MED15 mediates activation of lipid-related genes by WRI1 during seed maturation stages.
Abstract
The Mediator complex is known to be a master coordinator of transcription by RNA polymerase II, and this complex is recruited by transcription factors (TFs) to target promoters for gene activation or repression. The plant-specific TF WRINKLED1 (WRI1) activates glycolysis-related and fatty acid biosynthetic genes during embryogenesis. However, no Mediator subunit has yet been identified that mediates WRI1 transcriptional activity. Promoter-β-glucuronidase fusion experiments showed that MEDIATOR15 (MED15) is expressed in the same cells in the embryo as WRI1. We found that the Arabidopsis (Arabidopsis thaliana) MED15 subunit of the Mediator complex interacts directly with WRI1 in the nucleus. Overexpression of MED15 or WRI1 increased transcript levels of WRI1 target genes involved in glycolysis and fatty acid biosynthesis; these genes were down-regulated in wild-type or WRI1-overexpressing plants by silencing of MED15. However, overexpression of MED15 in the wri1 mutant also increased transcript levels of WRI1 target genes, suggesting that MED15 also may act with other TFs to activate downstream lipid-related genes. Chromatin immunoprecipitation assays confirmed the association of MED15 with six WRI1 target gene promoters. Additionally, silencing of MED15 resulted in reduced fatty acid content in seedlings and mature seeds, whereas MED15 overexpression increased fatty acid content in both developmental stages. Similar results were found in wri1 mutant and WRI1 overexpression lines. Together, our results indicate that the WRI1/MED15 complex transcriptionally regulates glycolysis-related and fatty acid biosynthetic genes during embryogenesis.
Seed development is a crucial process in the life cycle of flowering plants, and it can be roughly divided into two distinct stages: embryogenesis and seed maturation (Baud et al., 2002; Vicente-Carbajosa and Carbonero, 2005). In Arabidopsis (Arabidopsis thaliana), embryogenesis begins with a single embryogenic cell formed after sexual fertilization and ends at the heart stage of embryo development (6 d after pollination [DAP]). During this process, the activities of specific genes are required to generate the apical-basal axis (Vicente-Carbajosa and Carbonero, 2005), and seeds are white or pale yellow due to high water content (greater than 80%) and relatively low fatty acid content (Baud et al., 2008). The transition from embryogenesis to embryo maturation is characterized by an interruption of growth and specific gene expression. During the maturation phase (7–20 DAP), developing seeds initially accumulate starch, which is subsequently degraded to generate energy and carbon skeleton for the synthesis of lipids and seed storage proteins (Baud et al., 2002; Ruuska et al., 2002). Triacylglycerol (TAG), an ester derived from glycerol and fatty acids, is the major storage form of lipids in Arabidopsis embryos.
In Arabidopsis, three family members of the B3 domain transcription factors (TFs), LEAFY COTYLEDON2 (LEC2), ABSCISIC ACID INSENSITIVE3 (ABI3), and FUSCA3, as well as LEC1, which belongs to the CBF family, are involved in storage protein synthesis and seed maturation processes (Lotan et al., 1998; Gutierrez et al., 2007; Suzuki and McCarty, 2008). On the other hand, WRINKLED1 (WRI1), which is known as an important regulator in Arabidopsis seed development, specifies the activities of genes encoding most key enzymes involved in late stages of glycolysis and fatty acid biosynthesis (Focks and Benning, 1998; Baud et al., 2007; Maeo et al., 2009; Ma et al., 2013).
WRI1 is a plant-specific TF of the APETALA2 (AP2)/ethylene-responsive element-binding protein family. This factor binds to the AW box [CnTnG](n)7[CG] sequence that is enriched in promoters of fatty acid biosynthetic genes (Cernac and Benning, 2004; Baud et al., 2007; Maeo et al., 2009). Most AW boxes are located near the transcription start site and in the 5′ untranslated region of target genes (Maeo et al., 2009). The Arabidopsis wri1 mutant produces wrinkled seeds with only 20% of wild-type TAG content and increased starch levels, but otherwise it has no obvious phenotype during vegetative development (Baud et al., 2007). In addition, wri1 mutant seeds contain an altered fatty acid composition with higher levels of linolenic acid (C18:3) and erucic acid (C22:1) but lower levels of oleic (C18:1), linoleic (C18:2), and eicosenoic (C20:1) acids (Focks and Benning, 1998). Consistent with the results of the wri1 mutant, overexpression of WRI1 up-regulated a number of glycolytic and fatty acid biosynthetic genes in seedlings and increased the overall seed oil content without altering its fatty acid composition in Arabidopsis. Similar WRI1 overexpression results have been obtained in oilseed rape (Brassica napus), maize (Zea mays), and castor bean (Ricinus communis; Liu et al., 2010; Pouvreau et al., 2011; Sanjaya et al., 2011; Tajima et al., 2013), indicating functional conservation of WRI1 in both monocots and dicots.
In eukaryotes, gene expression is regulated by TFs that bind to specific cis-elements of promoters and enhancer sequences of target genes. After binding to the target DNA, the TF initiates the assembly of a large protein complex in order to alter chromatin structure and initiate transcription. Specifically, TFs are known to execute their functions through the Mediator complex, which is evolutionarily conserved from yeast to human (Taatjes, 2010). As a central coregulator of eukaryotic transcription, Mediator is a large complex comprising 25 to 30 different protein subunits, and it plays an essential role in transcriptional regulation by connecting DNA-binding TFs to the RNA polymerase II (Pol II) transcription machinery (Malik and Roeder, 2010; Bernecky et al., 2011). Depending on its associated protein components, Mediator can serve as either a transcriptional activator or a repressor (Conaway and Conaway, 2011). First identified in yeast and subsequently found in all eukaryotes, the Mediator complex is composed of four modules: head, middle, tail, and an additional detachable cyclin-dependent kinase module (Liu et al., 2001). The modular architecture and subunit composition also are highly conserved among eukaryotes from yeast to human (Bourbon, 2008). The head and middle modules bind to the C-terminal domain of Pol II (Asturias et al., 1999), whereas subunits of the tail module interact primarily with DNA-bound TFs (Malik and Roeder, 2005). The subunits of the cyclin-dependent kinase module are associated with negative regulation of a subset of genes by interacting with certain TFs (Holstege et al., 1998).
The Arabidopsis Mediator complex was initially characterized by Bäckström et al. (2007) to contain 27 subunits, but further analysis showed that it may contain up to 30 to 35 subunits (Mathur et al., 2011). MEDIATOR25 (MED25)/PHYTOCHROME AND FLOWERING TIME1 and MED14/STRUWWELPETER (SWP) were first identified as key regulators of flowering time and cell proliferation, respectively (Autran et al., 2002; Cerdán and Chory, 2003). Several mediators, MED8, MED16/SENSITIVE TO FREEZING6 (SFR6), and MED21, have been implicated in resistance to necrotrophic pathogens (Dhawan et al., 2009; Kidd et al., 2009; Knight et al., 2009; Hemsley et al., 2014). The production of small and long noncoding RNAs depends on the function of mediators such as MED8, MED17, MED18, and MED20a (Kim et al., 2011). Recently, MED18 was shown to have multiple functions through its interaction with several TFs, such as SUPPRESSOR OF FRIGIDA4, ABI4, and YIN YANG1, to determine flowering time and mediate plant responses to abscisic acid and bacterial infection, respectively (Lai et al., 2014).
Together with MED2, MED3, MED5, MED14/SWP, and MED16/SFR6, MED15 is a subunit of the tail module in yeast. The kinase-inducible domain-interacting (KIX) domain located at the N-terminal region of MED15 mediates protein-protein interaction through a hydrophobic pocket (Parker et al., 1996). MED15 plays a central role in fatty acid biosynthesis and stress metabolism by interaction with TFs such as Nuclear Hormone Receptor-49 and sterol regulatory element-binding proteins (SREBPs) in worms and mammals (Taubert et al., 2006; Yang et al., 2006). The yeast MED15, which also is known as Gal11p (Gal11p/MED15), is essential for the activation of fatty acid biosynthetic genes through its interaction with the TF Oaf1p (Thakur et al., 2009).
Among the three Arabidopsis MED15 orthologs identified by in silico analysis (Mathur et al., 2011), two genes, MED15_1 (At1g15780) and MED15_2 (At2g10440), have been characterized (Canet et al., 2012; Pasrija and Thakur, 2012; Thakur et al., 2013). MED15_1 has low sequence homology with MED15_2,with only about 30% amino acid identity. MED15_1, which is highly expressed in all tissues, is presumed to have a function in seed development (Thakur et al., 2013). Given the conserved function of many MED subunits among eukaryotes we hypothesized that MED15 may be involved in lipid homeostasis in Arabidopsis as well.
Here, we show that MED15 interacts directly with WRI1 in the nucleus. The expression patterns between MED15 and WRI1 are correlated during embryo development. Silencing of MED15 expression resulted in reduced fatty acid content in mature seeds. On the other hand, overexpression of MED15 was able to partially rescue wri1 mutant phenotypes. We found that MED15 was recruited to the promoters of WRI1 target genes. Our findings identify MED15 as a target of WRI1 and provide new insights into the role of MED15 in plant lipid metabolism.
