Abstract
The use of data-independent acquisition methods such as SWATH for mass spectrometry based proteomics is usually performed with peptide MS/MS assay libraries which enable identification and quantitation of peptide peak areas. Reference assay libraries can be generated locally through information dependent acquisition, or obtained from community data repositories for commonly studied organisms. However, there have been no studies performed to systematically evaluate how locally generated or repository-based assay libraries affect SWATH performance for proteomic studies. To undertake this analysis, we developed a software workflow, SwathXtend, which generates extended peptide assay libraries by integration with a local seed library and delivers statistical analysis of SWATH-quantitative comparisons. We designed test samples using peptides from a yeast extract spiked into peptides from human K562 cell lysates at three different ratios to simulate protein abundance change comparisons. SWATH-MS performance was assessed using local and external assay libraries of varying complexities and proteome compositions. These experiments demonstrated that local seed libraries integrated with external assay libraries achieve better performance than local assay libraries alone, in terms of the number of identified peptides and proteins and the specificity to detect differentially abundant proteins. Our findings show that the performance of extended assay libraries is influenced by the MS/MS feature similarity of the seed and external libraries, while statistical analysis using multiple testing corrections increases the statistical rigor needed when searching against large extended assay libraries.
Data Independent Acquisition (DIA)1 mass spectrometry workflows are gaining increasing use for proteomic analysis of model systems (1–8). The first integrated DIA and quantitative analysis protocol, termed SWATH (2) was shown to offer accurate, reproducible, and robust proteomic quantification (9–14). DIA offers advantages over conventional IDA methods (15) by overcoming the stochastic, intensity-based selection of peptide precursors—a problem which typically leads to inconsistent peptide detection and quantitation between replicate runs. By overcoming this problem, DIA is highly suited for large-scale comparative analyses as gaps in data points between samples are mostly eliminated. These digital, extensive proteome maps can be repeatedly mined for quantitative data by extracting ion chromatograms of defined peptides postacquisition, and yields fewer quantitative missing (NA) values than IDA. An important concept in DIA analysis is use of a LC-retention time referenced spectral ion assay library to enable peptide identification from DIA generated multiplexed MS/MS spectra (10, 13, 16). The depth and quality of this spectral reference library directly correlates with experimental outcome, therefore we consider it is essential to explore and understand this variable in detail.
The reference assay library should contain all the prior knowledge of the peptide components to be extracted from the SWATH data. Thus, assay library generation is one key challenge and limitation of this approach (17). Reference assay libraries must be species-specific and be of sufficient compositional depth to enable extensive peptide identification from DIA experiments. A common approach to establishing a reference assay library involves numerous IDA experiments, usually using fractionated samples to create library depth. It is acknowledged that library building is time consuming, and for some samples, such as plasma which have a large dynamic range of protein abundances, IDA fails to have the penetrance to detect less abundant proteins in the sample (14, 18). To underscore this point, it should be clearly recognized that a peptide must be present within an assay library for it to be detected and quantitated using the SWATH workflow with reference libraries. An alternate approach to sample-based, locally generated assay libraries is to use archived data available in-house or from external public data repositories. For many commonly examined species (e.g. human, yeast), extensive libraries are readily available in public data repositories (9, 10, 19–21). Recently, studies have demonstrated that combined assay libraries can be used for SWATH data extraction (22), and a few software tools and protocols have been proposed for creating combined assay libraries (17, 23, 24). Despite these developments there have been no studies performed to systematically evaluate the effect of local and extended assay libraries on SWATH proteomics quantification performance.
To undertake this systematic evaluation of assay library performance we developed a practical and easy-to-use software workflow, which we call SwathXtend. Used in combination with PeakView V2.1 SWATH app (SCIEX) (25) or OpenSWATH (23), this software covers all major components of in-silico processes in a SWATH workflow, from extended assay library building to final statistical analysis and reporting. Extended libraries are built from a locally generated seed library, which is combined with other libraries, including in-house archived assay libraries or externally acquired entire proteome repository libraries. As only some of the pre-existing repository libraries have spiked reference iRT peptides (26), SwathXtend encompasses alternative methods for automatic LC peptide retention time calibration by using supervised learning based retention time regression or hydrophobicity-based regression. Other features of SwathXtend include library cleaning by removing user-specified low confidence and low intensity spectra, optional inclusion of peptide modifications and enzymatic miss-cleavages, compatible library formats with commonly used DIA software including PeakView and OpenSWATH, and protein accession consolidation by merging duplicated protein accessions in heterogeneous formats. The statistical analysis part of the software automates the quantitative analysis of protein expression levels starting with the ion peak areas through to the identification of differentially expressed proteins using SWATH peak extraction results exported from PeakView. In this study, we used SwathXtend to build and assess the performance of various assay libraries using a set of purposefully designed human/yeast sample mixtures for quantitative assessment. These assessments included: (1) number of proteins and peptides extracted, (2) the ability to correctly detect differently expressed proteins, (3) detection consistency between locally generated and extended libraries, (4) the quantification accuracy, and (5) reporting thresholds for statistical analysis.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
Tryptic peptides of whole-cell protein extracts from yeast (Saccharomyces cerevisiae) and human (K562) cells were purchased from Promega, Madison, WI (Cat # V7461 and V6951). Both extracts had been reduced with dithiothreitol, alkylated with iodoacetamide and digested with trypsin/Lys-C mix by the manufacturer. The yeast and human samples were reconstituted in 0.1% formic acid at 0.1 μg/μl concentrations and stored in 10 μl aliquots. To make yeast-spiked-human samples, appropriate amounts of yeast protein digest were added into 1 μg human protein digest, making three groups of samples that contained 2%, 5%, and 10% yeast (Wyeast/Whuman × 100%) respectively in 0.05 μg/μl of human protein digest. Quantitative comparisons between the three yeast-spiked human samples yield theoretical ratios of ∼0.2, 0.4, and 0.5 (exact ratios 0.21, 0.41, 0.52 respectively). All SWATH mass spectrometry acquisitions and statistical analyses were conducted using three technical replicates for each group of samples.
Local IDA Data and SWATH Data Acquisition
Local IDA data and SWATH-MS data were acquired on a 5600 TripleTOF+ mass spectrometer (SCIEX, Framingham, MA) (termed 5600–1) coupled with an Eksigent Ultra-nanoLC-1D system (Eksigent, Dublin, CA) using identical chromatography conditions. Ten microliter peptide samples were injected onto a peptide trap (Bruker peptide Captrap) for preconcentration and desalted at 10 μl/min for 5 min with 0.1% formic acid and 2% acetonitrile. After desalting, the peptide trap was switched in-line with an in-house packed analytical column (75 μm × 10 cm of solid core Halo C18, 160 Å, 2.7 μm media (Bruker, Bruker Manning Park Billerica, MA)) and fused silica PicoTip emitter (New Objective, Woburn, MA). Peptides were eluted and separated from the column using the buffer B (99.9% (v/v) acetonitrile, 0.1% (v/v) formic acid) gradient starting from 2% and increasing to 30% over 100 min at a flow rate of 300 nL per minute. IDA data were acquired for the pure human sample, for pure yeast sample, and for 5% yeast-spiked human sample. In IDA mode, a TOFMS survey scan was acquired at m/z 350–1500 with 0.25 s accumulation time, with the ten most intense precursor ions (2+ - 5+; counts > 150) in the survey scan consecutively isolated for product ion scans. Dynamic exclusion was used for 20 s. Product ion spectra were accumulated for 200 milliseconds in the mass range m/z 100–1500 with rolling collision energy (0.0625 × m/z – 3 for z = 2, 0.0625 × m/z – 4 for z = 3, and 0.0625 × m/z – 5 for z = 4 and z = 5). SWATH data were acquired three times for each group of the yeast-spiked human samples. In SWATH mode, TOFMS survey scans were acquired (m/z 350–1500, 50 milliseconds) followed by 60 product ion scans with predefined consecutive variable Q1 windows from m/z 400 to m/z 1250 which were determined based on precursor m/z frequencies in the IDA data of human sample; details are included in supplemental Information S1. Product ion spectra were accumulated for 60 milliseconds in the mass range m/z 350–1500 with rolling collision energy for lowest m/z in Q1 window (assuming z = 2) +10%.
