Abstract
G protein-coupled receptors (GPCRs) are considered to represent the most promising drug targets; it has been repeatedly said that a large fraction of the currently marketed drugs elicit their actions by binding to GPCRs (with cited numbers varying from 30–50%). Closer scrutiny, however, shows that only a modest fraction of (~60) GPCRs are, in fact, exploited as drug targets, only ~20 of which are peptide-binding receptors. The vast majority of receptors in the humane genome have not yet been explored as sites of action for drugs. Given the drugability of this receptor class, it appears that opportunities for drug discovery abound. In addition, GPCRs provide for binding sites other than the ligand binding sites (referred to as the “orthosteric site”). These additional sites include (i) binding sites for ligands (referred to as “allosteric ligands”) that modulate the affinity and efficacy of orthosteric ligands, (ii) the interaction surface that recruits G proteins and arrestins, (iii) the interaction sites of additional proteins (GIPs, GPCR interacting proteins that regulate G protein signaling or give rise to G protein-independent signals). These sites can also be targeted by peptides. Combinatorial and natural peptide libraries are therefore likely to play a major role in identifying new GPCR ligands at each of these sites. In particular the diverse natural peptide libraries such as the venom peptides from marine cone-snails and plant cyclotides have been established as a rich source of drug leads. High-throughput screening and combinatorial chemistry approaches allow for progressing from these starting points to potential drug candidates. This will be illustrated by focusing on the ligand-based drug design of oxytocin (OT) and vasopressin (AVP) receptor ligands using natural peptide leads as starting points.
Keywords: GPCR, drug, ligand-based design, peptide, cyclotide, conotoxin, oxytocin, vasopressin
Introduction
G protein-coupled receptors (GPCRs) constitute probably the largest known protein families of cell surface receptors that regulate various cellular processes via signal transduction. The receptors consist of seven transmembrane-spanning α–helices, with an extracellular N-terminus, an intracellular C-terminus and three interhelical loops on each side of the membrane [1–3]. They recognize extracellular signaling molecules (ligands) of various nature (e.g., neurotransmitters, hormones, growth factors, odorant molecules and light) and size (from small molecules to peptides to large proteins) [4] that elicit a response inside the cell. The signal transmission leads to the activation or inactivation of a particular signaling pathway and hence a specific cellular response.
The human genome contains 791 - 865 genes coding for GPCRs [5–7]. This estimate is subject to continuous revision because of uncertainties and ambiguities in the nature of some genes: in the human genome, for instance, most pheromone receptors of the vomeronasal organ (VR1-class) are pseudogenes. There may also be pseudogenes in the abundant family of odorant receptors. More importantly from the perspective of drug discovery, at least ~150 are still “orphan receptors”: their endogenous ligand is not known; accordingly their physiological role is obscure [8–10]. The family of G protein-coupled receptors has been classified in various attempts according to the IUPHAR scheme [11] or the GPCRDB (database) [12] into four classes of GPCRs in humans; Class A: rhodopsin-like, with over 80% of all GPCRs in humans; Class B: secretin-like; Class C: metabotropic glutamate receptors; and the much smaller Class F: frizzled/smoothened family. Two additional classes (Class D: pheromone receptors; and Class E: cAMP receptors) are only found in non-mammalian species. These classes can be further divided, based on their function and their ligands.
In contrast to plants, animals are highly dependent on GPCRs to afford communication between cells. GPCRs have adapted to bind to a large variety of ligands; the range of endogenous agonists comprises lipids (e.g., prostanoids, lysophosphatidic acid, retinal), sugars (recognized by taste receptors), a bewildering array of volatile organic compounds (recognized by odorant receptors), amino acids (e.g., glutamate), organic acids (e.g., hydroxybutyrate, succinic acid), inorganic ions (e.g., Ca2+), nucleosides and nucleotides (e.g., adenosine, ATP, ADP, UTP, UDP), biogenic amines (e.g., dopamine, (nor-)epinephrine), peptides (e.g., secretin, gastrin, vasopressin, oxytocin), large proteins (e.g., TSH/thyrotropin, the gonadotropins). The mode of binding is highly divergent: binding may occur within the hydrophobic core of the receptor (the presumed binding mode for small molecular ligands other than GABA, glutamate and Ca2+), on the extracellular face of the receptor (for peptide ligands) or to an extended N-terminal domain (for receptors binding proteohormones, GABA, glutamate and Ca2+). In addition, even within topologically related binding sites, the structure of GPCRs accommodates distinct binding modes: X-ray crystal structures are available for two related group A (i.e., rhodopsin-like) receptors in the antagonist-bound state: within the binding pockets of the β1- and the β2-adrenergic receptors, the orientation of the antagonists cyanopindolol [13] and carazolol [14], respectively, is parallel to the membrane plane (and perpendicular to the axis of the transmembrane helices. In contrast, in the A2A-adenosine receptor the binding pocket is displaced towards helices VI and VII (relative to that of the β-adrenergic receptors) and the antagonist (ZM241385, a methylxanthine derivative) is bound in an extended conformation perpendicular to the plane of the membrane [15].
Typically, the “drugability” of GPCRs is highlighted by stressing that 30–50% of all currently registered drugs act on GPCRs [16–18], but these numbers obscure the untapped opportunities: regardless of whether these estimates are based on market share or on the percentage of the total currently approved chemical entities, the number of receptors that are targeted by these drugs is modest (~60 receptors, see Table 1). It is also evident from Table 1 that only a fraction (i.e., ~20) of those receptors are peptide-binding GPCRs [18]. In fact, “drugability” has not yet been explored for the vast majority of GPCRs. Last but not least, it is worth pointing out that GPCRs have binding sites other than the orthosteric binding site (i.e., the binding site of physiological agonists and competitive antagonists); these include an allosteric site for compounds that modulate agonist binding to the orthosteric site (for review see [19 20]). In addition, the interface between receptor and G protein may also be targeted [21, 22].
Table 1. Currently Targeted G Protein-Coupled Receptors.
| G Protein-Coupled Receptor(s) | Representative Targeting Drug(s) (Generic Name) |
|---|---|
| receptors for amines, lipid mediators and nucleosides/ nucleotides: | |
| A1 & A2A-adenosine receptor | caffeine, theophylline |
| α1A,B,C-adrenergic receptors | prazosin, doxazosin, terazosin, tamsulosin |
| α2A,B,C-adrenergic receptors | clonidine, moxonidine |
| β1-adrenergic receptor | bisoprolol, metoprolol, atenolol, esmolol |
| β2-adrenergic receptor | salbutamol, salmeterol, fenoterol, formoterol |
| CB1-cannabinoid receptor | rimonabant |
| D1,2(3-5) dopamine receptors | L-DOPA |
| D2-receptor | pramipexol, ropinirol; haloperidol, fluphenazine, sulpiride |
| D4-receptor | clozapine |
| GPR109a= HM74 = PUMA-G | nicotinic acid, acipimox |
| H1-histamine receptor | diphenhydramine, dimetindene, desloratadine |
| H2-histamine receptor | cimetidine, ranitidine |
| 5HT1a-serotonin receptor | buspirone |
| 5HT1b,d-receptors | sumatriptan, zolmitriptan |
| 5HT2A,C-receptors | risperidone, quetiapine, olanzapine |
| 5HT4-receptor | cisapride, metoclopramide |
| M1-muscarinic receptor | pirenzepine |
| M2,3,4-muscarinic receptors | atropine, ipratropium |
| M3-muscarinic receptor | tiotropium |
| MT1,2-melatonin-receptor | agomelatine |
| CysLT1-cysteinyl-leukotriene-receptor | montelukast, zafirlukast |
| EP3-prostanoid receptor | misoprostol |
| EP4-prostanoid receptor | alprostadil (PGE1) |
| FP(A,B)-prostanoid receptor | latanoprost, PGF2α |
| IP prostaglandin I2-receptor | treprostinil, epoprostenol |
| DP1 prostaglandin D2 receptor | laropripant |
| P2y12-purinergic receptor | clopidogrel |
| rhodopsin | retinal/vitamin A (for night blindness) |
| S1P1 (3, 4, 5)-sphingosin-1-phosphate receptors | fingolimod |
| “flytrap receptors”: | |
| Ca2+-sensing receptor | cinacalcet |
| GABAB-receptor (1/2) | baclofen |
| receptors for peptides /proteohormones: | |
| angiotensin-II-receptor type 1 | losartan, candesartan, telmisartan |
| B2-bradykinin receptor | Icatibant |
| calcitonin-receptor | calcitonin |
| ETA/B-endothelin receptor | bosentan |
| ETA-endothelin receptor | sitaxentan |
| GLP1-receptor | exenatide |
| glucagon-receptor | glucagon |
| FSH-receptor | FSH/follitropin, menotropin |
| GnRH-receptor | goserelin, buserelin; degarelix |
| LH-receptor | LH/lutropin, HCG |
| MC2-melanocortin-receptor | ACTH |
| NK1-Neurokinin-receptor | aprepitant |
| OT-oxytocin receptor | atosiban; oxytocin |
| µ-opioid receptor | morphine, fentanyl, buprenorphine |
| κ-opioid receptor | nalbuphine |
| PTH-receptor | PTH/parathyroid hormone |
| SSR2,5 (1,3,4)-somatostatin-receptor | somatostatin, octreotide, lanreotide |
| Vasopressin V1a+b-receptors | vasopressin, terlipressin |
| Vasopressin V2-receptor | desmopressin = DDAVP; tolvaptan |
Only receptors are listed, which are activated or inhibited by a drug that is approved for human pharmacotherapy. Subtypes in brackets indicate receptors that may be activated or blocked concomitantly by the drug, but that are not thought to contribute to the action for the approved indication.
Structure- and ligand-based design and the molecular diversity created by combinatorial peptide libraries are not mutually exclusive, but offer in combination the ability for the development of new specifically targeted peptide drugs [23]. Here, we focus on the use of ligand-based combinatorial peptide design for drug discovery to provide an overview on “peptides as ligands for GPCRs”. First, the reader is guided through the basic principles of GPCR signaling - explaining the G protein cycle and the role of GPCR accessory proteins. We will further present an overview of peptide-binding GPCRs and point out how peptides can interfere with GPCR signaling. We devote a section to discuss the role of combinatorial peptide libraries (including naturally-occurring peptide libraries) and ligand-based combinatorial peptide design for the purpose of drug discovery and the development of novel therapeutics. As an applied example we will review some recent work, which was carried out on the design of selective peptide agonists/antagonists for oxytocin and vasopressin receptors.
GPCR Signaling and Accessory Proteins - New Opportunities?
The eponymous term “G protein-coupled receptor” reflects the canonical signal transduction mechanism, namely the flow of information from an extracellular agonist, which activates a given receptor to engage its cognate G protein(s); this generates active subunits that regulate cellular effector systems and thus give rise to a biological response.
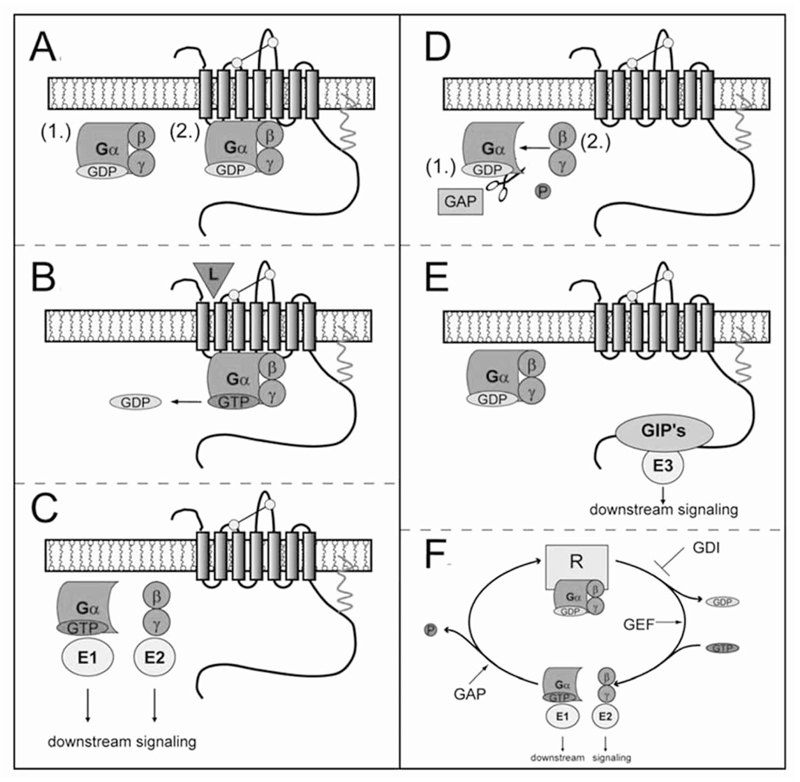
For the past 20 years, the G protein cycle has been understood in considerable detail [24]: the G protein cycles between an inactive GDP-bound conformation and an active GTP-bound conformation. Heterotrimeric G proteins consist of three subunits: α, β and γ. The G protein heterotrimer acts as a molecular switch in signal transduction pathways mediated by the GPCR [2] (Fig. 1). Upon receptor activation the GDP is released from the α–subunit; three models have been proposed to explain this GDP-exit mechanism, they are referred to as the (i) lever-arm, (ii) gear-shift and (iii) sequential fit models. The mechanistic details of these models explaining GDP-release have been recently reviewed [25] and the pertinent arguments will not be further considered. Suffice it to say that the GDP release is brought about by a conformational change of the receptor, which leads to a structural movement in the Gα subunit resulting in a change of the microenvironment of the GDP-binding pocket and the release of GDP. The three models differ in the molecular mechanism of these structural movements that lead to the release of GDP. Once the binding pocket of the nucleotide is empty, the G protein heterotrimer is in its activated conformation and forms a stable, high-affinity complex with the activated receptor. Binding of GTP to the Gα subunit destabilizes the complex, which leads to dissociation of Gα and Gβ/γ subunits from the receptor and their interaction with downstream effector proteins (E1 and E2). The signal is terminated by the intrinsic GTPase activity of the Gα subunit, which hydrolyses the GTP to GDP. This GTP-GDP “exchange” leads to re-formation of the heterotrimer Gαβγ and results in the inactive or basal state of the G protein. In summary, the G protein cycle is defined by a conformational switch of the receptor and hence G protein heterotrimer upon stimulation. The signal is initiated by GDP-GTP exchange and terminated by a conversion of GTP to GDP and the mutual inactivation of Gα and Gβγ. Thus the GDP/GTP-cycle drives the conformational cycle between inactive and active conformation of Gα, which results in a superimposed cycle of subunit association and dissociation.
Fig. (1).
GPCR signaling, G protein cycle and accessory proteins of signaling.
