Abstract
Regulatory variation in gene expression can be described by cis- and trans-genetic components. Here we used RNA-seq data from a population panel of Drosophila melanogaster test crosses to compare allelic imbalance (AI) in female head tissue between mated and virgin flies, an environmental change known to affect transcription. Indeed, 3048 exons (1610 genes) are differentially expressed in this study. A Bayesian model for AI, with an intersection test, controls type I error. There are ∼200 genes with AI exclusively in mated or virgin flies, indicating an environmental component of expression regulation. On average 34% of genes within a cross and 54% of all genes show evidence for genetic regulation of transcription. Nearly all differentially regulated genes are affected in cis, with an average of 63% of expression variation explained by the cis-effects. Trans-effects explain 8% of the variance in AI on average and the interaction between cis and trans explains an average of 11% of the total variance in AI. In both environments cis- and trans-effects are compensatory in their overall effect, with a negative association between cis- and trans-effects in 85% of the exons examined. We hypothesize that the gene expression level perturbed by cis-regulatory mutations is compensated through trans-regulatory mechanisms, e.g., trans and cis by trans-factors buffering cis-mutations. In addition, when AI is detected in both environments, cis-mated, cis-virgin, and trans-mated–trans-virgin estimates are highly concordant with 99% of all exons positively correlated with a median correlation of 0.83 for cis and 0.95 for trans. We conclude that the gene regulatory networks (GRNs) are robust and that trans-buffering explains robustness.
Keywords: transcription, genetic variation, cis- or trans-effects, allele-specific expression, Drosophila, regulatory mutations, purifying selection
DIFFERENCES in the level of expression between two alleles are widespread and play an important role in complex traits. They have been implicated in human disease phenotypes such as cancer and are an important component of variation in Drosophila, as well as yeast, plants, animals, and humans (Mendell and Dietz 2001; Brem et al. 2002; Cowles et al. 2002; Yan et al. 2002; Wittkopp et al. 2004, 2006, 2008b; Springer and Stupar 2007; Hutter et al. 2008; Smith and Kruglyak 2008; Tirosh et al. 2009; Emerson et al. 2010; McManus et al. 2010; Crowley et al. 2015). In Drosophila, variation in gene expression is heritable (Wayne et al. 2004, 2007) and evidence for both cis- and trans-regulatory polymorphisms is widespread (Wittkopp et al. 2004, 2008b; Hughes et al. 2006; Genissel et al. 2008; Wang et al. 2008; Graze et al. 2009, 2012, 2014) and evidence for cis- by trans-interactions has been reported (Wittkopp et al. 2004, 2008b; Wang et al. 2008; Graze et al. 2014). It has been argued that these interactions may also present evidence for coevolution of cis- and trans-regulation between species (Wittkopp et al. 2004).
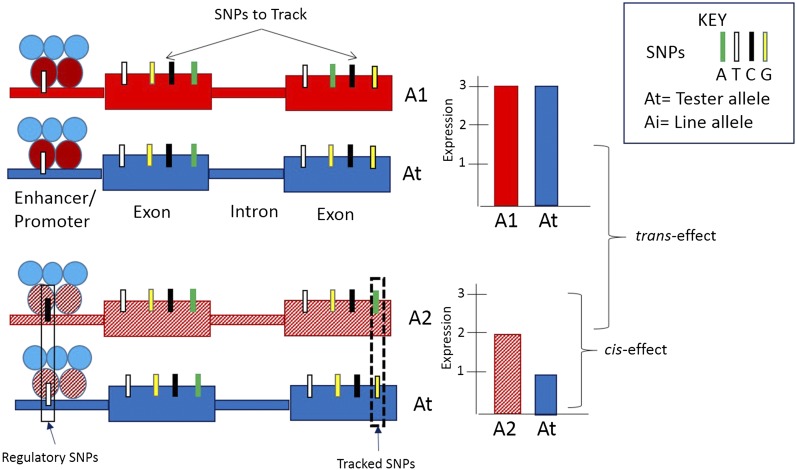
A variety of genetic designs have been used to identify cis- and trans-effects, including chromosome substitution (Hughes et al. 2006; Lemos et al. 2008; Wang et al. 2008; Wittkopp et al. 2008a; Graze et al. 2014), expression QTL (eQTL) (reviewed by Brem et al. 2002; Kirst et al. 2005; Genissel et al. 2008; Gilad et al. 2008; Mackay et al. 2009; King et al. 2012, 2014; Massouras et al. 2012), and allelic imbalance (reviewed by Yan et al. 2002; Lo et al. 2003; Wittkopp et al. 2004, 2008a,b; Ronald et al. 2005; Guo et al. 2008a; Graze et al. 2009, 2012; Tirosh et al. 2009; Zhang and Borevitz 2009; McManus et al. 2010; Pastinen 2010). While eQTL studies often refer to cis- and trans-effects, eQTL cis-effects are local effects and trans-effects are distal effects (Rockman and Kruglyak 2006). In contrast, the cis- and trans-effects in allelic imbalance (AI) studies refer to molecular effects in the regulatory region of the locus itself (cis) or variation in genes molecularly interacting with common regulatory elements (trans) (Figure 1) (Yan et al. 2002; Wittkopp et al. 2004; Graze et al. 2009, 2012; Wittkopp and Kalay 2012).
Figure 1.
Cartoon of cis- and trans-regulation. A1 and A2 are alleles from two different testcrosses of a parental line (A1, red; A2, cross-hatched red) with a tester allele (At, blue). Alleles are tracked by SNPs within the coding sequence. Note that these SNPs are not necessarily causal. For testcross 1, there is no allelic imbalance; allele-specific expression is equal between the A1 allele and the At allele. For testcross 2, the A2 allele is different from the At allele, indicating cis-regulation. In this cartoon, the expression of the exon is regulated by a regulatory SNP within the gene’s promoter. Trans-effects are identified by comparing the expression of the At allele in testcross 1 to the expression of the At allele in testcross 2.
Previous studies of AI in Drosophila examining interspecies hybrids (Michalak and Noor 2003, 2004; Ranz et al. 2004; Wittkopp et al. 2004; Ortiz-Barrientos et al. 2007; Graze et al. 2009; McManus et al. 2010, 2014; Graze et al. 2012) identified cis-effects in 20–90% of genes (Wittkopp et al. 2004; Graze et al. 2009, 2012; McManus et al. 2014; Meiklejohn et al. 2014). Most intraspecific studies have used chromosomal substitutions and these studies have found contributions of trans-effects ranging from 0.01% (McManus et al. 2014) to 40–50% of genes (Wittkopp et al. 2004; Meiklejohn et al. 2014) but sometimes reaching 90% of genes (Wang et al. 2008).
In a single hybrid, cis-regulatory variation is identified by comparing gene expression of two alleles at the same loci within that hybrid genotype (reviewed by Pastinen 2010) while trans-estimates are derived from comparing the same allele between hybrid genotypes (Figure 1). For a single hybrid, the estimates of cis are confounded with cis- by trans-interactions, and interactions are usually assumed to be negligible relative to the main effect of cis. When multiple genetically variable lines are crossed to a common tester line, cis- and trans-effects for each allele in the population can be estimated relative to the effects of the common tester allele (Nuzhdin et al. 2012). By modeling AI as a function of cis and trans and their interaction in the population, the contribution of these effects to AI can be estimated.
Identifying genes where AI occurs is a critically important biological problem, yet successfully identifying AI is technically challenging. Whereas mapping to a common reference strain is convenient, it introduces bias in the assessment of AI due to the difference in polymorphisms between the alleles interrogated and the reference (Degner et al. 2009; Graze et al. 2012). Masking SNPs and extensive filtering may ameliorate some sources of potential bias (Stevenson et al. 2013; van de Geijn et al. 2014), but bias may come from multiple sources and not all of these can be effectively filtered (Degner et al. 2009; Leon-Novelo et al. 2014; van de Geijn et al. 2014). There is mounting evidence that using personal genomes is more effective at reducing bias, particularly from structural variants (Degner et al. 2009; Rozowsky et al. 2011; Graze et al. 2012; Yuan and Qin 2012; Munger et al. 2014; Zou et al. 2014). Further, previously unidentified structural variation can be identified and the bias corrected using appropriate DNA controls (reviewed by Wittkopp et al. 2004; Pastinen 2010; Graze et al. 2012) and it is impossible to detect and filter such bias without DNA controls. This is often ignored, yet small amounts of unaccounted for bias lead to gross inflation of type I error rates (Leon-Novelo et al. 2014). Bias correction remains an important, and mostly ignored, feature of testing for AI.
The extent to which regulation of expression is sensitive to the environment is an interesting and not fully resolved issue. A frequent lack of concordance between eQTL across environments or genetic backgrounds has been previously interpreted as strong evidence for differential gene regulation in Drosophila (Mackay et al. 2009; Massouras et al. 2012; Huang et al. 2015). However, eQTL are reproduced between homozygous and heterozygous flies (Massouras et al. 2012), and regulatory variation measured using allelic imbalance appeared robust to environmental perturbations in Arabidopsis (Cubillos et al. 2014). Here, we have assayed the transcriptome in virgin and mated flies to test whether AI variation responds to mating.
