1) versus the wild type. Total numbers of DEGs; up- and down-regulated DEGs shown in magenta and green lettering, respectively. Red italic lettering indicates one gene (DEG30) down-regulated in C1 but up-regulated in C2. B, Heat map of clustering analysis of the 30 DEGs in the intersection of C1 and C2. Expression ratios shown as log2 values. Magenta represents increased expression; green represents decreased expression compared to control. C, Expression patterns of eight DEG candidates. Vertical axis shows fold enrichment of relative transcript levels between transgenic and wild-type plants. Data obtained with T3 generation seedlings. Error bars represent ± sd from three independent experiments. P value of Student’s t test: stressed plants compared with unstressed plants. *P < 0.05; **P < 0.01; ***P < 0.001
Validation of Differential Expression
To verify the expression pattern of the RNA-seq analysis, qRT-PCR was conducted on the 30 common DEGs with the same tissues used for RNA-seq. The expression pattern obtained was similar to the RNA-seq data for most of the genes except the fold change differed in a few instances (Supplemental Table S4). In the qRT-PCR ranking, eight DEGs among the top 10 DEGs that show the most dramatic change in gene expression (3.6- to 13.9-fold; Supplemental Table S4) are common in C1 and C2. They are DEG7 (AT5G26260), DEG11 (AT1G14960), DEG19 (AT5G48850), DEG20 (AT2G43535), DEG21 (AT4G13420), DEG23 (AT5G37990), DEG24 (AT5G48000), and DEG30 (AT5G39110).
WT(ZmOXS2b), WT(ZmO2L1), and wild-type control plants were again cultured on 0.5× Murashige and Skoog media without or with 75 μm CdCl2 for 11 d for qRT-PCR analysis of these eight specific DEGs. Even in the absence of Cd treatment, all but DEG30 in WT(ZmOXS2b) were up-regulated in the transgenic lines (Fig. 2C). All were induced by Cd treatment in the wild-type control (Supplemental Fig. S3), but the induction is more significant in the WT(ZmOXS2b) and WT(ZmO2L1) plants (Fig. 2C). This shows that ZmOXS2b and ZmO2L1 have further elevated the expression of at least seven genes that are normally up-regulated by Cd in Arabidopsis.
ZmOXS2b and ZmO2L1 Activate BOXS2-Containing Promoters
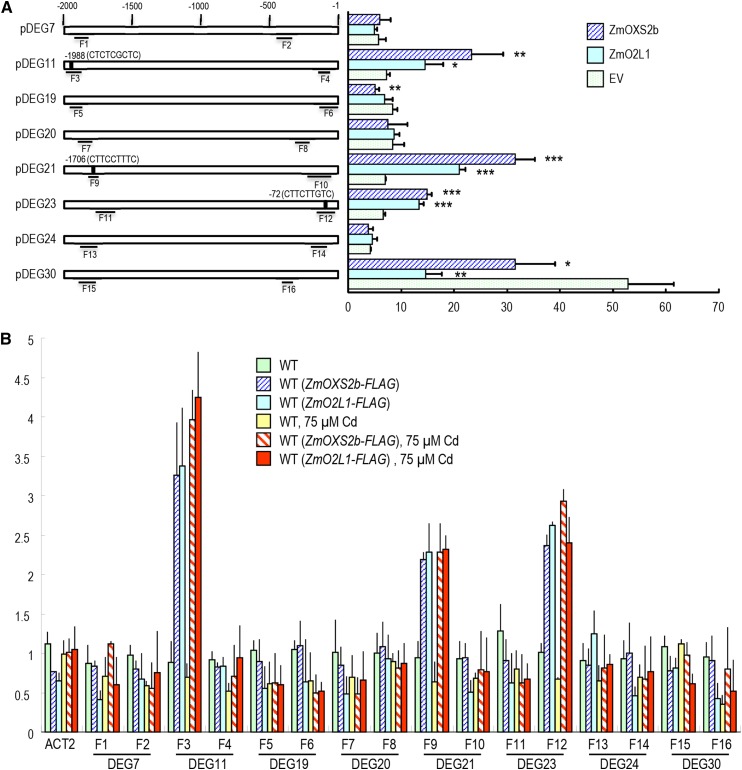
AtOXS2 has been shown previously to bind a 9-bp CT-rich motif named BOXS2 and that it can activate several BOXS2-containing promoters (Blanvillain et al., 2011). Among these eight genes, the promoters of DEG11, DEG21, and DEG23, but not the other five DEGs, contain sequences similar to a BOXS2 motif (Fig. 3A). We fused the firefly luciferase reporter gene (luc) to a 2-kb promoter including its 5′ untranslated region (UTR) fragment from each of the eight DEG candidates to generate all eight promoter-luc fusions. Each of these constructs was transiently introduced into tobacco leaf tissue by agroinfiltration, along with p35S::ZmOXS2b, p35S::ZmO2L1, or an empty vector control. In this transactivation assay, enhanced expression of pDEG-luc by p35S::ZmOXS2b and p35S::ZmO2L1 was seen only with the three BOXS2-containing constructs (Fig. 3A). None of the other five DEG promoters were affected, except that the DEG30 promoter showed repression by p35S::ZmOXS2b and p35S::ZmO2L1.