RESULTS
MED15 Interacts with WRI1 in Vitro and in Vivo
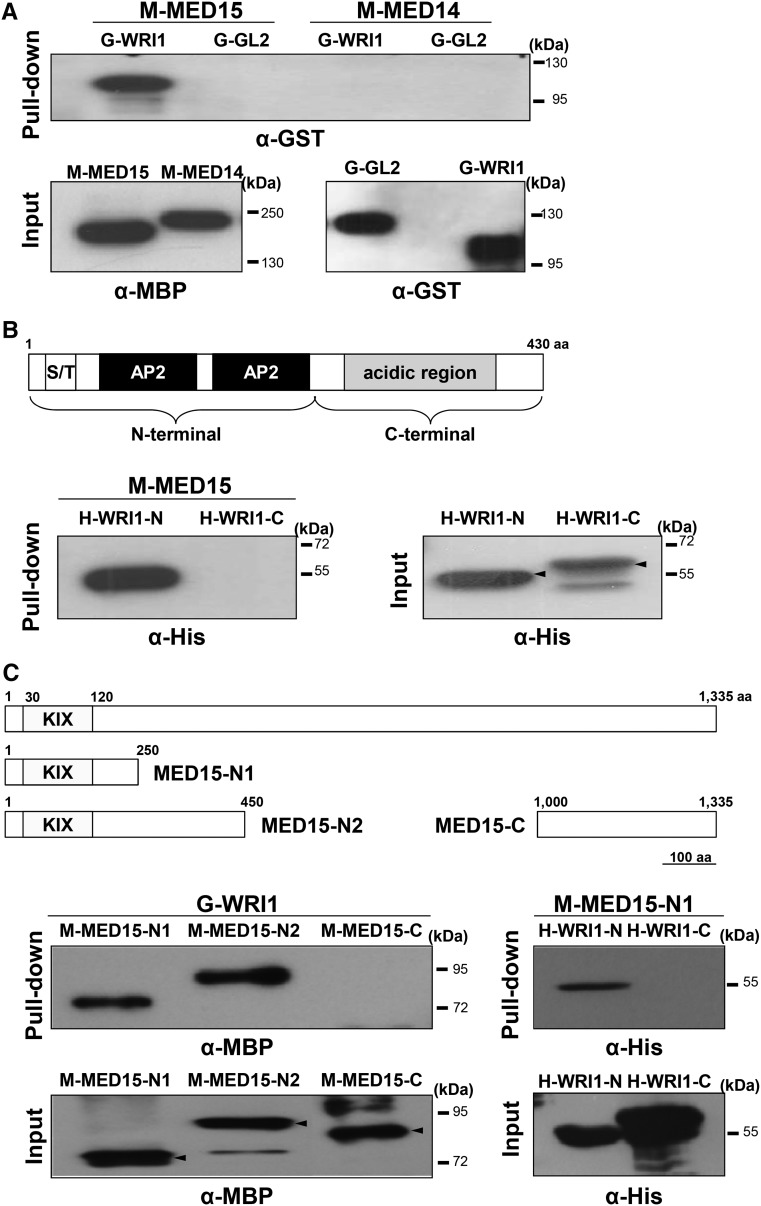
We purified MBP-MED15 (M-MED15) and glutathione S-transferase (GST)-WRI1 (G-WRI1) fusion proteins from Escherichia coli extracts and tested for possible interaction between them in vitro. GL2, another TF (Shen et al., 2006), was used as a negative control for WRI1, and MED14 was used as a negative control for MED15. Figure 1A shows that M-MED15 was able to bind to G-WRI1 but not G-GL2, whereas in similar assays, M-MED14 did not bind to G-WRI1 or G-GL2. These results show that the association between MED15 and WRI1 is specific.
Figure 1.
MED15 interacts with WRI1 in vitro. A, Top, In vitro pull-down assay showing direct interaction between WRI1 and MED15. M-MED15 and M-MED14 fusion proteins were used as bait proteins, and G-WRI1 and G-GL2 fusion proteins were used as target proteins. Proteins pulled down by M-MED15 or M-MED14 were detected by anti-GST antibody (α-GST). Bottom, Input amounts of purified recombinant M-MED15/M-MED14 and G-WRI1/G-GL2 proteins detected by anti-MBP antibody (α-MBP) and α-GST, respectively. B, In vitro pull-down assay showing interaction between MED15 and the N-terminal region of WRI1. Top, Schematic diagram of the WRI1 protein structure. S/T, Ser/Thr-rich domain; AP2, AP2 DNA-binding domain; aa, amino acids. Bottom left, His-tagged N-terminal fragment of WRI1 (H-WRI1-N; amino acids 1–250) and C-terminal fragment of WRI1 (H-WRI1-C; amino acids 251–430) were pull downed with M-MED15 and detected by anti-His antibody (α-His). Bottom right, Input amounts of purified H-WRI1-N and H-WRI1-C as detected by α-His. C, In vitro pull-down assay showing the interaction between WRI1 and the N-terminal region of MED15 including the KIX domain. Top, Schematic diagrams of full-length MED15 (amino acids 1–1,335), two overlapping N-terminal fragments of MED15, MED15-N1 (amino acids 1–250) and MED15-N2 (amino acids 1–450), and a C-terminal fragment of MED15, MED15-C (amino acids 1,000–1,335). KIX, The KIX domain (amino acids 30–120). Middle left, MBP-fused MED15 deletion proteins were pull downed with G-WRI1 and detected by α-MBP. Bottom left, Input amounts of purified M-MED15-N1, M-MED15-N2, and M-MED15-C detected by α-MBP. Middle right, His-tagged WRI1 deletion proteins were pull downed with M-MED15-N1 and detected by α-His. Bottom right, Input amounts of purified H-WRI1-N and H-WRI1-C detected by α-His.
Both MED15 and WRI1 are large proteins containing several putative domains. We used deletion derivatives of the two proteins to localize their interacting regions. M-MED15 associated with the N-terminal region (amino acids 1–250) but not the C-terminal region (amino acids 251–430) of WRI1 (Fig. 1B). Similar experiments showed that G-WRI1 bound to an N-terminal fragment of MED15 that contains the KIX domain (amino acids 30–120). Further experiments confirmed that this N-terminal fragment of MED15 was able to bind to the N-terminal fragment (amino acids 1–250) of WRI1 (Fig. 1C).
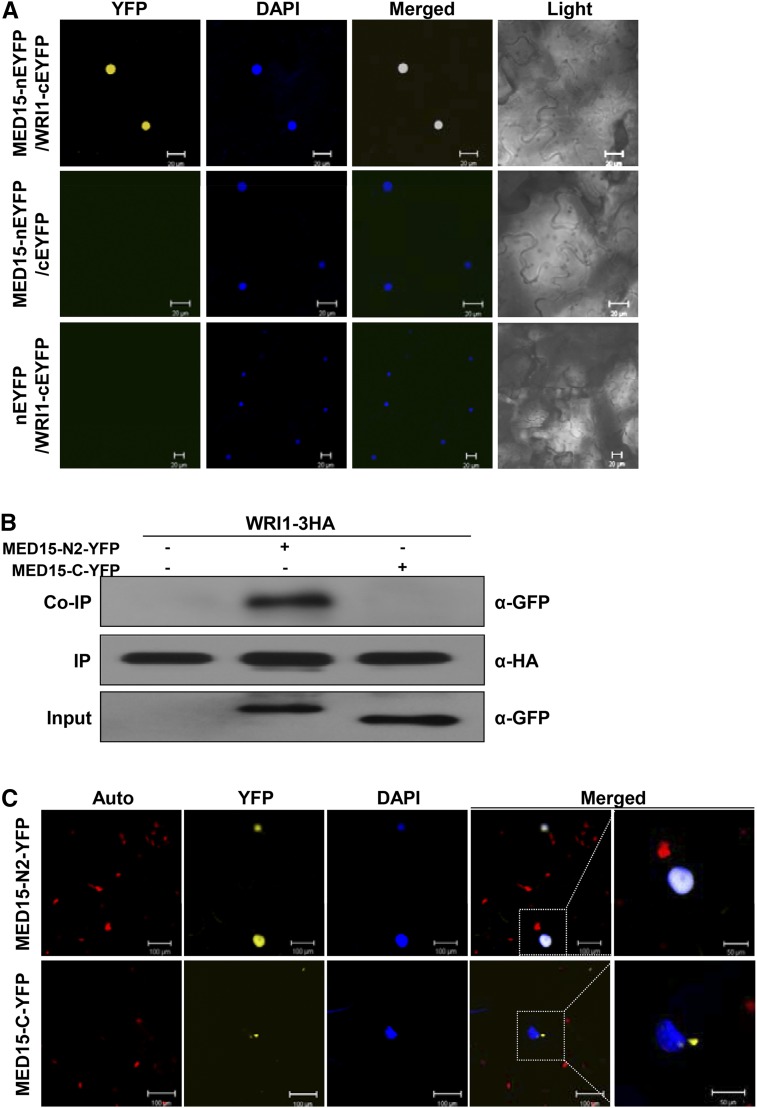
To see if the in vitro interaction of the two proteins can be recapitulated in vivo, we used the bimolecular fluorescence complementation (BiFC) assay. MED15 was fused to an N-terminal fragment of enhanced yellow fluorescent protein (nEYFP), whereas WRI1 was fused to the C-terminal fragment of EYFP (cEYFP). nEYFP and cEYFP alone were used as negative controls. Figure 2A shows that YFP fluorescence was reconstituted when the two proteins were coexpressed. No fluorescence was detected with MED15-nEYFP and cEYFP nor with nEYFP and WRI1-cEYFP. DAPI staining confirmed that the MED15/WRI1 complex was localized in the nucleus.
