SUMMARY
Eukaryotic initiation factor 3 (eIF3), an essential multi-protein complex involved in translation initiation, is composed of 12 tightly associated subunits in humans. While the overall structure of eIF3 is known, the mechanism of its assembly and structural consequences of dysregulation of eIF3 subunit expression seen in many cancers is largely unknown. Here we show that subunits in eIF3 assemble into eIF3 in an interdependent manner. Assembly of eIF3 is governed primarily by formation of a helical bundle, composed of helices extending C-terminally from PCI-MPN domains in eight subunits. We propose that, while the minimal subcomplex of human-like eIF3 functional for translation initiation in cells consists of subunits a, b, c, f, g, i, and m, numerous other eIF3 subcomplexes exist under circumstances of subunit over- or underexpression. Thus, eIF3 subcomplexes formed or “released” due to dysregulated subunit expression may be determining factors contributing to eIF3-related cancers.
Graphical abstract
INTRODUCTION
Regulating protein synthesis in eukaryotes occurs predominantly during translation initiation. Translational regulation is linked to many cellular processes including metabolism, proliferation, differentiation, and cell fate, and has been implicated in a number of diseases (Hershey, 2015; Hinnebusch, 2015). Translation initiation in eukaryotes is controlled by a number of eukaryotic initiation factors (eIFs), which are important for general translation (Hershey, 2010), but that can also regulate specific transcripts (Cao et al., 2015; Lee et al., 2015; Meyer et al., 2015; Pelletier et al., 2015; Wang et al., 2015; Zhou et al., 2015). In humans, eIF3 is the largest eIF and consists of 13 non-identical protein subunits named eIF3a to eIF3m (Damoc et al., 2007). Most other multicellular eukaroytes such as the filamentous fungus Neurospora crassa also have eIF3 complexes structurally and compositionally similar to that in humans (Smith et al., 2013).
During cap-dependent translation, eIF3 functions as a structural scaffold for other eIFs and is crucial in the formation of the translation preinitiation complex (Hinnebusch, 2014). Similarly, eIF3 is required for hepatitis C viral genomic RNA recruitment to the small ribosomal subunit during viral internal ribosome entry site (IRES)-dependent translation initiation (Fraser and Doudna, 2007; Gilbert, 2010; Hinnebusch, 2006; Lopez-Lastra et al., 2005). We recently found that eIF3 can bind directly to the 5′ UTRs of a number of mRNAs to activate or repress their translation (Lee et al., 2015). In addition, eIF3 may regulate translation of specific mRNAs during stress, mediated through binding of m6A modifications (Meyer et al., 2015; Wang et al., 2015).
In zebrafish and worms, knocking down certain eIF3 subunits results in specific developmental defects likely due to translational regulation of developmentally related mRNAs (Choudhuri et al., 2013; Curran and Ruvkun, 2007; Desai and Horvitz, 1989). A number of studies have linked the overexpression, truncation, or downregulation of eIF3 subunits to various cancers (Hershey, 2015), also suggesting that certain eIF3 subunits have direct functions in modulating cellular mechanisms such as cell fate. Despite the large body of evidence linking eIF3 subunit regulation to development, cancer, and disease, the consequences of misregulated eIF3 subunit expression on the overall structure and composition of eIF3, translation, and other cellular processes has not been examined.
The structural core of eIF3 is composed of eight subunits, six containing PCI (eIF3a, c, e, k, l, and m) and two containing MPN (eIF3f and h) domains, similar to the COP9 signalosome (CSN) and proteasome lid complexes (Querol-Audi et al., 2013). Structures of all three complexes show a helical bundle consisting of helices located C-terminal of the PCI-MPN domains (Beck et al., 2012; des Georges et al., 2015; Lander et al., 2012; Lingaraju et al., 2014). The contribution of these C-terminal helices to the assembly of the proteasome lid has been studied previously (Estrin et al., 2013). Although the overall structure of the proteasome lid and CSN are very similar, their proposed assembly pathways are distinct (Meister et al., 2015). Expression of recombinant human eIF3 in Escherichia coli requires all eight of the PCI-MPN-containing subunits, suggesting that the formation of the helical bundle is essential for eIF3 assembly and may be similar to either the proteasome lid or CSN (Sun et al., 2011). However, four of these eight subunits are completely dispensable in the N. crassa eIF3 complex (Smith et al., 2013), implying that eIF3 may assemble by alternative pathways that do not rely on the formation of the helical bundle in the same manner as the proteasome lid and CSN complexes. Therefore, despite recent cryo-electron microscopy (cryo-EM) reconstructions of human eIF3, the specific functions of its subunits and its cellular assembly pathway remain unclear (Aylett et al., 2015; des Georges et al., 2015; Hashem et al., 2013a; Querol-Audi et al., 2013).
Here we map the cellular assembly pathway of eIF3 in N. crassa and humans and find that removing certain non-essential subunits can have dramatic effects on the overall composition of the complex, in some cases liberating stable subcomplexes or subunits from eIF3. Furthermore, we investigate the importance of the helical bundle formed between helices located C-terminal to the PCI and MPN domains in eIF3 complex assembly. The interdependent nature of eIF3 subunit assembly provides a framework for understanding how dysregulation of eIF3 subunit expression impacts development and disease.
RESULTS
Subunit eIF3h Is Required for the Assembly of Four Other Subunits into eIF3
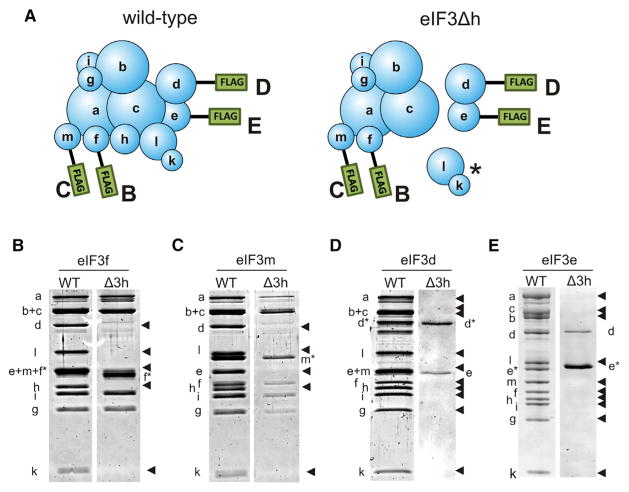
We previously showed that eIF3 subunits k and l in N. crassa assembled into the eIF3 complex as a dimer and that their assembly was dependent on eIF3h (Smith et al., 2013). Using a similar strategy we integrated FLAG-tagged eIF3 subunits d, e, f, or m in an N. crassa strain that expressed all eIF3 subunits (his-3) or in a strain with only eIF3h deleted (eIF3Δh his-3) (Figure 1A). As expected, affinity purification using any of the FLAG-tagged subunits in a wild-type (WT) eIF3 background pulled down every other subunit in the dodecamer in relatively stoichiometric quantities (Figures 1B–1E). However, complexes obtained from the eIF3Δh strains were missing a number of subunits, depending on which subunit was tagged. Using tagged eIF3f, we isolated an eIF3 subcomplex that contained subunits a, b, c, f, g, i, and m, but which lacked subunits k and l as previously observed (Smith et al., 2013), and also lacked subunits d and e in addition to the knocked-out h subunit (Figure 1B). We obtained similar results using tagged eIF3m (Figure 1C). Pull-downs using either tagged eIF3d or eIF3e expressed in the eIF3Δh strain yielded a stable dimer of subunits d and e (Figures 1D and 1E). These data clearly show that eIF3h is required for the assembly of up to four additional subunits into eIF3 (eIF3d, e, k, and l) and therefore plays a major role in eIF3 assembly in Neurospora.
Figure 1. Deletion of eIF3h Prevents Subunits d, e, k, and l from Associating with the eIF3 Complex.
(A) Cartoons of eIF3 showing subunits tagged with N-terminal FLAG tags used for purifying eIF3 complexes from wild-type (WT) (left) or eIF3Δh (right) Neurospora mycelia. Letters beside each FLAG tag signify the gels in (B–E) that correspond with eIF3 purifications using a FLAG-tagged subunit.
