Abstract
Human endometrium undergoes cyclic regeneration involving stem/progenitor cells, but the role of resident endometrial mesenchymal stem cells (eMSC) as progenitors of endometrial stromal fibroblasts (eSF) has not been definitively demonstrated. In endometriosis, eSF display progesterone (P4) resistance with impaired decidualization in vivo and in vitro. To investigate eMSC as precursors of eSF and whether endometriosis P4 resistance is inherited from eMSC, we analyzed transcriptomes of eutopic endometrium eMSC and eSF isolated by fluorescence-activated cell sorting (FACS) from endometriosis (eMSCendo, eSFendo) and controls (eMSCcontrol, eSFcontrol) and their derived primary cultures. Differentially expressed lineage-associated genes (LG) of FACS-isolated eMSC and eSF were largely conserved in endometriosis. In culture, eSFcontrol maintained in vitro expression of a subset of eSF LG and decidualized in vitro with P4. The eMSCcontrol cultures differentiated in vitro to eSF lineage, down-regulating eMSC LG and up-regulating eSF LG, showing minimal transcriptome differences versus eSFcontrol cultures and decidualizing in vitro. Cultured eSFendo displayed less in vitro LG stability and did not decidualize in vitro. In vitro, eMSCendo differentiated to eSF lineage but showed more differentially expressed genes versus eSFendo cultures, and did not decidualize in vitro, demonstrating P4 resistance inherited from eMSCendo. Compared to controls, cultures from tissue-derived eSFendo uniquely had a pro-inflammatory phenotype not present in eMSCendo differentiated to eSF in vitro, suggesting divergent niche effects for in vivo versus in vitro lineage differentiation. These findings substantiate eMSC as progenitors of eSF and reveal eSF in endometriosis as having P4 resistance inherited from eMSC and a pro-inflammatory phenotype acquired within the endometrial niche.
Keywords: differentiation, endometriosis, endometrium, fibroblasts, mesenchymal stem cells
INTRODUCTION
Human endometrium is a dynamic steroid hormone-dependent tissue that undergoes proliferation under the influence of estradiol (E2) and differentiation of its cellular constituents in response to progesterone (P4), preparing for embryo implantation. In the absence of pregnancy, it undergoes desquamation and subsequent regeneration without scarring [1], a process that involves epithelial, mesenchymal, and endothelial adult stem/progenitor cells [2–4]. Endometrial mesenchymal stem cells (eMSC) are clonogenic, multipotent pericytes that can differentiate into adipogenic, osteogenic, chondrogenic, and myogenic lineages [4–7]. Expression profiling, hierarchical clustering (HC), and principal component analyses (PCA) support a common lineage of eMSC and endometrial stromal fibroblasts (eSF) [4, 8]. However, whether eMSC are in fact progenitors of eSF has not been definitively demonstrated [4, 9] and was one of the aims of the current study.
Human eMSC display colony formation, side population phenotype, or stem cell surface marker expression (W5C5/SUSD2+, CD146+, PDGFRB+), and SUSD2+ eMSC give rise to endometrial stromal-like tissues in vivo [2, 10, 11]. Multiple groups have confirmed the presence of eMSC in human endometrium [2, 4, 6, 8, 9, 12, 13], and cultured SUSD2+ cells respond to medroxyprogesterone with activation of the PKA pathway [14], demonstrating the potential of eMSC differentiation to a cell type that is progestin responsive.
The eSF plays a central role in endometrial function. Its responsiveness to P4 is essential for establishment and maintenance of pregnancy [1], and the cell has a major role in tissue desquamation in the absence of pregnancy and overall tissue homeostasis [15, 16]. During the cycle, P4 induces eSF differentiation (decidualization) to an epithelial-like cell with characteristic morphology, transcriptome, and unique biomarkers, including IGFBP1 [17], mediated through PRA, PRB, MAPK, PKA, and other signaling pathways [18]. In response to P4, eSF create unique niches for angiogenesis and leukocyte recruitment, preparing for embryo implantation, immune tolerance, modulation of trophoblast invasion, and blood supply to the conceptus [19, 20]. Dysfunction of this differentiation process can lead to infertility or poor pregnancy outcome [21]. In endometriosis, an estrogen-dependent inflammatory disorder [22, 23], eSF are resistant to P4 [24–26], consistent with infertility and poor pregnancy outcomes observed in women with this disease [21, 22, 26, 27].
Herein, we investigated whether eMSC are precursors of eSF and if so, whether the observed P4 resistance in endometriosis is unique to eSF in the endometrial niche or is inherited from the eMSC progenitor. To address these fundamental questions, we studied eMSC and eSF freshly isolated from endometrium of women with and without endometriosis, and their respective short- and long-term cultures. The data substantiate eMSC as progenitors of eSF and reveal novel aspects of the eSF disease phenotype in endometriosis wherein P4 resistance is inherited from the eMSC and a pro-inflammatory component is acquired by the eSF lineage within the endometrial niche.
MATERIALS AND METHODS
Human Endometrial Tissues
Endometrial tissue samples were obtained through the National Institutes of Health (NIH)/University of California, San Francisco (UCSF) Human Endometrial Tissue and DNA Bank in accordance with the guidelines of the Declaration of Helsinki. The UCSF Committee on Human Research approved the study (protocol no. 10-02786, expiration 12/01/16), and written informed consent was obtained from all participants. Endometrial tissues were obtained prospectively by biopsy using a Pipelle catheter (Cooper Surgical) from 13 healthy volunteers and women undergoing benign gynecological surgery and documented to be free of endometriosis (controls) as well as from eight women with endometriosis (endo) diagnosed at laparoscopy and confirmed histologically. Participant characteristics are shown in Supplemental Table S1 (Supplemental Data are available online at www.biolreprod.org).
Endometrial Cell Isolation and Flow Cytometry
Pure populations of eMSC and eSF were isolated within 24 h of tissue collection by fluorescence-activated cell sorting (FACS) using fluorochrome-conjugated antibodies (BD Biosciences) as described previously [4, 8]. Briefly, fresh endometrial tissue samples were enzymatically digested and single cells separated through a 40 μm sieve, washed with phosphate buffered saline (PBS), treated with DNase, and contaminant erythrocytes lysed before processing for FACS. The FACS protocol used four-color sorting to exclude cluster of differentiation 45-positive (CD45+) leukocytes and epithelial cell adhesion molecule-positive (EPCAM+) endometrial epithelial cells, followed by sorting the CD45−/EPCAM− cells into eMSC co-expressing melanoma cell adhesion molecule (MCAM, CD146) and platelet-derived growth factor receptor β (PDGFRB, CD140b), and into eSF, which are CD146−/PDGFRB+. Dry cell pellets of freshly sorted CD146+/PDGFRB+ (eMSCFACS) and CD146−/PDGFRB+ (eSFFACS) were stored at −80°C until processed for RNA extraction. When FACS yields allowed, aliquots of the freshly sorted eMSC and eSF were immediately processed for cell culture as described below.
Cell Culture
Clonal growth, which typically spans longer culture times compared to high-density cultures, was chosen for the current experimental design (Fig. 1) based on previous studies defining clonal growth as selection/characterization criterion for eMSC in culture [5, 9, 28]. Freshly sorted eMSC and eSF were plated at clonal density (10–20 viable cells/cm2) and grown in culture medium composed of 75% high-glucose phenol red-free Dulbecco modified Eagle medium (Life Technologies) and 25% MCDB-105 (Sigma), supplemented with 10% charcoal-stripped fetal bovine serum (Gemini), 1 mM sodium pyruvate (Sigma), 1% antibiotic-antimycotic solution (Life Technologies), and 5 μg/ml insulin (Gemini). Primary eMSC and eSF clonal cultures were monitored for colony formation, and replicate cultures were harvested at early (2–3 wk, eMSCEarly, eSFEarly) and late (4–8 wk, eMSCLate, eSFLate) stages of colony development, according to individual culture colony-forming efficiency and growth. All colonies in individual primary clonal culture plates were pooled at the time of harvest. Cells were harvested with 0.25% trypsin and 1 mM ethylenediaminetetraacetic acid and cell aliquots frozen as dry pellets for RNA extraction or cryopreserved in culture medium containing 10% dimethylsulfoxide to be used for subsequent in vitro decidualization (see below).
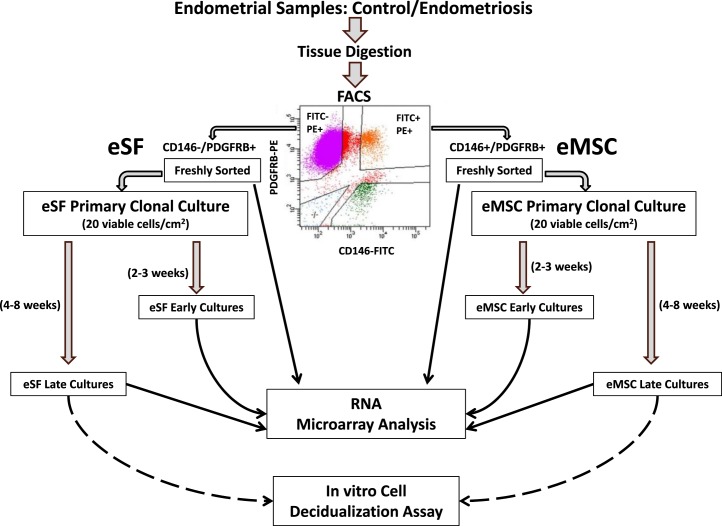
FIG. 1.
Experimental design. Pure populations of endometrial mesenchymal stem cells (eMSC) and endometrial stromal fibroblasts (eSF) were isolated by fluorescence-activated cell sorting (FACS) after enzymatic digestion of endometrial tissue. The four-color FACS excluded leukocytes and epithelial cells, sorting the remaining cells according to the binding of fluorescein isothiocyanate (FITC)-labeled antibody to cluster of differentiation 146 (CD146) and of phycoerythrin (PE)-labeled antibody to platelet-derived growth factor receptor beta (PDGFRB), isolating two populations: FITC+/PE+ eMSC co-expressing CD146 and PDGFR, and FITC−/PE+ eSF that express PDGFRB but not CD146. An aliquot of each freshly sorted population was processed for RNA extraction and microarrray analysis. When FACS yields were sufficient, the remainder of the sorted eMSC and eSF were established in primary culture at clonal density (10–20 viable cells/cm2). Replicates of primary eMSC and eSF clonal cultures were harvested at early (2–3 wk) and late (4–8 wk) stages of colony development and processed for RNA extraction and microarrray analysis. Cells harvested from late cultures were also used for in vitro decidualization assays.
RNA Isolation and cDNA Preparation for Microarray Analysis
Total cellular RNA was isolated using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems, Life Technologies) and subjected to DNase treatment using the RNase-Free DNase Set (Qiagen). Reverse transcription and amplification of isolated RNA into cDNA were performed using NuGEN Ovation V2 (NuGen). The integrity of the cDNA was assessed with the Agilent 2100 Bioanalyzer (Agilent Technologies), and individual samples meeting yield and quality standards were further processed and hybridized to Affymetrix Human Gene 1.0 ST arrays (Affymetrix), probing 36 079 genes. Arrays were scanned according to the protocol described in the WT Sense Target Labeling Assay Manual from Affymetrix (version 4, FS450_0007).
Microarray Gene Expression Data Analysis
Microarray data analysis was performed using GeneSpring 11.02 software (Agilent Technologies), essentially as previously described [4]. Briefly, intensity values of the probe sets (genes) were normalized and log to base 2 transformed using the robust multiarray analysis algorithm. Across-sample normalization was to the median of all samples. Differential expression analysis was conducted using one-way ANOVA with Benjamini-Hochberg multiple-testing correction for false discovery rate for a significance threshold of P < 0.05 and ≥1.5-fold change cutoff. The various experimental group comparisons used for differential expression analysis are summarized in Supplemental Table S2A. Unsupervised PCA algorithm was applied to all samples, using all 36 079 genes on the microarray, and HC analysis was conducted using only differentially expressed genes from all samples and among all experimental conditions. Raw data files have been uploaded to the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE73622. Biofunctional pathway analysis was conducted using Ingenuity Pathway Analysis (Qiagen), which identifies the activation states of biological pathways, networks, and cellular functions based on the differential gene expression analysis described above.
Validation of Microarray Data by Quantitative RT-PCR
Differentially expressed genes of select subsets of cell type and disease groups were validated by quantitative RT-PCR (Q-RT-PCR). A total of 28 cDNA samples from FACS-isolated endometrial cell populations, including freshly sorted cells from control (eMSCFACS.control n = 3; eSFFACS.control n = 3) and endometriosis groups (eMSCFACS.endo n = 3; eSFFACS.endo n = 3) groups and corresponding late stage primary clonal cultures (eMSCLate.control n = 5; eMSCLate.endo n = 3; eSFLate.control n = 5; eSFLate.endo n = 3) were assayed in duplicate by Q-RT-PCR using the Fluidigm (96.96 or 48.48) Dynamic Array Integrated Fluidic Circuits and the BioMark HD system (www.fluidigm.com/biomark-system.html) as previously described [4, 8]. Briefly, cDNA was preamplified to generate a pool of target genes using Taq-Man Pre-Amp master mix (Applied Biosystems), 100 ng cDNA, and 500 nM for each primer pair. Samples were then treated with exonuclease (Exonuclease I; New England BioLabs). Using previously generated optimal dilution curves, samples were diluted 1:5 in a Tris-ethylenediaminetetraacetic acid dilution buffer (TEKnova). Q-RT-PCR was performed using SsoFast Evagreen supermix with low ROX binding dye (Biotium Inc.) and a primer concentration of 5 μM. Data were processed by user-detected threshold settings and linear baseline correction using Biomark real-time PCR Analysis Software (version 3.0.4). Melt curves were assessed using the melting temperature threshold. The comparative cycle threshold (Ct) method was used as described [8] to obtain relative expression for each group comparison. Expression was normalized to an internal calibrator for cultured and sorted cells (ΔCt), then to the normalized controls (ΔΔCt). The ΔΔCt values were expressed as log 2 (2−ΔΔCt), which were used to calculate relative fold changes (docs.appliedbiosystems.com/pebiodocs/04303859.pdf).
