Abstract
Chaperones of the Hsp70 family interact with a multitude of newly synthesized polypeptides and prevent their aggregation. Saccharomyces cerevisiae cells lacking the Hsp70 homolog Ssb suffer from pleiotropic defects, among others a defect in glucose-repression. The highly conserved heterotrimeric kinase SNF1/AMPK (AMP-activated protein kinase) is required for the release from glucose-repression in yeast and is a key regulator of energy balance also in mammalian cells. When glucose is available the phosphatase Glc7 keeps SNF1 in its inactive, dephosphorylated state. Dephosphorylation depends on Reg1, which mediates targeting of Glc7 to its substrate SNF1. Here we show that the defect in glucose-repression in the absence of Ssb is due to the ability of the chaperone to bridge between the SNF1 and Glc7 complexes. Ssb performs this post-translational function in concert with the 14-3-3 protein Bmh, to which Ssb binds via its very C-terminus. Raising the intracellular concentration of Ssb or Bmh enabled Glc7 to dephosphorylate SNF1 even in the absence of Reg1. By that Ssb and Bmh efficiently suppressed transcriptional deregulation of Δreg1 cells. The findings reveal that Ssb and Bmh comprise a new chaperone module, which is involved in the fine tuning of a phosphorylation-dependent switch between respiration and fermentation.
INTRODUCTION
When the yeast Saccharomyces cerevisiae grows in the presence of glucose, energy is provided via fermentation and transcription of genes required for respiration is repressed (1–3). As soon as glucose becomes limiting, substantial transcriptional changes enable the cells to gain energy via respiration. The major regulator of the release from glucose repression is the kinase SNF1 (1–4). The heterotrimeric SNF1 complex consists of a catalytic α-subunit Snf1; one of three β-subunits, Sip1, Sip2 or Gal83; and the γ-subunit Snf4 (1 and see also ‘Discussion’ section). Activation of SNF1 requires phosphorylation at threonine 210 of the Snf1 subunit (5) (Supplementary Figure S1A). When active, SNF1 directly regulates the activity of key metabolic enzymes involved in cellular energy supply (1). Moreover, SNF1 phosphorylates important transcription factors including Mig1, Nrg1/Nrg2 and Cat8 (1–3,6) and is an important regulator of the transcription factor Adr1 (1–3,7). Via these and other, less well characterized transcription factors, SNF1 induces the expression of more than 400 genes, including 29 of the 40 most highly glucose-repressed genes (1,8). SNF1 activity is mainly regulated via dephosphorylation of Snf1-T210 (9), which is mediated by the essential protein phosphatase Glc7 in concert with its regulatory subunit Reg1. One function of Reg1 is to target Glc7 specifically to Snf1-T210 (1–3,10–11). As a consequence, Snf1-T210 is permanently hyperphosphorylated in Δreg1 cells and glucose repression cannot be established.
Bmh1 and Bmh2 (collectively termed Bmh) are the only yeast members of the highly conserved eukaryotic 14-3-3 protein family (12). The Δbmh1Δbmh2 double deletion is lethal in most strain backgrounds, indicating that Bmh1 and Bmh2 possess overlapping and essential functions (12). In general, 14-3-3 homologs form dimers, which interact with short consensus motifs, often localized within intrinsically disordered segments of their client proteins. 14-3-3 binding to clients can exert a variety of effects as for example regulation of enzyme activity, structure or complex assembly of proteins (12–14). Previous transcriptome studies revealed that the 14-3-3 proteins Bmh1 and Bmh2 play a pivotal role in SNF1/Glc7 signaling (15,16). A detailed study aimed on the identification of Reg1-interactors identified Bmh1 and demonstrated an effect of Bmh on glucose repression (17). Moreover, previous studies suggest that Snf1 interacts with Bmh directly or indirectly (18,19). How the Bmh proteins regulate SNF1/Glc7 function at a mechanistic level is currently not understood.
The Hsp70 homologs Ssb1 and Ssb2 (collectively termed Ssb) are encoded by two nearly identical and functionally redundant genes (20). Δssb1Δssb2 strains are viable, however, suffer from slow growth, cold sensitivity and hypersensitivity toward a variety of translational inhibitors (20). As a typical Hsp70 chaperone, Ssb interacts with unfolded polypeptide substrates via its peptide-binding domain (Supplementary Figure S1B). The cycle of substrate binding and release is modulated by the activity of the N-terminal ATPase domain, which is stimulated by the co-chaperone RAC (ribosome-associated complex) (20,21). Ssb can bind to the ribosome directly and then interacts with a variety of nascent polypeptides emerging from the ribosomal tunnel (20,22–23). However, at steady state, only approximately half of the Ssb molecules are bound to ribosomes, the other half is freely distributed in the cytosol (20). If the cytosolic pool of Ssb reflects dynamic ribosome binding and release, or serves ribosome-independent functions, is currently not understood. Accumulating evidence suggests that one function of cytosolic Ssb is connected to the SNF1/Glc7 signaling pathway. First, Δssb1Δssb2 strains display transcriptional deregulation reminiscent of glucose repression mutants (17,24). Second, Ssb is a strong multi-copy suppressor of Snf1-T210 hyperphosphorylation and growth defects of a Δreg1 strain (24). Third, a two-hybrid screen and TAP-tag purification of Reg1 revealed that Ssb interacts with Reg1 post-translationally (17,25). Moreover, data from a TAP-tag purification of Snf1 identified Ssb1 as a component of the SNF1 complex (19). However, the complex also contained Reg1 and it remained unclear if the interaction between Ssb and Snf1 was direct (19).
Here we show that the previously unexplained role of Ssb in SNF1/Glc7 signaling is intimately connected to the function of the Bmh proteins. We show that Ssb1 and Bmh1 have strikingly similar effects with respect to SNF1 activity. Moreover, we show that Ssb interacts with Bmh directly, and that this interaction requires the very C-terminus of Ssb. Our data reveal that Ssb was in direct contact with the essential components of the SNF1/Glc7 signaling module. Based on the findings we suggest that Ssb does not only work as a ribosome-bound chaperone in the de novo folding of newly synthesized polypeptides, but also, in concert with Bmh, post-translationally affects dynamic protein–protein interactions involved in the regulation of SNF1 activity.
MATERIALS AND METHODS
Yeast strains and plasmids
MH272–3f a/α (ura3/ura3, leu2/leu2, his3/his3, trp1/trp1, ade2/ade2) is the parental wild-type strain of the haploid derivatives used in this study (24,26). All strains employed in this study are summarized in Supplementary Table S1. Deletion strains lacking SSB1 and SSB2 (Δssb1Δssb2), REG1 (Δreg1), Δsnf1 and Δssb1Δssb2Δsnf1 were as described (24). Please note the frequent occurrence of spontaneous suppressors in the Δreg1 strain, which leads to fast-growing clones (24,27). Δsit4 was constructed by replacing SIT4 with the sit4::kanMX4 deletion cassette amplified by polymerase chain reaction (PCR) from strain YDL047w (Euroscarf). Δreg1Δsit4 + pYEplac195-SSB1 and Δreg1Δsit4 + pYEplac195-BMH1 were constructed by replacing SIT4 with the sit4::kanMX deletion cassette in the Δreg1 + pYEplac195-SSB1 or Δreg1 + pYEplac195-BMH1 strains. Transformation of Δreg1 with the sit4::kanMX deletion cassette without over-expression of Ssb1 or Bmh1 did not result in viable colonies. Lethality of Δreg1Δsit4 was previously reported (28). Δglc7 + GLC7 (glc7::TRP1) was constructed by replacing a 1175-base-pair (bp) ApaI/XbaI fragment in the GLC7 coding region with TRP1 +/−300 bp. Lethality of the Δglc7 strain was complemented with pYCplac111-GLC7 (cen, LEU2, GLC7 +/−300 bp) or pYCplac111-GLC7-T152K (cen, LEU2, GLC7-T152K +/−300 bp).
SSB1 and BMH1 were over-expressed under control of their own promoters from 2 μ plasmids, pYEplac195-SSB1 (2 μ, URA3, SSB1 +/−300), pYEplac195-BMH1 (2 μ, URA3, BMH1 +/−300 bp), or pYEplac112-BMH1 (2 μ, LEU2, BMH1 +/−300 bp). Crosslinking experiments were performed in a Δssb1Δssb2Δsnf1 background harboring pYCPlac22-FLAG-SNF1 (cen, TRP1, FLAG-SNF1 +/−300 bp) or pYCplac33-FLAG-SNF1 (cen, URA3, FLAG-SNF1 +/−300 bp). FLAG-SNF1 was generated by replacing the natural poly-histidine coding sequence within SNF1 (position 59–92 of the SNF1 orf, Supplementary Figure S1A) with a sequence encoding the FLAG-peptide (DYKDDDDK). Mutations within the potential 14-3-3 binding motifs of SNF1 (Supplementary Figure S1A) were introduced via PCR technology. The resulting triple mutant (S413A, R510A and S513A) termed Snf1-RSS was expressed in the Δsnf1 strain from the pYCplac33-Snf1-RSS plasmid. To generate an N-terminally His6-tagged version of Ssb a sequence encoding for GSSHHHHHHSS was inserted 3′ of the ATG start codon of Ssb1-A577K, which contains an alanine to lysine exchange at position 577 (Supplementary Figure S1B). The Ssb1-A577K mutant of Ssb was employed for crosslinking experiments, because the Ssb-specific antibody recognized Ssb1-A577K more effectively compared to wild-type Ssb (Supplementary Figure S1C). Δssb1Δssb2Δsnf1 + FLAG-SNF1 were transformed with pYEplac195-SSB1-A577K (2 μ, URA3, SSB1-A577K +/−300 bp) or pYEplac112-His6-SSB1-A577K (2 μ, TRP1, His6-SSB1-A577K +/−300 bp). FLAG-SNF1, SSB1-A577K and His6-SSB1-A577K (termed His-Ssb) fully complement growth defects of the Δsnf1 or Δssb1Δssb2 strains, respectively.
pTet111 (cen, LEU2, tetO) was constructed by introducing the tetracycline regulator region of pCM190 (29) via EcoRI–HindIII to pYCplac111. pTet111-Bmh1 and pCM190-Ssb1 were employed for expression of wild-type Bmh1 and Ssb1. pTet111-His8-Bmh1 was generated by exchanging the C-terminal AAEGEAPK sequence of Bmh1 for HHHHHHHH. The C-terminal region of Ssb1 is important for the recognition by the Ssb-specific antibody (Supplementary Figure S1B and C). In order to detect C-terminally truncated Ssb1 mutants we thus generated N-terminally Myc-tagged versions of Ssb1. To that end, a sequence coding for the Myc-tag peptide (EQKLISEEDL) was inserted 3′ of the ATG start codon of SSB1 in pCM190-Ssb1. C-terminal truncations of Ssb1 were then generated by introducing the TAA stop codon into the coding sequence of SSB1 in pCM190-Myc-Ssb1. The resulting plasmids, termed pCM190-Myc-Ssb1-Δ23 and pCM190-Myc-Ssb1-Δ40, give rise to Ssb1 versions C-terminally shortened by 23 or 40 residues, respectively (Supplementary Figure S1B). On glucose medium, Myc-Ssb1-Δ23 and Myc-Ssb1-Δ40 fully complemented growth defects of a Δssb1Δssb2 strain (Supplementary Figure S1D). For the generation of Δbmh1Δbmh2Δssb1Δssb2 strains expressing plasmid-borne versions of Ssb1 and Bmh1, a haploid Δbmh1Δbmh2 + pYCplac33-Bmh1 strain was crossed with a Δssb1Δssb2 strain and the resulting diploid was then sporulated and dissected to obtain a haploid Δbmh1Δbmh2Δssb1Δssb2 + pYCplac33-Bmh1. The haploid Δbmh1Δbmh2Δssb1Δssb2 + pYCplac33-Bmh1 was transformed with pTet111-Bmh1, or pTet111-His8Bmh1 and the URA3-plasmid pYCplac33-Bmh1 was subsequently removed by 5′-fluoro-orotic acid treatment (30). For expression of different versions of Ssb1 the strain was transformed with the respective pCM190 plasmids.
