Abstract
Trypanosoma brucei undergoes cytokinesis uni-directionally from the anterior tip of the new flagellum attachment zone (FAZ) toward the posterior end of the cell. We recently delineated a novel signaling pathway composed of polo-like kinase, cytokinesis initiation factor 1 (CIF1), and aurora B kinase that acts in concert at the new FAZ tip to regulate cytokinesis initiation. To identify new cytokinesis regulators, we carried out proximity-dependent biotin identification and identified many CIF1 binding partners and near neighbors. Here we report a novel CIF1-binding protein, named CIF2, and its mechanistic role in cytokinesis initiation. CIF2 interacts with CIF1 in vivo and co-localizes with CIF1 at the new FAZ tip during early cell cycle stages. RNAi of CIF2 inhibited the normal, anterior-to-posterior cytokinesis but activated an alternative, posterior-to-anterior cytokinesis. CIF2 depletion destabilized CIF1 and disrupted the localization of polo-like kinase and aurora B kinase to the new FAZ tip, thus revealing the mechanistic role of CIF2 in cytokinesis initiation. Surprisingly, overexpression of CIF2 also inhibited the normal, anterior-to-posterior cytokinesis and triggered the alternative, posterior-to-anterior cytokinesis, suggesting a tight control of CIF2 protein abundance. These results identified a new regulator in the cytokinesis regulatory pathway and reiterated that a backup cytokinesis pathway is activated by inhibiting the normal cytokinesis pathway.
Keywords: cell biology, cell cycle, cytokinesis, microbiology, Trypanosoma brucei, aurora B kinase, polo-like kinase, backup cytokinesis
Introduction
Trypanosoma brucei, an early divergent parasitic protozoan causing sleeping sickness in humans and nagana in cattle in sub-Saharan Africa, possesses a complex life cycle by alternating between the insect vector and the mammalian host. Within the insect midgut and the mammalian bloodstream, the parasite proliferates through binary fission along its longitudinal axis between the two flagella and their associated cytoskeletal structure termed flagellum attachment zone (FAZ)2 (1). The cell division plane in a dividing trypanosome is determined by the length of the elongating new flagellum/FAZ, and the anterior tip of the new FAZ constitutes the site from which cytokinesis cleavage furrow ingression is initiated (2, 3). Before cytokinesis cleavage furrow initiation, invagination of cell body occurs between the two flagella, leading to the formation of the so-called division fold (4). Subsequently, the anterior tip of the new flagellum is released from the old flagellum due to the dissolution of the flagella connector (5), and cleavage furrow ingression begins from the anterior tip of the new FAZ and proceeds along the division fold toward the posterior end of the cell (6). At the very late stage of cytokinesis, the two daughter cells are connected at the posterior ends via a thread of membrane termed the cytoplasmic bridge (4), which is finally severed to generate two uni-flagellate daughter cells.
Although the morphological events of cytokinesis in T. brucei have been well described (4), the mechanisms underlying this unusual mode of cytokinesis and the regulatory pathways remain poorly understood. Over the past decade, a number of proteins have been reported to be required for cytokinesis in T. brucei based on the defective cell division upon RNAi ablation of these proteins (for review, see Ref. 7). However, only a few of them localize to the new FAZ tip or both the new FAZ tip and the cleavage furrow and are directly implicated in cytokinesis (8–11). Among these bona fide cytokinesis regulators, two conserved protein kinases, the aurora B kinase TbAUK1 (12, 13) and the polo-like kinase TbPLK (9, 14, 15), have been extensively characterized, and their functions in cytokinesis appear to differ from their animal orthologs. In animals, the polo-like kinase and aurora B kinase cooperate at the central spindle to promote actomyosin contractile ring formation by recruiting the centralspindlin complex for the latter to further activate the small GTPase RhoA (for review, see Ref. 16). In contrast, TbPLK and TbAUK1 in trypanosomes act sequentially at the new FAZ tip to regulate cytokinesis, and they never co-localize at the new FAZ tip during the cell cycle (17). The new FAZ tip is the focal point in cytokinesis in T. brucei, as is the case of the central spindle in cytokinesis in animals, but our knowledge about the structural organization and the molecular composition of the new FAZ tip is still limited.
We recently delineated a novel cytokinesis signaling pathway, composed of TbPLK, TbAUK1, and a trypanosome-specific protein CIF1, that acts in concert at the new FAZ tip to promote cytokinesis initiation (11). TbPLK phosphorylates CIF1 during early cell cycle stages, and this phosphorylation is required for CIF1 localization to the new FAZ tip. When TbPLK disappears from the new FAZ tip at late anaphase, CIF1 recruits TbAUK1 to the new FAZ tip, and the two proteins then transfer to the cleavage furrow to promote cytokinesis. We also made an unexpected discovery that the CIF1 RNAi cells started to initiate a backup cytokinesis from the posterior end of the cell, and our findings indicate that T. brucei has evolved two distinct cytokinesis pathways that drive cell division in opposite directions.
In an attempt to identify new cytokinesis regulators that function in the TbPLK-CIF1-TbAUK1 pathway, we employed proximity-dependent biotin identification (BioID) (18, 19) to search for CIF1-interacting partners. Many CIF1-binding proteins and near neighbors were identified through affinity purification and mass spectrometry, including four known FAZ tip proteins, nine known FAZ filament proteins, and many hypothetical proteins of unknown function. Here we report the characterization of one of these hypothetical proteins, which we named CIF2 for cytokinesis initiation factor 2, and the functional interaction of CIF2 with CIF1 in regulating cytokinesis initiation in T. brucei. Our results identified the essential role of CIF2 in maintaining the stability of CIF1, thereby enabling CIF1 to recruit TbAUK1 to the new FAZ tip for cytokinesis initiation. Most importantly, our results further confirmed the activation of the backup cytokinesis pathway from the posterior end of the cell by inhibiting the typical anterior-to-posterior cytokinesis.
Results
Identification of CIF1-binding Proteins and Near Neighbors by BioID
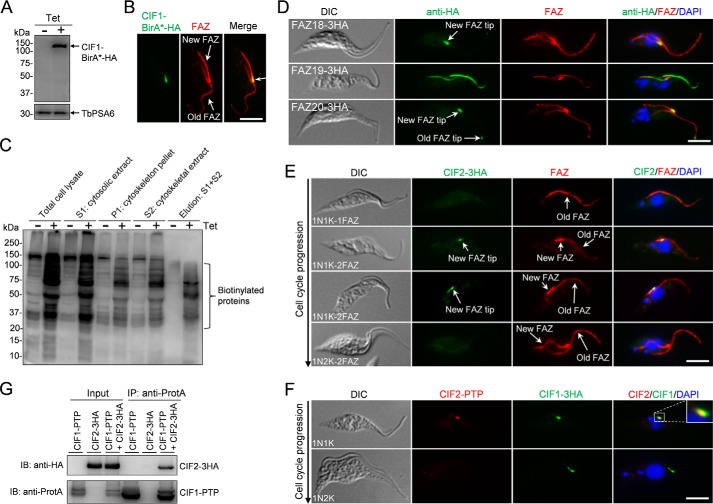
To identify CIF1-binding proteins and near neighbors, we carried out BioID. CIF1 was fused with a C-terminal BirA*-HA tag and overexpressed in procyclic trypanosomes, as confirmed by Western blotting with anti-HA antibody (Fig. 1A). Immunofluorescence microscopy confirmed that overexpressed CIF1-BirA*-HA was correctly localized to the new FAZ tip (Fig. 1B). Overexpression of CIF1-BirA*-HA did not affect cell growth (data not shown). We then carried out affinity purification of biotinylated proteins from CIF1-BirA*-HA overexpression cells and non-induced control cells, which showed that numerous biotinylated proteins were detected in CIF1-BirA*-HA overexpression cells (Fig. 1C). The eluted proteins from both the control cells and CIF1-BirA*-HA overexpression cells were digested with trypsin and analyzed by LC-MS/MS. The affinity purification and mass spectrometry were repeated three times, and the proteins that were only detectable in CIF1-BirA*-HA overexpression cells from all three experimental repeats were considered CIF1-binding proteins and near neighbors (supplemental Table S1). Among these proteins are nine known FAZ filament proteins, FAZ1-FAZ4 (20–22), FAZ9 (10, 20), FAZ10 (19), FAZ15 (10), CC2D (3), and KMP-11 (22), and four known FAZ tip proteins, FAZ6 (20), FAZ11 (19), FAZ14 (23), and TbSAS-4 (23) (Table 1).
FIGURE 1.
