Abstract
X-ray crystallography has shown that an antibody paratope typically binds 15–22 amino acids (aa) of an epitope, of which 2–5 randomly distributed amino acids contribute most of the binding energy. In contrast, researchers typically choose for B-cell epitope mapping short peptide antigens in antibody binding assays. Furthermore, short 6–11-aa epitopes, and in particular non-epitopes, are over-represented in published B-cell epitope datasets that are commonly used for development of B-cell epitope prediction approaches from protein antigen sequences. We hypothesized that such suboptimal length peptides result in weak antibody binding and cause false-negative results. We tested the influence of peptide antigen length on antibody binding by analyzing data on more than 900 peptides used for B-cell epitope mapping of immunodominant proteins of Chlamydia spp. We demonstrate that short 7–12-aa peptides of B-cell epitopes bind antibodies poorly; thus, epitope mapping with short peptide antigens falsely classifies many B-cell epitopes as non-epitopes. We also show in published datasets of confirmed epitopes and non-epitopes a direct correlation between length of peptide antigens and antibody binding. Elimination of short, ≤11-aa epitope/non-epitope sequences improved datasets for evaluation of in silico B-cell epitope prediction. Achieving up to 86% accuracy, protein disorder tendency is the best indicator of B-cell epitope regions for chlamydial and published datasets. For B-cell epitope prediction, the most effective approach is plotting disorder of protein sequences with the IUPred-L scale, followed by antibody reactivity testing of 16–30-aa peptides from peak regions. This strategy overcomes the well known inaccuracy of in silico B-cell epitope prediction from primary protein sequences.
Keywords: antibody, antigen, bioinformatics, epitope mapping, immunogenicity, protein motif, protein-protein interaction
Introduction
Knowledge of B-cell epitopes of proteins is essential in many fields of applied biomedical research, such as antibody diagnostics and therapeutics, vaccines, as well basic research. Laboratory methods for identification of such epitopes are time-consuming and labor-intensive. Hence, any reduction in the need for discovery and confirmatory wet-lab research by epitope prediction algorithms is highly desirable. Among in silico predictive methods from primary sequence information, epitope prediction algorithms are distinguished for their lack of reliability (1). This underperformance prompted us to examine current approaches to B-cell epitope prediction by use of extensive data on epitopes and confirmed non-epitope regions of the Chlamydia spp. proteome, accumulated in research on chlamydial molecular serology (2).
Recent three-dimensional antibody-antigen complex studies (3–7) show that about 15–22-aa2 antigen peptide residues are structurally involved in binding of epitopes to ∼17-aa residues in antibody complementarity-determining regions (CDRs; paratopes). Among these 15–22 structural epitope residues, about 2–5 aa, termed functional residues, contribute most of the total binding energy to antibodies (6). These functional residues lie only in a very small fraction of B-cell epitopes closely spaced to each other and embedded among the structural residues, representing the classical concept of continuous B-cell epitopes. In the vast majority (≥90%) of B-cell epitopes, functional as well as structural residues are randomly distributed within 15–150-aa linear antigen sequences, essentially representing discontinuous epitopes. Thus, a peptide antigen can effectively bind an antibody only if it contains the majority of the functional residues, and only a small fraction of the short peptides of 4–11 aa will contain sufficient functional residues for high affinity binding (6). Therefore, short peptide targets in B-cell epitope mapping and prediction may represent an inherent, unsolvable conundrum, because most of these short peptides, even from proven dominant epitope regions, will fail to bind antibodies strongly and therefore will give many false-negative (non-epitope) results.
Mammalian immune systems can be forced to generate antibodies against virtually any molecule, regardless of antigen origin, by using excessive amounts of adjuvants and antigens. However, the antibody response did evolve in response to infections that generate much lower antigen exposure, thus antibodies may be preferentially directed toward proteins and peptide regions with certain biological, structural, and physiochemical properties that determine optimal epitopes. Antibody formation during an immune response to any given epitope is inherently stochastic due to the random availability of a cognate B-cell receptor within the large pool of circulating B-cells, all with different B-cell receptors generated by recombination of the immunoglobulin gene (8). Another level of stochasticity in the antibody response to any given protein is the exposure of a protein to the immune system. Wang et al. (9) report that only 4.2% of about 900 Chlamydia trachomatis (Ctr) proteins induce natural antibody responses in ≥40% of human hosts. Therefore, any peptide of the remaining 95.8% non-immunodominant proteins is unlikely to elicit antibodies, regardless of its B-cell epitope properties. Hence, for accurate evaluation of epitope prediction methods, epitope/non-epitope data should be derived from testing of known immunodominant proteins, with multiple rather than single sera to account for the stochasticity of the antibody response.
B-cell epitope prediction has been first based on various properties of individual amino acids (aa) such as hydrophilicity, hydrophobicity, solvent accessibility, flexibility, or β-turn propensity, and combinations thereof (10–16). However, even the best combinations of aa propensity scales performed only marginally better than random sequence selection (1). With the availability of B-cell epitope databases, antigenicity scales (17, 18) and machine learning approaches (19–24) have been attempted, and improved prediction accuracy has been reported. Nevertheless, due to epitope redundancy (20), the predictive power may have been overestimated because these algorithms performed poorly on independent data (24). Therefore, B-cell epitope prediction algorithms must be evaluated on independent datasets that had not been used to train/develop the algorithms/scales.
The ever increasing number of solved three-dimensional protein structures has allowed the development and testing of numerous complex algorithms for prediction of physicochemical and structural properties of proteins directly from the aa sequences. Among these properties (scales), disorder tendency describes protein regions without defined three-dimensional structure that are inherently flexible, hydrophilic, solvent accessible, and thermally mobile (high B-factor) (25, 26). Incidentally, all of these properties are shared with B-cell epitopes (16); thus, protein disorder tendency is a prime candidate scale for B-cell epitope prediction due to its multifaceted properties (27).
This investigation is an extension of a comprehensive study that identified immunodominant B-cell epitopes of Chlamydia spp. (2). After encountering numerous failures of in silico B-cell epitope prediction, we used the first principles established above to analyze the shortcomings of B-cell prediction methodology. Using pools of hyperimmune mouse sera, we determined epitope/non-epitope regions of immunodominant Chlamydia spp. proteins by use of long 16–40-aa peptide antigens. These data created epitope/non-epitope datasets for accurate testing of numerous B-cell epitope prediction or aa/protein property algorithms/scales (henceforth termed scales). Subsequent testing revealed that public datasets were biased toward short epitope/non-epitope antigens, and removal of these short antigens dramatically increased prediction accuracy of most scales. We show that in general machine learning methods cannot predict epitopes with high accuracy; rather, many scales designed for prediction of protein properties, particularly disorder tendency, identify B-cell epitopes with better accuracy.
Experimental Procedures
B-cell Epitope Peptide Reactivity with Anti-chlamydial Hyperimmune Sera
Hyperimmune sera were raised in mice as described previously (2). Briefly, 9–50 mice were challenged three times with high but non-lethal intranasal chlamydial inocula, to mimic antibody production following natural infections. Bovine sera used were obtained from animals with PCR-confirmed natural Chlamydia spp. infection (2). Peptide antigens were chemically synthesized with N-terminal biotin, captured onto streptavidin-coated microtiter plates, and incubated with hyperimmune sera. Primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies in chemiluminescent ELISA, and data were expressed as relative light units/s (rlu/s) and for ease of display were divided by 1,000 (rlu/s × 10−3) (2). All peptides were analyzed on white microtiter plates by use of specific positive pooled hyperimmune sera as well as negative control sera in wells coated with specific peptides and in a non-coated well. For the final background-corrected results, 150% of the background signal (mean + 2 S.D.) in the non-coated well of each serum was subtracted from its specific peptide signals. To avoid false-positive results in quantitative evaluation of the reactivity of any peptide with individual mouse sera, and with bovine sera from naturally infected cattle, we used a more stringent cutoff of 10,000 rlu/s. Overall methods are described in detail by Rahman et al. (2).
B-cell Epitope/Non-epitope Datasets
These datasets are described briefly below, and a detailed description is provided in supplemental Table S1, and sequences are provided in the supplemental Appendix.
Concatenated Epitope/Non-epitope Virtual Proteins
Epitopes and non-epitopes of the Lbtope_Fixed_non_redundant and Lbtope_ Confirm datasets (24) were grouped by sequence length. Concatenated polyproteins of the sequences of each group were constructed by randomly combining all sequences. Similarly, concatenated polyproteins of Swiss-Prot sequences were constructed by randomly combining all sequences of the BCP12, BCP14, or BCP18 datasets (20).