Figure 2.
MED15 interacts with WRI1 in vivo. A, BiFC analysis showing the interaction between MED15 and WRI1. A complementary DNA (cDNA) encoding full-length MED15 or WRI1 was cloned into pSAT1 serial vectors containing DNA sequences encoding either N- or C-terminal fragments of EYFP. Fusion proteins were transiently expressed in Nicotiana benthamiana leaf cells by Agrobacterium tumefaciens-mediated infiltration. Fluorescence signals were captured using the YFP channel of a confocal microscope. Nuclei were visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining. YFP, YFP channel image; DAPI, DAPI channel image; Merged, merged image between YFP and DAPI; Light, light microscopic image. Bars = 20 µm. B, Coimmunoprecipitation (Co-IP) of MED15 and WRI1. Protein extracts of A. tumefaciens-infiltrated N. benthamiana leaves expressing WRI1-3HA, WRI1-3HA/MED15-N2-YFP, or WRI1-3HA/MED15-C-YFP were immunoprecipitated with anti-HA antibody (α-HA). Input proteins and the immunoprecipitates (IP) were detected by α-HA and anti-GFP antibody (α-GFP), respectively. C, Subcellular localization of YFP-fused MED15-N2 and YFP-fused MED15-C. Fluorescence signals were captured using the YFP channel of a confocal microscope. Nuclei were visualized by DAPI staining. The white dotted squares in the merged images are enlarged at right. Auto, Autofluorescence of chloroplasts; YFP, YFP channel image; DAPI, DAPI channel image; Merged, merged image between YFP and DAPI. Bars = 100 µm for all images except the enlarged images (50 µm).
We also tested if MED15 and WRI1 would interact in vivo using transient expression assays in leaf cells of Nicotiana benthamina. Indeed, MED15-N2-YFP but not MED15-C-YFP was coimmunoprecipitated along with WRI1-3X-hemagglutinin (HA; Fig. 2B). Figure 2C shows that the MED15-N2 fragment was able to localize to the nucleus. Note that the MED15-N1 fragment (amino acids 1–250), which contains the KIX domain, also was able to localize to the nucleus (Supplemental Fig. S1).
Spatiotemporal Expression of MED15 and WRI1
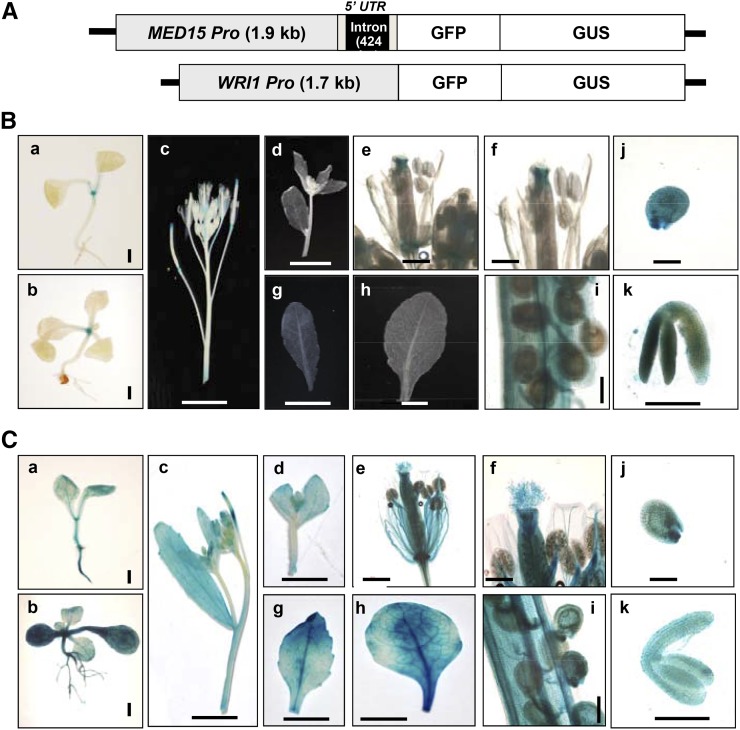
To investigate the expression patterns of MED15 and WRI1 during different stages of Arabidopsis growth and development, we generated independent transgenic plants expressing GFP-GUS fusion protein using either a MED15 promoter (2.3 kb) or a WRI1 promoter (1.7 kb; Fig. 3A). Figure 3B shows that WRI1 was highly expressed during seed maturation stages including globular stage embryo (j; 3–4 DAP) and mature green embryo (k; 9–10 DAP; Baud et al., 2007). In addition to seed maturation stages, WRI1 was expressed specifically in the shoot apical meristem (SAM) of 1-week-old (a) and 2-week-old (b) seedlings, young developing siliques (i), and carpels (e and f) but was not expressed in stamens (e and f), roots (a and b), cotyledons (a and b), and mature leaves (g and h; Fig. 3B). Similar gene expression results were obtained from the Arabidopsis eFP Browser 2.0 (http://bar.utoronto.ca/efp2/Arabidopsis/Arabidopsis_eFPBrowser2.html), which provides publicly available Arabidopsis gene expression data sets (Supplemental Fig. S2). However, WRI1 expression in SAM of 1- and 2-week-old seedlings was only detected in our experiments (Fig. 3B). On the other hand, MED15 was constitutively expressed in all tissues, from seedlings (a and b) to mature vegetative and reproductive stages (c–k). These observations were similar to the expression data derived from the Arabidopsis eFP Browser 2.0 (Fig. 3C; Supplemental Fig. S2). These results suggest that MED15 functions broadly during Arabidopsis growth and development and may act together with WRI1 in seed maturation stages.
Figure 3.
Expression patterns of MED15 promoter-GUS or WRI1 promoter-GUS in transgenic Arabidopsis plants. A, Schematic diagrams of MED15Pro::GFP-GUS and WRI1Pro::GFP-GUS constructs. The MED15 promoter contains the 5′ untranslated region (5′ UTR) with an intron (424 bp). B, Histochemical analysis of GUS activity in a WRI1Pro::GFP-GUS transgenic plant. C, Histochemical analysis of GUS activity in a MED15Pro::GFP-GUS transgenic plant. GUS assays were performed throughout different development stages, two stages of seedlings, 1 week old (a) and 2 weeks old (b), inflorescence (c and d), flower (e and f), cauline leaf (g), rosette leaf (h), young siliques with developing seeds (i), young seeds (3–4 DAP, globular embryo stage; j), and excised embryo from midstage seeds (9 to 10 DAP, mature embryo stage; k). Bars = 50 µm (a, b, e, f, and i–k) and 5 mm (c, d, g, and h).
MED15 Overexpression Induces WRI1 Target Genes
To investigate whether MED15 and WRI1 function together in seed maturation stages, we generated overexpression (OE) lines (WRI1-OE and MED15-OE) of each gene in the wild-type and wri1-4 backgrounds. In addition, we silenced MED15 expression by RNA interference (RNAi; MED15-Ri) and overexpressed WRI1 in the MED15-Ri background. Plants of all these genotypes were used for further analysis (Fig. 4).
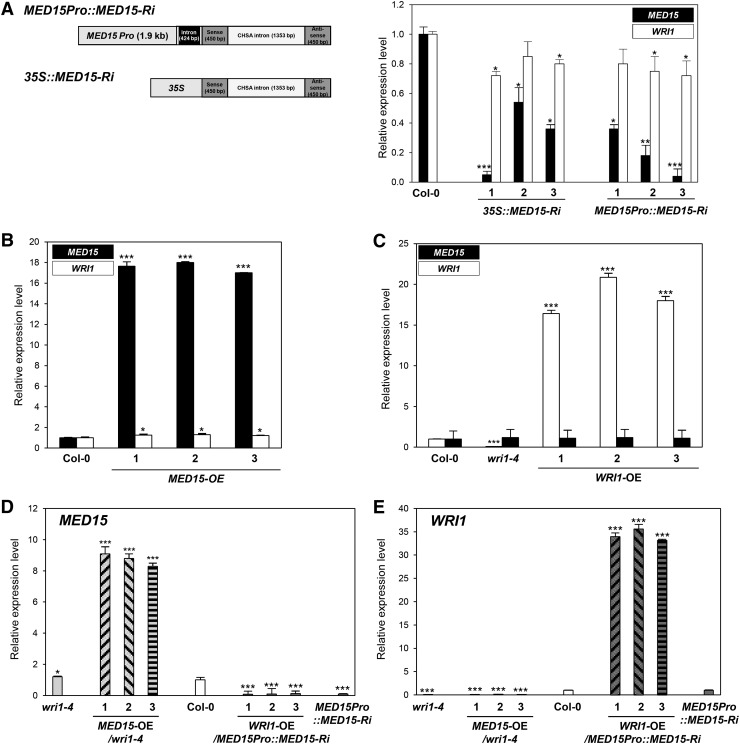
Figure 4.
