ABSTRACT
An intense period of chromatin remodeling takes place after fertilization in mammals, which is thought necessary for epigenetic reprogramming to start a new developmental program. While much attention has been given to the role of Polycomb Repressive Complex 2 (PRC2) and to canonical PRC1 complexes during this process, little is known as to whether there is any contribution of non-canonical PRC1 in shaping the chromatin landscape after fertilization. Here, we first describe in detail the temporal dynamics and abundance of H2A ubiquitylation (H2AK119ub), a histone modification catalyzed by PRC1, during pre-implantation mouse development. In addition, we have analyzed the presence of the 2 characteristic subunits of non-canonical PRC1 complexes, RYBP and its homolog YAF-2. Our results indicate that H2AK119ub is inherited from the sperm, rapidly removed from the paternal chromatin after fertilization, but detected again prior to the first mitosis, suggesting that PRC1 activity occurs as early as the zygotic stage. RYBP and YAF-2, together with the non-canonical subunit L3MBTL2, are all present during pre-implantation development but show different temporal dynamics. While RYBP is absent in the zygote, it is strongly induced from the 4-cell stage onwards. YAF-2 is inherited maternally and localizes to the pericentromeric regions in the zygote, is strongly induced between the 2- and 4-cell stages but then remains weak to undetectable subsequently. All together, our data suggest that non-canonical PRC1 is active during pre-implantation development and should be regarded as an additional component during epigenetic reprogramming and in the establishment of cellular plasticity of the early embryo.
KEYWORDS: Epigenetic reprogramming, histone modification, mouse embryo, PRC1
Introduction
Fertilization is the first step of embryonic development, whereby the paternal and the maternal genomic material get together to form a new organism. By implication, the process of fertilization triggers a number of molecular events on the parental genomes, including major chromatin remodeling. One of the main purposes of such remodeling is epigenetic reprogramming itself, which allows the reversion from a highly differentiated state in the gametes to a totipotent state in the zygote.35 The molecular mechanisms behind epigenetic reprogramming include histone variant exchange, changes in nuclear organization and DNA methylation, the activation of a large fraction of transposable elements, but also rapid and drastic changes in histone modifications.4,14
Among them, H3K27me3, catalyzed by the Polycomb Repressive Complex 2 (PRC2) and, in general, Polycomb group (PcG) proteins have been shown to play important roles from the earliest stages of development.8,25,27 At large, PcG repression is achieved through 2 major types of complexes that mediate various biochemical functions including histone modifying activities, recognition of covalent modifications, and physical compaction of chromatin. PRC2 displays its main catalytic activity toward H3K27me3 while the main catalytic activity of PRC1 is monoubiquitylation of H2A at lysine 119 (K119), although PRC1 has also been shown to be able to compact chromatin independently of H2AK119ub in vitro and in vivo.9,12,17 Genetically and biochemically, a role for PRC1 in transcriptional repression has been clearly demonstrated.33 The initial model for PcG repression posited that PRC1 recruitment depends upon previously established H3K27me3 domains by the action of PRC2. Subsequent recruitment of PRC1 then leads to H2AK119ub on those domains through the enzymatic activity of RING1A/RING1B, and to increased chromatin compaction. However, recent evidence supports an emerging view whereby PRC1 is indeed recruited to genomic regions occupied by PRC2 and in a manner dependent on H3K27, but additionally, PRC1 can be recruited onto chromatin independently of H3K27me3, presumably through alternative protein-protein interactions, via the Mel18 subunit for example 37, or through ncRNAs recruitment, such as the interaction between CBX7 and ANRIL.7,42 Thus, both PRC2 and PRC1 can mediate repression independently of each other.
In addition to RING1A/RING1B, the core subunits of canonical PRC1 include a CBX protein (2, 4, 6, 7 or 8) homolog of Polycomb, PHC proteins (1 to 3) and BMI1/PCGF4 or MEL18/PCGF2.12,20 However, a systematic proteomics approach in human cells identified additional non-canonical PRC1 complexes, revealing a greater complexity than previously anticipated.13 While RING1A/1B is a common theme for all PRC1 complexes, CBX and PHC proteins are replaced by either RYBP or YAF-2 in non-canonical PRC1 complexes. The incorporation of either RYBP or YAF-2 into PRC1 is presumably of functional relevance since—at least in vitro—RYBP can stimulate the ubiquitylation activity of PRC1 but YAF-2 does not.13 A further subdivision into functionally distinct families is provided by the presence of one of the 6 PCGF homologues.13 The high heterogeneity of PRC1 complexes is thought to allow a combinatorial assembly of multiple subunits to integrate additional biochemical activities including readers for other histone modifications—such as HP1s or the Malignant Brain Tumor (MBT) family members 26—as well as histone modifiers—such as HDACs.11
In the mouse embryo, PcG proteins support ‘facultative’ heterochromatin establishment in the male pronucleus 3,25,28, and a mutation in K27 within H3.3, the main histone variant incorporated in the male pronucleus, results in developmental arrest and heterochromatin defects.27 In addition, the combined deletion of both RING1 subunits, RING1A and RING1B, results in developmental arrest before the 2-cell stage due to defective regulation of the transcriptional program of the oocyte.23 Most studies addressing the function or the expression of PcG subunits in the early mammalian embryo have focused on PRC2 or on canonical PRC1 complexes, for example, the expression of most CBX proteins has been characterized in the early embryo.25,36 However, we still do not know whether non-canonical PRC1 can potentially contribute to the changes in chromatin remodeling that take place after fertilization. Indeed, the heterogeneous nature of PRC1 complexes makes it difficult to pinpoint the contribution of a specific complex and/or subunit to cellular plasticity and cell fate decisions. Here, we have first thoroughly analyzed the dynamic changes of the main catalytic activity of PRC1, H2AK119ub, during pre-implantation development. In addition, we have systematically characterized the main defining subunits of non-canonical PRC1 complexes, RYBP and YAF-2, and of additional non-canonical polycomb related proteins L3MBTL1 and L3MBTL2. Our results suggest that PRC1 activity is dynamic and takes place from the earliest stages of mouse development and suggest that non-canonical PRC1 activity contributes to the changes in chromatin remodeling that take place after fertilization.
