Abstract
Protein glycolysation is an essential posttranslational modification in eukaryotic cells. In pathogenic yeasts, it is involved in a large number of biological processes, such as protein folding quality control, cell viability and host/pathogen relationships. A link between protein glycosylation and apoptosis was established by the analysis of the phenotypes of oligosaccharyltransferase mutants in budding yeast. However, little is known about the contribution of glycosylation modifications to the adaptive response to apoptosis inducers. The cysteine protease metacaspase Mca1p plays a key role in the apoptotic response in Candida albicans triggered by the quorum sensing molecule farnesol. We subjected wild-type and mca1-deletion strains to farnesol stress and then studied the early phase of apoptosis release in quantitative glycoproteomics and glycomics experiments on cell-free extracts essentially devoid of cell walls. We identified and characterized 62 new glycosylated peptides with their glycan composition: 17 N-glycosylated, 45 O-glycosylated, and 81 additional sites of N-glycosylation. They were found to be involved in the control of protein folding, cell wall integrity and cell cycle regulation. We showed a general increase in the O-glycosylation of proteins in the mca1 deletion strain after farnesol challenge. We identified 44 new putative protein substrates of the metacaspase in the glycoprotein fraction enriched on concanavalin A. Most of these substrates are involved in protein folding or protein resolubilization and in mitochondrial functions. We show here that key Mca1p substrates, such as Cdc48p or Ssb1p, involved in degrading misfolded glycoproteins and in the protein quality control system, are themselves differentially glycosylated. We found putative substrates, such as Bgl2p (validated by immunoblot), Srb1p or Ugp1p, that are involved in the biogenesis of glycans. Our findings highlight a new role of the metacaspase in amplifying cell death processes by affecting several critical protein quality control systems through the alteration of the protein glycosylation machinery.
Data are available via ProteomeXchange with identifier PXD003677.
Protein glycosylation is the most common posttranslational modification in eukaryotic cells and glycosylation defects have been associated with severe developmental impairments and disease in humans (1, 2). Glycosylation mostly affects plasma membrane and secreted proteins, and it plays a critical role in regulating many cell-cell interactions (1, 3). In pathogenic yeasts, glycosylation has been implicated in host/pathogen interactions (3, 4) and particularly in virulence and immune recognition (5). As protein glycosylation is essential for cell viability in yeast, the glycosylation pathways is a potential target for new anti-fungal drugs. Two main types of glycosylation, N- and the O-glycosylation, have been described in Candida albicans, which has glycan biosynthesis pathways very similar to those of higher eukaryotes, but with glycan moieties of a simpler oligosaccharide composition. N-glycosylation affects the asparagine (Asn) residues of Asn-X-Ser/Thr motifs of proteins, in which X can be all amino acids except proline. The complex oligosaccharide moiety consists of two N-acetylglucosamine residues associated with a dolichol phosphate membrane-bound anchor and carrying nine mannose and three glucose residues. It is transferred “en bloc” to the nascent chain of newly synthesized proteins imported into the endoplasmic reticulum, by a complex machinery involving the OST complex (6). In yeast, this complex involves the proteins Ost1–6p, Wbp1p, Swp1p and the catalytic subunit Stt3p, which are encoded by essential genes. The core-glycosylated proteins are further modified by the addition of long oligomannoside outer chains, which make a major contribution to the antigenic variability of clinical isolates of Candida (7). N-glycans are essential structural components of the cell wall with a role in the folding of proteins targeted to membrane or cell wall and in the degradation of these proteins if they are incorrectly folded (8). O-glycosylation in yeast is somewhat much simpler, consisting of the sequential addition of mannose moieties to the serine and threonine residues of proteins in a process catalyzed by specific mannosyltransferases (9) with no particular recognition sequence other than a predominance of proline residues around the glycosylation site (10). Oligomannosylation occurs in the Golgi apparatus, but the first residue is added to the target protein in the endoplasmic reticulum. O-glycosylation has been described as a modification involved in the maintenance of cell wall integrity and protein secretion (11), providing some protection against proteolysis (12) and favoring protein stability (13, 14). It may also serve as a signal for protein degradation in the proteasome (15). The precise function of this modification thus remains unclear. O-glycosylation is mostly mediated by the proteins of the mannosyltransferase family (PMTs)1. Defects of the various PMTs result in changes in cell wall integrity, cell morphogenesis and adhesion to host cells in C. albicans (16). They also affect protein stability and transport to the endoplasmic reticulum in Saccharomyces cerevisiae (17). An interesting relationship between protein glycosylation and apoptosis has been described in S. cerevisiae, in which mutant strains lacking the Ost2 and Wbp1–1 proteins, essential oligosaccharyltransferases implicated in N-glycosylation, have been reported to display metacaspase-independent apoptotic phenotypes (18). We recently discovered new roles of the C. albicans metacaspase Mca1p in apoptosis, through the identification of a limited number of potential substrates of its trypsin-like proteolytic activity (19). Many of these substrates belong not only to a family of key proteins for maintenance of the mitochondria compartment including the AAA ATPase Cdc48p, but also to a vast group of proteins implicated in folding, such as the chaperones Hsp70p or Ssb1p, the degradation of which amplifies the cell death process. The ER-dependent folding processes were targeted, in particular, and direct targets of the Mca1p protease were also found in membrane extracellular and cell wall compartments. A second group of potential Mca1p substrates consists of mitochondrial proteins and proteins involved in mitochondrial biogenesis and maintenance. Several studies have reported the presence of glycosylated proteins in the mitochondrion (20–22), although the mitochondria itself as a site of protein glycosylation has recently been challenged (23). As protein glycosylation is clearly linked to cell viability, we wondered whether the early phase of apoptosis progression might also be connected to this key cellular process. We therefore decided to study the role of O- and N-glycosylation in farnesol-induced Mca1p-dependent apoptosis by quantitative glycoproteomics and glycomics with the wild-type (WT) and mca1Δ/Δ strains in the presence or absence of farnesol, a well characterized apoptosis inducer (24, 25).
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
C. albicans strains BWP17 (26) and BWP17-mca1Δ/Δ (mca1Δ::FRT/mca1Δ::FRT) (27) were used, as described in (19). The cells were grown in complete medium (YPD) until the mid-exponential growth phase. Farnesol (250 μm final concentration) was added to the culture and the cells were collected by centrifugation after 4 h of treatment. The cell/treatment combination defined four sets of conditions: two controls without farnesol treatment (WT and mca1Δ/Δ) and two sets of conditions with farnesol treatment (WT-F and mca1Δ/Δ-F). Cell-free extracts were prepared by disrupting the cells with a One-Shot apparatus (Constant Systems Ltd, Daventry, UK).
Enrichment in Glycoproteins and Glycopeptides by Lectin-based Affinity Chromatography
We used cell extracts (500 μg of protein) from each set of conditions directly for glycoprotein(/peptide) enrichment with concanavalin A (ConA) magnetic beads from the Pierce Glycosylation Isolation kit with ConA, according to the manufacturer's instructions (Thermo Scientific, Waltham, MA). We also digested 500 μg of cell protein from the various cell conditions overnight at 37 °C with sequencing-grade trypsin (12.5 μg/ml; Promega, Madison, Wi) in 200 μl of 25 mmol/L NH4HCO3. The resulting peptides were loaded onto a ConA column and processed as described above to enrich the samples in glycopeptides. In both experiments, two elutions were carried out and the proteins eluted from the beads were quantified with the Bradford protein assay. As a control, peptides from trypsin digests of crude samples were analyzed directly for protein quantification and glycopeptide identification. We obtained three fractions: (1) crude sample without enrichment, (2) fraction corresponding to proteins eluted from the ConA column, and (3) fractions corresponding to peptides eluted from the ConA column. The proteins eluted from the ConA column were also digested in solution, according to the same digestion protocol. Equal amounts of sample were used for the analysis of fraction (1), whereas similar volumes of eluate were analyzed for fractions (2) and (3).
LC-MS/MS Acquisition of Peptide Samples
We analyzed 2 μg of protein digests from crude samples and equal volumes of eluted proteins and peptides eluted from ConA columns from the different cell conditions in triplicate with an Orbitrap Fusion Tribrid coupled to a Nano-LC Proxeon 1000 equipped with an easy spray ion source (all from Thermo Scientific, San Jose, CA). Peptides were separated by chromatography with the following parameters: Acclaim PepMap100 C18 precolumn (2 cm, 75 μm i.d., 3 μm, 100 Å), Pepmap-RSLC Proxeon C18 column (50 cm, 75 μm i.d., 2 μm, 100 Å), 300 nl/min flow rate, gradient from 95% solvent A (water, 0.1% formic acid) to 35% solvent B (100% acetonitrile, 0.1% formic acid) over a period of 97 min, followed by column regeneration for 23 min, giving a total run time of 2 h. Peptides were analyzed in the Orbitrap cell, in full ion scan mode, at a resolution of 120,000 (at m/z 200), with a mass range of m/z 350–1550 (except for samples of isolated peptides from ConA columns with a mass range of 350–4000) and an AGC target of 2 × 105. Fragments were obtained by higher-energy C-trap dissociation (HCD) activation with a collisional energy of 30%, and a quadrupole isolation window of 1.6 Da. MS/MS data were acquired in the Orbitrap cell in top-speed mode, with a total cycle of 3 s at a resolution of 30,000, with an AGC target of 5 × 104, a dynamic exclusion of 50 s and an exclusion duration of 60 s. Precursor priority was highest charge state, followed by most intense. Peptides with charge states from 2 to 8 were selected for MS/MS acquisition. The maximum ion accumulation times were set to 250 ms for MS acquisition and 60 ms for MS/MS acquisition in parallelization mode.