(B–E) Denaturing polyacrylamide gels of eIF3 purified with FLAG-tagged (B) eIF3f, (C) eIF3m, (D) eIF3d, and (E) eIF3e. Tagged subunits are indicated over each pair of gels and to the side of the gels with an asterisk. WT or eIF3Δh genetic backgrounds are indicated over each gel. Missing subunits are indicated by arrowheads. The asterisk in (A) indicates the (k,l) subunit dimer previously shown to depend on eIF3h for assembly (Smith et al., 2013). See also Figures S1–S5.
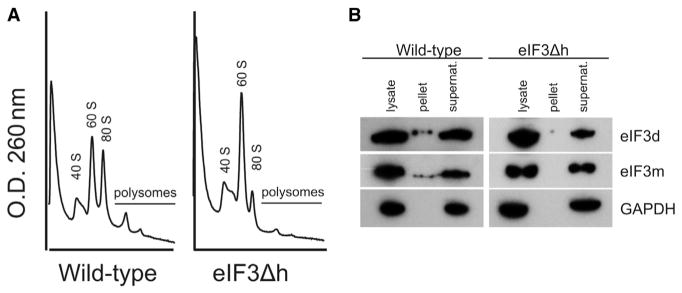
The structural impact of removing subunit h on eIF3 assembly has important implications for the functionality of eIF3 subcomplexes in the cell. The viability of the eIF3Δh strain indicates that one or more of the resulting subcomplexes (a,b,c,f,g,i,m), (d,e), and (k,l) (Smith et al., 2013) are functional in promoting translation initiation. However, compared with WT eIF3, the eIF3Δh strain has smaller polysomes indicating that initiation is less efficient (Figure 2A). Consistent with this interpretation, neither the seven-subunit nor the (d,e) subcomplexes can be pelleted with ribosomes in a sucrose cushion, whereas WT eIF3 was pelleted with ribosomes to some extent (Figure 2B). Interestingly, deletion of eIF3d is lethal to Neurospora suggesting that even though eIF3d appears to be separate from the rest of the complex in the eIF3Δh strain, it is still capable of its essential functions.
Figure 2. Deletion of eIF3h Affects Polysome Size and Affinity of eIF3 to Ribosomes.
(A) Polysomes are smaller and less abundant in eIF3Δh strains compared with WT N. crassa.
(B) Western blots showing FLAG-tagged eIF3d or eIF3m in WT or eIF3Δh N. crassa strains. Lysate, Pellet, and Supernatant indicate each fraction from a sucrose cushion. GAPDH was probed as a control for non-specific association with ribosomes.
Architecture of the eIF3Δh Subcomplex
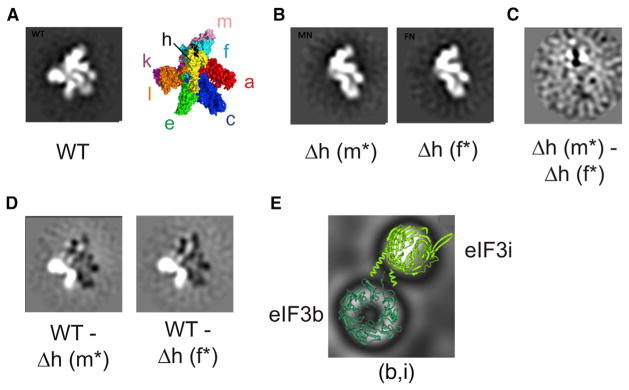
Subunits of eIF3 that are dispensable in N. crassa are clustered on one side of the complex (Querol-Audi et al., 2013; Smith et al., 2013). Structural models of eIF3 bound to the ribosome also show that the dispensable side of eIF3 is positioned away from the 40S ribosomal subunit (des Georges et al., 2015; Hashem et al., 2013a, 2013b). We previously postulated that this side of the eIF3 complex may be a regulatory platform that interacts with trans-acting factors or modulates eIF3 function through manipulation of a helical bundle within eIF3, proposed to be essential for its assembly (Smith et al., 2013). To test the latter possibility, we examined the structure of eIF3 isolated from eIF3Δh strains using negative-stain EM, to see if the overall conformation of eIF3 differed dramatically from the WT complex. We compared the structures of two eIF3Δh-derived subcomplexes, tagged either on the f or m subunit, with an eIF3 dodecamer tagged on the e subunit. The full dodecamer had the expected human-like eIF3 structure as previously published (Figures 3A and S1) (des Georges et al., 2015; Hashem et al., 2013a, 2013b; Querol-Audi et al., 2013). Both eIF3Δh-derived subcomplexes isolated using tagged f or m were entirely missing the right arm (eIF3e) and right leg (eIF3k and l) (Figure 3B), consistent with previous subunit mapping of human eIF3 (Querol-Audi et al., 2013). 2D difference maps between both eIF3Δh subcomplexes only showed slight variations proximal to the left leg (Figure 3C), likely due to the presence of the N-terminal FLAG tag on different subunits (eIF3f or m). Difference maps between the eIF3Δh-derived subcomplexes and the WT eIF3 dodecamer indicated that the subunits retained in the eIF3Δh-derived subcomplex (a,c,f,m) have no significant conformational differences compared with the dodecamer (Figure 3D). Thus, subunits d, e, h, k, and l have little or no influence on the conformation of remaining subunits at this resolution.
Figure 3. Negative-Stain 2D Class Averages ofNeurospora eIF3 and eIF3 Subcomplexes.
(A) Negative-stain image of WT eIF3 with an N-terminal FLAG tag on subunit e. The structural model of human eIF3 from cryo-EM reconstructions is included as a comparison (PDB: 5A5T) (des Georges et al., 2015).
(B) Negative-stain images of eIF3 purified from eIF3Δh strains, with N-terminal FLAG tags on either subunit m (Δh m*) or subunit f (Δh f*).
(C) 2D difference map between eIF3 complexes Δh m* and Δh f*.
(D) 2D difference maps between WT eIF3 and either Δh m* or Δh f*. White and black regions in difference maps correspond to regions of the structure with net positive or negative density, respectively. The top five class averages for each complex are depicted in Figure S1.
(E) Class average of solenoid images with crystal structures of the WD-repeat domains of eIF3b (dark green) and i (light green) modeled. See also Figure S1.
A number of 2D classes, particularly from the eIF3Δh-derived samples, consisted of two distinct solenoid structures adjacent to one another, suggesting they could represent the WD-repeat domains of eIF3b and eIF3i visualized in recent cryo-EM reconstructions of eIF3 (Aylett et al., 2015; des Georges et al., 2015). Crystal structures (Aylett et al., 2015) of the WD-repeat domains of eIF3b and i fit well into the solenoid-shaped density (Figure 3E), suggesting that (b,i) or (b,g,i) subcomplexes are distally bound to the eIF3 PCI-MPN subunits, consistent with cryo-EM reconstructions of eIF3 in isolation (Querol-Audi et al., 2013) and bound to the 40S ribosomal subunit (Aylett et al., 2015; des Georges et al., 2015). We also looked at the dimer formed between subunits d and e using negative-stain EM (Figure S1). The most populated class contains a single particle with a long tail structure, which we presume to be the isolated eIF3d subunit (Figure S1). A number of the remaining classes contain two distinct spots, with variable distances between them, which most likely represent the dimer formed between subunits d and e where the two spots are folded domains of e and d connected by flexible extensions in one or both subunits (Figure S1).
