Arabidopsis GOX1 and GOX2 have distinct roles under photorespiration-promoting conditions.
Abstract
The genes coding for the core metabolic enzymes of the photorespiratory pathway that allows plants with C3-type photosynthesis to survive in an oxygen-rich atmosphere, have been largely discovered in genetic screens aimed to isolate mutants that are unviable under ambient air. As an exception, glycolate oxidase (GOX) mutants with a photorespiratory phenotype have not been described yet in C3 species. Using Arabidopsis (Arabidopsis thaliana) mutants lacking the peroxisomal CATALASE2 (cat2-2) that display stunted growth and cell death lesions under ambient air, we isolated a second-site loss-of-function mutation in GLYCOLATE OXIDASE1 (GOX1) that attenuated the photorespiratory phenotype of cat2-2. Interestingly, knocking out the nearly identical GOX2 in the cat2-2 background did not affect the photorespiratory phenotype, indicating that GOX1 and GOX2 play distinct metabolic roles. We further investigated their individual functions in single gox1-1 and gox2-1 mutants and revealed that their phenotypes can be modulated by environmental conditions that increase the metabolic flux through the photorespiratory pathway. High light negatively affected the photosynthetic performance and growth of both gox1-1 and gox2-1 mutants, but the negative consequences of severe photorespiration were more pronounced in the absence of GOX1, which was accompanied with lesser ability to process glycolate. Taken together, our results point toward divergent functions of the two photorespiratory GOX isoforms in Arabidopsis and contribute to a better understanding of the photorespiratory pathway.
The increase in atmospheric CO2 levels linked to global warming, which entails unpredictable climate conditions, poses an unprecedented pressure on modern agriculture (Lobell and Gourdji, 2012; Wheeler and von Braun, 2013). However, apart from its greenhouse properties, CO2 and its photosynthetic assimilation into biomass are the primary foundation of life on Earth. Thus, atmospheric CO2 content has a direct effect on agricultural production, and even a modest CO2 increase might in theory result in higher crop yields, especially in plants with C3-type photosynthesis (Walker et al., 2016). In contrast to C4 and CAM-type plants, C3 plants do not possess carbon concentrating mechanisms, and the first stable CO2 assimilation product is 3-phosphoglycerate that is further processed in the Calvin-Benson cycle to fuel sugar synthesis. C4 plants initially incorporate CO2 into four-carbon acids that are subsequently decarboxylated in the vicinity of the primary CO2-assimilating enzyme Rubisco. The C4-type photosynthesis evolved independently over 60 times (Sage et al., 2011), reflecting the need to counteract the highly promiscuous nature of Rubisco that apart from carboxylation of ribulose-1,5-bisphosphate also catalyzes its oxygenation to 2-phosphoglycolate (PG). This oxygenation reaction initiates the photorespiratory pathway that recycles PG back to 3-phosphoglycerate with the release of CO2 in a series of enzymatic steps distributed between chloroplasts, peroxisomes, and mitochondria (Timm et al., 2008; Bauwe et al., 2012). The metabolic flux through the photorespiratory pathway increases substantially under adverse environmental conditions, such as drought and heat, which ultimately result in a loss of assimilated CO2 and a consequent yield reduction (Walker et al., 2016).
The photorespiratory pathway supports the second highest metabolic flux in photosynthesizing tissues after the Calvin-Benson cycle. Although the core photorespiratory pathway is now elucidated at the enzymatic level, its functional significance beyond carbon recycling and intimate interplay with primary metabolism remains a matter of debate (Bauwe et al., 2012; Hodges et al., 2016). Moreover, the existence of bypasses for core photorespiratory intermediates that play an important role during stress conditions imposes additional difficulties in elucidating the fine-tuning of the photorespiratory pathway (Timm et al., 2008, 2011). In the photorespiratory pathway, PG is first dephosphorylated by phosphoglycolate phosphatase to glycolate, which is subsequently exported from the chloroplasts via the plastidial glycolate/glycerate translocator 1 (Pick et al., 2013). Glycolate then undergoes oxidation in the peroxisomes catalyzed by the tetrameric FMN-dependent glycolate oxidase (GOX; EC 1.1.3.15), which generates glycoxylate and H2O2.
Arabidopsis (Arabidopsis thaliana) mutants lacking peroxisomal CATALASE2 (cat2-2) display around 10% wild-type catalase activity and have been instrumental in elucidating the impact of elevated photorespiratory H2O2 on the induction of defense responses and cell death (Queval et al., 2007; Kaurilind et al., 2015). The increase of H2O2 levels in cat2-2 mutants can be conditionally controlled by modulation of the growth environment. Restriction of the influx of fresh air to in vitro-grown plants, for example, decreases the available CO2 and ultimately results in enhanced photorespiration (Kerchev et al., 2015). Under such conditions, cat2-2 mutants display a rapid decline of photosynthetic activity and eventual cell death. In contrast, growing cat2-2 mutants under high CO2 atmosphere limits the rate of photorespiration and thus renders them indistinguishable from the wild type. Transfer of high CO2-grown cat2-2 mutants to photorespiration-promoting conditions, such as high light and ambient air, activates the photorespiratory pathway and triggers profound transcriptional and metabolic rearrangements and lesion formation (Queval et al., 2007).
The Arabidopsis genome contains five genes annotated as enzymes with GOX activity (Rojas et al., 2012; Hodges et al., 2013). They form the (l)-2-hydroxyacid-oxidases [(l)-2-HAOX] family that contains GOX1 (At3g14420), GOX2 (At3g14415), GOX3 (At4g18360), HAOX1 (At3g14130), and HAOX2 (At3g14150). GOX1 and GOX2 are the major isoforms found in leaf peroxisomes (Reumann et al., 2007; Foyer et al., 2009; Dellero et al., 2016). GOX3, on the other hand, is mainly expressed in roots where it converts glycolate and l-lactate with similar efficiency, making it an important player in lactate metabolism (Engqvist et al., 2015). HAOX1 and HAOX2 act predominantly on medium- and long-chain hydroxy acids, show a negligible activity toward glycolate (Esser et al., 2014), and are mainly expressed in seeds (Hodges et al., 2013).
Despite having only 30% residual GOX activity, single mutants lacking GOX1 or GOX2 show no visible phenotype implying a certain degree of functional redundancy (Rojas et al., 2012; Dellero et al., 2016). Moreover, recombinant GOX1 and GOX2 proteins exhibit similar kinetic parameters (Dellero et al., 2015). GOX1 and GOX2 reside in a tandem repeat on chromosome 3. The two homologs are separated by a mere 890 bp, which precludes the generation of double gox1 gox2 mutants through crossing of the available T-DNA insertion mutants. However, simultaneously knocking down GOX1 and GOX2 by artificial microRNA resulted in 5% residual GOX activity and stunted growth in ambient air (Dellero et al., 2016). Thus far, mutants affected in GOX activity have not been isolated in photorespiratory mutant screens in C3 species (Timm and Bauwe, 2013). The only example of a single GOX mutation that results in a photorespiratory phenotype is Zmgo1 in the C4 plant maize (Zea mays; Zelitch et al., 2009). In rice (Oryza sativa), an antisense-based strategy was employed to suppress the redundant GOX genes and to inhibit the flux through the photorespiratory pathway to levels negatively affecting photosynthesis and growth (Xu et al., 2009).
In a second-site suppressor screen for mutations attenuating the photorespiratory phenotype of Arabidopsis plants lacking peroxisomal catalase (cat2-2), we isolated a causative loss-of-function mutation in the GOX1 gene. Interestingly, the introduction of the gox2-1 allele into the cat2-2 background did not prevent the negative effects of photorespiration and the double cat2-2 gox2-1 mutants phenocopied the parental cat2-2 plants. Using a series of physiological, biochemical, and metabolomic approaches, we investigated the differential photorespiratory response of cat2-2 mutants in the absence of GOX1 and GOX2. Our results reveal the existence of nonredundant functions for GOX1 and GOX2 and contribute to a better understanding of the enzymes involved in the photorespiratory pathway.
RESULTS
Lack of GOX1, But Not GOX2, Attenuates the Photorespiratory Phenotype of Arabidopsis Plants Deficient in Peroxisomal Catalase
We previously conducted a high-throughput suppressor screen for second-site mutations that alleviate the photorespiratory phenotype of Arabidopsis plants lacking peroxisomal catalase (cat2-2; Waszczak et al., 2016). This screening strategy involved restriction of gas exchange to in vitro-grown plants and exposure to continuous light to avoid respiratory CO2 buildup. Under these conditions, cat2-2 mutants display a rapid decline of PSII maximum efficiency (Fv'/Fm') and cell death within 7 d from the onset of the treatment (Vanderauwera et al., 2011; Kerchev et al., 2015).
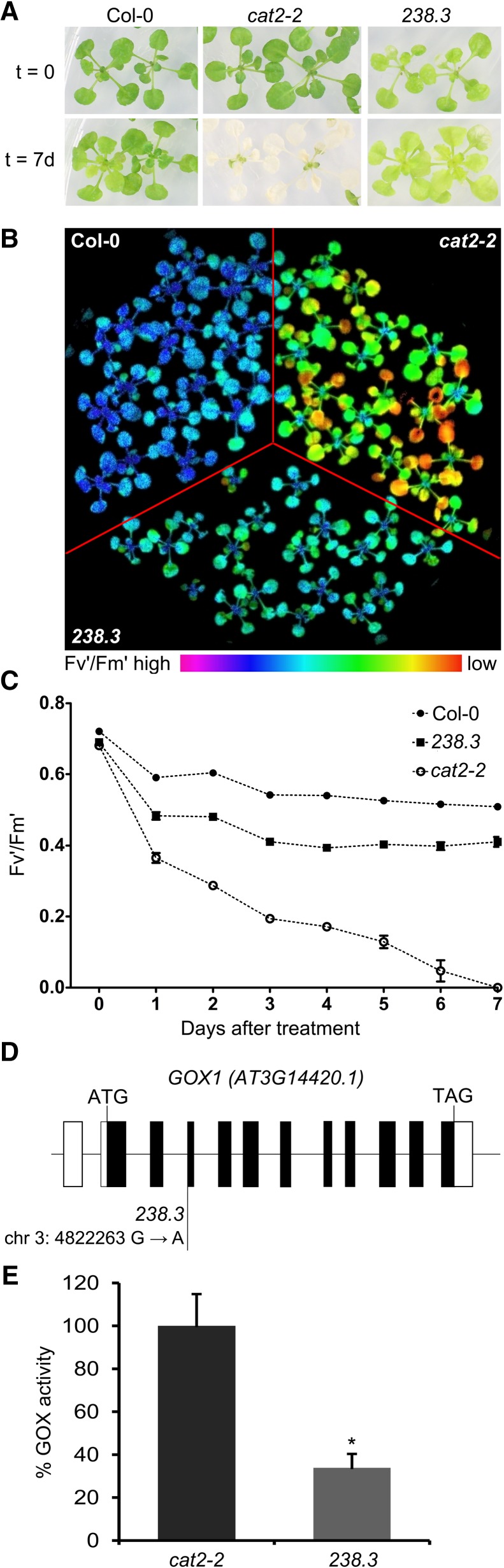
One of the mutants isolated in the screen, referred to here as 238.3, mitigated the characteristic Fv'/Fm' decline and cell death observed in single cat2-2 mutants under photorespiration-promoting conditions (Fig. 1, A–C). Using the SHOREmap backcross strategy (Hartwig et al., 2012; see “Materials and Methods”), the causative mutation was mapped to an intron-exon junction in the gene encoding GOX1 (Fig. 1D). Foliar GOX activity in 238.3 was reduced by more than 50% in comparison to single cat2-2 mutants (Fig. 1E), suggesting that the identified single-nucleotide polymorphism generates a loss-of-function mutation.
