Summary
Protein phosphorylation is a fundamental regulatory mechanism in many cellular processes and aberrant perturbation of phosphorylation has been implicated in various human diseases. Kinases and their cognate inhibitors have been considered as hotspots for drug development. Therefore, the emerging tools, which enable a system-wide quantitative profiling of phosphoproteome, would offer a powerful impetus in unveiling novel signaling pathways, drug targets and/or biomarkers for diseases of interest. This review highlights recent advances in phosphoproteomics, the current state of the art of the technologies and the challenges and future perspectives of this research area. Finally, some exemplary applications of phosphoproteomics in diabetes research are underscored.
Keywords: Phosphoproteomics, LC-MS/MS, phosphopeptide enrichment, quantification, targeted proteomics, phosphorylation, diabetes
Protein phosphorylation is a post-translational modification (PTM) on serine, threonine, and tyrosine residues that plays an essential role in signal transduction across many cellular processes, including cell proliferation, differentiation, and apoptosis, as well as metabolism. The dynamics of phosphorylation and the network of targets are delicately controlled and coordinated by the reciprocal actions of the arrays of protein kinases and phosphatases in response to stimuli [1]. In the context of biological significance of phosphorylation, the negatively charged phosphate group incorporated could either attract to or repel from a positively or negatively charged amino acid residue in close proximity, respectively. The protein conformation change resulting from phosphorylation in turn modifies its physical and chemical properties and functions, such as catalytic activity, interaction partners, and stability. Accordingly, aberrant protein phosphorylation is involved in many human diseases, such as Alzheimer’s disease [2,3], cancer [4–6], cardiovascular disease [7–9], as well as diabetes [10–12]. Ascribing to the biological impact of phosphorylation in the pathogenesis of human diseases, protein kinases and the cognate kinase inhibitors have been the quintessence in drug development [13–15], such as tyrosine kinase inhibitor imatinib (Gleevec®) for treating chronic myelogenous leukemia [16,17]; lipid kinase PI3K for solid tumors [18–20]; serine/threonine kinase BRAF for melanoma [21–23]; receptor tyrosine kinase EGFR for lung cancer [24,25]; and serine/threonine kinase mTOR for renal tumors [26].
The knowledge of the complexity of phosphorylation-mediated signaling networks has been greatly advanced in the last decade largely due to this emerging field of phosphoproteomics. Phosphoproteomics as a technology has become indispensable for biomedical research, which often enables quantitative profiling of site-specific phosphorylation across many different biological conditions with extensive coverage of the phosphoproteome [27]. The ability to identify >10,000 phosphorylation sites and to perform quantitative measurements of their levels of phosphorylation in such a broad scale allow investigators to systematically identify aberrant signaling activations, pathways, networks underlying disease conditions, thus providing a new level of mechanistic understanding of disease pathogenesis and potential therapeutic targets or biomarkers. Herein, we review the recent technological advances of phosphoproteomics with an emphasis on the quantitative approaches, including both global and targeted quantitative phosphoproteomics, highlight the current state-of-the-art of phosphoproteomics field, and discuss some recent applications to diabetes.
Global Phosphoproteomics
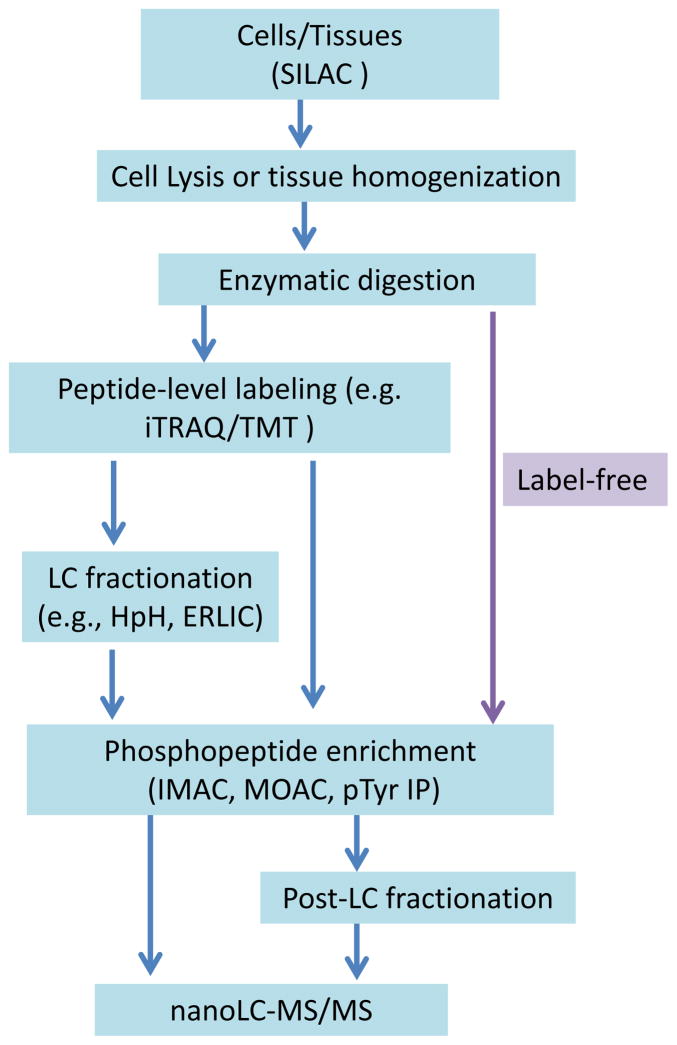
For large-scale profiling of the phosphoproteome, mass spectrometry (MS) has become the dominant tool due to its sensitivity and unique ability to identify site-specific PTMs. Traditionally, phosphoproteomics analysis by MS faces great challenges because of the low abundance of signaling proteins and low stoichiometry of phosphorylation events. However, recent advances in affinity enrichment approaches of phosphopeptides, liquid chromatography (LC) separations, and MS instrumentation has largely overcome this challenge by enabling large-scale proteome wide profiling of protein phosphorylation. The general workflow of MS-based quantitative phosphoproteomics is illustrated in Figure 1. While the detailed workflow will be varied depending on specific applications, it typically consists of four major steps: 1) protein extraction and enzymatic digestion, 2) isobaric labeling using reagents such as tandem mass tags (TMT) [28] or isobaric tags for relative and absolute quantification (iTRAQ) [29] to enabling multiplexed relative quantification, 3) enrichment of phosphopeptides by affinity chromatography, and 4) LC-MS/MS analyses. To further enhance the coverage of the phosphoproteome, LC fractionation strategies were often applied either prior to or after phosphopeptide enrichment [30]. Besides isobaric labeling, label-free or other stable isotope labeling approaches such as stable isotope labeling by amino acids in cell culture (SILAC) can also be incorporated to achieve quantitative measurements of protein phosphorylation [31,32].
Figure 1. The overall workflows of MS-based global quantitative phosphoproteomics.
Both label-free and stable isotope labeling based quantitative approaches can be applied. For labeling approach, SILAC can be performed at cell culture level and iTRAQ/TMT at peptide level. The peptides can either be subjected to LC fractionation prior to enrichment or directly subjected to phosphopeptide enrichment. LC-fractionation can also be applied after phosphopeptide enrichment.
Protein extraction and enzymatic digestion
Since phosphorylation is a labile PTM, preserving the integrity of in vivo phosphorylation status is critical. Therefore, it is pivotal to include appropriate phosphatase inhibitors during the steps of cell lysis and protein extractions. In shotgun proteomics, proteins are typically digested into peptides using trypsin. Other enzymes such as endoproteinase Lys-C or Glu-C can also be applied for digestion to enhance the coverage of the phosphoproteome due to the complementary specificity of enzymes. For example, in one study the integration of both Glu-C and trypsin digestion resulted in the identification of 8,507 phosphorylation sites compared to only 4,647 phosphorylation sites by trypsin alone [33]. Wisniewski et al. demonstrated that the consecutive use of Lys-C and trypsin enhanced both protein and phosphorylation site identification by 40% [34]. Despite the increase of sequence coverage adapting multiple enzyme digestion, the shortfall is the need of additional samples and MS instrument time. This caveat can be partially alleviated by using consecutive proteomic digestion with the implementation of filter aided sample preparation (FASP) as an enzyme reactor. In this way different populations of peptides can be obtained from a single sample without the need of an additional input material [34].
Quantitative Strategies
Elucidation of signaling networks requires quantification of the dynamic changes of protein phosphorylation. In principle most quantitative strategies are commonly applicable to both global proteomics and phosphoproteomics. Although recent advances in the robustness and reproducibility of LC-MS platforms have enabled label-free approaches to be more commonly employed in quantitative proteomics [35], the majority of quantitative phosphoproteomics studies to date are based on stable isotope labeling approaches. Among the isotope labeling approaches, SILAC [36] and isobaric labeling strategies are commonly employed in phosphoproteomics. In terms of quantification accuracy, SILAC typically performs better than isobaric labeling since the labeling process is conducted at a more upstream level (i.e., during cell culture) compared to peptide-level labeling for iTRAQ or TMT. Nonetheless, isobaric reagents offer several advantages in enabling quantitative analysis of multiple samples simultaneously (i.e., multiplexing), which is particularly useful for monitoring a biological system over multiple time points, and the universal applicability to all types of samples.