The Role of the PCI-MPN Helical Bundle in N. crassa eIF3 Assembly
Assembly of the proteasome lid is dictated by the formation of a helical bundle consisting of short stretches of α helices, C-terminal to the PCI or MPN domains (Estrin et al., 2013). Recombinant expression of an eIF3 octamer consisting of only PCI-MPN-containing subunits is possible, but absolutely requires the presence of all eight subunits (Sun et al., 2011). Of these eight subunits, four (e, h, k, and l) are dispensable in Neurospora, suggesting that either additional eIF3 subunits not in the proteasome lid-like architecture (e.g., subunits b, d, g, and i) or that other unknown factors aid eIF3 assembly in the cell (Smith et al., 2013). We were interested in the role of the helical bundle in eIF3 assembly in the cell and whether or not eIF3 has a similar subunit assembly pathway to the proteasome lid or CSN. We identified stretches of α helices just downstream of the PCI or MPN domains of subunits a, c, e, f, h, k, l, or m in Neurospora and made truncations to remove either the helices C-terminal of the PCI-MPN domains or regions of the subunit thought to be involved in interactions with other subunits in the complex (Figure S2). We inserted and expressed eIF3 subunit truncations from the his-3 locus in a series of N. crassa strains (Table 1) and isolated and identified the most stable subcomplexes of eIF3 using FLAG affinity purification. The identity of these subcomplexes allowed us to map the most likely assembly pathway of eIF3.
Table 1.
Neurospora Strains Created in This Study
| Strain | Genotype |
|---|---|
| WT (eIF3a 53–965) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3a 53-965) |
| WT (eIF3a 53–682) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3a 53-682) |
| WT (eIF3a 53–518) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3a 53-518) |
| WT (eIF3b) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3b) |
| WT (eIF3c) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3c) |
| WT (eIF3c 1–785) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3c 1-785) |
| WT (eIF3d) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3d) |
| WT (eIF3e) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3e) |
| WT (eIF3e 1–418) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3e 1-418) |
| WT (eIF3f) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3f) |
| WT (eIF3f 1–307) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3f 1-307) |
| WT (eIF3f 1–288) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3f 1-288) |
| WT (eIF3f 1–249) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3f 1-249) |
| WT (eIF3m) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3m) |
| WT (eIF3m 1–354) | mat A/a his-3(pccg1::N-FLAG::HAT::eIF3m 1-354) |
| Δ3h (eIF3b) | mat A his-3(pccg1::N-FLAG::HAT::eIF3b) |
| Δ3h (eIF3d) | mat A his-3(pccg1::N-FLAG::HAT::eIF3d) |
| Δ3h (eIF3e) | mat A his-3(pccg1::N-FLAG::HAT::eIF3e) |
| Δ3h (eIF3f) | mat A his-3(pccg1::N-FLAG::HAT::eIF3f) |
| Δ3h (eIF3h 1–310) | mat A his-3(pccg1::N-FLAG::HAT::eIF3h 1-310) |
| Δ3h (eIF3m) | mat A his-3(pccg1::N-FLAG::HAT::eIF3m) |
| Δ3k (eIF3k 65–215) | mat A his-3(pccg1::N-FLAG::HAT::eIF3k 65-215) |
| Δ3k (eIF3k 43–237) | mat A his-3(pccg1::N-FLAG::HAT::eIF3k 43-237) |
| Δ3k (eIF3k 43–215) | mat A his-3(pccg1::N-FLAG::HAT::eIF3k 43-215) |
| Δ3l (eIF3l 69–475) | mat A his-3(pccg1::C-Gly::HAT::FLAG:: eIF3l 69-475) |
Constructs labeled WT indicate that they contain all eIF3 subunits. mat A/a indicates that both mating types were made of the strain.
eIF3a and eIF3c
Subunits a and c are the largest in eIF3 and are thought to dictate the assembly of the complex (Smith et al., 2013; Sun et al., 2011; Wagner et al., 2014). Subunit a is the only subunit whose PCI-MPN domain is not located close to the C terminus. Removal of the region C-terminal to the coiled-coil/spectrin domain (eIF3a 53–964; Figure S2A) had no effect on complex assembly (Figure S3A), consistent with the fact that most of the C-terminal portion of eIF3a, downstream of the PCI domain, extends from the rest of the complex and is dispensable for the formation of the PCI-MPN octamer in recombinant human eIF3 expressed in E. coli (Sun et al., 2011). Removal of the majority of the coiled-coil/spectrin domain (eIF3a 53–682; Figure S2A) allowed assembly of a subcomplex that contained all subunits except for the (b,g,i) subcomplex (Figure S3A). Consistent with this result, biochemical experiments showed that the spectrin-like coiled-coil domain of eIF3a, downstream of the PCI domain, interacts with the (b,g,i) subcomplex (Dong et al., 2013), but that deletion of the spectrin domain results in weak association of subunit b with recombinant eIF3 assembled in vitro (Sun et al., 2011). Truncating eIF3a to remove its C-terminal helices that contribute to the helical bundle (eIF3a 53–518; Figure S2A) disrupted its association with all other eIF3 subunits (Figure S3A), despite eIF3a 53–518 having numerous conserved points of interaction upstream of the C-terminal helices in the helical bundle (Figure S4A). This suggests that incorporation of eIF3a into the helical bundle is the primary means of its assembly into eIF3 and that association of the (b,g,i) subcomplex appears to have little or no influence on the assembly of the PCI-MPN containing subunits (Sun et al., 2011).
Removing the C-terminal helices from eIF3c (eIF3c 1–785; Figure S2B) was sufficient for an entire eIF3 complex to assemble, albeit with substoichiometric quantities of the other subunits (Figure S3B) compared with full-length eIF3c (Figure S3I). These data suggest that the assembly of eIF3c likely involves stabilizing interactions between residues outside of the helical bundle with other subunits. Notably, eIF3c residues predicted to occur in interactions with subunits a and e, outside of the helical bundle, are highly conserved between N. crassa and multicellular organisms including humans, in contrast to prior reports (des Georges et al., 2015) (Figure S4B). Together with the fact that eIF3e is dispensable for eIF3 assembly (Figures 1B, 1C, and 3B) (Smith et al., 2013), these data suggest that the C-terminal helices of eIF3a play a more significant role than those of eIF3c in helical bundle formation and suggest that eIF3c assembles into the helical bundle downstream of or concurrently with eIF3a. However, conserved interactions between subunits a and c outside of the helical bundle may also make a significant contribution to assembly (Sun et al., 2011).
eIF3m and eIF3f
The m and f subunits are essential in Neurospora, and are nearly universally conserved in multicellular eukaryotes. The recent eIF3 structural models (des Georges et al., 2015) place the C-terminal helical region of eIF3m proximal to subunits f and h in the helical bundle. Removing the C-terminal helices in eIF3m just downstream of the PCI domain (eIF3m 1–354; Figure S2C) disrupted its assembly with other eIF3 subunits (Figure S3C). In eIF3f, removal of a C-terminal extension specific to N. crassa had no effect on eIF3 assembly (eIF3f 1–307; Figures S2D and S3D), whereas further deletion of the two C-terminal helices in the helical bundle, both of which have high sequence homology between N. crassa and humans (Figure S4D), prevented eIF3f binding to any other eIF3 subunit (eIF3f 1–249; Figure S3D). Interestingly, deletion of the more C-terminal helix (helix 2) permitted eIF3f assembly with subunits a, m, and the (b,g,i) subcomplex (eIF3f 1–288; Figure S3D), indicating that subunits a and m assemble with f exclusively via the more N-terminal helix (helix 1) in the helical bundle (Figure S3D). Furthermore, while these results do not rule out the possibility of subunits a and c assembling concurrently, they suggest that subunit c assembly into eIF3 via its C-terminal helix occurs subsequent to subunit a.