Culture conditions
Growth of strains was analyzed on rich media containing glucose (YPD: 1% yeast extract, 2% peptone, 2% glucose) or sucrose (YPSuc: 1% yeast extract, 2% peptone, 2% sucrose). To that end, 10-fold serial dilutions of early log-phase cultures containing the same number of cells were spotted on plates and were incubated as indicated. Glucose-repression was tested on YPSuc plates containing 200 μg/ml 2-deoxy-glucose (2DG). Sucrose-containing media were sterilized by filtration to avoid cleavage of the disaccharide during heat sterilization. Survival of Δreg1 or Δreg1Δsit4 deletion strains was tested on selective media containing 5′-fluoro-orotic acid (30). To that end, Δreg1 or Δreg1Δsit4 harboring the URA3-plasmids pYEplac195-Ssb1 or pYEplac195-Bmh1 were transferred to plates containing 5′-fluoro-orotic acid to select for those colonies, which were able to grow without pYEplac195-Ssb1 or pYEplac195-Bmh1.
Microarray analysis
Affymetrix microarrays and subsequent data analysis via the Genedata Expressionist software (version 5.1.2.) was performed as previously described (24) with 5 μg of three total RNA preparations, derived from independently grown cultures of wild-type, Δreg1 and Δreg1 over-expressing Ssb1 or Bmh1. To identify transcripts within the 5814 probe sets, which were differentially expressed in the mutant strains, we used the unpaired Bayes t-test (CyberT) (31) with the Bayes confidence estimate value set to 10 and a window size of 101 genes. Genes deregulated ≥ 1.5 (upregulated) or ≤0.66 (downregulated) with a false discovery rate (FDR) of ≤ 0.01 were selected for further analysis. Genes deregulated in the Δreg1 strain were compared to genes deregulated in the Δssb1Δssb2 strain selected by the same criteria (24). To analyze the effect of SSB1 or BMH1 over-expression in the Δreg1 background, analysis of variance over all samples (ANOVA) and principal component (PC) analysis was performed via the Analyst module implemented in the Expressionist Software. To control for the FDR the Benjamini–Hochberg q-value was calculated and set to ≤0.01 (32). By this, 1774 genes from the whole dataset were selected for further analysis. The N-fold regulation-activity of the Analyst module of the Expressionist software was employed to calculate the median ratio between the wild-type and mutant groups. Genes with deregulation values of ≥1.5 or ≤0.66 for Δreg1 were selected as significantly up (445 genes), or downregulated (307 genes). To assess the overall effect of Ssb1 or Bmh1 over-expression in the Δreg1 background, genes with at least 50% reregulation compared to the N-fold deregulation in the Δreg1 strain were selected (Supplementary Table S2). Microarray Gene Ontology (GO) analysis was performed with the ‘Generic GO Term Finder’ tool (33) with default settings to test for over- or under-representation of GO categories in selected gene lists (Supplementary Table S2).
Transcription factor analysis
Target gene lists from the YEASTRACT (34) database were employed to identify transcription factor regulons within the genes deregulated in Δreg1 or Δssb1Δssb2 cells (Supplementary Table S2). Significantly enriched TF regulons in the up and downregulated gene sets were identified via Fisher's exact test for over-representation. The sample frequency (i.e. number of TF targets in datasets of deregulated genes) was compared to the background frequency in the whole genome (5820 genes). A Bonferroni correction was applied to control the family-wise error rate (αBON = 0.05/n, n = number of tests, i.e. number of TFs in each analysis). Highly significant TFs (P ≤ 10−9) were considered for further analysis (Supplementary Table S3). Overlap between genes upregulated in batch culture cells and genes upregulated in Δreg1 or Δssb1Δssb2 was analyzed via Fisher's exact test for over-representation (Supplementary Table S3). The frequency of genes upregulated in batch culture cells, and of genes upregulated in Δreg1 or Δssb1Δssb2, respectively, was compared to the background frequency in the whole genome (5820 genes) for each OD600. Bonferroni correction was applied to control the family-wise error rate (αBON = 0.05/12 = 0.0042). Statistical calculations were performed with SAS® Studio.
Crosslinking and mass spectrometry
Δssb1Δssb2Δsnf1 + FLAG-Snf1 expressing either His-Ssb-A577K or Ssb1-A577K (Supplementary Table S1) were grown to mid-log phase on YPD at 30°C. Cells were harvested, rapidly frozen in liquid nitrogen and were disrupted with a cryo mill cooled with liquid nitrogen (MM400, Retsch, 35). For each reaction a volume of ∼25 μl frozen cell powder was mixed with 500 μl ice cold CL-buffer (20 mM HEPES-KOH pH 7.4, 80 mM potassium acetate, 2 mM magnesium acetate, 1 mM PMSF) containing 150 μM BS3 (Bis(sulfosuccinimidyl)-suberate: non-cleavable, homobifunctional, amino-reactive, spacer length 11.4 Å, Thermo Scientific). After incubation for 20 min on ice, reactions were quenched with Tris–HCl, pH 7.4 (final concentration of 10 mM) and were then cleared by centrifugation at 180000 × g, for 30 min. Crosslinking with DTSSP (3,3′-Dithiobis(sulfosuccinimidylpropionate), cleavable, homobifunctional, amino-reactive, spacer length 12.0 Å, Thermo Scientific) was performed as described for BS3 with a final concentration of 200 μM DTSSP. Supernatants of crosslinking reactions were diluted 1:4 in buffer A (final concentration: 40 mM HEPES-KOH pH 8.0, 100 mM potassium acetate, 5 mM imidazole, 5 mM magnesium acetate, 3.5 M urea, 1 mM PMSF, protease inhibitor mix (final concentration: leupeptin, 1.25 μg/ml; antipain 0.75 μg/ml; chymostatin 0.25 μg/ml; elastinal 0.25 μg/ml; pepstatin A 5.0 μg/ml)) and were then mixed with Ni-NTA beads (Qiagen, Ni-NTA Superflow; 40 μl beads/ml binding reaction). After 90 min of gentle agitation at 4°C Ni-NTA beads were collected and were washed once with 1 ml of buffer A, and three times with 1 ml buffer B (40 mM HEPES-KOH pH 8.0, 300 mM NaCl, 15 mM imidazole, 8M urea, 1 mM PMSF, protease inhibitor mix) at room temperature. Proteins bound to the beads were eluted in buffer B containing 300 mM imidazole adjusted to pH 7.4 and were subsequently precipitated with 5% trichloroacetic acid (TCA). Precipitated proteins dissolved in sample buffer containing 5% β-mercaptoethanol were separated on Tris-Tricine gels (36). Protein samples, crosslinked with DTSSP, were dissolved in sample buffer without reducing agent, or in sample buffer containing 5% β-mercaptoethanol plus 1% DTT.
For mass spectrometry, material from ∼10 parallel crosslinking reactions (∼250 μl cell powder) was combined and separated on 8% Tris-Tricine gels (36) which were subsequently stained with colloidal Coomassie Blue G-250 (37). Lanes were sliced above ∼90 kDa. Each gel slice was assigned to a maximum molecular mass (slicemass) employing a standard curve based on proteins with known molecular mass. Gel slices were subjected to in-gel digestion using trypsin and were analyzed by nano-HPLC-ESI-MS as described (38), except for the use of a 500 mm × 75 μm C18-column (Acclaim PepMap RSLC column, 2 μm particle size, 100Å pore size, Thermo Scientific) at a flow rate of 250 nl/min with a linear gradient ranging from 5 to 65% solvent B (0.1% (v/v) formic acid in 50% (v/v) MeOH, 30% (v/v) acetonitrile) in 50 min for the analytical HPLC. Proteins were identified by database searches against the protein translations of all ORFs of the Saccharomyces Genome database reference genome (www.yeastgenome.org, version R64) as described (38).
Filtering criteria for the selection of potential Ssb interactors
Proteins with a FDR of <0.01 on both the peptide and the protein level, with ≥2 peptides, and a sequence coverage of >15% were considered for further analysis (Supplementary Table S4). Abundance profiles for each protein were generated by plotting the number of the peptides identified in each gel slice against the corresponding slicemass. Candidate Ssb-crosslink partners were filtered under the assumption that the molecular mass of each crosslink product was at least the sum of the two individual protein masses (His-Ssb1 + identified protein). Candidates were rejected if the slicemass of the maximum peak was ≤85% of the calculated molecular mass of the crosslink product. An exception was made if an additional local maximum with higher slicemass was detected. Proteins carrying internal poly-His stretches, or with non-cytosolic localization were removed from the list of cytosolic Ssb-interactors (Supplementary Table S4).
Native pull-down experiments
Yeast cultures (200 ml) were grown to mid-log phase on YPD at 30°C and were subsequently harvested via centrifugation. A suspension consisting of equal volumes of cell pellet, glass beads and lysis buffer (20 mM HEPES-KOH pH 8.0, 120 mM potassium acetate, 2 mM magnesium acetate, 30 mM imidazole, 1 mM EDTA, 1 mM PMSF, protease inhibitor mix, 1× phosSTOP (Roche)) was prepared and glass bead disruption was performed by four cycles of vortexing for 30 s followed by 30 s incubation on ice and the resulting lysate was then cleared by centrifugation at 20000 × g for 10 min. For each pull-down reaction, 20 μl of Ni-NTA beads (Quiagen, Superflow) were pre-coated with 300 μg/ml trypsin inhibitor for 60 min to block unspecific binding. Cleared lysate and lysis buffer were mixed to a final protein concentration of ∼30 mg/ml and 1 ml of the mixture was added to the pre-coated Ni-NTA beads. Pull-down reactions were incubated at 4°C for 45 min with gentle agitation, Ni-NTA beads were then collected and were washed twice with 0.5 ml washing buffer (20 mM HEPES-KOH pH 8.0, 250 mM potassium acetate, 2 mM magnesium acetate, 100 mM imidazole, 1 mM PMSF, protease inhibitor mix). Bound proteins were released by boiling Ni-NTA beads at 95°C in sample buffer. Analysis was performed on 10% Tris-Tricine gels (36) followed by immunoblotting.
Miscellaneous
To assess the membrane potential of yeast strains, MitoTracker®Green-FM fluorescent dye (Invitrogen, M7514) was added to mid-log cultures at a final concentration of 200 nM (39). After 1h at 30°C on a shaker, aliquots were collected by centrifugation and immunofluorescence was analyzed in living cells (Axio Imager A2, Zeiss, 100× objective, fitted with a GFP filter set (Chroma Technology Corp., 41 017). Analysis of the images was performed with ImageJ software (National Institutes of Health, USA). Total yeast extract for immunoblot analysis was prepared as described (40). For the analysis of Snf1-T210 phosphorylation, cultures were boiled for 3 min prior to harvest. Immunoblots were developed using enhanced chemiluminescence (ECL) as described (24). Polyclonal antibodies were raised against peptides or purified proteins in rabbit (EUROGENTEC, Bel SA). Snf1-T210 phosphorylation was detected with anti-Thr(P)-172-AMPK (Cell Signalling Technologies, Beverly, MA, USA).