Decidualization In Vitro
Cells from late primary cultures of subject-paired eMSC and eSF from three control and two endometriosis subjects were used to assess in vitro decidualization. Cryopreserved cells from eMSC- and eSF-derived cultures (see above) were thawed, replated at 10–20 × 104 viable cells/cm2, and grown in serum-containing culture medium as described for primary cultures. Confluent replicate cultures were treated with 10 nM E2 plus 1 μM P4 (E2P4) or ethanol vehicle for 14 days in serum-free medium supplemented with epidermal growth factor, bovine serum albumin, ascorbic acid, and transferrin [29]. Decidualization was assessed by determining concentrations of the decidual biomarker IGFBP1 in conditioned media by enzyme-linked immunosorbent assay (ELISA) using kits from Alpha Diagnostic according to the manufacturer's instructions. All samples were assayed in duplicate, and a standard curve was run for each assay. Inter- and intra-assay coefficients of variation were 5.0%–7.4% and 2.4%–3.4%, respectively.
Statistical Analyses
Differences in relative expression by Q-RT-PCR were analyzed for each pairwise comparison using the t-test for equal/unequal variance as appropriate. Equality of variances was tested utilizing the F-test (Microsoft Excel). The Dixon Q-test was used to remove outliers [30]. Correlation between microarray and Q-RT-PCR data was evaluated with nonparametric Spearman and Kendall rank correlation, where positive rho and tau coefficients indicate agreement between microarray and Q-RT-PCR. P values were based on a two-tailed null hypothesis of no association [31]. ELISA data were analyzed by ANOVA and Sheffe post hoc test using StatView 5.0.1 (SAS Institute Inc.).
Ingenuity Pathways Analysis
Gene ontology and functional annotations were evaluated for the various experimental group comparisons from differential expression analysis of microarray data. RefSeq identifications and fold changes of differentially expressed genes in each comparison were imported into the Core Analysis function of Ingenuity Pathway Analysis. Inhibition or activation of pathways was predicted for functional groups of genes based on collective mRNA expression levels, and significance was determined using the right-tailed Fisher exact test. P values reflected the number of analysis-specific genes in a given pathway compared with the total number of occurrences of these genes in all pathways in the Ingenuity Knowledge Base. Results are shown for pathways with a bias-corrected Z score ≤ −2.0 or bias-corrected Z score ≥ 2.0 with P < 0.05 for inhibited or activated pathways, respectively.
RESULTS
A meaningful assessment of the potential of eMSC to give rise to progeny differentiated to the eSF lineage requires a clear definition of the corresponding phenotypes of these two closely related cell types, particularly in view of recent awareness of the similarities between mesenchymal stem cells and fibroblasts isolated from diverse human tissues [32–34], although studies are often confounded by unaccounted changes during ex vivo expansion in culture. In human endometrium, the distinct in vivo phenotypes of purified eMSC and eSF populations are well established [4], but the in vitro eSF phenotype vis-a-vis the eMSC is not clearly defined. Because this gap in knowledge is a major drawback for ex vivo studies on derivation of eSF lineage-differentiated cells from progenitors, it is critical to define, initially, the normal in vitro eSF phenotype compared to the corresponding in vivo eSF, serving as the cornerstone to address the fundamental questions of eMSC as eSF progenitor and the origin of the abnormal eSF phenotype in endometriosis. Thus, the experimental design of the current study (Fig. 1) involved the transcriptional profiling of freshly isolated eMSC and eSF obtained by FACS from subjects without or with endometriosis, obtaining short- and long-term primary clonal cultures from these cell populations (Figs. 2–5 and Tables 1–13), as well as assessing cultured cell responses to P4 (Fig. 6)—pathognomonic of the eSF.
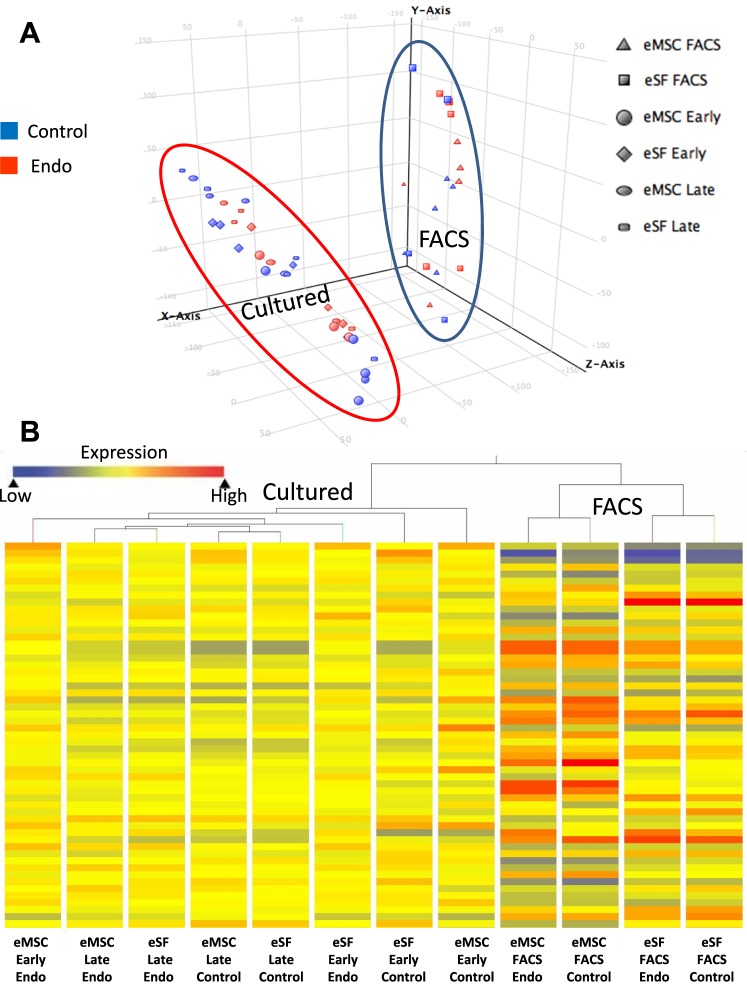
FIG. 2.
Clustering analyses. A) Principal component analysis (PCA) plot shows distribution of individual samples within the three-dimensional space defined by the three principal components accounting for the highest variances among probeset intensity values. In this two-dimensional representation of a three-dimensional image, the size of a symbol provides the perspective view of depth for its position, with smaller size symbols being closer to the origin and vice versa. The perspective shows two major clusters, one encircled by a blue line consisting of eMSC (triangles) and eSF (squares) analyzed immediately after FACS isolation; another cluster encircled by a red line corresponding to all cultured cells, including early eMSC (circles), early eSF (diamonds), late eMSC (ovals), and late eSF (rectangles), with blue symbols corresponding to control and red symbols to endo samples. Further subclustering by cell type is not clearly discernible in this particular perspective, but can be appreciated in a different orientation of the plot within the FACS cluster (not shown). B) Hierarchical clustering (HC) dendrogram, showing heat map bars representing average expression values in each experimental group indicated by the color spectrum shown from blue (low expression) to red (high expression). First branching segregates freshly FACS-isolated cells (right branch) and cultured cells (left branch), with further subbranching of FACS-isolated cells according to cell type (eMSC, eSF) and then by disease category (endo, control). Subbranching within cultured cells segregates successively first eMSC early control and second eSF early control from other groups, then the third and fourth branches separate eMSC early endo and eSF early endo, respectively, from all late cultures that cluster singularly first by disease (endo, control) and then by cell lineage (eMSC, eSF). Control, no endometriosis; Endo, endometriosis.
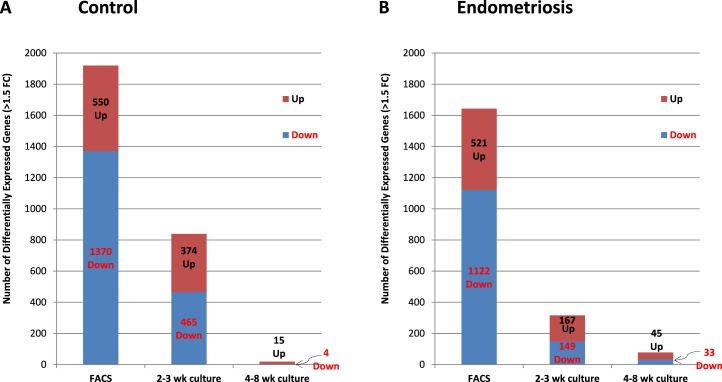
FIG. 3.
Number of differentially expressed genes in FACS-isolated and cultured eMSC versus eSF. Data represent average values for the number of differentially expressed genes (P < 0.05, >1.5-fold change) in each experimental group for eMSC versus eSF in freshly FACS-isolated samples (FACS) and in early (2–3 wk culture) and late (4–8 wk culture) primary clonal cultures from women without (A, control), or with endometriosis (B, endometriosis). Stacked bars show breakdown of the total number of differentially expressed genes into up-regulated (brown stack, black characters) and down-regulated (blue stack, red characters). Data show a progressive reduction in the number of eMSC versus eSF differentially expressed genes over time in culture in both control (A) and endometriosis (B).
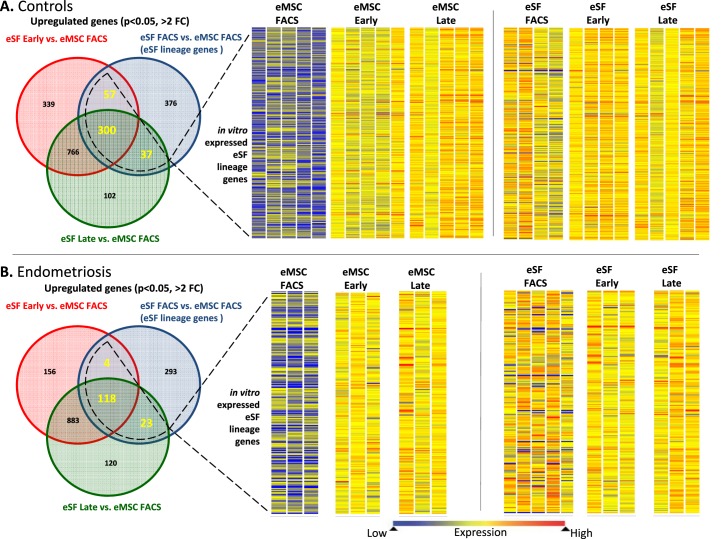
FIG. 4.
Expression of eSF lineage-associated genes in FACS-isolated and cultured eMSC and eSF. A) Controls. B) Endometriosis. Left panels show corresponding Venn diagrams of up-regulated genes (P < 0.05, >2-fold change) in eSF versus eMSC immediately after FACS isolation (eSF FACS vs. eMSC FACS, blue circles) representing eSF lineage genes in vivo and in eSF early (red circles) and eSF late (green circles) cultures versus eMSC FACS. Overlapping parts of the circles represent common up-regulated genes among the respective groups, with the corresponding number of common genes highlighted. The overall number of eSF lineage genes expressed in cultured eSF (surrounded by dotted line) represents the in vitro expressed eSF lineage genes whose expression levels in eMSC and eSF are shown as heat maps in the center and right panels, respectively. Heat map bars represent expression of in vitro expressed eSF lineage genes in individual samples from eMSC (center panel) or eSF (right panel) immediately after FACS isolation (eMSC FACS, eSF FACS) or from early (eMSC early, eSF early) or late (eMSC late, eSF late) cultures. Expression values are indicated by the color spectrum shown from blue (low expression) to red (high expression). Data shown in both controls (A) and endometriosis (B) eSF lineage gene expression by eMSC is low in freshly FACS-isolated cells (eMSC FACS) but increases in early (eMSC early) and late (eMSC late) cultures, while in eSF, the expression levels of freshly FACS-isolated cells (eSF FACS) are maintained in early (eSF early) and late (eSF late) cultures. Uneven numbers of samples per group in the eSF datasets (right panels) are the result of sample dropout due to failed quality control at either prehybridization (low/poor quality RNA amplification) or posthybridization (array quality control) or missing samples due to insufficient FACS yields for setting up primary cultures after allocating for analysis of freshly isolated cells.
FIG. 5.