The current commercially available isobaric reagents TMT and iTRAQ offer the options of 4-, 6-, 8-, and 10-plex labeling and quantification for both global proteomics and phosphoproteomics, which provides a great flexibility depending on the experimental designs in specific applications. The sample multiplexing ability also greatly increases the overall sample throughput for phosphoproteomics analysis especially when multi-dimensional LC separations are employed to enhance the coverage. The higher-energy collisional dissociation (HCD) performed on the new generation of Orbitrap mass spectrometers such as Orbitrap Velos or Q-Exactive has become the primary approach for analyzing isobaric labeled samples by producing excellent quality MS/MS data along with low m/z reporter ions for both identification and quantification [37]. One potential caveat related to isobaric labeling-based quantification is that the isolation window for selected precursors in the first stage MS, which is typically 3 Thomson, potentially include ions of multiple peptides, and such potential interferences could skew the quantification results of the identified peptides [38]. To address this potential interference issue, extensive multi-dimensional LC separations can be applied to at least partially alleviate the problem. More recently, triple-stage MS (MS3) strategy was reported to nearly completely eliminate interference [38], but with the expense of sensitivity. More recently, McAlister et al. described a MultiNotch MS3 method on an Orbitrap Fusion instrument that utilizes synchronous precursor selection for co-isolating and co-fragmenting multiple MS2 fragment ions to enhance the overall sensitivity [39].
In addition to labeling strategies, label-free quantification is also a commonly applied strategy in phosphoproteomics. Several software tools and strategies were reported for robust measurements of the levels of phosphopeptides in different samples using different strategies. For example, Schilling et al. demonstrated the use of MS1 extracted ion chromatograms using Skyline for quantification of phosphorylation [40]. Xue et al. introduced a library-assisted extracted ion chromatogram (LAXIC) approach which utilized a synthetic peptide library as internal standards for normalization [41]. Cox et al. reported the generic MaxLFQ approach using the Maxquant computational platform, which is applicable to phosphoproteomics [42]. However, one primary limitation of label-free quantification is its heavy reliance on the reproducibility of sample processing and instrument performance.
Phosphopeptide Enrichment
Due to the generally low-abundance of phosphopeptides, efficient enrichment of phosphorylated serine (pSer), threonine (pThr), and tyrosine (pTyr) containing peptides is a key step for phosphoproteomics analysis. Various affinity enrichment strategies based on either specific antibodies or chemical resins have been developed for effective isolation of phosphopeptides from complex mixtures. Anti-pTyr antibody-based immunoaffinity approaches have become the primary method for profiling the tyrosine phosphoproteome [43]. For global enrichment of pSer/pThr/pTyr peptides, immobilized metal affinity chromatography (IMAC) [44] and metal oxide affinity chromatography (MOAC) with TiO2 [45] have become two most popular methods.
pTyr immunoaffinity enrichment
Tyrosine phosphorylation is a prominent component of intracellular signaling involved in receptor tyrosine kinases activating and is essential for proliferation, differentiation, survival, and metabolism [46,47]. Specific anti-pTyr immunoaffinity strategies have proven highly successful in the comprehensive mapping of tyrosine phosphorylation [43,48–51]. However, phosphorylated tyrosine residues only constitute a very small fraction of the total amount of the cellular protein phosphorylation with an estimated relative abundance of 1800:200:1 for pSer/pThr/pTyr in vertebrate cells [52]. Therefore, a relatively large amount of proteins (in the levels of mg) is often required to achieve an adequate coverage of the tyrosine phosphoproteome. Recently, Boersema et al. has demonstrated the feasibility of in-depth quantitative profiling of the pTyr proteome using immunoaffinity enrichment coupled with stable isotope dimenthyl labeling, where more than 1,100 unique phosphopeptides were identified from 4 mg using single dimensional LC-MS/MS [53].
IMAC enrichment
The IMAC approach was first introduced in 1986 [54], leveraging the positively charged metal ions such as Fe3+, Ga3+, Ti4+, and Zr4+ [55–58], which are immobilized on resins to capture negatively charged phosphopeptides. The most commonly used resins are coated with iminodiacetic acid (IDA) and nitrilotriacetic acid (NTA). Despite being one the most extensively applied technique for phosphopeptide enrichment, IMAC has several caveats in terms of its low tolerance towards buffers or salts in the biological samples [59] and potential compromise of specificity during its unspecific binding towards acidic peptides. Initially, O-methylesterification was applied to derivatize carboxylic acid groups in acidic amino acid residues and the peptide C-terminal in order to enhance the specificity of IMAC [44]. A disadvantage of the esterification procedure is the occurrence of side reaction products (partial hydrolysis of peptides, deamidation of asparagine and glutamine residues) that can increase sample complexity.
Over the years, the commercially available IMAC resins have been improved for both its specificity and robustness through either the modification of the solid support or the chelating linker. For example, the availability of magnetic Ni-NTA agarose resins enabled high throughput automated isolation of phosphopeptides in 96-well format with relatively high specificity [60]. By coupling with different fractionation strategies, IMAC has been successfully applied to many large-scale studies to achieve extensive in-depth profiling of the global phosphoproteome where >10,000 phosphorylation sites were often identified [61,62]. Moreover, different metal ions can offer distinct enrichment efficiency and binding selectivity. For example, a sequential Ga3+- and Fe3+-IMAC enrichment was reported to significantly enhance the coverage of the phosphoproteome compared to a single IMAC enrichment [63].
MOAC enrichment
A number of metal oxides or hydroxides, including TiO2, Ga2O3, ZrO2, Fe3O4, Nb2O3, SnO2, HfO2, Ta2O5, and Al(OH)3, were reported to be adapted as matrix for enriching phosphopeptide based on the principles of complex formation between metal oxides and phosphopeptides [64]. TiO2 is by far the most widely employed owing to its remarkable sensitivity and selectivity in phosphopeptide enrichment. Pinkse et al. described the first implementation of TiO2 as a pre-column in tandem with LC-MS/MS for enrichment of phosphopeptide in 2004 [65]. One main limitation of MOAC is related to its specificity due to the potential binding to acidic peptides. Larsen et al. [73] in 2005 described an approach to improve the selectivity towards phosphopeptides by loading the sample in 2,5-dihydroxybenzoic acid (DHB) solution containing acetonitrile and TFA so that DHB competes with the binding of acidic peptides on TiO2 beads, thus achieving improved overall specificity for enriching phosphopeptides. Phosphopeptides were eluted using an alkaline solution at pH 10.5 using ammonium hydroxide. More recently, aliphatic hydroxyl acid modifiers such as lactic acid were proposed to improve selectivity and capacity of TiO2 towards phosphorylated peptides [66]. TiO2-based MOAC enrichment has been broadly applied in large-scale phosphoproteomics studies. For example, Paulo et al. were able to quantify 10,562 phosphorylation events on the mouse kidney, liver, and pancreas tissues treated with MEK inhibitors using MOAC coupled with 10-plex TMT labeling [67].
IMAC and MOAC have been reported as complementary in the aspects that IMAC favors multiply phosphorylated peptides while MOAC favors mono-phosphorylated peptides [59]. This complementary nature has led to the integration of both techniques into a single workflow termed sequential elution from IMAC (SIMAC) [68]. Briefly, this strategy involves multistage elution from IMAC: 1) the mono-phosphorylated peptides were initially eluted from IMAC under acidic condition (in 1% TFA, 20% acetonitrile at pH 1.0) after binding, and 2) the multiply phosphorylated peptides were subsequently eluted from the same IMAC resin using pH 11.30 (ammonia water). Both the IMAC flow-through and the 1% TFA acidic eluent were subjected to TiO2 chromatography for nriching the monophorylated peptides. Both Fe3+- and Ga3+-IMAC have been reported in the SIMAC strategy [69] and in one report a nearly 2-fold increase in phosphopeptide identification was observed from lysates of human mesenchymal stem cells compared to TiO2 alone [68]. This SIMAC strategy has been successfully applied to several large-scale phosphoproteomics studies [70,71]
In addition, polymer-based metal-ion affinity capture (PolyMAC) has emerged to as a novel chemical strategy based on water soluble, globular dentrimers multifunctionalized with metal ions (e.g., Ti4+, Fe3+) for capturing phosphate groups [72,73]. Comparing to TiO2, PolyMAC displayed excellent reproducibility, selectivity, and high recovery of phosphopeptides from complex mixtures.
Peptide-level LC fractionation
While the final enriched phosphopeptide samples are typically analyzed by LC-MS/MS, additional orthogonal LC fractionation strategies are often applied either prior to or after the enrichment of phosphopeptides to enhance the overall coverage. Traditionally, strong cation exchange (SCX) was widely adopted as the first dimensional fractionation [74]; however, SCX generally suffers from low resolution of separations and potential sample loss due to the need of additional desalting. Recent efforts have been converged onto better alternatives of SCX that can not only offer better separation but also mitigate the sample loss. HpH reversed phase LC [75] and electrostatic repulsion hydrophilic interaction chromatography (ERLIC) or hydrophilic interaction chromatography (HILIC) have been commonly applied for pre-fractionation prior to phosphopeptide enrichment. However, if the total protein amount is limited, the capillary-based LC fractionation was often applied after phosphopeptide enrichment to enhance the coverage of the phosphoproteome.