RESULTS
Gene expression patterns of Δreg1 and Δssb1Δssb2 cells correlate with distinct levels of glucose repression defects
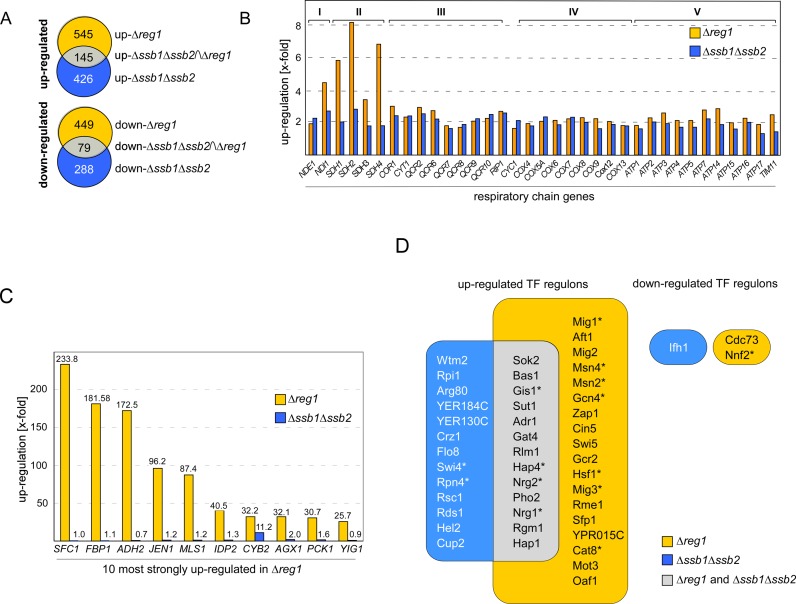
The Hsp70 homolog Ssb affects SNF1 activity, however, the mechanism is not understood (compare Introduction and 17,24). To get better insight into the role of Ssb with respect to SNF1 signaling we compared the transcriptome of a Δssb1Δssb2 strain (24) to the transcriptome of a Δreg1 strain in the same genetic background (Supplementary Table S2). In the combined datasets we first identified those genes deregulated ≥1.5-fold with a FDR of ≤0.01 (Supplementary Table S2). By these criteria, 426 genes were upregulated (up-Δssb1Δssb2) and 288 genes were downregulated (down-Δssb1Δssb2) in Δssb1Δssb2 cells, while 545 genes were upregulated (up-Δreg1) and 449 genes were downregulated (down-Δreg1) in Δreg1 cells (Figure 1A and Supplementary Table S2). The intersection groups up-Δssb1Δssb2∩Δreg1 and down-Δssb1Δssb2∩Δreg1 contained 145 and 79 genes, respectively (Figure 1A and Supplementary Table S2). The down-Δssb1Δssb2 group was enriched most significantly for genes connected to the GO term α-amino acid metabolic process (GO:1901605 P-value 8.37 × 10−11), the down-Δreg1 group for glycoprotein metabolic process (GO:0009100 P-value 5.86 × 10−12) (Supplementary Table S2, 41). Remarkably, genes in the down-Δssb1Δssb2∩Δreg1 intersection group were not enriched for any category of the GO domains ‘biological process’ or ‘molecular function’ (Supplementary Table S2). Thus, while 27% of the genes downregulated in Δssb1Δssb2 cells were also downregulated in Δreg1 cells (Figure 1A), these genes were diverse with respect to molecular function and biological process. In contrast, there was significant overlap with respect to the genes upregulated in Δssb1Δssb2 as well as Δreg1 cells. The up-Δssb1Δssb2∩Δreg1 group was most strongly enriched for the GO categories oxidative phosphorylation (GO:0006119 P-value 8.34 × 10−27, Supplementary Table S2) or ATP metabolic process (GO:0046034 P-value 2.12 × 10−25, Supplementary Table S2). The complete set of nuclear encoded subunits of complex I–V of the respiratory chain was upregulated in Δssb1Δssb2 as well as in Δreg1 cells (Figure 1B, GO:0070469 respiratory chain P-value 2.07 × 10−27, Supplementary Table S2). However, other genes under control of glucose repression were strongly upregulated in Δreg1, but not in Δssb1Δssb2 cells (Supplementary Table S2). For example, the 10 most highly upregulated genes in Δreg1 cells (Figure 1C, Supplementary Table S2), belong to the group of genes most strongly controlled by glucose repression (8). With the exception of CYB2, this group of genes was not, or only slightly, upregulated in Δssb1Δssb2 cells (Figure 1C, Supplementary Table S2).
Figure 1.
Transcriptional deregulation in Δssb1Δssb2 and Δreg1 cells. (A) Intersection set of genes deregulated ≥ 1.5-fold (FDR ≤ 0.01) in Δssb1Δssb2 (24) and Δreg1 cells (Supplementary Table S2). (B) Relative expression level of nuclear genes encoding for subunits complex I–V, (74)of the respiratory chain in Δreg1 and Δssb1Δssb2 cells. (C) Relative expression level of the 10 most highly upregulated genes in Δreg1 cells and of the same genes in Δssb1Δssb2 cells. (D) Enrichment of TF targets in the set of up or downregulated genes in Δreg1 or Δssb1Δssb2 cells. The frequency of TF targets in the dataset was compared to their background frequency in the whole genome using Fisher's exact test for over-representation (Supplementary Table S3). Shown are those TFs enriched in the genes up or downregulated in either Δssb1Δssb2 (blue) or Δreg1 (orange) with right-sided P-values ≤ 10−9. The overlap between the two groups is shown in gray. TFs which are direct targets of SNF1 are labeled with an asterisk.
Employing target gene lists from the YEASTRACT database (34) we identified 31 TF regulons highly enriched (P > 10−9) in the up-Δreg1, and 26 TF regulons in the up-Δssb1Δssb2 group, with an overlap of 13 TFs (Figure 1D, Supplementary Table S3). Only 1 TF regulon was enriched in the down-Δssb1Δssb2, and 2 TF regulons were enriched in the down-Δreg1 group (Figure 1D, Supplementary Table S3). Regulons enriched in up-Δreg1 as well as up-Δssb1Δssb2 included the transcriptional activator Hap4, which controls expression of respiratory chain genes (42,43, Figure 1D) providing an explanation for the similar regulation of this group of genes (Figure 1B). Both groups were also enriched for genes under control of the transcriptional repressors Nrg1, Nrg2 and the transcriptional activator Adr1, all of which are either directly or indirectly regulated by SNF1 and function in glucose repression of a specific subset of genes (6–8,44–46). Adr1 regulates the expression of many genes strictly controlled by glucose repression, as for example ADH2 (Figure 1C, 46). Interestingly, ADR1 itself was upregulated in both mutant strains, however, more significantly in Δreg1 (9.6-fold) when compared to Δssb1Δssb2 (2.3-fold) (Supplementary Table S2). The different levels of ADR1 expression are consistent with the more pronounced release from glucose repression in Δreg1 compared to Δssb1Δssb2 cells (Figure 1C, Supplementary Table S2).
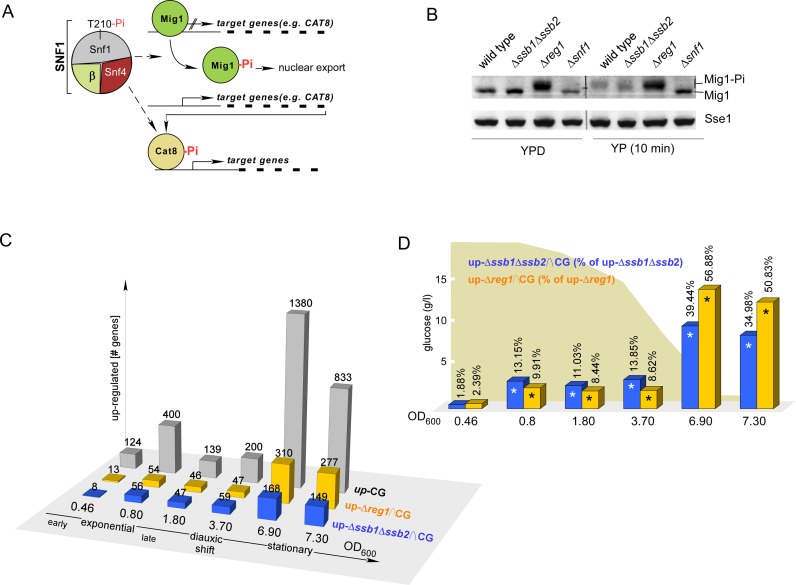
Besides Adr1, the hierarchically organized Mig1/Cat8 cascade, consisting of the transcriptional repressor Mig1 and the transcriptional activator Cat8, controls regulation of glucose repressed genes (Figure 2A, 1). Nuclear export of Mig1 correlates with the glucose level; at intermediate glucose concentrations Mig1 is partially distributed between nucleus and cytosol (47). Thus, the activity of Mig1/Cat8 is fine tuned with respect to glucose levels in an SNF1-dependent manner. As expected due to the hyperactivation of SNF1, Mig1-targets were significantly enriched in the up-Δreg1 group (Figure 1D, Supplementary Table S3). Mig1 targets were also enriched in the up-Δssb1Δssb2 group (Supplementary Table S3, P-value 9.11 × 10−7), however, the effect was less pronounced and did not pass the stringent criteria of the TF enrichment analysis (Figure 1D, Supplementary Table S3). Moreover, those Mig1 targets, which were upregulated in both mutant strains were more strongly affected in Δreg1 cells (Supplementary Table S2). For example CAT8, which is under direct control of Mig1 (Figure 2A), was upregulated 10.6-fold in Δreg1, and 2.4-fold in Δssb1Δssb2 cells (Supplementary Table S2). Targets of Cat8, in turn, were significantly enriched only in the up-Δreg1 group (P-value 6.94 × 10−10), but not in the up-Δssb1Δssb2 group (P-value 0.57) (Supplementary Table S3). Thus, transcription of CAT8 was induced, but Cat8 was not fully active in Δssb1Δssb2 cells, while Cat8 was seemingly fully active in Δreg1 cells. The data suggested that both the Δreg1 as well as the Δssb1Δssb2 mutation caused a release of Mig1 repression, however, the effect was significantly more pronounced in the Δreg1 strain.
Figure 2.
Transcriptional changes in Δssb1Δssb2 and Δreg1 cells resemble the transcriptional status of batch culture cells at distinct growth phases. (A) The transcriptional repressor Mig1 prevents transcription of many glucose-repressed genes among them CAT8. Cat8 is a major transcriptional activator of many glucose-repressed genes including those depicted in Figure 1C. Phosphorylation of Mig1 by SNF1 promotes nuclear export of Mig1 and releases transcriptional repression. In addition, Cat8 is activated by SNF1-dependent phosphorylation (1,8,75). (B) Mig1 is hyperphosphorylated in Δreg1 but not in Δssb1Δssb2 cells. Strains were grown to early logarithmic phase in the presence of glucose (YPD) followed by a 10 min incubation in the absence of glucose (YP). Total cell extracts were separated via sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and were subsequently analyzed via immunoblotting using an antibody recognizing Mig1 or Sse1 as a loading control. The hyperphosphorylated fraction of Mig1 (Mig-Pi) migrates with higher molecular mass. (C) Comparison of genes upregulated ≥ 1.5-fold during continuous growth in a batch culture (42) relative to the expression level at OD600 = 0.14, with genes upregulated in Δssb1Δssb2 or Δreg1 cells, respectively (24 and Supplementary Table S2). CG: number of genes upregulated in wild-type cells with increasing OD600 (42). Δssb1Δssb2∩CG and Δreg1∩CG indicate the intersecting gene sets. (D) Relative frequency of genes upregulated in batch culture, which are also upregulated in Δssb1Δssb2 or Δreg1 cells. Significant overlap (P < 0.0042, right-sided P-values of Fisher's exact test, Supplementary Table S3) is indicated with an asterisk.
Phosphorylation by SNF1 generates a complex mixture of hyperphosphorylated Mig1 species, which can be visualized due to their reduced mobility in sodium dodecyl sulphate (SDS) gels (5). Mig1 phosphorylation was readily detected in extracts derived from Δreg1 cells, but was not detected in extracts derived from Δssb1Δssb2 cells (Figure 2B). Thus, in Δssb1Δssb2 cells repression by Mig1 was partly released, even though Mig1 hyperphosphorylation was not detectable. Two possible scenarios might account for this observation. Either low level phosphorylation of Mig1, which only marginally affected mobility of Mig1 in SDS gels, caused partial release from glucose repression, or partial release from glucose repression occurred independent of Mig1 phosphorylation (compare also ‘Discussion’ section). Brief glucose depletion resulted in hyperphosphorylation of Mig1 in wild-type as well as Δssb1Δssb2 cells, indicating that Mig1 activation was not per se defective in the Δssb1Δssb2 strain (Figure 2B).