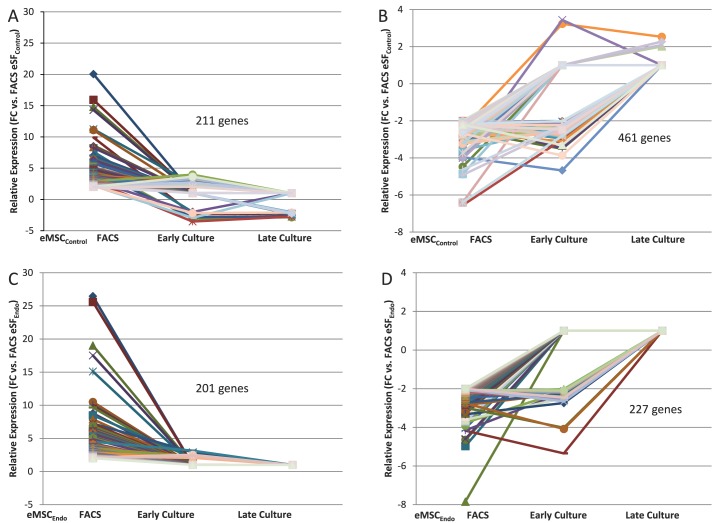
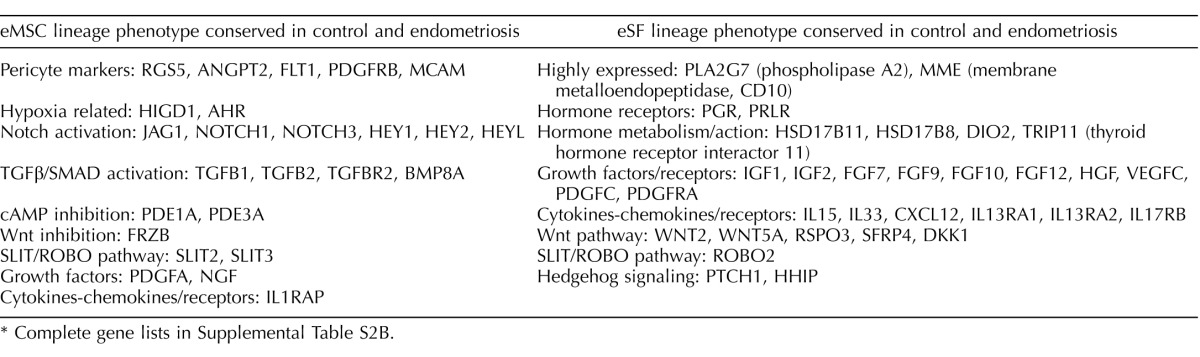
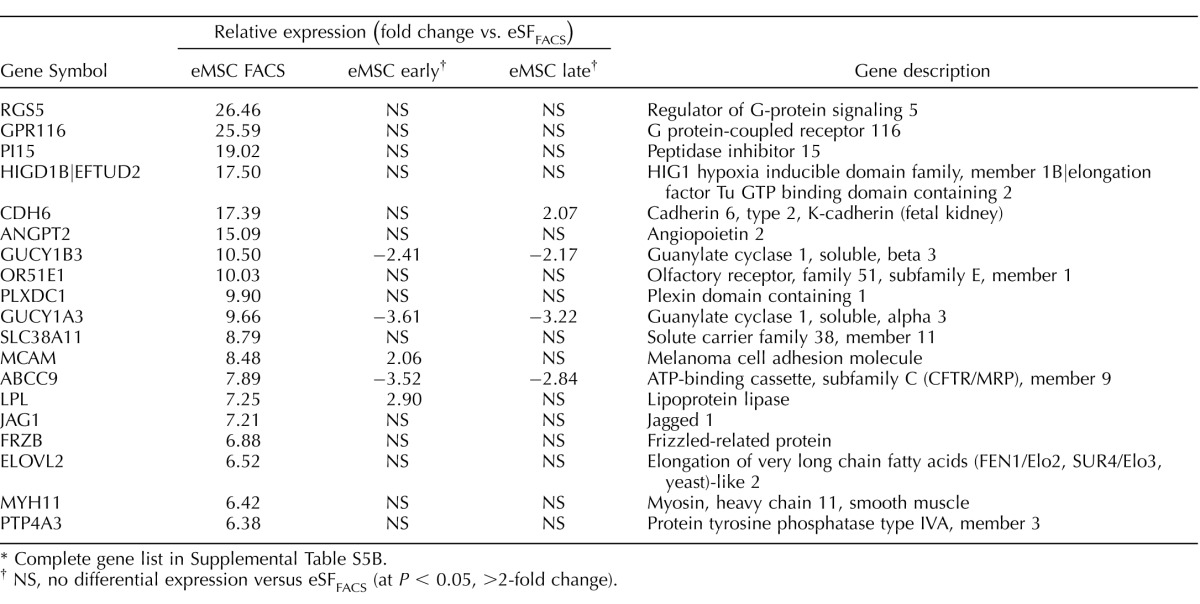
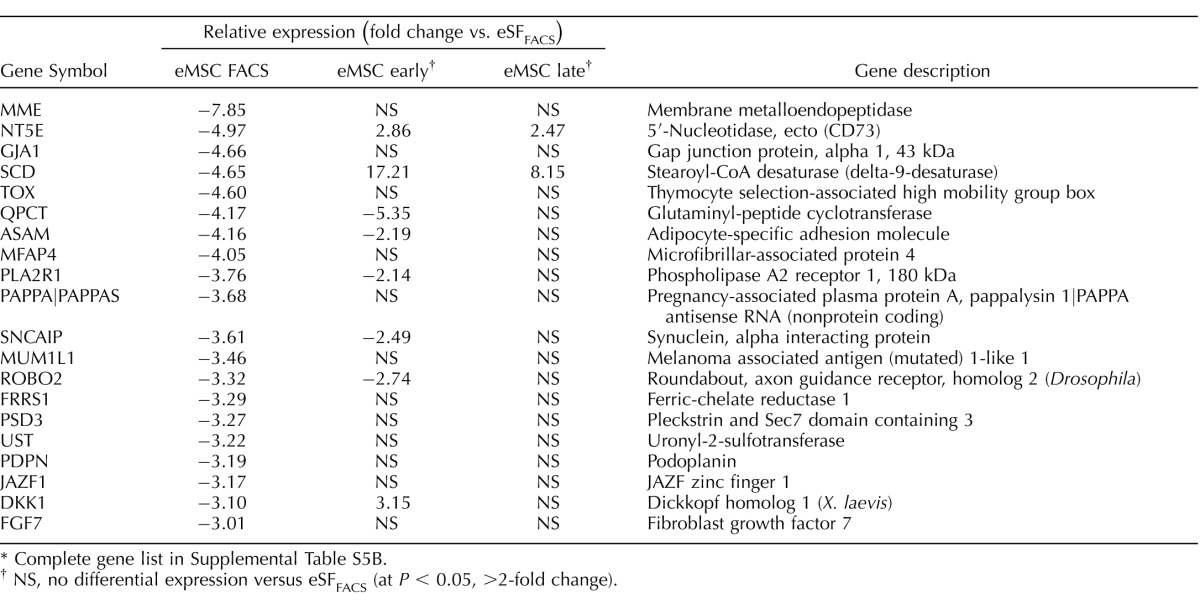
Expression of eMSC and eSF lineage-associated genes in FACS-isolated and cultured eMSC. Data correspond to differential expression analysis of the entire dataset consisting of n = 48 samples. Symbols/colors do not represent the same genes in different plots. Values represent the average relative expression (P < 0.05, >2-fold change) compared to freshly FACS-isolated eSF (eSF FACS) for each gene in the individual groups of eMSC from controls (A, B) or endometriosis (C, D): freshly FACS-isolated (eMSC FACS), early culture, and late culture. Genes up-regulated in eMSC FACS versus eSF FACS (A: control, 211 genes; C: endometriosis, 201 genes) correspond to in vivo eMSC lineage genes and are down-regulated in eMSC early and late cultures. Genes down-regulated in eMSC FACS versus eSF FACS (B: control, 461 genes; D: endometriosis, 227 genes) correspond to in vivo eSF lineage genes and are up-regulated in eMSC early and late cultures.
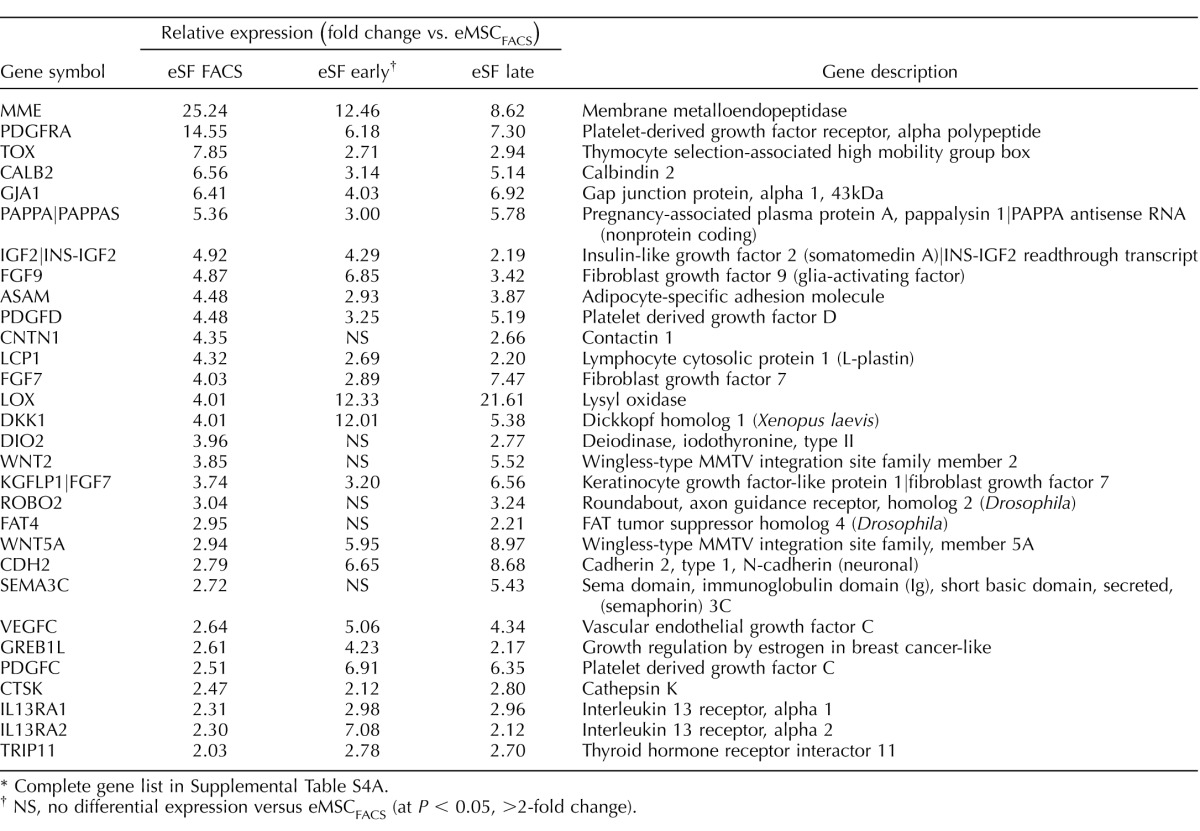
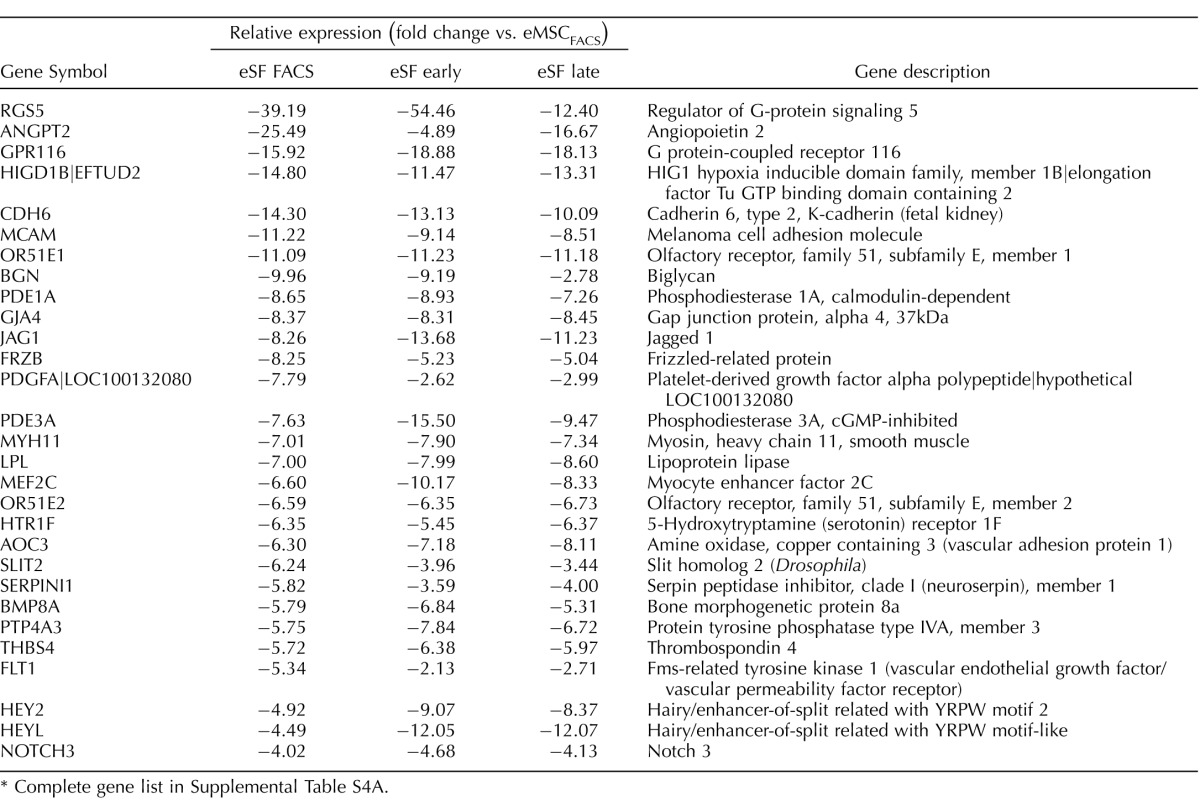
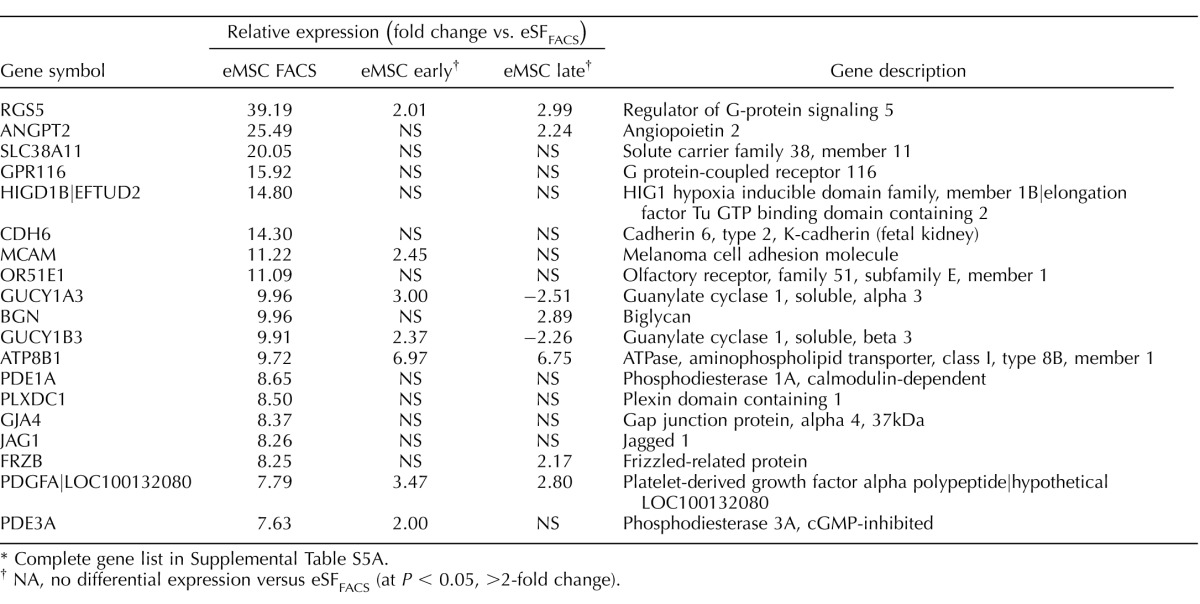
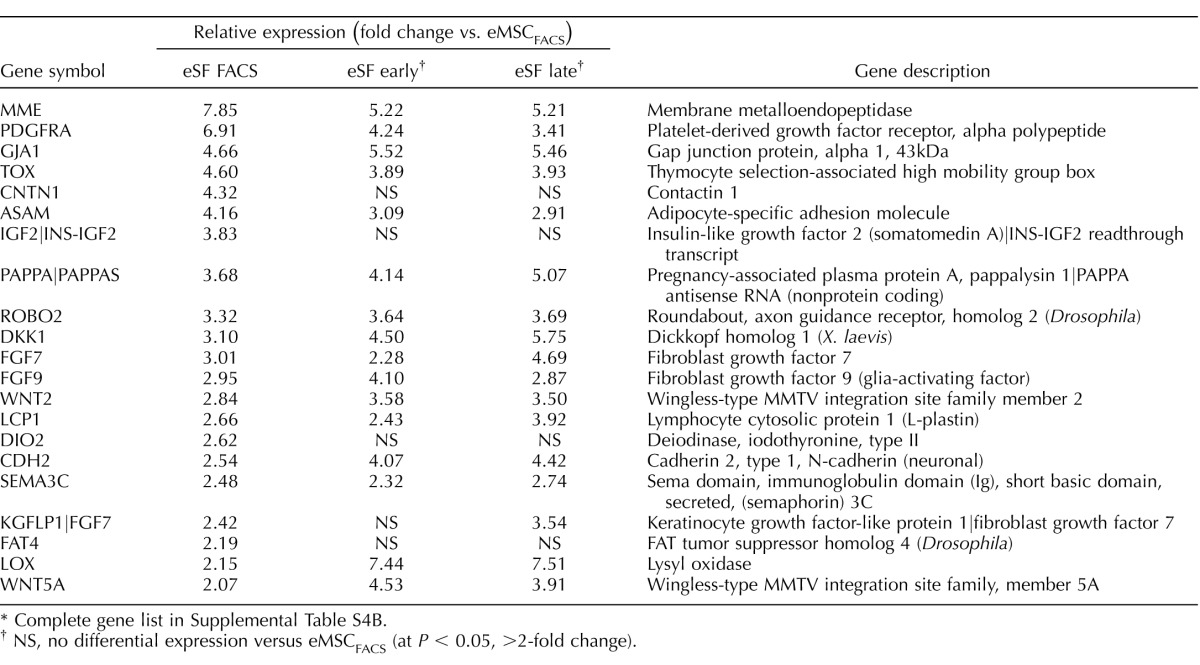
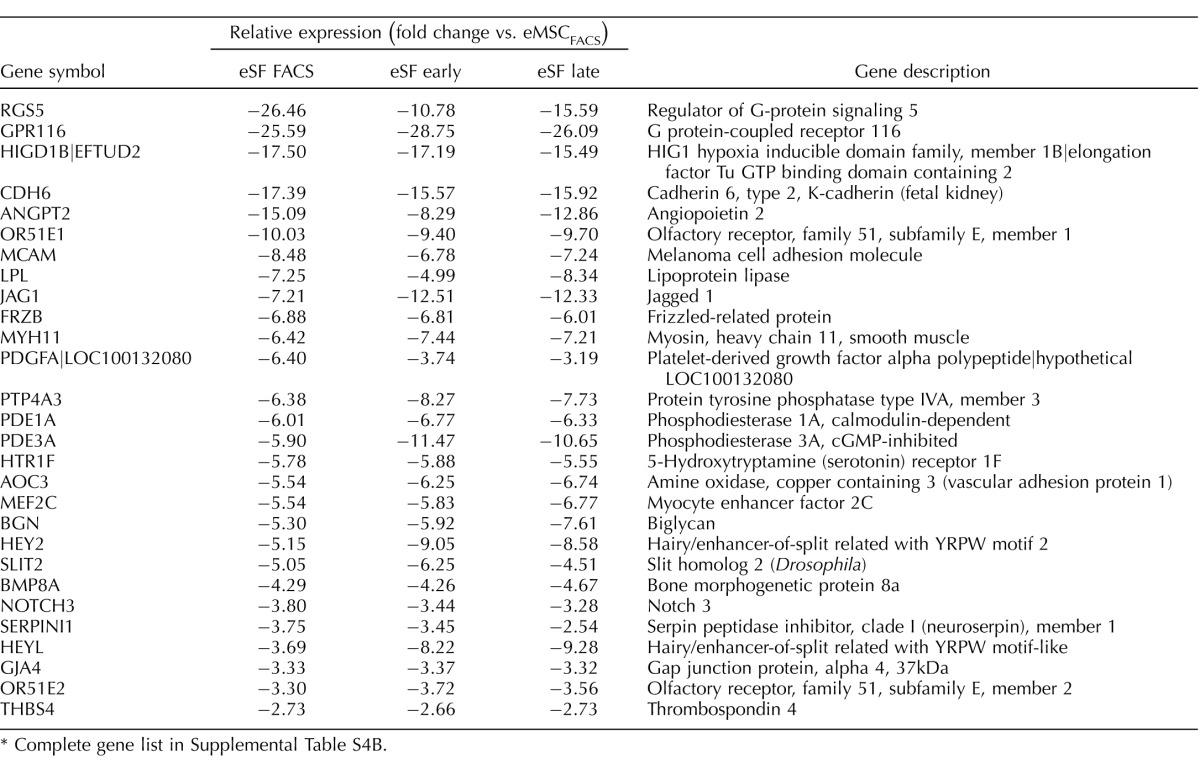
TABLE 1.
Highly conserved in vivo eMSC and eSF lineage-associated molecular phenotypes.*
Complete gene lists in Supplemental Table S2B.
FIG. 6.
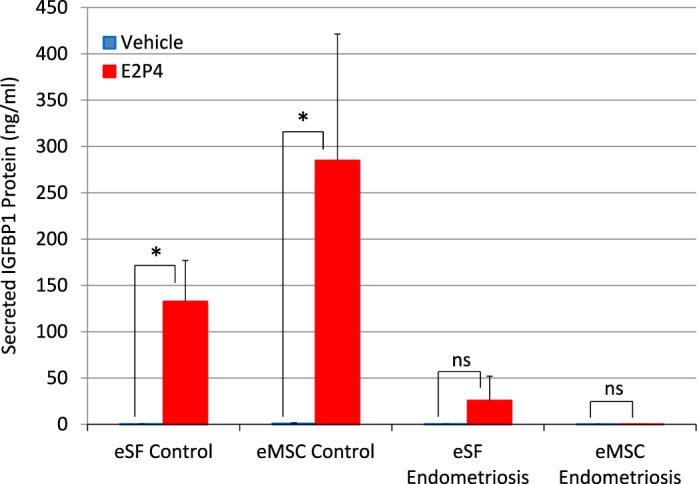
Impaired in vitro decidualization of eSF- and eMSC-derived cultures from women with endometriosis. Cells from late primary cultures of subject-paired eMSC and eSF from three control and two endometriosis subjects were subcultured and grown to confluence. Confluent replicate cultures were treated with estradiol plus progesterone (E2P4) or ethanol (vehicle) for 14 days. Decidualization was assessed measuring concentrations of the decidual biomarker insulin-like growth factor binding protein-1 (IGFBP-1) in conditioned media by ELISA. Values represent the mean ± SD of IGFBP1 concentration in conditioned media of eSF cultures from control (eSF control, n = 15) or endometriosis (eSF endometriosis, n = 6) treated with vehicle (blue bars) or E2P4 (red bars), and of subject-paired eMSC cultures from control (eMSC control, n = 15) or endometriosis (eMSC endometriosis, n = 6) treated with vehicle (blue bars) or E2P4 (red bars). Results of significance testing of the differences between E2P4-treated (red bars) and the corresponding vehicle-treated cultures (blue bars) are indicated: *P < 0.05 compared to vehicle; ns, not significant (P > 0.05) compared to vehicle. Mean IGFBP1 concentrations in all vehicle-treated groups (blue bars) and in the E2P4-treated endometriosis eMSC group (eMSC endometriosis, red bar) were <1 ng/ml. IGFBP1 concentrations in the E2P4-treated endometriosis eSF group (eSF endometriosis, red bar) were low and highly variable (25 ± 26 ng/ml), as frequently seen in P4-resistant eSF.
Gene Expression Profiling
Global gene expression differences among the various cell types isolated from endometrium of subjects without or with endometriosis by FACS and after short- and long-term primary cultures were evaluated after microarray analysis (Fig. 1). Differential gene expression comparisons among the different endometrial cell types and conditions are shown in Supplemental Table S2A (validation of select genes in Supplemental Table S3) and the corresponding differentially expressed genes (>1.5-fold change, P < 0.05) are shown in toto in Supplemental Tables S2B, S4, and S5.
The Transcriptome Segregates Samples by Culture Condition, Cell Type, and Disease Category
Microarray analysis revealed major differences in the transcriptomes of freshly isolated tissue-derived cells compared to their cultured counterparts (Supplemental Table S2B). PCA (Fig. 2A) and HC (Fig. 2B) of the data showed that groups clustered primarily by whether they were analyzed immediately after FACS isolation from the tissue or after being grown in primary culture (Fig. 2). Accordingly, the first branching in the HC dendrogram (Fig. 2B) segregated freshly isolated tissue-derived cells (right branch) from cultured cells (left branch). Further branching within tissue-derived cells was according to cell type (eMSC, eSF) and then by disease category (endometriosis, no endometriosis [controls]), reflecting strongly conserved gene expression by cell lineage followed by