Results
H2AK119 monoubiquitination is dynamic during pre-implantation development
We first analyzed the activity of the PRC1 complex by examining H2A monoubiquitylation on lysine 119 (H2AK119ub). We detected H2AK119ub throughout pre-implantation development at all stages analyzed (Fig. 1A). At the zygote stage, H2AK119ub was present in both female and male pronuclei and in the polar body (PB), which was thereafter used as an internal control for subsequent stages of development. Subsequently, H2AK119ub was detected throughout development, in 2-cell, 4-cell, 8-cell and morula stages (Fig. 1A). However, the pattern of distribution of H2AK119ub varied along these developmental times. H2AK119ub localized at the DAPI-dense regions surrounding the nucleolar-like bodies (NLBs) in zygotes at PN4 stage (ProNuclear4) (Fig. 1C) and in 2-cell stage embryos (Fig. 1A). These DAPI-rich regions are known to contain the pericentromeric chromatin.24 The pattern of localization of H2AK119ub changed at the 4-cell stage, which corresponds to the time when chromocenters are being established and the NLBs are lost.24 After the 4-cell stage, H2AK119ub did not colocalize anymore with the DAPI-dense regions (Fig. 1A and B), and showed instead a disperse distribution throughout the nucleoplasm. This localization persisted in the 8-cell stage, except that most blastomeres exhibited one large (arrows), single focus (n=5/7). One cell division later, H2AK119ub became localized in a more ‘dotty’ like pattern (Fig. 1A), reminiscent of Polycomb bodies observed in other cell types.29 Thus, H2AK119ub displays 3 different global patterns in the early embryo: i) an initial localization to DAPI-rich pericentromeric regions around the NLBs in the zygote and 2-cell stage embryos; ii) a wider nuclear localization with foci in 4- and 8-cell embryos; and iii) a foci-like distribution in the morula.
Figure 1.
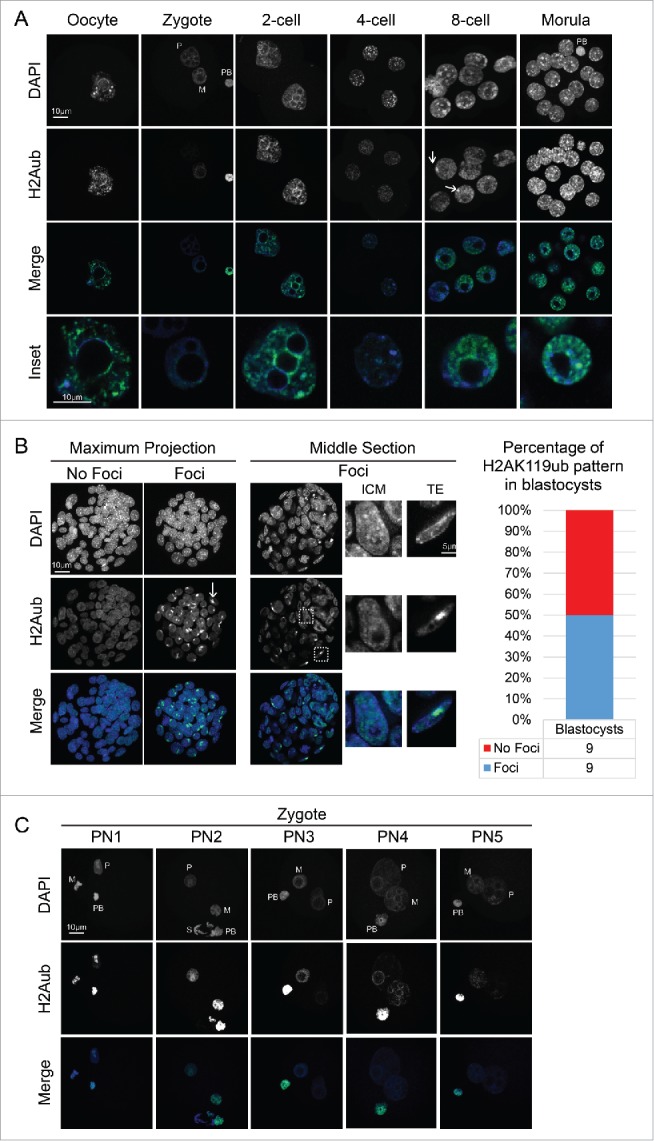
Dynamic establishment of H2AK119ub during mouse pre-implantation development. (A) Representative images showing full Z-projections of confocal sections of oocytes and embryos from the zygote to the morula stage stained with DAPI and H2AK119ub antibodies. White arrows at the 8-cell stage indicate accumulation of H2AK119ub at foci. Bottom panels are merged images of single sections showing insets of nuclei from the embryos above at a higher magnification. (B) Representative images of full Z-projections of confocal sections of blastocysts stained with DAPI and H2AK119ub antibody. A representative projection of a blastocyst showing H2AK119ub accumulation in foci is shown with 2 insets depicting one inner cell mass (ICM) nucleus and a trophectoderm (TE) nucleus. On the right, the percentage of embryos showing or not H2AK119ub foci, n =18. (C) Full projection of confocal Z-sections of H2AK119ub and DAPI at 5 different pronuclear (PN) stages in the zygote. Abbreviations: M: maternal, P: paternal, PB: polar body, S: sperm. Scale bars correspond to 10 µm.