IDA Data Acquisitions Using Different Instruments
To test SWATH extended libraries built on IDA data from different instruments, we further acquired IDA data for 1 μg pure human sample and 1 μg pure yeast sample on three other different mass spectrometers. The first one was a second, independent 5600 TripleTOF mass spectrometer (termed 5600–2) which had a different chromatography profile from 5600–1 (supplemental Information S2). The second instrument was a 6600 TripleTOF (SCIEX) coupled with a NanoLCTM ultra and cHiPLC® system (200 μm × 0.5 mm nano cHiPLC trap column and 15 cm × 200 μm nano cHiPLC columns ChromXPTM C18-CL 3 μm 120 Å; Eksigent, part of SCIEX). The same IDA data acquisition parameters were used for 5600–2 as used for 5600–1. For IDA data acquisition on 6600 TripleTOF, 10 μl sample was loaded onto the trap and desalted for 5 min at 10 μl/ml flow using loading buffer (2% acetonitrile and 0.1% formic acid). After desalting, the sample was subjected to a 120 min increasing acetonitrile gradients (5% to 40%; 99.9% acetonitrile 0.1% formic acid) at flow rate 600 nL/min for analytic separation and MS data acquisition. After a TOFMS survey scan from 350–1500 m/z, the 20 most intense m/z values exciding a threshold > 200 counts per second (cps) with charge states between 2+ and 4+ were selected for product ion scans (100–1500 m/z) with 100 msec accumulation time and 30 s dynamic exclusion. Rolling collision energy setting for product ion scans were 0.05 × m/z + 4 for z = 2, 0.05 × m/z + 3 for z = 3, and 0.05 × m/z + 2 for z = 4. The third instrument was an Orbitrap Elite coupled with Easy-nLC 1000 (Thermo Scientific, Waltham, MA). The mobile phase A consists of 0.1% formic acid and B consists of 99.9% acetonitrile/0.1% formic acid. Sample was loaded on a peptide trap (in house packed Halo® 2.7 μm 160 Å ES-C18, 100 μm × 3.5 cm) and separated with analytical column (in house packed Halo® 2.7 μm 160 Å ES-C18, 75 μm × 10 cm) at flow rate 550 nL/min and gradient from 3% B to 36% B in 100 min. MS/MS data were acquired in CID FTMS mode for 10 most intense precursor ions following a survey scan. Precursor isolation width was 2.00 m/z, normalized collision energy was 30.0, activation Q was 0.25 and activation time 10 ms.
Assay Library Generation
Seven single and five extended assay libraries were used in this study. Among the single libraries, five were in-house generated libraries and two were downloaded from public MS spectral repositories. The five extended libraries were generated by integrating two or more of these single libraries to serve our evaluation purpose. All libraries only contained b and y type ions with no water losses, precursor charge states of 2 to 5 and fragment ion charge states 1 and 2.
IDA Data Based SWATH Library Generation
IDA MS/MS data were subjected to database searches by ProteinPilot (V5.0, SCIEX) using the Paragon algorithm. Human (Homo sapiens) and yeast (Saccharomyces cerevisiae) reviewed protein databases were downloaded from UniProtKB (August 2014 version) and the two downloaded databases were merged making a new Yeast-Human database with 43,389 entries. This Yeast-Human database was used for database searches. The search parameters were as follows: sample type: identification; Cys alkylation: iodoacetamide; digestion: trypsin; special factors: none; ID focus: allow biological modifications. The group files from the database searches were loaded to PeakView (V2.1 with SWATH Quantitation plug-in) and exported as libraries in CSV format.
Extended Library Generation
We proposed a general workflow for local-library-based assay libraries integration. This workflow takes a seed library, usually a local spectral library which was generated with SWATH data using the same instrument and the same chromatography condition, and one or more add-on libraries as inputs to generate an extended library. All candidate assay libraries were first subject to a cleaning process which removes low confident peptides and low intensity ions by user-defined thresholds. Peptide confidence is based on the number of matches between the data and the theoretical fragment ions. Relative ion intensity is based on the fragment ion counts. These values are calculated by the protein identification search engine (ProteinPilot). The default values for these two thresholds are 99% for peptide confidence and 5 for ion intensity. This cleaning process not only improves the spectra quality that will be included in the final library but improves the efficiency of SWATH data extraction by significantly reducing the library size as well. The cleaned libraries undergo a quality checking to ensure that the fragment ion relative intensity match well between the add-on library and the seed library. Various measurement methods can be used to quantify the similarity of fragment ion spectra (27). In SwathXtend, we adopt Spearman rank correlation, ρ, as a measurement of the fragment ion relative intensity matching quality between two libraries (28). For those add-on libraries that pass the quality checking (median of ρ is greater than 0.7), their precursor retention time will be aligned to the seed library. We adopted two approaches for retention time alignment: retention-time based and hydrophobicity plus sequence based. The first approach requires (1) both the seed and the add-on libraries have existing retention times for all peptides; (2) the existence of a reasonable number of common peptides between the seed and add-on libraries; (3) good retention time correlation (R2 > 0.8) between the seed and add-on libraries. The second approach only uses peptide sequences and their hydrophobicity index in seed and candidate libraries. Hence, this approach can be applied when the add-on library has missing or inaccurate retention time. We use SSRCalc (29) to compute the peptide hydrophobicity index. The aligned add-on libraries are then merged with the seed library to form an integrated assay library. For peptides of conflicting or overlapping spectra between seed and add-on libraries, only seed library spectra are kept; in another word, if the same peptide appears in both seed and add-on libraries, only the spectra from the seed library will be kept in the extended library for this peptide. The function buildSpectraLibPair and buildSpectraLibTriple in the R package SwathXtend can be used to build an integrated assay library by using a seed and one or two add-on libraries, respectively. This package is available in supplemental SwathXtend package or downloadable from BioConductor (30).
SWATH Peak Extraction
PeakView V2.1 with SWATH quantitation plug-in (SCIEX) was used to extract SWATH MS peak areas with each of the libraries in our study. PeakView uses a set of processing settings to filter the ion library and determine which peptides or transitions should be used for quantification. These settings include the maximum number of peptides per protein to be included from the imported ion library; the number of transitions or fragment ions per peptide; a peptide confidence threshold in percentage which used to remove the peptides with a confidence below; a False Discovery Rate (FDR) in percentage, which is used to filter the SWATH extraction results and only export the peptide peak groups with a false positive rate below; XIC (Extracted Ion Chromatogram) retention time window and mass tolerance window for RT and m/z tolerance to pick the transitions. We evaluated these parameters based on the criterion of maximum number of peptide and protein identifications by using the gold-standard assay library, Lib1 (supplemental Table S1). Shared and modified peptides were excluded. The parameters that performed best in the tests were then applied to SWATH data extractions for all tested libraries. After SWATH peak extraction, the transition ion peak areas, peptide peak areas and protein peak areas were exported in Excel format for further statistical analysis.
Statistical Analysis
The ion peak areas exported by PeakView for each sample were normalized by total area normalization. Specifically, the total sum of all ion intensities for each sample was calculated, and the maximum of those totals determined. The ion intensities of each sample was divided by the ratio of (total ion intensities)/(maximum total ion intensity), ensuring that the normalized ion intensities sum up to the same amount for all samples. The normalized ion data was then summed by peptide and protein, which we refer to as peptide peak area and protein peak area. Two other types of data normalization were evaluated: median and MLR. For median normalization, each sample was scaled to have the median equal to the maximum median ion intensity. For MLR normalization the procedure used is described in detail in (31).
Two approaches were evaluated for determining differentially expressed proteins: the simple approach of working with the protein level quantitation only, and the second working with the peptide level quantitation separately for each peptide. For the protein level approach, differential expression was assessed by a two sample t test or ANOVA of the log transformed normalized protein peak areas. Natural logs (base e) were used throughout. The fold change (FC) ratio between any two conditions was calculated as the ratio of geometric means of the sample replicates, which corresponds to calculating the normal arithmetic ratio of log-transformed areas and back-transforming.
