Abstract
Many genomes display high levels of heterozygosity (i.e. presence of different alleles at the same loci in homologous chromosomes), being those of hybrid organisms an extreme such case. The assembly of highly heterozygous genomes from short sequencing reads is a challenging task because it is difficult to accurately recover the different haplotypes. When confronted with highly heterozygous genomes, the standard assembly process tends to collapse homozygous regions and reports heterozygous regions in alternative contigs. The boundaries between homozygous and heterozygous regions result in multiple assembly paths that are hard to resolve, which leads to highly fragmented assemblies with a total size larger than expected. This, in turn, causes numerous problems in downstream analyses such as fragmented gene models, wrong gene copy number, or broken synteny. To circumvent these caveats we have developed a pipeline that specifically deals with the assembly of heterozygous genomes by introducing a step to recognise and selectively remove alternative heterozygous contigs. We tested our pipeline on simulated and naturally-occurring heterozygous genomes and compared its accuracy to other existing tools. Our method is freely available at https://github.com/Gabaldonlab/redundans.
INTRODUCTION
The assembly of genomes from short sequencing reads is a complex computational problem. Numerous genome assemblers have been developed to address this task (1–5). Typically, when there is some heterogeneity in the sequence (e.g. non-haploid organisms, population of cells or individuals, etc.), a single reference sequence is recovered. In the particular case of non-haploid organisms that are highly polymorphic, the standard genome assemblers produce fragmented assemblies with a total size larger than expected (6,7). This is because short reads are generally not sufficient to accurately recover the different haplotypes in heterozygous regions, which are reported as alternative contigs. In contrast homozygous (or low heterozygosity) regions from the two homeologous chromosomes are collapsed into a single contig. The boundaries between these two types of contigs cannot be resolved by a unique path and, therefore, they are left unlinked. The final result is typically an assembly that is highly fragmented and contains redundant contigs (i.e. same region in homeologous chromosomes). Such assemblies mislead downstream analyses, from gene prediction (i.e. fragmented gene models, apparent paralogs) to comparative genome analysis (i.e. apparent duplicated blocks, synteny breaks).
Because heterozygous contigs represent the sequence of each haploid genome and homozygous contigs represent a consensus between two or more haploid genomes, these two categories of contigs can be recognized by similarity searches and differences in their depth-of-coverage. That is, heterozygous contigs should align to other heterozygous contigs originating from the same genomic region. In addition, when the reads are aligned back to the assembly, the consensus, homozygous contigs will have a higher number of reads aligned per a given length interval than haploid, heterozygous contigs (roughly double, for diploid organisms). We took advantage of these two properties to design a novel assembly strategy that is able to cope with highly heterozygous genomes. In brief our approach consists of three main steps: (i) detection and selectively removal of redundant contigs from an initial standard assembly, (ii) scaffolding of such non-redundant assembly using paired-end, mate-pair and/or fosmid-based reads and (iii) gap closing. The resulting assembly represents a chimeric reference genome in which each heterozygous region results from a random sorting of the haplotypes. Our strategy (and pipeline) is flexible and can be implemented on top of several software tools for the assembly, mapping, scaffolding, and gap closing steps. We have applied our methodology to both, real and simulated data sets, in order to evaluate its efficacy and accuracy.
MATERIALS AND METHODS
Genomes and short reads simulations
We used real data from Illumina and 454 whole genome shotgun sequencing of Candida orthopsilosis AY2 (NCBI accession: AMDC01) and MCO456 (6), Dekkera bruxellensis (AZMW01), and Wickerhamomyces anomalus (AEGI01). In addition, we simulated heterozygous genomes based on the small fungal genome (13 Mb C. parapsilosis CDC317 homozygous genome, which is organized in eight nuclear and one mitochondrial chromosomes) and the medium size plant genome (119 Mb Arabidopsis thaliana genome, which is organized in five nuclear chromosomes).
The simulations were performed in two complementary directions: (i) varying levels of heterozygosity and (ii) varying levels of divergence between heterozygous regions. At first, six genomes with 5% divergence between haploid genomes and increasing loss of heterozygosity (LOH, regions of the genome that lost heterozygosity through recombination) levels (0%, 20%, 40%, 60%, 80% and 100%) were generated using fasta2diverged.py v1.0 (see redundans github repository). Inserted LOH blocks sizes were modeled based on the real size distributions observed in C. orthopsilosis MCO456 (6) and C. metapsilosis PL429 (8). Secondly, 15 genomes with 40% loss of heterozygosity and increasing level of divergence between heterozygous regions (1%, 3%, 5%, 7%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55% and 60%) were generated. Subsequently, we simulated two Illumina libraries for each simulated genome: (i) 100 bp paired-end reads with 600 bp insert size (±50 bp) and 200X coverage, and (ii) 50 bp mate-pair reads with 5,000 bp insert size (±1200 bp) and 20× coverage using GemSIM v1.6 (9). In addition, we generated 50 bp fosmid-ends reads with 40 000 bp insert size (±10 000 bp) and 0.2× coverage for simulated heterozygous plant genomes. Finally, in order to run ALLPATHS-LG, we generated additional paired-end libraries: 2 × 100 bp with 150 bp insert size (so-called overlapping paired-end reads).
Heterozygous genome assembly pipeline
Three assemblers were used to assemble paired-end reads into contigs and scaffolds: Platanus v1.2.1 (10) with default parameters, SOAPdenovo v2.04 (1) with default parameters and K-mer ranging from 71 to 91 and SPAdes v3.1.0 (4) with default parameters. For SOAPdenovo we tried all -M possible settings (0, 1, 2, 3) in a subset of the data (5% divergence and 40% LOH simulated dataset), but no significant improvements were found and therefore this option was not used. In addition, dipSPAdes, an extension of SPAdes designed to handle polymorphic genomes was used for comparison with our pipeline (11). Finally, we also tested ALLPATHS-LG v.R44837 (12) in haploidify mode. ALLPATHS-LG was ran with three libraries: overlapping paired-end (2 × 100 bp with 150 bp insert size), paired-end with 600 bp insert size and mate-pair.