Figure 3.
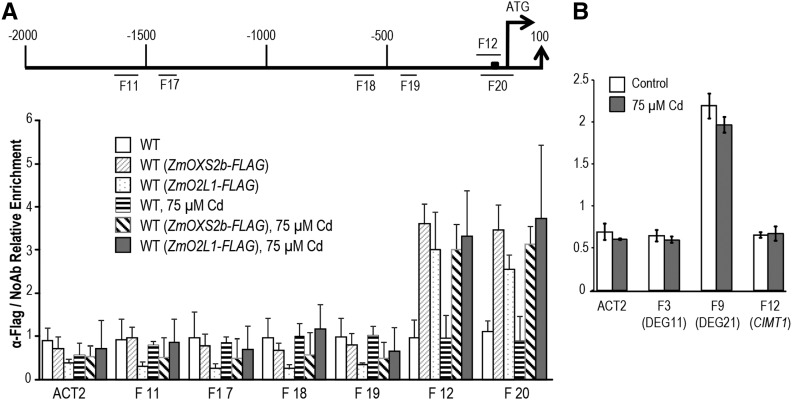
ZmOXS2b and ZmO2L1 activate and interact with BOXS2-containing promoters. A, ZmOXS2b and ZmO2L1 activation of DEG promoters (promoter fragment include 5′ UTR) determined by infiltration mediated transient expression assay. x axis is the ratio of LUC to rLUC activity 2 d after infiltration. Black boxes in promoters indicate putative BOXS2 motifs. Numbers indicate position of starting nucleotide of each BOXS2 relative to translation start. Error bars show ±sd from three independent experiments. P value of Student’s t test: ZmOXS2b or ZmO2L1 compared with empty vector. *P < 0.05; **P < 0.01; ***P < 0.001. B, ZmOXS2b and ZmO2L1 interact with BOXS2-containing promoters DEG11, DEG21, and DEG23. ChIP-qPCR to test in vivo interaction of promoters (including 5′ UTR) with ZmOXS2b or ZmO2L1 in tissues from the wild type, WT(ZmOXS2b-FLAG), and WT(ZmO2L1-FLAG) treated with or without 75 μm Cd. Promoter or 5′ UTR segments tested are labeled F1-F16 (shown in A). CP (crossing point) value of immunoprecipitated DNA fractions with α-FLAG or no antibody control (NoAb) normalized to CP value of input DNA fractions for the same qPCR assay. y axis is the ChIP signals calculated as the enrichment relative to the no antibody control (No Ab). Error bars indicate ±sd from three independent experiments on T2 seedlings.
A chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analysis was performed to test the in vivo interaction of these promoters in transgenic wild-type Arabidopsis producing FLAG-tagged ZmOXS2b or ZmO2L1, which showed similar Cd tolerance as the non-FLAG-tagged lines (data not shown). Following immunoprecipitation with anti-FLAG antibody, two pairs of primers were used for each promoter corresponding to fragments F1-F16 (Fig. 3, A and B). Positive interaction for ZmOXS2b and ZmO2L1 was found for F3, F9, and F12 (Fig. 3, A and B), but not for the other fragments, including the ACT2 (At3g18780) promoter used as negative control. Interestingly, only these three fragments span across DNA segments that encompass the BOXS2 element. It is tempting to suggest that ZmOXS2b and ZmO2L1 bind the BOXS2 element, as was found with AtOXS2.
Overexpression of DEG23 Enhances Cd Tolerance in Arabidopsis
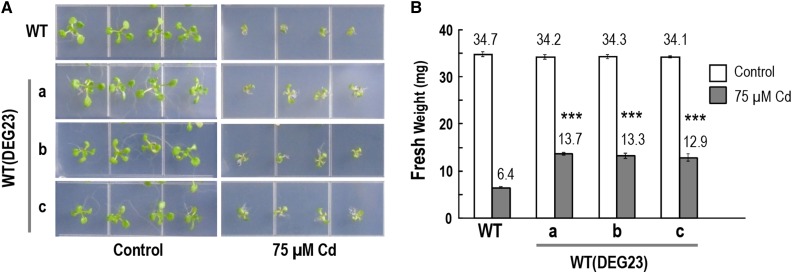
Each of the eight DEG candidates was fused to the Cauliflower mosaic virus 35S RNA promoter for expression in Arabidopsis. Approximately 10 independent transgenic lines were generated for each candidate. None showed an aberrant phenotype that differs from wild-type plants under normal growth conditions. Three independent lines of each genotype were randomly selected for testing. Under normal conditions, qRT-PCR analysis found 4- to 33-fold overexpression in these T2 generation lines compared to the wild-type control (Supplemental Fig. S4). When tested on Cd (75 µm), only plants overexpressing DEG23 (AT5G37990) grew stronger with more biomass than the wild-type control (Fig. 4, A and B; Supplemental Fig. S2D). However, the 75 µm concentration of Cd in synthetic media is too high for culturing the plants beyond the small plantlet stage. Therefore, we grew plants in soil for 3 weeks and treated them with 75 µm Cd for another week before harvest for Cd measurement. However, WT(DEG23) did not show a statistically significant difference in Cd content compared to wild-type, oxs2-1, o2l1-1, WT(ZmOXS2b), or WT(ZmO2L1) (Supplemental Fig. S5A). Previously, Huang et al. (2012) reported differences in Cd accumulation between wild type and transgenic Arabidopsis when grown in soil with 5 µm Cd. We also tried this same treatment, but as before, a difference in Cd accumulation in root, shoot, or seed from the wild-type level was not found among WT(DEG23), WT(ZmOXS2b), and WT(ZmO2L1) (Supplemental Fig. S5B). It does not appear that Cd tolerance mediated by DEG23, ZmOXS2b, and ZmO2L1 is due to a difference in Cd accumulation.