During successive depletion of glucose in a batch culture, cells continuously adapt their transcriptional program (42). Based on the above, we sought to test if transcriptional deregulation in Δssb1Δssb2 cells and Δreg1 cells resembled distinct growth phases of cells maturing in a batch culture. To test this, we employed the data from a microarray study analyzing global transcriptional changes during continuous growth (CG) (42). In this dataset we first identified those genes, which were ≥1.5-fold upregulated relative to the expression level at OD600 0.14 (Figure 2C up-CG, Supplementary Table S2). We then built the intersection groups between each up-CG group and the up-Δssb1Δssb2 or up-Δreg1 group, respectively (Figure 2C, up-Δssb1Δssb2∩CG and up-Δreg1∩CG). Starting from OD600 = 0.8 a significant number of genes upregulated in batch culture cells, was also upregulated in Δssb1Δssb2 or Δreg1 cells (Figure 2D, Supplementary Table S3). Prior to the diauxic shift the up-Δssb1Δssb2∩CG groups were more highly populated compared to the up-Δreg1∩CG groups (Figure 2D, OD600 0.8 - 3.7). Post diauxic shift the up-Δreg1∩CG groups were more highly populated (Figure 2B, OD600 6.9 - 7.3). The data suggested that the transcriptome of logarithmically growing Δssb1Δssb2 cells resembled the transcriptome of wild-type cells at the diauxic shift, while the transcriptome of Δreg1 cells more closely resembled wild-type cells in stationary phase. In summary the above data are consistent with the idea that the transcriptional program of logarithmically growing Δssb1Δssb2 cells resembled a time point close to the diauxic shift, with only partial activation of SNF1-dependent transcriptional activators, while logarithmically growing Δreg1 cells resembled a time point post-diauxic shift, with more pronounced activation of, for example, Adr1, Cat8 and their down-stream targets.
Ssb interacts with SNF1 and Glc7/Reg1 post-translationally
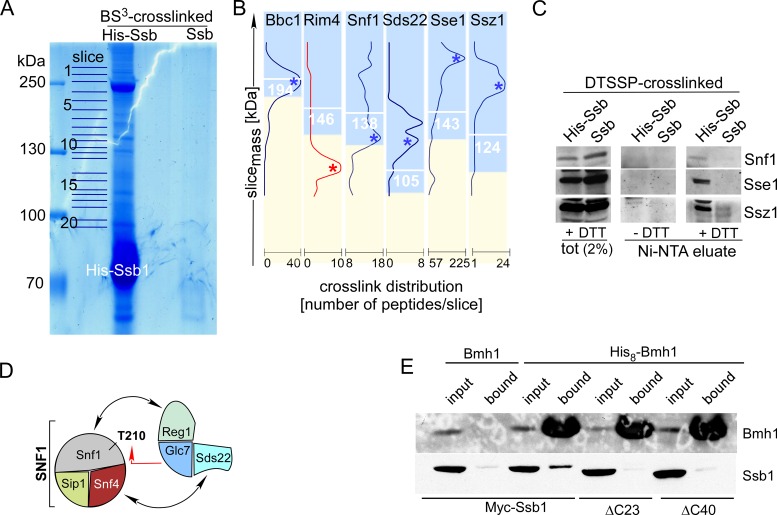
Based on the effect of the Δssb1Δssb2 mutation on SNF1-mediated transcriptional regulation and on previous observations (see ‘Introduction’ section) we speculated that Ssb might interact with SNF1 and/or Glc7/Reg1 directly. Experimental testing of this hypothesis was complicated by the fact that Ssb interacts with a large number of proteins cotranslationally, when they emerge from the ribosomal exit tunnel (20,22). By way of example a previous TAP-tag based protein–protein interaction approach identified more than 3000 potential Ssb interactors (48). To focus on those proteins, which interact with Ssb directly, however, post-translationally we revisited the Ssb interactome via a crosslinking approach. The procedure selected for proteins directly bound to His-Ssb, and, at the same time, trapped proteins interacting efficiently, however, too transiently to withstand conventional pull-down techniques (Supplementary Figure S2). Crosslinked proteins which passed the initial selection criteria (Supplementary Table S4, ‘Materials and Methods’ section) were assigned Ssb-interactors, if the maximum of identified peptides corresponded to an apparent molecular mass of ≥85% of the calculated molecular mass of a crosslink product (Figure 3B and Supplementary Table S4). As an example, Bbc1 was selected, because the maximum abundance of Bbc1 corresponded exactly to the calculated molecular mass of the Ssb1-Bbc1 crosslink (Figure 3B); Rim4 was excluded, because the maximum abundance of the Ssb1-Rim4 crosslink did not correspond to the expected molecular mass (Figure 3B). Based on these criteria, we identified 131 cytosolic proteins, which formed a crosslink to Ssb (Table 1). This rather small number of potential Ssb-interactors was consistent with a confinement to those proteins, which were in direct contact with Ssb post-translationally. The interaction of Ssb with the bulk of nascent chains was apparently below the detection limit of the LC/MS analysis. This is consistent with the low concentration of any protein in the nascent state combined with the inherently low efficiency of the crosslinking reaction.
Figure 3.
Ssb1 directly interacts with all subunits of the SNF1 complex, Glc7, Reg1 and Bmh1. (A) Purified His-Ssb crosslink products. His-Ssb was crosslinked with BS3 and was subsequently purified via Ni-NTA under denaturing conditions (‘Materials and Methods’ section). Untagged Ssb treated the same way is shown as a control. Slices of the Coomassie-stained gel employed for mass spec analysis are indicated. (B) Crosslink distribution profiles. Ssb-crosslinks were selected if a major peak of peptides was observed in the molecular mass range ≥85% of the calculated mass of the Ssb-crosslink in kDa (white numbers). Profiles of potential Ssb-interactors are shown in blue. An example profile (Rim4), which was discarded from the list of potential interactors (single maximum ≤ 85%) is shown in red (Supplementary Table S4). (C) Identification of representative Ssb-crosslinks via immunodetection. After crosslinking with the cleavable crosslinker DTSSP His-Ssb1 or, as a control, Ssb was purified under denaturing, non-reducing conditions via Ni-NTA. Aliquots of the total and of the affinity purified Ssb-crosslinks were separated via SDS-PAGE under reducing (+DTT) or non-reducing (−DTT) conditions. Subsequently immunoblots were decorated with antibodies directed against Snf1, Sse1 and Ssz1. Shown is the molecular mass range of the immunoblots at which the respective Ssb-interactor migrates after cleavage of the disulfide bond within DTSSP (+DTT). (D) Subunits of the SNF1 and Glc7 complexes, which formed crosslinks with Ssb. Interactions previously reported (see ‘Results’ section) are indicated by black arrows. The red arrow indicates the interaction between Snf1 and Glc7, which occurs during dephosphorylation of Snf1-T210. (E) Bmh1 interacts with the C-terminus of Ssb. His8-Bmh1 was purified via Ni-NTA under native conditions, untagged Bmh1 served as a control. Strains expressed N-terminally Myc-tagged Ssb, Ssb-Δ23 or Ssb-Δ40 as indicated. Input (0.66%) and the material bound to Ni-NTA (100%) was analyzed via immunoblotting using antibodies directed against Bmh1 or the Myc-tag (Supplementary Figure S1C). Background binding of Ssb was probably due to general stickiness of the chaperone, or was due to the association of Ssb with proteins like Snf1, which contain internal poly-histidine stretches (Supplementary Figures S1A and S4, and Table S4).
Table 1. Cytosolic proteins crosslinked to His-Ssb in total yeast extract.
| Cellular process | Proteins in close contact with Ssb |
|---|---|
| carbohydrate signaling | SNF1, SIP1, SNF4, GLC7, REG1, SDS22, SAP185 |
| ribosomal, large subunit | RPL2a, RPL3, RPL4a, RPL8b, RPL12b, RPL13a, RPL14b, RPL19b, RPL20a, RPL21a, RPL24a, RPL26b, RPL30, RPL31b, RPL32, RPL35b, RPL36b, RPP2b |
| ribosomal, small subunit | RPS1b, RPS3, RPS4b, RPS5, RPS7a, RPS8a, RPS11b, RPS14b, RPS16b, RPS18a, RPS19b, RPS20 |
| translation factors | TIF2, NPL3, PAB1, EFT2, YEF3, TEF4, EFB1, HYP2, STM1 |
| chaperones, general | SSA1, SSA2, SSE1, SSE2, FES1, HSC82, HSP42 |
| chaperones, specialized | NMA111, CCS1, BMH1 |
| chaperones, ribosome-bound | SSZ1, EGD1 |
| thioredoxin | TRX1, TRX2, TRR1 |
| glycolysis | GLK1, ENO1, ENO2, PFK1, PFK2, PGI1, TDH3, GPM1, FBA1, PYC2 |
| trehalose | UGP1, TSL1, TPS1, TPS2 |
| pentose phosphate | GND1, TKL1 |
| C2-utilization | ACH1, ALD6, ACS2, ADH1 |
| fatty acid biosynthesis | FAS1, FAS2 |
| tRNA synthetases | MES1, ILS1, THS1, DPS1, GRS1 |
| aa biosynthesis | SER33, SER1, SHM2, GLY1, ARO2, ARO4, TRP5, LYS20, CYS3, ASN1, ASN2, MET17, GLN1 |
| nucleotides and NAD | AMD1, ADE13, IMD3, GUA1, URA7, RNR2, PNC1, PSA1, NMA2 |
| cytoskeleton | ACT1, CRN1, BBC1, TUB1, TUB3 |
| miscellaneous | GAD1, GFA1, MRC1, DRE2, MDG1, SIN3, PEX14, RPA190, SMT3, ABF1, SPT6, ZDS1, NSR1, RPT4, ZEO1, YLR361C-A |
As a proof of principal, the crosslinking approach identified well-established co-chaperones of Ssb, like Sse1, Fes1 and Ssz1 (20) (Table 1 and Figure 3B). Direct interaction between Ssb and the highly expressed chaperones Sse1 and Ssz1 was readily confirmed employing the cleavable crosslinker DTSSP followed by immunoblotting (Figure 3C). As expected, Ssb was also in contact with components of the translational machinery, including 30 ribosomal proteins and 9 translation factors (Table 1). Four of the ribosomal proteins (Rpl35, Rpl19, Rpl26 and Rpl31) are components of the so-called exit platform, which surrounds the ribosomal polypeptide tunnel exit (Supplementary Figure S3), where Ssb gains access to nascent chains (20). Most relevant in the context of this study, five of the identified Ssb-interactors comprise the central components of the SNF1/Glc7 signaling pathway: Snf1, Sip1 and Snf4, which form a holo-SNF1 complex, Glc7 and Reg1 (Table 1 and Figure 3D) (2,3). Ssb also formed a crosslink to Sds22, an essential, however poorly characterized subunit of Glc7 (49), which also contacts Snf4 (50) and is phosphorylated by SNF1 (Yeast Kinase Interaction Database, 51). Employing a cleavable crosslinker, the interaction between Snf1 and Ssb was confirmed via immunoblotting (Figure 3C). Due to limited availability of antibodies, low expression levels of the proteins of interest, and low efficiency of crosslinking, attempts to confirm crosslinks to other components of the SNF1/Glc7 signaling pathway remained inconclusive. However, the specific interaction of Ssb with the complete set of SNF1 subunits, Glc7, and those Glc7-regulatory subunits involved in the dephosphorylation of SNF1 indicate that Ssb was in close contact with a significant fraction of the assembled SNF1 and also Glc7 complexes.
Bmh1 interacts with the C-terminal region of Ssb
Bmh1 was one of the Ssb interactors identified via the crosslinking approach (Table 1 and Supplementary Table S4). The finding corroborates previous studies, which identified Ssb as an interactor of affinity purified Bmh (18). To further characterize the interaction between Ssb and Bmh we employed a Δbmh1Δbmh2Δssb1Δssb2 strain complemented with Myc-Ssb1 and either Bmh1 (termed Myc-Ssb/Bmh strain) or His8-Bmh1 (termed Myc-Ssb/His8-Bmh1 strain). The tagged versions of Ssb1 and Bmh1 fully supported growth of the quadruple deletion strain (Supplementary Figure S1D). Ni-NTA pull-downs performed under native conditions revealed that a significant fraction of Myc-Ssb was associated with His8-Bmh1 (Figure 3E). 14-3-3 proteins bind to their clients via short internal motifs, which, most of the time, include a phosphoserine or phosphothreonine residue (13,52). More recently it was shown that in some cases 14-3-3 proteins can interact with short C-terminal segments of client proteins (53). Because Ssb contains such a potential C-terminal Bmh-binding motif (Supplementary Figure S1B, 54), we tested if the interaction of Ssb with Bmh was affected by C-terminal deletions of 23 or 40 amino acid (Ssb1-Δ23 and Ssb1-Δ40, Supplementary Figure S1B). The truncations within Ssb did not interfere with the substrate binding domain of Ssb and consistently were functional in vivo (Supplementary Figure S1D). Ni-NTA pull-downs revealed that neither Myc-Ssb1-Δ23 nor Myc-Ssb1-Δ40 was associated with His8-Bmh1 (Figure 3E). We conclude that the very C-terminus of Ssb was required for the interaction of the chaperone with Bmh (see also ‘Discussion’ section).