We applied redundans pipeline to assembled contigs and scaffolds. Redundans consists of three consecutive steps, in this order: (i) reduction, (ii) scaffolding and (iii) gap closing (Figure 1C). Firstly, contigs are aligned all versus all by BLAT (13) or LAST (14). Subsequently heterozygous contigs, those having identity and overlap above defined thresholds (by default 0.51 for identity and 0.66 for overlap), are identified from the alignments and shorter contigs are removed. The reduction steps consists of the identification and removal of heterozygous contigs, based on pair-wise sequence similarity searches. Contigs resulting from heterozygous regions are expected to have high sequence identity. By default, we cluster contigs with at least 85% identity and overlapping over 66% of the shorter sequence length from each pair-wise comparison. Only the longest contig representing each cluster is kept for further analysis. Subsequently, in the scaffolding step adjacent contigs connected by at least five read pairs are joined using SSPACE3 (15). Here, the parameters of each library are estimated automatically (including mean, median and standard deviation of insert size, and relative orientation of read pairs) and the sequencing libraries are used in order of increasing insert size. The reads are aligned using the fast and flexible short and long read mapper, BWA MEM (16), instead of the standalone SSPACE3 mapper (bowtie). Finally, in the final gap closing step remaining gaps are filled using GapCloser from the SOAPdenovo package (1). Noteworthy, in the last two steps only a subset of the reads in each library is processed in order to speed-up the scaffolding and gap closing. By default, redundans process 0.2 * ‘genome size in bp’ reads, for example up to 20 million read pairs from each library will be processed for 100 Mb genome (0.2 × 108 bp).
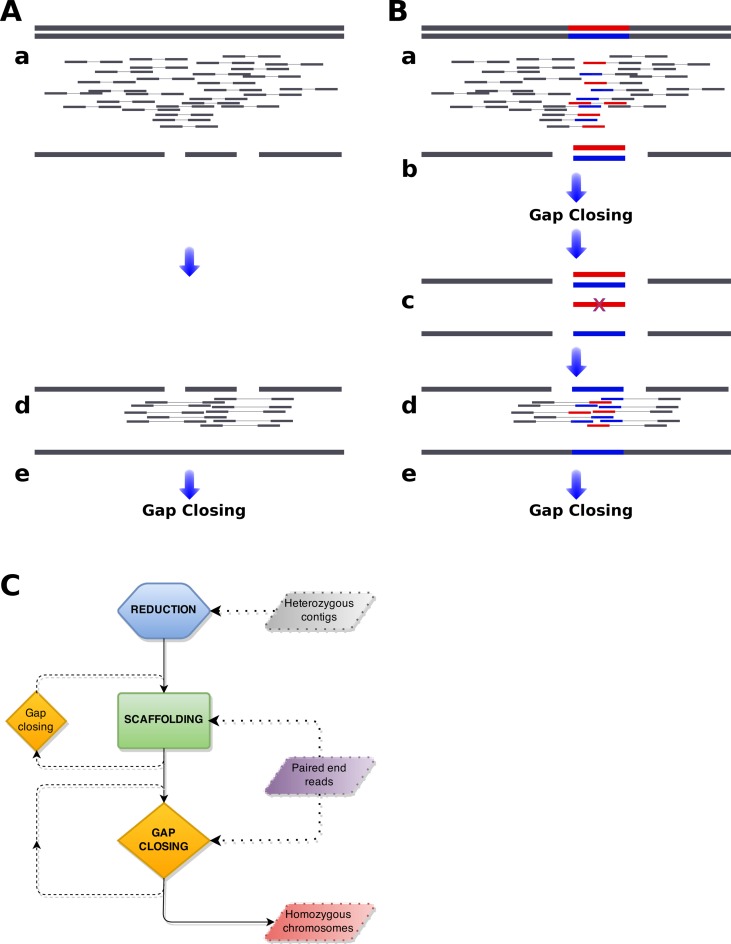
Figure 1.
Genome assembly from short reads. Standard (A) and heterozygous (B) genome assembly pipelines are compared. Diploid chromosomes are indicated as horizonal bars with heterozygous regions marked as red and blue. Paired-end reads produced from sequencing of those chromosomes are indicated as smaller bars linked by thin lines below the chromosomes. Assemblies are indicated as horizontal bars, in the same way as chromosomes, but a single reference is produced for diploid chromosomes. Heterozygous genome assembly pipeline consists of five steps. (a) Standard de novo assembly is performed and (b) optionally gaps are closed. Obtained assembly is larger than expected and fragmented because two alternative contigs are recovered from heterozygous regions (blue and red), while single contig is recovered from homozygous regions (gray). Further scaffolding of such assembly is impossible, as homozygous contigs can be joined to any of heterozygous contigs (blue and red).(c) To overcome this, redundant contigs from heterozygous regions are removed (here the red contig) and (d) homogenised assembly is further scaffolded. (e) Finally, gaps are closed.(C) Schematic representation of redundans mechanisms. Redundans pipeline consists of three steps: reduction, scaffolding and gap closing. Program takes as input assembled contigs, paired-end and/or mate pairs sequencing libraries and returns scaffolded homozygous genome assembly, that should be less fragmented and with total size smaller than the input contigs. Note, scaffolding and gap closing may be executed in multiple iterations. In the first step, only heterozygous contigs are used. Paired-end and/or mate pair libraries are used for scaffolding and gap closing. The latter steps can be repeated to achieve incremental assembly improvement. Redundans is very flexible, thus any of the above mentioned step can be omitted.