The above data suggest that subunits a, m, and f form the initial stable module in the helical bundle assembly. Sequence conservation of the C-terminal helical regions between human and Neurospora suggest that the main interactions governing the assembly of eIF3m into the helical bundle are through the helix 1 (via eIF3f) and not the helix 2 (via eIF3h) portion of the C-terminal helices (Figure S4D). Purified subcomplexes from eIF3Δh strains contain stoichiometric amounts of subunit m (Figure 1B), indicating that eIF3m assembles through the helical bundle and, like eIF3a, may participate early in the assembly pathway. Subunit c would have to assemble next to produce the minimal subcomplex that is functional in N. crassa (Figure 1B).
eIF3h
Like eIF3f, eIF3h has two C-terminal helices in the helical bundle (des Georges et al., 2015) supporting its importance in eIF3 assembly (Figures 1B and 1C). Although both eIF3h helices in the helical bundle have high sequence identity between Neurospora and humans (Figure S4E), the more C-terminal helix (helix 2) is nestled in a pocket of helices from several subunits (a, f, and m) in the helical bundle (des Georges et al., 2015). Helix 1 appears to be on the periphery of the helical bundle, possibly making contacts only with the C-terminal helix of eIF3c (des Georges et al., 2015). Removal of helix 2 (Figure S2E) revealed that helix 1 was insufficient to promote assembly of the eIF3 complex (eIF3h 1–310; Figure S3E). Surprisingly eIF3i bound to eIF3h 1–310 in stoichiometric amounts, in the complete absence of eIF3b and g (Figure S3E). Previous experiments (Fraser et al., 2004; Smith et al., 2013; Sun et al., 2011; Zhou et al., 2005) and the data presented above (Figure 1B) show that eIF3i and eIF3h can assemble into eIF3 complexes independently of each other. Thus it is not clear what functional role the interactions between eIF3i and eIF3h may play, if any, in eIF3 assembly. Substoichiometric amounts of subunits a, c, f, and m could assemble via eIF3h 1–310, consistent with subunits a, c, f, and m assembling a helical bundle prior to eIF3h.
To see if the weak interactions between eIF3h 1–310 with other PCI-MPN subunits or its binding to eIF3i were sufficient to restore the growth phenotype of the eIF3h knockout (KO) strain, we measured the linear growth of WT, eIF3Δh, and eIF3h 1–310 strains. Unlike full-length eIF3h, eIF3h 1–310 was not sufficient to rescue the growth phenotype (Figure S5) (Smith et al., 2013). Thus, neither the (h,i) subcomplex nor substoichiometric quantities of eIF3h 1–310 in an (a,c,f,i,m) subcomplex have an obvious functional relevance to growth. The phenotype of the eIF3Δh strain therefore reflects the role of C-terminal helix 2 in eIF3h in assembly of subunits h, e, d, k, and l into functional eIF3 complexes.
eIF3e
Removal of the C-terminal region of eIF3e allowed formation of a stable dimer with subunit d, but prevented eIF3e from integrating into the full eIF3 complex (eIF3e 1–418; Figures S2F and S3F). These results confirm the binary interaction between eIF3d and eIF3e (Figures 1D and 1E), in which eIF3d likely interacts with the N-terminal region of the PCI domain of eIF3e (Figure S4F), proximal to the region where eIF4G interacts with subunit e (Villa et al., 2013). Numerous potential points of interaction between eIF3e and subunits c and l are highly conserved between humans and Neurospora (Figure S4F). Thus, subunit e would assemble into eIF3 primarily through its highly conserved helical bundle (Figure S4F) and additional contacts with subunits c and l would form only after eIF3e is docked into the helical bundle. Based the data from eIF3Δh strains (Figure 1) and prior experiments (Smith et al., 2013), assembly of eIF3e in complex with subunit d into eIF3 would follow that of subunit h, but precede binding of subunits k and l.
eIF3k and eIF3l
Subunits k and l require each other to assemble into the rest of the eIF3 complex, contingent on the presence of eIF3h (Figures 1B and 1C) (Smith et al., 2013). The cryo-EM structural model of eIF3 shows that subunits k and l are tightly associated with each other, with the C-terminal helix of eIF3l making most of the contacts with the helical bundle (des Georges et al., 2015). We tested whether the interdependent assembly of subunits k and l is solely governed by the incorporation of their C-terminal helices into the helical bundle. Whereas deletion of an N-terminal 42 amino acid extension from eIF3k (eIF3k 43–237; Figures S2G and S3G) had no effect on assembly, additional removal of the C-terminal helix (eIF3k 43–217; Figure S2G) prevented subunit k incorporation into eIF3, but still allowed it to dimerize with subunit l (Figure S3G). Further truncation from the N terminus of eIF3k (eIF3k 65–217; Figure S2G), abolished the binary interaction with eIF3l (Figure S3G). N-terminal truncations of subunit l allowed the full eIF3 complex to assemble (eIF3l 69–475 or eIF3l 187–475; Figures S2H and S3H). However, the assembly of eIF3l 187–475 into eIF3 must not be efficient as suggested by its poor expression and/or stability (Smith et al., 2013). Furthermore, deletion of subunit k also leads to degradation of subunit l (Smith et al., 2013). Taken together, the deletion data suggest that efficient assembly of subunits k and l into eIF3 requires the C-terminal helices from both subunits combined with a stable interaction between their PCI domains.
Subcomplex of Subunits eIF3b, g, and i
Subunits eIF3b, g, and i form a stable subcomplex (Sun et al., 2011) (Figure 3E), which interacts with the spectrin/coiled-coil domain of eIF3a (Figure S3A), as well as the ribosome (Aylett et al., 2015; des Georges et al., 2015; Dong et al., 2013; Wagner et al., 2014). However, the point at which the (b,g,i) subcomplex assembles with eIF3a as part of intact eIF3 in cells is not well established. Our isolation of a subcomplex consisting of subunits eIF3f 1–288, a, and m, along with the (b,g,i) subcomplex, suggests that (b,g,i) assembles independently or after subunits m and f assemble with eIF3a (Figure S3D). Using an eIF3Δh strain, but not WT (Figure S3I), expressing an N-terminally tagged eIF3b subunit, we were able to isolate a subcomplex containing only subunits a, b, g, and i (Figure S3J), consistent with subcomplexes observed previously (Wagner et al., 2014; Zhou et al., 2008). This indicates that the (b,g,i) subunits can assemble with eIF3a prior to incorporation of subunits f and m into eIF3. However, the higher abundance of (b,i) complexes in the negative-stain EM class averages of samples isolated from eIF3Δh strains, compared with WT, suggests the (b,g,i) subcomplex is more labile in the absence of subunits d, e, h, k, and l (Figure S1), and may be stabilized in eIF3 subsequent to its initial integration.
Assembly of Human eIF3
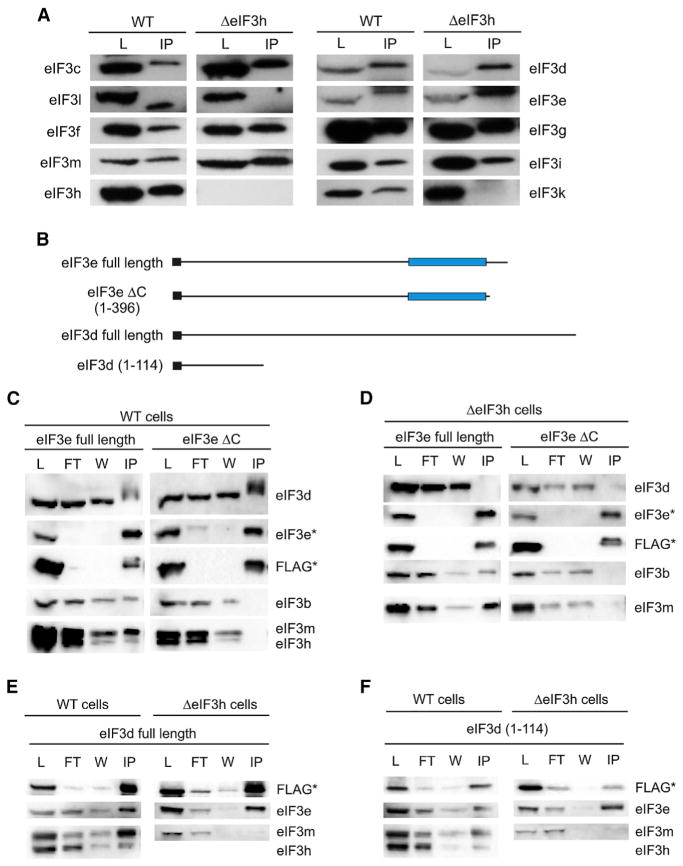
To test whether the eIF3 assembly pathway mapped in N. crassa is conserved in humans, we examined the assembly of subunits d, e, k, and l in human cells in which the eIF3h subunit gene was inactivated. HEK293T cells edited using CRISPR-Cas9 failed to produce eIF3h by western blot analysis (Figure 4A). We carried out immunoprecipitations (IPs) of eIF3 from lysates of either WT or EIF3H KO cell lines using an antibody for native eIF3b (Figure 4A). Consistent with results in N. crassa, removing eIF3h caused both the k and l subunits to dissociate from eIF3 (Figure 4A). However, unlike N. crassa, subunits d and e co-immunoprecipitated with eIF3b regardless of the presence or absence of eIF3h (Figure 4A).