Figure 4.
Overexpression of DEG23 (CIMT1) confers Cd Tolerance in Arabidopsis. A, Plants grown on 0.5× MS plates horizontally without or with 75 μm CdCl2 for 12 d. Three independent transgenic lines shown. Representative data from three independent experiments on T2 seedlings. B, Average fresh weight of 20 seedlings (12 d old) grown on 0.5× MS without or with 75 μm CdCl2 (measured in batches of five seedlings). Error bars indicate ±sd from three independent experiments. P value of Student’s t test: transgenic plants compared with wild-type plants. ***P < 0.001.
The fact that overexpression of DEG23 on its own is sufficient to enhance Cd tolerance indicates that this gene plays a role in ameliorating the toxic effects of Cd. Since DEG23 encodes a putative member of the superfamily of S-adenosyl-l-Met-dependent methyltransferases, we named the gene CIMT1 (Cd-Inducible Methyltransferase 1).
ZmOXS2 Family and AtOXS2 Interacts with Distinct and Overlapping BOXS2-Containing Promoters
To explore in greater detail of the interaction between the CIMT1 promoter and ZmOXS2b or ZmO2L1, four more pairs of primers were used against the CIMT1 (DEG23) promoter corresponding to fragments F17-F20 (Fig. 5A). Again, ChIP-qPCR showed positive signal for ZmOXS2b or ZmO2L1 interaction only with the BOXS2-containing fragments F12 and F20, and this interaction is independent on Cd treatment (Fig. 5A).
Figure 5.
Only one BOXS2-containing promoter interacts with all three proteins: ZmOXS2b, ZmO2L1, and AtOXS2. ChIP with FLAG-tagged ZmOXS2b, ZmO2L1, or AtOXS2 performed on tissues from plants treated without or with 75 μm Cd. Primers against ACT2 promoter used as negative control. A, ChIP with FLAG-tagged ZmOXS2b or ZmO2L1. F11, F17, F18, F19, F12, and F20 indicate CIMT1 (DEG23) promoter segments tested with qPCR. Black box indicates putative BOXS2 motif CTTCTTGTC. Numbers indicate position of starting nucleotide of BOXS2 relative to translation start. B, ChIP with FLAG-tagged AtOXS2 shows binding to BOXS2-containing fragment F9 in DEG21 promoter, but not those within F3 of DEG11 or F12 of CIMT1 (see Fig. 3A). Data show average ±sd of three independent experiments on T2 seedlings.
Previously, we reported that AtOXS2 activates BOXS2-containing promoters but overexpression of AtOXS2 did not confer Cd tolerance (Blanvillain et al., 2011). To examine whether AtOXS2 might interact with the three BOXS2-containing promoters (F3 in DEG11 promoter, F9 in DEG21 promoter, and F12 in CIMT1 promoter; Fig. 5B) that showed interaction with ZmOXS2b and ZmO2L1, a ChIP-qPCR was carried out with the oxs2-1 mutant transgenic for a 35S promoter-driven AtOXS2:FLAG construct. As shown in Figure 5B, interaction was not detected with fragments F3 or F12, but with F9, and the interaction was not affected by Cd stress (Fig. 5B). This suggests that DEG21 could be a target of AtOXS2. Indeed, expression of DEG21, but not the other DEGs, was elevated by overexpression of AtOXS2 (Fig. 6A). However, since overexpression of DEG21 alone could not enhance Cd tolerance, it is consistent with the previous finding that AtOXS2 overexpression could not enhance Cd tolerance (Blanvillain et al., 2011). The lack of AtOXS2 recognition of the CIMT1 F12 fragment is also consistent with the data that AtOXS2 could not elevate expression of CIMT1 (Fig. 6A).
Figure 6.