HpH reversed phase LC typically operates at either pH 10 using ammonium formate buffer or pH 7.5 using TEAB buffer [61]. In RPLC, it was observed that a change in the pH of the mobile phase from low to high will dramatically alter the overall charge distribution of a peptide, and its respective ion-pairing interactions with the LC stationary phase, thereby offering a distinct and at least partially orthogonal separation [76]. RPLC offers much higher resolution in separations and better sample recovery compared to traditional SCX, which makes it an ideal as the first dimensional fractionation strategy when coupled with a low pH LC-MS/MS [75]. Wang et al. first demonstrated the coupling of high- and low-pH RPLC offline for global proteome profiling using a so-called concatenation strategy to combined the early, middle and late eluting fractions from HpH RPLC for more effectively utilizing the partial orthogonality nature of the 2D separations [75]. Now offline HpH RPLC fractionation has been routinely applied prior to phosphopeptide enrichment by IMAC or MOAC for achieving extensively coverage of the phosphoproteome where the identification of >30,000 phosphopeptides were often reported in a given study [61,77,78].
ERLIC and HILIC are other types of promising orthogonal LC-based fractionation approaches for coupling with RPLC for phosphoproteomics [79,80]. HILIC is a high-resolution separation technique where the primary interaction between a peptide and the neutral, hydrophilic stationary phase is hydrogen bonding. In HILIC, retention increases with increasing polarity (hydrophilicity) of the peptide, opposite to the trends observed in RPLC [80]. In contrast, ERLIC is a hybrid-mode chromatography that makes use of the properties of HILIC and an anion exchange separation operated typically under a high percentage of organic solvent (e.g. 70% acetonitrile) at low pH [79]. In the absence of HILIC mode, phosphopeptides are electrostatically repulsed from the positive charged stationary column at low pH. However, in the presence of high content organic solvent, hydrophilic interaction dominates and outweighs the anion exchange interaction. Thereby, the phosphopeptides are less repulsed by (i.e., more retentive to) the column versus the non-phosphorylated counterparts [81]. In a comparison of ERLIC versus SCX-IMAC in human A431 epidermal cells, it was observed that ERLIC and SCX are complementary in phosphopeptide identifications [82].
Analysis of Phosphopeptides by MS/MS
Analysis of phosphopeptides by MS/MS has been traditionally challenging on ion trap instruments using collision-induced dissociation (CID) since the phosphorylated group on serine and threonine residues are very labile and a 98 Da neutral loss of phosphoric acid occurs preferentially over peptide backbone fragmentation [83]. The inefficient backbone fragmentation often resulted in MS/MS spectra that were essentially devoid of sequence information and hamper the phosphopeptide identification and phosphorylation site assignment, even with tedious manual examination of MS/MS spectra. The introduction of electron transfer dissociation (ETD) partially alleviates the challenge by successfully fragment the sequence backbone without little loss of the phosphate moiety on most fragment ions (complementary ions of types c and z) [84,85]. ETD is very useful for fragmentation of peptides containing labile modifications, such as phosphopeptides, as the resulting peptides fragments will retain their phosphate group, enabling direct identification of phosphorylation site. Drawbacks of ETD included its relatively low duty cycle due to the long reaction time compared to CID, and its low fragmentation inefficiency on doubly charged species, which constitutes the majority of peptides in a tryptic digest. While ETD is complementary and especially unique for site determination, these drawbacks prevent ETD being broadly applied in large-scale phosphoproteome profiling [86]. More recently, HCD has become more broadly available in the new generation of Orbitrap instruments (e.g., Orbitrap Velos and Q-Exactive). HCD deposits higher levels of energy into the precursor peptide ion on a shorter timescale, thus promoting the occurrence of much more prominent backbone fragmentation [87,88]. Recently, a new fragmentation scheme combining both ETD and HCD, namely EThcD, originally introduced by Frese et al. [89], has enabled dual fragment ion series of both b/y- and c/z-type fragment ions in a single spectrum. The EThcD, now available in the Orbitrap Fusion instrument, provides much richer fragmentation data, thus resulting in substantially enhanced phosphopeptide identification and site determination rate [90].
In the assignment of phosphorylation site, different probability-based algorithms have been developed to measure the probability of correct phosphorylation site localization based on the presence and intensity of site-determining ions in MS/MS spectra. These include PTM score [91], Ascore [92], PhosphoScore [93], PhosphoScan [94], and PhosCalc [95] for MS/MS spectra acquired using CID and SLoMo [96], Phosphinator [97], and PhosphoRS [98] for spectra acquired using both CID and ETD fragmentation modes.
Targeted Phosphoproteomics
In addition to global phosphoproteomics, there is increasing interest in pursuing accurate quantitative analyses of phosphorylation dynamics for a specific set of protein targets from particular signaling pathways or networks. In principle, both antibody-based and targeted MS approaches can be used to measure phosphorylation levels of specific proteins. Herein we briefly review antibody-based multiplex immunoassays, protein microarray, flow cytometry, and targeted MS approaches for targeted measurements of protein phosphorylation.
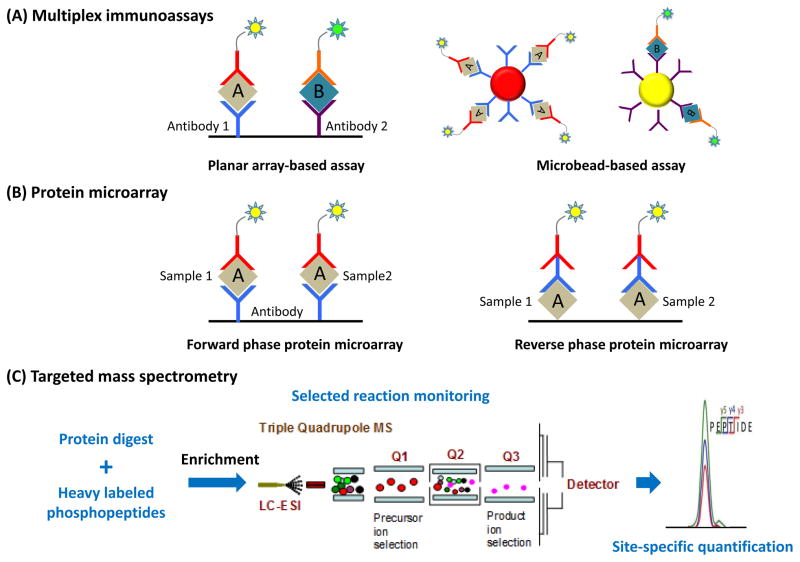
Multiplex immunoassays can simultaneously detect or quantify multiple analytes via an immunological reaction. This assay is carried out and presented in two major layouts: 1) planar array-based assay, also known as enzyme-linked immunosorbent assay (ELISA), for example, Meso-scale discovery (MSD); and 2) microbead-based assay, for instance, Luminex xMAP, and FlowCytomix [99]. In the setting of MSD, carbon electrode plate surface is implemented versus polystyrene ascribing to its 10-fold stronger anchoring capacity to the phospho-antibodies. In each 96-well, up to ten carbon electrodes could be immobilized; and each electrode is conjugated with a specific phospho-antibody. Therefore, it enables a 10-plex capability (i.e., up to 10 different phosphoproteins could be detected and analyzed per sample concurrently). In the platform of xMAP, polystyrene microspheres core is employed as the stationary matrix. Each batch of microspheres is coated and earmarked with both red and infrared fluorophores mixed at a distinct ratio, which are then coupled with the respective fluorescent R-phycoerythrin-labeled phospho-antibody. As one may anticipate, since there is an enormous possible combination of fluorophores in assigning distinct batches of microsphere conjugated with different phospho-antibodies, dozens to hundreds of phosphoprotein targets could be analyzed per sample at the same time [100,101] (Figure 2A). This innovation tremendously extends the capability of multiplexing, which is a critical merit in the application to large-scale clinical studies [102–105].
Figure 2. Overview of targeted approaches for phosphoproteomics.
(A) Multiplex immunoassays. In planar array-based assay, or ELISA, Specific primary phospho-antibodies are anchored to a carbon electrode plate (Antibody-1, and -2), which recognize phosphoproteins of interest. With the application of a fluorescent-tagged secondary antibody, the fluorescence signal measured corresponds to the expression level of that particular phosphoprotein in the sample. In the microbead-based array, primary antibodies are conjugated to the microspheres, and each microsphere batch is distinctly barcoded with different fluorophores mixed at a specific ratio (red, and yellow beads). (B) Protein microarray (or protein chip).In the forward-phase protein microarrays (FPPAs), phospho-antibodies are anchored on the chip to detect the phosphoproteins of interest; whereas in the reverse-phase protein microarrays (RPPAs), the clinical samples are directly spotted on the surface. With the sequential incubation of the cognate primary and secondary antibodies, phosphoprotein of interest could then be detected. (C) Targeted mass spectrometry. Heavy isotope-labeled synthetic peptides are typically spiked into protein digest as internal standards prior to IMAC/MOAC enrichment. The enriched phosphopeptides are subjected to SRM/PRM based targeted quantification, and the levels of phosphorylation are quantified based on light-to-heavy isotopic ratios.