subclustering within lineages by the effect of disease. In contrast, subclustering within culture-derived cells was more complex and indicative of a progressive change in phenotype of one or both cell types and/or widening differences between control and endometriosis cells in culture. The first two branches segregated successively the nonendometriosis early cultures from the other groups, and the third and fourth branches separated endometriosis early cultures from all late cultures. Late cultures clustered singularly first by disease (endometriosis, no endometriosis [controls]) and then by cell lineage (eMSC, eSF), suggesting a convergence of cell phenotypes in the late cultures and/or magnification of the disease effect.
Lineage-Associated Gene Expression In Vivo and In Vitro: Controls
Lineage-associated genes in vivo.
Genes differentially expressed in FACS-isolated, uncultured eMSC versus eSF from women without endometriosis (eMSCFACS.control, eSFFACS.control) defined the distinct phenotypes of these cell lineages. In the comparison of eMSCFACS.control versus eSFFACS.control (Fig. 3), there were 550 genes up-regulated >1.5-fold (P < 0.05) and 1370 genes down-regulated >1.5-fold (P < 0.05) (i.e., the latter being 1370 up-regulated in eSFFACS.control vs. eMSCFACS.control). Some of the most highly up-regulated genes are shown in Table 1, and the complete gene lists are found in Supplemental Table S2B. Expression of these genes is characteristic of eMSC from normal controls and also from endometriosis (see below). This highly conserved lineage-associated molecular phenotype that identifies eMSC is characterized by up-regulation of pericyte markers, hypoxia-related genes, and genes involved in Notch activation, Wnt inhibition, SLIT ligands, and growth factor-signaling pathways, as previously reported [4]. New in the current study are eSF lineage-associated genes that confer a highly conserved molecular phenotype of this cell type in vivo in normal controls and also in endometriosis (see below) (Table 1, select genes), including up-regulation of phospholipase A2 (PLA2S)and CD10 (MME, membrane metalloendopeptidase) as well as genes involving hormone receptors and hormone metabolism, growth factors, cytokines and chemokines and their receptors, Wnt ligands and inhibitors, ROBO receptors, and Hedgehog signaling. Herein, these lineage-specific genes served to guide assessment of lineage fidelity of eMSC and eSF in vitro. Select eMSC and eSF lineage-associated genes were validated by Q-RT-PCR, which demonstrated high and statistically significant positive associations (Supplemental Table S3, A and B).
The eSF demonstrate lineage-phenotype stability in vitro.
Because a goal of this study was to determine if eMSC differentiate to eSF, the question arose regarding in vivo (i.e., FACS isolated fresh from tissue) eSF lineage gene fidelity in short- and long-term primary culture, considering the potential confounding effect of cellular adaptation to the ex vivo culture environment. Predictably, major changes in gene expression occurred primarily during the in vivo to ex vivo transition from uncultured cells (FACS isolated) to early primary culture (Supplemental Table S2B). In the case of eSFcontrol, 1146 genes were down-regulated > 1.5-fold (P < 0.05) in early cultures compared to uncultured (FACS isolated) cells. More than half (581) of these corresponded to genes down-regulated in common by all early cultures regardless of cell type (eMSC/eSF) or disease (endometriosis/control), and thus represented nonspecific adaptational changes. Notwithstanding adaptation to the ex vivo culture environment, eSFcontrol-derived late cultures stably expressed a substantial subset (337, 44%) of the lineage genes defining the in vivo eSF phenotype (i.e., highly expressed by eSFcontrol vs. eMSCcontrol in vivo), and these provided an in vitro signature for the eSF lineage phenotype (Fig. 4A, left panel [Venn diagram], Table 2, and Supplemental Tables S4A and S3C, Q-RT-PCR validation). Thus, eSF demonstrated lineage phenotypic stability in vitro and notably did not increase expression of the majority of eMSC lineage genes. Specifically, 77% of lineage genes down-regulated in eSFcontrol versus eMSCcontrol in vivo were also down-regulated in cultured eSFcontrol (Table 3 and Supplemental Table S4A) despite up-regulating 1417 genes during the in vivo to ex vivo transition from uncultured cells (FACS isolated) to early primary culture.