Following mating in Drosophila melanogaster the fly experiences changes in metabolism (McGraw et al. 2004, 2008; Dalton et al. 2010), immune defense (reviewed by Lawniczak and Begun 2004; Peng et al. 2005; Lawniczak et al. 2007; Morrow and Innocenti 2012), life span (Chapman et al. 1995), detoxification products (McGraw et al. 2004), and gene expression (Smith et al. 2013; Zhou et al. 2014). Metabolic changes are found throughout the whole body (McGraw et al. 2004, 2008) as well as specific changes in the female head fat body (Dalton et al. 2010), including effects on nutritional state postmating.
This study explores whether an allelic imbalance in an interspecific population of D. melanogaster differs by environment and whether regulatory variation equally involves cis- and trans-effects. Consistent with the literature, this study finds 54% of all genes have evidence for AI. Coevolutionary processes between species result in cis- by trans-interactions, which have been clearly demonstrated in melanogaster/simulans hybrids (Wittkopp et al. 2004, 2008b; Landry et al. 2005; Graze et al. 2009, 2012; Fontanillas et al. 2010; Stevenson et al. 2013; McManus et al. 2014). However, coevolution is not a plausible explanation within species. We propose an alternative hypothesis for apparent cis–trans compensation, where the gene expression level perturbed by cis-regulatory mutations is partially compensated through trans-regulatory mechanisms. This is supported by recent experimental evidence of widespread molecular interactions as a result of cryptic genetic variation (Paaby and Rockman 2014).
Methods
All original data are attached to this article in File S1 and described in the Supplemental Material. All programs used for analysis, plus documentation, are at http://github.com/McIntyre-Lab/papers/tree/master/fear_ase_2016.
Sequencing data
RNA-seq data from a panel of 68 D. melanogaster F1 hybrids (Kurmangaliyev et al. 2015) were used to measure allele-specific expression. Briefly, females from two populations of naturally derived D. melanogaster strains [54 from the Drosophila Genetic Reference Panel (DGRP) (Mackay et al. 2012) and 14 from Winters, California] were crossed to males of a single D. melanogaster laboratory strain (w1118). Kurmangaliyev et al. (2015) isolated RNA from pools of ∼50 adult female heads from the F1 hybrid progeny. At least 3 independent biological replicates of each cross were evaluated. Libraries were constructed using standard protocols and pooled at equal molar concentrations. They were sequenced in eight or more lanes of Illumina HiSeq 2000. FlyBase v5.51 was used and exons were identified from this build. Exons that overlapped within a gene were collapsed as in Graze et al. (2012).
Genome ambiguity: simulation 1 random SNPs
Genome ambiguity is a major source of mapping bias (Degner et al. 2009), but it can be estimated via simulation (Degner et al. 2009; Leon-Novelo et al. 2014). To identify regions of genome ambiguity we simulated data similar to the intraspecific hybrid population studied here. On average, parental genotypes have ∼160,000 exonic SNPs compared to the reference (FlyBase v5.51). Parental genotypes for 101 lines were simulated by randomly incorporating 160,000 SNPs into 63,181 exons of the D. melanogaster reference (FlyBase v5.51). All possible 95-bp reads were created from each reference, using a sliding window. An F1 panel (n = 100) was constructed by taking simulated reads from each of 100 simulated genotypes (linei where i = 1–100) and mixing them with a single common simulated genotype (linei: tester). Reads were aligned to each “parental” reference (line101: tester), using Bowtie (v0.12.9, -m1, -v3) (Langmead et al. 2009) and Last (v247, -l 25) (Frith et al. 2010). Genotype-specific alignments were compared as in Graze et al. (2012) and reads were categorized as aligning best to the “line” or the “tester’ or aligning equally well to “both.” For each exon, bias (q) was estimated by taking the proportion of reads that aligned to the tester over the sum of the allele-specific reads (linei + tester). Because the simulated F1 panel was created by equally mixing reads from parental strains, q is expected to be 0.5. When q deviates from 0.5, the exons must contain sequence ambiguities that affect the mappability of this region. A total of 55,647 exons showed no sequence ambiguity, while 7534 have some evidence of ambiguity (Table S1). Of these there were 1372 that were ambiguous in >50% of simulated lines, and 807 were ambiguous in all 100 simulated F1 hybrids (Figure S1A). We removed the 807 exons that were ambiguous in all lines from the remainder of analyses.
Genotype-specific references
Genotype-specific references have been used to reduce map bias (Degner et al. 2009; Rozowsky et al. 2011; Graze et al. 2012; Yuan and Qin 2012; Stevenson et al. 2013; Leon-Novelo et al. 2014; Munger et al. 2014) by incorporating a filtered set of SNPs and indels that were identified in each parental strain. The GATK UnifiedGenotyper tool (ver. 2.1-8) was used to identify SNPs and indels in 68 genotypes (DGRP and Winters; line) and the laboratory strain (w1118; tester). To minimize biases from miscalled variants (Leon-Novelo et al. 2014), SNPs and indels were strictly filtered as follows. All variants were considered as a line–tester pair, and if a variant was heterozygous in either the line or the tester, it was removed. The remaining SNP locations were masked in the genomic reference (FlyBase r5.51) for each line–tester pair (n = 68), by replacing the current nucleotide with an “N”. RNA-seq data from each F1 hybrid were aligned to the masked reference, using Bowtie (v0.12.9, -a, -v3) (Langmead et al. 2009) followed by Last (v247, -l 25) (Frith et al. 2010). A custom python script summarized masked alignment files by counting the number of RNA-seq reads that supported each variant call (SNP or indel). A variant (SNP or indel) was retained if it had at least one supporting RNA-seq read or if the DNA coverage for the original variant call was five or more. Filtered variants (File S1) were used to create a genotype-specific exome reference by updating the D. melanogaster genomic reference (FlyBase r5.51) with SNPs and indels and removing redundant exons.
Genotype-specific bias estimation: simulation 2 (qSIM)
Previous studies have used DNA read counts to estimate inherent bias and then corrected these biases by including a random effect parameter in their allelic imbalance models (Wittkopp et al. 2004; Graze et al. 2012; Leon-Novelo et al. 2014). Here we do not have DNA read counts; however, we use read simulation to identify exons that contain ambiguity and to create a bias correction parameter (q = qSIM; see Leon-Novelo et al. 2014). To mimic the information provided by the DNA, we simulated all possible 95-bp DNA reads from each parental genotype-specific exome reference (DGRP, Winters, and w1118) (see Methods: Genotype-specific references). Similar to genome ambiguity simulation, a hybrid was created by mixing reads from each line (n = 49) with reads from the common laboratory strain (w1118). Simulated hybrid reads were aligned separately to the line and tester genotype-specific references, using Bowtie (v0.12.9, -m1, -v3) (Langmead et al. 2009) and Last (v247, -l 25) (Frith et al. 2010). Genotype-specific alignments were compared as in Graze et al. (2012) and reads were categorized as aligning best to the line or the tester or aligning equally well to both. For each exon, the bias term (qSIM) was calculated by taking the proportion of reads that aligned to the tester over the sum of the allele-specific reads (linei + tester). After removing 807 exons that are always biased in genome ambiguity simulations (see Methods: Genome ambiguity: simulation 1 random SNPs ), 53,923 exons showed no bias (qSIM = 0.5), while 8451 regions showed bias in at least 1 line–tester combination (Figure S1B and Table S1). The bias with the biggest difference from 0.5 was used as the estimate for bias for each exon.
Mapping
Genotype-specific references were created by updating the genome, using variants identified by global alignments of these and all other data for D. melanogaster lines simultaneously (Table S2). Data for the 68 lines were mapped to each parent and w1118 separately to the line and tester genotype-specific references, using Bowtie (v0.12.9, -m1, -v3) (Langmead et al. 2009) and Last (v247, -l 25) (Frith et al. 2010). Reads in exons were counted and classified as aligning to the line, to the tester, or to both. Several filtering steps were performed. Exons that always showed bias in the genome ambiguity simulation were removed. Exons with very low coverage (average per nucleotide coverage <25) were removed (n = 31,791). Exons that were not present in at least 10% of genotypes were removed (n = 11,382). Exons that were not present in both environments (virgin, mated) were removed (n = 1253; 5391 remaining). After filtering exons, genotypes whose median value across all lines (for the ratio of tester-specific to total allele-specific read counts) was extreme (≤0.4 or ≥0.6) were removed (n = 9) as these likely reflect poor variant detection and systematic bias. Finally, genotypes with <500 remaining exons were removed (n = 13). A total of 49 genotypes and 5391 exons were analyzed for allelic imbalance (File S2).
An intersection test for AI
We use the Poisson Gamma (PG) model developed by Leon-Novelo et al. (2014). Briefly, under the null hypothesis of no allelic imbalance vs. the alternative of allelic imbalance Let and be the line and tester RNA allele-specific read counts in biological replicate
and
Here μ is the overall mean, is the variation of biological replicates (), α is the effect of a read having AI, and q is a constant to incorporate bias information, where values >0.5 indicate bias toward the tester allele and values <0.5 are bias toward the line allele. If θ is the real proportion of reads from the tester allele, then
When there is no AI (), therefore The bias correction parameter q can either be a fixed constant (e.g., 0.5) or be a random variable. The PG model requires that exons have at least three biological replicates for a given genotype × mating status and that the exon is expressed in at least one of these replicates (i.e., average per nucleotide coverage >0). For those exons where qSIM captures the potential bias this value is used in the Bayesian model. The simulated value of qSIM, when different from 0.5, has been shown to reflect bias from DNA controls and to be close to the value of bias estimated from DNA (Leon-Novelo et al. 2014). Note that the input to this model is raw read counts and that no additional normalization is performed.