Concatenated Virtual Proteins of 50-Amino Acid-extended Epitopes/Non-epitopes Embedded in Random Sequences
All 16–20-aa epitopes of Lbtope_Confirm and fbcpred.pos.nr80 datasets were extended symmetrically to 50 aa with source protein sequences. These fragments were interspersed with random 150-aa Swiss-Prot sequences into a concatenated virtual polyprotein. Similar polyproteins were assembled from all 16–33-aa non-epitopes of the Lbtope_Confirm dataset and all epitopes/non-epitopes of the Chl_18Prot and Chl_43Prot datasets. For assembly of additional non-epitope datasets, random 50-aa peptides of the Bcpreds epitope source proteins (Bcpreds_Prot), Swiss-Prot proteins, and the C. trachomatis proteome (28) were similarly linked with 150-aa interspacing sequences into concatenated polyproteins.
B-cell Epitope/Non-epitope Annotation of Individual Chlamydia spp. Proteins
All residues of 18 immunodominant proteins of Chlamydia spp. were annotated in the Chl_18Prot dataset as Pos (positive, epitope), Neg (negative, non-epitope), or NT (not tested, unknown epitope status). The annotation is based on the reactivity of 16–20-aa peptide antigens with murine and/or bovine sera. For peptide datasets, 10-aa-spaced peptides of these proteins were used.
Computation of Amino Acid Residue Scores for Physicochemical, Structural, and Evolutionary Protein Properties
Website-based freeware algorithms/scales (10–15, 17–20, 22–24, 29–55) for protein properties were used to calculate individual residue scores for aa sequences of individual or polyproteins. Moving window scores were assigned to the center residue of the particular window. When required, missing scores for N- and C-terminal residues were inserted using scores of the adjacent residues. If algorithms/scales did not provide an output score for internal residues/windows, the minimum score of this protein was inserted. The polymorphism score was calculated by inverting the multiple sequence conservation score of the AACon algorithm in the Jalview freeware (35).
Comparison of Receiver Operating Characteristic (ROC) Curves of Protein Property Scales for B-cell Epitope Prediction
Bimodal epitope/non-epitope classification was achieved by F test classification based on the linear predictor variable in discriminant analysis with the software package JMP Pro 11 (SAS Institute Inc., Cary, NC). This software was also used to construct ROC curves and calculation of area under the ROC curve (AUC) for ranking of protein property scales for B-cell epitope prediction. Data formatting was performed in Microsoft Office Excel 2013, and all additional statistical analyses were performed by the Statistica 7.1 software package (Statsoft, Tulsa, OK). Differences between means of peptide reactivities and/or background were analyzed by one-tailed paired Student's t test, and p values ≤ 0.05 were considered significant. The significance of differences between B-cell epitope prediction scales was tested by one-tailed paired Student's t tests of AUCs of ROC analyses. If multiple independent test datasets were available, the mean AUC values of these scales for these datasets were compared. If multiple proteins of a single dataset were available, the mean AUC values for these proteins were compared. If ROC curves for different B-cell epitope prediction scales were analyzed for a single dataset, specificity was sampled in 0.05 increments for sensitivities from 0.05 to 0.85 and in 0.02 increments from 0.90 to 0.98, and the specificity means were compared, or the mean accuracy values at 40, 60, 80, 90, and 95% sensitivity were compared.
Results
Antibody Binding Increases with Length of Peptide Antigens
Within a peptide B-cell epitope, 15–22 residues are typically structurally involved in antibody binding (3–7). Sivalingam and Shepherd (6) reasoned that clustering or random distribution of the structural residues would determine the length of peptide antigens required for antibody binding. In this study, we tested length-dependent peptide antigen reactivity for previously identified epitope regions of the chlamydial outer membrane protein A, OmpA (2). Seven-12-aa peptide antigens invariably produced lower ELISA signals than longer ones (Fig. 1A). Occasionally, extensive elongation of peptide antigens may mask structural residues and reduce the signal relative to a slightly shorter peptide antigen of optimum length (Fig. 1A; 32- versus 24-aa Cpe peptides).
FIGURE 1.

Peptide reactivity increases with length. A, elongation of peptides around the center of two epitopes increases the ELISA signal; RLU, relative light units/s, mean of six repeats; %, percent signal of maximally reactive peptide; Ctr, C. trachomatis; numbers indicate peptide position on the OmpA protein; Cpe; Chlamydia pecorum. B, relative signal from 17 epitopes in dependence on peptide antigen length. Peptides for 17 epitopes were extended toward the C and N terminus by 12–20 residues around the epitope center (long 24–40-aa peptides), 8 residues (intermediate 16-aa peptides), or 3–6 residues (short 7–12-aa peptides) and tested with the respective epitope-positive pooled hyperimmune mouse sera. Peptide reactivities are represented by vertical lines in the same order for long, intermediate, and short peptides. C, relative reactivity with murine antisera of corresponding long and intermediate peptides of 55 epitopes. D, relative reactivity with bovine antisera of corresponding long and intermediate peptides of 45 epitopes. Mapping of epitopes and peptide antigens used for Fig. 1 are described in the supplemental Appendix.
To quantify the effect of peptide length on antibody binding, peptide antigens of different lengths from 17 epitope regions of OmpA and inclusion membrane A (IncA) proteins of Chlamydia spp. were tested (Fig. 1B). Compared with short 7–12-aa peptides, intermediate 16-aa peptides produced 1.8-fold ELISA signal intensity, and long 24–40-aa peptides produced 4.1-fold signal intensity (p value <10−2, one-tailed Student's t test, relative log-transformed signal). Importantly, the main 14.3-fold reactivity increase was achieved by elongation of the 13 lowest reactive short peptides from 7–12 to 24–40 aa (p value <10−4), whereas elongation of the 13 lowest reactive intermediate 16-aa peptides from 24 to 40 aa produced a moderate 3.3-fold increase (p value <10−4).
To identify optimum peptide antigen lengths, we tested the central 16- and 24–40-aa peptide antigens of 55 unique epitopes on 28 Chlamydia spp. proteins with pooled mouse sera (Fig. 1C). As observed before, ∼20% of elongated peptides produced a reduced signal, presumably due to epitope masking. However, long 24–40-aa peptides produced on average a 2.1-fold higher signal than the corresponding 16-aa peptides (p value <10−4). The reactivity of the 28 lowest reactive 16-aa peptides increased 9.1-fold for the respective long 24–40-aa peptides (p value <10−4). To confirm the host independence of length-dependent peptide reactivity, another set of chlamydial peptides yielded equivalent results with bovine sera (Fig. 1D).
Evaluation of B-cell Epitope Prediction Algorithms Confounded by Over-representation of Short False-negative Epitopes in Public Datasets
Many investigators typically use short peptide antigens of 4–11 aa for epitope mapping, with results added to public reference databases that are used for the development of B-cell epitope prediction algorithms/scales (10–24). These datasets may therefore be biased toward short epitopes and many false-negative epitope determinations due to the marginal antibody binding of short peptides. This may explain the poor, close to random, epitope prediction accuracy (1) that most epitope prediction scales show in practical application, even if they scored highly in evaluation with public datasets. We hypothesized that removing short epitope/non-epitope sequences from public datasets would allow correct performance ranking of B-cell epitope prediction scales. To test this hypothesis, we used the “Lbtope_Variable_non_redundant” dataset with 8,011 B-cell epitopes and 10,868 non-epitopes, retrieved by Singh et al. (24) from experimentally validated epitopes as well as non-epitopes from the Immune Epitope Database (IEDB). Importantly, 5–10-aa non-epitopes include ∼50% and 5–16-aa non-epitopes include ∼80% of all non-epitopes deposited in the parent IEDB (24). Among the 6–11-, 12–15-, 16–20-, and 21–30-aa sequences, the Lbtope_Variable_non_redundant dataset contains 2.12×, 1.49×, 0.93×, and 0.54× numbers of non-epitopes compared with epitopes. These data indicate that short non-epitope sequences are over-represented in the public knowledge base. For analysis shown in Fig. 2, epitopes and non-epitopes were grouped by length, and all sequences of each length group were randomly concatenated into a single virtual protein. Hydrophilicity of all consecutive non-overlapping 20-aa peptide windows of each concatenate was predicted by use of the Parker ProtScale (11) in ExPASy (29), a parameter used in B-cell epitope prediction. Results in Fig. 2A show that in the 12–15-, 16–20-, and 21–30-aa length concatenates, the hydrophilicity scores of epitope and non-epitope virtual proteins differ highly significantly (p value <10−6, Student's t test). In contrast, the hydrophilicity of epitope and non-epitope virtual proteins is not different for the 6–11-aa length concatenates (p value = 0.052). Thus, for peptides longer than 11 aa, the hydrophilicity scale discriminates between epitopes and non-epitopes but not for shorter peptides.
FIGURE 2.