Expression levels of MED15 and WRI1 in transgenic Arabidopsis plants for overexpression and reduction of MED15 and WRI1 transcripts. A, Left, Schematic diagrams of MED15-Ri constructs transcribed from a MED15 or a 35S promoter. Right, Quantitative reverse transcription (qRT)-PCR analysis showing MED15 and WRI1 expression levels in 35S::MED15-Ri (lines 1–3) and MED15Pro::MED15-Ri (lines 1–3). B and C, qRT-PCR analysis showing elevated MED15 (B) and WRI1 (C) expression levels in MED15-OE and WRI1-OE lines, respectively. D and E, qRT-PCR analysis showing MED15 (D) and WRI1 (E) expression levels in the wild type (Columbia-0 [Col-0]) and wri1-4, MED15-OE/wri1-4, WRI1-OE/MED15Pro::MED15-Ri, and MED15Pro::MED15-Ri lines. Two-week-old Arabidopsis seedlings were used for qRT-PCR analysis. The expression values were normalized using ACTIN1 as an internal control. Each experiment was repeated with three biological replicates. Student’s t test: *, P < 0.05, **, P < 0.01, or ***, P < 0.001 versus wild-type Col-0. Error bars represent sd.
As the homozygous mutant of MED15 (nrb4-4; non-recognition-of-BTH4-4) is sterile (Canet et al., 2012), we used two different promoters, cauliflower mosaic virus 35S and MED15, to silence MED15 expression in transgenic plants by RNAi technology (Fig. 4A). Figure 4A shows that RNAi was effective at reducing MED15 expression with either promoter, but the residual MED15 expression levels varied among independent lines. Irrespective of the promoter used, silencing of MED15 expression also moderately decreased WRI1 transcript levels in all the independent lines investigated. There was some correlation between the degree of MED15 suppression and the reduction levels of WRI1. Consistent with this observation, overexpression of MED15 led to slightly elevated WRI1 transcript levels (Fig. 4B). Figure 4, B to E, show overexpression levels of MED15 and WRI1 transcript in three independent lines of MED15-OE, WRI1-OE, MED15-OE/wri1-4 and WRI1-OE/MED15Pro::MED15-Ri.
To further understand how MED15 and WRI1 regulate fatty acid biosynthesis at the molecular level, we analyzed transcript levels of 11 key fatty acid biosynthetic genes, including glycolytic, late fatty acid biosynthetic, and storage-related genes, in 2-week-old seedlings (Fig. 5) and siliques bearing early-stage developing seeds (3–8 DAP; Supplemental Fig. S3) of various genotypes.
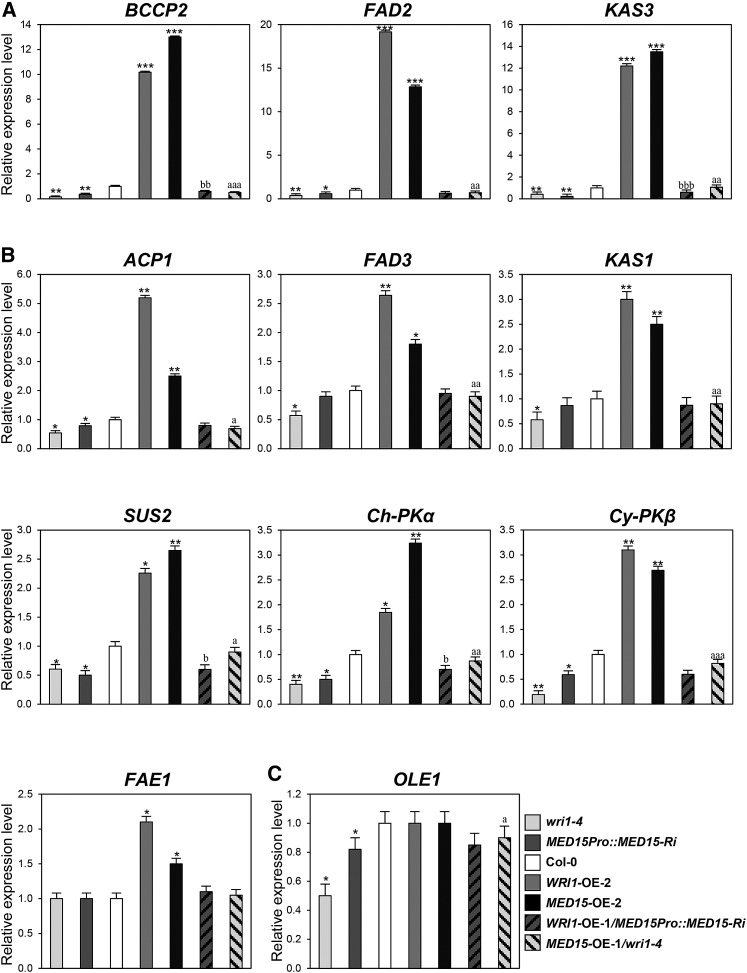
Figure 5.
Expression profiles of fatty acid biosynthetic genes in transgenic Arabidopsis plants with different MED15 and WRI1 transcript levels. A, Expression levels of BCCP2, FAD2, and KAS3 in wild-type Col-0, wri1-4, MED15Pro::MED15-Ri, WRI1-OE-2, MED15-OE-2, WRI1-OE-1/MED15Pro::MED15-Ri, and MED15-OE-1/wri1-4 transgenic plants. B, Expression levels ACP1, FAD3, KAS1, SUS2, Ch-PKα, Cy-PKβ, and FAE1 in wild-type Col-0, wri1-4, MED15Pro::MED15-Ri, WRI1-OE-2, MED15-OE-2, WRI1-OE-1/MED15Pro::MED15-Ri, and MED15-OE-1/wri1-4 transgenic plants. C, Expression levels of OLE1 in wild-type Col-0, wri1-4, MED15Pro::MED15-Ri, WRI1-OE-2, MED15-OE-2, WRI1-OE-1/MED15Pro::MED15-Ri, and MED15-OE-1/wri1-4 transgenic plants. Two-week-old Arabidopsis seedlings were used for qRT-PCR analysis. The expression values were normalized using ACTIN1 as an internal control. Each experiment was repeated with three biological replicates. Student’s t test: *, P < 0.05, **, P < 0.01, or ***, P < 0.001 versus wild-type Col-0; a, P < 0.05, aa, P < 0.01, or aaa, P < 0.001 versus wri1-4; b, P < 0.05, bb, P < 0.01, or bbb, P < 0.001 versus MED15Pro::MED15-Ri. Error bars represent sd.
Figure 5A shows that overexpression of MED15 and WRI1 in the wild-type background (WRI1-OE-2 and MED15-OE-2) increased transcript levels of BIOTIN CARBOXYL CARRIER PROTEIN2 (BCCP2), FATTY ACID DESATURASE2 (FAD2), and KETOACYL-ACP SYNTHASE3 (KAS3) by more than 10-fold compared with wild-type control plants. In addition, slight up-regulation of ACYL CARRIER PROTEIN1 (ACP1), FAD3, KAS1, SUCROSE SYNTHASE2 (SUS2), CHLOROPLAST PYRUVATE KINASE α (Ch-PKα), CYTOSOL PYRUVATE KINASE β (Cy-PKβ), and FATTY ACID ELONGASE1 (FAE1) was found in both lines, WRI1-OE-2 and MED15-OE-2, compared with the wild type (Fig. 5B). However, these WRI1 target genes, with the exception of FAE1, were down-regulated in wri1-4 and MED15Pro::MED15-Ri lines. Moreover, the reduced expression of these genes could be up-regulated by MED15 expression in the MED15-OE-1/wri1-4 line, but only BCCP2 and KAS3 were significantly up-regulated by WRI1 overexpression in the WRI1-OE-1/MED15Pro::MED15-Ri line. Expression of OLEOSIN1 (OLE1) was not changed notably in WRI1-OE-2 and MED15-OE-2 but was reduced significantly in wri1-4 and MED15Pro::MED15-Ri compared with the wild type (Fig. 5C). Its expression was increased by MED15 overexpression in the MED15-OE-1/wri1-4 line (Fig. 5C). OLE1 encodes TAG accumulation-related oil body protein. These results suggest that WRI1 along with MED15 transcriptionally regulate glycolysis-related and fatty acid biosynthetic genes.