All above steps are performed automatically by the wrapper (redundans.py). The wrapper takes as input assembled contigs, and paired-end and/or mate pairs reads. Runtime parameters, including library insert sizes and read orientations, are estimated automatically. The wrapper performs all necessary steps and returns scaffolded homozygous genome assembly, that should be less fragmented and with a total size smaller than the input contigs. All mentioned thresholds and parameters can be adjusted by the user. Redundans pipeline and all programs mentioned in the text can be access publicly (https://github.com/Gabaldonlab/redundans).
Comparison with heterozygosity reducing tools
The reduction step in redundans was compared with Haplomerger v20120810 (17). In these comparisons, we performed reduction of contigs recovered by SPAdes from each simulated heterozygous fungal genome. Because Haplomerger removed only a fraction of heterozygous contigs in the first iteration, we performed two additional iterations with this tool, while only one iteration of reduction was necessary for redundans. Subsequently, we used the resulting reduced assemblies from the redundans pipeline used while skipping the reduction step (–noreduction parameter). Redundans reduction method yielded less fragmented assemblies with more accurate total size, than Haplomerger in all but two simulated heterozygous genomes (divergence of 5% and LOH of 0% and 100%) (Supplementary Tables S8 and S9).
Estimation of assembly quality
We assessed the quality of the assemblies by using several parameters of general use (18). These include, number of contigs, N50, and genome completeness expressed as the ratio of the observed versus expected assembly size (18). For assemblies of simulated data the expected assembly size was known. We inferred the presence of redundant contigs/scaffolds if the assembly had a size larger than the reference. On the other hand, a smaller than expected assembly informed about the extent of missing reference genome regions. In addition, we analyzed the accuracy of each assembly by visual inspection of the alignments of its contigs/scaffolds and the reference chromosomes. The pairwise genome alignments were created and visualised using NUCmer v3.1 (19). The resulting alignments were filtered, keeping only the best alignment for each region from the query sequence, so called many-to-one mode. Subsequently, we counted large rearrangements, namely deletions, inversions or translocations, between every assembly and the respective reference genome. Additionally, we marked the reference sequences missing from each assembly. Finally, we checked by pairwise alignment of the de novo assemblies against the reference chromosomes, whether observed rearrangements originated from the original contigs/scaffolds (SPAdes or SOAPdenovo2) or were introduced during scaffolding. If a particular rearrangement was absent from the respective de novo assembly, we concluded it was introduced during the scaffolding step.
RESULTS
Rationale and design
In the course of our past and ongoing research in genomics, we have often encountered difficulties in producing high quality assemblies for highly heterozygous genomes. This problem is shared by many other colleagues, given the abundance in nature of highly heterozygous species, including hybrid species. For instance, in fungi, the number of reported hybrids has increased in the last years, of which many have been discovered in the process of genome sequencing (20). The assemblies of genomes from these highly heterozygous species are highly fragmented, which complicates downstream analyses. For instance the genome assemblies of recognized hybrids such as Dekkera bruxellensis LAMAP2480 (AZMW01) or Wickerhamomyces anomalus NRRL Y-366 (AEGI01), are highly fragmented (9167 contigs, and 3133 scaffolds, respectively) and larger (26.9 and 26.2 Mb, respectively) than those of closely related homozygous species or strains, i.e. D. bruxellensis AWRI1499 is 12.6 Mb in 324 contigs (AHIQ01) and Wickerhamomyces ciferrii is 15.9 Mb in 364 contigs (Supplementary Table S2). In the framework of the sequencing project of a hybrid strain from the emerging pathogen Candida orthopsilosis MCO456 (6), we obtained similar low quality initial assemblies. To solve this we devised an ad-hoc strategy to recognize and selectively remove one of the two haploid contigs from heterozygous regions. Subsequent scaffolding and gap closing steps yielded a high quality genome reducing from 5577 to 116 contigs and 14.4 Mb size to 13.2 Mb. Here, we describe the procedure in more detail and present a programmatic environment to facilitate its application. Although we have chosen a specific set of available tools to perform each step of the pipeline, it must be noted that the strategy itself is flexible and modular and can be implemented on top of different tools. In brief (see Materials and Methods for more details), our pipeline is similar to the standard de novo assembly methodology (Figure 1A): overlapping reads are assembled into contigs (a), contigs are subsequently joined into supercontigs using information from paired-end reads (d) and finally the remaining gaps are closed again utilizing paired-end reads (e). We recognised that the standard de novo assembly tools fail at the scaffolding step, as in the case of heterozygous genomes there are multiple redundant contigs that could be connected to any of the homozygous neighbours (see b in Figure 1B).
Our pipeline proceeds in three distinct steps: reduction, scaffolding and gap closing (Figure 1B and C). Firstly, the draft assembly is simplified by removing heterozygous contigs (c). These redundant contigs represent distinct haplotypes from polymorphic chromosomal regions. Such heterozygous contigs hinder the scaffolding step. To circumvent this, in our pipeline, the clusters of redundant contigs are recognised and only the longest contig from each cluster is kept. Such reduction of complexity allows for further scaffolding. This is conducted by our in-house solution, fasta2homozygous.py v1.0. We have implemented two similarity search algorithms for the reduction step: faster, but less sensitive BLAT (13); and slower but highly sensitive LAST (14). The similarity algorithm is chosen based on the identity stringency defined by the user. Such design guarantees very good performance for identification of less diverged heterozygous contigs and high sensitivity for contigs from more remote heterozygous regions.
In the second step, non-redundant contigs are joined using SSPACE3 (15) (d). We decided to replace the default SSPACE aligner, bowtie (21), with BWA MEM (16), because the latter is faster, more accurate and more flexible toward longer reads and various sequencing technologies. Finally, the gaps in the scaffolds are closed using GapCloser (1). Noteworthy, scaffolding and gap closing are iteratively repeated in order to improve scaffolding with another sequencing library or reduce the number of gaps. In order to improve the performance of redundans, only a subset of each sequencing library is processed, as, generally, the genome sequencing projects provide much larger depth of coverage that is needed for scaffolding and gap closing. We next assessed the accuracy of our pipeline by assembling simulated and naturally-occurring heterozygous genomes.