B-cell epitope prediction score and performance in dependence on peptide length. A, hydrophilicity scores of epitopes and non-epitopes are grouped by length in the Lbtope_Variable_non_redundant dataset (24). Hydrophilicity (Parker) (11) scores were obtained by using default settings in the ProtScale tools of the ExPASy server (29). Length-dependent hydrophilicity (±95% CI) of epitopes and non-epitopes is shown in green and red, respectively, and the p values for differences are shown in green. B, epitope length-dependent prediction performance (area under receiver operating characteristic curve) of different prediction scales in the Lbtope_Variable_non_redundant dataset. ***, p value <10−6 for comparison of any scale to any other scale of 6–11-aa epitopes versus longer epitopes. Hydrophobicity (Miyazawa, 30), a ProtScale (29) for hydrophobicity; Bepipred, a hidden Markov model combined with the Parker hydrophilicity scale (19); IUPred-L, an algorithm for protein disorder tendency (31). C, all 6–20-aa epitopes and non-epitopes of the Lbtope_Confirm dataset (24) grouped into 6–11-, 12–15-, and 16–20-aa peptides are compared with Swiss-Prot 12-, 14-, and 18-aa peptides of the Bcpreds BCP12, BCP14, and BCP18 datasets (20). The p value for hydrophilicity score differences between epitopes and non-epitopes is shown in green and between non-epitopes and Swiss-Prot random peptides in blue. All epitopes have higher hydrophilicity scores than Swiss-Prot random peptides (p value ≤ 10−5).
Similarly, with values of ∼0.50 for the AUC ROC curve, additional scales in Fig. 2B show random distribution of epitope versus non-epitope prediction for the 6–11-aa concatenates. In contrast, these scales show highly significantly increased prediction accuracy for concatenates of peptides longer than 11 amino acids, indicating significant discrimination between epitopes and non-epitopes (Fig. 2B). Analysis of the “Lbtope_Confirm” subset, composed of epitopes/non-epitopes that were at least twice independently experimentally validated, confirmed the result of the Lbtope_Variable_non_redundant dataset (Fig. 2C). An additional finding is that the 16–20-aa non-epitopes do not have a significantly higher score than the random Swiss-Prot peptides, although the 6–11- and 12–15-aa non-epitopes do (Fig. 2C). This suggests that the Lbtope_Confirm dataset may have a high frequency of incorrect identification of short 6–15-aa peptides, particularly 6–11-aa, as non-epitopes.
Evaluation Accuracy for B-cell Epitope Prediction Depends on Epitope/Non-epitope Discrimination in Test Datasets
Ideal algorithms/scales for prediction of B-cell epitopes should discriminate known epitopes from experimentally validated non-epitopes and identify epitopes within complete source proteins and proteomes. Since prediction accuracy should ideally be validated with multiple datasets, we evaluated the prediction performance by use of four positive datasets of experimentally validated B-cell epitopes and five negative datasets of experimentally validated non-epitopes or of random peptides from proteomes (Table 1). All 16–20-aa epitope/non-epitope sequences of the datasets were centered within their 50-aa source protein sequences, and these 50-aa sequences were randomly concatenated into a single virtual protein, interspersed with random 150-aa sequences from the Swiss-Prot database (supplemental Table S1 and Appendix). For evaluation of B-cell epitope prediction, we used the original score of each algorithm with default settings for each amino acid residue, thus each epitope/non-epitope sequence received individual scores for the 20 central residues. Correct or incorrect epitope prediction of all residues was evaluated by AUC in ROC analysis.
TABLE 1.
B-cell epitope prediction accuracy (AUC of ROC curves) in dependence on the evaluation dataset
a Datasets with experimentally identified epitopes/non-epitopes or random peptides are shown (see supplemental Table S1).
b AUC data are shown for the best performing scale of each category as defined for all scales tested (see supplemental Tables S2 and S3).
c Antigenicity scale (17), IEDB tool for antibody epitope prediction.
d Support vector machine model (SVM) trained on the Lbtope_Confirm dataset (24).
e Surface accessibility scale (13), IEDB tool for epitope prediction.
f β-turn scale (32), IEDB tool for epitope prediction.
g Average of seven propensity scales for epitope prediction (15).
h Flexibility scale (12), IEDB tool for epitope prediction.
i Quality of scales or datasets was ranked by AUC, with rank number determined by 1 for the highest AUC and addition of 1 for each 0.01 AUC reduction; antigenicity and Lbtope scales were excluded from ranking because antigenicity is a negative predictor and Lbtope was trained on the analysis dataset.
j Not applicable for quality ranking because the datasets served to train the Lbtope support vector machine model.
k Highest AUC in the compared dataset.
In Table 1, the column for each scale indicates epitope versus non-epitope discrimination (AUC in ROC analysis) of the scale for each compared combination of positive and negative datasets. The average AUC column, next to the rightmost column, indicates epitope/non-epitope discrimination averaged over all tested scales and therefore ranks the combined discrimination in both positive and negative datasets. The four positive datasets can be ranked by their AUC in comparison with the negative Swiss-Prot or Ctr-Proteome datasets, clearly showing significantly higher discrimination for both chlamydial datasets than for the fBcpreds and Lbtope_Confirm datasets (p value ≤0.02, one-tailed paired Student's t test).
The average AUC row, next to the bottom row in Table 1, indicates epitope/non-epitope discrimination averaged over the 12 tested pairs of datasets and shows that disorder tendency discriminated best of all algorithms tested (AUC = 0.75 for IUPred-L [31]), highly significantly better than Bepipred (AUC = 0.70), the next-best algorithm in the 12 AUC comparisons of positive and negative datasets (p value <10−3, one-tailed paired Student's t test). Since the machine learning Lbtope algorithm was trained on the Lbtope_Confirm dataset, it performed extremely well in this dataset (AUC = 0.97, 0.84, and 0.81), but very poorly, close to randomization, in all other datasets (average AUC = 0.57). In contrast, the protein disorder scale IUPred-L consistently discriminated best (Table 1), with AUCs depending on the datasets (0.58–0.88).
Wide Amino Acid Context and Standardized Scoring Maximize B-cell Epitope Prediction Accuracy
Most amino acid propensity and B-cell prediction scales score the context-dependent epitope likelihood for each amino acid residue by averaging the adjacent ±4 residues (10–19). In contrast, protein disorder prediction operates in a wider sequence context (31). To simulate the wider sequence context, we asked the question if scores for long peptides improved prediction accuracy of narrow-context scales and, if so, what the peptide length dependence of such an improvement was. In Table 2, we determined B-cell epitope prediction accuracy for single scores for the central 1-aa epitope/non-epitope residue and contrasted it to prediction by single average scores for the central 10-, 15-, 20-, 25-, or 30-aa residues. The mean AUC values of all 12 comparisons of four to five non-epitope datasets (as in Table 1) are shown in Table 2. The results indicate optimal B-cell epitope prediction for 20–30-aa peptide scores. Long peptide scoring improves prediction substantially for narrow-context scales such as hydrophilicity, hydrophobicity, flexibility, or Bepipred but not for wide-context protein disorder tendency scales such as IUPRed-L and VSL2B.
TABLE 2.
20–30-aa peptide scores optimally predict B-cell epitopes in virtual concatenated polyproteins
References 11, 12, 15, 19, 24, 30–32 are cited in the table.

a The average of 12 AUC values for the 12-way comparisons of four positive datasets with five negative datasets (Table 1) is shown. Average AUC values that differ by <0.01 from the maximum (bold red font) are shown in red font.
b A single score of the central residue was considered for each peptide in the datasets shown in Table 1.
c Scores of the central 10/15/20/25/30aa residues were averaged to a single peptide score.
d Comparison of 1-aa (single residue) versus 10-aa peptide scoring.
e Comparison of 10-aa versus 25-aa peptide scoring.
F/f AUC significantly higher for 10-aa peptide scoring than for 1-aa scoring (F, 10−6 ≤ p value < 10−3; 10−3≤ p value ≤ 10−2; one tailed paired Student's t-test with 12 AUC values).
G/g AUC significantly higher for 25-aa peptide scoring than for 10-aa peptide scoring (G, 10−4 ≤ p value <10−3; 10-3 ≤ P-value ≤ 0.023).
Table 2 shows prediction of epitopes/non-epitopes embedded in virtual concatenated polyproteins. In this approach, differences between the widely divergent average scores of source proteins cannot be offset by standardization (i.e. set to mean = 0 and S.D. = 1). To eliminate B-cell prediction bias induced by selection of epitope source proteins, we analyzed scores standardized for each protein of the 18 chlamydial protein datasets (Chl_18Prot; supplemental Table S1). Table 3 shows the results for comparison of experimentally validated epitopes of these proteins to experimentally validated non-epitopes (Pos versus Neg) or the total remaining protein (Pos versus Neg + NT). Standardization substantially improves B-cell epitope prediction accuracy for disorder and solvent accessibility scales, but not for individual amino acid propensity scales such as hydrophilicity or hydrophobicity. Similar to results for concatenated polyproteins (Table 2), standardized scores of long peptides improve performance of narrow-context but not of wide-context scales (Table 3).
TABLE 3.
Standardization of individual protein scores improves B-cell epitope prediction
References 11, 19, 24, 30, 31, 33–35 are cited in the table.