For the peptide level approach fold changes between the two categories were determined for each peptide separately as the ratio of average abundances in the two different categories, and then differential expression was assessed by a one sample t test of all log-transformed peptide fold changes corresponding to a particular protein. The advantage of using the peptide approach is that peptides of lower intensity can contribute without being dwarfed by the high intensity peptides; the disadvantage is that at least two different fold changes, hence two different peptides are necessary for the calculation of the one sample t test in this scenario, hence single peptide proteins cannot be considered as differentially expressed. The protein-level fold change was calculated as the geometric average of individual peptide-level fold changes.
For the pairwise comparisons, a grid of possible fold-change and p value cutoffs were considered for each library and each comparison, with possible fold change cutoffs ranging from 1.1 to 2, and p value cutoffs ranging from 0.01 to 0.2. For the case of the ANOVA protein-level analysis, a maximum fold change was calculated as the ratio of the largest group mean to the lowest group mean; in the context of an ANOVA analysis, whenever the term “fold change” is used it refers to the maximum fold change. Additionally, two multiple testing correction methods were evaluated, the Bonferroni correction and the Benjamini and Hochberg FDR-correction (32).
Two simple metrics were generated for across-library comparisons. The first metric took the top 100 proteins ranked in order of the increasing ANOVA p value and evaluated the quantification false discovery rate (qFDR) in this reduced list. The second metric identified the top N proteins yielding ∼10% qFDR using the same increasing ANOVA p value order.
RESULTS
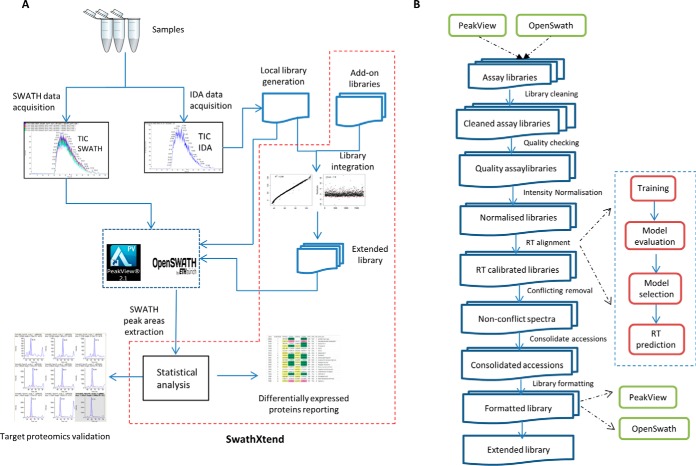
The purpose of this study was to use various peptide MS/MS assay libraries of different origins and complexities and assess how this impacts on SWATH-MS quantitative performance for analysis of complex biological samples. We examined the use of locally generated peptide MS/MS assay libraries and compared these to external reference libraries housed in public data repositories. To assess the quantitative performance of SWATH-MS to detect differentially abundant proteins among samples, we purposefully designed an experiment that mixed peptides of yeast extracts into trypsinized K562 human cell extracts in specific ratios to artificially create differentially abundant proteins in the sample mixture. Using this approach, we assessed: (1) the number of peptides and proteins extracted with each different peptide MS/MS reference assay library; (2) the quantitation false discovery rate (qFDR), which we define as the detection rate specificity for true/false assignment of differentially expressed proteins; (3) SWATH quantification accuracy (comparing the average quantitation obtained experimentally with the theoretical expected fold changes) for different library complexities. To undertake these analyses, we developed an R based software package called SwathXtend. This software integrates into SWATH analysis workflows as shown in Fig. 1A. The specific steps of SwathXtend for extending reference libraries are shown in Fig. 1B.
Fig. 1.
SWATH workflow integrating SwathXtend software package. A, Sample replicates undergo IDA and SWATH data acquisition separately. IDA data is used to generate a local spectral reference library. Extended assay libraries can be produced by combining the local library with archived IDA data or external assay libraries. SWATH acquisition data is extracted by using the local or extended assay libraries. Data extracted from matches of peptides in the library enable quantitation and statistical analysis to identify differentially abundant proteins. SWATH data can be re-used to perform target proteomics validation. B, SWATHXtend workflow takes a seed library, which is usually a locally generated spectra library, and one or more add-on libraries which are subjected to an optional cleaning process to remove low confidence and low intensity peptide ion spectra. The cleaned libraries undergo a quality checking to ensure that the add-on library and the seed library have good matching quality in terms of retention time and relative ion intensity. All quality libraries undertake ion intensity normalization and supervised learning based retention time alignment. For peptides of conflicting or overlapping spectra between seed and add-on libraries, only seed library spectra are kept. Protein accessions are consolidated by merging duplicated protein accessions in heterogeneous formats. The extended libraries can be output in PeakView or OpenSwath compatible format.
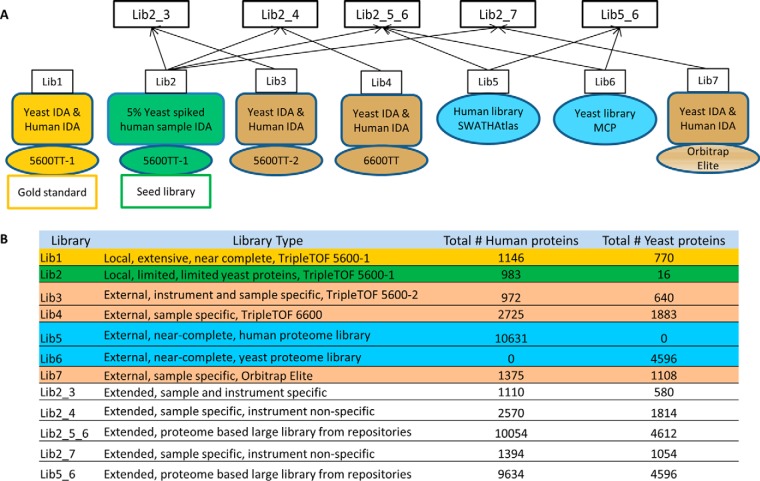
This study utilized seven peptide MS/MS assay libraries as shown in Fig. 2. These libraries can be broadly defined as locally generated or externally generated. Local libraries refer to those built from IDA data using the same chromatography and mass spectrometry conditions as we subsequently used for SWATH analysis. The library can be further described as being “extensive” or “limited” according to the number of yeast spectra contained within. The external libraries are classified into three categories: MS instrument specific and sample specific, sample specific only and near-complete species specific proteomes. To build the extensive local library, Lib1, we acquired one 1D nanoLC-MS/MS IDA analysis of the pure human K652 sample, and independently, one 1D nanoLC-MS/MS IDA analysis of the pure yeast sample on a TripleTOF 5600 + (5600 – 1). When merged together, Lib1 contained 1146 human proteins and 770 yeast proteins. Normally, such a library with extensive detection of proteins known to change in abundance between samples cannot be practically obtained as it specifically relied on obtaining spectra of the differentially expressed yeast proteins independently from the background human proteins. Lib 1 is therefore termed our “gold-standard” library to be used as a benchmark for comparisons as it contained a large number of yeast and human spectra despite the relative low abundance of yeast proteins in the mixed samples.
Fig. 2.
Assay library categories and composition. A, The assay library categories we used in this study. Lib1, the local extensive spectral library, serves as a “gold-standard” in this study; Lib2, a local limited spectral library serves as a seed library to generate extended assay libraries; Lib3, an external instrument and sample specific library; Lib4, an external sample specific library; Lib5 external, near-complete, human proteome library; Lib6, external, near-complete, yeast proteome library; Lib7, an external sample specific Orbitrap library. B, The number of human proteins and yeast proteins in each library.
Lib2 is a limited local library. This library simulates the common cases encountered in proteomics discovery projects in which most differentially expressed proteins are found in low abundances. For Lib2, two 1D nanoLC-MS/MS IDA data were acquired on the mixed sample that contained 5% (w/w) yeast spiked into the K562 human cell sample. In this library, due to the low relative abundance, only 16 yeast proteins were detected in a background of 983 human proteins. Clearly, the low number of yeast proteins contained in the library severely limits the capacity to detect the differentially abundant proteins. We used Lib2 as a seed library to generate more extensive libraries to evaluate how these might impact on SWATH quantitative performance.