Identification of CIF2 by CIF1 BioID and subcellular localization of CIF2 during the cell cycle. A, tetracycline (Tet)-induced overexpression of CIF1-BirA*-HA in procyclic trypanosomes was confirmed by Western blotting with anti-HA antibody. TbPSA6, the α-6 subunit of the 26S proteasome, served as the loading control. B, overexpressed CIF1-BirA*-HA localizes to the new FAZ tip. Cells were immunostained with FITC-conjugated anti-HA monoclonal antibody and anti-CC2D polyclonal antibody. Scale bar: 5 μm. C, affinity purification of biotinylated proteins from non-induced control cells and CIF1-BirA*-HA overexpression cells. Shown is the Western blot of the input samples and final elution samples detected by anti-HRP-streptavidin Western blotting. D, subcellular localization of FAZ18-FAZ20. Cells expressing endogenously 3HA-tagged FAZ18, FAZ19, or FAZ20 were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D pAb for FAZ filament. Scale bar: 5 μm. DIC, differential interference contrast. E, subcellular localization of CIF2 during the cell cycle. Cells expressing endogenously 3HA-tagged CIF2 were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D pAb and counterstained with DAPI for nucleus (N) and kinetoplast (K). Cell counts for this experiment: 201 1N1K cells with 1 FAZ, 186 1N1K cells with a short new FAZ, 136 1N2K cells. Scale bar: 5 μm. F, co-localization of CIF2 and CIF1. Cells expressing endogenously PTP-tagged CIF2 and 3HA-tagged CIF1 were co-immunostained with anti-protein A pAb and FITC-conjugated anti-HA mAb and counterstained with DAPI for nuclear (N) and kinetoplast (K) DNA. Scale bar: 5 μm. G, interaction of CIF1 and CIF2 in vivo in trypanosomes. Cells co-expressing PTP-tagged CIF1 and 3HA-tagged CIF2 at their respective endogenous locus were lysed by sonication for immunoprecipitation. The input samples and immunoprecipitated samples were analyzed by Western blotting with anti-HA antibody and anti-protein A antibody (anti-ProtA) to detect CIF2–3HA and CIF1-PTP, respectively.
TABLE 1.
FAZ-associated CIF1 binding partners and near neighbors identified by BioID
Shown are known FAZ filament proteins, known FAZ tip proteins, a new FAZ filament protein (FAZ19), and three new FAZ tip proteins (CIF2, FAZ18, and FAZ20). Additional CIF1 binding partners and near neighbors were listed in supplemental Table S1. MM, molecular mass; CC, coiled coil; WD40, tryptophan (W)-aspartic acid (D) motif; ZF, zinc finger; TCP10, T-complex protein 10; ARM, armadillo repeat.
| Protein name | GeneDB no. | Localization | Function | MM | Motifs | References |
|---|---|---|---|---|---|---|
| kDa | ||||||
| CIF1 | Tb927.11.15800 | New FAZ tip, cleavage furrow | Cytokinesis initiation | 89.6 | 2 × CC; 2 × ZF | Zhou et al. (11) |
| CIF2 | Tb927.9.14290 | New FAZ tip | Cytokinesis initiation | 49.8 | 1 × EF-hand | This work |
| FAZ1 | Tb927.4.3740 | FAZ filament | FAZ assembly | 192.6 | 5 × CC | Vaughan et al. (21) |
| FAZ2 | Tb927.1.4310 | FAZ filament | FAZ assembly | 183.8 | 4 × CC | Zhou et al. (22); Sunter et al. (20) |
| FAZ3 | Tb927.11.12530 | FAZ filament | Unknown | 90.7 | 2 × CC | Sunter et al. (20) |
| FAZ4 | Tb927.9.10530 | FAZ filament | Unknown | 118.7 | 6 × CC | Sunter et al. (20) |
| FAZ6 | Tb927.10.840 | FAZ tip | Unknown | 195.5 | 1 × CC; 2 × WD40 | Sunter et al. (20) |
| FAZ9 | Tb927.10.14320 | FAZ filament | Life cycle transition | 121.8 | 1 × CC; 4 × ARM | Sunter et al. (20); McAllaster et al. (10); Hu et al. (23) |
| FAZ10 | Tb927.7.3330 | FAZ filament | Unknown | 502.7 | >20 × CC | Morriswood et al. (19) |
| FAZ11 | Tb927.4.5340 | FAZ tip | Unknown | 94.7 | 8 × CC | Morriswood et al. (19) |
| FAZ14 | Tb927.8.6980 | FAZ tip | Unknown | 95.1 | 8 × CC | Hu et al. (23) |
| FAZ15 | Tb927.8.7070 | FAZ filament | Unknown | 35.4 | None | McAllaster et al. (10) |
| FAZ18 | Tb927.10.12920 | New FAZ tip | Unknown | 106.5 | 1 × CC | This work |
| FAZ19 | Tb927.3.3300 | FAZ filament | Unknown | 89.2 | None | This work |
| FAZ20 | Tb927.11.9290 | FAZ tip | Unknown | 85.4 | Protein kinase | This work |
| CC2D | Tb927.4.2080 | FAZ filament | FAZ assembly | 104.9 | 10 × CC; 1 × C2 | Zhou et al. (3) |
| KMP-11 | Tb927.9.13810 | FAZ filament, flagellum, etc. | FAZ assembly, etc | 11.1 | 1 × CC | Zhou et al. (22) |
| TbSAS-4 | Tb927.11.3300 | FAZ tip | Life cycle transition | 107.9 | TCP10 C-terminus | Hu et al. (23) |
To identify new proteins that co-localize with CIF1 at the new FAZ tip, we focused on those hypothetical proteins with more than five peptides detected by mass spectrometry (supplemental Table S1). A total of 18 hypothetical proteins were selected for epitope tagging at their respective endogenous locus, and 12 of them were successfully tagged (supplemental Table S1, highlighted in green). Among the 12 proteins, 4 proteins localize to the vicinity of CIF1, including two proteins (Tb927.9.14290 and Tb927.10.12920) that localize to the new FAZ tip, a putative protein kinase (Tb927.11.9290) that localizes to the tip of both the new and old FAZ structures, and one protein (Tb927.3.3300) that localizes along the entire length of the FAZ (Fig. 1, D and E). Tb927.10.12920, Tb927.3.3300, and Tb927.11.9290 were named FAZ18, FAZ19, and FAZ 20, respectively, and Tb927.9.14290 was named CIF2 for cytokinesis initiation factor 2 based on its essential role in cytokinesis initiation (see below). CIF2 contains a putative calcium binding EF-hand domain at its N terminus (amino acids 88–153), predicted by the HMMER algorithm (24), and is conserved in kinetoplastid parasites. CIF2 localizes to the new FAZ tip only during early cell cycle stages in 1N1K cells with a short new FAZ (Fig. 1E), during which it co-localizes with CIF1 at the new FAZ tip (Fig. 1F). However, given that the FAZ structure contains several domains (25), the precise location of CIF2 at the new FAZ tip remains to be determined.
To confirm that CIF2 interacts with CIF1, co-immunoprecipitation was carried out, and the results showed that PTP-tagged CIF1 was able to precipitated 3HA-tagged CIF2 from trypanosome cell lysate (Fig. 1G). Altogether, the results from BioID, immunofluorescence microscopy, and co-immunoprecipitation suggest that CIF2 and CIF1 form a complex at the new FAZ tip in the 1N1K cells with a short new FAZ. These cells very likely were at the S phase of the cell cycle.
Depletion of CIF2 Inhibited the Normal, Anterior-to-Posterior Cytokinesis but Activated the Alternative, Posterior-to-Anterior Cytokinesis
To investigate the function of CIF2, we carried out RNAi in the procyclic form. The efficiency of CIF2 RNAi was first examined by Western blotting to monitor the level of endogenously 3HA-tagged CIF2 upon RNAi induction. On the Western blot, CIF2–3HA was detected as a doublet band before RNAi induction and became undetectable after RNAi induction for only 1 day (Fig. 2A). This depletion of CIF2 protein slowed down cell growth but did not inhibit cell proliferation (Fig. 2B), similar to CIF1 RNAi (11). To examine the potential defects in cell cycle progression, cells with different numbers of nucleus and kinetoplast were counted in control and CIF2 RNAi cells induced for up to 4 days, and the results showed that there was a significant increase of cells with two nuclei and two kinetoplasts (2N2K) after RNAi for 2 days and subsequently an emergence of cells with multiple nuclei and multiple kinetoplasts (XNXK, X > 2) (Fig. 2C), which suggests a defect in cytokinesis.
FIGURE 2.
Depletion of CIF2 inhibited cytokinesis initiation from the anterior end and caused cytokinesis initiation from the posterior end. A, Western blotting to monitor the level of CIF2–3HA in CIF2 RNAi cells. CIF2 was endogenously tagged with a triple HA epitope in CIF2 RNAi cells, and RNAi was induced for 5 days. TbPSA6 served as the loading control. B, depletion of CIF2 caused growth defects. C, depletion of CIF2 caused initial accumulation of bi-nucleate (2N) cells and subsequent accumulation of multinucleate cells (XN, X > 2). A total of 300 cells were counted for each time point, and results are presented as the mean percentage ± S.D. (n = 3). N, nucleus; K, kinetoplast. ***, p < 0.001. D, RNAi of CIF2 abolished cleavage furrow ingression from the anterior tip of the new FAZ filament but caused furrow ingression from the posterior end of the cell. Cells were immunostained with anti-CC2D pAb to label the FAZ filament and counterstained with DAPI for nucleus (N) and kinetoplast (K). A, anterior; P, posterior. Scale bar: 5 μm. DIC, differential interference contrast. E, quantification of cells without or with visible cleavage furrow in the bi-nucleate cells from the control and CIF2 RNAi cells and multinucleate (>2 nuclei) cells from CIF2 RNAi cells. A total of 200 cells were counted for each time point and each cell type, and results are presented as the mean percentage ± S.D. (n = 3). ***, p < 0.001.