Bmh1 is a multicopy suppressor of the Δreg1 phenotype
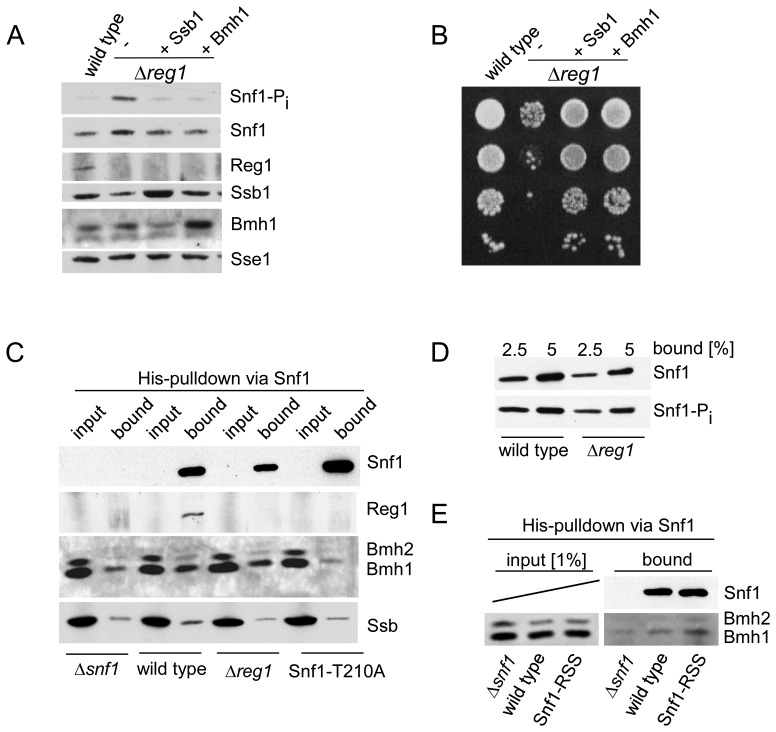
As previously shown (24), over-expression of Ssb1 abolished hyperphosphorylation of Snf1-T210 (Figure 4A) and fully suppressed growth defects (Figure 4B) of Δreg1 cells. Prompted by the similarities of Ssb and Bmh with respect to SNF1/Glc7 signaling (compare ‘Introduction’ section) and by the interaction between Ssb and Bmh, which was revealed via two independent approaches (Table 1 and Figure 3E), we tested the effect of Bmh1 over-expression in a Δreg1 strain. The result of the experiment was that Bmh1 efficiently suppressed hyperphosphorylation of Snf1-T210 and growth defects of Δreg1 cells (Figure 4A and B).
Figure 4.
Bmh suppresses growth defects of a Δreg1 strain and interacts with Snf1-T210-Pi. (A) Over-expression of Bmh1 suppresses growth defects of a Δreg1 strain. Phosphorylation of Snf1-T210 in glucose-grown wild-type, Δreg1 and Δreg1 over-expressing Ssb1 or Bmh1. Logarithmically growing yeast cultures were boiled prior to harvest and extract preparation to avoid harvest-induced phosphorylation of Snf1. Aliquots of total cell extracts were analyzed via immunoblotting with the antibodies indicated. Snf1 phosphorylation on Thr210 (Snf1-Pi) was analyzed using an antibody specifically recognizing the phosphorylated form of Snf1-T210. Sse1 served as a loading control. (B) Suppression of growth defects caused by the Δreg1 mutation. Serial 10-fold dilutions of log-phase cultures containing the same number of cells were spotted onto YPD plates and were incubated at 30°C for 2 days. (C) Bmh interacts with Snf1-T210-Pi. Cell extracts were prepared without boiling to maintain protein–protein interactions. Snf1, which contains a poly-histidine segment (Supplementary Figure S1A), was purified via Ni-NTA under native conditions, a Δsnf1 strain served as a control. Total extract (input) and affinity purified Snf1 (bound) from wild-type, Δreg1 and Snf1-T210A strains was analyzed via immunoblotting using antibodies directed against Snf1, Bmh, Reg1 and Ssb. The input corresponds to 0.33% of total extract added to the Ni-NTA pull down reaction. (see also Supplementary Figure S4). (D) Phosphorylation of Snf1-T210 occurs during cell harvest and extract preparation. Affinity purified Snf1 from wild-type and Δreg1 strains prepared without boiling (2.5 and 5% of the material shown in panel C) was analyzed for Snf1 and Snf1-Pi via immunoblotting. (E) Bmh interacts with the Snf1-RSS mutant. The experiment was performed and analyzed as described in panel C.
Binding of Bmh1 depends on the phosphorylation of Snf1-T210
Next we asked if the interaction of Bmh with Snf1 was affected by the phosphorylation level of Snf1-T210. To that end, we affinity purified Snf1 under native conditions via its internal poly-histidine segment (Supplementary Figure S1A, Figure 4C). With respect to the analysis of Bmh-binding the experimental system was challenging. First, in order to maintain native protein-protein interactions, cell cultures were not boiled prior to harvest in these experiments. However, Snf1 phosphorylation occurs during cell harvest (55) and therefore the Snf1-T210 phosphorylation level was high, even though cells were grown on glucose (Figure 4D, compare 4A for the Snf1-T210 phosphorylation level in extracts derived from boiled cells). To overcome this problem, we included the Snf1-T210A mutant in the analysis. Second, Reg1 interacts with Snf1 (56,57), Ssb and Bmh (17). Affinity purification of Snf1 from wild-type extract confirmed the interaction of Reg1 with Snf1 (Figure 4C). Moreover, analysis of Snf1-T210A (Figure 4C) confirmed that Reg1 only interacts with Snf1-T210-Pi, but does not interact with Snf1 lacking T210 phosphorylation (56,57). To distinguish between direct binding of Ssb/Bmh to Snf1 versus indirect binding via Reg1, we thus included a Δreg1 mutant in the analysis. Third, Snf1 is low abundant, while Bmh is highly abundant. Thus, only a minor fraction of Bmh can possibly co-purify with Snf1 (for a more detailed estimate refer to Supplementary Figure S4). Please note, that due to the latter problem and due to background binding (Figure 4C, Supplementary Figure S4) we were unable to analyze the binding of Ssb to Snf1 in this experimental setup (Supplementary Figure S4). The interaction between Snf1 and Ssb, however, was revealed via mass spectrometry and immunoblotting of crosslinked samples (see Figure 3B, C, Table 1 and Supplementary Table S4).
Bmh co-purified with wild-type Snf1, however, Bmh did not co-purify with Snf1-T210A (Figure 4C). As a rough estimate 30–50% of wild-type Snf1 was bound to Bmh under the conditions of the experiment (Supplementary Figure S4). Moreover, Bmh co-purified with Snf1 in a Δreg1 extract indicating that Bmh was not bound to Snf1 via Reg1 (see above). Snf1 contains a 14-3-3 consensus motif and an additional motif which closely resembles a 14-3-3 motif (Supplementary Figure S1A). To test if these motifs were required for the interaction of Bmh with Snf1 we generated Snf1-RSS, which lacks these motifs (Supplementary Figure S1A). Bmh binding to Snf1-RSS was not reduced, indicating the potential 14-3-3 motifs within Snf1 were not essential for the interaction with Bmh (Figure 4E). The combined data indicate that Bmh was bound to Snf1 in a Reg1-independent manner, however, like Reg1, displayed a strong binding preference for Snf1-T210-Pi. Whether or not Bmh directly binds to phosphorylated T210 in the activation loop is currently not clear. Alternatively, concurrent with T210 phosphorylation, other residues within the SNF1 complex might become phosphorylated leading to Bmh recruitment.
Ssb1 as well as Bmh1 act as efficient multicopy suppressors of the Δreg1 mutation at the transcriptional level
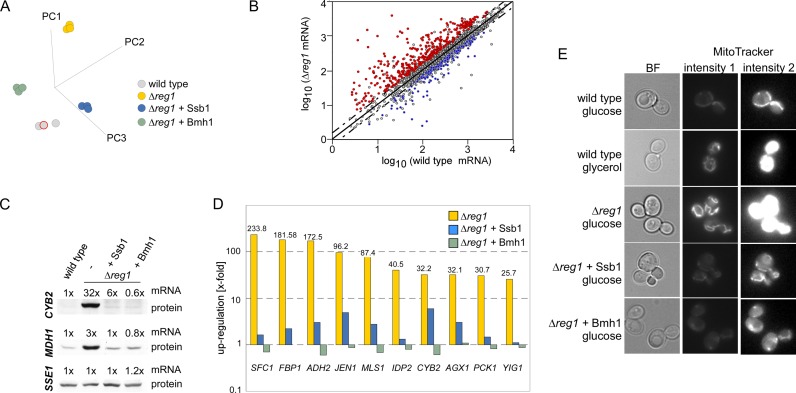
We next compared the effect of Ssb1 or Bmh1 over-expression with respect to transcriptional deregulation in the Δreg1 strain (Supplementary Table S2). PC analysis revealed that the variance between Δreg1 and the wild-type reference was significantly reduced by over-expression of Ssb1 or Bmh1 (Figure 5A). Analysis of variance (ANOVA) over the datasets (Figure 5A) identified 1774 genes, of which 752 were ≥1.5-fold deregulated in the Δreg1 strain (Supplementary Table S2). Of these genes, 71% were reregulated by at least 50% toward the wild-type expression level by over-expression of either Ssb1 or Bmh1 (Figure 5B, Supplementary Figure S5A, Table S2). As an example, mitochondrial malate dehydrogenase (MDH1) was upregulated 3-fold and cytochrome b2 (CYB2) was upregulated 32-fold in Δreg1 cells (Supplementary Table S2). Expression of both genes was efficiently reregulated by over-expression of either Ssb1 or Bmh1 at the level of the mRNA as well as at the protein level (Figure 5C and Supplementary Table S2). Over-expression of Ssb1 as well as Bmh1 reregulated transcript levels of genes under control of glucose repression (Figure 5D and Supplementary Table S2). With respect to 53 upregulated transcripts, including many of those under strong glucose repression, Bmh1 over-expression even led to a regulation overshoot (Figure 5D and Supplementary Table S2). Transcription factor ranking (34) within this group of genes revealed a strong preference for Adr1 (Supplementary Figure S5B), suggesting that the overshoot in regulation was likely due to the well characterized additional role of Bmh in the regulation of Adr1 in the nucleus (58). Over-expression of Bmh1 or Ssb1 also efficiently reregulated the expression level of genes required for respiration (Supplementary Figure S5C). The effect of Bmh1 and Ssb1 on respiration of Δreg1 cells was confirmed via in vivo uptake of MitoTracker fluorescent dye into mitochondria, which occurs in a membrane potential dependent manner (39). MitoTracker uptake increased in the order: glucose-grown wild-type cells < glycerol grown wild-type cells < glucose-grown Δreg1 cells (Figure 5E). This indicated that respiratory chain genes were not only upregulated in Δreg1 cells (Figure 1B and Supplementary Table S2), but were assembled into functional respiratory chain complexes, resulting in a high membrane potential across the inner mitochondrial membrane. Over-expression of Ssb1 or Bmh1 suppressed the effect of the Δreg1 mutation with respect to MitoTracker uptake (Figure 5E). The combined data indicate that over-expression of Bmh1 or Ssb1 exerted a very similar effect in Δreg1 cells, and that this effect was connected to the normalization of Snf1-T210 phosphorylation. One possible explanation was that Ssb1 and Bmh1 facilitated the Glc7-dependent dephosphorylation of Snf1-T210 in the absence of Reg1.
Figure 5.