a The Chl-18Prot dataset was analyzed. Pos, Positive (epitopes); Neg, negative (non-epitope); NT, not tested (epitope or non-epitope status is unknown). Pos versus Neg indicates epitopes were compared to non-epitopes; and Pos vs Neg+NT indicates epitopes were compared to the total remaining protein. Average AUC values that differ by <0.01 from the maximum (bold red font) are shown in red font.
b Solvent accessibility (ASA_Spine-X) residue solvent accessibility (34); polymorphism is sequence divergence in multiple sequence alignment, calculated by inverting the conservation score of AACon in the Jalview freeware (35).
c Original non-standardized score for central 1-aa residue in the peptides. These scores were obtained with individual protein sequences as input.
d Original scores were standardized (mean = 0 and S.D. = 1) for each of the 18 chlamydial proteins, and the standardized score for the central 1-aa residue in the peptides is shown.
e Difference in AUC values between standardized and non-standardized scores.
f Sensitivity at a given specificity is significantly higher in ROC curves for standardized versus non-standardized scores (f, 10−6 ≤ p value≤ 0.01; one-tailed paired Student's t test).
g Peptide scores were calculated using the average of standardized scores for the central 5-, 9-, 17-, 25-, 33-, 41- or 49-aa residues.
h Difference in AUC values between standardized scores of 25- and 9-aa peptides.
i Sensitivity at given specificity is significantly higher in ROC curves for 25-aa versus 9-aa standardized scores (i, 10-6 ≤ p value ≤0.01; one-tailed paired Student's t test).
The polymorphism scale for the 18 chlamydial protein dataset was derived by inverting the Jalview AACon conservation score (35) calculated from multiple sequence alignments of these proteins with the available homologous Chlamydia sequences. Thus, it is completely independent of individual amino acid properties and quantifies only evolutionary sequence change at each residue. Standardization of polymorphism scores in Table 3 improves prediction because chlamydial proteins have widely divergent rates of evolution (2). Importantly, averaging over 25-aa residues again provides maximum prediction accuracy, suggesting that wide-context properties in general are the best predictors of B-cell epitopes from their primary amino acid sequences.
Protein Disorder Most Accurately Predicts B-cell Epitopes
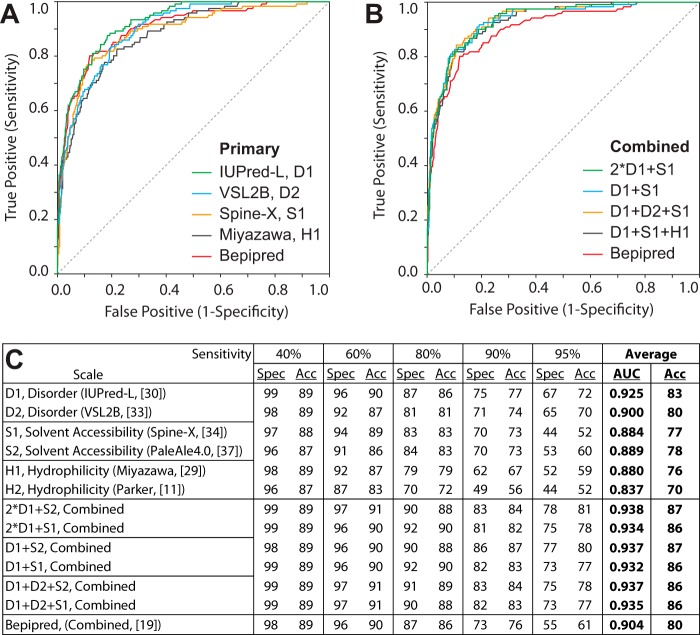
For epitope/non-epitope discrimination in the ROC curve in Fig. 3A, sensitivity at given specificity (or specificity at given sensitivity) of the IUPred-L disorder or the combined scale is higher than that of Bepipred (19) or LBTope (24) (p value <10−4, one-tailed paired Student's t test). Similarly, when epitopes were discriminated from the complete remaining proteins in Fig. 3B, IUPred-L scale performed significantly better than Bepipred or LBTope. In final testing of the overall prediction approach applied to the 18 individual proteins of the Chl_18Prot dataset, IUPred-L scale also best discriminated individual epitope residues from the whole remaining protein (average AUC of IUPred-L = 0.91, minimum = 0.74, maximum = 1.00, S.D. = 0.08; Table 4).
FIGURE 3.

Comparison of ROC curves for prediction of 25-aa epitopes (Table 3). Plots of epitope-positive rate versus false-positive rate for the 18 chlamydial protein dataset are shown. A, prediction of epitopes from confirmed non-epitopes (25-aa epitopes/non-epitopes spaced 10 aa). The combined scale represents the arithmetic mean of two disorder scales, IUPred-L (31) and VSL2B (33), and one solvent accessibility scale, Accessible Surface Area, Spine-X (34). B, prediction of epitopes from the total remaining proteins (non-epitope plus non-tested regions). In both datasets (A and B), the combined scale and the single disorder (IUPred-L) scale performed best (highest sensitivity at given specificity or vice versa), significantly better than Bepipred or LBTope (one-tailed paired Student's t test, p value <10−4).
TABLE 4.
Epitope prediction accuracy (AUC) averaged for individual proteins of the 18-chlamydial protein dataseta

a Original scores obtained with default options for the algorithm/scale were smoothed by a sliding window method in which the score for each residue was averaged for the adjacent ± 12 residues (25-aa moving window). Smoothed scores of residues were standardized for each of the 18 chlamydial proteins and discrimination of epitope residues from remaining total residues was tested for each of the 18 proteins individually.
b Coils (Spine-X) indicate the coils predicted in secondary structure (36).
c A indicates 0.05 ≤ p value > 0.01; B indicates 0.01 ≤p value > 0.001; C indicates 0.001 > p value.
Marginal Improvement in B-cell Epitope Prediction by Combinations of Multiple Scales
In analyses shown in Tables 1 and 2, most epitope/non-epitope sequences were derived from public datasets of variable and largely unknown discrimination accuracy. For maximum accuracy, we therefore selected the 18 chlamydial protein datasets (Chl_18Prot; supplemental Table S1) with extensively validated epitopes as well as non-epitopes on each protein, all identified in a single investigation (2). For the Chl_18Prot dataset, 151 standardized primary scales for B-cell epitope prediction were evaluated (supplemental Tables S2 and S3). To improve B-cell epitope prediction, investigators frequently combine scales (16). To test this concept, we evaluated 126 combined scales that were derived by linear combination of 2–14 standardized primary scales (Fig. 4 and Tables S2 and S3). In Fig. 4, we asked whether the combined scales, derived from 25-aa moving averages of the primary scales, improve B-cell epitope prediction. Results show that the combination of scales only incrementally improves B-cell epitope prediction (Fig. 4). The best combination of the primary scales provides only a 3.8% improvement of prediction accuracy over IUPred-L protein disorder in five tests at 40, 60, 80, 90, and 95% sensitivities (Fig. 4C, p value <0.049, paired Student's t test, with five accuracy values). Collectively, the dominant conclusion is that the main improvement for B-cell epitope prediction comes from using the optimal IUPred-L primary scale (Tables 3, supplemental Tables S2 and S3, and Fig. 4).
FIGURE 4.
Combined scales provide only marginal improvement for B-cell epitope prediction. A, prediction by use of primary scales or B, combined scales. 2·D1 + S1, D1 score weighted 2×. Plots of true positive versus false-positive (ROC curve) are shown. C, prediction performance with 25-aa moving average scores of the Chl-18Prot dataset. At five specified sensitivities, B-cell epitope prediction specificities (Spec) and the corresponding accuracies (Acc) are shown.
Underperformance of Machine-learning B-cell Prediction Algorithms
In evaluation of B-cell epitope prediction algorithms, scores of most physicochemical, structural, and evolutionary protein properties are higher than those of machine learning algorithms (Fig. 5). In addition, the discrimination power of all scales is higher when epitopes are tested against the remaining protein than against experimentally validated non-epitopes (Table 3 and Fig. 5A). As a consequence, the prediction performance against the remaining total protein sequences is also consistently higher for all scales. An explanation for this counterintuitive observation is that non-epitopes had initially been selected as candidate epitopes by high scores in prediction scales (Fig. 2) but failed to react with antibodies. The higher scores for tested non-epitopes thus induced a pre-selection bias that makes evaluation of B-cell epitope prediction scales more difficult.
FIGURE 5.
Comparative discriminatory power of protein property scales and machine learning algorithms, and dominant properties of B-cell epitope regions. Discrimination of proven epitopes, non-epitopes, and untested remaining total protein regions was evaluated in the Chl_18Prot dataset of 18 tested chlamydial protein. A, primary scales. Prediction scores (±95% CI) of protein property scales for epitope, non-epitope, and not tested datasets. B, combined scales and machine learning algorithms. Prediction scores (±95% CI) shown for combined scales derived from primary scales and for machine learning algorithms developed for B-cell epitope prediction. Combined scales are derived from primary scales (supplemental Table S2). C, comparative amino acid frequencies of B-cell epitope and non-epitope regions.