Lib3, Lib4, Lib5, Lib6, and Lib7 are external libraries. Lib3 refers to an instrument and sample specific external library. Like Lib1, it was generated from one 1D nanoLC-MS/MS IDA run of the pure human sample and one run of the pure yeast sample, but used a different TripleTOF 5600+ instrument (5600–2). Lib 3 contained 972 human and 640 yeast proteins. This represents a situation where the extensive IDA data used to build the library (for example 2D LC IDA data or longer gradient IDA data) were acquired with the intention to use it to mine pre-existing SWATH data. Thus, the peptide LC retention time in Lib3 is different from those obtained with the SWATH data. Lib4 is a sample specific external library. It was made from IDA of one independent run of yeast, and separately, the human cell lysate using a TripleTOF 6600 instrument with ChipLC (2725 human and 1883 yeast proteins, respectively). This instrument provides greater sensitivity than the TripleTOF 5600+. Lib5 and Lib6 represent proteome wide large external libraries. Lib5 is a recently reported extensive human proteome assay library of ∼10000 proteins (10), while Lib6 is a near-complete yeast proteome assay library which contained ∼ 83% of the predicted yeast proteome (9). Lib7 is a sample specific external library generated using IDA on an Orbitrap Elite instrument with CID fragmentation from an independent acquisition of yeast and human K652 cell lysate. Using the local limited library, Lib2, as a seed, we merged the external libraries and created extended libraries Lib2_3, Lib2_4, Lib2_5_6, and Lib2_7. Lib6 has a median of four fragment ions per peptide, whereas the other six libraries have a median of 13 to 22 ions per peptide depending on the specific library (supplemental Information S3). We also evaluated another utility of an external library only library, Lib5_6 (no locally generated seed library). This library was made by combining Lib5 and Lib6 using Lib5 as the seed library.
We evaluated the time-based and hydrophobicity-based retention time alignment methods and the results are reported in supplemental Information S4. Our evaluation results show, for this study, the time-based method outperforms the hydrophobicity-based method. Therefore, we used the time based method for retention time alignment throughout this study.
Peptides and Proteins Extracted Using Various Reference Libraries
We first performed thorough testing of various PeakView parameter settings for SWATH data extraction by using the gold-standard library, Lib1 (supplemental Table S1). The evaluation criteria included the total number of peptides extracted, total number of proteins extracted and the number of yeast proteins extracted. We concluded that the optimal parameter settings for SWATH extraction using this library was: (1) the maximum number of peptide per protein to be included from the imported library as 100, (2) the number of fragment ions per peptide as 6, (3) peptide identification confidence setting as 99%, (4) SWATH FDR for exported peak group detection, calculated by using a decoy strategy tailored to the targeted analysis of DIA (2, 33) as 1%, (5) XIC RT window for picking the peptide transitions as 10 min, and (6) XIC mass window, a tolerance m/z for the targeted transition, as 75 ppm. Shared and modified peptides were excluded.
Using this optimal parameter set, SWATH data was extracted using local libraries, Lib1 and Lib2, and extended libraries, Lib2_3, Lib2_4, Lib2_5_6, Lib2_7, and Lib5_6. For all local or local-seeded libraries, i.e. Lib1, Lib2, Lib2_3, Lib2_4, Lib2_5_6, and Lib2_7, the SWATH data extraction process was facile and rapid because of the pre-aligned library retention time. However, when the external standalone library (Lib5_6) was used for SWATH data extraction, this required peptide retention time alignment as an initial step. As iRT peptides were not included in our SWATH data, this required choosing appropriate retention time calibration peptides. This was carried out by extracting SWATH data for one sample (10% yeast) using a 50 min retention time window. Twenty-two peptides that had 0 FDR, peptide peak area greater than 1.5 × 106, and unique retention times across the chromatogram gradient were manually checked and selected as calibration references. The retention time calibration was then applied to all peptides in the library and used for extraction of all SWATH raw data files.
Table I shows the number of peptides and proteins obtained from these extractions for all test samples. Compared with the local limited library, Lib2, all extended libraries increased total number of proteins extracted. For human proteins there was a near threefold improvement when extended libraries were used, from 962 (Lib2) to ∼2940 (Lib2_5_6 and Lib5_6). For detecting yeast proteins which are present at a maximum of 10% in the human-yeast mixed samples, dramatic improvements were seen, from only 16 proteins detected using the library generated by IDA with the mixed sample, to ∼400 proteins using the extended library that contained many more yeast protein spectra. Compared with the “gold-standard” locally generated IDA-based library, Lib1, the extended libraries showed a comparable and often greater extraction performance. Because the local extensive library, Lib1, was generated by a single1D nanoLC-MS/MS IDA run of human and yeast samples separately, it does not contain a comprehensive list of human proteins, but contains most yeast proteins that were detectable from the SWATH data which were acquired for yeast-human mixed samples. Increasing the library size dramatically increased the number of human peptides extracted, however, the extraction of yeast proteins plateaued at ∼400 proteins even though extended libraries Lib2_4, Lib2_5_6, and Lib5_6 possess many more yeast peptide reference spectra. We attribute this detection plateau to the low relative abundance of yeast proteins in the test samples (2%, 5 and 10% w/w), where many of these proteins could not be confidently detected in the mixed sample because of low relative signal intensity compared with the far greater number of human peptides that are in higher molar abundance. The extracted results including protein identification and peak intensity can be found in supplemental SwathXtend package.
Table I. Comparison of peptide and protein extractions using local and extended assay libraries. Lib1, the local extensive library, serving as the “gold-standard”; Lib2, the local limited library, serving as the seed library; Lib2_3, Lib2_4, Lib2_5_6, and Lib2_7 are extended assay libraries by combining the local limited library and various types ofexternal libraries; Lib5_6 is a combined near-complete proteome external standalone library.
| Peptides in library (extracted) | Human proteins in library | Yeast proteins in library | Human proteins extracted | Yeast proteins extracted | Percentage of human protein extraction | Percentage of yeast protein extraction | |
|---|---|---|---|---|---|---|---|
| Lib1 | 12097 (5718) | 1146 | 770 | 1062 | 390 | 92.7 | 50.6 |
| Lib2 | 7463 (5088) | 983 | 16 | 962 | 16 | 97.9 | 100.0 |
| Lib2_3 | 7919 (5566) | 1110 | 580 | 1059 | 308 | 95.4 | 53.1 |
| Lib2_4 | 20414 (8665) | 2570 | 1814 | 2006 | 349 | 78.1 | 19.2 |
| Lib2_5_6 | 111741 (9603) | 10054 | 4612 | 2931 | 390 | 29.2 | 8.5 |
| Lib2_7 | 10805 (5853) | 1394 | 1054 | 1240 | 335 | 89.0 | 31.8 |
| Lib5_6 | 108767 (7903) | 9634 | 4596 | 2948 | 408 | 30.6 | 8.9 |
Specificity to Detect Differentially Abundant Proteins
In the context of this benchmarked dataset we can evaluate with precision the performance of different statistical approaches to correctly detect differentially abundant proteins from SWATH data sets. We determined the number of true positives (TP, i.e. yeast proteins identified as differentially expressed), false positives (FP, i.e. human proteins identified as differentially expressed) and computed the quantitative FDR (qFDR) (qFDR = FP/(TP + FP)). Note, qFDR as defined is distinct from identification FDR commonly used in 'omics data analysis. We first evaluated the overall change across the three experimental conditions (2% versus 5%, 2% versus 10%, 5% versus 10% yeast spiked samples) by an analysis of variance (ANOVA) conducted separately for each individual protein using the total area normalized data, for all assay libraries (Table II). Some additional multivariate analyses were also undertaken for all libraries to examine the overall data set quality. Boxplots and density plots showed the overall data distribution, and PCA plots were used to assess variability among replicates. The differentially expressed proteins identified by ANOVA were clustered and revealed the expected patterns, for both the locally generated and extended libraries (supplemental Fig. S1). Full results of ANOVA tests for each library are available in supplemental Table S2.