To investigate whether CIF2 RNAi also inhibited cytokinesis initiation from the anterior tip of the new FAZ and caused cytokinesis initiation from the posterior end of the cell, as in the case of CIF1 RNAi (11), we examined the cleavage furrow in control and CIF2 RNAi cells by light microscopy (Fig. 2, D and E). Like CIF1 RNAi, depletion of CIF2 resulted in a significant decrease of bi-nucleate cells with a visible anterior cleavage furrow from ∼19% to ∼3% after RNAi for 48 h and an emergence of bi-nucleate cells with a visible posterior cleavage furrow to ∼21% and multinucleate cells with one or more visible posterior cleavage furrows to ∼60% (Fig. 2E).
We next examined the cytokinesis cleavage furrow in control and CIF2 RNAi cells by scanning electron microscopy. The non-induced control cells established a division fold between the flagella/FAZs (Fig. 3A) and initiated cytokinesis from the anterior tip of the new flagellum/FAZ toward the posterior end of the cell by forming an anterior cleavage furrow (Fig. 3B), as reported previously (4, 6, 11). At the late cytokinesis stage, the two daughter cells were connected by a cytoplasmic bridge at the posterior end of the cell (Fig. 3C). CIF2 RNAi cells established a similar division fold as the control cells (Fig. 3D), but no cleavage furrow from the anterior tip of the new flagellum/FAZ was observed, confirming that cytokinesis initiation from the anterior was inhibited. In the majority (>95%) of these cells, the tip of the new flagellum was detached (Fig. 3D, arrowhead), indicating the dissolution of the flagella connector, a typical event before cytokinesis initiation in T. brucei (4, 5). Moreover, in agreement with the results obtained with light microscopy (Fig. 2, D and E), cleavage furrow formation at the posterior end of the CIF2 RNAi cells was detected (Fig. 3E), indicating that CIF2 RNAi cells initiated an alternative cytokinesis from the posterior end of the cell toward the anterior. Additionally, dividing CIF2-deficient cells that were connected by a cytoplasmic bridge at the anterior end were identified (Fig. 3F), suggesting that these cells were at the late stage of the alternative cytokinesis. These cells always contained four flagella and thus were likely 4N4K cells. As reported in our previous publication (11), this alternative, posterior-to-anterior cytokinesis is slower than the normal, anterior-to-posterior cytokinesis. Because mitosis and organelle duplication/segregation were not affected by CIF2 RNAi, this may explain why CIF2 RNAi cells gradually became multinucleated (Fig. 2, C–E).
FIGURE 3.
Scanning electron microscopic analysis of cleavage furrow ingression in control and CIF2 RNAi cells. A–C, cytokinesis in control cells occurs from the anterior toward the posterior. A, a division fold was formed before cytokinesis furrow ingression in a biflagellate cell. B, cleavage furrow was formed at the anterior end of a biflagellate cell. C, at the late cytokinesis stage, the two daughter cells were connected with a cytoplasmic bridge at the posterior end. D–I, CIF2 RNAi cells underwent an alternative cytokinesis from the posterior toward the anterior. D, a division fold was formed in a biflagellate cell. The arrowhead indicates the detached flagellum tip. E, cleavage furrow was formed at the posterior end in a biflagellate cell. F, at the late stage of the alternative cytokinesis, the two daughter cells were connected by a cytoplasmic bridge at the anterior end. This cell has four flagella and is likely a 4N4K cell. G, posterior cleavage furrow ingression in a cell with two full-length flagella and two elongating 2nd new flagella (NF). H, a quadri-flagellate cell that has a posterior cleavage furrow started to form new (2nd) division folds in the two daughter cells. I, a quadri-flagellate cell that has a posterior cleavage furrow (1st furrow) started to form additional posterior cleavage furrows (2nd furrow) from the posterior ends of the two daughter cells. A, anterior; P, posterior. Scale bar: 5 μm.
Scanning electron microscopy also showed that the alternative cytokinesis cleavage furrow ingression from the posterior end of CIF2 RNAi cells was initiated in biflagellate cells (Fig. 3E). A posterior cleavage furrow was also observed in cells containing two full-length flagella and two elongating 2nd new flagella (Fig. 3G), suggesting that this cell entered the next round of organelle duplication and mitosis. In the CIF2 RNAi cells that contain four full-length flagella and a posterior cleavage furrow, two new (2nd) division folds (Fig. 3H) or two new (2nd) posterior cleavage furrows (Fig. 3I) were formed in the two daughter cells. These observations suggest that before the completion of the first round of the alternative, posterior-to-anterior cytokinesis, an additional round of alternative, posterior-to-anterior cytokinesis was initiated in the two dividing daughter cells.
CIF2 and CIF1 Are Interdependent for Stability
Given that CIF2 forms a complex with CIF1 at the new FAZ tip (Fig. 1, F and G), we examined the effect of CIF2 depletion on the localization of CIF1 to the new FAZ tip. CIF1 was endogenously tagged with a triple HA epitope in CIF2 RNAi cell line, and immunofluorescence microscopy showed that CIF1 was undetectable in >80% of the bi-nucleate cells after CIF2 RNAi for 48 h (Fig. 4, A and B), indicating that CIF2 depletion disrupted CIF1 localization to the new FAZ tip. To further investigate whether CIF1 protein level was affected, Western blotting was carried out. As reported previously (11), CIF1 was detected as a doublet band, a phosphorylated form and non-phosphorylated form, in non-induced control cell (Fig. 4C). Upon CIF2 RNAi induction, the phosphorylated form of CIF1 (the upper band) disappeared after RNAi induction for 24 h, whereas the non-phosphorylated form of CIF1 (the lower band) gradually decreased (Fig. 4C). To test whether CIF1 was degraded in CIF2 RNAi cells, the proteasome inhibitor MG-132 was added for 8 h, and Western blotting showed that CIF1 protein was stabilized (Fig. 4C). These results suggest that when CIF2 was depleted, CIF1 protein was destabilized. Disappearance of the phosphorylated form of CIF1 within 24 h of CIF2 RNAi was due to the fact that CIF1 was not localized to the new FAZ tip and thus was not phosphorylated by the polo-like kinase TbPLK.
FIGURE 4.
CIF2 and CIF1 are interdependent for protein stability. A, depletion of CIF2 disrupted the localization of CIF1 to the new FAZ tip. CIF1 was endogenously tagged with a triple HA epitope in CIF2 RNAi cells. Cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D pAb and counterstained with DAPI for nuclear DNA (N) and kinetoplast DNA (K). Scale bar: 5 μm. DIC, differential interference contrast. B, quantification of cells with different CIF1 localization patterns. A total of 300 cells were counted for each cell line, and results are presented as the mean percentage ± S.D. (n = 3). ***, p < 0.001. C, effect of CIF2 depletion on CIF1 protein level. CIF2 RNAi was induced for 72 h, and time course samples were collected for Western blotting. The proteasome inhibitor MG-132 was added after CIF1 RNAi was induced for 64 h and incubated for 8 h. TbPSA6 was detected with anti-TbPSA6 and served as the loading control. D, depletion of CIF1 disrupted the localization of CIF2 to the new FAZ tip in S-phase cells (1N1K cells containing a short new FAZ filament). CIF2 was endogenously tagged with a triple HA epitope in CIF1 RNAi cells. Cells were co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D pAb and counterstained with DAPI for nuclear and kinetoplast DNA. Scale bar: 5 μm. E, quantification of the 2N2K cells with different CIF2 localization patterns in S-phase cells. A total of 300 cells were counted for each cell line, and results are presented as the mean percentage ± S.D. (n = 3). ***, p < 0.001. F, effect of CIF1 depletion on CIF2 protein levels. CIF1 RNAi was induced for 72 h, and time course samples were collected for Western blotting. The proteasome inhibitor MG-132 was added after CIF1 RNAi was induced for 64 h and incubated for 8 h. CIF2–3HA was detected with anti-HA mAb. TbPSA6 was detected with anti-TbPSA6 pAb and served as the loading control.
Conversely, we investigated the effect of CIF1 depletion on CIF2 localization and stability. Similar to the effect of CIF2 depletion on CIF1, depletion of CIF1 also disrupted CIF2 localization (Fig. 4, D and E) and severely destabilized CIF2 protein (Fig. 4F). Together, these results suggest that CIF1 and CIF2 are interdependent for maintaining their stability.
Depletion of CIF2 Disrupted the Localization of TbPLK and TbAUK1 to the New FAZ Tip
We previously showed that depletion of CIF1 disrupted the localization of TbPLK to the new FAZ tip (11). Because CIF2 depletion destabilized CIF1 (Fig. 4C), we speculated that CIF2 depletion would also disrupt the localization of TbPLK to the new FAZ tip. Indeed, RNAi of CIF2 abolished TbPLK localization to the new FAZ tip, and TbPLK was instead detected in the basal body and the bilobe structure in ∼40% of the bi-nucleate cells or was undetectable in the remaining bi-nucleate cells (Fig. 5, A and B), similar to the effect caused by CIF1 RNAi (11). Western blotting showed that in CIF2 RNAi cells, TbPLK protein level was moderately reduced, but TbPLK was stabilized in the presence of MG-132 (Fig. 4C), indicating that CIF2 RNAi also partially destabilized TbPLK.