Protein microarray or protein chip is another form of multiplex immunoassay albeit carried out on a glass slide or membrane. Specifically, nitrocellulose-coated slides, nylon or silanized silica are employed owing to its high binding affinity to proteins [106]. Proteome Profiler Antibody Arrays (R&D system), PathScan, and RayBioTech Phosphorylation Assay are the exemplary commercially available microarrays. The protein microarray could be subdivided into two groups based on the molecules being immobilized. In the context of forward-phase protein microarrays (FPPAs) or the antibody microarrays, phospho-antibodies are fabricated on the slide, whereas in the reverse-phase protein microarrays (RPPAs), sample of interest is glazed on the surface. In the regard of multiplexing, each array could probe for a specific phosphoprotein target across hundreds of samples in just one single run [107,108] (Figure 2B). Moreover, protein microarray is compatible with a broad spectrum of clinical samples, including biopsies, laser capture microdissection, and multiple biological fluids.
Flow cytometry is another technology that couples cell-sorting capability with the multiplexing capability of the fluorescent cell barcoding system to generate cell-type specific phosphor-proteomic fingerprints of a heterogeneous cell population [109,110]. More recently, multiplexed mass cytometry was developed by coupling flow cytometry, mass-tag cellular barcoding of antibodies, and mass spectrometry to offer single cell analyses of many proteins including phosphoproteins [111,112].
Besides immunoassays, targeted quantification of site-specific phosphorylation dynamics using SRM/PRM or data-independent acquisitions (DIA) for specific signaling proteins or pathways has become an emerging technique [113–117]. While there have been significant advances in antibody-based assays in the format of multiplexed immunoassays or protein microarrays, the major limitations for antibody-based assays are their limited specificity and often reproducibility in analyte detection due to non-specific binding and the lack of site-specific phosphorylation information. In contrast, targeted mass spectrometry based on selected reaction monitoring (SRM, also called multiple reaction monitoring MRM) or parallel reaction monitoring (PRM) provides high specificity, site-specific, and reproducible measurements of phosphorylation levels on targeted proteins [118]. The general concept of targeted MS quantification of site-specific phosphorylation is illustrated in Figure 2C. Heavy-isotope labeled phosphopeptides for the phosphorylation sites of interest will be synthesized and spiked into the peptide samples. The samples containing internal standards can be subjected to phosphopeptide enrichment using IMAC or MOAC prior to LC-SRM analyses. Both the dynamics of site-specific phosphorylation and the phosphorylation stoichiometry can be accurately quantified [113]. Direct quantification of phosphorylation dynamics is feasible without affinity enrichment by applying alternative more sensitive targeted proteomics workflow [115]. Several recent studies have demonstrated targeted quantification of phosphorylation for the entire signaling pathway such as DNA damage pathway [117] and PI3K-mTOR/MAPK pathway [116].
The Current State-of-the-Art of Phosphoproteomics
Given the technology advances in phosphoproteomics in its ability to comprehensive quantitative profiling of the phosphoproteome, we have seen an explosion of both phosphoproteomics data and literature. Table 1 summarizes some representative studies and their workflows that are reflecting the current state-of-the-art of phosphoproteomics in terms of the depth of coverage, sensitivity, and quantification dynamics. In many of these global studies, an enrichment specificity of >90% was typically obtained. By coupling phosphopeptide enrichment technique such as TiO2-MOAC or Ti4+-based IMAC with single dimensional LC-MS/MS, ~10,000 phosphorylation sites were identified and quantified from mouse liver tissue [119] or Jurkat T-cells [120]. Moreover, the phosphoproteome coverage could be further enhanced with the incorporation of different proteolytic enzymes such as AspN, Chymotrypsin, GluC, Trypsin, and LysC where 18,430 phosphosites were identified [121].
Table 1.
Selected recent studies that are representative of the current state-of-the-art of phosphoproteomics
| Workflow | Sample type | Starting sample amount | Coveragea | Quantitative strategies | Reference |
|---|---|---|---|---|---|
| Global Phosphoproteomics | |||||
| b TiO2-LC-MS/MS (Q-Exactive) | Mouse liver | 1 mg protein per analysis | ~10,000 sites | Label-free | Humphrey et al., 2015 [119] |
| Ti4+-IMAC-LC-MS/MS (Orbitrap Elite) | Jurkat T cells | 1 mg protein per condition | 12,799 sites quantified | Label-free | De Graaf et al., 2014 [120] |
| c Multiple protease- Ti4+-IMAC-LC-MS/MS (Orbitrap Elite or Fusion) | Jurkat T cells | 1 mg protein per condition | 18,430 sites | N/A | Giansanti et al., 2015 [121] |
| SCX (24 fractions)-IMAC-LC-MS/MS (Orbitrap) | 9 mouse tissues | ~10 mg protein per tissue | ~36,000 sites from 12,000 proteins; 10,000 sites per tissue | N/A | Huttlin et al., 2010 [62] |
| SCX (24 fractions)-TiO2 and anti-pTyr IP-LC-MS/MS (Q-Exactive) | Hela S3 Cells | 12.5 mg protein per experiment | 38,000 sites | Label-free | Sharma et al., 2014 [122] |
| iTRAQ-HpH (12 fractions)-IMAC-LC- MS/MS (Orbitrap and Q-Exactive) | Human ovarian tumor and breast cancer xenograft | ~2–5 mg protein per condition | 23,000–38,000 sites quantified per tissue | 4-plex iTRAQ | Mertins et al., 2014 [61] |
| iTRAQ-Online 3D-HpH RP-SAX-low pH RPLC-MS/MS (Orbitrap) | Mouse CD8+ T cells | ~100 μg peptide per condition | ~7,700 sites | 4 plex iTRAQ | Ficarro et al., 2011 [123] |
| SIMAC-HILIC-LC-MS/MS (Orbitrap velos) | Mouse INS-1 cells | 300 μg peptide per condition | ~6,600 sites | Dimethyl labeling | Engholm-Keller, et al., 2012 [70] |
| TiO2-miniatuated LC (25 μm ID)-MS/MS (Orbitrap) | Human cancer cell lines | 10,000 cells or ~1 μg protein | 1011 sites | N/A | Masuda, et al., 2011 [124] |
| Targeted Phosphoproteomics | |||||
| Ti4+-IMAC and LC-SRM (TSQ Vantage) | Human diploid fibroblast (HDF) cell line Tig3 | ~200 μg peptides | 89 phosphopeptides from PI3K- mTOR/MAPK pathway targeted | Heavy isotope- labeled internal standards | De Graaf et al., 2015 [116] |
| IMAC and LC-SRM (5500 QTRAP) | Human cell line MCF10A and human PBMCs | ~200 μg peptides | 107 DNA damage-responsive phosphosites targeted | Heavy isotope-labeled internal standards | Kennedy, et al., 2015 [117] |
The false discovery rate for all global studies was typically controlled at <1% at the peptide level.
Long gradient (2–4 hrs) separations were applied.
The multiple proteases include AspN, chymotrypsin, GluC, LysC, and trypsin.
To achieve more in-depth coverage, multi-dimensional separations were often coupled with phosphopeptide enrichment when the starting sample amounts are not limited. For example, SCX fractionation has been coupled with IMAC enrichment or TiO2 enrichment as well as anti-pTyr IP, followed by LC-MS/MS, where >30,000 phosphosites were identified [62,122]. More recently, the integration of isobaric labeling (e.g., iTRAQ) with HpH RPLC fractionation followed by IMAC enrichment and LC-MS/MS has become an increasingly popular quantitative phosphoproteomics workflow, which provides quantification of typically 20–40K phosphosites [61,78]. With this workflow, 4 to 10 biological samples can be multiplexed for quantification depending on the type of iTRAQ or TMT reagents to be used. One caveat of all the current in-depth phosphoproteome profiling workflows is the requirement of at least several mg of proteins per sample. Recently, there are several developments designed to start with a relative small amounts of proteins. For example, Ficarro et al. reported an online nanoflow 3D-LC-MS/MS workflow integrating HpH, strong anion exchange, and low pH RPLC to achieve the identification of 7,700 unique phosphopeptides from a total of only 400 μg peptides from mouse CD8+ T cells [123]. Engholm-Keller et al. reported the integration of TiO2-based pre-fractionation followed by SIMAC fractionation and capillary HILIC fractionation, which resulted in an average of ~6,600 unique phosphopeptides from 300 μg peptides/condition in a duplex dimethyl labeling experiment [70]. The most sensitive phosphoproteomics work reported to date is the coupling of hydroxy acid-modified MOAC and miniaturized analytical column (25 μm I.D. capillary LC column) for successful identification of ~1000 phosphosites from 10,000 human cancer cells [124].
Besides global phosphoproteomics, targeted quantifications of hundreds of phosphorylation sites from specific pathways have also become feasible. For example, De Graaf et al. reported the Ti4+-IMAC-SRM based quantification of 89 phosphosites from PI3K-mTOR/MAPK pathway [116], and Kennedy et al., reported IMAC-SRM based quantification of 107 phosphosites from DNA repair pathway [117]. The IMAC-SRM workflow typically only requires ~200 μg peptides per sample due to the overall higher sensitivity of LC-SRM based targeted measurements compared to conventional LC-MS/MS global measurements.
Besides the technological advances, we are also witnessing an explosion of the amount data on phosphorylation and other PTMs. The public available data on site-specific phosphorylation and other PTMs represent a hugely valuable resource to the research community at large. For instance, a number of comprehensive database resources of phosphosites as well as sites for other PTMs have been made available to the public during the past decade. These include the Phopspho.ELM [125], PhosphoSitePlus [126,127], Phosida [128], SysPTM [129], and dbPTM [130]. The SysPTM and dbPTM databases are curated databases of different types of site-specific PTMs from public resources. The PhosphoSitePlus is currently the most comprehensive database on key types of PTMs for human and mouse, containing over 330,000 non-redundant PTMs, including phospho, acetyl, ubiquityl, and methyl groups [127].