MED15 Is Associated with Promoters of WRI1 Target Genes
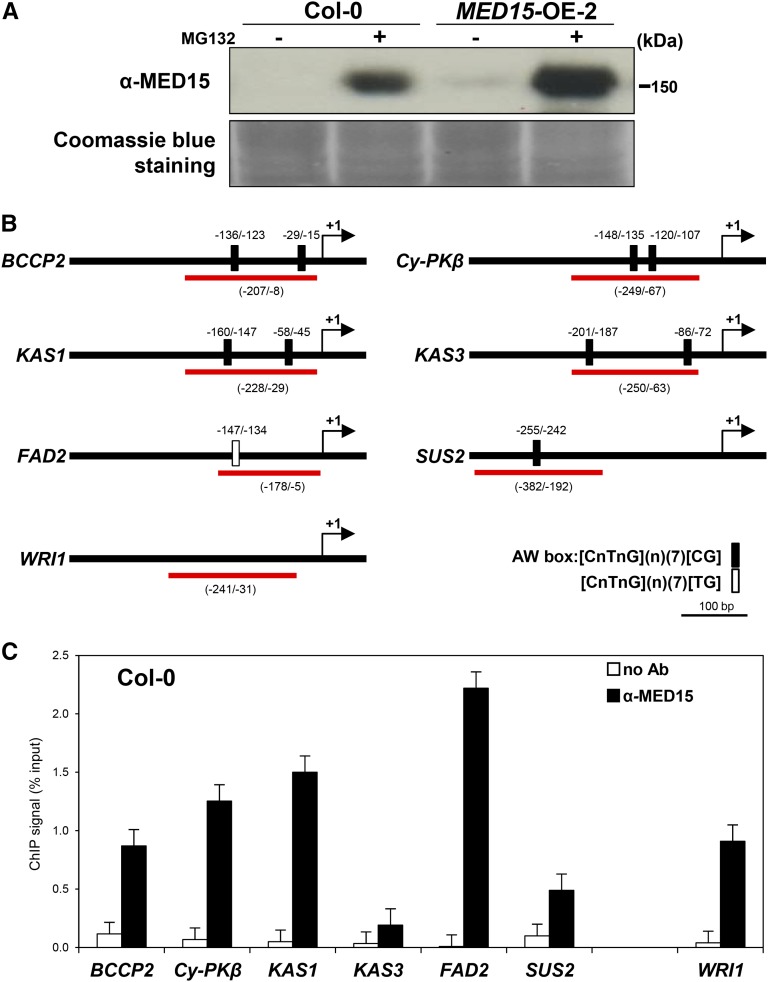
The interaction of MED15 with WRI1 in vitro and in vivo is consistent with the role of this Mediator subunit in regulating WRI1 target genes. One possible scenario is that MED15 is recruited to the promoters of target genes. To explore this possibility, we performed chromatin immunoprecipitation (ChIP) assays using rabbit antibody against MED15. Figure 6A shows that MED15 was undetectable in wild-type seedlings unless the materials were pretreated with MG132, which blocks protein degradation. This result suggests that MED15 is an unstable protein. Figure 6A shows that MED15 could be detected in the MED15-OE line and that its levels were highly increased with MG132 treatment.
Figure 6.
MED15 is recruited to promoter regions of WRI1 target genes. A, Immunoblot analysis showing MED15 protein levels in Arabidopsis. Two-week-old wild-type Col-0 and MED15-OE-2 seedlings were treated with (+) or without (−) MG132 (50 µm) in Murashige and Skoog liquid medium for 16 h. MED15 proteins were detected by anti-MED15 rabbit antibodies. The Coomassie Blue staining pattern was used as a loading control. B, Schematic diagram of the promoter regions of WRI1 target genes, BCCP2, Cy-PKβ, KAS1, KAS3, FAD2 and SUS2, and WRI1. Black boxes and a white box on the line indicate the putative WRI1-binding AW box, [CnTnG](n)(7)[CG] and [CnTnG](n)(7)[TG], respectively. The red lines indicate the DNA fragments used for ChIP-qPCR. The translational start sites (ATG) are marked as +1. C, ChIP-qPCR analysis for MED15 occupancy on the promoters of six WRI1 target genes and WRI1. ChIP assays were performed with chromatin prepared from wild-type Col-0 Arabidopsis seedlings after MG132 treatment for 16 h. Black and white bars represent the ChIP signals (% input) with and without MED15 antibody, respectively. ChIP results are presented as percentages of input DNA. Error bars represent sd (n = 3 biological replicates).
For the quantitative PCR (qPCR) of ChIP products, we designed primers to amplify the region including the WRI1-binding consensus site AW box in the proximal upstream regions of representative WRI1 target genes, BCCP2, Cy-PKβ, KAS1, KAS3, FAD2, and SUS2 (Fig. 6B). Note that BCCP2, Cy-PKβ, KAS1, and KAS3 contain two AW boxes [CnTnG](n)7[CG] in the promoter regions, whereas the SUS2 and FAD2 promoters had one AW box and one AW box-like sequence, [CnTnG](n)7[TG], respectively. Figure 6C shows that the promoters of all six WRI1 target genes tested were enriched by anti-MED15 antibody. Interestingly, the promoter of WRI1 also could be enriched by anti-MED15 antibody, suggesting that MED15 also mediates WRI1 expression. Consistent with this notion, WRI1 expression was indeed reduced in MED15-Ri lines (Fig. 4A). The ACTIN7 promoter was not enriched and was used as a negative control.
Overexpression of MED15 Increases Total Fatty Acid Content
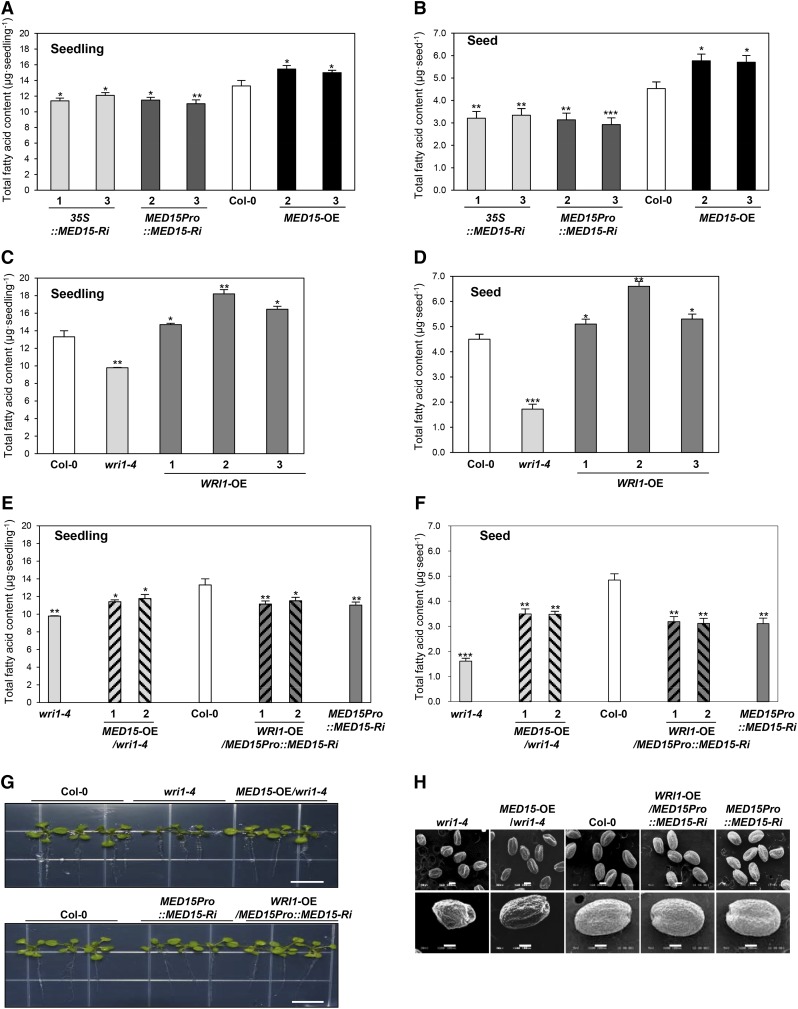
To further explore the role of MED15 in fatty acid biosynthesis, we analyzed total fatty acid content in seedlings and seed stages of wild-type, wri1-4, and transgenic plants in various genotypic backgrounds. Figure 7, A and B, show the total fatty acid content of two independent lines of MED15-Ri and MED15-OE. In MED15-Ri lines, the total fatty acid content was decreased by about 10% to 15% in seedlings (Fig. 7A) and about 30% in seeds (Fig. 7B). On the other hand, overexpression of MED15 (MED15-OE) resulted in about 15% increase in total fatty acid content in both stages. With respect to fatty acid profile, the C18:2 level relative to total fatty acid levels was decreased in both 35S::MED15-Ri (lines 1 and 3) and MED15Pro::MED15-Ri (lines 2 and 3) lines compared with the wild type, whereas the relative amount of C18:3 was increased in the same lines (Supplemental Fig. S4A). However, we were unable to find any significant changes of fatty acid composition in MED15-OE (lines 2 and 3) lines (Supplemental Fig. S4A).
Figure 7.
Total fatty acid content in seedlings and mature dried seeds of transgenic Arabidopsis plants with different MED15 and WRI1 expression levels. A and B, Total fatty acid content in 2-week-old seedlings (A) and seeds (B) of wild-type Col-0, 35S::MED15-Ri (lines 1 and 3), MED15Pro::MED15-Ri (lines 2 and 3), and MED15-OE (lines 2 and 3). C and D, Total fatty acid content in 2-week-old seedlings (C) and seeds (D) of wild-type Col-0, wri1-4, and WRI1-OE (lines 1–3). E and F, Total fatty acid content in 2-week-old seedlings (E) and seeds (F) of wild-type Col-0, wri1-4, MED15-OE/wri1-4 (lines 1 and 2), WRI1-OE /MED15Pro::MED15-Ri (lines 1 and 2), and MED15Pro::MED15-Ri. G, Phenotypes of lines shown in E. Bars = 1 cm. H, Scanning electron microscopy analysis of lines shown in F. Bars = 200 µm (top row) and 100 µm (bottom row). In A to F, experiments were performed with 100 seedlings or 100 seeds per line with five biological replicates. Student’s t test: *, P < 0.05, **, P < 0.01, or ***, P < 0.001 versus wild-type Col-0. n = 500. Error bars represent sd.
In confirmation of previous results (Baud et al., 2007), we found reduction of total fatty acid content of about 30% in seedlings (Fig. 7C) and about 56% in seeds of wri1-4 (Fig. 7D) compared with the wild type. This reduction of total fatty acid content was accompanied by significant changes of the relative amounts of fatty acids, with considerable increases of C18:3, C20:0, and C22:1 and decreases of C18:1 and C18:2 (Supplemental Fig. S4B). Moreover, overexpression of WRI1 increased total fatty acids by about 10% to 35% in seedlings and about 20% to 45% in seeds compared with the wild type, without any changes in fatty acid composition (Fig. 7, C and D; Supplemental Fig. S4B).