(A) In its basal (non-activated) state the G protein (shown in green) is a heterotrimer composed of three subunits (α, β and γ) and binds the guanine nucleotide diphosphate (shown in pale orange). The G protein(s) interact with their receptors (shown as 7-transmembrane spanning model in grey, with commonly observed motifs pointed out, i.e., disulfide bond in yellow and palmytoylation anchor in red) by (restricted/unrestricted) collision coupling (1→2). (B) Upon activation (i.e., agonist stimulation), the GDP is released from the α–subunit and GTP (red) is bound (see text for explanation). (C) Binding of GTP to the Gα subunit destabilizes the complex, which leads to dissociation of Gα and Gβ/γ subunits from the receptor and their interaction with downstream effector proteins (E1 and E2, shown in yellow). (D) The signal is terminated by the intrinsic GTPase (pale blue) activity of the Gα subunit, which hydrolyses the GTP to GDP. This GTP-GDP “exchange” leads to re-formation (1→2) of the heterotrimer Gαβγ and results in the inactive or basal state of the G protein. (E) In addition, GPCRs can interact with numerous proteins other than their cognate G protein(s). These so-called GPCR interacting proteins (GIPs) can elicit specific cellular responses, independently of the G protein and/or the arrestin-mediated cellular signaling. (F) Several proteins impinge on the G protein cycle by interacting directly with individual G proteins: (i) GTPase activating proteins (GAPs), (ii) non-receptor guanine nucleotide exchange factors (GEFs) and (iii) guanine nucleotide dissociation inhibitors (GDIs).
Several proteins impinge on the G protein cycle by interacting directly with individual G proteins [26]: (i) the intrinsic GTPase of the G protein α–subunit functions as the timed turn-off switch; the rate of GTP-cleavage (kcat) of the intrinsic GTPase is in the range of 2 - 15 min-1 and thus too slow to allow for rapid signaling events (following receptor activation, the turn-off reaction becomes rate-limiting in the cycle; the turn-off reaction, however, also determines the speed at which the agonist-induced response reaches its plateau). Thus, the turn-off reaction is - in most instances - assisted by the eponymous action of GAPs (GTPase activating proteins) (see Fig. 1F). For historical reasons (the proteins were originally identified in functional screens in Saccharomyces cervisiae and Caenorhabditis elegans), these GAPs are referred to as RGS proteins (regulators of G proteins signaling) [27, 28]. Additional proteins are (ii) non-receptor guanine nucleotide exchange factors (GEFs) and (iii) guanine nucleotide dissociation inhibitors (GDIs) [26, 29], which modulate the G protein cycle in opposite ways (Fig. 1F).
There are many different isoforms or closely related subtypes of G proteins. In mammalian organisms, for example, there are 20 Gα, 5 Gβ and 12 Gγ isoforms assembled in various combinations to generate a diverse population of G protein heterotrimers [26, 30]. In spite of this combinatorial diversity, G proteins are classified according to their Gα–subunit because this highlights the principal signaling mechanism (e.g., the isoforms of Gαs and Gαi stimulate and inhibit, respectively, adenylyl cyclases and the production of cAMP, those of Gαq/11 stimulates phospholipase C and the breakdown of PIP2, Gα12/13 Rho-mediated cytoskeletal rearrangements). It is evident that this approach is an oversimplification, because it ignores the contribution of Gβγ.
However, GPCRs do not only trigger cellular response via the canonical signaling pathway, i.e., via the G-protein activation/inactivation cycle. The agonist-liganded GPCR is subject to phosphorylation by regulatory kinases (GRK1-6, G protein-coupled receptor kinases). Phosphorylation triggers recruitment of arrestins, which associate with the phosphorylated receptor. This interaction precludes the recruitment of G protein and thus leads to desensitization of G protein-dependent signaling [31]. During the past decade, it was appreciated that upon internalization, the complex of GPCR and arrestin triggers a second round of signals that involves nonreceptor tyrosine kinases of the SRC-family, MAP kinase family members (ERK1/2, jun-N-terminal kinase, p38 MAP kinase etc.) and regulators of small G proteins [32]. In this context, it is interesting to note that (partial) agonists can be identified that bias the receptor conformation; i.e., they promote recruitment of arrestins although they do not stimulate the canonical (i.e., G protein-dependent signaling) pathway [33]. These observations were generated by using the β2-adrenergic receptor, which has a rich pharmacology: thus a long list of compounds is available that can systematically be tested for their ability to raise cAMP and to support stimulation of ERK1/2 phosphorylation. However, it is very likely that similar bias for agonists will be discovered with many other receptors. In fact, desensitization of μ-opioid receptor is an instructive example: peptide agonists act as full agonists that efficaciously trigger signaling, desensitization and internalization; morphine and several semi-synthetic alkaloids are agonists that also give rise to desensitization, but they are poorly effective in triggering internalization. While these observations were originally made in transfected cells, they have been recapitulated ex vivo (i.e., in slice preparation) in neurons indicating that these differences also exist in the native environment [34].
In addition, GPCRs can interact with numerous proteins other than their cognate G protein(s), GRKs and arrestins [35–37]. These so-called GPCR interacting proteins (GIPs) can elicit specific cellular responses, independently of the G protein and/or the arrestinmediated cellular signaling. A case in point is the A2A-adenosine receptor, where stimulation of mitogen-activated protein kinase/ERK1/2 is independent of the cognate G protein Gs [38] and of any other heterotrimeric G protein [39], but requires ARNO (ARF-nucleotide binding site opener, the guanine nucleotide exchange factor of the monomeric G protein ARF6) and ARF6 [40, 41]; ARNO binds directly to the C-terminus of the receptor. However, in many instances, the significance of the interaction between a given receptor and the GIPs is not fully understood. It is specifically not clear to what extent these interactions contribute to the signal transduction elicited by the receptor and what the relative role of each interaction is. There have only been a few attempts to characterize the interactome of GPCRs, in particular at the receptors C-terminus. Two strategies have been reported: (i) using the isolated C-terminus as bait in pull-down experiments coupled to LC-MS/MS identification; e.g., for the interactomes of the 5- hydroxytryptamine receptor [42] and the melatonin receptors [43] or (ii) tandem affinity purification of tagged receptor from recombinant cells coupled to MS identification [44]. These surveys enabled a first glimpse of possible GIPs for two GPCRs. The “interactome” of other GPCRs as well as the mechanism of these interactions, their regulation and their exact physiological role is not yet understood. It is therefore of interest to characterize the (i) list of accessory proteins of GPCRs, in particular in vivo, (ii) how the interactome of the receptor changes in the presence of agonists/antagonists and within various tissues and (iii) identify the signaling pathways that are regulated by these additional interactors.
In principle, G proteins – like any other protein interacting with multiple partners – can act (i) as signal integrators (two or more competing signals impinge on the receptor and the output is determined by the relative strength of each input), (ii) coincidence detectors (two or more signals must be present simultaneously to elicit a response) and (iii) molecular switches (in the presence of interactor A, signal 1 is generated; in the presence of interactor B, a different effector is activated). Charting the interactome of individual GPCRs and its functional network may translate into opportunities for chemical biology and drug discovery. It is obvious that blockage of the protein-protein interaction surfaces will elicit biological effects that are distinct from those that can be triggered by agonists or antagonists acting at the receptor ligand binding site.
Peptides as Ligands for GPCRs and Modulators of Signaling
Mechanisms by which Peptides can Interfere with GPCR Signaling
Generally there are at least five mechanisms by which peptides can interfere with GPCR signaling.
These are:
-
(I)
as orthosteric ligands of GPCRs,
-
(II)
as allosteric ligands of GPCRs that enhance or block the action of the orthosteric ligand (for review see [19 20]),
-
(III)
at the receptor-G protein interface,
-
(IV)
at the interface of the receptor with other accessory proteins and
-
(V)
by direct interaction with G proteins.
As mentioned above, there are numerous examples of peptides that act as natural ligands for GPCRs (see Tables 1 and 2). These peptide ligands can either stimulate the receptor and thus act as agonists or inhibit receptor-induced signaling and thus act as antagonists. Besides the direct interaction of the peptides with the receptor as ligands on the extracellular side, peptides can interfere with the receptor on the intracellular side. By this interaction they may block the recruitment of the G protein and hence inhibit signaling. This has been shown, for instance, for synthetic peptides mimicking the C-terminal tail of Gα subunits and hence blocking an efficient binding of the G proteins to the receptor (summarized in [21, 22, 29]). In similar fashion peptides could block interactions of the receptor with other accessory proteins. GPCR interacting proteins (GIPs) and their respective receptors are dynamic signaling complexes that form differentially, depending on the cellular signal status, thus allowing therapeutic intervention. This intervention can be accomplished with peptides (from natural or synthetic origin) that specifically block or mimic the receptor interaction site and hence compete with the original GIP [45]. This approach has for example been used to inhibit protein interactions of the metabotropic glutamate receptor with the aim of preventing neurodegenerative diseases [46, 47]. Last but not least there are many possibilities how peptides/ligands can directly interfere with the G protein side of GPCR signaling (reviewed in [21, 22, 29]), for instance by binding to either G protein subunit (Gα, β and γ) and inhibiting the formation of the heterotrimer or by acting as GEFs, GAPs or GDIs. Obviously, the above-mentioned mechanisms of signal interception are only viable for in vivo pharmacological applications if the peptides can cross cellular membranes. In instances where this is not feasible, the use of small organic molecules may be the better alternative [48, 49]. It is likely that a collection of new ligands will emerge because high-throughput assays have been developed to screen for peptides that bind selectively to different conformations of G protein subunits [50] or target the interface with a specific subset of RGS proteins [51].
Table 2. G Protein-Coupled Receptors for Peptides and Proteins.
| Receptor Family | Receptor Subtypes | Cellular/Biological Function | Natural Ligand | Nature of Ligands |
|---|---|---|---|---|
| Angiotensin receptors | AT1, AT2 | Blood pressure, water and electrolyte homeostasis; tissue development and repair | Angiotensin | Peptides (8-22 amino acids) |
| Apelin R | APJ | Cardiovascular, fluid homeostasis | Apelin | Peptides (13/36 aa) |
| Bombesin R | BB1, BB2, BB3 | Wide range of physiological and pathological functions | neuromedin B, gastrinreleasing peptide | Peptides |
| Bradykinin R | B1, B2 | Blood vessel vasodilatation | Bradykinin | Small peptide (9 aa) |
| Calcitonin R | AMY1-3, AM1,2, CT, CALCRL, CGRP | Calcium and phosphorus metabolism | amylin, adrenomedullin, CGRP | Peptide (32 aa) |
| Chemokine R | CXCR1-5, CCR1-10, CX3CR1, XCR1, CXCR6 | Immunity | CXCLl-3, 5-13,16, CCL …, CX3CL1, XLC1,2, macrophage derived lectin | Proteins |
| Cholecystokinin R | CCK1,2 | Gastrointestinal tract hormone, digestion | cholesystokinin, gastrin | Peptides (17/33 aa) |
| Corticotropin releasing factor R | CRF1,2 | Stress response hormone, CNS | Peptide (41 aa, α-helical) | |
| Endothelin R | ETA, B | Vasoconstriction of smooth muscle cells | endothelin 1-3 | Peptides (21 aa) |
| Formylpeptide R | FPR, FPRL1,2 | Chemotaxis | N-formyl peptides | Amino acids and peptides with modifications (N-formyl) |
| Frizzled R | - | Development, cell proliferation | Wnt-family lipoglycoproteins | Proteins |
| Galanin R | GAL1-3 | Neuropeptide, regulation of neurotransmitter release | galanin | Peptides (30 aa) |
| Ghrelin R | GRLN | Peptide hormone, involved in digestion and obesity (see ghrelin) | ghrelin | Peptides (modified) |
| Glucagon R | GHRH, GIP, GLP1,2, Glucagon, Secretin | Peptide hormone, blood-sugar regulation | glucagon, secretin | Peptides |
| Glycoprotein hormone R | FSH, LH, TSH | Reproduction, Development | follicle-stimulating hormone (FSH), luteinizing hormone (LH), chorionic gonadotropin, thyroid-stimulating hormone | Proteins |
| Gonadotropin releasing hormone R | GNRH, GNRH2 | Neuropeptide hormone, release of FSH and LH | gonadotropin releasing hormone | Peptide/protein |
| KiSS1 derived peptide R | KiSS1 | Regulation of endocrine function | kisspeptin | Peptide |
| Melanocortin R | MC1-5 | Peptide hormone; appetite and sexual arousal | α, β, γ-melanocyte stimulating hormone | Peptide (39 aa) |
| Motilin R | MTLN | Peptide hormone, digestion (see ghrelin) | motilin | Peptide (22 aa) |
| Neurotensin R | NTS1,2 | Peptide hormone, digestion, CNS | neurotensin | Peptide (13 aa) |
| Neuromedin U R | NMU1,2 | Bombesin-related function | neuromedin | Peptide (8/25 aa) |
| Neuropeptide S R | NPS | Behavioral functions, linked to asthma suseptibility | neuropeptide S | Peptide |
| Neuropeptide Y R | NPY1,2,4,5 | neurotransmitter | neuropeptide Y, pancreatic polypeptide | Peptide (36 aa) |
| Neuropeptide B/W R | NPBW1,2 | Regulation of feeding, obesity | neuropeptide W, B | Peptides |
| Neuropeptide FF/AF | NPFF1,2 | Several physiological functions | neropeptide FF, AF | Peptides |
| Opioid R | μ–, κ–, δ-OP, NOP | Analgesic | β-endorphin, dynorphin A, nociceptin/orphanin FQ | Opioid peptides |
| Orexin R | OX1,2 | Neuropeptide hormone, food intake, energy metabolism | orexin A, B | Peptides (28/33 aa) |
| Parathyroid hormone R | PTH1,2 | Calcium metabolism | parathyroid hormone, TIP-39 | Peptide/protein (84 aa) |
| Peptide P518 R | QRFP | Orphan receptor (neuroendocrine system) | RF-amide P518 gene product | Amidated peptides |
| Prokineticin R | PK1,2 | Contraction of gastro-intestinal smooth muscle | prokineticin | Cysteine-rich proteins (81/86 aa) |
| Prolactin releasing peptide R (GPR10) | PRRP | processing of nociceptive signals in the brain | prolactin releasing peptide | Peptides (20/31 aa) |
| Relaxin family peptide R | RXFP1-4 | Reproduction | relaxin | Peptides |
| Somatostatin R | sst1-5 | inhibition of growth hormone release and modulation of neuronal activity | somatostatin, cortistatin | Cyclic peptides 14/28 aa (somtostatin); 17 aa (cortistatin) |
| Tachykinin R | NK1-3 | behavioral responses; potent vasodilators; contraction (directly or indirectly) of many smooth muscles | substance P, neurokinin A, neurokinin B | Peptides (10-12 aa) |
| Thyrotropin releasing hormone R | TRH1 | release of thyroid-stimulating hormone and prolactin by the anterior pituitary | thyrotropin releasing hormone | Tripeptide |
| Urotensin R | UT | Neurosecretory system | urotensin II | Cyclic dodecapeptide |
| Vasopressin and oxytocin R | V1a, V1b, V2, OT | See text | vasopressin, oxytocin | Small peptides (cyclic, disulfide-bonds) |
| VIP and PACAP R | PAC1, VPAC1,2 | circadian rhythms, pancreatic insulin secretion, control of immunity and inflammation | vasoactive intestinal peptide (VIP) and pituitary adenylate cyclase-activating polypeptide (PACAP) | Peptides (VIP 28 aa, PACAP 27/38 aa) |
Information extracted from [11]; without consideration of orphan GPCRs
GPCRs that Bind Peptides and Proteins
Of the ~800 distinct GPCRs, the list of known peptide- or protein-targeted GPCRs is readily comprehensible with “only” a few hundred [18]. Nevertheless peptide- and protein-GPCRs are involved in the regulation of basic and indispensible physiological and cellular processes. Table 2 summarizes the currently known GPCRs, their cognate physiological agonist, which are peptides and proteins (as extracted from the IUPHAR database [11]). For the sake of simplicity, the 19 identified chemokine receptors and their 44 endogenous ligands were not included. It is emphasized that peptide-binding GPCRs are spread over all GPCR subfamilies, but the majorities are members of the rhodopsin and secretin classes.