Fig. 5B compares B-cell epitope prediction scales that were among the best combinations of the primary scales in our study with several publicly available algorithms/scales that almost uniformly perform poorly. This poor discriminatory power of machine learning algorithms most likely results from suboptimal training datasets with an over-representation of short non-epitopes. For example, Lbtope was trained on 80% short 6–16-aa confirmed non-epitopes. In contrast, Bcpreds was trained by use of random Swiss-Prot peptides as non-epitopes, equal in length to confirmed epitopes, and they performed better than Lbtope (0.06–0.10 AUC value difference between Bcpreds and Lbtope; Fig. 5B). Among the published combined B-cell epitope prediction scales, only Bepipred showed acceptable performance, better than the accurate Parker hydrophilicity scale (0.06–0.09 ΔAUC compared with Parker hydrophilicity; see Table 3). Bepipred nevertheless requires long peptide scores for optimal performance (0.07–0.10 ΔAUC between 25-aa peptide and default scoring; Table 3), and it is not a pure machine learning algorithm because it combines a protein property scale, Parker hydrophilicity with a hidden Markov model (19).
Dominant Properties of B-cell Epitope Regions
Our evaluation of the discriminatory power of B-cell prediction algorithms in the extensively experimentally confirmed Chl_18Prot dataset allowed us to deduce some critical global properties that define natural B-cell epitope regions. Clearly, the dominant property is the propensity for a disordered state of amino acids in B-cell epitopes. This property is linearly correlated to hydrophilicity (inverted Miyazawa hydrophobicity scale (30); R2 = 0.66, p value <10−6; linear regression analysis of the 25-aa peptide scores centered around each residue of the Chl_18Prot dataset), flexibility (Karplus and Schulz (12); R2 = 0.56, p value <10−6; solvent accessibility (Spine-X (34); R2 = 0.49, p value <10−6), evolutionary mutation rate (R2 = 0.43, p value <10−6), coils in secondary structure (PSIPRED (52); R2 = 0.42, p value <10−6), and β-turns (Levitt (54); R2 = 0.40, p value <10−6). Thus, due to multi-collinearity, the multifaceted properties of protein disorder tendency synthesizes all of these properties into a single descriptor (Fig. 5). The physicochemical, structural, and evolutionary properties of B-cell epitope regions discriminate them sufficiently to translate into significant differences in amino acid composition to the remaining total proteins. B-cell epitopes are enriched for proline, followed by glutamic and aspartic acids, asparagine, threonine, alanine, and serine (Fig. 5C). Epitopes are also relatively depleted of leucine, isoleucine, tryptophan, phenylalanine, tyrosine, and cysteine.
Proposed B-cell Epitope Prediction
As a result of the preceding analyses, an easily implemented approach for accurate B-cell epitope prediction has emerged that should be useful for investigators in many fields of antibody research. Fig. 6 demonstrates the application of the previous findings for B-cell epitope prediction in an actual example for which we generated epitope scanning data of the complete chlamydial protein IncA. In Fig. 6A, the default IUPred-L and VSL2B disorder scores of the IncA protein are plotted against IncA residue number. Solvent accessibility and hydrophilicity are shown in Fig. 6, B and C. In Fig. 6D, noise was reduced by smoothing the scores as 25-aa moving averages, and comparison was improved by standardizing the data. Comparison of these IncA epitope prediction plots with actual IncA peptide reactivity in Fig. 6E clearly shows that IUPred-L predicted scores best match experimental observations and confirm the superior B-cell epitope discriminatory power of protein disorder tendency as calculated by IUPred-L.
FIGURE 6.
Optimal B-cell epitope prediction. A, disorder scores plotted against the C. pecorum IncA protein residues. Scores were obtained at default settings from IUPred-L (31) and VSL2B (33) web servers. B, default ASA_Spine-X solvent accessibility scores (34). C, hydrophilicity scores (inverted default Miyazawa hydrophobicity (29, 30)). D, standardized 25-aa moving average smoothed scores of scales shown in A–C. E, IUPred-L and combined scale scores compared with IncA peptide antigen reactivity with mouse sera. The combined scale is derived from the unweighted mean of standardized smoothed scores of scales shown in D.
Fig. 6E displays optimal prediction approaches by the combined scores of scales shown in Fig. 6D, and smoothed and original default IUPred-L scores. While combined scores have marginally better discriminatory power (Fig. 5), for practical purposes we consider the accuracy of IUPred-L sufficient. Also, given the wide-context nature of protein disorder scales, scores are sufficiently stable to even render smoothing unnecessary, allowing direct use of default plots obtained from the IUPred-L webserver for B-cell epitope prediction. Therefore, 16–30-aa peptide antigens for laboratory testing can be selected directly from peak disorder regions of the IUPred-L plot.
Discussion
The results of this study suggest strategies for B-cell epitope identification that deviate from current approaches that many investigators use. Our approach initially identifies protein regions that harbor B-cell epitopes rather than immediately focusing on identifying peptide antigens of specified length. B-cell epitope regions can be predicted with high accuracy simply by selection of the peak regions from the IUPred-L disorder plot (31) of a protein antigen (Fig. 6E). Next, these high probability epitope regions should be confirmed with 16–30-aa-long peptide antigens using pooled antisera. Fine mapping of highly reactive regions with overlapping 16-aa peptides, using the individually reactive antisera of the pool, identifies regions with several functional aa residues embedded among structural epitope residues (6). Further reduction in peptide antigen length entails mapping with very short 6–12-aa peptides. Success at this stage relies on stochastic identification (Fig. 2) of closely spaced randomly distributed functional residues that maximally contribute to antibody binding. Antibody binding of such short peptides is, however, typically low (Fig. 1), most likely because antigens of less than 16 aa will not bind to the complete CDR of an antibody (3–7).
This approach is derived from the conclusive evidence that short 7–12-aa peptide antigens of confirmed Chlamydia spp. epitopes bind antibodies poorly (Fig. 1), and therefore many of these epitopes would be falsely classified as non-epitopes if they were identified by short peptide mapping. The poor reactivity of short peptide antigens combined with data in Fig. 2 strongly suggest that many of the short non-epitopes in public B-cell epitope datasets are likely to be actual epitopes. Most investigators who develop B-cell epitope prediction algorithms/scales draw training and test datasets from public databases such as IEDB. These reference datasets are suboptimal due to over-representation of short non-epitope peptides and inherently compromise the performance (supplemental Tables S2 and S3 and Fig. 5B) of machine learning algorithms (CBTope, LBTope, COBEpro, BCPreds (20, 22–24)) or antigenicity scales (Chen AAP, Kolaskar antigenicity, BcePred (15, 17, 18)). For instance, the LBTope algorithms perform optimally in the IEDB-derived LBTope datasets (AUC = 0.81–0.97) but poorly in independent datasets (average AUC = 0.57, Table 1). In contrast, many untrained protein property scales, such as protein disorder tendency, that were developed for different reasons nevertheless predict B-cell epitopes with higher accuracy than specifically developed B-cell epitope prediction scales/machine learning algorithms (supplemental Tables S2 and S3 and Fig. 5B).
A fundamental conundrum in B-cell epitope prediction is the conceptual and methodological approach that leads to the eventual identification of a B-cell epitope. Vastly preferable is the use of x-ray crystallography-solved three-dimensional structures of antigen-antibody complexes. Such data define precisely the actual determinants of a protein antigen that specifically contact an antibody, in essence the set of protein residues that are buried under a cognate antibody in the antibody-antigen complex (3–7). However, only 26–107 non-identical three-dimensional structures of antigen-antibody complexes have been generated by different investigators from the Protein Data Bank crystallographic database (3–7, 56–62). Such data were used for training and development of several B-cell epitope prediction methods such as CEP, DiscoTope, Rapberger's method, Ellipro, PEPITO, and Epitopia (56–62). The major shortcoming is the requirement for the three-dimensional structure of the protein antigen. In practice, this limitation is currently insurmountable because we do not know the three-dimensional structure of most proteins.
In practice, B-cell epitopes are commonly determined by use of peptide antigens and their ability to capture antibodies. This approach does not identify which residues of the peptide are in binding contact with antibody CDR residues and which actually contribute to the antigen-antibody complex formation. Nonetheless, antibody-reactive peptide sequences, particularly those identified by systematic mapping with overlapping peptides, are commonly referred to as B-cell epitopes (16, 63–65). This terminology is justified because even non-binding residues are specifically required to provide the structural context for binding residues, thus the linear peptide sequence is still an indispensable, if not complete, characterization of a B-cell epitope. It is important, however, to understand that epitope prediction from linear peptide sequences will weigh the total combined contributions of binding (functional) and spacer (structural) amino acids to an epitope. Nevertheless, B-cell epitope prediction from the primary amino acid sequence of a protein is a valid and, for practical purposes, highly desirable approach. In addition, tens of thousands of B-cell epitope/non-epitope sequences have been deposited in IEDB (24). Thus, the sequence-based B-cell epitope datasets provide a viable basis for training and development of B-cell epitope prediction algorithms (10–22).