The type I error rate for this test is <5% only when there is modest misspecification of bias; however, error rate increases as the percentage of misspecification increases (Leon-Novelo et al. 2014). For the commonly used binomial procedure in the test for AI type I error rates are large (Degner et al. 2009; Fontanillas et al. 2010; Nothnagel et al. 2011; Yuan and Qin 2012; Leon-Novelo et al. 2014; van de Geijn et al. 2014; Zou et al. 2014; Castel et al. 2015). Using DNA controls to estimate bias reveals that most bias (95%) is <20% (Graze et al. 2012). This translates to values of the parameter q between 0.4 and 0.6. Evaluating the posterior at three values of q, 0.4, 0.5, and 0.6, provides an estimate of the behavior of the posterior for the most likely set of conditions. Using an intersection test approach (Stell et al. 1980; Berger 1997; Berger and Boos 1999; Coffman et al. 2003), AI is declared if the credible interval excludes 0.5 for all three values of q.
The type I error behavior of the proposed intersection test was evaluated using simulation. Read counts for 10,000 genes were simulated using a Poisson distribution. The Poisson model was parameterized similarly to the above AI PG model: with mean read counts estimated from the population panel for three classes of coverage (low, medium, and high), the bias varied from 0.35 to 0.65 (19 values altogether), and all simulations were the null case (no AI). For each level of bias simulated, the simulated bias was used in the model and several deviations from the simulated bias, misspecification, were evaluated (1%, 2%, 3%, 5%, 10%, and 20%). For each combination of bias and misspecification (19 × 6) the posterior distribution of θ was estimated using the above AI PG model and intersection test (q = [0.4, 0.5, 0.6]). Note that the simulation is performed in relative bias amounts and not absolute. For example, 10% of 0.4 is 0.04 and 10% of 0.6 is 0.06. This was done for demonstration purposes, as the direction of the AI is arbitrary and this reveals more about the behavior of the bias.
Estimation of cis- and trans-effects
With the tester-cross design, allele-specific read counts can be used to separate cis- and trans-effects (Nuzhdin et al. 2012). For the sake of clarity in this article the reasoning in Nuzhdin et al. (2012) is reproduced here. In the testcross, the expression of the population allele i and the tester allele t in genotype i can be expressed as
whereis the population mean,is the cis-effects of allele i,is the trans-effects of the allele i, and is the trans-effects of the tester allele. Several constraints are necessary to solve this system. For simplicity assume and The expected difference between the alleles over the population is which is equivalent to The tester trans-effects are estimated by which is The cis-effects for each allele are estimated as The line trans-effects are estimated by This formulation assumes equal coverage across lines. To account for the varying number of reads and the varying specificity of each cross, allelic expression was scaled to the number of non-allele-specific reads in the same region. That is, Eii is estimated as the number of reads mapping uniquely to allele i divided by the number of reads mapping to the region but not assignable to allele i or the tester allele. Without scaling, the estimation is reflective only of the relative number of reads for that cross. In addition, this is a system of four unknowns and two equations, meaning there is not a unique solution for all four effects. The effects of Ct and Tt in this design are confounded with each other and will vary, depending on the exact estimation strategy for the population mean. However, the estimation of the effects Ci and Ti is not sensitive to the strategy for estimation of the population mean.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Allele-specific expression was compared between 49 testcrosses of females that were either mated or virgin. F1 hybrids were produced between a w1118 tester strain and genotypes from the DGRP and a Winters collection (Campo et al. 2013). We use a Bayesian test for AI that accounts for bias and controls type I errors (Figure S2). The ability to detect AI is related to coverage. Exons with allelic imbalance were underrepresented in genotypes with low levels of coverage (Table S2) of fewer than three reads per nucleotide. AI is concordant across the length of the transcript with very few (n = 82) exceptions (Table S3). These genes show AI in two or more exons in different directions, with at least one exon with more expression in the line and at least one other exon with more expression from the tester allele. The few differences across the gene may represent a frequency difference in isoform usage between alleles (Kurmangaliyev et al. 2015) and are consistent with data observed in other species (Skelly et al. 2011; Anders et al. 2012; Trapnell et al. 2013).
As with previous studies (Lawniczak and Begun 2004; McGraw et al. 2004, 2008; Mack et al. 2006; Kocher et al. 2008; Dalton et al. 2010; Smith et al. 2013) there is substantial difference in expression between virgin and mated flies. We observed 3048 exons from 1610 genes exhibiting differential expression at a false discovery rate of 0.05 (Benjamini and Hochberg 1995). In addition, 7 of the top 10 genes were reported previously in an analysis of gene expression in the heads of virgin and mated flies, which provides support for the present results (Dalton et al. 2010).
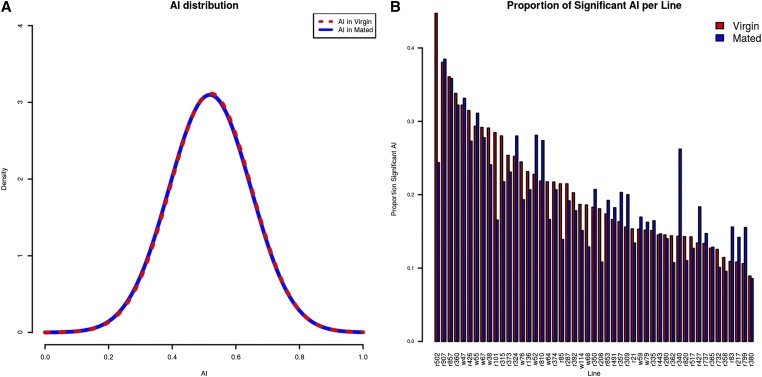
Of the 5391 exons analyzed, 4090 (76%) showed evidence of allelic imbalance in at least 1 cross. Fourteen regions had AI in >40 crosses (of 49 total), and 31 regions had AI in 100% of the crosses measured. The few that are different in all crosses likely result from regulatory mutations unique to the w1118 strain. There were 3004 exons that had AI in ≤5 crosses, and 570 exons showed AI in <5 crosses. The distribution of θ is centered on 0.5 for both mated and virgin environments (Figure 2A). An average of 19% of exons show evidence for AI in any one cross. The proportion of exons with significant AI ranged from 8% of exons tested to 39% (Figure 2B and Table S3). Of the exons with allelic imbalance, one-third are also differentially expressed between environments.
Figure 2.
Distribution of AI. (A) Distribution of for mated (blue) and virgin (red) environments for all data. The distribution is centered at 0.5, indicating that there is no overall bias toward the line or the tester allele. (B) The proportion (y-axis) of exons significant for AI, determined by the intersection test, for each line (x-axis) in mated (blue) and virgin (red) environments.
Environmental effects on allelic imbalance
We looked to see whether there were any differences in AI between mated and virgin flies. The exons that show differences in estimates of allelic imbalance between mated and virgin flies are listed in Table S4. There were 93 genes that were significant in AI in mated flies only. Several genes associated with reproduction were significant for AI in mated flies only, including Arpc1, CG10433, CG17919, fry, heph, Sec5, Cg25C, and Shark. Mating has been shown to reduce life span (Chapman et al. 1995; Flatt 2011; Landis et al. 2015) and several genes reported to affect life span were also significant for AI in mated flies only (α-Man-Ia, bmm, mt:ATPase6, sNPF, and sm). Postmating immunity is also compromised in female fruit flies (reviewed by Peng et al. 2005; Lawniczak et al. 2007; McGraw et al. 2008) and the genes CG10433, CG17652, CG17919, Gs1l, α-Man-Ia, mtd, Sec5, and Shark are involved in processes such as apoptosis, phagocytosis, and general immune responses. In addition, Cyp4ac1, bmm, Gs1l, and sNPF are associated with metabolism, which is also altered by mating in fruit flies (McGraw et al. 2004, 2008; Dalton et al. 2010; Zhou et al. 2014).
Interestingly, there were two genes that have been shown to be directly involved in regulatory regions of the genome that were significant for AI in mated flies. cha is involved in cholinergic neuronal regulation and cis-regulation in the brain (Yasuyama et al. 1995). cnc encodes the Drosophila homolog of the Nrf2 transcription factor that is directly linked to metabolic regulation, stress response (Misra et al. 2011; Sykiotis et al. 2011; Pickering et al. 2013), and life span (Obata and Miura 2015). cnc has a direct influence on the oocyte nucleus and gurken, signaling (Guichet et al. 2001) innate immunity and lipid metabolism (Karim et al. 2015), and its overexpression can restore movement in a Drosophila model of Parkinson’s disease (Barone et al. 2011).
In virgin flies, there were 99 genes that were significant for AI. Reproductive-related activities were also represented in virgins as they were in mated flies (Cdc42, Cys, dnc, fl(2)d, mei-P26, qm, and zip). In addition, Tim10, a negative regulator of innate immune response, was also significant in AI in virgin females only, suggesting that upon mating this gene is differentially regulated in virgin but not in mated flies.