FIGURE 5.
Depletion of CIF2 disrupted the localization of TbPLK and TbAUK1 to the new FAZ tip. A, depletion of CIF2 disrupted TbPLK localization to the new FAZ tip in bi-nucleate (2N2K) cells. Control and CIF2 RNAi cells were co-immunostained with anti-TbPLK pAb and 20H5, which labels basal body (BB) and bilobe. Scale bar: 5 μm. DIC, differential interference contrast. B, quantification of TbPLK localization in control and CIF2-depleted cells. A total of 300 bi-nucleate cells were counted for each cell line, and results are presented as the mean percentage ± S.D. (n = 3). ***, p < 0.001. C, effect of CIF2 depletion on TbPLK protein level. CIF2 RNAi was induced for 72 h, and time course samples were collected for Western blotting. The proteasome inhibitor MG-132 was added after CIF1 RNAi was induced for 64 h and incubated for 8 h. TbPLK was detected with anti-TbPLK pAb, and TbPSA6 was detected with anti-TbPSA6 to serve as the loading control. D, CIF2 depletion abolished TbAUK1 localization to the new FAZ tip. TbAUK1 was endogenously tagged with a triple HA epitope in cells harboring the CIF2 RNAi construct. RNAi was induced for 48 h, and the non-induced control cells and RNAi-induced cells were co-immunostained with FITC-conjugated anti-HA mAb to stain TbAUK1–3HA and anti-CC2D pAb to label the FAZ filament. Scale bar: 5 μm. E, quantification of TbAUK1 localization in control and CIF2-depleted cells. A total of 300 bi-nucleate cells were counted for each cell line, and results are presented as the mean percentage ± S.D. (n = 3). F, TbAUK1 protein level was not changed in CIF2 RNAi cells. TbAUK1–3HA in non-induced control and CIF2 RNAi-induced cells was detected by Western blotting with anti-HA antibody. TbPSA6 served as the loading control.
TbAUK1 functions downstream of CIF1 in the cytokinesis regulatory pathway and is targeted to the new FAZ tip by CIF1 (11). We investigated whether depletion of CIF2 also disrupted TbAUK1 localization to the new FAZ tip. TbAUK1 has a dynamic subcellular localization during the cell cycle by localizing to kinetochores, central spindle, the new FAZ tip, and the cleavage furrow at different cell cycle stages (8). In the non-induced control bi-nucleate cells, TbAUK1 was detected at the central spindle (early anaphase), the central spindle, and the new FAZ tip (late anaphase), or the new FAZ tip (telophase). However, in CIF2-depleted bi-nucleate cells, ∼90% of them had no detectable TbAUK1 signal (Fig. 5, D and E), suggesting that depletion of CIF2 disrupted TbAUK1 localization to the new FAZ tip. Taken together, these results provided additional evidence to support the notion that CIF2 functions in the TbPLK-CIF1-TbAUK1 pathway to promote cytokinesis initiation from the anterior tip of the new FAZ.
It should be noted that the lack of TbAUK1 on the central spindle in CIF2-depleted cells was not a direct effect of CIF2 depletion because CIF2 deficiency arrested cells before cytokinesis initiation (see below), and therefore, these bi-nucleate cells have already disassembled the spindle and exited mitosis. These CIF2-deficient bi-nucleate cells were equivalent to the control telophase cells in which TbAUK1 only localizes to the new FAZ tip.
To confirm that CIF2 RNAi did not abolish TbAUK1 localization to the central spindle, we examined TbAUK1 localization in the next cell cycle of CIF2 RNAi cells. As reported previously (6, 8), in non-induced control cells, TbAUK1 was detected at kinetochores from S phase to metaphase, central spindle at early anaphase, central spindle and new FAZ tip at late anaphase, new FAZ tip at telophase, and cleavage furrow during cytokinesis (Fig. 6A). CIF2 RNAi cells were inhibited before cytokinesis initiation but underwent an additional round of mitosis and organelle duplication/segregation, generating 2N2K cells with two elongating FAZs and two full-length FAZs, 2N4K cells with two elongating FAZs and two full-length FAZs, and 4N4K cells with four full-length FAZs (Fig. 6B). In these cells, TbAUK1 was first detected in kinetochores in 2N2K-4FAZ and 2N4K-4FAZ cells, which were likely at the 2nd S phase to the 2nd metaphase, and the central spindle in some 4N4K-4FAZ cells, which were likely at the 2nd anaphase (Fig. 6B). TbAUK1 was not detectable in some other 4N4K-4FAZ cells, which were likely at the 2nd telophase (Fig. 6B). These data demonstrated that CIF2 depletion did not affect TbAUK1 localization to the central spindle.
FIGURE 6.
Localization of TbAUK1 in control cells and CIF2 RNAi cells undergoing the next round of mitosis. A, subcellular localization of TbAUK1 in non-induced control cells during the cell cycle. Cells expressing endogenously 3HA-tagged TbAUK1 were co-immunostained with FITC-conjugated anti-HA mAb to stain TbAUK1–3HA and anti-CC2D pAb to label the FAZ filament and counterstained with DAPI for nucleus (N) and kinetoplast (K). Scale bar: 5 μm. DIC, differential interference contrast. B, subcellular localization of TbAUK1 in CIF2 RNAi cells undergoing the second round of mitosis. CIF2 RNAi cells harboring 3HA-tagged TbAUK1 were induced for RNAi for 48 h, co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D pAb, and counterstained with DAPI for nucleus (N) and kinetoplast (K). Shown are the cells at different stages of the second round of mitosis, 2N2K-4FAZ (2nd S phase), 2N4K-4FAZ (2nd G2 phase to 2nd metaphase), 4N4K-4FAZ (2nd anaphase to 2nd telophase). The cell cycle stages were estimated according to the numbers of nucleus and kinetoplast and the length of the 2nd new FAZ filaments. Scale bar: 5 μm.
Overexpression of CIF2 Inhibited Anterior-to-Posterior Cytokinesis and Triggered the Alternative, Posterior-to-Anterior Cytokinesis
The observations that CIF2 localizes to the new FAZ tip only during the early cell cycle stages led us to hypothesize that CIF2 protein level is strictly controlled, and hence, constitutive expression of CIF2 may be deleterious to cells. We, therefore, investigated the potential effect of CIF2 overexpression on cell proliferation and cytokinesis. CIF2 was tagged with a C-terminal triple HA epitope and overexpressed in the procyclic form of T. brucei. Immunofluorescence microscopy showed that overexpressed CIF2–3HA was correctly localized to the new FAZ tip in 1N1K cells with a short new FAZ, but surprisingly, two CIF2–3HA foci were detected in the basal body region in 1N2K and 2N2K cells (Fig. 7A). Western blotting with anti-HA antibody detected a doublet band of CIF2–3HA throughout the course of tetracycline induction without leaky expression in the non-induced control cells (Fig. 7B). To estimate the level of CIF2–3HA overexpression, we tagged the endogenous CIF2 with a triple HA epitope at one of the two loci in cells containing the CIF2–3HA overexpression construct and induced CIF2–3HA overexpression with tetracycline for Western blotting with anti-HA antibody. CIF2–3HA was estimated to be overexpressed ∼2-fold over the endogenous CIF2 protein that was also tagged with a triple HA epitope (Fig. 7C). Overexpression of CIF2–3HA slowed down cell growth but did not inhibit cell proliferation (Fig. 7D). Tabulation of cells with different numbers of nucleus and kinetoplast showed that there was a significant increase of bi-nucleate (2N2K) cells and subsequently an emergence of multinucleate (XNXK, X > 2) cells (Fig. 7E), suggesting a cytokinesis defect.
FIGURE 7.
Overexpression of CIF2 inhibited cytokinesis initiation from the anterior and led to alternative cytokinesis from the posterior. A, localization of overexpressed CIF2–3HA during the cell cycle. Expression of CIF2–3HA was induced with 1 μg/ml tetracycline, and cells were then co-immunostained with FITC-conjugated anti-HA mAb and anti-CC2D pAb and counterstained with DAPI for nucleus (N) and kinetoplast (K). The white arrowheads indicate the CIF2–3HA foci localizing to the basal body region. Scale bar: 5 μm. B, Western blotting with anti-HA antibody to monitor the overexpression of CIF2–3HA. TbPSA6 served as the loading control. Tet, tetracycline. C, Western blotting to monitor the level of CIF2–3HA overexpression. CIF2 was endogenously tagged with a triple HA epitope in cells harboring the CIF2–3HA overexpression construct. Western blotting was carried out with anti-HA antibody. TbPSA6 served as the loading control. D, overexpression (OE) of CIF2 caused growth defects in the procyclic form. E, overexpression of CIF2 caused initial accumulation of bi-nucleate (2N) cells and subsequent accumulation of multinucleate cells (XN, X > 2). A total of 300 cells were counted for each time point, and results are presented as the mean percentage ± S.D. (n = 3). N, nucleus; K, kinetoplast. ***, p < 0.001. F–K, scanning electron microscopic analysis of the cleavage furrow in control and CIF2-overexpression cells. A, anterior; P, posterior. Scale bar: 5 μm. L, quantification of cells without or with visible cleavage furrow in control and CIF2-overexpression cells. A total of 300 bi-nucleate cells and 300 multinucleate (>2 nuclei) cells were counted for each time point, and results are presented as the mean percentage ± S.D. (n = 3). ***, p < 0.001.