A recent review has examined ~120 GPCRs and their respective peptide ligands with respect to structural properties of ligand binding [18]. This comparison has highlighted common ligand binding motifs allowing receptor recognition: these GPCRs apparently recognize (broadly defined) peptide turn conformations in their diverse peptide ligands. This information may help to better understand peptide-mediated binding and activation of GPCRs and will be discussed in more detail below.
Combinatorial Peptide Libraries and Design of Peptides as Ligands for GPCRs
Common Structural Motifs of GPCR-Binding Peptides
The majority of registered pharmaceuticals that act on GPCRs are derived from ligand-based drug design strategies that were originally based on structure-activity relationships (SARs) of the endogenous ligand and derivatives thereof that interacted with the receptor. Future drugs are still likely to rely on ligand-based drug design due to the limited availability of structural data on GPCRs. It is unlikely that structural information will be available for the majority of GPCRs, regardless if the number is limited to the nonsensory GPCRs with known ligands (180 out of 210) that have not been targeted yet or whether the > 150 “orphan” receptors are included. For these latter receptors different strategies have been devised, which generally involve the screening of large combinatorial libraries and in silico molecular modeling.
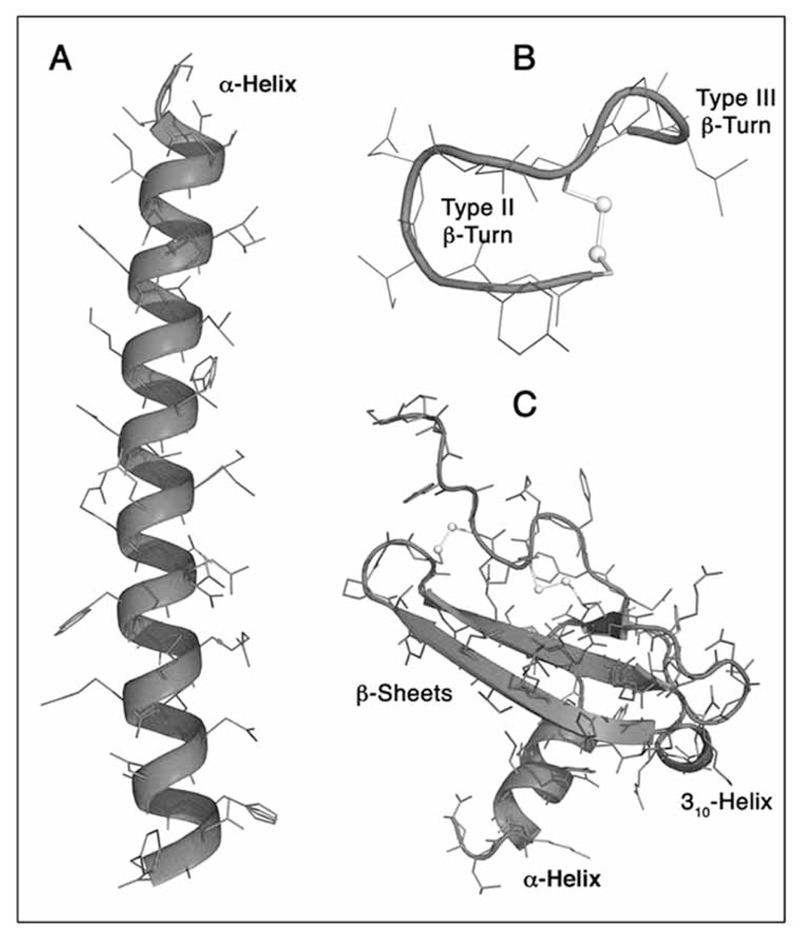
Many of the known endogenous ligands have been studied extensively and common structural binding motifs have been identified [18, 52, 53]. At the primary structure level similar pattern of amino acid sequences are found in secretin, glucagons, growth hormone-releasing hormone, glucose-dependent insulinotropic polypeptide, glucagon-like-peptide 1 and 2 [54]. An even richer source of recognition motifs can be found at the secondary structure level, particularly considering that information content in proteins/peptides is evolutionary more conserved through threedimensional structures rather than through linear amino acid sequences [55]. The main structural motif identified is the turn [18]. A turn may be defined by 3 residues (γ-turn), 4 residues (β-turn) and 5 residues (α-turn) (see Fig. 2). These can form 7-, 10- and 13- membered hydrogen bonded rings, respectively. Many of these turn structures are found to be stabilized by cyclic ring and loop moieties, in particular in the case of smaller and more flexible peptides that require conformational stabilization to maintain a rigid threedimensional structure. Examples of such cyclic peptides targeting GPCRs are the calcitonins, chemokines, endothelins, melaninconcentrating hormone, oxytocin, relaxins, somatostatin, vasopressin and urotensin II. Recognition of turn motifs generally only involves interactions of the spatially-orientated side chain residues of the ligand with the receptor and they can therefore be considered as scaffolds, which could theoretically be substituted by alternative rigid non-peptidic scaffolds that maintain the functional side chains in the right conformation. This field of peptidomimetics has been thoroughly reviewed and the reader is directed to some key articles [56–59]. Certainly, these common recognition motifs (Fig. 2) can be used as well-defined starting points for ligand-based drug design that can lead with the help of combinatorial chemistry to novel bioactive peptides as well as non-peptidic entities.
Fig. (2).
Common structural recognition motifs of peptides targeting GPCRs.
(A) α-Helix of the human parathyroid hormone [199]. (B) Type II β-turn of deamino-oxytocin [180]. (C) Stromal cell-derived factor-1α (SDF-1α), a member of the chemokine superfamily, exhibiting β-sheets and helices as structural motifs [200].
Similar to the peptide recognition motifs, so called privileged structures have been identified within small molecule drugs. A privileged structure is considered to be “a single molecular framework able to provide ligands for diverse receptors” [60]. The concept originated from the study of 1,4-benzodiazepin-2-ones that bind to cholecystokinin, gastrin and central benzodiazepine receptors. In addition, the benzodiazepine substructure is found in neurokinin-1 antagonists, κ-secretase inhibitors, farnesyl: protein transferase inhibitor and ion channel ligands. Privileged structures are generally identified by virtual screening of the structures of approved drugs employing various filters and algorithms. Once a privileged structure has been identified, it can be utilized as a scaffold for the design of drug-like libraries that can be screened against other GPCR targets. In this way many small organic privileged substructures have been identified and their use in library design has been extensively reviewed [53, 61–64]. Interestingly, some of them, including benzodiazepines have been reported to be β-turn mimetics [65, 66] and it is speculated that the multiple receptor binding properties are mainly due to the specific orientation of the attached side chains similar to above discussed peptide turn motifs [53]. It can therefore be argued that the term privileged structure can be extended to common structural recognition motifs, such as the turn structures [67, 68].
Ligand-Based Peptide Design and Combinatorial Approaches
Over the last two decades the production of efficient and high quality combinatorial libraries has been one of the most rapidly developing fields in the pharmaceutical industry driven by the unmet compound supply for high throughput screening and the vast number of novel drug targets derived from genome projects across the world [69]. The promise of a seemingly inexhaustible supply of compound libraries with drug-like properties resulted in global downsizing of the more tedious (time intensive, expensive, structurally and synthetically challenging, difficult to scale-up, unclear intellectual property, etc.) natural product discovery divisions in most pharmaceutical companies [69]. It is now considered to have been a premature decision: the vast, random compound libraries did not produce the results initially anticipated; in fact, there is only one FDA-approved drug that emerged from this approach (Sunitinib™ for renal carcinoma in 2005 [69–71]). Nonetheless, the techniques developed in the field of combinatorial chemistry certainly revolutionized drug development. Highly automated solid-phase synthesis, high yield reactions, convergent scaffold approaches in combination with split and pool or parallel techniques increased the efficacy of SAR studies and optimization of lead compounds drastically. It has been estimated that 1063 small drug-like molecule compounds or 2050 (1065) 50-residue peptides (not including different folds and conformers) could in principle be synthesized [72–74]. Realistically, this chemical space can neither be covered nor characterized systematically, with the largest libraries at present only containing up to 106 compounds. Conversely, biological relevant space is restricted to biosynthesis and natural amino acids, and functional domains are found to be conserved in their three-dimensional structure rather than through their underlying sequences [55, 75]. Consequently, biologically relevant space explored by nature during evolution is highly conserved and strongly limited in size compared to the possible chemical space [74, 76, 77]. It is therefore not surprising that random libraries based on commercially available reagents do not often overlap with structurally complex biological-relevant space. This realization resulted in a paradigm shift in library design to more focused, rational designed and smaller libraries that have an enhanced likelihood of obtaining biologically active compounds.
Two main synthetic approaches emerged: biology-orientated synthesis (BIOS) and diversity-orientated synthesis (DOS) [77]. BIOS libraries primarily employ combinatorial techniques to build compounds around privileged structures or known pharmacophores. In contrast, DOS libraries rely on methods that generate structurally complex and natural product-like compounds: the aim is to probe chemical space by structural and functional diversity. Both approaches are not mutually exclusive. DOS libraries can be based on privileged structures and BIOS libraries can be generated around scaffolds derived from DOS. The compounds of both library designs can be screened against a wide range of biological targets, considering that privileged structures interact with multiple proteins differently. These two interactive drug discovery platforms in combination with the rapid advances in computational modeling and in silico screening are powerful tools in the quest for novel GPCR modulators or in general new bioactive entities.
Implementing recognition motifs in library designs has already yielded novel leads. For example, cyclic heterochiral penta- and hexapeptides that adopt a βII’/γ-turn motif were used as conformational scaffolds for probing receptor recognition, where a recognition motif (such as Arg-Gly-Asp) was systematically shifted around the cyclic peptide backbone structure to spatially sample various conformations, leading to the identification of a vitronectin binding inhibitor to the αvβ3-integrin [78, 79]. Micromolar non-peptidic heterocyclic agonists associated with the melanocortin-1 receptor were discovered by screening a library of 951 compounds based upon a β-turn motif [80, 81].
Another interesting approach takes advantage of “turn-scans”: an initial library of 2 x 107 peptides with a β-turn constrained mimetic block in their centre was screened against the human opioid receptors μ, δ, and κ, and the opioid receptor like ORL1 [82]. The initial hits underwent optimization towards affinity and selectivity by exchanging the original β-turn mimetic moiety with other turn mimetics (“turn-scan”), which resulted in identification of a novel class of opioid ligands that act as inverse agonists as well as peptide antagonist on the κ-receptor and possess different selectivity profiles. Considering bioavailability shortcoming of peptides as drugs, the advantages of this approach towards drug development are evident: incorporation of peptidomimetics design within the library cuts out the challenging process of transferring the biological relevant peptide/protein topology into a small molecule structure.
The value of any drug discovery strategy can be evaluated by the quality and quantity of drug leads produced, with “quality” being associated to the drug-like properties of the hits (such as pharmacokinetics, toxicity, ADME, i.e., Absorption-Distribution- Metabolism-Excretion), their specificity, potency, synthetic availability, intellectual property and the target novelty, while “quantity” depends on the hit success rate and how rapidly a hit can be moved into the clinic. No doubt that combinatorial chemistry has changed the drug discovery landscape and time will tell whether these new approaches can use combinatorial chemistry in combination with rational library design to their advantage to overcome the early setbacks of random libraries and to provide the new generation of pharmaceuticals.
Peptide Libraries
Attempts to indentify privileged structures of GPCR ligands - as pointed out above - can lead to some success in the development of new and optimized peptide drugs. The lack of three-dimensional receptor structures was a major limitation to structure-based drug design. Recently several X-ray crystal structures of GPCRs with their bound ligands have been reported [13, 15, 83–87]. Although these reports only include small-molecule but not peptide ligands, the structural information can be used in bioinformatics and molecular modeling approaches to advance the field of structure- and ligand-based peptide design as targets for GPCRs [88].
Alternatively, peptide libraries offer a unique way to screen the natural diversity of peptide-protein interactions for the modification of GPCR signaling response and efficiency [23, 89] and drug lead discovery. Natural product libraries have been applied to oligonucleotides, synthetic oligomers, oligosaccharides, small molecules proteins and peptides (summarized in [90]). Naturally occurring peptides and synthetic peptides derived from portions of receptors, G proteins and effector proteins have been used in the past to study protein-peptide and receptor-peptide interactions and how these peptides can interfere with GPCR signaling [29, 91]. Several approaches are known to generate molecular diversity through synthetic or biological (encoded) peptide libraries [16, 23, 29, 89, 92, 93]. A combinatorial library approach generally involves three main steps: (i) preparation of the library, (ii) screening of the components (peptides) and (iii) identification and characterization of the active components [90]. The following section gives a brief general introduction to combinatorial peptide library approaches.
Encoded and Synthetic Approaches
Biological (also known as encoded or DNA-based libraries) include mainly mRNA display, phage display and ribosome display techniques (summarized in [89, 94]). A typical experimental library setup includes: (i) construction of a DNA library, (ii) expression of the library using the above mentioned methods, (iii) affinity selection against an immobilized target and (iv) amplification of the recovered nucleotide sequence to identify the sequence corresponding to the functional peptide. In vitro methods that do not require an in vivo transformation step, such as ribosome display, exhibit the greatest library sizes (>1013 unique peptides). Typical DNA-based (selection) libraries such as phage, bacteria, yeast, insect and mammalian cells have smaller library sizes (108 − 109), but exhibit higher biosynthetic capabilities [29, 94]. Furthermore, mRNA display has several advantages, such as higher complexity of the library and the possibility to identify high affinity sequences and is therefore the preferred biological library method [29, 95].