Performance on simulated heterozygous fungal genomes
The underlying difficulty of heterozygous genome reconstruction is the lack of a ‘golden’ reference that would allow the identification of possible pitfalls of the genome reconstruction process. To circumvent this, we simulated six diploid genomes in which the two haploid sequences had 5% sequence divergence and which presented varying levels of loss of heterozygosity (LOH). LOH is a recombination event that renders homozygous regions with the sequence of only one of the two haplotypes (22). Additionally, we simulated fifteen diploid genomes with increasing divergence between two haploid sequences. Subsequently, we simulated short reads from these genomes, which included typical Illumina-related errors (see Materials and Methods). Then we assembled these genomes from the simulated short reads with either a standard pipeline and the redundans pipeline. These were performed for a small fungal genome (13 Mb) and a medium-size plant genome (119 Mb).
Loss of heterozygosity
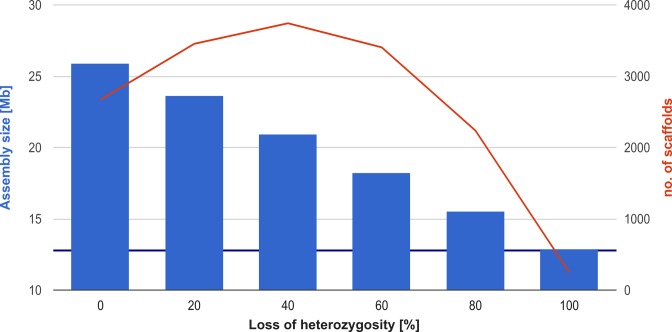
As expected, standard de novo assembly approaches (SPAdes and SOAPdenovo) obtained very fragmented genome assemblies (2237–3743 scaffolds) with increased size (119–198% of the original genome) for five heterozygous fungal genomes (Figure 2). In contrast, a fully homozygous genome (i.e. LOH of 100%) was recovered in 250 scaffolds with roughly the expected assembly size (99% of the original assembly). Interestingly, the size of the genome assembly was negatively correlated to the amount of LOH (Pearson r = −0.9939) (Supplementary Table S2). We next applied the redundans pipeline to the contigs (Supplementary Table S3) and scaffolds (Supplementary Table S4) reconstructed by SPAdes from the simulated reads. Firstly, we removed heterozygous contigs from all assemblies (reduction). The resulting non-redundant assemblies based on SPAdes contigs and scaffolds were very close to the expected size (99–104%). The non-redundant assemblies based on contigs and scaffolds from SOAPdenovo2 were slightly larger than expected (99–125%), suggesting SOAPdenovo2 may report wrongly resolved or not overlapping heterozygous contigs/scaffolds that cannot be recognised as heterozygous by our program. Because of this observation, we decided to use SPAdes assemblies for further analysis. Subsequently, the non-redundant contigs/scaffolds were further scaffolded using paired-end (two iterations) and mate-pairs (three iterations) reads. While a single pass of heterozygous reduction provides sufficiently contiguous assemblies, additional iterations of scaffolding further decreased the fragmentation of the assembly (Supplementary Table S3). Finally, the gaps were closed. Redundas was able to decrease the fragmentation of the assemblies from several thousands of contigs to <110.
Figure 2.
Heterozygous genome assemblies characteristics. The total SOAPdenovo2 assembly size (blue bars) as well as number of scaffolds longer than 1 kb (red plot) are given for reconstructed genome assemblies: one homozygous (LOH of 100%) and five heterozygous (with 5% divergence between haplomes and varying loss of heterozygosity level: 0%, 20%, 40%, 60%, 80%). Expected genome size is marked with purple baseline. The assemblies recovered for heterozygous genomes are much more fragmented (2237–3743 scaffolds) than those recovered for homozygous genome (250 scaffolds). The assembly reconstructed for homozygous genome (100% LOH) has expected size, while the remaining assemblies have size larger than expected (119.20–198.85% of expected size). The size of the genome assembly is negatively correlated with LOH level (Pearson coefficient of -0.9996).
Interestingly, the final redundans assemblies which started from contigs were less fragmented (17–40 contigs) and more similar in size to the target simulated genome (99–101%, except 0% LOH) than those which started from scaffolds (25–225 contigs and 101%-104% of the expected size). Such an observation suggests, that de novo assemblers may produce some wrongly resolved scaffolds for the heterozygous genomes, as every homozygous region (usually recovered as single consensus contig) can be joined ambiguously with two or more polymorphic neighbour regions on both ends (see b in Figure 1B). Unexpectedly, the redundans pipeline that started from thousands of contigs/scaffolds, returned full size chromosomes in nearly all reconstructions (Supplementary Figure S1). In the case of a simulated genome with 0% LOH (heterozygous over the entire length), the full size chromosomes were reconstructed for three reference chromosomes (including the longest 3 Mb chromosome), while the remaining five were reconstructed in two scaffolds. This was true for the reconstructions starting from both, contigs and scaffolds. The simulated genome with 20% LOH was reconstructed with six full or nearly full size chromosomes and the remaining two were represented in 2–4 scaffolds. On the other hand, the assemblies reconstructed for simulated genomes with a higher level of LOH had fewer full size chromosomes (1,2), if any.