Quantitative Analysis, in Label-free Experiments, of Proteins and Peptides from the Different Data Sets
All MS and MS/MS data for protein samples were processed with Progenesis QI 2.0 (Nonlinear Dynamics, Newcastle, UK) for label-free quantification and with the Byonic search engine (Protein Metrics, San Carlos, CA, version 2.3.5) (28). The abundances of the peptides obtained by the trypsin digestion of crude sample were normalized with Progenesis software, a procedure not directly applicable to samples obtained by ConA chromatography because the amount of bound material varied to the sample origin. The mass tolerance was set to 10 ppm for precursor ions and 20 ppm for fragments. The peptide identifications obtained with the search engine Byonic were validated if the two-dimensional Post Error Probabilities which include PSM FDR and protein FDR (2D PEP) (29) were lower than 0.01 (log 2D PEP >2). The following variable modifications were allowed: oxidation (M), phosphorylation (ST), acetylation (K) (2 by peptide maximum in the parameter rare modifications). An additional glycan database was used, containing the N-glycosylations (N) and O-glycosylations (ST) described in yeast and making it possible to investigate 12 O-glycosylation subtypes (from 1 to 12 mannose residues) and 14 N-glycosylation subtypes (1 N-acetylhexosamine or 2 N-acetylhexosamine with 0 to 12 mannose residues) simultaneously in the various data sets (one modification by peptide maximum in common modifications). The maximum number of missed cleavages was limited to two for all the proteases used. MS/MS data were used to search a C. albicans ORF database (C_albicans_SC5314_A21_current_orf_trans_all.fasta, 6221 entries, 06–09-2014) retrieved from the Candida Genome Database website (30) (http://www.candidagenome.org/). Between-subject analyses were carried out on the various data sets. Nonnormalized abundances of the proteins eluted form ConA columns and normalized abundances of proteins from trypsin digests of crude sample with similar normalized abundance variations (ANOVA p value < 0.05) were classified together by the AutoClass Bayesian clustering system (webserver Autoclass@IJM, http://ytat2.ijm.univ-paris-diderot.fr/) (31) and visualized with Javatreeview (http://jtreeview.sourceforge.net/). Proteins were considered to be putatively glycosylated if they were more abundant in the ConA column eluate than in the crude sample. Gene Ontology analyses were performed on each cluster in which putative glycosylated proteins were overrepresented and more abundant in mca1Δ/Δ-F conditions than in other conditions. These Gene Ontology analyses were carried out with the GO Slim mapper tool from the Candida Genome Database website. Similar GO analyses were performed for all the glycosylated proteins found in the various data sets.
Determination of N-glycosylation Sites After Treatment With the Endoglycosidase H (Endo H)
We digested 20 μg of crude protein samples from the different conditions (WT, WT-farnesol, mca1Δ/Δ and mca1Δ/Δ-farnesol) by the Endo H (2500 U for 20 μg; Promega) overnight at pH7.8 in 20 μl of a 50 mm NH4HCO3 buffer according to the manufacturer's instructions. Then deglycosylated proteins were digested by trypsin (12.5 μg/ml; Promega) overnight. Peptides obtained were analyzed in LC-MS/MS with an Orbitrap Fusion Tribrid mass spectrometer coupled to a Nano-LC Proxeon 1000 equipped with an easy spray ion source (all from Thermo Scientific). Parameters used for the chromatographic separation and for the mass spectrometry analyses were identical to those described in the method tab LC-MS/MS acquisition of peptide samples. The Proteome Discoverer 1.4 node of the Byonic software was used to analyze the data. The mass tolerance was set to 10 ppm for precursor ions and 20 ppm for fragments. The peptide identifications obtained with the search engine Byonic were validated if the 2D PEP were lower than 0.01 (log 2D PEP >2). We searched for variable modifications: oxidation (Met), phosphorylation (Ser, Thr, Tyr), acetylation (Lys) and HexNAc (Asn). This last modification allowed us to identify N-glycosylation sites if just one Asn or one consensus site was present in a peptide.
Evaluation of Consensus Sites for N-glycosylation
The N-glycosylation site contexts of the identified glycosylated peptides from the different data sets (trypsin digests of crude samples, released proteins, and peptides released from ConA columns, crude samples digested by Endo H and trypsin) were investigated according to the different cell conditions. O-glycosylations site contexts were not investigated because most of the site localization were ambiguous because of the HCD fragmentation mode used. Five amino-acids on the N- and C-terminal sides of the modification sites were selected. Extended modification specificities other than the consensus motif Asn-X-Ser/Thr were investigated with IceLogo (https://code.google.com/p/icelogo/) (32), using the natural abundance of amino acids in the C. albicans protein sequence database (C_albicans_SC5314_A21_current_orf_trans_all.fasta, 6221 entries, 06–09-2014).
Search for New Potential Substrates of Mca1p
We digested 10 μg of proteins after glycopeptide enrichment by ConA chromatography, for each set of conditions, with the GluC protease (12.5 μg/ml; Sigma Aldrich, St-Louis, MO) overnight at 37 °C in 20 μl of 20 mm phosphate buffer pH7.8. MS/MS data were processed with the Byonic search engine and Proteome Discoverer 1.4 (Thermo Scientific), using the same parameters as above, except for the enzyme parameter which was set to GluC and semi-specific. Peptides identified exclusively in WT-F conditions, in at least two of three replicates, were selected for further analysis. For substrate validation, label-free, between-subject approaches were applied to the GluC data set, with Progenesis QI. The Mca1p-specific cleavage products identified were considered validated if more abundant in WT-F conditions than in the other conditions. ANOVA p values below 0.05 after label-free analysis were used as a criterion for the selection of the best potential substrates. GO analyses were performed on the list of potential substrates with the GO Slim Mapper tool from the Candida Genome Database website. Variation probabilities were calculated with R software for each GO term.
SDS-PAGE
We subjected 20 μg of crude protein samples from the different conditions to SDS-PAGE in a 4–12% NUPAGE gel (Invitrogen, Carlsbad, CA), which were then stained with Gel code Coomassie blue solution (Thermo Scientific). For each set of conditions, the fractions (equal volumes loaded) enriched in glycosylated protein by ConA chromatography support were run on the same gel.
Western Blot Analyses
We subjected 30 μg of crude lysate proteins digested or not by Endo H (2500 U for 20 μg; Promega) from the different conditions (WT, WT-F, mca1Δ/Δ, mca1Δ/Δ -F) to electrophoresis in a NUPAGE 4–12% acrylamide gel (Invitrogen) and transferred the protein bands obtained to a nitrocellulose membrane (Protran BA85, GE Healthcare Life Sciences). The abundance and glycosylation status of the Ssb1p and Cdc48p proteins were assessed by immunoblotting with anti-Ssb1p and anti-Cdc48p polyclonal rabbit primary antibodies. We also used an anti-Bgl2p antibody (33) to probe metacaspase-dependent degradation events on this protein. An anti-3-Pgk1p antibody (rabbit polyclonal IgG fractions, Nordic Immunology, Tillburg, The Netherlands) was used to control protein load in the different lanes. All the primary antibodies were raised against the proteins from S. cerevisiae. We used an anti-rabbit horseradish peroxidase-conjugated secondary antibody (Sigma Aldrich) and enhanced chemiluminescence reagents (West Pico and Femto, Thermo Scientific) for detection of immunoreactive material. ConA-HRP (Sigma Aldrich) was used on the same extracts (5 μg each) processed with the same conditions of migration, transfer and detection.
Quantitative Analysis of the Released Glycans by TMT-labeling
Glycan release and labeling with TMT reporters were performed in accordance with the general instructions for the aminoxy-TMT sixplex kit (Thermo Scientific). We digested 1 mg of sample from each set of conditions with trypsin in 100 μl of 20 mm triethylammonium bicarbonate (TEAB) buffer overnight at 37 °C. We added 5 μl of the glycosidase PNGase F (Prozyme, Velizy-Villacoublay, France) and the samples were incubated for 16 h at 37 °C with continuous shaking. A conditioned Oasis (Waters, Milford, MA) column was then used to separate peptides from the released glycans. Glycan fractions for the WT, WT-F, mca1Δ/Δ and mca1Δ/Δ-F conditions were labeled with 126 (monoisotopic m/z = 126.12773), 127 (m/z = 127.1248 Da), 128 (m/z = 128.1344 Da), and 129 (m/z = 129.1315 Da) labeling reagents, respectively. Labeled glycans were washed in 95% methanol, in 10% acetone and then combined in a 1:1 by volume ratio. A new Oasis column was used to discard excess aminoxy-TMT reagents. The released glycans were analyzed on an Orbitrap Fusion Tribrid mass spectrometer coupled to a Nano-LC Proxeon 1000 and equipped with an easy spray source (all from Thermo Scientific). Peptides were separated by chromatography with the following parameters: Accucore Amide HILIC column (15 cm, 75 μm i.d., 2.6 μm, 150 Å), 300 nl/min flow rate, gradient from 95% solvent B (100% acetonitrile, 0.1% formic acid) to 40% solvent A (water, 0.1% formic acid) over 100 min followed by column regeneration for 20 min, giving a total run time of 2 h. Glycans were analyzed in two independent experiments, depending on the acquisition mass range (mass ranges of m/z 270–1000 and m/z 850–4000). In both cases, the instrument parameters were set as follows: full ion scan mode in the Orbitrap cell, at a resolution of 60,000 (at m/z 200) in positive mode, with an AGC target of 4 × 105. Fragments were obtained by higher-energy C-trap dissociation (HCD) activation with a collisional energy of 35% and a quadrupole isolation window of 1.6 Da. MS/MS data were acquired in the Orbitrap cell, in top-speed mode, with a total cycle of 3 s, a resolution of 15,000, an AGC target of 5 × 104, a dynamic exclusion of 50 s and an exclusion duration of 60 s. Precursor priority for MS/MS was highest charge state followed by most intense. Glycans with charge states from 1 to 8 were selected for MS/MS acquisition. The maximum ion accumulation times were set to 250 ms for MS acquisition and 120 ms for MS/MS acquisition in parallelization mode. Glycans were quantified and identified manually.
Data Availability
The complete sets of data are available as .raw files, Proteome Discoverer 1.4 .msf files and pep.xml files associated, label-free reports generated by Progenesis QI and from the Candida albicans protein sequence database, provided as a multiple fasta, in the PRIDE partner repository (34) under the identification number: PXD003677.