There were several genes involved in regulatory regions that were significant for AI in virgin flies only. Act57B contributes to developmental processes and cytoskeleton dynamics but has also been shown to play a role in courtship conditioning (Winbush et al. 2012). Kank, a gene that affects muscle tendon development, has been shown to be differentially expressed downstream of dsx (Lebo et al. 2009), suggesting AI regulation in the sex determination pathway in virgins only. p24-1 has sex-specific expression (Boltz et al. 2007) and has been shown to play a role in the neural signaling involved in female egg-laying defects and reproductive behavior (Saleem et al. 2012). In addition, ζCOP is involved in lipid storage and regulation of lipid droplets (Guo et al. 2008b) consistent with regulatory changes in lipid metabolism after mating in the fruit fly.
Cis- and trans-effects
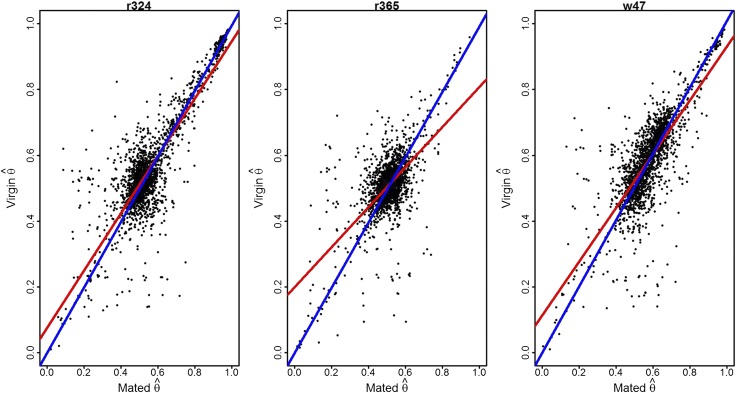
We compared all estimates of AI in both environments (Figure 3). AI is largely concordant between environments (Figure 3 and Figure 5), a somewhat surprising result given the magnitude of expression differences reported for these conditions previously (Dalton et al. 2010) and in these data (see above) and the presence of ∼200 differentially regulated genes. We hypothesize that the large degree of concordance in expression regulation between environments is due to a robust gene regulatory network (Cubillos et al. 2014). To explicitly test this hypothesis, we can estimate cis- and trans-effects for each exon within the population based on the F1-hybrid tester design (Nuzhdin et al. 2012). For exons significant for AI in at least 10 lines (n = 879) cis- and trans-effects were estimated for all lines in mated and virgin conditions separately (Figure 4 and Table S3).
Figure 3.
Estimates of AI in the mated vs. virgin environment. Three lines (r324, r365, and w47) were selected at random. for the mated environment (x-axis) and the virgin environment (y-axis) for all exons was evaluated. Each exon is a point on the graph. Estimates of AI are remarkably similar between mated and virgin environments. The red line in the panels is a regression while the blue line has an intercept of (0, 0).
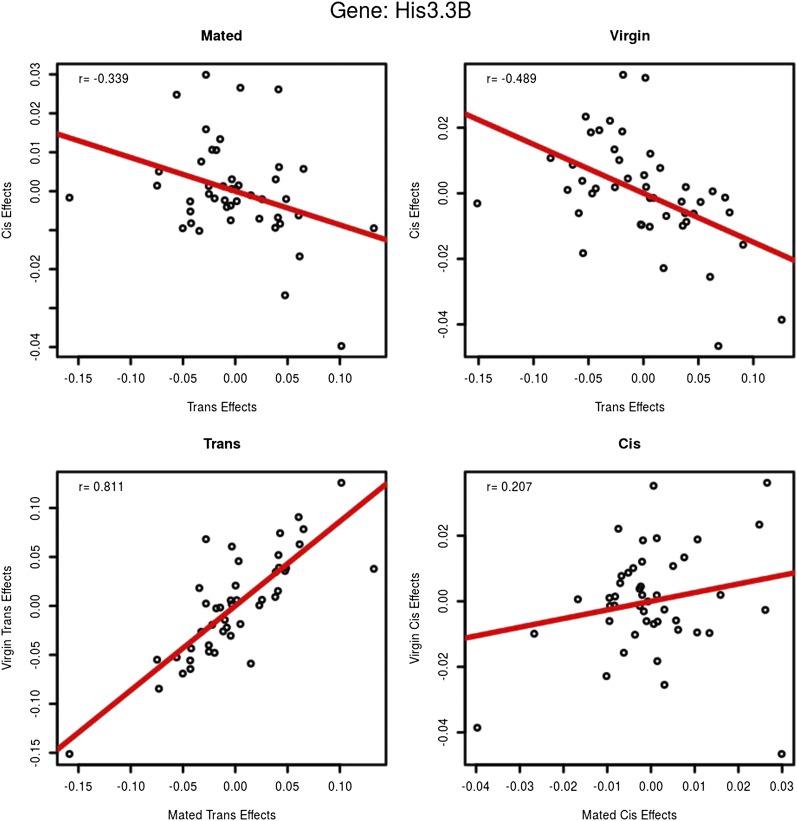
Figure 5.
His3.3B: cis- and trans-estimates. This gene was chosen at random as an example to demonstrate how cis- and trans-effects were compared. Each circle represents one testcross/F1-hybrid line. The top row compares cis- and trans-estimates within environment. The x-axis is the estimated trans-effect and the y-axis is the cis-effect. Cis- and trans-effects are negatively correlated for this exon (85% of exons show negative associations in both environments). The bottom row compares cis- and trans-effects across environments. The x-axis is the mated environment and the y-axis is the virgin environment. In this example, trans-effects are positively associated between mated and virgin flies. Cis effects are less correlated in this example but the correlation is positive (99% of all exons are positively correlated for both cis- and trans-effects across environments with a median correlation of 0.83 for cis and 0.95 for trans). The red line is the regression. Figure S4 shows the same relationships between cis- and trans-effects across all crosses and exons.
Figure 4.
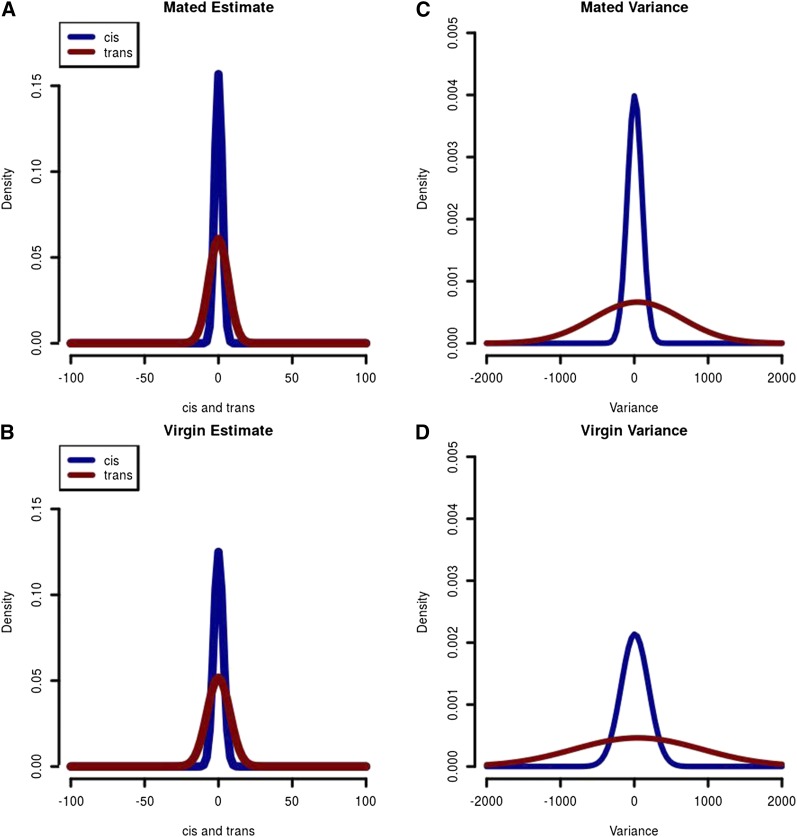
Distribution of cis and trans for exons with significant AI. (A) Distribution of estimates of cis and trans for the mated environment. Trans-estimates appear to have a larger variance than the cis-estimates. Cis and trans are centered at zero, as expected. (B) Distribution of estimates of cis and trans for the virgin environment. Trans-estimates appear to have a larger variance than the cis-estimates. Cis and trans are centered at zero, as expected. (C) Distribution of the variance for cis- and trans-estimates for the mated environment. For each exon, the variance of the estimate is calculated. The variance in trans-estimates can be much larger than the variance in cis-estimates. (D) Distribution of the variance for cis- and trans-estimates for the virgin environment. For each exon, the variance of the estimate is calculated. The variance in trans-estimates can be much larger than the variance in cis-estimates.
The distribution of estimates (Figure 4, A and B) shows a larger range for trans-estimates than for cis-estimates. For each exon in mated and virgin environments we calculated the variance of cis- and trans-estimates (Figure 4, C and D). The variance of trans-estimates is much larger than the variance of cis-estimates in both environments, consistent with previous studies (Genissel et al. 2008). The direction of the estimates for cis and trans for a particular cross/environment is often different (63%, Table 1) and the association between estimates for cis and trans for a particular exon across crosses is negative 85% of the time (Figure 5, gene His3.3B; and Figure S4). In contrast, when comparing trans-effects for the same exon across environments the results are strikingly concordant (Figure 5 and Figure S4). Cis-effects are likewise similar across environments (Figure 5 and Figure S4).