TABLE 2.
Expression of eSF lineage-associated genes in FACS-isolated control eSF and derived early and late cultures.*
Complete gene list in Supplemental Table S4A.
NS, no differential expression versus eMSCFACS (at P < 0.05, >2-fold change).
TABLE 3.
Expression of eMSC lineage-associated genes in FACS-isolated control eSF and derived early and late cultures.*
Complete gene list in Supplemental Table S4A.
The eMSC differentiate in vitro to eSF-like cells.
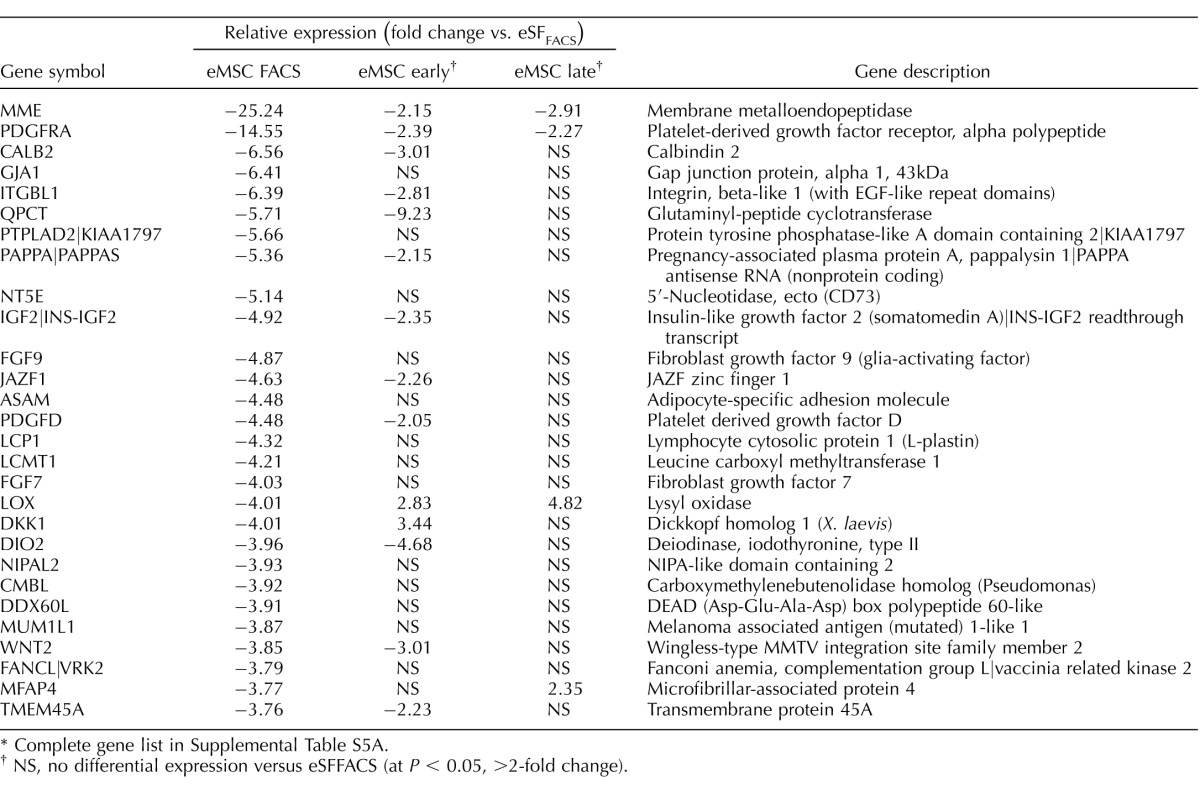
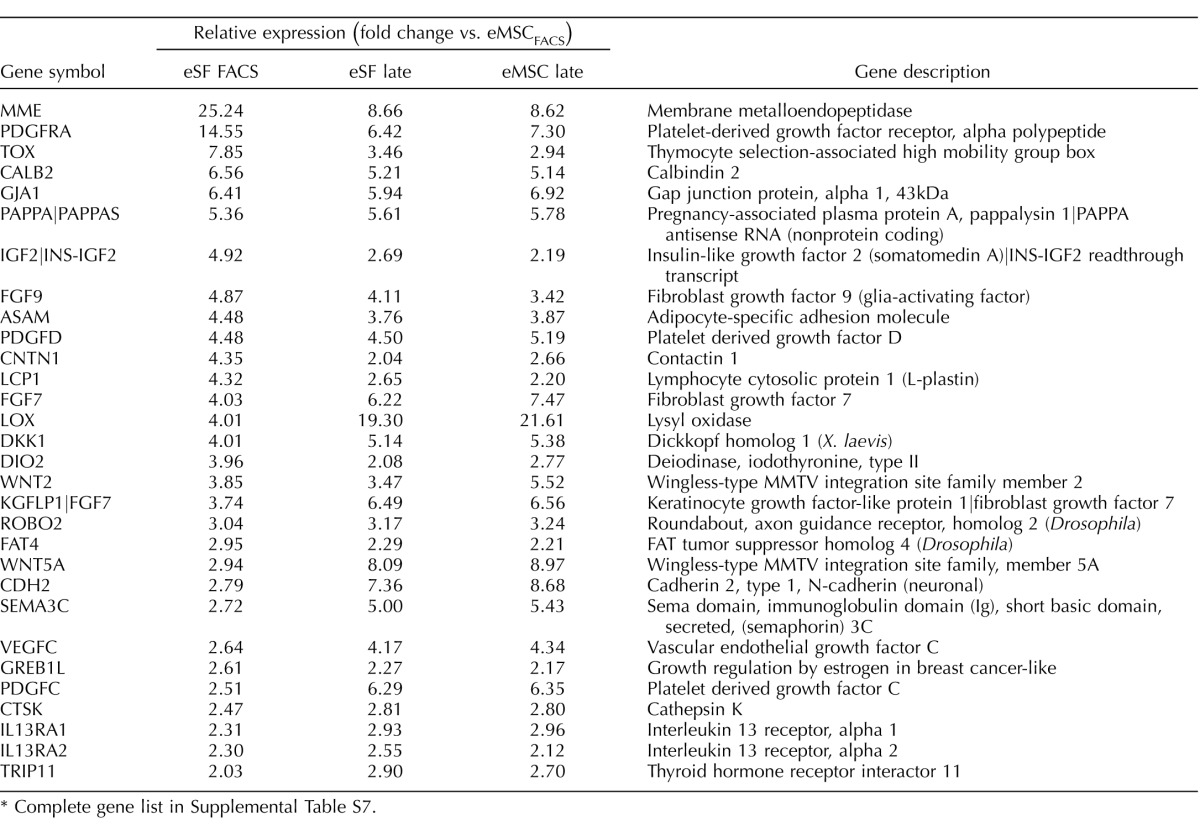
In contrast to the in vitro phenotypic stability observed in eSFcontrol, cultured eMSCcontrol displayed marked changes in expression of lineage genes (Figs. 4A, center panel, and 5, A and B, Tables 2 and 3, and Supplemental Table S5A). There was down-regulation of 211 eMSC lineage genes (Fig. 5A and Table 4) and up-regulation of 461 eSF lineage genes (Fig. 5B and Table 5) (complete gene list in Supplemental Table S5A). In culture, 81% of eMSC lineage-associated genes were down-regulated, suggesting that eMSC lose in vitro most of their stemness regarding lineage genes. Importantly, they concomitantly up-regulated 55% of eSF lineage genes (Figs. 4A and 5B, Tables 4 and 5, and Supplemental Tables S4A and S3D, Q-RT-PCR validation). Thus, changes in expression of lineage-associated genes in eMSC-derived cultures suggested a transition to an eSF-like phenotype. Consistent with this finding was a progressive reduction in the number of cell lineage differentially expressed genes (eMSCcontrol vs. eSFcontrol) through early and late cultures, resulting in minimal transcriptome differences (19 genes differentially expressed ranging from −1.64-fold change to +1.75-fold change, P < 0.05) in late cultures (4–8 wk) of eMSCcontrol versus eSFcontrol (Supplemental Table S2B and Figs. 3A and 4A). In addition, late cultures of eMSCcontrol displayed polygonal cell morphology typical of postconfluent eSF, consistent with eMSC differentiation to the eSF lineage (data not shown). Table 6 (Supplemental Table S7, full list) shows select co-expressed eSF lineage genes in eSFcontrol- and eMSCcontrol-derived late cultures, demonstrating that eSF and eMSC converge in vitro to an eSF molecular phenotype. Supplemental Table S3D shows high concordance of genes validated by Q-RT-PCR.
TABLE 4.
Expression of eMSC lineage-associated genes in FACS-isolated control eMSC and derived early and late cultures.*
Complete gene list in Supplemental Table S5A.
NA, no differential expression versus eSFFACS (at P < 0.05, >2-fold change).
TABLE 5.
Expression of eSF lineage-associated genes in FACS-isolated control eMSC and derived early and late cultures.*
Complete gene list in Supplemental Table S5A.
NS, no differential expression versus eSFFACS (at P < 0.05, >2-fold change).
TABLE 6.
Select co-expressed eSF lineage genes in control eSF- and eMSC-derived late cultures.*
Complete gene list in Supplemental Table S7.
Lineage-Associated Gene Expression In Vivo and In Vitro: Endometriosis
Lineage-associated genes and cellular functions in vivo.
While the in vivo eMSC and eSF molecular phenotypes were largely conserved in endometriosis (>95% of the 200 most highly expressed eMSC and eSF lineage genes; Table 1, select genes), overall there were fewer differentially expressed genes (521 up-regulated >1.5-fold change, P < 0.05) in eMSC isolated by FACS from women with endometriosis (eMSCFACS.endo) versus eSFFACS.endo compared to 550 genes in control eMSCFACS.control versus eSFFACS.control (Fig. 3B, Supplemental Table S2B, full list, and Table 1, select lineage-associated genes). Similarly, there were fewer differentially expressed genes in eSFFACS.endo versus eMSCFACS.endo (1122 genes up-regulated >1.5-fold change, P < 0.05) from women with disease, compared to 1370 genes in control eSFFACS.control versus eMSCFACS.control (Fig. 3B, Table 1, and Supplemental Table S2B). These data suggest that in endometriosis the endometrial mesenchymal lineage cells retain their molecular barcode phenotype in vivo, albeit with somewhat less fidelity than their counterparts in endometrium from women without disease.
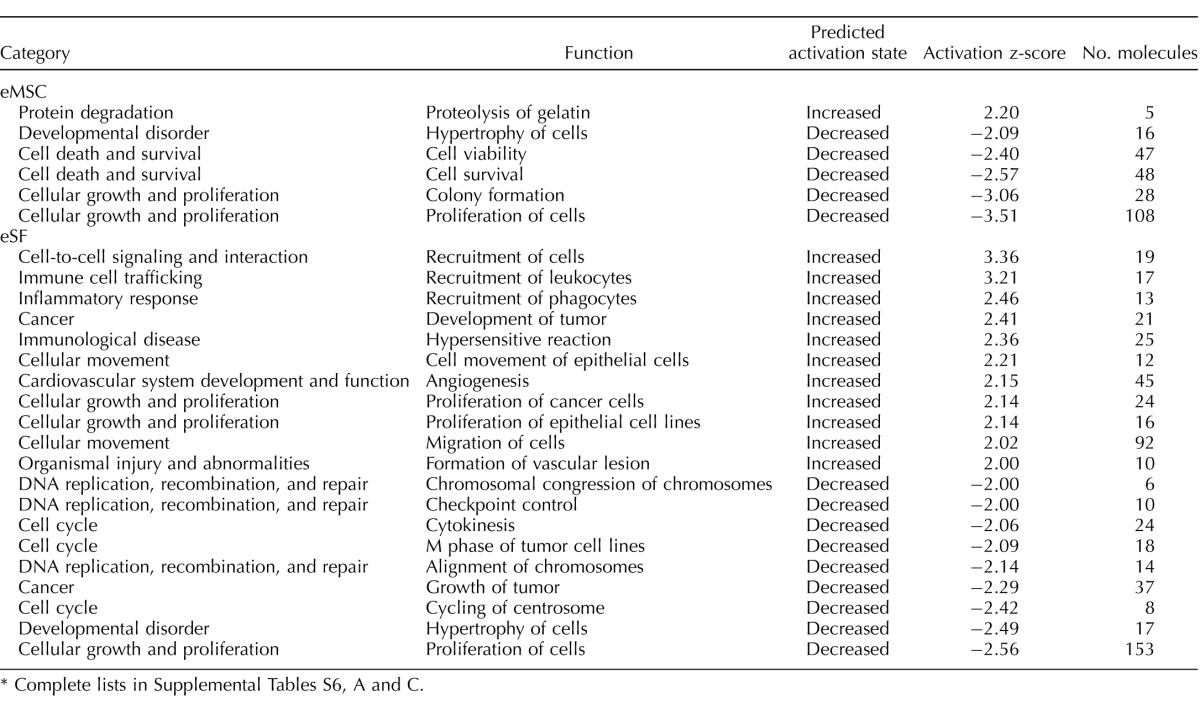
Head-to-head comparisons of FACS-isolated eMSC and eSF from women with versus without endometriosis were conducted (Supplemental Table S2B). In eMSCFACS.endo versus eMSCFACS.control, there were 188 genes up-regulated and 132 genes down-regulated. Of note are up-regulation of numerous SNORDs, INHBA, and 61 eSF lineage genes including DIO2, the chaperone HSPA6, PDGFRA, MME, PDGFC, LOX, and WNT5A. Among down-regulated genes were 22 eMSC lineage genes including GJD4, SLC38A11, THBS4, OR51E2, and FLT1. Pathway analysis revealed altered eMSC cellular functions (Table 7 and Supplemental Table S6A), with activation of protein degradation and inhibition of cell viability, colony formation, and proliferation.
TABLE 7.
Pathway analysis: altered cellular functions in endometriosis versus control in vivo.*
Complete lists in Supplemental Tables S6, A and C.