Figure 1.
Characterization of line 238.3 that displays attenuated cell death in the absence on peroxisomal catalase under photorespiratory stress. A, Representative bright-field images of 3-week-old Col-0, cat2-2, and 238.3 plants before (top panel) and after 7 d (bottom panel) of photorespiratory stress (restricted gas exchange and continuous light). B, Color-coded image of PSII maximum efficiency (Fv'/Fm') of 3-week-old Col-0, 238.3, and cat2-2 plants exposed to photorespiratory stress for 48 h. C, Fv'/Fm' decrease during the course of the photorespiratory treatment. Data points represent means of three biological replicates ± se. D, GOX1 gene model together with the position of the causative EMS-induced mutation. E, Extractable leaf glycolate oxidase activity from 2-week-old plants grown in vitro expressed as a percentage of the cat2-2 value. Bars represent means of three biological replicates ± se. Asterisks indicate significant differences according to Student’s t test (*P < 0.05).
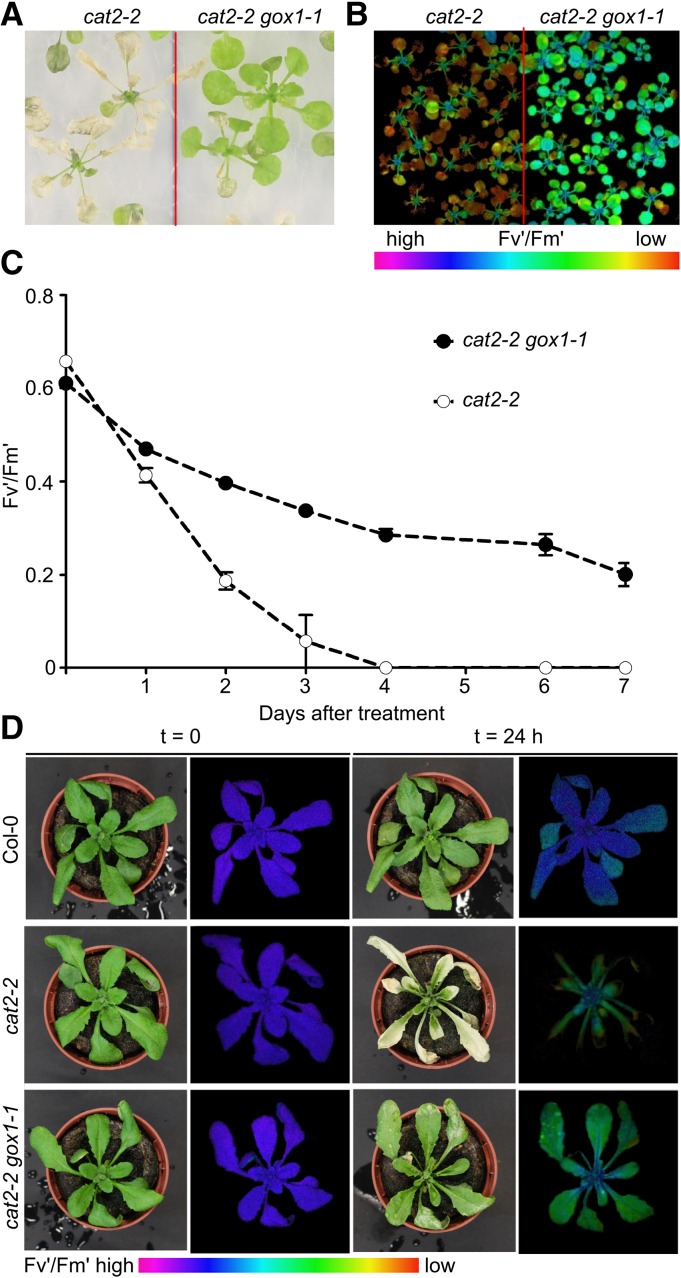
To confirm that lack of GOX1 is responsible for the attenuation of the photorespiratory phenotype observed in 238.3, we introduced the T-DNA insertion allele gox1-1 (Supplemental Fig. S1) into the cat2-2 background and exposed the resulting cat2-2 gox1-1 double mutants (Supplemental Fig. S2) to the same photorespiratory stress as applied in the original high-throughput screen. Under these conditions, cat2-2 gox1-1 mutants retained higher Fv'/Fm' values and exhibited limited cell death in comparison to single cat2-2 mutants (Fig. 2, A–C). We further tested whether the protective effect of gox1-1 can be similarly observed in soil-grown plants exposed to severe photorespiratory conditions. To this end, 3-week-old plants grown under high CO2 atmosphere (3,000 μL L−1), aimed to limit photorespiration, were exposed to high light (∼1,000 μmol m−2 s−1) and ambient air. The transfer resulted in cell death lesion formation in cat2-2 mutants within 24 h, whereas cat2-2 gox1-1 double mutants were largely unaffected and survived the photorespiratory stress (Fig. 2D).
Figure 2.
Attenuation of the photorespiratory phenotype of cat2-2 mutants in the absence of GOX1. A, Representative bright-field image of 3-week-old in vitro-grown cat2-2 and cat2-2 gox1-1 mutant plants exposed to photorespiratory stress for 4 d (restricted gas exchange and continuous light). B, Color-coded Fv′/Fm′ images of plants exposed to photorespiratory stress for 48 h. C, PSII maximum efficiency (Fv′/Fm′) decrease in the course of the photorespiratory treatment. Data points represent means of three biological replicates ± se. D, Representative bright-field and color-coded Fv′/Fm′ images of 3-week-old Col-0, cat2-2, and cat2-2 gox1-1 plants grown in soil under high CO2 (3,000 μL L−1) atmosphere and low light (100 μmol m−2 s−1) before (T=0 h) and after (T=24 h) transfer to ambient air and high light (1,000 μmol m−2 s−1).
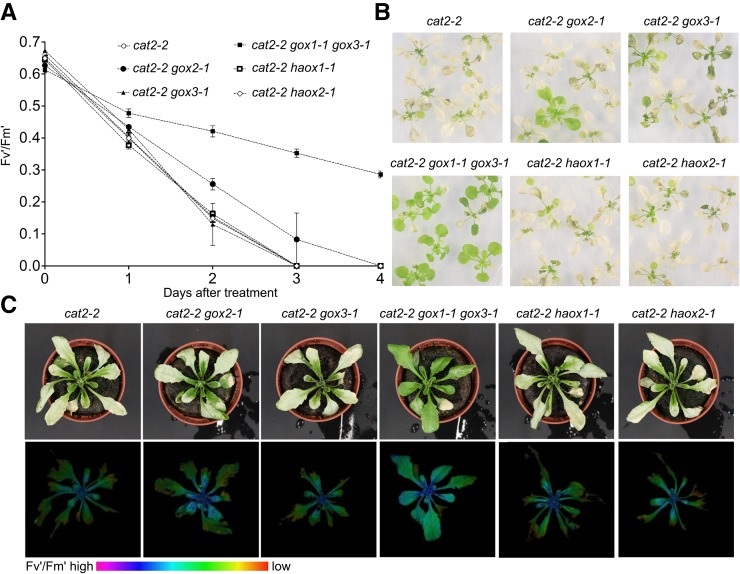
To test whether the loss of any other glycolate-metabolizing enzyme from the (l)-2-HAOX family, apart from GOX1, might attenuate of the photorespiratory phenotype of cat2-2, we generated a series of double (cat2-2 gox2-1, cat2-2 gox3-1, cat2-2 haox1-1, and cat2-2 haox2-1) and triple (cat2-2 gox1-1 gox3-1) mutants and assessed their phenotype under photorespiratory stress. Surprisingly, knocking out GOX2, the other predicted photorespiratory GOX isoform, in the cat2-2 background did not alleviate the cell death of cat2-2 gox2-1 double mutants under in vitro and in soil photorespiration-promoting conditions (Fig. 3, A–C). Similarly, introduction of either haox1-1 or haox2-1 alleles into the cat2-2 background had no effect on the photorespiratory phenotype (Fig. 3, A–C). The cat2-2 gox3-1 double mutants were indistinguishable from the parental cat2-2 plants under photorespiration-promoting conditions (Fig. 3, A–C), which is in line with the notion that GOX3 is active predominantly in roots. Moreover, the negative effects of photorespiration in cat2-2 gox1-1 gox3-1 triple mutants were attenuated to the same extent as in cat2-2 gox1-1 double mutants (Fig. 3, A–C), implying that the combined loss of GOX3 and GOX1 does not have an additive effect.
Figure 3.
Contribution of (l)-2-hydroxyacid-oxidase family members to the photorespiratory phenotype of cat2-2 mutants. A, Decrease of PSII maximum efficiency (Fv′/Fm′) in 3-week-old in vitro-grown plants during the course of the photorespiratory stress (restricted gas exchange and continuous light). Data points represent means of three biological replicates ± se. B, Representative bright-field images of 3-week-old plants exposed photorespiratory stress for 4 d (restricted gas exchange and continuous light). C, Representative bright-field (top panel) and color-coded Fv′/Fm′ (bottom panel) images of 3-week-old plants grown in soil under high CO2 (3,000 μL L−1) atmosphere and low light (100 μmol m−2 s−1) after transfer to ambient air and high light (1,000 μmol m−2 s−1) for 24 h.
Our genetic analysis indicated that the lack of GOX3, HAOX1, and HAOX2 does not influence the photorespiratory phenotype of cat2-2, which is in line with their substrate preferences and tissue-specific expression (Hodges et al., 2013; Esser et al., 2014; Engqvist et al., 2015). GOX1 and GOX2, on the other hand, metabolize glycolate in vitro with similar kinetic parameters and have been assumed to equally support the photorespiratory pathway. We therefore set out to investigate the differential response of cat2-2 gox1-1 and cat2-2 gox2-1 mutants under photorespiratory stress.