Table 1. Direction of cis- and trans-effects.
| Direction trans | |||
|---|---|---|---|
| Direction cis | + | − | Total |
| + | 9,090 | 22,300 | 31,390 |
| − | 11,869 | 10,621 | 22,490 |
| Total | 20,959 | 32,921 | 53,880 |
If the value of the estimate is positive, the direction is recorded as (+); similarly negative values are scored as (−). The direction of cis-effects is compared to the direction of trans-effects. There are (9090 + 10,621) cis- and trans-estimates that are concordant and (22,300 + 11,869) cis- and trans-estimates that are compensatory.
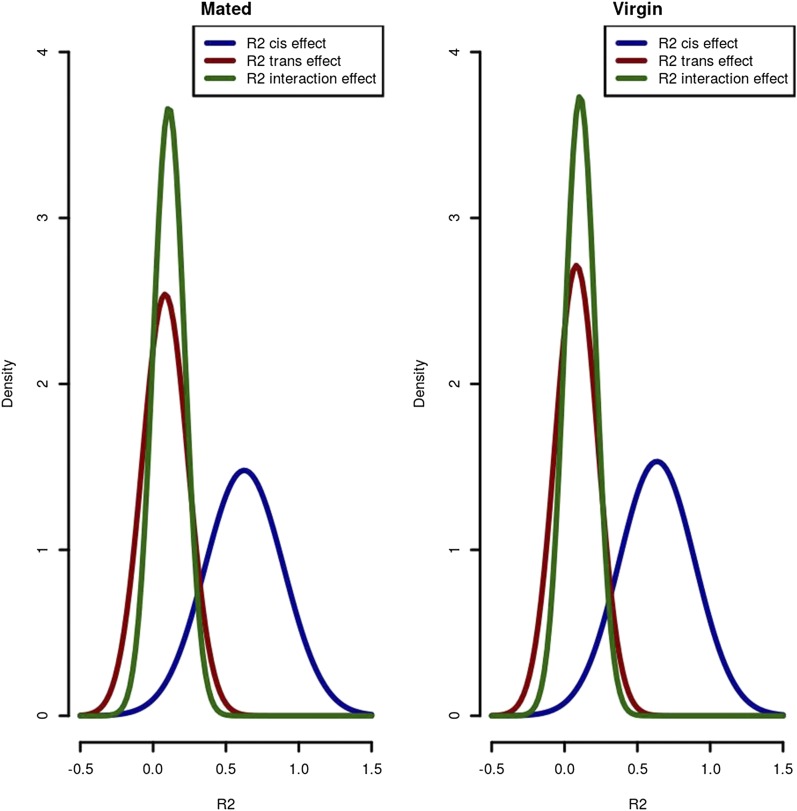
AI is a result of both cis and trans and the interaction between them (Figure 1). For each environment we modeled the effect of AI as a function of cis- and trans-effects and their interaction, using a linear model: Yi = μ + Ci +Ti + Ci × Ti, where Y is AI, Ci is the estimate of cis, and Ti is trans for line i (i = 1–nj). The association between cis-effects and AI was nearly always significant. For 75% of the exons tested, cis-effects explained at least 45% of the variation in expression (Figure 6 and Table S5). For 25% of exons, cis-effects explained >85% of the variance. Trans-effects added on average 8% to the proportion of variance explained. We found that 25% of exons had at least 10% of their variance explained by trans. Even after accounting for both cis and trans, the interaction between cis and trans explains an additional 11% on average. The full model explained at least 40% of the variance for 95% of the exons modeled and 90% of the variance for 50% of the exons (Figure 6, Figure 7, and Table S5).
Figure 6.
The distribution of R2 for the regression Yij = μ + Ci + Ti + Ci × Ti, where Y is Ci is the estimate of cis, and Ti is trans for line i for each exon j. R2 is an estimate of the contribution of cis-interactions, trans-interactions, and cis- by trans-interactions to AI. Distributions of R2 are similar in the mated and virgin environments. Cis-effects explain at least 45% of the variation in expression for 75% of exons tested. Trans-effects explain 8% of the variance on average. The cis- by trans-interaction explains an additional 11% of the variance on average.
Figure 7.
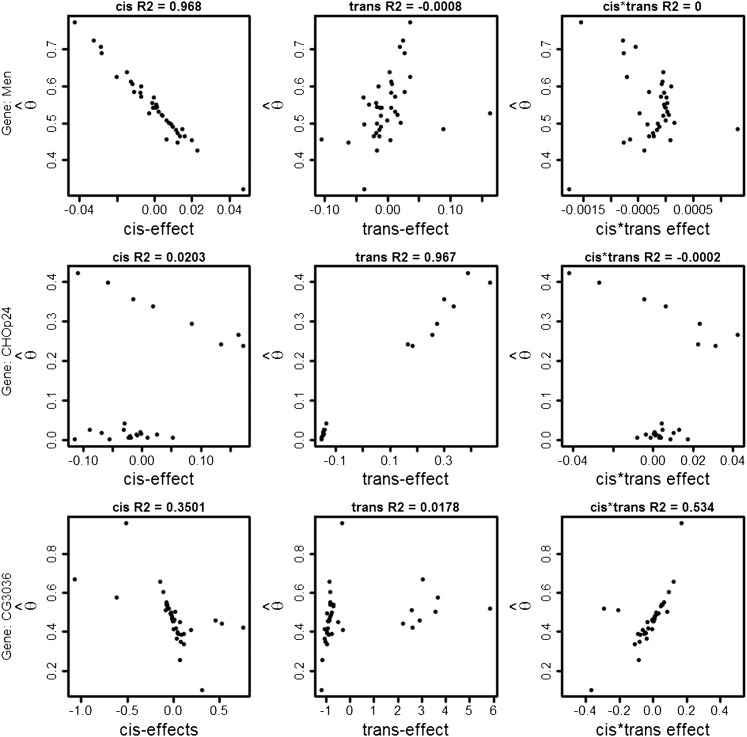
Three exons displaying different patterns of association between cis-interactions, trans-interactions, and cis- by trans-interactions and Each point is a testcross/F1 hybrid. On the y-axis is the estimated AI () and on the x-axis in left, center, and right columns are cis, trans, and cis by trans. The top row represents the gene Men. This gene has ∼97% of the variance explained by the cis-effect and a negative association between cis and AI. The middle row represents the gene CHOp24. This gene has ∼97% of the variance explained by trans-effects. The bottom row represents the gene CG3036. This gene has ∼53% of variance explained by the cis- by trans-interaction. Figure S3 depicts lack of association between cis- and trans-effects with when all crosses and all exons are considered simultaneously.
We identified genes that are regulated completely by cis (R2 > 0.90, n = 170), have a large trans-effect (R2 due to trans >0.40, n = 50), or have a large cis- by trans-interaction effect (R2 due to the interaction >0.40). Genes regulated primarily by cis were enriched for biological Gene Ontology processes involved in centrosome organization (P = 0.001439, 12 genes), centrosome cycle (P < 0.0001, 11 genes), and centrosome duplication (P = 0.001839, 10 genes). In addition, phototransduction (P = 0.004142, 8 genes), detection of light stimulus (P = 0.00949, 8 genes), response to light stimulus (P = 0.017, 11 genes), and detection of visible light (P = 0.046, 6 genes) were also enriched. Phototransduction genes are known to be altered by mating in Drosophila (Gioti et al. 2012; Landis et al. 2015).
Of the genes with a large trans-effect on AI, Chmp1 is involved in the negative regulation of the epidermal growth signaling pathway as well as involved in chromatin structure and cell cycle progression (Stauffer et al. 2001). HPS4 is also involved with chromatin, but in the silencing and negative regulation of gene silencing (Lee et al. 2009). jim is also involved in chromatin silencing (Mugat et al. 2015) and PIWI-interacting RNA processes in follicle cells (Saito et al. 2009). Finally, ps is spliced sex specifically (Telonis-Scott et al. 2009) and is a splicing factor (Seshaiah et al. 2001) and Vmat is involved in transmembrane transport as a splice variant that is involved in dopamine, serotonin, and octopamine transport (Greer et al. 2005). Both dopamine signaling and octopamine signaling are implicated in mediating the postmating response (Rezaval et al. 2014; Landis et al. 2015).
Discussion
This study supports previous findings of differences in expression between mated and virgin flies and goes further in identifying ∼200 genes with AI differences between environments. Yet, when AI is detected in both environments, it is concordant and cis- and trans-effects are concordant across the environments. When AI is detected, cis is almost always significant and in this first population-scale study of natural variation within a species, trans-effects and cis- by trans-interactions are sizable. Trans-effects are evidence of molecular connections across loci and cis- by trans-interactions are evidence of genetic variation across loci influencing the phenotype of gene expression. In stark contrast to the cisM–cisV and transM–transV comparison across environments the cis- and trans-effects within an environment (cisM–transM; cisV–transV) are markedly not concordant in this study (Table S6). The cis/trans combinations +/+ and −/− are concordant while +/− and −/+ are compensatory. Here 63% of the effects within a line/exon are compensatory and across lines 85% of the associations between cis- and trans-effects have a negative slope, leaving us with an unambiguous finding of large-scale compensatory effects in expression regulation. Large-scale compensatory effects within the same species are also supported by a recent elegant experimental study in Caenorhabditis elegans (Paaby and Rockman 2014).