RESULTS
Strategy for the Analysis of Glycoproteome Variations and Glycan Composition
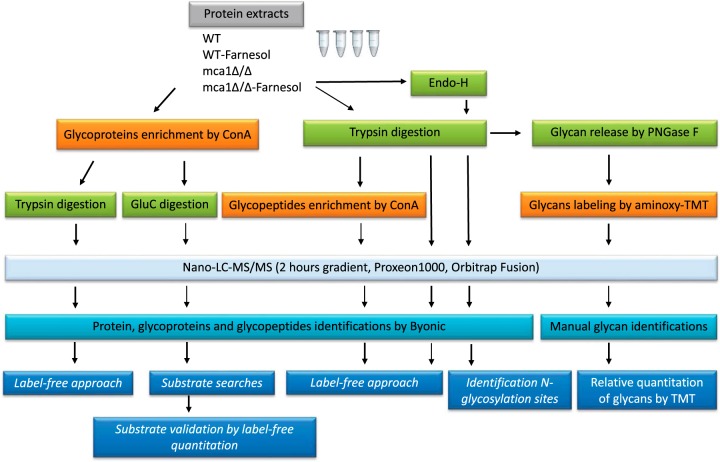
The general workflow used to generate the data sets for this study is shown in Fig. 1. Three data sets were used to characterize and quantify glycosylation, and one data set was used for the characterization of glycans. The complete data sets are provided in supplemental Files S1 to S4.
Fig. 1.
Study workflow.
Analysis of Putative Glycosylated Proteins After Enrichment by ConA Chromatography
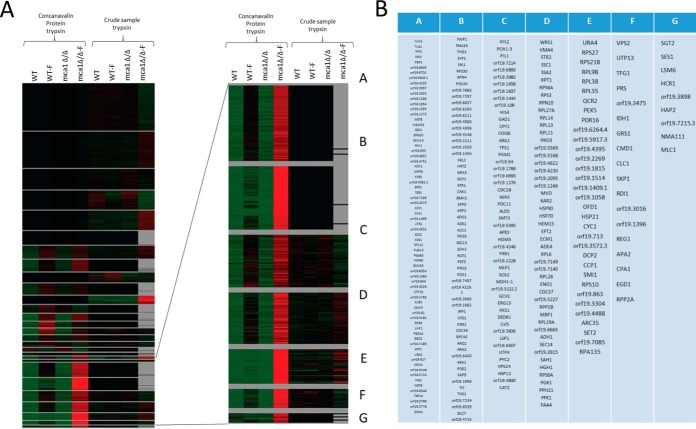
Quantitative label-free analyses were performed on two sets of samples digested with trypsin (samples enriched in glycosylated proteins by chromatography on ConA columns and crude samples), for set of cell conditions (WT, WT-F, mca1Δ/Δ, and mca1Δ/Δ-F). All the proteins identified in both sets and quantified with a significant (p < 0.05) ANOVA were selected. Relative abundances were then used to classify the proteins on the basis of abundance variations. We used AutoClass@ijm to cluster the proteins and Javatreeview to visualize the clusters obtained (Fig. 2). Proteins were identified as potentially glycosylated if they were more abundant in the sample from ConA columns than in the crude samples. An analysis of Gene Ontology terms showed that O-glycosylated proteins were more abundant than N-glycosylated proteins in the cell wall and that both types of glycosylated proteins were overrepresented in the plasma membrane. The “endoplasmic reticulum” term was more frequent for N-glycosylated proteins only in comparisons with the total data set. This finding is consistent with the occurrence of early steps of N-glycan biosynthesis in the endoplasmic reticulum, whereas O-glycosylation biosynthesis occurs mostly within the Golgi compartment (35). Cell cycle proteins were found to have particularly high levels of O- and N-glycosylation. All the proteins identified and quantified by LC-MS/MS are described in supplemental File S1 for proteins eluted from ConA columns and in supplemental File S2 for digested proteins from the crude sample.
Fig. 2.
Determination of putative glycosylated proteins by label-free proteomics and enrichment by ConA chromatography. A, Normalized abundances obtained by label-free analyses of trypsin digests of crude proteins and proteins eluted from ConA columns were used to classify proteins (with an ANOVA p value < 0.05) from the two data sets by the AutoClass Bayesian clustering system (webserver Autoclass@ijm). A higher abundance of the protein in the eluate from the ConA column than in the crude sample was considered to indicate that the protein was probably glycosylated. B, Putative glycosylated proteins from clusters defined in (A) with higher abundances in mca1Δ/Δ-F conditions.
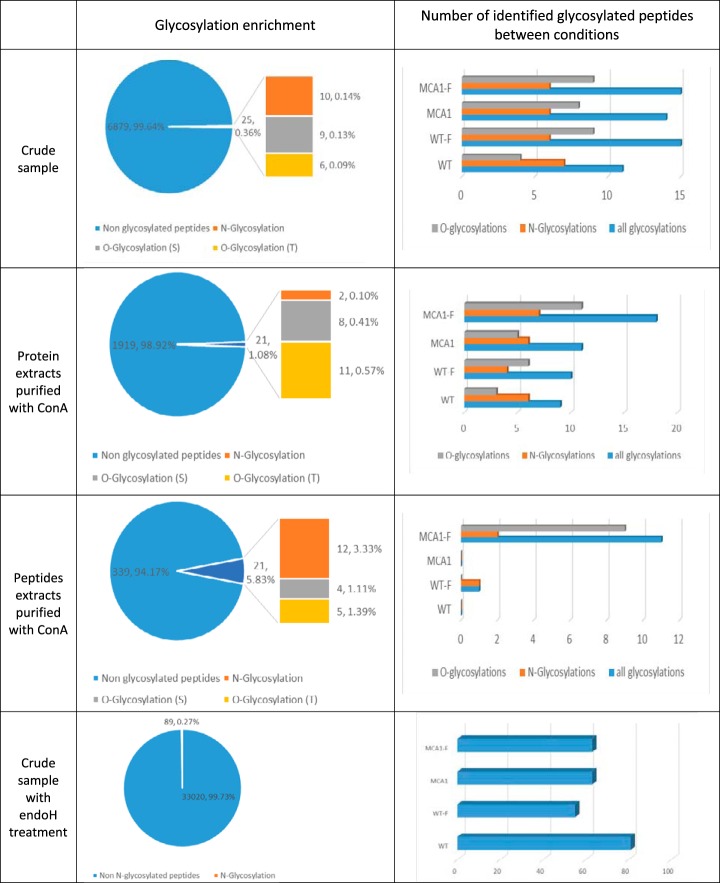
Evaluation and Comparison of Glycosylation Enrichment Methods
We compared three data sets for glycosylation enrichment (Fig. 3). In samples without enrichment, only 0.36% of the peptides (25 out of 6879) were identified as glycosylated by the Byonic search engine. The ConA chromatography of protein samples resulted in a 1.08% enrichment in glycosylated peptides with respect to the total peptide sample (21 out of 1919 peptides). It corresponded to 2 N-glycosylations, and 8 O-glycosylations on serine residues and 11 on threonine residues. Higher levels of enrichment were observed when ConA chromatography was used on samples digested with trypsin. This led to the identification of 21 glycosylated peptides (5.83%) among a total of 339 identified peptides. We detected 12 N-glycosylations, and 4 O-glycosylations on serine and 5 on threonine residues. Using the three data sets, we identified a total of 62 different glycosylated peptides (from 45 proteins), corresponding to 45 O-glycosylations and 17 N-glycosylations (supplemental Table S1 and supplemental Table S2). We identified also 81 additional sites of N-glycosylation with the data set using crude samples digested by Endo H and trypsin (supplemental Table S3). Among the 17 N-glycosylated peptides identified directly (without using Endo H) 8 were identified in the data set from the crude sample treated with Endo H with the same sites of N-glycosylation. This is a good indicator of the relevance of the proposed identifications. For crude samples, the number of glycosylated proteins was greater for WT and WT-F samples than for mca1Δ/Δ samples, consistent with the ConA-HRP signal detected on Western blots of proteins from the crude lysates. An increase in the number of O-glycosylated proteins in the presence of farnesol (WT and mca1Δ/Δ) was observed with proteins eluted from ConA columns. The ConA-HRP signal and Coomassie blue staining confirmed these changes (supplemental Fig. S1). An increase of the number of O-glycosylated proteins was also observed for peptides eluted from ConA columns for mca1Δ/Δ-F cells. Surprisingly, more N-glycosylations than O-glycosylations were observed with eluted proteins from ConA column in contrast to what is measured with eluted peptides from ConA columns. The proteins released from ConA columns were identified and quantified, and the data are presented in the supplemental File S3.
Fig. 3.
Evaluation of enrichment methods for glycosylated peptides and proteins according to the type of glycosylation and cell conditions. Glycosylation was compared between proteins from crude samples, proteins and peptides eluted from ConA columns and crude samples digested by Endo H, according to the nature of the glycosylation (N- or O-glycosylations on Asn and Ser or Thr residues, respectively) and the cell conditions (WT, WT-F, mca1Δ/Δ and mca1Δ/Δ-F). Glycosylated peptides were identified with the Byonic search engine.