Applications in Diabetes
Phosphoproteomics has become an indispensable technology for biomedical research. Herein, we highlight some recent applications in diabetes, an important research area where the study of protein phosphorylation is pivotal and increasing its role. Protein phosphorylation is essential in the orchestration of pancreatic β-cell function, including β-cell proliferation, apoptosis, and insulin secretion, as well as insulin resistance of peripheral tissues. The challenge in applying phosphoproteomics to diabetes research is associated with the very limited sample availability of pancreatic islets samples, especially human samples. To date, most studies were carried out with cell lines or samples from mouse models. For example, Engholm-Keller et al. identified ~6,600 unique phosphopeptides from an insulin-secreting rat INS-cell line using the SIMAC-HILIC approach [70]. Li et al. reported the phosphoproteome dynamics during glucose-stimulated insulin secretion (GSIS) using rat pancreatic islets by implementing TiO2 enrichment with SILAC and LC-MS/MS [131]. Their work represents the first study focusing on phosphorylation dynamics during GSIS in islets. Despite the very small starting protein amounts (20–47 μg) from each rat, they managed to identify 8,539 phosphosites from 2,487 proteins, revealing phosphorylation responses on many different pathways such as insulin secretion related pathways, cytoskeleton dynamics, protein processing in ER and Golgi, transcription, and translation. Ca2+-dependent kinases such as Camk2b, L-type Ca2+ channel-activity-related protein Rem2, and Ca2+ transporters Atp2b1 and Slc24a2 were among those regulated proteins by the short-term high glucose stimulation. This confirms the essential role of Ca2+ in exocytosis. In another recent study of insulin signaling by Zhang et al., 3,876 phosphorylation sites were identified and a large number of potential new substrates of protein phosphatase 1 regulatory subunit 12A were revealed by quantitative phosphoproteomics [132]. A total of 698 phosphosites were responsive to PPP1R12A knockdown at both basal and insulin stimulated states and 295 of which were implicated in insulin signaling-relevant pathways such as insulin receptor signaling, mTOR signaling, and ERK/MAPK signaling. This study unveils new mechanistic evidence of the role of PPP1R12A in the skeletal muscle insulin signaling, and its implications in skeletal muscle insulin resistance and type 2 diabetes. More recently, El Ouaamari et al. reported SerpinB1 as a novel as a novel factor regulating pancreatic β-cell proliferation through the canonical growth factor pathways based on the quantitative phosphoproteomics data obtained by applying TMT-IMAC-LC-MS/MS to mouse islets and Western blotting validation [133].
In the case of type 1 diabetes (T1D), it involves the autoimmune attack specifically on the pancreatic β-cells mediated by the autoreactive T lymophocytes. Iwai et al. tested the hypothesis that the downstream signaling cascade of the antigen-specific T cell receptor (TCR) is a key factor instrumental in orchestrating the T cell response by carrying out quantitative phosphoproteomics of the primary CD4+ T cells derived from the diabetes-prone NOD versus –resistant B6.H2g7 mice [134]. With the anti-pTyr IP and IMAC along with iTRAQ labeling, they were able to quantify 77 tyrosine phosphorylation events from 54 unique proteins downstream of TCR stimulation [134]. Moreover, differentially orchestrated phosphorylation sites, including TXK, CD5, PAG1, and ZAP-70, underscore the key signaling molecules involved in the CD4+ T cell activation in NOD mice. In another study related to the complications of T1D, quantitative phosphoproteomics was applied to examine the mechanism of complications using induced pluripotent stem cells (iPSC) from patients with longstanding T1D, which allowed the observation of many key phosphorylation events in the DNA repair pathway related to T1D complications [135].
Expert Commentary
The recent advances in the field of phosphoproteomics confer us with an unprecedented capability and exciting opportunities for systems-wide study of signaling pathways and their regulation for various biological systems to gain fundamental understanding of cellular processes such as cell proliferation and apoptosis and disease pathogenesis. The powerful tools to study phosphorylation dynamics for a large number of phosphorylation sites (e.g., >10,000) are extremely valuable in identifying novel pathways, new drug targets and/or mechanistic biomarkers for the disease of interest. In the last decade we witnessed an explosion of phosphoproteomics applications, we anticipate the impact will only be accelerating now that the current phosphoproteomics workflows have been well disseminated to many different laboratories around the world based on standard enrichment protocols such as IMAC and MOAC and commercially available instrumentation for separations and MS measurements. We also anticipate that the integration of global quantitative phosphoproteomics for discovery and targeted phosphoproteomics for specific pathways will become a more prevailing tool in future applications.
In spite of the advances we recapitulate above and the great potential for applications, the phosphoproteomics field in general still faces a number of challenges for broad applications in biological and clinical research. First, most phosphoproteomics workflows require a relatively large amount of proteins (e.g., mg or more proteins are required for in-depth profiling). However, many clinical applications only have very limited amounts of samples available. For example, it is difficult to obtain even ~100 μg proteins from laser-capture microdissection (LCM) samples or pancreatic islet samples. Future advances in microscale phosphoproteomics will be important to enable this important area of clinical applications [124]. Second, although many recent studies reported very impressive coverage of the phosphoproteome (e.g., >30,000 phosphosites) using extensive fractionation [61,62,122], this workflows offer a low throughput for sample analyses, which is not suitable for large-scale profiling of clinical samples. Further advances in MS instrumentation and separations will likely lead to enhanced throughput in the near future. Third, there is also a bioinformatics challenge in identifying functionally important phosphorylation sites and regulatory networks among the extensive coverage of the whole phosphoproteome. Towards this direction, improvements in bioinformatics tools along with more accurate quantification, especially the dynamics of phosphorylation stoichiometry [115,136], will enable more effectively identify novel functional sites and regulatory networks. Finally, the targeted MS-based quantification of PTMs including phosphorylation is still a relative new area, which offers significant advantages in terms of quantification accuracy, precision and reproducibility. We envision significant efforts will be dedicated to enhance the sensitivity and multiplexing of targeted quantification of PTMs to enable broad applications of this technology for accurately quantification of potentially multiple types of PTMs such as phosphorylation, acetylation, ubiquitination, redox modifications in specific pathways, thus allowing the study of signaling crosstalk through different PTMs.
Five-year view
While phosphoproteomics has been broadly applied in many biological applications, there are still many hurdles on the way to apply the technologies to specific types of applications as discussed above. In the next 5 years, we anticipate the phosphoproteomics technologies will become more mature with further advance in sensitivity, throughput, reproducibility, and accuracy of quantification to enable even broader applications in both normal physiology (e.g., the effect of physical activity [137]) and disease pathogenesis. Specifically, the sensitivity for handling microscale samples will potentially to be a key technology advance to enable phosphoproteome profiling of extremely small clinical samples such as LCM-tissue samples. Further advances in the multiplexed quantification (20-plex or more) [138] and the overall sample throughput will likely be achieved through the must faster scanning rate of MS instrumentation and advanced separations. Such advances should enable in-depth quantitative profiling of tissue phosphoproteome in large-scale clinical studies such as those performed for the global proteome [139]. Moreover, we anticipate significant advances in MS-based targeted quantification of phosphorylation and other types of PTMs in terms of sensitivity, multiplexing capacity, and throughput to enable detailed time course studies or large-scale population studies of specific pathways and signaling networks. With these advances, phosphoproteomics studies will become more integrative for both discovery and functional verification, and be more routinely applied in broad areas of biomedical research.
Key Issues.
The current global quantitative phosphoproteomics workflows integrating stable isotope labeling, LC fractionation, IMAC/MOAC phosphopeptide enrichment, and LC-MS/MS enable quantitative profiling of the phosphoproteome with in-depth coverage (e.g., >30,000 phosphosites)
MS-based targeted quantification of site-specific phosphorylation is an emerging technology and further advances in sensitivity and multiplexing capacity are anticipated in the near future.
The current limitations for phosphoproteomics include the relatively low overall sample throughput, and the requirement of relatively large amount of starting protein materials.
Advances in phosphoproteomics towards micro-scale clinical samples (e.g., LCM samples) will be key for broad clinical applications.
Acknowledgments
This work is partially supported by National Institutes of Health grants UC4 DK104167 (to WJ Qian) and P41 GM103493 (to RD Smith).