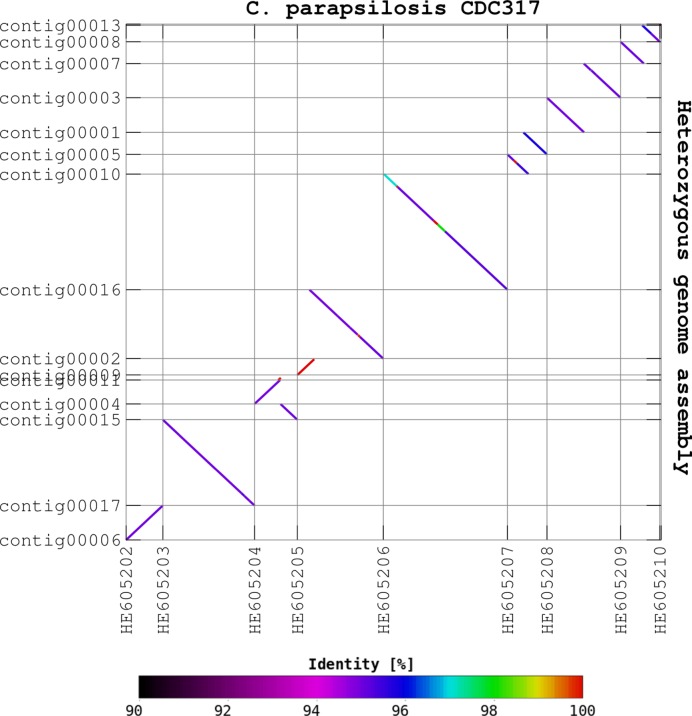
In order to evaluate the correctness of each assembly, we aligned the obtained contigs/scaffolds onto C. parapsilosis CDC317 chromosomes, as this genome was used to simulate the heterozygous genomes and short reads (Figure 3). Notably, four assemblies for the most heterozygous simulated genomes, these with LOH of 0% and 20%, were resolved correctly (Supplementary Figure S1). The remaining eight assemblies with larger LOH levels were carrying between 1 to 4 translocations as compared to the reference (Supplementary Figure S1). No large inversions and deletions were observed. We were interested to know whether the above mentioned translocations were present in the original SPAdes contigs/scaffolds or rather they were introduced during the scaffolding process. To test this we aligned the SPAdes contigs and scaffolds onto the C. parapsilosis CDC317 chromosomes (Supplementary Figure S2). As standard de novo assemblies were highly fragmented, it was difficult to trace large rearrangements. For this reason, we assumed that most of the observed translocations were introduced during the scaffolding step in our pipeline. Nevertheless, two deletions or translocations are present in the scaffolds from the assembly with 100% LOH, suggesting that at least some of the observed incongruencies may be attributed to errors in the de novo contigs or scaffolds from SPAdes.
Figure 3.
The assembly of simulated heterozygous genome. Pairwise genome alignment of the final assembly for simulated heterozygous genome with 0% LOH and its reference, C. parapsilosis CDC317. Synteny blocks have been coloured accordingly to the identity level between pair of query and target sequences. The assembled genome represent a mixture of two haplomes: 5% diverged (blue or violet) and identical (red) to reference genome. In addition, two short regions with divergence of 2–3% (green and cyan) are present in HE605206. The regions with intermediate divergence were likely assembled from very short contigs from both haplomes.
Sequence divergence
Similar results were obtained from fifteen simulated diploid genomes with 40% LOH and varying level of sequence divergence. Standard de novo assemblers produced assemblies proportionally larger to the level of heterozygosity of the genome under investigation for the entire spectrum of simulated divergences between haploid sequences (Supplementary Table S5). Interestingly, heterozygous genomes with sequence divergence close to expected sequencing error (1%, 3%) produced more fragmented assemblies (98 658 and 54 664 contigs, respectively), than genomes with higher divergence between haploid sequences. Redundans reduced correctly heterozygous regions with up to 45% divergence between haploid genomes. Higher fragmentation and slightly larger assembly was obtained for the genome with 1% divergence. We suspect that extremely high fragmentation of the initial assembly hindered reduction and scaffolding in this case (Supplementary Table S5). Although, our pipeline returns superior assemblies compared to standard de novo assemblers, we need to stress that the resulting scaffolds are chimeric, representing a mixture of both haplotypes (Figure 3).
Performance on simulated heterozygous plant genome
Arabidopsis thaliana is a model plant having a compact genome (119 Mb) that contains 28.9 Mb (24.3%) of repetitive sequences (23). One quarter of repetitive sequence space has identity above 95%, another quarter has identity between 85% and 95%, while the remaining half has identity below 85%. These make A. thaliana genome a very good model for testing the usability of bioinformatics tools in plant genomics. To make this model even more challenging, we have simulated heterozygous genomes based on A. thaliana chromosomes by introducing various level of divergence (1–20%) and LOH (0–100%).
As expected, the initial assemblies, recovered using SPAdes and Platanus, were highly fragmented and much larger than expected (Supplementary Table S7). Similarly to what we observed in the fungal dataset, redundans correctly recognised and removed heterozygous contigs from the heterozygous plant genome. Here, it is important to note, that during the reduction step redundans removes heterozygous contigs having identity over 51% and overlap over 66% by default. Because literally all A. thaliana repeats have identity above 60%, it may be expected that they will be removed from the initial assembly resulting in a final assembly that is reduced in excess. Although the reduced assemblies had smaller sizes than expected (Supplementary Table S9), subsequent scaffolding and gap closing resulted in very good assemblies, with a total size very close to expected (92–102%), organised in 127–1471 scaffolds and having N50 and N90 of 811–2910 and 52–612 kb, respectively (Supplementary Table S7). Interestingly, when scaffolds returned by Platanus were used with redundans, the final assemblies were even better; 97–864 scaffolds were recovered with total size very close (98–101%) to expected and having N50 and N90 of 1796–13 602 and 163–4371 kb, respectively (Supplementary Table S7).