Chemical (or synthetic) combinatorial peptide library approaches include positional scanning libraries, mixture based oriented peptide libraries and one bead-one peptide (or split pool) solid phase libraries [23, 89, 90, 96]. Synthetic libraries have been successful to identify bioactive peptides, including antimicrobial peptides, ligands for cell surface receptors, protein kinase inhibitors, peptide binding to MHC molecules, peptide mimetics of receptor binding sites (summarized in [23]). Furthermore there are some examples to use this approach for the identification of GPCR ligands, in particular α-MSH antagonists [90, 97], opioid receptor antagonists [98] and ligands for the D2 dopamine receptor [90, 99].
Both approaches, biological and chemical libraries, are equally powerful tools in drug discovery, but they differ specifically in the diversity (number of different molecules, library size), incorporation of modified and non-proteinogenic amino acids (only possible in chemical libraries), codon degeneracy and hence bias towards amino acids (in biological libraries), higher synthesis capabilities and cost efficiency (for biological libraries) [94]. Overall, encoded and synthetic peptide library approaches play an important role in the identification of drug leads and drug development for GPCRs and will remain a powerful tool for basic research and molecular recognition studies [23, 90].
Natural Peptide Libraries
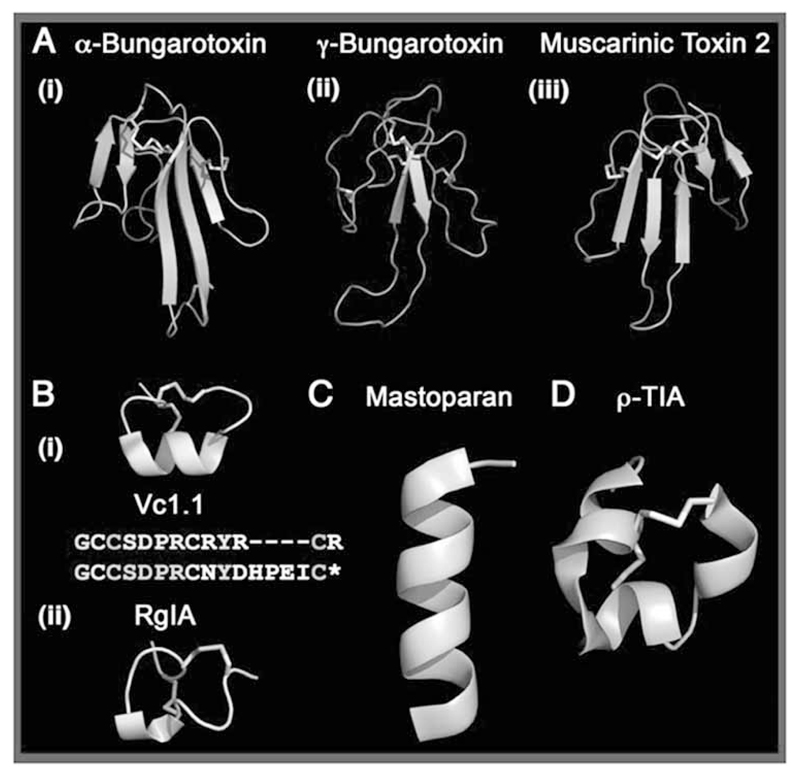
The diversity in nature has long been and still is one of the biggest resources of pharmacological lead compounds. Many natural products often exhibit biological activity against unrelated biological targets, thus providing researchers with starting points for drug development [69, 70, 100, 101]. Natural peptides of great number and diversity occur in all organisms from microbes to man (see Table 3). One such rich and yet largely untapped library of bioactive compounds can be found in venom peptides. Rich sources of venom peptides can be found in spiders [102, 103], scorpions [104], snakes [105–107] and marine animals, in particular cone-snails [108–110], to name only a few. Evolutionary pressures have afforded a pre-optimized, structurally sophisticated collection of disulfide-rich peptide toxins that have been produced in a combinatorial fashion and fine-tuned over million of years. Hence it does not come as a surprise that within the venom of these animals structural scaffolds are found that give rise to a very large number of agonists and antagonists, which act on functionally diverse targets, e.g., ion channels, transporters and GPCRs (see Fig. 3). In particular the well-studied “three-finger” scaffold found within the snake neurotoxins (Fig. 3A) highlights the functional versatility of this defined fold: it is able to act on unrelated receptor families with high potency. Another group of extensively studied peptides can be found in the venom of the predatory marine cone snail (genus Conus). These conotoxins have become a rich source for highly potent and subtype selective molecular probes for neuroscience [108,110, 111]. Conotoxins are between 6 and 50 amino acids in length, they possess a highly conserved cysteine framework. The latter gives rise to well-defined three-dimensional structures containing many of the structural motifs present in proteins, including α-helices, β-sheets and β-turns. In total only approximately 100 conotoxins have been characterized so far - a mere 0.2 % of the estimated 50,000 biologically active peptides. Nevertheless, this rather small sample has already afforded a drug of proven clinical utility (ziconotide, a derivative of ω-conopeptide MVIIA marketed under the name Pri- alt®), several pre-clinical leads for CNS disorders and various valuable tools to probe receptor subtypes [112]. Although they are thought to mainly target ligand-gated ion channels, some of these peptides have also been shown to modulate GPCRs [113–117]. The case of the α-conotoxins Vc1.1 and RgIA (Fig. 3B) is in particular interesting: these bicyclic peptides block the neuronal α9α10 subtype of the nicotinic acetylcholine receptor with nanomolar affinity and are also able to bind to the GABAB receptor. In order to extract information for the design of peptidomimetics and strategic scaffolds for libraries, it certainly would be of interest to address to questions: (i) Is the helix in the highly conserved first loop the motif responsible for high affinity interaction with both receptors? (ii) What is the role of the second loop?
Table 3. Bioactive Peptides from Natural Sources.
| Source | Peptide Class | Estimated Number of Peptides | Activity/Targets | References |
|---|---|---|---|---|
| Bacteria, plants, animals (invertebrates, vertebrates) humans | Anti-microbial peptides (AMPs), defensins | -* | Immunity, anti-microbial, insecticidal, antifungal, enzyme inhibitors | [190–193] |
| Animals/venom toxins | Bees, wasps and ants | Hundreds to thousands§ | Hemolytic, anti-microbial, cytolytic, receptor/G-protein interface/receptomimetic (e.g., mastoparan) | [107 194–196] |
| Cone-snails, Conotoxins | 50,000 | Anti-pain (ion-channels, acetylcholine receptor), neurotensin R, entdothelin R, adreno R, 5-HT R, OT/AVP R, and many more | [109 197] | |
| Scorpions | 100,000 | Ion-channels, cytolytic | [104 197] | |
| Snakes | Many thousands# | Neurotoxins; muscarinic-, α-adrenergic- and neurotensin R | [106, 107] | |
| Spiders | 1.5 – 16 million | Ion channels, cytolytic | [103 197] | |
| Plants | Cyclotides | >19,000-59,000 | Insecticidal, anti-HIV, cytotoxic, uterotonic, neurotensin R | [122, 124, 125 198] |
Total number has not been reliably been determined, but due to the wide distribution and diversity, these peptides make up large natural peptide libraries
Only few peptides have been reported from individual species, although some venom components have been studies in great detail (e.g., melittin and mastoparan); generally arthropod venom peptides are thought to have great pharmacological potential
Including peptides and larger proteins
Fig. (3).
Examples of versatile venom peptide scaffolds.
(A) The “three-finger” scaffold of snake neurotoxins highlight the target diversity of venom peptides with (i) α-bungaratoxin acting on the nAChR, (ii) γ-bungaratoxin targeting both, nicotinic and muscarinic acetylcholine receptors, and (iii) muscarinic toxin 2 modulating the muscarinic acetylcholine and α-adrenergic receptors. (B) RgIA and Vc1.1, two α-conotxin that are α9α10 nicotinic acetylcholine receptor antagonists, but act also via the GABAB receptor. (C) Mastoparan, a G-protein activating wasp venom peptide that uses its α-helical secondary structure to penetrate cell membranes and to mimic the G protein-activating portion of GPCRs. (D) TIA, an example of the ρ-conotoxin superfamily that targets α1–adrenergic receptors.
Another intriguing example are the conopressins [114–117]. These cyclic nonapeptides act on the closely related vasotocin/vasopressin/oxytocin GPCRs. These receptors are present in many different species, including mollusc, non-mammalian vertebrates, annelids, fish, mammals and humans [118] and they are characterized by a high degree of interspecies conservation among orthologues. It is not clear, which evolutionary advantage is conferred by the presence of these peptides in the venom of the cone snail. However, the discovery and characterization of conopressin-T in comparison with the human neuropeptides vasopressin and oxytocin led to the identification of an agonist/antagonist switch, which is currently investigated regarding novel antagonist design for the human receptors [117], which will be discussed in more detail below.
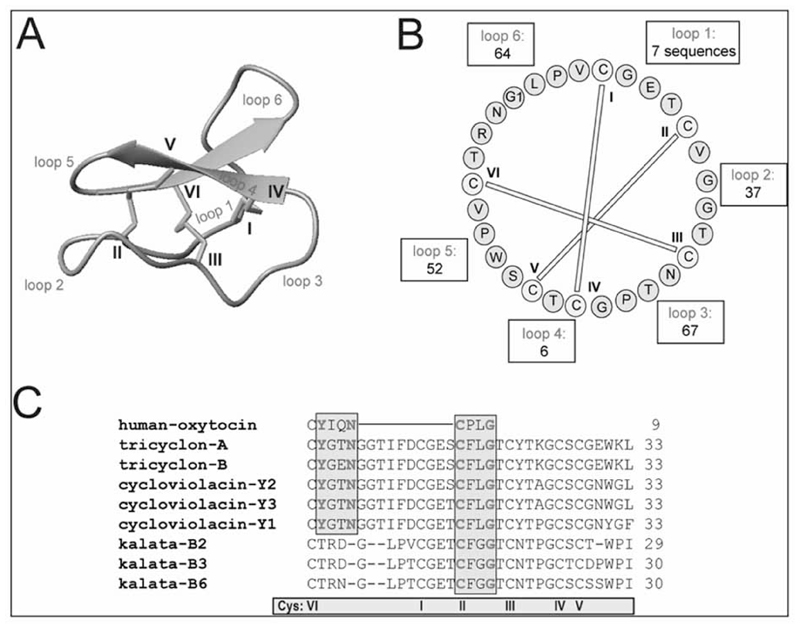
Another abundant group of naturally-occurring peptides, yet less explored in particular with respect to possible receptor targets, are bioactive plant peptides, especially circular peptides, so-called cyclotides. They have recently attracted much attention due to their impressive stability, their intrinsic bioactivity and their natural abundance and sequence diversity [119]. Cyclotides are disulfide-rich peptides discovered in plants of the Violaceae, Rubiaceae and Apocynaceae families [120–122]. They are about 30 amino acids in size and have the unique structural features of a head-to-tail cyclized backbone and a knotted arrangement of three-disulfide bonds, referred to as a cyclic cystine knot (CCK) motif [120] (see Fig. 4A). The compact CCK motif makes cyclotides exceptionally resistant to thermal, chemical or enzymatic degradation [121, 123].
Fig. (4).
Structure, diversity and oxytocin-like sequence motif of plant cyclotides.
(A) Cyclotides comprise the typical structural cyclic-cystine-knot motif, characterized by three disulfide bonds (shown in yellow) in a knotted arrangement (two disulfide bonds and the adjacent backbone segments from a ring that is threaded by the third disulfide bond). Cys-residues are numbered with Roman numerals, loops are indicated in red. The characteristic anti-parallel beta sheets are colored in cyan. (B) The diversity of cyclotides is pointed out by the number of to sequence permutations discovered to date. The wheel diagram shows the sequence of the typical cyclotide kalata B1; residues, disulfide bonds, and loops are colored/labeled according to (A). (C) Sequence alignment of human oxytocin with selected cyclotides (www.cybase.com.au). Some cyclotides and oxytocin share one or both of the activity-bearing sequence elements YxxN and CxxG.
Recent studies on the evolution, distribution and abundance of cyclotides in plants has led to the conclusion that there are at least 9,000 novel cyclotides to be discovered in the violet family (Violaceae) [124] and an even greater number (up to 50,000) in the coffee family (Rubiaceae) [122]. The discovery of cyclotides and cyclotide-like proteins in other monocotyledon [125] and dicotyledon plant families suggests that cyclotides are much more numerous than originally anticipated and it is likely that cyclotides will ultimately comprise one of the largest protein families within the plant kingdom [122] and hence constitute an immense “library of natural peptides”. The family of cyclotides impresses not only by their sheer number, but also by their diversity. Typically one species can express 15–60 different, unique cyclotides [122, 124, 126, 127] and a recent report suggests this number may be increased by varying growth conditions [128]. Their diversity is a result of the conserved cysteines framework and the backbone chains (loops) between those cysteines, which are tolerant to various amino acid combinations (see Fig. 4B). The plasticity of the cyclotide framework and its tolerance to substitution [129] have been recently used as a tool for the development of novel anti-cancer [130] and anti-infectious agents [131]. These examples underpin the application of cyclotides as combinatorial peptide scaffolds in pharmaceutical drug design [132–134] and it makes the cyclotides a large naturally occurring combinatorial peptide library.
Apart from their unique structural framework, abundance and sequence diversity, cyclotides make interesting starting points for pharmacological analysis. They exhibit a range of biological activities, including uterotonic [135], hemolytic [136], anti-neurotensin [137], anti-HIV [138], cytotoxic [139], anti-bacterial [140], anti-fouling [141] and insecticidal properties [142–144]. Only little is known about the mechanism of how they elicit these activities, but this probably involves a combination of “non-specific” membrane disruption and directed, specific receptor interaction [145].
The proposed uterotonic and anti-neurotensin activities of cyclotides are of particular interest in the present context as this would be the first example of cyclotides acting as ligands on GPCRs. Gran et al. studied the pharmacological properties of kalata B1 and found that the peptides induce oxytocin-like uterine muscle contractions in vitro (isolated rat and rabbit uteri) at concentrations of 20 μg/mL and similar effects in vivo (rabbit) at concentrations of 100 μg/kg. However, doses of > 1 mg/kg, were found to be toxic (increased blood pressure and ventricular tachycardia) and eventually lethal (ventricular fibrillation) [135, 146, 147]. Nevertheless, it is intriguing to further explore the uterotonic effect of cyclotides in molecular detail, in particular because kalata B1 [148], and other cyclotides (for instance tricyclons A and B from the violet plant Viola tricolor [149]), contain sequence- and structural-motifs (β-turns) that are similar to the presumed activity-bearing motifs in the oxytocin peptide [18] (see Fig. 4C). Another interesting cyclotide is cyclopsychotride A, which reportedly inhibits the activity (antagonistic) of the neuro-peptide neurotensin, which is the natural ligand of the neurotensin receptor and of interest as a target in neurodegenerative disease [150]. The cyclotide was originally isolated from the tropical tree Psychotria vellosiana (formerly known as Psychotria longipes) and inhibits the binding of neurotensin to its receptor with an IC50 of 3 μM and also was found to increase the intracellular Ca2+ concentration in a dose-dependent manner [137]. The neurotensin receptor is also targeted by the conotoxin contulakin-G, which is currently undergoing clinical trials for the treatment of neuropathic pain [151].