There is a strong consensus emerging from the analyses of a variety of model organisms: in yeast (Kvitek et al. 2008; Artieri and Fraser 2014), fly (Wang et al. 2008; Coolon et al. 2014, 2015; Graze et al. 2014), and mouse (Goncalves et al. 2012; Crowley et al. 2015; Pinter et al. 2015) that cis- and trans-effects are compensatory. In intraspecific D. melanogaster F1’s, compensatory interactions were observed in 79% of cases, and in interspecific D. simulans/D. sechelia and D. melanogaster/D. simulans crosses, they corresponded to 73% and 87% of cases (see summary in Coolon et al. 2014). How does one explain this strong and phylogenetically ubiquitous pattern?
One frequent explanation for compensatory effects is that trans- and cis-factors coevolve, as there is a stabilizing selection for overall transcript level (Wittkopp et al. 2004, 2008b; Landry et al. 2005; Graze et al. 2009, 2012; Fontanillas et al. 2010; Stevenson et al. 2013; McManus et al. 2014). A neutral trans-mutation is envisioned that spreads jointly with a slightly deleterious cis-mutation (see Takahasi et al. 2011 for a helpful summary). Takahasi et al. (2011, p. 15,279) demonstrate via extensive simulation that “compensatory cis-trans interactions gradually accumulated over time.” Also of note, in their simulations (Takahasi et al. 2011, p. 15,279) “background dependency of relative allelic expression was not observed within species.” Within species, cis- and trans-effects cannot coevolve as they are not cotransmitted. In this experiment in Drosophila the likelihood of long-range LD in flies accounting for ∼85% compensatory vs. ∼15% concordant cis–trans combinations is vanishingly small as the scale of this effect is known to be very small (Pool et al. 2012). What is then the evolutionary explanation of such compensation within species?
Gene expression evolves under a house-of-cards model of stabilizing selection (Hodgins-Davis et al. 2015), maintaining population genetic variation (Jin et al. 2001; Gibson and Dworkin 2004; Nuzhdin et al. 2004). This variation contributes to phenotypic diversity and is widespread. If instead of thinking about individual loci we focus on gene regulatory networks (GRNs), then logically there must be variation among loci. How then would that variation be measured in the context of cis- and trans-effects? If each locus contained variation that affected expression at that locus, we would expect a large number of cis-effects, as observed here. Yet, GRNs are inherently models of trans-effects, as the connections among genes in the pathway are trans. For these data a pathway analysis of the virgin environment modeled regulation of the sex hierarchy, using structural equations (Fear et al. 2015). This analysis suggested that there were 754 genes that were linked to the core sex hierarchy. We conducted a similar path analysis of the mated environment and found 380 (50%) were in common, suggesting a large overlap in the trans-regulation of genes upstream and downstream of the sex hierarchy in both environments.
In thinking about cis- and trans-effects in the context of a GRN the relative directions of the cis- and trans-effects must be considered carefully. For example: If an allele 1 in genotype 1 is upregulated due to cis-regulatory mutation (+ effect) and there is also a significant trans-effect estimated for the same allele, what direction—upregulating or downregulating (− effect)—would it more frequently have? An alternate effect of the allele in cis and trans suggests that the overall regulation of the GRN is constrained. Compensatory effects of the allele within a GRN may explain widespread observations of GRN robustness.
GRN robustness has been explicitly formulated to account for the prevalence of compensatory cis–trans interactions (Denby et al. 2012; Bader et al. 2015). Denby et al. (2012) have proposed that negative feedback controlling the level of RNA expression could be a common mechanism to buffer effects of regulatory variants in yeast. Screening for autoregulated transcription factors in yeast, Denby et al. (2012) found ROX1 to be under strong negative feedback. Mutant experiments showed that this negative feedback confers robustness to the expression of ROX1 in the face of naturally occurring allelic variants present in a set of divergent yeast strains. This study demonstrated for a single gene that negative feedback could act as a buffering mechanism for regulatory variants. Bader et al. (2015) quantified buffering by feedback against naturally occurring regulatory variants genome-wide to be ∼15%. Note that when buffering arises from feedback, it is not necessary to invoke additional genetic variation across the genome to explain the trans-effects.
If GRNs are generally robust, they have implications in human health research. Analysis of the structure of GRNs in the K562 cancer cell line suggests poor buffering compared to the noncancer cell line GM12878 (Albergante et al. 2014). Most of the genome-wide association study associations are between noncoding regions and disease (reviewed in Zhang and Lupski 2015). One potential explanation is that genes are misregulated in disease (reviewed in Lee and Young 2013). As GRN robustness should buffer against misregulation, it implies that disease may be a result of lack of effective buffering. This may make the hunt for genes underlying disease a little more straightforward, as rather than looking for direct links with phenotype, lack of robust regulation can be used to identify potential candidate pathways.
Acknowledgments
We thank Jeremy Newman for his help in the revisions. We thank the editor and anonymous reviewers for insightful comments. This work was supported by National Institutes of Health (NIH) grants GMS102227 (to L.M.M. and S.V.N.), R01MH091561 (to S.V.N. and L.M.M.), 5P50HG002790 (to S.V.N. and J.T.), and AG011833 (to J.T.). The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Communicating editor: J. A. Birchler
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.188797/-/DC1.
Literature Cited
- Albergante L., Blow J. J., Newman T. J., 2014. Buffered qualitative stability explains the robustness and evolvability of transcriptional networks. eLife 3: e02863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Reyes A., Huber W., 2012. Detecting differential usage of exons from RNA-seq data. Genome Res. 22: 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artieri C. G., Fraser H. B., 2014. Evolution at two levels of gene expression in yeast. Genome Res. 24: 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader D. M., Wilkening S., Lin G., Tekkedil M. M., Dietrich K., et al. , 2015. Negative feedback buffers effects of regulatory variants. Mol. Syst. Biol. 11: 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone M. C., Sykiotis G. P., Bohmann D., 2011. Genetic activation of Nrf2 signaling is sufficient to ameliorate neurodegenerative phenotypes in a Drosophila model of Parkinson’s disease. Dis. Model. Mech. 4: 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Berger, R. L., 1997 Likelihood Ratio Tests and Intersection-Union Tests, pp. 225–237 in Advances in Statistical Decision Theory and Applications, edited by S. Panchapakesan and N. Balakrishnan. Birkhäuser Boston, Boston, MA. [Google Scholar]
- Berger R. L., Boos D. D., 1999. Confidence limits for the onset and duration of treatment effect. Biom. J. 41: 517–531. [Google Scholar]
- Boltz K. A., Ellis L. L., Carney G. E., 2007. Drosophila melanogaster p24 genes have developmental, tissue‐specific, and sex‐specific expression patterns and functions. Dev. Dyn. 236: 544–555. [DOI] [PubMed] [Google Scholar]
- Brem R. B., Yvert G., Clinton R., Kruglyak L., 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. [DOI] [PubMed] [Google Scholar]
- Campo D., Lehmann K., Fjeldsted C., Souaiaia T., Kao J., et al. , 2013. Whole-genome sequencing of two North American Drosophila melanogaster populations reveals genetic differentiation and positive selection. Mol. Ecol. 22: 5084–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel S. E., Levy-Moonshine A., Mohammadi P., Banks E., Lappalainen T., 2015. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 16: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T., Liddle L. F., Kalb J. M., Wolfner M. F., Partridge L., 1995. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature 373: 241–244. [DOI] [PubMed] [Google Scholar]
- Coffman C. J., Doerge R. W., Wayne M. L., McIntyre L. M., 2003. Intersection tests for single marker QTL analysis can be more powerful than two marker QTL analysis. BMC Genet. 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon J. D., McManus C. J., Stevenson K. R., Graveley B. R., Wittkopp P. J., 2014. Tempo and mode of regulatory evolution in Drosophila. Genome Res. 24: 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon J. D., Stevenson K. R., McManus C. J., Yang B., Graveley B. R., et al. , 2015. Molecular mechanisms and evolutionary processes contributing to accelerated divergence of gene expression on the Drosophila X chromosome. Mol. Biol. Evol. 32: 2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. R., Hirschhorn J. N., Altshuler D., Lander E. S., 2002. Detection of regulatory variation in mouse genes. Nat. Genet. 32: 432–437. [DOI] [PubMed] [Google Scholar]
- Crowley J. J., Zhabotynsky V., Sun W., Huang S., Pakatci I. K., et al. , 2015. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat. Genet. 47: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F. A., Stegle O., Grondin C., Canut M., Tisne S., et al. , 2014. Extensive cis-regulatory variation robust to environmental perturbation in Arabidopsis. Plant Cell 26: 4298–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton J. E., Kacheria T. S., Knott S. R., Lebo M. S., Nishitani A., et al. , 2010. Dynamic, mating-induced gene expression changes in female head and brain tissues of Drosophila melanogaster. BMC Genomics 11: 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner J. F., Marioni J. C., Pai A. A., Pickrell J. K., Nkadori E., et al. , 2009. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25: 3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby, C. M., J. H. Im, R. C. Yu, C. G. Pesce and R. B. Brem, 2012 Negative feedback confers mutational robustness in yeast transcription factor regulation. Proc. Natl. Acad. Sci. USA 109: 3874–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J. J., Hsieh L. C., Sung H. M., Wang T. Y., Huang C. J., et al. , 2010. Natural selection on cis and trans regulation in yeasts. Genome Res. 20: 826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fear J. M., Arbeitman M. N., Salomon M. P., Dalton J. E., Tower J., et al. , 2015. The Wright stuff: reimagining path analysis reveals novel components of the sex determination hierarchy in Drosophila melanogaster. BMC Syst. Biol. 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T., 2011. Survival costs of reproduction in Drosophila. Exp. Gerontol. 46: 369–375. [DOI] [PubMed] [Google Scholar]