At the blastocyst stage we observed 2 types of patterns for H2AK119ub (Fig. 1B). Half of the embryos analyzed (n=18) displayed intense foci labeled with H2AK119ub (Fig. 1B), similarly to the appearance of foci of RING1B/RNF2 during X inactivation.6 Furthermore, these foci were not detected in the inner cell mass (ICM), but were detected exclusively in the trophectoderm (Fig. 1B). This pattern is reminiscent of previous observations of colocalization between H2AK119ub, RING1 (RING1A and RING1B/RNF2) and the inactive X that have been shown to occur in female trophoblast stem (TS) and differentiating female embryonic stem (ES) cells.6,10 Thus, we conclude that H2AK119ub likely marks sites of imprinted X chromosome inactivation in mouse blastocyst in vivo. While this pattern may be set at an earlier developmental time, we did not find a correlation with the foci of H2AK119ub that we observed, e.g., at the morula stage (Fig. 1A). Indeed, while we observed foci in morula stage embryos, we observed this pattern in more than half of the embryos and, therefore, it is likely not to be related to embryonic X inactivation.
Next, to better understand the establishment of H2AK119ub after fertilization, we carefully analyzed the presence of H2AK119ub at different stages of zygotic development (PN1 to PN5) (Fig. 1C). We first detected H2AK119ub in both pronuclei immediately after fertilization (PN1) (Fig. 1C), suggesting that the sperm chromatin already contains H2AK119ub. H2AK119ub retained the same pattern until PN2, where it was enriched in the central part of the decondensing male pronucleus, which corresponds to the region where pericentromeric chromatin localizes at these stages.18,24 Thereafter, H2AK119ub levels were drastically reduced in the male pronucleus at PN3 (Fig. 1C). At PN4, the levels of H2AK119ub became stronger again, and localized at the NLBs on both pronuclei, in agreement with a previous report.36 H2AK119ub levels were further reduced as the embryo reached PN5 and approached the first mitotic division (Fig. 1C). Thus, H2AK119ub is detected in both, oocyte and sperm chromatin at fertilization, but is barely detectable in the male pronucleus at mid-zygotic stages, suggesting that PRC1 activity is dynamic and takes place from the earliest stages of mouse development.
RYBP is expressed in pre-implantation mouse embryos
PRC1 complexes can mediate H2AK119ub independently of H3K27me3 and PRC2 proteins.22,40 The dynamics and localization pattern of H3K27me3 in conjunction with that of PRC2 components have been well studied in the mouse embryo.25,27,28 Therefore, having established that H2AK119ub was present in pre-implantation development, we wondered whether non-canonical PRC1 complexes contribute also to H2AK119 ubiquitylation. We thus next analyzed the expression of members of the non-canonical PRC1 complexes that can mediate H2AK119ub independently of PRC2 activity. When assembled with RYBP, non-canonical PRC1 mediates H2AK119ub without the need of pre-existing H3K27me3.37 We first examined the localization of RING1 and YY1 Binding Protein (RYBP). Albeit weakly, in the oocyte RYBP was distributed both in the nucleus and the cytoplasm. This observation was surprising given the expected nuclear localization of RYBP based on findings in other cell types.15,37 However, immediately after fertilization, we did not detect RYBP at the zygote stage, neither in the cytoplasm nor in the nucleoplasm of any of the 2 pronuclei (Fig. 2A). At the 2-cell stage, RYBP was weakly detected in the nuclei of both blastomeres, where it showed a slight accumulation around the NLBs at DAPI dense regions (Fig. 2A, arrows). Although the immunofluorescence signal for RYBP was weak, RYBP remained present at all stages analyzed until the blastocyst stage (Fig. 2A). Apart from the slight accumulation around the NLBs in 2-cell stage embryos, we did not observe any particular nuclear accumulation when the protein was detected. However, we noticed that, besides the nuclear localization, RYBP also was present in the mid-body after nuclear division (Fig. 2A). We also observed that RYBP was present during mitosis and localized to the spindle (data not shown). These observations indicate that RYBP may not play a role in H2AK119ub establishment or maintenance at the zygote stage, but could do so after the 2-cell stage.
Figure 2.
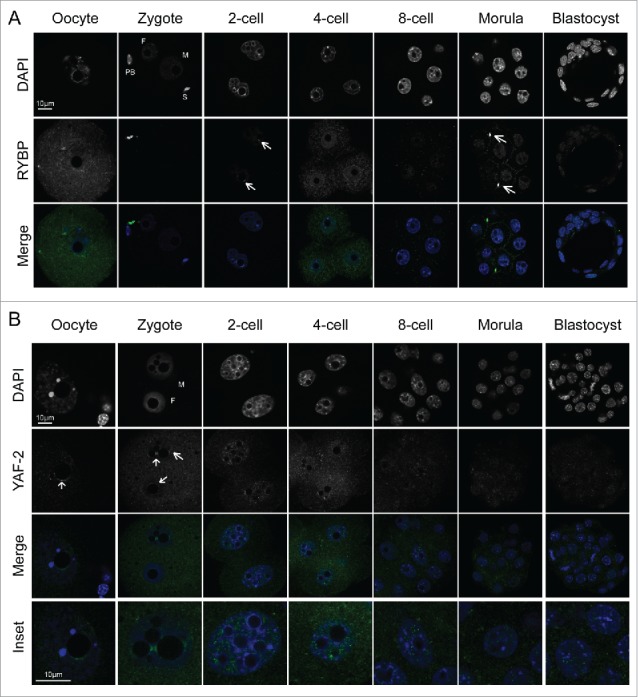
Changes in non-canonical PRC1 components during pre-implantation development. A –B. Confocal single projection images of DAPI and RYBP (A), or YAF-2 (B), are shown. White arrows indicate the accumulation of the protein. Merge insets of nuclei are shown for YAF-2 at the bottom. Abbreviations: M: maternal, P: paternal, PB: polar body, S: sperm. Scale bar is shown in white and corresponds to 10 µm for both stainings.