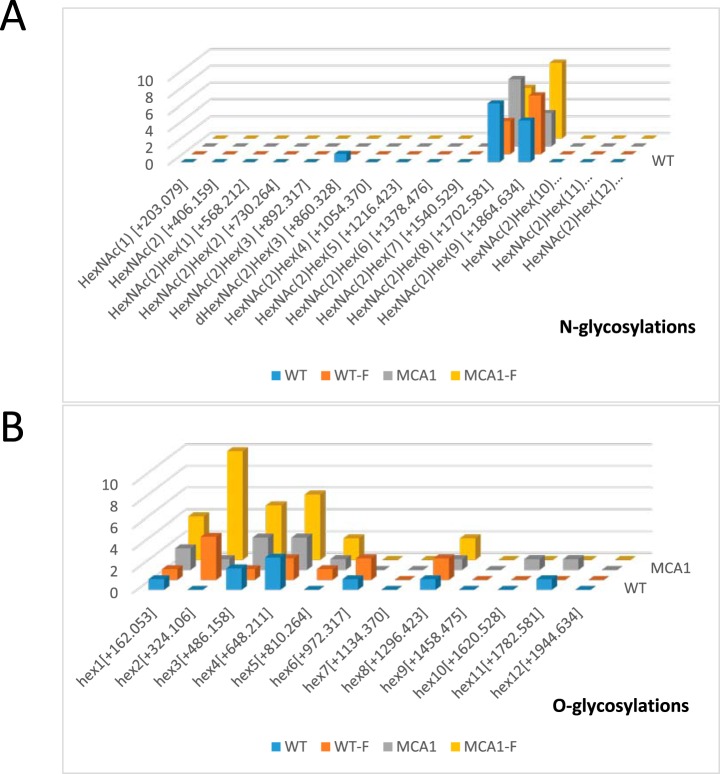
Determination of the Occurrence Rates of the Identified N- and O-glycosylations
The O- and N-glycosylations identified in all data sets were compiled as a function of their mannan composition. We observed no increase in any particular subtype for N-linked glycans (Fig. 4A). No change in Man7-(HexNAc)2 content was observed, even though this compound has been identified as a degradation signal in the ERAD system (36, 37). This molecule does not therefore appear to be a signal inducing apoptosis in response to farnesol. We observed a large increase in O-glycosylation in mca1Δ/Δ-F extracts. The oligomannans characterized were significantly enriched in Man2, Man3, and Man4 chains (Fig. 4B).
Fig. 4.
Occurrence of N- and O-glycosylation subtypes as a function of cell conditions. Occurrence of glycosylation subtypes (N-glycosylations in A and O-glycosylations in B), identified with the Byonic search engine, in all data sets (proteins from crude samples, proteins and peptides eluted from ConA columns) were pooled and compared as a function of cell conditions (WT, WT-F, mca1Δ/Δ and mca1Δ/Δ-F).
Evaluation of Consensus Sites for N-glycosylations
The sites of N-glycosylation were investigated by analyzing the sequences surrounding the glycosylated amino acid residues (supplemental Fig. S2). For N-glycosylation, the Asn-X-Ser/Thr motif was imposed by the use of the Byonic search engine. This motif is relevant, given that 97% to 99% of N-glycosylation sites contain this canonical motif (23). We observed the same consensus when the data sets from the Endo H treated samples were searched using Mascot, and only the modification of an Asparagine residue by HexNAc specified. Overall, these results indicated that there was no modification to the profile of N-glycosylation sites, indicating that there was no aberrant glycosylation under farnesol treatment or apoptosis release.
Analysis and Quantitative Assessment of the Glycosylated Peptides Identified in the Various Data Sets
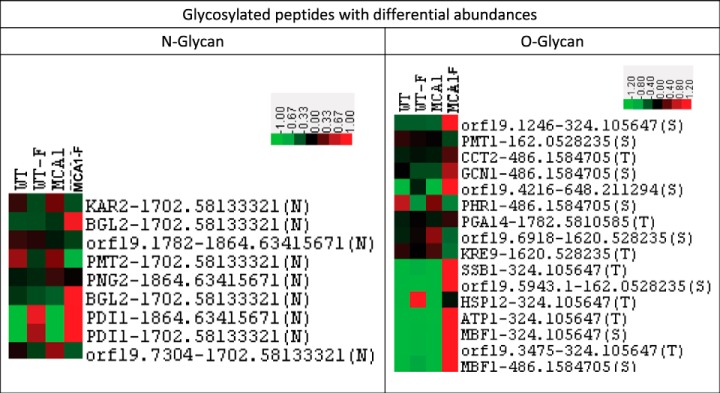
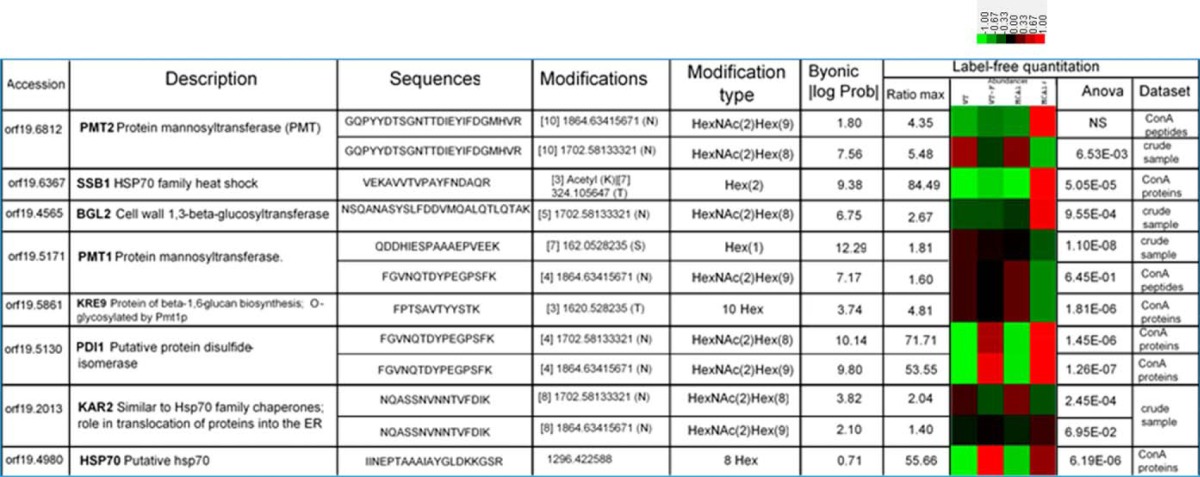
We also identified glycosylated peptides directly in our three data sets (supplemental Table S4). Quantitative label-free analyses were performed on the three data sets for which trypsin was used (glycosylated peptide and protein enrichments by ConA chromatography and crude samples), for the four sets of cell conditions (WT, WT-F, mca1Δ/Δ, and mca1Δ/Δ-F). All the identified and quantified glycosylated peptides with ANOVA p values below 0.05 were selected and the changes in their abundance were visualized with Javatreeview software. The O-glycosylated peptides with differential abundances in the three data sets were mostly overrepresented in mca1Δ/Δ-F cells (Fig. 5). This pattern was not observed for N-glycosylated peptides, which had less homogeneous abundances variations. Many glycosylated proteins, and O-glycosylated proteins in particular, were implicated in the cell cycle (Table I). The Gene Ontology term “transport” was underrepresented in N-glycosylated proteins, whereas the term “protein folding” was overrepresented, consistent with the involvement of this type of glycosylation in the ERAD system of glycosylated protein folding. Most of the O-glycosylated proteins overexpressed in mca1Δ/Δ-F cells were present in the plasma membrane or cell wall. Proteins involved in glycosylation processes, such as Pmt1p, Pmt2p, Phr2p and Kre9p, were themselves glycosylated. N-glycosylations of the protein mannosyltransferases Pmt1p and Pmt2p, which are involved in the O-glycosylation process, were less abundant in mca1Δ/Δ-F cells (Table II). Pdi1p, a major protein of the ERAD system for degrading misfolded glycoproteins, was also N-glycosylated and farnesol increased the abundance of its glycosylated peptides in a Mca1p-independent manner.
Fig. 5.
Label-free relative quantification of N- and O-glycosylated peptides as a function of cell conditions. Abundances of quantified glycosylated peptides for which ANOVA p values below 0.05 were obtained were represented with the Javatreeview tool, for all data sets (proteins from crude samples, proteins and peptides eluted from ConA columns) and cell conditions (WT, WT-F, mca1Δ/Δ and mca1Δ/Δ-F).
Table I. Gene ontology term enrichment in the different data sets, computed for (1) the type of glycosylation (N- or O-glycosylation) (2) proteins with differentially glycosylated peptides (according to the label-free experiment) and (3) proteins with higher abundances of O-glycosylated peptides in mca1Δ/Δ-F conditions than in the other conditions.
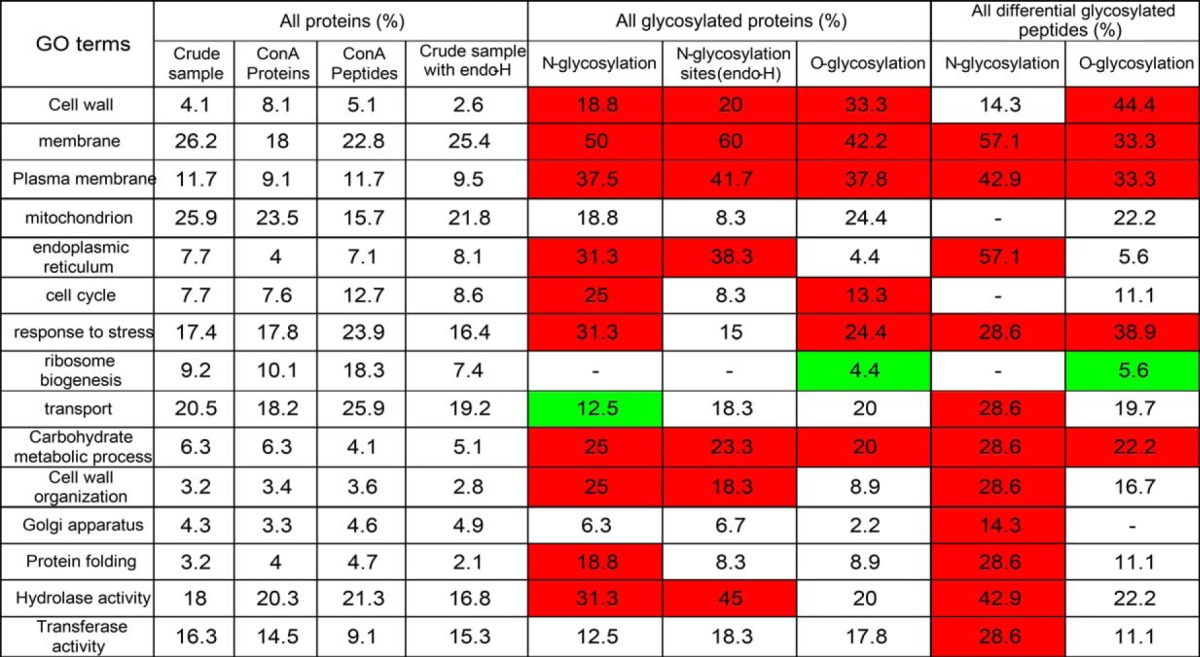
Table II. Examples of the glycosylated peptides identified and their label-free quantification, for the different cell conditions. Glycosylated peptides were identified with the Byonic search engine and differences in peptide levels for all datasets were quantified in label-free experiments. All the glycosylated peptides identified are listed in supplemental Table S1 and S2 and their quantification in the various cell conditions is detailed in the supplemental Table S4.