Table II. Differentially detected proteins with local and extended assay libraries using ANOVA tests. Six criteria were tested including ANOVA p value, Bonferoni adjusted p value, FDR-adjusted p value using the multiple testing correction of Benjamini and Hochberg[32], maximum fold change (max FC) among the three comparisons, and the combination of p value and max FC. The results are shown in format TP|FP(qFDR). Additionally, two simple metrics for library comparisons are shown: performance for the top 100 proteins in order of Anova p value, and performance for the top N proteins achieving approximately 10% qFDR.
| Lib1 | Lib2 | Lib2_3 | Lib2_4 | Lib2_5_6 | Lib2_7 | Lib5_6 | |
|---|---|---|---|---|---|---|---|
| max FC > 1.5 only | 324|94 (22.5%) | 12|86 (88%) | 277|119 (30.1%) | 279|375 (57.3%) | 251|806 (76.3%) | 290|221 (43.2%) | 273|836 (75.4%) |
| Anova p value < 0.05 | 173|133 (43.5%) | 11|76 (87%) | 162|142 (46.7%) | 128|160 (55.6%) | 141|258 (64.7%) | 111|120 (51.9%) | 146|198 (57.6%) |
| Anova p value < 0.05 and max FC > 1.5 | 167|13 (7.2%) | 11|6 (35%) | 160|13 (7.5%) | 125|32 (20.4%) | 130|71 (35.3%) | 111|14 (11.2%) | 131|57 (30.3%) |
| Anova Bonferoni-adjusted p value < 0.05 | 29|1 (3.3%) | 5|0 (0%) | 22|0 (0.0%) | 11|1 (8.3%) | 9|0 (0.0%) | 11|0 (0.0%) | 21|0 (0.0%) |
| Anova BH fdr-adjusted p value < 0.05 | 123|21 (14.6%) | 11|0 (0%) | 98|11 (10.1%) | 63|6 (8.7%) | 73|7 (8.8%) | 56|10 (15.2%) | 60|6 (9.1%) |
| Anova BH fdr-adjusted p value < 0.05 and max FC > 1.5 | 121|5 (4.0%) | 11|0 (0%) | 98|3 (3.0%) | 62|2 (3.1%) | 72|3 (4.0%) | 56|2 (3.4%) | 60|1 (1.6%) |
| Top 100 ranked by increasing p value | 92|8 (8.0%) | 11|76 (87.4%) | 92|8 (8.0%) | 80|20 (20.0%) | 86|14 (14.0%) | 78|22 (22.0%) | 80|20 (20.0%) |
| Top N ranked by increasing p value yielding approximately 10% qFDR | 107|12 (10%) | 11|2 (15.4%) | 98|11 (10.1%) | 67|8 (10.7%) | 79|9 (10.2%) | 42|5 (10.6%) | 62|7 (10.1%) |
In the context of an ANOVA analysis, we assessed the impact of using the multiple testing corrections methods most commonly used in proteomics (34) (Benjamini and Hochberg FDR-correction and Bonferroni; the Storey Q-value was also assessed and behaved similarly to BH - supplemental Table Qvalue) and incorporating a fold-change threshold as a requirement for differential expression (Table II). Overall, all criteria have similar effects for all libraries. Though the multiple testing corrected p values yields very low qFDR, it is too stringent at the conventional 0.05 cutoff resulting in significant loss of TP proteins. Using uncorrected p values coupled with a fold change requirement achieves a much higher number of TPs for this data set with an acceptably low qFDR. The results also show that, for the local extensive library, multiple testing p value correction has little effect on decreasing qFDR, whereas for extended libraries, it does. This is not surprising since the extended libraries have a much larger proportion of proteins with no change in expression levels (human proteins) and the p value corrections work as expected to limit the chance of FPs there.
The criterion of ANOVA BH FDR-adjusted p value < 0.05 and FC > 1.5 yields a very low qFDR of less than 5% for all libraries considered, though at the expense of having fewer TPs. However, for this study the simple criterion of uncorrected p value < 0.05 coupled with FC > 1.5 provides low qFDR for the local extensive library and extended instrument and sample specific library, and a larger set of TPs identified (Table II).
In addition, Table II includes a simple ranking-based evaluation of the libraries: the performance of the top 100 proteins in increasing p value ranking, and the top proteins guaranteeing a qFDR of ∼10%. Although such criteria cannot be applied to other experiments where the true positives are not known, in the current experiment they serve to show the best performance is achieved using with the gold-standard library and this is comparable to that of Lib 2_3, followed by the external library Lib2_5_6, Lib2_4, Lib5_6, Lib2_7, and lastly Lib2. These results are consistent with the qFDR method based on fold change and p value cutoffs. Interestingly, this shows that at a fixed 10% qFDR, the external standalone library (Lib 5_6) detected ∼ 60% of the number of TPs found in the “gold-standard” local library Lib1, however this can be enhanced to 74%, by simply incorporating a locally generated seed library (Lib2_5_6). The assay library generated with an Orbitrap instrument by CID performed poorly when used to match against TripleTOF SWATH data (<40% of the TPs found in Lib1).
Detection Consistency for Differentially Abundant Proteins
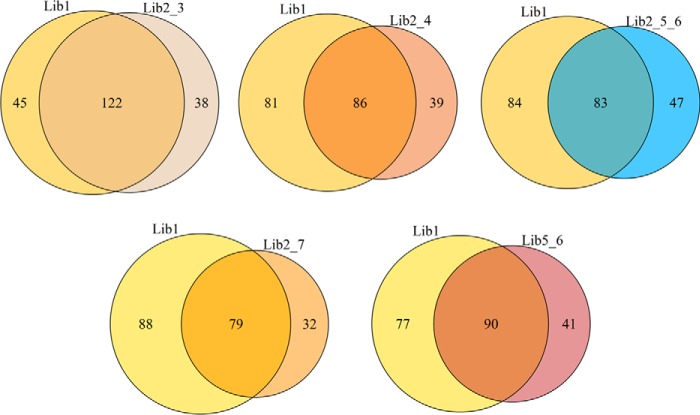
We further investigated the consistency of detecting differentially abundant proteins for each reference assay library. This evaluation was based on the set of TP proteins (yeast proteins classified as differentially expressed based on ANOVA p value < 0.05 and FC > 1.5 at the protein level) detected using local (Lib1) and extended assay libraries (Lib 2_3, Lib 2_4, Lib 2_5_6, Lib2_7, and Lib5_6). As shown by Venn diagrams in Fig. 3 there is a high overlap of TP proteins detected using the local extensive library and extended libraries, confirming that use of extended libraries are feasible to correctly detect many of the differentially expressed proteins that are discovered by using the local extensive library, Lib1. Nonetheless, it is also evident that as the library size increases there is concomitant loss of some TP proteins, as much as 25% decrease in TP detection compared with using the “gold-standard” local library, Lib1. The heat maps and examples of ion chromatographic data in supplemental Fig. S2 show the protein expression levels for the exclusive TP proteins detected using extended libraries. For most proteins, the abundance levels agree with the expected change patterns for these sample comparisons. This trend supports the view that most of the exclusive TP proteins detected by the extended libraries are valid differentially abundant proteins.
Fig. 3.
True positive consistency of differentially expressed proteins detected by local and extended libraries. Venn diagrams of TP differentially expressed proteins by using “gold-standard” spectral library, Lib1, and extended libraries, Lib2_3, Lib2_4,Lib2_5_6, Lib2_7 and Lib5_6.
Pairwise Comparisons
In the previous sections, we examined the detection of changes in protein levels for all three spiked samples in one analysis by using ANOVA. Here, we consider in more detail three pairwise comparisons, 2% to 5%, 2% to 10 and 5% to 10%, for various libraries (Table III and supplemental Fig. S3). In each case we undertook both protein level analyses and peptide level analyses, where a fold change is generated for each peptide identified for each protein separately and the results from multiple peptides belonging to the same protein are combined. The peptide level approach is stricter because it requires a minimum of two peptides per protein, but should better account for contributions from lower intensity peptides. We allowed a range of p values and fold change thresholds to be considered, and also examined the effect of BH-FDR multiple testing corrections for all libraries, where we highlight and regard 10% qFDR as an acceptable false quantitation level that would be found in contemporary proteomic data sets using the reporting thresholds we tested (A subset of these results are shown in Table III and the full results are in supplemental Table S3). It should be pointed out that qFDR is undefined in proteomic discovery experiments as TP and FP are unknown. By using the reporting thresholds described below we can extrapolate qFDR reported here to prospective proteomic studies.