Abbreviations
- CID
Collision-induced dissociation
- DHB
2,5-dihydroxybenzoic acid
- DIA
Data-independent acquisitions
- ELISA
Enzyme-linked immunosorbent assay
- ERLIC
Electrostatic repulsion hydrophilic interaction chromatography
- ETD
Electron transfer dissociation
- FASP
Filter aided sample preparation
- FPPAs
Forward-phase protein microarrays
- GSIS
Glucose stimulated insulin secretion
- HCD
Higher-energy collisional dissociation
- HILIC
Hydrophilic interaction chromatography
- HpH
High pH
- IDA
Iminodiacetic acid
- IMAC
Immobilized metal affinity chromatography
- iPSC
Induced pluripotent stem cells
- iTRAQ
Isobaric tags for relative and absolute quantification
- LC
Liquid chromatography
- LCM
Laser-capture microdissection
- LIRKO
Liver-specific insulin receptor knockout
- MOAC
Metal oxide affinity chromatography
- MRM
Multiple reaction monitoring
- MS
Mass spectrometry
- MS3
Triple-stage MS
- MSD
Meso-scale discovery
- NTA
Nitrilotriacetic acid
- PolyMAC
Polymer-based metal-ion affinity capture
- PRM
Parallel reaction monitoring
- pSer
Phosphorylated serine
- pThr
Phosphorylated threonine
- PTM
Post-translational modification
- pTyr
Phosphorylated tyrosine
- RPPAs
Reverse-phase protein microarrays
- SCX
Strong cation exchange
- SILAC
Stable isotope labeling by amino acids in cell culture
- SIMAC
Sequential elution from IMAC
- SRM
Selected reaction monitoring
- T1D
Type 1 diabetes
- TCR
Antigen-specific T cell receptor
Footnotes
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Hunter T. Signaling--2000 and beyond. Cell. 2000;100(1):113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Ohno M. Roles of eIF2alpha kinases in the pathogenesis of Alzheimer's disease. Frontiers in molecular neuroscience. 2014;7:22. doi: 10.3389/fnmol.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tepper K, Biernat J, Kumar S, et al. Oligomer formation of tau protein hyperphosphorylated in cells. The Journal of biological chemistry. 2014;289(49):34389–34407. doi: 10.1074/jbc.M114.611368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411(6835):355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 5.Zanivan S, Meves A, Behrendt K, et al. In vivo SILAC-based proteomics reveals phosphoproteome changes during mouse skin carcinogenesis. Cell reports. 2013;3(2):552–566. doi: 10.1016/j.celrep.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Huelsemann MF, Patz M, Beckmann L, et al. Hypoxia-induced p38 MAPK activation reduces Mcl-1 expression and facilitates sensitivity towards BH3 mimetics in chronic lymphocytic leukemia. Leukemia. 2015;29(4):981–984. doi: 10.1038/leu.2014.320. [DOI] [PubMed] [Google Scholar]
- 7.Decker RS, Decker ML, Kulikovskaya I, et al. Myosin-binding protein C phosphorylation, myofibril structure, and contractile function during low-flow ischemia. Circulation. 2005;111(7):906–912. doi: 10.1161/01.CIR.0000155609.95618.75. [DOI] [PubMed] [Google Scholar]
- 8.El-Armouche A, Pohlmann L, Schlossarek S, et al. Decreased phosphorylation levels of cardiac myosin-binding protein-C in human and experimental heart failure. J Mol Cell Cardiol. 2007;43(2):223–229. doi: 10.1016/j.yjmcc.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Schoutsen B, Blom JJ, Verdouw PD, Lamers JM. Calcium transport and phospholamban in sarcoplasmic reticulum of ischemic myocardium. J Mol Cell Cardiol. 1989;21(7):719–727. doi: 10.1016/0022-2828(89)90613-5. [DOI] [PubMed] [Google Scholar]
- 10.Scheuner D, Vander Mierde D, Song B, et al. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nature medicine. 2005;11(7):757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 11.Ortsater H, Grankvist N, Honkanen RE, Sjoholm A. Protein phosphatases in pancreatic islets. J Endocrinol. 2014;221(3):R121–144. doi: 10.1530/JOE-14-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyerovitch J, Backer JM, Kahn CR. Hepatic phosphotyrosine phosphatase activity and its alterations in diabetic rats. J Clin Invest. 1989;84(3):976–983. doi: 10.1172/JCI114261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez de Castro D, Clarke PA, Al-Lazikani B, Workman P. Personalized cancer medicine: molecular diagnostics, predictive biomarkers, and drug resistance. Clin Pharmacol Ther. 2013;93(3):252–259. doi: 10.1038/clpt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Dar AC, Shokat KM. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu Rev Biochem. 2011;80:769–795. doi: 10.1146/annurev-biochem-090308-173656. [DOI] [PubMed] [Google Scholar]
- 16.Sawyers CL, Hochhaus A, Feldman E, et al. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a phase II study. Blood. 2002;99(10):3530–3539. doi: 10.1182/blood.v99.10.3530. [DOI] [PubMed] [Google Scholar]
- 17.Sawyers CL. Shifting paradigms: the seeds of oncogene addiction. Nature medicine. 2009;15(10):1158–1161. doi: 10.1038/nm1009-1158. [DOI] [PubMed] [Google Scholar]
- 18.Bachman KE, Argani P, Samuels Y, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3(8):772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 19.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 20.Serra V, Markman B, Scaltriti M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68(19):8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 21.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62(23):6997–7000. [PubMed] [Google Scholar]
- 22.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 23.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 25.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 26.Motzer RJ, Escudier B, Oudard S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372(9637):449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 27.Roux PP, Thibault P. The coming of age of phosphoproteomics--from large data sets to inference of protein functions. Mol Cell Proteomics. 2013;12(12):3453–3464. doi: 10.1074/mcp.R113.032862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson A, Schafer J, Kuhn K, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical Chemistry. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 29.Ross PL, Huang YLN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & Cellular Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Engholm-Keller K, Larsen MR. Technologies and challenges in large-scale phosphoproteomics. Proteomics. 2013;13(6):910–931. doi: 10.1002/pmic.201200484. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson CL. Advances in quantitative phosphoproteomics. Anal Chem. 2012;84(2):735–746. doi: 10.1021/ac202877y. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Belkina N, Jacob HK, et al. Identifying novel targets of oncogenic EGF receptor signaling in lung cancer through global phosphoproteomics. Proteomics. 2015;15(2–3):340–355. doi: 10.1002/pmic.201400315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bian Y, Ye M, Song C, et al. Improve the coverage for the analysis of phosphoproteome of HeLa cells by a tandem digestion approach. J Proteome Res. 2012;11(5):2828–2837. doi: 10.1021/pr300242w. [DOI] [PubMed] [Google Scholar]
- 34.Wisniewski JR, Mann M. Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Anal Chem. 2012;84(6):2631–2637. doi: 10.1021/ac300006b. [DOI] [PubMed] [Google Scholar]
- 35.Neilson KA, Ali NA, Muralidharan S, et al. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11(4):535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 36.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & cellular proteomics : MCP. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 37.Kocher T, Pichler P, Schutzbier M, et al. High precision quantitative proteomics using iTRAQ on an LTQ Orbitrap: a new mass spectrometric method combining the benefits of all. J Proteome Res. 2009;8(10):4743–4752. doi: 10.1021/pr900451u. [DOI] [PubMed] [Google Scholar]
- 38.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8(11):937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McAlister GC, Nusinow DP, Jedrychowski MP, et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem. 2014;86(14):7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schilling B, Rardin MJ, MacLean BX, et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics. 2012;11(5):202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue L, Wang P, Wang L, et al. Quantitative measurement of phosphoproteome response to osmotic stress in arabidopsis based on Library-Assisted eXtracted Ion Chromatogram (LAXIC) Mol Cell Proteomics. 2013;12(8):2354–2369. doi: 10.1074/mcp.O113.027284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding SJ, Qian WJ, Smith RD. Quantitative proteomic approaches for studying phosphotyrosine signaling. Expert Rev Proteomics. 2007;4(1):13–23. doi: 10.1586/14789450.4.1.13. [DOI] [PubMed] [Google Scholar]
- 44*.Ficarro SB, McCleland ML, Stukenberg PT, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20(3):301–305. doi: 10.1038/nbt0302-301. The first report of global phosphoproteome profiling using IMAC. [DOI] [PubMed] [Google Scholar]