The Potential Substrates of Mca1p are Proteins Involved in the Regulation of Glycosylation
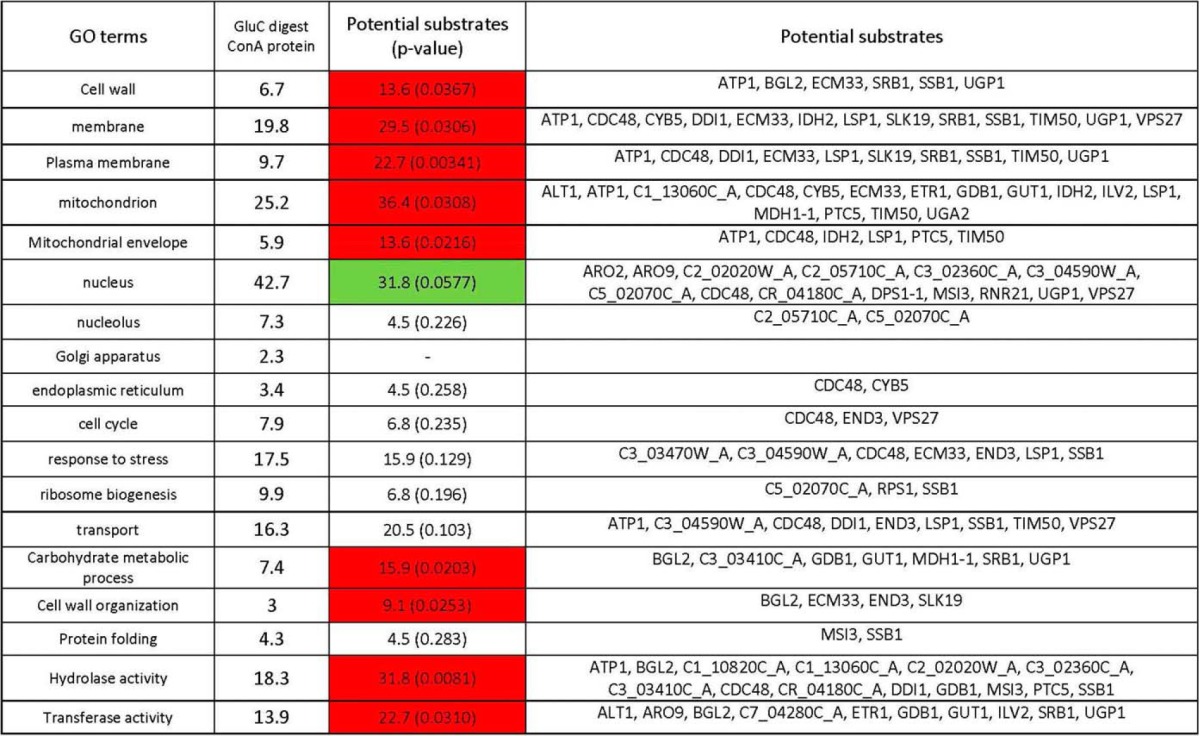
Proteins eluted from ConA columns were digested with the GluC protease. Peptides with K or R ends identified in at least two of three replicates in WT-F conditions only were considered to be potential substrates of Mca1p. Potential substrates were validated by label-free quantification. Potential substrates with the abundance ratio of cleavage signature peptide greater than 2 in WT-F condition relative to the other conditions for which a p value ANOVA below 0.05 was obtained were considered to be potential substrates with a high level of confidence. In these conditions, we identified 49 highly probable substrates for 44 proteins (supplemental Table S5), some of which, including cdc48p and the chaperone Ssb1p, had already been identified in a previous study (19). We identified other potential substrates involved in glycoprotein biosynthesis and regulation, such as Srb1p, Bgl2p, Ugp1p, Ecm33p, Gdp1p, and orf19.338 (putative glycoside hydrolase), providing a possible explanation for the direct involvement of the metacaspase in the regulation of glycosylation (Table III). An analysis of Gene Ontology terms for potential substrates showed an increase in mitochondrial proteins (Tim50p, Cdc48p, Atp1p, Idh2p), as in our previous study, but also enrichments in the terms “cell wall,” “membrane,” “plasma membrane,” or “cell wall organization,” indicating a direct action of the metacaspase on these particular compartments (Table IV). The metacaspase targeted proteins involved in carbohydrate metabolism in particular. All the peptides identified and quantified after the GluC digestion of proteins eluted from ConA columns are described in supplemental File S4.
Table III. Examples of candidate substrates identified by the analysis of proteins eluted from ConA columns, digested with the GluC enzyme and validated by the label-free approach. Peptides were considered to be putative substrates if they were identified in at least two of the three replicates in WT-F conditions only and had a K or R amino-acid residue at one end. Putative substrates were validated by a label-free approach and those with an abundance higher in WT-F conditions than in other conditions (with a ratio greater than 2) and an ANOVA p value < 0.05 were considered to be highly probable substrates.
| Accession | Description | Sequences | Log Prob Byonic | Identifications (3 replicates) Byonic scores | Label-free quantitation |
|||
|---|---|---|---|---|---|---|---|---|
| Ratio WT-F/WT | Ratio WT-F/mca1 | Ratio WT-F/mca1-F | Anova | |||||
| orf19.6190 | SRB1; Essential GDP-mannose pyrophosphorylase; makes GDP-mannose for protein glycosylation | MKGLILVGGYGTR | 4.42 | 116.2, 232.4 | infinity | infinity | 26.4 | 0.0014 |
| orf19.4565 | BGL2; Cell wall 1,3-beta-glucosyltransferase; mutant has cell-wall and growth defects, but wild-type 1,3- or 1,6-beta-glucan content | GDLDFLKSHSK | 9.27 | 480.5, 391.7, 532.1 | 96.6 | 900.2 | 11.9 | 0.0009 |
| LASKINDIKDLVK | 8.2 | 497.5, 257.9, 547.9 | 96.5 | 29 | 6.1 | 0.015 | ||
| orf19.1738 | UGP1; UTP-glucose-1-phosphaturidyl transferase; localizes to yeast, not hyphal cell surface; Hog1-repressed | HGALVLDPTRDGFSNPLIKLGSHFK | 10.07 | 447.9, 340.3, 546.1 | 277.5 | 140 | 16.7 | 0.008 |
| orf19.3010.1 | ECM33; GPI-anchored cell wall protein; mutants show cell-wall defects and reduced adhesion, host cell damage, and endocytosis | RVSGGFILKTD | 3.36 | 321, 219.4, 229.8 | Infinity | Infinity | 18 | 1.1E-08 |
| LTGVQQIYGSLHVK | 7.83 | 313.1, 435, 363.2 | 31.1 | 21.8 | 13.9 | 0.0002 | ||
| orf19.744 | GDB1; Putative glycogen debranching enzyme | KALGAIIIADK | 7.04 | 399.5, 403.4 | 22.3 | 46.3 | 5.3 | 6.9E-5 |
| orf19.338 | Uncharacterized ORF; Putative glycoside hydrolase; stationary phase enriched protein; Hog1p-downregulated | YFDGDSGRGLGASHQTGWTSLVTK | 6.49 | 39.1.3, 356.9, 375.1 | 103.6 | 28.6 | 5.4 | 1.42E-6 |
| orf19.2340 | CDC48; Putative microsomal ATPase; plasma membrane-localized | LVELPLRHPQLFK | 8.06 | 470.3, 458.1, 473.4 | Infinity | 237.5 | 15.1 | 0.0003 |
| orf19.6367 | SSB1; HSP70 family heat shock protein | VVLVGGSTRIPKVQKLLSDFFDGK | 3.52 | 120.8, 174.3, 87.3 | 316.5 | 33.4 | 8.1 | 0.003 |
Table IV. Gene ontology term enrichment of potential substrates relative to all identified proteins obtained by GluC digestion of the proteins eluted from ConA columns. A hypergeometric distribution was calculated for each GO term.
Validation of the Induction of Ssb1p and Cdc48p Glycosylation in the mca1Δ/Δ-F Extract
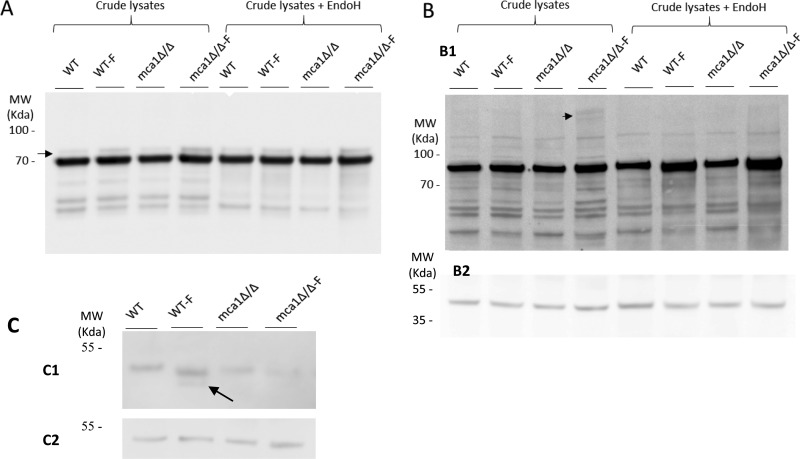
The glycosylation of the Cdc48p and Ssb1p proteins was demonstrated indirectly, based on the increase in their abundances following ConA chromatography (in mca1Δ/Δ-F conditions) with respect to the crude sample, and directly for Ssb1p, through the identification and quantification of glycosylated peptides. Immunoblotting with antibodies directed against Ssb1p and Cdc48p revealed an increase in the abundance of these proteins in mca1Δ/Δ-F conditions, as already reported in our previous study for Ssb1p only (19). Bands with higher molecular weights than Ssb1p and Cdc48p were observed, indicating a possible increase in glycosylation in mca1Δ/Δ-F condition (Fig. 6A and 6B), consistent with the results of the LC-MS/MS study presented here. A much larger mass shift was observed with Cdc48p than with Ssb1p. This is consistent with our indirect demonstration of putative N-glycosylation for Cdc48p and our direct demonstration of Ssb1p O-glycosylation by LC-MS/MS. These findings suggest that the glycan moiety associated with Cdc48p is probably very large and out of the mass detection range of the mass spectrometer used.