To investigate possible coregulatory roles of MED15 and WRI1 in fatty acid biosynthesis, we also overexpressed MED15 in the wri1 mutant background. Figure 7, G and H, show that MED15 overexpression was able to partially restore the wrinkled seed and seedling phenotypes of the mutant to the wild-type phenotype. This incomplete restoration to wild-type levels also was seen with total fatty acid content. Overexpression of MED15 in wri1-4 (MED15-OE/wri1-4 lines 1 and 2) increased total fatty acid content by 10% in seedlings and 45% in seeds compared with wri1-4, but the levels were still lower than wild-type levels (Fig. 7, E and F). Seeds of wri1-4 showed increases in C18:3, C20:1, and C22:1 but decreases in C18:1 and C18:2. These changes in relative fatty acid levels were incompletely reversed by MED15 overexpression (Supplemental Fig. S4C).
We also performed the reciprocal experiment by overexpressing WRI1 in transgenic plants deficient in MED15 (MED15Pro::MED15-Ri). WRI1-OE/MED15Pro::MED15-Ri lines 1 and 2 showed no significant change in total fatty acid content in seedlings and seeds compared with the parental MED15Pro::MED15-Ri line (Fig. 7, E and F). Moreover, the fatty acid profile in WRI1-OE/MED15Pro::MED15-Ri was similar to that of MED15Pro::MED15-Ri. These results show that MED15 overexpression can increase fatty acid content and alter fatty acid composition in the wri1-4 mutant, suggesting that MED15 is able to mediate the positive action of other TFs in fatty acid biosynthesis.
DISCUSSION
WRI1 is an AP2 domain transcription factor specific to plants, and it plays a central role in carbon partitioning between carbohydrate and lipids during plant embryogenesis (Focks and Benning, 1998; Cernac and Benning, 2004). Mutants deficient in WRI1 accumulate only 20% of the normal seed lipid content, and mutant seed appears wrinkled; hence, the mutant is called wri1. Despite the cloning and characterization of the WRI1 gene more than a decade ago (Cernac and Benning, 2004) interacting proteins of the encoded transcription factor have not yet been described, and how its transcriptional activity is executed has not been explored.
It is generally believed that TFs such as WRI1 recognize specific DNA sequences on target promoters. Upon binding to the latter, TFs recruit the Mediator complex and, in turn, the Pol II complex to initiate or block transcription (Malik and Roeder, 2010). Results in the last decade indicate that specific TFs bind to one or more subunits of the Mediator complex. Here, we identify the MED15 subunit of the Mediator complex as an interacting partner of WRI1.
The following lines of evidence support this claim. (1) WRI1 binds to MED15 but not MED14 in vitro. Experiments using deletion derivations showed that the N-terminal region of WRI1, which contains the AP2 domain, associates with the N-terminal fragment of MED15. (2) The association between WRI1 and MED15 was confirmed in vivo by complementation of YFP fluorescence using split YFP fusion proteins as well as by coimmunoprecipitation in transient expression experiments. (3) Promoter-GUS fusion experiments showed that WRI1 and MED15 are indeed expressed in embryos, although the expression of MED15 also extends to other plant tissues and organs, suggesting its roles in other developmental processes and signaling pathways as well.
Consistent with the hypothesis that MED15 mediates the transcription activity of WRI1, we found that MED15 can bind to the promoter regions of six WRI1 target genes that harbor a conserved AW sequence motif in their proximal upstream regions. In the case of the FAD2 gene, MED15 enrichment was found in the −5 to −179 region, which contains the first intron (1,134 bp from −5 to −1,139). This may not be surprising, as some intronic regions are known to enhance transcription. In addition to the above genes, we also found that MED15 binds to the promoter region of WRI1.
The binding of MED15 to WRI1 target promoters provides preliminary evidence that MED15 mediates the activity of WRI1. Direct evidence supporting this view comes from comparative experiments overexpressing WRI1 in the wild type and in MED15-Ri lines. We found that overexpression of WRI1 in the wild type elevates the expression levels of seven genes related to glycolysis and fatty acid biosynthesis (ACP1, FAD3, KAS1, SUS2, Ch-PKα, Cy-PKβ, and FAE1); however, increased expression levels of these genes were reversed to levels close to that of the wri1 mutant when MED15 was silenced by RNAi. These results provide evidence that WRI1 transcriptional activity is largely dependent on the integrity of MED15.
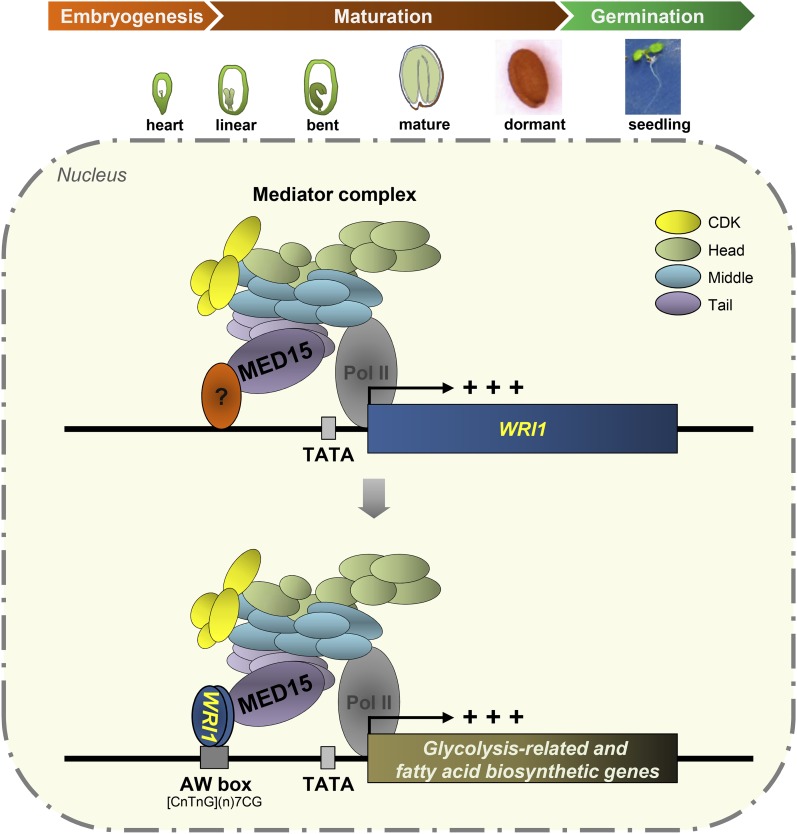
We also performed the reciprocal experiment by overexpressing MED15 in the wild type and in the wri1 mutant background. Overexpression of MED15 in the wild type also increased the expression levels of the seven glycolysis-related and fatty acid biosynthetic genes tested. Here, again, the enhancing effect of MED15 overexpression was negated by the wri1 mutation to levels close to those in the wri1 mutant itself. These results, together with those obtained with WRI1 overexpression, support the claim that WRI1/MED15 function as a complex in promoting the transcription of this gene set. Here, we propose a working model of how WRI1/MED15 activates the transcription of WRI1 target genes for fatty acid metabolism from late embryo development to the seedling stage in Arabidopsis (Fig. 8).
Figure 8.
Working model of WRI1/MED15 interaction and its influence on the transcriptional regulation of genes involved in glycolysis and fatty acid biosynthesis in Arabidopsis. A subunit of the tail module of the Mediator complex, MED15, mediates WRI1 expression through its possible interaction with an unidentified TF. Once WRI1 is expressed during late embryonic development (heart stage) and seed maturation (linear cotyledon stage to dormant stage) or in the SAM in the seedling stage in Arabidopsis, it binds to the AW box sequence in the 5′ upstream promoter region of WRI1 target genes, which are involved in glycolysis and fatty acid biosynthesis. Upon binding to the target promoters, WRI1 recruits the Mediator complex and Pol II through its direct interaction with MED15 to initiate the transcription of WRI1 target genes. CDK, Cyclin-dependent kinase module.
The effects of MED15 and WRI1 on gene expression are reflected to a certain extent in fatty acid accumulation in seeds and seedlings, although the effects obtained with MED15 are more modest compared with those with WRI1. The enhancing effect of WRI1 overexpression is abrogated when MED15 expression is silenced by RNAi. On the other hand, the increase in fatty acid accumulation caused by MED15 overexpression is only partially reversed by the wri1-4 mutation.
This result may be explained in part by the presence of three WRI1-like genes (WRI2, At2g41710; WRI3, At1g16060; and WRI4, At1g79700) in Arabidopsis (Maeo et al., 2009). These genes encode proteins with high sequence similarity in the AP2 DNA-binding domain but very low sequence similarity in the C terminus of the proteins. Recent work shows that WRI3 and WRI4 can each compensate for the low fatty acid content of the wri1-4 mutant (To et al., 2012), suggesting partial functional redundancy.