Performance on real data sets
As mentioned above, we had earlier applied our pipeline to the C. orthopsilosis MCO456 genome, an intraspecies hybrid with 4.5% divergence between parental genomes and over 80% LOH (6). To test the generality of our approach we here applied our pipeline to improve the assemblies of the highly heterozygous genomes of another C. orthopsilosis heterozygous strain AY2 (NCBI Assembly ID: AMDC01), D. bruxellensis LAMAP2480 (AZMW01) and W. anomalus (AEGI01). In the case of Candida orthopsilosis AY2 with a 14.5 Mb genome assembled in 4152 contigs represents a heterozygous genome with a high level of LOH. 1293 contigs representing 1.7 Mb were found to be heterozygous in this genome. Again, the haploid assembly of AY2 (12.6 Mb in 255 contigs) is very similar in size to the genome of the highly homozygous strain of the same species, C. orthopsilosis 90–125 (12.6 Mb in eight chromosomes) (Supplementary Table S1). In the case of W. anomalus, the first, standard assembly contained 26.2 Mb of sequence in 3133 scaffolds. This represents a heterozygous genome with low level of LOH. Nearly half of this assembly (11 Mb in 1247 scaffolds) was redundant with an average divergence of 8.6%, suggesting that it is a heterozygous genome with approximately 15% LOH (Supplementary Table S1). Similar numbers were obtained for the second version of the W. anomalus assembly (AEGI02). Our pipeline identified and removed 11 Mb of sequence in redundant contigs (Supplementary Table S6). The remaining 2801 non-redundant contigs were further scaffolded using publicly available paired-end (SRR072086, SRR072088) and mate-pair (SRR073582, SRR073583, SRR073584) libraries. Noteworthy, the mate-pair libraries were generated by Roche 454, but redundans was able to use these data transparently, showing the flexibility of our pipeline. We obtained a well resolved assembly (85 scaffolds), with 41 times larger N50 and 80 times larger N90 than in the original contigs (Supplementary Table S6). Interestingly, the haploid assembly of W. anomalus (15.1 Mb) obtained after reduction of heterozygous scaffolds is similar in size to the genome of the closely-related and homozygous Wickerhamomyces ciferrii (15.9 Mb) (24). Importantly, our improved W. anomalus assembly is less fragmented, than that of W. ciferii (Supplementary Tables S1 and S6, Supplementary Figure S4). Similarly, D. bruxellensis LAMAP2480 (AZMW01) assembly was improved from 26.9 Mb in 9167 contigs in initial assembly to 13.6 Mb in 146 contigs using just two mate-pair libraries (SRR1222155, SRR1222162).
Comparison with heterozygosity-aware tools
Recently, the developers of SPAdes implemented a mode (dipSPAdes) specialised in the assembly of polymorphic genomes. We ran dipSPAdes on our simulated datasets. The resulting assemblies were neither very fragmented nor larger than expected (Supplementary Table S2). Surprisingly, the most heterozygous genome (0% LOH) was the least fragmented with 161 contigs, while the least heterozygous genomes (80% and 100% of LOH) were the most fragmented with 282 and 281 contigs, respectively. Importantly, dipSPAdes produced genome assemblies from 8% to 12% smaller than expected for the genomes that are not 100% heterozygous (Supplementary Table S2). In addition, dipSPAdes failed to recognise heterozygous regions in simulated genomes with sequence divergence of 1% and above 10%, reporting only 79% or below 60% of the genome, respectively (Supplementary Table S5). Moreover, the assemblies returned by dipSPAdes are more fragmented than those from redundans. In line with this, dipSPAdes assembled only the fully heterozygous (0% LOH) simulated genome correctly, while the remaining genomes were fragmented and contained several incongruencies as compared to the reference (Supplementary Figure S3). This is an important obstacle as, due to the existence of recombination, in hybrid genomes the heterozygosity rarely reaches 100% (Supplementary Table S1). Importantly, although our pipeline is as fast as dipSPAdes (Supplementary Table S3), redundans returned more complete and less fragmented assemblies than dipSPAdes. Typical computation and memory requirements of our pipeline including complexity reduction, scaffolding and gap closing is just a fraction of those necessary for de novo assembly. Thus, contig assembly is the most time and memory consuming step.
Another recently published method, Platanus, deals with heterozygous genome assembly by simplifying the assembly graph structures not only during contig assembly, but also during scaffolding (10). Thus, at least conceptually, Platanus uses a similar approach to redundans. We have tested Platanus on our simulated datasets. While Platanus deals well over the full spectrum of loss of heterozygosity, it fails at divergences above 10% (Supplementary Table S2 and S5). Moreover, the assemblies returned by Platanus are more fragmented than those produced by redundans. Support for polymorphic genomes have been also implemented in ALLPATHS-LG, as so-called 'haploidify' mode. We have tested this mode on the simulated dataset with 40% LOH of heterozygosity and 5% of divergence. ALLPATHS-LG requires an additional overlapping paired-end library in order to run, so we needed to simulate such library (2 × 100 bp with 150 bp insert size). ALLPATHS-LG produced an assembly with the size close to expected (13.0 Mb), but fragmented (401 scaffolds) (Supplementary Table S2). Importantly, ALLPATHS-LG assembly process took over 6 h using 32 cores (over five CPU days) and over 111GB of RAM. Moreover, nearly 102 GB of output files were created. In contrast, redundans dealt with the same data in less than two hours using eight cores and less than 16 GB of RAM (including contigs assembly). Importantly, the assembly obtained with our pipeline was less fragmented (57 scaffolds) than the one from ALLPATHS-LG. Such high computational demands prevented us from evaluating ALLPATHS-LG on the entire simulated dataset.