The profound conundrum for epitope prediction by use of linear peptide sequence-based methods, however, is the fact that more than 90% of all B-cell epitopes are not linear, composed of immediately neighboring binding residues, but they are discontinuous. In almost all B-cell epitopes, the typical 2–5 dominant binding residues will be discontinuously arranged randomly in the linear epitope sequence (3, 6, 7). Nevertheless, in the majority of epitopes these binding residues are still closely spaced. For instance, Sivalingam and Shepherd (6) show that 30-aa peptides will encompass the functional residues of 75% of all B-cell epitopes. Thus, increased lengths of peptide antigens will increase the probability of capturing more of the residues of any epitope that are required for high affinity antibody binding (Fig. 1). In addition, long peptides may increase the probability of capturing different antibody clones that may bind the same epitope region differently (65). For instance, C. trachomatis OmpA serovar-specific peptide serology has used 6–10-aa peptides, with inconsistent results (66–70). In our study, we observed strong but completely serovar-specific antibody reactivity by use of ≥16-aa peptide antigens (2). Importantly, inclusion of conserved adjacent residues shared among chlamydial species, in addition to the 7–10 central polymorphic serovar-determinant OmpA residues, was required for strong, yet specific, antibody binding (2).
Conceptually, a peptide antigen captures antibodies if it can fold to complement the binding region of the cognate antibody (65). Because of such structural constraints, the length of peptide antigens may also negatively influence antibody binding. For instance, if the few randomly spaced dominant binding residues are obstructed by structural constraints such as misfolding, masking by non-epitope residues, or peptide aggregation (65), antibody binding may be compromised. Our study clearly shows that moderate elongation of peptide antigens strongly enhances antibody binding, while more extensive elongation reduces antibody binding again in 20% of B-cell epitopes, presumably by masking epitope residues (Fig. 1). The implication of this fact is that an optimal sequence length exists that most reliably discriminates between true epitopes and non-epitopes and that sequences of that length should be used to generate datasets for the development of B-cell prediction methods.
A protein surface can be thought of as a continuous landscape of epitopic regions, and any region of this landscape may be identified as an epitope under specific conditions (56, 63–65). For instance, Singh et al. (24) reported that all non-epitopes in the LBTope_Confirm dataset have been reported as “non-epitopes” in at least two studies. Yet 8.3% of these non-epitopes are reported as “epitopes” in the fBcpreds dataset (21). Thus, binary classification of antigen regions into epitopes or non-epitopes is problematic because all epitopes of most antigens are not known, and defining B-cell epitopes and non-epitopes is a challenging task due to the variability in epitope discovery assays (71) and the stochastic antibody responses to protein antigens (9) and their epitopes (2). Muller et al. (71) found almost the entire histone 2A protein antigenic when they forced highest B-cell stimulation and antibody reactivity by excessive use of adjuvants and high antigen doses. In contrast, raising antisera in our study by experimental infection rather than by forced immunization very likely resulted in much lower adjuvantation and lower antigenic stimulus by physiologically processed native protein antigens (2). Thus, antibodies likely were generated mainly against exposed antibody-binding regions of highly expressed proteins. In addition, targeting known immunodominant proteins by the use of antisera pooled from multiple individuals maximized correct epitope/non-epitope discrimination by offsetting the inherent stochasticity of antibody formation in individuals and by minimizing false-negative results. We observed a clear trend that certain protein regions are a more frequent source of B-cell epitopes than others, and we think that our study identified the distinctive properties of such preferentially antibody-recognized regions.
Kringelum et al. (7) determined by x-ray crystallography that hydrophobic amino acids of epitopes located closest to the antibody, and charged amino acids most distant, but that the amino acid composition of equally surface-exposed non-epitopes did not differ significantly from epitopes. However, the amino acid composition of epitopes deviated significantly from the whole protein (7). We compared properties of B-cell epitope regions with experimentally confirmed non-epitope regions or the remaining protein regions, but we do not know about surface exposure. Similarly, we report that many protein properties of B-cell epitope regions differ substantially from the total remaining proteins (Fig. 5), making these properties candidates for B-cell epitope prediction. Accessibility of the antigen by cognate B-cell receptors or antibodies is the central concept in molecular recognition of epitope by the paratope, and thus highly surface-exposed hydrophilic/charged epitope residues will first interact with the antibody (Fig. 5C). Although hydrophobic amino acids except for alanine, the smallest one, are under-represented in epitopes, those that are present may “provide the glue” in the final stabilization of the antigen-antibody complex by hydrophobic interaction. All non-covalent antigen-antibody interactions are thought to be driven by shape complementarities in the complex formation (57). Thus, paratopes may interact preferentially with flexible regions of an antigen rather than with highly structured regions (72). Precisely because of relaxed structural constraints, such protein regions should accommodate higher amino acid substitution rates, favoring under immunoselective pressure the emergence of escape mutants.
B-cell epitopes have historically been recognized as hydrophilic (10, 11), flexible (12), mobile (high B factor; 73, 74), surface-exposed or solvent-accessible (3, 5, 13, 57, 75), enriched with β-turns (14) or coils/loops (3–4, 7), and highly sequence-polymorphic (3–5). Recent three-dimensionally based studies (3–7) also show that epitopes compared with non-epitope regions are (i) enriched for polar and charged amino acids and depleted of hydrophobic amino acids, (ii) more surface-exposed than the remaining protein, (iii) more sequence polymorphic, and (iv) enriched with unorganized secondary structure elements and depleted of strands and helices (3–7). In our best characterized 18 chlamydial proteins (Chl_18Prot dataset), hydrophilicity, solvent accessibility/surface-exposed tendency, coils in secondary structure, and evolutionary mutation rate are all collinear and highly predictive of B-cell epitopes. Protein disorder tendency synthesizes these properties into a single descriptor, rendering IUPred-L disorder scores the single best predictor of epitope regions (Fig. 5). Hence, our findings regarding B-cell epitope properties are in agreement with modern three-dimensional structure-based studies (3–7) or classical peptide sequence-based studies (10–14), and protein disorder is the unifying concept behind them.
Important antigenic regions of viral and bacterial proteins have been identified as disordered regions of these protein antigens (72). However, x-ray crystallography studies have not specifically reported the localization of B-cell epitopes in disordered protein regions (3–7, 56–65). The most likely explanation for this discrepancy to our results is the fact that the Protein Data Bank database is biased toward proteins of common interest that are easy to produce and crystallize. Many expressed proteins cannot be crystallized, and among the main factors for this failure is the presence of even small numbers (1–10 aa) of disordered residues that are well known to have deleterious effects on crystallization (76, 77). For convenient determination of three-dimensional structures, disordered protein regions are removed from expressed proteins (78). As a result, disordered proteins or protein regions are rare in the Protein Data Bank database compared with whole proteomes (79–81). Moreover, crystal packing is thought to enforce certain disordered regions to become ordered (31), resulting in incorrect characterization of disordered protein residues. In addition, disordered segments crystallized together with binding antibodies are usually classified as ordered structure in the antigen-antibody complex, despite their lack of ordered structure in the unbound state. Thus, datasets generated by crystallography may inherently under-represent B-cell epitopes with high disorder tendency.
Protein disorder tendency has also not been proposed for B-cell epitope prediction from primary amino acid sequences, although many protein property scales, particularly aa propensity scales, have been tested and recommended for B-cell epitope prediction (16, 63–65). In our study, the IUPred-L disorder scale has the highest epitope discriminatory power in all datasets. We explain this discrepancy by the typical experimental approach with which investigators test sequence-based epitope prediction methods as follows: wide-context disorder properties of proteins will not be correctly determined by solely analyzing the typically short peptide sequences of databases. To achieve correct results, we elongated test peptides with source protein sequences and embedded them in a wider context of random Swiss-Prot sequences (supplemental Table S1 and supplemental Appendix).
As a norm in investigations addressing protein disorder, protein residues are binary-classified as either “ordered” or “disordered.” In contrast, disorder prediction algorithms quantify the probability of protein disorder, and binary classification converts the prediction scores by using an arbitrary cutoff at a pre-determined threshold. By these criteria, many epitopes would not classify as disordered. However, relative to the moving average score of the whole source protein, B-cell epitopes consistently score highest for protein disorder tendency. For actual B-cell epitope prediction, the IUPred-L protein disorder scale consistently performs best (87% specificity at 80% sensitivity, 86% accuracy; Fig. 4). However, if a 25-aa moving average score is used, several other protein property scales such as hydrophilicity (Parker), hydrophobicity (Miyazawa), solvent accessibility (Spine-X), or Bepipred perform similarly. In fact, scoring by narrow-context scales for long 20–30-aa peptides reflects protein disorder tendency such as the Globplot-2 algorithm predicts protein disorder tendency by a wide-context hydrophilicity score (38). It is noteworthy that even the best combination of top performing scales does not substantially increase prediction performance (Fig. 4), due to multi-collinearity of these scales. The best performing combined scale (Figs. 4 and 5 and supplemental Tables S2 and S3), derived from smoothed and standardized 25-aa peptide scores of three primary scales, improves prediction accuracy only marginally (90–92% specificity at 80% sensitivity, 88–90% accuracy; Fig. 4).