Fig. 6.
Validation of the induction of Ssb1p and Cdc48p glycosylation in the mca1Δ/Δ-F extract and immunocharacterization of a putative substrate of Mca1p. The abundances of Ssb1p (A) and Cdc48p (B) were assessed in immunoassays, as a function of cell conditions (WT, WT-F, mca1Δ/Δ and mca1Δ/Δ-F) in samples treated or not by Endo H. Arrows indicate immunoreactive material of the higher mass, attributed to some glycosylated forms of the proteins. Those were Endo H insensitive (Ssb1p, panel A) or Endo H sensitive (Cdc48p, panel B). Protein loading are probed using anti-Pgk1 antibodies (B2). C, Immunodetection of Bgl2p in the different experimental conditions. The arrow points to a degradation product observed only in the WT-F condition. Control of protein loading using the anti-Pgk1p antibody is presented in C2.
Immunocharacterization of a Putative Substrate of Mca1p
A degradation product of the Bgl2p protein was highlighted by immunoblot (Fig. 6C) only in the WT-F condition, the sole condition with Mca1p activated and with apoptosis release. The electrophoretic mobility of the immunodetected protein (apparent Mr 45 kDa versus 33.6 kDa theoretical) is suggestive of some post-translational modification of the protein. Indeed, the Bgl2p protein is known to be N-glycosylated, and we observed a N-glycosylated peptide from Bgl2p in our proteomics data of sequence K.NSQAN[+1702.581]ASYSLFDDVMQALQTLQTAK (supplemental Table S1).
Changes in Glycan Composition are Mediated by farnesol and mca1 Disruption
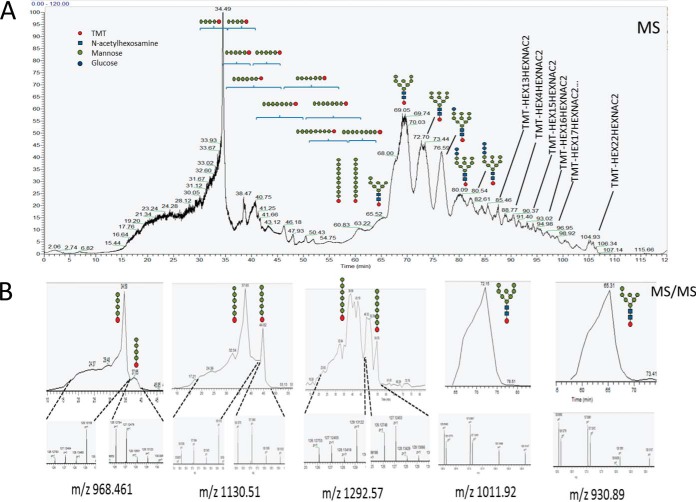
Glycans released from protein samples in the four sets of conditions (WT, WT-F, mca1Δ/Δ, mca1Δ/Δ-F) were labeled with aminoxy-TMT reagents and analyzed by LC-MS/MS. N-glycans were detected, as expected, by a combination of PGNase F glycosidase and trypsin digestion. However, TMT-labeled O-glycans were also found in the samples. We investigated whether these unexpected O-glycans were free glycans or whether they were released from the proteins by the PGNase F or trypsin, or by the TMT reagents. Protein extracts from mca1Δ/Δ-F cells were incubated with PNGase F after trypsin digestion, with trypsin alone and in the absence of these two enzymes. The glycans were then purified and analyzed by LC-MS/MS, as described above. High levels of enrichment in oligomannosides were found only after combined digestion with PNGAse F and trypsin, indicating that this treatment released O-glycans efficiently from proteins (File S5). By contrast to what was observed for O-glycans, N-glycan levels differed little between the various conditions. Indeed, all the O-glycan subtypes identified displayed similar variations in abundance. Structural analyses of O-glycan subtypes revealed a small increase in a certain subpopulation in WT-F conditions and a large increase in that subpopulation in mca1Δ/Δ-F conditions, whereas a small decrease, similar to that for N-glycans, was found for another population in the mca1 disruption context (Fig. 7 and supplemental Fig. S3). The variations of O-glycans abundancies were similar to those found for those of the glycosylated peptides, with an increase in mca1Δ/Δ-F for both glycans and glycopeptides. The oligomannosides molecules detected would therefore probably correspond to glycans released from serine or threonine residues. All the representative annotated spectra obtained for the glycans identified and TMT-quantified are presented in supplemental Table S6.
Fig. 7.
Relative quantification by TMT-labeling of the glycans released, for the four experimental conditions. Glycans were released from trypsin digests of cell lysates for the four sets of conditions (WT, WT-F, mca1Δ/Δ, and mca1Δ/Δ-F), by incubation with the englycosidase PNGase F. A, Total Ion current (TIC) chromatogram and localization of the major glycans identified in the separation gradient for the amide-HILIC phase. B, Examples of glycans quantified with the aminoxy-TMT reagents. Labels 126, 127, 128, and 129 correspond to WT, WT-F, mca1Δ/Δ, and mca1Δ/Δ-F conditions, respectively. Mass spectrometry acquisition was performed in triplicate with a mass range in MS of m/z 250 to 1000 and for m/z 850 to 4000. The results for the second mass range are shown in supplemental Fig. S2. Details of the quantification are provided in supplemental Table S6.
DISCUSSION
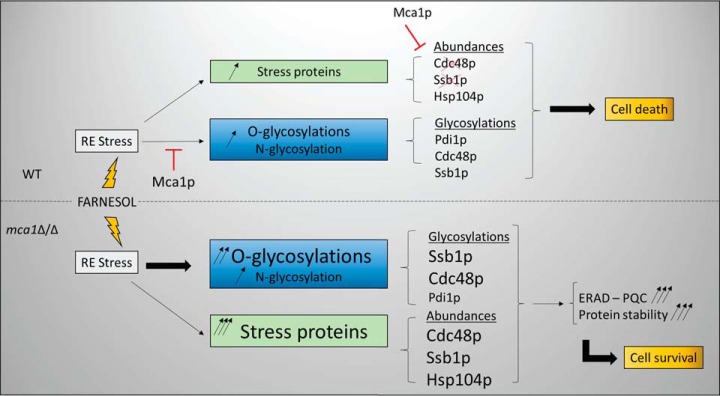
Recent proteomics studies have identified many N-glycosylation sites in diverse species, including budding yeast. These studies generally make use of enrichment procedures based on sugar-specific lectin chromatography, the cleavage specificity of various glycosidases, including PNGase F with H2 18O (23) and EndoH (21) and/or chemical reagents, such as boronic acid (38). These approaches provide valuable information about glycopeptides but little insight into structural aspects of protein glycosylation, concerning glycan composition and the stereospecificity of sugar bonding, for example. Such information is important, to determine the role of these specific modifications in various biological processes, such as apoptosis. For example, we lack precise informations about the nature of the transition from Man8-(HexNAc)2 to Man7-(HexNAc)2, which has been recognized as a signal for the clearance of unfolded proteins from the endoplasmic reticulum (8). We investigated O-glycosylated and N-glycosylated protein content by quantitative glycoproteomics approaches, with the aim of deciphering the mechanisms underlying metacaspase-dependent farnesol-induced apoptosis in C. albicans. One limitation of this study concerned the sizes of the glycan moieties accessible to mass spectrometry measurement. This made it impossible to address changes in the complexity of the large, branched oligomannosyl outer chains described for many exocellular glycoproteins. Our sample preparation method was not designed to conserve the parietal components in the cell-free extracts (most of the cell-wall proteins were removed by the low-speed centrifugation used to eliminate unbroken cells and large debris from the samples). Most of the proteins analyzed were therefore derived from the internal soluble and membrane-bound compartments. Using the Byonic search engine (28), we were able to carry out several different analyses for 26 glycosylation subtypes simultaneously. This would not have been possible with most standard database search engines. This approach proved effective, as we were able to identify 62 glycosylated peptides (in 45 proteins), 17 of which were N-glycosylated and 45 of which were O-glycosylated. We identified also 81 additional sites of N-glycosylation in crude samples digested by Endo H and trypsin. Previous proteomics studies characterized only N-glycosylation sites, 135 (39) and 516 (23) of which were identified in S. cerevisiae. Interestingly, we observed differential N- or O-glycosylation of several proteins involved in the general machinery of protein glycosylation, such as Pmt1p, Pmt2p, suggesting that feedback control loops occur in this process. We investigated whether the apoptosis process or a lack of Mca1p could reveal aberrant glycosylation sites. We found that glycosylation sites displayed an enrichment in proline residues in the P1′ position, and also in the P2′ and P1 positions for O-glycosylation; this finding is consistent with those deduced from the statistical analysis of glycosylation sites in a database for O-glycosylated proteins (10). For N-glycosylation, we observed no particular enrichment in specific amino acids around the glycosylation sites. Characterization of the specific substrates of the metacaspase Mca1p remains limited. We used a glycoprotein enrichment procedure to identify new potential substrates. An analysis of peptides eluted from ConA columns and digested with the GluC endoprotease identified 49 peptides for 44 proteins as potential substrates of the metacaspase, based on the most stringent selection parameters. These new substrates can be added to the list of 77 putative substrates (14 with the most stringent filters) that we identified in previous studies (19). Some of these substrates were differentially expressed in the samples analyzed. It was therefore important to distinguish between differences in protein abundance and differences in glycosylation status between samples. We carried out label-free protein quantification and glycan quantification to address this question. We observed an increase in O-glycosylation with mannose moieties with low levels of branching in apoptotic conditions (WT-F condition), this increase being accentuated in the context of mca1 disruption. The label-free quantification of O-glycosylated peptides from our various data sets revealed differences in peptide abundant, with some peptides more abundant in the mca1Δ/Δ-F protein extract. As the signal triggering apoptosis is abolished in mca1Δ/Δ-F cells, this increase in O-glycosylation may correspond to an attempt to improve protein folding to escape apoptosis. It may also be interpreted as a