These are just a few of many examples that show how nature’s vast libraries can be a source of diverse, biologically pre-validated and evolutionary fine-tuned scaffolds that can be used as biological relevant starting points in the quest of GPCR modulators. One could argue that natural products optimized to defend against and prey on a large selection of species would have an enhanced likelihood to preserve common structural motifs that are active on more than one target. It can therefore be hoped that, in times of high-throughput screening, such libraries will be screened also against unrelated target receptors to identify such interesting privileged motifs systematically rather than by serendipity.
Design of Selective Peptide Agonists and Antagonists for Oxytocin and Vasopressin Receptors
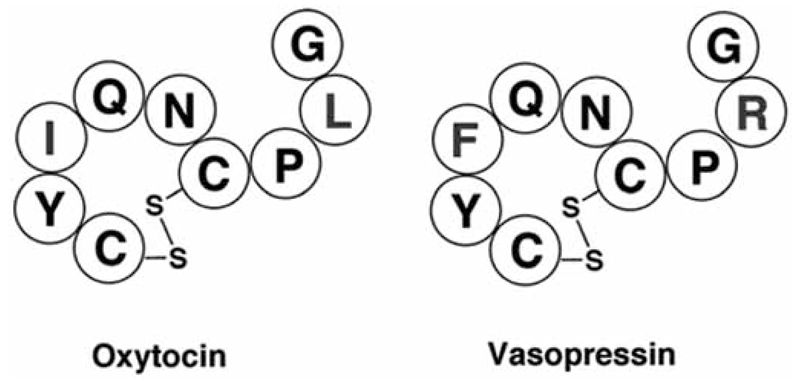
Oxytocin (OT) and vasopressin (AVP, arginine-vasopressin) are closely related, highly conserved, multifunctional nonapeptides with an N-terminal cyclic 6-residue ring structure stabilized by an intramolecular disulfide bond, and a flexible C-terminal 3-residue tail. They are structurally very similar: they differ only by two amino acids in position 3 and 8 (see Fig. 5), however their physiological functions are quite distinct. In the periphery, OT is involved in uterine smooth muscle contraction during parturition, ejaculation, and milk ejection during lactation [152]. In the central nervous system, OT functions as a neurotransmitter involved in complex social behaviors [153–157], maternal care [158, 159], bonding [160], stress and anxiety [96, 161]. AVP regulates peripheral fluid balance and blood pressure [162, 163] and is centrally implicated in memory and learning [164, 165], stress related disorders [166] and aggressive behavior [167, 168]. Both peptides act on GPCRs, OT via one oxytocin receptor (OTR) and AVP via three vasopressin receptors (AVPRs, vasopressor V1aR, pituitary V1bR, renal V2R) [169]. Their distinct physiological effects can be accounted for by the low sequence homology of the intracellular domains of the OTR and the AVPRs. In contrast, the transmembrane portion and extracellular domains have high sequence homology. The related docking sites and the structural similarity of OT and AVP create a problem of specificity and result in significant cross reactivity [170, 171]. In fact, receptor specificity is not only controlled by (modest) ligand selectivity, but rather by a complex interplay comprising the enzymes oxytocinase and vasopressinase and by cell-specific up- and down-regulation of individual receptor expression [172]. Interspecies variability further complicates the quest for selective agonist/antagonists: many compounds selective in rat turned out to be unspecific in humans [173]. These factors constitute a major challenge and although thousands of OT and AVP analogues have been synthesized, there remains still a shortage of receptor selective agonists and antagonists, in particular for the human receptors [174–176]. Clinically, OT is administrated intravenously to induce labor and treat post-partum hemorrhage, and intra-nasally to elicit milk letdown; the antagonist atosiban is employed during premature labor. Vasopressin is administrated orally in the form of deamino-D-Arg8-vasopressin (desmopressin) for the treatment of diabetes insipidus and bedwetting; it is also used in coagulation disorders such as von Willebrand disease and mild forms of factor VIII-deficiency [174, 177, 178]. However, all of these ligands have been shown to be non-selective at the human OT and AVP receptors [174, 175]. Other potential applications for selective OT/AVP agonists and antagonists include treatment of congestive heart failure, blood pressure, anxiety, stress, anger, depression, hypotension, hypo-osmolality, hyponatremia and bleeding disorders [174, 179]. Understanding the mechanism of ligand-receptor interaction and the structural motifs involved in binding and GPCR activation is essential for the library design of such a delicate and exigent system. There are significant differences in the design of peptide versus small molecule modulators and the focus will be on peptide library design. However, selective peptide agonist/antagonists represent excellent starting point for further small molecule drug designs.
Fig. (5).
Amino acid sequences of oxytocin and vasopressin.
The amino acid of mature oxytocin and vasopressin peptide hormones is presented. The two differences in each of the peptides are highlighted in blue. Disulfide bonds are indicated.
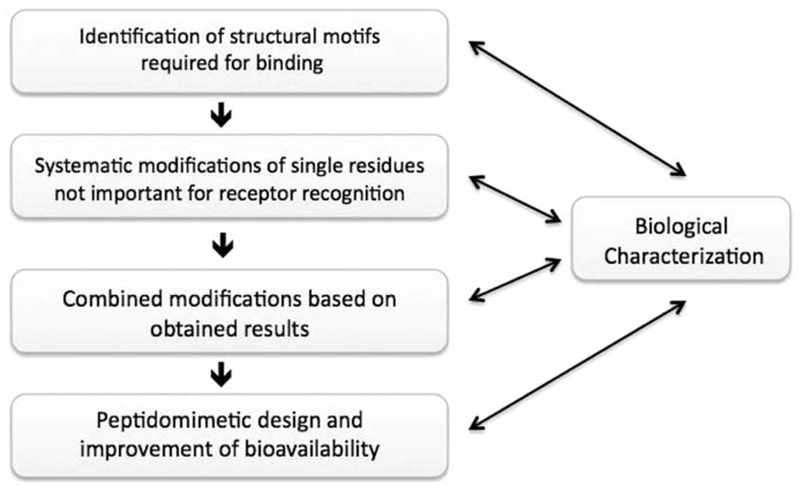
The crystal structure of deamino-OT and NMR structures of OT and AVP showed secondary structure motifs in form of a Type-II β-turn between the backbone of −NH of Tyr 2 and the carbonyl of Asn5 and a Type-III β-turn between the Cys 6 and the Gly 9 imino group [180–183]. Structure-activity relationship and NMR studies further revealed that the disulfide bond is crucial for agonistic activity because it acts as a conformational constraint to limit the flexibility and to align tyrosine in position 2 and the carboxylic tripeptide tail [181, 184]. These observations indicate that binding might occur via a receptor-induced fit, which is further reinforced by the comparison of the deamino-OT crystal structure, (two β-turns) [180] with the crystal structure of OT bound to its carrier-protein neurophysin, where no β-turns were observed [185]. To obtain receptor-specific ligand selectivity it is hence essential for the agonist library design to preserve the recognition motifs and the conformational flexibility of the ring structure. The emphasis is on agonist design as subtle differences exist between the mechanism of agonist and antagonist. To act as an agonist, the ligand needs to undergo receptor recognition followed by binding and conformational changes in both, the ligand and the receptor, which results in a functional response. In contrast, receptor inhibition can result from simple blockage of the agonist binding pocket or by binding to an allosteric site that changes the conformation of the receptor suppressing the functional response. Hence SAR studies relevant for agonists do not necessarily yield results that are predictive for the SAR of antagonists. This has also been realized during the antagonist development for oxytocin and vasopressin receptors [186–188]. Nevertheless the overall methodology of finding selective peptide agonists as well as antagonists remains the same (see Fig. 6).
Fig. (6).
Workflow for the search for GPCR agonists and antagonists.
The identification of agonists and antagonists works generally via the same methodological workflow (1) Identification of the structural motifs required for binding, (2) systematic modifications of single residues not important for receptor recognition, (3) biological characterization, (4) combined modifications guided by the results obtained, (5) biological characterization and (6) peptidomimetic design and improvement of bioavailability.
Modifications include changes in hydrophobicity, charge states, conformational constraints, polarity and aromaticity to extract information on their effect on potency and selectivity to guide the direction of subsequent efforts. For selectivity optimization it is essential to screen against all four receptors of the same species. The information obtained from the single residue mutations series and from the literature is the basis for the next series that incorporates additive changes in more than one position. This cycle of analysis and design is repeated until the desired parameters have been optimized. Use of synthetic/unnatural building blocks is of benefit not only because it significantly extends the chemical diversity around the structural framework important for selectivity, but also because it often results in analogues with improved bioavailability.
High-throughput synthesis and biological screening is required to drive this type of approach. A strategy in combinatorial chemistry was originally developed for the drug discovery pipeline of venom peptides that gives rapid synthetic access to a large quantity of disulfide-rich peptides [189]. This has been adapted for the oxytocin/vasopressin library design. It utilizes a safety-catch acid labile linker (SCAL) that allows complete side-chain deprotection of the assembled peptides without cleaving the peptide from the solid support. The “naked” peptides are folded on-resin and a chemoselective reaction activates the SCAL linker, which releases the folded peptides from the solid support. Depending on the quality of assembly and analogue separation, the folded peptides are either screened directly or purified prior as mixtures by reverse phase HPLC. When combined with highly automated solid-phase peptide synthesis, this approach allows for rapid production of a large number of analogues. Biological screening occurs via radioligand binding assays or functional assays that analyze the intracellular secondary messengers (accumulation of PIP2 or cAMP; Ca2+-ion transients) signal. In this way, hundreds of peptide ligands can be synthesized and characterized efficiently. Selectivity and other pharmacological properties should improve significantly within a few iterative cycles. Once the structural features for binding and selectivity have been identified, it might be possible to further modify the structure to enhance stability against proteolytic degradation, increase the half-life of the peptide in the circulation, improve membrane permeability and/or create a prodrug. There are various well-established approaches such as implementation of peptidomimetic design, truncations, cyclizations and PEGylation that can be applied to improve the bioavailability of the peptide and improve its drug properties.
Summary and Future Perspective
There is a school of thoughts that assumes that all therapeutically useful, readily drugable GPCRs have already been discovered. Future investigations in these areas are then argued to be futile because they are subject to the law of diminishing returns. However, it is evident from Table 1 that several compounds (agomelatine, aprepitant, cinacalcet, exenatide, fingolimod, icatibant, laropripant. maraviroc, rimonabant, sitaxentane) have been approved for clinical pharmacotherapy in the past 5-6 years. In most instances, the cognate receptors of these drugs had previously not been exploited as drug targets. Thus > 10% of the extant drugable GPCRs were discovered recently. It is therefore likely that many more GPCRs will prove useful as therapeutic targets in the future. In addition, GPCRs can be targeted at sites other than the binding site proper, i.e., the orthosteric pocket for the physiological endogenous agonist. These additional sites include (i) binding sites for allosteric modulators of the orthosteric site, (ii) interaction surface with the G proteinsubunits and with the arrestins, (iii) binding sites for accessory proteins (GIPs) that regulate receptor sorting and targeting and (iv) binding sites for pharmacochaperones that promote folding of mutated receptors. In all instances, screening systems can be devised that afford medium- to high-throughput and natural peptide libraries may be a starting point to identify drug candidates and to extract a pharmacophore, followed by optimization of the drugs candidate by combinatorial approaches. We are therefore confident that the family of GPCRs will remain fruitful and prolific for drug discovery.
Acknowledgements
CWG is supported by a “Lise-Meitner” fellowship from the Austrian science fund/FWF. Screening efforts in the authors’ laboratories are supported by the PLACEBO-consortium supported by the Gen-AU program of the Austrian Federal Ministry of Science and Research.
References
References 201-203 are related articles recently published.