Figure 4. The N Terminus of Subunit d Interacts with Subunit e Independently of Other Subunits in Human eIF3.
(A) Western blots of human eIF3 subunits in crude lysates (L) or immunoprecipitate (IP) with eIF3b primary antibody from either WT or EIF3H KO (ΔeIF3h) HEK293T cells.
(B) Schematics depicting N-terminally FLAG-tagged eIF3d or e constructs used for FLAG IPs in (C–F). PCI domains are represented with blue rectangles and FLAG tags are black rectangles. The amino acids included in truncated constructs are also shown.
(C–F) Western blots of anti-FLAG immunoprecipitations using transiently transfected FLAG-tagged constructs, as indicated. The cell lines, WT or EIF3H KO (ΔeIF3h), used for expression are indicated over each pair of western blots. The probed eIF3 subunit is indicated to the left or right of each blot. Subunits marked with an asterisk indicate the bands of the FLAG-tagged subunit. (C–F) Contain samples from crude L, unbound by anti-FLAG antibody flow through (FT), wash (W), and FLAG IP.
We next tested for the dimerization of subunits d and e, and whether their dimerization depends on assembly mediated by the helical bundle. We transiently expressed N-terminal FLAG-tagged eIF3e, either full length or with the C-terminal α helices deleted (eIF3e-ΔC) to prevent assembly into the helical bundle, in either WT or EIF3H KO cell lines (Figure 4B). In cells transiently co-expressing full-length eIF3d and full-length eIF3e, full-length eIF3e pulled down subunit d as well as subunits b, h, and m, in WT cells (Figure 4C). By contrast, truncated subunit eIF3e-ΔC interacted with subunit d, but not subunits b, h, and m. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis confirmed the absence of any other eIF3 or ribosomal subunit pulled down by eIF3e-ΔC (Table S1) indicating that the interaction between eIF3d and e is binary. These results are identical to those observed in N. crassa, in which the interaction between subunits d and e occurs independently of the rest of the eIF3 complex and requires the C-terminal helices of eIF3e for assembly into eIF3.
Surprisingly, in the EIF3H KO cells the interaction between eIF3d and either eIF3e construct was abolished (Figure 4D). However, transiently expressed full-length eIF3e co-immunoprecipitated with subunits b and m, consistent with results in WT cells (Figure 4C). To test whether transient expression eIF3e individually may be responsible for the difference between N. crassa and humans, transiently expressed FLAG-tagged eIF3d was able to pull down endogenous eIF3e in IPs (Figure 4E). Furthermore, as observed in N. crassa, the absence of eIF3h prevented the (d,e) dimer from interacting with rest of the eIF3 complex (Figure 4E). Taken together, these data suggest that, similar to the case in N. crassa, eIF3h influences the assembly of subunits d, e, k, and l into human eIF3. However, the interactions governing the assembly of subunits d and e into eIF3 may be highly sensitive to the expression levels of the individual subunits.
Although subunits eIF3d and e interact with each other biochemically and independently of the rest of the eIF3 complex (Figures 1D, 1E, 4C, 4E, and S3F), the positions of subunits d and e in the recent cryo-EM reconstruction of eIF3 are modeled far apart, with discontinuous density between the globular domain of eIF3d and subunit e (des Georges et al., 2015). It was previously shown that the N-terminal 114 amino acid region of human eIF3d remains bound to the eIF3 complex after HIV protease cleavage (Jager et al., 2012). We therefore tested whether eIF3e could directly interact with the N-terminal region of eIF3d (eIF3d 1–114). In WT cells, FLAG-tagged eIF3d 1–114 pulled down the same eIF3 subunits as observed with full-length eIF3d (Figures 4E and 4F). Furthermore, in the EIF3H KO cell lines, eIF3d 1–114 interacted with eIF3e independently of the rest of the eIF3 complex, as did full-length eIF3d (Figures 4E and 4F).
The cryo-EM reconstruction of eIF3 bound to the 40S subunit and the observed binding of eIF3d 1–114 to eIF3e suggests that eIF3d bridges eIF3e to the 40S subunit in an extended conformation. Amino acids 1–114 of human eIF3d are predicted to be mostly unstructured or in β-sheet conformations, consistent with an extended interaction between eIF3e and the eIF3d (1–114) suggested by cryo-EM difference maps between eIF3 with and without eIF3d (Sun et al., 2011). The distances observed between subunits d and e in negative-stain images of the (d,e) dimer (Figure S1, Table 2), which range from 67 to 173 Å, with the most populated class being 144 Å and a weighted average of classes yielding 111 ± 33 Å (Table 2), also agree well with the measured distance between the approximate center of masses (154 Å) of subunits d and e in the cryo-EM reconstruction of eIF3 bound to the human 40S ribosomal subunit (des Georges et al., 2015).
Table 2.
Measured Distances between d and e Subunits and Their Class Particle Counts
| Class | Particle Count | Distance (Å)a | Weighted Average |
|---|---|---|---|
| 3 | 329 | 144 | 0.21 |
| 4 | 283 | 77 | 0.18 |
| 7 | 159 | 67 | 0.10 |
| 10 | 156 | 104 | 0.10 |
| 11 | 151 | 173 | 0.10 |
| 12 | 133 | 102 | 0.08 |
| 13 | 107 | 73 | 0.07 |
| 15 | 98 | 125 | 0.06 |
| 16 | 80 | 116 | 0.05 |
| 17 | 77 | 130 | 0.05 |
Average distance between subunits d and e in the above classes is 111 ± 34 Å, with the average and SD calculated using values weighted by particle counts.
DISCUSSION
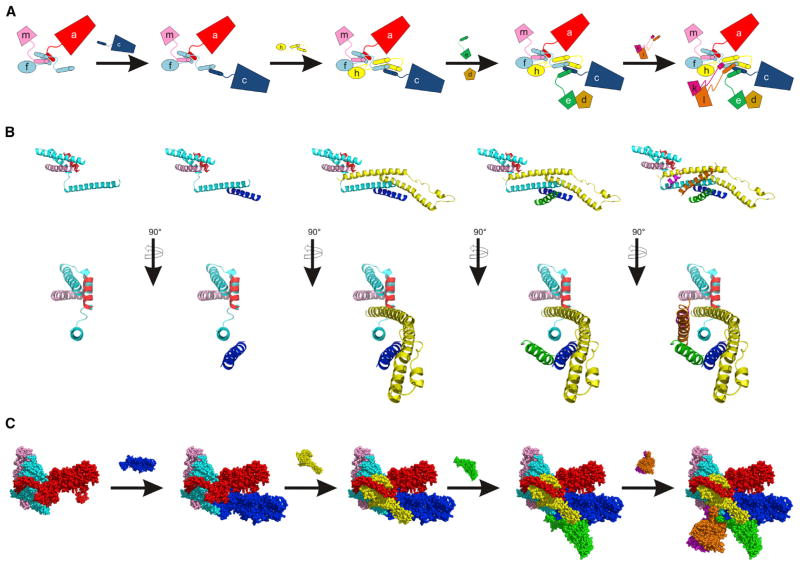
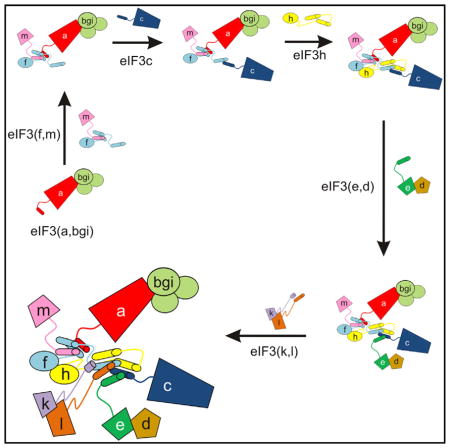
While the compositional complexity of eIF3 raises the possibility of the coexistence of eIF3 subcomplexes (Zhou et al., 2008) and assembly pathways, using subunit deletion and truncation experiments in N. crassa and human cells we propose a predominant pathway for eIF3 assembly in cells. The assembly pathway of eIF3 likely nucleates with subunit a, forming a subcomplex with subunits b, g, and i (Figures 5 and S3J). In parallel with or subsequent to this nucleation event, subunits a and m assemble with eIF3f through its first helical segment that contributes to the core helical bundle (Figures 5 and S3D). Subunit c then joins the complex through interactions with the second helix of eIF3f in the helical bundle, followed by subunit h (Figure 5). Subunit e, which binds the N-terminal segment of subunit d (Figure 4F), can then assemble with eIF3, either individually or as a dimer with d (Figures 5, 1B–1E, 4D, and S3F). Finally, subunits k and l assemble as a dimer to complete the assembly of the helical bundle and entire complex (Figures 5, S3G, and S3H) (Smith et al., 2013).