Expression pattern of DEGs and organ-specific analysis of CIMT1. Transcript abundance in Arabidopsis is determined by qRT-PCR. A, Whole seedlings grown for 11 d without or with 75 µm CdCl2 were assayed for fold change of DEG transcription between transgenic plants over wild-type control. P value of Student’s t test: stressed plants compared with unstressed plants. B, Shoot and root expression of CIMT1 (relative to ACT1 control) in 11-d-old Arabidopsis seedlings exposed to 0 or 25 µm CdCl2; T5 for WT(ZmOXS2b) and WT(ZmO2L1) and T3 for WT(CIMT1). Error bars indicate ±sd from three independent experiments. P value of Student’s t test: WT(ZmOXS2b), WT(ZmO2L1), and WT(CIMT1) compared with the wild type. *P < 0.05; **P < 0.01; ***P < 0.001.
CIMT1 Expression Is Cd Inducible Only in Shoot
To check the tissue-specific expression of CIMT1, the plant lines WT(ZmOXS2), WT(ZmO2L1), WT(CIMT1), and wild-type control were grown without or with CdCl2. With 75 µm Cd, the root lengths were rather short, especially with the wild-type control, and this made it difficult to separate the roots from the shoots. Hence, we grew the plants at 25 µm Cd. The wild-type control shows that Cd up-regulates CIMT1 mRNA abundance (∼20×) in shoot but down-regulates its abundance (∼30%) in root (Fig. 6B). WT(ZmOXS2), WT(ZmO2L1), and WT(CIMT1) all showed a similar pattern of regulation, although compared to the wild type, higher mRNA abundance is found in the absence or presence of Cd treatment. This suggests that overexpression did not alter the overall pattern, but strengthened transcriptional output. Given that CIMT1 mRNA in wild-type plants is down-regulated in root in response to Cd, it is possible that the higher root mRNA abundance in the transgenic plants may not be physiologically relevant, as compared to the elevated shoot transcript level, which were up-regulated by as much as 2.5-fold in WT(CIMT1). Alternatively, or additionally, it is possible that the higher shoot CIMT1 transcript abundance in the absence of Cd might prime the plant for a more robust stress response.
DISCUSSION
Previous research in Arabidopsis has led to our current model on OXS2 regulation of oxidative stress (Blanvillain et al., 2011). In this working model, stress induces OXS2 translocation from the cytoplasm to the nucleus. Nuclear OXS2 along with other family members activate the stress tolerance pathway to alleviate the stress. At a higher stress level, nuclear OXS2 autoactivates its own promoter to produce higher levels of the transcription factor commensurate to the stress challenge. Upon relief of stress, OXS2 accumulates in the cytoplasm where it is needed to resume vegetable growth. In the case of a very high level of stress, where tolerance pathways are unable to cope with the damage, nuclear OXS2 activates the stress escape response, inducing reproduction to insure survival of the species over that of the individual. Much has been learned of the molecular details of this stress escape pathway. In transactivation assays, OXS2 has been shown to activate several flowering genes: the floral integrators SOC1 and LFY, and the floral identity gene AP1. In ChIP-qPCR assays, OXS2 binds to the SOC1 promoter and to a fragment that encompasses a BOXS2 motif. Mutation analysis shows that stress-induced expression of SOC1 depends on OXS2 and other family members.
In contrast, not much is known on how OXS2 activates the stress tolerance pathway. Presumably, there are target genes that OXS2 acts on, and a goal of this study was to find these stress tolerance pathway targets. The most logical route is to continue with the research on the Arabidopsis OXS2 family of proteins. However, since we were eager to embrace translational research to a major crop plant, we isolated the two maize homologs. Identifying target genes in maize would be the ultimate goal, but since generating transgenic maize takes much greater effort, we regressed back to using transgenic Arabidopsis for a first analysis. Given that OXS2 family members of Arabidopsis, maize, and rice (Oryza sativa) all can enhance stress tolerance in the heterologous organism Schizosaccharomyces pombe (data not shown), we considered it possible that all of them recognize similar target genes in evolutionarily conserved stress tolerance pathways.
Indeed, the RNA-seq analysis of overexpressed maize OXS2 genes revealed a number of DEGs in the Arabidopsis transgenic lines, among which at least 88 are up-regulated. Finding many genes is consistent with an expectation that stress tolerance requires the activity of numerous proteins to alleviate the stress challenge. It would also justify the need for upstream regulators to orchestrate their expression. Therefore, it was quite surprising to find that expression of a single target gene, CIMT1, was sufficient to enhance Cd tolerance to a level similar to that from expression of ZmOXS2b or ZmO2L1. This effect may be due to the much higher dosage of expression. In ZmOXS2b and ZmO2L1 expressing lines, CIMT1 was activated by as much as 5-fold (Fig. 2C; Supplemental Table S4), whereas in the CIMT1 overexpression lines, this gene transcript was elevated by 14- to 17-fold (Supplemental Fig. S4).