The RYBP homolog YAF-2 is inherited maternally and its levels decrease during pre-implantation development
YY1 Associated Factor 2 (YAF-2), an RYBP homolog, is another component of non-canonical PRC1 complexes. The presence of YAF-2 is mutually exclusive with that of RYBP and, therefore, defines different complexes.13 Thus, we next addressed whether YAF-2 could potentially play a role in mediating H2AK119ub establishment and maintenance in early mouse development. We detected YAF-2, albeit weakly, in germinal vesical stage (GV) oocytes around the NLB and nuclear periphery (Fig 2B). However, YAF-2 was below the detection limit immediately after fertilization (data not shown) and it only started getting detected at PN4 in the zygote (Fig. 2B, arrows), where it shows some association to the NLBs. YAF-2 was present in the 2-cell embryos, where it showed some enrichment around the NLBs, but its localization appeared rather punctuate throughout the nucleoplasm, without exclusive localization with DAPI-rich regions (Fig. 2B). Subsequently, the levels of YAF-2 were reduced throughout pre-implantation development after the 4-cell and till the blastocyst stage, but remained visible albeit weak (Fig. 2B). This data leads us to conclude that YAF-2 is present in the early mouse embryo and that it could potentially contribute the ubiquitylation of H2A via a non-canonical form of PRC1 in which YAF-2 is part of. However, the patterns of YAF-2 at the morula and blastocyst stages do not indicate a specific accumulation at foci (Fig. 2B) like H2AK119ub does (Fig. 1A-B), suggesting that YAF-2 may globally not be involved in the subsequent establishment of H2AK119ub on the X-chromosome.
L3MBTL2, a specific subunit of non-canonical PRC1.6 complex is expressed in pre-implantation embryos
Considering the diversity of non-canonical PRC1 complexes, we next focused more specifically on one of them, PRC1.6. Our reasoning behind this choice was 2-fold. Firstly, PRC1.6 is the only PRC1 complex that, in addition to catalytic activity toward H2AK119, possesses histone deacetylation activity 13 and, therefore, could also contribute to the rapid changes in histone acetylation that characterize pre-implantation development.1,2,4 Secondly, PRC1.6 contains a recognition module, through L3MBTL2, for H4K20me1, which is essential for pre-implantation development, as deletion of PR-Set7 results in lethality by the 8-cell stage, in a manner dependent of its methyltransferase activity.13,21 Since L3MBTL2 is a defining, unique feature of PRC1.6, we therefore assessed its localization through pre-implantation development. L3MBTL2 was readily detected in the mature GV oocyte, where it exhibited a rather uniform nuclear localization, but was excluded from the DAPI-rich regions (Fig. 3A). After fertilization, L3MBTL2 was present on both pronuclei at the zygote stage. The levels of L3MBTL2 decreased significantly at the 2-cell stage, and gradually increased between the 4-cell and morula stages (Fig. 3A). In blastocysts, L3MBTL2 displayed a more diffuse signal, with high background. However, some weak accumulation was visible across the nuclei of both the ICM and the trophectoderm. Thus, L3MBTL2 is inherited maternally but strongly reduced at the 2-cell stage, becoming then present again after the 4-cell stage. It is possible that the low levels of L3MBTL2 between the zygote and 2-cell stage help to relieve a repressive chromatin state globally, thereby helping to promote zygotic genome activation.
Figure 3.