Global Transcriptome and Metabolome Rearrangements Accompanying the Increased Survival of cat2-2 gox1-1 Mutants under Photorespiratory Stress
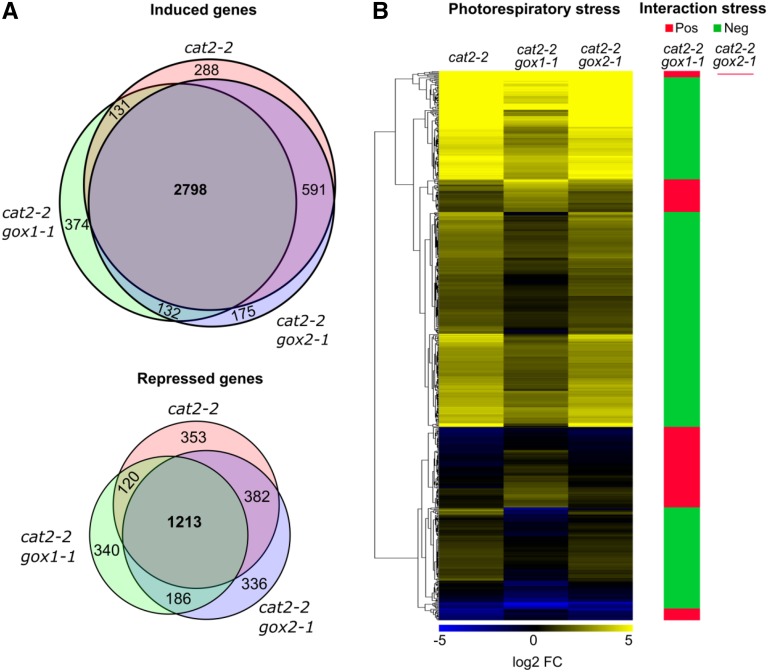
To gain insight into the early events underlying the attenuation of the photorespiratory phenotype of cat2-2 in the absence of GOX1, we performed global transcriptome (RNA-seq) and metabolome (gas chromatography-mass spectrometry [GC-MS]) profiling of high-CO2-grown plants before and after exposure to photorespiratory stress. Only 14 transcripts showed differential regulation (|log2 FC| > 1, FDR < 0.05) between cat2-2 gox1-1 and cat2-2 gox2-1 mutants relative to cat2-2 under control high CO2 conditions, implying that all genotypes are comparable (Supplemental Fig. S3). The transfer to photorespiratory stress (high light and ambient air) triggered a profound transcriptional response with 5,876 differentially regulated genes (|log2 FC| > 1, FDR < 0.01) after 3 h in cat2-2 mutants (Fig. 4A; Supplemental Data Set 1). The majority of the transcripts (3,808) were induced, while 2,068 were repressed. A significant transcriptional overlap (2,798 common up- and 1,213 down-regulated transcripts) was observed between the tested genotypes (Fig. 4A). Nevertheless, 608 transcripts responded to the photorespiratory treatment in a genotype-specific manner according to two-factor (genotype and photorespiratory stress) ANOVA (Fig. 4B; Supplemental Data Set 2). In contrast to cat2-2 gox1-1, the cat2-2 gox2-1 transcriptional profile was largely comparable to that of cat2-2 mutants with only one gene being more responsive to photorespiratory stress (Supplemental Data Set 2). Genes that exhibited higher induction in cat2-2 gox1-1 in comparison to cat2-2 mutants were predominantly involved in glycosinolate biosynthesis, jasmonic acid biosynthesis, oxylipin metabolism, and wounding (Supplemental Data Set 3). In contrast, transcripts that were less induced by the photorespiratory treatment in cat2-2 gox1-1 could be associated with a range of processes such as response to high light, plant-pathogen interaction, salicylic acid-mediated signaling, ethylene biosynthesis, and regulation of programmed cell death (Supplemental Data Set 3).
Figure 4.
Global transcriptome changes triggered by photorespiratory stress in cat2-2, cat2-2 gox1-1, and cat2-2 gox2-1 mutants. Three-week-old plants grown in soil under high CO2 (3,000 μL L−1) atmosphere and low light (100 μmol m−2 s−1) were transferred to ambient air and high light (1,000 μmol m−2 s−1) for 3 h. Genome-wide gene expression levels were quantified in mature rosette leaves before and after exposure to photorespiratory stress using RNA-seq. A, Venn diagrams showing the number of induced (top panel) or repressed (bottom panel) transcripts (|log2 FC| > 1, FDR < 0.01) upon photorespiratory stress. B, Heat map of transcripts that responded significantly (FDR < 0.05) to the photorespiratory stress in a genotype-specific manner according to a two-way ANOVA. Clusters of transcripts with higher or lower expression in the double mutants relative to cat2-2 mutants are marked with red or green, respectively.
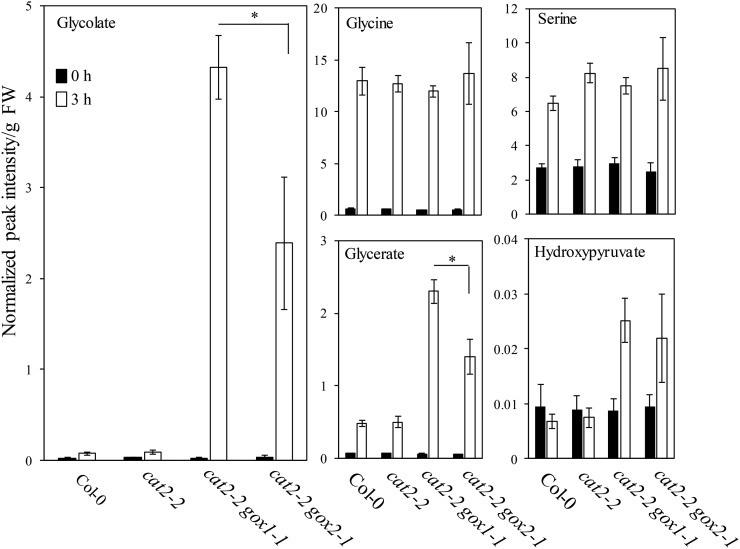
Parallel analysis of photorespiratory intermediates demonstrated that exposure to photorespiratory stress led to a modest increase (∼3-fold) of glycolate in cat2-2 and Col-0 plants, whereas cat2-2 gox1-1 and cat2-2 gox2-1 mutants accumulated 168 and 93 times more glycolate in comparison to their high CO2-grown controls, respectively (Fig. 5). In contrast, transfer to photorespiration-promoting conditions boosted the Gly (∼20-fold) and Ser (∼3-fold) content to a comparable level in all genotypes (Fig. 5). Although the steady-state glycerate levels were elevated in all genotypes, its content was higher in cat2-2 gox1-1 and cat2-2 gox2-1 double mutants (Fig. 5). Photorespiratory treatment increased the hydroxypyruvate content approximately 3-fold in cat2-2 gox1-1 and cat2-2 gox2-1 mutants, while it did not affect Col-0 and cat2-2 (Fig. 5).
Figure 5.
Changes of photorespiratory intermediates triggered by photorespiratory stress. Three-week-old Col-0, cat2-2, cat2-2 gox1-1, and cat2-2 gox2-1 plants grown in soil under high CO2 (3,000 μL L−1) atmosphere and low light (100 μmol m−2 s−1) were transferred to ambient air and high light (1,000 μmol m−2 s−1) for 3 h. Metabolites were extracted from rosette leaves before (0 h) and after exposure to photorespiratory stress (3 h). Bars represents averages of 5 to 6 replicates ± sd. Data were analyzed with two-way ANOVA using photorespiratory stress (photorespiratory conditions versus control conditions) and genotype as main factors. Asterisks indicate significant differences between genotypes under photorespiratory stress (P < 0.05).
Taken together, these results suggest that the lack of either GOX1 or GOX2 results in inability to process photorespiratory glycolate in a catalase-deficient background. However, the effect is much more pronounced in the absence of GOX1 and cat2-2 gox1-1 mutants accumulate nearly 2 times more glycolate than cat2-2 gox2-1 mutants.
Redox Homeostasis in cat2-2 gox1-1 and cat2-2 gox2-1 Mutants under Photorespiration-Promoting Conditions
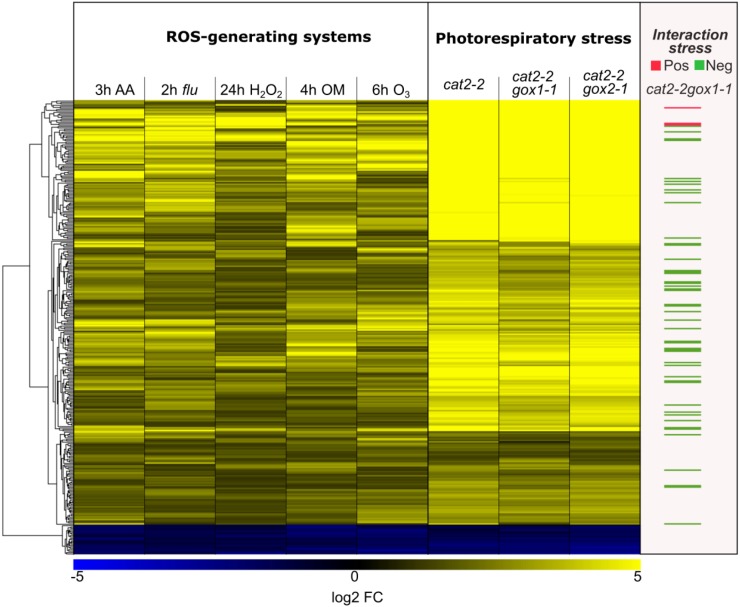
GOX activity is the primary source of H2O2 production in photosynthesizing tissues (Noctor et al., 2002). Decreased GOX activity thus might limit H2O2 accumulation and attenuate photorespiration-induced cell death in the absence of peroxisomal catalase. Therefore, we assessed the impact of photorespiration-promoting conditions on a common set of differentially regulated genes retrieved from six microarray studies featuring various ROS generating conditions (Willems et al., 2016; see “Materials and Methods”). This ROS-induced transcriptome imprint comprises 322 genes, which are robustly regulated upon perturbation of the redox homeostasis, and was used here as a proxy for H2O2 accumulation. The majority of these transcripts responded upon transfer from high CO2 to photorespiratory stress (high light and ambient air) in all tested genotypes (cat2-2 gox1-1, cat2-2 gox2-1, and cat2-2; Fig. 6). However, the abundance of 48 transcripts was significantly lower in cat2-2 gox1-1 mutants (Fig. 6), implying an impaired H2O2 production and a more stringent regulation of the redox homeostasis.
Figure 6.
Cellular redox homeostasis upon exposure to photorespiratory stress in cat2-2, cat2-2 gox1-1, and cat2-2 gox2-1 mutants. Comparison between the expression patterns of a subset of ROS-responsive genes, obtained by meta-analysis of ROS-generating systems, found in the transcriptome signatures of cat2-2, cat2-2 gox1-1, and cat2-2 gox2-1 mutants. The following experiments were used in the meta-analysis: reillumination of the conditional flu mutant for 2 h after a dark acclimation (2 h flu, GSE10812); treatment of seedlings with oligomycin for 4 h (4 h OM, GSE38965); treatment of seedlings with 50 µM antimycin A for 3 h (3 h AA, GSE41136); exposure of seedlings to ozone for 6 h (6 h O3, E-MEXP-342); treatment of seedlings with 10 mm H2O2 for 24 h (24 h H2O2).