degradation signal for the efficient removal of unfolded proteins, thereby preventing the accumulation of misfolded proteins systematically associated with apoptosis. In N-glycosylated proteins, the occurrence of a glycan moiety with fewer mannose residues than in Man8-(HexNAc)2 could be considered as a signal for the targeting of the protein to degradation pathways, or as a reflection of a kinetic process of protein modification occurring during degradation. However, as we did not observe the induction of large numbers of N-glycosylation subtypes by farnesol treatment and mca1 disruption, this signaling pathway appears to be marginal in the experimental conditions used (early phase of apoptosis induction). As O-glycosylation is a sequential process, the occurrence of oligomannosides with small numbers of mannose residues may also reflect the kinetics of mannan transfer to targeted proteins. These forms may be intermediates in the synthesis of longer oligomannoside chains. The ConA enrichment procedure for glycopeptides clearly revealed differences in the abundances of specific peptides correlated with growth conditions or genetic background. However, these differences may reflect differences in protein abundance, or glycan composition and/or abundance. We therefore performed a quantitative analysis of the N-linked glycan moieties released by PNGase F and labeled with the aminoxy-TMT reagents. The quantification of TMT reporters highlighted the dependence of the abundances of some glycans on the biological conditions tested. An analysis of the fragmentation spectra of the glycans separated with a hydrophilic chromatographic support made it possible to distinguish a number of structural variants of oligosaccharides with the same mass and very similar retention times. Some of these variants were branched oligomannosides. Specific structure-dependent differences in the abundances of these oligosaccharides were observed, with higher abundances in WT-F conditions than in WT or mca1Δ/Δ conditions, and much more abundant in mca1Δ/Δ-F conditions. It is puzzling to label and quantify these oligomannans chains because the aminoxy-TMT reagent has been described as reacting with the reducing end of the sugar released by PNGase-F treatment. We quantified the N-glycans released by this enzyme and found that unexpectedly large amounts of O-mannosides were released during the experiment. The control experiments excluded the possibility of these O-glycans being free glycans present in the initial samples. The precise mechanisms involved in this atypical reactivity of aminoxy-TMT are currently being investigated. Our data indicate that the activity of the key enzymes involved in the synthesis of the various types of oligomannoside chains is subject to differential control, and we provide evidence that the metacaspase Mca1p plays a direct role in determining the glycan composition and glycosylation profile. Indeed, Mca1p directly targeted proteins involved in glycan biosynthesis, such as the GDP-mannose pyrophosphorylase Srb1p, the deletion of which causes N- and O-glycosylation deficiency in C. albicans (40), and impairs cell wall integrity (41–43) and cell growth in S. cerevisiae (as Psa1p) (40); the UDP-glucose pyrophosphorylase Ugp1p, which has been implicated in N-linked glycan biosynthesis and the formation of beta-glucans and glucomannoproteins in S. cerevisiae (44) and the 1,3-beta-glucosyltransferase Bgl2p that is validated here as a substrate of Mca1p by immunoblot. The Bgl2p protein is essential to the maintenance of the cell wall integrity and is involved in the incorporation of newly synthetized mannoproteins into the cell wall (45). The phosphomannomutase Pmm1p (46), which is homologous to the Sec53p protein in S. cerevisiae, was also identified as a putative substrate of Mca1p in our previous study; this enzyme converts mannose-6-phosphate into mannose-1-phosphate. This last glycan is essential for the generation of GDP-mannose, which is required for the elongation of mannose chains and, thus, for O- and N-glycosylations (47). We previously showed that Mca1p degrades Cdc48p, Ssb1p and other chaperones involved in protein folding in apoptotic conditions, resulting in an increase in the abundance of these proteins in mca1Δ/Δ-F conditions. This study provides the new finding that both proteins are more glycosylated in mca1Δ/Δ-F conditions. This was confirmed by both the LC-MS/MS analysis (indicating a high level of O-glycosylation of Ssb1p) and by immunoblotting. The ConA-affinity enrichment step led to the identification of new putative substrates of the metacaspase, such as Msi3p, another chaperone involved in the cell cycle, the deletion of which results in spindle elongation during S phase. Cdc48 has also been implicated in mitotic spindle disassembly (48), and Ssb1p is involved in the regulation of the mitotic cell cycle in S. pombe (as sks2 protein) (49). These results suggest that the metacaspase Mca1p plays a critical role in the regulation of cell cycle progression, as reported for nonapoptotic S. cerevisiae cells (50), in which the disturbance of cell cycle progression may lead to cell death. Many of the putative substrates of the metacaspase were located in mitochondria and in the mitochondrial envelope, as demonstrated in both this and our previous study, but some substrates were also found in the cell wall, membrane and plasma membrane. As expected, the glycosylated proteins identified and quantified were most present in or targeted to the cell wall compartment and the plasma membrane. More N-glycosylated proteins than O-glycosylated proteins were associated with the endoplasmic reticulum, as expected, because N-glycosylation is initiated in this compartment (51). We also found N-glycosylated proteins in the mitochondrial compartment, as previously reported (20–22). We hypothesize that the presence of these proteins in this compartment may result from the activity of the OST complex at the sites of contact between the mitochondria and the endoplasmic reticulum. We are currently testing this hypothesis by analyzing the proteome of MAM domains. We found that that the protein disulfide isomerase Pdi1p, another key component of the ERAD system, was hyper-N-glycosylated in the presence of farnesol, both with and without mca1 disruption. The N-glycosylation of this protein was reported in a previous proteomics study, but information was provided only about the glycosylation site, and in the absence of stress (39). The N-glycosylation of Pdia2, a protein disulfide isomerase, has been observed in human cells and was found to be essential for the dimerization of this protein (52). This modification may activate this protein, which connects control of the protein glycosylation pathways to the apoptotic stress response. These findings demonstrate the involvement of Mca1p in glycan and glycosylation regulation and show how cells abolish apoptosis in the absence of Mca1p and in stress conditions, by maintaining the correct functioning of several key mechanisms of the stress response, such as the ERAD, PQC or mitochondrial integrity pathways. Our results suggest that the high levels of O-glycosylation observed are instrumental to these processes (Fig. 8), and may significantly contribute to protection against the proteolytic activities amplified during apoptosis. These findings open up unexpected new possibilities for developing targeted inhibitors of C. albicans growth.
Fig. 8.
Proposed model for the interaction between Mca1p and the glycosylation machineries, ERAD and PQC systems, in the apoptosis release.
Supplementary Material
Acknowledgments
We thank Dr Véronique Albanese (IJM, Paris) for providing the anti-Ssb1p antibody, Dr Alexander Buchberger (Univ. Würzburg, Germany) for providing the anti-Cdc48p antibody, Pr Wei Guo (University of Pennsylvania, Philadelphia) for the gift of the anti-Bgl2p antibody, Junie Hovsepian and Sébastien Léon (IJM, Paris) for the anti-Pgk1p primary antibody. We are grateful to Sergei Snovida (Thermo Scientific) and Sanjib Meitei (Premier Biosoft) for helpful discussions and advices in MS-based glycan analyses. We thank Alex Edelman & Associates for correcting the English version of this text.
Footnotes
Author contributions: Conceived and designed the experiments: TL, JMC. Performed the experiments: TL, CM, JMC. Analyzed the data: TL, CM, JMC. Wrote the paper: TL, JMC.
* This work was supported by NSF grant MCB 1517986 to YFN-M, and NIEHS grant R01-ES024478 to YNF-M. A portion of this work was performed at the National High Magnetic Field Laboratory, which is supported by National Science Foundation Cooperative Agreement # DMR-1157490 and the State of Florida.
 This article contains supplemental material.
This article contains supplemental material.
Authors declare no conflict of interest.
1 The abbreviations used are:
- PMT
- protein mannosyltransferase
- ERAD
- ER-associated degradation
- ConA
- concanavalin A
- PQC
- protein quality control
- WT
- wild-type.