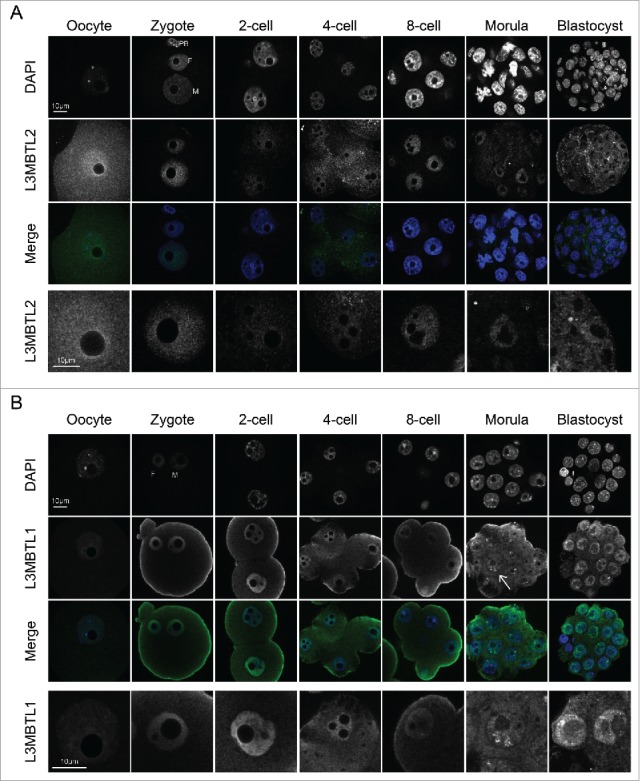
MBT family proteins are present in early mammalian development. (A) Representative confocal sections of immunofluorescence analysis of oocytes and embryos at the indicated stages of development analyzed with the L3MBTL2 antibody. DNA was stained with DAPI (top panel). A merged, single section is also shown (L3MBTL2 in green, DAPI in blue). Insets showing a higher magnification of nuclei from the same embryos, but with the L3MBTL2 signal in gray scale are shown at the bottom. (B) Representative single confocal sections of oocytes or embryos stained with the L3MBTL1 antibody, as indicated. The DAPI (top panel) and the L3MBTL1 channels are shown in gray scale. In the merge image L3MBTL1 is shown in green and DAPI in blue. The bottom panel shows higher magnifications of nuclei of the same representative embryos, with the L3MBTL1 staining is depicted in gray scale. M: maternal, P: paternal, PB: polar body. Note that the L3MBTL1 antibody used gave some non-specific staining around the cell membrane in some of the embryos analyzed. Scale bar as indicated.
In addition to L3MBTL2, another MBT family member and polycomb-related protein, L3MBTL1, can also bind to methyl-lysines, including H4K20me1/2.34,39 Thus, we finally addressed the protein levels of L3MBTL1, the mouse homolog of the Drosophila PcG protein l(3)mbt.19,41 The fully-grown GV oocyte displayed a clear nuclear localization of L3MBTL1, which appeared dispersed, without any obvious enrichment in either DAPI-rich foci or the regions surrounding the NLB (Fig. 3B). We detected L3MBTL1 at all stages of pre-implantation development that we analyzed, from the zygote to the blastocyst stage (Fig. 3B). In the zygote, L3MBTL1 localized to both pronuclei and had no particular enrichment in DAPI-dense regions. L3MBTL1 retained the same localization pattern until the morula stage, where all nuclei analyzed displayed L3MBTL1 foci in association with the nucleoli, but excluded from DAPI-rich regions (Fig. 3B, arrows). In all the blastocysts analyzed, L3MBTL1 was present in both the ICM and the TE lineages (Fig. 3B). Like in the morula stage, L3MBTL1 displayed condensed foci in proximity to the nucleoli in the blastocyst (Fig. 3B). Because this localization was detected in all blastocysts analyzed, it suggests that these foci may not be linked to X inactivation, as in the case of H2AK119ub.6 Thus, while L3MBTL2 shows temporal changes in its expression pattern during pre-implantation development, L3MBTL1 remained present throughout all stages analyzed.
Discussion
Our observations above suggest that different types of PRC1 complexes 32 might be at play during early mouse development. While expression analysis does not imply that a complex is formed biochemically, it does provide an indication of the components present in the developmental time window studied. Given the temporal changes in the expression of the subunits that we analyzed (Fig. 4), it is possible that different complexes play different functions during this period, but also that their activities and assembly change dynamically during these first divisions. For example, CBX2, a member of the canonical PRC1, is expressed between the zygote and 8-cell stages and later at the blastocyst stage.25 Thus, one could postulate that CBX2 is involved in the targeting of canonical PRC1 activity at these stages. While we did not detect RYBP in zygotes, we detected YAF-2 enriched around the NLBs, suggesting that CBX-2 directed PRC1 activity is not alone in promoting H2AK119ub at these regions. Indeed, Ring1a/Ring1b double knockout results in derepression of major satellites transcription from the paternal chromatin.25 Since the Ring1a/Ring1b double knockout abolishes PRC1 activity as a whole, additional experiments will be necessary to address whether there is a differential contribution between those 2 potential PRC1 complexes in mediating heterochromatin silencing. This can be further assessed by the depletion of YAF-2 in the zygote or a complete YAF-2 knockout model to study the different roles that the non-canonical mediated H2AK119ub compared to the canonical ones during early mouse development.
Figure 4.
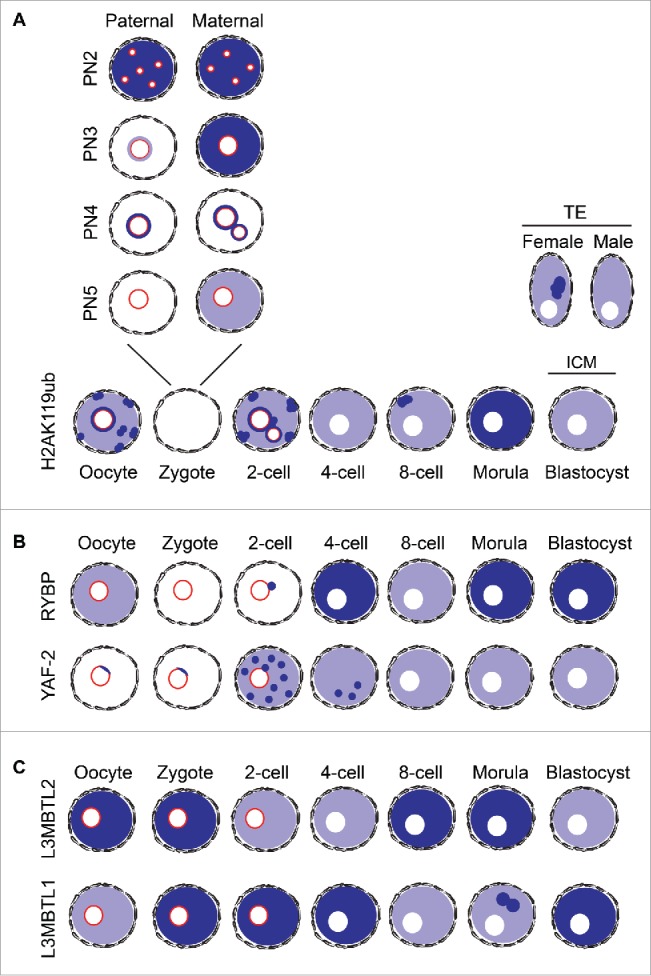
Schematic representation of the distribution of H2AK119ub and the proteins studied across developmental stages. Nuclear pattern distribution of H2AK119ub (A), RYBP, YAF-2 (B), L3MBTL2 and L3MBTL1 (C) at all stages of pre-implantation development. Levels H2AK119ub (A) at the zygote stage are subdivided into 4 ProNuclear (PN) stages to reflect dynamic changes in the pattern and levels of the mark. Color representation: Light blue indicates low levels of enrichment whereas darker blue is representative of high levels and specific localization in foci (dots in the diagram). Nucleolar-like bodies (NLBs) in the oocyte, zygote, and 2-cell stage embryos are shown as red circles in the nucleus.