Our data show that wide-context disorder scores or long 20–30-aa peptide scores of narrow-context scales are optimal for B-cell epitope prediction (Tables 2 and 3), consistent with the higher antibody binding of 16–30-aa peptide antigens (Fig. 1). Compared with highly structured protein regions, disordered regions may have several functional advantages for efficient interactions with partner molecules (26, 27), such as the capacity of initiating binding by long range electrostatic interactions, high flexibility, binding plasticity and speed, minimal steric restrictions in binding, and the ability to form very stable intertwined complexes (26, 27, 82–87). Hence, our investigation merges theoretical advances in protein biophysics with very practical aspects of protein interaction, the identification of peptide sequences best suited for recognition by CDRs of antibodies.
Author Contributions
K. S. R. and B. K. planned the experiments; K. S. R. and E. U. C. performed the experiments; K. S. R. and B. K. analyzed the data; K. S. R., B. K., and K. S. contributed reagents and essential material; and K. S. R. and B. K. wrote the paper.
Supplementary Material
This work was supported by the Molecular Diagnostics Laboratory at the Department of Pathobiology, College of Veterinary Medicine at Auburn University. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1–S3 and Appendix.
- aa
- amino acid
- CDR
- complementarity-determining region
- AUC
- area under the curve
- ROC
- receiver operating characteristic
- IEDB
- Immune Epitope Database
- rlu
- relative light unit
- NT
- not tested
- Pos
- positive
- Neg
- negative
- CI
- confidence interval.
References
- 1.Blythe M. J., and Flower D. R. (2005) Benchmarking B-cell epitope prediction: underperformance of existing methods. Protein Sci. 14, 246–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahman K. S., Chowdhury E. U., Poudel A., Ruettger A., Sachse K., and Kaltenboeck B. (2015) Defining species-specific immunodominant B-cell epitopes for molecular serology of Chlamydia species. Clin. Vaccine Immunol. 22, 539–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinstein N. D., Mayrose I., Halperin D., Yekutieli D., Gershoni J. M., and Pupko T. (2008) Computational characterization of B-cell epitopes. Mol. Immunol. 45, 3477–3489 [DOI] [PubMed] [Google Scholar]
- 4.Ofran Y., Schlessinger A., and Rost B. (2008) Automated identification of complementarity determining regions (CDRs) reveals peculiar characteristics of CDRs and B-cell epitopes. J. Immunol. 181, 6230–6235 [DOI] [PubMed] [Google Scholar]
- 5.Sun J., Xu T., Wang S., Li G., Wu D., and Cao Z. (2011) Does difference exist between epitope and non-epitope residues? Immunome Res. 201, 1–11 [Google Scholar]
- 6.Sivalingam G. N., and Shepherd A. J. (2012) An analysis of B-cell epitope discontinuity. Mol. Immunol. 51, 304–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kringelum J. V., Nielsen M., Padkjær S. B., and Lund O. (2013) Structural analysis of B-cell epitopes in antibody: protein complexes. Mol. Immunol. 53, 24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brack C., Hirama M., Lenhard-Schuller R., and Tonegawa S. (1978) A complete immunoglobulin gene is created by somatic recombination. Cell 15, 1–14 [DOI] [PubMed] [Google Scholar]
- 9.Wang J., Zhang Y., Lu C., Lei L., Yu P., and Zhong G. (2010) A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J. Immunol. 185, 1670–1680 [DOI] [PubMed] [Google Scholar]
- 10.Hopp T. P., and Woods K. R. (1981) Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. U.S.A. 78, 3824–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker J. M., Guo D., and Hodges R. S. (1986) New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and x-ray-derived accessible sites. Biochemistry 25, 5425–5432 [DOI] [PubMed] [Google Scholar]
- 12.Karplus P. A., and Schulz G. E. (1985) Prediction of chain flexibility in proteins–a tool for the selection of peptide antigens. Naturwissenschaften 72, 212–213 [Google Scholar]
- 13.Emini E. A., Hughes J. V., Perlow D. S., and Boger J. (1985) Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 55, 836–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellequer J. L., Westhof E., and Van Regenmortel M. H. (1993) Correlation between the location of antigenic sites and the prediction of turns in proteins. Immunol. Lett. 36, 83–99 [DOI] [PubMed] [Google Scholar]
- 15.Saha S., and Raghava G. P. (2004) in Artificial Immune Systems (Nicosia G., Cutello V., Bentley P. J., and Timmis J. I., eds) pp. 197–204, Springer, Heidelberg, Germany [Google Scholar]
- 16.Ponomarenko J. V., and Van Regenmortel M. H. (2009) in Structural Bioinformatics (Bourne P. E., and Gu J., eds) 2nd Ed., pp. 849–879, John Wiley, Hoboken, NJ [Google Scholar]
- 17.Kolaskar A. S., and Tongaonkar P. C. (1990) A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 276, 172–174 [DOI] [PubMed] [Google Scholar]
- 18.Chen J., Liu H., Yang J., and Chou K. C. (2007) Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 33, 423–428 [DOI] [PubMed] [Google Scholar]
- 19.Larsen J. E., Lund O., and Nielsen M. (2006) Improved method for predicting linear B-cell epitopes. Immunome Res. 2, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Manzalawy Y., Dobbs D., and Honavar V. (2008) Predicting linear B-cell epitopes using string kernels. J. Mol. Recognit. 21, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Manzalawy Y., Dobbs D., and Honavar V. (2008) Predicting flexible length linear B-cell epitopes. Comput. Syst. Bioinformatics Conf. 7, 121–132 [PMC free article] [PubMed] [Google Scholar]
- 22.Sweredoski M. J., and Baldi P. (2009) COBEpro: a novel system for predicting continuous B-cell epitopes. Protein Eng. Des. Sel. 22, 113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ansari H. R., and Raghava G. P. (2010) Identification of conformational B-cell epitopes in an antigen from its primary sequence. Immunome Res. 6, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh H., Ansari H. R., and Raghava G. P. (2013) Improved method for linear B-cell epitope prediction using antigen's primary sequence. PLoS ONE 8, e62216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uversky V. N. (2013) Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta 1834, 932–951 [DOI] [PubMed] [Google Scholar]
- 26.Uversky V. N. (2013) A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 22, 693–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z., and Huang Y. (2014) Advantages of proteins being disordered. Protein Sci. 23, 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens R. S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R. L., Zhao Q., Koonin E. V., and Davis R. W. (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759 [DOI] [PubMed] [Google Scholar]
- 29.Gasteiger E., Hoogland C., Gattiker A., Wilkins M. R., Appel R. D., and Bairoch A. (2005) in Proteomics Protocols Handbook (Walker J. M., ed) pp 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 30.Miyazawa S., and Jernigan R. L. (1985) Estimation of effective interresidue contact energies from protein crystal structures: quasi-chemical approximation. Macromolecules 18, 534–552 [Google Scholar]
- 31.Dosztányi Z., Csizmok V., Tompa P., and Simon I. (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 [DOI] [PubMed] [Google Scholar]
- 32.Chou P. Y., and Fasman G. D. (1978) Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 47, 45–148 [DOI] [PubMed] [Google Scholar]
- 33.Peng K., Radivojac P., Vucetic S., Dunker A. K., and Obradovic Z. (2006) Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics 7, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faraggi E., Xue B., and Zhou Y. (2009) Improving the prediction accuracy of residue solvent accessibility and real-value backbone torsion angles of proteins by guided-learning through a two-layer neural network. Proteins 74, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faraggi E., Yang Y., Zhang S., and Zhou Y. (2009) Predicting continuous local structure and the effect of its substitution for secondary structure in fragment-free protein structure prediction. Structure 17, 1515–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirabello C., and Pollastri G. (2013) Porter, PaleAle 4.0: high-accuracy prediction of protein secondary structure and relative solvent accessibility. Bioinformatics 29, 2056–2058 [DOI] [PubMed] [Google Scholar]
- 38.Linding R., Russell R. B., Neduva V., and Gibson T. J. (2003) GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31, 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galzitskaya O. V., Garbuzynskiy S. O., and Lobanov M. Y. (2006) FoldUnfold: web server for the prediction of disordered regions in protein chain. Bioinformatics 22, 2948–2949 [DOI] [PubMed] [Google Scholar]
- 40.Ishida T., and Kinoshita K. (2007) PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35, W460–W464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., and Russell R. B. (2003) Protein disorder prediction: implications for structural proteomics. Structure 11, 1453–1459 [DOI] [PubMed] [Google Scholar]