As shown by ChIP-qPCR, CIMT1 is indeed a target of ZmOXS2b and ZmO2L1. Moreover, these proteins interact with a segment of the CIMT1 promoter that contains a BOXS2 motif. However, a follow-up ChIP-qPCR experiment failed to show AtOXS2 interaction with this promoter (Fig. 5B), nor was CIMT1 mRNA up-regulated in the AtOXS2-overexpressing line (Fig. 6A). This at least partially explains why overexpressing AtOXS2 fails to confer Cd tolerance in Arabidopsis, since it cannot activate this gene. However, this does not rule out CIMT1 as a target of other members of the AtOXS2 family.
CIMT1 is a root-specific protein under normal growth condition (Baerenfaller et al., 2008), but its shoot mRNA can be induced by Cd treatment (Fig. 6B). CIMT1 is one of 25 SAM-dependent methyltransferases in Arabidopsis. Highly conserved in all branches of life, SAM-dependent methyltransferases catalyze the transfer of methyl groups from SAM to a broad range of substrates including DNA, proteins, and small metabolites. In plants, stress hormones such as salicylic acid and jasmonic acid are both substrates of these enzymes (Lee et al., 2007; Seo et al., 2001). With a Rossmann-like superfold, CIMT1 belongs to the Class I methyltransferases, the largest group of methyltransferases catalyzing the majority of methylation reactions in all kinds of organisms. As far as we are aware, this particular member has not been functionally characterized in Arabidopsis. Hence, this research provides direct evidence of its involvement in plant heavy metal tolerance. We presume that a similar homolog would exist in maize. However, with over 20 homologs in the database, some effort is needed to sort out which maize homolog(s) might play a similar role in response to Cd stress. Perhaps we could follow the clue that its promoter would likely be regulated by ZmOXS2b and ZmO2L1, and thus through ChIP-qPCR with these proteins, we may be able to fish out the true ortholog(s).
As reported previously (Blanvillain et al., 2011), subcellular localization patterns differ among members of AtOXS2 family proteins. Only AtOXS2 and AtO2L1 translocate from the cytoplasmic to the nucleus during stress. AtO2L2 is largely cytoplasmic, while AtO2L3 and AtO2L4 are mainly in the nucleus. As for the subcellular localization of ZmOXS2b, ZmO2L1, and AtCIMT1, a protoplast transient expression assay of GFP-fused proteins in Arabidopsis showed that all three proteins were found in both the cytoplasm and the nucleus, and this pattern is not altered by Cd treatment (Supplemental Fig. S6). Nuclear ZmOXS2b and ZmO2L1 fit their role in transcription, but whether there is also a cytoplasmic role is not clear. For AtCIMT1, as a putative methyl transferase, its subcellular localization pattern could raise the possibility of substrate(s) in both the nucleus and cytoplasm.
To conclude, we have shown that maize OXS2 family proteins can enhance Cd tolerance in Arabidopsis, resulting in the differential expression of a large number of genes. From this large collection of leads, we have at least narrowed down one gene, CIMT1, as a target of the maize transcription factors. More importantly, from a biotechnology point of view, its expression alone can enhance Cd tolerance. This redirects our focus to finding a similar CIMT1 in maize that may someday be engineered for higher stress tolerance in a major crop.
MATERIALS AND METHODS
Plant Treatments
Arabidopsis (Arabidopsis thaliana) wild-type Col-0 (SALK_6000) and mutants oxs2-1 (SALK_037470) and o2l1-1 (SALK_020612) have been described (Blanvillain et al., 2011). Arabidopsis plants were grown in a controlled environment at 22°C in a 16-h-light/8-h-dark photoperiod.
Plant transgene expression constructs were transformed into Agrobacterium tumefaciens strain GV3101 by the floral dip procedure. Transgenic plants were selected with phosphinothricin (10 µg/mL) or hygromycin (50 µg/mL). Seeds used for phenotypic assays were harvested at the same time. The growth assay was performed on plates with 0.5× MS solid media without or with diamide (1 and 2 mm), NaCl (100 and 150 mm), Cd (25 and 75 µm), mannitol (100 and 200 mm), and abscisic acid (1.5 and 3.0 µm). Heat shock treatment was conducted in a 37°C chamber for 3 h and cold treatment at 4°C for 3 h with 10-d-old plants.
Sweet corn (cv FengTian1) was germinated (5 d in water, 4 d without water) and grown in MS hydroponic cultures with 30 min aeration twice per day in a controlled environment at 22°C with a 16-h-light/8-h-dark photoperiod. After 5 d of growth in hydroponic culture, plantlets were placed into fresh culture without or with 200 µm Cd. Leaf samples were collected at 0, 3, 6, 12, 24, and 48 h. Three leaves from independent plants were mixed as one sample, flash frozen with liquid nitrogen, and stored at −80°C.