Reregulation of transcriptional deregulation in a Δreg1 strain by over-expression of Ssb1 or Bmh1. (A) Principal component (PC) analysis of wild-type, Δreg1 and Δreg1 over-expressing Ssb1 or Bmh1. (B) mRNA abundance in wild-type and Δreg1 cells. Mean log values from three independent microarray datasets were plotted with wild-type on the x-axis and Δreg1 on the y-axis. Genes ≥ 1.5-fold deregulated in the in Δreg1 strain are above and below the dotted cut-off lines. Genes deregulated in Δreg1 and reregulated by over-expression of Ssb1 as well as Bmh1 are shown in red (upregulated in Δreg1) or blue (downregulated in Δreg1). (C) Levels of CYB2, MDH1 and SSE1 transcripts and of the corresponding proteins in wild-type, Δreg1 and Δreg1 over-expressing Ssb1 or Bmh1. (D) Relative expression level of the 10 most highly upregulated genes in Δreg1 cells and in Δreg1 cells over-expressing either Ssb1 or Bmh1. (E) The mitochondrial membrane potential in wild-type, Δreg1 and Δreg1 cells over-expressing Ssb1 or Bmh1 was monitored with MitoTracker®Green-FM fluorescent dye. Strains were grown on glucose (YPD) or glycerol (YPGly) as indicated. Shown are two different brightness settings (intensity 1 and 2) for the same set of images. Intensity 2 was adjusted to visualize the weak fluorescence of mitochondria in fermenting strains.
Suppression of Snf1-T210-Pi phosphorylation by high levels of Ssb1 or Bmh1 requires functional Glc7
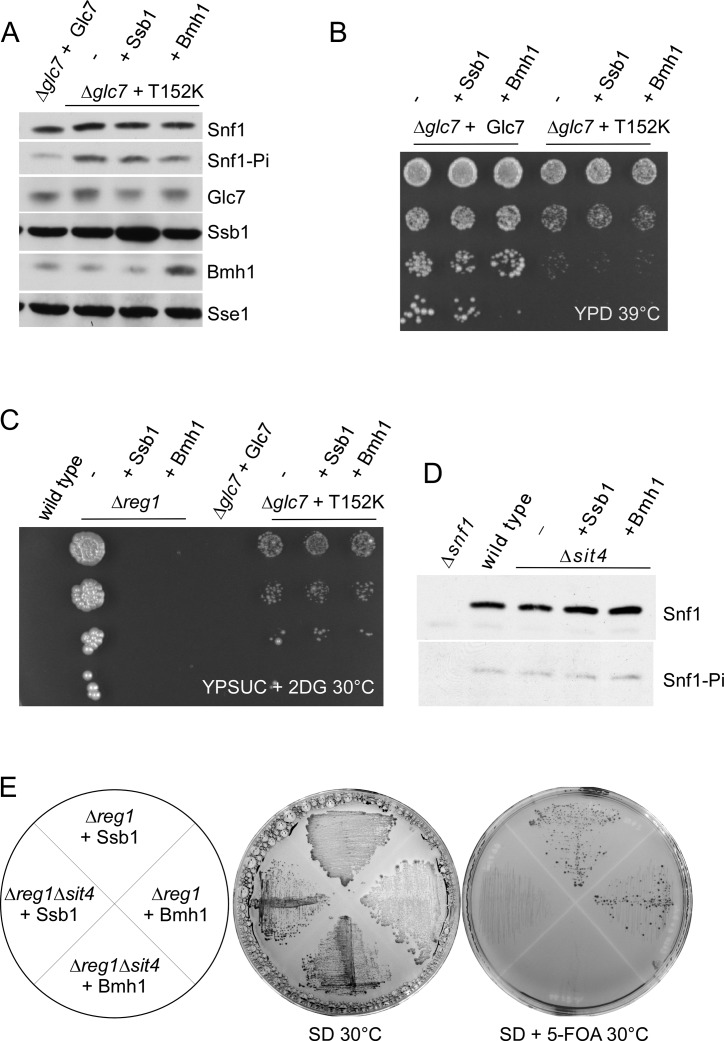
To test for this possibility, we employed the conditional mutant Glc7-T152K, which causes a release from glucose repression (59). As expected, Snf1-T210 was hyperphosphorylated in the Δglc7 + Glc7-T152K strain (Glc7-T152K strain) (Figure 6A). In contrast to the Δreg1 strain (Figure 4A), over-expression of Ssb1 or Bmh1 in the Glc7-T152K strain did not prevent hyperphosphorylation of Snf1-T210 (Figure 6A). Moreover, the temperature-sensitive phenotype of Glc7-T152K cells was unaffected by over-expression of Ssb1 or Bmh1 (Figure 6B). We also tested the ability of Ssb1 or Bmh1 to restore glucose-repression in Glc7-T152K cells directly. Wild-type yeast cannot utilize sucrose in the presence of glucose, because invertase (SUC2), which is required for sucrose utilization, is strongly repressed by glucose. 2-deoxy-glucose (2DG) mediates glucose-repression, however, cannot be utilized as an energy substrate. Therefore, 2DG strongly inhibits growth of wild-type yeast on sucrose (Figure 6C) (60). In the Δreg1 strain SUC2 was upregulated 12.2-fold (Supplementary Table S2) and thus 2DG did not inhibit growth of Δreg1 on sucrose (Figure 6C). As expected based on the above results, expression of SUC2 in Δreg1 was efficiently reregulated by over-expression of Ssb1 (upregulated 1.5-fold) or Bmh1 (same as wild-type) (Supplementary Table S2). Consistently, over-expression of Ssb1 or Bmh1 prevented growth of Δreg1 on sucrose supplemented with 2DG (Figure 6C). The Glc7-T152K mutation conferred moderate resistance to 2DG, indicating that glucose-repression was indeed partly released (Figure 6C). However, in this case, over-expression of Ssb1 or Bmh1 did not affect 2DG sensitivity (Figure 6C). Besides conditional glc7 mutants and the Δreg1 mutation, Δsit4 is the only phosphatase deletion mutant, for which a role in dephosphorylation of Snf1-T210 was previously demonstrated (61). We thus aimed to test if over-expression of Ssb1 or Bmh1 affected Snf1-T210 phosphorylation in a Δsit4 strain. However, Snf1-T210 phosphorylation was not enhanced by the Δsit4 deletion in our strain background and consequently was not affected by over-expression of Ssb1 or Bmh1 (Figure 6D). It was previously reported that the Δreg1Δsit4 double deletion is lethal (61). This enabled us to test if Ssb and/or Bmh were able to suppress the lethal phenotype of the Δreg1Δsit4 double deletion. To that end, we introduced the Δsit4 mutation into Δreg1 strains over-expressing plasmid-borne Ssb1 or Bmh1, respectively. Indeed the Δreg1Δsit4 + Ssb1 and Δreg1Δsit4 + Bmh1 strains were viable (Figure 6E). Viability was critically dependent on the over-expression of Ssb or Bmh, because cells did not survive curing of the respective plasmids (Figure 6E). Thus, the ability of Ssb or Bmh to enhance dephosphorylation of Snf1-T210-Pi in the absence of Reg1 is independent of Sit4. The combined data are consistent with a model in which high levels of Ssb1 or Bmh1 enabled Glc7 to dephosphorylate Snf1-T210 in the absence of the regulatory subunit Reg1.
Figure 6.
Suppression of Snf1-T210 hyperphosphorylation requires functional Glc7. (A) Effect of Ssb1 and Bmh1 over-expression on the phosphorylation status of Snf1-T210 in a GLC7-T152K mutant strain. To induce the Glc7-T152K defect, cultures were shifted to 39°C for 1h prior to harvest. Aliquots of total cell extracts were analyzed via immunoblotting using the indicated antibodies. Sse1 served as a loading control. (B and C) Suppression of growth defects caused by the Glc7-T152K mutation. Serial 10-fold dilutions of the strains indicated were spotted onto YPD plates and were grown for 3 days at 39°C (B) or were spotted onto YPSuc plates containing 200 μg/ml 2-deoxy-glucose (2DG) and were incubated for 8 days at 30°C (C). (D) Snf1-T210 is not significantly hyperphosphorylated in Δsit4 cells. Aliquots of total cell extracts were analyzed via immunoblotting using the indicated antibodies. (E) Over-expression of Ssb or Bmh rescues lethality of a Δreg1Δsit4 strain. The strains indicated were grown on SD for 5 days, or on SD supplemented with 5-FOA for 6 days. For details compare ‘Materials and Methods’ section.
DISCUSSION
Recent observations revealed that release from glucose-repression does not occur uniformly upon SNF1 activation. Rather, genes under control of SNF1 fall into distinct groups, which require different levels of SNF1 activity for efficient transcription (62). These findings fit with our observation that the Δssb1Δssb2 and Δreg1 mutations, which both increase the level of Snf1-T210 phosphorylation, though to a different extent (24), led to distinct patterns of transcriptional deregulation. One striking difference between the Δssb1Δssb2 and Δreg1 strains was that the transcriptional repressor Mig1, which is a direct target of SNF1 (1), was hyperphosphorylated in Δreg1, but not in Δssb1Δssb2 cells. Activation of SNF1 without subsequent Mig1 hyperphosphorylation is not without precedence. When cells are stressed with 2DG (60) or high sodium (5,63) Snf1-T210 becomes hyperphosphorylated, however, Mig1 phosphorylation remains unaffected. Thus, Δssb1Δssb2 cells resembled 2DG or sodium stress conditions with respect to Snf1-T210 and Mig1 phosphorylation. However, even though Mig1 was not phosphorylated in Δssb1Δssb2 cells, transcript levels of the Mig1-target CAT8 were moderately increased. Of note, sodium stress conditions did not affect CAT8 expression levels (64). The combined data support a model in which activation of SNF1 by Snf1-T210 phosphorylation does not result in uniform downstream effects but is seemingly modulated by additional factors, such as, for example, Ssb.
How could a chaperone, which is known to interact with nascent polypeptides (20) perform such a function? Based on our data we suggest that this role of Ssb is independent of its well characterized role as a cotranslational folding helper. First, in a Δssb1Δssb2 strain the expression levels of Snf1, and also Reg1 and Glc7, are not reduced compared to the wild-type (24). Thus even though Ssb interacts e.g. with Snf1 cotranslationally (22), Ssb is seemingly not essential to maintain a normal steady state level of these proteins. Second, the identification of the whole SNF1/Glc7/Reg1 module via crosslinking/mass spectrometry strongly argues against an exclusively cotranslational binding mode of Ssb, because the steady state expression level of the SNF1 subunits and of Glc7/Reg1 (65) is low, crosslinking via amino-groups is inefficient, and only a minor fraction of each protein is in the nascent state. Consistently only 131 proteins were identified via the crosslinking approach, which is way below the number of cotranslational Ssb interactors (22,48). In this context it is interesting to note that an unexpected theme of the post-translational Ssb-interactors was that many of them comprised subunits of the same complexes and pathways (Table 1). For example, Ssb formed crosslinks with components of the thioredoxin system, consisting of thioredoxin reductase Trr1 and its two substrates Trx1 and Trx2, both subunits of the fatty acid synthase (Fas1 and Fas2), 10 enzymes of the glycolysis/gluconeogenesis pathway, and additional cytosolic enzymes directly connected to glycolysis, as for example the subunits (Tsl1, Tps1, Tps2) of the trehalose-6-P synthase/phosphatase complex (Table 1). These findings suggest that Ssb was not only in contact with the folded and assembled SNF1 and Glc7-Reg1 complexes but also with a defined set of other heterooligomeric complexes.