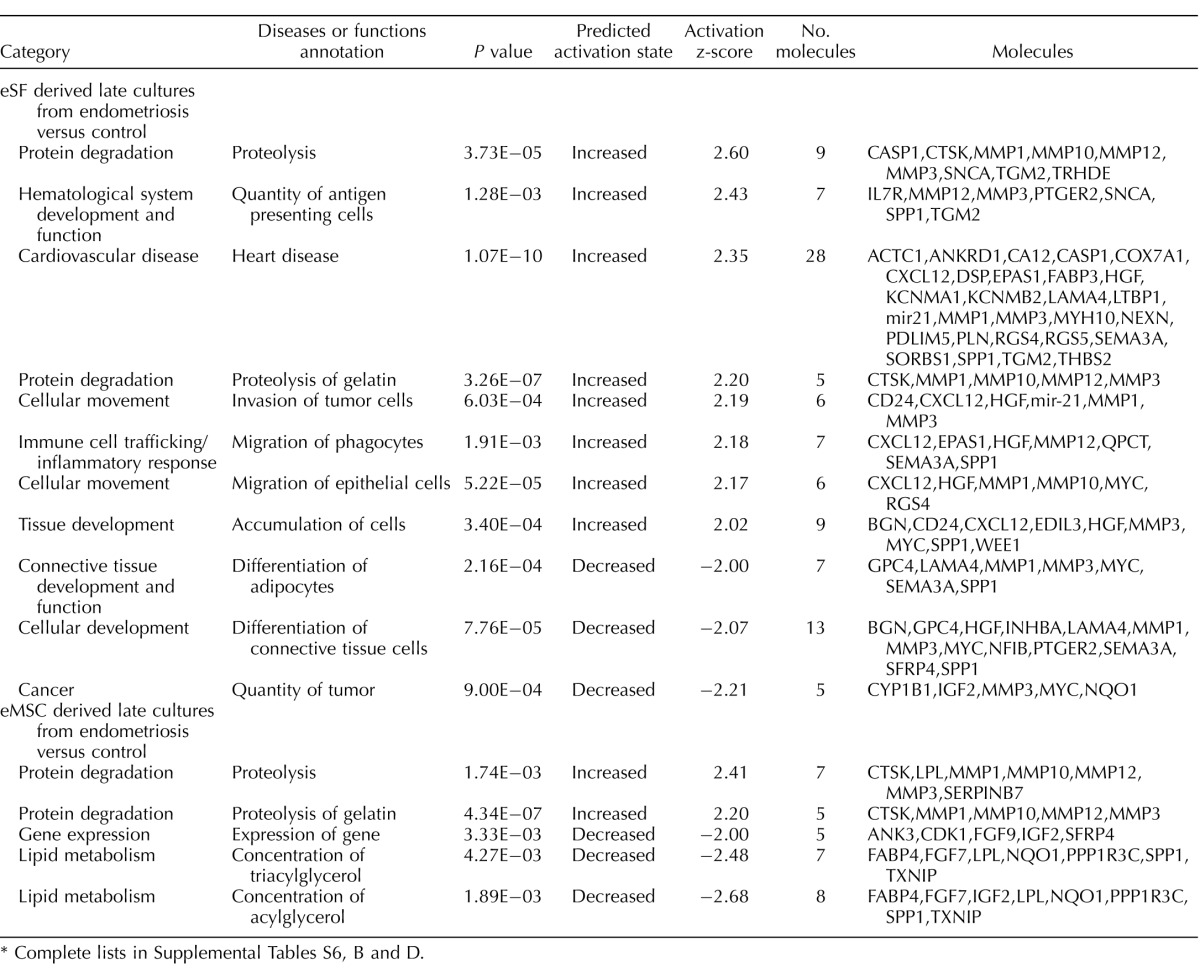
In eSFFACS.endo versus eSFFACS.control, there were 170 up-regulated genes, 22 of them eMSC lineage genes, including HAS2, ANGPT2, BGN, FRZB, AHR, and TGFB2, as well as genes associated with inflammation (CXCL2, IL8, C3, NFKB1, matrix metalloproteinases). There were 243 genes down-regulated, including 62 eSF lineage genes (PLA2G7, TOX, OMD, LCP1, DKK1, FGF9, IL17RB, GREB1L) as well as several genes involved in cell cycle control and cytokinesis. Pathway analysis (Table 7 and Supplemental Table S6C) revealed increased recruitment of cells, including leukocytes and phagocytes, tumor development/proliferation of cancer cells and epithelial cells, hypersensitive reaction, epithelial cell movement and cell migration, angiogenesis, and formation of vascular lesions. Decreased cellular functions (Table 7 and Supplemental Table S6C) included cell proliferation, hypertrophy, tumor growth, chromosome alignment, cytokinesis, and checkpoint control and chromosomal congression in DNA replication, recombination, and repair.
The eSFendo Demonstrate Less Robust Lineage-Phenotypic Stability In Vitro
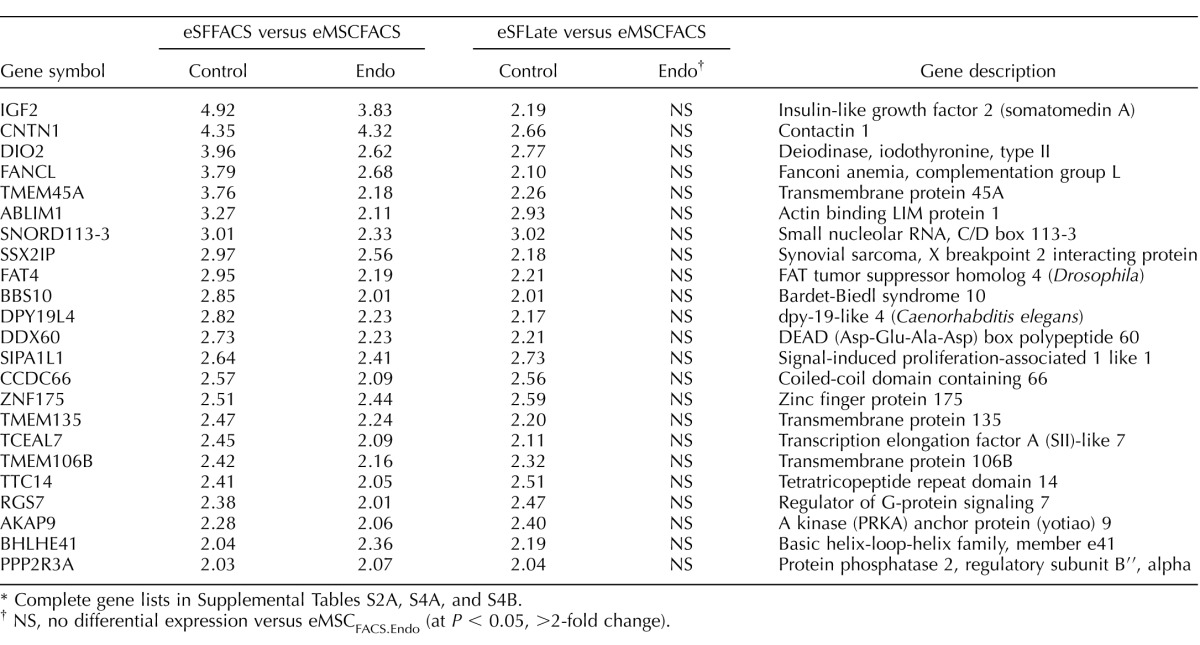
The eSFendo-derived cultures showed less robust eSF phenotypic stability compared to eSFcontrol-derived cultures (Fig. 4B, left panel). Specifically, eSFendo stably expressed a smaller subset (136, 33%) of eSF lineage genes compared to eSF lineage genes stably expressed in eSFcontrol (332, 44%) and maintained down-regulation of fewer (64%) eMSC lineage genes compared to eSFcontrol (77%) (Tables 8 and 9 and Supplemental Table S4B). In addition, eSFendo lost in vitro expression of some eSF lineage genes stably expressed by eSFcontrol (Table 10 and Supplemental Tables S2A and S4, A and B).
TABLE 8.
Expression of eSF lineage-associated genes in FACS-isolated endometrosis eSF and derived early and late cultures.*
Complete gene list in Supplemental Table S4B.
NS, no differential expression versus eMSCFACS (at P < 0.05, >2-fold change).
TABLE 9.
Expression of eMSC lineage-associated genes in FACS-isolated endometrosis eSF and derived early and late cultures.*
Complete gene list in Supplemental Table S4B.
The eMSCendo Differentiate In Vitro to eSF-Like Cells Expressing a Limited Subset of Lineage-Associated Genes in eMSCendo-Derived Cultures
The eMSC from women with endometriosis underwent major transcriptome changes during primary clonal culture (Figs. 3B, 4B, and 5, C and D), similar to those without disease (Figs. 3A, 4A, and 5, A and B). There was a progressive reduction in the overall number of differentially expressed genes in eMSCendo versus eSFendo in early and late cultures (Fig. 3B), while eSF lineage genes were up-regulated in cultured eMSC and stably expressed in cultured eSF (Fig. 4B); however, there was greater transcriptome differences in late eMSCendo cultures versus late eSFendo cultures (78 genes differentially expressed ranging from −2.73-fold change to +2.81-fold change, P < 0.05) compared with 19 genes differently expressed in late cultures from controls (Supplemental Table S2B). Moreover, while late cultures of eMSCendo demonstrated similar numbers of down-regulated eMSC lineage genes (201) compared to late culture eMSCcontrol (211), eMSCendo had notably fewer up-regulated eSF lineage genes (227) compared to eMSCcontrol (461) during in vitro differentiation (Tables 11, 12 [select genes]; Supplemental Table 5B [full gene list]). These observations suggest that eMSCLate.endo have impaired lineage differentiation compared with eMSCLate.control because the latter differentiate to eSF phenotype with more fidelity to both tissue-derived and late culture eSFcontrol.
The eSF- and eMSC-Derived Late Cultures from Endometriosis Versus Controls
In comparing eSFLate.endo to eSFLate.control, there were 60 genes up-regulated and 67 genes down-regulated (>1.5-fold change, P < 0.05) (Supplemental Table S2B). Pathway analysis (Table 13 and Supplemental Table S6D) revealed increased proteolysis, antigen presenting cells, cell invasion, migration of phagocytes and epithelial cells, and decreased differentiation of adipocytes and connective tissue and tumor size. Similarly, in eMSCLate.endo versus eMSCLate.control, there were 60 genes up-regulated and 78 genes down-regulated (>1.5-foldchange, Supplemental Table S2B). However, pathway analysis (Table 13 and Supplemental Table S6B) showed a more muted pathway activation profile in eMSC-derived late cultures from endometriosis versus control women, with only activation of pathways involved in protein degradation (especially matrix degradation) and decreased pathway activation involving genes expressed in lipid metabolism.
Decidualization In Vitro
The eSF uniquely differentiate to a decidual phenotype in response to P4 both in vivo and in vitro. This process, involving complex functional, biochemical, and morphological changes, with concomitant secretion of biomarkers, for example, IGFBP-1 [18], is compromised in women with endometriosis [24, 25]. To determine if late eMSC cultures that have acquired in vitro an eSF molecular phenotype respond to P4 as do eSF derived from the same tissue and cultured in parallel, cells from subject-matched eMSC- and eSF-derived late cultures from women without and with endometriosis were treated with P4 (and E2), and secreted IGFBP1 was measured in the conditioned media. The eMSCcontrol-derived cultures showed robust decidualization in response to E2P4, secreting equal or higher amounts of IGFBP1 (285 ± 35 ng/ml) as E2P4-treated eSFcontrol-derived cultures (132 ± 11 ng/ml) and significantly different (P < 0.0001) from vehicle-treated cultures (Fig. 6). As anticipated, eSFendo-derived cultures showed P4 resistance with inability to decidualize as evidenced by low/undetectable (25 ± 11 ng/ml) secreted IGFBP1 after 14 days of treatment with E2P4 that was not significantly different (p = 0.99) from vehicle-treated cultures. Remarkably, the in vitro differentiated progeny of eMSC → eSF from women with endometriosis also failed to decidualize in response to E2P4 with low/undetectable secreted IGFBP1 (0.12 ± 0.05 ng/ml) and not significantly different from vehicle-treated cultures (P > 0.99) or from the low/undetectable IGFBP1 secretion in endometriosis subject-paired eSF cultures (P = 0.96).
DISCUSSION
The eMSC: Progenitors of Stromal Fibroblasts
We have previously identified evidence supporting a common lineage of eMSC and eSF, based mainly on expression profiling and cluster analyses (PCA and HC) of differentially expressed genes [4, 8]. Herein, we present data that confirm that eMSC are bona fide progenitors of eSF, differentiating in vitro to cells that share similar eSF lineage-associated genes compared with eSF freshly isolated from endometrium and also eSF cultured in parallel under identical conditions. In addition these eMSC-derived eSF display hallmark features of human eSF, namely, characteristic culture morphology and the ability to respond to P4 with induction of decidualization markers (e.g., IGFBP1) and morphological changes [29]. Based on the findings of the current study, we propose the model of eMSC → eSF → decidualized eSF shown in Figure 7.
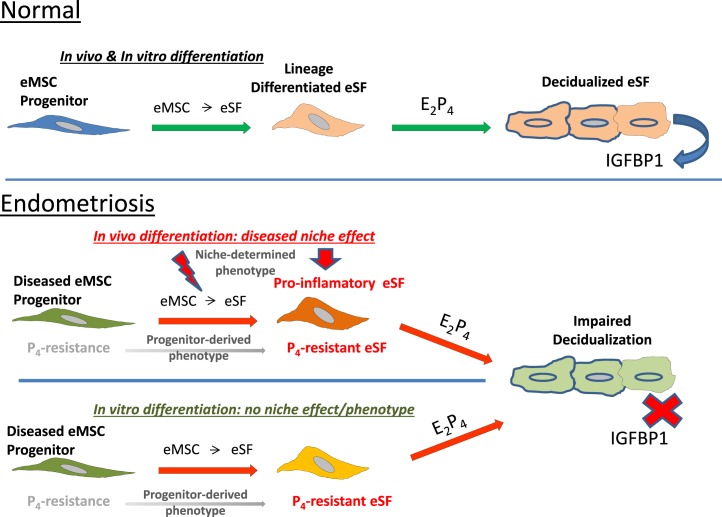
FIG. 7.