- 45.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4(7):873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Gnad F, Forner F, Zielinska DF, Birney E, Gunawardena J, Mann M. Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol Cell Proteomics. 2010;9(12):2642–2653. doi: 10.1074/mcp.M110.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller M, Donat S, Rakette S, et al. Staphylococcal PknB as the first prokaryotic representative of the proline-directed kinases. PLoS One. 2010;5(2):e9057. doi: 10.1371/journal.pone.0009057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol. 2004;22(9):1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 49.Rush J, Moritz A, Lee KA, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23(1):94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 50.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 51.Heibeck TH, Ding SJ, Opresko LK, et al. An extensive survey of tyrosine phosphorylation revealing new sites in human mammary epithelial cells. J Proteome Res. 2009;8(8):3852–3861. doi: 10.1021/pr900044c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunter T. The Croonian Lecture 1997. The phosphorylation of proteins on tyrosine: its role in cell growth and disease. Philos Trans R Soc Lond B Biol Sci. 1998;353(1368):583–605. doi: 10.1098/rstb.1998.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boersema PJ, Foong LY, Ding VM, et al. In-depth qualitative and quantitative profiling of tyrosine phosphorylation using a combination of phosphopeptide immunoaffinity purification and stable isotope dimethyl labeling. Mol Cell Proteomics. 2010;9(1):84–99. doi: 10.1074/mcp.M900291-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal Biochem. 1986;154(1):250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 55.Neville DC, Rozanas CR, Price EM, Gruis DB, Verkman AS, Townsend RR. Evidence for phosphorylation of serine 753 in CFTR using a novel metal-ion affinity resin and matrix-assisted laser desorption mass spectrometry. Protein Sci. 1997;6(11):2436–2445. doi: 10.1002/pro.5560061117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou H, Ye M, Dong J, et al. Specific phosphopeptide enrichment with immobilized titanium ion affinity chromatography adsorbent for phosphoproteome analysis. J Proteome Res. 2008;7(9):3957–3967. doi: 10.1021/pr800223m. [DOI] [PubMed] [Google Scholar]
- 57.Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal Chem. 1999;71(14):2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 58.Feng S, Ye M, Zhou H, et al. Immobilized zirconium ion affinity chromatography for specific enrichment of phosphopeptides in phosphoproteome analysis. Mol Cell Proteomics. 2007;6(9):1656–1665. doi: 10.1074/mcp.T600071-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Jensen SS, Larsen MR. Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun Mass Spectrom. 2007;21(22):3635–3645. doi: 10.1002/rcm.3254. [DOI] [PubMed] [Google Scholar]
- 60.Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, Marto JA. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal Chem. 2009;81(11):4566–4575. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61**.Mertins P, Yang F, Liu T, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics. 2014;13(7):1690–1704. doi: 10.1074/mcp.M113.036392. Multi-laboratory large-scale quantitative phosphoproteome profiling for tumor tissues using iTRAQ-HpH fractionation-IMAC-LC-MS/MS with coverage more than 20,000 phosphosites per tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huttlin EL, Jedrychowski MP, Elias JE, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsai CF, Hsu CC, Hung JN, et al. Sequential phosphoproteomic enrichment through complementary metal-directed immobilized metal ion affinity chromatography. Anal Chem. 2014;86(1):685–693. doi: 10.1021/ac4031175. [DOI] [PubMed] [Google Scholar]
- 64.Leitner A. Phosphopeptide enrichment using metal oxide affinity chromatography. Trends in Analytical Chemistry. 2010;29(2):177–185. [Google Scholar]
- 65*.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem. 2004;76(14):3935–3943. doi: 10.1021/ac0498617. The first report of phosphopeptide enrichment using TiO2 for LC-MS/MS analysis. [DOI] [PubMed] [Google Scholar]
- 66.Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics. 2007;6(6):1103–1109. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
- 67.Paulo JA, McAllister FE, Everley RA, Beausoleil SA, Banks AS, Gygi SP. Effects of MEK inhibitors GSK1120212 and PD0325901 in vivo using 10-plex quantitative proteomics and phosphoproteomics. Proteomics. 2015;15(2–3):462–473. doi: 10.1002/pmic.201400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol Cell Proteomics. 2008;7(4):661–671. doi: 10.1074/mcp.M700362-MCP200. [DOI] [PubMed] [Google Scholar]
- 69.Sun Z, Hamilton KL, Reardon KF. Evaluation of quantitative performance of sequential immobilized metal affinity chromatographic enrichment for phosphopeptides. Anal Biochem. 2014;445:30–37. doi: 10.1016/j.ab.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70**.Engholm-Keller K, Birck P, Storling J, Pociot F, Mandrup-Poulsen T, Larsen MR. TiSH--a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J Proteomics. 2012;75(18):5749–5761. doi: 10.1016/j.jprot.2012.08.007. A multi-dimensional phosphoproteomics strategy combining TiO2, SIMAC, and HILIC (collectively abbreviated "TiSH") was applied in the study of interferon-gamma signaling. [DOI] [PubMed] [Google Scholar]
- 71.Engholm-Keller K, Hansen TA, Palmisano G, Larsen MR. Multidimensional strategy for sensitive phosphoproteomics incorporating protein prefractionation combined with SIMAC, HILIC, and TiO(2) chromatography applied to proximal EGF signaling. J Proteome Res. 2011;10(12):5383–5397. doi: 10.1021/pr200641x. [DOI] [PubMed] [Google Scholar]
- 72.Iliuk AB, Martin VA, Alicie BM, Geahlen RL, Tao WA. In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol Cell Proteomics. 2010;9(10):2162–2172. doi: 10.1074/mcp.M110.000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iliuk A, Jayasundera K, Wang WH, Schluttenhofer R, Geahlen RL, Tao WA. In-Depth Analyses of B Cell Signaling Through Tandem Mass Spectrometry of Phosphopeptides Enriched by PolyMAC. Int J Mass Spectrom. 2015;377:744–753. doi: 10.1016/j.ijms.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81(11):4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- 75.Wang Y, Yang F, Gritsenko MA, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11(10):2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dwivedi RC, Spicer V, Harder M, et al. Practical implementation of 2D HPLC scheme with accurate peptide retention prediction in both dimensions for high-throughput bottom-up proteomics. Anal Chem. 2008;80(18):7036–7042. doi: 10.1021/ac800984n. [DOI] [PubMed] [Google Scholar]
- 77.Batth TS, Francavilla C, Olsen JV. Off-line high-pH reversed-phase fractionation for in-depth phosphoproteomics. J Proteome Res. 2014;13(12):6176–6186. doi: 10.1021/pr500893m. [DOI] [PubMed] [Google Scholar]
- 78**.Mertins P, Qiao JW, Patel J, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10(7):634–637. doi: 10.1038/nmeth.2518. The first report of serial enrichments of different PTMs (SEPTM) for generating large-scale profiling of phosphorylation, ubiquitination, and acetylation sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alpert AJ. Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal Chem. 2008;80(1):62–76. doi: 10.1021/ac070997p. [DOI] [PubMed] [Google Scholar]
- 80.McNulty DE, Annan RS. Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol Cell Proteomics. 2008;7(5):971–980. doi: 10.1074/mcp.M700543-MCP200. [DOI] [PubMed] [Google Scholar]
- 81.Chien KY, Liu HC, Goshe MB. Development and application of a phosphoproteomic method using electrostatic repulsion-hydrophilic interaction chromatography (ERLIC), IMAC, and LC-MS/MS analysis to study Marek's Disease Virus infection. J Proteome Res. 2011;10(9):4041–4053. doi: 10.1021/pr2002403. [DOI] [PubMed] [Google Scholar]
- 82.Gan CS, Guo T, Zhang H, Lim SK, Sze SK. A comparative study of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) versus SCX-IMAC-based methods for phosphopeptide isolation/enrichment. J Proteome Res. 2008;7(11):4869–4877. doi: 10.1021/pr800473j. [DOI] [PubMed] [Google Scholar]
- 83.Annan RS, Carr SA. Phosphopeptide analysis by matrix-assisted laser desorption time-of-flight mass spectrometry. Anal Chem. 1996;68(19):3413–3421. doi: 10.1021/ac960221g. [DOI] [PubMed] [Google Scholar]
- 84.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(7):2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2007;6(11):1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 87.Lehmann WD, Kruger R, Salek M, Hung CW, Wolschin F, Weckwerth W. Neutral loss-based phosphopeptide recognition: a collection of caveats. J Proteome Res. 2007;6(7):2866–2873. doi: 10.1021/pr060573w. [DOI] [PubMed] [Google Scholar]
- 88.Jedrychowski MP, Huttlin EL, Haas W, Sowa ME, Rad R, Gygi SP. Evaluation of HCD- and CID-type fragmentation within their respective detection platforms for murine phosphoproteomics. Mol Cell Proteomics. 2011;10(12) doi: 10.1074/mcp.M111.009910. M111 009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frese CK, Altelaar AF, van den Toorn H, et al. Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal Chem. 2012;84(22):9668–9673. doi: 10.1021/ac3025366. [DOI] [PubMed] [Google Scholar]
- 90.Frese CK, Zhou H, Taus T, et al. Unambiguous phosphosite localization using electron-transfer/higher-energy collision dissociation (EThcD) J Proteome Res. 2013;12(3):1520–1525. doi: 10.1021/pr301130k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Olsen JV, Blagoev B, Gnad F, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 92.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 93.Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD. PhosphoScore: an open-source phosphorylation site assignment tool for MSn data. J Proteome Res. 2008;7(7):3054–3059. doi: 10.1021/pr800169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wan Y, Cripps D, Thomas S, et al. PhosphoScan: a probability-based method for phosphorylation site prediction using MS2/MS3 pair information. J Proteome Res. 2008;7(7):2803–2811. doi: 10.1021/pr700773p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.MacLean D, Burrell MA, Studholme DJ, Jones AM. PhosCalc: a tool for evaluating the sites of peptide phosphorylation from mass spectrometer data. BMC research notes. 2008;1:30. doi: 10.1186/1756-0500-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bailey CM, Sweet SM, Cunningham DL, Zeller M, Heath JK, Cooper HJ. SLoMo: automated site localization of modifications from ETD/ECD mass spectra. J Proteome Res. 2009;8(4):1965–1971. doi: 10.1021/pr800917p. [DOI] [PubMed] [Google Scholar]