We further assessed whether the absence of GOX1 could similarly affect the redox poise by quantifying the key redox buffer glutathione/glutathione disulfide (GSH/GSSG) and in situ visualization of H2O2 levels with 3,3′-diaminobenzidine (DAB). In 3-week-old plants grown continuously under ambient air, the total glutathione pool in cat2-2 rosette leaves was significantly larger and more oxidized in comparison to the wild type. Whereas the introduction of gox2-1 in cat2-2 background did not alter the total glutathione content and its oxidation status, cat2-2 gox1-1 mutants accumulated less total glutathione (Supplemental Fig. S4A). In addition, DAB staining revealed an intense signal in cat2-2 and cat2-2 gox2-1 mutants, which was largely absent in cat2-2 gox1-1 mutants and the wild type (Supplemental Fig. S4B).
gox1-1 and gox2-1 Single Mutants Are Morphologically Indistinguishable under Moderate Light Intensities But, Display Distinct Metabolic Phenotypes
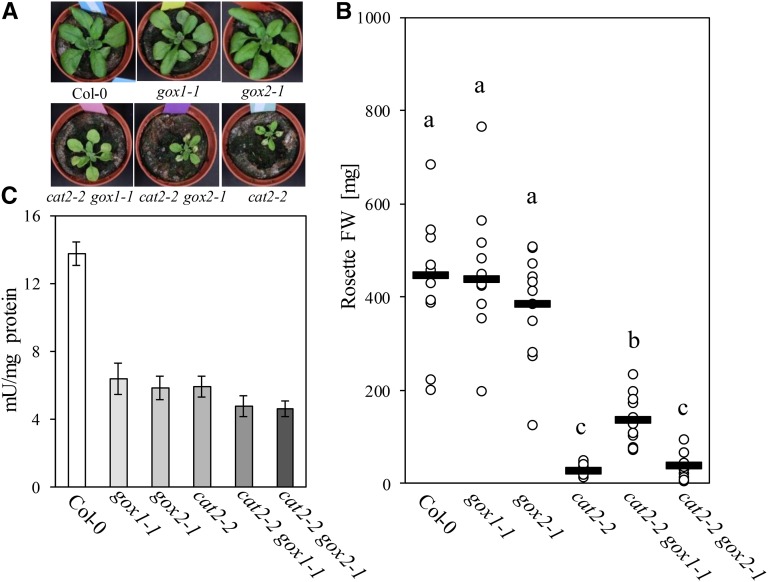
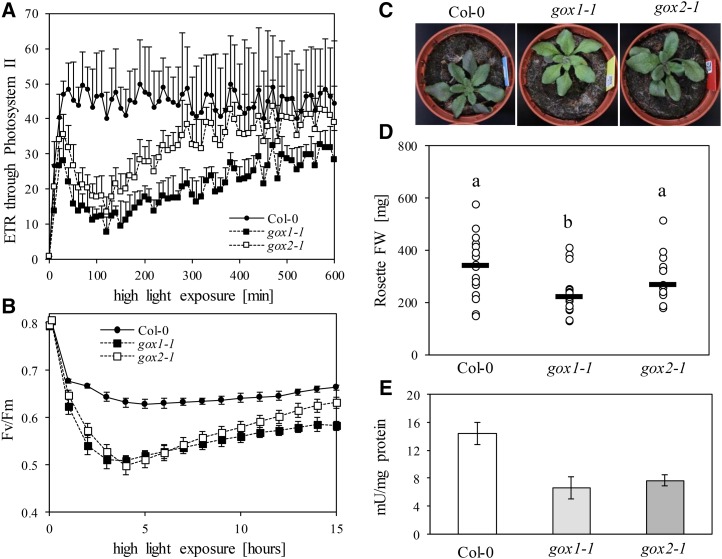
The fact that mutants affected in GOX activity have never been isolated in photorespiratory mutant screens (Timm and Bauwe, 2013), together with the similar enzymatic properties of recombinant GOX1 and GOX2 (Dellero et al., 2015), point toward a strong functional redundancy. Indeed, under moderate light intensities (300 μmol m−2 s−1), gox1-1 and gox2-1 single mutants were indistinguishable from the wild type and accumulated similar rosette biomass (Fig. 7, A and B), despite displaying only half of the wild-type foliar GOX activity (Fig. 7C). In contrast, under such growth conditions, cat2-2 and cat2-2 gox2-1 mutants were stunted and developed leaf lesions. Both of these phenotypes were largely abolished in cat2-2 gox1-1 mutants (Fig. 7A). Interestingly, GOX activity from cat2-2 leaves was comparable to that extracted from single gox mutants (Fig. 7C). Introduction of gox1-1 or gox2-1 into the cat2-2 background further reduced leaf GOX activity to levels below those found in single cat2-2 mutants (Fig. 7C).
Figure 7.
Phenotypic characteristics of Col-0, gox1-1, gox2-1, cat2-2, cat2-2 gox1-1, and cat2-2 gox2-1 plants grown under moderate light intensity (300 µmol m−2 s−1). A, Representative bright-field images of 3-week-old plants. B, Rosette fresh weight (FW) of 3-week-old plants. Black lines represent medians from at least 10 replicates and dots individual measurements. Different letters denote homogenous subsets according to a one-way ANOVA with Tukey post hoc test. C, Extractable leaf GOX activity from 3-week-old plants. Bars represent averages from three biological replicates ± sd.
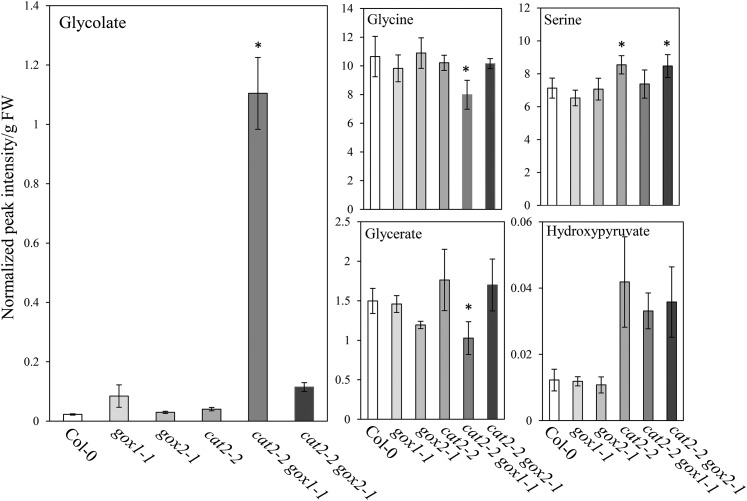
We further tested whether, despite the lack of growth phenotype, the low foliar GOX activity found in gox1-1 and gox2-1 mutants affected the steady-state levels of photorespiratory metabolites. Interestingly, gox1-1 mutants accumulated more glycolate (3.7-fold) than gox2-1 mutants (1.3-fold) relative to the wild type (Fig. 8). Moreover, cat2-2 gox1-1 and cat2-2 gox2-1 mutants contained approximately 50 and 5 times more glycolate than the wild type, respectively. Apart from glycerate, which was lower in gox2-1 single mutants, the levels of the other quantified photorespiratory intermediates (Gly, Ser, and hydroxypyruvate) were similar between gox1-1, gox2-1, and wild-type plants (Fig. 8).
Figure 8.
Levels of photorespiratory intermediates in rosettes of Col-0, gox1-1, gox2-1, cat2-2, cat2-2 gox1-1, and cat2-2 gox2-1 plants grown under moderate light intensity (300 µmol m−2 s−1). Bars represents averages of 5 to 6 replicates ± sd. Asterisks indicate significant differences (P < 0.05) in comparison to Col-0 according to one-way ANOVA and Tukey post hoc test.
High Light Conditions Elicit Differential Responses in gox1-1 and gox2-1 Mutants
The differential photorespiratory response of cat2-2 gox1-1 and cat2-2 gox2-1 double mutants indicates that GOX1 and GOX2 are not entirely redundant. To further deconvolute the individual roles of the two isoforms, 3-week-old gox1-1 and gox2-1 mutants grown under light intensities limiting photorespiration (100 µmol m−2 s−1), which showed no apparent differences in morphology and photosynthetic light reactions in comparison to the wild type, were exposed to high-intensity blue light (700 µmol m−2 s−1) inside an imaging PAM (pulse amplitude modulation), which allowed to follow their photosynthetic performance. Data for steady-state chlorophyll fluorescence (Fs) and maximal fluorescence under light (Fm') were collected over 10 h of exposure to high light and used to calculate the apparent rate of electron transfer (ETR) through PSII and nonphotochemical quenching (NPQ). Initially, high light exposure negatively affected the ETR through PSII in both gox1-1 and gox2-1 mutants in comparison to the wild type (Fig. 9A). However, after 2 h of high light treatment, ETR started to recover. The recovery was more pronounced in gox2-1 mutants and they eventually reached wild-type ETR levels, whereas gox1-1 mutants did not fully recover. The high light treatment increased the NPQ levels in both gox1-1 and gox2-1 mutants above those recorded for the wild type (Supplemental Fig. S5A).
Figure 9.
Phenotypic characteristics of Col-0, gox1-1, and gox2-1 single mutants under high light. A and B, Changes in photosynthetic electron transfer reactions after shift from a nonphotorespiratory growth environment to high light. Changes in apparent ETR through PSII (A) and maximal quantum yield of PSII (Fv/Fm; B) are plotted against the incubation time under high light. C, Representative bright-field images of 3-week-old plants grown under high light intensities (1,000 µmol m−2 s−1). D, Rosette biomass of 3-week-old plants grown under high light intensities (1,000 µmol m−2 s−1). Black lines represent medians from at least 15 replicates and dots individual measurements. Different letters denote statistical differences according to a one-way ANOVA with Tukey post hoc test. E, Extractable leaf GOX activity from 3-week-old plants grown under high light intensities (1000 µmol m−2 s−1). Bars represent averages from three biological replicates ± sd.
To explore the underlying reason for the diminished ETR and increased NPQ in both gox lines, we collected maximal quantum yield of PSII (Fv/Fm) data over time by subjecting the plants to consecutive periods of high-intensity blue light (900 µmol m−2 s−1) and darkness (see “Materials and Methods”). Such treatment negatively affected the Fv/Fm ratio, indicative for PSII photoinhibition, in all tested genotypes, but the decline was especially pronounced in gox1-1 and gox2-1 single mutants (Fig. 9B). Both mutants entered the recovery phase simultaneously; however, the recovery rate was significantly higher in gox2-1 than in gox1-1. Moreover, gox2-1 mutants nearly reached wild-type Fv/Fm levels by the end of the treatment. These findings suggest that the most likely explanation for the transient drop in ETR upon exposure to continuous high-intensity blue light is inhibition of PSII activity.
To probe whether the PSII photoinhibition observed in the gox mutants exposed to high light is accompanied with an altered redox status of the plastoquinone pool, the phosphorylation state of light-harvesting antennae of PSII (LHCBII) was assessed with an anti-phospho-Thr antibody. Overreduction of the plastoquinone pool activates the thylakoid-associated kinase STN7 that phosphorylates LHCBII, thereby increasing their affinity to PSI (Rochaix, 2014). The exposure to high light led to a time-dependent dephosphorylation of LHCBII in all tested genotypes, pointing toward a uniform impact on the plastoquinone pool (Supplemental Fig. S5B).
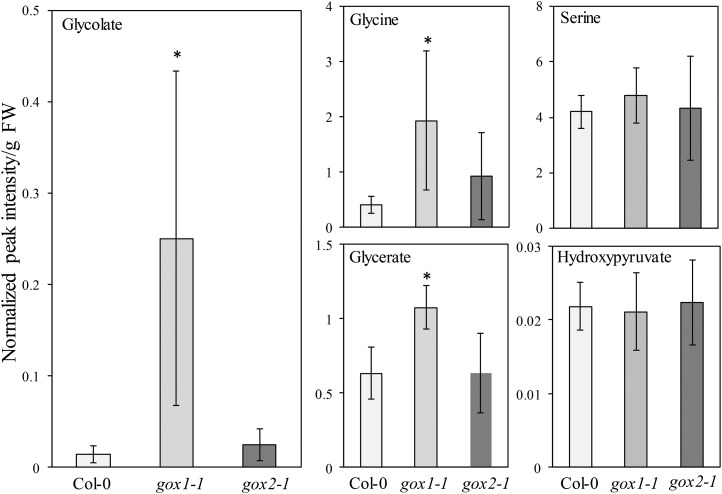
To further corroborate specific functionalities for GOX1 and GOX2, we grew gox1-1 and gox2-1 mutants under high light intensity (∼1,000 µmol m−2 s−1). Under these conditions, gox1-1 mutants accumulated less rosette biomass than gox2-1 mutants, whereas both gox1-1 and gox2-1 were smaller than the wild type (Fig. 9, C and D). The foliar GOX activity was similar in both mutants and nearly half of that found in the wild type (Fig. 9E). The lower biomass of gox1-1 mutants correlated with elevated glycolate levels (∼18-fold) relative to the wild type (Fig. 10). In contrast, the glycolate content of gox2-1 mutants was not significantly different from that of the wild type. Similarly to glycolate, Gly and glycerate levels were elevated in gox1-1, but not in gox2-1 mutants under these growth conditions (Fig. 10).