Table III. Pairwise t-test detailed results for the three comparisons and each library, under three scenarios for determining differential expression. A, p values < 0.05 and fold change > 1.5; B, Benjamini and Hochberg FDR-corrected p values< 0.05 and fold change > 1.5, and C, p values < 0.05 and fold change > 2. In each case the protein level analyses and peptide level analyses are presented side by side. The results are shown in format TP|FP(qFDR); cells with qFDR < 10% are highlighted. A subset of these results is shown in picture form in supplemental Fig. S3, for ease of interpretation. Panel A shows that low qFDR is maintained for the local library Lib1 for all comparisons, and for some of the extended libraries provided the peptide level analysis is used. Panel B shows the multiple testing corrections enforce low false discovery rates, but at the expense of having almost no true positives for the comparisons with smaller effects, 2% to 5% and 5% to 10%. Panel C shows that increasing the fold change cutoff to 2 yields low qFDR of less than 10% for all extended libraries provided the peptide level analysis is used. Note: NaN stands for an undefined value and here it represents 0/0.
We now find that for the local extensive library, Lib 1, and extended instrument and sample specific library, Lib 2_3, using a reporting threshold commonly used in proteomics (uncorrected t test p value < 0.05 and FC > 1.5) still ensures qFDR <10% for all comparisons at the protein level, except the more challenging situation of 2–5% spiked yeast comparison (Table III). We find that the peptide level analysis generates a lower qFDR in all cases, but at the expense of significantly fewer TPs. For most of the extended libraries, qFDR < 10% and maximizing TPs can still be achieved by using the peptide level and by raising the fold change threshold to 2.
Quantification Accuracy
Our experimental design allowed for the assessment of SWATH quantitation accuracy. All human proteins were expected to be detected at an approximate 1:1 ratio in each of the samples, whereas the yeast proteins were expected to be detected at approximate ratios of 0.2 (fivefold), 0.4 (2.5-fold), and 0.5 (twofold) respectively for each of the comparisons. The mean detected fold changes and standard deviations (S.D.) for the human proteins were very close to the expected and are tabulated in supplemental Table S4. The quantitation means and S.D. for the TP yeast identifications is tabulated in Table IV for each of the libraries in this experiment (based on the protein level analysis, p value < 0.05 and FC > 1.5).
Table IV. Quantitative accuracy for all libraries using TP yeast proteins. For each pairwise comparison the mean listed represents the geometric mean of the abundance ratios between the two conditions calculated using protein peak areas for all yeast proteins identified as differentially expressed; the standard deviation (S.D.) of the same ratios is shown in brackets.
| TP RESULTS | Comparison | Comparison | Comparison |
|---|---|---|---|
| 2% to 5% | 2% to 10% | 5% to 10% | |
| Expect 0.41 | Expect 0.21 | Expect 0.52 | |
| Mean (S.D.) | Mean (S.D.) | Mean (S.D.) | |
| Lib1 | 0.45 (0.11) | 0.27 (0.11) | 0.43 (0.11) |
| Lib2 | 0.47 (0.06) | 0.23 (0.06) | 0.49 (0.07) |
| Lib2_3 | 0.47 (0.11) | 0.26 (0.10) | 0.44 (0.11) |
| Lib2_4 | 0.37 (0.16) | 0.25 (0.12) | 0.43 (0.10) |
| Lib2_5_6 | 0.37 (0.14) | 0.20 (0.12) | 0.41 (0.11) |
| Lib2_7 | 0.40 (0.14) | 0.23 (0.11) | 0.36 (0.13) |
| Lib5_6 | 0.44(0.10) | 0.22 (0.11) | 0.43 (0.10) |
The volcano plots in Fig. 4 demonstrate the quantification accuracy for different classes (TP, FP, TN, FN) of yeast and human proteins for three pairwise comparisons by using the data extracted with Lib1. The bold purple vertical line in each plot shows the expected intensity ratios for each comparison, i.e. ∼0.4 (2% to 5%), 0.2 (2% to 10%), and 0.5 (5% to 10%), respectively. For comparison 1 (2% to 5%) and comparison 2 (2% to 10%), most of the yeast protein intensity ratios are all compressed, i.e. closer to 1 than the expected ratios. However, for comparison 3 (5% to 10%), most of the ratios are closer to the expected ratio. This can be explained that, in the case of one of the samples in the comparison has low abundance, the SWATH quantification ratios will be reduced due to contribution of background noise in the extracted peak areas; when samples in the comparison have a relatively high abundance, the quantification ratios are very close to the expected ratios. Volcano plots for the extracted data with the extended libraries can be found in supplemental Fig. S4.
Fig. 4.
Volcano plots for yeast and human proteins in three comparisons with extracted data of Lib1. Classification based on protein level analysis, FC > 1.5 and pval < 0.05. Purple dots represent true positive (TP) proteins, i.e. yeast proteins detected as differentially expressed; blue represents true negatives (TN), i.e. human proteins not detected as differentially expressed; green represents false positives (FP), i.e. human proteins detected as differentially expressed; and red represent false negatives (FN), i.e. yeast proteins not detected as differentially expressed. The numbers inside the circles show the number of peptides identified for the respective protein.
DISCUSSION
SWATH-MS is demonstrated to provide accurate, robust and reproducible proteomic quantification. In the most widely deployed SWATH workflow a high quality reference assay library is required to deconvolute the mixed MS/MS spectra to enable peptide identifications. Establishing such a comprehensive peptide assay library in a local environment can require numerous IDA runs to build sufficient depth. Re-using archived peptide assay libraries to build new assay libraries for a specific SWATH experiment has been proposed (17, 23). However, no previous studies have been carried out to systematically evaluate the quantification performance of SWATH dependent upon assay library characteristics. A recent study by Muntel et al. (35) made some progress by evaluating spectral library performance based on DIA analysis of human urine samples focusing on the number and reproducibility of identified peptides and proteins. Their libraries included in-house IDA libraries generated from TripleTOF 5600 and Q Exactive instruments and a comprehensive human external repository library downloaded from SWATHAtlas(10). Unlike Muntel et al., our study using known quantities of spiked-in proteins enabled an evaluation of the quantitation accuracy and detection of differentially expressed proteins. Both our report and that of Muntel et al. demonstrated the utility of in house generated spectral libraries for high peptide identification success. Our study using a local library as a seed for integrating with the comprehensive external repository library led to the largest gain in protein identifications. However, this was not the case in Muntel et al. where the comprehensive repository library based on human cell lines and tissues was poorly representative of the composition of urine, hence limiting the value of this library. We observed a similar outcome for the analysis of human sperm using the Rosenberger et al. extensive human library (data not shown). These observations clearly highlight the requirement for compositional alignment between the spectral library and experimental samples.
Though some previous studies (17, 23) have performed assay libraries generation in silico by integrating archived assay libraries, they were all based on use of pre-spiked iRT peptides (26) for LC retention time calibration. The iRT method requires these peptides to be prespiked into the protein samples at the sample preparation stage. Although the iRT strategy enables excellent peptide retention time alignment among multiple IDA runs and SWATH data, it also poses limitations as most pre-existing IDA data does not contain any iRT peptides. For example, among the six original assay libraries in this study, five of them do not have any iRT peptides, and only one has five of the 11 iRT peptides. One of the complicating aspects of using iRTs is the issue of ensuring an appropriate quantity of iRT peptides is spiked to samples. The amount has to be sufficiently high enough for the iRT peptide signals to be detectable, but as low as possible to cause minimal interferences to the analytes. The recent CiRT (36) methods alleviate the requirement of spiked peptides but instead require a set of predetermined internal common peptides to be established for alignment. Some other retention time calibration methods require manual selection of a set of reference peptides, which can be tedious and time consuming especially for large data sets, which was demonstrated in our study in the case of using the external standalone library, Lib5_6. SwathXtend does not rely on iRTs or user selected peptides for LC-retention time calibration, but rather uses supervised learning based methods with model optimization and quality control for retention time alignment, so is suitable as a generic workflow for any assay library.