- Fontanillas P., Landry C. R., Wittkopp P. J., Russ C., Gruber J. D., et al. , 2010. Key considerations for measuring allelic expression on a genomic scale using high‐throughput sequencing. Mol. Ecol. 19: 212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M. C., Wan R., Horton P., 2010. Incorporating sequence quality data into alignment improves DNA read mapping. Nucleic Acids Res. 38: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genissel A., McIntyre L. M., Wayne M. L., Nuzhdin S. V., 2008. Cis and trans regulatory effects contribute to natural variation in transcriptome of Drosophila melanogaster. Mol. Biol. Evol. 25: 101–110. [DOI] [PubMed] [Google Scholar]
- Gibson G., Dworkin I., 2004. Uncovering cryptic genetic variation. Nat. Rev. Genet. 5: 681–690. [DOI] [PubMed] [Google Scholar]
- Gilad Y., Rifkin S. A., Pritchard J. K., 2008. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 24: 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioti A., Wigby S., Wertheim B., Schuster E., Martinez P., et al. , 2012. Sex peptide of Drosophila melanogaster males is a global regulator of reproductive processes in females. Proc. Biol. Sci. 279: 4423–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves A., Leigh-Brown S., Thybert D., Stefflova K., Turro E., et al. , 2012. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 22: 2376–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graze, R. M., L. M. McIntyre, B. J. Main, M. L. Wayne and S. V. Nuzhdin, 2009 Regulatory divergence in Drosophila melanogaster and D. simulans, a genome-wide analysis of allele-specific expression. Genetics 183: 547–561. [DOI] [PMC free article] [PubMed]
- Graze R. M., Novelo L. L., Amin V., Fear J. M., Casella G., et al. , 2012. Allelic imbalance in Drosophila hybrid heads: exons, isoforms, and evolution. Mol. Biol. Evol. 29: 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graze R. M., McIntyre L. M., Morse A. M., Boyd B. M., Nuzhdin S. V., et al. , 2014. What the X has to do with it: differences in regulatory variability between the sexes in Drosophila simulans. Genome Biol. Evol. 6: 818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer C. L., Grygoruk A., Patton D. E., Ley B., Romero-Calderon R., et al. , 2005. A splice variant of the Drosophila vesicular monoamine transporter contains a conserved trafficking domain and functions in the storage of dopamine, serotonin, and octopamine. J. Neurobiol. 64: 239–258. [DOI] [PubMed] [Google Scholar]
- Guichet A., Peri F., Roth S., 2001. Stable anterior anchoring of the oocyte nucleus is required to establish dorsoventral polarity of the Drosophila egg. Dev. Biol. 237: 93–106. [DOI] [PubMed] [Google Scholar]
- Guo M., Yang S., Rupe M., Hu B., Bickel D. R., et al. , 2008a Genome-wide allele-specific expression analysis using Massively Parallel Signature Sequencing (MPSS) reveals cis- and trans-effects on gene expression in maize hybrid meristem tissue. Plant Mol. Biol. 66: 551–563. [DOI] [PubMed] [Google Scholar]
- Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., et al. , 2008b Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453: 657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins-Davis A., Rice D. P., Townsend J. P., 2015. Gene expression evolves under a house-of-cards model of stabilizing selection. Mol. Biol. Evol. 32: 2130–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Carbone M. A., Magwire M. M., Peiffer J. A., Lyman R. F., et al. , 2015. Genetic basis of transcriptome diversity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 112: E6010–E6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. A., Ayroles J. F., Reedy M. M., Drnevich J. M., Rowe K. C., et al. , 2006. Segregating variation in the transcriptome: cis regulation and additivity of effects. Genetics 173: 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter S., Saminadin-Peter S. S., Stephan W., Parsch J., 2008. Gene expression variation in African and European populations of Drosophila melanogaster. Genome Biol. 9: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Riley R. M., Wolfinger R. D., White K. P., Passador-Gurgel G., et al. , 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29: 389–395. [DOI] [PubMed] [Google Scholar]
- Karim M. R., Taniguchi H., Kobayashi A., 2015. Constitutive activation of Drosophila CncC transcription factor reduces lipid formation in the fat body. Biochem. Biophys. Res. Commun. 463: 693–698. [DOI] [PubMed] [Google Scholar]
- King E. G., Merkes C. M., McNeil C. L., Hoofer S. R., Sen S., et al. , 2012. Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 22: 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Sanderson B. J., McNeil C. L., Long A. D., Macdonald S. J., 2014. Genetic dissection of the Drosophila melanogaster female head transcriptome reveals widespread allelic heterogeneity. PLoS Genet. 10: e1004322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst M., Basten C. J., Myburg A. A., Zeng Z. B., Sederoff R. R., 2005. Genetic architecture of transcript-level variation in differentiating xylem of a eucalyptus hybrid. Genetics 169: 2295–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher S. D., Richard F. J., Tarpy D. R., Grozinger C. M., 2008. Genomic analysis of post-mating changes in the honey bee queen (Apis mellifera). BMC Genomics 9: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmangaliyev Y. Z., Favorov A. V., Osman N. M., Lehmann K. V., Campo D., et al. , 2015. Natural variation of gene models in Drosophila melanogaster. BMC Genomics 16: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek D. J., Will J. L., Gasch A. P., 2008. Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet. 4: e1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis G. N., Salomon M. P., Keroles D., Brookes N., Sekimura T., et al. , 2015. The progesterone antagonist mifepristone/RU486 blocks the negative effect on life span caused by mating in female Drosophila. Aging 7: 53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry C. R., Wittkopp P. J., Taubes C. H., Ranz J. M., Clark A. G., et al. , 2005. Compensatory cis–trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171: 1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak M. K., Begun D. J., 2004. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome 47: 900–910. [DOI] [PubMed] [Google Scholar]
- Lawniczak M. K., Barnes A. I., Linklater J. R., Boone J. M., Wigby S., et al. , 2007. Mating and immunity in invertebrates. Trends Ecol. Evol. 22: 48–55. [DOI] [PubMed] [Google Scholar]
- Lebo M. S., Sanders L. E., Sun F., Arbeitman M. N., 2009. Somatic, germline and sex hierarchy regulated gene expression during Drosophila metamorphosis. BMC Genomics 10: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Young R. A., 2013. Transcriptional regulation and its misregulation in disease. Cell 152: 1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. S., Pressman S., Andress A. P., Kim K., White J. L., et al. , 2009. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 11: 1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B., Araripe L. O., Fontanillas P., Hartl D. L., 2008. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc. Natl. Acad. Sci. USA 105: 14471–14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Novelo L. G., McIntyre L. M., Fear J. M., Graze R. M., 2014. A flexible Bayesian method for detecting allelic imbalance in RNA-seq data. BMC Genomics 15: 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H. S., Wang Z., Hu Y., Yang H. H., Gere S., et al. , 2003. Allelic variation in gene expression is common in the human genome. Genome Res. 13: 1855–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack P. D., Kapelnikov A., Heifetz Y., Bender M., 2006. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103: 10358–10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Stone E. A., Ayroles J. F., 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10: 565–577. [DOI] [PubMed] [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massouras A., Waszak S. M., Albarca-Aguilera M., Hens K., Holcombe W., et al. , 2012. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet. 8: e1003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw L. A., Gibson G., Clark A. G., Wolfner M. F., 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 14: 1509–1514. [DOI] [PubMed] [Google Scholar]
- McGraw L. A., Clark A. G., Wolfner M. F., 2008. Postmating gene expression profiles of female Drosophila melanogaster in response to time and to four male accessory gland proteins. Genetics 179: 1395–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus C. J., Coolon J. D., Duff M. O., Eipper-Mains J., Graveley B. R., et al. , 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 20: 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus C. J., Coolon J. D., Eipper-Mains J., Wittkopp P. J., Graveley B. R., 2014. Evolution of splicing regulatory networks in Drosophila. Genome Res. 24: 786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Coolon J. D., Hartl D. L., Wittkopp P. J., 2014. The roles of cis- and trans-regulation in the evolution of regulatory incompatibilities and sexually dimorphic gene expression. Genome Res. 24: 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. T., Dietz H. C., 2001. When the message goes awry: disease-producing mutations that influence mRNA content and performance. Cell 107: 411–414. [DOI] [PubMed] [Google Scholar]