The advantages of using redundans are even more clear on simulated heterozygous plant genomes. SPAdes returned very fragmented assemblies (7261–45 769 scaffolds) and much larger than expected (84–191% of expected size). In contrast, dipSPAdes assemblies were less fragmented (978–4305 scaffolds), while being severely smaller than expected (58–82% of expected size). Assemblies produced by Platanus were fragmented (1225–29 856 scaffolds), but very close to the expected size in genomes with divergence below 15% (97–105%). Redundans returned 127–1471 scaffolds, but smaller than expected (92–102%) when SPAdes contigs were used as input. Interestingly, even less fragmented assemblies (97–864 scaffolds) and with more accurate size (98–101%) were produced by redundans when Platanus scaffolds were used as input (Table 1).
Table 1. Genome and assembly statistics.
| No. of scaffolds | N50 | N90 | Ns | Longest scaffold | Size [%] | ||
|---|---|---|---|---|---|---|---|
| C. parapsilosis | ref | 9 | 2 091 826 | 957 321 | 0 | 3 023 470 | - |
| SPAdes | min | 188 | 2607 | 111 | 0 | 35 992 | 102.61 |
| max | 3861 | 183 185 | 38 399 | 469 611 | 975 662 | 199.59 | |
| dipSPAdes | min | 47 | 140 841 | 10 876 | 0 | 369 644 | 44.06 |
| max | 201 | 443 228 | 168 647 | 0 | 1 277 709 | 100.67 | |
| Platanus | min | 97 | 8525 | 2240 | 7816 | 62 024 | 99.30 |
| max | 3503 | 314 901 | 105 072 | 115 364 | 1 233 366 | 156.29 | |
| Redundansa | min | 16 | 294 087 | 88 621 | 277 | 870 098 | 99.58 |
| max | 109 | 1 880 160 | 557 645 | 73 286 | 3 104 871 | 106.17 | |
| Redundansb | min | 13 | 432 772 | 179 116 | 3053 | 1 026 161 | 99.39 |
| max | 49 | 1 599 252 | 651 286 | 38 113 | 3 144 038 | 100.35 | |
| A. thaliana | ref | 5 | 23 459 830 | 18 585 056 | 185 738 | 30 427 671 | – |
| SPAdes | min | 7261 | 1242 | 111 | 0 | 14 802 | 84.00 |
| max | 45 769 | 91 641 | 6562 | 0 | 786 328 | 190.87 | |
| dipSPAdes | min | 978 | 38 818 | 4393 | 0 | 378 250 | 57.99 |
| max | 4305 | 217 604 | 41 794 | 0 | 1 120 742 | 81.89 | |
| Platanus | min | 1225 | 9078 | 2304 | 300 250 | 89 082 | 96.80 |
| max | 29 856 | 330 677 | 63 136 | 1 907 392 | 2 741 383 | 152.32 | |
| Redundansa | min | 127 | 810 991 | 51 721 | 191 751 | 2 162 015 | 92.30 |
| max | 1471 | 2 909 961 | 612 329 | 1 830 833 | 9 950 963 | 101.68 | |
| Redundansb | min | 97 | 1 796 239 | 163 909 | 151 424 | 7 206 143 | 97.67 |
| max | 864 | 13 602 130 | 4 371 075 | 1 162 308 | 28 354 754 | 101.29 |
All genomes used in this study are listed. For respective reference genomes, we provide it's accession together with number of scaffolds/chromosomes, N50 and N90, cumulative size of gaps and the length of the longest scaffold / chromosome. For assemblies produced in this study based on simulated heterozygous genomes with various level of divergence between heterozygous regions (1–40%) and level of loss of heterozygosity (0–100%), we provide minimum and maximum value for each of these metrics obtained. In addition, minimum and maximum percentage of expected assembly size is given.
aRedundans reconstruction started with SPAdes contigs.
bRedundans reconstruction started with Platanus scaffolds.
Comparison with heterozygosity reducing tools
The first step of redundans pipeline consists of heterozygous contigs reduction. To the best of our knowledge, there are no tools developed exactly for that purpose. Nevertheless, the tools that separate haplotypes from the assembly, i.e. Haplomerger (17), could be used for heterozygous contigs reduction. Thus we compared our assembly reducing implementation (fasta2homozygous.py) with Haplomerger. In brief, our solution is faster and more robust toward wide range of divergence and LOH (Supplementary Tables S8 and S9). Multiple iterations of Haplomerger (typically 2–3) are needed in order to remove heterozygous contigs. Haplomerger has problems recognizing heterozygous contigs in assemblies with 40–60% LOH and with low (1%) or high (above 20%) divergence between heterozygous regions. Notably, Haplomerger crashed on nine simulated heterozygous plant genomes, while in the three remaining cases only part of heterozygous contigs were correctly recognised and removed (Supplementary Table S9). In addition, Haplomerger is limited to diploid genomes, while our method could be used with polyploid genomes. These findings put into question the applicability of Haplomerger on larger genomes. Here, we need to stress, however, that our methodology performs only the reduction of the assembly, while Haplomerger reports contigs representing alternative haplotypes.
DISCUSSION
We have introduced redundans, a pipeline that improves the genome assembly of heterozygous genomes. Redundans is fast, lightweight, modular, and flexible toward many sequencing technologies and library types. We show that our approach reduces the heterozygous regions with substantial divergence from the genomes under various levels of loss of heterozygosity and sequence divergence in both, simulated and real data sets. Moreover, we showed that such reduced assembly can be further scaffolded with success, resulting in full size chromosomes if mate-pair libraries are available. Redundans is superior to existing tools, resolving complete and correct assemblies by using fewer resources. Redundans is equipped with two similarity search algorithms: very fast, but less sensitive (BLAT) and slower, but very sensitive (LAST). Such design guarantees high performance in identification of highly similar heterozygous contigs and high sensitivity for identification of more remote heterozygous contigs. To provide further performance boost, the LAST alignment step is performed in multiple threads.