- [1].Kroeze WK, Sheffler DJ, Roth BL. G-protein-coupled receptors at a glance. J Cell Sci. 2003;116:4867–9. doi: 10.1242/jcs.00902. [DOI] [PubMed] [Google Scholar]
- [2].Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- [3].Perez DM. From plants to man: the GPCR “tree of life”. Mol Pharmacol. 2005;67:1383–4. doi: 10.1124/mol.105.011890. [DOI] [PubMed] [Google Scholar]
- [4].Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol Sci. 2001;22:368–76. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- [5].Bjarnadottir TK, Gloriam DE, Hellstrand SH, et al. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics. 2006;88:263–73. doi: 10.1016/j.ygeno.2006.04.001. [DOI] [PubMed] [Google Scholar]
- [6].Jacoby E, Bouhelal R, Gerspacher M, et al. The 7TM G-protein-coupled receptor target family. ChemMedChem. 2006;1:760–82. doi: 10.1002/cmdc.200600134. [DOI] [PubMed] [Google Scholar]
- [7].Fredriksson R, Schioth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–25. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- [8].Civelli O, Saito Y, Wang ZW, et al. Orphan GPCRs and their ligands. Pharmacol Ther. 2006;110:525–32. doi: 10.1016/j.pharmthera.2005.10.001. [DOI] [PubMed] [Google Scholar]
- [9].Robas N, O’Reilly M, Katugampola S, et al. Maximizing serendipity: strategies for identifying ligands for orphan G-protein-coupled receptors. Curr Opin Pharmacol. 2003;3:121–6. doi: 10.1016/s1471-4892(03)00010-9. [DOI] [PubMed] [Google Scholar]
- [10].Wise A, Jupe SC, Rees S. The identification of ligands at orphan G-protein coupled receptors. Annu Rev Pharmacol Toxicol. 2004;44:43–66. doi: 10.1146/annurev.pharmtox.44.101802.121419. [DOI] [PubMed] [Google Scholar]
- [11].Harmar AJ, Hills RA, Rosser EM, et al. IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res. 2009;37:D680–D5. doi: 10.1093/nar/gkn728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Horn F, Weare J, Beukers MW, et al. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 1998;26:275–9. doi: 10.1093/nar/26.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Warne T, Serrano-Vega MJ, Baker JG, et al. Structure of a beta(1)-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–U2. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cherezov V, Rosenbaum DM, Hanson MA, et al. High-resolution crystal structure of an engineered human beta(2)-adrenergic G protein-coupled receptor. Science. 2007;318:1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jaakola VP, Griffith MT, Hanson MA, et al. The 2.6 angstrom crystal structure of a human A(2A) adenosine receptor bound to an antagonist. Science. 2008;322:1211–7. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jimonet P, Jaeger R. Strategies for designing GPCR-focused libraries and screening sets. Curr Opin Drug Discov Develop. 2004;7:325–33. [PubMed] [Google Scholar]
- [17].Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. ChemBioChem. 2002;3:928–44. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- [18].Tyndall JDA, Pfeiffer B, Abbenante G, et al. Over One Hundred Peptide-Activated G Protein-Coupled Receptors Recognize Ligands with Turn Structure. Chem Rev. 2005;105:793–826. doi: 10.1021/cr040689g. [DOI] [PubMed] [Google Scholar]
- [19].Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov. 2009;8:41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Leach K, Sexton PM, Christopoulos A. Allosteric GPCR modulators: taking advantage of permissive receptor pharmacology. Trends Pharmacol Sci. 2007;28:382–9. doi: 10.1016/j.tips.2007.06.004. [DOI] [PubMed] [Google Scholar]
- [21].Freissmuth M, Waldhoer M, Bofill-Cardona E, et al. G protein antagonists. Trends Pharmacol Sci. 1999;20:237–45. doi: 10.1016/s0165-6147(99)01337-1. [DOI] [PubMed] [Google Scholar]
- [22].Holler C, Freissmuth M, Nanoff C. G proteins as drug targets. Cell Mol Life Sci. 1999;55:257–70. doi: 10.1007/s000180050288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Falciani C, Lozzi L, Pini A, et al. Bioactive peptides from libraries. Chem Biol. 2005;12:417–26. doi: 10.1016/j.chembiol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- [24].Freissmuth M, Casey PJ, Gilman AG. G-Pproteins control diverse pathways of transmembrane signaling. FASEB J. 1989;3:2125–31. [PubMed] [Google Scholar]
- [25].Gsandtner I, Gruber CW, Freissmuth M. In: Shifting Paradigms in GPCRs. Gilchrist A, editor. 2010. in press. [Google Scholar]
- [26].Sato M, Blumer JB, Simon V, et al. Accessory proteins for G proteins: Partners in signaling. Annu Rev Pharmacol Toxicol. 2006;46:151–87. doi: 10.1146/annurev.pharmtox.46.120604.141115. [DOI] [PubMed] [Google Scholar]
- [27].Hollinger S, Hepler JR. Cellular regulation of RGS proteins: Modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–59. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- [28].Siderovski D, Willard F. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ja WW, Roberts RW. G-protein-directed ligand discovery with peptide combinatorial libraries. Trends Biochem Sci. 2005;30:318–24. doi: 10.1016/j.tibs.2005.04.001. [DOI] [PubMed] [Google Scholar]
- [30].Hildebrandt JD. Role of subunit diversity in signaling by hetero-trimeric G proteins. Biochem Pharmacol. 1997;54:325–39. doi: 10.1016/s0006-2952(97)00269-4. [DOI] [PubMed] [Google Scholar]
- [31].Moore CAC, Milano SK, Benovic JL. Regulation of receptor trafficking by GRKs and arrestins. Annu Rev Physiol. 2007;69:451–82. doi: 10.1146/annurev.physiol.69.022405.154712. [DOI] [PubMed] [Google Scholar]
- [32].Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–7. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- [33].Drake MT, Violin JD, Whalen EJ, et al. beta-Arrestin-biased Agonism at the beta 2-Adrenergic Receptor. J Biol Chem. 2008;283:5669–76. doi: 10.1074/jbc.M708118200. [DOI] [PubMed] [Google Scholar]
- [34].Arttamangkul S, Quillinan N, Low MJ, et al. Differential activation and trafficking of mu-opioid receptors in brain slices. Mol Pharmacol. 2008;74:972–9. doi: 10.1124/mol.108.048512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bockaert J, Dumuis A, Fagni L, et al. GPCR-GIP networks: a first step in the discovery of new therapeutic drugs? Curr Opin Drug Discov Develop. 2004;7:649–57. [PubMed] [Google Scholar]
- [36].Bockaert J, Marin P, Dumuis A, et al. The ‘magic tail’ of G protein-coupled receptors: an anchorage for functional protein networks. FEBS Lett. 2003;546:65–72. doi: 10.1016/s0014-5793(03)00453-8. [DOI] [PubMed] [Google Scholar]
- [37].Bockaert J, Roussignol G, Becamel C, et al. GPCR-interacting proteins (GIPs): nature and functions. Biochem Soc Trans. 2004;32:851–5. doi: 10.1042/BST0320851. [DOI] [PubMed] [Google Scholar]
- [38].Seidel MG, Klinger M, Freissmuth M, et al. Activation of mitogen-activated protein kinase by the A(2A)-adenosine receptor via a rap1-dependent and via a p21(ras)-dependent pathway. J Biol Chem. 1999;274:25833–41. doi: 10.1074/jbc.274.36.25833. [DOI] [PubMed] [Google Scholar]
- [39].Klinger M, Kuhn M, Just H, et al. Removal of the carboxy terminus of the A(2A)-adenosine receptor blunts constitutive activity: differential effect on cAMP accumulation and MAP kinase stimulation. Naunyn-Schmiedeberg’s Arch Pharmacol. 2002;366:287–98. doi: 10.1007/s00210-002-0617-z. [DOI] [PubMed] [Google Scholar]
- [40].Charalambous C, Gsandtner I, Keuerleber E, et al. Restricted collision coupling of the A(2A) receptor revisited - Evidence for physical separation of two signaling cascades. J Biol Chem. 2008;283:9276–88. doi: 10.1074/jbc.M706275200. [DOI] [PubMed] [Google Scholar]
- [41].Gsandtner I, Charalambous C, Stefan E, et al. Heterotrimeric G protein-independent signaling of a G protein-coupled receptor. J Biol Chem. 2005;280:31898–905. doi: 10.1074/jbc.M506515200. [DOI] [PubMed] [Google Scholar]
- [42].Becamel C, Alonso G, Galeotti N, et al. Synaptic multiprotein complexes associated with 5-HT(2C) receptors: a proteomic approach. EMBO J. 2002;21:2332–42. doi: 10.1093/emboj/21.10.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Maurice P, Daulat AM, Broussard C, et al. A generic approach for the purification of signaling complexes that specifically interact with the carboxyl-terminal domain of G protein-coupled receptors. Mol Cell Proteomics. 2008;7:1556–69. doi: 10.1074/mcp.M700435-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Daulat AM, Maurice P, Froment C, et al. Purification and identification of G protein-coupled receptor protein complexes under native conditions. Mol Cell Proteomics. 2007;6:835–44. doi: 10.1074/mcp.M600298-MCP200. [DOI] [PubMed] [Google Scholar]
- [45].Enz R. The trick of the tail: protein-protein interactions of metabotropic glutamate receptors. BioEssays. 2007;29:60–73. doi: 10.1002/bies.20518. [DOI] [PubMed] [Google Scholar]
- [46].El-Agnaf OMA, Paleologou KE, Greer B, et al. A strategy for designing inhibitors of alpha-synuclein aggregation and toxicity as a novel treatment for Parkinson’s disease and related disorders. FASEB J. 2004;18:1315–+. doi: 10.1096/fj.03-1346fje. [DOI] [PubMed] [Google Scholar]
- [47].Nagai Y, Fujikake N, Ohno K, et al. Prevention of polyglutamine oligomerization and neurodegeneration by the peptide inhibitor QBP1 in Drosophila. Hum Mol Genet. 2003;12:1253–9. doi: 10.1093/hmg/ddg144. [DOI] [PubMed] [Google Scholar]
- [48].Ayoub MA, Damian M, Gespach C, et al. Inhibition of heterotrimeric G protein signaling by a small molecule acting on Galpha subunit. J Biol Chem. 2009;284:29136–45. doi: 10.1074/jbc.M109.042333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Breitweg-Lehmann E, Czupalla C, Storm R, et al. Activation and inhibition of G proteins by lipoamines. Mol Pharmacol. 2002;61:628–36. doi: 10.1124/mol.61.3.628. [DOI] [PubMed] [Google Scholar]
- [50].Johnston CA, Willard FS, Ramer JK, et al. State-selective binding peptides for heterotrimeric G-protein subunits: novel tools for investigating G-protein signaling dynamics. Comb Chem High Throughput Scr. 2008;11:370–81. doi: 10.2174/138620708784534798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kimple AJ, Yasgar A, Hughes M, et al. A high throughput fluorescence polarization assay for inhibitors of the GoLoco motif/G-alpha interaction. Comb Chem High Throughput Scr. 2008;11:396–409. doi: 10.2174/138620708784534770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Marshall GR. Peptide interactions with G-protein coupled receptors. Biopolymers. 2001;60:246–77. doi: 10.1002/1097-0282(2001)60:3<246::AID-BIP10044>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- [53].Horton DA, Bourne GT, Smythe ML. The combinatorial synthesis of bicyclic privileged structures or privileged substructures. Chem Rev. 2003;103:893–930. doi: 10.1021/cr020033s. [DOI] [PubMed] [Google Scholar]
- [54].Sherwood NM, Krueckl SL, McRory JE. The origin and function of the pituitary adenylate cyclase-activating polypeptide (PACAP)/glucagon superfamily. Endocr Rev. 2000;21:619–70. doi: 10.1210/edrv.21.6.0414. [DOI] [PubMed] [Google Scholar]
- [55].Grishin NV. Fold change in evolution of protein structures. J Struct Biol. 2001;134:167–85. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- [56].Gante J. Peptide mimetics - tailor-made enzyme inhibitors. Angew Chem Int Ed Engl. 1994;1733:1699–720. [Google Scholar]
- [57].Hanessian S, McNaughton-Smith G, Lombart H-G, et al. Design and synthesis of conformationally constrained amino acids as versatile scaffolds and peptide mimetics. Tetrahedron. 1997;53:12789–854. [Google Scholar]
- [58].Hruby VJ, Li G, Haskell-Luevano C, et al. Design of peptides, proteins, and peptidomimetics in chi space. Biopolymers. 1997;43:219–66. doi: 10.1002/(SICI)1097-0282(1997)43:3<219::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- [59].Fairlie DP. 223rd ACS National Meeting Orlando; FL, United States. 2002. pp. ORGN–411. [Google Scholar]
- [60].Evans BE, Rittle KE, Bock MG, et al. Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J Med Chem. 1988;31:2235–46. doi: 10.1021/jm00120a002. [DOI] [PubMed] [Google Scholar]
- [61].Horton DA, Bourne GT, Smythe ML. Exploring privileged structures: The combinatorial synthesis of cyclic peptides. Mol Divers. 2002;5:289–304. doi: 10.1023/a:1021365402751. [DOI] [PubMed] [Google Scholar]
- [62].DeSimone RW, Currie KS, Mitchell SA, et al. Privileged structures: Applications in drug discovery. Comb Chem High Throughput Scr. 2004;7:473–93. doi: 10.2174/1386207043328544. [DOI] [PubMed] [Google Scholar]
- [63].Guo T, Hobbs DW. Privileged structure-based combinatorial libraries targeting G protein-coupled receptors. Assay Drug Dev Technol. 2003;1:579–92. doi: 10.1089/154065803322302835. [DOI] [PubMed] [Google Scholar]
- [64].Costantino L, Barlocco D. Privileged structures as leads in medicinal chemistry. Curr Med Chem. 2006;13:65–85. [PubMed] [Google Scholar]
- [65].Fecik RA, Frank KE, Gentry EJ, et al. The search for orally active medications through combinatorial chemistry. Med Res Rev. 1998;18:149–85. doi: 10.1002/(sici)1098-1128(199805)18:3<149::aid-med2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- [66].Ripka WC, De Lucca GV, Bach AC, II, et al. Protein beta -turn mimetics. I. Design, synthesis, and evaluation in model cyclic peptides. Tetrahedron. 1993;49:3593–608. [Google Scholar]
- [67].Newman DJ. Natural Products as Leads to Potential Drugs: An Old Process or the New Hope for Drug Discovery? J Med Chem. 2008;51:2589–99. doi: 10.1021/jm0704090. [DOI] [PubMed] [Google Scholar]
- [68].Che Y, Marshall GR. Privileged scaffolds targeting reverse-turn and helix recognition. Expert Opin Ther Targets. 2008;12:101–14. doi: 10.1517/14728222.12.1.101. [DOI] [PubMed] [Google Scholar]
- [69].Ortholand JY, Ganesan A. Natural products and combinatorial chemistry: back to the future. Curr Opin Chem Biol. 2004;8:271–80. doi: 10.1016/j.cbpa.2004.04.011. [DOI] [PubMed] [Google Scholar]