For Cd accumulation measurement, Arabidopsis plants were grown in soil for 3 weeks (for 75 µm CdCl2 treatment) and 4 weeks (for 5 µm CdCl2 treatment) and then bottom flooded once with 0.4 liters per pot (pot size 0.4 liters) of CdCl2 with indicated concentrations of CdCl2 solution followed by normal watering without Cd. Whole seedlings were collected 1 week after 75 µm CdCl2 treatment. As for plants treated with 5 µm CdCl2 shoots, roots and seeds were collected for sampling. Cd2+ content was measured with the 7700X ICP-MS (Agilent).
Protoplast of Arabidopsis were prepared and transfected according to standard protocol by Dr. Jen Sheen’s lab (http://molbio.mgh.harvard.edu/sheenweb/protocols_reg.html).
Molecular Constructs
For expression constructs ZmOXS2b and ZmO2L1, DNA was PCR amplified from sweet corn (cv FengTian1) genomic DNA with primer sets ZmOXS2b-1F/ZmOXS2b-1R and ZmO2L1-1F/ZmO2L1-1R, respectively (all primers listed in Supplemental Table S5). Seven of the eight DEGs were PCR cloned with one pair of primers against Arabidopsis genomic DNA, with 5′ and 3′ UTRs included. For DEG21 (AT4G13420), which is over 5,500 bp (genomic DNA, UTRs included), we divided the PCR amplification into three parts and fused them together using In-Fusion HD cloning kit (catalog no. 011614; Clontech). All primers were designed according to information from the TAIR website (http://www.arabidopsis.org). Each was inserted into the XbaΙ site of binary vector pCambia3300 (http://www.cambia.org) to yield p35S::ZmOXS2b, p35S::ZmO2L1, and p35S::DEGs. For binary vector with FLAG, ZmOXS2b and ZmO2L1 open reading frame (ORF) fragments without stop codon were PCR amplified with primers ZmOXS2b-2F and ZmOXS2b-2R or ZmO2L1-2F and ZmO2L1-2R. The fragments were cloned into binary vector pCambia1305 that contains a FLAG tag after the XbaI site. To make the 35S::AtOXS2-FLAG construct, AtOXS2 coding region without its stop codon was PCR amplified from genomic DNA with primers AtOXS2-F and AtOXS2-R and inserted between KpnI and PstI sites on pCambia1305. Promoters including 5′ UTR of the eight DEG candidates (2,000 bp upstream of ATG) were PCR cloned and fused to a firefly luciferase ORF. Likewise, a double enhancer 35S promoter was fused to the Renilla luciferase ORF. Each of the luciferase fusions was inserted into the XbaΙ site of pCambia3300 using In-Fusion HD cloning kit (catalog no. 011614; Clontech).
For subcellular localization vectors, ZmOXS2b and ZmO2L1 ORF fragments without stop codon were PCR amplified with primers ZmOXS2b-3F and ZmOXS2b-3R or ZmO2L1-3F and ZmO2L1-3R. ZmOXS2b and ZmO2L1 were inserted between KpnI and SpeI or XbaI and BglII in pGFP vectors, respectively. CIMT1 ORF without stop codon was PCR amplified with primers CIMT1 F and CIMT1 R and cloned into SacI and BamHI in frame with eGFP in vector pCambia3300.
qRT-PCR
RNA extraction was conducted using a plant RNA kit (catalog no. R5105; GBCBIO Technologies). Reverse transcription was conducted using PrimeScript RT reagent kit with gDNA Eraser (catalog no. RR047A; TaKaRa). qPCR was conducted with SYBR Premix Ex Taq (catalog no. DRR820A; TaKaRa) on LightCycler 480 II (Roche). Maize (Zea mays) elongation factor 1-α (EF1-α; NM_001112117) and Arabidopsis ACTIN1 (ACT1, AT2G37620) were used as internal controls.
Transactivation Assay
Nicotiana benthamiana plants were grown in soil in a controlled environment at 28°C, with a 14-h-light/10-h-dark photoperiod. Infiltration was done on 5- to 6-week-old plants. Single clones of GV3101 carrying different vectors were inoculated to LB medium containing 10 μg/mL rifampicin and 50 μg/mL kanamycin and grown for more than 24 h at 28°C. One hundred microliters of near-saturation Agrobacterium tumefaciens was inoculated to 5 mL fresh LB medium containing 10 μg/mL rifampicin, 50 μg/mL kanamycin, 10 mm MES (pH 5.6) buffer, and 20 μm acetosyringone and grown for more than 8 h at 28°C. Cells were collected by centrifugation (4,000 rpm, 10 min), resuspended to an OD600 of 0.8 infiltration medium (10 mm MgSO4, 200 μm acetosyringone, and 10 mm MES), and incubated at room temperature for 3 h. Infiltration medium contains three A. tumefaciens strains: 1 mL of transcription activator strain (p35S::ZmOXS2b, p35S::ZmO2L1, or empty vector), 100 µL promoter strain (DEG promoter-luc), and 5 µL reference strain (35S-rLUC). Infiltration carried out with healthy N. benthamiana leaves using a 1-mL syringe without needle. After infiltration, plants were kept in a dark chamber with high humidity for one night and then put back to a normal growth room for 2 d. Luciferase values measured with Dual-Luciferase Reporter Assay System (catalog no. E1910; Promega).