Figure 5. Main Assembly Pathway for Human-like eIF3.
(A) Cartoon scheme showing the ordered assembly of eIF3. Helical bundle formation is guided by the C-terminal helices of the indicated subunits. The (b,g,i) subcomplex assembles with eIF3a, independently of assembly of eIF3f and m with eIF3a, and is not depicted. Assembly of eIF3d with eIF3e is indicated, but is not dependent on the C-terminal helices of eIF3e.
(B) Two views of eIF3 helical bundle assembly. Subunits are added as in (A), except eIF3d is not included.
(C) Main assembly pathway of the human eIF3 octamer according to helical bundle assembly in (B), showing the available structural models for the subunits. Images in (B) and (C) are from the cryo-EM structure of human eIF3 (PDB: 5A5T) (des Georges et al., 2015). Subunits in all panels are colored as follows: eIF3a (red), eIF3c (blue), eIF3d (brown), eIF3e (green), eIF3f (cyan), eIF3h (yellow), eIF3k (magenta), eIF3l (orange), and eIF3m (light pink). See also Figures S2–S4.
The proposed cellular assembly pathway of eIF3 can be compared with that of recombinantly expressed human eIF3 (Masutani et al., 2007; Sun et al., 2011) and eIF3 assembly in human cells with subunits eIF3a or c partially depleted by siRNAs (Wagner et al., 2014). With respect to recombinant assembly, there are differences in subunit composition proposed for the minimally functional eIF3 from multicellular organisms. Masutani et al. (2007) found that subunits d, k, and l were structurally dispensable for assembling the remaining subunits, consistent with our results. However, contrary to our findings, they also found that subunits e and h were essential in assembling recombinant human eIF3 isolated from insect cells (Figures 1, 3, and 4) (Smith et al., 2013). This is possibly due to the fact that eIF3m had not yet been identified as a component eIF3 and was not included in their study (Masutani et al., 2007). Knock down of eIF3m in mouse embryonic fibroblasts (Zeng et al., 2013) caused depletion of eIF3c, f, and h—subunits that assemble concurrently or after eIF3m in our model (Figure 5)—but not subunits eIF3a, b, and i, which we propose are upstream or independent of eIF3m in the assembly pathway (Figure 5). A later study by Masutani et al. (2013) showed that in vitro reconstitution of human eIF3 lacking eIF3l or eIF3e also resulted in the absence of subunits eIF3k or eIF3d, respectively. Furthermore, removal of eIF3h resulted in loss or reduction of eIF3e, d, k, and l, also consistent with our model for cellular assembly.
Sun et al., (2013) could form a stable eIF3 octameric core of the PCI-MPN domain containing subunits (a,c,e,f,h,k,l,m) expressed in E. coli. However, to obtain functional preinitiation complex formation on the hepatitis C viral IRES, heterologously expressed and purified recombinant eIF3 in addition required subunits b, d, g, and i. In human cells in which eIF3c was depleted, Wagner et al. (2014) identified (a,b,g,i) and (f,h,m) sub-complexes by chemical crosslinking, and proposed that these interact independently with the 40S subunit. Based on the assembly pathway we derived by comparing N. crassa and human cell experiments, in which subunits m, f, and h assemble with subunit a in that order (Figure 5), we propose that knock down of eIF3c in human cells results in predominantly (a,b,g,i), with more labile subunits forming an (f,h,m) complex, in which the h subunit is stabilized by crosslinking (Wagner et al., 2014). Future experiments with recombinant eIF3 using core subunits identified in vivo and in cells (a,b,c,f,g,i,m) will be needed to test human eIF3 activity requirements in vitro.
Comparison with the Proteasome Lid or CSN
The proteasome lid and CSN are complexes compositionally and structurally similar to eIF3 (des Georges et al., 2015; Lander et al., 2012; Lingaraju et al., 2014; Querol-Audi et al., 2013). All three complexes contain eight subunits with either PCI or MPN domains and C-terminal helices that form helical bundles analogous to eIF3 (des Georges et al., 2015; Estrin et al., 2013; Lingaraju et al., 2014), but their proposed assembly pathways differ (Meister et al., 2015). The current model for assembly of the proteasome lid is similar to that of the eIF3 assembly pathway we propose here, although Rpn11 (the paralog of eIF3h) assembles much earlier into the proteasome lid compared with eIF3h into eIF3. The other major difference is the assembly of Rpn7 and 3 (paralogs of eIF3e and l) as a dimer, followed by Rpn8 (eIF3k paralog), rather than Rpn3 and 8 (eIF3k and l) as we see with eIF3.
The assembly of the CSN complex is not as well understood compared with the proteasome lid and eIF3. However, consistent with our model of eIF3 assembly, N. crassa KOs of Csn4 or 6 (paralogs of eIF3a or f) are lethal, suggesting they assemble prior to dispensable subunits Csn1, 2, 3, and 5 (paralogs of eIF3e, c, l, and h) (Colot et al., 2006). However, one notable difference is that Csn5 (eIF3h paralog) is thought to be the final subunit to be incorporated (Beckmann et al., 2015; Enchev et al., 2012), affecting the activity of the CSN, but not its structure (Zhou et al., 2012).
Implications for Cancer and Disease
Dysregulation of many eIF3 subunits has been implicated in a number of cancers, impaired development, and other diseases (Choudhuri et al., 2013; Hershey, 2015). Using our model for eIF3 assembly, we can now propose a framework for how eIF3 subunit over-, underexpression, and truncation affects the composition of eIF3 subcomplexes that can co-exist in cells, and possibly contribute to cancer and disease.
Unlike the proteasome lid and CSN, eIF3 is functional prior to the assembly of a number of subunits (d,e,h,k,l), and retains its ribosome binding surface (Figure 3) while leaving the helical bundle partially assembled. It is possible that the role of the helical bundle is to maintain a level of structural plasticity, allowing subcomplexes of eIF3 to form and fine-tune translation rates by, for example, affecting interactions with other eIFs such as eIF4G (Villa et al., 2013) or reinitiation following upstream open reading frames (Roy et al., 2010), without completely shutting off canonical translation. Some evidence for these possibilities was presented by Wagner et al. (2014). Alternatively, certain eIF3 subcomplexes might independently recruit specific mRNAs to the translation machinery or block their activity (Lee et al., 2015). Finally, some eIF3 subunits may have cellular functions apart from eIF3’s known roles in translation initiation. This idea is supported by the lethality of the eIF3d KO in N. crassa, which contrasts with the viability of the eIF3h KO strain, in which a (d,e) dimer persists separate from the rest of eIF3 and must support viability in some way.
Overexpression of a number of eIF3 subunits has been linked to various cancers (reviewed in Hershey, 2015). Although over-expression could lead to an excess of unincorporated subunits, overexpression may also coordinately upregulate the expression of other eIF3 subunits or the entire eIF3 complex. For example, overexpression of subunits a, b, or c leads to upregulation of multiple eIF3 subunits (Zhang et al., 2007). Alternatively, overexpression could create a “sink” for other subunits assembled downstream, leading to the generation of specific eIF3 subcomplexes such as (d,e) or (k,l). Subcomplexes with different subunit compositions could change the comparative rates of cap-dependent versus cap-independent translation initiation, which can lead to cancer (Silvera et al., 2010). The formation of eIF3 subcomplexes could also affect the rate in which other eIF3 subunits are turned over in the cell. For example, eIF3l is stabilized in the cell by eIF3k (Smith et al., 2013).