REFERENCES
- 1. Ohtsubo K., and Marth J. D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 2. Matthijs G., Schollen E., Pardon E., Veiga-Da-Cunha M., Jaeken J., Cassiman J. J., and Van Schaftingen E. (1997) Mutations in PMM2, a phosphomannomutase gene on chromosome 16p13, in carbohydrate-deficient glycoprotein type I syndrome (Jaeken syndrome). Nat. Genet. 16, 88–92 [DOI] [PubMed] [Google Scholar]
- 3. Mora-Montes H. M., Bates S., Netea M. G., Diaz-Jimenez D. F., Lopez-Romero E., Zinker S., Ponce-Noyola P., Kullberg B. J., Brown A. J., Odds F. C., Flores-Carreon A., and Gow N. A. (2007) Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot. Cell 6, 2184–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Albrecht A., Felk A., Pichova I., Naglik J. R., Schaller M., de Groot P., Maccallum D., Odds F. C., Schafer W., Klis F., Monod M., and Hube B. (2006) Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281, 688–694 [DOI] [PubMed] [Google Scholar]
- 5. Hall R. A., Bates S., Lenardon M. D., Maccallum D. M., Wagener J., Lowman D. W., Kruppa M. D., Williams D. L., Odds F. C., Brown A. J., and Gow N. A. (2013) The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans. PLoS Pathog. 9, e1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwarz F., and Aebi M. (2011) Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582 [DOI] [PubMed] [Google Scholar]
- 7. Poulain D., and Jouault T. (2004) Candida albicans cell wall glycans, host receptors and responses: elements for a decisive crosstalk. Curr. Opin. Microbiol. 7, 342–349 [DOI] [PubMed] [Google Scholar]
- 8. Gauss R., Kanehara K., Carvalho P., Ng D. T., and Aebi M. (2011) A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol. Cell 42, 782–793 [DOI] [PubMed] [Google Scholar]
- 9. Hall R. A., and Gow N. A. (2013) Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol. Microbiol. 90, 1147–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thanka Christlet T. H., and Veluraja K. (2001) Database analysis of O-glycosylation sites in proteins. Biophys. J. 80, 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gentzsch M., and Tanner W. (1996) The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 15, 5752–5759 [PMC free article] [PubMed] [Google Scholar]
- 12. Hart G. W. (1992) Glycosylation. Curr. Opin. Cell Biol. 4, 1017–1023 [DOI] [PubMed] [Google Scholar]
- 13. Petkova M. I., Pujol-Carrion N., and de la Torre-Ruiz M. A. (2012) Mtl1 O-mannosylation mediated by both Pmt1 and Pmt2 is important for cell survival under oxidative conditions and TOR blockade. Fungal Genet. Biol. 49, 903–914 [DOI] [PubMed] [Google Scholar]
- 14. Lommel M., Bagnat M., and Strahl S. (2004) Aberrant processing of the WSC family and Mid2p cell surface sensors results in cell death of Saccharomyces cerevisiae O-mannosylation mutants. Mol. Cell. Biol. 24, 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hirayama H., Fujita M., Yoko-o T., and Jigami Y. (2008) O-mannosylation is required for degradation of the endoplasmic reticulum-associated degradation substrate Gas1*p via the ubiquitin/proteasome pathway in Saccharomyces cerevisiae. J. Biochem. 143, 555–567 [DOI] [PubMed] [Google Scholar]
- 16. Prill S. K., Klinkert B., Timpel C., Gale C. A., Schroppel K., and Ernst J. F. (2005) PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55, 546–560 [DOI] [PubMed] [Google Scholar]
- 17. Harty C., Strahl S., and Romisch K. (2001) O-mannosylation protects mutant alpha-factor precursor from endoplasmic reticulum-associated degradation. Mol. Biol. Cell 12, 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hauptmann P., Riel C., Kunz-Schughart L. A., Frohlich K. U., Madeo F., and Lehle L. (2006) Defects in N-glycosylation induce apoptosis in yeast. Mol. Microbiol. 59, 765–778 [DOI] [PubMed] [Google Scholar]
- 19. Leger T., Garcia C., Ounissi M., Lelandais G., and Camadro J. M. (2015) The metacaspase (Mca1p) has a dual role in farnesol-induced apoptosis in Candida albicans. Mol. Cell. Proteomics 14, 93–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chandra N. C., Spiro M. J., and Spiro R. G. (1998) Identification of a glycoprotein from rat liver mitochondrial inner membrane and demonstration of its origin in the endoplasmic reticulum. J. Biol. Chem. 273, 19715–19721 [DOI] [PubMed] [Google Scholar]
- 21. Kung L. A., Tao S. C., Qian J., Smith M. G., Snyder M., and Zhu H. (2009) Global analysis of the glycoproteome in Saccharomyces cerevisiae reveals new roles for protein glycosylation in eukaryotes. Mol. Syst. Biol. 5, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burnham-Marusich A. R., and Berninsone P. M. (2012) Multiple proteins with essential mitochondrial functions have glycosylated isoforms. Mitochondrion 12, 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zielinska D. F., Gnad F., Schropp K., Wisniewski J. R., and Mann M. (2012) Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol. Cell 46, 542–548 [DOI] [PubMed] [Google Scholar]
- 24. Shirtliff M. E., Krom B. P., Meijering R. A., Peters B. M., Zhu J., Scheper M. A., Harris M. L., and Jabra-Rizk M. A. (2009) Farnesol-induced apoptosis in Candida albicans. Antimicrob. Agents Chemother. 53, 2392–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu J., Krom B. P., Sanglard D., Intapa C., Dawson C. C., Peters B. M., Shirtliff M. E., and Jabra-Rizk M. A. (2011) Farnesol-induced apoptosis in Candida albicans is mediated by Cdr1-p extrusion and depletion of intracellular glutathione. PloS One 6, e28830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilson R. B., Davis D., and Mitchell A. P. (1999) Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aerts A. M., Carmona-Gutierrez D., Lefevre S., Govaert G., Francois I. E., Madeo F., Santos R., Cammue B. P., and Thevissen K. (2009) The antifungal plant defensin RsAFP2 from radish induces apoptosis in a metacaspase independent way in Candida albicans. FEBS Lett. 583, 2513–2516 [DOI] [PubMed] [Google Scholar]
- 28. Bern M., Kil Y. J., and Becker C. (2012) Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics Chapter 13, Unit13 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bern M. W., and Kil Y. J. (2011) Two-dimensional target decoy strategy for shotgun proteomics. J. Proteome Res. 10, 5296–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Inglis D. O., Arnaud M. B., Binkley J., Shah P., Skrzypek M. S., Wymore F., Binkley G., Miyasato S. R., Simison M., and Sherlock G. (2012) The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res. 40, D667–D674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Achcar F., Camadro J. M., and Mestivier D. (2009) AutoClass@IJM: a powerful tool for Bayesian classification of heterogeneous data in biology. Nucleic Acids Res. 37, W63–W67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Colaert N., Helsens K., Martens L., Vandekerckhove J., and Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 33. He B., Xi F., Zhang J., TerBush D., Zhang X., and Guo W. (2007) Exo70p mediates the secretion of specific exocytic vesicles at early stages of the cell cycle for polarized cell growth. J. Cell Biol. 176, 771–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vizcaino J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Herscovics A., and Orlean P. (1993) Glycoprotein biosynthesis in yeast. FASEB J. 7, 540–550 [DOI] [PubMed] [Google Scholar]
- 36. Brodsky J. L. (2012) Cleaning up: ER-associated degradation to the rescue. Cell 151, 1163–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Molinari M. (2007) N-glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 3, 313–320 [DOI] [PubMed] [Google Scholar]
- 38. Chen W., Smeekens J. M., and Wu R. (2014) A universal chemical enrichment method for mapping the yeast N-glycoproteome by mass spectrometry (MS). Mol. Cell. Proteomics 13, 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cao L., Yu L., Guo Z., Shen A., Guo Y., and Liang X. (2014) N-Glycosylation site analysis of proteins from Saccharomyces cerevisiae by using hydrophilic interaction liquid chromatography-based enrichment, parallel deglycosylation, and mass spectrometry. J. Proteome Res. 13, 1485–1493 [DOI] [PubMed] [Google Scholar]
- 40. Warit S., Zhang N., Short A., Walmsley R. M., Oliver S. G., and Stateva L. I. (2000) Glycosylation deficiency phenotypes resulting from depletion of GDP-mannose pyrophosphorylase in two yeast species. Mol. Microbiol. 36, 1156–1166 [DOI] [PubMed] [Google Scholar]
- 41. Hashimoto H., Sakakibara A., Yamasaki M., and Yoda K. (1997) Saccharomyces cerevisiae VIG9 encodes GDP-mannose pyrophosphorylase, which is essential for protein glycosylation. J. Biol. Chem. 272, 16308–16314 [DOI] [PubMed] [Google Scholar]
- 42. Yoda K., Kawada T., Kaibara C., Fujie A., Abe M., Hitoshi, Hashimoto, Shimizu J., Tomishige N., Noda Y., and Yamasaki M. (2000) Defect in cell wall integrity of the yeast Saccharomyces cerevisiae caused by a mutation of the GDP-mannose pyrophosphorylase gene VIG9. Biosci. Biotechnol. Biochem. 64, 1937–1941 [DOI] [PubMed] [Google Scholar]
- 43. Janik A., Sosnowska M., Kruszewska J., Krotkiewski H., Lehle L., and Palamarczyk G. (2003) Overexpression of GDP-mannose pyrophosphorylase in Saccharomyces cerevisiae corrects defects in dolichol-linked saccharide formation and protein glycosylation. Biochim. Biophys. Acta 1621, 22–30 [DOI] [PubMed] [Google Scholar]
- 44. Daran J. M., Dallies N., Thines-Sempoux D., Paquet V., and Francois J. (1995) Genetic and biochemical characterization of the UGP1 gene encoding the UDP-glucose pyrophosphorylase from Saccharomyces cerevisiae. Eur. J. Biochem. 233, 520–530 [DOI] [PubMed] [Google Scholar]
- 45. Sarthy A. V., McGonigal T., Coen M., Frost D. J., Meulbroek J. A., and Goldman R. C. (1997) Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-beta-glucosyltransferase. Microbiology 143, 367–376 [DOI] [PubMed] [Google Scholar]
- 46. Smith D. J., Cooper M., DeTiani M., Losberger C., and Payton M. A. (1992) The Candida albicans PMM1 gene encoding phosphomannomutase complements a Saccharomyces cerevisiae sec 53–6 mutation. Current Genet. 22, 501–503 [DOI] [PubMed] [Google Scholar]
- 47. Feldman R. I., Bernstein M., and Schekman R. (1987) Product of SEC53 is required for folding and glycosylation of secretory proteins in the lumen of the yeast endoplasmic reticulum. J. Biol. Chem. 262, 9332–9339 [PubMed] [Google Scholar]
- 48. Cao K., Nakajima R., Meyer H. H., and Zheng Y. (2003) The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115, 355–367 [DOI] [PubMed] [Google Scholar]
- 49. Usui T., Yoshida M., Kasahara K., Honda A., Beppu T., and Horinouchi S. (1997) A novel HSP70 gene of Schizosaccharomyces pombe that confers K-252a resistance. Gene 189, 43–47 [DOI] [PubMed] [Google Scholar]
- 50. Lee R. E., Puente L. G., Kaern M., and Megeney L. A. (2008) A nondeath role of the yeast metacaspase: Yca1p alters cell cycle dynamics. PloS One 3, e2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aebi M. (2013) N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 [DOI] [PubMed] [Google Scholar]
- 52. Walker A. K., Soo K. Y., Levina V., Talbo G. H., and Atkin J. D. (2013) N-linked glycosylation modulates dimerization of protein disulfide isomerase family A member 2 (PDIA2). FEBS J. 280, 233–243 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete sets of data are available as .raw files, Proteome Discoverer 1.4 .msf files and pep.xml files associated, label-free reports generated by Progenesis QI and from the Candida albicans protein sequence database, provided as a multiple fasta, in the PRIDE partner repository (34) under the identification number: PXD003677.