The effect of CIF2 overexpression on cytokinesis furrow ingression was investigated by scanning electron microscopy (Fig. 7, F–K). CIF2-overexpressing cells formed a division fold between the two flagella (Fig. 7I), similar to that in the non-induced control cells (Fig. 7F). However, cleavage furrow ingression from the anterior tip of the new flagellum/FAZ was inhibited, but furrow ingression from the posterior end of the cell was detected (Fig. 7, H–L). These results demonstrated that CIF2 overexpression exerted almost identical cytokinesis defects as CIF2 RNAi (compare Figs. 2 and 3 with Fig. 7).
Overexpression of CIF2 Disrupted CIF1 Localization to the New FAZ Tip
The unexpected cytokinesis defects by CIF2 overexpression prompted us to investigate the underlying mechanism. Given that CIF1 forms a complex with CIF2 (Fig. 1G) and that overexpressed CIF2 was mis-localized to the basal body region in 1N2K and 2N2K cells (Fig. 7A), we reasoned that CIF2 overexpression might disrupt CIF1 localization to the new FAZ tip in these cells. To test this possibility, we examined the localization of CIF1 in CIF2-overexpressing cells. The results showed that endogenously PTP-tagged CIF1 was not localized to the new FAZ tip in CIF2-overexpressing 1N2K cells (data not shown) and 2N2K cells but was detected as two foci near the proximal end of the FAZ, where it co-localized with the overexpressed CIF2–3HA (Fig. 8, A and B). Co-immunostaining with the 20H5 antibody, which labels the basal body and the bilobe in T. brucei (26), showed that CIF1-PTP was mis-localized in the basal body region in 1N2K cells (data not shown) and 2N2K cells (Fig. 8A). Western blotting showed that the level of PTP-tagged CIF1 was not altered upon CIF2 overexpression (Fig. 8C). It is likely that the two CIF1-PTP foci detected in the basal body region were the aggregate of CIF1-PTP and CIF2–3HA. These results suggest that CIF2 overexpression caused CIF1 mis-localization to the basal body region in 2N2K cells.
FIGURE 8.

Overexpression of CIF2 disrupted the localization of CIF1 to the new FAZ tip. A, Localization of CIF1 in control and CIF2-overexpressing cells. CIF1 was endogenously tagged with the PTP epitope in cells harboring the pLew100-CIF2–3HA construct. Overexpression of CIF2 was induced with 1.0 μg/ml tetracycline for 24 h. Cells were co-immunostained with anti-protein A pAb and anti-FAZ1 (L3B2) mAb, with anti-protein A pAb and FITC-conjugated anti-HA mAb, or with anti-protein A pAb and 20H5 mAb and counterstained with DAPI for nucleus (N) and kinetoplast (K). The solid arrowheads indicate the CIF1-PTP foci, whereas the open arrowheads show the CIF2–3HA foci in 2N2K cells. BB, basal body. Scale bar: 5 μm. DIC, differential interference contrast. B, quantification of cells with different CIF1 localization patterns in bi-nucleate cells from control and CIF2-overexpressing (OE) cells. A total of 200 bi-nucleate cells were counted for each cell line, and data are presented as the mean percentage ± S.D. (n = 3). ***, p < 0.001. C, overexpression of CIF2 did not affect the protein level of CIF1. CIF1 was endogenously tagged with the PTP epitope in cells containing the pLew100-CIF2–3HA construct. CIF2–3HA overexpression was induced with 1.0 μg/ml tetracycline (Tet). CIF2–3HA was detected by anti-HA mAb, and CIF1-PTP was detected by anti-Protein A pAb. The level of TbPSA6 served as the loading control.
Discussion
Cytokinesis in T. brucei is unusual in that it occurs uni-directionally along the longitudinal axis of the cell from the anterior toward the posterior and is regulated by a novel signaling pathway at the anterior tip of the new FAZ, the cell's cytokinesis initiation site. The new FAZ tip appears to comprise many proteins, which likely play distinct functions, including cytokinesis initiation (8, 11) and life cycle transition (23). The cytokinesis regulators, including the three components of the chromosomal passenger complex (TbAUK1, TbCPC1, and TbCPC2), TbPLK, and CIF1 (aka TOEFAZ1), localize to the new FAZ tip only (8–11), whereas TbSAS-4, which controls life cycle transition from the epimastigote form to the trypomastigote form, and its associated proteins, FAZ6, FAZ11, FAZ13, and FAZ14, are enriched at the anterior tip of both the new and old FAZs (23). Despite the functional distinction between CIF1 and TbSAS-4, BioID with CIF1 as the bait was able to detect TbSAS-4 and its associated FAZ tip proteins (Table 1), indicating that TbSAS-4 and its partner proteins are located at the new FAZ tip in close proximity to CIF1. Moreover, CIF1 BioID also identified three new FAZ tip proteins, CIF2, FAZ18, and FAZ20 (Table 1). CIF2 is involved in cytokinesis initiation, but the biological functions of FAZ18 and FAZ20 remain to be explored. Given the distinct localization patterns of FAZ18 and FAZ20, they likely play different functions. Finally, it should be noted that >100 hypothetical proteins were identified as CIF1 near neighbors (supplemental Table S1), and the subcellular localizations of most of these proteins have not been determined. Epitope tagging and immunofluorescence microscopy are currently being carried out to determine their subcellular localizations, and additional FAZ tip proteins that function in cytokinesis initiation may be identified.
We have expanded the inventory of cytokinesis regulators in T. brucei by identifying CIF2 as a CIF1 binding partner, which maintains CIF1 protein stability (Figs. 1 and 4, A–C). Conversely, CIF1 depletion also destabilized CIF2 (Fig. 4, D–F), indicating that formation of the CIF1·CIF2 complex is essential to maintain the stability of both proteins. The interdependence of individual subunit proteins in a protein complex for their stability was also observed in other protein complexes in T. brucei and other organisms, such as the TbKIN-C·TbKIN-D complex (27), the TbCentrin3·TbIAD5-1 complex (28), the CC2D·FAZ2·KMP11 complex (22), the γ-tubulin complex (29), and the CRK9·CYC12·CRK9AP complex (30) in T. brucei, the kinetochore protein complex in Candida albicans (31), multiple DNA repair protein complexes (32) and the Laforin-Malin complex (33) in humans, and the Pes1·Bop1·WDR12 complex in animals (34). It appears that such control of protein stability provides an effective means to regulate the function of the individual subunits in the protein complex.
The fact that CIF2 is detectable only during early cell cycle stages in the 1N1K cells with a short new FAZ (Fig. 1E) and is required to maintain CIF1 stability (Fig. 4, A–C) raises an intriguing question of how CIF1 stability is maintained after CIF2 disappears from the new FAZ tip in 1N2K and 2N2K cells. Given that CIF1 and CIF2 localize to the new FAZ tip concurrently in the 1N1K cells with a short new FAZ (Fig. 1F), the two proteins may form a complex in the cytosol before being targeted to the new FAZ tip or are simultaneously and independently recruited to the new FAZ tip, where they form a stable complex. In the first scenario we speculate that formation of the CIF1·CIF2 complex in the cytosol prevents them from being degraded by the proteasome, and once the complex is targeted to the new FAZ tip, CIF2 is no longer needed and thus is removed from the new FAZ tip. In the second scenario, we postulate that formation of the CIF1·CIF2 complex is essential for maintaining them at the new FAZ tip during early cell cycle stages, but thereafter CIF2 is no longer needed or is replaced by another FAZ tip protein(s). Although the precise mechanism behind the control of CIF1 and CIF2 stability remains unknown, it is clear that CIF2 is tightly regulated during the cell cycle, and its function at the new FAZ tip is restricted to early cell cycle stages. The mechanism underlying the control of CIF2 protein abundance will be further explored in the future.
CIF2 is a new player in the recently discovered cytokinesis regulatory pathway in T. brucei (11), and its implication in cytokinesis initiation was demonstrated by several lines of evidence. First, CIF2 RNAi caused a significant increase in bi-nucleate cell population and then an emergence of multinucleate (>2 nuclei) cell population, without compromising FAZ elongation and organelle duplication/segregation (Fig. 2, C and D). Depletion of CIF2 also inhibited cleavage furrow ingression at the anterior end of the new FAZ (Figs. 2, D and E, and 3, D–I), which provided direct evidence for its requirement for cytokinesis initiation. Second, ablation of CIF2 by RNAi impaired the localization of TbPLK and CIF1, which cooperate to regulate cytokinesis initiation (11), to the new FAZ tip (Figs. 4, A–C, and 5, A–C). Finally, CIF2 deficiency disrupted the localization of TbAUK1, a crucial cytokinesis regulator that functions directly at the cytokinesis initiation site and the cleavage furrow (6, 8), to the new FAZ tip (Fig. 5, D–F). These findings placed CIF2 upstream of TbAUK1 in the TbPLK-CIF1-TbAUK1 pathway and highlighted the cooperative actions of multiple regulators (TbPLK, CIF1, and CIF2) at the new FAZ tip during early cell cycle stages to regulate cytokinesis.