Our model for eIF3 assembly suggests that subunit underexpression, truncation, or expression of alternative isoforms could also lead to the coexistence of predominant eIF3 subcomplexes in cells. Underexpression of eIF3h causes defects in zebrafish embryo development, suggesting that eIF3h could be directly involved in translation initiation of specific transcripts during embryogenesis (Choudhuri et al., 2013). However, our assembly data suggest that these defects could be caused by a loss of function from the inability of downstream subunits to assemble (e.g., eIF3d, e, k, and/or l; Figure 1) into eIF3, which may also be functionally equivalent to downregulating the “unassembled” subunits directly (e.g., eIF3d or e) (Desnoyers et al., 2015; Gao et al., 2015; Li et al., 2015). Alternatively, the defects could be caused by aberrant function of subcomplexes (d,e) or (k,l) unable to assemble in the absence of eIF3h (Figures 1 and 3) (Smith et al., 2013). For example, eIF3d and e interact with CDC48 in fission yeast and could alter cell-cycle progression (Otero et al., 2010). The underexpression or truncation of eIF3e has been identified in many breast cancers (Desnoyers et al., 2015; Gillis and Lewis, 2013; Mayeur and Hershey, 2002). In our model, reduction of eIF3e levels in the cell would prevent eIF3d from assembling with eIF3, where it might localize to the nucleus as seen in certain cancers (Uhlen et al., 2005) or activate or repress the translation of certain cell-proliferation transcripts (Lee et al., 2015). Alternative splicing of eIF3 subunits might serve to activate specialized or aberrant functions of certain eIF3 subunits by preventing their assembly into the complex. For example, an isoform of human eIF3k, which lacks a region critical for its interaction with eIF3l (amino acids 21–53; Figures S2G and S3G) and thus the rest of eIF3, has high affinity for promyelocytic leukemia protein and co-localizes with its associated nuclear bodies (Salsman et al., 2013). In addition, there may be cellular mechanisms to prevent subcomplexes of eIF3 from forming when subunits of eIF3 are underexpressed. Translational down-regulation of subunits a, e, f, h, and l occurs in response to mammalian target of rapamycin inhibition (Hsieh et al., 2012; Olshen et al., 2013; Thoreen et al., 2012), and may serve to shut down eIF3-dependent translation initiation by lowering the expression of subunits that are involved in subcomplexes throughout the entire eIF3 assembly pathway (Figure 5).
Taken together, our model for human-like eIF3 assembly should enable future experiments to probe the role of eIF3 in development and disease. It will now be useful to explore which eIF3 subcomplexes co-exist in cells when expression of individual subunits is changed, as these subcomplexes may be causative for disease and cancer.
EXPERIMENTAL PROCEDURES
Neurospora eIF3 Expression Constructs, Strain Creation, and Complex Isolation
All eIF3 constructs were cloned into the AscI and PacI restriction sites of plasmid pCCG::N-FLAG::HAT (N-terminally tagged; GenBank: FJ457007) as previously described (Honda and Selker, 2009). The construct (eIF3l 69–475) utilized a C-terminal FLAG tag to match previously created strains (Smith et al., 2013).
All strains created in this study are summarized in Table 1. The his-3 auxotrophs of eIF3h, eIF3k, or eIF3l KOs have been previously described (Smith et al., 2013). Strains expressing FLAG-tagged eIF3 constructs were generated previously (Smith et al., 2013) or as described in the Supplemental Experimental Procedures.
Strains containing tagged eIF3 subunits were grown and purified as previously described (Smith et al., 2013). Additional purification steps were carried out to enhance lysis and prepare samples for MS or EM and are described in the Supplemental Experimental Procedures.
EM
Purified eIF3 complexes: WT (FLAG-eIF3e), Δ3h (FLAG-eIF3d), Δ3h (FLAG-eIF3f), and Δ3h (FLAG-eIF3m); (see Table 1) were diluted to ~50 nM in Tris-buffered saline (TBS) with 5% glycerol. Sample aliquots of 4 μl were pipetted onto 400 mesh continuous carbon grids, plasma cleaned in air for 10 s in a Solarus plasma cleaner (Gatan), and negatively stained with a 2% uranyl acetate solution. Data were acquired using a Philips CM200F electron microscope operating at 200 keV equipped with an UltraScan 1000 (Gatan) at a nominal magnification of 38,000× (2.8 Å/pixel). 2D data processing was carried out using the Relion and EMAN1.9 software packages (Ludtke et al., 1999; Scheres, 2012). Particles were picked manually within Relion, with a box size of 120 × 120 pixels. The contrast transfer function (CTF) was estimated using CTFFind4 (Rohou and Grigorieff, 2015). Reference-free 2D classification was performed with CTFs ignored until the first peak for 25 rounds. Selected class averages were subsequently aligned and difference maps were created within EMAN1.9.
Human eIF3 IPs
FLAG-tagged and un-tagged eIF3 subunits were all cloned into the plasmid nLv-103 (Addgene) using In-Fusion cloning (Clonetech). Constructs with N-terminal FLAG tags had tag sequences identical to the Neurospora constructs created in this study, which were added by PCR.
HEK293T cell lines (WT or eIF3h KO) were transiently transfected with eIF3 subunits using Lipofectamine 2000 (Thermo Fisher Scientific). Cells were harvested 20–30 hr after transfection. Cell pellets were lysed using 250 μl of lysis buffer (TBS, 0.5% Triton X-100, 10% glycerol, 2 mM EDTA) on ice. Complexes of eIF3 were immunoprecipitated using anti-FLAG affinity beads or Dynabeads (Thermo Fisher Scientific) with protein-G-coupled eIF3b primary antibody (Bethyl). Detailed experimental procedures, including the creation of the eIF3h KO strain, are outlined in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The assembly of many subunits into eIF3 is interdependent
Assembly of eIF3 is ordered and depends on C-terminal helices in PCI-MPN subunits
Dysregulated eIF3 assembly could play important roles in cancer and disease
Acknowledgments
We thank Anthony Iavarone of the QB3/Chemistry Mass Spectrometry Facility, University of California, Berkeley, for conducting the LC-MS/MS analyses. We thank Zoltan Metlagel for electron microscopy support and Tom Houweling for computer support, Starlynn Clarke and Charlie Craik for providing us with the FLAG-tagged constructs for eIF3d and eIF3d (1–114), Angel Cabral for help with cloning N. crassa subunit k and l truncations, and Luke Gilbert and Jonathan Weissman for the sgRNA plasmid backbone. This work was funded by NIH grants R01-GM65050 and P50-GM102706 to J.H.D.C. and the NIH Bridges to Baccalaureate Grant R25 GM095401 to A.C., and Pfizer for funding for E.M. L.A.-T. is supported by a CAPES fellowship.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.str.2016.02.024.
AUTHOR CONTRIBUTIONS
M.D.S., E.M., and J.H.D.C. designed the experiments; M.D.S., L.A.-T., A.N., E.M., and A.P carried out the experiments; M.D.S. and J.H.D.C wrote the paper; all authors edited the paper.