Interestingly, all 3 YAF-2, RYBP (this work), and CBX2 36 are expressed in 2-cell stage embryos, suggesting that at this stage, at least 3 types of PRC1 complexes could potentially form to enable widespread PRC1 activity. This is supported by the strong H2AK119ub signal observed at this stage, where H2AK119ub is not only restricted to the heterochromatic regions surrounding the NLBs where RYBP is enriched, but clearly extends to the nucleoplasm. Indeed, RYBP containing PRC1 exhibit stronger H2AK119ub activity than those containing CBX2/CBX8 in vitro.13 The 2-cell stage coincides with embryonic genome activation (EGA), which also corresponds with the structural changes that start to take shape between the 2- and 4-cell stages when the NLBs are replaced by the chromocenters as the organizing centers of chromatin structure. Thus, the 2-cell stage embryo may require a large deal of chromatin remodeling and changes, to which increased PRC1 activity may contribute. Conditional knockout of Rybp in ES cells leads to derepression of genes related to pre-implantation development during EGA, such as Eif1a, Zscan4, and some retrotransposons. Of special interest is the upregulation of MuERVs among the retrotransposons upon Rybp deletion. Double knockout of Rybp and Yaf2 in ES cells showed no synergistic effects, suggesting potential independent effects for these 2 forms of non-canonical PRC1 complexes.15
At later stages of development, at the blastocyst stage, previous data suggested a lack of enrichment of CBX2 at RING1B/RNF2 foci in the blastocyst.25 We observed foci of H2AK119ub at this stage, which are presumably linked to X inactivation.6 However, neither RYBP nor YAF-2 displays a strictly similar pattern to that of H2AK119ub at this stage. Thus, a different PRC1 complex might be at play when RING1 is recruited by Xist to the X inactivation center.31 One should keep in mind, however, that mechanisms other than H2A ubiquitylation could also contribute to PRC1 function. This has been clearly demonstrated for the case of L3MBTL2, which does not seem to stimulate RING1A/RING1B activity in vitro, and was therefore suggested to act as a recruiter exclusively.40 Because L3MBTL2 has its own biochemical activities by which it can compact chromatin in vitro 40, recruitment of L3MBTL2 and PRC1 could facilitate local chromatin compaction in a histone modification independent manner. Indeed, our observations that both L3MBTL2 and L3MBTL1 are expressed throughout pre-implantation development could provide more versatile actions of polycomb-related function at specific genome locations at a given developmental time. While it is known that the catalytic, ubiquitylation activity of PRC1 is not necessary to compact chromatin at the time of gastrulation in vivo 9,17 whether catalytic activity per se of PRC1 is necessary after fertilization, has not been addressed.
The dynamic expression profiling of the PRC1 subunits RYBP, YAF-2, and L3MBTL2 suggests that different variants of PRC1 complexes might play a role during early development. In addition, and without excluding this possibility, it is conceivable that the proteins studied play additional roles via the interaction with partners other than polycomb proteins. Indeed, RYBP has been documented to interact with MDM2 and the transcription factors E2F2 and E2F3.5,30 Likewise, the NuRD complex associates with L3MBTL2 in ES cells.26
In sum, our work contributes to the molecular characterization of the embryonic chromatin after fertilization, and places non-canonical PRC1 activity as a potential important player to consider when understanding the molecular definition of epigenetic reprogramming in vivo.
Materials & Methods
Embryo collection
Mice were bred in a 12 h light cycle. Embryos were collected as described 16 from CD1 X CD1 natural mating. Zygotes and cleavage stage embryos were collected at the indicated developmental stages upon puncturing of the ampullaswollen and the oviduct. Blastocysts were obtained by flushing the uterus with M2 medium (Sigma). All embryos were fixed immediately after collection. Pronuclear stages (PN) were classified according to Adenot et al. 1. Experiments with animals were carried out according to valid legislation in France and under the authorization of the Com'eth ethical committee.