Chromatin Immunoprecipitation
Fresh tissue (1 g, whole seedlings) was infiltrated in 25 mL of 1% formaldehyde MC buffer (10 mm potassium phosphate, pH 7.0, 50 mm NaCl, and 0.1 m Suc) for 1 h at 20 p.s.i. on ice and placed on ice for another 1 h. The reaction was stopped by adding Gly powder to 0.15 m final concentration and incubated at 4°C for 40 min. Samples were washed twice with MC buffer at 4°C for 40 min. Tissues were ground in a cold mortar to make a relatively thick slurry with M1 buffer (10 mm potassium phosphate, pH 7.0, 0.1 m NaCl, 10 mm β-mercaptoethanol, 1 m hexylene glycol, and 1 mm PMSF), centrifuged at top speed at 4°C for 3 min, and the pellet washed thoroughly four times with 1 mL M2 buffer (10 mm potassium phosphate, pH 7.0, 0.1 m NaCl, 10 mm β-mercaptoethanol, 1 m hexylene glycol, 10 mm MgCl2, and 0.5% Triton X-100) supplemented with protease inhibitor cocktail (catalog no. 78410; Thermo), then washed with 1 mL M3 buffer (10 mm potassium phosphate, pH 7.0, 0.1 m NaCl, and 10 mm β-mercaptoethanol). The pellet was resuspended in 0.75 mL of sonication buffer (10 mm potassium phosphate, pH 7.0, 0.1 m NaCl, 10 mm EDTA, pH 8.0, and 0.5% sarkosyl) and vortexed for 30 s. Chromatin complexes were sonicated to DNA fragment sizes 250 to 500 bp (cycles of 5-s sonication/5-s pause, 9 min) and centrifuged at top speed for 5 min at 4°C. Chromatin supernatants were kept in IP buffer (50 mm HEPES, pH 7.5, 150 mm KCl, 5 mm MgCl2, 1% Triton X-100, and 0.05% SDS) and incubated without or with anti-FLAG (1:500) and Dynabeads Protein A (catalog no. 10002D; Life Technologies)/protein G (catalog no. 10004D; Life Technologies) mixtures (1:1) overnight at 4°C with gentle agitation. After several rounds of washing with IP buffer, the pellets were eluted using elution buffer (50 mm Tris, pH 8.0, 1% SDS, and 10 mm EDTA) followed by reverse cross-link and purification. DNA fragments were quantified by qPCR with SYBR Premix ExTaq Mix (catalog no. DRR820A; TaKaRa). The CP (crossing point) value of immunoprecipitated DNA fractions with α-FLAG or no antibody control (NoAb) normalized to CP value of input DNA fractions for the same qPCR assay. The ChIP signals were calculated as the relative enrichment in signal relative to the NoAb control. All experiments were repeated three times.
RNA-Seq Library Construction and Sequencing
The wild type, WT(ZmOXS2b), and WT(ZmO2L1) were grown on 0.5× MS plates supplemented with 75 μm CdCl2 for 11 d. Whole seedlings were collected and sent to BGI-Tech for RNA-seq analysis. Total RNA isolation, library construction, sequencing, and basic data analysis were carried out by BGI-Tech.
Screening of DEGs and Expression Pattern Analysis of DEGs (Heat Map)
Expression level calculated as RPKM (reads per kilobase per million reads) according to Mortazavi et al. (2008) was compared between transgenic plants (with either ZmOXS2b or ZmO2L1) and wild-type control. NOISeq approach was used to evaluate the significance of the gene expression differences (Tarazona et al., 2011). Clustering software was used to perform cluster analysis of gene expression patterns (de Hoon et al., 2004). Assessment of RNA-seq quality, screening of DEGs, and expression pattern analysis of DEGs were carried out by BGI-Tech.
Other Customized Data Analysis for RNA-Seq
Gene Ontology analysis was carried out by BGI-Tech. BOXS2 analysis in Arabidopsis genome was conducted by GENE DENOVO with transcription factor binding site software (http://tfbs.genereg.net).
Accession Numbers
Sequence data for the RNA-seq samples can be found in the NCBI database under the following accession numbers: SAMN04259680, SAMN04259681, SAMN04259682, SAMN04259683, SAMN04259684, and SAMN04259685.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. OXS2 homolog proteins in maize.
Supplemental Figure S2. Overexpression of ZmOXS2s or DEG23(CIMT1) enhances cadmium tolerance.
Supplemental Figure S3. Expression of eight DEG candidates in response to Cd treatment.
Supplemental Figure S4. Relative transcription of DEGs in WT(DEGs)/wild type.
Supplemental Figure S5. Cd accumulation in different genetic backgrounds
Supplemental Figure S6. Subcellular localization of AtCIMT1, ZmOXS2b, and ZmO2L1 fused to GFP and transiently expressed in Arabidopsis protoplasts.