References
- Aylett CH, Boehringer D, Erzberger JP, Schaefer T, Ban N. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat Struct Mol Biol. 2015;22:269–271. doi: 10.1038/nsmb.2963. [DOI] [PubMed] [Google Scholar]
- Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci USA. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann EA, Kohler AM, Meister C, Christmann M, Draht OW, Rakebrandt N, Valerius O, Braus GH. Integration of the catalytic subunit activates deneddylase activity in vivo as final step in fungal COP9 signalosome assembly. Mol Microbiol. 2015;97:110–124. doi: 10.1111/mmi.13017. [DOI] [PubMed] [Google Scholar]
- Cao R, Gkogkas CG, de Zavalia N, Blum ID, Yanagiya A, Tsukumo Y, Xu H, Lee C, Storch KF, Liu AC, et al. Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nat Neurosci. 2015;18:855–862. doi: 10.1038/nn.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri A, Maitra U, Evans T. Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proc Natl Acad Sci USA. 2013;110:9818–9823. doi: 10.1073/pnas.1302934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoc E, Fraser CS, Zhou M, Videler H, Mayeur GL, Hershey JW, Doudna JA, Robinson CV, Leary JA. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol Cell Proteomics. 2007;6:1135–1146. doi: 10.1074/mcp.M600399-MCP200. [DOI] [PubMed] [Google Scholar]
- des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491–495. doi: 10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C, Horvitz HR. Caenorhabditis elegans mutants defective in the functioning of the motor neurons responsible for egg laying. Genetics. 1989;121:703–721. doi: 10.1093/genetics/121.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Frost LD, Courteau L, Wall ML, Lewis SM. Decreased eIF3e expression can mediate epithelial-to-mesenchymal transition through activation of the TGFbeta signaling pathway. Mol Cancer Res. 2015;13:1421–1430. doi: 10.1158/1541-7786.MCR-14-0645. [DOI] [PubMed] [Google Scholar]
- Dong Z, Qi J, Peng H, Liu J, Zhang JT. Spectrin domain of eukaryotic initiation factor 3a is the docking site for formation of the a:b:i:g subcomplex. J Biol Chem. 2013;288:27951–27959. doi: 10.1074/jbc.M113.483164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enchev RI, Scott DC, da Fonseca PC, Schreiber A, Monda JK, Schulman BA, Peter M, Morris EP. Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep. 2012;2:616–627. doi: 10.1016/j.celrep.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrin E, Lopez-Blanco JR, Chacon P, Martin A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure. 2013;21:1624–1635. doi: 10.1016/j.str.2013.06.023. [DOI] [PubMed] [Google Scholar]
- Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- Fraser CS, Lee JY, Mayeur GL, Bushell M, Doudna JA, Hershey JW. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J Biol Chem. 2004;279:8946–8956. doi: 10.1074/jbc.M312745200. [DOI] [PubMed] [Google Scholar]
- Gao Y, Teng J, Hong Y, Qu F, Ren J, Li L, Pan X, Chen L, Yin L, Xu D, et al. The oncogenic role of EIF3D is associated with increased cell cycle progression and motility in prostate cancer. Med Oncol. 2015;32:518. doi: 10.1007/s12032-015-0518-x. [DOI] [PubMed] [Google Scholar]
- Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem. 2010;285:29033–29038. doi: 10.1074/jbc.R110.150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis LD, Lewis SM. Decreased eIF3e/Int6 expression causes epithelial-to-mesenchymal transition in breast epithelial cells. Oncogene. 2013;32:3598–3605. doi: 10.1038/onc.2012.371. [DOI] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013a;503:539–543. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CUT, Pestova TV, Frank J. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013b;153:1108–1119. doi: 10.1016/j.cell.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JW. Regulation of protein synthesis and the role of eIF3 in cancer. Braz J Med Biol Res. 2010;43:920–930. doi: 10.1590/s0100-879x2010007500098. [DOI] [PubMed] [Google Scholar]
- Hershey JW. The role of eIF3 and its individual subunits in cancer. Biochim Biophys Acta. 2015;1849:792–800. doi: 10.1016/j.bbagrm.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational control 1995–2015: unveiling molecular underpinnings and roles in human biology. RNA. 2015;21:636–639. doi: 10.1261/rna.049957.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Selker EU. Tools for fungal proteomics: multifunctional neurospora vectors for gene replacement, protein expression and protein purification. Genetics. 2009;182:11–23. doi: 10.1534/genetics.108.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Cate JH. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhou F, Wang H, Lin D, Chen G, Zuo X, Sun L, Zhang X, Yang S. Knockdown of EIF3D suppresses proliferation of human melanoma cells through G2/M phase arrest. Biotechnol Appl Biochem. 2015;62:615–620. doi: 10.1002/bab.1305. [DOI] [PubMed] [Google Scholar]
- Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, Renatus M, Fischer ES, Thoma NH. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–165. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- Lopez-Lastra M, Rivas A, Barria MI. Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation. Biol Res. 2005;38:121–146. doi: 10.4067/s0716-97602005000200003. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani M, Machida K, Kobayashi T, Yokoyama S, Imataka H. Reconstitution of eukaryotic translation initiation factor 3 by co-expression of the subunits in a human cell-derived in vitro protein synthesis system. Protein Expr Purif. 2013;87:5–10. doi: 10.1016/j.pep.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Mayeur GL, Hershey JW. Malignant transformation by the eukaryotic translation initiation factor 3 subunit p48 (eIF3e) FEBS Lett. 2002;514:49–54. doi: 10.1016/s0014-5793(02)02307-4. [DOI] [PubMed] [Google Scholar]
- Meister C, Kolog Gulko M, Kohler AM, Braus GH. The devil is in the details: comparison between COP9 signalosome (CSN) and the LID of the 26S proteasome. Curr Genet. 2015;62:129–136. doi: 10.1007/s00294-015-0525-7. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Hsieh AC, Stumpf CR, Olshen RA, Ruggero D, Taylor BS. Assessing gene-level translational control from ribosome profiling. Bioinformatics. 2013;29:2995–3002. doi: 10.1093/bioinformatics/btt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero JH, Suo J, Gordon C, Chang EC. Int6 and Moe1 interact with Cdc48 to regulate ERAD and proper chromosome segregation. Cell Cycle. 2010;9:147–161. doi: 10.4161/cc.9.1.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015;75:250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querol-Audi J, Sun C, Vogan JM, Smith MD, Gu Y, Cate JH, Nogales E. Architecture of human translation initiation factor 3. Structure. 2013;21:920–928. doi: 10.1016/j.str.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Vaughn JN, Kim BH, Zhou F, Gilchrist MA, Von Arnim AG. The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA. 2010;16:748–761. doi: 10.1261/rna.2056010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsman J, Pinder J, Tse B, Corkery D, Dellaire G. The translation initiation factor 3 subunit eIF3K interacts with PML and associates with PML nuclear bodies. Exp Cell Res. 2013;319:2554–2565. doi: 10.1016/j.yexcr.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- Smith MD, Gu Y, Querol-Audi J, Vogan JM, Nitido A, Cate JH. Human-like eukaryotic translation initiation factor 3 from Neurospora crassa. PLoS One. 2013;8:e78715. doi: 10.1371/journal.pone.0078715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Todorovic A, Querol-Audi J, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis CS, Hartland A, Gradia S, et al. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3) Proc Natl Acad Sci USA. 2011;108:20473–20478. doi: 10.1073/pnas.1116821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Querol-Audi J, Mortimer SA, Arias-Palomo E, Doudna JA, Nogales E, Cate JH. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res. 2013;41:7512–7521. doi: 10.1093/nar/gkt510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- Villa N, Do A, Hershey JW, Fraser CS. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem. 2013;288:32932–32940. doi: 10.1074/jbc.M113.517011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Herrmannova A, Malik R, Peclinovska L, Valasek LS. Functional and biochemical characterization of human eukaryotic translation initiation factor 3 in living cells. Mol Cell Biol. 2014;34:3041–3052. doi: 10.1128/MCB.00663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Wan Y, Li D, Wu J, Shao M, Chen J, Hui L, Ji H, Zhu X. The m subunit of murine translation initiation factor eIF3 maintains the integrity of the eIF3 complex and is required for embryonic development, homeostasis, and organ size control. J Biol Chem. 2013;288:30087–30093. doi: 10.1074/jbc.M113.506147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2007;282:5790–5800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- Zhou C, Arslan F, Wee S, Krishnan S, Ivanov AR, Oliva A, Leatherwood J, Wolf DA. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 2005;3:14. doi: 10.1186/1741-7007-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci USA. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang Y, Cai G, He Q. Neurospora COP9 signalosome integrity plays major roles for hyphal growth, conidial development, and circadian function. PLoS Genet. 2012;8:e1002712. doi: 10.1371/journal.pgen.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.