Model of disease phenotype of eMSC and eSF in endometriosis. Top panel: normal eMSC progenitors differentiate both in vivo and in vitro to eSF that decidualize in response to P4. Bottom panel: in endometriosis, in vivo differentiated eSF inherit P4 resistance from eMSC progenitors failing to decidualize, and also acquire a pro-inflammatory phenotype within the endometrial niche. The eMSC from endometriosis differentiate in vitro to eSF that inherit P4 resistance with impaired decidualization, but eMSC differentiated to eSF outside of the endometrial niche do not acquire the pro-inflammatory phenotype displayed by their in vivo differentiated counterparts.
The gene expression profiles identified herein have enabled lineage-associated gene assignments for eMSC and for eSF. For the former, our findings are consistent with previously published work [4]. For the latter, the unique eSF signature displayed considerable fidelity when eSF were cultured in vitro and when eMSC were differentiated in vitro to eSF. The case has been argued for close similarities between mesenchymal stem cells and fibroblasts isolated from diverse human tissues [32–34] even though studies are often confounded by unaccounted changes during ex vivo expansion in culture. Mindful of these broader outstanding issues, in the current study, the definitions of in vitro lineage phenotypes were anchored in the gold standard of the in vivo lineage phenotypes as established in our previous studies [4, 8] and extended herein. The current data show a distinct in vivo eSF lineage molecular phenotype that, notwithstanding adaptation to ex vivo culture, is substantially recapitulated in vitro by the surface marker-selected homologous population. In contrast, surface marker-selected eMSC showed a progressive shift in vitro, arguably not random and demonstrably specific to an eSF phenotype. Indeed, the two eMSC/pericyte markers MCAM (CD146) and SUSD2 were down-regulated in early eMSC cultures (Supplemental Table S2B), and late eMSC cultures analyzed by flow cytometry (n = 3, data not shown) showed a shift toward an eSF (CD146−/PDGFRB+) phenotype, with >90% of the population exhibiting reduced CD146 protein expression while remaining PDGFRB+. This is consistent with a recent study by Murakami et al. [14] showing down-regulation of SUSD2 expression in primary cultures of purified SUSD2+/perivascular cells from human endometrium. However, that study also found up-regulation of SUSD2 expression in primary cultures of endometrial SUSD2−/nonperivascular cells. This is in contrast with our current data from control eSF cultures showing no change in SUSD2 expression (Supplemental Table S4A). These divergent results likely reflect the differences in experimental conditions between the two studies and also imply an inherent plasticity of these endometrial cell populations, which can respond to shifting environmental cues with distinct changes in cell fate/lineage commitment. Such phenotypic plasticity would be of particular significance in cyclic endometrium wherein changes in the hormonal milieu and tissue microenvironment would introduce distinct environmental cues and impose significant demands on acute cellular responses to maintain tissue homeostasis. Of note, in the current study, in contrast to controls, primary cultures of endometriosis eSF showed up-regulation of SUSD2 gene expression (Supplemental Table S4B). This is consistent with the subpar overall in vitro phenotypic stability observed in endometriosis versus control eSF in the current study and underscores that the presence of endometriosis can be a confounder if not properly controlled for when procuring tissues for in vitro studies of endometrial cell populations.
In a broader context, our findings are consistent with the reported similarity of mesenchymal stem cells and fibroblasts isolated from nonendometrial tissues [32–34]. However, our data show that for endometrial (eMSC) cells, the shared in vitro phenotype conformed to the tissue eSF, not the eMSC. This may reflect an inherent plasticity of the eMSC and/or a preexisting commitment of the isolated tissue eMSC to the eSF lineage. On the other hand, the demonstrated phenotypic stability of eSF in vitro may relate to the highly specialized nature of the eSF compared to fibroblasts in other tissues as they uniquely undergo decidual differentiation, thus playing a key role in the establishment and maintenance of pregnancy [18]. Indeed, while many eSF lineage-associated genes reflect key functions shared by cultured fibroblasts from various human tissues, including immune modulation (e.g., IDO, PGE2, IGFs, specific interleukins, and cytokines), and specific fibroblast-derived factors (e.g., FGFs) [32], more specific to human endometrium are the expression of CD10, a cell surface marker/zinc-dependent endopeptidase [15, 24] and genes involved in hormone action and metabolism and growth factor signaling, including Wnt, SLIT/ROBO, and Hedgehog pathways, which are involved in the steroid hormone response of the tissue across the cycle [35–37].
In the current study, FACS-isolated eMSC readily differentiated to the eSF lineage under standard conditions used to culture eSF. This may reflect the absence in this particular culture environment of key elements of the eMSC niche required to maintain the stem/progenitor phenotype and/or the presence of factors promoting lineage differentiation [38]. The requirement of specific factors to maintain stemness in vitro as well as the use of inducers for specific lineage differentiation in vitro are well documented in the literature [39]. Therefore, in the case of eMSC, it is possible that changes in the culture environment such as extracellular matrix, bioactive molecules (e.g., growth factors/cytokines, hormones, and agonists/antagonists of cellular-signaling pathways), and/or oxygen concentration may influence stability of the stem/progenitor phenotype and/or specific lineage differentiation. In this regard, in a recent study, Gurung et al. [40] identified TGFβ signaling as a factor involved in the loss of eMSC stemness during culture. The cells in that study were serially passaged eMSC in serum-free medium with bFGF/EGF, which implies involvement of autocrine TGFβ. In this context, our data show that FACS-isolated eMSC express higher TGFβ ligands (TGFB1, 2) and receptors (TGFBR2) versus FACS-isolated eSF (Table 1, Supplemental Table S2B), consistent with autocrine TGFβ signaling in PDGFRB+/CD146+/SUSD2+ cells, potentially implying active exiting from the stem cell compartment within this population in vivo. This would be consistent with the spontaneous differentiation of eMSC cultures observed in our study, where primary eMSC cultures show overall down-regulation of TGFβ ligands (TGFB1, 2, 3) and receptors (TGFBR1, 3) versus FACS-isolated eMSC, indicating a shift toward an eSF-like, low TGFβ-signaling phenotype. While many other relevant biological questions remain to be addressed in future studies, the fact that eSF derive from eMSC underscores the importance of the endometrial niche in proper eMSC functioning and lineage differentiation to functional fibroblasts as well as the inherent niche environment effect on the differentiated eSF as they assume their important roles within the endometrium.
A key outstanding question is whether eMSC have unique properties in comparison to other sources of mesenchymal stem cells (MSC) (e.g., bone marrow, adipose, placental, cord). In this regard, an earlier study from our group [41] found that bone marrow MSC did not decidualize in response to P4 in contrast to eSF and expressed a limited subset of decidualization-associated genes (20 of 353 genes expressed by eSF) only after prolonged exposure to cAMP (14–21 days) as compared to eSF that decidualized after 2–4 days with cAMP. These data indicate MSC isolated from human endometrium have unique properties compared to MSC from this other tissue source, and potentially also compared to bone marrow-derived MSC in the systemic circulation.
The eMSC and eSF in Endometriosis
Endometriosis is an estrogen-dependent, P4-resistant disorder wherein eutopic endometrial tissue refluxed at menses seeds the peritoneum and surrounding organs and adheres to and establishes a blood supply, proliferates, and elicits an inflammatory response resulting in pelvic pain and infertility [22]. The latter is believed due to the inflammatory milieu's effects on oocyte quality and sperm and embryo viability, and on impaired implantation due to the marked pro-inflammatory phenotype within the eutopic endometrium [37]. Endometrial tissue and stromal fibroblast gene expression, proteome profiles, hormone responsiveness, cellular differentiation, and cell-signaling pathways differ in women with endometriosis versus unaffected women [25, 42, 43] due in part to P4 receptor mutations, P4 receptor co-regulator and epigenetic abnormalities, as well as local synthesis of E2 and epigenetic alterations [44–46]. The pathogenesis of these abnormalities has largely eluded definition, although a role for inflammation has been suggested [23]. In the setting of endometriosis, our data demonstrate that eSF freshly isolated from the tissue have a marked pro-inflammatory transcriptional profile with activation of pathways involved in cell-cell signaling, immune cell trafficking, the inflammatory response, epithelial migration, angiogenesis, and formation of vascular lesions. Part of this pro-inflammatory phenotype is retained in vitro after eSF are established in culture together with P4 resistance. This is consistent with whole tissue transcriptomic analysis revealing marked immune activation within the endometrium of women with endometriosis [37]. Of note when eMSC are differentiated to eSF in vitro, the latter, while displaying P4 resistance, do not exhibit a comparable pro-inflammatory phenotype, suggesting that in vivo eSF acquire their pro-inflammatory characteristics within the endometrial niche (model in Fig. 7). What contributes to this phenotype is unclear, although mediators of systemic inflammation in endometriosis [22] may be candidates.
The eMSC are pericytes believed to facilitate tissue repair and regeneration after menses [9, 47]. It is of interest that eMSCendo compared with eMSCcontrol are transcriptionally characterized by reduced cell growth and proliferation, clonogenic potential and cell survival, and increased ECM proteolysis. Whether and how eMSC acquire these characteristics within the endometrial niche is uncertain, and whether the systemic inflammatory component of endometriosis plays a role in affecting eMSC functionality warrants further investigation.
Roles of eMSC and eSF in the pathogenesis of endometriosis when refluxed into the peritoneal cavity have been proposed [18, 47, 48]. The persistence of altered transcriptomic signatures of eSF and eMSC in eutopic endometrium and eSF P4 resistance in endometrium and peritoneal lesions coupled with the potential of eMSC to differentiate into lineage cells refractory to P4 in the setting of endometriosis gain further importance in understanding the pathogenesis and pathophysiology of this disorder.
Clinical Implications of Molecular Disease Phenotypes
Given the central role of decidualization in pregnancy establishment and maintenance, abnormalities in decidualization have been invoked as underlying causes for pregnancy complications in women with endometriosis wherein there is P4 resistance in the eSF differentiation process. Higher rates of implantation-related disorders (placental abnormalities, miscarriage, preterm birth, postpartum hemorrhage) have been reported in pregnancies achieved in women with endometriosis, either in spontaneous conceptions or through assisted reproductive technologies [21, 49–52]. Deficient spiral artery remodeling during pregnancy has also been considered causative of poor pregnancy outcomes in women with endometriosis, a function facilitated by decidualized eSF [53]. Which regions, pathways, and biological functions of the decidua (basalis, parietalis, capsularis) during implantation and pregnancy are affected in women with endometriosis remains an area of active investigation. In addition, endometriosis has been identified in the decidua in contact with the fetal membranes during pregnancy [54], and whether this involves inherited phenotypes from eMSC and eSF or other mechanisms is not clear. Furthermore, the conventional wisdom has been that eSF play a central role in endometrial tissue homeostasis in the nonpregnant state, including tissue integrity, epithelial functionality, menstruation, and regeneration. Given that eSF are daughters of eMSC, how eMSC may be affected by organismal changes is an important consideration for endometrial tissue homeostasis as well as pregnancy success and disease pathogenesis. Thus, how upstream abnormalities in eMSC progenitors are acquired and affect these processes are important questions to be answered in future studies.
Our data are consistent with eMSC as the progenitor of eSF with the latter inheriting the P4-resistance phenotype from their eMSC progenitors in endometriosis. In addition, there was a pronounced pro-inflammatory phenotype not evident in the eMSC progenitors in vivo and not manifested in their in vitro differentiated progeny, indicating acquisition during/after eSF lineage differentiation within the endometrial niche in the setting of endometriosis. This study raises questions about mechanisms underlying eMSC abnormalities in endometriosis predisposing to transmitting the propensity to P4 resistance to their progeny eSF along with how and why eSF acquire a pro-inflammatory phenotype in the endometrial niche in endometriosis. The strengths of our study include the use of in vitro culture models allowing functional assessment of the hormone responsive phenotype. Limitations of the study include a relatively small number of samples for analysis and the inherent changes introduced by ex vivo cellular adaptation. Indeed, the latter is an unavoidable compromise with most in vitro model systems, and herein we have addressed this shortcoming by including the relevant data from noncultured freshly isolated cells to provide the in vivo reference standards and further thoroughly assessing the transcriptomic changes during the in vivo to ex vivo transition to identify potential liabilities of this experimental confounder.
TABLE 10.
Impaired in vitro expression by eSFEndo cultures of eSF lineage genes stably expressed by eSFControl.*
Complete gene lists in Supplemental Tables S2A, S4A, and S4B.
NS, no differential expression versus eMSCFACS.Endo (at P < 0.05, >2-fold change).
TABLE 11.
Expression of eMSC lineage-associated genes in FACS-isolated endometriosis eMSC and derived early and late cultures.*
Complete gene list in Supplemental Table S5B.
NS, no differential expression versus eSFFACS (at P < 0.05, >2-fold change).
TABLE 12.
Expression of eSF lineage-associated genes in FACS-isolated endometriosis eMSC and derived early and late cultures.*
Complete gene list in Supplemental Table S5B.
NS, no differential expression versus eSFFACS (at P < 0.05, >2-fold change).
TABLE 13.
Pathway analysis: altered cellular functions in late cultures derived from eMSCEndo and eSFEndo versus eMSCControl and eSFControl.*
Complete lists in Supplemental Tables S6, B and D.
Supplementary Material
ACKNOWLEDGMENT
We thank the participants who gave informed consent for the use of their tissue in this study; Drs. A Jacoby, M Goldman, M Autry, and S Knight of the University of California, San Francisco, for collecting some of the endometrial samples; and the nurses and clinical, laboratory, and administrative staff who made this study possible. We also acknowledge the UCSF NIH Human Endometrial Tissue and DNA Bank through which samples were obtained, and Ms. Kim Chi Vo for her participation in specimen collection and processing.
Footnotes
This research was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)/NIH National Centers for Translational Research in Reproduction and Infertility P50 HD 055764-09 (L.C.G.).
These authors contributed equally to this work and are considered co-first authors.