Figure 10.
Levels of photorespiratory intermediates in rosettes of 3-week-old Col-0, gox1-1, and gox2-1 plants grown under high light intensity (1,000 µmol m−2 s−1). Bars represent averages of 5 to 6 replicates ± sd. Asterisks indicate significant difference (P < 0.05) in comparison to Col-0 according to one-way ANOVA with Tukey post hoc test.
Evolutionary Analysis of GOX1 and GOX2
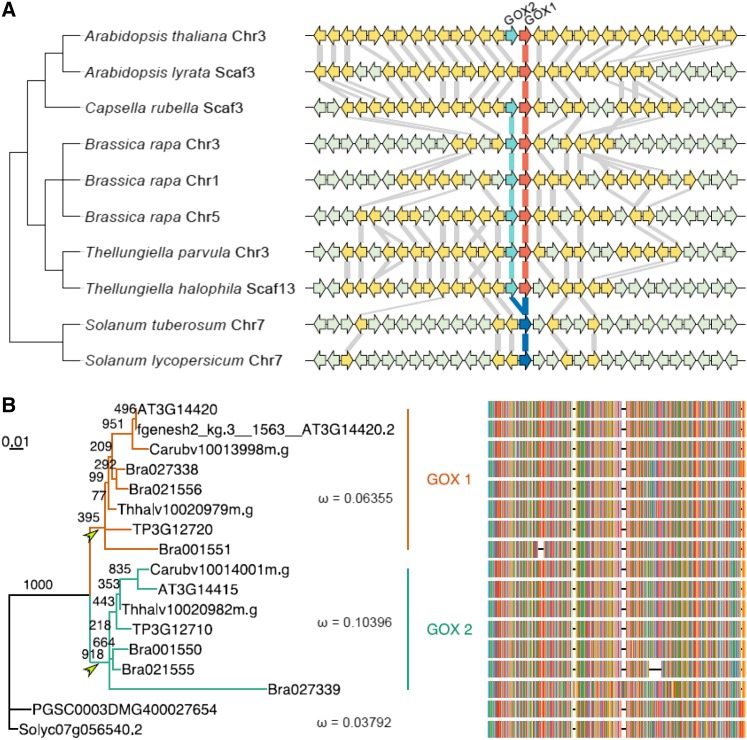
To further investigate the origin and evolution of the two highly similar protein-coding GOX1 and GOX2 sequences in Arabidopsis, we selected their orthologs in close relatives based on conserved synteny blocks that flank GOX1 and GOX2 (Louis et al., 2015). GOX1 and GOX2 are present as “tandem” duplicates in all Brassicaceae species with the exception of Arabidopsis lyrata, which retained only GOX1 (Fig. 11A). The only other plant species for which statistically significant colinearity or synteny could be found (our criterion to decide on true orthology) were the Solanaceae species Solanum tuberosum and Solanum lycopersicum, which contain only one GOX gene (Fig. 11A). This, together with the fact that GOX1 and GOX2 genes in Brassicaceae form a monophyletic cluster when compared with genes from more close relatives (Supplemental Fig. S6), would suggest that the tandem duplication that gave rise to GOX1 and GOX2 occurred in the most recent common ancestor of the Brassicaceae and is therefore Brassicaceae specific.
Figure 11.
Evolutionary analysis of GOX1 and GOX2. A, Conserved synteny surrounding GOX1 and GOX2 as retrieved from Genomicus (v16.03) and visualized with genoPlotR. Arrowed blocks indicate genes, with yellow and gray denoting homologous and nonhomologous genes to Arabidopsis, respectively. Red and green arrows indicate GOX1 and GOX2 in Brassicaceae, respectively, and blue arrows are GOX genes in Solanaceae. Gray lines connect homologous genes between neighbors in the tree. Chromosome or scaffold numbers on which the conserved synteny blocks were found are also indicated. B, Unrooted maximum likelihood tree and multiple sequence alignment for GOX genes. The numbers along branches show bootstrap support. Green and red indicate the GOX1 and GOX2 clade, respectively. Two arrowheads denote two postduplication branches. ω is the ratio of the nonsynonymous substitution rate over the synonymous substitution rate for the indicated clades as calculated with PAML under a three-ratio model.
Using the GOX1 and GOX2 orthologs, we tested whether they had experienced different selection forces during their evolutionary history following the duplication event. Such differences might explain the different phenotypes observed in single gox1-1 and gox2-1 mutants. To this end, we estimated the ratio of nonsynonymous substitutions rates to synonymous substitution rates (ω) over time using the branch models implemented in PAML (Yang, 2007). We further reconstructed a maximum likelihood tree based on amino acid sequences (see “Materials and Methods”) that also reflected the duplication event prior to the divergence of Brassicaceae (Fig. 11B). In a three-ratio model using the Solanaceae gene clade as a background, the obtained ω values for the GOX1 and GOX2 clades were 0.064 and 0.104, respectively. Both of them were higher than the ω value for the Solanaceae clade: 0.038. The three-ratio model was significantly better than a one-ratio model with one ω value for all three clades (2Δℓ = 18.4962; df = 2; P = 9.63 E−5), which implies that the GOX1 and GOX2 clades experienced different selection forces following duplication.
Because most Brassicaceae have retained both GOX1 and GOX2 genes (Fig. 11A), different selection pressures might have acted in both postduplication branches before the divergence of the Brassicaceae. We thus compared ω values on the two postduplication branches, indicated by arrows in Figure 11B, using the rest of the branches as a background (Supplemental Tables S1 and S2). Apparently, both early postduplication branches underwent purifying selection with an ω value significantly less than 1 (P = 0.02, D–H in Supplemental Table S2). The ω value for the branch leading to the GOX1 clade was indistinguishable from the background (P = 0.16 for A–C and P = 0.57 for B–E in Supplemental Table S2), whereas the one leading to the GOX2 clade was significantly higher than the background (P = 0.001 for A and B and P = 0.003 for C–E in Supplemental Table S2), but not significantly over 1 (P = 0.67, E–I in Supplemental Table S2). To further examine whether positive selection acted on GOX2 in Arabidopsis, we carried out PAML with the modified branch-site model (Zhang et al., 2005). No signal of positive selection was detected (Supplemental Table S3).
Our evolutionary analysis implies that although both GOX1 and GOX2 underwent purifying selection following their duplication, GOX2 did experience a more relaxed selection pressure than GOX1, which might underlie the differential phenotypes observed in single gox1-1 and gox2-1 mutants.
DISCUSSION
In contrast to the majority of the photorespiratory enzymes, gox mutants have never been isolated in photorespiratory mutant screens pointing toward a genetic redundancy among the GOX family members (Timm and Bauwe, 2013). Indeed, the Arabidopsis genome contains five genes annotated as glycolate oxidases (GOX1, GOX2, GOX3, HAOX1, and HAOX2). The data presented here provide new insights into the GOX functional redundancy and reveal distinct functions for the photorespiratory GOX isoforms in Arabidopsis.
In a second-site suppressor screen, we identified a causative loss-of-function mutation in the gene encoding GOX1 that attenuated the photorespiratory phenotype of Arabidopsis mutants lacking peroxisomal catalase. Interestingly, the highly similar GOX2 did not alleviate the photorespiratory phenotype of cat2-2 mutants, implying that GOX1 and GOX2 are functionally nonidentical. The absence of none of the other Arabidopsis enzymes predicted to metabolize glycolate could not counteract the characteristic photosynthetic decline and cell death observed in cat2-2 mutants exposed to photorespiratory stress (Fig. 3). This could be largely explained by their substrate specificities and expression patterns. HAOX1 and HAOX2 preferentially metabolize medium- and long-chain hydroxyacids (Esser et al., 2014), and their absence did not significantly affect the extractable leaf GOX activity (Supplemental Fig. S7). Similarly, the lack of GOX3, which is predominantly expressed in roots and senescing leaves and uses glycolate and l-lactate with comparable efficiency (Engqvist et al., 2015), did not reduce the GOX activity (Supplemental Fig. S7). Interestingly, haox1-1, haox2-1, and gox3-1 mutants have been previously reported to possess only 30% of the wild-type GOX activity (Rojas et al., 2012). The discrepancy with our results, which revealed no differences in GOX activity between haox1-1, haox2-1, and gox3-1 mutants and the wild type, might be explained by plant age and growth conditions.
The photorespiratory GOX activity produces approximately 70% of the total H2O2 pool and is thus the most important H2O2 source in photosynthesizing C3 leaves (Noctor et al., 2002). The absence of peroxisomal catalase (CAT2) in cat2-2 mutants results in perturbed redox homeostasis and cell death formation under photorespiration-promoting conditions (Queval et al., 2007). Given that the majority of H2O2 in photosynthesizing leaves is generated by the GOX activity, the most straightforward explanation for the alleviated photorespiratory phenotype of cat2-2 mutants in the absence of GOX is a limited H2O2 production. This assumption implies that GOX1 and GOX2 are functionally divergent and that GOX1 produces more H2O2, thus explaining the alleviated cell death of cat2-2 gox1-1 but not cat2-2 gox2-1 mutants. Indeed, DAB staining indicated lower H2O2 levels in cat2-2 gox1-1 mutants in comparison to cat2-2 gox2-1 and cat2-2 mutants, which were accompanied with a more stringent regulation of glutathione homeostasis and higher accumulation of glycolate (Supplemental Fig. S4A; Fig. 8). Moreover, ROS-related transcripts were less induced in cat2-2 gox1-1 mutants in comparison to cat2-2 and cat2-2 gox2-1 mutants, which were largely identical. Taken together, these results reveal that the seemingly redundant GOX1 and GOX2 are not identical and the H2O2 levels produced by the residual GOX1 in cat2-2 gox2-1 mutants are sufficient to trigger cell death under photorespiration-promoting conditions.
Activation of antioxidant systems that can compensate for the lack of CAT2 and scavenge excess H2O2 could also underlie the alleviated cell death of cat2-2 gox1-1 mutants. Ascorbate peroxidases (APXs) are key enzymes in the ascorbate-glutathione cycle that remove H2O2 using ascorbate as an electron donor (Dietz, 2016). Nine APX isoforms distributed between chloroplasts, mitochondria, cytosol, and microsomes, including peroxisomes, have been described in Arabidopsis (Mittler et al., 2004). APX transcripts levels are responsive to adverse environmental conditions and total APX activity generally increases under stress (Caverzan et al., 2012). Knocking out the most abundant cytosolic and chloroplastic APXs negatively affects plant growth, development, and susceptibility to stresses, such as high light (Pnueli et al., 2003; Maruta et al., 2010). In contrast, apx3 mutants lacking peroxisomal APX are phenotypically not affected under optimal or stress conditions (Narendra et al., 2006). The transcript abundance of the H2O2-responsive cytosolic APX2 was lower in cat2-2 gox1-1 mutants, whereas APX1, another cytosolic isoform, was slightly more induced in comparison to cat2-2 and cat2-2 gox2-1 mutants (Supplemental Data Set 2). The absence of APX1 results in H2O2 accumulation and degradation of the chloroplastic antioxidant machinery under moderate light intensities, suggesting a protective role of the cytosol on the chloroplast function (Davletova et al., 2005). The moderate induction of APX1 in cat2-2 gox1-1 mutants might therefore contribute to the alleviated cell death phenotype in the absence of peroxisomal catalase.