Immunostaining
After removal of the zona pellucida with acid Tyrode's solution (Sigma), embryos were washed 3 times in PBS and fixed as described.38 Embryos were permeabilized, washed with PBS-T (0.1% Tween in PBS), blocked and incubated with the primary antibodies. The primary antibodies were: H2AK119ub (Cell Signaling D27C4, rabbit monoclonal; characterized in 13, YAF-2 (ThermoFisher Scientific PA5-30359, rabbit polyclonal, http://www.thermofisher.com/YAF2-Antibody-Polyclonal/PA5-30359.html), RYBP (Abcam ab5976, rabbit polyclonal, characterized in 37, L3MBTL1 and L3MBTL2 (from D. Reinberg, rabbit polyclonal; characterized in 40 and 39). A dilution of 1/250 was used for all the primary antibodies. After O.N. incubation in the primary antibody, embryos were washed twice in PBS-T, blocked for additional 20 minutes and incubated for 3 h with the corresponding secondary antibodies at RT. The secondary antibody used in this work is AlexaFluor 488 (Life Technologies, goat anti-rabbit). After washing, embryos were mounted in Vectashield (Vector Laboratories) supplemented with 4'-6-Diamidino-2-phenylindole (DAPI) for visualization of the DNA.
Confocal analysis
Confocal microscopy was performed using a 63x oil objective in a Leica SP5 or SP8 inverted microscope. Confocal sections were acquired every 0.5 µm and images were analyzed using Lecica LAS AF Lite 2.6.3 build 8173. Images for this manuscript were formatted using Adobe Illustrator CS6. The number of embryos analyzed in this work are shown in the table S1 as follows: A) Number of embryos collected per stage for each antibodies; B) Number of zygotes stained with H2AK119ub at the different ProNuclear stage (PN); and C) Number of blastocysts stained with H2AK119ub and pattern of foci. Each staining was repeated at least in 2 independent biological replicates.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Jin Zhang, Patrick Trojer and Danny Reinberg for providing the L3MBTL1 and L3MBTL2 antibodies and to Celine Ziegler-Birling for support with mouse handling and embryo collection. M.E.T.-P. acknowledges funding from EpiGeneSys NoE, ERC-Stg ‘NuclearPotency’, EMBO Young Investigator Program and the Schlumberger Foundation for Research and Education. A.E is a recipient of a doctoral fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche and from the Fondation pour la Recherche Médicale (FDT20150532012).
References
- 1.Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 1997; 124:4615-4625; PMID:9409678 [DOI] [PubMed] [Google Scholar]
- 2.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the pre-implantation mouse embryo. Dev Biol 1997; 181:296-307; PMID:9013938; http://dx.doi.org/ 10.1006/dbio.1996.8466 [DOI] [PubMed] [Google Scholar]
- 3.Arney KL, Bao S, Bannister AJ, Kouzarides T, Surani MA. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int J Dev Biol 2002; 46:317-320; PMID:12068953 [PubMed] [Google Scholar]
- 4.Burton A, Torres-Padilla ME. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol 2014; 15:723-734; PMID:25303116; http://dx.doi.org/ 10.1038/nrm3885 [DOI] [PubMed] [Google Scholar]
- 5.Chen D, Zhang J, Li M, Rayburn ER, Wang H, Zhang R. RYBP stabilizes p53 by modulating MDM2. EMBO Rep 2009; 10:166-172; PMID:19098711; http://dx.doi.org/ 10.1038/embor.2008.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 2004; 7:663-676; PMID:15525528; http://dx.doi.org/ 10.1016/j.devcel.2004.10.005 [DOI] [PubMed] [Google Scholar]
- 7.El Messaoudi-Aubert S, Nicholls J, Maertens GN, Brookes S, Bernstein E, Peters G. Role for the MOV10 RNA helicase in polycomb-mediated repression of the INK4a tumor suppressor. Nat Struct Mol Biol 2010; 17:862-868; PMID:20543829; http://dx.doi.org/ 10.1038/nsmb.1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during pre-implantation mouse development. Development 2003; 130:4235-4248; PMID:12900441; http://dx.doi.org/ 10.1242/dev.00625 [DOI] [PubMed] [Google Scholar]
- 9.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 2010; 38:452-464; PMID:20471950; http://dx.doi.org/ 10.1016/j.molcel.2010.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem 2004; 279:52812-52815; PMID:15509584; http://dx.doi.org/ 10.1074/jbc.C400493200 [DOI] [PubMed] [Google Scholar]
- 11.Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 2012; 1:e00205; PMID:23256043; http://dx.doi.org/ 10.7554/eLife.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science 2004; 306:1574-1577; PMID:15567868; http://dx.doi.org/ 10.1126/science.1100576 [DOI] [PubMed] [Google Scholar]
- 13.Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 2012; 45:344-356; PMID:22325352; http://dx.doi.org/ 10.1016/j.molcel.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol 2009; 10:526-537; PMID:19603040; http://dx.doi.org/ 10.1038/nrm2727 [DOI] [PubMed] [Google Scholar]
- 15.Hisada K, Sanchez C, Endo TA, Endoh M, Roman-Trufero M, Sharif J, Koseki H, Vidal M. RYBP represses endogenous retroviruses and pre-implantation- and germ line-specific genes in mouse embryonic stem cells. Mol Cell Biol 2012; 32:1139-1149; PMID:22269950; http://dx.doi.org/ 10.1128/MCB.06441-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan BL, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo, 2nd. edn (Cold Spring Harbor Laboratory Press; ) 1994; http://dx.doi.org/ 10.1017/s0016672300034571 [DOI] [Google Scholar]