Multiple lines of evidence suggest that Ssb fine tunes SNF1 activity in concert with the Bmh proteins. First, Bmh interacts with the core components of the SNF1/Glc7 signaling module (17–19). Our data confirm these earlier observations and reveal that the interaction of Bmh with Snf1 is independent of Reg1, however, depends on the phosphorylation of Snf1-T210. Second, transcriptional analysis employing conditional mutants of Bmh revealed defects in glucose repression, indicating a role of the Bmh proteins in the regulation of SNF1 activity (15,16). Third, we found that the Δreg1 mutation was not only efficiently suppressed by over-expression of Ssb (24) but also by Bmh. Multicopy suppression of the Δreg1 mutation by Ssb1 or Bmh1 had surprisingly similar consequences: (i) the Snf1-T210 phosphorylation level was normalized in a Glc7-dependent manner, (ii) wild-type growth was restored and (iii) transcriptional deregulation was normalized. Fourth, Bmh and Ssb interact with each other. This was previously suggested by the results of high throughput studies analyzing the interactome of Bmh (18,54). Here we confirm these predictions and provide experimental evidence for a direct interaction between Ssb and Bmh. We find that specifically the C-terminus of Ssb is required for binding to Bmh, leaving the substrate binding domain of Ssb free to interact with client proteins (20,21). Interestingly, binding sites of 14-3-3 proteins often localize to intrinsically unfolded regions of client proteins (13). One may thus speculate that Ssb, which also preferentially interacts with unfolded protein segments (20,21) interacts with the same intrinsically unfolded domains, which also interact with Bmh.
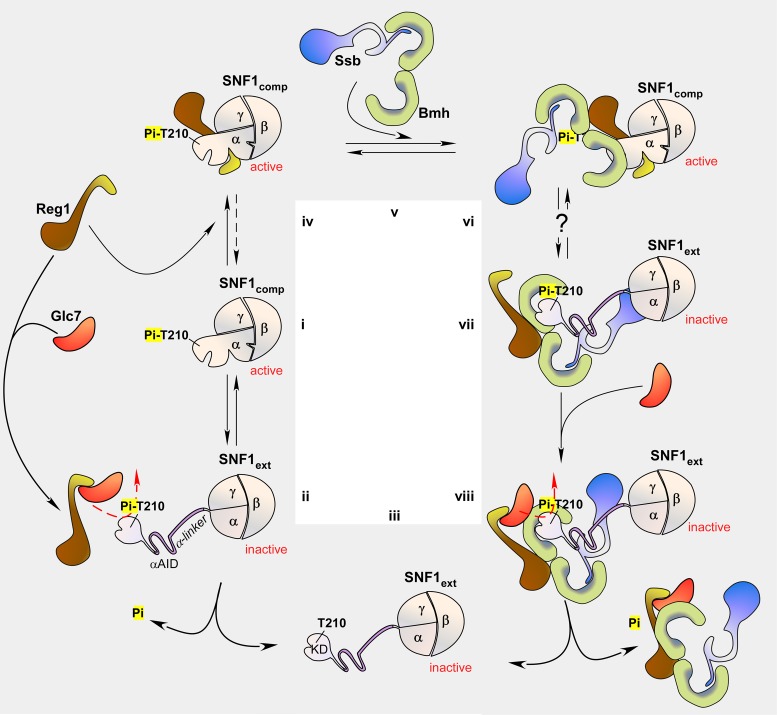
How could Bmh and Ssb together affect the phosphorylation level of Snf1-T210? We suggest a mechanism, which incorporates recent models of the regulation of SNF1 (57,66) (Figure 7i–iii). Structural and biochemical studies of the last years revealed that the regulation of SNF1 not only depends on phosphorylation of Snf1-T210, but also on a conformational switch between a compact conformation, which is active (Figure 7i, SNF1comp) and an extended conformation (Figure 7ii, SNF1ext), which is inactive (3,66–70). The switch between SNF1comp and SNF1ext involves structural rearrangements in all subunits of the heterotrimeric complex. Central for the regulation is a conformational change within the α-subunit Snf1, consisting of the N-terminal kinase domain followed by the autoinhibitory domain (αAID), regulatory α-linker and C-terminal domains (Supplementary Figure S1A). In SNF1comp the αAID is recruited to the γ-subunit, which holds it apart from the kinase domain (67–71) (Figure 7i). Biochemical studies support a model in which SNF1comp is not only active but also protects Snf1-T210-Pi from dephosphorylation (66). When SNF1 adopts the extended conformation, the Snf1 α-linker becomes unstructured allowing the αAID to interact and inhibit the kinase domain (66–70) (Figure 7ii). SNF1ext is highly susceptible to dephosphorylation by Glc7-Reg1 (10,57,72) (Figure 7ii), resulting in the fully inactive, dephosphorylated kinase complex (3,66) (Figure 7iii). Recent findings suggest that ADP binding to Snf1 and/or Snf4 may be involved in regulating the conformational switch between the compact and extended conformation (3,66,73).
Figure 7.
Schematic model of SNF1 regulation via Glc7-dependent dephosphorylation and of the potential role of Bmh/Ssb. (i–iii) Regulation of SNF1 involves phosphorylation of T210 and a conformational switch. (i) The C-terminal domain of the α-subunit Snf1, one β-subunit and the γ-subunit Snf4 form the heterotrimeric core of SNF1. In its active state (SNF1comp), residue T210 within the N-terminal kinase domain (KD) is phosphorylated and tethered closely to the core of the complex. (ii) In its inactive state (SNF1ext), the regulatory α-linker of Snf1 adopts an extended conformation, which allows the autoinhibitory domain (αAID) to inhibit kinase activity. SNF1ext is susceptible to dephosphorylation via Glc7-Reg1, (iii) which results in the dephosphorylated, inactive state of SNF1. (iv–viii) Hypothetical model for the role of Bmh/Ssb in the regulated dephosphorylation of Snf1-T210. (iv) Reg1 can bind to SNF1comp and then protects Snf1-T210-Pi from dephosphorylation. Please note that binding of Reg1 to Snf1 or Glc7 is mutually exclusive. (v) Ssb interacts with Bmh via its C-terminal domain. (vi) Because the Bmh-binding region within Reg1 is distinct from the region required for Snf1-binding, Reg1 can recruit Bmh/Ssb to SNF1comp. According to the model Bmh/Ssb does not directly bind to SNF1comp. (vii) When Snf1 adopts the extended conformation, Reg1 remains bound to Bmh/Ssb, however, is released from SNF1ext. At the same time, Bmh/Ssb interact and stabilize the extended α-linker of Snf1. (viii) Reg1 can now recruit Glc7, which dephosphorylates Snf1-T210-Pi. For details and references compare ‘Discussion’ section. The model takes into account the results from structural and biochemical studies on the regulation of SNF1 and its mammalian homolog AMPK. While some aspects remain speculative, it is consistent with the ring-a-ring-a-roses between the subunits of SNF1, Glc7, Reg1, Bmh and Ssb observed in this and many previous studies (see ‘Discussion’ section). The αβγ-heterotrimeric SNF1 complex is shown in beige. The α-subunit corresponds to Snf1. The domain structure of Snf1 is shown in Supplementary Figure S1A. Reg1 is shown in brown, the Reg1 domain, which interacts with either Glc7 or Snf1 is shown in in yellow. Ssb is shown in blue, Bmh in green. Bmh/Ssb is regarded as a single functional complex.
Reg1 performs a central role in the regulation of SNF1 activity, however, its exact function is not yet understood. On the one hand Reg1 binds to active SNF1 (56,57, and this work) (Figure 7iv), thereby protecting Snf1-T210-Pi from dephosphorylation (57), while on the other hand Reg1 binds to Glc7 and is required for the Glc7-dependent dephosphorylation of Snf1-T210-Pi (1,5, and this work) (Figure 7ii). Because Reg1 interacts with Snf1 and Glc7 via the same, short motif, binding of Reg1 to Snf1 and Glc7 is mutually exclusive (57) (Figure 7ii and iv). What determines if Reg1 associates with Snf1 or Glc7 is currently not understood.
Based on our data we propose that Reg1 functions together with Bmh and Ssb (Bmh/Ssb) as a module, which is required for the switch from SNF1comp to SNF1ext (Figure 7v–viii). We speculate that Reg1 has a high affinity for SNF1comp and a low affinity for SNF1ext, while Bmh/Ssb has a low affinity for SNF1comp, but a high affinity SNF1ext. The model is supported by the following observations. Ssb interacts with Bmh via its C-terminal domain (this work and see prediction in 54) (Figure 7v). Bmh/Ssb and Reg1 form a complex, which contains approximately stoichiometric amounts of Reg1, Bmh and Ssb, however, does not contain Glc7 (17). Because Reg1 interacts with Bmh via a domain distinct from the domain which interacts with Snf1/Glc7 (17), we speculate that Reg1 can recruit Bmh/Ssb to SNF1comp, resulting in a complex, in which Reg1 bridges between SNF1comp and Bmh/Ssb. In this situation Bmh/Ssb does not directly interact with SNF1comp, because the α-linker is folded (Figure 7vi). Only after the conformational rearrangement to SNF1ext (3,66) the α-linker becomes extended (70) providing a binding site for Bmh/Ssb (Figure 7vii). We speculate that Bmh/Ssb-binding may stabilize the extended conformation of Snf1, because it prevents folding of the α-linker. Reg1 is released from SNF1ext, however, is not released from Bmh/Ssb and thus remains in close proximity of SNF1ext (Figure 7vii). In this situation the Snf1/Glc7-binding site within Reg1 is free (57) and can recruit Glc7 which then dephosphorylates Snf1-T210-Pi (Figure 7viii). As a consequence, Reg1 and Bmh/Ssb, which both possess a low affinity for the dephosphorylated SNF1ext complex (this work) are released.
The model provides a possible explanation also for the effect of over-expression of Bmh or Ssb in the Δreg1 strain. Bmh/Ssb would not be recruited to SNF1comp in the absence of Reg1. However, in the absence of Reg1, SNF1comp is less stable and the equilibrium is shifted to SNF1ext (3,57,66). This may allow Bmh/Ssb to bind and stabilize SNF1ext. By the law of mass action higher levels of Bmh/Ssb will trap more SNF1ext in the extended conformation allowing Glc7 to dephosphorylate Snf1-T210-Pi even in the absence of Reg1. Enhanced phosphorylation of Snf1 in Δssb1Δssb2 (24) suggests that also in the wild-type binding of Bmh/Ssb to SNF1ext contributes to efficient dephosphorylation. However, the data presented here and earlier (17,24) indicate that Ssb only modulates SNF1 activity. Likely, Bmh is the central component of the Bmh/Ssb module (see also above and ‘Introduction’ section). Due to the lethality of the Δbmh1Δbmh2 strain this has not been tested directly. However, such a model is consistent also with the observation that in a Δreg1 strain conditional Bmh mutants cause a synthetic lethal phenotype (58).