We need to emphasize, however, that the resulting assembly does not represent individual haploid genomes, but it is a mosaic of segments from each of the haploid genomes present in a polyploid organism (i.e. haplome). Thus, one of many haplomes is randomly chosen to fill a given heterozygous region. This is a common feature of all heterozygosity-aware methods. In order to identify individual haplomes, sequencing reads need to be realigned onto the genome assembly and re-analyzed. Although, such an assembly is somewhat chimeric, in the same way as genomes derived from several individuals as the reference human genome (25), it simplifies downstream analysis. In contrast, the typical heterozygous genome assembly is a mixture of consensus and haploid-contigs, which misleads subsequent analyses. Emerging new technologies that produce longer reads will certainly help in fully resolving the haplotype structure of heterozygous genomes. Summarizing, redundans simplifies genome assembly of polymorphic genomes, and we show that in the tested fungal genomes it is superior to other existing methods both in terms of performance and sensitivity. Our tool has been designed with heterozygous diploid genomes in mind. In principle, however, it could be applied to polyploid genomes. Redundans is freely available at https://github.com/Gabaldonlab/redundans.
Supplementary Material
Acknowledgments
We would like to thank for fruitful discussion to Darek Kędra, Marina Marcet-Houben, Ken Wolfe and the entire community of BioStars forum (www.biostars.org). Finally, we would like to acknowledge the beta testers for their engagement and useful feedback.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness grants, ‘Centro de Excelencia Severo Ochoa [2013–2017’ SEV-2012-0208, BFU2015-67107 to TG group] cofounded by European Regional Development Fund (ERDF); European Union and ERC Seventh Framework Programme [FP7/2007-2013] under grant agreements [FP7-PEOPLE-2013-ITN-606786 and ERC-2012-StG-310325]; European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie [H2020-MSCA-ITN-2014-642095]; La Caixa-CRG International Fellowship Program (to L.P.P.). Funding for open access charge: ERC Seventh Framework Programme [FP7/2007-2013] under grant agreements [FP7-PEOPLE-2013-ITN-606786 and ERC-2012-StG-310325]; European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie [H2020-MSCA-ITN-2014-642095]; Spanish Ministry of Economy and Competitiveness grants, ‘Centro de Excelencia Severo Ochoa [2013–2017’ SEV-2012-0208, BFU2015-67107] cofounded by ERDF; Catalan Research Agency (AGAUR) [SGR857].
Conflict of interest statement. None declared.
REFERENCES
- 1.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J.M., Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zerbino D.R., McEwen G.K., Margulies E.H., Birney E. Pebble and rock band: heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler. PLoS One. 2009;4:e8407. doi: 10.1371/journal.pone.0008407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weisenfeld N.I., Yin S., Sharpe T., Lau B., Hegarty R., Holmes L., Sogoloff B., Tabbaa D., Williams L., Russ C., et al. Comprehensive variation discovery in single human genomes. Nat. Genet. 2014;46:1350–1355. doi: 10.1038/ng.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pryszcz L.P., Németh T., Gácser A., Gabaldón T. Genome comparison of Candida orthopsilosis clinical strains reveals the existence of hybrids between two distinct subspecies. Genome Biol. Evol. 2014;6:1069–1078. doi: 10.1093/gbe/evu082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Small K.S., Brudno M., Hill M.M., Sidow A. A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biol. 2007;8:R41. doi: 10.1186/gb-2007-8-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pryszcz L.P., Németh T., Saus E., Ksiezopolska E., Hegedűsová E., Nosek J., Wolfe K.H., Gacser A., Gabaldón T. The genomic aftermath of hybridization in the opportunistic pathogen Candida metapsilosis. PLoS Genet. 2015;11:e1005626. doi: 10.1371/journal.pgen.1005626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McElroy K.E., Luciani F., Thomas T. GemSIM: general, error-model based simulator of next-generation sequencing data. BMC Genomics. 2012;13:74. doi: 10.1186/1471-2164-13-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., Okuno M., Yabana M., Harada M., Nagayasu E., Maruyama H., et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safonova Y., Bankevich A., Pevzner P.A. dipSPAdes?: Assembler for Highly Polymorphic Diploid Genomes. Res. Comput. Mol. Biol. 2014;8394:265–279. doi: 10.1089/cmb.2014.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnerre S., Maccallum I., Przybylski D., Ribeiro F.J., Burton J.N., Walker B.J., Sharpe T., Hall G., Shea T.P., Sykes S., et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frith M.C., Hamada M., Horton P. Parameters for accurate genome alignment. BMC Bioinformatics. 2010;11:80. doi: 10.1186/1471-2105-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boetzer M., Henkel C. V., Jansen H.J., Butler D., Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 16.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang S., Chen Z., Huang G., Yu T., Yang P., Li J., Fu Y., Yuan S., Chen S., Xu A. HaploMerger: reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res. 2012;22:1581–1588. doi: 10.1101/gr.133652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradnam K.R., Fass J.N., Alexandrov A., Baranay P., Bechner M., Birol I., Boisvert S., Chapman J. a, Chapuis G., Chikhi R., et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2013;2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales L., Dujon B. Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiol. Mol. Biol. Rev. 2012;76:721–739. doi: 10.1128/MMBR.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett R.J., Forche A., Berman J. Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb. Perspect. Med. 2014;4:a019604. doi: 10.1101/cshperspect.a019604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maumus F., Quesneville H. Ancestral repeats have shaped epigenome and genome composition for millions of years in Arabidopsis thaliana. Nat. Commun. 2014;5:4104. doi: 10.1038/ncomms5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider J., Andrea H., Blom J., Jaenicke S., Rückert C., Schorsch C., Szczepanowski R., Farwick M., Goesmann A., Pühler A., et al. Draft genome sequence of Wickerhamomyces ciferrii NRRL Y-1031 F-60-10. Eukaryot. Cell. 2012;11:1582–1583. doi: 10.1128/EC.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.