The physiological role of CIF2 is well understood, but the biochemical function of CIF2 remains unknown. CIF2 contains a putative EF-hand domain at its N terminus and presumably should be capable of binding calcium ions. The presence of an EF-hand domain in CIF2 raises the questions of whether binding to calcium is necessary for CIF2 function in cytokinesis initiation and whether calcium signaling is involved in cytokinesis in T. brucei. Calcium signaling has been demonstrated to play important roles in cytokinesis in other eukaryotes (35, 36) and is mediated by the actin-binding protein cofilin, which is required for cytokinesis (37), and by calmodulin, which localizes to the spindle midzone, the cleavage furrow, and the intercellular bridge during cytokinesis (38–40) and plays an essential role in cytokinesis (40–43). However, nothing is known about the potential involvement of calcium in cytokinesis in T. brucei, and no other protein with calcium binding capacity has been found to play a role in cytokinesis in T. brucei. Thus, the identification of CIF2 provides an excellent start point to dissect the calcium signaling pathway in regulating cytokinesis in T. brucei.
Although we postulate that overexpression of CIF2 might be toxic to cells based on its short stint at the new FAZ tip during early cell cycle stages (Fig. 1E), we did not expect that overexpression of CIF2 would cause cytokinesis defects identical to that caused by CIF2 depletion (Figs. 2, 3, and 7). Intriguingly, the mechanisms underlying the cytokinesis defects in CIF2 RNAi cells and CIF2-overexpressing cells are different. CIF2 RNAi destabilized CIF1, whereas CIF2 overexpression caused CIF1 mis-localization from the new FAZ tip to the basal body region without destabilizing CIF1 (Figs. 4, A–C, and 8). Given that overexpressed CIF2 was mis-localized to the basal body region in 1N2K and 2N2K cells (Fig. 7A), it titrated CIF1 away from the new FAZ tip to the basal body region (Fig. 8, A and B). Because CIF1 localization to the new FAZ tip is necessary for cytokinesis initiation (11), these results revealed the underlying mechanism for the cytokinesis defects caused by CIF2 overexpression.
Our current work on CIF2 provided additional evidence to support our previous discovery that inhibition of the normal cytokinesis regulatory pathway can trigger a backup cytokinesis pathway to initiate cytokinesis from the opposite cell end (11). This alternative pathway is cryptic under normal growth conditions but is activated when the normal cytokinesis signaling cascade is defective. Our work thus suggests the existence of redundant mechanisms that T. brucei employs to ensure cell division and survival. An alternative explanation for furrow ingression at the posterior end observed in CIF2 RNAi cells is that remodeling (reorganization) of cytoskeleton and membrane caused furrow ingression. Although furrow ingression involves extensive remodeling of the microtubule cytoskeleton and membrane, there is still the requirement of regulatory protein(s) to promote the initiation of furrow ingression. In fungi and animals, initiation of furrow ingression, i.e. the assembly of the actomyosin contractile ring, is regulated by the aurora B kinase- and RhoA-mediated signaling pathway (44). In T. brucei, no actomyosin ring is formed at the cleavage furrow, and how cleavage furrow ingression is regulated remains elusive. However, given that CIF2 depletion as well as CIF1 depletion (11) and TbAUK1 depletion (8) inhibited cleavage furrow ingression from the anterior tip of the new FAZ (Fig. 3, D–I), it suggests that furrow ingression at the anterior end and likely the remodeling of cytoskeleton and membrane is activated by the TbAUK1-mediated pathway. By the same token, cleavage furrow ingression and remodeling of cytoskeleton and membrane at the posterior end of the cell in CIF2 RNAi cells likely also require certain regulatory proteins.
One may argue that the alternative cytokinesis pathway does not contribute to normal cytokinesis or is not relevant to the natural process of cytokinesis in T. brucei. However, one should be aware that genetic redundancy not only occurs on individual genes but also on the level of cellular pathways. Additional examples of a backup pathway have been reported on several important cellular processes, including cytokinesis in humans (45). The backup cytokinesis pathway in humans, termed cytofission, can be activated in interphase cells and does not require components of the canonical cytokinesis machinery, such as the polo-like kinase Plk1, the chromosomal passenger protein INCENP, and the centralspindlin subunit MKLP1, but it still requires the actomyosin contractile ring (45).
The backup cytokinesis in T. brucei is also independent of the canonical cytokinesis regulators, including TbPLK and TbAUK1, which were not redirected to the posterior end of the cell in CIF2 RNAi cells (Fig. 5). However, given that the backup cytokinesis uses the same division fold as the normal cytokinesis (Fig. 3, A and D) and still needs to bisect the membrane at the cytoplasmic bridge, it is likely that the backup cytokinesis may use the same membrane abscission machinery as the normal cytokinesis. T. brucei possesses homologs of the ESCRT-III (endosomal sorting complex required for transport-III) machinery, which is regulated by aurora B kinase to promote the final membrane abscission step at the end of cytokinesis in other eukaryotes (46), but the potential role of the ESCRT-III machinery in T. brucei cytokinesis has not been explored. Future work will be directed to investigate the involvement of the membrane abscission machinery in both the normal cytokinesis and the backup cytokinesis. Efforts will also be directed to identify the regulators involved in the backup cytokinesis.
Experimental Procedures
Trypanosome Cell Culture
The procyclic form of T. brucei strain 29-13 (47) was cultured in SDM-79 medium supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Inc.), 15 μg/ml G418, and 50 μg/ml hygromycin at 27 °C. The procyclic form of T. brucei strain 427 was grown in SDM-79 medium containing 10% heat-inactivated fetal bovine serum at 27 °C.
Identification of CIF1 Binding Partners and Near Neighbors by BioID and LC-MS/MS
To overexpress CIF1-BirA*-HA for identification of CIF1-binding proteins and near neighbors, the full-length coding sequence of CIF1 was PCR-amplified and cloned into the pLew100-BirA*-HA vector (23). The resulting plasmid was then linearized with NotI and transfected into the 29-13 strain. Transfectants were selected under 2.5 μg/ml phleomycin and cloned by limiting dilution in a 96-well plate. Expression of CIF1-BirA*-HA was induced with 0.5 μg/ml tetracycline and was confirmed by Western blotting with anti-HA antibody and immunofluorescence microscopy with FITC-conjugated anti-HA antibody.
Affinity purification of biotinylated proteins was performed according to our previous publication (23). CIF1-BirA*-HA was overexpressed by induction with 0.5 μg/ml tetracycline for 24 h, and cells (∼2.5 × 109) were incubated with 50 μm biotin for an additional 24 h. Cells were washed 3 times with PBS and treated with PEME buffer (100 mm PIPES, pH 6.9, 2 mm EGTA, 0.1 mm EDTA, 1 mm MgSO4) containing 0.5% Nonidet P-40. Cytosolic (soluble) and cytoskeletal (pellet) fractions were separated, and the cytoskeletal fraction was further extracted with lysis buffer (0.4% SDS, 500 mm NaCl, 5 mm EDTA, 1 mm DTT, 50 mm Tris-HCl, pH 7.4). Solubilized cytoskeletal materials were collected by centrifugation. The cytosolic extract and the cytoskeletal extract were combined and then incubated with 500 μl of pre-washed streptavidin-coated Dynabeads (Invitrogen) at 4 °C for 4 h. The Dynabeads were washed extensively with PBS. As the negative control, non-induced control cells (∼2.5 × 109) were similarly treated and subject to purification with the same amount of Dynabeads. Three independent purifications were performed for both CIF1-BirA*-HA and the negative control.
The Dynabeads were washed 5 times with 50 mm ammonium bicarbonate and then resuspended in 100 mm ammonium bicarbonate. 10% DTT was then added to reduce the disulfide bond, and subsequently 50% iodoacetamide was added for alkylation. 5% DTT was then added to the solution, and proteins were digested with trypsin overnight at 37 °C. Digestion was stopped by adding trifluoroacetic acid to approximately pH 2.0. The protein digests were desalted and analyzed on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) interfaced with an Eksgent nano-LC 2D plus chipLC system (Eksigent Technologies) at the Proteomics Core Facility of the University of Texas Health Science Center at Houston.
Data analysis was performed according to our published methods (22, 28). Raw data files were searched against the T. brucei genome database using the Mascot search engine. The search conditions used peptide tolerance of 10 ppm and MS/MS tolerance of 0.8 Da with the enzyme trypsin and two missed cleavages.
RNA Interference
To generate the CIF2 RNAi cell line, a 555-bp DNA fragment (nucleotides 154–708) corresponding to the N-terminal coding region of CIF2 was cloned into the pZJM vector (48). The pZJM-CIF2 plasmid was then linearized with NotI and transfected into the 29-13 strain by electroporation. Transfectants were selected with 2.5 μg/ml phleomycin in SDM-79 medium containing 15 μg/ml G418 and 50 μg/ml hygromycin and cloned by limiting dilution in a 96-well plate. The CIF1 RNAi cell line has been reported previously (11). To induce RNAi, cells were cultured in medium containing 1.0 μg/ml tetracycline, and cell growth was monitored daily by counting the cells with a hemacytometer.