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [SFB 746, DFG RO 1028/5-1]; Excellence Initiative of the German federal and state governments [BIOSS-2]. Funding for open access charge: Deutsche Forschungsgemeinschaft [SFB 746].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hedbacker K., Carlson M. SNF1/AMPK pathways in yeast. Front. Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broach J.R. Nutritional control of growth and development in yeast. Genetics. 2012;192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad M., Schothorst J., Kankipati H.N., Zeebroeck G., Rubio-Texeira M., Thevelein J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014;38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J., Vaga S., Chumnanpuen P., Kumar R., Vemuri G.N., Aebersold R., Nielsen J. Mapping the interaction of Snf1 with TORC1 in Saccharomyces cerevisiae. Mol. Syst. Biol. 2011;7:545. doi: 10.1038/msb.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCartney R.R., Schmidt M.C. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 2001;276:36460–36466. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- 6.Braun K.A., Vaga S., Dombek K.M., Fang F., Palmisano S., Aebersold R., Young E.T. Phosphoproteomic analysis identifies proteins involved in transcription-coupled mRNA decay as targets of Snf1 signaling. Sci. Signal. 2014;7:ra64. doi: 10.1126/scisignal.2005000. [DOI] [PubMed] [Google Scholar]
- 7.Ratnakumar S., Kacherovsky N., Arms E., Young E.T. Snf1 controls the activity of Adr1 through dephosphorylation of Ser230. Genetics. 2009;182:735–745. doi: 10.1534/genetics.109.103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young E.T., Dombek K.M., Tachibana C., Ideker T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 2003;278:26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- 9.Rubenstein E.M., McCartney R.R., Zhang C., Shokat K.M., Shirra M.K., Arndt K.M., Schmidt M.C. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J. Biol. Chem. 2008;283:222–230. doi: 10.1074/jbc.M707957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu J., Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dombek K.M., Voronkova V., Raney A., Young E.T. Functional analysis of the yeast Glc7-binding protein Reg1 identifies a protein phosphatase type 1-binding motif as essential for repression of ADH2 expression. Mol. Cell. Biol. 1999;19:6029–6040. doi: 10.1128/mcb.19.9.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Heusden G.P. 14-3-3 Proteins: insights from genome-wide studies in yeast. Genomics. 2009;94:287–293. doi: 10.1016/j.ygeno.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Obsil T., Obsilova V. Structural basis of 14-3-3 protein functions. Semin. Cell Dev. Biol. 2011;22:663–672. doi: 10.1016/j.semcdb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Kleppe R., Martinez A., Doskeland S.O., Haavik J. The 14-3-3 proteins in regulation of cellular metabolism. Semin. Cell Dev. Biol. 2011;22:713–719. doi: 10.1016/j.semcdb.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Bruckmann A., Steensma H.Y., Teixeira De Mattos M.J., Van Heusden G.P. Regulation of transcription by Saccharomyces cerevisiae 14-3-3 proteins. Biochem. J. 2004;382:867–875. doi: 10.1042/BJ20031885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichimura T., Kubota H., Goma T., Mizushima N., Ohsumi Y., Iwago M., Kakiuchi K., Shekhar H.U., Shinkawa T., Taoka M., et al. Transcriptomic and proteomic analysis of a 14-3-3 gene-deficient yeast. Biochemistry. 2004;43:6149–6158. doi: 10.1021/bi035421i. [DOI] [PubMed] [Google Scholar]
- 17.Dombek K.M., Kacherovsky N., Young E.T. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:39165–39174. doi: 10.1074/jbc.M400433200. [DOI] [PubMed] [Google Scholar]
- 18.Kakiuchi K., Yamauchi Y., Taoka M., Iwago M., Fujita T., Ito T., Song S.Y., Sakai A., Isobe T., Ichimura T. Proteomic analysis of in vivo 14-3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry. 2007;46:7781–7792. doi: 10.1021/bi700501t. [DOI] [PubMed] [Google Scholar]
- 19.Elbing K., McCartney R.R., Schmidt M.C. Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem. J. 2006;393:797–805. doi: 10.1042/BJ20051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peisker K., Chiabudini M., Rospert S. The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2010;1803:662–672. doi: 10.1016/j.bbamcr.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Mayer M.P., Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willmund F., Del Alamo M., Pechmann S., Chen T., Albanese V., Dammer E.B., Peng J., Frydman J. The cotranslational function of ribosome-associated hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pechmann S., Willmund F., Frydman J. The ribosome as a hub for protein quality control. Mol. Cell. 2013;49:411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Plehwe U., Berndt U., Conz C., Chiabudini M., Fitzke E., Sickmann A., Petersen A., Pfeifer D., Rospert S. The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev. 2009;23:2102–2115. doi: 10.1101/gad.529409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayordomo I., Regelmann J., Horak J., Sanz P. Saccharomyces cerevisiae 14-3-3 proteins Bmh1 and Bmh2 participate in the process of catabolite inactivation of maltose permease. FEBS Lett. 2003;544:160–164. doi: 10.1016/s0014-5793(03)00498-8. [DOI] [PubMed] [Google Scholar]
- 26.Heitman J., Movva N.R., Hiestand P.C., Hall M.N. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1991;88:1948–1952. doi: 10.1073/pnas.88.5.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett L., Orlova M., Maziarz M., Kuchin S. Protein kinase A contributes to the negative control of Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot. Cell. 2012;11:119–128. doi: 10.1128/EC.05061-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz A., Liu Y., Xu X., Carlson M. Heterotrimer-independent regulation of activation-loop phosphorylation of Snf1 protein kinase involves two protein phosphatases. Proc. Natl. Acad. Sci. U.S.A. 2012;109:8652–8657. doi: 10.1073/pnas.1206280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gari E., Piedrafita L., Aldea M., Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 30.Boeke J.D., LaCroute F., Fink G.R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 31.Baldi P., Long A.D. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–800. [Google Scholar]
- 33.Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teixeira M.C., Monteiro P.T., Guerreiro J.F., Goncalves J.P., Mira N.P., dos Santos S.C., Cabrito T.R., Palma M., Costa C., Francisco A.P., et al. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:D161–D166. doi: 10.1093/nar/gkt1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens S.W., Abelson J. Yeast pre-mRNA splicing: methods, mechanisms, and machinery. Methods Enzymol. 2002;351:200–220. doi: 10.1016/s0076-6879(02)51849-8. [DOI] [PubMed] [Google Scholar]
- 36.Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 37.Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G.M., Carnemolla B., Orecchia P., Zardi L., Righetti P.G. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 38.Cristodero M., Mani J., Oeljeklaus S., Aeberhard L., Hashimi H., Ramrath D.J., Lukes J., Warscheid B., Schneider A. Mitochondrial translation factors of Trypanosoma brucei: elongation factor-Tu has a unique subdomain that is essential for its function. Mol. Microbiol. 2013;90:744–755. doi: 10.1111/mmi.12397. [DOI] [PubMed] [Google Scholar]
- 39.Poot M., Zhang Y.Z., Kramer J.A., Wells K.S., Jones L.J., Hanzel D.K., Lugade A.G., Singer V.L., Haugland R.P. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J. Histochem. Cytochem. 1996;44:1363–1372. doi: 10.1177/44.12.8985128. [DOI] [PubMed] [Google Scholar]
- 40.Kushnirov V.V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 41.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeRisi J.L., Iyer V.R., Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 43.Schüller H.J. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 2003;43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- 44.Vyas V.K., Kuchin S., Carlson M. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics. 2001;158:563–572. doi: 10.1093/genetics/158.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkey C.D., Vyas V.K., Carlson M. Nrg1 and Nrg2 transcriptional repressors are differently regulated in response to carbon source. Eukaryot. Cell. 2004;3:311–317. doi: 10.1128/EC.3.2.311-317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young E.T., Kacherovsky N., Riper K. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J. Biol. Chem. 2002;277:38095–38103. doi: 10.1074/jbc.M206158200. [DOI] [PubMed] [Google Scholar]
- 47.De Vit M.J., Waddle J.A., Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong Y., Kakihara Y., Krogan N., Greenblatt J., Emili A., Zhang Z., Houry W.A. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol. Syst. Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong G., Trumbly R.J., Reimann E.M., Schlender K.K. Sds22p is a subunit of a stable isolatable form of protein phosphatase 1 (Glc7p) from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2000;376:288–298. doi: 10.1006/abbi.2000.1715. [DOI] [PubMed] [Google Scholar]
- 50.Ghosh A., Cannon J.F. Analysis of protein phosphatase-1 and aurora protein kinase suppressors reveals new aspects of regulatory protein function in Saccharomyces cerevisiae. PLoS One. 2013;8:e69133. doi: 10.1371/journal.pone.0069133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharifpoor S., Nguyen Ba A.N., Youn J.Y., van Dyk D., Friesen H., Douglas A.C., Kurat C.F., Chong Y.T., Founk K., Moses A.M., et al. A quantitative literature-curated gold standard for kinase-substrate pairs. Genome Biol. 2011;12:R39. doi: 10.1186/gb-2011-12-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muslin A.J., Tanner J.W., Allen P.M., Shaw A.S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 53.Coblitz B., Wu M., Shikano S., Li M. C-terminal binding: an expanded repertoire and function of 14-3-3 proteins. FEBS Lett. 2006;580:1531–1535. doi: 10.1016/j.febslet.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 54.Panni S., Montecchi-Palazzi L., Kiemer L., Cabibbo A., Paoluzi S., Santonico E., Landgraf C., Volkmer-Engert R., Bachi A., Castagnoli L., et al. Combining peptide recognition specificity and context information for the prediction of the 14-3-3-mediated interactome in S. cerevisiae and H. sapiens. Proteomics. 2011;11:128–143. doi: 10.1002/pmic.201000030. [DOI] [PubMed] [Google Scholar]
- 55.Wilson W.A., Hawley S.A., Hardie D.G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 56.Ludin K., Jiang R., Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabba S., Mangat S., McCartney R., Schmidt M.C. PP1 phosphatase-binding motif in Reg1 protein of Saccharomyces cerevisiae is required for interaction with both the PP1 phosphatase Glc7 and the Snf1 protein kinase. Cell Signal. 2010;22:1013–1021. doi: 10.1016/j.cellsig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Braun K.A., Parua P.K., Dombek K.M., Miner G.E., Young E.T. 14-3-3 (Bmh) proteins regulate combinatorial transcription following RNA polymerase II recruitment by binding at Adr1-dependent promoters in Saccharomyces cerevisiae. Mol. Cell. Biol. 2013;33:712–724. doi: 10.1128/MCB.01226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tu J., Carlson M. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:6789–6796. doi: 10.1128/mcb.14.10.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCartney R.R., Chandrashekarappa D.G., Zhang B.B., Schmidt M.C. Genetic analysis of resistance and sensitivity to 2-deoxyglucose in Saccharomyces cerevisiae. Genetics. 2014;198:635–646. doi: 10.1534/genetics.114.169060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruiz A., Xu X., Carlson M. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6349–6354. doi: 10.1073/pnas.1102758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young E.T., Zhang C., Shokat K.M., Parua P.K., Braun K.A. The AMP-activated protein kinase Snf1 regulates transcription factor binding, RNA polymerase II activity, and mRNA stability of glucose-repressed genes in Saccharomyces cerevisiae. J. Biol. Chem. 2012;287:29021–29034. doi: 10.1074/jbc.M112.380147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye T., Elbing K., Hohmann S. The pathway by which the yeast protein kinase Snf1p controls acquisition of sodium tolerance is different from that mediating glucose regulation. Microbiology. 2008;154:2814–2826. doi: 10.1099/mic.0.2008/020149-0. [DOI] [PubMed] [Google Scholar]
- 64.Yale J., Bohnert H.J. Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 2001;276:15996–16007. doi: 10.1074/jbc.M008209200. [DOI] [PubMed] [Google Scholar]
- 65.Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 66.Chandrashekarappa D.G., McCartney R.R., Schmidt M.C. Ligand binding to the AMP-activated protein kinase active site mediates protection of the activation loop from dephosphorylation. J. Biol. Chem. 2013;288:89–98. doi: 10.1074/jbc.M112.422659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen L., Jiao Z.H., Zheng L.S., Zhang Y.Y., Xie S.T., Wang Z.X., Wu J.W. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature. 2009;459:1146–1149. doi: 10.1038/nature08075. [DOI] [PubMed] [Google Scholar]
- 68.Chen L., Wang J., Zhang Y.Y., Yan S.F., Neumann D., Schlattner U., Wang Z.X., Wu J.W. AMP-activated protein kinase undergoes nucleotide-dependent conformational changes. Nat. Struct. Mol. Biol. 2012;19:716–718. doi: 10.1038/nsmb.2319. [DOI] [PubMed] [Google Scholar]
- 69.Chen L., Xin F.-J., Wang J., Hu J., Zhang Y.-Y., Wan S., Cao L.-S., Lu C., Li P., Yan S.F., et al. Conserved regulatory elements in AMPK. Nature. 2013;498:E8–E10. doi: 10.1038/nature12189. [DOI] [PubMed] [Google Scholar]
- 70.Xin F.-J., Wang J., Zhao R.-Q., Wang Z.-X., Wu J.-W. Coordinated regulation of AMPK activity by multiple elements in the [alpha]-subunit. Cell Res. 2013;23:1237–1240. doi: 10.1038/cr.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao B., Sanders M.J., Underwood E., Heath R., Mayer F.V., Carmena D., Jing C., Walker P.A., Eccleston J.F., Haire L.F., et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanz P., Alms G.R., Haystead T.A., Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 2000;20:1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mayer F.V., Heath R., Underwood E., Sanders M.J., Carmena D., McCartney R.R., Leiper F.C., Xiao B., Jing C., Walker P.A., et al. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 2011;14:707–714. doi: 10.1016/j.cmet.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joseph-Horne T., Hollomon D.W., Wood P.M. Fungal respiration: a fusion of standard and alternative components. Biochim. Biophys. Acta. 2001;1504:179–195. doi: 10.1016/s0005-2728(00)00251-6. [DOI] [PubMed] [Google Scholar]
- 75.Hedges D., Proft M., Entian K.D. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1915–1922. doi: 10.1128/mcb.15.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.