REFERENCES
- Hess N, Giudice L. Oviduct and endometrium: cyclic changes in primate oviduct and endometrium. Knobil and Neill's Physiology of Reproduction 2005:337–381. In. Neill JD, Plant DM, Pfaff DW, Challis JRG, de Kretser DM, Richards JS, Wassarman PM (eds.), , vol. 1. St. Louis, MO: Academic Press; [Google Scholar]
- Gargett C, Masuda H. Adult stem cells in the endometrium. Mol Hum Reprod. 2010;16:818–834. doi: 10.1093/molehr/gaq061. [DOI] [PubMed] [Google Scholar]
- Gargett C, Ye L. Endometrial reconstruction from stem cells. Fertil Steril. 2012;98:11–20. doi: 10.1016/j.fertnstert.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Spitzer T, Rojas A, Zelenko Z, Aghajanova L, Erikson D, Barragan F, Meyer M, Tamaresis J, Hamilton A, Irwin J, Giudice L. Perivascular human endometrial mesenchymal stem cells express pathways relevant to self-renewal, lineage specification, and functional phenotype. Biol Reprod. 2012;86:58. doi: 10.1095/biolreprod.111.095885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R, Schwab K, Gargett C. Clonogenicity of human endometrial epithelial and stromal cells. Biol Reprod. 2004;70:1738–1750. doi: 10.1095/biolreprod.103.024109. [DOI] [PubMed] [Google Scholar]
- Wolff E, Wolff A, Hongling D, Taylor H. Demonstration of multipotent stem cells in the adult human endometrium by in vitro chondrogenesis. Reprod Sci. 2007;14:524–533. doi: 10.1177/1933719107306896. [DOI] [PubMed] [Google Scholar]
- Mutlu L, Hufnagel D, Taylor H. The endometrium as a source of mesenchymal stem cells for regenerative medicine. Biol Reprod. 2015;92:138. doi: 10.1095/biolreprod.114.126771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piltonen T, Chen J, Erikson D, Spitzer T, Barragan F, Rabban J, Huddleston H, Irwin J, Giudice L. Mesenchymal stem/progenitors and other endometrial cell types from women with polycystic ovary syndrome (PCOS) display inflammatory and oncogenic potential. J Clin Endocrinol Metab. 2013;98:3765–3775. doi: 10.1210/jc.2013-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab K, Gargett C. Co-expression of two perivascular cell markers isolates mesenchymal stem-like cells from human endometrium. Hum Reprod. 2007;22:2903–2911. doi: 10.1093/humrep/dem265. [DOI] [PubMed] [Google Scholar]
- Masuda H, Anwar S, Bühring H-J, Rao J, Gargett C. A novel marker of human endometrial mesenchymal stem-like cells. Cell Transplant. 2012;21:2201–2214. doi: 10.3727/096368911X637362. [DOI] [PubMed] [Google Scholar]
- Ulrich D, Tan K, Deane J, Schwab K, Cheong A, Rosamilia A, Gargett C. Mesenchymal stem/stromal cells in post-menopausal endometrium. Hum Reprod. 2014;29:1895–1905. doi: 10.1093/humrep/deu159. [DOI] [PubMed] [Google Scholar]
- Dimitrov R, Timeva T, Kyurkchiev D, Stamenova M, Shterev A, Kostova P, Zlatkov V, Kehayov I, Kyurkchiev S. Characterization of clonogenic stromal cells isolated from human endometrium. Reproduction. 2008;135:551–558. doi: 10.1530/REP-07-0428. [DOI] [PubMed] [Google Scholar]
- Cervelló I, Mas A, Gil-Sanchis C, Peris L, Faus A, Saunders P, Critchley H, Simón C. Reconstruction of endometrium from human endometrial side population cell lines. Plos One. 2011;6:e21221. doi: 10.1371/journal.pone.0021221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Lee Y, Lucas E, Chan Y-W, Durairaj R, Takeda S, Moore J, Tan B, Quenby S, Chan J, Gargett C, Brosens J. Decidualization induces a secretome switch in perivascular niche cells of the human endometrium. Endocrinology. 2014;155:4542–4553. doi: 10.1210/en.2014-1370. [DOI] [PubMed] [Google Scholar]
- Dunn C, Kelly R, Critchley H. Decidualization of the human endometrial stromal cell: an enigmatic transformation. Reprod Biomed Online. 2003;7:151–161. doi: 10.1016/s1472-6483(10)61745-2. [DOI] [PubMed] [Google Scholar]
- Henriet P. Gaide Chevronnay H, Marbaix E. The endocrine and paracrine control of menstruation. Mol Cell Endocrinol. 2012;358:197–207. doi: 10.1016/j.mce.2011.07.042. [DOI] [PubMed] [Google Scholar]
- Irwin J, Suen L, Martina N, Mark S, Giudice L. Role of the IGF system in trophoblast invasion and pre-eclampsia Hum Reprod 1999. 14 (Suppl 2): 90 96 [DOI] [PubMed] [Google Scholar]
- Gellersen B, Brosens J. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr Rev. 2014;35:851–905. doi: 10.1210/er.2014-1045. [DOI] [PubMed] [Google Scholar]
- Du M-R, Wang S-C, Li D-J. The integrative roles of chemokines at the maternal-fetal interface in early pregnancy. Cell Mol Immunol. 2014;11:438–448. doi: 10.1038/cmi.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-X, Jin L-P, Xu B, Liang S-S, Li D-J. Decidual stromal cells recruit Th17 cells into decidua to promote proliferation and invasion of human trophoblast cells by secreting IL-17. Cell Mol Immunol. 2014;11:253–262. doi: 10.1038/cmi.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannuccini SCV, Fraser IS, Taylor HS, Critchley H, Giudice LC, Petraglia F. Infertility and reproductive disorders: impact of hormonal and inflammatory mechanisms on pregnancy outcomes. Hum Reprod Update. 2016;22:104–115. doi: 10.1093/humupd/dmv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice L. Clinical practice. Endometriosis. N Engl J Med. 2010;362:2389–2398. doi: 10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney R, Giudice L. Pathogenesis and pathophysiology of endometriosis. Fertil Steril. 2012;98:511–519. doi: 10.1016/j.fertnstert.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemmt P, Carver J, Kennedy S, Koninckx P, Mardon H. Stromal cells from endometriotic lesions and endometrium from women with endometriosis have reduced decidualization capacity. Fertil Steril. 2006;85:564–572. doi: 10.1016/j.fertnstert.2005.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanova L, Tatsumi K, Horcajadas J, Zamah A, Esteban F, Herndon C, Conti M, Giudice L. Unique transcriptome, pathways, and networks in the human endometrial fibroblast response to progesterone in endometriosis. Biol Reprod. 2011;84:801–815. doi: 10.1095/biolreprod.110.086181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanova L, Velarde M, Giudice L. The progesterone receptor coactivator Hic-5 is involved in the pathophysiology of endometriosis. Endocrinology. 2009;150:3863–3870. doi: 10.1210/en.2009-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosens I, Brosens J, Fusi L, Al-Sabbagh M, Kuroda K, Benagiano G. Risks of adverse pregnancy outcome in endometriosis. Fertil Steril. 2012;98:30–35. doi: 10.1016/j.fertnstert.2012.02.024. [DOI] [PubMed] [Google Scholar]
- Gargett C, Schwab K, Zillwood R, Nguyen H, Wu D. Isolation and culture of epithelial progenitors and mesenchymal stem cells from human endometrium. Biol Reprod. 2009;80:1136–1145. doi: 10.1095/biolreprod.108.075226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J, Suen L, Faessen G, Popovici R, Giudice L. Insulin-like growth factor (IGF)-II inhibition of endometrial stromal cell tissue inhibitor of metalloproteinase-3 and IGF-binding protein-1 suggests paracrine interactions at the decidua:trophoblast interface during human implantation. J Clin Endocrinol Metab. 2001;86:2060–2064. doi: 10.1210/jcem.86.5.7451. [DOI] [PubMed] [Google Scholar]
- Dixon W. Analysis of extreme values. Ann Math Stat. 1950;21:488–506. [Google Scholar]
- Zar J. Biostatistical Analysis Upper Saddle River, NJ: Prentice Hall; 1999. [Google Scholar]
- Haniffa M, Collin M, Buckley C, Dazzi F. Mesenchymal stem cells: the fibroblasts' new clothes? Haematologica. 2009;94:258–263. doi: 10.3324/haematol.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hematti P. Mesenchymal stromal cells and fibroblasts: a case of mistaken identity? Cytotherapy. 2012;14:516–521. doi: 10.3109/14653249.2012.677822. [DOI] [PubMed] [Google Scholar]
- Chang Y, Li H, Guo Z. Mesenchymal stem cell-like properties in fibroblasts. Cell Physiol Biochem. 2014;34:703–714. doi: 10.1159/000363035. [DOI] [PubMed] [Google Scholar]
- Talbi S, Hamilton A, Vo K, Tulac S, Overgaard M, Dosiou C, Le Shay N, Nezhat C, Kempson R, Lessey B, Nayak N, Giudice L. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology. 2006;147:1097–1121. doi: 10.1210/en.2005-1076. [DOI] [PubMed] [Google Scholar]
- Wei Q, Levens E, Stefansson L, Nieman L. Indian Hedgehog and its targets in human endometrium: menstrual cycle expression and response to CDB-2914. J Clin Endocrinol Metab. 2010;95:5330–5337. doi: 10.1210/jc.2010-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaresis J, Irwin J, Goldfien G, Rabban J, Burney R, Nezhat C, DePaolo L, Giudice L. Molecular classification of endometriosis and disease stage using high-dimensional genomic data. Endocrinology. 2014;155:4986–4999. doi: 10.1210/en.2014-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezza A, Sennett R, Rendl M. Adult stem cell niches: cellular and molecular components. Curr Top Dev Biol. 2014;107:333–372. doi: 10.1016/B978-0-12-416022-4.00012-3. [DOI] [PubMed] [Google Scholar]
- Kolf C, Cho E, Tuan R. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther. 2007;9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung S, Werkmeister J, Gargett C. Inhibition of transforming growth factor-β receptor signaling promotes culture expansion of undifferentiated human endometrial mesenchymal stem/stromal cells. Sci Rep. 2015;5:15042. doi: 10.1038/srep15042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanova L, Horcajadas J, Esteban F, Giudice L. The bone marrow-derived human mesenchymal stem cell: potential progenitor of the endometrial stromal fibroblast. Biol Reprod. 2010;82:1076–1087. doi: 10.1095/biolreprod.109.082867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney R, Talbi S, Hamilton A, Vo K, Nyegaard M, Nezhat C, Lessey B, Giudice L. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007;148:3814–3826. doi: 10.1210/en.2006-1692. [DOI] [PubMed] [Google Scholar]
- Bulun S. Endometriosis. N Engl J Med. 2009;360:268–279. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- Al-Sabbagh M, Lam E, Brosens J. Mechanisms of endometrial progesterone resistance. Mol Cell Endocrinol. 2012;358:208–215. doi: 10.1016/j.mce.2011.10.035. [DOI] [PubMed] [Google Scholar]
- Guo S-W. The endometrial epigenome and its response to steroid hormones. Mol Cell Endocrinol. 2012;358:185–196. doi: 10.1016/j.mce.2011.10.025. [DOI] [PubMed] [Google Scholar]
- Huhtinen K, Saloniemi-Heinonen T, Keski-Rahkonen P, Desai R, Laajala D, Ståhle M, Häkkinen M, Awosanya M, Suvitie P, Kujari H, Aittokallio T, Handelsman D, et al. Intra-tissue steroid profiling indicates differential progesterone and testosterone metabolism in the endometrium and endometriosis lesions. J Clin Endocrinol Metab. 2014;99:97. doi: 10.1210/jc.2014-1913. [DOI] [PubMed] [Google Scholar]
- Gargett C, Nguyen H, Ye L. Endometrial regeneration and endometrial stem/progenitor cells. Rev Endocr Metab Disord. 2012;13:235–251. doi: 10.1007/s11154-012-9221-9. [DOI] [PubMed] [Google Scholar]
- Gargett C, Schwab K, Brosens J, Puttemans P, Benagiano G, Brosens I. Potential role of endometrial stem/progenitor cells in the pathogenesis of early-onset endometriosis. Mol Hum Reprod. 2014;20:591–598. doi: 10.1093/molehr/gau025. [DOI] [PubMed] [Google Scholar]
- Stephansson O, Kieler H, Granath F, Falconer H. Endometriosis, assisted reproduction technology, and risk of adverse pregnancy outcome. Hum Reprod. 2009;24:2341–2347. doi: 10.1093/humrep/dep186. [DOI] [PubMed] [Google Scholar]
- Healy D, Breheny S, Halliday J, Jaques A, Rushford D, Garrett C, Talbot J, Baker H. Prevalence and risk factors for obstetric haemorrhage in 6730 singleton births after assisted reproductive technology in Victoria Australia. Hum Reprod. 2010;25:265–274. doi: 10.1093/humrep/dep376. [DOI] [PubMed] [Google Scholar]
- Vercellini P, Parazzini F, Pietropaolo G, Cipriani S, Frattaruolo M, Fedele L. Pregnancy outcome in women with peritoneal, ovarian and rectovaginal endometriosis: a retrospective cohort study. BJOG. 2012;119:1538–1543. doi: 10.1111/j.1471-0528.2012.03466.x. [DOI] [PubMed] [Google Scholar]
- Takemura Y, Osuga Y, Fujimoto A, Oi N, Tsutsumi R, Koizumi M, Yano T, Taketani Y. Increased risk of placenta previa is associated with endometriosis and tubal factor infertility in assisted reproductive technology pregnancy. Gynecol Endocrinol. 2013;29:113–115. doi: 10.3109/09513590.2012.706669. [DOI] [PubMed] [Google Scholar]
- Brosens I, Pijnenborg R, Benagiano G. Defective myometrial spiral artery remodelling as a cause of major obstetrical syndromes in endometriosis and adenomyosis. Placenta. 2013;34:100–105. doi: 10.1016/j.placenta.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Marcellin L, Santulli P, Gogusev J, Lesaffre C, Jacques S, Chapron C, Goffinet F, Vaiman D, Méhats C. Endometriosis also affects the decidua in contact with the fetal membranes during pregnancy. Hum Reprod. 2015;30:392–405. doi: 10.1093/humrep/deu321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.