- 97.Swaney DL, Wenger CD, Thomson JA, Coon JJ. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(4):995–1000. doi: 10.1073/pnas.0811964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taus T, Kocher T, Pichler P, et al. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res. 2011;10(12):5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 99.Chowdhury F, Williams A, Johnson P. Validation and comparison of two multiplex technologies, Luminex and Mesoscale Discovery, for human cytokine profiling. J Immunol Methods. 2009;340(1):55–64. doi: 10.1016/j.jim.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 100.Du J, Bernasconi P, Clauser KR, et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat Biotechnol. 2009;27(1):77–83. doi: 10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schwenk JM, Nilsson P. Antibody suspension bead arrays. Methods Mol Biol. 2011;723:29–36. doi: 10.1007/978-1-61779-043-0_3. [DOI] [PubMed] [Google Scholar]
- 102.Dahut WL, Scripture C, Posadas E, et al. A phase II clinical trial of sorafenib in androgen-independent prostate cancer. Clin Cancer Res. 2008;14(1):209–214. doi: 10.1158/1078-0432.CCR-07-1355. [DOI] [PubMed] [Google Scholar]
- 103.Yap TA, Yan L, Patnaik A, et al. First-in-man clinical trial of the oral pan-AKT inhibitor MK-2206 in patients with advanced solid tumors. J Clin Oncol. 2011;29(35):4688–4695. doi: 10.1200/JCO.2011.35.5263. [DOI] [PubMed] [Google Scholar]
- 104.Angevin E, Lopez-Martin JA, Lin CC, et al. Phase I study of dovitinib (TKI258), an oral FGFR, VEGFR, and PDGFR inhibitor, in advanced or metastatic renal cell carcinoma. Clin Cancer Res. 2013;19(5):1257–1268. doi: 10.1158/1078-0432.CCR-12-2885. [DOI] [PubMed] [Google Scholar]
- 105.Haura EB, Tanvetyanon T, Chiappori A, et al. Phase I/II study of the Src inhibitor dasatinib in combination with erlotinib in advanced non-small-cell lung cancer. J Clin Oncol. 2010;28(8):1387–1394. doi: 10.1200/JCO.2009.25.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.VanMeter A, Signore M, Pierobon M, Espina V, Liotta LA, Petricoin EF., 3rd Reverse-phase protein microarrays: application to biomarker discovery and translational medicine. Expert Rev Mol Diagn. 2007;7(5):625–633. doi: 10.1586/14737159.7.5.625. [DOI] [PubMed] [Google Scholar]
- 107.Mueller C, Liotta LA, Espina V. Reverse phase protein microarrays advance to use in clinical trials. Molecular oncology. 2010;4(6):461–481. doi: 10.1016/j.molonc.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.VanMeter AJ, Rodriguez AS, Bowman ED, et al. Laser capture microdissection and protein microarray analysis of human non-small cell lung cancer: differential epidermal growth factor receptor (EGPR) phosphorylation events associated with mutated EGFR compared with wild type. Mol Cell Proteomics. 2008;7(10):1902–1924. doi: 10.1074/mcp.M800204-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krutzik PO, Trejo A, Schulz KR, Nolan GP. Phospho flow cytometry methods for the analysis of kinase signaling in cell lines and primary human blood samples. Methods Mol Biol. 2011;699:179–202. doi: 10.1007/978-1-61737-950-5_9. [DOI] [PubMed] [Google Scholar]
- 110.Krutzik PO, Clutter MR, Trejo A, Nolan GP. Fluorescent cell barcoding for multiplex flow cytometry. In: Paul Robinson J managing editor, et al., editors. Current protocols in cytometry. Unit 6. Chapter 6. 2011. p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bodenmiller B, Zunder ER, Finck R, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30(9):858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Simmons AJ, Banerjee A, McKinley ET, et al. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-alpha-induced apoptosis in vivo. Mol Syst Biol. 2015;11(10):835. doi: 10.15252/msb.20156282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113**.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci U S A. 2007;104(14):5860–5865. doi: 10.1073/pnas.0608638104. The first proof-of-concept of applying targeted mass spectrometry for quantification of site-specific phosphorylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Parker BL, Yang G, Humphrey SJ, et al. Targeted phosphoproteomics of insulin signaling using data-independent acquisition mass spectrometry. Sci Signal. 2015;8(380):rs6. doi: 10.1126/scisignal.aaa3139. [DOI] [PubMed] [Google Scholar]
- 115*.Shi T, Gao Y, Gaffrey MJ, et al. Sensitive targeted quantification of ERK phosphorylation dynamics and stoichiometry in human cells without affinity enrichment. Anal Chem. 2015;87(2):1103–1110. doi: 10.1021/ac503797x. Demonstrated the direct quantification of phosphorylation stoichiometry without affinity enrichment by ultrasensitive targeted proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.de Graaf EL, Kaplon J, Mohammed S, et al. Signal Transduction Reaction Monitoring Deciphers Site-Specific PI3K-mTOR/MAPK Pathway Dynamics in Oncogene-Induced Senescence. J Proteome Res. 2015;14(7):2906–2914. doi: 10.1021/acs.jproteome.5b00236. [DOI] [PubMed] [Google Scholar]
- 117**.Kennedy JJ, Yan P, Zhao L, et al. Immobilized metal affinity chromatography coupled to multiple reaction monitoring enables reproducible quantification of phospho-signaling. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.O115.054940. Demonstrated the ability to perform relatively broad targeted quantification for 107 phosphosites by IMAC-MRM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Addona TA, Abbatiello SE, Schilling B, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27(7):633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Humphrey SJ, Azimifar SB, Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat Biotechnol. 2015;33(9):990–995. doi: 10.1038/nbt.3327. [DOI] [PubMed] [Google Scholar]
- 120.de Graaf EL, Giansanti P, Altelaar AF, Heck AJ. Single-step enrichment by Ti4+-IMAC and label-free quantitation enables in-depth monitoring of phosphorylation dynamics with high reproducibility and temporal resolution. Mol Cell Proteomics. 2014;13(9):2426–2434. doi: 10.1074/mcp.O113.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Giansanti P, Aye TT, van den Toorn H, Peng M, van Breukelen B, Heck AJ. An Augmented Multiple-Protease-Based Human Phosphopeptide Atlas. Cell reports. 2015;11(11):1834–1843. doi: 10.1016/j.celrep.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 122.Sharma K, D'Souza RC, Tyanova S, et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell reports. 2014;8(5):1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 123.Ficarro SB, Zhang Y, Carrasco-Alfonso MJ, et al. Online nanoflow multidimensional fractionation for high efficiency phosphopeptide analysis. Mol Cell Proteomics. 2011;10(11) doi: 10.1074/mcp.O111.011064. O111 011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124*.Masuda T, Sugiyama N, Tomita M, Ishihama Y. Microscale phosphoproteome analysis of 10,000 cells from human cancer cell lines. Anal Chem. 2011;83(20):7698–7703. doi: 10.1021/ac201093g. The most sensisitive phosphoproteomics work reported to date starting with only 10,000 cells. [DOI] [PubMed] [Google Scholar]
- 125.Dinkel H, Chica C, Via A, et al. Phospho.ELM: a database of phosphorylation sites--update 2011. Nucleic Acids Res. 2011;39(Database issue):D261–267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hornbeck PV, Kornhauser JM, Tkachev S, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40(Database issue):D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127*.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512–520. doi: 10.1093/nar/gku1267. PhosphositePlus: the most comprehensive PTM database for mammals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gnad F, Ren S, Cox J, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome biology. 2007;8(11):R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li H, Xing X, Ding G, et al. SysPTM: a systematic resource for proteomic research on post-translational modifications. Mol Cell Proteomics. 2009;8(8):1839–1849. doi: 10.1074/mcp.M900030-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lu CT, Huang KY, Su MG, et al. DbPTM 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications. Nucleic Acids Res. 2013;41(Database issue):D295–305. doi: 10.1093/nar/gks1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li J, Li Q, Tang J, Xia F, Wu J, Zeng R. Quantitative Phosphoproteomics Revealed Glucose-Stimulated Responses of Islet Associated with Insulin Secretion. J Proteome Res. 2015;14(11):4635–4646. doi: 10.1021/acs.jproteome.5b00507. [DOI] [PubMed] [Google Scholar]
- 132.Zhang X, Ma D, Caruso M, Lewis M, Qi Y, Yi Z. Quantitative phosphoproteomics reveals novel phosphorylation events in insulin signaling regulated by protein phosphatase 1 regulatory subunit 12A. J Proteomics. 2014;109:63–75. doi: 10.1016/j.jprot.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.El Ouaamari A, Dirice E, Gedeon N, et al. SerpinB1 Promotes Pancreatic beta Cell Proliferation. Cell metabolism. 2015 doi: 10.1016/j.cmet.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Iwai LK, Benoist C, Mathis D, White FM. Quantitative phosphoproteomic analysis of T cell receptor signaling in diabetes prone and resistant mice. J Proteome Res. 2010;9(6):3135–3145. doi: 10.1021/pr100035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bhatt S, Gupta MK, Khamaisi M, et al. Preserved DNA Damage Checkpoint Pathway Protects against Complications in Long-Standing Type 1 Diabetes. Cell metabolism. 2015;22(2):239–252. doi: 10.1016/j.cmet.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu R, Haas W, Dephoure N, et al. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat Methods. 2011;8(8):677–683. doi: 10.1038/nmeth.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Neufer PD, Bamman MM, Muoio DM, et al. Understanding the Cellular and Molecular Mechanisms of Physical Activity-Induced Health Benefits. Cell metabolism. 2015;22(1):4–11. doi: 10.1016/j.cmet.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 138*.Everley RA, Kunz RC, McAllister FE, Gygi SP. Increasing throughput in targeted proteomics assays: 54-plex quantitation in a single mass spectrometry run. Anal Chem. 2013;85(11):5340–5346. doi: 10.1021/ac400845e. A 54-plex approach was applied to measure the inhibition of PKA activity of MCF cells by nine compounds. This platform demonstrates a ~50-fold improvement in acquisition throughput. [DOI] [PubMed] [Google Scholar]
- 139.Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513(7518):382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]