Supplemental Table S1. Sequence differences found between ZmOXS2 homologs from sweet corn and information from NCBI.
Supplemental Table S2. Summary of differentially expressed genes discovered by RNA-seq.
Supplemental Table S3. Significantly enriched Gene Ontology terms of DEGs.
Supplemental Table S4. RNA-seq and qRT-PCR values of 30 DEGs in intersection of C1 and C2.
Supplemental Table S5 Primer sequences.
Supplementary Material
Acknowledgments
We thank R. Blanvillain for Arabidopsis mutant seeds, M. Liang for oxs2-1(AtOXS2-FLAG) transgenic plants and technical advice on ChIP and western blots, X. Pan for pGFP vectors, and Z. Han and C. Wang for reading the manuscript.
Glossary
- DEG
differentially expressed gene
- UTR
untranslated region
- ChIP-qPCR
chromatin immunoprecipitation-quantitative PCR
- SAM
S-adenosyl-l-Met
- ORF
open reading frame
References
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Blanvillain R, Wei S, Wei P, Kim JH, Ow DW (2011) Stress tolerance to stress escape in plants: role of the OXS2 zinc-finger transcription factor family. EMBO J 30: 3812–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YX, He YF, Luo YM, Yu YL, Lin Q, Wong MH (2003) Physiological mechanism of plant roots exposed to cadmium. Chemosphere 50: 789–793 [DOI] [PubMed] [Google Scholar]
- Clemens S, Kim EJ, Neumann D, Schroeder JI (1999) Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J 18: 3325–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Hartwig A, Schwerdtle T (2002) Interactions by carcinogenic metal compounds with DNA repair processes: toxicological implications. Toxicol Lett 127: 47–54 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang Y, Peng JS, Zhong C, Yi HY, Ow DW, Gong JM (2012) Fission yeast HMT1 lowers seed cadmium through phytochelatin-dependent vacuolar sequestration in Arabidopsis. Plant Physiol 158: 1779–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y (2007) The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J 50: 207–218 [DOI] [PubMed] [Google Scholar]
- Küpper H, Parameswaran A, Leitenmaier B, Trtílek M, Setlík I (2007) Cadmium-induced inhibition of photosynthesis and long-term acclimation to cadmium stress in the hyperaccumulator Thlaspi caerulescens. New Phytol 175: 655–674 [DOI] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA (1997) A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci USA 94: 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin MJ, Parker DR, Clarke JM (1999) Metals and micronutrients - food safety issues. Field Crops Res 60: 143–163 [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nriagu JO, Pacyna JM (1988) Quantitative assessment of worldwide contamination of air, water and soils by trace metals. Nature 333: 134–139 [DOI] [PubMed] [Google Scholar]
- Patra M, Bhowmik N, Bandopadhyay B, Sharma A (2004) Comparison of mercury, lead and arsenic with respect to genotoxic effects on plant systems and the development of genetic tolerance. Environ Exp Bot 52: 199–223 [Google Scholar]
- Perfus-Barbeoch L, Leonhardt N, Vavasseur A, Forestier C (2002) Heavy metal toxicity: cadmium permeates through calcium channels and disturbs the plant water status. Plant J 32: 539–548 [DOI] [PubMed] [Google Scholar]
- Prasad MNV. (1995) Cadmium toxicity and tolerance in vascular plants. Environ Exp Bot 35: 525–545 [Google Scholar]
- Sandalio LM, Dalurzo HC, Gómez M, Romero-Puertas MC, del Río LA (2001) Cadmium-induced changes in the growth and oxidative metabolism of pea plants. J Exp Bot 52: 2115–2126 [DOI] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytar O, Kumar A, Latowski D, Kuczynska P, Strzałka K, Prasad MNV (2012) Heavy metal-induced oxidative damage, defense reactions, and detoxification mechanisms in plants. Acta Physiol Plant 35: 985–999 [Google Scholar]
- Takiguchi M, Achanzar WE, Qu W, Li G, Waalkes MP (2003) Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp Cell Res 286: 355–365 [DOI] [PubMed] [Google Scholar]
- Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A (2011) Differential expression in RNA-seq: a matter of depth. Genome Res 21: 2213–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalkes MP. (2003) Cadmium carcinogenesis. Mutat Res 533: 107–120 [DOI] [PubMed] [Google Scholar]
- Wagner GJ. (1993) Accumulation of cadmium in crop plants and its consequences to human health. Adv Agron 51: 173–212 [Google Scholar]
- Waisberg M, Joseph P, Hale B, Beyersmann D (2003) Molecular and cellular mechanisms of cadmium carcinogenesis. Toxicology 192: 95–117 [DOI] [PubMed] [Google Scholar]
- Yadav SK. (2010) Heavy metals toxicity in plants: An overview on the role of glutathione and phytochelatins in heavy metal stress tolerance of plants. S Afr J Bot 76: 167–179 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.