The structural and functional characterization of the plant Mediator complex has been a subject of considerable research interest, and recent investigations have focused mainly on the functions of various plant Mediator subunits in a number of abiotic and biotic signaling pathways (Kidd et al., 2009; Elfving et al., 2011; Canet et al., 2012; Zhang et al., 2012) and developmental processes (Autran et al., 2002; Wang and Chen, 2004; Ito et al., 2011). Along with MED14/SWP and MED16/SFR6 (Wathugala et al., 2012; Zhang et al., 2012, 2013), MED15 has been shown to be a regulator of salicylic acid signaling (Canet et al., 2012). Here, we have extended the role of this Mediator subunit to lipid metabolism. Sequence comparison between Arabidopsis MED15 and that of yeast (Gal11p/MED15) and human (ARC105/MED15) reveals little homology other than the conserved KIX domain. Nevertheless, it is surprising that the function of MED15 in lipid metabolism appears to be conserved in evolution. TFs Gcn4 and Gal4 in yeast (Fishburn et al., 2005; Reeves and Hahn, 2005), SREBPs in worms (Taubert et al., 2006; Yang et al., 2006), and Smad4 in Xenopus spp. (Kato et al., 2002) activate the transcription of lipid-related genes, and they all interact with MED15. Future work should be directed toward understanding how the WRI1/MED15 complex recruits other factors to alter the chromatin structure of target genes to activate transcription.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 was used as the wild type. Mutant wri1-4 (SALK_008559) was obtained from the Arabidopsis Biological Resource Center. Plants were grown at 22°C under long-day conditions (16 h of light/8 h of dark).
Vector Construction and Plant Transformation
Full-length open reading frame encoding MED15 or WRI1 was PCR amplified with the appropriate primer sets listed in Supplemental Table S1. After cloning into pENTR/D-TOPO (Invitrogen), DNA sequences were recombined into the Gateway destination vector, pBA-DC (Zhang et al., 2006), to generate 35S::MED15 and 35S::WRI1 constructs.
For MED15 deletion derivatives, DNA sequences coding for two overlapping N-terminal fragments of MED15, MED15-N1 (+1 to +750; 250 amino acids) and MED15-N2 (+1 to +1,350; 450 amino acids), and a C-terminal fragment of MED15, MED15-C (+3,000 to +4,005; 335 amino acids), were PCR amplified with appropriate primers and cloned into pBA-DC-YFP to generate 35S::MED15-N1-YFP, 35S::MED15-N2-YFP, and 35S::MED15-C-YFP constructs.
For the promoter-GUS experiment, the MED15 promoter sequence (2,377 bp; from −1 to −2,377, with +1 being the translational start site) with a 5′ untranslated region containing one intron (424 bp; from −9 to −432) and the WRI1 promoter sequence (1,739 bp; from −1 to −1,739, with +1 being the translational start site) was PCR amplified from genomic DNA using specific primer sets (Supplemental Table S1). Promoter sequences were then cloned into pHGWFS7 (Invitrogen), which carries both GFP and GUS reporter genes. For MED15-Ri constructs, we inserted a 450-bp fragment (+1,801 to +2,250) of MED15 cDNA into pFGC5941 RNAi vector (GenBank accession no. AY310901) through a two-step cloning procedure. The final vector, 35S::MED15-Ri, contained MED15 cDNA fragments in opposite orientations on either side of the 1,352-bp intron of the petunia (Petunia hybrida) CHALCONE SYNTHASE A (CHSA) gene. The 35S promoter in 35S::MED15-Ri was replaced with the MED15 promoter (2,377 bp) to produce MED15Pro::MED15-Ri. All constructs were transformed into Agrobacterium tumefaciens GV3101 using the floral dip method (Clough and Bent, 1998). Primers used in this study are listed in Supplemental Table S1.
In Vitro Pull-Down Assays
To produce MBP- or GST-tagged proteins, full-length open reading frames of MED15 or MED14 and WRI1 or GL2 were cloned into pMAL-c2 DC and pGEX-6P-1, respectively. All constructs were transformed into Escherichia coli BL21 (Novagen), and the expression of recombinant proteins was induced by isopropyl β-D-1-thiogalactopyranoside. MBP fusion proteins were bound to amylose resin and GST fusion proteins to glutathione-Sepharose 4B. Beads were washed, and bound proteins were eluted from the column using a buffer containing 10 mm maltose for MBP fusion proteins or 10 mm reduced glutathione in 50 mm Tris-HCl (pH 8) buffer for GST fusion proteins.
For pull-down assays, 2 µg of GST-WRI1 or GST-GL2 was incubated with immobilized MBP-MED15 or MBP-MED14 fusion proteins at 4°C for 3 h in binding buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1% Triton X-100, and 0.5 mm β-mercaptoethanol). After washing with a buffer (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, and 1% Triton X-100) five times, pulled-down proteins were separated on 10% SDS-PAGE gels and detected by immunoblotting using anti-GST antibody.
To define the interacting region between WRI1 and MED15, 6His-tagged N terminus (+1 to +750; 250 amino acids) and C terminus (+751 to +1,290; 180 amino acids) or MBP-tagged MED15-N1, MED15-N2, and MED15-C were used as target proteins for in vitro pull-down assays.
Subcellular Localization of WRI1 and MED15
YFP-fused MED15 deletion derivatives were infiltrated into Nicotiana benthamiana leaves, and the infiltrated plants were incubated at 24°C for 3 d. To visualize nuclei, DAPI (5 µg mL−1) solution dissolved in water containing 0.1% Silwet L-77 was infiltrated into leaves, and the images were monitored by confocal microscopy. YFP and DAPI signals were detected with a confocal scanning laser microscope with standard filter sets. Images were recorded and processed using the Zeiss LSM Image Browser (Carl Zeiss).
BiFC Assays
DNA fragments encoding full-length MED15 and WRI1 were cloned into serial pSAT1 vectors (Tzfira et al., 2005) containing DNA sequences for either the N or C terminus of EYFP and a 35S promoter. The resulting fusion genes with a 35S promoter were transferred to pGreen binary vector HY105. These vectors were transformed into A. tumefaciens strain C58C1 containing a pSOUP helper plasmid (Hellens et al., 2000). For BiFC experiments, A. tumefaciens cultures containing each plasmid were coinfiltrated into N. benthamiana leaves using a needleless syringe. YFP signals were detected by confocal scanning laser microscopy using a standard filter set.
Coimmunoprecipitation
N. benthamiana leaves were infiltrated with an A. tumefaciens strain harboring genes encoding 3HA-tagged WRI1 and YFP-fused MED15 deletion proteins, MED15-N2 and MED15-C. The infiltrated plants were grown at 24°C for 3 d. To increase protein stability, MG132, a proteasome inhibitor, was added to 10 mm MgCl2, 10 mm MES (pH 5.6), and 100 µm acetosyringone infiltration medium. For coimmunoprecipitation analysis, total proteins from 2 g of N. benthamiana leaves were extracted in prechilled lysis buffer (50 mm Tris-HCl [pH 8], 10% [v/v] glycerol, 2 mm EDTA, 150 mm NaCl, 1 mm dithiothreitol, 1% [w/v] Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail). Approximately 2 mg of protein extracts was incubated with anti-HA antibody (Santa Cruz Biotechnology) for 3 h at 4°C followed by the addition of protein A agarose beads (Santa Cruz Biotechnology) equilibrated with the extraction buffer. After an overnight incubation, the beads were collected by centrifugation and then washed four times with ice-cold immunoprecipitation buffer at 4°C. Immunoprecipitants were separated on 7.5% SDS-PAGE gels, blotted onto membranes, and detected with anti-GFP antibody (Santa Cruz Biotechnology).
Histochemical GUS Staining
Organs/tissues derived from transgenic plants carrying MED15Pro::GFP-GUS and WRI1Pro::GFP-GUS were incubated in GUS staining buffer (0.1 m sodium phosphate, pH 7, 1 mm 5-bromo-4-chloro-3-indolyl-d-glucuronide [Sigma], 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, 10 mm Na2EDTA, and 0.1% Triton X-100) for 20 h at 37°C. After staining, the plant materials were rinsed with 70% ethanol to remove chlorophyll (Jefferson et al., 1987) before visualization by light microscopy.
RNA Isolation and qRT-PCR Analysis
Total RNA was extracted from Arabidopsis seedlings (2 weeks old) or siliques bearing early-stage developing seeds (3–8 DAP) using TRIzol reagent (Invitrogen). One microgram of total RNA treated with DNase I (Promega) was used for cDNA synthesis using Moloney murine leukemia virus SuperScript II (Promega). The cDNA was quantified using Power SYBR Green PCR Master Mix (Life Technologies) with gene-specific primers in the ABI7900 Sequence Detection System (Life Technologies). ACTIN1 was used as an internal control. All reactions were performed with three biological replicates, and each sample was tested in triplicate by PCR.
Fatty Acid Analysis in Arabidopsis
To analyze the fatty acid content of seedlings or seeds in plants of various genotypes, 2-week-old seedlings or mature seeds of homozygous lines were used. One hundred seedlings or dry seeds were weighed on an analytical balance. Each sample was transmethylated at 85°C for 2 h in 1 mL of 3 m HCl-methanol, 0.01% butylated hydroxytoluene solution as an antioxidant, and 300 µL of toluene as a cosolvent. Fifty micrograms of pentadecanoic acid (C15:0) was added to each sample as an internal standard. After cooling the reaction mixture to room temperature, 1.5 mL of 0.9% (w/v) NaCl was added. Fatty acid methyl esters (FAMEs) were extracted two times with 1 mL of hexane. Pooled extracts were evaporated under nitrogen gas and then dissolved in 100 µL of hexane. FAME extracts were analyzed using an Agilent 6890 gas chromatograph with a flame ionization detector on a DB23 column (30 m length × 0.25 mm i.d. × 0.25 µm film thickness) employing helium as the carrier gas. The fatty acid content was estimated by comparing the total FAME peak area with that of the C15:0 that was used as an internal standard. Total fatty acid content was calculated as the sum of each detected fatty acid within the sample, and the relative amount of each fatty acid was determined by dividing the concentration of an individual fatty acid by the total fatty acid content.