Overexpression of CIF2 in the Procyclic Form of T. brucei
To overexpress CIF2 in T. brucei, the full-length coding sequence of CIF2 was cloned into pLew100–3HA vector. The resulting plasmid, pLew100-CIF2–3HA, was linearized with NotI and transfected into 29-13 strain by electroporation. Transfectants were selected under 2.5 μg/ml phleomycin in addition to 15 μg/ml G418 and 50 μg/ml hygromycin and then cloned by limiting dilution in a 96-well plate.
In Situ Epitope Tagging of Proteins
To tag the CIF1 binding partners and near neighbors for determining their subcellular localizations, 19 hypothetical proteins were selected (see supplemental Table S1 for the accession numbers). The DNA fragment corresponding to their C-terminal coding sequence was cloned into pC-3HA-PAC vector. The resulting plasmids were each linearized with appropriate restriction enzymes and transfected into the 427 cell line by electroporation. Transfectants were selected with 1 μg/ml puromycin and cloned by limiting dilution in a 96-well plate.
For C-terminal epitope tagging of CIF2 at one of its endogenous loci, a 984-bp fragment corresponding to the C-terminal coding region of CIF2 was cloned into pC-3HA-PAC and pC-PTP-NEO vectors. The two plasmids were each linearized with XhoI and electroporated into the wild-type 427 strain, whereas the pC-3HA-PAC was transfected into the cell lines harboring pZJM-CIF2 or pZJM-CIF1. Transfectants were selected with 1 μg/ml puromycin and cloned by limiting dilution.
For co-localization of CIF2-PTP and CIF1–3HA, the cell line harboring the pC-CIF2-PTP-NEO vector was transfected with pC-CIF1–3HA-PAC, which was linearized by XcmI digestion (11). Transfectants were selected with 40 μg/ml G418 in addition to 1.0 μg/ml puromycin and cloned by limiting dilution.
For endogenous epitope tagging of TbAUK1 in CIF2 RNAi cell line, the pC-TbAUK1–3HA-PAC vector (11) was linearized with SphI and transfected into the cell line harboring the pZJM-CIF2 construct. Transfectants were selected under 1 μg/ml puromycin in addition to 15 μg/ml G418, 50 μg/ml hygromycin, and 2.5 μg/ml phleomycin, and clonal cell lines were obtained by limiting dilution.
For endogenous epitope tagging of CIF1 in CIF2–3HA overexpression cell line, a 1358-bp DNA fragment (nucleotides 1021–2378) corresponding to the C-terminal coding region of CIF1 was cloned into the pC-PTP-PAC vector, linearized with XcmI, and transfected into the cell line harboring the pLew100-CIF2–3HA construct. For endogenous epitope tagging of CIF2 in CIF2–3HA overexpression cell line, the pC-CIF2–3HA-PAC vector was linearized with XhoI and electroporated into the cell line containing the pLew100-CIF2–3HA construct. Transfectants were selected with 1 μg/ml puromycin in addition to 15 μg/ml G418, 50 μg/ml hygromycin, and 2.5 μg/ml phleomycin and cloned by limiting dilution.
Co-immunoprecipitation
Cells expressing endogenously PTP-tagged CIF1 and 3HA-tagged CIF2 were lysed by sonication in 1 ml of immunoprecipitation buffer (25 mm Tris-HCl, pH 7.6, 500 mm NaCl, 1 mm DTT, 1% Nonidet P-40, and protease inhibitor mixture). The lysate was cleared by centrifugation at the highest speed in a microcentrifuge, and the supernatant was incubated with 50 μl of settled IgG-Sepharose 6 fast flow beads (GE Healthcare) at 4 °C for 1 h. The beads were then washed six times with the immunoprecipitation buffer. Bound proteins were eluted with 10% SDS, separated on SDS-PAGE, transferred onto a PVDF membrane, and immunoblotted with anti-HA mAb and anti-protein A pAb to detect CIF2–3HA and CIF1-PTP, respectively. Cells expressing CIF1-PTP alone and CIF2–3HA alone were used as the controls.
Immunofluorescence Microscopy
Cells were adhered to the coverslips for 30 min, fixed with cold methanol (−20 °C) for 30 min, and then rehydrated with PBS for 10 min. Cells were blocked with 3% BSA in PBS for 1 h at room temperature and then incubated with the primary antibody for 1 h at room temperature. The following primary antibodies were used: FITC-conjugated anti-HA monoclonal antibody (1:400 dilution, Sigma), anti-protein A polyclonal antibody (1:400 dilution, Sigma), anti-CC2D polyclonal antibody for the FAZ filament (1:1000 dilution) (3), anti-FAZ1 monoclonal antibody (L3B2, 1:50 dilution) (49), 20H5 monoclonal antibody for basal body and bilobe (1:400 dilution, EMD Millipore) (26), and anti-TbPLK antibody (1:400 dilution) (50). Cells were washed 3 times with PBS and then incubated with FITC-conjugated anti-mouse IgG (1:400 dilution, Sigma) or Cy3-conjugated anti-rabbit IgG (1:400 dilution, Sigma) for 1 h at room temperature. Cells on the coverslips were washed 3 times with PBS, mounted with DAPI-containing VectaShield mounting medium (Vector Laboratories), and imaged using an inverted fluorescence microscope (Olympus IX71) equipped with a cooled CCD camera (model Orca-ER, Hamamatsu) and a PlanApo N 60 × 1.42-NA lens. Images were acquired using the Slidebook 5 software.
Scanning Electron Microscopy
Scanning electron microscopy was performed essentially as described in our previous publication (11). Briefly, cells were settled onto coverslips and fixed with 2.5% (v/v) glutaraldehyde in PBS for 30 min at room temperature. After washing the cells three times with PBS, cells were dehydrated in a series of alcohol (30%, 50%, 70%, 90%, and 100%) for 10 min each. After critical point drying, samples were coated with a 5-nm metal film (Pt:Pd 80:20, Ted Pella Inc.) using a sputter-coater (Cressington Sputter Coater 208 HR, Ted Pella Inc.) and imaged using Nova NanoSEM 230 (FEI). The scanning work distance was at 5 mm, and the accelerating high voltage was at 8 kV.
Statistical Analysis
Statistical analysis was performed using the t test in the Microsoft Excel software. Detailed n values for each panel in the figures were stated in the corresponding legends. For immunofluorescence microscopy, images were randomly taken, and all cells in each image were counted.
Author Contributions
Q. Z., H. H., and Z. L. conceived and designed the research. Q. Z. and H. H. performed the research. Q. Z., H. H., and Z. L. analyzed the data. Q. Z. and Z. L. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Drs. Arthur Günzl, Cynthia Y. He, Keith Gull, and Kyle Roux for providing epitope tagging vectors, anti-CC2D antibody, anti-FAZ1 (L3B2) antibody, and BirA*-HA plasmid (from Addgene), respectively. We also thank Dr. Jianhua Gu for assistance with scanning electron microscopy, and Li Li for LC-MS/MS.
This work was supported by National Institutes of Health Grant R01 AI101437 (to Z. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1.
- FAZ
- flagellum attachment zone
- CIF1
- cytokinesis initiation factor 1
- CIF2
- cytokinesis initiation factor 2
- TbPLK
- T. brucei polo-like kinase
- TbAUK1
- T. brucei aurora B kinase
- BioID
- proximity-dependent biotin identification
- N
- nuclear DNA
- K
- kinetoplast DNA
- PTP
- protein A/tobacco etch virus protease site/protein C epitope.