- [70].Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–77. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- [71].Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci. 2001;41:856–64. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- [72].Lipinski C, Hopkins A. Navigating chemical space for biology and medicine. Nature. 2004;432:855–61. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- [73].Dobson CM. Chemical space and biology. Nature. 2004;432:824–8. doi: 10.1038/nature03192. [DOI] [PubMed] [Google Scholar]
- [74].Wilks HM, Cortes A, Emery DC, et al. Opportunities and limits in creating new enzymes: experiences with the NAD-dependent lactate dehydrogenase frameworks of humans and bacteria. Ann N Y Acad Sci. 1992;672:80–93. doi: 10.1111/j.1749-6632.1992.tb32662.x. [DOI] [PubMed] [Google Scholar]
- [75].Smith JM. Natural selection and the concept of a protein space. Nature. 1970;225:563–4. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- [76].Russell RB, Sasieni PD, Sternberg MJE. Supersites within superfolds. Binding site similarity in the absence of homology. J Mol Biol. 1998;282:903–18. doi: 10.1006/jmbi.1998.2043. [DOI] [PubMed] [Google Scholar]
- [77].Kaiser M, Wetzel S, Kumar K, et al. Biology-inspired synthesis of compound libraries. Cell Mol Life Sci. 2008;65:1186–201. doi: 10.1007/s00018-007-7492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Haubner R, Schmitt W, Hoelzemann G, et al. Cyclic RGD peptides containing beta -turn mimetics. J Am Chem Soc. 1996;118:7881–91. [Google Scholar]
- [79].Pfaff M, Tangemann K, Mueller B, et al. Selective recognition of cyclic RGD peptides of NMR defined conformation by alpha IIb-beta 3, alpha Vbeta 3, and alpha 5beta 1 integrins. J Biol Chem. 1994;269:20233–8. [PubMed] [Google Scholar]
- [80].Porcelli M, Casu M, Lai A, et al. Cyclic pentapeptides of chiral sequence DLDDL as scaffold for antagonism of G-protein coupled receptors: synthesis, activity and conformational analysis by NMR and molecular dynamics of ITF 1565, a substance P inhibitor. Biopolymers. 1999;50:211–9. doi: 10.1002/(SICI)1097-0282(199908)50:2<211::AID-BIP10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- [81].Haskell-Luevano C, Rosenquist A, Souers A, et al. Compounds that activate the mouse melanocortin-1 receptor identified by screening a small molecule library based upon the beta -turn. J Med Chem. 1999;42:4380–7. doi: 10.1021/jm990190s. [DOI] [PubMed] [Google Scholar]
- [82].Becker JAJ, Wallace A, Garzon A, et al. Ligands for κ-opioid and ORL1 receptors identified from a conformationally constrained peptide combinatorial library. J Biol Chem. 1999;274:27513–22. doi: 10.1074/jbc.274.39.27513. [DOI] [PubMed] [Google Scholar]
- [83].Cherezov V, Rosenbaum DM, Hanson MA, et al. High-resolution crystal structure of an engineered human beta(2)-adrenergic G protein-coupled receptor. Science. 2007;318:1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Nygaard R, Frimurer TM, Holst B, et al. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol Sci. 2009;30:249–59. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- [85].Palczewski K, Kumasaka T, Hori T, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- [86].Rasmussen SGF, Choi HJ, Rosenbaum DM, et al. Crystal structure of the human beta(2) adrenergic G-protein-coupled receptor. Nature. 2007;450:383–4. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- [87].Rosenbaum DM, Cherezov V, Hanson MA, et al. GPCR engineering yields high-resolution structural insights into beta(2)-adrenergic receptor function. Science. 2007;318:1266–73. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- [88].Michino M, Abola E, Participants GD, et al. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat Rev Drug Discov. 2009 doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Turk BE, Cantley LC. Peptide libraries: at the crossroads of proteomics and bioinformatics. Curr Opin Chem Biol. 2003;7:84–90. doi: 10.1016/s1367-5931(02)00004-2. [DOI] [PubMed] [Google Scholar]
- [90].Lam KS, Lebl M, Krchnak V. The “one-bead-one-compound” combinatorial library method. Chem Rev. 1997;97:411–48. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- [91].Taylor JM, Neubig RR. Peptides as probes for G protein signal transduction. Cell Signal. 1994;6:841–9. doi: 10.1016/0898-6568(94)90017-5. [DOI] [PubMed] [Google Scholar]
- [92].Houghten RA, Pinilla C, Blondelle SE, et al. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature. 1991;354:84–6. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- [93].Lam KS, Salmon SE, Hersh EM, et al. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–4. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- [94].Mersich C, Jungbauer A. International symposium for biochromatography and nanotechnologies; Vellore, INDIA. 2007. pp. 160–70. [Google Scholar]
- [95].Takahashi TT, Austin RJ, Roberts RW. mRNA display: ligand discovery, interaction analysis and beyond. Trends Biochem Sci. 2003;28:159–65. doi: 10.1016/S0968-0004(03)00036-7. [DOI] [PubMed] [Google Scholar]
- [96].Jezova D, Skultetyova I, Tokarev DI, et al. Vasopressin and oxytocin in stress. Ann N Y Acad Sci. 1995;771:192–203. doi: 10.1111/j.1749-6632.1995.tb44681.x. [DOI] [PubMed] [Google Scholar]
- [97].Jayawickreme CK, Quillan JM, Graminski GF, et al. Discovery and structure-function analysis of alpha-melanocyte-stimulating hormone antagonists. J Biol Chem. 1994;269:29846–54. [PubMed] [Google Scholar]
- [98].Dooley CT, Ny P, Bidlack JM, et al. Selective ligands for the mu, delta, and Kappa opioid receptors identified from a single mixture based tetrapeptide positional scanning combinatorial library. J Biol Chem. 1998;273:18848–56. doi: 10.1074/jbc.273.30.18848. [DOI] [PubMed] [Google Scholar]
- [99].Sasaki S, Takagi M, Tanaka Y, et al. A new application of a peptide library to identify selective interaction between small peptides in an attempt to develop recognition molecules toward protein surfaces. Tetrahedron Lett. 1996;37:85–8. [Google Scholar]
- [100].Lewis RJ, Garcia ML. Therapeutic potential of venom peptides. Nat Rev Drug Discov. 2003;2:790–802. doi: 10.1038/nrd1197. [DOI] [PubMed] [Google Scholar]
- [101].Ganesan A. The impact of natural products upon modern drug discovery. Curr Opin Chem Biol. 2008;12:306–17. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- [102].Escoubas P. Molecular diversification in spider venoms: A web of combinatorial peptide libraries. Mol Divers. 2006;10:545–54. doi: 10.1007/s11030-006-9050-4. [DOI] [PubMed] [Google Scholar]
- [103].Escoubas P, Sollod B, King GF. Venom landscapes: Mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon. 2006;47:650–63. doi: 10.1016/j.toxicon.2006.01.018. [DOI] [PubMed] [Google Scholar]
- [104].Possani LD, Becerril B, Delepierre M, et al. Scorpion toxins specific for Na+-channels. Eur J Biochem. 1999;264:287–300. doi: 10.1046/j.1432-1327.1999.00625.x. [DOI] [PubMed] [Google Scholar]
- [105].Fox JW, Serrano SMT. Approaching the golden age of natural product pharmaceuticals from venom libraries: An overview of toxins and toxin-derivatives currently involved in therapeutic or diagnostic applications. Curr Pharm Des. 2007;13:2927–34. doi: 10.2174/138161207782023739. [DOI] [PubMed] [Google Scholar]
- [106].Tsetlin V. Snake venom alpha-neurotoxins and other ‘three-finger’ proteins. Eur J Biochem. 1999;264:281–6. doi: 10.1046/j.1432-1327.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- [107].Theakston RD, Kamiguti AS. A list of animal toxins and some other natural products with biological activity. Toxicon. 2002;40:579–651. doi: 10.1016/s0041-0101(01)00244-6. [DOI] [PubMed] [Google Scholar]
- [108].Alewood P, Hopping G, Armishaw C. Marine toxins as sources of drug leads. Aust J Chem. 2003;56:769–74. [Google Scholar]
- [109].Norton RS, Olivera BM. In 6th asia/pacific congress on animals, plant and microbial toxins/11th annual scientific meeting of the australiasian-college-of-tropical-medicine; Cairns, Australia. 2002. pp. 780–98. [Google Scholar]
- [110].Olivera BM, Rivier J, Clark C, et al. Diversity of conus neuropeptides. Science. 1990;249:257–63. doi: 10.1126/science.2165278. [DOI] [PubMed] [Google Scholar]
- [111].Armishaw CJ, Alewood PF. Conotoxins as research tools and drug leads. Curr Protein Pept Sci. 2005;6:221–40. doi: 10.2174/1389203054065437. [DOI] [PubMed] [Google Scholar]
- [112].Livett BG, Sandall DW, Keays D, et al. Therapeutic applications of conotoxins that target the neuronal nicotinic acetylcholine receptor. Toxicon. 2006;48:810–29. doi: 10.1016/j.toxicon.2006.07.023. [DOI] [PubMed] [Google Scholar]
- [113].Callaghan B, Haythornthwaite A, Berecki G, et al. Analgesic alpha -conotoxins Vc1.1 and Rg1A inhibit N-type calcium channels in rat sensory neurons via GABAB receptor activation. J Neurosci. 2008;28:10943–51. doi: 10.1523/JNEUROSCI.3594-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Nielsen DB, Dykert J, Rivier JE, et al. Isolation of Lys-conopressin-G from the venom of the worm-hunting snail, Conus imperialis. Toxicon. 1994;32:845–8. doi: 10.1016/0041-0101(94)90009-4. [DOI] [PubMed] [Google Scholar]
- [115].Moeller C, Mari F. A vasopressin/oxytocin-related conopeptide with gamma -carboxyglutamate at position 8. Biochem J. 2007;404:413–9. doi: 10.1042/BJ20061480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Cruz LJ, de Santos V, Zafaralla GC, et al. Invertebrate vasopressin/oxytocin homologs. Characterization of peptides from Conus geographus and Conus straitus venoms. J Biol Chem. 1987;262:15821–4. [PubMed] [Google Scholar]
- [117].Dutertre S, Croker D, Daly NL, et al. Conopressin-T from conus tulipa reveals an antagonist switch in vasopressin-like peptides. J Biol Chem. 2008;283:7100–8. doi: 10.1074/jbc.M706477200. [DOI] [PubMed] [Google Scholar]
- [118].Hoyle CHV. Neuropeptide families and their receptors: evolutionary perspectives. Brain Res. 1999;848:1–25. doi: 10.1016/s0006-8993(99)01975-7. [DOI] [PubMed] [Google Scholar]
- [119].Craik DJ. Chemistry. Seamless proteins tie up their loose ends. Science. 2006;311:1563–4. doi: 10.1126/science.1125248. [DOI] [PubMed] [Google Scholar]
- [120].Craik DJ, Daly NL, Bond T, et al. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294:1327–36. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- [121].Craik DJ, Daly NL, Mulvenna J, et al. Discovery, structure and biological activities of the cyclotides. Curr Protein Pept Sci. 2004;5:297–315. doi: 10.2174/1389203043379512. [DOI] [PubMed] [Google Scholar]
- [122].Gruber CW, Elliott AG, Ireland DC, et al. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell. 2008;20:2471–83. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Colgrave ML, Craik DJ. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–75. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- [124].Simonsen SM, Sando L, Ireland DC, et al. A Continent of plant defense peptide diversity: cyclotides in australian hybanthus (Violaceae) Plant Cell. 2005;17:3176–89. doi: 10.1105/tpc.105.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Mulvenna JP, Mylne JS, Bharathi R, et al. Discovery of cyclotide-like protein sequences in graminaceous crop plants: ancestral precursors of circular proteins? Plant Cell. 2006;18:2134–44. doi: 10.1105/tpc.106.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Trabi M, Svangard E, Herrmann A, et al. Variations in cyclotide expression in viola species. J Nat Prod. 2004;67:806–10. doi: 10.1021/np034068e. [DOI] [PubMed] [Google Scholar]
- [127].Plan MRR, Göransson U, Clark RJ, et al. The cyclotide fingerprint in oldenlandia affinis: elucidation of chemically modified, linear and novel macrocyclic peptides. ChemBioChem. 2007;8:1001–11. doi: 10.1002/cbic.200700097. [DOI] [PubMed] [Google Scholar]
- [128].Seydel P, Gruber CW, Craik DJ, et al. Formation of cyclotides and variations in cyclotide expression in oldenlandia affinis suspension cultures. Appl Microbiol Biotechnol. 2007;77:275–84. doi: 10.1007/s00253-007-1159-6. [DOI] [PubMed] [Google Scholar]
- [129].Clark RJ, Daly NL, Craik DJ. Structural plasticity of the cyclic-cystine-knot framework: implications for biological activity and drug design. Biochem J. 2006;394:85–93. doi: 10.1042/BJ20051691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Gunasekera S, Foley FM, Clark RJ, et al. Engineering stabilized vascular endothelial growth factor-A antagonists: synthesis, structural characterization, and bioactivity of grafted analogues of cyclotides. J Med Chem. 2008;51:7697–704. doi: 10.1021/jm800704e. [DOI] [PubMed] [Google Scholar]
- [131].Thongyoo P, Roque-Rosell N, Leatherbarrow RJ, et al. Chemical and biomimetic total syntheses of natural and engineered MCoTI cyclotides. Org Biomol Chem. 2008;6:1462–70. doi: 10.1039/b801667d. [DOI] [PubMed] [Google Scholar]
- [132].Craik DJ, Cemazar M, Daly NL. The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr Opin Drug Discov Develop. 2006;9:251–60. [PubMed] [Google Scholar]
- [133].Craik DJ, Mylne JS, Daly NL. Cyclotides: macrocyclic peptides with applications in drug design and agriculture. Cell Mol Life Sci. 2009 doi: 10.1007/s00018-009-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Henriques ST, Craik DJ. Cyclotides as templates in drug design. Drug Discov Today. 2009 doi: 10.1016/j.drudis.2009.10.007. [DOI] [PubMed] [Google Scholar]
- [135].Gran L, Sandberg F, Sletten K. Oldenlandia affinis (R&S) DC. A plant containing uteroactive peptides used in African traditional medicine. J Ethnopharmacol. 2000;70:197–203. doi: 10.1016/s0378-8741(99)00175-0. [DOI] [PubMed] [Google Scholar]
- [136].Schöpke T, Hasan Agha MI, Kraft R, et al. Hämolytisch aktive komponenten aus viola tricolor 1. und viola arvensis murray. Sci Pharm. 1993;61:145–53. [Google Scholar]
- [137].Witherup KM, Bogusky MJ, Anderson PS, et al. Cyclopsychotride A, A biologically active, 31-residue cyclic peptide isolated from psychotria longipes. J Nat Prod. 1994;57:1619–25. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- [138].Gustafson KR, McKee TC, Bokesch HR. Anti-HIV cyclotides. Curr Protein Pept Sci. 2004;5:331–40. doi: 10.2174/1389203043379468. [DOI] [PubMed] [Google Scholar]
- [139].Lindholm P, Goransson U, Johansson S, et al. Cyclotides: A novel type of cytotoxic agents. Mol Cancer Ther. 2002;1:365–9. [PubMed] [Google Scholar]