In this study we demonstrate that the quality of the extended assay library is heavily influenced by the quality of matching between the seed library and add-on libraries. A seed library is usually a peptide assay library generated locally using the same instrument and settings as the SWATH-MS experiments. The seed library can be readily generated by an IDA run, which is normally done as a sample preparation and instrument condition check. The IDA run data are used to define SWATH-MS Q1 windows. Besides being easy to acquire, a seed library is also sample and instrument specific therefore has the best retention time matching and ion pattern matching to the SWATH data. The seed library alone can be used to extract SWATH data for high or medium abundant proteins quantification, but to quantitate less abundant proteins it will normally be necessary to add further library depth through library extension. The add-on libraries should be organism specific with high quality matching to the seed library. As demonstrated in supplemental Fig. S5, highest quality matching will be achieved when the same instrument architecture (or at least a similar fragmentation technique) is used for the seed library and add-on library, where MS/MS features are most consistent.
The matching quality refers to the similarity of the spectra in the seed and add-on assay library, and is dictated by mass spectrometer type and acquisition parameter settings. We used two parameters to assess the matching quality of two assay libraries: retention time correlation and relative ion intensity correlation (Supplemental Fig. S5). In this study, Lib2 and Lib3 have the greatest matching quality because they were acquired with two different TripleTOF 5600+ mass spectrometers using the same acquisition settings. They showed high correlations for both retention time (R2) and relative ion intensity (Spearman rank correlation median ρ = 0.88). Lib4 IDA data were acquired using a TripleTOF 6600 system which provides higher sensitivity and greater dynamic range compared with the TripleTOF 5600+. However, it is interesting to note that the default collision energy (CE) settings on these two systems are not uniform, resulting in some differences in MS/MS features and ion intensities populating the libraries. We observed the matching quality of Lib2 and Lib4 is lower than that of Lib2 and Lib3 (peptide retention time correlation and relative ion intensity correlation are 0.97 and 0.79, respectively). It should be noted that most of the published assay reference libraries useful for SWATH have been generated on TripleTOF 5600+ instruments, and our data illustrates that fewer TP differentially abundant peptides will be identified in attempting to match TripleTOF 6600 SWATH acquisition to TripleTOF 5600+ reference assay libraries, unless TripleTOF 6600 CE is adjusted to enable uniform relative ion intensity. Lib5 and Lib6 are repository based proteome-wide assay libraries established using TripleTOF 5600+ instruments for human and yeast, respectively. The extended reference assay library generated from Lib5 and Lib6, Lib2_5_6, has comparable matching quality, (i.e. retention time correlations are R2 = 0.99 and 0.98 respectively, and the median relative ion intensity correlations are 0.74 and 0.8 respectively). Lib7 IDA data were acquired using an Orbitrap system with CID fragmentation which yields different MS/MS ions patterns to the TripleTOF-based seed library. Thus, it is no surprise that the matching quality between Lib2 and Lib7 is relatively low compared with the other TripleTOF-based external libraries, with a median relative ion intensity correlation of 0.55. As shown in Table II, the quantitative performance of Lib2_7, was inferior to other extended libraries tested in this study.
Using a very large combined library such as, Lib2_5_6 and Lib5_6 can results in high qFDR for differentially abundant protein detection compared with smaller sized libraries (i.e. Lib2_3 and Lib2_4). We attribute this to the significantly larger size of Lib2_5_6 and Lib5_6 (∼110,000 peptides) which is 14 times and 6 times larger than Lib2_3 and Lib2_4, respectively. In our study design, where differential detection of yeast proteins are TPs and human proteins are FPs, adding a large number of human proteins (Lib5) to the extended library is equivalent to adding noise (FPs). Therefore, to maintain low qFDR, it is recommended that extended assay libraries should be relevant to the sample being studied (species and tissue type) and should not be excessively large. It is especially important to control qFDR in extended libraries by applying multiple testing corrections or more stringent criteria for differential expression (Table II and Table III), as discussed later.
ANOVA and pairwise T-Tests were used to identify differentially abundant proteins after data normalization. Statistical analysis showed that the test p value and fold change magnitude should always be used in combination to set the reporting threshold. In most cases, p value less than 0.05 and fold change greater than 1.5 were a satisfactory reporting threshold to identify differentially expressed proteins in the context of this study, where the true expected changes were at least twofold. Depending on the intended follow up applications of the SWATH results, one might choose to use more relaxed criteria which would allow more TP at the expense of additional FP, or more stringent criteria (for example, peptide T-Test or FDR corrected p value or FC greater than 2) which would enable lower qFDR at the expense of some TPs. We found the use of p value FDR correction was particularly useful to reduce FP reports in ANOVA tests when the library size is very large, such as in using proteome-wide libraries (Table II). However, the p value FDR correction with the conventional cutoff is clearly too stringent for pairwise comparisons, where the statistical analysis power is lower, resulting in very few TP identifications (Table III). It is therefore desirable to use higher FC thresholds or peptide-level tests to reduce qFDR. One of the advantages of SWATH is that the data can be re-analyzed using targeted MRM peak extraction techniques to verify initial findings. This is compatible with using more relaxed cut off criteria to find protein targets during the initial stage of data processing, followed by targeted MRM analysis using the same SWATH data with manual inspection to minimize FP reports.
In conclusion, this study shows that by using high matching quality extended assay libraries, the number of peptides and proteins extracted as well as the number of correctly identified differentially abundant proteins can be increased while minimizing qFDR. The set of proteins detected as differentially expressed by using the extended assay library and the local comprehensive library are mostly consistent, and the new set of differentially expressed proteins detected by the extended libraries typically show the expected expression pattern. The quantification accuracy of the SWATH data extracted using the extended libraries is similar to that obtained when using the local comprehensive library. It is also noted that an active area of research is to develop bioinformatics approaches that eliminate the use of reference libraries for the interpretation of mixed MS/MS spectra from DIA experiments (17, 37). It remains to be determined whether these methodologies will replace the need to use reference libraries for SWATH analyses.
Data and package access
The mass spectrometry proteomics data and PeakView annotated SWATH result files have been deposited into the ProteomeXchange Consortium (38) via PRIDE partner repository with the dataset identifier PXD002836. Interested readers can download and open the SWATH result files using PeakView with SWATH plugin. The SwathXtend R package is downloadable from BioConductor (30) (April 2016 release) at bioconductor.org.
Supplementary Material
Acknowledgments
We thank Ardeshir Amirkhani for providing the Orbitrap IDA data. We thank Dr Ludovic Gillett for helpful discussions.
Footnotes
Author contributions: X.S. and M.P.M. designed research; J.X.W., T.Z., N.C., and C.K. performed research; J.X.W. and D.P. contributed new reagents or analytic tools; J.X.W. and D.P. analyzed data; J.X.W., X.S., D.P., T.Z., N.C., C.K., and M.P.M. wrote the paper.
* This work was supported by BioPlatforms Australia through the Australian Government's National Collaborative Research Infrastructure Scheme. MPM acknowledges support from the Cancer Institute NSW through Research Equipment grants and funding of Sydney Vital translational cancer research center.
 This article contains supplemental material.
This article contains supplemental material.
The authors have no disclosures to make.
1 The abbreviations used are:
- DIA
- Data-Independent Acquisition
- CiRT
- Common internal Retention Time standard peptides
- FC
- Fold Change
- FDR
- False Discovery Rate
- FP
- False Positive
- FN
- False Negative
- IDA
- Information Dependent Acquisition
- iRT
- Normalized Retention time
- MLR
- Most Likely Ratio
- m/z
- mass/charge ratio
- NA
- Not Available
- qFDR
- Quantification False Discovery Rate
- RT
- Retention Time
- SWATH
- Sequential Window Acquisition of all Theoretical fragment-ion spectra
- TN
- True Negative
- TP
- True Positive
- XIC
- Extracted Ion Chromatogram.