References
- 1.Vaughan S., and Gull K. (2008) The structural mechanics of cell division in Trypanosoma brucei. Biochem. Soc. Trans. 36, 421–424 [DOI] [PubMed] [Google Scholar]
- 2.Kohl L., Robinson D., and Bastin P. (2003) Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22, 5336–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Q., Liu B., Sun Y., and He C. Y. (2011) A coiled-coil- and C2-domain-containing protein is required for FAZ assembly and cell morphology in Trypanosoma brucei. J. Cell Sci. 124, 3848–3858 [DOI] [PubMed] [Google Scholar]
- 4.Wheeler R. J., Scheumann N., Wickstead B., Gull K., and Vaughan S. (2013) Cytokinesis in Trypanosoma brucei differs between bloodstream and tsetse trypomastigote forms: implications for microtubule-based morphogenesis and mutant analysis. Mol. Microbiol. 90, 1339–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs L. J., McKean P. G., Baines A., Moreira-Leite F., Davidge J., Vaughan S., and Gull K. (2004) The flagella connector of Trypanosoma brucei: an unusual mobile transmembrane junction. J. Cell Sci. 117, 1641–1651 [DOI] [PubMed] [Google Scholar]
- 6.Li Z., Umeyama T., and Wang C. C. (2009) The Aurora Kinase in Trypanosoma brucei plays distinctive roles in metaphase-anaphase transition and cytokinetic initiation. PLoS Pathog. 5, e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z. (2012) Regulation of the cell division cycle in Trypanosoma brucei. Eukaryot. Cell 11, 1180–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z., Lee J. H., Chu F., Burlingame A. L., Günzl A., and Wang C. C. (2008) Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS ONE 3, e2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Graffenried C. L., Ho H. H., and Warren G. (2008) Polo-like kinase is required for Golgi and bilobe biogenesis in Trypanosoma brucei. J. Cell Biol. 181, 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAllaster M. R., Ikeda K. N., Lozano-Núñez A., Anrather D., Unterwurzacher V., Gossenreiter T., Perry J. A., Crickley R., Mercadante C. J., Vaughan S., and de Graffenried C. L. (2015) Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in Trypanosoma brucei. Mol. Biol. Cell 26, 3013–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q., Gu J., Lun Z. R., Ayala F. J., and Li Z. (2016) Two distinct cytokinesis pathways drive trypanosome cell division initiation from opposite cell ends. Proc. Natl. Acad. Sci. U.S.A. 113, 3287–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z., and Wang C. C. (2006) Changing roles of aurora-B kinase in two life cycle stages of Trypanosoma brucei. Eukaryot. Cell 5, 1026–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu X., Kumar P., Li Z., and Wang C. C. (2006) An aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J. Biol. Chem. 281, 9677–9687 [DOI] [PubMed] [Google Scholar]
- 14.Kumar P., and Wang C. C. (2006) Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryot. Cell 5, 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammarton T. C., Kramer S., Tetley L., Boshart M., and Mottram J. C. (2007) Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation, and cytokinesis. Mol. Microbiol. 65, 1229–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carmena M. (2008) Cytokinesis: the final stop for the chromosomal passengers. Biochem. Soc. Trans. 36, 367–370 [DOI] [PubMed] [Google Scholar]
- 17.Li Z., Umeyama T., Li Z., and Wang C. C. (2010) Polo-like kinase guides cytokinesis in Trypanosoma brucei through an indirect means. Eukaryot. Cell 9, 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morriswood B., Havlicek K., Demmel L., Yavuz S., Sealey-Cardona M., Vidilaseris K., Anrather D., Kostan J., Djinovic-Carugo K., Roux K. J., and Warren G. (2013) Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot. Cell 12, 356–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sunter J. D., Varga V., Dean S., and Gull K. (2015) A dynamic coordination of flagellum and cytoplasmic cytoskeleton assembly specifies cell morphogenesis in trypanosomes. J. Cell Sci. 128, 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaughan S., Kohl L., Ngai I., Wheeler R. J., and Gull K. (2008) A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist 159, 127–136 [DOI] [PubMed] [Google Scholar]
- 22.Zhou Q., Hu H., He C. Y., and Li Z. (2015) Assembly and maintenance of the flagellum attachment zone filament in Trypanosoma brucei. J. Cell Sci. 128, 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu H., Zhou Q., and Li Z. (2015) SAS-4 Protein in Trypanosoma brucei controls life cycle transitions by modulating the length of the flagellum attachment zone filament. J. Biol. Chem. 290, 30453–30463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., Bateman A., and Eddy S. R. (2015) HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sunter J. D., and Gull K. (2016) The flagellum attachment zone: “the cellular ruler” of trypanosome morphology. Trends Parasitol. 32, 309–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He C. Y., Pypaert M., and Warren G. (2005) Golgi duplication in Trypanosoma brucei requires Centrin2. Science 310, 1196–1198 [DOI] [PubMed] [Google Scholar]
- 27.Wei Y., Hu H., Lun Z. R., and Li Z. (2013) The cooperative roles of two kinetoplastid-specific kinesins in cytokinesis and in maintaining cell morphology in bloodstream trypanosomes. PLoS ONE 8, e73869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei Y., Hu H., Lun Z. R., and Li Z. (2014) Centrin3 in trypanosomes maintains the stability of a flagellar inner-arm dynein for cell motility. Nat. Commun. 5, 4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Q., and Li Z. (2015) γ-Tubulin complex in Trypanosoma brucei: molecular composition, subunit interdependence, and requirement for axonemal central pair protein assembly. Mol. Microbiol. 98, 667–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badjatia N., Park S. H., Ambrósio D. L., Kirkham J. K., and Günzl A. (2016) Cyclin-dependent kinase CRK9, required for spliced leader trans splicing of pre-mRNA in trypanosomes, functions in a complex with a new L-type cyclin and a kinetoplastid-specific protein. PLoS Pathog. 12, e1005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thakur J., and Sanyal K. (2012) A coordinated interdependent protein circuitry stabilizes the kinetochore ensemble to protect CENP-A in the human pathogenic yeast Candida albicans. PLoS Genet. 8, e1002661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe R., Ui A., Kanno S., Ogiwara H., Nagase T., Kohno T., and Yasui A. (2014) SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 74, 2465–2475 [DOI] [PubMed] [Google Scholar]
- 33.Mittal S., Upadhyay M., Singh P. K., Parihar R., and Ganesh S. (2015) Interdependence of laforin and malin proteins for their stability and functions could underlie the molecular basis of locus heterogeneity in Lafora disease. J. Biosci. 40, 863–871 [DOI] [PubMed] [Google Scholar]
- 34.Rohrmoser M., Hölzel M., Grimm T., Malamoussi A., Harasim T., Orban M., Pfisterer I., Gruber-Eber A., Kremmer E., and Eick D. (2007) Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol. Cell Biol. 27, 3682–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webb S. E., Li W. M., and Miller A. L. (2008) Calcium signalling during the cleavage period of zebrafish development. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 363, 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong R., Hadjiyanni I., Wei H. C., Polevoy G., McBride R., Sem K. P., and Brill J. A. (2005) PIP2 hydrolysis and calcium release are required for cytokinesis in Drosophila spermatocytes. Curr. Biol. 15, 1401–1406 [DOI] [PubMed] [Google Scholar]
- 37.Nusco G. A., Chun J. T., Ercolano E., Lim D., Gragnaniello G., Kyozuka K., and Santella L. (2006) Modulation of calcium signalling by the actin-binding protein cofilin. Biochem. Biophys. Res. Commun. 348, 109–114 [DOI] [PubMed] [Google Scholar]
- 38.Yu Y. Y., Chen Y., Dai G., Chen J., Sun X. M., Wen C. J., Zhao D. H., Chang D. C., and Li C. J. (2004) The association of calmodulin with central spindle regulates the initiation of cytokinesis in HeLa cells. Int. J. Biochem. Cell Biol. 36, 1562–1572 [DOI] [PubMed] [Google Scholar]
- 39.Li C. J., Heim R., Lu P., Pu Y., Tsien R. Y., and Chang D. C. (1999) Dynamic redistribution of calmodulin in HeLa cells during cell division as revealed by a GFP-calmodulin fusion protein technique. J. Cell Sci. 112, 1567–1577 [DOI] [PubMed] [Google Scholar]
- 40.Gonda K., Katoh M., Hanyu K., Watanabe Y., and Numata O. (1999) Ca2+/calmodulin and p85 cooperatively regulate an initiation of cytokinesis in Tetrahymena. J. Cell Sci. 112, 3619–3626 [DOI] [PubMed] [Google Scholar]
- 41.Liu T., Williams J. G., and Clarke M. (1992) Inducible expression of calmodulin antisense RNA in Dictyostelium cells inhibits the completion of cytokinesis. Mol. Biol. Cell 3, 1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Y. Y., Dai G., Pan F. Y., Chen J., and Li C. J. (2005) Calmodulin regulates the post-anaphase reposition of centrioles during cytokinesis. Cell Res. 15, 548–552 [DOI] [PubMed] [Google Scholar]
- 43.Tsang W. Y., Spektor A., Luciano D. J., Indjeian V. B., Chen Z., Salisbury J. L., Sánchez I., and Dynlacht B. D. (2006) CP110 cooperates with two calcium-binding proteins to regulate cytokinesis and genome stability. Mol. Biol. Cell 17, 3423–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mierzwa B., and Gerlich D. W. (2014) Cytokinetic abscission: molecular mechanisms and temporal control. Dev. Cell 31, 525–538 [DOI] [PubMed] [Google Scholar]
- 45.Choudhary A., Lera R. F., Martowicz M. L., Oxendine K., Laffin J. J., Weaver B. A., and Burkard M. E. (2013) Interphase cytofission maintains genomic integrity of human cells after failed cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 110, 13026–13031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt O., and Teis D. (2012) The ESCRT machinery. Curr. Biol. 22, R116–R120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wirtz E., Leal S., Ochatt C., and Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 48.Wang Z., Morris J. C., Drew M. E., and Englund P. T. (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275, 40174–40179 [DOI] [PubMed] [Google Scholar]
- 49.Kohl L., Sherwin T., and Gull K. (1999) Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46, 105–109 [DOI] [PubMed] [Google Scholar]
- 50.Hu H., Zhou Q., and Li Z. (2015) A novel basal body protein that is a polo-like kinase substrate is required for basal body segregation and flagellum adhesion in Trypanosoma brucei. J. Biol. Chem. 290, 25012–25022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.