Functional catalase is crucial for the protection of GOX, which is highly susceptible to oxidative damage (Schäfer and Feierabend, 2000). In support of this notion, we found that the leaf GOX activity in cat2-2 mutants is significantly lower than in the wild type (Fig. 7). The low GOX activity observed in cat2-2 mutants can be attributed to the preventive role played by the peroxisomal catalase in limiting the degradation of the redox vulnerable GOX cofactor FMN which in its reduced form (FMNH2) contributes to ROS generation (Massey, 1994). Chemically inhibiting catalase with 3-amino-1,2,4-triazole, similarly to the cat2-2 mutation, negatively affects GOX activity (Schäfer and Feierabend, 2000). The crucial role of catalase in maintaining GOX activity seems to be also dependent on the presence of GSH because pharmacologically blocking GSH biosynthesis inactivated GOX even in the presence of catalase (Schäfer and Feierabend, 2000). Both GOX1 and GOX2 have a single, highly conserved Cys residue (Proost et al., 2015) that potentially could serve as a site for redox-dependent posttranslational modification governing protein function and stability under oxidative conditions. The redox activity of the Cys residue was corroborated by Rouhier et al. (2005) who isolated GOX1 following affinity chromatography purification with poplar GLUTAREDOXIN C4, indicating that it could be subjected to S-glutathionylation. Moreover, GOX from Kalanchoe pinnata (Abat et al., 2008) and Pisum sativum (Ortega-Galisteo et al., 2012) were described as targets for S-nitrosylation.
Mutants lacking GOX1 and GOX2 have been previously shown to be morphologically and metabolically indistinguishable from wild-type plants under light intensity of 200 µmol m−2 s−1, implying a redundant role for both isoforms (Dellero et al., 2016). Similarly, under our experimental conditions, light intensity of 300 µmol m−2 s−1 did not provoke any visible difference between single gox1-1 and gox2-1 mutants and the wild type. However, we observed a trend toward increased glycolate levels in the absence of GOX1 (Fig. 8), suggesting the existence of distinct metabolic roles for GOX1 and GOX2. Indeed, when grown under higher light intensities (1,000 µmol m−2 s−1), gox1-1 mutants accumulated nearly 18 times more glycolate than gox2-1 mutants and the wild type (Fig. 10). The functional difference between the two isoforms is further supported by the faster PSII recovery rate observed in gox2-1 mutants under high light in comparison to gox1-1 (Fig. 9A). Nevertheless, both gox1-1 and gox2-1 mutants displayed similar initial rates of PSII photodamage upon exposure to high light (Fig. 9A; Supplemental Fig. S5), implying that the abrupt increase in photosynthetic electron flow against the background of restricted photorespiratory pathway creates similar imbalance in electron transfer chains. This notion is further supported by the increased phosphorylation of LHCBII in both mutant lines in comparison to the wild type (Supplemental Fig. S5), which is indicative of overreduction of the plastoquinone pool. The predominant metabolic role of GOX1 evident from the above described experiments is also substantiated by the higher transcriptional abundance of GOX1 (Supplemental Fig. S8; Dellero et al., 2016). The transcriptional differences, however, were not reflected in the extractable leaf GOX activity (Fig. 7). Thus, it would be important to quantitatively evaluate the individual GOX isoforms at the protein level. The gox1-1 mutants are only slightly affected under high light, suggesting that the residual GOX2 can partially compensate for the loss of GOX1. Only the simultaneous knock down of GOX1 and GOX2 by artificial microRNA, which results in 5% residual GOX activity, leads to a severely stunted growth phenotype (Dellero et al., 2016).
The duplication event that gave rise to GOX1 and GOX2 occurred before the divergence of Brassicaceae, which has been dated approximately 40 million years ago (Bailey et al., 2006). The high similarity of the two genes suggests that both of them have been under a purifying selection, which minimized the generation of deleterious mutations in their coding sequences, for the last 40 million years. However, our evolutionary analysis revealed more relaxed selection pressure on GOX2 that might underlie the differential phenotypes observed in the absence of GOX2. Although the differences might be due to a positive selection acting on a few sites, the branch-site model did not support this hypothesis. The relaxed purifying selection on GOX2 was also reflected in the evolutionary forces acting on the six GOX paralogs from Brassica rapa that originated in the well-documented paleohexaploidization (Wang et al., 2011). Not only four of the six genes have already started degrading, but also two GOX2 paralogs accumulated larger insertions and deletions in comparison to the GOX1 paralogs (Fig. 11B). This is in accordance with the different ω values for the two ancestral branches and the loss of GOX2 from a conserved synteny block in A. lyrata, which together support a major role for GOX1 in photorespiratory metabolism.
In conclusion, our results provide evidence for distinct roles of the two photorespiratory GOX isoforms in Arabidopsis. Lack of GOX1, but not GOX2, attenuates the cell death phenotype of cat2-2 mutants under photorespiratory conditions. This is accompanied by higher levels of accumulated glycolate in the absence of GOX1, suggesting that under conditions promoting photorespiration, GOX1 is the isoform that predominantly metabolizes glycolate and produces H2O2.
MATERIALS AND METHODS
Plant Material
All mutants used in this study are in the Col-0 background and were described before: cat2-2 (Queval et al., 2007); gox1-1, gox2-1, gox3-1, haox1-1, haox2-1, and gox1-1 gox3-1 (Rojas et al., 2012). Double and triple knockout lines cat2-2 gox1-1, cat2-2 gox2-1, cat2-2 gox3-1, cat2-2 haox1-1, cat2-2 haox2-1, and cat2-2 gox1-1 gox3-1 were generated by crossing cat2-2 plants (pollen acceptors) with the respective mutant lines (pollen donors). Double mutant plants were identified in F2 segregating populations by PCR genotyping with respective primers (Supplemental Table S4).
Growth Conditions and Stress Treatments
To impose photorespiratory stress in vitro, seeds were surfaced-sterilized by fumigation and cold-treated at 4°C for 3 to 4 d. Plants were germinated and grown on full-strength Murashige and Skoog (MS) medium (1% [w/v] Suc and 0.8% [w/v] agar) under controlled environmental conditions (16 h/8 h light/dark, 100 μmol m−2 s−1 light intensity, 21°C). After 3 weeks, plates were sealed with multiple layers of Parafilm M (Bemis Flexible Packaging) in order to reduce gas exchange and transferred to continuous light (100 μmol m−2 s−1 light intensity, 21°C). Data for PSII maximum efficiency (Fv'/Fm') were recorded daily using an Imaging-PAM-Series chlorophyll fluorescence system (Heinz Walz).
To impose severe photorespiration, plants were initially germinated in soil and grown under high CO2 atmosphere (3000 μL L−1) in a controlled climate chamber (Vötsch Industrietechnik; 16 h/8 h light/dark, 21°C, 50% relative humidity). Three-week-old plants were transferred to high light (∼1,000 μmol m−2 s−1) and ambient air for the specified time periods.
To investigate the effect of light intensity, plants were grown in soil under long-day conditions (16 h/8 h light/dark) in a controlled climate chamber (Sanyo Fitotron; 21°C and 50% relative humidity). Light intensity was set to either 300 or 1000 μmol m−2 s−1.
Identification of a Causative Mutation in 238.3
In order to identify the causative mutation of line 238.3, we applied the SHOREmap backcross pipeline (Hartwig et al., 2012). For this, we analyzed an F2 mapping population derived from a cross between cat2-2 and 238.3 mutant lines. In total, 1,794 F2 individuals were subjected to photorespiratory assay conditions in vitro and about 13% of plants (234) exhibited reversion of the cell death phenotype. This segregation did not fit the Mendelian 1:3 ratio (χ2 = 137.319, P < 0.0001) presumably due to the limited penetrance of the revertant phenotype and/or presence of independent EMS-induced mutations negatively affecting the seedling performance. Surviving plants were harvested and used for preparation of nuclear DNA (Schneeberger et al., 2009). This sample was used to prepare an Illumina library, which was sequenced to an average 53.6-fold genome coverage. Backcrossing of a mutant to its parent line does not introduce prespecified markers into the mapping population. Therefore, we applied a SHOREmap backcross strategy (Hartwig et al., 2012) that uses EMS-induced markers as novel markers in order to determine the mapping interval. Using a concordance threshold set to 0.8, we identified 10 EMS-specific SNPs (Supplemental Table S5) significantly enriched in plants displaying a revertant phenotype. All polymorphisms localized to the upper arm of chromosome 3 within 3.3 to 7.4 Mb physical interval. The most frequent SNP was localized within the GOX1 (At3g14420) coding sequence at position 1 of the third exon. Analysis of the GOX1 mutant sequence performed with NetGene2 software (http://www.cbs.dtu.dk/services/NetGene2/) indicated that this G → A substitution could potentially disrupt the splicing site between the second intron and the third exon.
Photosynthesis-Related Measurements
Wild-type (Col-0), gox1-1, and gox2-1 lines were grown in soil under 60 to 100 µmol photons m−2 s−1 light intensity and a 12 h/12 h light/dark photoperiod. Following a dark adaptation period of 20 min, 3-week-old plants were transferred to an Imaging-PAM (M-series; Heinz Walz), and the minimal (Fo) and maximal (Fm) fluorescence was determined for the dark-adapted state. In one set of experiments, plants were exposed to blue light (700 µmol photons m−2 s−1) for 10 h. The measuring beam was applied once every minute throughout the experiment to probe steady-state fluorescence under light (Fs). Saturating flashes were triggered every 10 min to assess maximal fluorescence under light (Fm′). The apparent ETR through PSII was calculated as follows: ETR = 0.5 × ([Fm′−Fs]/Fm′) × PAR × 0.84. NPQ was calculated as: NPQ = (Fm−Fm')/Fm'. In another set of experiments, plants were exposed to 15 consecutive 1-h periods of blue light (900 µmol m−2 s−1) each followed by 23 min of darkness. Maximal quantum yield of PSII was calculated as Fv/Fm = (Fm−Fo)/Fm.
To probe the dephosphorylation state of the light-harvesting antennae of PSII (LHCBII), plants treated with continuous blue light (700 µmol photons m−2 s−1) were harvested at 0, 45, 120, and 300 min from the onset of the light treatment. Total protein extracts were separated by SDS-PAGE and immunoblotted with an anti-phospho-Thr antibody (Cell Signaling).
Evolutionary Analysis
To obtain conserved synteny blocks containing GOX1 and GOX2, we queried the synteny browser GenomicusPlants (v16.03; Louis et al., 2015) with GOX1. The synteny blocks were then visualized using the R package “genoPlotR” (Guy et al., 2010). To further confirm the loss of GOX2 in Arabidopsis lyrata, we extracted the corresponding genomics sequence that, according to the conserved synteny block, is supposed to contain GOX2. We then predicted a gene model by aligning the protein sequence of GOX2 from Arabidopsis thaliana with GeneWise (Birney et al., 2004). No gene showing similarity to GOX2 was predicted in that genomic region.