- 42.Cilia E., Pancsa R., Tompa P., Lenaerts T., and Vranken W. F. (2014) The DynaMine webserver: predicting protein dynamics from sequence. Nucleic Acids Res. 42, W264–W270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozlowski L. P., and Bujnicki J. M. (2012) MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics 13, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizianty M. J., Stach W., Chen K., Kedarisetti K. D., Disfani F. M., and Kurgan L. (2010) Improved sequence-based prediction of disordered regions with multilayer fusion of multiple information sources. Bioinformatics 26, i489–i496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishida T., and Kinoshita K. (2008) Prediction of disordered regions in proteins based on the meta approach. Bioinformatics 24, 1344–1348 [DOI] [PubMed] [Google Scholar]
- 46.Guy H. R. (1985) Amino acid side-chain partition energies and distribution of residues in soluble proteins. Biophys. J. 47, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sweet R. M., and Eisenberg D. (1983) Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J. Mol. Biol. 171, 479–488 [DOI] [PubMed] [Google Scholar]
- 48.Joo K., Lee S. J., and Lee J. (2012) SANN: Solvent accessibility prediction of proteins by nearest neighbor method. Proteins 80, 1791–1797 [DOI] [PubMed] [Google Scholar]
- 49.Petersen B., Petersen T. N., Andersen P., Nielsen M., and Lundegaard C. (2009) A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magnan C. N., and Baldi P. (2014) SSpro/ACCpro 5: almost perfect prediction of protein secondary structure and relative solvent accessibility using profiles, machine learning and structural similarity. Bioinformatics 30, 2592–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhaskaran R., and Ponnuswamy P. K. (1988) Positional flexibilities of amino acid residues in globular proteins. Int. J. Pept. Protein Res. 32, 241–255 [DOI] [PubMed] [Google Scholar]
- 52.McGuffin L. J., Bryson K., and Jones D. T. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 53.Rose G. D., Geselowitz A. R., Lesser G. J., Lee R. H., and Zehfus M. H. (1985) Hydrophobicity of amino acid residues in globular proteins. Science 229, 834–838 [DOI] [PubMed] [Google Scholar]
- 54.Levitt M. (1978) Conformational preferences of amino acids in globular proteins. Biochemistry 17, 4277–4285 [DOI] [PubMed] [Google Scholar]
- 55.Deléage G., and Roux B. (1987) An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1, 289–294 [DOI] [PubMed] [Google Scholar]
- 56.Ponomarenko J. V., and Bourne P. E. (2007) Antibody-protein interactions: benchmark datasets and prediction tools evaluation. BMC Struct. Biol. 7, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rapberger R., Lukas A., and Mayer B. (2007) Identification of discontinuous antigenic determinants on proteins based on shape complementarities. J. Mol. Recognit. 20, 113–121 [DOI] [PubMed] [Google Scholar]
- 58.Kulkarni-Kale U., Bhosle S., and Kolaskar A. S. (2005) CEP: a conformational epitope prediction server. Nucleic Acids Res. 33, W168–W171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haste Andersen P., Nielsen M., and Lund O. (2006) Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 15, 2558–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ponomarenko J., Bui H. H., Li W., Fusseder N., Bourne P. E., Sette A., and Peters B. (2008) ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics 9, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sweredoski M. J., and Baldi P. (2008) PEPITO: improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure. Bioinformatics 24, 1459–1460 [DOI] [PubMed] [Google Scholar]
- 62.Rubinstein N. D., Mayrose I., Martz E., and Pupko T. (2009) Epitopia: a web-server for predicting B-cell epitopes. BMC Bioinformatics 10, 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.El-Manzalawy Y., and Honavar V. (2010) Recent advances in B-cell epitope prediction methods. Immunome Res. 6, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greenbaum J. A., Andersen P. H., Blythe M., Bui H. H., Cachau R. E., Crowe J., Davies M., Kolaskar A. S., Lund O., Morrison S., Mumey B., Ofran Y., Pellequer J. L., Pinilla C., Ponomarenko J. V., et al. (2007) Towards a consensus on datasets and evaluation metrics for developing B-cell epitope prediction tools. J. Mol. Recognit. 20, 75–82 [DOI] [PubMed] [Google Scholar]
- 65.Van Regenmortel M. H. (2009) What is a B-cell epitope? Methods Mol. Biol. 524, 3–20 [DOI] [PubMed] [Google Scholar]
- 66.Zhong G. M., Reid R. E., and Brunham R. C. (1990) Mapping antigenic sites on the major outer membrane protein of Chlamydia trachomatis with synthetic peptides. Infect. Immun. 58, 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conlan J. W., Clarke I. N., and Ward M. E. (1988) Epitope mapping with solid-phase peptides: identification of type-, subspecies-, species-and genus-reactive antibody binding domains on the major outer membrane protein of Chlamydia trachomatis. Mol. Microbiol. 2, 673–679 [DOI] [PubMed] [Google Scholar]
- 68.Pal S., Cheng X., Peterson E. M., and de la Maza L. M. (1993) Mapping of a surface-exposed B-cell epitope to the variable sequent 3 of the major outer-membrane protein of Chlamydia trachomatis. J. Gen. Microbiol. 139, 1565–1570 [DOI] [PubMed] [Google Scholar]
- 69.Villeneuve A., Brossay L., Paradis G., and Hébert J. (1994) Determination of neutralizing epitopes in variable domains I and IV of the major outer-membrane protein from Chlamydia trachomatis serovar K. Microbiology 140, 2481–2487 [DOI] [PubMed] [Google Scholar]
- 70.Batteiger B. E. (1996) The major outer membrane protein of a single Chlamydia trachomatis serovar can possess more than one serovar-specific epitope. Infect. Immun. 64, 542–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muller S., Plaue S., Couppez M., and Van Regenmortel M. H. (1986) Comparison of different methods for localizing antigenic regions in histone H2A. Mol. Immunol. 23, 593–601 [DOI] [PubMed] [Google Scholar]
- 72.Uversky V. N., Oldfield C. J., and Dunker A. K. (2005) Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 18, 343–384 [DOI] [PubMed] [Google Scholar]
- 73.Westhof E., Altschuh D., Moras D., Bloomer A. C., Mondragon A., Klug A., and Van Regenmortel M. H. (1984) Correlation between segmental mobility and the location of antigenic determinants in proteins. Nature 311, 123–126 [DOI] [PubMed] [Google Scholar]
- 74.Tainer J. A., Getzoff E. D., Alexander H., Houghten R. A., Olson A. J., Lerner R. A., and Hendrickson W. A. (1984) The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature 312, 127–134 [DOI] [PubMed] [Google Scholar]
- 75.Novotný J., Handschumacher M., Haber E., Bruccoleri R. E., Carlson W. B., Fanning D. W., Smith J. A., and Rose G. D. (1986) Antigenic determinants in proteins coincide with surface regions accessible to large probes (antibody domains). Proc. Natl. Acad. Sci. U.S.A. 83, 226–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Canaves J. M., Page R., Wilson I. A., and Stevens R. C. (2004) Protein biophysical properties that correlate with crystallization success in Thermotoga maritima: maximum clustering strategy for structural genomics. J. Mol. Biol. 344, 977–991 [DOI] [PubMed] [Google Scholar]
- 77.Price W. N. 2nd., Chen Y., Handelman S. K., Neely H., Manor P., Karlin R., Nair R., Liu J., Baran M., Everett J., Tong S. N., Forouhar F., Swaminathan S. S., Acton T., Xiao R., et al (2009) Understanding the physical properties that control protein crystallization by analysis of large-scale experimental data. Nat. Biotechnol. 27, 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oldfield C. J., Ulrich E. L., Cheng Y., Dunker A. K., and Markley J. L. (2005) Addressing the intrinsic disorder bottleneck in structural proteomics. Protein 59, 444–453 [DOI] [PubMed] [Google Scholar]
- 79.Huntley M. A., and Golding G. B. (2002) Simple sequences are rare in the Protein Data Bank. Proteins 48, 134–140 [DOI] [PubMed] [Google Scholar]
- 80.Le Gall T., Romero P. R., Cortese M. S., Uversky V. N., and Dunker A. K. (2007) Intrinsic disorder in the protein data bank. J. Biomol. Struct. Dyn. 24, 325–342 [DOI] [PubMed] [Google Scholar]
- 81.Xue B., Dunker A. K., and Uversky V. N. (2012) Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 30, 137–149 [DOI] [PubMed] [Google Scholar]
- 82.Kriwacki R. W., Hengst L., Tennant L., Reed S. I., and Wright P. E. (1996) Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. U.S.A. 93, 11504–11509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wright P. E., and Dyson H. J. (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 293, 321–331 [DOI] [PubMed] [Google Scholar]
- 84.Shoemaker B. A., Portman J. J., and Wolynes P. G. (2000) Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl. Acad. Sci. U.S.A. 97, 8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausio J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., Bailey R. W., Griswold M. D., Chiu W., Garner E. C., and Obradovic Z. (2001) Intrinsically disordered protein. J. Mol. Graph. Model. 9, 26–59 [DOI] [PubMed] [Google Scholar]
- 86.Dyson H. J., and Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 87.Sugase K., Dyson H. J., and Wright P. E. (2007) Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.