REFERENCES
- 1. Weisbrod CR, Eng JK, Hoopmann MR, Baker T and Bruce JE (2012) Accurate peptide fragment mass analysis: multiplexed peptide identification and quantification. Journal of proteome research 11, 1621–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gillet Ludovic C., et al. “Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis.” Molecular & Cellular Proteomics 11.6 (2012): O111–016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Venable J. D., Dong M. Q., Wohlschlegel J., Dillin A., and Yates J. R. (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 4. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., and Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 5. Panchaud A., Scherl A., Shaffer S. A., von Haller P. D., Kulasekara H. D., Miller S. I., and Goodlett D. R. (2009) Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean. Anal. Chem. 81, 6481–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carvalho P. C., Han X., Xu T., Cociorva D., da Gloria, Carvalho M., Barbosa V. C., and Yates J. R. (2010) XDIA: improving on the label-free data-independent analysis. Bioinformatics 26, 847–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geiger T., Cox J., and Mann M. (2010) Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol. Cell. Proteomics 9, 2252–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kapp E., and Schütz F. (2007) Overview of tandem mass spectrometry (MS/MS) database search algorithms. Current Protocols Protein Sci. 25, 1–19 [DOI] [PubMed] [Google Scholar]
- 9. Selevsek N., Chang C. Y., Gillet L. C., Navarro P., Bernhardt O. M., Reiter L., Cheng L. Y., Vitek O., and Aebersold R. (2015) Reproducible and Consistent Quantification of the Saccharomyces cerevisiae Proteome by SWATH-mass spectrometry. Mol. Cell. Proteomics 14, 739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosenberger G, Koh CC, Guo T, et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Scientific Data. 2014;1:140031. doi: 10.1038/sdata.2014.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y., Chen J., Sethi A., Li Q. K., Chen L., Collins B., Gillet L. C., Wollscheid B., Zhang H., and Aebersold R. (2014) Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers N-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Mol. Cell. Proteomics 13, 1753–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Y., Hüttenhain R., Surinova S., Gillet L. C., Mouritsen J., Brunner R., Navarro P., and Aebersold R. (2013) Quantitative measurements of N-linked glycoproteins in human plasma by SWATH-MS. Proteomics 13, 1247–1256 [DOI] [PubMed] [Google Scholar]
- 13. Bruderer R., Bernhardt O. M., Gandhi T., Miladinović S. M., Cheng L. Y., Messner S., Ehrenberger T., Zanotelli V., Butscheid Y., and Escher C. (2015) Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics 14, 1400–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Y., Buil A., Collins B. C., Gillet L. C., Blum L. C., Cheng L. Y., Vitek O., Mouritsen J, Lachance G., and Spector T. D. (2015) Quantitative variability of 342 plasma proteins in a human twin population. Molecular systems biology 11, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Domon B., and Aebersold R. (2006) Mass spectrometry and protein analysis. science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 16. Panchaud A., Jung S., Shaffer S. A., Aitchison J. D., and Goodlett D. R. (2011) Faster, quantitative, and accurate precursor acquisition independent from ion count. Analytical chemistry 83, 2250–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsou C. C., Avtonomov D., Larsen B., Tucholska M., Choi H., Gingras A. C., and Nesvizhskii A. I. (2015) DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Meth 12, 258–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Farrah T., Deutsch E. W., Omenn G. S., Campbell D. S., Sun Z., Bletz J. A., Mallick P., Katz J. E., Malmström J., and Ossola R. (2011) A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol. Cell. Proteomics 10, M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharma V., Eckels J., Taylor G. K. Shulman N. J., Stergachis A. B., Joyner S. A., Yan P., Whiteaker J. R., Halusa G. N., Schilling B., Gibson B. W., Colangelo C. M., Paulovich A. G., Carr S. A., Jaffe J. D., MacCoss M. J., and MacLean B. (2014) Panorama: A Targeted Proteomics Knowledge Base. Journal of Proteome Research 13(9), 4205–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Biology I. f. S. (2014) SWATHAtlas. SWATHAtlas database http://www.swathatlas.org/
- 21. Schubert O. T., Ludwig C., Kogadeeva M., Zimmermann M., Rosenberger G., Gengenbacher M., Gillet L. C., Collins B. C., Röst H. L., and Kaufmann S. H. (2015) Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis. Cell Host Microbe 18, 96–108 [DOI] [PubMed] [Google Scholar]
- 22. Zi J., Zhang S., Zhou R., Zhou B., Xu S., Hou G., Tan F., Wen B., Wang Q., Lin L., and Liu S. (2014) Expansion of the ion library for mining swath-ms data through fractionation proteomics. Anal. Chem. 86, 7242–7246 [DOI] [PubMed] [Google Scholar]
- 23. Rost H. L., Rosenberger G., Navarro P., Gillet L., Miladinovic S. M., Schubert O. T., Wolski W., Collins B. C., Malmstrom J., Malmstrom L., and Aebersold R. (2014) OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32:219–23. doi: 10.1038/nbt.2841 [DOI] [PubMed] [Google Scholar]
- 24. Schubert O. T., Gillet L. C., Collins B. C., Navarro P., Rosenberger G., Wolski W. E., Lam H., Amodei D., Mallick P., and MacLean B. (2015) Building high-quality assay libraries for targeted analysis of SWATH MS data. Nature protocols 10, 426–441 [DOI] [PubMed] [Google Scholar]
- 25. Schreiber A., and Cox D. (2011) Using PeakView® software with the XIC manager for screening and identification with high confidence based on high resolution and accurate mass LC-MS/MS. Application Note. AB SCIEX, 2170811–03 [Google Scholar]
- 26. Escher C., Reiter L., MacLean B., Ossola R., Herzog F., Chilton J., MacCoss M.J., and Rinner O. (2012) Using iRT, a normalized retention time for more targeted measurement of peptides. PROTEOMICS 12, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Toprak U. H., Gillet L. C., Maiolica A., Navarro P., Leitner A., and Aebersold R. (2014) Conserved peptide fragmentation as a benchmarking tool for mass spectrometers and a discriminating feature for targeted proteomics. Mol. Cell. Proteomics 13, 2056–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim S and Zhang X (2013) Comparative analysis of mass spectral similarity measures on peak alignment for comprehensive two-dimensional gas chromatography mass spectrometry. Computational and mathematical methods in medicine 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krokhin O. V. (2006) Sequence-specific retention calculator. Algorithm for peptide retention prediction in ion-pair RP-HPLC: application to 300-and 100-Å pore size C18 sorbents. Anal. Chem. 78, 7785–7795 [DOI] [PubMed] [Google Scholar]
- 30. Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., and Gentry J. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lambert J. P., Ivosev G., Couzens A. L., Larsen B., Taipale M., Lin Z. Y., Zhong Q., Lindquist S., Vidal M., and Aebersold R. (2013) Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods 10, 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Statist. Soc. Series B (Methodological), 57, 289–300 [Google Scholar]
- 33. Reiter L., Rinner O., Picotti P., Hüttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., and Aebersold R. (2011) mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 34. Diz A. P, Carvajal-Rodríguez A., and Skibinski D. O. (2011) Multiple hypothesis testing in proteomics: a strategy for experimental work. Mol. Cell. Proteomics 10, M110.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muntel J., Xuan Y., Berger S. T., Reiter L., Bachur R., Kentsis A., and Steen H. (2015) Advancing urinary protein biomarker discovery by data-independent acquisition on a quadrupole-orbitrap mass spectrometer. J. Proteome Res. 14, 4752–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parker S. J., Rost H., Rosenberger G., Collins B. C., Malmström L., Amodei D., Venkatraman V., Raedschelders K., Van Eyk J. E., and Aebersold R. (2015) Identification of a set of conserved eukaryotic internal retention time standards for data-independent acquisition mass spectrometry. Mol. Cell. Proteomics 14, 2800–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J., Tucholska M., Knight J. D., Lambert J. P., Tate S., Larsen B., Gingras A. C., and Bandeira N. (2015) MSPLIT-DIA: sensitive peptide identification for data-independent acquisition. Nat. Methods 12, 1106–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., and Bandeira N. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnology 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.