Scanning Electron Microscopy Analysis
Seeds were mounted directly on the microscope stage and observed using a JSM-6360LV device with an acceleration voltage of 20 kV (JEOL).
MED15 Antibody
A 1,350-bp DNA fragment (+601 to + 1,950) encoding 450 amino acids (amino acids 200–650) of the MED15 protein was cloned into pET28a (Invitrogen). Purified His-tagged partial MED15 protein was sent to AbFrontier for the production of anti-MED15 rabbit antibody. The final serum was purified using the Amino Link Plus Immobilization Kit (Pierce).
ChIP-qPCR Assay
ChIP assays were performed according to Saleh et al. (2008). Two-week-old seedlings were transferred to Murashige and Skoog liquid medium containing 50 µm MG132 and incubated for 16 h at 22°C under long-day conditions (16 h of light/8 h of dark). Approximately 3 g of Arabidopsis seedlings (2 weeks old) treated with MG132 was fixed with 1% formaldehyde and cross-linked. The chromatin fraction was sonicated to a fragment size of around 0.3 to 1 kb. Rabbit antibody specific for Arabidopsis MED15 was used to immunoprecipitate DNA/protein complex from the chromatin preparation. The DNA recovered by reverse cross-linking from the chromatin complex was purified using a PCR purification kit (Qiagen) and then used for qPCR. To ensure the reliability of ChIP data, the input sample and no-antibody control sample were analyzed with each primer set.
Accession Numbers
Sequence data are available in the GenBank databases under the following accession numbers: At3g54320 (WRI1), At1g15780 (MED15), At5g15530 (BCCP2), At3g22960 (Ch-PKα), At5g52920 (Cy-PKβ), At5g49190 (SUS2), At5g46290 (KAS1), At1g62640 (KAS3), At4g25140 (OLE1), At3g12120 (FAD2), At2g29980 (FAD3), At4g34520 (FAE1), At3g05020 (ACP1), At1g16060 (WRI3), At1g79700 (WRI4), and At2g37620 (ACTIN1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Subcellular localization of MED15-N1-YFP.
Supplemental Figure S2. Organ/tissue expression pattern of MED15 and WRI1 derived from the Arabidopsis eFP Browser 2.0.
Supplemental Figure S3. Expression profiles of fatty acid biosynthetic genes in developing young seeds with siliques of transgenic Arabidopsis plants with different MED15 and WRI1 expression levels.
Supplemental Figure S4. Fatty acid profiles in mature seeds of transgenic Arabidopsis plants with different MED15 and WRI1 expression levels.
Supplemental Table S1. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Sheshayan Gayathri and Sivaramakrishnan P. Veena for technical assistance and the Temasek Life Sciences Laboratory central facilities for support in scanning electron microscopy and confocal microscopy.
Glossary
- DAP
days after pollination
- TAG
triacylglycerol
- TF
transcription factor
- KIX
kinase-inducible domain-interacting
- BiFC
bimolecular fluorescence complementation
- DAPI
4′,6-diamidino-2-phenylindole
- SAM
shoot apical meristem
- RNAi
RNA interference
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative PCR
- Col-0
Columbia-0
- cDNA
complementary DNA
- qRT
quantitative reverse transcription
- FAME
fatty acid methyl ester
Footnotes
This work was supported by the Temasek Life Sciences Laboratory.
Articles can be viewed without a subscription.
References
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD (1999) Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283: 985–987 [DOI] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L (2008) Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. The Arabidopsis Book 6: e0113 doi: 10.1199/tab.0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Bernecky C, Grob P, Ebmeier CC, Nogales E, Taatjes DJ (2011) Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS Biol 9: e1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon HM. (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res 36: 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet JV, Dobón A, Tornero P (2012) Non-recognition-of-BTH4, an Arabidopsis mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24: 4220–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW (2011) Origins and activity of the Mediator complex. Semin Cell Dev Biol 22: 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan A, Bajpayee M, Parmar D (2009) Comet assay: a reliable tool for the assessment of DNA damage in different models. Cell Biol Toxicol 25: 5–32 [DOI] [PubMed] [Google Scholar]
- Elfving N, Davoine C, Benlloch R, Blomberg J, Brännström K, Müller D, Nilsson A, Ulfstedt M, Ronne H, Wingsle G, et al. (2011) The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc Natl Acad Sci USA 108: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J, Mohibullah N, Hahn S (2005) Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell 18: 369–378 [DOI] [PubMed] [Google Scholar]
- Focks N, Benning C (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C (2007) Combined networks regulating seed maturation. Trends Plant Sci 12: 294–300 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hemsley PA, Hurst CH, Kaliyadasa E, Lamb R, Knight MR, De Cothi EA, Steele JF, Knight H (2014) The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26: 465–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Ito J, Sono T, Tasaka M, Furutani M (2011) MACCHI-BOU 2 is required for early embryo patterning and cotyledon organogenesis in Arabidopsis. Plant Cell Physiol 52: 539–552 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Habas R, Katsuyama Y, Näär AM, He X (2002) A component of the ARC/Mediator complex required for TGF beta/Nodal signalling. Nature 418: 641–646 [DOI] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Mugford SG, Ulker B, Gao D, Thorlby G, Knight MR (2009) Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J 58: 97–108 [DOI] [PubMed] [Google Scholar]
- Lai Z, Schluttenhofer CM, Bhide K, Shreve J, Thimmapuram J, Lee SY, Yun DJ, Mengiste T (2014) MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat Commun 5: 3064. [DOI] [PubMed] [Google Scholar]
- Liu J, Hua W, Zhan G, Wei F, Wang X, Liu G, Wang H (2010) Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol Biochem 48: 9–15 [DOI] [PubMed] [Google Scholar]
- Liu Y, Ranish JA, Aebersold R, Hahn S (2001) Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J Biol Chem 276: 7169–7175 [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Ma W, Kong Q, Arondel V, Kilaru A, Bates PD, Thrower NA, Benning C, Ohlrogge JB (2013) Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS ONE 8: e68887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG (2005) Dynamic regulation of Pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 30: 256–263 [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur S, Vyas S, Kapoor S, Tyagi AK (2011) The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol 157: 1609–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Ferreri K, Nakajima T, LaMorte VJ, Evans R, Koerber SC, Hoeger C, Montminy MR (1996) Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol 16: 694–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasrija R, Thakur JK (2012) Analysis of differential expression of Mediator subunit genes in Arabidopsis. Plant Signal Behav 7: 1676–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouvreau B, Baud S, Vernoud V, Morin V, Py C, Gendrot G, Pichon JP, Rouster J, Paul W, Rogowsky PM (2011) Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol 156: 674–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves WM, Hahn S (2005) Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol 25: 9092–9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Sanjaya, Durrett TP, Weise SE, Benning C (2011) Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol J 9: 874–883 [DOI] [PubMed] [Google Scholar]
- Shen B, Sinkevicius KW, Selinger DA, Tarczynski MC (2006) The homeobox gene GLABRA2 affects seed oil content in Arabidopsis. Plant Mol Biol 60: 377–387 [DOI] [PubMed] [Google Scholar]
- Suzuki M, McCarty DR (2008) Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol 11: 548–553 [DOI] [PubMed] [Google Scholar]
- Taatjes DJ. (2010) The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci 35: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima D, Kaneko A, Sakamoto M, Ito Y, Hue N, Miyazaki M, Ishibashi Y, Yuasa T, Iwaya-Inoue M (2013) Wrinkled 1 (WRI1) homologs, AP2-type transcription factors involving master regulation of seed storage oil synthesis in castor bean (Ricinus communis L.). Am J Plant Sci 4: 333–339 [Google Scholar]
- Taubert S, Van Gilst MR, Hansen M, Yamamoto KR (2006) A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev 20: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur JK, Agarwal P, Parida S, Bajaj D, Pasrija R (2013) Sequence and expression analyses of KIX domain proteins suggest their importance in seed development and determination of seed size in rice, and genome stability in Arabidopsis. Mol Genet Genomics 288: 329–346 [DOI] [PubMed] [Google Scholar]
- Thakur JK, Arthanari H, Yang F, Chau KH, Wagner G, Näär AM (2009) Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J Biol Chem 284: 4422–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Joubès J, Barthole G, Lécureuil A, Scagnelli A, Jasinski S, Lepiniec L, Baud S (2012) WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24: 5007–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Carbonero P (2005) Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol 49: 645–651 [DOI] [PubMed] [Google Scholar]
- Wang W, Chen X (2004) HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131: 3147–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathugala DL, Hemsley PA, Moffat CS, Cremelie P, Knight MR, Knight H (2012) The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol 195: 217–230 [DOI] [PubMed] [Google Scholar]
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, et al. (2006) An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442: 700–704 [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang C, Zhang Y, Sun Y, Mou Z (2012) The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24: 4294–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yao J, Zhang Y, Sun Y, Mou Z (2013) The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J 75: 484–497 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.