- Michalak P., Noor M. A., 2003. Genome-wide patterns of expression in Drosophila pure species and hybrid males. Mol. Biol. Evol. 20: 1070–1076. [DOI] [PubMed] [Google Scholar]
- Michalak P., Noor M. A., 2004. Association of misexpression with sterility in hybrids of Drosophila simulans and D. mauritiana. J. Mol. Evol. 59: 277–282. [DOI] [PubMed] [Google Scholar]
- Misra J. R., Horner M. A., Lam G., Thummel C. S., 2011. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 25: 1796–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow E. H., Innocenti P., 2012. Female postmating immune responses, immune system evolution and immunogenic males. Biol. Rev. Camb. Philos. Soc. 87: 631–638. [DOI] [PubMed] [Google Scholar]
- Mugat B., Akkouche A., Serrano V., Armenise C., Li B., et al. , 2015. MicroRNA-dependent transcriptional silencing of transposable elements in Drosophila follicle cells. PLoS Genet. 11: e1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger S. C., Raghupathy N., Choi K., Simons A. K., Gatti D. M., et al. , 2014. RNA-Seq alignment to individualized genomes improves transcript abundance estimates in multiparent populations. Genetics 198: 59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothnagel M., Wolf A., Herrmann A., Szafranski K., Vater I., et al. , 2011. Statistical inference of allelic imbalance from transcriptome data. Hum. Mutat. 32: 98–106. [DOI] [PubMed] [Google Scholar]
- Nuzhdin S. V., Wayne M. L., Harmon K. L., McIntyre L. M., 2004. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol. Biol. Evol. 21: 1308–1317. [DOI] [PubMed] [Google Scholar]
- Nuzhdin S. V., Friesen M. L., McIntyre L. M., 2012. Genotype-phenotype mapping in a post-GWAS world. Trends Genet. 28: 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata F., Miura M., 2015. Enhancing S-adenosyl-methionine catabolism extends Drosophila lifespan. Nat. Commun. 6: 8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D., Counterman B. A., Noor M. A., 2007. Gene expression divergence and the origin of hybrid dysfunctions. Genetica 129: 71–81. [DOI] [PubMed] [Google Scholar]
- Paaby A. B., Rockman M. V., 2014. Cryptic genetic variation: evolution’s hidden substrate. Nat. Rev. Genet. 15: 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastinen T., 2010. Genome-wide allele-specific analysis: insights into regulatory variation. Nat. Rev. Genet. 11: 533–538. [DOI] [PubMed] [Google Scholar]
- Peng J., Zipperlen P., Kubli E., 2005. Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr. Biol. 15: 1690–1694. [DOI] [PubMed] [Google Scholar]
- Pickering A. M., Staab T. A., Tower J., Sieburth D., Davies K. J., 2013. A conserved role for the 20S proteasome and Nrf2 transcription factor in oxidative stress adaptation in mammals, Caenorhabditis elegans and Drosophila melanogaster. J. Exp. Biol. 216: 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter S. F., Colognori D., Beliveau B. J., Sadreyev R. I., Payer B., et al. , 2015. Allelic imbalance is a prevalent and tissue-specific feature of the mouse transcriptome. Genetics 200: 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., et al. , 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8: e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz J. M., Namgyal K., Gibson G., Hartl D. L., 2004. Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res. 14: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaval C., Nojima T., Neville M. C., Lin A. C., Goodwin S. F., 2014. Sexually dimorphic octopaminergic neurons modulate female postmating behaviors in Drosophila. Curr. Biol. 24: 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L., 2006. Genetics of global gene expression. Nat. Rev. Genet. 7: 862–872. [DOI] [PubMed] [Google Scholar]
- Ronald J., Akey J. M., Whittle J., Smith E. N., Yvert G., et al. , 2005. Simultaneous genotyping, gene-expression measurement, and detection of allele-specific expression with oligonucleotide arrays. Genome Res. 15: 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J., Abyzov A., Wang J., Alves P., Raha D., et al. , 2011. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol. Syst. Biol. 7: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., et al. , 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299. [DOI] [PubMed] [Google Scholar]
- Saleem S., Schwedes C. C., Ellis L. L., Grady S. T., Adams R. L., et al. , 2012. Drosophila melanogaster p24 trafficking proteins have vital roles in development and reproduction. Mech. Dev. 129: 177–191. [DOI] [PubMed] [Google Scholar]
- Seshaiah P., Miller B., Myat M. M., Andrew D. J., 2001. pasilla, the Drosophila homologue of the human Nova-1 and Nova-2 proteins, is required for normal secretion in the salivary gland. Dev. Biol. 239: 309–322. [DOI] [PubMed] [Google Scholar]
- Skelly D. A., Johansson M., Madeoy J., Wakefield J., Akey J. M., 2011. A powerful and flexible statistical framework for testing hypotheses of allele-specific gene expression from RNA-seq data. Genome Res. 21: 1728–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. N., Kruglyak L., 2008. Gene-environment interaction in yeast gene expression. PLoS Biol. 6: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G., Fang Y., Liu X., Kenny J., Cossins A. R., et al. , 2013. Transcriptome-wide expression variation associated with environmental plasticity and mating success in cactophilic Drosophila mojavensis. Evolution 67: 1950–1963. [DOI] [PubMed] [Google Scholar]
- Springer N. M., Stupar R. M., 2007. Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19: 2391–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer D. R., Howard T. L., Nyun T., Hollenberg S. M., 2001. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J. Cell Sci. 114: 2383–2393. [DOI] [PubMed] [Google Scholar]
- Stell R., Torrie J., Dickey D., 1980. Principles and Procedures of Statistics: A Biometrical Approach. MacGraw-Hill, New York. [Google Scholar]
- Stevenson K. R., Coolon J. D., Wittkopp P. J., 2013. Sources of bias in measures of allele-specific expression derived from RNA-sequence data aligned to a single reference genome. BMC Genomics 14: 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis G. P., Habeos I. G., Samuelson A. V., Bohmann D., 2011. The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr. Opin. Clin. Nutr. Metab. Care 14: 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K. R., Matsuo T., Takano-Shimizu-Kouno T., 2011. Two types of cis-trans compensation in the evolution of transcriptional regulation. Proc. Natl. Acad. Sci. USA 108: 15276–15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telonis-Scott M., Kopp A., Wayne M. L., Nuzhdin S. V., McIntyre L. M., 2009. Sex-specific splicing in Drosophila: widespread occurrence, tissue specificity and evolutionary conservation. Genetics 181: 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Reikhav S., Levy A. A., Barkai N., 2009. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., et al. , 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Geijn B., McVicker G., Gilad Y., Pritchard J., 2015. WASP: allele-specific software for robust discovery of molecular quantitative trait loci. Nat. Methods 12: 1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-Y., Fu Y., McPeek M. S., Lu X., Nuzhdin S., et al. , 2008. Complex genetic interactions underlying expression differences between Drosophila races: analysis of chromosome substitutions. Proc. Natl. Acad. Sci. USA 105: 6362–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne M. L., Pan Y. J., Nuzhdin S. V., McIntyre L. M., 2004. Additivity and trans-acting effects on gene expression in male Drosophila simulans. Genetics 168: 1413–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne M. L., Telonis-Scott M., Bono L. M., Harshman L., Kopp A., et al. , 2007. Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104: 18577–18582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winbush A., Reed D., Chang P. L., Nuzhdin S. V., Lyons L. C., et al. , 2012. Identification of gene expression changes associated with long-term memory of courtship rejection in Drosophila males. G3 2: 1437–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., Kalay G., 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13: 59–69. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2006. Parent-of-origin effects on mRNA expression in Drosophila melanogaster not caused by genomic imprinting. Genetics 173: 1817–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2008a Independent effects of cis- and trans-regulatory variation on gene expression in Drosophila melanogaster. Genetics 178: 1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2008b Regulatory changes underlying expression differences within and between Drosophila species. Nat. Genet. 40: 346–350. [DOI] [PubMed] [Google Scholar]
- Yan H., Yuan W., Velculescu V. E., Vogelstein B., Kinzler K. W., 2002. Allelic variation in human gene expression. Science 297: 1143. [DOI] [PubMed] [Google Scholar]
- Yasuyama K., Kitamoto T., Salvaterra P. M., 1995. Immunocytochemical study of choline acetyltransferase in Drosophila melanogaster: an analysis of cis-regulatory regions controlling expression in the brain of cDNA-transformed flies. J. Comp. Neurol. 361: 25–37. [DOI] [PubMed] [Google Scholar]
- Yuan S., Qin Z., 2012. Read-mapping using personalized diploid reference genome for RNA sequencing data reduced bias for detecting allele-specific expression. IEEE Int. Conf. Bioinform. Biomed. Workshops 2012: 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Lupski J. R., 2015. Non-coding genetic variants in human disease. Hum. Mol. Genet. 24: R102–R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Borevitz J. O., 2009. Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics 182: 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Mackay T., Anholt R. R., 2014. Transcriptional and epigenetic responses to mating and aging in Drosophila melanogaster. BMC Genomics 15: 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F., Sun W., Crowley J. J., Zhabotynsky V., Sullivan P. F., et al. , 2014. A novel statistical approach for jointly analyzing RNA-Seq data from F1 reciprocal crosses and inbred lines. Genetics 197: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.