- 17.Illingworth RS, Moffat M, Mann AR, Read D, Hunter CJ, Pradeepa MM, Adams IR, Bickmore WA. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev 2015; 29:1897-1902; PMID:26385961; http://dx.doi.org/ 10.1101/gad.268151.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jachowicz JW, Santenard A, Bender A, Muller J, Torres-Padilla ME. Heterochromatin establishment at pericentromeres depends on nuclear position. Genes Dev 2013; 27:2427-2432; PMID:24240232; http://dx.doi.org/ 10.1101/gad.224550.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koga H, Matsui S, Hirota T, Takebayashi S, Okumura K, Saya H. A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene 1999; 18:3799-3809; PMID:10445843; http://dx.doi.org/ 10.1038/sj.onc.1202732 [DOI] [PubMed] [Google Scholar]
- 20.Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol 2002; 22:6070-6078; PMID:12167701; http://dx.doi.org/ 10.1128/MCB.22.17.6070-6078.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 2009; 29:2278-2295; PMID:19223465; http://dx.doi.org/ 10.1128/MCB.01768-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 2007; 27:3769-3779; PMID:17339329; http://dx.doi.org/ 10.1128/MCB.01432-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posfai E, Kunzmann R, Brochard V, Salvaing J, Cabuy E, Roloff TC, Liu Z, Tardat M, van Lohuizen M, Vidal M, et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev 2012; 26:920-932; PMID:22499591; http://dx.doi.org/ 10.1101/gad.188094.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Probst AV, Santos F, Reik W, Almouzni G, Dean W. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma 2007; 116:403-415; PMID:17447080; http://dx.doi.org/ 10.1007/s00412-007-0106-8 [DOI] [PubMed] [Google Scholar]
- 25.Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet 2008; 40:411-420; PMID:18311137; http://dx.doi.org/ 10.1038/ng.99 [DOI] [PubMed] [Google Scholar]
- 26.Qin J, Whyte WA, Anderssen E, Apostolou E, Chen HH, Akbarian S, Bronson RT, Hochedlinger K, Ramaswamy S, Young RA, et al. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell 2012; 11:319-332; PMID:22770845; http://dx.doi.org/ 10.1016/j.stem.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol 2010; 12:853-862; PMID:20676102; http://dx.doi.org/ 10.1038/ncb2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol 2005; 280:225-236; PMID:15766761; http://dx.doi.org/ 10.1016/j.ydbio.2005.01.025 [DOI] [PubMed] [Google Scholar]
- 29.Saurin AJ, Shiels C, Williamson J, Satijn DP, Otte AP, Sheer D, Freemont PS. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol 1998; 142:887-898; PMID:9722603; http://dx.doi.org/ 10.1083/jcb.142.4.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. Embo J 2002; 21:5775-5786; PMID:12411495; http://dx.doi.org/ 10.1093/emboj/cdf577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. Embo J 2006; 25:3110-3122; PMID:16763550; http://dx.doi.org/ 10.1038/sj.emboj.7601187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet 2013; 14:853-864; PMID:24217316; http://dx.doi.org/ 10.1038/nrg3603 [DOI] [PubMed] [Google Scholar]
- 33.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 2009; 10:697-708; PMID:19738629; http://dx.doi.org/ 10.1038/nrn2731 [DOI] [PubMed] [Google Scholar]
- 34.Stielow C, Stielow B, Finkernagel F, Scharfe M, Jarek M, Suske G. SUMOylation of the polycomb group protein L3MBTL2 facilitates repression of its target genes. Nucleic Acids Res 2014; 42:3044-3058; PMID:24369422; http://dx.doi.org/ 10.1093/nar/gkt1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell 2007; 128:747-762; PMID:17320511; http://dx.doi.org/ 10.1016/j.cell.2007.02.010 [DOI] [PubMed] [Google Scholar]
- 36.Tardat M, Albert M, Kunzmann R, Liu Z, Kaustov L, Thierry R, Duan S, Brykczynska U, Arrowsmith CH, Peters AH. Cbx2 targets PRC1 to constitutive heterochromatin in mouse zygotes in a parent-of-origin-dependent manner. Mol Cell 2015; 58:157-171; PMID:25801166; http://dx.doi.org/ 10.1016/j.molcel.2015.02.013 [DOI] [PubMed] [Google Scholar]
- 37.Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012; 148:664-678; PMID:22325148; http://dx.doi.org/ 10.1016/j.cell.2011.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and pre-implantation embryos. Int J Dev Biol 2006; 50:455-461; PMID:16586346; http://dx.doi.org/ 10.1387/ijdb.052073mt [DOI] [PubMed] [Google Scholar]
- 39.Trojer P, Li G, Sims RJ, III, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, Wang YH, Reinberg D. L3MBTL1, a Histone-Methylation-Dependent Chromatin Lock. Cell 2007; 129:915-928; PMID:17540172; http://dx.doi.org/ 10.1016/j.cell.2007.03.048 [DOI] [PubMed] [Google Scholar]
- 40.Trojer P, Cao AR, Gao Z, Li Y, Zhang J, Xu X, Li G, Losson R, Erdjument-Bromage H, Tempst P, et al. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol Cell 2011; 42:438-450; PMID:21596310; http://dx.doi.org/ 10.1016/j.molcel.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wismar J, Loffler T, Habtemichael N, Vef O, Geissen M, Zirwes R, Altmeyer W, Sass H, Gateff E. The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech Dev 1995; 53:141-154; PMID:8555106; http://dx.doi.org/ 10.1016/0925-4773(95)00431-9 [DOI] [PubMed] [Google Scholar]
- 42.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010; 38:662-674; PMID:20541999; http://dx.doi.org/ 10.1016/j.molcel.2010.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]