We then reconstructed an amino acid-based phylogeny of the GOX genes present in the above identified conserved synteny blocks (Supplemental Table S6). To this end, we performed a multiple sequence alignment with T-Coffee and used trimAl 1.4 (Capella-Gutiérrez et al., 2009) to remove low-quality aligned regions in a heuristic approach (automated1). After the first round of multiple sequence alignment, a gene in Brassica rapa (Bra027339) exhibited large insertions in comparison to other sequences, indicating an annotation error. We hence used the protein sequence of GOX1 from Arabidopsis to predict a gene model in the genomic sequence of Bra027339 by GeneWise and identified a gene with a similar length to the other GOX genes. After a new round of multiple sequence alignment with the predicted gene, we tested all available amino acid substitution matrices using ProtTest3 (Darriba et al., 2011) and selected the best-fit model according to Akaike Information Criterion (AIC), Bayesian Information Criterion (BIC) score and corrected AIC (AICc). The LG+GAMMA model outperformed the others and was used in PhyML 3.0 (Guindon et al., 2010) to search the maximum likelihood tree. PhyML made use of a neighbor-joining tree as the initial tree and optimized tree topology, branch lengths, and rate parameters. The best tree produced from either Nearest Neighbor Interchange or Subtree Pruning and Regrafting was retained as the maximum likelihood tree. Branch support values were obtained in a bootstrap analysis with 1,000 replicates. The “ggtree” package implemented in Bioconductor was used to visualize the obtained maximum likelihood tree.
To explore the evolutionary forces experienced from the GOX1 and GOX2 clades, we used “codeml” implemented in PAML (Yang, 2007). After back-translating amino acid alignment into nucleotide sequence alignment with trimAl 1.4, we compared a one-ratio model, in which all clades in the tree have the same ω value, with a three-ratio model, in which the GOX1 and GOX2 clades can have different ω values using the Solanaceae clade as a background. To further test whether the evolutionary pressures on the two postduplication branches were different after the duplication event but before the divergence of Brassicaceae, we performed a series of analyses with branch models and compared ω values on the two postduplication branches using the rest of the branches as a background (Supplemental Table S1). Then, a group of likelihood ratio tests was carried out to compare different models (Supplemental Table S2). To screen possible positive selections on sites along specific branches in Arabidopsis GOX2, we applied the modified branch-site model in PAML to each of the six branches leading to GOX2 in Arabidopsis after the duplication event. The ω values in the modified branch model were allowed to vary across branches on the tree as well as among sites in the sequence (Zhang et al., 2005).
GC-MS Analysis
Metabolites were extracted from 50 mg fresh weight material ground under liquid nitrogen in 600 µL ice-cold N,N-dimethiylformamide in the presence of ribitol (30 µL from 0.2 mg/mL aqueous solution) used as an internal standard. After the addition of 400 µL water, the samples were shaken for 10 min at 4°C and subsequently centrifuged for 8 min at 14,000g. The aqueous phase was transferred to a new tube and vigorously mixed with 600 µL xylene for 10 min before centrifugation for 3 min at 14,000g. Aliquots (300 µL) from the lower aqueous phase were dried under vacuum, and the residue was derivatized for 120 min at 37°C (in 40 µL of 20 mg mL−1 methoxyamine hydrochloride in pyridine) followed by a 30-min treatment at 37°C with 70 µL of MSTFA. The GC-MS system used was a gas chromatograph coupled to a time-of-flight mass spectrometer (Leco Pegasus HT TOF-MS). An autosampler Gerstel Multi Purpose system injected the samples. Helium was used as carrier gas at a constant flow rate of 2 mL s−1, and gas chromatography was performed on a 30-m DB-35 column. The injection temperature was 230°C, and the transfer line and ion source were set to 250°C. The initial temperature of the oven (85°C) increased at a rate of 15°C/min up to a final temperature of 360°C. After a solvent delay of 180 s, mass spectra were recorded at 20 scans s−1 with m/z 70 to 600 scanning range. Chromatograms and mass spectra were evaluated by using Chroma TOF 4.5 (Leco) and TagFinder 4.2 software as described by Lisec et al. (2006).
RNA-Seq Analysis
cat2-2, cat2-2 gox1-1, and cat2-2 gox2-1 mutant plants were grown under elevated CO2 concentrations (3,000 μL L−1) to impair photorespiration. Individual plants were organized according to a completely randomized block design in a controlled climate chamber (Votsch Industrietechnik) in a 16-h/8-h light/dark regime (relative humidity of 75%, 21°C, and an irradiance of 120 µmol m−2 s−1). For high light treatments, 3-week-old plants were transferred to a Sanyo Fitotron growth chamber (Weiss Technik) in ambient air (∼400 μL L−1 CO2, relative humidity of 55%, 21°C) and irradiation of 1,000 µmol m−2 s−1 for 3 h. Middle aged leaves of 15 individual plants per line were sampled, pooled, and frozen in liquid nitrogen. This entire setup was repeated to obtain three biological repeats. Shoot tissue was sampled in three biological replicates before (t = 0) and after 3 h of treatment. RNA was extracted using a combination of TRIzol and RNeasy kit (Qiagen) according to the manufacturer’s instructions. Library preparation and sequencing was performed at the VIB Nucleomics Core. Sequencing libraries were constructed with using the TruSeq Stranded mRNA Library Preparation Kit (Illumina). Three biological replicates were sequenced on Illumina NextSEquation 500, resulting in approximately 20 million 75-bp single-end reads per sample. Adapter sequences and low quality base pairs (Q < 20) were trimmed using Trim Galore (v0.3.3; http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), retaining high-quality reads of at least 50 bp length. Quality-filtered reads were aligned to the TAIR10 Arabidopsis reference genome using the spliced aligner TopHat2 (v2.1.0). The number of reads per gene was quantified using featureCounts function as implemented in the Subread package v1.4.6. Reads mapping to genes annotated as microRNA, pseudogenes, and protein coding genes (TAIR10) were retained for further downstream analysis (28,517 genes in total).
Differentially expressed genes were identified using the R (v3.1.2) software package edgeR. For the analysis genes with expression values higher than 0.21 cpm (corresponding to five read counts) in at least three samples were retained (19,719 genes). TMM normalization was applied using the “calcNormFactors” function. Variability in the data set was assessed with a MDSplot employing the 3,000 top genes to calculate pairwise distances (top = 3000). There was a clear separation according to photorespiratory treatment; however, the third replicate also showed separation, especially under control conditions. Trended negative binomial dispersion parameters were estimated based on a model with main effects of genotype, treatment, replicate, and an interaction term between genotype and treatment using the “estimateGLMTrendedDisp” function, followed by the estimation of the empirical Bayes dispersion for each transcript. A negative binomial regression model was then used to model the overdispersed counts for each gene separately with fixed values for the dispersion parameter as implemented in the function glmFit using the above described model. A likelihood ratio test was performed to compare this model with a model without replicate to assess possible replicate (batch) effects. After FDR adjustments of the P values with the Benjamini and Hochberg method, 4,931 genes were found to have a significant batch effect as was expected by the MDS plot. Hence, we omitted the third replicate and used solely the first and second replicates for which 19,712 genes had expression values higher than 0.21 cpm (corresponding to five read counts) in at least two samples. The MDS plot showed a more pronounced effect in the absence of GOX1 under stress than the previous one including all three replicates. Following the same procedure as described above, a neglectable batch effect was observed. The estimate of the dispersions and the fitting of the model was repeated with only the main effects of stress treatment and genotype (cat2-2 gox1-1 and cat2-2 gox2-1) and treatment and their interaction. A likelihood ratio test followed by FDR adjustment showed 608 genes and 1 gene to be affected (FDR < 0.05) under stress due the lack of GOX1 or GOX2, respectively, which was in line with the MDS plot. To test user-defined hypotheses, a no intercept single-factor model was defined combining genotype and treatment factors (e.g. cat2-2_3h). Dispersions were estimated using the “estimateGLMRobustDisp” function. A negative binomial regression model was then used to model the overdispersed counts for each gene separately with fixed values for the dispersion parameter as outlined in McCarthy et al. (2012) and as implemented in the function “glmFit” using the above described model. Hypothesis testing was based on likelihood ratio tests. Contrasts of interest were the response between different genotypes under control conditions (t = 0 h) and the effect of photorespiratory stress in each genotype.
Identification of ROS-Responsive Genes
In order to generate a robust ROS signature the raw intensity (.CEL), files from five studies featuring ROS-generating conditions were obtained from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/). The following experiments were considered: reillumination of the conditional flu mutant for 2 h following a dark acclimation (GSE10812); treatment of seedlings with oligomycin for 4 h (GSE38965); treatment of seedlings with 50 µM antimycin A for 3 h (GSE41136); exposure of seedlings to ozone for 6 h (E-MEXP-342); treatment of seedlings with 10 mm H2O2 for 24 h. All raw intensity files were normalized by robust multiarray averaging using the Bioconductor package affy (v1.40.0). Probe sets were up to date using the TAIR10 CDF annotation retrieved from BrainArray (TAIRG v18.0.0; http://brainarray.mbni.med.umich.edu/). Differential gene expression was analyzed with the limma package (v3.18.13) using empirical Bayes moderated t-statistics. Differentially expressed genes were selected for each transcriptomic response, using a P value of 0.01 as a significance threshold and absolute log2 fold change greater than 1. Differential gene expression values were imported into the TM4 Microarray Software suite Multi Experiment Viewer v4.9.0 (TIGR). Genes were hierarchically clustered using Euclidean distance with average linkage.
Quantitative RT-PCR
For quantification of expression levels, total RNA was extracted from 3-week- old rosettes with the RNeasy Plant Mini Kit (Qiagen). First-strand cDNA synthesis was performed with iScript cDNA Synthesis Kit with 1 µg of total RNA used as input material. Five microliters of the 1:8 diluted first-strand cDNA was used as a template in subsequent PCR performed on the iCycler iQ (Bio-Rad) with respective gene specific primers (Supplemental Table S3). Reactions were performed in three technical repeats with the SYBR Green I Master Kit according to the manufacturer’s instructions. Expression analysis was performed with qBASEPlus software (Biogazelle) with ELONGATION FACTOR1α (EF-1α) and POLYUBIQUTIN5 (UBIQ5) used for data normalization.
All experiments were performed with four biological replicates. Statistical analysis was carried out with one-way ANOVA followed by the Tukey-Kramer post-hoc test.
GOX Enzyme Assays
The glycolate oxidase activity was measured using the o-Dianisidine method described before (Rojas et al., 2012) downscaled for 200 mg ground tissue. The shoot tissue was collected and frozen in liquid nitrogen and ground with ball mill (Retsch). The material was then mixed with 1 mL of extraction buffer by vortexing and centrifuged at 16,100g for 30 min at 4°C. The protein concentration in soluble phase was determined with Bradford Assay (Bradford, 1976). Next, 10 µL supernatant was used in spectrophotometric glycolate oxidase activity assay conducted in VersaMax microplate reader (Molecular Devices) at room temperature for 1 h. Specific activity was calculated by dividing ΔA by time and amount of protein present in the sample and expressed as percentage of control. All enzymatic assays were performed on three biological replicates with three technical repeats.
Alternatively, GOX activity was assessed as described by Yamaguchi and Nishimura (2000) with minor modifications. Briefly, 100 mg plant material ground under liquid nitrogen was homogenized in 100 mm HEPES (pH 7.2) containing 1 mm EDTA and 10 mm 2-mercaptoethanol. Following centrifugation, 10 µL supernatant was introduced in 180 µL reaction medium (100 mm triethanolamine, pH 7.8, 3 mm EDTA, 0.75 mm oxidized glutathione, and 4 mm phenylhydrazine), and the reaction was started with 2.3 mm glycolate. The amount of enzyme catalyzing the production of 1 μmol glyoxylate-phenylhydrazone min−1 based on the extinction coefficient for phenylhydrazone at 324 nm (16.8 mM−1 cm−1) was defined as one unit. The enzyme activity was expressed as mU per mg protein determined as described above.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GSE77170 and GSE77171.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GOX1 and GOX2 gene models.