- [140].Tam JP, Lu YA, Yang JL, et al. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc Natl Acad Sci USA. 1999;96:8913–8. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Göransson U, Sjogren M, Svangard E, et al. Reversible antifouling effect of the cyclotide cycloviolacin O2 against barnacles. J Nat Prod. 2004;67:1287–90. doi: 10.1021/np0499719. [DOI] [PubMed] [Google Scholar]
- [142].Gruber CW, Cemazar M, Anderson MA, et al. Insecticidal plant cyclotides and related cystine knot toxins. Toxicon. 2007;49:561–75. doi: 10.1016/j.toxicon.2006.11.018. [DOI] [PubMed] [Google Scholar]
- [143].Jennings C, West J, Waine C, et al. Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Old-enlandia affinis. Proc Natl Acad Sci USA. 2001;98:10614–9. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Jennings CV, Rosengren KJ, Daly NL, et al. Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist: do Mobius strips exist in nature? Biochemistry. 2005;44:851–60. doi: 10.1021/bi047837h. [DOI] [PubMed] [Google Scholar]
- [145].Barbeta BL, Marshall AT, Gillon AD, et al. Plant cyclotides disrupt epithelial cells in the midgut of lepidopteran larvae. Proc Natl Acad Sci USA. 2008;105:1221–5. doi: 10.1073/pnas.0710338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Gran L. On the effect of a polypeptide isolated from “Kalata-Kalata” (Oldenlandia affinis DC) on the oestrogen dominated uterus. Acta Pharmacol Toxicol (Copenh) 1973;33:400–8. doi: 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- [147].Gran L, Sletten K, Skjeldal L. Cyclic peptides from oldenlandia affinis DC. Molecular and biological properties. Chem Biodivers. 2008;5:2014–22. doi: 10.1002/cbdv.200890184. [DOI] [PubMed] [Google Scholar]
- [148].Rosengren KJ, Daly NL, Plan MR, et al. Twists, knots, and rings in proteins - structural definition of the cyclotide framework. J Biol Chem. 2003;278:8606–16. doi: 10.1074/jbc.M211147200. [DOI] [PubMed] [Google Scholar]
- [149].Mulvenna JP, Sando L, Craik DJ. Processing of a 22 kDa precursor protein to produce the circular protein tricyclon A. Structure. 2005;13:691–701. doi: 10.1016/j.str.2005.02.013. [DOI] [PubMed] [Google Scholar]
- [150].Antonelli T, Fuxe K, Tomasini MC, et al. Neurotensin receptor mechanisms and its modulation of glutamate transmission in the brain relevance for neurodegenerative diseases and their treatment. Prog Neurobiol. 2007;83:92–109. doi: 10.1016/j.pneurobio.2007.06.006. [DOI] [PubMed] [Google Scholar]
- [151].Craig AG, Norberg T, Griffin D, et al. Contulakin-G, an O-glycosylated invertebrate neurotensin. J Biol Chem. 1999;274:13752–9. doi: 10.1074/jbc.274.20.13752. [DOI] [PubMed] [Google Scholar]
- [152].Nishimori K, Young LJ, Guo Q, et al. Oxytocin is required for nursing but is not essential for parturition or reproductive behavior. Proc Natl Acad Sci USA. 1996;93:11699–704. doi: 10.1073/pnas.93.21.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Veenema AH, Neumann ID. Central vasopressin and oxytocin release: regulation of complex social behaviours. Prog Brain Res. 2008;170:261–76. doi: 10.1016/S0079-6123(08)00422-6. [DOI] [PubMed] [Google Scholar]
- [154].Zak PJ, Stanton AA, Ahmadi S. Oxytocin increases generosity in humans. PLoS One. 2007;2:e1128. doi: 10.1371/journal.pone.0001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Kosfeld M, Heinrichs M, Zak PJ, et al. Oxytocin increases trust in humans. Nature. 2005;435:673–6. doi: 10.1038/nature03701. [DOI] [PubMed] [Google Scholar]
- [156].Neumann ID. Brain oxytocin: a key regulator of emotional and social behaviours in both females and males. J Neuroendocrinol. 2008;20:858–65. doi: 10.1111/j.1365-2826.2008.01726.x. [DOI] [PubMed] [Google Scholar]
- [157].Donaldson ZR, Young LJ. Oxytocin, vasopressin, and the neurogenetics of sociality. Science. 2008;322:900–4. doi: 10.1126/science.1158668. [DOI] [PubMed] [Google Scholar]
- [158].Gimpl G, Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol Rev. 2001;81:629–83. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- [159].Argiolas A, Gessa GL. Central functions of oxytocin. Neurosci Biobehav Rev. 1991;15:217–31. doi: 10.1016/s0149-7634(05)80002-8. [DOI] [PubMed] [Google Scholar]
- [160].Bielsky IF, Young LJ. Oxytocin, vasopressin, and social recognition in mammals. Peptides. 2004;25:1565–74. doi: 10.1016/j.peptides.2004.05.019. [DOI] [PubMed] [Google Scholar]
- [161].McCarthy MM, McDonald CH, Brooks PJ, et al. An anxiolytic action of oxytocin is enhanced by estrogen in the mouse. Physiol Behav. 1996;60:1209–15. doi: 10.1016/s0031-9384(96)00212-0. [DOI] [PubMed] [Google Scholar]
- [162].Holmes Cheryl L, Landry Donald W, Granton John T. Science Review: Vasopressin and the cardiovascular system part 2 - clinical physiology. Crit Care. 2004;8:15–23. doi: 10.1186/cc2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Holmes Cheryl L, Landry Donald W, Granton John T. Science review: Vasopressin and the cardiovascular system part 1--receptor physiology. Crit Care. 2003;7:427–34. doi: 10.1186/cc2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Alescio-Lautier B, Soumireu-Mourat B. Role of vasopressin in learning and memory in the hippocampus. Prog Brain Res. 1998;119:501–21. doi: 10.1016/s0079-6123(08)61590-3. [DOI] [PubMed] [Google Scholar]
- [165].Guelpinar MA, Yegen BC. The physiology of learning and memory: Role of peptides and stress. Curr Prot Peptide Sci. 2004;5:457–73. doi: 10.2174/1389203043379341. [DOI] [PubMed] [Google Scholar]
- [166].Landgraf R. The involvement of the vasopressin system in stress-related disorders. CNS Neurol Disord: Drug Targets. 2006;5:167–79. doi: 10.2174/187152706776359664. [DOI] [PubMed] [Google Scholar]
- [167].Wersinger SR, Ginns EI, O’Carroll AM, et al. Vasopressin V1b receptor knockout reduces aggressive behavior in male mice. Mol Psychiatry. 2002;7:975–84. doi: 10.1038/sj.mp.4001195. [DOI] [PubMed] [Google Scholar]
- [168].Ferris CF, Melloni RH, Jr, Koppel G, et al. Vasopressin/serotonin interactions in the anterior hypothalamus control aggressive behavior in golden hamsters. J Neurosci. 1997;17:4331–40. doi: 10.1523/JNEUROSCI.17-11-04331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Barberis C, Mouillac B, Durroux T. Structural bases of vaso-pressin/oxytocin receptor function. J Endocrinol. 1998;156:223–9. doi: 10.1677/joe.0.1560223. [DOI] [PubMed] [Google Scholar]
- [170].Chini B, Mouillac B, Balestre M-N, et al. Two aromatic residues regulate the response of the human oxytocin receptor to the partial agonist arginine vasopressin. FEBS Lett. 1996;397:201–6. doi: 10.1016/s0014-5793(96)01135-0. [DOI] [PubMed] [Google Scholar]
- [171].Kimura T, Makino Y, Saji F, et al. Molecular characterization of a cloned human oxytocin receptor. Eur J Endocrinol. 1994;131:385–90. doi: 10.1530/eje.0.1310385. [DOI] [PubMed] [Google Scholar]
- [172].Mitchell BF, Fang X, Wong S. Oxytocin: a paracrine hormone in the regulation of parturition? Rev Reprod. 1998;3:113–22. doi: 10.1530/ror.0.0030113. [DOI] [PubMed] [Google Scholar]
- [173].Saito M, Tahara A, Sugimoto T. 1-desamino-8-D-arginine vasopressin (DDAVP) as an agonist on V1b vasopressin receptor. Biochem Pharmacol. 1997;53:1711–7. doi: 10.1016/s0006-2952(97)00070-1. [DOI] [PubMed] [Google Scholar]
- [174].Manning M, Stoev S, Chini B, et al. Peptide and non-peptide agonists and antagonists for the vasopressin and oxytocin V1a, V1b, V2 and OT receptors: research tools and potential therapeutic agents. Prog Brain Res. 2008;170:473–512. doi: 10.1016/S0079-6123(08)00437-8. [DOI] [PubMed] [Google Scholar]
- [175].Chini B, Manning M, Guillon G. Affinity and efficacy of selective agonists and antagonists for vasopressin and oxytocin receptors: an “easy guide” to receptor pharmacology. Prog Brain Res. 2008;170:513–7. doi: 10.1016/S0079-6123(08)00438-X. [DOI] [PubMed] [Google Scholar]
- [176].Chini B, Manning M. Agonist selectivity in the oxytocin/vasopressin receptor family: new insights and challenges. Biochem Soc Trans. 2007;35:737–41. doi: 10.1042/BST0350737. [DOI] [PubMed] [Google Scholar]
- [177].Fewtrell MS, Loh KL, Blake A, et al. Randomised, double blind trial of oxytocin nasal spray in mothers expressing breast milk for preterm infants. Arch Dis Child Fetal Neonatal Ed. 2006;91:F169–74. doi: 10.1136/adc.2005.081265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Janknegt RA, Zweers MM, Delaere KPJ, et al. Oral desmopressin as a new treatment modality for primary nocturnal enuresis in adolescents and adults: a double-blind, randomized, multicenter study. J Urol. 1997;157:513–7. [PubMed] [Google Scholar]
- [179].Treschan TA, Peters J. The vasopressin system: physiology and clinical strategies. Anesthesiology. 2006;105:599–612. doi: 10.1097/00000542-200609000-00026. [DOI] [PubMed] [Google Scholar]
- [180].Wood SP, Tickle IJ, Treharne AM, et al. Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding. Science. 1986;232:633–6. doi: 10.1126/science.3008332. [DOI] [PubMed] [Google Scholar]
- [181].Bhaskaran R, Chuang LC, Yu C. Conformational properties of oxytocin in dimethyl sulfoxide solution: NMR and restrained molecular dynamics studies. Biopolymers. 1992;32:1599–608. doi: 10.1002/bip.360321203. [DOI] [PubMed] [Google Scholar]
- [182].Iwadate M, Nagao E, Williamson MP, et al. Structure determination of [Arg8]vasopressin methylenedithioether in dimethyl sulfoxide using NMR. Eur J Biochem. 2000;267:4504–10. doi: 10.1046/j.1432-1327.2000.01500.x. [DOI] [PubMed] [Google Scholar]
- [183].Schmidt JM, Ohlenschlaeger O, Rueterjans H, et al. Conformation of [8-arginine]vasopressin and V1 antagonists in dimethyl sulfoxide solution derived from two-dimensional NMR spectroscopy and molecular dynamics simulation. Eur J Biochem. 1991;201:355–71. doi: 10.1111/j.1432-1033.1991.tb16293.x. [DOI] [PubMed] [Google Scholar]
- [184].Meraldi JP, Hruby VJ, Brewster AIR. Relative conformational rigidity in oxytocin and [1-penicillamine]oxytocin: A proposal for the relation of conformational flexibility to peptide hormone agonism and antagonism. Proc Natl Acad Sci USA. 1977;74:1373–7. doi: 10.1073/pnas.74.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Rose JP, Wu C-K, Hsiao C-D, et al. Crystal structure of the neurophysin-oxytocin complex. Nat Struct Biol. 1996;3:163–9. doi: 10.1038/nsb0296-163. [DOI] [PubMed] [Google Scholar]
- [186].Hruby VJ. Implications of the x-ray structure of deamino-oxytocin to agonist/antagonist-receptor interactions. Trends Pharmacol Sci. 1987;8:336–9. [Google Scholar]
- [187].Hruby VJ. Conformational and topographical considerations in the design of biologically active peptides. Biopolymers. 1993;33:1073–82. doi: 10.1002/bip.360330709. [DOI] [PubMed] [Google Scholar]
- [188].Hruby VJ. Designing peptide receptor agonists and antagonists. Nat Rev Drug Discov. 2002;1:847–58. doi: 10.1038/nrd939. [DOI] [PubMed] [Google Scholar]
- [189].Brust A, Tickle AE. High-throughput synthesis of conopeptides: a safety-catch linker approach enabling disulfide formation in 96-well format. J Pept Sci. 2007;13:133–41. doi: 10.1002/psc.825. [DOI] [PubMed] [Google Scholar]
- [190].Adermann K, John H, Standker L, et al. Exploiting natural peptide diversity: novel research tools and drug leads. Curr Opin Biotechnol. 2004;15:599–606. doi: 10.1016/j.copbio.2004.10.007. [DOI] [PubMed] [Google Scholar]
- [191].Brown KL, Hancock REW. Cationic host defense (antimicrobial) peptides. Curr Opin Immunol. 2006;18:24–30. doi: 10.1016/j.coi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- [192].Bulet P, Stocklin R, Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol Rev. 2004;198:169–84. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- [193].Lay FT, Anderson MA. Defensins - Components of the innate immune system in plants. Curr Prot Peptide Sci. 2005;6:85–101. doi: 10.2174/1389203053027575. [DOI] [PubMed] [Google Scholar]
- [194].Gauldie J, Hanson JM, Rumjanek FD, et al. Peptide components of bee venom. Eur J Biochem. 1976;61:369–76. doi: 10.1111/j.1432-1033.1976.tb10030.x. [DOI] [PubMed] [Google Scholar]
- [195].Monteiro MC, Romao PR, Soares AM. Pharmacological perspectives of wasp venom. Protein Pept Lett. 2009;16:944–52. doi: 10.2174/092986609788923275. [DOI] [PubMed] [Google Scholar]
- [196].Pimenta AMC, De Lima ME. Small peptides, big world: biotechnological potential in neglected bioactive peptides from arthropod venoms. J Pept Sci. 2005;11:670–6. doi: 10.1002/psc.701. [DOI] [PubMed] [Google Scholar]
- [197].King GF, Gentz MC, Escoubas P, et al. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon. 2008;52:264–76. doi: 10.1016/j.toxicon.2008.05.020. [DOI] [PubMed] [Google Scholar]
- [198].Craik DJ. Circling the enemy: cyclic proteins in plant defense. Trends Plant Sci. 2009;14:328–35. doi: 10.1016/j.tplants.2009.03.003. [DOI] [PubMed] [Google Scholar]
- [199].Jin L, Briggs SL, Chandrasekhar S, et al. Crystal structure of human parathyroid hormone 1-34 at 0.9-angstrom resolution. J Biol Chem. 2000;275:27238–44. doi: 10.1074/jbc.M001134200. [DOI] [PubMed] [Google Scholar]
- [200].Dealwis C, Fernandez EJ, Thompson DA, et al. Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1 alpha, a potent ligand for the HIV-1 “fusin” coreceptor. Proc Natl Acad Sci USA. 1998;95:6941–6. doi: 10.1073/pnas.95.12.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Liebmann C. G protein-coupled receptors and their signaling pathways: classical therapeutical targets susceptible to novel therapeutic concepts. Curr Pharm Des. 2004;10(16):1937–58. doi: 10.2174/1381612043384367. [DOI] [PubMed] [Google Scholar]
- [202].Kostenis E. G proteins in drug screening: from analysis of receptor-G protein specificity to manipulation of GPCR-mediated signalling pathways. Curr Pharm Des. 2006;12(14):1703–15. doi: 10.2174/138161206776873734. [DOI] [PubMed] [Google Scholar]
- [203].Bortolato A, Mobarec JC, Provasi D, Filizola M. Progress in elucidating the structural and dynamic character of G Protein-Coupled Receptor oligomers for use in drug discovery. Curr Pharm Des. 2009;15(35):4017–25. doi: 10.2174/138161209789824768. [DOI] [PMC free article] [PubMed] [Google Scholar]