Supplemental Figure S2. Read coverage of GOX1 and GOX2 loci from RNA-seq analysis of cat2-2 gox1-1, cat2-2, and cat2-2 gox2-1 mutants.
Supplemental Figure S3. Heat map of differentially expressed transcripts (|log2 FC| > 1, FDR < 0.05) between cat2-2 gox1-1, cat2-2 gox2-1, and cat2-2 mutants under control high CO2 conditions aimed at inhibiting the photorespiratory flux.
Supplemental Figure S4. Redox status of Col-0, cat2-2, cat2-2 gox1-1, and cat2-2 gox2-1 plants grown under ambient air and moderate light intensity (300 μmol m−2 s−1).
Supplemental Figure S5. Nonphotochemical quenching and phosphorylation of LHCBII upon exposure of Col-0, gox1-1, and gox2-1 plants grown under conditions limiting photorespiration to high light.
Supplemental Figure S6. GOX1 and GOX2 genes in Brassicaceae are clustered as monophyly in the gene tree of ORTHO03D000507 from PLAZA 3.0 dicots.
Supplemental Figure S7. Extractable leaf GOX activity.
Supplemental Figure S8. GOX1 and GOX2 transcript abundance.
Supplemental Table S1. Maximum likelihood estimates of parameters under branch models on both postduplication branches leading to the GOX1 and GOX2 clades.
Supplemental Table S2. Likelihood ratio tests for comparing different branch models applied to both postduplication branches leading to the GOX1 and GOX2 clades.
Supplemental Table S3. The modified branch-site models for detecting positive selection on six branches after the duplication event leading to GOX2 in Arabidopsis.
Supplemental Table S4. List of primers used in this study.
Supplemental Table S5. Mutations enriched in line 238.3.
Supplemental Table S6. Data sources and sequence accessions.
Supplemental Data Set 1. RNA-seq results.
Supplemental Data Set 2. List of transcripts that responded to the photorespiratory stress treatment in a genotype-specific manner.
Supplemental Data Set 3. Gene Set Enrichment Analysis (http://structuralbiology.cau.edu.cn/PlantGSEA) of the transcripts that responded to the photorespiratory stress treatment in a genotype-specific manner.
Supplementary Material
Glossary
- PG
2-phosphoglycolate
- GC-MS
gas chromatography-mass spectrometry
- DAB
3,3′-diaminobenzidine
- ETR
rate of electron transfer
- NPQ
nonphotochemical quenching
- APX
ascorbate peroxidase
Footnotes
Articles can be viewed without a subscription.
References
- Abat JK, Mattoo AK, Deswal R (2008) S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata- ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J 275: 2862–2872 [DOI] [PubMed] [Google Scholar]
- Bailey CD, Koch MA, Mayer M, Mummenhoff K, O’Kane SL Jr., Warwick SI, Windham MD, Al-Shehbaz IA (2006) Toward a global phylogeny of the Brassicaceae. Mol Biol Evol 23: 2142–2160 [DOI] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Kern R, Timm S (2012) Photorespiration has a dual origin and manifold links to central metabolism. Curr Opin Plant Biol 15: 269–275 [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R (2004) GeneWise and Genomewise. Genome Res 14: 988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caverzan A, Passaia G, Rosa SB, Ribeiro CW, Lazzarotto F, Margis-Pinheiro M (2012) Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet Mol Biol 35: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellero Y, Jossier M, Glab N, Oury C, Tcherkez G, Hodges M (2016) Decreased glycolate oxidase activity leads to altered carbon allocation and leaf senescence after a transfer from high CO2 to ambient air in Arabidopsis thaliana. J Exp Bot 67: 3149–3163 [DOI] [PubMed] [Google Scholar]
- Dellero Y, Mauve C, Boex-Fontvieille E, Flesch V, Jossier M, Tcherkez G, Hodges M (2015) Experimental evidence for a hydride transfer mechanism in plant glycolate oxidase catalysis. J Biol Chem 290: 1689–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ. (2016) Thiol-based peroxidases and ascorbate peroxidases: why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? Mol Cells 39: 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist MKM, Schmitz J, Gertzmann A, Florian A, Jaspert N, Arif M, Balazadeh S, Mueller-Roeber B, Fernie AR, Maurino VG (2015) GLYCOLATE OXIDASE3, a glycolate oxidase homolog of yeast l-lactate Cytochrome c Oxidoreductase, supports l-lactate oxidation in roots of Arabidopsis. Plant Physiol 169: 1042–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, Kuhn A, Groth G, Lercher MJ, Maurino VG (2014) Plant and animal glycolate oxidases have a common eukaryotic ancestor and convergently duplicated to evolve long-chain 2-hydroxy acid oxidases. Mol Biol Evol 31: 1089–1101 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Bloom AJ, Queval G, Noctor G (2009) Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu Rev Plant Biol 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Guy L, Kultima JR, Andersson SG (2010) genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26: 2334–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig B, James GV, Konrad K, Schneeberger K, Turck F (2012) Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol 160: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M, Dellero Y, Keech O, Betti M, Raghavendra AS, Sage R, Zhu XG, Allen DK, Weber AP (2016) Perspectives for a better understanding of the metabolic integration of photorespiration within a complex plant primary metabolism network. J Exp Bot 67: 3015–3026 [DOI] [PubMed] [Google Scholar]
- Hodges M, Jossier M, Boex-Fontvieille E, Tcherkez G (2013) Protein phosphorylation and photorespiration. Plant Biol (Stuttg) 15: 694–706 [DOI] [PubMed] [Google Scholar]
- Kaurilind E, Xu E, Brosché M (2015) A genetic framework for H2O2 induced cell death in Arabidopsis thaliana. BMC Genomics 16: 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev P, Mühlenbock P, Denecker J, Morreel K, Hoeberichts FA, Van Der Kelen K, Vandorpe M, Nguyen L, Audenaert D, Van Breusegem F (2015) Activation of auxin signalling counteracts photorespiratory H2O2-dependent cell death. Plant Cell Environ 38: 253–265 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Lobell DB, Gourdji SM (2012) The influence of climate change on global crop productivity. Plant Physiol 160: 1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis A, Murat F, Salse J, Crollius HR (2015) GenomicusPlants: a web resource to study genome evolution in flowering plants. Plant Cell Physiol 56: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T, Tanouchi A, Tamoi M, Yabuta Y, Yoshimura K, Ishikawa T, Shigeoka S (2010) Arabidopsis chloroplastic ascorbate peroxidase isoenzymes play a dual role in photoprotection and gene regulation under photooxidative stress. Plant Cell Physiol 51: 190–200 [DOI] [PubMed] [Google Scholar]
- Massey V. (1994) Activation of molecular oxygen by flavins and flavoproteins. J Biol Chem 269: 22459–22462 [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Narendra S, Venkataramani S, Shen G, Wang J, Pasapula V, Lin Y, Kornyeyev D, Holaday AS, Zhang H (2006) The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J Exp Bot 57: 3033–3042 [DOI] [PubMed] [Google Scholar]
- Noctor G, Veljovic-Jovanovic S, Driscoll S, Novitskaya L, Foyer CH (2002) Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Ann Bot 89: 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Rodríguez-Serrano M, Pazmiño DM, Gupta DK, Sandalio LM, Romero-Puertas MC (2012) S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. J Exp Bot 63: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schulz MA, Obata T, Fernie AR, Weber APM (2013) PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc Natl Acad Sci USA 110: 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res 43: D974–D981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Liang H, Rozenberg M, Mittler R (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34: 187–203 [DOI] [PubMed] [Google Scholar]
- Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakière B, Vanacker H, Miginiac-Maslow M, Van Breusegem F, Noctor G (2007) Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J 52: 640–657 [DOI] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lüder F, Weckwerth W, Jahn O (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD. (2014) Regulation and dynamics of the light-harvesting system. Annu Rev Plant Biol 65: 287–309 [DOI] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Wang K, Ryu C-M, Kaundal A, Mysore KS (2012) Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24: 336–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Villarejo A, Srivastava M, Gelhaye E, Keech O, Droux M, Finkemeier I, Samuelsson G, Dietz KJ, Jacquot JP, Wingsle G (2005) Identification of plant glutaredoxin targets. Antioxid Redox Signal 7: 919–929 [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin PA, Edwards EJ (2011) The C(4) plant lineages of planet Earth. J Exp Bot 62: 3155–3169 [DOI] [PubMed] [Google Scholar]
- Schäfer L, Feierabend J (2000) Photoinactivation and protection of glycolate oxidase in vitro and in leaves. Z Naturforsch C 55: 361–372 [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen J-E, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Timm S, Bauwe H (2013) The variety of photorespiratory phenotypes - employing the current status for future research directions on photorespiration. Plant Biol (Stuttg) 15: 737–747 [DOI] [PubMed] [Google Scholar]
- Timm S, Florian A, Jahnke K, Nunes-Nesi A, Fernie AR, Bauwe H (2011) The hydroxypyruvate-reducing system in Arabidopsis: multiple enzymes for the same end. Plant Physiol 155: 694–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S, Nunes-Nesi A, Pärnik T, Morgenthal K, Wienkoop S, Keerberg O, Weckwerth W, Kleczkowski LA, Fernie AR, Bauwe H (2008) A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell 20: 2848–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, Suzuki N, Miller G, van de Cotte B, Morsa S, Ravanat J-L, Hegie A, Triantaphylidès C, Shulaev V, Van Montagu MCE, Van Breusegem F, Mittler R (2011) Extranuclear protection of chromosomal DNA from oxidative stress. Proc Natl Acad Sci USA 108: 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR (2016) The costs of photorespiration to food production now and in the future. Annu Rev Plant Biol 67: 107–129 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, et al. ; Brassica rapa Genome Sequencing Project Consortium (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Waszczak C, Kerchev K, Mühlenbock P, Frank A. Hoeberichts, Van Der Kelen K, Mhamdi A, Willems P, Denecker J, Kumpf R, Noctor G, Messens J, Van Breusegem F (2016) SHORT-ROOT deficiency alleviates the cell death phenotype of the Arabidopsis catalase2 mutant under photorespiration-promoting conditions. Plant Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler T, von Braun J (2013) Climate change impacts on global food security. Science 341: 508–513 [DOI] [PubMed] [Google Scholar]
- Willems P, Mhamdi A, Stael S, Storme V, Kerchev PI, Noctor G, Gevaert K, Van Breusegem F (2016) The ROS Wheel: refining ROS transcriptional footprints in Arabidopsis. Plant Physiol 171: 1720–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhang J, Zeng J, Jiang L, Liu E, Peng C, He Z, Peng X (2009) Inducible antisense suppression of glycolate oxidase reveals its strong regulation over photosynthesis in rice. J Exp Bot 60: 1799–1809 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Nishimura M (2000) Reduction to below threshold levels of glycolate oxidase activities in transgenic tobacco enhances photoinhibition during irradiation. Plant Cell Physiol 41: 1397–1406 [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z (2005) Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22: 2472–2479 [DOI] [PubMed] [Google Scholar]
- Zelitch I, Schultes NP, Peterson RB, Brown P, Brutnell TP (2009) High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol 149: 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.